

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137079.15 + phase: 0 /pseudo
(864 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
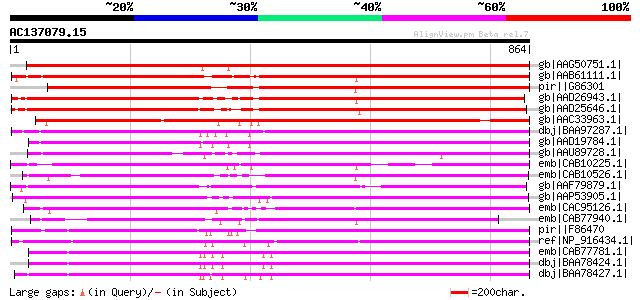
Score E
Sequences producing significant alignments: (bits) Value
gb|AAG50751.1| polyprotein, putative [Arabidopsis thaliana] gi|2... 780 0.0
gb|AAB61111.1| Strong similarity to Zea mays retrotransposon Hop... 768 0.0
pir||G86301 probable retroelement polyprotein [imported] - Arabi... 765 0.0
gb|AAD26943.1| putative retroelement pol polyprotein [Arabidopsi... 753 0.0
gb|AAD25646.1| putative retroelement pol polyprotein [Arabidopsi... 744 0.0
gb|AAC33963.1| contains similarity to reverse transcriptases (Pf... 729 0.0
dbj|BAA97287.1| retroelement pol polyprotein-like [Arabidopsis t... 725 0.0
gb|AAD19784.1| putative retroelement pol polyprotein [Arabidopsi... 725 0.0
gb|AAU89728.1| putative retroelement pol polyprotein-like [Solan... 695 0.0
emb|CAB10225.1| retrovirus-related like polyprotein [Arabidopsis... 688 0.0
emb|CAB10526.1| retrotransposon like protein [Arabidopsis thalia... 682 0.0
gb|AAF79879.1| T7N9.5 [Arabidopsis thaliana] 665 0.0
gb|AAP53905.1| putative pol polyprotein [Oryza sativa (japonica ... 625 e-177
emb|CAC95126.1| gag-pol polyprotein [Populus deltoides] 620 e-176
emb|CAB77940.1| putative polyprotein [Arabidopsis thaliana] gi|4... 615 e-174
pir||F86470 probable retroelement polyprotein [imported] - Arabi... 601 e-170
ref|NP_916434.1| putative gag/pol polyprotein [Oryza sativa (jap... 583 e-164
emb|CAB77781.1| putative polyprotein of LTR transposon [Arabidop... 580 e-164
dbj|BAA78424.1| polyprotein [Arabidopsis thaliana] 580 e-164
dbj|BAA78427.1| polyprotein [Arabidopsis thaliana] 579 e-163
>gb|AAG50751.1| polyprotein, putative [Arabidopsis thaliana] gi|25301686|pir||F96610
probable polyprotein T8L23.26 [imported] - Arabidopsis
thaliana
Length = 1468
Score = 780 bits (2014), Expect = 0.0
Identities = 401/868 (46%), Positives = 570/868 (65%), Gaps = 33/868 (3%)
Query: 29 FDVWHMRLGHVSSSGLSVISKQFPFIPCIKNAPPCDACHYAKQKRLPFPHSSIKSSAPFD 88
FD+WH RLGH S ++++ ++ CD C AKQ R FP S +S F
Sbjct: 513 FDLWHRRLGHASDKIVNLLPRELLSSGKEILENVCDTCMRAKQTRDTFPLSDNRSMDSFQ 572
Query: 89 LLHADLWGPYSTPSFLGHKYFLTLVDDYSRFTWVIFLKTKDETQKHLKHFISYVENQFHT 148
L+H D+WGPY PS+ G +YFLT+VDDYSR WV + K ETQKHLK FI+ VE QF T
Sbjct: 573 LIHCDVWGPYRAPSYSGARYFLTIVDDYSRGVWVYLMTDKSETQKHLKDFIALVERQFDT 632
Query: 149 TLKCLRSDNGSEFIAMTSFLLSKGIIHHKTCVETPQQNGVVERKHQHILNVARSLAFHSH 208
+K +RSDNG+EF+ M + L KGI H +CV TP QNG VERKH+HILN+AR+L F S+
Sbjct: 633 EIKIVRSDNGTEFLCMREYFLHKGIAHETSCVGTPHQNGRVERKHRHILNIARALRFQSY 692
Query: 209 VPITMWNFTVQHAVHIINRIPSPLLKFKSPFELLHKEPPSIIHLKVFGCLAYASTLQAHR 268
+PI W + A ++INR PS LL+ KSP+E+L+K P HL+VFG L YA
Sbjct: 693 LPIQFWGECILSAAYLINRTPSMLLQGKSPYEMLYKTAPKYSHLRVFGSLCYAHNQNHKG 752
Query: 269 TKFNPRARKTIFLGFKEGTKGSILYDLNSNELFVSRNVIFYENHFPFTLAT--------- 319
KF R+R+ +F+G+ G KG L+DL + FVSR+VIF E FP++ +
Sbjct: 753 DKFAARSRRCVFVGYPHGQKGWRLFDLEEQKFFVSRDVIFQETEFPYSKMSCNEEDERVL 812
Query: 320 ---------KQANIPTTSSHIDLGDPIT--DLSPHPISAPEFQLTSTPPSQYVS------ 362
++A P T ++G+ +++ PI PE S+ PS++VS
Sbjct: 813 VDCVGPPFIEEAIGPRTIIGRNIGEATVGPNVATGPI-IPEINQESSSPSEFVSLSSLDP 871
Query: 363 ---APAVQHA-IPVTDSISEPT-VRKSTRISQRPSYLADYHCNLPS-KSCSNVSSGISSY 416
+ VQ A +P++ + P +R+S+R +Q+P L ++ N S +S S +S S Y
Sbjct: 872 FLASSTVQTADLPLSSTTPAPIQLRRSSRQTQKPMKLKNFVTNTVSVESISPEASSSSLY 931
Query: 417 PLSSFLSYDNCSPTYTHFCCTISSINEPKTFAQANKSECWREAMTTELNALAKNNTWSVV 476
P+ ++ + ++ F +++ EP T+ +A + WREAM+ E+ +L N T+S+V
Sbjct: 932 PIEKYVDCHRFTSSHKAFLAAVTAGMEPTTYNEAMVDKAWREAMSAEIESLRVNQTFSIV 991
Query: 477 TLPPGKVPIGCKWVYKVKYHANGSIERYKARLVAQGYTQTEGVDYFDTFSPVAKLTTIRV 536
LPPGK +G KWVYK+KY ++G+IERYKARLV G Q EGVDY +TF+PVAK++T+R+
Sbjct: 992 NLPPGKRALGNKWVYKIKYRSDGAIERYKARLVVLGNCQKEGVDYDETFAPVAKMSTVRL 1051
Query: 537 LLSLAAIKGWHLEQLDVNNAFLHGDLHEEVYMALPPGYPTINSSQVCKLNKSLYGLKQAS 596
L +AA + WH+ Q+DV+NAFLHGDL EEVYM LP G+ + S+VC+L+KSLYGLKQA
Sbjct: 1052 FLGVAAARDWHVHQMDVHNAFLHGDLKEEVYMKLPQGFQCDDPSKVCRLHKSLYGLKQAP 1111
Query: 597 RQWYSKLSTSLISFGYTQSLADYSLFVKVSGASFTALLVYVDDIVLAGNCISEIKSVKTF 656
R W+SKLS++L +G+TQSL+DYSLF + F +LVYVDD++++G+C + K++
Sbjct: 1112 RCWFSKLSSALKQYGFTQSLSDYSLFSYNNDGIFVHVLVYVDDLIISGSCPDAVAQFKSY 1171
Query: 657 LDNKFCIKDLGQLRYFLGFEIARSKSGILLNQRKYTLELLEDAGTLGSKPAATPFDPSTK 716
L++ F +KDLG L+YFLG E++R+ G L+QRKY L+++ + G LG++P+A P + + K
Sbjct: 1172 LESCFHMKDLGLLKYFLGIEVSRNAQGFYLSQRKYVLDIISEMGLLGARPSAFPLEQNHK 1231
Query: 717 LGATTGTPFTDASSYRRLIGRLLYLTNTRPDISYSVQNLSQFVSRPMVPHYQAAQRILKY 776
L +T +D+S YRRL+GRL+YL TRP++SYSV L+QF+ P H+ AA R+++Y
Sbjct: 1232 LSLSTSPLLSDSSRYRRLVGRLIYLVVTRPELSYSVHTLAQFMQNPRQDHWNAAIRVVRY 1291
Query: 777 LKSSPAKGLFFSSSSELKLHGFADSDWACCPDTRRSVTGYCVLLGSSLISWKSKKQSTVS 836
LKS+P +G+ SS+S L+++G+ DSD+A CP TRRS+TGY V LG + ISWK+KKQ TVS
Sbjct: 1292 LKSNPGQGILLSSTSTLQINGWCDSDYAACPLTRRSLTGYFVQLGDTPISWKTKKQPTVS 1351
Query: 837 RSSTEAEYRALAHLTCELQWLNYLFHDL 864
RSS EAEYRA+A LT EL WL + +DL
Sbjct: 1352 RSSAEAEYRAMAFLTQELMWLKRVLYDL 1379
>gb|AAB61111.1| Strong similarity to Zea mays retrotransposon Hopscotch polyprotein
(gb|U12626). [Arabidopsis thaliana]
gi|25301690|pir||G96722 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana
Length = 1315
Score = 768 bits (1982), Expect = 0.0
Identities = 404/876 (46%), Positives = 563/876 (64%), Gaps = 39/876 (4%)
Query: 3 VGGLYLI----AAGPSLANKLSCNSVFTDCFDVWHMRLGHVSSSGLSVISKQFPFIPCIK 58
V LY++ + P + ++ SV + D+WH RLGH S L +S F P K
Sbjct: 376 VANLYIVDLDSLSHPGTDSSITVASVTSH--DLWHKRLGHPSVQKLQPMSSLLSF-PKQK 432
Query: 59 NAPP--CDACHYAKQKRLPFPHSSIKSSAPFDLLHADLWGPYSTPSFLGHKYFLTLVDDY 116
N C CH +KQK LPF + KSS PFDL+H D WGP+S + G++YFLT+VDDY
Sbjct: 433 NNTDFHCRVCHISKQKHLPFVSHNNKSSRPFDLIHIDTWGPFSVQTHDGYRYFLTIVDDY 492
Query: 117 SRFTWVIFLKTKDETQKHLKHFISYVENQFHTTLKCLRSDNGSEFIAMTSFLLSKGIIHH 176
SR TWV L+ K + + F++ VENQF TT+K +RSDN E + T F SKGI+ +
Sbjct: 493 SRATWVYLLRNKSDVLTVIPTFVTMVENQFETTIKGVRSDNAPE-LNFTQFYHSKGIVPY 551
Query: 177 KTCVETPQQNGVVERKHQHILNVARSLAFHSHVPITMWNFTVQHAVHIINRIPSPLLKFK 236
+C ETPQQN VVERKHQHILNVARSL F SH+PI+ W + AV++INR+P+P+L+ K
Sbjct: 552 HSCPETPQQNSVVERKHQHILNVARSLFFQSHIPISYWGDCILTAVYLINRLPAPILEDK 611
Query: 237 SPFELLHKEPPSIIHLKVFGCLAYASTLQAHRTKFNPRARKTIFLGFKEGTKGSILYDLN 296
PFE+L K P+ H+KVFGCL YAST R KF+PRA+ F+G+ G KG L DL
Sbjct: 612 CPFEVLTKTVPTYDHIKVFGCLCYASTSPKDRHKFSPRAKACAFIGYPSGFKGYKLLDLE 671
Query: 297 SNELFVSRNVIFYENHFPFT---LATKQANIPTTSSHIDLGDPITDLSPHPISAPEFQLT 353
++ + VSR+V+F+E FPF L+ ++ N DL+P P +
Sbjct: 672 THSIIVSRHVVFHEELFPFLGSDLSQEEQNF------------FPDLNPTPPMQRQSSDH 719
Query: 354 STPPSQYVSAPAVQHAIPVTDSISEPTVRKSTRISQRPSYLADYHCNLPSKSCSNVSSGI 413
P S + A P T+++ EP+V+ S R +++P+YL DY+C+ S VSS
Sbjct: 720 VNPSDSSSSVEILPSANP-TNNVPEPSVQTSHRKAKKPAYLQDYYCH------SVVSS-- 770
Query: 414 SSYPLSSFLSYDNCSPTYTHFCCTISSINEPKTFAQANKSECWREAMTTELNALAKNNTW 473
+ + + FLSYD + Y F + EP + +A K + WR+AM E + L +TW
Sbjct: 771 TPHEIRKFLSYDRINDPYLTFLACLDKTKEPSNYTEAEKLQVWRDAMGAEFDFLEGTHTW 830
Query: 474 SVVTLPPGKVPIGCKWVYKVKYHANGSIERYKARLVAQGYTQTEGVDYFDTFSPVAKLTT 533
V +LP K IGC+W++K+KY+++GS+ERYKARLVAQGYTQ EG+DY +TFSPVAKL +
Sbjct: 831 EVCSLPADKRCIGCRWIFKIKYNSDGSVERYKARLVAQGYTQKEGIDYNETFSPVAKLNS 890
Query: 534 IRVLLSLAAIKGWHLEQLDVNNAFLHGDLHEEVYMALPPGYPT-----INSSQVCKLNKS 588
+++LL +AA L QLD++NAFL+GDL EE+YM LP GY + + + VC+L KS
Sbjct: 891 VKLLLGVAARFKLSLTQLDISNAFLNGDLDEEIYMRLPQGYASRQGDSLPPNAVCRLKKS 950
Query: 589 LYGLKQASRQWYSKLSTSLISFGYTQSLADYSLFVKVSGASFTALLVYVDDIVLAGNCIS 648
LYGLKQASRQWY K S++L+ G+ QS D++ F+K+S F +LVY+DDI++A N +
Sbjct: 951 LYGLKQASRQWYLKFSSTLLGLGFIQSYCDHTCFLKISDGIFLCVLVYIDDIIIASNNDA 1010
Query: 649 EIKSVKTFLDNKFCIKDLGQLRYFLGFEIARSKSGILLNQRKYTLELLEDAGTLGSKPAA 708
+ +K+ + + F ++DLG+L+YFLG EI RS GI ++QRKY L+LL++ G LG KP++
Sbjct: 1011 AVDILKSQMKSFFKLRDLGELKYFLGLEIVRSDKGIHISQRKYALDLLDETGQLGCKPSS 1070
Query: 709 TPFDPSTKLGATTGTPFTDASSYRRLIGRLLYLTNTRPDISYSVQNLSQFVSRPMVPHYQ 768
P DPS +G F + YRRLIGRL+YL TRPDI+++V L+QF P H Q
Sbjct: 1071 IPMDPSMVFAHDSGGDFVEVGPYRRLIGRLMYLNITRPDITFAVNKLAQFSMAPRKAHLQ 1130
Query: 769 AAQRILKYLKSSPAKGLFFSSSSELKLHGFADSDWACCPDTRRSVTGYCVLLGSSLISWK 828
A +IL+Y+K + +GLF+S++SEL+L +A++D+ C D+RRS +GYC+ LG SLI WK
Sbjct: 1131 AVYKILQYIKGTIGQGLFYSATSELQLKVYANADYNSCRDSRRSTSGYCMFLGDSLICWK 1190
Query: 829 SKKQSTVSRSSTEAEYRALAHLTCELQWLNYLFHDL 864
S+KQ VS+SS EAEYR+L+ T EL WL +L
Sbjct: 1191 SRKQDVVSKSSAEAEYRSLSVATDELVWLTNFLKEL 1226
>pir||G86301 probable retroelement polyprotein [imported] - Arabidopsis thaliana
gi|9989054|gb|AAG10817.1| Putative retroelement
polyprotein [Arabidopsis thaliana]
Length = 1413
Score = 765 bits (1976), Expect = 0.0
Identities = 389/810 (48%), Positives = 527/810 (65%), Gaps = 43/810 (5%)
Query: 63 CDACHYAKQKRLPFPHSSIKSSAPFDLLHADLWGPYSTPSFLGHKYFLTLVDDYSRFTWV 122
CD C AKQK+L +P APFDLLH D+WGP+S P+ G+ YFLT+VDD++R TWV
Sbjct: 566 CDICQRAKQKKLTYPSRHNICLAPFDLLHIDVWGPFSEPTQEGYHYFLTIVDDHTRVTWV 625
Query: 123 IFLKTKDETQKHLKHFISYVENQFHTTLKCLRSDNGSEFIAMTSFLLSKGIIHHKTCVET 182
+K K + FI+ VE Q+ T +K +RSDN E + KGI+ + +C ET
Sbjct: 626 YLMKYKSDVLTIFPDFITMVETQYDTKVKAVRSDNAPE-LKFEELYRRKGIVAYHSCPET 684
Query: 183 PQQNGVVERKHQHILNVARSLAFHSHVPITMWNFTVQHAVHIINRIPSPLLKFKSPFELL 242
P+QN VVERKHQHILNVAR+L F S +P++ W + AV IINR PSP++ K+ FE+L
Sbjct: 685 PEQNSVVERKHQHILNVARALLFQSQIPLSYWGDCILTAVFIINRTPSPVISNKTLFEML 744
Query: 243 HKEPPSIIHLKVFGCLAYASTLQAHRTKFNPRARKTIFLGFKEGTKGSILYDLNSNELFV 302
K+ P HLK FGCL YAST R KF RAR FLG+ G KG L DL S+ +F+
Sbjct: 745 TKKVPDYTHLKSFGCLCYASTSPKQRHKFEDRARTCAFLGYPSGYKGYKLLDLESHTIFI 804
Query: 303 SRNVIFYENHFPF---TLATKQANIPTTSSHIDLGDPITDLSPHPISAPEFQLTSTPPSQ 359
SRNV+FYE+ FPF +++++ ++D D
Sbjct: 805 SRNVVFYEDLFPFKTKPAENEESSVFFPHIYVDRND------------------------ 840
Query: 360 YVSAPAVQHAIPVTDSISEPTVRKSTRISQRPSYLADYHCNLPSKSCSNVSSGISSYPLS 419
S P+ + T + + P ++++R+S+ P+YL DYHCN + S + +P+S
Sbjct: 841 --SHPSQPLPVQETSASNVPAEKQNSRVSRPPAYLKDYHCNSVTSS--------TDHPIS 890
Query: 420 SFLSYDNCSPTYTHFCCTISSINEPKTFAQANKSECWREAMTTELNALAKNNTWSVVTLP 479
LSY + S Y F ++ I EP T+AQA + + W +AM E+ AL N TW V +LP
Sbjct: 891 EVLSYSSLSDPYMIFINAVNKIPEPHTYAQARQIKEWCDAMGMEITALEDNGTWVVCSLP 950
Query: 480 PGKVPIGCKWVYKVKYHANGSIERYKARLVAQGYTQTEGVDYFDTFSPVAKLTTIRVLLS 539
GK +GCKWVYK+K +A+GS+ERYKARLVA+GYTQTEG+DY DTFSPVAKLTT+++L++
Sbjct: 951 VGKKAVGCKWVYKIKLNADGSLERYKARLVAKGYTQTEGLDYVDTFSPVAKLTTVKLLIA 1010
Query: 540 LAAIKGWHLEQLDVNNAFLHGDLHEEVYMALPPGY-----PTINSSQVCKLNKSLYGLKQ 594
+AA KGW L QLD++NAFL+G L EE+YM LPPGY + + VC+L KSLYGLKQ
Sbjct: 1011 VAAAKGWSLSQLDISNAFLNGSLDEEIYMTLPPGYSPRQGDSFPPNAVCRLKKSLYGLKQ 1070
Query: 595 ASRQWYSKLSTSLISFGYTQSLADYSLFVKVSGASFTALLVYVDDIVLAGNCISEIKSVK 654
ASRQWY K S SL + G+TQS D++LF + S S+ A+LVYVDDI++A +C E + ++
Sbjct: 1071 ASRQWYLKFSESLKALGFTQSSGDHTLFTRKSKNSYMAVLVYVDDIIIASSCDRETELLR 1130
Query: 655 TFLDNKFCIKDLGQLRYFLGFEIARSKSGILLNQRKYTLELLEDAGTLGSKPAATPFDPS 714
L ++DLG LRYFLG EIAR+ GI + QRKYTLELL + G LG K ++ P +P+
Sbjct: 1131 DALQRSSKLRDLGTLRYFLGLEIARNTDGISICQRKYTLELLAETGLLGCKSSSVPMEPN 1190
Query: 715 TKLGATTGTPFTDASSYRRLIGRLLYLTNTRPDISYSVQNLSQFVSRPMVPHYQAAQRIL 774
KL G DA YR+L+G+L+YLT TRPDI+Y+V L QF S P VPH +A +I+
Sbjct: 1191 QKLSQEDGELIDDAEHYRKLVGKLMYLTFTRPDITYAVHRLCQFTSAPRVPHLKAVYKII 1250
Query: 775 KYLKSSPAKGLFFSSSSELKLHGFADSDWACCPDTRRSVTGYCVLLGSSLISWKSKKQST 834
YLK + +GLF+S++ +LKL GFADSD++ C D+R+ TGYC+ LG+SL++WKSKKQ
Sbjct: 1251 YYLKGTVGQGLFYSANVDLKLSGFADSDFSSCSDSRKLTTGYCMFLGTSLVAWKSKKQEV 1310
Query: 835 VSRSSTEAEYRALAHLTCELQWLNYLFHDL 864
+S SS EAEY+A++ E+ WL +L DL
Sbjct: 1311 ISMSSAEAEYKAMSMAVREMMWLRFLLEDL 1340
>gb|AAD26943.1| putative retroelement pol polyprotein [Arabidopsis thaliana]
gi|25301694|pir||E84535 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana
Length = 1454
Score = 753 bits (1943), Expect = 0.0
Identities = 401/861 (46%), Positives = 539/861 (62%), Gaps = 42/861 (4%)
Query: 3 VGGLYLIAAGPSLANKLSCNSVFTDCFDVWHMRLGHVSSSGLSVISKQFPFIPCI-KNAP 61
V LYL+ G +S N+V +WH RLGH S L IS K +
Sbjct: 534 VANLYLLDVGDQ---SISVNAVVD--ISMWHRRLGHASLQRLDAISDSLGTTRHKNKGSD 588
Query: 62 PCDACHYAKQKRLPFPHSSIKSSAPFDLLHADLWGPYSTPSFLGHKYFLTLVDDYSRFTW 121
C CH AKQ++L FP S+ FDLLH D+WGP+S + G+KYFLT+VDD+SR TW
Sbjct: 589 FCHVCHLAKQRKLSFPTSNKVCKEIFDLLHIDVWGPFSVETVEGYKYFLTIVDDHSRATW 648
Query: 122 VIFLKTKDETQKHLKHFISYVENQFHTTLKCLRSDNGSEFIAMTSFLLSKGIIHHKTCVE 181
+ LKTK E FI VENQ+ +K +RSDN E + TSF KGI+ +C E
Sbjct: 649 MYLLKTKSEVLTVFPAFIQQVENQYKVKVKAVRSDNAPE-LKFTSFYAEKGIVSFHSCPE 707
Query: 182 TPQQNGVVERKHQHILNVARSLAFHSHVPITMWNFTVQHAVHIINRIPSPLLKFKSPFEL 241
TP+QN VVERKHQHILNVAR+L F S VP+++W V AV +INR PS LL K+P+E+
Sbjct: 708 TPEQNSVVERKHQHILNVARALMFQSQVPLSLWGDCVLTAVFLINRTPSQLLMNKTPYEI 767
Query: 242 LHKEPPSIIHLKVFGCLAYASTLQAHRTKFNPRARKTIFLGFKEGTKGSILYDLNSNELF 301
L P L+ FGCL Y+ST R KF PR+R +FLG+ G KG L DL SN +F
Sbjct: 768 LTGTAPVYEQLRTFGCLCYSSTSPKQRHKFQPRSRACLFLGYPSGYKGYKLMDLESNTVF 827
Query: 302 VSRNVIFYENHFPFTLATKQANIPTTSSHIDLGDPITDLSPHPISAPEFQLTSTPPSQYV 361
+SRNV F+E FP A P + S + L P+ P+S+ T+ PS
Sbjct: 828 ISRNVQFHEEVFPL------AKNPGSESSLKLFTPMV-----PVSSGIISDTTHSPS--- 873
Query: 362 SAPAVQHAIPVTDSISEPTVRKSTRISQRPSYLADYHCNLPSKSCSNVSSGISSYPLSSF 421
++P S P + S R+ + P++L DYHCN YP+SS
Sbjct: 874 -------SLPSQISDLPPQI-SSQRVRKPPAHLNDYHCNTMQSD--------HKYPISST 917
Query: 422 LSYDNCSPTYTHFCCTISSINEPKTFAQANKSECWREAMTTELNALAKNNTWSVVTLPPG 481
+SY SP++ + I+ I P +A+A ++ W EA+ E+ A+ K NTW + TLP G
Sbjct: 918 ISYSKISPSHMCYINNITKIPIPTNYAEAQDTKEWCEAVDAEIGAMEKTNTWEITTLPKG 977
Query: 482 KVPIGCKWVYKVKYHANGSIERYKARLVAQGYTQTEGVDYFDTFSPVAKLTTIRVLLSLA 541
K +GCKWV+ +K+ A+G++ERYKARLVA+GYTQ EG+DY DTFSPVAK+TTI++LL ++
Sbjct: 978 KKAVGCKWVFTLKFLADGNLERYKARLVAKGYTQKEGLDYTDTFSPVAKMTTIKLLLKVS 1037
Query: 542 AIKGWHLEQLDVNNAFLHGDLHEEVYMALPPGYP-----TINSSQVCKLNKSLYGLKQAS 596
A K W L+QLDV+NAFL+G+L EE++M +P GY + S+ V +L +S+YGLKQAS
Sbjct: 1038 ASKKWFLKQLDVSNAFLNGELEEEIFMKIPEGYAERKGIVLPSNVVLRLKRSIYGLKQAS 1097
Query: 597 RQWYSKLSTSLISFGYTQSLADYSLFVKVSGASFTALLVYVDDIVLAGNCISEIKSVKTF 656
RQW+ K S+SL+S G+ ++ D++LF+K+ F +LVYVDDIV+A + +
Sbjct: 1098 RQWFKKFSSSLLSLGFKKTHGDHTLFLKMYDGEFVIVLVYVDDIVIASTSEAAAAQLTEE 1157
Query: 657 LDNKFCIKDLGQLRYFLGFEIARSKSGILLNQRKYTLELLEDAGTLGSKPAATPFDPSTK 716
LD +F ++DLG L+YFLG E+AR+ +GI + QRKY LELL+ G L KP + P P+ K
Sbjct: 1158 LDQRFKLRDLGDLKYFLGLEVARTTAGISICQRKYALELLQSTGMLACKPVSVPMIPNLK 1217
Query: 717 LGATTGTPFTDASSYRRLIGRLLYLTNTRPDISYSVQNLSQFVSRPMVPHYQAAQRILKY 776
+ G D YRR++G+L+YLT TRPDI+++V L QF S P H AA R+L+Y
Sbjct: 1218 MRKDDGDLIEDIEQYRRIVGKLMYLTITRPDITFAVNKLCQFSSAPRTTHLTAAYRVLQY 1277
Query: 777 LKSSPAKGLFFSSSSELKLHGFADSDWACCPDTRRSVTGYCVLLGSSLISWKSKKQSTVS 836
+K + +GLF+S+SS+L L GFADSDWA C D+RRS T + + +G SLISW+SKKQ TVS
Sbjct: 1278 IKGTVGQGLFYSASSDLTLKGFADSDWASCQDSRRSTTSFTMFVGDSLISWRSKKQHTVS 1337
Query: 837 RSSTEAEYRALAHLTCELQWL 857
RSS EAEYRALA TCE+ WL
Sbjct: 1338 RSSAEAEYRALALATCEMVWL 1358
>gb|AAD25646.1| putative retroelement pol polyprotein [Arabidopsis thaliana]
gi|25301701|pir||E84589 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana
Length = 1461
Score = 744 bits (1922), Expect = 0.0
Identities = 395/864 (45%), Positives = 546/864 (62%), Gaps = 46/864 (5%)
Query: 3 VGGLYLIAAGPSLANKLSCNSVFTDCFDVWHMRLGHVSSSGLSVISKQFPFIPCI-KNAP 61
+G LY++ + + +S N+V VWH RLGH S S L +S+ K +
Sbjct: 545 IGNLYVL---DTQSPAISVNAVVD--VSVWHKRLGHPSFSRLDSLSEVLGTTRHKNKKSA 599
Query: 62 PCDACHYAKQKRLPFPHSSIKSSAPFDLLHADLWGPYSTPSFLGHKYFLTLVDDYSRFTW 121
C CH AKQK+L FP ++ ++ F+LLH D+WGP+S + G+KYFLT+VDD+SR TW
Sbjct: 600 YCHVCHLAKQKKLSFPSANNICNSTFELLHIDVWGPFSVETVEGYKYFLTIVDDHSRATW 659
Query: 122 VIFLKTKDETQKHLKHFISYVENQFHTTLKCLRSDNGSEFIAMTSFLLSKGIIHHKTCVE 181
+ LK+K + FI VENQ+ T +K +RSDN E +A T F +KGI+ +C E
Sbjct: 660 IYLLKSKSDVLTVFPAFIDLVENQYDTRVKSVRSDNAKE-LAFTEFYKAKGIVSFHSCPE 718
Query: 182 TPQQNGVVERKHQHILNVARSLAFHSHVPITMWNFTVQHAVHIINRIPSPLLKFKSPFEL 241
TP+QN VVERKHQHILNVAR+L F S++ + W V AV +INR PS LL K+PFE+
Sbjct: 719 TPEQNSVVERKHQHILNVARALMFQSNMSLPYWGDCVLTAVFLINRTPSALLSNKTPFEV 778
Query: 242 LHKEPPSIIHLKVFGCLAYASTLQAHRTKFNPRARKTIFLGFKEGTKGSILYDLNSNELF 301
L + P LK FGCL Y+ST R KF PR+R +FLG+ G KG L DL SN +
Sbjct: 779 LTGKLPDYSQLKTFGCLCYSSTSSKQRHKFLPRSRACVFLGYPFGFKGYKLLDLESNVVH 838
Query: 302 VSRNVIFYENHFPFTLATKQANIPTTSSHIDLGDPITDLSPHPISAPEFQLTSTPPSQYV 361
+SRNV F+E FP LA+ Q + T S DP++ + +TS PS +
Sbjct: 839 ISRNVEFHEELFP--LASSQQSATTASDVFTPMDPLSSGN---------SITSHLPSPQI 887
Query: 362 SAPAVQHAIPVTDSISEPTVRKSTRISQRPSYLADYHCNLPSKSCSNVSSGISSYPLSSF 421
S P+ Q + RI++ P++L DYHC +K S+P+SS
Sbjct: 888 S-PSTQIS--------------KRRITKFPAHLQDYHCYFVNKD--------DSHPISSS 924
Query: 422 LSYDNCSPTYTHFCCTISSINEPKTFAQANKSECWREAMTTELNALAKNNTWSVVTLPPG 481
LSY SP++ + IS I P+++ +A S+ W A+ E+ A+ + +TW + +LPPG
Sbjct: 925 LSYSQISPSHMLYINNISKIPIPQSYHEAKDSKEWCGAIDQEIGAMERTDTWEITSLPPG 984
Query: 482 KVPIGCKWVYKVKYHANGSIERYKARLVAQGYTQTEGVDYFDTFSPVAKLTTIRVLLSLA 541
K +GCKWV+ VK+HA+GS+ER+KAR+VA+GYTQ EG+DY +TFSPVAK+ T+++LL ++
Sbjct: 985 KKAVGCKWVFTVKFHADGSLERFKARIVAKGYTQKEGLDYTETFSPVAKMATVKLLLKVS 1044
Query: 542 AIKGWHLEQLDVNNAFLHGDLHEEVYMALPPGYPTINSSQ-----VCKLNKSLYGLKQAS 596
A K W+L QLD++NAFL+GDL E +YM LP GY I + VC+L KS+YGLKQAS
Sbjct: 1045 ASKKWYLNQLDISNAFLNGDLEETIYMKLPDGYADIKGTSLPPNVVCRLKKSIYGLKQAS 1104
Query: 597 RQWYSKLSTSLISFGYTQSLADYSLFVKVSGASFTALLVYVDDIVLAGNCISEIKSVKTF 656
RQW+ K S SL++ G+ + D++LFV+ G+ F LLVYVDDIV+A +S+
Sbjct: 1105 RQWFLKFSNSLLALGFEKQHGDHTLFVRCIGSEFIVLLVYVDDIVIASTTEQAAQSLTEA 1164
Query: 657 LDNKFCIKDLGQLRYFLGFEIARSKSGILLNQRKYTLELLEDAGTLGSKPAATPFDPSTK 716
L F +++LG L+YFLG E+AR+ GI L+QRKY LELL A L KP++ P P+ +
Sbjct: 1165 LKASFKLRELGPLKYFLGLEVARTSEGISLSQRKYALELLTSADMLDCKPSSIPMTPNIR 1224
Query: 717 LGATTGTPFTDASSYRRLIGRLLYLTNTRPDISYSVQNLSQFVSRPMVPHYQAAQRILKY 776
L G D YRRL+G+L+YLT TRPDI+++V L QF S P H A ++L+Y
Sbjct: 1225 LSKNDGLLLEDKEMYRRLVGKLMYLTITRPDITFAVNKLCQFSSAPRTAHLAAVYKVLQY 1284
Query: 777 LKSSPAKGLFFSSSSELKLHGFADSDWACCPDTRRSVTGYCVLLGSSLISWKSKKQSTVS 836
+K + +GLF+S+ +L L G+ D+DW CPD+RRS TG+ + +GSSLISW+SKKQ TVS
Sbjct: 1285 IKGTVGQGLFYSAEDDLTLKGYTDADWGTCPDSRRSTTGFTMFVGSSLISWRSKKQPTVS 1344
Query: 837 RSSTEAEYRALAHLTCELQWLNYL 860
RSS EAEYRALA +CE+ WL+ L
Sbjct: 1345 RSSAEAEYRALALASCEMAWLSTL 1368
>gb|AAC33963.1| contains similarity to reverse transcriptases (Pfam; rvt.hmm, score:
11.19) [Arabidopsis thaliana] gi|7486705|pir||T01879
hypothetical protein F8M12.17 - Arabidopsis thaliana
Length = 1633
Score = 729 bits (1883), Expect = 0.0
Identities = 400/859 (46%), Positives = 536/859 (61%), Gaps = 57/859 (6%)
Query: 44 LSVISKQFPFIPCIKN----APPCDACHYAKQKRLPFPHSSIKSSAPFDLLHADLWGPYS 99
L + K IP +K+ A C AKQKRL + + +S+PFDL+H D+WGP+S
Sbjct: 509 LPALQKLVSSIPSLKSVSSTASHCRISPLAKQKRLAYVSHNNLASSPFDLIHLDIWGPFS 568
Query: 100 TPSFLGHKYFLTLVDDYSRFTWVIFLKTKDETQKHLKHFISYVENQFHTTLKCLRSDNGS 159
S G +YFLTLVDD +R TWV +K K E F+ + Q++ +K +RSDN
Sbjct: 569 IESVDGFRYFLTLVDDCTRTTWVYMMKNKSEVSNIFPVFVKLIFTQYNAKIKAIRSDNVK 628
Query: 160 EFIAMTSFLLSKGIIHHKTCVETPQQNGVVERKHQHILNVARSLAFHSHVPITMWNFTVQ 219
E +A T F+ +G+IH +C TPQQN VVERKHQH+LN+ARSL F S+VP+ W+ V
Sbjct: 629 E-LAFTKFVKEQGMIHQFSCAYTPQQNSVVERKHQHLLNIARSLLFQSNVPLQYWSDCVL 687
Query: 220 HAVHIINRIPSPLLKFKSPFELLHKEPPSIIHLKVFGCLAYASTLQAHRTKFNPRARKTI 279
A ++INR+PSPLL K+PFELL K+ P LK CL YAST R KF+PRAR +
Sbjct: 688 TAAYLINRLPSPLLDNKTPFELLLKKIPDYTLLK--SCLCYASTNVHDRNKFSPRARPCV 745
Query: 280 FLGFKEGTKGSILYDLNSNELFVSRNVIFYENHFPFTLAT--KQANIPTTSSHIDLGDPI 337
FLG+ G KG + DL S+ + ++RNV+F+E FPF + K++ +S + L P+
Sbjct: 746 FLGYPSGYKGYKVLDLESHSISITRNVVFHETKFPFKTSKFLKESVDMFPNSILPLPAPL 805
Query: 338 TDLSPHPI-----SAPEFQLTSTPPSQYVSAPAVQHAIPV--TDSISEPT----VRKSTR 386
+ P+ + TS S S P + + TD++ T + + R
Sbjct: 806 HFVESMPLDDDLRADDNNASTSNSASSASSIPPLPSTVNTQNTDALDIDTNSVPIARPKR 865
Query: 387 ISQRPSYLADYHCN----------LPSKSCSNVSSGI------SSYPLSSFLSYDNCSPT 430
++ P+YL++YHCN S S SS I + YP+S+ +SYD +P
Sbjct: 866 NAKAPAYLSEYHCNSVPFLSSLSPTTSTSIETPSSSIPPKKITTPYPMSTAISYDKLTPL 925
Query: 431 YTHFCCTISSINEPKTFAQANKSECWREAMTTELNALAKNNTWSVVTLPPGKVPIGCKWV 490
+ + C + EPK F QA KSE W A EL+AL +N TW V +L GK +GCKWV
Sbjct: 926 FHSYICAYNVETEPKAFTQAMKSEKWTRAANEELHALEQNKTWIVESLTEGKNVVGCKWV 985
Query: 491 YKVKYHANGSIERYKARLVAQGYTQTEGVDYFDTFSPVAKLTTIRVLLSLAAIKGWHLEQ 550
+ +KY+ +GSIERYKARLVAQG+TQ EG+DY +TFSPVAK ++++LL LAA GW L Q
Sbjct: 986 FTIKYNPDGSIERYKARLVAQGFTQQEGIDYMETFSPVAKFGSVKLLLGLAAATGWSLTQ 1045
Query: 551 LDVNNAFLHGDLHEEVYMALPPGY--PT---INSSQVCKLNKSLYGLKQASRQWYSKLST 605
+DV+NAFLHG+L EE+YM+LP GY PT + S VC+L KSLYGLKQASRQWY +LS+
Sbjct: 1046 MDVSNAFLHGELDEEIYMSLPQGYTPPTGISLPSKPVCRLLKSLYGLKQASRQWYKRLSS 1105
Query: 606 SLISFGYTQSLADYSLFVKVSGASFTALLVYVDDIVLAGNCISEIKSVKTFLDNKFCIKD 665
+ + QS AD ++FVKVS S +LVYVDD+++A N S ++++K L ++F IKD
Sbjct: 1106 VFLGANFIQSPADNTMFVKVSCTSIIVVLVYVDDLMIASNDSSAVENLKELLRSEFKIKD 1165
Query: 666 LGQLRYFLGFEIARSKSGILLNQRKYTLELLEDAGTLGSKPAATPFDPSTKLGATTGTPF 725
LG R+FLG EIARS GI + QRKY LLED G G KP++ P DP+ L GT
Sbjct: 1166 LGPARFFLGLEIARSSEGISVCQRKYAQNLLEDVGLSGCKPSSIPMDPNLHLTKEMGTLL 1225
Query: 726 TDASSYRRLIGRLLYLTNTRPDISYSVQNLSQFVSRPMVPHYQAAQRILKYLKSSPAKGL 785
+A+SYR L+GRLLYL TRPDI+++V LSQF+S P H QAA ++L+YLK +P +
Sbjct: 1226 PNATSYRELVGRLLYLCITRPDITFAVHTLSQFLSAPTDIHMQAAHKVLRYLKGNPGQ-- 1283
Query: 786 FFSSSSELKLHGFADSDWACCPDTRRSVTGYCVLLGSSLISWKSKKQSTVSRSSTEAEYR 845
D+DW C D+RRSVTG+C+ LG+SLI+WKSKKQS VSRSSTE+EYR
Sbjct: 1284 --------------DADWGTCKDSRRSVTGFCIYLGTSLITWKSKKQSVVSRSSTESEYR 1329
Query: 846 ALAHLTCELQWLNYLFHDL 864
+LA TCE+ WL L DL
Sbjct: 1330 SLAQATCEIIWLQQLLKDL 1348
>dbj|BAA97287.1| retroelement pol polyprotein-like [Arabidopsis thaliana]
Length = 1491
Score = 725 bits (1872), Expect = 0.0
Identities = 407/920 (44%), Positives = 548/920 (59%), Gaps = 65/920 (7%)
Query: 4 GGLYLIAAGPSLANKLSCNSVFTDCFDVWHMRLGHVSSSGLSVISKQFPFIPCIKNAPPC 63
G YL A + +K+ V TD +WH RLGH S S LS + F C ++ C
Sbjct: 489 GVYYLTDAATTTVHKVD---VTTD-HALWHQRLGHPSFSVLSSLPL-FSGSSCSVSSRSC 543
Query: 64 DACHYAKQKRLPFPHSSIKSSAPFDLLHADLWGPYSTPSFLGHKYFLTLVDDYSRFTWVI 123
D C AKQ R FP SS KS+ F L+H D+WGPY PS G YFLT+VDD+SR W
Sbjct: 544 DVCFRAKQTREVFPDSSNKSTDCFSLIHCDVWGPYRVPSSCGAVYFLTIVDDFSRSVWTY 603
Query: 124 FLKTKDETQKHLKHFISYVENQFHTTLKCLRSDNGSEFIAMTSFLLSKGIIHHKTCVETP 183
L K E + L +F++Y E QF ++K +RSDNG+EF+ ++S+ +GI+H +CV TP
Sbjct: 604 LLLAKSEVRSVLTNFLAYTEKQFGKSVKIIRSDNGTEFMCLSSYFKEQGIVHQTSCVGTP 663
Query: 184 QQNGVVERKHQHILNVARSLAFHSHVPITMWNFTVQHAVHIINRIPSPLLKFKSPFELLH 243
QQNG VERKH+HILNV+R+L F + +PI W V A ++INR PS + SP+ELLH
Sbjct: 664 QQNGRVERKHRHILNVSRALLFQASLPIKFWGEAVMTAAYLINRTPSSIHNGLSPYELLH 723
Query: 244 KEPPSIIHLKVFGCLAYASTLQAHRTKFNPRARKTIFLGFKEGTKGSILYDLNSNELFVS 303
P L+VFG YA + + KF R+R IF+G+ G KG +YDL++NE VS
Sbjct: 724 GCKPDYDQLRVFGSACYAHRVTRDKDKFGERSRLCIFVGYPFGQKGWKVYDLSTNEFIVS 783
Query: 304 RNVIFYENHFPF------TLATKQANIPTT---------------SSHIDLGDP---ITD 339
R+V+F EN FP+ T+ T P T S L DP +TD
Sbjct: 784 RDVVFRENVFPYATNEGDTIYTPPVTCPITYDEDWLPFTTLEDRGSDENSLSDPPVCVTD 843
Query: 340 LS------------PHPISAPEFQLTSTPPSQ------YVSAPAVQHAIPVTDS---ISE 378
+S P P+ P TS P+Q ++P+ + P D+ I
Sbjct: 844 VSESDTEHDTPQSLPTPVDDPLSPSTSVTPTQTPTNSSSSTSPSTNVSPPQQDTTPIIEN 903
Query: 379 PTVRKSTRISQRPSYLADY------------HCNLPSKSCSNVS-SGISSYPLSSFLSYD 425
R+ R Q+ + L DY H PS S S+ S G S YPL+ ++ +D
Sbjct: 904 TPPRQGKRQVQQLARLKDYILYNASCTPNTPHVLSPSTSQSSSSIQGNSQYPLTDYI-FD 962
Query: 426 NC-SPTYTHFCCTISSINEPKTFAQANKSECWREAMTTELNALAKNNTWSVVTLPPGKVP 484
C S + F I++ +EPK F +A K + W +AM E++AL N TW +V LP GKV
Sbjct: 963 ECFSAGHKVFLAAITANDEPKHFKEAVKVKVWNDAMYKEVDALEVNKTWDIVDLPTGKVA 1022
Query: 485 IGCKWVYKVKYHANGSIERYKARLVAQGYTQTEGVDYFDTFSPVAKLTTIRVLLSLAAIK 544
IG +WVYK K++A+G++ERYKARLV QG Q EG DY +TF+PV K+TT+R LL L A
Sbjct: 1023 IGSQWVYKTKFNADGTVERYKARLVVQGNNQIEGEDYTETFAPVVKMTTVRTLLRLVAAN 1082
Query: 545 GWHLEQLDVNNAFLHGDLHEEVYMALPPGYPTINSSQVCKLNKSLYGLKQASRQWYSKLS 604
W + Q+DV+NAFLHGDL EEVYM LPPG+ + +VC+L KSLYGLKQA R W+ KLS
Sbjct: 1083 QWEVYQMDVHNAFLHGDLEEEVYMKLPPGFRHSHPDKVCRLRKSLYGLKQAPRCWFKKLS 1142
Query: 605 TSLISFGYTQSLADYSLFVKVSGASFTALLVYVDDIVLAGNCISEIKSVKTFLDNKFCIK 664
+L FG+ Q DYS F +LVYVDD+++ GN ++ K +L F +K
Sbjct: 1143 DALKRFGFIQGYEDYSFFSYSCKGIELRVLVYVDDLIICGNDEYMVQKFKEYLGRCFSMK 1202
Query: 665 DLGQLRYFLGFEIARSKSGILLNQRKYTLELLEDAGTLGSKPAATPFDPSTKLGATTGTP 724
DLG+L+YFLG E++R GI L+QRKY L+++ D+GTLG++PA TP + + L + G
Sbjct: 1203 DLGKLKYFLGIEVSRGPDGIFLSQRKYALDIISDSGTLGARPAYTPLEQNHHLASDDGPL 1262
Query: 725 FTDASSYRRLIGRLLYLTNTRPDISYSVQNLSQFVSRPMVPHYQAAQRILKYLKSSPAKG 784
D +RRL+GRLLYL +TRP++SYSV LSQF+ P H +AA RI++YLK SP +G
Sbjct: 1263 LQDPKPFRRLVGRLLYLLHTRPELSYSVHVLSQFMQAPREAHLEAAMRIVRYLKGSPGQG 1322
Query: 785 LFFSSSSELKLHGFADSDWACCPDTRRSVTGYCVLLGSSLISWKSKKQSTVSRSSTEAEY 844
+ SS+ +L L + DSD+ CP TRRS++ Y VLLG S ISWK+KKQ TVS SS EAEY
Sbjct: 1323 ILLSSNKDLTLEVYCDSDFQSCPLTRRSLSAYVVLLGGSPISWKTKKQDTVSHSSAEAEY 1382
Query: 845 RALAHLTCELQWLNYLFHDL 864
RA++ E++WLN L +L
Sbjct: 1383 RAMSVALKEIKWLNKLLKEL 1402
>gb|AAD19784.1| putative retroelement pol polyprotein [Arabidopsis thaliana]
gi|25301698|pir||C84512 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana
Length = 1501
Score = 725 bits (1871), Expect = 0.0
Identities = 390/887 (43%), Positives = 526/887 (58%), Gaps = 56/887 (6%)
Query: 31 VWHMRLGHVSSSGLSVISKQFPFIPCIKNAPPCDACHYAKQKRLPFPHSSIKSSAPFDLL 90
+WH RLGH S S LS + F + CD C AKQ R FP S K+ F L+
Sbjct: 529 LWHQRLGHPSFSVLSSLPL-FSKTSSTVTSHSCDVCFRAKQTREVFPESINKTEECFSLI 587
Query: 91 HADLWGPYSTPSFLGHKYFLTLVDDYSRFTWVIFLKTKDETQKHLKHFISYVENQFHTTL 150
H D+WGPY P+ G YFLT+VDDYSR W L K E ++ L +F+ Y E QF T+
Sbjct: 588 HCDVWGPYRVPASCGAVYFLTIVDDYSRAVWTYLLLEKSEVRQVLTNFLKYAEKQFGKTV 647
Query: 151 KCLRSDNGSEFIAMTSFLLSKGIIHHKTCVETPQQNGVVERKHQHILNVARSLAFHSHVP 210
K +RSDNG+EF+ ++S+ GIIH +CV TPQQNG VERKH+HILNVAR+L F + +P
Sbjct: 648 KMVRSDNGTEFMCLSSYFRENGIIHQTSCVGTPQQNGRVERKHRHILNVARALLFQASLP 707
Query: 211 ITMWNFTVQHAVHIINRIPSPLLKFKSPFELLHKEPPSIIHLKVFGCLAYASTLQAHRTK 270
I W ++ A ++INR PS +L ++P+E+LH P L+VFG Y + + K
Sbjct: 708 IKFWGESILTAAYLINRTPSSILSGRTPYEVLHGSKPVYSQLRVFGSACYVHRVTRDKDK 767
Query: 271 FNPRARKTIFLGFKEGTKGSILYDLNSNELFVSRNVIFYENHFPF------TLATKQANI 324
F R+R IF+G+ G KG +YD+ NE VSR+VIF E FP+ TLA+ ++
Sbjct: 768 FGQRSRSCIFVGYPFGKKGWKVYDIERNEFLVSRDVIFREEVFPYAGVNSSTLAS--TSL 825
Query: 325 PTTSSHIDLGDP-------------------------ITDLSPHPISAPEFQLTSTPPSQ 359
PT S D P T +S I EF TPPS
Sbjct: 826 PTVSEDDDWAIPPLEVRGSIDSVETERVVCTTDEVVLDTSVSDSEIPNQEFVPDDTPPSS 885
Query: 360 YVS--------APAVQHAIPVTDSI--SEPTVRKSTRISQRPSYLADY------------ 397
+S P +PV I S P RKS R + P L DY
Sbjct: 886 PLSVSPSGSPNTPTTPIVVPVASPIPVSPPKQRKSKRATHPPPKLNDYVLYNAMYTPSSI 945
Query: 398 HCNLPSKSCSNVSSGISSYPLSSFLSYDNCSPTYTHFCCTISSINEPKTFAQANKSECWR 457
H S S+ G S +PL+ ++S S ++ + I+ EPK F +A + + W
Sbjct: 946 HALPADPSQSSTVPGKSLFPLTDYVSDAAFSSSHRAYLAAITDNVEPKHFKEAVQIKVWN 1005
Query: 458 EAMTTELNALAKNNTWSVVTLPPGKVPIGCKWVYKVKYHANGSIERYKARLVAQGYTQTE 517
+AM TE++AL N TW +V LPPGKV IG +WV+K KY+++G++ERYKARLV QG Q E
Sbjct: 1006 DAMFTEVDALEINKTWDIVDLPPGKVAIGSQWVFKTKYNSDGTVERYKARLVVQGNKQVE 1065
Query: 518 GVDYFDTFSPVAKLTTIRVLLSLAAIKGWHLEQLDVNNAFLHGDLHEEVYMALPPGYPTI 577
G DY +TF+PV ++TT+R LL A W + Q+DV+NAFLHGDL EEVYM LPPG+
Sbjct: 1066 GEDYKETFAPVVRMTTVRTLLRNVAANQWEVYQMDVHNAFLHGDLEEEVYMKLPPGFRHS 1125
Query: 578 NSSQVCKLNKSLYGLKQASRQWYSKLSTSLISFGYTQSLADYSLFVKVSGASFTALLVYV 637
+ +VC+L KSLYGLKQA R W+ KLS SL+ FG+ QS DYSLF +L+YV
Sbjct: 1126 HPDKVCRLRKSLYGLKQAPRCWFKKLSDSLLRFGFVQSYEDYSLFSYTRNNIELRVLIYV 1185
Query: 638 DDIVLAGNCISEIKSVKTFLDNKFCIKDLGQLRYFLGFEIARSKSGILLNQRKYTLELLE 697
DD+++ GN ++ K +L F +KDLG+L+YFLG E++R GI L+QRKY L+++
Sbjct: 1186 DDLLICGNDGYMLQKFKDYLSRCFSMKDLGKLKYFLGIEVSRGPEGIFLSQRKYALDVIA 1245
Query: 698 DAGTLGSKPAATPFDPSTKLGATTGTPFTDASSYRRLIGRLLYLTNTRPDISYSVQNLSQ 757
D+G LGS+PA TP + + L + G +D YRRL+GRLLYL +TRP++SYSV L+Q
Sbjct: 1246 DSGNLGSRPAHTPLEQNHHLASDDGPLLSDPKPYRRLVGRLLYLLHTRPELSYSVHVLAQ 1305
Query: 758 FVSRPMVPHYQAAQRILKYLKSSPAKGLFFSSSSELKLHGFADSDWACCPDTRRSVTGYC 817
F+ P H+ AA R+++YLK SP +G+ ++ +L L + DSDW CP TRRS++ Y
Sbjct: 1306 FMQNPREAHFDAALRVVRYLKGSPGQGILLNADPDLTLEVYCDSDWQSCPLTRRSISAYV 1365
Query: 818 VLLGSSLISWKSKKQSTVSRSSTEAEYRALAHLTCELQWLNYLFHDL 864
VLLG S ISWK+KKQ TVS SS EAEYRA+++ E++WL L +L
Sbjct: 1366 VLLGGSPISWKTKKQDTVSHSSAEAEYRAMSYALKEIKWLRKLLKEL 1412
>gb|AAU89728.1| putative retroelement pol polyprotein-like [Solanum tuberosum]
Length = 1476
Score = 695 bits (1794), Expect = 0.0
Identities = 387/857 (45%), Positives = 512/857 (59%), Gaps = 60/857 (7%)
Query: 30 DVWHMRLGHVSSSGLSVISKQFPFIPCIKNAPPCDACHYAKQKRLPFPHSSIKSSAPFDL 89
++WH RLGH+ S L I K F P P CD C A+Q RLPFP S +S FDL
Sbjct: 551 EMWHKRLGHIPMSVLRKI-KMFDS-PQKLVLPSCDVCPLARQVRLPFPISQSRSENCFDL 608
Query: 90 LHADLWGPYSTPSFLGHKYFLTLVDDYSRFTWVIFLKTKDETQKHLKHFISYVENQFHTT 149
+H D+WGPY + +YFLT+VDD+SR+TW+ + K + L++FI ++ QF
Sbjct: 609 IHLDVWGPYKAATHNKMRYFLTVVDDHSRWTWIFLMHLKSDVSTVLQNFILMIDTQFGQK 668
Query: 150 LKCLRSDNGSEFI--AMTSFLLSKGIIHHKTCVETPQQNGVVERKHQHILNVARSLAFHS 207
+K RSDNG+EF S GI+H +C TPQQNGVVER+H+HIL AR+L F
Sbjct: 669 IKIFRSDNGTEFFNAQCDGLFKSHGIVHQSSCPHTPQQNGVVERRHKHILETARALRFQG 728
Query: 208 HVPITMWNFTVQHAVHIINRIPSPLLKFKSPFELLHKEPPSIIHLKVFGCLAYASTLQAH 267
H+PI W V AVHIINRIPS +L KSPFEL++K P + +++V GCL +A+ L
Sbjct: 729 HLPIRFWGECVLSAVHIINRIPSSVLHNKSPFELMYKRSPDLSYMRVIGCLCHATNLVNT 788
Query: 268 RTKFNPRARKTIFLGFKEGTKGSILYDLNSNELFVSRNVIFYENHFPF---TLATKQAN- 323
T+ KG LYDL FVSR+++F E FPF LA
Sbjct: 789 STQ-----------------KGYKLYDLEHQHFFVSRDMVFNEAVFPFQSPALADPHDTP 831
Query: 324 ----IPTTSSHIDLGDPITDLSPHPISAPEFQLTSTPPSQYVSAPAVQHAIPVTDSISEP 379
P SSH + D + P I++ E ++PPS A + H P P
Sbjct: 832 VFLASPPCSSHTEDADAV---QPAIITSEEIIPVASPPS----AVSDDHLHP------PP 878
Query: 380 TVRKSTRISQRPSYLADYHCNLPSKSCSNVSSGISSYPLSSFLSYDNCSPTYTHFCCTIS 439
R+S R + P + D+ S+S + YP+S + Y S TY + + S
Sbjct: 879 ERRRSYRTGKPPIWQKDFITTSTSRSNHCL------YPISDNIDYSCLSSTYQCYIASSS 932
Query: 440 SINEPKTFAQANKSECWREAMTTELNALAKNNTWSVVTLPPGKVPIGCKWVYKVKYHANG 499
EP+ + QA W AM E+ AL N TW VV+LP GK IGCKWVYK+KY A+G
Sbjct: 933 VETEPQFYYQAANDCRWVHAMKEEIQALEDNKTWEVVSLPKGKKAIGCKWVYKIKYKASG 992
Query: 500 SIERYKARLVAQGYTQTEGVDYFDTFSPVAKLTTIRVLLSLAAIKGWHLEQLDVNNAFLH 559
IER+KARLVA+GY Q EG+DY +TFSPV K+ T+R +L+LA KGW ++Q+DV NAFL
Sbjct: 993 EIERFKARLVAKGYNQKEGLDYQETFSPVVKMVTLRTVLTLAVSKGWDIQQMDVYNAFLQ 1052
Query: 560 GDLHEEVYMALPPG--YPTINSSQVCKLNKSLYGLKQASRQWYSKLSTSLISFGYTQSLA 617
GDL EEVYM LP G Y +VC+L KSLYGLKQASRQW KL+T+L++ G+ QS
Sbjct: 1053 GDLIEEVYMQLPQGFQYDKTGDPKVCRLLKSLYGLKQASRQWNVKLTTALLAAGFQQSHL 1112
Query: 618 DYSLFVKVSGASFTALLVYVDDIVLAGNCISEIKSVKTFLDNKFCIKDLGQLRYFLGFEI 677
DYSL +K + +L+YVDD+++ G+ + I K L F IKDLG LRYFLG E
Sbjct: 1113 DYSLMLKRTADGIVIVLIYVDDLLITGSSLQLIDDAKQVLKANFKIKDLGTLRYFLGMEF 1172
Query: 678 ARSKSGILLNQRKYTLELLEDAGTLGSKPAATPFDPSTKL----------GATTGTPFTD 727
AR+ SG+L++QRKY LEL+ D G GSKP+ TP + KL + + D
Sbjct: 1173 ARNASGMLMHQRKYALELISDLGLGGSKPSVTPVELHLKLTTREFDLHVGSSGADSLLAD 1232
Query: 728 ASSYRRLIGRLLYLTNTRPDISYSVQNLSQFVSRPMVPHYQAAQRILKYLKSSPAKGLFF 787
+ Y+RL+GRLLYLT TRPDIS++VQ+LSQF+ P V H +AA R++KY+K +P GL+
Sbjct: 1233 PTEYQRLVGRLLYLTITRPDISFAVQHLSQFMHAPKVSHMEAAIRVVKYVKQAPGLGLYM 1292
Query: 788 SSSSELKLHGFADSDWACCPDTRRSVTGYCVLLGSSLISWKSKKQSTVSRSSTEAEYRAL 847
+ + L + D+DW C +TR+S+TGY + GS+L+SWKSKKQ T+SRSS EAEYR+L
Sbjct: 1293 AVQTADTLQAYCDADWGSCINTRKSITGYMIQFGSALLSWKSKKQPTISRSSAEAEYRSL 1352
Query: 848 AHLTCELQWLNYLFHDL 864
A EL WL LF +L
Sbjct: 1353 ASTVAELVWLTGLFKEL 1369
>emb|CAB10225.1| retrovirus-related like polyprotein [Arabidopsis thaliana]
gi|7268152|emb|CAB78488.1| retrovirus-related like
polyprotein [Arabidopsis thaliana] gi|7488175|pir||G71406
probable retrovirus-related polyprotein - Arabidopsis
thaliana
Length = 1489
Score = 688 bits (1775), Expect = 0.0
Identities = 386/909 (42%), Positives = 530/909 (57%), Gaps = 126/909 (13%)
Query: 2 LVGGLYLIAA---GPSLANKLSC---NSVFTDCFDVWHMRLGHVSSSGLSVISKQFPFIP 55
L LY++ PS + +C SV D +WH RLGH SS L
Sbjct: 567 LYHNLYILETENTSPSTSTPAACLFTGSVLNDGH-LWHQRLGHPSSVVLQ---------- 615
Query: 56 CIKNAPPCDACHYAKQKRLPFPHSSIKSSAPFDLLHADLWGPYSTPSFLGHKYFLTLVDD 115
K KRL + + +S PFDL+H D+WGP+S S G +YFLT+VDD
Sbjct: 616 --------------KLKRLAYISHNNLASNPFDLVHLDIWGPFSIESIEGFRYFLTVVDD 661
Query: 116 YSRFTWVIFLKTKDETQKHLKHFISYVENQFHTTLKCLRSDNGSEFIAMTSFLLSKGIIH 175
+R TWV L+ K + FI V QF+ +K +RSDN E + T + G++H
Sbjct: 662 CTRTTWVYMLRNKKDVSSVFPEFIKLVSTQFNAKIKAIRSDNAPE-LGFTEIVKEHGMLH 720
Query: 176 HKTCVETPQQNGVVERKHQHILNVARSLAFHSHVPITMWNFTVQHAVHIINRIPSPLLKF 235
H +C TPQQN VVERKHQHILNVAR+L F S++P+ W+ V AV +INR+PSPLL
Sbjct: 721 HFSCAYTPQQNSVVERKHQHILNVARALLFQSNIPMQYWSDCVTTAVFLINRLPSPLLNN 780
Query: 236 KSPFELLHKEPPSIIHLKVFGCLAYASTLQAHRTKFNPRARKTIFLGFKEGTKGSILYDL 295
KSP+EL+ + P LK FGCL + ST RTKF PRAR +FLG+ G KG + DL
Sbjct: 781 KSPYELILNKQPDYSLLKNFGCLCFVSTNAHERTKFTPRARACVFLGYPSGYKGYKVLDL 840
Query: 296 NSNELFVSRNVIFYENHFPFTLAT--KQANIPTTSSHIDLGDPITDLSPHPISAPEFQLT 353
S+ + VSRNV+F E+ FPF + +A +S + L P+ + P+ + +
Sbjct: 841 ESHSVTVSRNVVFKEHVFPFKTSELLNKAVDMFPNSILPLPAPLHFVETMPLIDEDSLIP 900
Query: 354 STPPSQYV------SAPAVQHAIPVT--------DSISEPTVRKSTRISQRPSYLADYHC 399
+T S+ S+ A+ IP + DS + P R S R ++ PSYL++YHC
Sbjct: 901 TTTDSRTADNHASSSSSALPSIIPPSSNTETQDIDSNAVPITR-SKRTTRAPSYLSEYHC 959
Query: 400 NL-------------------PSKSCSNVSSGISSYPLSSFLSYDNCSPTYTHFCCTISS 440
+L P ++ + YP+S+ +SYD +P + ++
Sbjct: 960 SLVPSISTLPPTDSSIPIHPLPEIFTASSPKKTTPYPISTVVSYDKYTPLCQSYIFAYNT 1019
Query: 441 INEPKTFAQANKSECWREAMTTELNALAKNNTWSVVTLPPGKVPIGCKWVYKVKYHANGS 500
EPKTF+QA KSE W EL A+ N TWSV +LPP K +GCKWV+ +KY+ +G+
Sbjct: 1020 ETEPKTFSQAMKSEKWIRVAVEELQAMELNKTWSVESLPPDKNVVGCKWVFTIKYNPDGT 1079
Query: 501 IERYKARLVAQGYTQTEGVDYFDTFSPVAKLTTIRVLLSLAAIKGWHLEQLDVNNAFLHG 560
+ERYKARLVAQG+TQ EG+D+ DTFSPVAKLT+ +++L LAAI GW L Q+DV++AFLHG
Sbjct: 1080 VERYKARLVAQGFTQQEGIDFLDTFSPVAKLTSAKMMLGLAAITGWTLTQMDVSDAFLHG 1139
Query: 561 DLHEEVYMALPPGYPT-----INSSQVCKLNKSLYGLKQASRQWYSKLSTSLISFGYTQS 615
DL EE++M+LP GY + + VC+L KS+YGLKQASRQWY +
Sbjct: 1140 DLDEEIFMSLPQGYTPPAGTILPPNPVCRLLKSIYGLKQASRQWYKR------------- 1186
Query: 616 LADYSLFVKVSGASFTALLVYVDDIVLAGNCISEIKSVKTFLDNKFCIKDLGQLRYFLGF 675
F A LVY+DDI++A N +E++++K L ++F IKDLG R+FLG
Sbjct: 1187 --------------FVAALVYIDDIMIASNNDAEVENLKALLRSEFKIKDLGPARFFLGL 1232
Query: 676 EIARSKSGILLNQRKYTLELLEDAGTLGSKPAATPFDPSTKLGATTGTPFTDASSYRRLI 735
LG KP++ P DP+ L GTP + ++YR+LI
Sbjct: 1233 --------------------------LGCKPSSIPMDPTLHLVRDMGTPLPNPTAYRKLI 1266
Query: 736 GRLLYLTNTRPDISYSVQNLSQFVSRPMVPHYQAAQRILKYLKSSPAKGLFFSSSSELKL 795
GRLLYLT TRPDI+Y+V LSQF+S P H QAA ++L+Y+K++P +GL +S+ E+ L
Sbjct: 1267 GRLLYLTITRPDITYAVHQLSQFISAPSDIHLQAAHKVLRYIKANPGQGLMYSADYEICL 1326
Query: 796 HGFADSDWACCPDTRRSVTGYCVLLGSSLISWKSKKQSTVSRSSTEAEYRALAHLTCELQ 855
+GF+D+DWA C DTRRS++G+C+ LG+SLISWKSKKQ+ SRSSTE+EYR++A TCE+
Sbjct: 1327 NGFSDADWAACKDTRRSISGFCIYLGTSLISWKSKKQAVASRSSTESEYRSMAQATCEII 1386
Query: 856 WLNYLFHDL 864
WL L DL
Sbjct: 1387 WLQQLLKDL 1395
>emb|CAB10526.1| retrotransposon like protein [Arabidopsis thaliana]
gi|7268497|emb|CAB78748.1| retrotransposon like protein
[Arabidopsis thaliana] gi|7444421|pir||A71444 probable
LTR retrotransposon - Arabidopsis thaliana
Length = 1433
Score = 682 bits (1761), Expect = 0.0
Identities = 369/854 (43%), Positives = 510/854 (59%), Gaps = 66/854 (7%)
Query: 21 CNSVFTDCFDVWHMRLGHVSSSGLSVISK--QFPFIPCIKNAPP----CDACHYAKQKRL 74
C+SV D WH RLGH + S + ++S K P C CH +KQK L
Sbjct: 543 CSSVVVDSV-TWHKRLGHPAYSKIDLLSDVLNLKVKKINKEHSPVCHVCHVCHLSKQKHL 601
Query: 75 PFPHSSIKSSAPFDLLHADLWGPYSTPSFLGHKYFLTLVDDYSRFTWVIFLKTKDETQKH 134
F SA FDL+H D WGP+S P+ + TW+ LK K +
Sbjct: 602 SFQSRQNMCSAAFDLVHIDTWGPFSVPT--------------NDATWIYLLKNKSDVLHV 647
Query: 135 LKHFISYVENQFHTTLKCLRSDNGSEFIAMTSFLLSKGIIHHKTCVETPQQNGVVERKHQ 194
FI+ V Q+ T LK +RSDN E + T + GI+ + +C ETP+QN VVERKHQ
Sbjct: 648 FPAFINMVHTQYQTKLKSVRSDNAHE-LKFTDLFAAHGIVAYHSCPETPEQNSVVERKHQ 706
Query: 195 HILNVARSLAFHSHVPITMWNFTVQHAVHIINRIPSPLLKFKSPFELLHKEPPSIIHLKV 254
HILNVAR+L F S++P+ W V AV +INR+P+P+L KSP+E L PP+ LK
Sbjct: 707 HILNVARALLFQSNIPLEFWGDCVLTAVFLINRLPTPVLNNKSPYEKLKNIPPAYESLKT 766
Query: 255 FGCLAYASTLQAHRTKFNPRARKTIFLGFKEGTKGSILYDLNSNELFVSRNVIFYENHFP 314
FGCL Y+ST R KF PRAR +FLG+ G KG L D+ ++ + +SR+VIF+E+ FP
Sbjct: 767 FGCLCYSSTSPKQRHKFEPRARACVFLGYPLGYKGYKLLDIETHAVSISRHVIFHEDIFP 826
Query: 315 FTLATKQANIPTTSSHIDLGDPITDLSPHPISAPEFQLTSTPPSQYVSAPAVQHAIPVTD 374
F +T + +I + DL P+ + + T P Q VS+ A+ D
Sbjct: 827 FISSTIKDDIKDFFPLLQFPARTDDL---PLE--QTSIIDTHPHQDVSS---SKALVPFD 878
Query: 375 SISEPTVRKSTRISQRPSYLADYHCNLPSKSCSNVSSGISSYPLSSFLSYDNCSPTYTHF 434
+S+ R + P +L D+HC Y+N + + F
Sbjct: 879 PLSK-------RQKKPPKHLQDFHC------------------------YNNTTEPFHAF 907
Query: 435 CCTISSINEPKTFAQANKSECWREAMTTELNALAKNNTWSVVTLPPGKVPIGCKWVYKVK 494
I++ P+ +++A + W +AM E+ A+ + NTWSVV+LPP K IGCKWV+ +K
Sbjct: 908 INNITNAVIPQRYSEAKDFKAWCDAMKEEIGAMVRTNTWSVVSLPPNKKAIGCKWVFTIK 967
Query: 495 YHANGSIERYKARLVAQGYTQTEGVDYFDTFSPVAKLTTIRVLLSLAAIKGWHLEQLDVN 554
++A+GSIERYKARLVA+GYTQ EG+DY +TFSPVAKLT++R++L LAA W + QLD++
Sbjct: 968 HNADGSIERYKARLVAKGYTQEEGLDYEETFSPVAKLTSVRMMLLLAAKMKWSVHQLDIS 1027
Query: 555 NAFLHGDLHEEVYMALPPGY-----PTINSSQVCKLNKSLYGLKQASRQWYSKLSTSLIS 609
NAFL+GDL EE+YM +PPGY + +C+L+KS+YGLKQASRQWY KLS +L
Sbjct: 1028 NAFLNGDLDEEIYMKIPPGYADLVGEALPPHAICRLHKSIYGLKQASRQWYLKLSNTLKG 1087
Query: 610 FGYTQSLADYSLFVKVSGASFTALLVYVDDIVLAGNCISEIKSVKTFLDNKFCIKDLGQL 669
G+ +S AD++LF+K + +LVYVDDI++ N + L + F ++DLG
Sbjct: 1088 MGFQKSNADHTLFIKYANGVLMGVLVYVDDIMIVSNSDDAVAQFTAELKSYFKLRDLGAA 1147
Query: 670 RYFLGFEIARSKSGILLNQRKYTLELLEDAGTLGSKPAATPFDPSTKLGATTGTPFTDAS 729
+YFLG EIARS+ GI + QRKY LELL G LGSKP++ P DPS KL G P TD++
Sbjct: 1148 KYFLGIEIARSEKGISICQRKYILELLSTTGFLGSKPSSIPLDPSVKLNKEDGVPLTDST 1207
Query: 730 SYRRLIGRLLYLTNTRPDISYSVQNLSQFVSRPMVPHYQAAQRILKYLKSSPAKGLFFSS 789
SYR+L+G+L+YL TRPDI+Y+V L QF P H A ++L+YLK + +GLF+S+
Sbjct: 1208 SYRKLVGKLMYLQITRPDIAYAVNTLCQFSHAPTSVHLSAVHKVLRYLKGTVGQGLFYSA 1267
Query: 790 SSELKLHGFADSDWACCPDTRRSVTGYCVLLGSSLISWKSKKQSTVSRSSTEAEYRALAH 849
+ L G+ DSD+ C D+RR V YC+ +G L+SWKSKKQ TVS S+ EAE+RA++
Sbjct: 1268 DDKFDLRGYTDSDFGSCTDSRRCVAAYCMFIGDYLVSWKSKKQDTVSMSTAEAEFRAMSQ 1327
Query: 850 LTCELQWLNYLFHD 863
T E+ WL+ LF D
Sbjct: 1328 GTKEMIWLSRLFDD 1341
>gb|AAF79879.1| T7N9.5 [Arabidopsis thaliana]
Length = 1436
Score = 665 bits (1717), Expect = 0.0
Identities = 366/870 (42%), Positives = 511/870 (58%), Gaps = 59/870 (6%)
Query: 1 SLVGGLYLIAAGPSLAN------KLSCNSVFTDCFDVWHMRLGHVSSSGLSVISK--QFP 52
S VG LY++ SL + K C+SV + ++WH RLGH S + + +S P
Sbjct: 522 SQVGNLYILNLDKSLVDVSSFPGKSVCSSVKNES-EMWHKRLGHPSFAKIDTLSDVLMLP 580
Query: 53 FIPCIKNAPPCDACHYAKQKRLPFPHSSIKSSAPFDLLHADLWGPYSTPSFLGHKYFLTL 112
K++ C CH +KQK LPF + F+L+H D WGP+S P+ ++YFLT+
Sbjct: 581 KQKINKDSSHCHVCHLSKQKHLPFKSVNHIREKAFELVHIDTWGPFSVPTVDSYRYFLTI 640
Query: 113 VDDYSRFTWVIFLKTKDETQKHLKHFISYVENQFHTTLKCLRSDNGSEFIAMTSFLLSKG 172
VDD+SR TW+ LK K + F+ VE Q+HT + +RSDN E + +G
Sbjct: 641 VDDFSRATWIYLLKQKSDVLTVFPSFLKMVETQYHTKVCSVRSDNAHE-LKFNELFAKEG 699
Query: 173 IIHHKTCVETPQQNGVVERKHQHILNVARSLAFHSHVPITMWNFTVQHAVHIINRIPSPL 232
I C ETP+QN VVERKHQH+LNVAR+L F S +P+ W V AV +INR+ SP+
Sbjct: 700 IKADHPCPETPEQNFVVERKHQHLLNVARALMFQSGIPLEYWGDCVLTAVFLINRLLSPV 759
Query: 233 LKFKSPFELLHKEPPSIIHLKVFGCLAYASTLQAHRTKFNPRARKTIFLGFKEGTKGSIL 292
+ ++P+E L K P LK FGCL Y ST RTKF+PRA+ IFLG+ G KG L
Sbjct: 760 INNETPYERLTKGKPDYSSLKAFGCLCYCSTSPKSRTKFDPRAKACIFLGYPMGYKGYKL 819
Query: 293 YDLNSNELFVSRNVIFYENHFPFTLATKQANIPTTSSHIDLGDPITDLSPHPISAPEFQL 352
D+ + + +SR+VIFYE+ FPF SS+I D D PH I P
Sbjct: 820 LDIETYSVSISRHVIFYEDIFPFA-----------SSNIT--DAAKDFFPH-IYLPAPNN 865
Query: 353 TSTPPSQYVSAPAVQHAIPVTDSISEPTVRKSTRISQRPSYLADYHC--NLPSKSCSNVS 410
P S+ A + + I P+ KSTR + PS+L D+HC N P+ +
Sbjct: 866 DEHLPLVQSSSDAPHNHDESSSMIFVPSEPKSTRQRKLPSHLQDFHCYNNTPT------T 919
Query: 411 SGISSYPLSSFLSYDNCSPTYTHFCCTISSINEPKTFAQANKSECWREAMTTELNALAKN 470
+ S YPL++++SY S + F I++ P+ +++A + W +AM E++A +
Sbjct: 920 TKTSPYPLTNYISYSYLSEPFGAFINIITATKLPQKYSEARLDKVWNDAMGKEISAFVRT 979
Query: 471 NTWSVVTLPPGKVPIGCKWVYKVKYHANGSIERYKARLVAQGYTQTEGVDYFDTFSPVAK 530
TWS+ LP GKV +GCKW+ +K+ A+GSIER+KARLVA+GYTQ EG+D+F+TFSPVAK
Sbjct: 980 GTWSICDLPAGKVAVGCKWIITIKFLADGSIERHKARLVAKGYTQQEGIDFFNTFSPVAK 1039
Query: 531 LTTIRVLLSLAAIKGWHLEQLDVNNAFLHGDLHEEVYMALPPGYPTINSSQVCKLNKSLY 590
+ T++VLLSLA W+L QLD++NA L+GDL EE+YM LPPGY I +V N +
Sbjct: 1040 MVTVKVLLSLAPKMKWYLHQLDISNALLNGDLEEEIYMKLPPGYSEIQGQEVSP-NAKCH 1098
Query: 591 GLKQASRQWYSKLSTSLISFGYTQSLADYSLFVKVSGASFTALLVYVDDIVLAGNCISEI 650
G D++LFVK F +LVYVDDI++A +
Sbjct: 1099 G--------------------------DHTLFVKAQDGFFLVVLVYVDDILIASTTEAAS 1132
Query: 651 KSVKTFLDNKFCIKDLGQLRYFLGFEIARSKSGILLNQRKYTLELLEDAGTLGSKPAATP 710
+ + L + F ++DLG+ ++FLG EIAR+ GI L QRKY L+LL + KP++ P
Sbjct: 1133 AELTSQLSSFFQLRDLGEPKFFLGIEIARNADGISLCQRKYVLDLLASSDFSDCKPSSIP 1192
Query: 711 FDPSTKLGATTGTPFTDASSYRRLIGRLLYLTNTRPDISYSVQNLSQFVSRPMVPHYQAA 770
+P+ KL TGT D YRR++G+L YL TRPDI+++V L+Q+ S P H QA
Sbjct: 1193 MEPNQKLSKDTGTLLEDGKQYRRILGKLQYLCLTRPDINFAVSKLAQYSSAPTDIHLQAL 1252
Query: 771 QRILKYLKSSPAKGLFFSSSSELKLHGFADSDWACCPDTRRSVTGYCVLLGSSLISWKSK 830
+IL+YLK + +GLF+ + + L GF+DSDW CPDTRR VTG+ + +G+SL+SW+SK
Sbjct: 1253 HKILRYLKGTIGQGLFYGADTNFDLRGFSDSDWQTCPDTRRCVTGFAIFVGNSLVSWRSK 1312
Query: 831 KQSTVSRSSTEAEYRALAHLTCELQWLNYL 860
KQ VS SS EAEYRA++ T EL WL Y+
Sbjct: 1313 KQDVVSMSSAEAEYRAMSVATKELIWLGYI 1342
>gb|AAP53905.1| putative pol polyprotein [Oryza sativa (japonica cultivar-group)]
gi|37534632|ref|NP_921618.1| putative pol polyprotein
[Oryza sativa (japonica cultivar-group)]
Length = 1688
Score = 625 bits (1613), Expect = e-177
Identities = 363/908 (39%), Positives = 511/908 (55%), Gaps = 70/908 (7%)
Query: 5 GLYLIAAGPSLANKLSCNSVF-----TDC--FDVWHMRLGHVSSSGLSVISKQ--FPFIP 55
GLY++ + ++ + SV+ T C F WH RLGH+ S L+ + Q +P
Sbjct: 291 GLYILDSLSLPSSSTNTPSVYSPMCSTACKSFPQWHHRLGHLCGSRLATLINQGVLGSVP 350
Query: 56 CIKNAPPCDACHYAKQKRLPFPHSSIKSSAPFDLLHADLWGPYSTPSFLGHKYFLTLVDD 115
+ C C KQ +LP+P S+ +SS PFDL+H+D+WG PS GH Y++ VDD
Sbjct: 351 -VDTTFVCKGCKLGKQVQLPYPSSTSRSSRPFDLVHSDVWGKSPFPSKGGHNYYVIFVDD 409
Query: 116 YSRFTWVIFLKTKDETQKHLKHFISYVENQFHTTLKCLRSDNGSEFI--AMTSFLLSKGI 173
YSR+TW+ F+K + + + F + QF + ++ RSD+G E++ A FL+S+G
Sbjct: 410 YSRYTWIYFMKHRSQLISIYQSFAQMIHTQFSSAIRIFRSDSGGEYMSNAFREFLVSQGT 469
Query: 174 IHHKTCVETPQQNGVVERKHQHILNVARSLAFHSHVPITMWNFTVQHAVHIINRIPSPLL 233
+ +C QNGV ERKH+HI+ AR+L S VP W + AV++IN PS L
Sbjct: 470 LPQLSCPGAHAQNGVAERKHRHIIETARTLLIASFVPAHFWAEAISTAVYLINMQPSSSL 529
Query: 234 KFKSPFELLHKEPPSIIHLKVFGCLAYASTLQAHRTKFNPRARKTIFLGFKEGTKGSILY 293
+ +SP E+L PP HL+VFGC Y RTK ++ + +FLG+ KG Y
Sbjct: 530 QGRSPGEVLFGSPPRYDHLRVFGCTCYVLLAPRERTKLTAQSVECVFLGYSLEHKGYRCY 589
Query: 294 DLNSNELFVSRNVIFYENHFPFTLATKQANIPTTSSHIDLGDPITDLSPHPISAPEFQLT 353
D ++ + +SR+V F EN F +T Q + P S I+ L PI +PE
Sbjct: 590 DPSARRIRISRDVTFDENKPFFYSSTNQPSSPENS--------ISFLYLPPIPSPE---- 637
Query: 354 STPPSQYVSAPAVQHAIPVTDSISEPTVRKSTRISQRPSYLADYHCNLP-SKSCSNVSSG 412
S P S +P+ P+ S+ PT S PS ++ ++P S S +V S
Sbjct: 638 SLPSSPITPSPS-----PIPPSVPSPTYVPPPPPSPSPSPVSPPPSHIPASSSPPHVPST 692
Query: 413 IS-----------------SYPLSSFLSYDNCS--------------------PTYTHFC 435
I+ S P L CS P F
Sbjct: 693 ITLDTFPFHYSRRPKIPNESQPSQPTLEDPTCSVDDSSPAPRYNLRARDALRAPNRDDF- 751
Query: 436 CTISSINEPKTFAQANKSECWREAMTTELNALAKNNTWSVVTLPPGKVPIGCKWVYKVKY 495
+ + EP T+ +A W+ AM+ EL AL + NTW VV LP VPI CKWVYKVK
Sbjct: 752 -VVGVVFEPSTYQEAIVLPHWKLAMSEELAALERTNTWDVVPLPSHAVPITCKWVYKVKT 810
Query: 496 HANGSIERYKARLVAQGYTQTEGVDYFDTFSPVAKLTTIRVLLSLAAIKGWHLEQLDVNN 555
++G +ERYKARLVA+G+ Q G DY +TF+PVA +TT+R L+++AA + W + Q+DV N
Sbjct: 811 KSDGQVERYKARLVARGFQQAHGRDYDETFAPVAHMTTVRTLIAVAATRSWTISQMDVKN 870
Query: 556 AFLHGDLHEEVYMALPPGYPTINSSQVCKLNKSLYGLKQASRQWYSKLSTSLISFGYTQS 615
AFLHGDLHEEVYM PPG V +L ++LYGLKQA R W+++ S+ +++ G++ S
Sbjct: 871 AFLHGDLHEEVYMHPPPGVEA-PPGHVFRLRRALYGLKQAPRAWFARFSSVVLAAGFSPS 929
Query: 616 LADYSLFVKVSGASFTALLVYVDDIVLAGNCISEIKSVKTFLDNKFCIKDLGQLRYFLGF 675
D +LF+ S T LL+YVDD+++ G+ + I VK L +F + DLG L YFLG
Sbjct: 930 DHDPALFIHTSSRGRTLLLLYVDDMLITGDDLEYIAFVKGKLSEQFMMSDLGPLSYFLGI 989
Query: 676 EIARSKSGILLNQRKYTLELLEDAGTLGSKPAATPFDPSTKLGATTGTPFTDASSYRRLI 735
E+ + G L+Q +Y +LL +G S+ TP + +L +T GTP D S YR L+
Sbjct: 990 EVTSTVDGYYLSQHRYIEDLLAQSGLTDSRTTTTPMELHVRLRSTDGTPLDDPSRYRHLV 1049
Query: 736 GRLLYLTNTRPDISYSVQNLSQFVSRPMVPHYQAAQRILKYLKSSPAKGLFFSSSSELKL 795
G L+YLT TRPDI+Y+V LSQFVS P+ HY R+L+YL+ + + LF+++SS L+L
Sbjct: 1050 GSLVYLTVTRPDIAYAVHILSQFVSAPISVHYGHLLRVLRYLRGTTTQCLFYAASSPLQL 1109
Query: 796 HGFADSDWACCPDTRRSVTGYCVLLGSSLISWKSKKQSTVSRSSTEAEYRALAHLTCELQ 855
F+DS WA P RRSVTGYC+ LG+SL++WKSKKQ+ VSRSSTEAE RALA T E+
Sbjct: 1110 RAFSDSTWASDPIDRRSVTGYCIFLGTSLLTWKSKKQTAVSRSSTEAELRALATTTSEIV 1169
Query: 856 WLNYLFHD 863
WL +L D
Sbjct: 1170 WLRWLLAD 1177
>emb|CAC95126.1| gag-pol polyprotein [Populus deltoides]
Length = 1382
Score = 620 bits (1599), Expect = e-176
Identities = 354/860 (41%), Positives = 501/860 (58%), Gaps = 55/860 (6%)
Query: 23 SVFTDCFDVWHMRLGHVSSSGLSVISKQFPFIPCIKNAPPCD-----ACHYAKQKRLPFP 77
S+ + F +WH RLGHVSSS L ++ + N CD C AK LPF
Sbjct: 472 SLSSSSFYLWHSRLGHVSSSRLRFLAST----GALGNLKTCDISDCSGCKLAKFSALPFN 527
Query: 78 HSSIKSSAPFDLLHADLWGPYSTPSFLGHKYFLTLVDDYSRFTWVIFLKTKDETQKHLKH 137
S+ SS+PFDL+H+D+WGP + G +Y+++ +DD++R+ WV +K + E +
Sbjct: 528 RSTSVSSSPFDLIHSDVWGPSPVSTKGGSRYYVSFIDDHTRYCWVYLMKHRSEFFEIYAA 587
Query: 138 FISYVENQFHTTLKCLRSDNGSEFIA--MTSFLLSKGIIHHKTCVETPQQNGVVERKHQH 195
F + ++ Q +KC R D G E+ + L G IH +C +TP+QNGV ERKH+H
Sbjct: 588 FRALIKTQHSAVIKCFRCDLGGEYTSNKFCQMLALDGTIHQTSCTDTPEQNGVAERKHRH 647
Query: 196 ILNVARSLAFHSHVPITMWNFTVQHAVHIINRIPSPLLKFKSPFELLHKEPPSIIHLKVF 255
I+ ARSL + V W V AV +IN IPS SPFE L+ P +VF
Sbjct: 648 IVETARSLLLSAFVLSEFWGEAVLTAVSLINTIPSSHSSGLSPFEKLYGHVPDYSSFRVF 707
Query: 256 GCLAYASTLQAHRTKFNPRARKTIFLGFKEGTKGSILYDLNSNELFVSRNVIFYENHFPF 315
GC + R K + R+ +FLG+ EG KG +D + +L+VS +V+F E H PF
Sbjct: 708 GCTYFVLHPHVERNKLSSRSAICVFLGYGEGKKGYRCFDPITQKLYVSHHVVFLE-HIPF 766
Query: 316 TLATKQANIPTTSS--HID--LGDPITDLSP-------HPISAPEFQLTSTPPSQYVSAP 364
+ T S HID D D SP H + L+ TP + + S
Sbjct: 767 FSIPSTTHSLTKSDLIHIDPFSEDSGNDTSPYVRSICTHNSAGTGTLLSGTPEASFSST- 825
Query: 365 AVQHAIPVTDSISEPTVRKSTRISQRPSYLADYHCNLPSKSCSNVSSGISSYPLSSFLSY 424
A + I +P R+S RI ++ + L D+ + SC + S +SFL+Y
Sbjct: 826 ----APQASSEIVDPPPRQSIRI-RKSTKLPDF-----AYSCYSSS-------FTSFLAY 868
Query: 425 DNCSPTYTHFCCTISSINEPKTFAQANKSECWREAMTTELNALAKNNTWSVVTLPPGKVP 484
+C + EP ++ +A ++AM EL+AL K +TW +V LPPGK
Sbjct: 869 IHC-------------LFEPSSYKEAILDPLGQQAMDEELSALHKTDTWDLVPLPPGKSV 915
Query: 485 IGCKWVYKVKYHANGSIERYKARLVAQGYTQTEGVDYFDTFSPVAKLTTIRVLLSLAAIK 544
+GC+WVYK+K +++GSIERYKARLVA+GY+Q G+DY +TF+P+AK+TTIR L+++A+I+
Sbjct: 916 VGCRWVYKIKTNSDGSIERYKARLVAKGYSQQYGMDYEETFAPIAKMTTIRTLIAVASIR 975
Query: 545 GWHLEQLDVNNAFLHGDLHEEVYMALPPGYPTINSSQVCKLNKSLYGLKQASRQWYSKLS 604
WH+ QLDV NAFL+GDL EEVYMA PPG + +S VCKL K+LYGLKQA R W+ K S
Sbjct: 976 QWHISQLDVKNAFLNGDLQEEVYMAPPPGI-SHDSGYVCKLKKALYGLKQAPRAWFEKFS 1034
Query: 605 TSLISFGYTQSLADYSLFVKVSGASFTALLVYVDDIVLAGNCISEIKSVKTFLDNKFCIK 664
+ S G+ S D +LF+K + A L +YVDD+++ G+ I I +KT L +F +K
Sbjct: 1035 IVISSLGFVSSSHDSALFIKCTDAGRIILSLYVDDMIITGDDIDGISVLKTELARRFEMK 1094
Query: 665 DLGQLRYFLGFEIARSKSGILLNQRKYTLELLEDAGTLGSKPAATPFDPSTKLGATTGTP 724
DLG LRYFLG E+A S G LL+Q KY +LE A +K TP + + + ++ G P
Sbjct: 1095 DLGYLRYFLGIEVAYSPRGYLLSQSKYVANILERARLTDNKTVDTPIEVNARYSSSDGLP 1154
Query: 725 FTDASSYRRLIGRLLYLTNTRPDISYSVQNLSQFVSRPMVPHYQAAQRILKYLKSSPAKG 784
D + YR ++G L+YLT T PDI+Y+V +SQFV+ P H+ A RIL+YL+ + +
Sbjct: 1155 LIDPTLYRTIVGSLVYLTITHPDIAYAVHVVSQFVASPTTIHWAAVLRILRYLRGTVFQS 1214
Query: 785 LFFSSSSELKLHGFADSDWACCPDTRRSVTGYCVLLGSSLISWKSKKQSTVSRSSTEAEY 844
L SS+S L+L ++D+D P R+SVTG+C+ LG SLISWKSKKQS VS+SSTEAEY
Sbjct: 1215 LLLSSTSSLELRAYSDADHGSDPTDRKSVTGFCIFLGDSLISWKSKKQSIVSQSSTEAEY 1274
Query: 845 RALAHLTCELQWLNYLFHDL 864
A+A T E+ W +L D+
Sbjct: 1275 CAMASTTKEIVWSRWLLADM 1294
>emb|CAB77940.1| putative polyprotein [Arabidopsis thaliana] gi|4325355|gb|AAD17352.1|
contains similarity to retrovirus-related polyproteins
[Arabidopsis thaliana] gi|25301678|pir||C85077 probable
polyprotein [imported] - Arabidopsis thaliana
Length = 1366
Score = 615 bits (1585), Expect = e-174
Identities = 356/811 (43%), Positives = 466/811 (56%), Gaps = 83/811 (10%)
Query: 35 RLGHVSSSGLSVISKQFPFIPCIKNAPPCDACHYAKQKRLPFPHSSIKSSAPFDLLHADL 94
RLGH S S + +S IP + C CH +KQK L F ++ PF L+H D
Sbjct: 506 RLGHPSMSRVQALSSNL-HIPQKLSEFHCKICHLSKQKCLSFVSNNKIYEEPFPLIHID- 563
Query: 95 WGPYSTPSFLGHKYFLTLVDDYSRFTWVIFLKTKDETQKHLKHFISYVENQFHTTLKCLR 154
+ F+ V+ QF T+K +R
Sbjct: 564 ----------------------------------SDVTTIFPEFLKLVQTQFGCTVKSIR 589
Query: 155 SDNGSEFIAMTSFLLSKGIIHHKTCVETPQQNGVVERKHQHILNVARSLAFHSHVPITMW 214
SDN E + L + GI H+ +C TPQQN VVER HQH+LNVARSL F S++P+ W
Sbjct: 590 SDNAPE-LQFKDLLATFGIFHYHSCAYTPQQNYVVERNHQHLLNVARSLYFQSNIPLAYW 648
Query: 215 NFTVQHAVHIINRIPSPLLKFKSPFELLHKEPPSIIHLKVFGCLAYASTLQAHRTKFNPR 274
V A +INR P+P L+ KSP+E+L+K+ P L+VF CL YAST Q R KF R
Sbjct: 649 PECVSTAAFLINRTPTPNLEHKSPYEVLYKKLPDYNSLRVFCCLCYASTHQHERHKFTER 708
Query: 275 ARKTIFLGFKEGTKGSILYDLNSNELFVSRNVIFYENHFPFTLATKQANIPTTSSHIDLG 334
A +F+G++ G KG + DL SN + V+RNV+F+E FPF N+
Sbjct: 709 ATSCVFIGYESGFKGYKILDLESNTVSVTRNVVFHETIFPFIDKHSTQNVS--------- 759
Query: 335 DPITDLSPHPISAPE-------------FQLTSTPPSQYVSAPAVQHA---IPVTDSISE 378
D S PIS + L P + + PA H P++ +++
Sbjct: 760 --FFDDSVLPISEKQKENRFQIYDYFNVLNLEVCPVIEPTTVPAHTHTRSLAPLSTTVTN 817
Query: 379 PTV----------RKSTRISQRPSYLADYHCNLPSKSCSNVSSGISSYPLSSFLSYDNCS 428
RK TR PSYL+ YHC+ K S+ G +++ LSS LSYD S
Sbjct: 818 DQFGNDMDNTLMPRKETRA---PSYLSQYHCSNVLKEPSSSLHG-TAHSLSSHLSYDKLS 873
Query: 429 PTYTHFCCTISSINEPKTFAQANKSECWREAMTTELNALAKNNTWSVVTLPPGKVPIGCK 488
Y FC I + EP TF +A + W +AM EL+AL +T + +L GK IGCK
Sbjct: 874 NEYRLFCFAIIAEKEPTTFKEAALLQKWLDAMNVELDALVSTSTREICSLHDGKRAIGCK 933
Query: 489 WVYKVKYHANGSIERYKARLVAQGYTQTEGVDYFDTFSPVAKLTTIRVLLSLAAIKGWHL 548
WV+K+KY ++G+IERYKARLVA GYTQ EGVDY DTFSP+AKLT++R++L+LAAI W +
Sbjct: 934 WVFKIKYKSDGTIERYKARLVANGYTQQEGVDYIDTFSPIAKLTSVRLILALAAIHNWSI 993
Query: 549 EQLDVNNAFLHGDLHEEVYMALPPGYPT-----INSSQVCKLNKSLYGLKQASRQWYSKL 603
Q+DV NAFLHGD EE+YM LP GY + VC+L KSLYGLKQASRQW+ K
Sbjct: 994 SQMDVTNAFLHGDFEEEIYMQLPQGYTPRKGELLPKRPVCRLVKSLYGLKQASRQWFHKF 1053
Query: 604 STSLISFGYTQSLADYSLFVKVSGASFTALLVYVDDIVLAGNCISEIKSVKTFLDNKFCI 663
S LI G+ QSL D +LFV+V +F ALLVYVDDI+L N S + VK L +F +
Sbjct: 1054 SGVLIQNGFMQSLFDPTLFVRVREDTFLALLVYVDDIMLVSNKDSAVIEVKQILAKEFKL 1113
Query: 664 KDLGQLRYFLGFEIARSKSGILLNQRKYTLELLEDAGTLGSKPAATPFDPSTKLGATTGT 723
KDLGQ RYFLG EIARSK GI ++QRKY LELLE+ G LG KP TP + + KL G
Sbjct: 1114 KDLGQKRYFLGLEIARSKEGISISQRKYALELLEEFGFLGCKPVPTPMELNLKLSQEDGA 1173
Query: 724 PFTDASSYRRLIGRLLYLTNTRPDISYSVQNLSQFVSRPMVPHYQAAQRILKYLKSSPAK 783
DAS YR+LIGRL+YLT TRPDI ++V L+Q++S P PH AA+RIL+YLK+ P +
Sbjct: 1174 LLLDASHYRKLIGRLVYLTVTRPDICFAVNKLNQYMSAPREPHLMAARRILRYLKNDPGQ 1233
Query: 784 GLFFSSSSELKLHGFADSDWACCPDTRRSVT 814
G+F+ +SS L FAD+DW+ CP++ S++
Sbjct: 1234 GVFYPASSTLTFRAFADADWSNCPESSISIS 1264
>pir||F86470 probable retroelement polyprotein [imported] - Arabidopsis thaliana
gi|9989049|gb|AAG10812.1| Putative retroelement
polyprotein [Arabidopsis thaliana]
Length = 1404
Score = 601 bits (1550), Expect = e-170
Identities = 347/911 (38%), Positives = 512/911 (56%), Gaps = 74/911 (8%)
Query: 4 GGLYLIA-AGPSLANKLSCNSVFTDCFD-VWHMRLGHVSSSGLSVISKQFPFIPCIKNAP 61
G LY++ P+ ++ S S F+ +WH RLGH + L ++ F +
Sbjct: 430 GELYVLEDLSPNSSSCFSSKSHLGISFNTLWHARLGHPHTRALKLMLPNISF-----DHT 484
Query: 62 PCDACHYAKQKRLPFPHSSIKSSAPFDLLHADLWGPYSTP--SFLGHKYFLTLVDDYSRF 119
C+AC K + FP S FDL+H+D+W ++P S +KYF+T +++ S++
Sbjct: 485 SCEACILGKHCKSVFPKSLTIYEKCFDLVHSDVW---TSPCVSRDNNKYFVTFINEKSKY 541
Query: 120 TWVIFLKTKDETQKHLKHFISYVENQFHTTLKCLRSDNGSEFIAMT--SFLLSKGIIHHK 177
TW+ L +KD + +F +YV NQF+ +K R+DNG E+ + L +GIIH
Sbjct: 542 TWITLLPSKDRVFEAFTNFETYVTNQFNAKIKVFRTDNGGEYTSQKFRDHLAKRGIIHQT 601
Query: 178 TCVETPQQNGVVERKHQHILNVARSLAFHSHVPITMWNFTVQHAVHIINRIPSPLLKFKS 237
+C TPQQNGV ERK++H++ VARS+ FH+ VP W V A ++INR P+ +L S
Sbjct: 602 SCPYTPQQNGVAERKNRHLMEVARSMMFHTSVPKRFWGDAVLTACYLINRTPTKVLSDLS 661
Query: 238 PFELLHKEPPSIIHLKVFGCLAYASTLQAHRTKFNPRARKTIFLGFKEGTKGSILYDLNS 297
PFE+L+ P I HL+VFGC+ + R+K + ++ K +FLG+ KG +D
Sbjct: 662 PFEVLNNTKPFIDHLRVFGCVCFVLIPGEQRSKLDAKSTKCMFLGYSTTQKGYKCFDPTK 721
Query: 298 NELFVSRNVIFYENHFPFTLATKQANIP----TTSSHID--------LGDPITDLSPHPI 345
N F+SR+V F EN + N+ +TS ++ LG+ T + H
Sbjct: 722 NRTFISRDVKFLENQ-DYNNKKDWENLKDLTHSTSDRVETLKFLLDHLGNDSTSTTQHQP 780
Query: 346 SAPEFQLTSTPPSQYVSA-------------PAVQ----HAIPVTDSISE---------- 378
+ Q ++ VS P Q H + D SE
Sbjct: 781 EMTQDQEDLNQENEEVSLQHQENLTHVQEDPPNTQEHSEHVQEIQDDSSEDEEPTQVLPP 840
Query: 379 -PTVRKSTRISQRPSYLADYHCNLPSKSCSNVSSGISSYPLSSFLSYDNCSPTYTHFCCT 437
P +R+STRI ++ + +S ++P + S + F
Sbjct: 841 PPPLRRSTRIRRKKEFF---------------NSNAVAHPFQATCSLALVPLDHQAFLSK 885
Query: 438 ISSINEPKTFAQANKSECWREAMTTELNALAKNNTWSVVTLPPGKVPIGCKWVYKVKYHA 497
IS P+T+ +A + + WR+A+ E+NA+ +N+TW LP GK + +WV+ +KY +
Sbjct: 886 ISEHWIPQTYEEAMEVKEWRDAIADEINAMKRNHTWDEDDLPKGKKTVSSRWVFTIKYKS 945
Query: 498 NGSIERYKARLVAQGYTQTEGVDYFDTFSPVAKLTTIRVLLSLAAIKGWHLEQLDVNNAF 557
NG IERYK RLVA+G+TQT G DY +TF+PVAKL T+RV+L+LA W L Q+DV NAF
Sbjct: 946 NGDIERYKTRLVARGFTQTYGSDYMETFAPVAKLHTVRVVLALATNLSWGLWQMDVKNAF 1005
Query: 558 LHGDLHEEVYMALPPGY-PTINSSQVCKLNKSLYGLKQASRQWYSKLSTSLISFGYTQSL 616
L G+L ++VYM PPG TI +V +L K++YGLKQ+ R WY KLS +L G+ +S
Sbjct: 1006 LQGELEDDVYMTPPPGLEDTIPCDKVLRLRKAIYGLKQSPRAWYHKLSRTLKDHGFKKSE 1065
Query: 617 ADYSLFVKVSGASFTALLVYVDDIVLAGNCISEIKSVKTFLDNKFCIKDLGQLRYFLGFE 676
+D++LF S +L+YVDD+++ G+ I S KTFL + F IKDLG+L+YFLG E
Sbjct: 1066 SDHTLFTLQSPQGIVVVLIYVDDLIITGDNKDGIDSTKTFLKSCFDIKDLGELKYFLGIE 1125
Query: 677 IARSKSGILLNQRKYTLELLEDAGTLGSKPAATPFDPSTKL---GATTGTPFTDASSYRR 733
+ RS +G+ L+QRKYTL+LL + G + +KPA TP + K+ G F DA YR+
Sbjct: 1126 VCRSNAGLFLSQRKYTLDLLNETGFMDAKPARTPLEDGYKVNRKGEKEDEKFGDAPLYRK 1185
Query: 734 LIGRLLYLTNTRPDISYSVQNLSQFVSRPMVPHYQAAQRILKYLKSSPAKGLFFSSSSEL 793
L+G+L+YLTNTRPDI ++V +SQ + PMV H+ +RIL+YLK S +G++ +S
Sbjct: 1186 LVGKLIYLTNTRPDICFAVNQVSQHMKVPMVYHWNMVERILRYLKGSSGQGIWMGKNSST 1245
Query: 794 KLHGFADSDWACCPDTRRSVTGYCVLLGSSLISWKSKKQSTVSRSSTEAEYRALAHLTCE 853
++ G+ D+D+A RRS TGYC +G +L +WK+KKQ VS SS E+EYRA+ LT E
Sbjct: 1246 EIVGYCDADYAGDRGDRRSKTGYCTFIGGNLATWKTKKQKVVSCSSAESEYRAMRKLTNE 1305
Query: 854 LQWLNYLFHDL 864
L WL L DL
Sbjct: 1306 LTWLKALLKDL 1316
>ref|NP_916434.1| putative gag/pol polyprotein [Oryza sativa (japonica
cultivar-group)]
Length = 1090
Score = 583 bits (1502), Expect = e-164
Identities = 337/896 (37%), Positives = 486/896 (53%), Gaps = 45/896 (5%)
Query: 4 GGLYLI-AAGPSLANKLSCNSVFTDCFDVWHMRLGHVSSSGLSVISKQFPFIPCIK-NAP 61
G LY + AA PS A + + +WH RLGH + + + + I C K +
Sbjct: 100 GELYTLPAATPSSA----AHGLLATSSTLWHCRLGHPGPAAIHGL-RNIASISCNKIDTS 154
Query: 62 PCDACHYAKQKRLPFPHSSIKSSAPFDLLHADLW-GPYSTPSFLGHKYFLTLVDDYSRFT 120
C AC K RLPF +SS ++S PF+L+H D+W P + S G KY+L ++DD+S F
Sbjct: 155 LCHACQLGKHTRLPFHNSSSRTSVPFELVHCDVWTSPVMSTS--GFKYYLVVLDDFSHFC 212
Query: 121 WVIFLKTKDETQKHLKHFISYVENQFHTTLKCLRSDNGSEFI--AMTSFLLSKGIIHHKT 178
W L+ K + +H+ F+ YV QF LK ++DNG EF+ A+T+FL S+G +
Sbjct: 213 WTFLLRLKSDVHRHIVEFVEYVSTQFGLPLKSFQADNGREFVNTAITTFLASRGTQLRLS 272
Query: 179 CVETPQQNGVVERKHQHILNVARSLAFHSHVPITMWNFTVQHAVHIINRIPSPLLKFKSP 238
C T QNG ER + I N R+L + +P + W + A +++NR PS + P
Sbjct: 273 CPYTSPQNGKAERMLRTINNSIRTLLIQASMPPSYWAEALATATYLLNRRPSSSIHQSLP 332
Query: 239 FELLHKEPPSIIHLKVFGCLAYASTLQAHRTKFNPRARKTIFLGFKEGTKGSILYDLNSN 298
F+LLH+ P HL+VFGCL Y + K +PR+ +FLG+ KG DL+++
Sbjct: 333 FQLLHRTIPDFSHLRVFGCLCYPNLSATTPHKLSPRSTACVFLGYPTSHKGYRCLDLSTH 392
Query: 299 ELFVSRNVIFYENHFPFTLATKQANI---------PTTSSHIDLGDP-------ITDLSP 342
+ +SR+V+F E+ FPF A+ P + +++ P T++
Sbjct: 393 RIIISRHVVFDESQFPFAATPPAASSFDFLLQGLSPADAPSLEVEQPRPLTVAPSTEVEQ 452
Query: 343 HPISAPEFQLTSTPPSQYVSAPAVQHAIPVTDSISEPTVRKSTRISQ-----RPSYLADY 397
+ P +L++ + AP+ + T S +TR S R Y
Sbjct: 453 PYLPLPSRRLSAGTVTVASEAPSAGAPLVGTSSADATPPGSATRASTIVSPFRHVYTRRP 512
Query: 398 HCNLPSKSCSNVSSGISSYPLSSFLSYDNCSP-------TYTHFCCTISSINEPKTFAQA 450
+P S + V++ +++ S ++ TYT S + P + A
Sbjct: 513 VTTVPPSSSTAVTNAVAAPQPHSMVTRSQSGSLRPVDRLTYTATQAAASPV--PANYHSA 570
Query: 451 NKSECWREAMTTELNALAKNNTWSVVTLPPGKVPIGCKWVYKVKYHANGSIERYKARLVA 510
WR AM E L N TW +V+ PP KW++K K+H++GS+ RYKAR V
Sbjct: 571 LADPNWRAAMADEYKELVDNGTWRLVSRPPRANIATGKWIFKHKFHSDGSLARYKARWVV 630
Query: 511 QGYTQTEGVDYFDTFSPVAKLTTIRVLLSLAAIKGWHLEQLDVNNAFLHGDLHEEVYMAL 570
+GY+Q G+DY +TFSPV KL TIRV+LS+AA + W + QLDV NAFLHG L E VY
Sbjct: 631 RGYSQQHGIDYDETFSPVVKLATIRVVLSIAASRAWPIHQLDVKNAFLHGHLKETVYCQQ 690
Query: 571 PPGY--PTINSSQVCKLNKSLYGLKQASRQWYSKLSTSLISFGYTQSLADYSLFVKVSGA 628
P G+ PT + VC L KSLYGLKQA R WY + +T + G+ S +D SLFV G
Sbjct: 691 PSGFVDPTAPDA-VCLLQKSLYGLKQAPRAWYQRFATYIRQMGFMPSASDTSLFVYKDGD 749
Query: 629 SFTALLVYVDDIVLAGNCISEIKSVKTFLDNKFCIKDLGQLRYFLGFEIARSKSGILLNQ 688
LL+YVDDI+L + + ++ + L ++F + DLG L +FLG + RS G+ L+Q
Sbjct: 750 RIAYLLLYVDDIILTASTTTLLQQLTARLHSEFAMTDLGDLHFFLGISVKRSPDGLFLSQ 809
Query: 689 RKYTLELLEDAGTLGSKPAATPFDPSTKLGATTGTPFTDASSYRRLIGRLLYLTNTRPDI 748
R+Y ++LL+ AG +TP D KL AT G P D S+YR + G L YLT TRPD+
Sbjct: 810 RQYAVDLLQRAGMAECHSTSTPVDTHAKLSATDGLPVADPSAYRSIAGALQYLTLTRPDL 869
Query: 749 SYSVQNLSQFVSRPMVPHYQAAQRILKYLKSSPAKGLFFSSSSELKLHGFADSDWACCPD 808
+Y+VQ + F+ P PH +RIL+Y+K S + GL S L ++D+DWA CP+
Sbjct: 870 AYAVQQVCLFMHDPREPHLALVKRILRYVKGSLSIGLHIGSGPIQSLTAYSDADWAGCPN 929
Query: 809 TRRSVTGYCVLLGSSLISWKSKKQSTVSRSSTEAEYRALAHLTCELQWLNYLFHDL 864
+RRS +GYCV LG +L+SW SK+Q+TVSRSS EAEYRA+AH E WL L +L
Sbjct: 930 SRRSTSGYCVYLGDNLVSWSSKRQTTVSRSSAEAEYRAVAHAVAECCWLRQLLQEL 985
>emb|CAB77781.1| putative polyprotein of LTR transposon [Arabidopsis thaliana]
gi|3924609|gb|AAC79110.1| putative polyprotein of LTR
transposon [Arabidopsis thaliana] gi|7444420|pir||T01397
LTR gag/pol polyprotein homolog T4I9.16 - Arabidopsis
thaliana
Length = 1456
Score = 580 bits (1496), Expect = e-164
Identities = 340/919 (36%), Positives = 500/919 (53%), Gaps = 89/919 (9%)
Query: 32 WHMRLGHVSSSGL-SVISKQ-FPFIPCIKNAPPCDACHYAKQKRLPFPHSSIKSSAPFDL 89
WH RLGH S + L SVIS P + C C K ++PF +S+I SS P +
Sbjct: 446 WHSRLGHPSLAILNSVISNHSLPVLNPSHKLLSCSDCFINKSHKVPFSNSTITSSKPLEY 505
Query: 90 LHADLWGPYSTP--SFLGHKYFLTLVDDYSRFTWVIFLKTKDETQKHLKHFISYVENQFH 147
+++D+W S+P S ++Y++ VD ++R+TW+ LK K + + F S VEN+F
Sbjct: 506 IYSDVW---SSPILSIDNYRYYVIFVDHFTRYTWLYPLKQKSQVKDTFIIFKSLVENRFQ 562
Query: 148 TTLKCLRSDNGSEFIAMTSFLLSKGIIHHKTCVETPQQNGVVERKHQHILNVARSLAFHS 207
T + L SDNG EF+ + +L GI H + TP+ NG+ ERKH+HI+ + +L H+
Sbjct: 563 TRIGTLYSDNGGEFVVLRDYLSQHGISHFTSPPHTPEHNGLSERKHRHIVEMGLTLLSHA 622
Query: 208 HVPITMWNFTVQHAVHIINRIPSPLLKFKSPFELLHKEPPSIIHLKVFGCLAYASTLQAH 267
VP T W + AV++INR+P+PLL+ +SPF+ L +PP+ LKVFGC Y +
Sbjct: 623 SVPKTYWPYAFSVAVYLINRLPTPLLQLQSPFQKLFGQPPNYEKLKVFGCACYPWLRPYN 682
Query: 268 RTKFNPRARKTIFLGFKEGTKGSILYDLNSNELFVSRNVIFYENHFPFT-----LATKQA 322
R K ++++ F+G+ + + + L+ SR+V F E FPF+ ++T Q
Sbjct: 683 RHKLEDKSKQCAFMGYSLTQSAYLCLHIPTGRLYTSRHVQFDERCFPFSTTNFGVSTSQE 742
Query: 323 ----NIPTTSSHIDLGD------------PITDLSPHPISAPEFQLT-----STPPSQYV 361
+ P SH L P D SP P S+P T S PS +
Sbjct: 743 QRSDSAPNWPSHTTLPTTPLVLPAPPCLGPHLDTSPRPPSSPSPLCTTQVSSSNLPSSSI 802
Query: 362 SAPAVQHAIPVTDSISEPTVR--KSTRISQRPSYLADYHCNLPSKSCSNVSSGISSYPLS 419
S+P+ + + +PT + ++ + L + + N PS + N +S + P+S
Sbjct: 803 SSPSSSEPTAPSHNGPQPTAQPHQTQNSNSNSPILNNPNPNSPSPNSPNQNSPLPQSPIS 862
Query: 420 S----------------------------------FLSYDNCSPTYTH------------ 433
S + + +P TH
Sbjct: 863 SPHIPTPSTSISEPNSPSSSSTSTPPLPPVLPAPPIIQVNAQAPVNTHSMATRAKDGIRK 922
Query: 434 ------FCCTISSINEPKTFAQANKSECWREAMTTELNALAKNNTWSVVTLPPGKVPI-G 486
+ ++++ +EP+T QA K + WR+AM +E+NA N+TW +V PP V I G
Sbjct: 923 PNQKYSYATSLAANSEPRTAIQAMKDDRWRQAMGSEINAQIGNHTWDLVPPPPPSVTIVG 982
Query: 487 CKWVYKVKYHANGSIERYKARLVAQGYTQTEGVDYFDTFSPVAKLTTIRVLLSLAAIKGW 546
C+W++ K++++GS+ RYKARLVA+GY Q G+DY +TFSPV K T+IR++L +A + W
Sbjct: 983 CRWIFTKKFNSDGSLNRYKARLVAKGYNQRPGLDYAETFSPVIKSTSIRIVLGVAVDRSW 1042
Query: 547 HLEQLDVNNAFLHGDLHEEVYMALPPGYPTINSSQ-VCKLNKSLYGLKQASRQWYSKLST 605
+ QLDVNNAFL G L +EVYM+ PPG+ + VC+L K++YGLKQA R WY +L T
Sbjct: 1043 PIRQLDVNNAFLQGTLTDEVYMSQPPGFVDKDRPDYVCRLRKAIYGLKQAPRAWYVELRT 1102
Query: 606 SLISFGYTQSLADYSLFVKVSGASFTALLVYVDDIVLAGNCISEIKSVKTFLDNKFCIKD 665
L++ G+ S++D SLFV G S +LVYVDDI++ GN +K L +F +K+
Sbjct: 1103 YLLTVGFVNSISDTSLFVLQRGRSIIYMLVYVDDILITGNDTVLLKHTLDALSQRFSVKE 1162
Query: 666 LGQLRYFLGFEIARSKSGILLNQRKYTLELLEDAGTLGSKPAATPFDPSTKLGATTGTPF 725
L YFLG E R G+ L+QR+YTL+LL L +KP ATP S KL +GT
Sbjct: 1163 HEDLHYFLGIEAKRVPQGLHLSQRRYTLDLLARTNMLTAKPVATPMATSPKLTLHSGTKL 1222
Query: 726 TDASSYRRLIGRLLYLTNTRPDISYSVQNLSQFVSRPMVPHYQAAQRILKYLKSSPAKGL 785
D + YR ++G L YL TRPD+SY+V LSQ++ P H+ A +R+L+YL +P G+
Sbjct: 1223 PDPTEYRGIVGSLQYLAFTRPDLSYAVNRLSQYMHMPTDDHWNALKRVLRYLAGTPDHGI 1282
Query: 786 FFSSSSELKLHGFADSDWACCPDTRRSVTGYCVLLGSSLISWKSKKQSTVSRSSTEAEYR 845
F + L LH ++D+DWA D S GY V LG ISW SKKQ V RSSTEAEYR
Sbjct: 1283 FLKKGNTLSLHAYSDADWAGDTDDYVSTNGYIVYLGHHPISWSSKKQKGVVRSSTEAEYR 1342
Query: 846 ALAHLTCELQWLNYLFHDL 864
++A+ + ELQW+ L +L
Sbjct: 1343 SVANTSSELQWICSLLTEL 1361
>dbj|BAA78424.1| polyprotein [Arabidopsis thaliana]
Length = 1330
Score = 580 bits (1496), Expect = e-164
Identities = 340/919 (36%), Positives = 500/919 (53%), Gaps = 89/919 (9%)
Query: 32 WHMRLGHVSSSGL-SVISKQ-FPFIPCIKNAPPCDACHYAKQKRLPFPHSSIKSSAPFDL 89
WH RLGH S + L SVIS P + C C K ++PF +S+I SS P +
Sbjct: 320 WHSRLGHPSLAILNSVISNHSLPVLNPSHKLLSCSDCFINKSHKVPFSNSTITSSKPLEY 379
Query: 90 LHADLWGPYSTP--SFLGHKYFLTLVDDYSRFTWVIFLKTKDETQKHLKHFISYVENQFH 147
+++D+W S+P S ++Y++ VD ++R+TW+ LK K + + F S VEN+F
Sbjct: 380 IYSDVW---SSPILSIDNYRYYVIFVDHFTRYTWLYPLKQKSQVKDTFIIFKSLVENRFQ 436
Query: 148 TTLKCLRSDNGSEFIAMTSFLLSKGIIHHKTCVETPQQNGVVERKHQHILNVARSLAFHS 207
T + L SDNG EF+ + +L GI H + TP+ NG+ ERKH+HI+ + +L H+
Sbjct: 437 TRIGTLYSDNGGEFVVLRDYLSQHGISHFTSPPHTPEHNGLSERKHRHIVEMGLTLLSHA 496
Query: 208 HVPITMWNFTVQHAVHIINRIPSPLLKFKSPFELLHKEPPSIIHLKVFGCLAYASTLQAH 267
VP T W + AV++INR+P+PLL+ +SPF+ L +PP+ LKVFGC Y +
Sbjct: 497 SVPKTYWPYAFSVAVYLINRLPTPLLQLQSPFQKLFGQPPNYEKLKVFGCACYPWLRPYN 556
Query: 268 RTKFNPRARKTIFLGFKEGTKGSILYDLNSNELFVSRNVIFYENHFPFT-----LATKQA 322
R K ++++ F+G+ + + + L+ SR+V F E FPF+ ++T Q
Sbjct: 557 RHKLEDKSKQCAFMGYSLTQSAYLCLHIPTGRLYTSRHVQFDERCFPFSTTNFGVSTSQE 616
Query: 323 ----NIPTTSSHIDLGD------------PITDLSPHPISAPEFQLT-----STPPSQYV 361
+ P SH L P D SP P S+P T S PS +
Sbjct: 617 QRSDSAPNWPSHTTLPTTPLVLPAPPCLGPHLDTSPRPPSSPSPLCTTQVSSSNLPSSSI 676
Query: 362 SAPAVQHAIPVTDSISEPTVR--KSTRISQRPSYLADYHCNLPSKSCSNVSSGISSYPLS 419
S+P+ + + +PT + ++ + L + + N PS + N +S + P+S
Sbjct: 677 SSPSSSEPTAPSHNGPQPTAQPHQTQNSNSNSPILNNPNPNSPSPNSPNQNSPLPQSPIS 736
Query: 420 S----------------------------------FLSYDNCSPTYTH------------ 433
S + + +P TH
Sbjct: 737 SPHIPTPSTSISEPNSPSSSSTSTPPLPPVLPAPPIIQVNAQAPVNTHSMATRAKDGIRK 796
Query: 434 ------FCCTISSINEPKTFAQANKSECWREAMTTELNALAKNNTWSVVTLPPGKVPI-G 486
+ ++++ +EP+T QA K + WR+AM +E+NA N+TW +V PP V I G
Sbjct: 797 PNQKYSYATSLAANSEPRTAIQAMKDDRWRQAMGSEINAQIGNHTWDLVPPPPPSVTIVG 856
Query: 487 CKWVYKVKYHANGSIERYKARLVAQGYTQTEGVDYFDTFSPVAKLTTIRVLLSLAAIKGW 546
C+W++ K++++GS+ RYKARLVA+GY Q G+DY +TFSPV K T+IR++L +A + W
Sbjct: 857 CRWIFTKKFNSDGSLNRYKARLVAKGYNQRPGLDYAETFSPVIKSTSIRIVLGVAVDRSW 916
Query: 547 HLEQLDVNNAFLHGDLHEEVYMALPPGYPTINSSQ-VCKLNKSLYGLKQASRQWYSKLST 605
+ QLDVNNAFL G L +EVYM+ PPG+ + VC+L K++YGLKQA R WY +L T
Sbjct: 917 PIRQLDVNNAFLQGTLTDEVYMSQPPGFVDKDRPDYVCRLRKAIYGLKQAPRAWYVELRT 976
Query: 606 SLISFGYTQSLADYSLFVKVSGASFTALLVYVDDIVLAGNCISEIKSVKTFLDNKFCIKD 665
L++ G+ S++D SLFV G S +LVYVDDI++ GN +K L +F +K+
Sbjct: 977 YLLTVGFVNSISDTSLFVLQRGRSIIYMLVYVDDILITGNDTVLLKHTLDALSQRFSVKE 1036
Query: 666 LGQLRYFLGFEIARSKSGILLNQRKYTLELLEDAGTLGSKPAATPFDPSTKLGATTGTPF 725
L YFLG E R G+ L+QR+YTL+LL L +KP ATP S KL +GT
Sbjct: 1037 HEDLHYFLGIEAKRVPQGLHLSQRRYTLDLLARTNMLTAKPVATPMATSPKLTLHSGTKL 1096
Query: 726 TDASSYRRLIGRLLYLTNTRPDISYSVQNLSQFVSRPMVPHYQAAQRILKYLKSSPAKGL 785
D + YR ++G L YL TRPD+SY+V LSQ++ P H+ A +R+L+YL +P G+
Sbjct: 1097 PDPTEYRGIVGSLQYLAFTRPDLSYAVNRLSQYMHMPTDDHWNALKRVLRYLAGTPDHGI 1156
Query: 786 FFSSSSELKLHGFADSDWACCPDTRRSVTGYCVLLGSSLISWKSKKQSTVSRSSTEAEYR 845
F + L LH ++D+DWA D S GY V LG ISW SKKQ V RSSTEAEYR
Sbjct: 1157 FLKKGNTLSLHAYSDADWAGDTDDYVSTNGYIVYLGHHPISWSSKKQKGVVRSSTEAEYR 1216
Query: 846 ALAHLTCELQWLNYLFHDL 864
++A+ + ELQW+ L +L
Sbjct: 1217 SVANTSSELQWICSLLTEL 1235
>dbj|BAA78427.1| polyprotein [Arabidopsis thaliana]
Length = 1421
Score = 579 bits (1492), Expect = e-163
Identities = 349/943 (37%), Positives = 506/943 (53%), Gaps = 93/943 (9%)
Query: 9 IAAGPSLANKLSCNSVFTDCFDVWHMRLGHVSSSGL-SVISKQ-FPFIPCIKNAPPCDAC 66
IA+ +++ S S T C WH RLGH S + L SVIS P + C C
Sbjct: 444 IASSQAVSMFASPCSKATHCS--WHSRLGHPSLAILNSVISNHSLPVLNPSHKLLSCSDC 501
Query: 67 HYAKQKRLPFPHSSIKSSAPFDLLHADLWGPYSTP--SFLGHKYFLTLVDDYSRFTWVIF 124
K ++PF +S+I SS P + +++D+W S+P S ++Y++ VD ++R+TW+
Sbjct: 502 FINKSHKVPFSNSTITSSKPLEYIYSDVW---SSPILSIDNYRYYVIFVDHFTRYTWLYP 558
Query: 125 LKTKDETQKHLKHFISYVENQFHTTLKCLRSDNGSEFIAMTSFLLSKGIIHHKTCVETPQ 184
LK K + + F S VEN+F T + L SDNG EF+ + +L GI H + TP+
Sbjct: 559 LKQKSQVKDTFIIFKSLVENRFQTRIGTLYSDNGGEFVVLRDYLSQHGISHFTSPPHTPE 618
Query: 185 QNGVVERKHQHILNVARSLAFHSHVPITMWNFTVQHAVHIINRIPSPLLKFKSPFELLHK 244
NG+ ERKH+HI+ + +L H+ VP T W + AV++INR+P+PLL+ +SPF+ L
Sbjct: 619 HNGLSERKHRHIVEMGLTLLSHASVPKTYWLYAFSVAVYLINRLPTPLLQLQSPFQKLFG 678
Query: 245 EPPSIIHLKVFGCLAYASTLQAHRTKFNPRARKTIFLGFKEGTKGSILYDLNSNELFVSR 304
+PP+ LKVFGC Y +R K ++++ F+G+ + + + L+ SR
Sbjct: 679 QPPNYEKLKVFGCACYPWLRPYNRHKLEDKSKQCAFMGYSLTQSAYLCLHIPTGRLYTSR 738
Query: 305 NVIFYENHFPFT-----LATKQ-------------ANIPTTSSHID----LGDPITDLSP 342
+V F E FPF+ ++T Q +PTT + LG P D SP
Sbjct: 739 HVQFDERCFPFSTTNFGVSTSQEQRSDSASNWPSHTTLPTTPLVLPAPPCLG-PHLDTSP 797
Query: 343 HPISAPEFQLT-------------STPPSQYVSAPAVQHAIPVTDSISEPTVRKSTRISQ 389
P S+P T S+P S +AP+ P T ++ I
Sbjct: 798 RPPSSPSPLCTTQVSSSNLPSSSISSPSSSEPTAPSYNGPQPTTQPHQTQNSNSNSPILN 857
Query: 390 RP------------------SYLADYHCNLPSKSCSNVSSGISS----------YPLSSF 421
P S ++ H PS S S +S SS P
Sbjct: 858 NPNPNSPSPNSPNQNSPLPQSPISSPHIPTPSTSISEPNSPSSSSTSTPPLPPVLPAPPI 917
Query: 422 LSYDNCSPTYTH------------------FCCTISSINEPKTFAQANKSECWREAMTTE 463
+ + +P TH + ++++ +EP+T QA K + WR+AM +E
Sbjct: 918 IQVNAQAPVNTHSMATRAKDGIRKPNQKYSYATSLAANSEPRTAIQAMKDDRWRQAMGSE 977
Query: 464 LNALAKNNTWSVVTLPPGKVPI-GCKWVYKVKYHANGSIERYKARLVAQGYTQTEGVDYF 522
+NA N+TW +V PP V I GC+W++ K++++GS+ RYKARLVA+GY Q G+DY
Sbjct: 978 INAQIGNHTWDLVPPPPPSVTIVGCRWIFTKKFNSDGSLNRYKARLVAKGYNQRPGLDYA 1037
Query: 523 DTFSPVAKLTTIRVLLSLAAIKGWHLEQLDVNNAFLHGDLHEEVYMALPPGYPTINSSQV 582
+TFSPV K T+IR++L +A + W + QLDVNNAFL G L +E+YM+ PPG+ N
Sbjct: 1038 ETFSPVIKSTSIRIVLGVAVDRSWPIRQLDVNNAFLQGTLTDELYMSQPPGFVDKNRPDY 1097
Query: 583 -CKLNKSLYGLKQASRQWYSKLSTSLISFGYTQSLADYSLFVKVSGASFTALLVYVDDIV 641
C+L K++YGLKQA R WY +L T L++ G+ S++D SLFV G S +LVYVDDI+
Sbjct: 1098 FCRLKKAIYGLKQAPRAWYVELQTYLLTVGFVNSISDTSLFVLQRGRSIIYMLVYVDDIL 1157
Query: 642 LAGNCISEIKSVKTFLDNKFCIKDLGQLRYFLGFEIARSKSGILLNQRKYTLELLEDAGT 701
+ GN +K L +F +K+ L YFLG E R G+ L+QR+YTL+LL
Sbjct: 1158 ITGNDTVLLKHTLDALSQRFSVKEHEDLHYFLGIEAKRVPQGLHLSQRRYTLDLLARTNM 1217
Query: 702 LGSKPAATPFDPSTKLGATTGTPFTDASSYRRLIGRLLYLTNTRPDISYSVQNLSQFVSR 761
L +KP ATP S KL +GT D + YR ++G L YL TRPD+SY+V LSQ++
Sbjct: 1218 LTAKPVATPMATSPKLTLHSGTKLPDPTEYRGIVGSLQYLAFTRPDLSYAVNRLSQYMHM 1277
Query: 762 PMVPHYQAAQRILKYLKSSPAKGLFFSSSSELKLHGFADSDWACCPDTRRSVTGYCVLLG 821
P H+ A +R+L+YL +P G+F + L LH ++D+DWA D S GY V LG
Sbjct: 1278 PTDDHWNALKRVLRYLAGTPDHGIFLKKGNTLSLHAYSDADWAGDTDDYVSTNGYIVYLG 1337
Query: 822 SSLISWKSKKQSTVSRSSTEAEYRALAHLTCELQWLNYLFHDL 864
ISW SKKQ V RSSTEAEYR++A+ + ELQW+ L +L
Sbjct: 1338 HHPISWSSKKQKGVVRSSTEAEYRSVANTSSELQWICSLLTEL 1380
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.320 0.134 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,490,047,578
Number of Sequences: 2540612
Number of extensions: 63126646
Number of successful extensions: 155730
Number of sequences better than 10.0: 2207
Number of HSP's better than 10.0 without gapping: 1663
Number of HSP's successfully gapped in prelim test: 545
Number of HSP's that attempted gapping in prelim test: 148030
Number of HSP's gapped (non-prelim): 3618
length of query: 864
length of database: 863,360,394
effective HSP length: 137
effective length of query: 727
effective length of database: 515,296,550
effective search space: 374620591850
effective search space used: 374620591850
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 80 (35.4 bits)
Medicago: description of AC137079.15


