

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137553.2 - phase: 0
(264 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
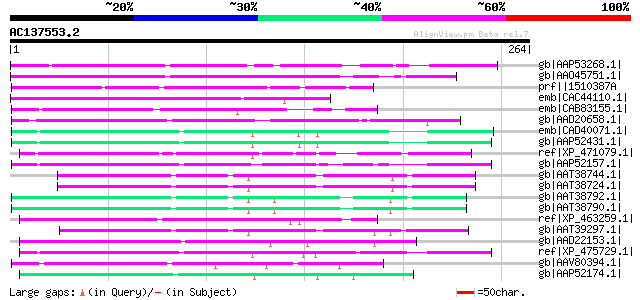
Score E
Sequences producing significant alignments: (bits) Value
gb|AAP53268.1| putative 22 kDa kafirin cluster; Ty3-Gypsy type [... 110 5e-23
gb|AAO45751.1| gag-protease polyprotein [Cucumis melo] 90 6e-17
prf||1510387A retrotransposon del1-46 83 7e-15
emb|CAC44110.1| gag polyprotein [Cicer arietinum] 83 7e-15
emb|CAB83155.1| putative protein [Arabidopsis thaliana] gi|11358... 78 2e-13
gb|AAD20658.1| putative retroelement pol polyprotein [Arabidopsi... 78 3e-13
emb|CAD40071.1| OSJNBa0085C10.24 [Oryza sativa (japonica cultiva... 78 3e-13
gb|AAP52431.1| putative retroelement [Oryza sativa (japonica cul... 77 4e-13
ref|XP_471079.1| OSJNBb0089K24.5 [Oryza sativa (japonica cultiva... 77 4e-13
gb|AAP52157.1| putative polyprotein [Oryza sativa (japonica cult... 77 6e-13
gb|AAT38744.1| putative gag-pol polyprotein [Solanum demissum] 77 6e-13
gb|AAT38724.1| putative retrotransposon protein [Solanum demissum] 77 6e-13
gb|AAT38792.1| putative gag-pol polyprotein [Solanum demissum] g... 75 1e-12
gb|AAT38790.1| putative gag-pol polyprotein [Solanum demissum] 75 1e-12
ref|XP_463259.1| putative polyprotein [Oryza sativa] gi|16924104... 75 2e-12
gb|AAT39297.1| putative gag-pol protein [Solanum demissum] 75 2e-12
gb|AAD22153.1| polyprotein [Sorghum bicolor] 75 2e-12
ref|XP_475729.1| hypothetical protein [Oryza sativa (japonica cu... 74 4e-12
gb|AAV80394.1| polyprotein [Hordeum vulgare subsp. vulgare] 74 4e-12
gb|AAP52174.1| putative polyprotein [Oryza sativa (japonica cult... 74 5e-12
>gb|AAP53268.1| putative 22 kDa kafirin cluster; Ty3-Gypsy type [Oryza sativa
(japonica cultivar-group)] gi|37533358|ref|NP_920981.1|
putative 22 kDa kafirin cluster; Ty3-Gypsy type [Oryza
sativa (japonica cultivar-group)]
gi|21327374|gb|AAM48279.1| Putative 22 kDa kafirin
cluster; Ty3-Gypsy type [Oryza sativa (japonica
cultivar-group)] gi|18767374|gb|AAL79340.1| Putative 22
kDa kafirin cluster; Ty3-Gypsy type [Oryza sativa]
Length = 1230
Score = 110 bits (274), Expect = 5e-23
Identities = 78/248 (31%), Positives = 115/248 (45%), Gaps = 29/248 (11%)
Query: 1 MLAEEADDWWKSLLPVLEQDGVVVTWAVFRREFLNRYFPEDVRGKKEIEFLELKQGDMSV 60
ML A +WW + + + +TW +F+ F +YFPE V+ KE EFLELKQG+ SV
Sbjct: 119 MLQSSAFEWWDAHKKSYSER-IFITWELFKEAFYKKYFPESVKRMKEKEFLELKQGNKSV 177
Query: 61 TEYAAKFVELAKFYPHYSAETAEFSKCIKFENGLRADIKRAIGYQKIRNFSELVSSYMIY 120
EY +F LA+F P + + SK +FE+GLR +KR + ++ F E+VS +
Sbjct: 178 AEYEIEFSRLARFAPEFVQ--TDGSKARRFESGLRQPLKRRVEAFELTIFREVVSKAQLL 235
Query: 121 EEDTKAHYKIRNERRNKGQQGRPKPYSAPVNKGEQRLNDERRPKRRDDPAEIVCYKCGEK 180
E K +++ R E GQ + + P N+G R N + +R+ +
Sbjct: 236 E---KGYHEQRIEH---GQPQKKFKTNNPQNQGRFRGNYSGQMQRKSSE---------NQ 280
Query: 181 GHKSNVCNGEEKKCFRCGKKWHTIAECKRGDIVCFNCDEEGHLTSQCKKPKKGQASVYDS 240
G K +C G C W CF C E GH QC +KG+ V +
Sbjct: 281 GRKCPICQGSHVPSI-CPNCWGR----------CFECGEAGHTRYQCPLLQKGKNRVSST 329
Query: 241 DYVLTKVM 248
TKV+
Sbjct: 330 TQPNTKVL 337
>gb|AAO45751.1| gag-protease polyprotein [Cucumis melo]
Length = 429
Score = 90.1 bits (222), Expect = 6e-17
Identities = 62/228 (27%), Positives = 100/228 (43%), Gaps = 18/228 (7%)
Query: 1 MLAEEADDWWKSLLPVLEQDGVVVTWAVFRREFLNRYFPEDVRGKKEIEFLELKQGDMSV 60
ML + WW++ +L D +TW F+ F ++F +R K EFL L+QGDM+V
Sbjct: 109 MLTDRGTAWWETTERMLGGDVSQITWQQFKESFYAKFFSASLRDAKRQEFLNLEQGDMTV 168
Query: 61 TEYAAKFVELAKFYPHYSAETAEFSKCIKFENGLRADIKRAI-GYQKIRNFSELVSSYMI 119
+Y A+F L++F P A E ++ KF GLR DI+ + ++ + L + +
Sbjct: 169 EQYDAEFDMLSRFAPEMIA--TEAARADKFVRGLRLDIQGLVRAFRPATHADALRLAVDL 226
Query: 120 YEEDTKAHYKIRNERRNKGQQGRPKPYSAPVNKGEQRLNDERRPKRRDDPAEIVCYKCGE 179
++ K GQ+ + + PV + R E R ++ ++ GE
Sbjct: 227 SLQERANSSKTAGRGSTSGQKRKAEQQPVPVPQRNFRPGGEFRSFQQKP------FEAGE 280
Query: 180 KGHKSNVCNGEEKKCFRCGKKWHTIAECKRGDIVCFNCDEEGHLTSQC 227
+C CGK H + C G CF C +EGH +C
Sbjct: 281 AARGKPLCT-------TCGK--HHLGRCLFGTRTCFKCRQEGHTADRC 319
>prf||1510387A retrotransposon del1-46
Length = 1443
Score = 83.2 bits (204), Expect = 7e-15
Identities = 54/185 (29%), Positives = 94/185 (50%), Gaps = 8/185 (4%)
Query: 2 LAEEADDWWKSLLPVLEQDGVVVTWAVFRREFLNRYFPEDVRGKKEIEFLELKQGDMSVT 61
L EA+ WW + + E D + W F F ++FP+ VR E F+ L QG ++V
Sbjct: 106 LQGEAEFWWDAKVRSREDDTTQIKWDEFVEVFTEKFFPDTVRDDLE-RFMTLVQGSLTVA 164
Query: 62 EYAAKFVELAKFYPHYSAETAEFSKCIKFENGLRADIKRAIGYQKIRNFSELVSSYMIYE 121
+Y AKF EL+++ P+ K +FE GL+ I + + I+++ E+++ + E
Sbjct: 165 QYEAKFEELSRYAPY--QVDINIRKVKRFEQGLKLSITKQLSSHLIKDYREVITRALSVE 222
Query: 122 EDTKAHYKIRNERRNKGQQGRPKPYSAPVNKGEQRLNDERRPKRRDDPAEIVCYKCGEKG 181
+ + +I +R+K +G PY+ + ++ ND+ + P + VCY G+ G
Sbjct: 223 KREQREAQIM-AKRSKKNRGHNHPYTR--KEPHRQSNDKSQCPNPLSPGK-VCYSYGQPG 278
Query: 182 H-KSN 185
H KSN
Sbjct: 279 HFKSN 283
>emb|CAC44110.1| gag polyprotein [Cicer arietinum]
Length = 277
Score = 83.2 bits (204), Expect = 7e-15
Identities = 54/169 (31%), Positives = 83/169 (48%), Gaps = 7/169 (4%)
Query: 1 MLAEEADDWWKSLLPVLEQDGVVVTWAVFRREFLNRYFPEDVRGKKEIEFLELKQGDMSV 60
ML +A+ WW+S ++ V W F+R+FL++YFP R K +FL+L QG ++V
Sbjct: 104 MLLGDAEYWWRSARLLMGAAHEEVNWESFKRKFLDKYFPMSTRTKLGDDFLKLHQGSLTV 163
Query: 61 TEYAAKFVELAKFYPHYSAETAEFSKCIKFENGLRADIKRAIGYQKIRNFSELVSSYMIY 120
EYAAKF L++ + + E E C +F++GL+ +I+ ++ I+ F LV
Sbjct: 164 GEYAAKFESLSRHFRFFREEIDEPFMCHRFQDGLKYEIQDSVLPLGIQRFQALVEKCR-E 222
Query: 121 EEDTKAHYKIRNERRNKG------QQGRPKPYSAPVNKGEQRLNDERRP 163
ED K R N G Q R K + P N+ + RP
Sbjct: 223 VEDMKNKRASRVGNFNSGGPSRATYQNRGKQVAKPYNRPQNNNGGPSRP 271
>emb|CAB83155.1| putative protein [Arabidopsis thaliana] gi|11358258|pir||T47419
hypothetical protein T28A8.120 - Arabidopsis thaliana
Length = 329
Score = 78.2 bits (191), Expect = 2e-13
Identities = 57/189 (30%), Positives = 86/189 (45%), Gaps = 26/189 (13%)
Query: 2 LAEEADDWWKSLLPVLEQDGVVVTWAVFRREFLNRYFPEDVRGKKEIEFLELKQGDMSVT 61
L EA +WW+ + EQ G V W+ F+ EF RY PE+ E++FL L+QG +V
Sbjct: 154 LRGEAQEWWERVKQ-REQVGCVDQWSFFKEEFTRRYLPEETIDDLEMKFLRLQQGTKTVR 212
Query: 62 EYAAKFVELAKFYPHYSAETAEFSKCIKFENGLRADIKRAIGYQKIRNFSELV---SSYM 118
+Y +F L +F + E KF +GLR DI+ + N +LV +S
Sbjct: 213 KYEKEFHSLERF---ERRKRGEHELIHKFISGLRVDIRLCCHVRDFDNMIDLVEKAASLE 269
Query: 119 IYEEDTKAHYKIRNERRNKGQQGRPKPYSAPVNKGEQRLNDERRPKRRDDPAEIVCYKCG 178
I E+ + +I E+ G G Q + +RR + E+ CY+CG
Sbjct: 270 IGLEEEARYKRIAQEKEAMGSYG-------------QTGHSKRRCQ------EVTCYRCG 310
Query: 179 EKGHKSNVC 187
GH + C
Sbjct: 311 VAGHIARDC 319
>gb|AAD20658.1| putative retroelement pol polyprotein [Arabidopsis thaliana]
gi|25411368|pir||G84493 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana
Length = 1611
Score = 77.8 bits (190), Expect = 3e-13
Identities = 65/240 (27%), Positives = 103/240 (42%), Gaps = 26/240 (10%)
Query: 2 LAEEADDWWKSLLPVLEQDGVVV-TWAVFRREFLNRYFPEDVRGKKEIEFLELKQGDMSV 60
L +A +WW L V ++ G V ++A F EF +YFP + + E +L+L QG+ +V
Sbjct: 220 LEGDAHNWW---LTVEKRRGDEVRSFADFEDEFNKKYFPPEAWDRLECAYLDLVQGNRTV 276
Query: 61 TEYAAKFVELAKFYPHYSAETAEFSKCIKFENGLRADIKRAIGYQKIRNFSELVSSYMIY 120
EY +F L ++ E E ++ +F GLR +I+ + + SELV +
Sbjct: 277 REYDEEFNRLRRYVGRELEE--EQAQLRRFIRGLRIEIRNHCLVRTFNSVSELVERAAMI 334
Query: 121 EEDTKAHYKIRNERRNKGQQGRPKPYSAPVNKGEQRLNDERRPKRRDDPAEIVCYKCGEK 180
EE + E R ++ P + +++ ++ + D C CG K
Sbjct: 335 EEGIE-------EERYLNREKAPIRNNQSTKPADKKRKFDKVDNTKSDAKTGECVTCG-K 386
Query: 181 GHKSNVCNGEEKKCFRCGKKWHTIAECKRGD-----------IVCFNCDEEGHLTSQCKK 229
H S C C RCG K H I C + + CF C + GHL +C K
Sbjct: 387 NH-SGTCWKAIGACGRCGSKDHAIQSCPKMEPGQSKVLGEETRTCFYCGKTGHLKRECPK 445
>emb|CAD40071.1| OSJNBa0085C10.24 [Oryza sativa (japonica cultivar-group)]
Length = 369
Score = 77.8 bits (190), Expect = 3e-13
Identities = 68/263 (25%), Positives = 106/263 (39%), Gaps = 40/263 (15%)
Query: 2 LAEEADDWWKSLLPVLEQDGVVVTWAVFRREFLNRYFPEDVRGKKEIEFLELKQGDMSVT 61
L A +WW + +G +TWA F F + P V K+ EF LKQ D +VT
Sbjct: 105 LQGSASEWWDHFR-MNRAEGQPITWAEFTEAFKKTHIPAGVVALKKREFRALKQKDRTVT 163
Query: 62 EYAAKFVELAKFYPHYSAETAEFSKCIKFENGLRADIKRAIGYQKIRNFSELVSSYMIYE 121
EY +F LA++ P E + KF GL+ ++ + +F ELV + E
Sbjct: 164 EYLHEFNRLARYAPEDVRTDEERQE--KFLEGLKDELSVMLISHDYEDFQELVDKAIRLE 221
Query: 122 E------DTKAHYKIRNERRNKGQQGRPKP------YSAPVNKGEQ-----RLNDERRPK 164
+ + K + E + Q+ R KP +SA + +Q +N E
Sbjct: 222 DKKNRMDNRKRGRIVFQEAQGSSQRQRIKPLQIGESFSAGQEQSQQLGISGEINSETNAS 281
Query: 165 RRDDPAEIV-CYKCGEKGHKSNVCNGEEKKCFRCGKKWHTIAECKRGDIVCFNCDEEGHL 223
D+ + + + + NG E++ VCFNC E GH
Sbjct: 282 NEDNDEQATQSVSFQQDQSQEDNSNGREQR-------------------VCFNCYEPGHF 322
Query: 224 TSQCKKPKKGQASVYDSDYVLTK 246
+C KPK+ Q ++ VLT+
Sbjct: 323 VKECPKPKRQQPQGQVNNLVLTE 345
>gb|AAP52431.1| putative retroelement [Oryza sativa (japonica cultivar-group)]
gi|37531684|ref|NP_920144.1| putative retroelement
[Oryza sativa (japonica cultivar-group)]
gi|21671934|gb|AAM74296.1| Putative retroelement [Oryza
sativa (japonica cultivar-group)]
Length = 365
Score = 77.4 bits (189), Expect = 4e-13
Identities = 68/262 (25%), Positives = 105/262 (39%), Gaps = 40/262 (15%)
Query: 2 LAEEADDWWKSLLPVLEQDGVVVTWAVFRREFLNRYFPEDVRGKKEIEFLELKQGDMSVT 61
L A +WW + +G +TWA F F + P V K+ EF LKQ D +VT
Sbjct: 105 LQGSASEWWDHFR-MNRAEGQPITWAEFTEAFKKTHIPAGVVALKKREFRALKQKDRTVT 163
Query: 62 EYAAKFVELAKFYPHYSAETAEFSKCIKFENGLRADIKRAIGYQKIRNFSELVSSYMIYE 121
EY +F LA++ P E + KF GL+ ++ + +F ELV + E
Sbjct: 164 EYLHEFNRLARYAPEDVRTDEERQE--KFLEGLKDELSVMLISHDYEDFQELVDKAIRLE 221
Query: 122 E------DTKAHYKIRNERRNKGQQGRPKPY------SAPVNKGEQ-----RLNDERRPK 164
+ + K + E ++ Q+ R KP SA + +Q +N E
Sbjct: 222 DKKNRMDNRKRGRIVFQEAQDSSQRQRIKPLQIGEPSSASQGQSQQLNIAGEINSETNAS 281
Query: 165 RRDDPAEIV-CYKCGEKGHKSNVCNGEEKKCFRCGKKWHTIAECKRGDIVCFNCDEEGHL 223
+D+ + + + N NG E++ VCFNC E GH
Sbjct: 282 NKDNDEQATQSVSVQQDQSQVNNSNGREQR-------------------VCFNCYEPGHF 322
Query: 224 TSQCKKPKKGQASVYDSDYVLT 245
+C KPK+ Q ++ V T
Sbjct: 323 VKECPKPKRQQPQGQVNNIVFT 344
>ref|XP_471079.1| OSJNBb0089K24.5 [Oryza sativa (japonica cultivar-group)]
gi|38344099|emb|CAD39395.2| OSJNBb0089K24.5 [Oryza
sativa (japonica cultivar-group)]
Length = 1365
Score = 77.4 bits (189), Expect = 4e-13
Identities = 67/235 (28%), Positives = 100/235 (42%), Gaps = 24/235 (10%)
Query: 6 ADDWWKSLLPVLEQDGVVVTWAVFRREFLNRYFPEDVRGKKEIEFLELKQGDMSVTEYAA 65
A +WW + G V+TWA F F + P V K+ EF ELKQ +VTE+
Sbjct: 96 ASEWWDHF-QATQPAGQVITWACFSTAFRRAHVPAGVVALKKKEFRELKQDGRTVTEFLH 154
Query: 66 KFVELAKFYPHYSAETAEFSKCIKFENGLRADIKRAIGYQKIRNFSELVSSYMIYEE--- 122
+F A++ P T E K KF GL ++ + +F +LV + E+
Sbjct: 155 EFNRFARYAPE-DVRTDE-EKQEKFLEGLNDELSVQLISGDYEDFQKLVDKAIRLEDKHR 212
Query: 123 --DTKAHYKIRNERRNKGQQGRPKPYSAPVNKGEQRLNDERRPKRRDDPAEIVCYKCGEK 180
++K H K+ N R +G +P+ Y P G +R P+ + K
Sbjct: 213 KMESKKH-KMANARMFQGPSQKPR-YVFPPQVGSSS-TAQRAPRPQ----------MNNK 259
Query: 181 GHKSNVCNGEEKKCFRCGKKWHTIAECKRGDIVCFNCDEEGHLTSQCKKPKKGQA 235
S NG+ + G T + +VC+NC E GH +C KPK+ QA
Sbjct: 260 LQNSGWNNGQRSTTAQGGS---TRKDSSGKILVCYNCYEPGHFADKCPKPKRPQA 311
>gb|AAP52157.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
gi|37531136|ref|NP_919870.1| putative polyprotein [Oryza
sativa (japonica cultivar-group)]
gi|22725908|gb|AAN04918.1| Putative polyprotein [Oryza
sativa] gi|18425259|gb|AAL69437.1| Putative polyprotein
[Oryza sativa]
Length = 333
Score = 76.6 bits (187), Expect = 6e-13
Identities = 66/244 (27%), Positives = 101/244 (41%), Gaps = 37/244 (15%)
Query: 2 LAEEADDWWKSLLPVLEQDGVVVTWAVFRREFLNRYFPEDVRGKKEIEFLELKQGDMSVT 61
L A +WW + +G +TWA F F + P V K+ EF LKQ D +VT
Sbjct: 105 LQGRASEWWDHFR-MNRAEGQPITWAEFTEAFKKTHIPTGVVSLKKREFRALKQKDRTVT 163
Query: 62 EYAAKFVELAKFYPHYSAETAEFSKCIKFENGLRADIKRAIGYQKIRNFSELVSSYMIYE 121
EY +F LA++ P E + KF GL+ ++ + +F ELV + E
Sbjct: 164 EYLHEFNRLARYAPEDVRTDEERQE--KFLEGLKDELSVMLISHDYEDFQELVDKAIRLE 221
Query: 122 EDTKAHYKIRNERRNKGQQGRPKPYSAPVNKGEQRLNDERRPKRRDDPAEIVCYKCGEKG 181
+ K R + R Q G ++ N + NDE+ + V ++ +
Sbjct: 222 DK-----KNRMDNRQSQQLGISGEINSETNASNEG-NDEQA-------TQSVSFQQDQSQ 268
Query: 182 HKSNVCNGEEKKCFRCGKKWHTIAECKRGDIVCFNCDEEGHLTSQCKKPKKGQASVYDSD 241
++ NG E++ VCFNC E GH +C KPK+ Q ++
Sbjct: 269 EDNS--NGREQR-------------------VCFNCYEPGHFVKECPKPKRQQPQGQVNN 307
Query: 242 YVLT 245
VLT
Sbjct: 308 IVLT 311
>gb|AAT38744.1| putative gag-pol polyprotein [Solanum demissum]
Length = 1515
Score = 76.6 bits (187), Expect = 6e-13
Identities = 61/233 (26%), Positives = 98/233 (41%), Gaps = 29/233 (12%)
Query: 25 TWAVFRREFLNRYFPEDVRGKKEIEFLELKQGDMSVTEYAAKFVELAKFYPHYSAETAEF 84
+WA F FL +FP +++ K EFL LKQ +SV EY+ KF +L+++ P A+
Sbjct: 194 SWANFEEAFLGHFFPRELKEAKVREFLTLKQEPLSVREYSLKFTQLSRYAPEMVADMR-- 251
Query: 85 SKCIKFENGL-RADIKRAIGYQKIRNFSELVSSYMIY-----EEDTKAHYKIRNERRNKG 138
++ F GL R K I + +S M+Y EE + + R++R G
Sbjct: 252 NRMSLFVAGLSRLSSKEGRAAMLIGDMD--ISRLMVYVQQVEEEKLRDREEFRSKRAKTG 309
Query: 139 QQGRPKPYSAPVNKGEQRLNDERRPKRRDDPAEIVCYKCGEKGHKSNVCNGEEKK----- 193
+ + +A ++R ++ P A Y+ G G S +
Sbjct: 310 NEPGQQKGNANRPSFQER---QKGPAPSSVSAPAPRYRGGHNGQNSKDFKARPVQSSGSV 366
Query: 194 ---------CFRCGKKWHTIAECKRGDIVCFNCDEEGHLTSQCKKPKKGQASV 237
C RCG+ +C+ G CF C +EGH +C K +G S+
Sbjct: 367 AQRSSLFPACARCGRTHP--GKCRDGQTGCFKCGQEGHFVKECPKNNQGSGSL 417
>gb|AAT38724.1| putative retrotransposon protein [Solanum demissum]
Length = 1602
Score = 76.6 bits (187), Expect = 6e-13
Identities = 61/233 (26%), Positives = 98/233 (41%), Gaps = 29/233 (12%)
Query: 25 TWAVFRREFLNRYFPEDVRGKKEIEFLELKQGDMSVTEYAAKFVELAKFYPHYSAETAEF 84
+WA F FL +FP +++ K EFL LKQ +SV EY+ KF +L+++ P A+
Sbjct: 200 SWANFEEAFLGHFFPRELKEAKVREFLTLKQEPLSVREYSLKFTQLSRYAPEMVADMR-- 257
Query: 85 SKCIKFENGL-RADIKRAIGYQKIRNFSELVSSYMIY-----EEDTKAHYKIRNERRNKG 138
++ F GL R K I + +S M+Y EE + + R++R G
Sbjct: 258 NRMSLFVAGLSRLSSKEGRAAMLIGDMD--ISRLMVYVQQVEEEKLRDREEFRSKRAKTG 315
Query: 139 QQGRPKPYSAPVNKGEQRLNDERRPKRRDDPAEIVCYKCGEKGHKSNVCNGEEKK----- 193
+ + +A ++R ++ P A Y+ G G S +
Sbjct: 316 NEPGQQKGNANRPSFQER---QKGPAPSSVSAPAPRYRGGHNGQNSKDFKARPVQSSGSV 372
Query: 194 ---------CFRCGKKWHTIAECKRGDIVCFNCDEEGHLTSQCKKPKKGQASV 237
C RCG+ +C+ G CF C +EGH +C K +G S+
Sbjct: 373 AQRSSLFPACARCGRTHP--GKCRDGQTGCFKCGQEGHFVKECPKNNQGSGSL 423
>gb|AAT38792.1| putative gag-pol polyprotein [Solanum demissum]
gi|47825020|gb|AAT38791.1| putative gag-pol polyprotein
[Solanum demissum]
Length = 1351
Score = 75.5 bits (184), Expect = 1e-12
Identities = 65/254 (25%), Positives = 101/254 (39%), Gaps = 35/254 (13%)
Query: 2 LAEEADDWWKSLLPVLEQDGVVVTWAVFRREFLNRYFPEDVRGKKEIEFLELKQGDMSVT 61
L + A W+ ++ +WA F FL +FP +++ K EFL LKQ +SV
Sbjct: 75 LKDVARTWFDQWKGGRVENAPPASWACFEEAFLGHFFPRELKEVKVREFLTLKQESLSVH 134
Query: 62 EYAAKFVELAKFYPHYSAETAEFSKCIKFENGL-RADIKRAIGYQKIRNFSELVSSYMIY 120
EY+ KF +L+++ P A+ ++ F GL R K I + +S M+Y
Sbjct: 135 EYSLKFTQLSRYAPEMVADMR--NRMSLFVAGLSRLSSKEGRAAMLIGDMD--ISRLMVY 190
Query: 121 ------------EEDTKAHYKIRNE--------RRNKGQQGRPKPYSAPVNKGEQRLNDE 160
EE K RNE R+ QQ + P ++ R E
Sbjct: 191 VQQVEEEKLRDREEFRNKRVKTRNESGQQRGNSNRSSFQQRQKGPATSSARAPAPRYRGE 250
Query: 161 RRPKRRDDPAEIVCYKCGEKGHKSNVCNGEEK--KCFRCGKKWHTIAECKRGDIVCFNCD 218
+ D +K +V G C +CG+ +C++G CF C
Sbjct: 251 HNVQNSKD------FKVTPAQSSGSVVRGGSSFPACAKCGRVHP--GKCRQGQTCCFRCG 302
Query: 219 EEGHLTSQCKKPKK 232
+EGH +C K K+
Sbjct: 303 QEGHFMKECPKNKQ 316
>gb|AAT38790.1| putative gag-pol polyprotein [Solanum demissum]
Length = 1351
Score = 75.5 bits (184), Expect = 1e-12
Identities = 65/254 (25%), Positives = 101/254 (39%), Gaps = 35/254 (13%)
Query: 2 LAEEADDWWKSLLPVLEQDGVVVTWAVFRREFLNRYFPEDVRGKKEIEFLELKQGDMSVT 61
L + A W+ ++ +WA F FL +FP +++ K EFL LKQ +SV
Sbjct: 75 LKDVARTWFDQWKGGRVENAPPASWACFEEAFLGHFFPRELKEVKVREFLTLKQESLSVH 134
Query: 62 EYAAKFVELAKFYPHYSAETAEFSKCIKFENGL-RADIKRAIGYQKIRNFSELVSSYMIY 120
EY+ KF +L+++ P A+ ++ F GL R K I + +S M+Y
Sbjct: 135 EYSLKFTQLSRYAPEMVADMR--NRMSLFVAGLSRLSSKEGRAAMLIGDMD--ISRLMVY 190
Query: 121 ------------EEDTKAHYKIRNE--------RRNKGQQGRPKPYSAPVNKGEQRLNDE 160
EE K RNE R+ QQ + P ++ R E
Sbjct: 191 VQQVEEEKLRDREEFRNKRVKTRNESGQQRGNSNRSSFQQRQKGPATSSARAPAPRYRGE 250
Query: 161 RRPKRRDDPAEIVCYKCGEKGHKSNVCNGEEK--KCFRCGKKWHTIAECKRGDIVCFNCD 218
+ D +K +V G C +CG+ +C++G CF C
Sbjct: 251 HNVQNSKD------FKVTPAQSSGSVVRGGSSFPACAKCGRVHP--GKCRQGQTCCFRCG 302
Query: 219 EEGHLTSQCKKPKK 232
+EGH +C K K+
Sbjct: 303 QEGHFMKECPKNKQ 316
>ref|XP_463259.1| putative polyprotein [Oryza sativa] gi|16924104|gb|AAL31683.1|
putative polyprotein [Oryza sativa]
Length = 962
Score = 75.1 bits (183), Expect = 2e-12
Identities = 57/194 (29%), Positives = 87/194 (44%), Gaps = 18/194 (9%)
Query: 6 ADDWWKSLLPVLEQDGVVVTWAVFRREFLNRYFPEDVRGKKEIEFLELKQGDMSVTEYAA 65
A++WW++ +E ++TW F F + + P V K+ EF L+QG M+ EY
Sbjct: 36 ANEWWEAYQASIEDPTHIITWMEFTTAFRDAFIPTAVIRMKKNEFRRLRQGFMTGQEYLN 95
Query: 66 KFVELAKFYPHYSAETAEFSKCIKFENGLRADIKRAIGYQKIRNFSELVSSYMIYEEDTK 125
KF +LA++ S E K +F GL +++ ++ Q NF LV+ + E D +
Sbjct: 96 KFTQLARYTA--SDIQDEKEKIERFIEGLHDELQGSMISQDHENFQSLVNKVVRLENDRR 153
Query: 126 AHYKIRNERRNKGQQG---------RPKPY--SAPVNK-GEQRLNDERRPKRRDDPAEIV 173
+R R N +Q RP P+ S P K G + K D P
Sbjct: 154 MVEGLRKRRMNLQRQSPTRSSLQVRRPYPFKPSLPNPKPGMPPILPRPGLKMEDSP---- 209
Query: 174 CYKCGEKGHKSNVC 187
CY CG GH + +C
Sbjct: 210 CYVCGNIGHFARLC 223
>gb|AAT39297.1| putative gag-pol protein [Solanum demissum]
Length = 1438
Score = 74.7 bits (182), Expect = 2e-12
Identities = 62/228 (27%), Positives = 97/228 (42%), Gaps = 29/228 (12%)
Query: 26 WAVFRREFLNRYFPEDVRGKKEIEFLELKQGDMSVTEYAAKFVELAKFYPHYSAETAEFS 85
WA F FL +FP ++R K EFL LKQ +SV EY+ KF +L+++ P A+ +
Sbjct: 142 WACFEEAFLGHFFPRELREAKVREFLTLKQESLSVHEYSLKFTQLSRYAPEMVADMR--N 199
Query: 86 KCIKFENGL-RADIKRAIGYQKIRNFSELVSSYMIY-----EEDTKAHYKIRNERRNKGQ 139
+ F GL R K I + +S M+Y EE + + +N+R G
Sbjct: 200 RMSLFVAGLSRLSSKEGRAAMLIGDMD--ISRLMVYVQQVEEEKLRDREEFKNKRAKSGS 257
Query: 140 QGRPKPYSAPVNKGEQRLNDERRPKRRDDPAEIVCYKCGEKGHKS------------NVC 187
+ + + +QR +R P A Y+ G S +V
Sbjct: 258 ESGHQKGNVNRPSFQQR---QRGPAPSSARAPAPRYRGEFNGQNSKDFKARPAQSSGSVA 314
Query: 188 NGEEK--KCFRCGKKWHTIAECKRGDIVCFNCDEEGHLTSQCKKPKKG 233
G K +CG+ + + C+ G I CF C + GH +C K ++G
Sbjct: 315 QGSSKPPAYAKCGR--NHLGICREGSIGCFKCGQNGHFMRECPKNRQG 360
>gb|AAD22153.1| polyprotein [Sorghum bicolor]
Length = 1484
Score = 74.7 bits (182), Expect = 2e-12
Identities = 56/229 (24%), Positives = 95/229 (41%), Gaps = 29/229 (12%)
Query: 6 ADDWWKSLLPVLEQDGVVVTWAVFRREFLNRYFPEDVRGKKEIEFLELKQGDMSVTEYAA 65
A WW+S + VTW F R+F R+ E V K+ EF L+ G M+V EY
Sbjct: 124 AQTWWESYQAARPNNASPVTWLEFCRDFRARHINEGVMELKQEEFRSLRMGSMTVAEYHD 183
Query: 66 KFVELAKFYPHYSAETAEFSKCIKFENGLRADIKRAIGYQKIRNFSELVSSYMIYEEDTK 125
F +LA++ P+ E A+ + F GL +++ + F LV+ ++ + K
Sbjct: 184 TFEQLARYAPNDVREDADKQRL--FMKGLYYELRLQLAGNTYPTFQALVNRAIVLDNMRK 241
Query: 126 AHYKIR----------NERRNKGQQGRPKPYSAPV-----NKGEQRLNDERRPKRRDDPA 170
+ K R N +R QG + + PV N+ QR ++ + + P
Sbjct: 242 SQDKKRRMLAQGSGSNNRQRFSSHQGHQQRFQGPVSQWNRNQQPQRFQNQSQSGNQRPPF 301
Query: 171 EIVCYKCGEKGHKS------------NVCNGEEKKCFRCGKKWHTIAEC 207
+ + + G ++ + +CFRCGK+ H +C
Sbjct: 302 QQRNHNQQQHGQQAPRPGNPNANSTKRSASNTPTRCFRCGKEGHMSYDC 350
>ref|XP_475729.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|50080334|gb|AAT69668.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 371
Score = 73.9 bits (180), Expect = 4e-12
Identities = 67/251 (26%), Positives = 104/251 (40%), Gaps = 26/251 (10%)
Query: 6 ADDWWKSLLPVLEQDGVVVTWAVFRREFLNRYFPEDVRGKKEIEFLELKQGDMSVTEYAA 65
A +WW + +G +TWA F F + P V K+ EF LKQ D +VTEY
Sbjct: 109 ASEWWDHFR-MNRAEGQPITWAEFTEAFKKTHIPAGVVALKKREFRALKQKDRTVTEYLH 167
Query: 66 KFVELAKFYPHYSAETAEFSKCIKFENGLRADIKRAIGYQKIRNFSELVSSYMIYEE--- 122
+F LA++ P E + KF GL+ ++ + +F ELV + E+
Sbjct: 168 EFNRLARYAPEDVRTDEERQE--KFLEGLKDELSVMLISHDYEDFQELVDKAIRLEDKKN 225
Query: 123 ---DTKAHYKIRNERRNKGQQGRPKPYSA--PVNKGE---QRLNDERRPKRRDDPAEIVC 174
+ K + E + Q+ R KP P + G+ Q+LN K + + V
Sbjct: 226 RMDNRKRKMTVFQEAQGSSQRQRIKPLQIGEPFSAGQGQSQQLNIGGEIKSETNESNEVN 285
Query: 175 YKCGEKGHKSNVCNGEEKKCFRCGKKWHTIAECKRGDIVCFNCDEEGHLTSQCKKPKKGQ 234
K + V + ++ G+K VCFNC + GH +C KPK Q
Sbjct: 286 IK--QANQPVPVQQDQSQENNSSGRKQQ----------VCFNCYKPGHFARECPKPKHQQ 333
Query: 235 ASVYDSDYVLT 245
++ V+T
Sbjct: 334 PQGQVNNIVVT 344
>gb|AAV80394.1| polyprotein [Hordeum vulgare subsp. vulgare]
Length = 1486
Score = 73.9 bits (180), Expect = 4e-12
Identities = 57/222 (25%), Positives = 101/222 (44%), Gaps = 39/222 (17%)
Query: 2 LAEEADDWWKSLLPVLEQDGVVVTWAVFRREFLNRYFPEDVRGKKEIEFLELKQGDMSVT 61
L +A WW+ L + G V++W F R+F + Y P + +F LKQG+ SV
Sbjct: 126 LKGQASIWWQHLKD--SRGGRVMSWDEFCRDFRSHYIPSSFVEEMREKFRRLKQGNNSVY 183
Query: 62 EYAAKFVELAKFYPHYSAETAEFSKCIKFENGLRADIKRAIG------YQKIRNFSELVS 115
+Y +F ELA++ + + SK +F GL+ D++ A+ + K N +
Sbjct: 184 KYNVEFHELARYALQDVPD--QKSKIYQFRGGLKEDLQLALALHDPEEFDKFYNLALRAE 241
Query: 116 SYMIYEEDTKAHYK-----------------------IRNERRNKGQQGRPKPYSAPVNK 152
+ ++ E++K ++ R+ ++ K GR S P N
Sbjct: 242 AALLRVENSKKRFRDSSSSSTQVVQKQQRFWVSSPPPPRHSQQPKHSGGRGS--SRPPNP 299
Query: 153 G-EQRLNDERRPKRR---DDPAEIVCYKCGEKGHKSNVCNGE 190
G +QR +++ + PA++ C+KCG+KGH +N C +
Sbjct: 300 GFQQRYQQQKQGPPQIQFRQPADVTCHKCGQKGHYANKCTSQ 341
>gb|AAP52174.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
gi|37531170|ref|NP_919887.1| putative polyprotein [Oryza
sativa (japonica cultivar-group)]
gi|22725924|gb|AAN04934.1| Putative polyprotein [Oryza
sativa] gi|20177629|gb|AAM14684.1| Putative polyprotein
[Oryza sativa (japonica cultivar-group)]
Length = 1569
Score = 73.6 bits (179), Expect = 5e-12
Identities = 59/224 (26%), Positives = 89/224 (39%), Gaps = 26/224 (11%)
Query: 6 ADDWWKSLLPVLEQDGVVVTWAVFRREFLNRYFPEDVRGKKEIEFLELKQGDMSVTEYAA 65
A DWW + E+D TW F F + P V K+ EF L+QG+ +V EY
Sbjct: 312 AGDWWDTYKEAREEDAGEPTWEEFTAAFHENFVPAAVMRMKKNEFRRLQQGNTTVQEYLN 371
Query: 66 KFVELAKFYPHYSAETAEFSKCIKFENGLRADIKRAIGYQKIRNFSELVSSYMIYEED-- 123
+F +LA++ A E K KF GL +++ + Q +F L++ + E D
Sbjct: 372 RFTQLARYAIGDLANEEE--KIDKFIEGLNDELRGPMIGQDHESFRSLINKVVRLENDQR 429
Query: 124 TKAHYKIRNERRNKGQQGRPKPYSAPVNKGEQ----RLNDERRPKRRDDPAEIV------ 173
T H + R N QG P+ + + G + N P+ + P I
Sbjct: 430 TVGHNRKRRLAMNCPPQGVPQRHKGATSSGWKPPIVATNRPAAPRNFNRPVAIQNRTPTP 489
Query: 174 ------------CYKCGEKGHKSNVCNGEEKKCFRCGKKWHTIA 205
C+ CGE GH +N C K G T++
Sbjct: 490 TLAAPGAKKNVDCFNCGEYGHYANNCPHPRKTPVHTGANAMTVS 533
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.137 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 462,929,177
Number of Sequences: 2540612
Number of extensions: 20381632
Number of successful extensions: 81194
Number of sequences better than 10.0: 4834
Number of HSP's better than 10.0 without gapping: 3796
Number of HSP's successfully gapped in prelim test: 1049
Number of HSP's that attempted gapping in prelim test: 59250
Number of HSP's gapped (non-prelim): 16023
length of query: 264
length of database: 863,360,394
effective HSP length: 126
effective length of query: 138
effective length of database: 543,243,282
effective search space: 74967572916
effective search space used: 74967572916
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 74 (33.1 bits)
Medicago: description of AC137553.2


