

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137510.8 - phase: 0 /pseudo
(254 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
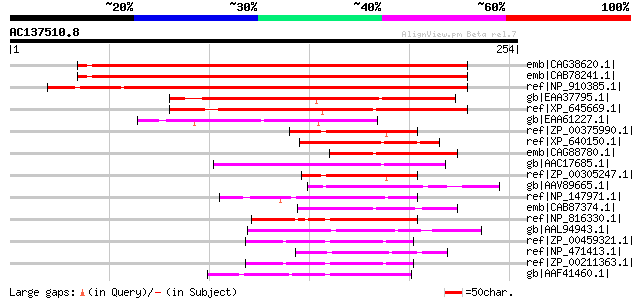
emb|CAG38620.1| ADP-sugar diphosphatase [Arabidopsis thaliana] g... 292 5e-78
emb|CAB78241.1| putative protein [Arabidopsis thaliana] gi|45861... 276 3e-73
ref|NP_910385.1| ESTs AU029348(E30206),C74035(E30206) correspond... 263 5e-69
gb|EAA37795.1| GLP_549_31342_32085 [Giardia lamblia ATCC 50803] 121 2e-26
ref|XP_645669.1| hypothetical protein DDB0216797 [Dictyostelium ... 119 8e-26
gb|EAA61227.1| hypothetical protein AN7712.2 [Aspergillus nidula... 72 2e-11
ref|ZP_00375990.1| NTP pyrophosphohydrolase [Erythrobacter litor... 60 6e-08
ref|XP_640150.1| hypothetical protein DDB0204862 [Dictyostelium ... 54 4e-06
emb|CAG88780.1| unnamed protein product [Debaryomyces hansenii C... 53 9e-06
gb|AAC17685.1| Nudix family protein 2 [Caenorhabditis elegans] g... 53 9e-06
ref|ZP_00305247.1| COG0494: NTP pyrophosphohydrolases including ... 49 2e-04
gb|AAV89665.1| NTP pyrophosphohydrolase [Zymomonas mobilis subsp... 49 2e-04
ref|NP_147971.1| hypothetical protein APE1481 [Aeropyrum pernix ... 48 2e-04
emb|CAB87374.1| SPBP35G2.12 [Schizosaccharomyces pombe] gi|19112... 48 2e-04
ref|NP_816330.1| MutT/nudix family protein [Enterococcus faecali... 48 3e-04
gb|AAL94943.1| Phosphohydrolase (MUTT/NUDIX family protein) [Fus... 48 3e-04
ref|ZP_00459321.1| NUDIX hydrolase [Burkholderia cenocepacia HI2... 47 5e-04
ref|NP_471413.1| hypothetical protein lin2079 [Listeria innocua ... 47 5e-04
ref|ZP_00211363.1| COG0494: NTP pyrophosphohydrolases including ... 47 5e-04
gb|AAF41460.1| conserved hypothetical protein [Neisseria meningi... 47 7e-04
>emb|CAG38620.1| ADP-sugar diphosphatase [Arabidopsis thaliana]
gi|15450341|gb|AAK96464.1| AT4g11980/F16J13_50
[Arabidopsis thaliana] gi|18413722|ref|NP_567384.1|
MutT/nudix family protein [Arabidopsis thaliana]
gi|24797050|gb|AAN64537.1| At4g11980/F16J13_50
[Arabidopsis thaliana]
Length = 309
Score = 292 bits (748), Expect = 5e-78
Identities = 152/195 (77%), Positives = 169/195 (85%), Gaps = 2/195 (1%)
Query: 35 KMSSSTESPSLTHSITLPSKQSEPVHILAAPGVSSSDFWSAIDSSLFKQWLHNLQTENGI 94
KMSSS S SLT SITLPS+ +EPV + A G+SSSDF AIDSSLF+ WL NL++E+GI
Sbjct: 35 KMSSS--SSSLTQSITLPSQPNEPVLVSATAGISSSDFRDAIDSSLFRNWLRNLESESGI 92
Query: 95 LANDTMTLRQVLIQGVDMFGKRIGFLKFIAEIIDKETGNKVPGIVFARGPAVAMLILLES 154
LA+ +MTL+QVLIQGVDMFGKRIGFLKF A+I DKETG KVPGIVFARGPAVA+LILLES
Sbjct: 93 LADGSMTLKQVLIQGVDMFGKRIGFLKFKADIFDKETGQKVPGIVFARGPAVAVLILLES 152
Query: 155 EGETYAVLTEQARVPVGRIILELPAGMLDDDKGDIVGTAVREVEEETGIKLNVEDMVDLT 214
+GETYAVLTEQ RVP G+I+LELPAGMLDDDKGD VGTAVREVEEE GIKL EDMVDLT
Sbjct: 153 DGETYAVLTEQVRVPTGKIVLELPAGMLDDDKGDFVGTAVREVEEEIGIKLKKEDMVDLT 212
Query: 215 AFLDSSTGSTVFPSP 229
AFLD STG +FPSP
Sbjct: 213 AFLDPSTGYRIFPSP 227
>emb|CAB78241.1| putative protein [Arabidopsis thaliana] gi|4586103|emb|CAB40939.1|
putative protein [Arabidopsis thaliana]
gi|7485580|pir||T06605 hypothetical protein F16J13.50 -
Arabidopsis thaliana
Length = 310
Score = 276 bits (707), Expect = 3e-73
Identities = 147/196 (75%), Positives = 164/196 (83%), Gaps = 3/196 (1%)
Query: 35 KMSSSTESPSLTHSITLPSKQSEPVHILAAPGVSSSDFWSAIDSSLFK-QWLHNLQTENG 93
KMSSS S SLT SITLPS+ +EPV + A G+SSSDF +D F WL NL++E+G
Sbjct: 35 KMSSS--SSSLTQSITLPSQPNEPVLVSATAGISSSDFRRVLDIRGFPLNWLRNLESESG 92
Query: 94 ILANDTMTLRQVLIQGVDMFGKRIGFLKFIAEIIDKETGNKVPGIVFARGPAVAMLILLE 153
ILA+ +MTL+QVLIQGVDMFGKRIGFLKF A+I DKETG KVPGIVFARGPAVA+LILLE
Sbjct: 93 ILADGSMTLKQVLIQGVDMFGKRIGFLKFKADIFDKETGQKVPGIVFARGPAVAVLILLE 152
Query: 154 SEGETYAVLTEQARVPVGRIILELPAGMLDDDKGDIVGTAVREVEEETGIKLNVEDMVDL 213
S+GETYAVLTEQ RVP G+I+LELPAGMLDDDKGD VGTAVREVEEE GIKL EDMVDL
Sbjct: 153 SDGETYAVLTEQVRVPTGKIVLELPAGMLDDDKGDFVGTAVREVEEEIGIKLKKEDMVDL 212
Query: 214 TAFLDSSTGSTVFPSP 229
TAFLD STG +FPSP
Sbjct: 213 TAFLDPSTGYRIFPSP 228
>ref|NP_910385.1| ESTs AU029348(E30206),C74035(E30206) correspond to a region of the
predicted gene.~Similar to lipase (U38916) [Oryza sativa
(japonica cultivar-group)] gi|5734637|dbj|BAA83368.1|
MutT/nudix protein-like [Oryza sativa (japonica
cultivar-group)]
Length = 325
Score = 263 bits (671), Expect = 5e-69
Identities = 132/210 (62%), Positives = 166/210 (78%), Gaps = 3/210 (1%)
Query: 20 APNLIPKKKKNGFFYKMSSSTESPSLTHSITLPSKQSEPVHILAAPGVSSSDFWSAIDSS 79
AP+ P ++ SS +P L+ ++ +P + PV ++AAPG++ +DF SA++SS
Sbjct: 28 APSPAPSSRRGARM--ASSGDHAPQLSTAVAVPGAGA-PVRVVAAPGLTEADFTSAVESS 84
Query: 80 LFKQWLHNLQTENGILANDTMTLRQVLIQGVDMFGKRIGFLKFIAEIIDKETGNKVPGIV 139
LF+QWL NLQ E G+L + LRQ+LIQGVDMFGKR+GF+KF A+IID+ET K+PGIV
Sbjct: 85 LFRQWLKNLQEEKGVLTYGRLNLRQILIQGVDMFGKRVGFVKFKADIIDEETKAKIPGIV 144
Query: 140 FARGPAVAMLILLESEGETYAVLTEQARVPVGRIILELPAGMLDDDKGDIVGTAVREVEE 199
FARGPAVA+LILLES+G+TYAVLTEQ RVPVG+ ILELPAGMLDD+KGD VGTAVREVEE
Sbjct: 145 FARGPAVAVLILLESKGQTYAVLTEQVRVPVGKFILELPAGMLDDEKGDFVGTAVREVEE 204
Query: 200 ETGIKLNVEDMVDLTAFLDSSTGSTVFPSP 229
ETGIKLN+EDM+DLTA L+ TG + PSP
Sbjct: 205 ETGIKLNLEDMIDLTALLNPDTGCRMLPSP 234
>gb|EAA37795.1| GLP_549_31342_32085 [Giardia lamblia ATCC 50803]
Length = 247
Score = 121 bits (303), Expect = 2e-26
Identities = 70/146 (47%), Positives = 93/146 (62%), Gaps = 12/146 (8%)
Query: 81 FKQWLHNLQTENGILANDTMTLRQVLIQGVDMFGKRIGFLKFIAEIIDKETGNKVPGIVF 140
F++W + ++ + +R + +Q VD FG RIGFLKF AE K G +VPGIVF
Sbjct: 36 FREWCEEI--------DENLDVRGLTVQSVDYFGARIGFLKFSAEAYSKIHGQRVPGIVF 87
Query: 141 ARGPAVAMLILL--ESEGETYAVLTEQARVPVGRI-ILELPAGMLDDDKGDIVGTAVREV 197
RG +V +L +L E E Y VLTEQARVPVG+ LE+PAGMLD++ GD+VG AV+E+
Sbjct: 88 MRGGSVGVLPVLIDEKTAEKYIVLTEQARVPVGKASFLEIPAGMLDEN-GDVVGVAVQEM 146
Query: 198 EEETGIKLNVEDMVDLTAFLDSSTGS 223
EETGI L D+ L ++ S GS
Sbjct: 147 AEETGISLKRSDLCSLGSYYTSPGGS 172
>ref|XP_645669.1| hypothetical protein DDB0216797 [Dictyostelium discoideum]
gi|60473879|gb|EAL71818.1| hypothetical protein
DDB0216797 [Dictyostelium discoideum]
Length = 253
Score = 119 bits (298), Expect = 8e-26
Identities = 63/152 (41%), Positives = 97/152 (63%), Gaps = 10/152 (6%)
Query: 81 FKQWLHNLQTENGILANDTMTLRQVLIQGVDMFGKRIGFLKFIAEIIDKETGNKVPGIVF 140
F +W+ ++ E + N + +Q VDMFGK +GFLKF A+++ + G VPGI+F
Sbjct: 36 FNKWIKKMELEKELKVNS------ISVQSVDMFGKNVGFLKFKADVVTVKEGRVVPGIIF 89
Query: 141 ARGPAVAMLILLESE--GETYAVLTEQARVPVGRI-ILELPAGMLDDDKGDIVGTAVREV 197
RG +VA+L++L+S+ G+ Y+VLT Q RVPV E+PAGML D G VG A +E+
Sbjct: 90 CRGGSVAILVILKSKETGKEYSVLTVQTRVPVASFQYSEIPAGML-DGSGHFVGVAAKEL 148
Query: 198 EEETGIKLNVEDMVDLTAFLDSSTGSTVFPSP 229
+EETG++++ + ++DLT + V+PSP
Sbjct: 149 KEETGLEVSEDKLIDLTKLAYGTEVDGVYPSP 180
>gb|EAA61227.1| hypothetical protein AN7712.2 [Aspergillus nidulans FGSC A4]
gi|67901450|ref|XP_680981.1| hypothetical protein
AN7712_2 [Aspergillus nidulans FGSC A4]
gi|49111949|ref|XP_411849.1| hypothetical protein
AN7712.2 [Aspergillus nidulans FGSC A4]
Length = 332
Score = 71.6 bits (174), Expect = 2e-11
Identities = 52/136 (38%), Positives = 69/136 (50%), Gaps = 20/136 (14%)
Query: 65 PGVSSSDFWSAIDSSLFKQWLHNLQTE-------NGILANDTMTLRQVLIQGVDMF-GKR 116
PG+S D F+ W LQ + D LR++ +Q VD F G R
Sbjct: 160 PGLSKEDLSRF---PAFRVWFATLQRSLSRQKDPSHEFHKDPYLLRKIEVQAVDFFQGGR 216
Query: 117 IGFLKFIAEIIDKETGNKVPGIVFARGPAVAMLILLE-------SEGETYAVLTEQARVP 169
+GF+K AEI G +PG VF RG +V ML+LL+ E + +AVLT Q R+P
Sbjct: 217 LGFVKLRAEI-SNAGGESLPGSVFLRGGSVGMLLLLQPHDVPSTEEDDKWAVLTVQPRIP 275
Query: 170 VGRIIL-ELPAGMLDD 184
G + E+PAGMLDD
Sbjct: 276 AGSLAFSEIPAGMLDD 291
>ref|ZP_00375990.1| NTP pyrophosphohydrolase [Erythrobacter litoralis HTCC2594]
gi|60737211|gb|EAL75468.1| NTP pyrophosphohydrolase
[Erythrobacter litoralis HTCC2594]
Length = 183
Score = 60.1 bits (144), Expect = 6e-08
Identities = 30/68 (44%), Positives = 46/68 (67%), Gaps = 6/68 (8%)
Query: 141 ARGPAVAMLILLESEGETYAVLTEQARVPVGRIILELPAGMLDDDKG----DIVGTAVRE 196
+RG A ++ ++ EG + +L EQ RVP+GR+ LE+PAG++ D +G D V A+RE
Sbjct: 33 SRGIRAAAIVAIDDEG--HVLLVEQCRVPLGRVCLEIPAGLIGDHEGQEDEDAVEAAIRE 90
Query: 197 VEEETGIK 204
+EEETG +
Sbjct: 91 LEEETGYR 98
>ref|XP_640150.1| hypothetical protein DDB0204862 [Dictyostelium discoideum]
gi|60468151|gb|EAL66161.1| hypothetical protein
DDB0204862 [Dictyostelium discoideum]
Length = 199
Score = 53.9 bits (128), Expect = 4e-06
Identities = 27/70 (38%), Positives = 48/70 (68%), Gaps = 2/70 (2%)
Query: 146 VAMLILLESEGETYAVLTEQARVPVGRIILELPAGMLDDDKGDIVGTAVREVEEETGIKL 205
V +L ++ +G+ Y ++ Q RVPV +++E PAG++D+D+ + V +A+RE++EETG +
Sbjct: 52 VDILATVKKDGKKYLIVVVQYRVPVDNLVIEFPAGLVDNDE-NFVNSAIRELKEETGYR- 109
Query: 206 NVEDMVDLTA 215
D V LT+
Sbjct: 110 TTPDKVLLTS 119
>emb|CAG88780.1| unnamed protein product [Debaryomyces hansenii CBS767]
gi|50423777|ref|XP_460473.1| unnamed protein product
[Debaryomyces hansenii]
Length = 248
Score = 52.8 bits (125), Expect = 9e-06
Identities = 30/64 (46%), Positives = 39/64 (60%), Gaps = 1/64 (1%)
Query: 161 VLTEQARVPVGRIILELPAGMLDDDKGDIVGTAVREVEEETGIKLNVEDMVDLTAFLDSS 220
VLT+Q R PVG +++ELPAG++ D K + TAVRE+ EETG + D A L S
Sbjct: 113 VLTKQFRPPVGAVVIELPAGLV-DPKESVESTAVRELIEETGYHSTFNHLTDSMADLVSD 171
Query: 221 TGST 224
G T
Sbjct: 172 PGLT 175
>gb|AAC17685.1| Nudix family protein 2 [Caenorhabditis elegans]
gi|17564994|ref|NP_503726.1| NuDiX hydrolase (ndx-2)
[Caenorhabditis elegans] gi|7508854|pir||T33225
hypothetical protein W02G9.1 - Caenorhabditis elegans
Length = 223
Score = 52.8 bits (125), Expect = 9e-06
Identities = 33/116 (28%), Positives = 62/116 (53%), Gaps = 1/116 (0%)
Query: 103 RQVLIQGVDMFGKRIGFLKFIAEIIDKETGNKVPGIVFARGPAVAMLILLESEGETYAVL 162
++V+ G + +++GF ++ ++ ++ V A V+++ + +G+ Y VL
Sbjct: 34 QEVVWNGKWIQTRQVGFKTHTGQVGVWQSVHRNTKPVEASADGVSIIARVRKQGKLYIVL 93
Query: 163 TEQARVPVGRIILELPAGMLDDDKGDIVGTAVREVEEETGIKLNVEDMVDLTAFLD 218
+Q R+P G++ LELPAG++D + A+RE++EETG M FLD
Sbjct: 94 VKQYRIPCGKLCLELPAGLIDAGE-TAQQAAIRELKEETGYVSGKVVMESKLCFLD 148
>ref|ZP_00305247.1| COG0494: NTP pyrophosphohydrolases including oxidative damage
repair enzymes [Novosphingobium aromaticivorans DSM
12444]
Length = 180
Score = 48.5 bits (114), Expect = 2e-04
Identities = 28/62 (45%), Positives = 39/62 (62%), Gaps = 6/62 (9%)
Query: 147 AMLILLESEGETYAVLTEQARVPVGRIILELPAGMLDDDKG----DIVGTAVREVEEETG 202
A +IL +G + +L EQ RVP+GR +ELPAG++ D+ G D A RE+EEETG
Sbjct: 40 AAVILAIDDG--HVLLVEQFRVPLGRPCIELPAGLIGDEAGAENEDAATAASRELEEETG 97
Query: 203 IK 204
+
Sbjct: 98 YR 99
>gb|AAV89665.1| NTP pyrophosphohydrolase [Zymomonas mobilis subsp. mobilis ZM4]
gi|56551937|ref|YP_162776.1| NTP pyrophosphohydrolase
[Zymomonas mobilis subsp. mobilis ZM4]
Length = 185
Score = 48.5 bits (114), Expect = 2e-04
Identities = 33/98 (33%), Positives = 57/98 (57%), Gaps = 12/98 (12%)
Query: 150 ILLESEGETYAVLTEQARVPVGRIILELPAGML-DDDKGDIVGT-AVREVEEETGIKLNV 207
++L +GE Y +L EQ R+P G +ELPAG++ D D + V T A RE+ EETG + ++
Sbjct: 44 VILALDGE-YVILVEQPRIPFGAHTIELPAGLIGDTDSHESVETAAARELIEETGYQADI 102
Query: 208 EDMVDLTAFLDSSTGSTVFPSPSAESVLIKRSSHISKV 245
+ +L F S P ++ES + R+ +++++
Sbjct: 103 --IENLGQFASS-------PGMTSESFFLVRAQNLTRI 131
>ref|NP_147971.1| hypothetical protein APE1481 [Aeropyrum pernix K1]
gi|5105165|dbj|BAA80479.1| 188aa long hypothetical
protein [Aeropyrum pernix K1] gi|7516527|pir||A72628
hypothetical protein APE1481 - Aeropyrum pernix (strain
K1)
Length = 188
Score = 48.1 bits (113), Expect = 2e-04
Identities = 37/101 (36%), Positives = 54/101 (52%), Gaps = 8/101 (7%)
Query: 106 LIQGVDMFGKRIGFLKFIAEIIDKETGNK--VPGIVFARGPAVAMLILLESEGETYAVLT 163
L V+ G+R+ KF A + G + V +VF +VA+L L+E +GE + VL
Sbjct: 7 LALNVECRGRRV---KFEARMETLPNGRQILVDRVVFP--DSVAVLPLVEKDGEWHVVLV 61
Query: 164 EQARVPVGRIILELPAGMLDDDKGDIVGTAVREVEEETGIK 204
Q R +G+ LE PAG L + + A RE+EEE G+K
Sbjct: 62 RQFRPSIGKWTLEAPAGTLKEGETP-EAAAARELEEEAGLK 101
>emb|CAB87374.1| SPBP35G2.12 [Schizosaccharomyces pombe]
gi|19112179|ref|NP_595387.1| hypothetical protein
SPBP35G2.12 [Schizosaccharomyces pombe 972h-]
Length = 205
Score = 48.1 bits (113), Expect = 2e-04
Identities = 27/80 (33%), Positives = 46/80 (56%), Gaps = 4/80 (5%)
Query: 145 AVAMLILLESEGETYAVLTEQARVPVGRIILELPAGMLDDDKGDIVGTAVREVEEETGIK 204
AVA+L ++ +G + + +Q R P+G+ +E+PAG++ D K A+RE+ EETG
Sbjct: 55 AVAILAIVPIDGSPHVLCQKQFRPPIGKFCIEIPAGLV-DSKESCEDAAIRELREETGY- 112
Query: 205 LNVEDMVDLTAFLDSSTGST 224
V ++D T + + G T
Sbjct: 113 --VGTVMDSTTVMYNDPGLT 130
>ref|NP_816330.1| MutT/nudix family protein [Enterococcus faecalis V583]
gi|29344642|gb|AAO82400.1| MutT/nudix family protein
[Enterococcus faecalis V583]
Length = 198
Score = 47.8 bits (112), Expect = 3e-04
Identities = 34/84 (40%), Positives = 52/84 (61%), Gaps = 5/84 (5%)
Query: 122 FIAEIIDKETGNKVPGIVFARGPAVAMLILLESEGETYAVLTEQARVPVGRIILELPAGM 181
F+ ++ G +VF G AVAM I L +EG+ VL +Q R P+ ++ILE+PAG
Sbjct: 30 FLDDVALPTGGTAKRELVFHSG-AVAM-IPLTAEGKI--VLVKQFRKPLEQVILEIPAGK 85
Query: 182 LD-DDKGDIVGTAVREVEEETGIK 204
+D ++ + TA+RE+EEETG +
Sbjct: 86 IDPGEENQLETTAMRELEEETGYR 109
>gb|AAL94943.1| Phosphohydrolase (MUTT/NUDIX family protein) [Fusobacterium
nucleatum subsp. nucleatum ATCC 25586]
gi|19704082|ref|NP_603644.1| Phosphohydrolase
(MUTT/NUDIX family protein) [Fusobacterium nucleatum
subsp. nucleatum ATCC 25586]
Length = 171
Score = 47.8 bits (112), Expect = 3e-04
Identities = 40/117 (34%), Positives = 55/117 (46%), Gaps = 8/117 (6%)
Query: 120 LKFIAEIIDKETGNKVPGIVFARGPAVAMLILLESEGETYAVLTEQARVPVGRIILELPA 179
LKF+ +D + N + A+A LIL S + V Q R V I E+PA
Sbjct: 9 LKFLKVGVDTDPLNNHNLEYLEKQNAIAALILNHSGDKVLFV--NQYRAGVHNYIYEVPA 66
Query: 180 GMLDDDKGDIVGTAVREVEEETGIKLNVEDMVDLTAFLDSSTGSTVFPSPSAESVLI 236
G++++D+ IV REV EETG K D DS+TG V P + E + I
Sbjct: 67 GLIENDEKPIVALE-REVREETGYKRE-----DYDILYDSNTGFLVSPGYTTEKIYI 117
>ref|ZP_00459321.1| NUDIX hydrolase [Burkholderia cenocepacia HI2424]
gi|67660957|ref|ZP_00458290.1| NUDIX hydrolase
[Burkholderia cenocepacia AU 1054]
gi|67104615|gb|EAM21734.1| NUDIX hydrolase [Burkholderia
cenocepacia HI2424] gi|67091458|gb|EAM09034.1| NUDIX
hydrolase [Burkholderia cenocepacia AU 1054]
Length = 196
Score = 47.0 bits (110), Expect = 5e-04
Identities = 29/84 (34%), Positives = 45/84 (53%), Gaps = 4/84 (4%)
Query: 119 FLKFIAEIIDKETGNKVPGIVFARGPAVAMLILLESEGETYAVLTEQARVPVGRIILELP 178
FLK + + G K + + P M+I L +G ++ Q R P+G+++ E P
Sbjct: 26 FLKLKRDTVRLPDGKKATR-EYVQHPGAVMVIPLFDDGRV--LMESQFRYPIGKVMAEFP 82
Query: 179 AGMLDDDKGDIVGTAVREVEEETG 202
AG LD ++G + AVRE+ EETG
Sbjct: 83 AGKLDPNEG-ALACAVRELREETG 105
>ref|NP_471413.1| hypothetical protein lin2079 [Listeria innocua Clip11262]
gi|16414580|emb|CAC97309.1| lin2079 [Listeria innocua]
gi|25303390|pir||AE1692 hypothetical protein homolog
lin2079 [imported] - Listeria innocua (strain Clip11262)
Length = 185
Score = 47.0 bits (110), Expect = 5e-04
Identities = 29/76 (38%), Positives = 45/76 (59%), Gaps = 5/76 (6%)
Query: 144 PAVAMLILLESEGETYAVLTEQARVPVGRIILELPAGMLDDDKGDIVGTAVREVEEETGI 203
P +I ++G Y L EQ R P+ + I+E+PAG ++ + +V TA RE+EEETG
Sbjct: 43 PGAVAIIPFSADGRMY--LVEQFRKPLEKNIIEIPAGKMEPGEDPLV-TAKRELEEETGF 99
Query: 204 KLNVEDMVDLTAFLDS 219
+ +D+ LT+F S
Sbjct: 100 Q--SDDLTYLTSFYTS 113
>ref|ZP_00211363.1| COG0494: NTP pyrophosphohydrolases including oxidative damage
repair enzymes [Burkholderia cepacia R18194]
Length = 196
Score = 47.0 bits (110), Expect = 5e-04
Identities = 29/84 (34%), Positives = 45/84 (53%), Gaps = 4/84 (4%)
Query: 119 FLKFIAEIIDKETGNKVPGIVFARGPAVAMLILLESEGETYAVLTEQARVPVGRIILELP 178
FLK + + G K + + P M+I L +G ++ Q R P+G+++ E P
Sbjct: 26 FLKLKRDTVRLPDGKKATR-EYVQHPGAVMVIPLFDDGRV--LMESQYRYPIGKVMAEFP 82
Query: 179 AGMLDDDKGDIVGTAVREVEEETG 202
AG LD ++G + AVRE+ EETG
Sbjct: 83 AGKLDPNEG-ALACAVRELREETG 105
>gb|AAF41460.1| conserved hypothetical protein [Neisseria meningitidis MC58]
gi|11278608|pir||B81127 conserved hypothetical protein
NMB1064 [imported] - Neisseria meningitidis (strain MC58
serogroup B) gi|15676948|ref|NP_274097.1| hypothetical
protein NMB1064 [Neisseria meningitidis MC58]
Length = 178
Score = 46.6 bits (109), Expect = 7e-04
Identities = 35/102 (34%), Positives = 51/102 (49%), Gaps = 5/102 (4%)
Query: 100 MTLRQVLIQGVDMFGKRIGFLKFIAEIIDKETGNKVPGIVFARGPAVAMLILLESEGETY 159
M LR+V + G ++ GF+ + + GN+ IV R P A ++ + EG+
Sbjct: 1 MDLREVKLGGETIYEG--GFVSISRDKVRLPNGNEGQRIVI-RHPGAACVLAVTDEGKV- 56
Query: 160 AVLTEQARVPVGRIILELPAGMLDDDKGDIVGTAVREVEEET 201
VL Q R + LELPAG LD D+ A+RE+ EET
Sbjct: 57 -VLVRQWRYAANQATLELPAGKLDVAGEDMAACALRELAEET 97
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.317 0.133 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 407,681,897
Number of Sequences: 2540612
Number of extensions: 16696676
Number of successful extensions: 46330
Number of sequences better than 10.0: 272
Number of HSP's better than 10.0 without gapping: 63
Number of HSP's successfully gapped in prelim test: 209
Number of HSP's that attempted gapping in prelim test: 46172
Number of HSP's gapped (non-prelim): 276
length of query: 254
length of database: 863,360,394
effective HSP length: 125
effective length of query: 129
effective length of database: 545,783,894
effective search space: 70406122326
effective search space used: 70406122326
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 73 (32.7 bits)
Medicago: description of AC137510.8


