

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137670.4 + phase: 0
(324 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
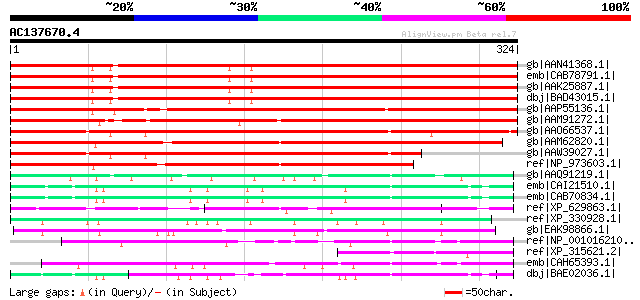
Score E
Sequences producing significant alignments: (bits) Value
gb|AAN41368.1| unknown protein [Arabidopsis thaliana] gi|6232009... 384 e-105
emb|CAB78791.1| putative protein [Arabidopsis thaliana] gi|28945... 384 e-105
gb|AAK25887.1| unknown protein [Arabidopsis thaliana] 383 e-105
dbj|BAD43015.1| unknown protein [Arabidopsis thaliana] 383 e-105
gb|AAP55136.1| unknown protein [Oryza sativa (japonica cultivar-... 353 5e-96
gb|AAM91272.1| zinc finger protein Glo3-like [Arabidopsis thalia... 350 3e-95
gb|AAO66537.1| putative zinc finger protein [Oryza sativa (japon... 344 2e-93
gb|AAM62820.1| zinc finger protein Glo3-like [Arabidopsis thalia... 311 1e-83
gb|AAW39027.1| putative zinc finger protein [Oryza sativa (japon... 261 3e-68
ref|NP_973603.1| human Rev interacting-like family protein / hRI... 241 3e-62
gb|AAQ91219.1| ADP-ribosylation factor GTPase activating protein... 92 2e-17
emb|CAI21510.1| OTTHUMP00000028526 [Homo sapiens] gi|56203507|em... 87 7e-16
emb|CAB70834.1| hypothetical protein [Homo sapiens] 87 7e-16
ref|XP_629863.1| hypothetical protein DDB0184259 [Dictyostelium ... 83 1e-14
ref|XP_330928.1| hypothetical protein [Neurospora crassa] gi|289... 79 1e-13
gb|EAK98866.1| potential ARF GAP [Candida albicans SC5314] gi|46... 72 2e-11
ref|NP_001016210.1| hypothetical protein LOC548964 [Xenopus trop... 72 2e-11
ref|XP_315621.2| ENSANGP00000007262 [Anopheles gambiae str. PEST... 71 4e-11
emb|CAH65393.1| hypothetical protein [Gallus gallus] gi|60302730... 70 6e-11
dbj|BAE02036.1| unnamed protein product [Macaca fascicularis] 70 6e-11
>gb|AAN41368.1| unknown protein [Arabidopsis thaliana] gi|62320091|dbj|BAD94263.1|
hypothetical protein [Arabidopsis thaliana]
gi|62319827|dbj|BAD93852.1| hypothetical protein
[Arabidopsis thaliana] gi|18414983|ref|NP_567543.1|
human Rev interacting-like family protein / hRIP family
protein [Arabidopsis thaliana]
gi|51971433|dbj|BAD44381.1| unknown protein [Arabidopsis
thaliana] gi|51970716|dbj|BAD44050.1| unknown protein
[Arabidopsis thaliana]
Length = 413
Score = 384 bits (987), Expect = e-105
Identities = 211/336 (62%), Positives = 253/336 (74%), Gaps = 15/336 (4%)
Query: 1 MSFGGNSRAQIFFKQHGWTDGGKIEAKYTSRAAELYRQILTKEVAKSMALE--KGLPSSP 58
M FGGN+RAQ+FFKQHGWTDGGKIEAKYTSRAA+LYRQIL KEVAK++A E GL SSP
Sbjct: 81 MMFGGNNRAQVFFKQHGWTDGGKIEAKYTSRAADLYRQILAKEVAKAIAEETNSGLLSSP 140
Query: 59 VASQS----SNGFLDVRTSEVLKENTLDKAEKLESTSSPRASHTSASNNLKKSIGGKKPG 114
VA+ SNG V + V +E L K E +TSSP+AS+T + KK IG K+ G
Sbjct: 141 VATSQLPEVSNG---VSSYSVKEELPLSKHEATSATSSPKASNTVVPSTFKKPIGAKRTG 197
Query: 115 KSGGLGARKLNKKPSESFYEQKPEE--PPAPVPSTTNNNVS---ARPSMTSRFEYVDNVQ 169
K+GGLGARKL KP ++ YEQKPEE P P S+TNN S A S SRFEY D++Q
Sbjct: 198 KTGGLGARKLTTKPKDNLYEQKPEEVAPVIPAVSSTNNGESKSSAGSSFASRFEYNDDLQ 257
Query: 170 SSELDSRGSNTFNHVSVPKSSNFFADFGMDSGFPKKFGSNTSKVQIEESDEARKKFSNAK 229
S G+ NHV+ PKSS+FF+DFGMDS FPKK SN+SK Q+EESDEARKKF+NAK
Sbjct: 258 SGGQSVGGTQVLNHVAPPKSSSFFSDFGMDSSFPKKSSSNSSKSQVEESDEARKKFTNAK 317
Query: 230 SISSSQFFGDQNKARDAETRATLSKFSSSSAISSADFFG-DSADSSIDLAASDLINRLSF 288
SISS+Q+FGDQNK D E++ATL KF+ S++ISSADF+G D DS+ID+ ASDLINRLSF
Sbjct: 318 SISSAQYFGDQNKNADLESKATLQKFAGSASISSADFYGHDQDDSNIDITASDLINRLSF 377
Query: 289 QAQQDISSLKNIAGETGKKLSSLASSLMTDLQDRIL 324
QAQQD+SSL NIAGET KKL +LAS + +D+QDR+L
Sbjct: 378 QAQQDLSSLVNIAGETKKKLGTLASGIFSDIQDRML 413
>emb|CAB78791.1| putative protein [Arabidopsis thaliana] gi|2894598|emb|CAA17132.1|
putative protein [Arabidopsis thaliana]
gi|7487780|pir||T05075 hypothetical protein T6K21.70 -
Arabidopsis thaliana
Length = 1082
Score = 384 bits (987), Expect = e-105
Identities = 211/336 (62%), Positives = 253/336 (74%), Gaps = 15/336 (4%)
Query: 1 MSFGGNSRAQIFFKQHGWTDGGKIEAKYTSRAAELYRQILTKEVAKSMALE--KGLPSSP 58
M FGGN+RAQ+FFKQHGWTDGGKIEAKYTSRAA+LYRQIL KEVAK++A E GL SSP
Sbjct: 81 MMFGGNNRAQVFFKQHGWTDGGKIEAKYTSRAADLYRQILAKEVAKAIAEETNSGLLSSP 140
Query: 59 VASQS----SNGFLDVRTSEVLKENTLDKAEKLESTSSPRASHTSASNNLKKSIGGKKPG 114
VA+ SNG V + V +E L K E +TSSP+AS+T + KK IG K+ G
Sbjct: 141 VATSQLPEVSNG---VSSYSVKEELPLSKHEATSATSSPKASNTVVPSTFKKPIGAKRTG 197
Query: 115 KSGGLGARKLNKKPSESFYEQKPEE--PPAPVPSTTNNNVS---ARPSMTSRFEYVDNVQ 169
K+GGLGARKL KP ++ YEQKPEE P P S+TNN S A S SRFEY D++Q
Sbjct: 198 KTGGLGARKLTTKPKDNLYEQKPEEVAPVIPAVSSTNNGESKSSAGSSFASRFEYNDDLQ 257
Query: 170 SSELDSRGSNTFNHVSVPKSSNFFADFGMDSGFPKKFGSNTSKVQIEESDEARKKFSNAK 229
S G+ NHV+ PKSS+FF+DFGMDS FPKK SN+SK Q+EESDEARKKF+NAK
Sbjct: 258 SGGQSVGGTQVLNHVAPPKSSSFFSDFGMDSSFPKKSSSNSSKSQVEESDEARKKFTNAK 317
Query: 230 SISSSQFFGDQNKARDAETRATLSKFSSSSAISSADFFG-DSADSSIDLAASDLINRLSF 288
SISS+Q+FGDQNK D E++ATL KF+ S++ISSADF+G D DS+ID+ ASDLINRLSF
Sbjct: 318 SISSAQYFGDQNKNADLESKATLQKFAGSASISSADFYGHDQDDSNIDITASDLINRLSF 377
Query: 289 QAQQDISSLKNIAGETGKKLSSLASSLMTDLQDRIL 324
QAQQD+SSL NIAGET KKL +LAS + +D+QDR+L
Sbjct: 378 QAQQDLSSLVNIAGETKKKLGTLASGIFSDIQDRML 413
>gb|AAK25887.1| unknown protein [Arabidopsis thaliana]
Length = 413
Score = 383 bits (983), Expect = e-105
Identities = 210/336 (62%), Positives = 253/336 (74%), Gaps = 15/336 (4%)
Query: 1 MSFGGNSRAQIFFKQHGWTDGGKIEAKYTSRAAELYRQILTKEVAKSMALE--KGLPSSP 58
M FGGN+RAQ+FFKQHGWTDGGKIEAKYTSRAA+LYRQIL KEVAK++A E GL SSP
Sbjct: 81 MMFGGNNRAQVFFKQHGWTDGGKIEAKYTSRAADLYRQILAKEVAKAIAEETNSGLLSSP 140
Query: 59 VASQS----SNGFLDVRTSEVLKENTLDKAEKLESTSSPRASHTSASNNLKKSIGGKKPG 114
VA+ SNG V + V +E L K E +TSSP+AS+T + KK IG K+ G
Sbjct: 141 VATSQLPEVSNG---VSSYSVKEELPLSKHEATSATSSPKASNTVVPSTFKKPIGAKRTG 197
Query: 115 KSGGLGARKLNKKPSESFYEQKPEE--PPAPVPSTTNNNVS---ARPSMTSRFEYVDNVQ 169
K+GGLGARKL KP ++ YEQKPEE P P S+TNN S A S SRFEY D++Q
Sbjct: 198 KTGGLGARKLTTKPKDNLYEQKPEEVAPVIPAVSSTNNGESKSSAGSSFASRFEYNDDLQ 257
Query: 170 SSELDSRGSNTFNHVSVPKSSNFFADFGMDSGFPKKFGSNTSKVQIEESDEARKKFSNAK 229
S G+ NHV+ PKSS+FF+DFGMDS FPKK SN+SK Q+EESDEAR+KF+NAK
Sbjct: 258 SGGQSVGGTQVLNHVAPPKSSSFFSDFGMDSSFPKKSSSNSSKSQVEESDEAREKFTNAK 317
Query: 230 SISSSQFFGDQNKARDAETRATLSKFSSSSAISSADFFG-DSADSSIDLAASDLINRLSF 288
SISS+Q+FGDQNK D E++ATL KF+ S++ISSADF+G D DS+ID+ ASDLINRLSF
Sbjct: 318 SISSAQYFGDQNKNADLESKATLQKFAGSASISSADFYGHDQDDSNIDITASDLINRLSF 377
Query: 289 QAQQDISSLKNIAGETGKKLSSLASSLMTDLQDRIL 324
QAQQD+SSL NIAGET KKL +LAS + +D+QDR+L
Sbjct: 378 QAQQDLSSLVNIAGETKKKLGTLASGIFSDIQDRML 413
>dbj|BAD43015.1| unknown protein [Arabidopsis thaliana]
Length = 413
Score = 383 bits (983), Expect = e-105
Identities = 211/336 (62%), Positives = 252/336 (74%), Gaps = 15/336 (4%)
Query: 1 MSFGGNSRAQIFFKQHGWTDGGKIEAKYTSRAAELYRQILTKEVAKSMALE--KGLPSSP 58
M FGGN+RAQ+FFKQHGWTDGGKIEAKYTSRAA+LYRQIL KEVAK++A E GL SSP
Sbjct: 81 MMFGGNNRAQVFFKQHGWTDGGKIEAKYTSRAADLYRQILAKEVAKAIAEETNSGLLSSP 140
Query: 59 VASQS----SNGFLDVRTSEVLKENTLDKAEKLESTSSPRASHTSASNNLKKSIGGKKPG 114
VA+ SNG V + V +E L K E +TSSP+AS+T + KK IG K+ G
Sbjct: 141 VATSQLPEVSNG---VSSYSVKEELPLSKHEATSATSSPKASNTVVPSTFKKPIGAKRTG 197
Query: 115 KSGGLGARKLNKKPSESFYEQKPEE--PPAPVPSTTNNNVS---ARPSMTSRFEYVDNVQ 169
K+GGLGARKL KP ++ YEQKPEE P P S+TNN S A S SRFEY D++Q
Sbjct: 198 KTGGLGARKLTTKPKDNLYEQKPEEVAPVIPAVSSTNNGESKSSAGSSFASRFEYNDDLQ 257
Query: 170 SSELDSRGSNTFNHVSVPKSSNFFADFGMDSGFPKKFGSNTSKVQIEESDEARKKFSNAK 229
S G+ NHV+ PKSS+FF+DFGMDS FPKK SN+SK Q+EESDEARKKF+NAK
Sbjct: 258 SGGQSVGGTQVLNHVAPPKSSSFFSDFGMDSSFPKKSSSNSSKSQVEESDEARKKFTNAK 317
Query: 230 SISSSQFFGDQNKARDAETRATLSKFSSSSAISSADFFG-DSADSSIDLAASDLINRLSF 288
SISS+Q+FGDQNK D E++ATL KF+ S++ISSADF G D DS+ID+ ASDLINRLSF
Sbjct: 318 SISSAQYFGDQNKNADLESKATLQKFAGSASISSADFHGHDQDDSNIDITASDLINRLSF 377
Query: 289 QAQQDISSLKNIAGETGKKLSSLASSLMTDLQDRIL 324
QAQQD+SSL NIAGET KKL +LAS + +D+QDR+L
Sbjct: 378 QAQQDLSSLVNIAGETKKKLGTLASGIFSDIQDRML 413
>gb|AAP55136.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|37537094|ref|NP_922849.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
gi|12643061|gb|AAK00450.1| unknown protein [Oryza
sativa]
Length = 407
Score = 353 bits (905), Expect = 5e-96
Identities = 201/332 (60%), Positives = 243/332 (72%), Gaps = 14/332 (4%)
Query: 1 MSFGGNSRAQIFFKQHGWTDGGKIEAKYTSRAAELYRQILTKEVAKSMALE--KGLPSSP 58
M +GGN+RAQ FFKQHGWTDGGKIEAKYTSRAA+LYRQ+L K+VAK+ + PSSP
Sbjct: 82 MVYGGNNRAQAFFKQHGWTDGGKIEAKYTSRAADLYRQLLAKDVAKNSTEDGNNSWPSSP 141
Query: 59 VA-SQSSN---GFLDVRTSEVLKENTLDKAEKLESTSSPRASHTSASNNLKKSIGGKKPG 114
VA SQ +N D++ +E KE +K E E SPRA +++ KK I KKPG
Sbjct: 142 VAASQPTNQADAIPDLKLAEASKEVANEKTEP-EVIRSPRAP----THSFKKPIVAKKPG 196
Query: 115 -KSGGLGARKLNKKPSESFYEQKPEEPPAPVPSTTNNNVSARPSMTSRFEYVDNVQSSEL 173
K+GGLGARKL KP+ES YEQKPEE +P T N+ + S TSRFEYV+N S+
Sbjct: 197 NKTGGLGARKLTSKPNESLYEQKPEELAPALPPVTENSTAKSKSHTSRFEYVENTPSAGS 256
Query: 174 DSRGSNTFNHVSVPKSSNFFADFGMDSGFPKKFGSNTSKVQIEESDEARKKFSNAKSISS 233
+S + HV+ PKSSNFF +FGMDSG+ KK SKVQIEES EAR+KFSNAKSISS
Sbjct: 257 NSEENQVIGHVAPPKSSNFFGEFGMDSGYHKKSAPGPSKVQIEESSEARQKFSNAKSISS 316
Query: 234 SQFFGDQNKARDAETRATLSKFSSSSAISSADFFGDSADSS-IDLAASDLINRLSFQAQQ 292
SQFFGDQ + + E + +L KFS SSAISSAD FG +SS +DL+ASDLINRLSFQA Q
Sbjct: 317 SQFFGDQ-ASFEKEAQVSLQKFSGSSAISSADLFGHPTNSSNVDLSASDLINRLSFQASQ 375
Query: 293 DISSLKNIAGETGKKLSSLASSLMTDLQDRIL 324
D+SS+KN+AGETGKKL+SLAS++M+DLQDRIL
Sbjct: 376 DLSSIKNMAGETGKKLTSLASNIMSDLQDRIL 407
>gb|AAM91272.1| zinc finger protein Glo3-like [Arabidopsis thaliana]
gi|9758511|dbj|BAB08919.1| zinc finger protein Glo3-like
[Arabidopsis thaliana] gi|20466454|gb|AAM20544.1| zinc
finger protein Glo3-like [Arabidopsis thaliana]
gi|15237500|ref|NP_199487.1| human Rev interacting-like
family protein / hRIP family protein [Arabidopsis
thaliana]
Length = 402
Score = 350 bits (898), Expect = 3e-95
Identities = 194/334 (58%), Positives = 241/334 (72%), Gaps = 19/334 (5%)
Query: 1 MSFGGNSRAQIFFKQHGWTDGGKIEAKYTSRAAELYRQILTKEVAKSMALEKGLPS---- 56
M FGGN+RAQ+FFKQHGW DGGKIEAKYTSRAA++YRQ L KEVAK+MA E LPS
Sbjct: 78 MMFGGNNRAQVFFKQHGWNDGGKIEAKYTSRAADMYRQTLAKEVAKAMAEETVLPSLSSV 137
Query: 57 ---SPVASQSSNGFLDVRTSEVLKENTLDKAEKLESTSSPRASHTSASNNLKKSIGGKKP 113
PV S S NGF TSE KE++L + + SSP+AS ++ KK + +K
Sbjct: 138 ATSQPVES-SENGF----TSESPKESSLKQEAAV--VSSPKASQKVVASTFKKPLVSRKS 190
Query: 114 GKSGGLGARKLNKKPSESFYEQKPEEPPAPVP--STTNNNVSARPSMTSRFEYVDNVQSS 171
GK+GGLGARKL K ++ YEQKPEEP +P S TN+ +A S SRFEY D+ QS
Sbjct: 191 GKTGGLGARKLTTKSKDNLYEQKPEEPVPVIPAASPTNDTSAAGSSFASRFEYFDDEQSG 250
Query: 172 ELDSRGSNTFNHVSVPKSSNFFADFGMDSGFPKKFGSNTSKVQIEESDEARKKFSNAKSI 231
G+ +HV+ PKSSNFF +FGMDS FPKK S++SK Q+EE+DEARKKFSNAKSI
Sbjct: 251 --GQSGTRVLSHVAPPKSSNFFNEFGMDSAFPKKSSSSSSKAQVEETDEARKKFSNAKSI 308
Query: 232 SSSQFFGDQNKARDAETRATLSKFSSSSAISSADFFGDSA-DSSIDLAASDLINRLSFQA 290
SS+QFFG+QN+ D +++ATL KFS S+AISS+D FG DS+ID+ ASDLINR+SFQA
Sbjct: 309 SSAQFFGNQNRDADLDSKATLQKFSGSAAISSSDLFGHGPDDSNIDITASDLINRISFQA 368
Query: 291 QQDISSLKNIAGETGKKLSSLASSLMTDLQDRIL 324
QQD+SS+ N+A ET KL + ASS+ +DLQDR+L
Sbjct: 369 QQDMSSIANLAEETKNKLGTFASSIFSDLQDRML 402
>gb|AAO66537.1| putative zinc finger protein [Oryza sativa (japonica
cultivar-group)] gi|50920175|ref|XP_470448.1| putative
zinc finger protein [Oryza sativa (japonica
cultivar-group)]
Length = 412
Score = 344 bits (883), Expect = 2e-93
Identities = 190/338 (56%), Positives = 248/338 (73%), Gaps = 17/338 (5%)
Query: 1 MSFGGNSRAQIFFKQHGWTDGGKIEAKYTSRAAELYRQILTKEVAKSMALEKGLPSSPVA 60
M+FGGN+RA FFKQHGWTDGGK++AKYTSRAAELYRQIL KEVAKS A + LPSSPVA
Sbjct: 78 MAFGGNNRAHAFFKQHGWTDGGKVDAKYTSRAAELYRQILQKEVAKSSA-DNVLPSSPVA 136
Query: 61 SQS----SNGFLDVRTSEVLKENTLDKAE------KLESTSSPRA-SHTSASNNLKKSIG 109
+ S+ F + + E ENT K E + T +P+A +H + + ++KKSIG
Sbjct: 137 ASQPQNPSDDFPEFKLPEAPAENTNGKQEPDVTNSQKAPTQTPKAPTHPTFATSVKKSIG 196
Query: 110 GKK-PGKSGGLGARKLNKKPSESFYEQKPEEPPAPVPSTTNNNVSARPSMTSRFEYVDNV 168
KK GK+GGLG +KL KPSES Y+QKPEEP P T + + PS+ SRFEYV+N
Sbjct: 197 AKKIGGKTGGLGVKKLTTKPSESLYDQKPEEPKPAAPVMTTSTTKSGPSLHSRFEYVENE 256
Query: 169 QSSELDSRGSNTFNHVSVPKSSNFFADFGMDSGFPKKFGSNTSKVQIEESDEARKKFSNA 228
+ + + G+ HV+ PKSSNFF ++GMD+GF KK + +K QI+E+DEARKKFSNA
Sbjct: 257 PAVDSRNGGTQMTGHVAPPKSSNFFQEYGMDNGFQKKTSTAATKTQIQETDEARKKFSNA 316
Query: 229 KSISSSQFFGDQNKARDAETRATLSKFSSSSAISSADFFG--DSADSSIDLAASDLINRL 286
K+ISSSQFFG+Q++ + + + +L KF+ SS+ISSAD FG D DS++DL+A+DLINR+
Sbjct: 317 KAISSSQFFGNQSR-EEKDAQMSLQKFAGSSSISSADLFGRRDMDDSNLDLSAADLINRI 375
Query: 287 SFQAQQDISSLKNIAGETGKKLSSLASSLMTDLQDRIL 324
SFQA QD+SSLKN+AGETGKKL+S+AS+ ++DL DRIL
Sbjct: 376 SFQASQDLSSLKNMAGETGKKLTSIASNFISDL-DRIL 412
>gb|AAM62820.1| zinc finger protein Glo3-like [Arabidopsis thaliana]
gi|3668084|gb|AAC61816.1| expressed protein [Arabidopsis
thaliana] gi|25408361|pir||H84765 hypothetical protein
At2g35210 [imported] - Arabidopsis thaliana
gi|18403775|ref|NP_565801.1| human Rev interacting-like
family protein / hRIP family protein [Arabidopsis
thaliana]
Length = 395
Score = 311 bits (798), Expect = 1e-83
Identities = 182/322 (56%), Positives = 230/322 (70%), Gaps = 13/322 (4%)
Query: 1 MSFGGNSRAQIFFKQHGWTDGGKIEAKYTSRAAELYRQILTKEVAKSMALEKG--LPSSP 58
M +GGN+RAQ+FFKQ+GW+DGGK EAKYTSRAA+LY+QIL KEVAKS A E+ PS P
Sbjct: 78 MIYGGNNRAQVFFKQYGWSDGGKTEAKYTSRAADLYKQILAKEVAKSKAEEELDLPPSPP 137
Query: 59 VASQSSNGFLDVRTSEVLKE-NTLDKAEKLESTS-SPRASHTSASNNLKKSIGGKKPGKS 116
++Q NG ++TSE LKE NTL + EK + SPR S + +KK +G KK GK+
Sbjct: 138 DSTQVPNGLSSIKTSEALKESNTLKQQEKPDVVPVSPRISRS-----VKKPLGAKKTGKT 192
Query: 117 GGLGARKLNKKPSESFYEQKPEEPPAPVPSTTNNNVSARPSMTSRFEYVDNVQSSELDSR 176
GGLGARKL K S + Y+QKPEE ++ + SAR S +SRF+Y DNVQ+ E D
Sbjct: 193 GGLGARKLTTKSSGTLYDQKPEESVIIQATSPVSAKSARSSFSSRFDYADNVQNRE-DYM 251
Query: 177 GSNTFNHVSVPKSSNFFAD-FGMDSG-FPKKFGSNTSKVQIEESDEARKKFSNAKSISSS 234
+HV+ PKSS FF + M+ G F KK +++SK+QI+E+DEARKKF+NAKSISS+
Sbjct: 252 SPQVVSHVAPPKSSGFFEEELEMNGGRFQKKPITSSSKLQIQETDEARKKFTNAKSISSA 311
Query: 235 QFFGDQNKARDAETRATLSKFSSSSAISSADFFGD-SADSSIDLAASDLINRLSFQAQQD 293
Q+FG+ N + D E +++L KFS SSAISSAD FGD D +DL A DL+NRLS QAQQD
Sbjct: 312 QYFGNDNNSADLEAKSSLKKFSGSSAISSADLFGDGDGDFPLDLTAGDLLNRLSLQAQQD 371
Query: 294 ISSLKNIAGETGKKLSSLASSL 315
ISSLKN+A ET KKL S+ASSL
Sbjct: 372 ISSLKNMAEETKKKLGSVASSL 393
>gb|AAW39027.1| putative zinc finger protein [Oryza sativa (japonica
cultivar-group)]
Length = 384
Score = 261 bits (666), Expect = 3e-68
Identities = 143/275 (52%), Positives = 191/275 (69%), Gaps = 14/275 (5%)
Query: 1 MSFGGNSRAQIFFKQHGWTDGGKIEAKYTSRAAELYRQILTKEVAKSMALEKGLPSSPVA 60
M+FGGN+RA FFKQHGWTDGGK++AKYTSRAAELYRQIL KEVAKS A + LPSSPVA
Sbjct: 78 MAFGGNNRAHAFFKQHGWTDGGKVDAKYTSRAAELYRQILQKEVAKSSA-DNVLPSSPVA 136
Query: 61 SQS----SNGFLDVRTSEVLKENTLDKAE------KLESTSSPRA-SHTSASNNLKKSIG 109
+ S+ F + + E ENT K E + T +P+A +H + + ++KKSIG
Sbjct: 137 ASQPQNPSDDFPEFKLPEAPAENTNGKQEPDVTNSQKAPTQTPKAPTHPTFATSVKKSIG 196
Query: 110 GKK-PGKSGGLGARKLNKKPSESFYEQKPEEPPAPVPSTTNNNVSARPSMTSRFEYVDNV 168
KK GK+GGLG +KL KPSES Y+QKPEEP P T + + PS+ SRFEYV+N
Sbjct: 197 AKKIGGKTGGLGVKKLTTKPSESLYDQKPEEPKPAAPVMTTSTTKSGPSLHSRFEYVENE 256
Query: 169 QSSELDSRGSNTFNHVSVPKSSNFFADFGMDSGFPKKFGSNTSKVQIEESDEARKKFSNA 228
+ + + G+ HV+ PKSSNFF ++GMD+GF KK + +K QI+E+DEARKKFSNA
Sbjct: 257 PAVDSRNGGTQMTGHVAPPKSSNFFQEYGMDNGFQKKTSTAATKTQIQETDEARKKFSNA 316
Query: 229 KSISSSQFFGDQNKARDAETRATLSKFSSSSAISS 263
K+ISSSQFFG+Q++ + + + +L KF+ +++S
Sbjct: 317 KAISSSQFFGNQSR-EEKDAQMSLQKFAVRISLAS 350
>ref|NP_973603.1| human Rev interacting-like family protein / hRIP family protein
[Arabidopsis thaliana]
Length = 371
Score = 241 bits (614), Expect = 3e-62
Identities = 140/264 (53%), Positives = 184/264 (69%), Gaps = 12/264 (4%)
Query: 1 MSFGGNSRAQIFFKQHGWTDGGKIEAKYTSRAAELYRQILTKEVAKSMALEK--GLPSSP 58
M +GGN+RAQ+FFKQ+GW+DGGK EAKYTSRAA+LY+QIL KEVAKS A E+ PS P
Sbjct: 78 MIYGGNNRAQVFFKQYGWSDGGKTEAKYTSRAADLYKQILAKEVAKSKAEEELDLPPSPP 137
Query: 59 VASQSSNGFLDVRTSEVLKE-NTLDKAEKLESTS-SPRASHTSASNNLKKSIGGKKPGKS 116
++Q NG ++TSE LKE NTL + EK + SPR S ++KK +G KK GK+
Sbjct: 138 DSTQVPNGLSSIKTSEALKESNTLKQQEKPDVVPVSPR-----ISRSVKKPLGAKKTGKT 192
Query: 117 GGLGARKLNKKPSESFYEQKPEEPPAPVPSTTNNNVSARPSMTSRFEYVDNVQSSELDSR 176
GGLGARKL K S + Y+QKPEE ++ + SAR S +SRF+Y DNVQ+ E D
Sbjct: 193 GGLGARKLTTKSSGTLYDQKPEESVIIQATSPVSAKSARSSFSSRFDYADNVQNRE-DYM 251
Query: 177 GSNTFNHVSVPKSSNFF-ADFGMDSG-FPKKFGSNTSKVQIEESDEARKKFSNAKSISSS 234
+HV+ PKSS FF + M+ G F KK +++SK+QI+E+DEARKKF+NAKSISS+
Sbjct: 252 SPQVVSHVAPPKSSGFFEEELEMNGGRFQKKPITSSSKLQIQETDEARKKFTNAKSISSA 311
Query: 235 QFFGDQNKARDAETRATLSKFSSS 258
Q+FG+ N + D E +++L KFS S
Sbjct: 312 QYFGNDNNSADLEAKSSLKKFSQS 335
>gb|AAQ91219.1| ADP-ribosylation factor GTPase activating protein 3 [Danio rerio]
gi|45387621|ref|NP_991160.1| ADP-ribosylation factor
GTPase activating protein 3 [Danio rerio]
Length = 498
Score = 92.0 bits (227), Expect = 2e-17
Identities = 111/431 (25%), Positives = 169/431 (38%), Gaps = 123/431 (28%)
Query: 1 MSFGGNSRAQIFFKQHGWTDGGKIEAKYTSRAAELYR---QILTKEVAKSMALEKGL--- 54
M GGN+ A FF QHG + AKY+SRAA LYR + L + + E L
Sbjct: 79 MQVGGNASANAFFSQHGCSSSSAANAKYSSRAAALYRDKIRALANQATRQHGTELWLDAQ 138
Query: 55 ----PSSPVASQSSNGFLDVRTSEVL-----------KENTLDKAEKLESTSSPRASHTS 99
PSSP+ Q F T L ++ D +S + P S
Sbjct: 139 APLSPSSPLDKQED--FFTQHTQSALPDTVQLNISQSQKRVQDNNNNAKSEAGPSVEQLS 196
Query: 100 ASN----------------NLKKSIGGKKPGKSGGLGARKLNKK---------------- 127
S ++KS GG G+ GLGA+K++ +
Sbjct: 197 VSPVQSAAEPLSLLKKKPAAVRKSAGG---GRRAGLGAQKVSSQSFTLQQRRAQAEDRLT 253
Query: 128 -------PSESFYEQKPEEPPAPVPSTTNNNVSAR------------------------- 155
P S + + V + ++ S+R
Sbjct: 254 EQRTSTAPDHSVWSVSRVQQAVDVQRSAQDSSSSRMKQQQEERLGMGLSGRSGLSHSVTE 313
Query: 156 ----------PSMTSRFEYVDNVQSSEL--------DSRGSNT--FNHVSVPKSSNFF-- 193
PS++SR Y ++ Q + + RGS+ F+ S K S
Sbjct: 314 DMQTIIQQDCPSLSSRRVYEEDEQQEDTHFCSRALDEGRGSSDLFFSQWSSEKQSRMKPE 373
Query: 194 ADFGMDSGFPKKFGSNTSKVQIEESDEARKKFSNAKSISSSQFFGDQNKARDAETRATLS 253
+D+ ++ + S + K D+A++KF +AK+ISS FFG Q+++ + E RA L
Sbjct: 374 SDYCVED---EHRASASRKEPQSVCDDAQRKFGDAKAISSDMFFGTQDRS-EYEVRARLE 429
Query: 254 KFSSSSAISSADFFGDSADSSIDLAASDLINRLS--FQAQQDISSLKNIAGETGKKLSSL 311
FSSSSAISSAD F + AA RLS + D++ L++ KLS +
Sbjct: 430 NFSSSSAISSADLFDEQKK-----AAGSSSYRLSSVLSSVPDMTQLRSGVRSVAGKLSGM 484
Query: 312 ASSLMTDLQDR 322
AS +++ +QDR
Sbjct: 485 ASGVVSTIQDR 495
>emb|CAI21510.1| OTTHUMP00000028526 [Homo sapiens] gi|56203507|emb|CAI20950.1|
OTTHUMP00000028526 [Homo sapiens]
gi|7208833|emb|CAB76901.1| hypothetical protein [Homo
sapiens] gi|21263420|sp|Q9NP61|ARFG3_HUMAN
ADP-ribosylation factor GTPase-activating protein 3 (ARF
GAP 3) gi|7211442|gb|AAF40310.1| ARFGAP1 protein [Homo
sapiens]
Length = 516
Score = 87.0 bits (214), Expect = 7e-16
Identities = 114/443 (25%), Positives = 171/443 (37%), Gaps = 129/443 (29%)
Query: 1 MSFGGNSRAQIFFKQHGWTDGGKIEAKYTSRAAELYRQILTKEVAKSMALEKGL------ 54
M GGN+ A FF QHG + AKY SRAA+LYR+ + K +A + G
Sbjct: 79 MQVGGNASASSFFHQHGCSTNDT-NAKYNSRAAQLYREKI-KSLASQATRKHGTDLWLDS 136
Query: 55 ----PSSP------------------------VASQSSNGFLDVRTSEVLKENTLDKAEK 86
P SP +A SS V T+ E ++
Sbjct: 137 CVVPPLSPPPKEEDFFASHVSPEVSDTAWASAIAEPSSLTSRPVETTLENNEGGQEQGPS 196
Query: 87 LESTSSPRASHTSASNNLKKSIGGKKPG---KSGGLGARKLN-------KKPSESFYEQK 136
+E + P + S+ +KK K G K G LGA+KL +K +++ + K
Sbjct: 197 VEGLNVPTKATLEVSSIIKKKPNQAKKGLGAKKGSLGAQKLANTCFNEIEKQAQAADKMK 256
Query: 137 PEEPPAPVPSTTNN----------------------NVSARPSMTS-------------- 160
+E A V S + N+S + ++ S
Sbjct: 257 EQEDLAKVVSKEESIVSSLRLAYKDLEIQMKKDEKMNISGKKNVDSDRLGMGFGNCRSVI 316
Query: 161 ---------------------RFEYVDNVQSSELDSRGSNTFNHVSVPKSSNFFADFGMD 199
R +Y D+ S S S V + SS D D
Sbjct: 317 SHSVTSDMQTIEQESPIMAKPRKKYNDDSDDSYFTSSSSYFDEPVELRSSSFSSWDDSSD 376
Query: 200 SGFPKKFGSNTSKV-------------------QIEESDEARKKFSNAKSISSSQFFGDQ 240
S + K+ +T V +E +DEA+KKF N K+ISS +FG Q
Sbjct: 377 SYWKKETSKDTETVLKTTGYSDRPTARRKPDYEPVENTDEAQKKFGNVKAISSDMYFGRQ 436
Query: 241 NKARDAETRATLSKFSSSSAISSADFFGDSADSSI-DLAASDLINRLSFQAQQDISSLKN 299
++A D ETRA L + S+SS+ISSAD F + + + S ++ AQ +++
Sbjct: 437 SQA-DYETRARLERLSASSSISSADLFEEPRKQPAGNYSLSSVLPNAPDMAQFK-QGVRS 494
Query: 300 IAGETGKKLSSLASSLMTDLQDR 322
+AG KLS A+ ++T +QDR
Sbjct: 495 VAG----KLSVFANGVVTSIQDR 513
>emb|CAB70834.1| hypothetical protein [Homo sapiens]
Length = 473
Score = 87.0 bits (214), Expect = 7e-16
Identities = 114/443 (25%), Positives = 171/443 (37%), Gaps = 129/443 (29%)
Query: 1 MSFGGNSRAQIFFKQHGWTDGGKIEAKYTSRAAELYRQILTKEVAKSMALEKGL------ 54
M GGN+ A FF QHG + AKY SRAA+LYR+ + K +A + G
Sbjct: 36 MQVGGNASASSFFHQHGCSTNDT-NAKYNSRAAQLYREKI-KSLASQATRKHGTDLWLDS 93
Query: 55 ----PSSP------------------------VASQSSNGFLDVRTSEVLKENTLDKAEK 86
P SP +A SS V T+ E ++
Sbjct: 94 CVVPPLSPPPKEEDFFASHVSPEVSDTAWASAIAEPSSLTSRPVETTLENNEGGQEQGPS 153
Query: 87 LESTSSPRASHTSASNNLKKSIGGKKPG---KSGGLGARKLN-------KKPSESFYEQK 136
+E + P + S+ +KK K G K G LGA+KL +K +++ + K
Sbjct: 154 VEGLNVPTKATLEVSSIIKKKPNQAKKGLGAKKGSLGAQKLANTCFNEIEKQAQAADKMK 213
Query: 137 PEEPPAPVPSTTNN----------------------NVSARPSMTS-------------- 160
+E A V S + N+S + ++ S
Sbjct: 214 EQEDLAKVVSKEESIVSSLRLAYKDLEIQMKKDEKMNISGKKNVDSDRLGMGFGNCRSVI 273
Query: 161 ---------------------RFEYVDNVQSSELDSRGSNTFNHVSVPKSSNFFADFGMD 199
R +Y D+ S S S V + SS D D
Sbjct: 274 SHSVTSDMQTIEQESPIMAKPRKKYNDDSDDSYFTSSSSYFDEPVELRSSSFSSWDDSSD 333
Query: 200 SGFPKKFGSNTSKV-------------------QIEESDEARKKFSNAKSISSSQFFGDQ 240
S + K+ +T V +E +DEA+KKF N K+ISS +FG Q
Sbjct: 334 SYWKKETSKDTETVLKTTGYSDRPTARRKPDYEPVENTDEAQKKFGNVKAISSDMYFGRQ 393
Query: 241 NKARDAETRATLSKFSSSSAISSADFFGDSADSSI-DLAASDLINRLSFQAQQDISSLKN 299
++A D ETRA L + S+SS+ISSAD F + + + S ++ AQ +++
Sbjct: 394 SQA-DYETRARLERLSASSSISSADLFEEPRKQPAGNYSLSSVLPNAPDMAQFK-QGVRS 451
Query: 300 IAGETGKKLSSLASSLMTDLQDR 322
+AG KLS A+ ++T +QDR
Sbjct: 452 VAG----KLSVFANGVVTSIQDR 470
>ref|XP_629863.1| hypothetical protein DDB0184259 [Dictyostelium discoideum]
gi|60463237|gb|EAL61430.1| hypothetical protein
DDB0184259 [Dictyostelium discoideum]
Length = 522
Score = 83.2 bits (204), Expect = 1e-14
Identities = 58/200 (29%), Positives = 106/200 (53%), Gaps = 15/200 (7%)
Query: 125 NKKPSES-FYEQKPEEPPAPVPSTTNNNVSARPSMTSRFEYVDNVQSSELDSRGSNTFNH 183
NK+ S S Y QK + NNN + + + +N ++ + SN N+
Sbjct: 334 NKQDSSSPLYSQKYNNSNNSNNNNNNNNNNNNNNNNNNNNSYNNNNNNN-NKNYSNNSNN 392
Query: 184 VSVPKSSNFFADFGMDSGFPKKFGSNTSKVQIEESDEARKKFSNAKSISSSQFFGDQNKA 243
S ++ ++ P+ N + E+D ARK F+NAKSISSS ++G+ +
Sbjct: 393 NSYNNNNR-----SPNNQSPRNNNENKNYQHQNETDYARKNFTNAKSISSSTYYGEDKEK 447
Query: 244 RDAETRATLSKFSSSSAISSADFFG-DSADSSIDLAASDLINRLSFQAQQDISSLKNIAG 302
D++ + +SKF++S +ISSA ++ D S + +AS++ L++ A+ D++S+ N
Sbjct: 448 VDSDKQQRISKFTNSKSISSAQYYDRDETPSFSERSASNIARDLAYNARSDLTSISN--- 504
Query: 303 ETGKKLSSLASSLMTDLQDR 322
K+S++A++++ DLQDR
Sbjct: 505 ----KISNIANNIINDLQDR 520
Score = 49.7 bits (117), Expect = 1e-04
Identities = 71/285 (24%), Positives = 117/285 (40%), Gaps = 42/285 (14%)
Query: 1 MSFGGNSRAQIFFKQHGWTDGGKIEAKYTSRAAELYRQILTKEVAKSMALEKGLPSSPVA 60
M GGN A+ +F++HG D E+KY S+ Y+QIL V K++ SP +
Sbjct: 79 MELGGNQVAKQYFQEHG-GDIRDTESKYQSQVGINYKQILDARVKKALK-----EISPFS 132
Query: 61 SQSSNGFLDVRTSEVLKENTLDKAEKLESTSSPRASHTSASNNLKKSIGGKKPGKSGGLG 120
S SS T L +T++ + +TSSP S S+S S G G
Sbjct: 133 STSSTPLQAPTTPSPLSISTIN-SNPSTTTSSPMFSTNSSS--------------SSGSG 177
Query: 121 ARKLNKKPSESFYEQKPEEPPAPVPSTTNNNVSARPSMTSRFEYVDNVQSSELDS----- 175
+S +Q+ ++ +P S ++ VS++ T+ S + DS
Sbjct: 178 I-------LQSKQQQQQQQQQSPTTSFSDKKVSSQQKSTTTTTTTTKNDSDDFDSFFSSS 230
Query: 176 -RGSNTFNHVSVPKSSNFFADFGMDSGFPK---KFGSNTSKVQIEESDEARKKFSNAKSI 231
S + V+ K D DS K KF +N + +KK ++
Sbjct: 231 PPKSTSTKKVATKKIDQKSFDDDWDSLPDKKEEKFDNNNNNNNNNNYKNNKKKGGSSFKF 290
Query: 232 SSSQFFGDQNKARDAETRATLSKFSSSSAISSADFFGDSADSSID 276
++ D++++ D+ R+ LS S +S+ F S D S+D
Sbjct: 291 NNEYIENDEDES-DSNNRSDLSP-ELRSLYTSSKF---SMDFSVD 330
>ref|XP_330928.1| hypothetical protein [Neurospora crassa] gi|28923588|gb|EAA32762.1|
hypothetical protein [Neurospora crassa]
Length = 496
Score = 79.3 bits (194), Expect = 1e-13
Identities = 100/408 (24%), Positives = 156/408 (37%), Gaps = 100/408 (24%)
Query: 1 MSFGGNSRAQIFFKQHGWT---DGGKIEAKYTSRAAELYRQILTKEVAKSM--------- 48
M GGN A FF+Q+G + + + KY S AA Y++ L K A+
Sbjct: 79 MKVGGNESATKFFQQNGGSAALNSKDPKTKYQSAAATKYKEELKKRAARDAREYPEEVVI 138
Query: 49 ---------------------------ALEKGLP------SSPVASQSSNGFLDVRTSEV 75
A++K P + PV ++++ L
Sbjct: 139 TDGDDATSSNTPAGEPDDDFFSSWDKPAIKKPTPPISRTSTPPVIGRTASPLLGNGKDIQ 198
Query: 76 LKENTLDKAEKLESTSSPRASHTSASNNLKKSIGGKK----PGKSGG-----------LG 120
+ L K + T +P AS + S L+K+ G P K GG LG
Sbjct: 199 RTSSPLSKTDSDAPTPAPAASRITTSAALRKTTPGSSTTGGPRKVGGGILGAKKPAAKLG 258
Query: 121 ARKLN----------KKPSES--------FYEQKPEEPPAPVPSTTNNNV----SARPSM 158
+K++ KK E + + EE A T NN+ +A S
Sbjct: 259 VKKISADLIDFDEAEKKAKEEAERIEKLGYDPEAEEEKAAKKAETKTNNIITPAAAPVSS 318
Query: 159 TSRFEYVDNVQSSELDSRGSNT----FNHVSVPKSSNFFADFGMDSGFPKKFGS--NTSK 212
+S ++E++ G F V P + A G + K G +
Sbjct: 319 SSSRSAAQEKSAAEVERLGMGVRKLGFGMVGKPGGAGAAAGAGGAAAAKKNAGGFGSVGP 378
Query: 213 VQIEESDE---ARKKFSNAKSISSSQFF--GDQNKARDAETRATLSKFSSSSAISSADFF 267
++ E+DE AR KF+N K+ISS +FF G+ + + AET+A L F + AISS +F
Sbjct: 379 IKASEADEEQYARNKFANQKAISSDEFFNKGNYDPSVKAETKARLQGFEGAQAISSNAYF 438
Query: 268 GDSADSS-------IDLAASDLINRLSFQAQQDISSLKNIAGETGKKL 308
G D + ++ AA D I + A D+ +L + GE +L
Sbjct: 439 GRPEDDAPAEDYGDLESAAKDFIRKFGITASDDLENLTQMVGEGAGRL 486
>gb|EAK98866.1| potential ARF GAP [Candida albicans SC5314]
gi|46439448|gb|EAK98766.1| potential ARF GAP [Candida
albicans SC5314]
Length = 451
Score = 72.4 bits (176), Expect = 2e-11
Identities = 89/366 (24%), Positives = 153/366 (41%), Gaps = 61/366 (16%)
Query: 3 FGGNSRAQIFFKQHGWT------DGGKIEAKYTSRAAELYRQILTKEVAKSMALEKGLPS 56
FGGN +A+ FF ++G + +G AKYTS A Y++ L ++ A+ A + +
Sbjct: 82 FGGNQQAKDFFLKNGGSQFVNNKNGVDATAKYTSPCANKYKEKLKQKAAQDAAKHPDIVT 141
Query: 57 ----------SPVASQSSNGFLDVRTSEVLKENTLDKAEKLESTSSP------------R 94
S S+S++ F V NT +T + R
Sbjct: 142 LDDVTDVMSLSDSPSESTDDFFSNWNKPVNNSNTASPLSSRAATPNASTDDLTKKKPVVR 201
Query: 95 ASHTSA---SNN--LKKSI-GGKKPG-KSGGLGARKLNKKPSESFYEQKPEEPPAPVPST 147
+ TSA SNN +KKSI GK G ++ L A+++NK + +++ E A +
Sbjct: 202 TTTTSARLKSNNTAVKKSILSGKGNGPRTSRLAAKRINKTEDDIDFDEI--EKKAKQEAE 259
Query: 148 TNNNVSARPSMTSRFEYVDNVQSSELDSRGSNT---FNHVSVPKSSNFFAD--FGMDSGF 202
+ +P+ T S+ L + N + +++ F FGM G
Sbjct: 260 EAKKLGYKPTETVEPTVKKQPTSTSLSLKKENEEVKLTPAPIQETTQQFQKLGFGMTQG- 318
Query: 203 PKKFGSNTSKV-QIEESDEARKKFSNAKSISSSQFFGDQ---NKARDAETRATLSKFSSS 258
GS+T K +++ + E K+ K ISS +FFG ++ E R L F+ +
Sbjct: 319 DNIVGSSTKKYKEVKYTGEVSNKYGTQKGISSDEFFGRGPRFDEQAKTEARTKLQAFNGA 378
Query: 259 SAISSADFFGDSADSS--------------IDLAASDLINRLSFQAQQDISSLKNIAGET 304
+ISS+ +FG+ + ++ +A + ++ S A QD+ LK+ +
Sbjct: 379 QSISSSSYFGEEEGGARGGRSNSGAGGLGDLEASAREFASKFSGNANQDLEVLKDALEDG 438
Query: 305 GKKLSS 310
KL S
Sbjct: 439 ATKLGS 444
>ref|NP_001016210.1| hypothetical protein LOC548964 [Xenopus tropicalis]
Length = 535
Score = 72.0 bits (175), Expect = 2e-11
Identities = 81/303 (26%), Positives = 133/303 (43%), Gaps = 43/303 (14%)
Query: 34 ELYRQILTKEVAKSMALEKGLPSSPVASQSSNGFLDV-----RTSEVLKENTLDKAEKLE 88
E Q + K+ + A PV + + D+ + E LK KA + E
Sbjct: 259 ERQAQAVDKQKEQEAATVTKRAEEPVVASLRLAYKDLEIQKQKDEEKLKNLPPKKAAEAE 318
Query: 89 STSSPRASHTSASNNLKKSIGGKKPGKSGGLGARKL-NKKPSESFYEQKP-----EEPPA 142
+ + S+++ + + + A+KL ++ +E FY P E+PP
Sbjct: 319 RLGMGFGARSGISHSVLSEMTTIEQEAPKSVKAKKLYQEEEAEDFYFSAPRSRYREDPPE 378
Query: 143 PVPSTTNNNVSARPSMTSRFEYVDNVQSSELDSRGSNTFNHVSVPKSSNFFADFGMDSGF 202
PV S S S E D+ ++DS G++T V++ + + D
Sbjct: 379 PVTS----------SFLSWDERSDDYW--KMDSPGADT--EVALTSKTTSYDDRP----- 419
Query: 203 PKKFGSNTSKVQIE---ESDEARKKFSNAKSISSSQFFGDQNKARDAETRATLSKFSSSS 259
+ K +E +D+A+KKF NAK+ISS FFG Q+ A D E R+ L + S +S
Sbjct: 420 -----ATRRKHDLEPAPSTDDAQKKFGNAKAISSDMFFGKQDSA-DYEARSRLDRLSGNS 473
Query: 260 AISSADFFGDSADSSIDLAASDLINRLSFQAQQDISSLKNIAGETGKKLSSLASSLMTDL 319
+ISSAD F + D + + R+ A D+++ K +LS LA+ +MT +
Sbjct: 474 SISSADLFDEQKK---DPTGNYTLTRV-LPAAPDMANFKQGVKSVAGRLSVLANGVMTTI 529
Query: 320 QDR 322
QDR
Sbjct: 530 QDR 532
>ref|XP_315621.2| ENSANGP00000007262 [Anopheles gambiae str. PEST]
gi|55238411|gb|EAA11857.2| ENSANGP00000007262 [Anopheles
gambiae str. PEST]
Length = 519
Score = 71.2 bits (173), Expect = 4e-11
Identities = 45/116 (38%), Positives = 68/116 (57%), Gaps = 6/116 (5%)
Query: 210 TSKVQIEESDEARKKFSNAKSISSSQFFGDQNKARDAETRATLSKFSSSSAISSADFFGD 269
+S + ++ SD A+KKF +AK ISS QFFGD+ + E A LSKF S++ISSAD+FG
Sbjct: 404 SSSISVDSSDVAQKKFGSAKGISSDQFFGDEQSS--YERSANLSKFQGSTSISSADYFGH 461
Query: 270 SADSSIDLAASDLINRLSFQAQ--QDI-SSLKNIAGETGKKLSSLASSLMTDLQDR 322
+ SS+ L + +D+ S++ + +LSSLAS +M +QD+
Sbjct: 462 GS-SSMGSGGGPRGPALQYNGPDLEDVRESVRQGVTKVAGRLSSLASDVMNSIQDK 516
Score = 44.3 bits (103), Expect = 0.005
Identities = 84/382 (21%), Positives = 132/382 (33%), Gaps = 94/382 (24%)
Query: 1 MSFGGNSRAQIFFKQHGW--TDGGKIEAKYTSRAAELYRQILTKEVAKSMAL-------E 51
M GGN+ A FF+QH TD + KY SRAA+LY+ L + +S+ L +
Sbjct: 82 MQVGGNANAAQFFRQHNCNTTDA---QQKYNSRAAQLYKDKLLNKAQQSLQLYGTTLHID 138
Query: 52 KGLPSSPVASQSSN-------GFLDVRTSEVLKENTLDKAEKL----------ESTSSPR 94
+ AS+ D T+ V + N K+ + T+ PR
Sbjct: 139 NAHDLNTTASEKKEMDFFADCDNFDNDTATVERNNNPSSGPKIPKLASLSATVDPTAGPR 198
Query: 95 ASHTSAS---NNLKKSIGGKK-PGKSGGLGAR--------------------------KL 124
++ +K +IG +K K GGLGA+ KL
Sbjct: 199 VDFLTSEVPVEPVKSTIGVRKIQPKKGGLGAKKGGLGATRVKTNFAEIEERANMADKLKL 258
Query: 125 NKKPSESFYEQKPEEPPAPVPSTTNNNVSARPSMTSRFEYVDNVQSSELDSRGSNTFNHV 184
P + E++ E A V + + + + +D ++ +++ G
Sbjct: 259 APAPEKPITEEEKAETLASVRLAYQDLSIKQHREEEKLKAIDPNKAKQIERLGMGFGRAA 318
Query: 185 SVPKS-----------------------SNFFADFGMDSGFP-KKFGSNTSKV------Q 214
V S + FF D+ + GF G++ S+
Sbjct: 319 GVSHSALTDMKTLTPEEATRSSAASLSKALFFDDYSIIYGFSGSGKGADMSEATQMGFDT 378
Query: 215 IEESDEARKKFSNAKSISSSQFFGDQNKARDAETRATLSKFSSSSAISSADFFGDSADSS 274
+E D R S + G + + KF S+ ISS FFGD SS
Sbjct: 379 LEPIDSKRPTVKTMFSPAGGSLKGGSSSISVDSSDVAQKKFGSAKGISSDQFFGDE-QSS 437
Query: 275 IDLAASDLINRLSFQAQQDISS 296
+ +A N FQ ISS
Sbjct: 438 YERSA----NLSKFQGSTSISS 455
>emb|CAH65393.1| hypothetical protein [Gallus gallus]
gi|60302730|ref|NP_001012557.1| similar to
ADP-ribosylation factor GTPase-activating protein 3 (ARF
GAP 3) [Gallus gallus]
Length = 440
Score = 70.5 bits (171), Expect = 6e-11
Identities = 97/333 (29%), Positives = 146/333 (43%), Gaps = 40/333 (12%)
Query: 21 GGKIEAKYTSRAAELYRQILTK----EVAKSMALEKG-LPSSPVASQSSNGF-LDVRTSE 74
G ++ TS A L K +V K + +KG L + V+SQS N + +
Sbjct: 114 GPSVDCLSTSPKATLENTSFIKKKPNQVKKGLGAKKGGLGAQKVSSQSFNEIEKQAQAVD 173
Query: 75 VLKENT-LDKAEKLESTSSPRASHTSASNNL-------KKSIGGKKPGK----SGGLGAR 122
+KE L + K+E +S A +L K ++ GKK + G G+
Sbjct: 174 KMKEQEDLHSSRKIEMEEPLVSSLRLAYRDLDIKTKEEKLNLSGKKKNELERLGMGFGSN 233
Query: 123 K--LNKKPSESFYEQKPEEPPAPVPSTT-NNNVSARPSMTSRFEYVDNVQSSELDSRGSN 179
+ ++ + E P P ++V +S Y D SS+L S +
Sbjct: 234 RSGISHSVLSDMQTIEQETPTIGKPKKKYTDDVEDSYFSSSSSRYYD---SSDLRSNTFS 290
Query: 180 TFNHVSVP----KSSNFFADFGM---DSGFPKKFGSNTSKVQIEES---DEARKKFSNAK 229
++ S P +S+N D + ++GF + S K + E S DEA+KKF N K
Sbjct: 291 KWDDNSDPFWKKESNNRDVDVILTPKNTGFSDRPASRR-KPEYEPSLSTDEAQKKFGNVK 349
Query: 230 SISSSQFFGDQNKARDAETRATLSKFSSSSAISSADFFGDSADSSIDLAASDLINRLSFQ 289
+ISS +FG Q+ A D E RA L + S SS+ISSAD F D ++ N L
Sbjct: 350 AISSDMYFGRQDHA-DYEARARLERLSGSSSISSADLFEDQKKQ--PAGGYNITNVL--P 404
Query: 290 AQQDISSLKNIAGETGKKLSSLASSLMTDLQDR 322
+ DI+ K KLS LA+ +MT +QDR
Sbjct: 405 SAPDIAQFKQGVKSVAGKLSVLANGVMTSIQDR 437
>dbj|BAE02036.1| unnamed protein product [Macaca fascicularis]
Length = 516
Score = 70.5 bits (171), Expect = 6e-11
Identities = 82/275 (29%), Positives = 128/275 (45%), Gaps = 48/275 (17%)
Query: 77 KENTLDKAEKLES-TSSPRASHTSASNNLKK----SIGGKKPGKSGGLGARKLNKKPSES 131
+E+ A K ES SS R ++ +KK +I GKK S LG N + S
Sbjct: 258 QEDLAKAAPKEESIVSSLRLAYKDLEIQMKKDEKLNISGKKNVDSDRLGMGFGNCRSGIS 317
Query: 132 FY---EQKPEEPPAPVPSTTNNNVSARPSMTSRFEYVDNVQSSELDSRGSNTFNHVSVPK 188
+ + E +P+ A+P R +Y D+ S S S F+ +
Sbjct: 318 HSVTSDMQTIEQESPI--------MAKP----RKKYNDDSDDSYFTS-SSRYFDEAVELR 364
Query: 189 SSNFFA-DFGMDSGFPKKFGSNTSKV-------------------QIEESDEARKKFSNA 228
SS+F + D DS + K+ +T + +E +DEA+KKF N
Sbjct: 365 SSSFSSWDDSSDSYWKKETSKDTETILKTTGYSDRPTARRKPDYEPVENTDEAQKKFGNV 424
Query: 229 KSISSSQFFGDQNKARDAETRATLSKFSSSSAISSADFFGDSADSSI-DLAASDLINRLS 287
K+ISS +FG Q +A D ETRA L + S+SS+ISSAD F + + + + S+++
Sbjct: 425 KAISSDMYFGRQAQA-DYETRARLERLSASSSISSADLFEEQRKQAAGNYSLSNVLPSAP 483
Query: 288 FQAQQDISSLKNIAGETGKKLSSLASSLMTDLQDR 322
AQ ++++AG KLS A+ ++T +QDR
Sbjct: 484 NMAQFK-QGVRSVAG----KLSVFANGVVTSIQDR 513
Score = 47.0 bits (110), Expect = 8e-04
Identities = 86/364 (23%), Positives = 141/364 (38%), Gaps = 71/364 (19%)
Query: 1 MSFGGNSRAQIFFKQHGWTDGGKIEAKYTSRAAELYRQILTKEVAKSMALEKGL------ 54
M GGN+ A FF QHG + AKY SRAA+LYR+ + K +A + G
Sbjct: 79 MQVGGNANASSFFHQHGCST-SDTNAKYNSRAAQLYREKI-KSLASQATRKHGTDLWLDS 136
Query: 55 ----PSSP------------------------VASQSSNGFLDVRTSEVLKENTLDKAEK 86
P SP +A SS V T+ E ++
Sbjct: 137 CVVPPLSPPPKEEDFFASHVSPEVSDTAWASAIAEPSSLTSRPVETTLENNEGGQEQGPS 196
Query: 87 LESTSSPRASHTSASNNLKKSIGGKKPG---KSGGLGARKLN-------KKPSESFYEQK 136
+E + P + S+ +KK K G K G LGA+KL +K +++ + K
Sbjct: 197 VEGLNVPSKAALEVSSIIKKKPNQAKKGLGAKKGSLGAQKLANTCFNEIEKQAQAADKMK 256
Query: 137 PEEPPAPVPSTTNNNVSARPSMTSRFEYVDNVQSSELDSRGSNTFNHVSVPKSSNFFAD- 195
+E A + VS + R Y D + D + +++ N +D
Sbjct: 257 EQEDLAKAAPKEESIVS-----SLRLAYKDLEIQMKKDEK-------LNISGKKNVDSDR 304
Query: 196 FGMDSGFPKKFGSN--TSKVQIEESD-----EARKKFSNAKSISSSQFFGDQNKARDAET 248
GM G + S+ TS +Q E + + RKK+++ S +F ++ D
Sbjct: 305 LGMGFGNCRSGISHSVTSDMQTIEQESPIMAKPRKKYNDD---SDDSYFTSSSRYFDEAV 361
Query: 249 RATLSKFSSSSAISSADFFGD-SADSSIDLAASDLINRLSFQAQQDISSLKNIAGETGKK 307
S FSS S + + + S D+ L + +R + + + D ++N E KK
Sbjct: 362 ELRSSSFSSWDDSSDSYWKKETSKDTETILKTTGYSDRPTARRKPDYEPVEN-TDEAQKK 420
Query: 308 LSSL 311
++
Sbjct: 421 FGNV 424
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.306 0.123 0.324
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 494,563,845
Number of Sequences: 2540612
Number of extensions: 19898221
Number of successful extensions: 86266
Number of sequences better than 10.0: 751
Number of HSP's better than 10.0 without gapping: 118
Number of HSP's successfully gapped in prelim test: 652
Number of HSP's that attempted gapping in prelim test: 80359
Number of HSP's gapped (non-prelim): 2706
length of query: 324
length of database: 863,360,394
effective HSP length: 128
effective length of query: 196
effective length of database: 538,162,058
effective search space: 105479763368
effective search space used: 105479763368
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 75 (33.5 bits)
Medicago: description of AC137670.4


