

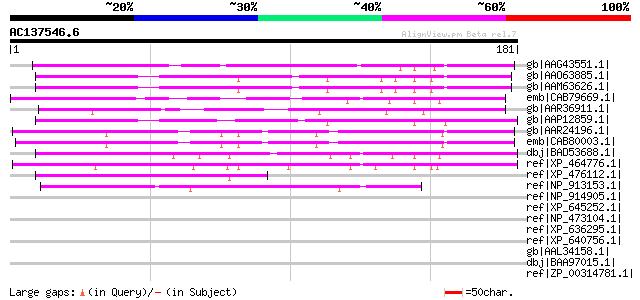
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137546.6 - phase: 0
(181 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAG43551.1| Avr9/Cf-9 rapidly elicited protein 146 [Nicotiana... 112 4e-24
gb|AAO63885.1| unknown protein [Arabidopsis thaliana] gi|2839341... 81 2e-14
gb|AAM63626.1| unknown [Arabidopsis thaliana] 80 3e-14
emb|CAB79669.1| putative protein [Arabidopsis thaliana] gi|49720... 75 6e-13
gb|AAR36911.1| disease resistance gene [Pinus sylvestris] 74 2e-12
gb|AAP12859.1| At3g16330 [Arabidopsis thaliana] gi|26453104|dbj|... 70 3e-11
gb|AAR24196.1| At4g32860 [Arabidopsis thaliana] gi|40824060|gb|A... 68 1e-10
emb|CAB80003.1| putative protein [Arabidopsis thaliana] gi|15233... 67 2e-10
dbj|BAD53688.1| hypothetical protein [Oryza sativa (japonica cul... 67 2e-10
ref|XP_464776.1| hypothetical protein [Oryza sativa (japonica cu... 60 3e-08
ref|XP_476112.1| unknown protein [Oryza sativa (japonica cultiva... 57 2e-07
ref|NP_913153.1| P0419B01.19 [Oryza sativa (japonica cultivar-gr... 46 4e-04
ref|NP_914905.1| OSJNBa0052O12.23 [Oryza sativa (japonica cultiv... 40 0.030
ref|XP_645252.1| hypothetical protein DDB0168763 [Dictyostelium ... 37 0.33
ref|NP_473104.1| hypothetical protein PFB0880w [Plasmodium falci... 36 0.57
ref|XP_636295.1| hypothetical protein DDB0219424 [Dictyostelium ... 35 0.97
ref|XP_640756.1| hypothetical protein DDB0203942 [Dictyostelium ... 35 1.3
gb|AAL34158.1| unknown protein [Arabidopsis thaliana] gi|1387818... 34 1.6
dbj|BAA97015.1| unnamed protein product [Arabidopsis thaliana] 34 1.6
ref|ZP_00314781.1| hypothetical protein Mdeg02003956 [Microbulbi... 34 2.1
>gb|AAG43551.1| Avr9/Cf-9 rapidly elicited protein 146 [Nicotiana tabacum]
Length = 182
Score = 112 bits (281), Expect = 4e-24
Identities = 75/184 (40%), Positives = 103/184 (55%), Gaps = 20/184 (10%)
Query: 9 VVAKKLWNMVRVLLFIIRKGIAKSKTMVHLNLILKRGKLAGKALINTLMLNHQLYHSSFT 68
V AKK W +VRV F++RKG++K K M LNL++KRGK+AGKA I LM +H++ T
Sbjct: 7 VTAKKFWKIVRVAFFMLRKGLSKRKLMFDLNLLMKRGKIAGKAAIQNLM----FHHNNNT 62
Query: 69 CRSDNNSFISPCEYEFSCRNTPANPLRHSSRRFSKSRRQRHNDFSIMNNNIAVQKVFIEM 128
C S +S S YEFSC N+PA L + + SK R D ++ N AV K +EM
Sbjct: 63 CPSSTSS--SKEYYEFSCSNSPAFHLPFNLNKRSKHNRHHETDGDVLMVNAAVLKA-LEM 119
Query: 129 MLNNEKVEAV---ANSPL--------APFTLED-EGNCHQVDIAAEEFINNFYKELNRQN 176
+ + A+ +P +PF + D EGN H VD A+EFI+ FYK+L R+
Sbjct: 120 IQSETASPALPGFGRTPTVRQLRVTDSPFPIRDGEGNSH-VDEKADEFISRFYKDLRREA 178
Query: 177 RTIA 180
A
Sbjct: 179 SAFA 182
>gb|AAO63885.1| unknown protein [Arabidopsis thaliana] gi|28393418|gb|AAO42131.1|
unknown protein [Arabidopsis thaliana]
gi|18403871|ref|NP_564601.1| expressed protein
[Arabidopsis thaliana] gi|4220461|gb|AAD12688.1| ESTs
gb|T75642 and gb|AA650997 come from this gene.
[Arabidopsis thaliana] gi|25405529|pir||D96561
hypothetical protein F5F19.20 [imported] - Arabidopsis
thaliana
Length = 208
Score = 80.9 bits (198), Expect = 2e-14
Identities = 66/209 (31%), Positives = 99/209 (46%), Gaps = 49/209 (23%)
Query: 10 VAKKLWNMVRVLLFIIRKGIAKSKTMVHLNLILKRGKLAGKALINTLMLNHQLYHSSFTC 69
++KKLWN+VR LL++IRKG++K+K + N LKRGK N + + H+ T
Sbjct: 7 ISKKLWNIVRFLLYMIRKGVSKNKLIADFNATLKRGK-------NLMFHQRRRVHAGSTA 59
Query: 70 RSDNNSFISPC----EYEFSCRNTPANPLRHSSRRFSKSRRQRHNDF-------SIMNNN 118
+ N+ + EYEFSC NTP S+ F R++ HN+ ++++
Sbjct: 60 SAALNATSATASSRQEYEFSCSNTPNYSFPFSNMAF--MRKKSHNNLFTCGQTPQTLDDD 117
Query: 119 IAVQKVFIEMMLN-NEKVE------AVANSPL-----------------APFTLE----D 150
+A + +E++ EK VA SP +PF L D
Sbjct: 118 VAAARAVLELLNGVGEKGNVTPADLTVALSPYFPGFGQTPLVRPLRVTDSPFPLTPENGD 177
Query: 151 EGNCHQVDIAAEEFINNFYKELNRQNRTI 179
N H VD AA++FI FYK LN+Q + I
Sbjct: 178 VANGH-VDKAADDFIKKFYKNLNQQKKMI 205
>gb|AAM63626.1| unknown [Arabidopsis thaliana]
Length = 208
Score = 79.7 bits (195), Expect = 3e-14
Identities = 65/209 (31%), Positives = 99/209 (47%), Gaps = 49/209 (23%)
Query: 10 VAKKLWNMVRVLLFIIRKGIAKSKTMVHLNLILKRGKLAGKALINTLMLNHQLYHSSFTC 69
++KKLWN+VR LL++IRKG++K+K + N LKRGK N + + H+ T
Sbjct: 7 ISKKLWNIVRFLLYMIRKGVSKNKLIADFNATLKRGK-------NLMFHQRRRVHAGSTA 59
Query: 70 RSDNNSFISPC----EYEFSCRNTPANPLRHSSRRFSKSRRQRHNDF-------SIMNNN 118
+ N+ + EYEFSC NTP S+ F R++ HN+ ++++
Sbjct: 60 SAALNATSATASSRQEYEFSCSNTPNYSFPFSNMAF--MRKKSHNNLFTCGQTPQTLDDD 117
Query: 119 IAVQKVFIEMMLN-NEKVE------AVANSPL-----------------APFTLE----D 150
+A + +E++ +K VA SP +PF L D
Sbjct: 118 VAAARAVLELLNGVGDKGNVTPADLTVALSPYFPGFGQTPLVRPLRVTDSPFPLTPENGD 177
Query: 151 EGNCHQVDIAAEEFINNFYKELNRQNRTI 179
N H VD AA++FI FYK LN+Q + I
Sbjct: 178 VANGH-VDKAADDFIKKFYKNLNQQKKMI 205
>emb|CAB79669.1| putative protein [Arabidopsis thaliana] gi|4972057|emb|CAB43925.1|
putative protein [Arabidopsis thaliana]
gi|60547867|gb|AAX23897.1| hypothetical protein
At4g29110 [Arabidopsis thaliana]
gi|15233454|ref|NP_194640.1| hypothetical protein
[Arabidopsis thaliana] gi|7459218|pir||T08966
hypothetical protein F19B15.140 - Arabidopsis thaliana
Length = 200
Score = 75.5 bits (184), Expect = 6e-13
Identities = 67/197 (34%), Positives = 96/197 (48%), Gaps = 43/197 (21%)
Query: 1 MEIEARQGVVAKKLWNMVRVLLFIIRKGIAKSKTMVHLNLILKRGKLAGKALINTLMLNH 60
ME+E V AK+LW +VR++ +++ G K+K M+ LNL+LKRG KA+ N
Sbjct: 12 MEMEQNAQVAAKRLWKVVRIVFCVLKTGTVKNKLMLDLNLMLKRG---NKAITNLRR--- 65
Query: 61 QLYHSSFTCRSDNNSFISPCEYEFSCRNTPANPLRHSSRRFSKSRRQRHNDFSIMNNNI- 119
SS T D +S S R +P S+R +R+ H + + +
Sbjct: 66 ---RSSSTGSHDVSS---------SSRVRDYDPFAFISKR----KRRVHGGYDNEEDAVE 109
Query: 120 -AVQKVFIEMMLNNEK----VEAVANSPL-------------APFTLEDEG-NCHQVDIA 160
AV+KVF E++ N+K E+ SPL +PF L+D G + H VD A
Sbjct: 110 AAVKKVF-ELLGENDKKTVATESARESPLIMSPAVRQLRVTDSPFPLDDGGDHDHVVDKA 168
Query: 161 AEEFINNFYKELNRQNR 177
AEEFI FYK L Q +
Sbjct: 169 AEEFIKKFYKNLKLQKK 185
>gb|AAR36911.1| disease resistance gene [Pinus sylvestris]
Length = 207
Score = 73.9 bits (180), Expect = 2e-12
Identities = 58/198 (29%), Positives = 97/198 (48%), Gaps = 48/198 (24%)
Query: 11 AKKLWNMVRVLLFIIRKG-IAKSKTMVHLNLILKRGKLAGKALINTLMLNHQLYHSSFTC 69
AK+ WN++R+ LF+IRKG I+K K ++ ++L+++RGK+ G++L N L+ +H
Sbjct: 12 AKRFWNILRIALFMIRKGLISKRKMLMDMHLMMERGKVYGRSLRN-LVFHHS-------- 62
Query: 70 RSDNNSFISPCEYEFSCRNTPANPLRHSSRRFSKSRRQRH------------NDFSIMNN 117
R +N+ +YEFSC N+PA F ++R+ H + + +
Sbjct: 63 RGNNHGGFGLQDYEFSCSNSPA--------IFHMTKRKHHYFPTHILHFPCIHPHQVEDK 114
Query: 118 NIAVQKVFIEMMLNNE--------KVEAVANSPLAPF----------TLEDEGNCHQVDI 159
VF ++ +NE + + L+P + ED+ HQVD
Sbjct: 115 EEPSTPVFPQLDYSNEYFSKDCLDQNDLPVLQKLSPLLSPLCRRISNSGEDQAYDHQVDR 174
Query: 160 AAEEFINNFYKELNRQNR 177
A+EFI FY++L QNR
Sbjct: 175 RADEFIAKFYEQLRLQNR 192
>gb|AAP12859.1| At3g16330 [Arabidopsis thaliana] gi|26453104|dbj|BAC43628.1|
unknown protein [Arabidopsis thaliana]
gi|9279715|dbj|BAB01272.1| unnamed protein product
[Arabidopsis thaliana] gi|15228179|ref|NP_188254.1|
expressed protein [Arabidopsis thaliana]
gi|2062170|gb|AAB63644.1| unknown protein [Arabidopsis
thaliana]
Length = 205
Score = 70.1 bits (170), Expect = 3e-11
Identities = 57/211 (27%), Positives = 90/211 (42%), Gaps = 54/211 (25%)
Query: 10 VAKKLWNMVRVLLFIIRKGIAKSKTMVHLNLILKRGKLAGKALINTLMLNHQLYHSSFTC 69
++KKL N+VR +L+++ KGI+K K + N LKRGK N + N + S
Sbjct: 7 ISKKLGNIVRFVLYMLHKGISKQKLLADFNATLKRGK-------NLMFHNRRRVPGSAVA 59
Query: 70 RSDNNSFISPCEYEFSCRNTPANPLRHSSRRFSKSRRQRHNDF-------SIMNNNIAVQ 122
N EYEFSC +TP + F K + HN ++++ +V
Sbjct: 60 SHPQN------EYEFSCSDTPNYTFPFNMAAFKK--KSHHNSLFSCGQAPPTLDDDTSVS 111
Query: 123 KVFIEMMLNNEKVEAVANSPL------------------------------APFTLEDEG 152
+ +E++ + + +N+P +PF L +EG
Sbjct: 112 RAVLELLNSGGDHDQGSNTPAFSIEALTALSPYLPGFGRSTSSVRPLRVTDSPFPLREEG 171
Query: 153 NC--HQVDIAAEEFINNFYKELNRQNRTIAS 181
+ VD AA+EFI FYK L +Q + I S
Sbjct: 172 DVANGHVDKAADEFIKKFYKNLYQQKKMIES 202
>gb|AAR24196.1| At4g32860 [Arabidopsis thaliana] gi|40824060|gb|AAR92334.1|
At4g32860 [Arabidopsis thaliana]
Length = 199
Score = 67.8 bits (164), Expect = 1e-10
Identities = 64/201 (31%), Positives = 98/201 (47%), Gaps = 32/201 (15%)
Query: 2 EIEARQGVVAKKLWNMVRVLLFIIRKGIAKSK----TMVHLNLILKRGKLAGKALINTLM 57
++E V KKL ++ +++LF I+K S+ T + +L+ KRGK+ K+L +
Sbjct: 6 DMEVCSTVTTKKLSSLAKLILFTIQKVSDASRHKLLTTLDPHLLAKRGKILRKSLNEAVS 65
Query: 58 LNHQLYHSSFTCRSDNN-----SFISPC----EYEFSCRNTPANPLRHSSRRFSKSRRQR 108
+H S TCR ++ SFISP EYEFSC +TP P R + SK RR
Sbjct: 66 TSH----SRITCRPSDHQDVRSSFISPVPLQLEYEFSCSSTP--PRRSYATTVSKGRRSN 119
Query: 109 --HNDFSIMNNNIAVQKVFIEMMLNNEKVEAVANSPLAPFTLEDEGN-------CHQVDI 159
HN I N ++ +I + +++ + +A D + CH VD
Sbjct: 120 GSHNRPLI---NKRQRQAYIRYNTLPKVRDSIWDRHVAAAVFPDVASSTGTMESCH-VDR 175
Query: 160 AAEEFINNFYKELNRQNRTIA 180
AAEEFI +FY++L Q +A
Sbjct: 176 AAEEFIQSFYRQLRLQKWMMA 196
>emb|CAB80003.1| putative protein [Arabidopsis thaliana]
gi|15233964|ref|NP_195012.1| expressed protein
[Arabidopsis thaliana] gi|7487079|pir||T10693
hypothetical protein T16I18.70 - Arabidopsis thaliana
Length = 193
Score = 67.0 bits (162), Expect = 2e-10
Identities = 64/200 (32%), Positives = 97/200 (48%), Gaps = 32/200 (16%)
Query: 3 IEARQGVVAKKLWNMVRVLLFIIRKGIAKSK----TMVHLNLILKRGKLAGKALINTLML 58
+E V KKL ++ +++LF I+K S+ T + +L+ KRGK+ K+L +
Sbjct: 1 MEVCSTVTTKKLSSLAKLILFTIQKVSDASRHKLLTTLDPHLLAKRGKILRKSLNEAVST 60
Query: 59 NHQLYHSSFTCRSDNN-----SFISPC----EYEFSCRNTPANPLRHSSRRFSKSRRQR- 108
+H S TCR ++ SFISP EYEFSC +TP P R + SK RR
Sbjct: 61 SH----SRITCRPSDHQDVRSSFISPVPLQLEYEFSCSSTP--PRRSYATTVSKGRRSNG 114
Query: 109 -HNDFSIMNNNIAVQKVFIEMMLNNEKVEAVANSPLAPFTLEDEGN-------CHQVDIA 160
HN I N ++ +I + +++ + +A D + CH VD A
Sbjct: 115 SHNRPLI---NKRQRQAYIRYNTLPKVRDSIWDRHVAAAVFPDVASSTGTMESCH-VDRA 170
Query: 161 AEEFINNFYKELNRQNRTIA 180
AEEFI +FY++L Q +A
Sbjct: 171 AEEFIQSFYRQLRLQKWMMA 190
>dbj|BAD53688.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 266
Score = 67.0 bits (162), Expect = 2e-10
Identities = 66/251 (26%), Positives = 114/251 (45%), Gaps = 82/251 (32%)
Query: 10 VAKKLWNMVRVLLFIIRKGIAKSKTMVHLNLILKRGKLAGKALINTLM------LNHQLY 63
+A++LW++VR +LF++RKG++K K + L+L+L RGK+AG + +M +H
Sbjct: 1 MARRLWHVVRAVLFMLRKGMSKRKLAMDLHLLLHRGKIAGNKALGKIMNTTTATASHGHG 60
Query: 64 HSS---------------FTCRSDNNSFI-------SPCEYEFSCRNTPANPLRHSSRRF 101
H++ F+C + + + E EFSC NTP+ P S
Sbjct: 61 HAADAASTAAGEAAAAAPFSCGRALDPALAVYDPRGAGLEVEFSCSNTPSYP--SSFHLI 118
Query: 102 SKSRRQRHNDFS------------------IMNNNIA-VQKVFIEMMLNNEKV-----EA 137
RR+R+N+ S N + A + +VF E++ +++++ A
Sbjct: 119 PTKRRRRNNNGSNGRRRGGGGRGANGGEPGWYNYDAADIARVF-EILNSSDQLLGDGGAA 177
Query: 138 VANSPL-----------------------APFTLEDEG----NCHQVDIAAEEFINNFYK 170
VA +P +PF + D+G VD+ AEEFIN FY+
Sbjct: 178 VAATPSPALWRTSFGGRSPAPVRQLRITDSPFPIRDDGGEDAGAGLVDLEAEEFINKFYE 237
Query: 171 ELNRQNRTIAS 181
+L Q +++A+
Sbjct: 238 QLRTQQQSLAT 248
>ref|XP_464776.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|49388946|dbj|BAD26166.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 295
Score = 60.1 bits (144), Expect = 3e-08
Identities = 70/266 (26%), Positives = 112/266 (41%), Gaps = 87/266 (32%)
Query: 2 EIEARQGVVAKKLWNMVRVLLFIIRKGI---AKSKTMVHLNLILKRGKLAGKALINTLML 58
++ A + VA++LW +VR +L+++R+G+ + K + L+L+L+RGK+AGKAL + +
Sbjct: 8 KVVAAEPGVARRLWRVVRAVLYMLRRGLQAPSGRKLAMDLHLLLRRGKIAGKALGHLVTF 67
Query: 59 NHQLYH--------------SSFTCRSDNNSF--ISPC-----EYEFSCRNTPANPLRHS 97
+H ++ SS +CR + + P E EFSC NTP++
Sbjct: 68 HHHHHNHGHGFSASAAAAGSSSLSCRGIDPALAVYEPSRGRRREVEFSCSNTPSSTTGGG 127
Query: 98 S-----RRFSKSRRQRHNDFSIMNNNIA--------------VQKVFIEMM-----LNNE 133
R ++R R +D+ N+ A V +VF EM+ L N+
Sbjct: 128 GGGGLLGRRRRNRHHRRDDYGFSNDAGAGGSGYYDHGYDAAYVARVF-EMLNDSEHLFND 186
Query: 134 KVEAVANSPL---------------------------------APFTLE--DE---GNCH 155
AVA +P +PF DE G
Sbjct: 187 DDAAVAVAPATAETTPLWTPARSHHSHSPAPAAPSRHRGRTTDSPFAASNGDEAGGGAQQ 246
Query: 156 QVDIAAEEFINNFYKELNRQNRTIAS 181
QVD A+EFI FY++L Q A+
Sbjct: 247 QVDRKADEFIRRFYEQLRAQRSVAAT 272
>ref|XP_476112.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|48475243|gb|AAT44312.1| unknown protein [Oryza sativa
(japonica cultivar-group)]
Length = 250
Score = 57.0 bits (136), Expect = 2e-07
Identities = 31/90 (34%), Positives = 51/90 (56%), Gaps = 7/90 (7%)
Query: 10 VAKKLWNMVRVLLFIIRKGIAKSKTMVHLNLILKRGKLAGKALINTLMLNHQLYHSSFTC 69
+A++LW++V + ++R+G+ + + MV L+++L RGKLAGKAL L + H
Sbjct: 16 MARRLWHVVLAVCHMLRRGLCRKRLMVDLHVLLGRGKLAGKALRGLLAHHAAAGHGHHLA 75
Query: 70 RSDNNSFI-------SPCEYEFSCRNTPAN 92
S ++S P E EFSC TP++
Sbjct: 76 ASSSSSAALASFYGRRPREVEFSCTTTPSS 105
>ref|NP_913153.1| P0419B01.19 [Oryza sativa (japonica cultivar-group)]
gi|20160462|dbj|BAB89415.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
gi|14209608|dbj|BAB56103.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 237
Score = 46.2 bits (108), Expect = 4e-04
Identities = 44/159 (27%), Positives = 71/159 (43%), Gaps = 26/159 (16%)
Query: 12 KKLWNMVRVLLFIIRKGIAKSKTMVHLNLILKRGKLAGKALINTLMLNHQLY-------- 63
K+L +++R + ++R+G+ + + M+ L+L+L RGKLAG+AL ++L HQ +
Sbjct: 7 KRLMHVLRAVYHMLRRGLCRKRLMMDLHLLLGRGKLAGRAL-RDVLLAHQPHGGAAAVAV 65
Query: 64 -----------HSSFTCRSDNNSFISPCEYEFSCRNTPANPLRHSSRRFSKSRRQRHNDF 112
SS + S + +P + EFSC TP+ RF RH
Sbjct: 66 MGGGAGVARGGDSSSSPLSASFFHHNPRDVEFSCTTTPSYAPGVFPFRFRGRGGSRHAGG 125
Query: 113 SIMN----NNIAVQKVFIEMMLNNEKVEAVANSPLAPFT 147
N + AV +VF MLN + A P +
Sbjct: 126 GASNYGGLDASAVARVF--EMLNADAAAAAGAGGETPLS 162
>ref|NP_914905.1| OSJNBa0052O12.23 [Oryza sativa (japonica cultivar-group)]
gi|20161851|dbj|BAB90765.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
gi|15623873|dbj|BAB67931.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 232
Score = 40.0 bits (92), Expect = 0.030
Identities = 26/93 (27%), Positives = 45/93 (47%), Gaps = 14/93 (15%)
Query: 1 MEIEARQGVVAKKLWNMVRVLLFIIRKGIAKSKTM---VHLNLILKRGKLAGKALINTLM 57
ME+E+ A+++W +R + F++RKG+ + + + L + LKR A + +L+
Sbjct: 1 MEVES----AARRMWGYLRAVFFMVRKGVISKRRLLLGMQLAMRLKRRNRAVARSVASLL 56
Query: 58 LNHQLYHSSFTCRSDNNSFISPCEYEFSCRNTP 90
+H R EYEFSC N+P
Sbjct: 57 SHHHGGGGGGALRRRR-------EYEFSCSNSP 82
>ref|XP_645252.1| hypothetical protein DDB0168763 [Dictyostelium discoideum]
gi|42761515|gb|AAS45333.1| similar to coiled-coil
protein involved in spindle-assembly checkpoint; Mad1p
[Saccharomyces cerevisiae] [Dictyostelium discoideum]
gi|60473284|gb|EAL71230.1| hypothetical protein
DDB0168763 [Dictyostelium discoideum]
Length = 540
Score = 36.6 bits (83), Expect = 0.33
Identities = 38/153 (24%), Positives = 67/153 (42%), Gaps = 14/153 (9%)
Query: 35 MVHLNLILKRGKLAGKALINTLMLNHQLYHSSFTCRSDNNSFISPCEYEFSC-RNTPANP 93
++ +NLIL+ + L M+N +L +S ++NN+ IS +YE N +
Sbjct: 200 LISINLILQFQNINDCKLCFYEMINKKLIYSESLSNNNNNNIISIIDYEILIDENNYFSF 259
Query: 94 LRHSSRRFSK----SRRQRHNDFSIMNNNIAVQKVFIEMMLNNEKVE-----AVANSPLA 144
R+F K R + + I+ N+ ++ IE EK+E V N +A
Sbjct: 260 ENIRKRKFEKELLLKREKERKEKIIIENDKKIKNEEIERQ-RKEKIEQDMKLKVKNQLIA 318
Query: 145 PFTLEDEGNCHQVDIAAEEFINNFYKELNRQNR 177
+E+E ++I E+ +KE +Q R
Sbjct: 319 ---IEEEMRLKMIEIENEKLKEQQFKEKEKQER 348
>ref|NP_473104.1| hypothetical protein PFB0880w [Plasmodium falciparum 3D7]
gi|3845300|gb|AAC71965.1| hypothetical protein,
conserved [Plasmodium falciparum 3D7]
gi|7494169|pir||H71603 FAD-dependent oxidoreductase (OO)
PFB0880w - malaria parasite (Plasmodium falciparum)
Length = 426
Score = 35.8 bits (81), Expect = 0.57
Identities = 22/56 (39%), Positives = 27/56 (47%), Gaps = 9/56 (16%)
Query: 127 EMMLNNEKVEAVANSPLAPFTLED--------EGNCHQVDIAAE-EFINNFYKELN 173
E +LN K+ V S + P+ L EG CH VD+ E FI N YKE N
Sbjct: 79 ESILNCCKISKVVISTIGPYILYGYNIVKACVEGGCHYVDVCGEHNFILNIYKEFN 134
>ref|XP_636295.1| hypothetical protein DDB0219424 [Dictyostelium discoideum]
gi|60464650|gb|EAL62783.1| hypothetical protein
DDB0219424 [Dictyostelium discoideum]
Length = 915
Score = 35.0 bits (79), Expect = 0.97
Identities = 18/62 (29%), Positives = 32/62 (51%), Gaps = 3/62 (4%)
Query: 60 HQLYHSSFTCRSDNNSFISPCEYEFSC---RNTPANPLRHSSRRFSKSRRQRHNDFSIMN 116
HQ +H + ++NN+ SPC+ E S N+P +P +S+ ++ + ND N
Sbjct: 461 HQYHHQIQSSHNNNNNPSSPCDSEISSNGGNNSPTSPQEYSTPMETEDNLENENDNENDN 520
Query: 117 NN 118
+N
Sbjct: 521 DN 522
>ref|XP_640756.1| hypothetical protein DDB0203942 [Dictyostelium discoideum]
gi|60468792|gb|EAL66792.1| hypothetical protein
DDB0203942 [Dictyostelium discoideum]
Length = 591
Score = 34.7 bits (78), Expect = 1.3
Identities = 21/76 (27%), Positives = 38/76 (49%), Gaps = 3/76 (3%)
Query: 59 NHQLYHSSFTCRSDNNSFISPCEYEFSCRNTPANPLRHSSRRFSKSRRQRHNDFSIMNNN 118
N+ + +++ +DN +++ EY SC + + S +N+ I NNN
Sbjct: 273 NNNGFQNNYNNNNDNYHYVNN-EYISSCNSINITVSNNDEEIISGDNNNNNNN--INNNN 329
Query: 119 IAVQKVFIEMMLNNEK 134
IAV K E+++NN+K
Sbjct: 330 IAVSKTDEEVIINNDK 345
>gb|AAL34158.1| unknown protein [Arabidopsis thaliana] gi|13878181|gb|AAK44168.1|
unknown protein [Arabidopsis thaliana]
gi|26452324|dbj|BAC43248.1| unknown protein [Arabidopsis
thaliana] gi|18423084|ref|NP_568715.1| nitrogen fixation
NifU-like family protein [Arabidopsis thaliana]
gi|16226434|gb|AAL16167.1| AT5g49940/K9P8_8 [Arabidopsis
thaliana] gi|28207818|emb|CAD55559.1| NFU2 protein
[Arabidopsis thaliana]
Length = 235
Score = 34.3 bits (77), Expect = 1.6
Identities = 23/71 (32%), Positives = 35/71 (48%), Gaps = 5/71 (7%)
Query: 90 PANPLRHSSRRFSKSRRQRHNDFSIMNNNIAVQKVFIEMMLNNEKVEAVANSPLAPFTLE 149
P NPLR RF SR+ S + +A +E+ L E VE+V + + P+ +
Sbjct: 47 PCNPLRRGLSRFLSSRQLFRR--SKVVKAVATPDPILEVPLTEENVESVLDE-IRPYLMS 103
Query: 150 DEGN--CHQVD 158
D GN H++D
Sbjct: 104 DGGNVALHEID 114
>dbj|BAA97015.1| unnamed protein product [Arabidopsis thaliana]
Length = 684
Score = 34.3 bits (77), Expect = 1.6
Identities = 23/71 (32%), Positives = 35/71 (48%), Gaps = 5/71 (7%)
Query: 90 PANPLRHSSRRFSKSRRQRHNDFSIMNNNIAVQKVFIEMMLNNEKVEAVANSPLAPFTLE 149
P NPLR RF SR+ S + +A +E+ L E VE+V + + P+ +
Sbjct: 47 PCNPLRRGLSRFLSSRQLFRR--SKVVKAVATPDPILEVPLTEENVESVLDE-IRPYLMS 103
Query: 150 DEGN--CHQVD 158
D GN H++D
Sbjct: 104 DGGNVALHEID 114
>ref|ZP_00314781.1| hypothetical protein Mdeg02003956 [Microbulbifer degradans 2-40]
Length = 209
Score = 33.9 bits (76), Expect = 2.1
Identities = 26/135 (19%), Positives = 55/135 (40%), Gaps = 19/135 (14%)
Query: 58 LNHQLYHSSFTCRSDNNSFISPCEYEFSCRNTPANPLRHS--------SRRFSKSRRQRH 109
L +L + ++ ++ N +++ +Y S R +P RR+S++
Sbjct: 64 LEEKLKQAGYSVQTGNTQYVARLDYNVSRREAEQSPSSQIVVATGVGFHRRYSRTGLYFS 123
Query: 110 NDFSIMNNNIAVQKVFIEMMLNNEKVEAVANSPLA--------PFTLEDEGNCHQVDIAA 161
++F +++ + ++ + K+ A +P A T GNC + I
Sbjct: 124 DNFE---KRFEFERLLVLVIADAGKLAAAKANPDAVDETINVLEITARSVGNCESLPIVY 180
Query: 162 EEFINNFYKELNRQN 176
EE + +K+L R N
Sbjct: 181 EEMLEAIFKDLKRPN 195
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.322 0.133 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 277,543,197
Number of Sequences: 2540612
Number of extensions: 10561819
Number of successful extensions: 24077
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 27
Number of HSP's that attempted gapping in prelim test: 24042
Number of HSP's gapped (non-prelim): 45
length of query: 181
length of database: 863,360,394
effective HSP length: 120
effective length of query: 61
effective length of database: 558,486,954
effective search space: 34067704194
effective search space used: 34067704194
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 71 (32.0 bits)
Medicago: description of AC137546.6


