

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
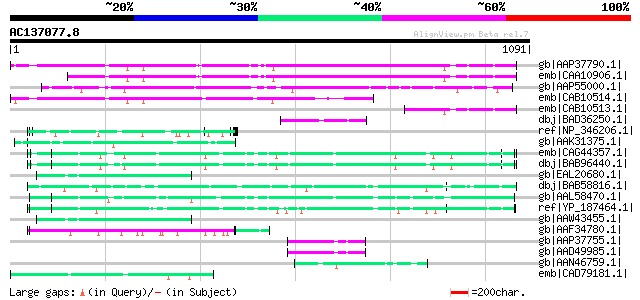
Query= AC137077.8 - phase: 0
(1091 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAP37790.1| At4g17330 [Arabidopsis thaliana] gi|20466528|gb|A... 747 0.0
emb|CAA10906.1| G2484-1 [Arabidopsis thaliana] 687 0.0
gb|AAP55000.1| unknown protein [Oryza sativa (japonica cultivar-... 480 e-133
emb|CAB10514.1| hypothetical protein [Arabidopsis thaliana] gi|7... 424 e-116
emb|CAB10513.1| hypothetical protein [Arabidopsis thaliana] gi|7... 167 2e-39
dbj|BAD36250.1| agenet domain-containing protein / bromo-adjacen... 73 6e-11
ref|NP_346206.1| cell wall surface anchor family protein [Strept... 68 1e-09
gb|AAK31375.1| ppg3 [Leishmania major] 67 4e-09
emb|CAG44357.1| putative cell wall-anchored protein [Staphylococ... 65 1e-08
dbj|BAB96440.1| MW2575 [Staphylococcus aureus subsp. aureus MW2]... 65 1e-08
gb|EAL20680.1| hypothetical protein CNBE0450 [Cryptococcus neofo... 64 2e-08
dbj|BAB58816.1| serine-threoinine rich antigen [Staphylococcus a... 64 3e-08
gb|AAL58470.1| serine-threonine rich antigen [Staphylococcus aur... 64 4e-08
ref|YP_187464.1| LPXTG cell wall surface anchor family protein [... 63 5e-08
gb|AAW43455.1| hypothetical protein CNE00530 [Cryptococcus neofo... 62 1e-07
gb|AAF34780.1| srpA [Streptococcus cristatus] 61 2e-07
gb|AAP37755.1| At1g68580 [Arabidopsis thaliana] gi|22135896|gb|A... 61 2e-07
gb|AAD49985.1| Contains PF|01426 BAH (bromo-adjacent homology) d... 61 2e-07
gb|AAN46759.1| At5g55600/MDF20_4 [Arabidopsis thaliana] gi|18087... 59 7e-07
emb|CAD79181.1| hypothetical protein-signal peptide prediction [... 59 7e-07
>gb|AAP37790.1| At4g17330 [Arabidopsis thaliana] gi|20466528|gb|AAM20581.1| G2484-1
protein [Arabidopsis thaliana]
gi|22328747|ref|NP_193464.2| agenet domain-containing
protein [Arabidopsis thaliana]
Length = 1058
Score = 747 bits (1929), Expect = 0.0
Identities = 467/1108 (42%), Positives = 657/1108 (59%), Gaps = 101/1108 (9%)
Query: 1 MISAYGGTGTCGRMSGVSAWKGSVV---KNLILIPQKHLCSQDLAARTSDSTVKQSVLQG 57
MISA+GG G+ S +W+ VV K+L+ P+ L S+ G
Sbjct: 1 MISAFGGADG-GKGSWEKSWRTCVVRAQKSLVATPETPLQSRP----------------G 43
Query: 58 KGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSALARGSVVDYSQALTPL 117
K + G +SK + NP+IPLSSPLWSL T S D+LQSS++ RGS + L+
Sbjct: 44 KTETPSAGHTNSKESSG-TNPMIPLSSPLWSLST-SVDTLQSSSVQRGSAATHQPLLSAS 101
Query: 118 HPYQSPSPRNFLGHSTSWISQAPLRGPWIGSP-TPAPDNNTHLSASPSSDTIKLASVKGS 176
H +Q+P +N +GH+T W+S P R W+ S T D + P +D +KL +K S
Sbjct: 102 HAHQTPPTQNIVGHNTPWMSPLPFRNAWLASQQTSGFDVGSRFPVYPITDPVKLTPMKES 161
Query: 177 LPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQSSGPKAKKRKKDVLSE 236
S K V G ++ S + T L+ + V PAQ S+ K++KRKK +S
Sbjct: 162 SMTLSGAKHVQSGTSSNVSKVTPT-------LEPTSTVVAPAQHSTRVKSRKRKKMPVSV 214
Query: 237 DHGQKLLQSL----------TPAVASRASTSVSAATPVGNVPMSSVEKSVVSV------- 279
+ G +L SL P + A+ +A T V M++V +VS
Sbjct: 215 ESGPNILNSLKQTELAASPLVPFTPTPANLGYNAGTLPSVVSMTAVPMDLVSTFPGKKIK 274
Query: 280 ----SPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDK 335
SP+ + ++ +LS++++ K+KEA++HAE+ASAL+ AAV+HS +W Q+++
Sbjct: 275 SSFPSPIFGGNLVREVKQRSVLSEDTIEKLKEAKMHAEDASALATAAVSHSEYVWKQIEQ 334
Query: 336 HKNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYEN 395
++G + + +LASAAVAIAAAAAVAKAAAAAANVA+NAA QAKLMA+EA + +
Sbjct: 335 QSHAGLQPETQDRLASAAVAIAAAAAVAKAAAAAANVAANAALQAKLMAEEASLPNA--- 391
Query: 396 TSQGNNTFLPEGTSNL--GQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKR 453
+ QG LP+ ++ GQ TPASILKG + S ++AA+EA ++RVEAA+AATKR
Sbjct: 392 SDQG----LPKSYDSILPGQGTPASILKGEGAVVNSSSVLIAAREASKKRVEAATAATKR 447
Query: 454 AENMDAILKAAELAAEAVSQAGKIVTMGDPLPLIELIEAGPEGCWKASRESSREVGLLKD 513
AENMD+I+KAAELA+EAVSQAG +V+MG P L +L+EAGP W+ ++ES +EV K
Sbjct: 448 AENMDSIVKAAELASEAVSQAGILVSMGHPPLLNKLVEAGPSNYWRQAQES-QEVQPCKT 506
Query: 514 MTRDLVNIDMVRDIPETSHAQNRDILSSEISASIMINEK---------NTRGQQARTVSD 564
+ + + E + A + I+ +E S+ + +GQ+ +D
Sbjct: 507 VVLEKETVST----SEGTFAGPK-IVQTEFGGSVNTADGVSGPVPATGKLKGQEGDKYAD 561
Query: 565 LVKPVDMVLGSESETQDPSFTVRNGSENLEENTFKEGSLVEVFKDEEGHKAAWFMGNILS 624
L K D+V E ++ S + + + KEGS VEVFK+E G + AW+ N+LS
Sbjct: 562 LAKNNDVVFEPEVGSKF-SIDAQQTIKATKNEDIKEGSNVEVFKEEPGLRTAWYSANVLS 620
Query: 625 LKDGKVYVCYTSLVAVEGP--LKEWVSLECEGDKPPRIRTARPLTSLQHEGTRKRRRAAM 682
L+D K YV ++ L +G LKEWV+L+ EGD+ P+IR AR +T+L +EGTRKRRRAA+
Sbjct: 621 LEDDKAYVLFSDLSVEQGTDKLKEWVALKGEGDQAPKIRPARSVTALPYEGTRKRRRAAL 680
Query: 683 GDYAWSVGDRVDAWIQESWREGVITEKNKKDETTLTVHIPASGETSVLRAWNLRPSLIWK 742
GD+ W +GDRVD+W+ +SW EGVITEKNKKDE T+TVH PA ET ++AWNLRPSL+WK
Sbjct: 681 GDHIWKIGDRVDSWVHDSWLEGVITEKNKKDENTVTVHFPAEEETLTIKAWNLRPSLVWK 740
Query: 743 DGQWLDFSKVGANDSSTHKGDTPHEKRPKLGSNAVEVKGKDKMSKNIDAAESANPDEMRS 802
DG+W++ S G SS+H+GDTP EKRP+LG+ A+ + KD K +D + P +
Sbjct: 741 DGKWIECSSSGETISSSHEGDTPKEKRPRLGTPALVAEVKDTSMKIVDDPDLGKPPQTGV 800
Query: 803 LNLTENEIVFNIGKSSTNESKQDPQRQVRSGLQKEGSKVIFGVPKPGKKRKFMEVSKHYV 862
LNL +E FNIGKS+ E+K DP R R+GLQK+GSKVIFGVPKPGKKRKFM+VSKHYV
Sbjct: 801 LNLGVSENTFNIGKSTREENKPDPLRMKRTGLQKQGSKVIFGVPKPGKKRKFMDVSKHYV 860
Query: 863 AHGSSKVNDKNDSVKIANFSMPQGSELRGWRNSSKNDSKEKLGADSKPKT-----KFGKP 917
+ S+K ++ + VK +PQ S + W+ SK S EK S+PKT K +
Sbjct: 861 SEASTKTQERKEPVKPVRSIVPQNSGIGSWKMPSKTISIEKQTTISRPKTFKPAPKPKEK 920
Query: 918 PGVLGRVNPPRNTSVSNTEMNKDSSNHTKNASQS-ESRVERAPYSTTDGATQVPIVFSSQ 976
PG R+ P KDS N T + +S ES R P S G + V
Sbjct: 921 PGATARIIP-----------RKDSRNTTASDMESDESAENRGPGS---GVSFKGTVEEQT 966
Query: 977 ATSTNTLPTKRTFTSRASKGKLAPASDKLRKGGGGKALNDKPTTSTSEPDALEPRRSNRR 1036
+S++ +K + + +KG++AP + +L K KAL + S+ + +EPRRS RR
Sbjct: 967 TSSSHDTGSKNSSSLSTNKGRVAPTAGRLAKIEEDKALAE---NSSKTSEGMEPRRSIRR 1023
Query: 1037 IQPTSRLLEGLQSSLMVSKIPSVSHNRN 1064
IQPTSRLLEGLQ+S+M SKIPSVSH+++
Sbjct: 1024 IQPTSRLLEGLQTSMMTSKIPSVSHSKS 1051
>emb|CAA10906.1| G2484-1 [Arabidopsis thaliana]
Length = 954
Score = 687 bits (1773), Expect = 0.0
Identities = 419/985 (42%), Positives = 590/985 (59%), Gaps = 79/985 (8%)
Query: 121 QSPSPRNFLGHSTSWISQAPLRGPWIGSP-TPAPDNNTHLSASPSSDTIKLASVKGSLPP 179
Q+P +N +GH+T W+S P R W+ S T D + P +D +KL +K S
Sbjct: 1 QTPPTQNIVGHNTPWMSPLPFRNAWLASQQTSGFDVGSRFPVYPITDPVKLTPMKESSMT 60
Query: 180 SSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQSSGPKAKKRKKDVLSEDHG 239
S K V G ++ S + T L+ + V PAQ S+ K++KRKK +S + G
Sbjct: 61 LSGAKHVQSGTSSNVSKVTPT-------LEPTSTVVAPAQHSTRVKSRKRKKMPVSVESG 113
Query: 240 QKLLQSL----------TPAVASRASTSVSAATPVGNVPMSSVEKSVVSV---------- 279
+L SL P + A+ +A T V M++V +VS
Sbjct: 114 PNILNSLKQTELAASPLVPFTPTPANLGYNAGTLPSVVSMTAVPMDLVSTFPGKKIKSSF 173
Query: 280 -SPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHKN 338
SP+ + ++ +LS++++ K+KEA++HAE+ASAL+ AAV+HS +W Q+++ +
Sbjct: 174 PSPIFGGNLVREVKQRSVLSEDTIEKLKEAKMHAEDASALATAAVSHSEYVWKQIEQQSH 233
Query: 339 SGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENTSQ 398
+G + + +L SAAVAIA AAAVAKAAAAAANVA+N A QAKLMA+EA + + + Q
Sbjct: 234 AGLQPETQDRLGSAAVAIAGAAAVAKAAAAAANVAANDALQAKLMAEEASLPNA---SDQ 290
Query: 399 GNNTFLPEGTSNL--GQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAEN 456
G LP+ ++ GQ TPASILKG + S ++AA+EA ++RVEAA+AATKRAEN
Sbjct: 291 G----LPKSYDSILPGQGTPASILKGEGAVVNSSSVLIAAREASKKRVEAATAATKRAEN 346
Query: 457 MDAILKAAELAAEAVSQAGKIVTMGDPLPLIELIEAGPEGCWKASRESSREVGLLKDMTR 516
MD+I+KAAELA+EAVSQAG +V+MG P L +L+EAGP W+ + ES +EV K +
Sbjct: 347 MDSIVKAAELASEAVSQAGILVSMGHPPLLNKLVEAGPSNYWRQAHES-QEVQPCKTVVL 405
Query: 517 DLVNIDMVRDIPETSHAQNRDILSSEISASIMINEK---------NTRGQQARTVSDLVK 567
+ + E + A + I+ +E S+ + +GQ+ +DL K
Sbjct: 406 EKETVST----SEGTFAGPK-IVQTEFGGSVNTADGVSGPVPATGKLKGQEGDKYADLAK 460
Query: 568 PVDMVLGSESETQDPSFTVRNGSENLEENTFKEGSLVEVFKDEEGHKAAWFMGNILSLKD 627
D+V E ++ S + + + KEGS VEVFK+E G + AW+ N+LSL+D
Sbjct: 461 NNDVVFEPEVGSKF-SIDAQQTIKATKNEDIKEGSNVEVFKEEPGLRTAWYSANVLSLED 519
Query: 628 GKVYVCYTSLVAVEGP--LKEWVSLECEGDKPPRIRTARPLTSLQHEGTRKRRRAAMGDY 685
K YV ++ L +G LKEWV+L+ EGD+ P+IR AR +T+L +EGTRKRRRAA+GD+
Sbjct: 520 DKAYVLFSDLSVEQGTDKLKEWVALKGEGDQAPKIRPARSVTALPYEGTRKRRRAALGDH 579
Query: 686 AWSVGDRVDAWIQESWREGVITEKNKKDETTLTVHIPASGETSVLRAWNLRPSLIWKDGQ 745
W +GDRVD+W+ +SW EGVITEKNKKDE T+TVH PA ET ++AWNLRPSL+WKDG+
Sbjct: 580 IWKIGDRVDSWVHDSWLEGVITEKNKKDENTVTVHFPAEEETLTIKAWNLRPSLVWKDGK 639
Query: 746 WLDFSKVGANDSSTHKGDTPHEKRPKLGSNAVEVKGKDKMSKNIDAAESANPDEMRSLNL 805
W++ S G SS+H+GDTP EKRP+LG+ A+ + KD K +D + P + LNL
Sbjct: 640 WIECSSSGETISSSHEGDTPKEKRPRLGTPALVAEVKDTSMKIVDDPDLGKPPQTGVLNL 699
Query: 806 TENEIVFNIGKSSTNESKQDPQRQVRSGLQKEGSKVIFGVPKPGKKRKFMEVSKHYVAHG 865
+E FNIGKS+ E+K DP R R+GLQK+GSKVIFGVPKPGKKRKFM+VSKHYV+
Sbjct: 700 GVSENTFNIGKSTREENKPDPLRMKRTGLQKQGSKVIFGVPKPGKKRKFMDVSKHYVSEA 759
Query: 866 SSKVNDKNDSVKIANFSMPQGSELRGWRNSSKNDSKEKLGADSKPKT-----KFGKPPGV 920
S+K ++ + VK +PQ S + W+ SK S EK S+PKT K + PG
Sbjct: 760 STKTQERKEPVKPVRSIVPQNSGIGSWKMPSKTISIEKQTTISRPKTFKPAPKPKEKPGA 819
Query: 921 LGRVNPPRNTSVSNTEMNKDSSNHTKNASQS-ESRVERAPYSTTDGATQVPIVFSSQATS 979
R+ P KDS N T + +S ES R P S G + V +S
Sbjct: 820 TARIIP-----------RKDSRNTTASDMESDESAENRGPGS---GVSFKGTVEEQTTSS 865
Query: 980 TNTLPTKRTFTSRASKGKLAPASDKLRKGGGGKALNDKPTTSTSEPDALEPRRSNRRIQP 1039
++ +K + + +KG++AP + +L K KAL + S+ + +EPRRS RRIQP
Sbjct: 866 SHDTGSKNSSSLSTNKGRVAPTAGRLAKIEEDKALAE---NSSKTSEGMEPRRSIRRIQP 922
Query: 1040 TSRLLEGLQSSLMVSKIPSVSHNRN 1064
TSRLLEGLQ+S+M SKIPSVSH+++
Sbjct: 923 TSRLLEGLQTSMMTSKIPSVSHSKS 947
>gb|AAP55000.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|37536822|ref|NP_922713.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
gi|18873854|gb|AAL79800.1| unknown protein [Oryza sativa]
Length = 2036
Score = 480 bits (1235), Expect = e-133
Identities = 366/1051 (34%), Positives = 547/1051 (51%), Gaps = 125/1051 (11%)
Query: 67 ASSKATPTI--ANPLIPLSSPLWSLPTLSADSLQSSALA--RGSVVDYSQALTPLHPYQS 122
AS K T+ A+ +PL LPTL+ L SSAL+ RG+ +D+ QA++P+ PY S
Sbjct: 1031 ASKKGGKTVLPAHTAVPLH-----LPTLNMSPLGSSALSLPRGTHLDFGQAVSPVFPYSS 1085
Query: 123 PSPRNFLGHSTSWISQAP--LRGPWIGSP------------TPAPDNNTHLSASPSSDTI 168
+ + G + SW Q+P PW+ P PA N T AS + +I
Sbjct: 1086 QTRQPTSGVA-SWFPQSPGGRAAPWLVQPQNLIFDSSMKPPVPASANETAKGASSKNISI 1144
Query: 169 KLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQSSGP-KAK 227
A + PP+ + ++P + + Q +V +++ + P K++
Sbjct: 1145 SQAVSPVAFPPNQAPSTISP----------LAVIPEEKQ----KASVSTSKRGATPQKSR 1190
Query: 228 KRKKDVLSEDHG------QKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVVSVSP 281
KRKK S + + + S+TPA ++S +P N+ S + + V+P
Sbjct: 1191 KRKKAPASPEQPIIAPLLKTDIASVTPATQHTPGFTLSTHSP-SNILASGLVSNTGLVTP 1249
Query: 282 LAD-QPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHKNSG 340
+ + Q + E+RI S++ ++++ A+ A + AV H+ +W+ L +
Sbjct: 1250 VPNYQITGIKDAEQRIFSEQISGAIEQSMGQAKGAGVHAMDAVRHAEGIWSHLSTNSKGK 1309
Query: 341 FMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENTSQGN 400
+++E KL SAA A +AA +VAKAAA AA +AS AA QAK+MA+E L SS Y N+SQ +
Sbjct: 1310 LPAEVEEKLTSAAAAASAAVSVAKAAAEAAKMASEAALQAKMMAEEVL-SSTYANSSQKH 1368
Query: 401 NTFLPEGTSNLGQ---ATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAENM 457
+ + ++NL TP S K S GS I A+E R+RVE A+AA KRAEN+
Sbjct: 1369 DAGEFKVSNNLASFSSLTPTSSWK-TKDDISKGSIISVAREVARKRVEEAAAAAKRAENL 1427
Query: 458 DAILKAAELAAEAVSQAGKIVTMGDPLP--LIELIEAGPEGCWKASRESSREVGLLKDMT 515
DAILKAAELAAEAV +AG I+ MG+PLP L EL+EAGP+G WK+ + +++
Sbjct: 1428 DAILKAAELAAEAVFKAGTIIGMGEPLPFTLSELLEAGPDGYWKSDQVRNKKA------- 1480
Query: 516 RDLVNIDMVRDIPETSHAQNRDILSSEISASIMINEKNTRGQQARTVSDLVKPVDMVLGS 575
+++ D ++ + ++ +I E ++ G+ + +D+V
Sbjct: 1481 ---TKLELPTDFSKSGRKRGG---KAKHDHAIQNLEPSSSGKGLQ--------LDVVHSG 1526
Query: 576 ESETQDPSFTVRNGSEN-----LEENTFKEGSLVEVFKDEEGHKAAWFMGNILSLKDGKV 630
P+ NG+ N + N ++GS VEV + AWF +L + +
Sbjct: 1527 NVAEDVPTIAPVNGNRNDAAPNIIWNGIEKGSAVEVLVHKGESGVAWFSAKVLDINNDSA 1586
Query: 631 YVCYTSLVAVEGPLKEWVSLECEGDKPPRIRTARPLTSLQHEGTRKRRRAAMGDYAWSVG 690
+ Y S G KEWV L EG+K P+IR A P T + +GTRKRRR G+Y+W++G
Sbjct: 1587 CISYDSHTEETGLRKEWVPLRQEGEKAPQIRLAHPATVSRFKGTRKRRRDTSGNYSWAIG 1646
Query: 691 DRVDAWIQESWREGVITEKNKKDETTLTVHIPA-SGETSVLRAWNLRPSLIWKDGQWLDF 749
D VD I++SWREG+I+ DET LTV A + ++ V+ AWNLRPSL+W DGQW+++
Sbjct: 1647 DHVDVLIEDSWREGIISRNRDGDETKLTVQFSAGTSDSLVVDAWNLRPSLVWNDGQWIEW 1706
Query: 750 SKVGANDSSTHKGDTPHEKRPKL-GSNAVEVKGK---DKMSKNIDAAESANPDEMRSLNL 805
S+ D +KGD+PHEKR + G++ V + G M + +AA A P+E + L L
Sbjct: 1707 SRGKTVD--CNKGDSPHEKRQRTKGNDHVPIGGAAAGPSMDTSTNAA--AKPEEPKPLAL 1762
Query: 806 TENEIVFNIGKSSTNESKQDPQRQVRSGLQKEGSKVIFGVPKPGKKRKFMEVSKHYVAHG 865
++ ++VFNIGK ESK D R GL+KEGS+V+ GVPKPGKK+KFMEVSKHY A
Sbjct: 1763 SDRDMVFNIGKRVV-ESKTDGVAFKRPGLRKEGSRVV-GVPKPGKKKKFMEVSKHYDADQ 1820
Query: 866 SSKVNDKNDSVKIANFSMPQGSELRGWRNSSKNDSK-EKLGADSKPKTKFGKPPGVLGRV 924
+ K+++ N S + +P R +SK D K +++G K K +
Sbjct: 1821 ADKISEGNASTRPVKHLVPNVPRPR--EGTSKVDQKGKRIGEMRSRVPKSTKSQDGATNI 1878
Query: 925 NP---PRNTSVSNTEMNKD----------SSNHTKNASQSESRVERAPYSTTDGATQVPI 971
P P + S +T + + SSN+ + + S V + D + P
Sbjct: 1879 IPGKGPLSMSAPSTGVFESSHTFIGSTIGSSNNMNLSVEKNSSVHGVGLRSEDSSVSEPH 1938
Query: 972 VFSSQATSTNTLPTKRTFTSRASKGKLAPASDKLRKGGGGKALNDKPTTSTSE-----PD 1026
+ QA S K T+ +K KL P+ D +++ T TSE D
Sbjct: 1939 I---QAASAAPTSRKNLTTTDRAKRKLVPSMDN----------SNRTTNKTSEIPGKSAD 1985
Query: 1027 ALEPRRSNRRIQPTSRLLEGLQSSLMVSKIP 1057
+ EPRRSNRRIQPTSRLLEGLQSSL+VSK+P
Sbjct: 1986 STEPRRSNRRIQPTSRLLEGLQSSLIVSKVP 2016
>emb|CAB10514.1| hypothetical protein [Arabidopsis thaliana]
gi|7268485|emb|CAB78736.1| hypothetical protein
[Arabidopsis thaliana] gi|7485136|pir||E71442
hypothetical protein - Arabidopsis thaliana
Length = 1732
Score = 424 bits (1089), Expect = e-116
Identities = 297/809 (36%), Positives = 430/809 (52%), Gaps = 135/809 (16%)
Query: 1 MISAYGGT--GTC-----GRMSGVSAWKGSVV---KNLILIPQKHLCSQDLAARTSDSTV 50
MISA+GG C G+ S +W+ VV K+L+ P+ L S+
Sbjct: 1000 MISAFGGAVCSICISLDGGKGSWEKSWRTCVVRAQKSLVATPETPLQSRP---------- 1049
Query: 51 KQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSALARGSVVDY 110
GK + G +SK + NP+IPLSSPLWSL T S D+LQSS++ RGS +
Sbjct: 1050 ------GKTETPSAGHTNSKESSG-TNPMIPLSSPLWSLST-SVDTLQSSSVQRGSAATH 1101
Query: 111 SQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSP-TPAPDNNTHLSASPSSDTIK 169
L+ H +Q+P +N +GH+T W+S P R W+ S T D + P +D +K
Sbjct: 1102 QPLLSASHAHQTPPTQNIVGHNTPWMSPLPFRNAWLASQQTSGFDVGSRFPVYPITDPVK 1161
Query: 170 LASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQSSGPKAKKR 229
L +K S S K V G ++ S + T L+ + V PAQ S+ K++KR
Sbjct: 1162 LTPMKESSMTLSGAKHVQSGTSSNVSKVTPT-------LEPTSTVVAPAQHSTRVKSRKR 1214
Query: 230 KKDVLSEDHGQKLLQSL----------TPAVASRASTSVSAATPVGNVPMSSVEKSVVSV 279
KK +S + G +L SL P + A+ +A T V M++V +VS
Sbjct: 1215 KKMPVSVESGPNILNSLKQTELAASPLVPFTPTPANLGYNAGTLPSVVSMTAVPMDLVST 1274
Query: 280 -----------SPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLE 328
SP+ + ++ +LS++++ K+KEA++HAE+ASAL+ AAV+HS
Sbjct: 1275 FPGKKIKSSFPSPIFGGNLVREVKQRSVLSEDTIEKLKEAKMHAEDASALATAAVSHSEY 1334
Query: 329 LWNQLDKHKNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEA- 387
+W Q+++ ++G + + +LASAAVAIAAAAAVAKAAAAAANVA+NAA QAKLMA+EA
Sbjct: 1335 VWKQIEQQSHAGLQPETQDRLASAAVAIAAAAAVAKAAAAAANVAANAALQAKLMAEEAS 1394
Query: 388 LISSGYENTSQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAA 447
L ++ + + ++ LP GQ TPASILKG + S ++AA+EA ++RVEAA
Sbjct: 1395 LPNASDQGLPKSYDSILP------GQGTPASILKGEGAVVNSSSVLIAAREASKKRVEAA 1448
Query: 448 SAATKRAENMDAILKAAELAAEAVSQAGKIVTMGDPLPLIELIEAGPEGCWKASRESSRE 507
+AATKRAENMD+I+KAAELA+EAVSQAG +V+MG P L +L+EAGP W+ ++E S+E
Sbjct: 1449 TAATKRAENMDSIVKAAELASEAVSQAGILVSMGHPPLLNKLVEAGPSNYWRQAQE-SQE 1507
Query: 508 VGLLKDMTRDLVNIDMVRDIPETSHA--QNRDILSSEISASIMINE---------KNTRG 556
V K + + ++ TS I+ +E S+ + +G
Sbjct: 1508 VQPCK-------TVVLEKETVSTSEGTFAGPKIVQTEFGGSVNTADGVSGPVPATGKLKG 1560
Query: 557 QQARTVSDLVKPVDMVLGSESETQDPSFTVRNGSENLEENTFKEGSLVEVFKDEEGHKAA 616
Q+ +DL K D+V E ++ S + + + KEGS VEVFK+E G + A
Sbjct: 1561 QEGDKYADLAKNNDVVFEPEVGSKF-SIDAQQTIKATKNEDIKEGSNVEVFKEEPGLRTA 1619
Query: 617 WFMGNILSLKDGKVYVCYTSLVAVEGPLKEWVSLECEGDKPPRIRTARPLTSLQHEGTRK 676
W+ N+LSL+D K YV ++ L +G + H+
Sbjct: 1620 WYSANVLSLEDDKAYVLFSDLSVEQG------------------------FCIYHQ---- 1651
Query: 677 RRRAAMGDYAWSVGDRVDAWIQESWREGVITEKNKKDETTLTVHIPASGETSVLRAWNLR 736
W+ EGVITEKNKKDE T+TVH PA ET ++AWNLR
Sbjct: 1652 ------------------CWL-----EGVITEKNKKDENTVTVHFPAEEETLTIKAWNLR 1688
Query: 737 PSLIWKDGQWLDFSKVGANDSSTHKGDTP 765
PSL+WKDG+W++ S G SS+H+ +P
Sbjct: 1689 PSLVWKDGKWIECSSSGETISSSHEVFSP 1717
>emb|CAB10513.1| hypothetical protein [Arabidopsis thaliana]
gi|7268484|emb|CAB78735.1| hypothetical protein
[Arabidopsis thaliana] gi|7485109|pir||D71442
hypothetical protein - Arabidopsis thaliana
Length = 232
Score = 167 bits (422), Expect = 2e-39
Identities = 105/240 (43%), Positives = 142/240 (58%), Gaps = 23/240 (9%)
Query: 831 RSGLQKEGSKVIFGVPKPGKKRKFMEVSKHYVAHGSSKVNDKNDSVKIANFSMPQGSELR 890
R+GLQK+GSKVIFGVPKPGKKRKFM+VSKHYV+ S+K ++ + VK +PQ S +
Sbjct: 3 RTGLQKQGSKVIFGVPKPGKKRKFMDVSKHYVSEASTKTQERKEPVKPVRSIVPQNSGIG 62
Query: 891 GWRNSSKNDSKEKLGADSKPKT-----KFGKPPGVLGRVNPPRNTSVSNTEMNKDSSNHT 945
W+ SK S EK S+PKT K + PG R+ P KDS N T
Sbjct: 63 SWKMPSKTISIEKQTTISRPKTFKPAPKPKEKPGATARIIP-----------RKDSRNTT 111
Query: 946 KNASQS-ESRVERAPYSTTDGATQVPIVFSSQATSTNTLPTKRTFTSRASKGKLAPASDK 1004
+ +S ES R P S G + V +S++ +K + + +KG++AP + +
Sbjct: 112 ASDMESDESAENRGPGS---GVSFKGTVEEQTTSSSHDTGSKNSSSLSTNKGRVAPTAGR 168
Query: 1005 LRKGGGGKALNDKPTTSTSEPDALEPRRSNRRIQPTSRLLEGLQSSLMVSKIPSVSHNRN 1064
L K KAL + S+ + +EPRRS RRIQPTSRLLEGLQ+S+M SKIPSVSH+++
Sbjct: 169 LAKIEEDKALAE---NSSKTSEGMEPRRSIRRIQPTSRLLEGLQTSMMTSKIPSVSHSKS 225
>dbj|BAD36250.1| agenet domain-containing protein / bromo-adjacent homology (BAH)
domain-containing protein-like [Oryza sativa (japonica
cultivar-group)]
Length = 956
Score = 72.8 bits (177), Expect = 6e-11
Identities = 50/190 (26%), Positives = 87/190 (45%), Gaps = 19/190 (10%)
Query: 569 VDMVLGSESETQDPSFTVRNGSENLEENTFKEGSLVEVFKDEEGHKAAWFMGNILSLKDG 628
++++ + + T+ S + + E E F G LVE + G + WF+G+++
Sbjct: 425 INVLARNAAPTESASALINSALEKYLEQYFSHGCLVECLSQDSGIRGCWFIGSVIRRCGD 484
Query: 629 KVYVCYTSLVAVEGP---LKEWVSLECEGDKPPRIRTARPLT-SLQHEGTRKRRRAAMGD 684
++ V Y L E P L+EW+ + RTA P T ++ G + R M +
Sbjct: 485 RIKVRYQHLQDPETPRANLEEWLLV---------TRTANPDTLRIRLSGRTRIRPHNMSE 535
Query: 685 ----YAWSVGDRVDAWIQESWREGVITEKNKKDETTLTVHIPASGETSVLRAWNLRPSLI 740
SVG +D W+ + W EG++ + N D L ++P + + R LR SL
Sbjct: 536 RENPSTISVGTVIDGWLYDGWWEGIVLKVN--DARRLLAYLPGEKKMVLFRKDQLRHSLE 593
Query: 741 WKDGQWLDFS 750
W D +W +F+
Sbjct: 594 WIDNEWKNFA 603
>ref|NP_346206.1| cell wall surface anchor family protein [Streptococcus pneumoniae
TIGR4] gi|14973269|gb|AAK75846.1| cell wall surface
anchor family protein [Streptococcus pneumoniae TIGR4]
gi|25389341|pir||E95206 cell wall surface anchor family
protein [imported] - Streptococcus pneumoniae (strain
TIGR4)
Length = 4776
Score = 68.2 bits (165), Expect = 1e-09
Identities = 92/442 (20%), Positives = 165/442 (36%), Gaps = 14/442 (3%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S +A TS S + +S AS+ A+ + + +S S ++ S
Sbjct: 1382 SASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSASA 1441
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
+S A S + A + +S S STS + A S + + +T
Sbjct: 1442 SASTSASASASTSASASASISASESASTSASASASTSASASASTSASVSASTSASASAST 1501
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPP 217
SAS S + AS S S+S + S+S ST + + A+
Sbjct: 1502 SASASASISASESASTSASASASTS---ASASASTSASASASTSASASASISASESASTS 1558
Query: 218 AQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVV 277
A S+ A S S + + ++ ASTS SA+ +S S
Sbjct: 1559 ASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSAS-A 1617
Query: 278 SVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHK 337
S S A + E S + + + ASA ++ + + S
Sbjct: 1618 SASTSASASASTSASESASTSASASASTSASASASTSASASASTSASVSA---------S 1668
Query: 338 NSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENTS 397
S S + ASA+ + +A+A+ + +A+A+ + + +A+ A A + +S + S
Sbjct: 1669 TSASESASTSASASASTSASASASTSASASASTSASESASTSASASASTSASASASTSAS 1728
Query: 398 QGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAENM 457
+ +T S A+ AS A+ S + A+ A +ASA+ + +
Sbjct: 1729 ESASTSASASASTSASAS-ASTSASASASTSASASASASTSASASASTSASASASTSASA 1787
Query: 458 DAILKAAELAAEAVSQAGKIVT 479
A + A+E A+ + S++ T
Sbjct: 1788 SASISASESASTSASESASTST 1809
Score = 68.2 bits (165), Expect = 1e-09
Identities = 88/430 (20%), Positives = 165/430 (37%), Gaps = 23/430 (5%)
Query: 48 STVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSAD-----SLQSSAL 102
ST QS+ Q K +S +++S + T A+ S+ + + SA S+ +S
Sbjct: 377 STTSQSLSQSKSLSVSASQSASASASTSASASASTSASASASTSASASASTSASVSASTS 436
Query: 103 ARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSAS 162
A S + A +S S STS + A S + + +T SAS
Sbjct: 437 ASASASTSASASASTSASESASTSASASASTSASASASTSASASASTSASESASTSASAS 496
Query: 163 PSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQSS 222
S+ + AS S S+S + S+SG ST + A+ A S+
Sbjct: 497 ASTSASESASTSASASASTS---ASASASTSASGSASTSTSASASTSASASASTSASASA 553
Query: 223 GPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVVSVSPL 282
A + SE S + + + ASTS SA+ +S S + +
Sbjct: 554 SISASESASTSASESASTSTSASASTSASESASTSASASASTSASASASTSASASASTSA 613
Query: 283 ADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHKNSGFM 342
+ ++ + S A A +++ SA+A S +
Sbjct: 614 SASTSASESASTSASASASTSASASASTSASASASTSASA------------SASTSASV 661
Query: 343 SDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENTSQGNNT 402
S + ASA+ + +A+A+ + + +A+ + +++A+ A A + +S + S +T
Sbjct: 662 SASTSASASASTSASASASTSASESASTSASASASTSASASASTSASASASTSASASAST 721
Query: 403 FLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAENMDAILK 462
+++ + S A+ S + A+ A +ASA+ + + A +
Sbjct: 722 ---SASASASTSASESASTSASASASTSASASASTSASASASTSASASASTSASASASIS 778
Query: 463 AAELAAEAVS 472
A+E A+ + S
Sbjct: 779 ASESASTSAS 788
Score = 62.4 bits (150), Expect = 8e-08
Identities = 92/435 (21%), Positives = 165/435 (37%), Gaps = 26/435 (5%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S +A TS S + +S AS+ A+ + + +S S ++ S
Sbjct: 698 SASASASTSASASASTSASASASTSASASASTSASESASTSASASASTSASASASTSASA 757
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
+S A S + A + +S S STS + A S + + +T
Sbjct: 758 SASTSASASASTSASASASISASESASTSASASASTSASASASTSASASASTSASESAST 817
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPP 217
SAS S+ AS S S+S AS+S S T + A+
Sbjct: 818 SASASASTSASASASTSASASASTSAS-------ASASTSASASASTSASASAST----S 866
Query: 218 AQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVV 277
A +S+ A SE S + + ++ ASTS SA+ +S S
Sbjct: 867 ASESASTSASASASTSASESASTSASASASTSASASASTSASASASTSASASASTSAS-A 925
Query: 278 SVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHK 337
S S A + S+ + + + ASA ++A+ + S
Sbjct: 926 SASTSASASASTSASASTSASESASTSASASASTSASASASTSASASASTS--------- 976
Query: 338 NSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENTS 397
S+ + ASA+ + +A+A+ + +A+A+A+ +++A+ A ++ +S +TS
Sbjct: 977 ----ASESASTSASASASTSASASASTSASASASTSASASASTSASASASISASESASTS 1032
Query: 398 QGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAENM 457
+ S A+ AS A+ S + A+ A +ASA+ + +
Sbjct: 1033 ASASASTSASVSASTSAS-ASASTSASESASTSASASASTSASESASTSASASASTSASA 1091
Query: 458 DAILKAAELAAEAVS 472
A + A+E A+ + S
Sbjct: 1092 SASISASESASTSAS 1106
Score = 62.4 bits (150), Expect = 8e-08
Identities = 89/435 (20%), Positives = 159/435 (36%), Gaps = 10/435 (2%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S +A S S + +S AS+ A+ + + +S S ++ S
Sbjct: 2402 STSASASASTSASASASTSASASASTSASASTSASESASTSASASASTSASASASTSASA 2461
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
+S A S + A S S STS + A S + + +T
Sbjct: 2462 SASTSASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSASASAST 2521
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPP 217
SAS S+ + AS S S+S + S+S ST + A+
Sbjct: 2522 SASASASTSASESASTSASASASTS---ASASASTSASASASTSASVSASTSASESASTS 2578
Query: 218 AQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVV 277
A S+ A S +S + + ++ ASTS SA+ S+ E +
Sbjct: 2579 ASASASTSASASASTSASASASTSASESASTSASASASTSASASAST-----SASESAST 2633
Query: 278 SVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHK 337
S S A + S + + + ASA ++A+ + S
Sbjct: 2634 SASASASTSASASASTSASASASTSASASASASTSASASASTSASASASTSASASASISA 2693
Query: 338 NSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENTS 397
+ + A ASA+ + +A+A+ + +A+A+ + +++A+ A A + S +TS
Sbjct: 2694 SESASTSASAS-ASASTSASASASTSASASASTSASASASISASESASTSASESASTSTS 2752
Query: 398 QGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAENM 457
+T E S A+ AS A+ S + + A E+AS + + +
Sbjct: 2753 ASASTSASESASTSASAS-ASTSASASASTSASASASTSASASTSASESASTSASASAST 2811
Query: 458 DAILKAAELAAEAVS 472
A A+ A+ + S
Sbjct: 2812 SASASASTSASASAS 2826
Score = 62.4 bits (150), Expect = 8e-08
Identities = 84/440 (19%), Positives = 162/440 (36%), Gaps = 7/440 (1%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S +A TS S + +S AS+ A+ + + +S S ++ S
Sbjct: 2426 STSASASTSASESASTSASASASTSASASASTSASASASTSASASASTSASASASTSASA 2485
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
+S A S + A S S STS + A S + + +T
Sbjct: 2486 SASTSASASASTSASASASTSASASASTSASASASTSASASASTSASESASTSASASAST 2545
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPP 217
SAS S+ AS S+ S+S + S+S ST + A+
Sbjct: 2546 SASASASTSASASASTSASVSASTSASE---SASTSASASASTSASASASTSASASASTS 2602
Query: 218 AQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSV- 276
A +S+ A S +S + + ++ ASTS SA+ +S S
Sbjct: 2603 ASESASTSASASASTSASASASTSASESASTSASASASTSASASASTSASASASTSASAS 2662
Query: 277 --VSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLD 334
S S A + S + + E+ + ASA ++ + + S
Sbjct: 2663 ASASTSASASASTSASASASTSASASASISASESASTSASASASASTSASASASTSASAS 2722
Query: 335 KHKNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYE 394
++ + I A SA+ + + +A+ + +A+A+ + + +A+ A A + +S
Sbjct: 2723 ASTSASASASISAS-ESASTSASESASTSTSASASTSASESASTSASASASTSASASAST 2781
Query: 395 NTSQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRA 454
+ S +T TS A+ ++ + ++ S +A + A+++A+ A
Sbjct: 2782 SASASASTSASASTSASESASTSASASASTSASASASTSASASASTSASASASTSASASA 2841
Query: 455 ENMDAILKAAELAAEAVSQA 474
++ + +A A + A
Sbjct: 2842 STSASVSASTSASASASTSA 2861
Score = 62.4 bits (150), Expect = 8e-08
Identities = 92/435 (21%), Positives = 165/435 (37%), Gaps = 26/435 (5%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S +A TS S + +S AS+ A+ + + +S S ++ S
Sbjct: 3184 SASASASTSASASASTSASASASTSASASASTSASESASTSASASASTSASASASTSASA 3243
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
+S A S + A + +S S STS + A S + + +T
Sbjct: 3244 SASTSASASASTSASASASISASESASTSASASASTSASASASTSASASASTSASESAST 3303
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPP 217
SAS S+ AS S S+S AS+S S T + A+
Sbjct: 3304 SASASASTSASASASTSASASASTSAS-------ASASTSASASASTSASASAST----S 3352
Query: 218 AQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVV 277
A +S+ A SE S + + ++ ASTS SA+ +S S
Sbjct: 3353 ASESASTSASASASTSASESASTSASASASTSASASASTSASASASTSASASASTSAS-A 3411
Query: 278 SVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHK 337
S S A + S+ + + + ASA ++A+ + S
Sbjct: 3412 SASTSASASASTSASASTSASESASTSASASASTSASASASTSASASASTS--------- 3462
Query: 338 NSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENTS 397
S+ + ASA+ + +A+A+ + +A+A+A+ +++A+ A ++ +S +TS
Sbjct: 3463 ----ASESASTSASASASTSASASASTSASASASTSASASASTSASASASISASESASTS 3518
Query: 398 QGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAENM 457
+ S A+ AS A+ S + A+ A +ASA+ + +
Sbjct: 3519 ASASASTSASVSASTSAS-ASASTSASESASTSASASASTSASESASTSASASASTSASA 3577
Query: 458 DAILKAAELAAEAVS 472
A + A+E A+ + S
Sbjct: 3578 SASISASESASTSAS 3592
Score = 62.4 bits (150), Expect = 8e-08
Identities = 92/435 (21%), Positives = 165/435 (37%), Gaps = 26/435 (5%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S +A TS S + +S AS+ A+ + + +S S ++ S
Sbjct: 1942 SASASASTSASASASTSASASASTSASASASTSASESASTSASASASTSASASASTSASA 2001
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
+S A S + A + +S S STS + A S + + +T
Sbjct: 2002 SASTSASASASTSASASASISASESASTSASASASTSASASASTSASASASTSASESAST 2061
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPP 217
SAS S+ AS S S+S AS+S S T + A+
Sbjct: 2062 SASASASTSASASASTSASASASTSAS-------ASASTSASASASTSASASAST----S 2110
Query: 218 AQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVV 277
A +S+ A SE S + + ++ ASTS SA+ +S S
Sbjct: 2111 ASESASTSASASASTSASESASTSASASASTSASASASTSASASASTSASASASTSAS-A 2169
Query: 278 SVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHK 337
S S A + S+ + + + ASA ++A+ + S
Sbjct: 2170 SASTSASASASTSASASTSASESASTSASASASTSASASASTSASASASTS--------- 2220
Query: 338 NSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENTS 397
S+ + ASA+ + +A+A+ + +A+A+A+ +++A+ A ++ +S +TS
Sbjct: 2221 ----ASESASTSASASASTSASASASTSASASASTSASASASTSASASASISASESASTS 2276
Query: 398 QGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAENM 457
+ S A+ AS A+ S + A+ A +ASA+ + +
Sbjct: 2277 ASASASTSASVSASTSAS-ASASTSASESASTSASASASTSASESASTSASASASTSASA 2335
Query: 458 DAILKAAELAAEAVS 472
A + A+E A+ + S
Sbjct: 2336 SASISASESASTSAS 2350
Score = 62.0 bits (149), Expect = 1e-07
Identities = 90/446 (20%), Positives = 165/446 (36%), Gaps = 12/446 (2%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S +A TS S + +S AS+ A+ + + +S S ++ S
Sbjct: 3868 SASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSASA 3927
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
+S A S + A + +S S STS + A S + + +T
Sbjct: 3928 SASTSASASASTSASASASISASESASTSASASASTSASASASTSASVSASTSASASAST 3987
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPP 217
SAS S + AS S S+S + S+S ST + + A+
Sbjct: 3988 SASASASISASESASTSASASASTS---ASASASTSASASASTSASASASISASESASTS 4044
Query: 218 AQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSV- 276
A S+ A S S + + ++ ASTS SA+ +S S
Sbjct: 4045 ASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSASAS 4104
Query: 277 --VSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLD 334
S S A ++ S + + +E AS ++A+ + S
Sbjct: 4105 ASTSASASASTSASESASTSASASASTSASASASISASESASTSASASASTSASASASTS 4164
Query: 335 KHKN-SGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGY 393
+ S S+ + SA+ + +A+ + + +A+A+A+ +++A+ A + +S
Sbjct: 4165 ASASASTSASESASTSTSASASTSASESASTSASASASTSASASASTSASASASTSASAS 4224
Query: 394 ENTSQGNNTFLPEGTSNLGQAT-----PASILKGANGPNSPGSFIVAAKEAIRRRVEAAS 448
+TS +T E S A+ AS A+ S + A+ A +AS
Sbjct: 4225 ASTSASASTSASESASTSASASASTSASASASTSASASASTSASASASTSASASASTSAS 4284
Query: 449 AATKRAENMDAILKAAELAAEAVSQA 474
A+ + + A A+ A+ + S++
Sbjct: 4285 ASASTSASASASTSASASASTSASES 4310
Score = 61.2 bits (147), Expect = 2e-07
Identities = 92/439 (20%), Positives = 165/439 (36%), Gaps = 37/439 (8%)
Query: 42 AARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSA 101
A+ ++ ++ S S+ + S +T T A+ S S T ++ S +SA
Sbjct: 2349 ASASASTSASASASTSASASASTSASESASTSTSASASTSASE---SASTSASASASTSA 2405
Query: 102 LARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSA 161
A S + A T S S STS A S + + +T SA
Sbjct: 2406 SASASTSASASASTSA----SASASTSASASTSASESASTSASASASTSASASASTSASA 2461
Query: 162 SPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQS 221
S S+ AS S S+S + S+S ST + A+ A S
Sbjct: 2462 SASTSASASASTSASASASTS---ASASASTSASASASTSASASASTSASASASTSASAS 2518
Query: 222 SGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSA-ATPVGNVPMSSVEKSVVSVS 280
+ A SE S + + ++ ASTS SA A+ +V S+ S S
Sbjct: 2519 ASTSASASASTSASESASTSASASASTSASASASTSASASASTSASVSASTSASESASTS 2578
Query: 281 PLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHKNSG 340
A + S + E+ + ASA ++A+ + S
Sbjct: 2579 ASASASTSASASASTSASASASTSASESASTSASASASTSASASASTSA----------- 2627
Query: 341 FMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENTSQGN 400
S+ + ASA+ + +A+A+ + +A+A+A+ +++A+ A A + +S
Sbjct: 2628 --SESASTSASASASTSASASASTSASASASTSASASASASTSASASASTSA-------- 2677
Query: 401 NTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAENMDAI 460
+++ + ASI + S + A+ A +ASA+ + + A
Sbjct: 2678 -----SASASTSASASASISASESASTSASASASASTSASASASTSASASASTSASASAS 2732
Query: 461 LKAAELAAEAVSQAGKIVT 479
+ A+E A+ + S++ T
Sbjct: 2733 ISASESASTSASESASTST 2751
Score = 60.8 bits (146), Expect = 2e-07
Identities = 89/436 (20%), Positives = 163/436 (36%), Gaps = 28/436 (6%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S +A TS S + +S AS+ A+ + + +S S T +++S
Sbjct: 2828 SASASASTSASASASTSASVSASTSASASASTSASASASTSASESASTSASASTSASESA 2887
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
+SA A S + A STS + A S + + +T
Sbjct: 2888 STSASASASTSASASA------------------STSASASASTSASESASTSASASAST 2929
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPP 217
SAS S+ + AS S S+S + S+S ST + A+
Sbjct: 2930 SASASASTSASESASTSASASASTS---ASASASTSASASASTSASESASTSASASASTS 2986
Query: 218 AQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVV 277
A +S+ A S S + + ++ ASTS SA+ +S+ S
Sbjct: 2987 ASESASTSASASASTSASASASTSASGSASTSTSASASTSASASASTSASASASISASES 3046
Query: 278 SVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAE-EASALSAAAVNHSLELWNQLDKH 336
+ + ++ + + ES A ASA ++A+ + S
Sbjct: 3047 ASTSASESASTSTSASASTSASESASTSASASASTSASASASTSASASASTSA-----SA 3101
Query: 337 KNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENT 396
S S + ASA+ + +A+A+ + +A+A+ + +++A+ A + A + +S +
Sbjct: 3102 STSASESASTSASASASTSASASASTSASASASTSASASASTSASVSASTSASASASTSA 3161
Query: 397 SQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAEN 456
S +T E S A+ AS A+ S + + A +ASA+T +E+
Sbjct: 3162 SASASTSASESASTSASAS-ASTSASASASTSASASASTSASASASTSASASASTSASES 3220
Query: 457 MDAILKAAELAAEAVS 472
A+ + + S
Sbjct: 3221 ASTSASASASTSASAS 3236
Score = 60.5 bits (145), Expect = 3e-07
Identities = 92/438 (21%), Positives = 171/438 (39%), Gaps = 34/438 (7%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S ++A TS S + +S AS+ A+ + + +S S T +++S
Sbjct: 1302 SASVSASTSASESASTSASASASTSASASASTSASESAS------TSASASASTSASESA 1355
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
+SA A S + A T S S STS + A S + + +T
Sbjct: 1356 STSASASASTSASASASTSASASASTSAS--ASASTSASASASTSASASASTSASASAST 1413
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPP 217
SAS S+ AS S S+S + S+S ST + + A+
Sbjct: 1414 SASASASTSASASASTSASASASTS---ASASASTSASASASTSASASASISASESASTS 1470
Query: 218 AQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSV- 276
A S+ A S S + + ++ ASTS SA+ + +S S
Sbjct: 1471 ASASASTSA--------SASASTSASVSASTSASASASTSASASASISASESASTSASAS 1522
Query: 277 VSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKH 336
S S A + S + + E+ + ASA ++A+ + S
Sbjct: 1523 ASTSASASASTSASASASTSASASASISASESASTSASASASTSASASAS---------- 1572
Query: 337 KNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENT 396
S S + ASA+ + +A+A+ + +A+A+ + +++A+ A A + +S +
Sbjct: 1573 -TSASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSA 1631
Query: 397 SQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAEN 456
S+ +T +++ + AS A+ S + + A+ A +ASA+ + +
Sbjct: 1632 SESAST---SASASASTSASASASTSASASASTSASVSASTSASESASTSASASASTSAS 1688
Query: 457 MDAILKAAELAAEAVSQA 474
A A+ A+ + S++
Sbjct: 1689 ASASTSASASASTSASES 1706
Score = 60.5 bits (145), Expect = 3e-07
Identities = 87/439 (19%), Positives = 161/439 (35%), Gaps = 6/439 (1%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S +A TS S + +S AS+ A+ + + +S S ++ S
Sbjct: 1558 SASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSASA 1617
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
+S A S + S S STS + A S + + +T
Sbjct: 1618 SASTSASASASTSASESASTSASASASTSASASASTSASASASTSASVSASTSASESAST 1677
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPP 217
SAS S+ AS S S+S + + AS+S S + + T
Sbjct: 1678 SASASASTSASASASTSASASASTSASE-SASTSASASASTSASASASTSASESASTSAS 1736
Query: 218 AQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVV 277
A S+ A S S + + ++ ASTS SA+ +S+ S
Sbjct: 1737 ASASTSASASASTSASASASTSASASASASTSASASASTSASASASTSASASASISASES 1796
Query: 278 SVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAE-EASALSAAAVNHSLELWNQLDKH 336
+ + ++ + + ES A ASA ++A+ + S
Sbjct: 1797 ASTSASESASTSTSASASTSASESASTSASASASTSASASASTSASASASTSASASTSAS 1856
Query: 337 KNSGFMSDIEAKLASAAVAIAAAAAVAK---AAAAAANVASNAAFQAKLMADEALISSGY 393
+++ + A +++A A +A+A A +A+A+ + +++A+ A + A + +S
Sbjct: 1857 ESASTSASASASTSASASASTSASASASTSASASASTSASASASTSASVSASTSASASAS 1916
Query: 394 ENTSQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKR 453
+ S +T E S A+ AS A+ S + + A +ASA+T
Sbjct: 1917 TSASASASTSASESASTSASAS-ASTSASASASTSASASASTSASASASTSASASASTSA 1975
Query: 454 AENMDAILKAAELAAEAVS 472
+E+ A+ + + S
Sbjct: 1976 SESASTSASASASTSASAS 1994
Score = 60.1 bits (144), Expect = 4e-07
Identities = 88/431 (20%), Positives = 161/431 (36%), Gaps = 26/431 (6%)
Query: 42 AARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSA 101
+A TS S + +S AS+ A+ + + +S S T +++S +SA
Sbjct: 1804 SASTSTSASASTSASESASTSASASASTSASASASTSASASASTSASASTSASESASTSA 1863
Query: 102 LARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSA 161
A S + A T S S STS + A S + + +T SA
Sbjct: 1864 SASASTSASASASTSASASASTSAS--ASASTSASASASTSASVSASTSASASASTSASA 1921
Query: 162 SPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQS 221
S S+ + AS S S+S + S+S ST + A+ A +S
Sbjct: 1922 SASTSASESASTSASASASTS---ASASASTSASASASTSASASASTSASASASTSASES 1978
Query: 222 SGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVVSVSP 281
+ A S S + + ++ ASTS SA+ + +S S + +
Sbjct: 1979 ASTSASASASTSASASASTSASASASTSASASASTSASASASISASESASTSASASASTS 2038
Query: 282 LADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHKNSGF 341
+ + + ES A A +++ SA+A S
Sbjct: 2039 ASASASTSASASASTSASES------ASTSASASASTSASA------------SASTSAS 2080
Query: 342 MSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENTSQGNN 401
S + ASA+ + +A+A+ + +A+A+ + + +A+ A A + S + S +
Sbjct: 2081 ASASTSASASASTSASASASTSASASASTSASESASTSASASASTSASESASTSASASAS 2140
Query: 402 TFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAENMDAIL 461
T +++ + AS A+ S + A+ A +ASA+T +E+
Sbjct: 2141 T---SASASASTSASASASTSASASASTSASASASTSASASASTSASASTSASESASTSA 2197
Query: 462 KAAELAAEAVS 472
A+ + + S
Sbjct: 2198 SASASTSASAS 2208
Score = 59.3 bits (142), Expect = 7e-07
Identities = 85/436 (19%), Positives = 162/436 (36%), Gaps = 18/436 (4%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S +A TS S + S AS+ A+ + + +S S ++ S
Sbjct: 1526 SASASASTSASASASTSASASASISASESASTSASASASTSASASASTSASASASTSASA 1585
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
+S A S + A S S STS + A S + + +T
Sbjct: 1586 SASTSASASASTSASASASTSASASASTSASASASTSASASASTSASESASTSASASAST 1645
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIKD-VTPGPPASSSGLQSTFVGTDSQLDANNVTVP 216
SAS S+ AS S+ S+S + + AS+S S T + A+
Sbjct: 1646 SASASASTSASASASTSASVSASTSASESASTSASASASTSASASASTSASASASTSASE 1705
Query: 217 PAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSV 276
A S+ A S + S + + ++ AS S S + +S S
Sbjct: 1706 SASTSASASASTSASASASTSASESASTSASASASTSASASASTSASASASTSASASASA 1765
Query: 277 VSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKH 336
+ + + + + S+ + A A E+++ S +A
Sbjct: 1766 STSASASASTSASASASTSASASASISASESASTSASESASTSTSA-------------- 1811
Query: 337 KNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENT 396
S S+ + ASA+ + +A+A+ + +A+A+A+ +++A+ A A + +S +
Sbjct: 1812 SASTSASESASTSASASASTSASASASTSASASASTSASASTSASESASTSASASASTSA 1871
Query: 397 SQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAEN 456
S +T +++ + AS A+ S + + A+ A +ASA+ + +
Sbjct: 1872 SASAST---SASASASTSASASASTSASASASTSASVSASTSASASASTSASASASTSAS 1928
Query: 457 MDAILKAAELAAEAVS 472
A A+ A+ + S
Sbjct: 1929 ESASTSASASASTSAS 1944
Score = 59.3 bits (142), Expect = 7e-07
Identities = 84/434 (19%), Positives = 158/434 (36%), Gaps = 9/434 (2%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S +A TS S + +S AS+ A+ + + +S S ++ S
Sbjct: 420 SASASASTSASVSASTSASASASTSASASASTSASESASTSASASASTSASASASTSASA 479
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
+S A S + A +S S STS + A S + + +T
Sbjct: 480 SASTSASESASTSASASASTSASESASTSASASASTSASASASTSASGSASTSTSASAST 539
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIKD-VTPGPPASSSGLQSTFVGTDSQLDANNVTVP 216
SAS S+ AS+ S S+S + + AS+S S T + A+
Sbjct: 540 SASASASTSASASASISASESASTSASESASTSTSASASTSASESASTSASASASTSASA 599
Query: 217 PAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSV 276
A S+ A S S + + ++ AS S SA+ +S S
Sbjct: 600 SASTSASASASTSASASTSASESASTSASASASTSASASASTSASASASTSASASASTS- 658
Query: 277 VSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKH 336
SVS + T S + + + SA ++A+ + S
Sbjct: 659 ASVSASTSASASASTSASASASTSASESASTSASASASTSASASASTSASASA------- 711
Query: 337 KNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENT 396
S S + ASA+ + + +A+ + +A+A+ + +++A+ A A + +S +
Sbjct: 712 STSASASASTSASASASTSASESASTSASASASTSASASASTSASASASTSASASASTSA 771
Query: 397 SQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAEN 456
S + E S A+ ++ + ++ S +A E+ A+++ + A
Sbjct: 772 SASASISASESASTSASASASTSASASASTSASASASTSASESASTSASASASTSASASA 831
Query: 457 MDAILKAAELAAEA 470
+ +A +A A
Sbjct: 832 STSASASASTSASA 845
Score = 58.5 bits (140), Expect = 1e-06
Identities = 93/443 (20%), Positives = 164/443 (36%), Gaps = 22/443 (4%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S +A TS S + +S AS+ A+ + + +S S ++ S
Sbjct: 460 SASASASTSASASASTSASASASTSASESASTSASASASTSASESASTSASASASTSASA 519
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
+S A GS + A S S STS + A + S + + +T
Sbjct: 520 SASTSASGSASTSTSA--------SASTSASASASTSASASASISASESASTSASESAST 571
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIK---DVTPGPPASSSGLQSTFVGTDSQLDANNVT 214
SAS S+ + AS S S+S + AS+S ST + A+
Sbjct: 572 STSASASTSASESASTSASASASTSASASASTSASASASTSASASTSASESASTSASASA 631
Query: 215 VPPAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEK 274
A S+ A S S + + ++ ASTS SA+ +S
Sbjct: 632 STSASASASTSASASASTSASASASTSASVSASTSASASASTSASASASTSASESASTSA 691
Query: 275 SV---VSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWN 331
S S S A + S + + +E AS ++A+ + S
Sbjct: 692 SASASTSASASASTSASASASTSASASASTSASASASTSASESASTSASASASTSASA-- 749
Query: 332 QLDKHKNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISS 391
S S + ASA+ + +A+A+++ + +A+ + +++A+ A A + +S
Sbjct: 750 ---SASTSASASASTSASASASTSASASASISASESASTSASASASTSASASASTSASAS 806
Query: 392 GYENTSQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAAT 451
+ S+ +T S A+ AS A+ S + + A +ASA+T
Sbjct: 807 ASTSASESASTSASASASTSASAS-ASTSASASASTSASASASTSASASASTSASASAST 865
Query: 452 KRAE--NMDAILKAAELAAEAVS 472
+E + A A+ A+E+ S
Sbjct: 866 SASESASTSASASASTSASESAS 888
Score = 58.5 bits (140), Expect = 1e-06
Identities = 84/434 (19%), Positives = 161/434 (36%), Gaps = 15/434 (3%)
Query: 42 AARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSA 101
+A TS S + +S AS+ A+ + + +S S ++ S +S
Sbjct: 3840 SASTSASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSASASAST 3899
Query: 102 LARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSA 161
A S + A S S STS + A S + + +T SA
Sbjct: 3900 SASASASTSASASASTSASASASTSASASASTSASASASTSASASASISASESASTSASA 3959
Query: 162 SPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQS 221
S S+ AS S+ S+ S+S ST + + A+ A S
Sbjct: 3960 SASTSASASASTSASVSAST-----------SASASASTSASASASISASESASTSASAS 4008
Query: 222 SGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSA-ATPVGNVPMSSVEKSVVSVS 280
+ A S S + + + ASTS SA A+ + S+ + S S
Sbjct: 4009 ASTSASASASTSASASASTSASASASISASESASTSASASASTSASASASTSASASASTS 4068
Query: 281 PLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHKNSG 340
A + S + + + ASA ++A+ + S + + S
Sbjct: 4069 ASASASTSASASASTSASASASTSASASASTSASASASTSASASAST---SASESASTSA 4125
Query: 341 FMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENTSQGN 400
S + ASA+++ + +A+ + +A+A+ + +++A+ A A + S +TS
Sbjct: 4126 SASASTSASASASISASESASTSASASASTSASASASTSASASASTSASESASTSTSASA 4185
Query: 401 NTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAENMDAI 460
+T E S A+ ++ + ++ S +A + A+++A++ A +
Sbjct: 4186 STSASESASTSASASASTSASASASTSASASASTSASASASTSASASTSASESASTSASA 4245
Query: 461 LKAAELAAEAVSQA 474
+ +A A + A
Sbjct: 4246 SASTSASASASTSA 4259
Score = 58.5 bits (140), Expect = 1e-06
Identities = 93/443 (20%), Positives = 164/443 (36%), Gaps = 22/443 (4%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S +A TS S + +S AS+ A+ + + +S S ++ S
Sbjct: 2946 SASASASTSASASASTSASASASTSASESASTSASASASTSASESASTSASASASTSASA 3005
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
+S A GS + A S S STS + A + S + + +T
Sbjct: 3006 SASTSASGSASTSTSA--------SASTSASASASTSASASASISASESASTSASESAST 3057
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIK---DVTPGPPASSSGLQSTFVGTDSQLDANNVT 214
SAS S+ + AS S S+S + AS+S ST + A+
Sbjct: 3058 STSASASTSASESASTSASASASTSASASASTSASASASTSASASTSASESASTSASASA 3117
Query: 215 VPPAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEK 274
A S+ A S S + + ++ ASTS SA+ +S
Sbjct: 3118 STSASASASTSASASASTSASASASTSASVSASTSASASASTSASASASTSASESASTSA 3177
Query: 275 SV---VSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWN 331
S S S A + S + + +E AS ++A+ + S
Sbjct: 3178 SASASTSASASASTSASASASTSASASASTSASASASTSASESASTSASASASTSASA-- 3235
Query: 332 QLDKHKNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISS 391
S S + ASA+ + +A+A+++ + +A+ + +++A+ A A + +S
Sbjct: 3236 ---SASTSASASASTSASASASTSASASASISASESASTSASASASTSASASASTSASAS 3292
Query: 392 GYENTSQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAAT 451
+ S+ +T S A+ AS A+ S + + A +ASA+T
Sbjct: 3293 ASTSASESASTSASASASTSASAS-ASTSASASASTSASASASTSASASASTSASASAST 3351
Query: 452 KRAE--NMDAILKAAELAAEAVS 472
+E + A A+ A+E+ S
Sbjct: 3352 SASESASTSASASASTSASESAS 3374
Score = 58.2 bits (139), Expect = 2e-06
Identities = 83/423 (19%), Positives = 158/423 (36%), Gaps = 13/423 (3%)
Query: 42 AARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSA 101
A+ ++ ++ S S+ ++S +T A+ S+ S +++S +SA
Sbjct: 1975 ASESASTSASASASTSASASASTSASASASTSASASASTSASA---SASISASESASTSA 2031
Query: 102 LARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSA 161
A S + A T S S STS + A S + + +T SA
Sbjct: 2032 SASASTSASASASTSASASASTSASE--SASTSASASASTSASASASTSASASASTSASA 2089
Query: 162 SPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQS 221
S S+ AS S S+S + S+S ST + A+ A S
Sbjct: 2090 SASTSASASASTSASASASTSASE---SASTSASASASTSASESASTSASASASTSASAS 2146
Query: 222 SGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVVSVSP 281
+ A S S + + ++ ASTS SA+T +S S + +
Sbjct: 2147 ASTSASASASTSASASASTSASASASTSASASASTSASASTSASESASTSASASASTSAS 2206
Query: 282 LADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHKNSGF 341
+ + S A A +++ SA+A + + S
Sbjct: 2207 ASASTSASASASTSASESASTSASASASTSASASASTSASASASTSASASASTSASASAS 2266
Query: 342 MSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENTSQGNN 401
+S E+ SA+ + + +A+V+ + +A+A+ +++A+ A A + +S E+ S +
Sbjct: 2267 ISASESASTSASASASTSASVSASTSASASASTSASESASTSASASASTSASESASTSAS 2326
Query: 402 TFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAENMDAIL 461
+++ + ASI + S + + A +ASA+T +E+
Sbjct: 2327 A-----SASTSASASASISASESASTSASASASTSASASASTSASASASTSASESASTST 2381
Query: 462 KAA 464
A+
Sbjct: 2382 SAS 2384
Score = 58.2 bits (139), Expect = 2e-06
Identities = 92/428 (21%), Positives = 163/428 (37%), Gaps = 32/428 (7%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S ++A TS S + +S AS+ A+ + + +S S T +++S
Sbjct: 3788 SASVSASTSASESASTSASASASTSASASASTSASESAS------TSASASASTSASESA 3841
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
+SA A S + A T S S STS + A S + + +T
Sbjct: 3842 STSASASASTSASASASTSASASASTSAS--ASASTSASASASTSASASASTSASASAST 3899
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPP 217
SAS S+ AS S S+S + S+S ST + + A+
Sbjct: 3900 SASASASTSASASASTSASASASTS---ASASASTSASASASTSASASASISASESASTS 3956
Query: 218 AQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSV- 276
A S+ A S S + + ++ ASTS SA+ + +S S
Sbjct: 3957 ASASASTSA--------SASASTSASVSASTSASASASTSASASASISASESASTSASAS 4008
Query: 277 VSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKH 336
S S A + S + + E+ + ASA ++A+ + S
Sbjct: 4009 ASTSASASASTSASASASTSASASASISASESASTSASASASTSASASAS---------- 4058
Query: 337 KNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENT 396
S S + ASA+ + +A+A+ + +A+A+ + +++A+ A A + +S +
Sbjct: 4059 -TSASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSA 4117
Query: 397 SQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAEN 456
S+ +T S A+ ASI + S + + A +ASA+T +E+
Sbjct: 4118 SESASTSASASASTSASAS-ASISASESASTSASASASTSASASASTSASASASTSASES 4176
Query: 457 MDAILKAA 464
A+
Sbjct: 4177 ASTSTSAS 4184
Score = 58.2 bits (139), Expect = 2e-06
Identities = 83/423 (19%), Positives = 158/423 (36%), Gaps = 13/423 (3%)
Query: 42 AARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSA 101
A+ ++ ++ S S+ ++S +T A+ S+ S +++S +SA
Sbjct: 3217 ASESASTSASASASTSASASASTSASASASTSASASASTSASA---SASISASESASTSA 3273
Query: 102 LARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSA 161
A S + A T S S STS + A S + + +T SA
Sbjct: 3274 SASASTSASASASTSASASASTSASE--SASTSASASASTSASASASTSASASASTSASA 3331
Query: 162 SPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQS 221
S S+ AS S S+S + S+S ST + A+ A S
Sbjct: 3332 SASTSASASASTSASASASTSASE---SASTSASASASTSASESASTSASASASTSASAS 3388
Query: 222 SGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVVSVSP 281
+ A S S + + ++ ASTS SA+T +S S + +
Sbjct: 3389 ASTSASASASTSASASASTSASASASTSASASASTSASASTSASESASTSASASASTSAS 3448
Query: 282 LADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHKNSGF 341
+ + S A A +++ SA+A + + S
Sbjct: 3449 ASASTSASASASTSASESASTSASASASTSASASASTSASASASTSASASASTSASASAS 3508
Query: 342 MSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENTSQGNN 401
+S E+ SA+ + + +A+V+ + +A+A+ +++A+ A A + +S E+ S +
Sbjct: 3509 ISASESASTSASASASTSASVSASTSASASASTSASESASTSASASASTSASESASTSAS 3568
Query: 402 TFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAENMDAIL 461
+++ + ASI + S + + A +ASA+T +E+
Sbjct: 3569 A-----SASTSASASASISASESASTSASASASTSASASASTSASASASTSASESASTST 3623
Query: 462 KAA 464
A+
Sbjct: 3624 SAS 3626
Score = 58.2 bits (139), Expect = 2e-06
Identities = 83/423 (19%), Positives = 158/423 (36%), Gaps = 13/423 (3%)
Query: 42 AARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSA 101
A+ ++ ++ S S+ ++S +T A+ S+ S +++S +SA
Sbjct: 731 ASESASTSASASASTSASASASTSASASASTSASASASTSASA---SASISASESASTSA 787
Query: 102 LARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSA 161
A S + A T S S STS + A S + + +T SA
Sbjct: 788 SASASTSASASASTSASASASTSASE--SASTSASASASTSASASASTSASASASTSASA 845
Query: 162 SPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQS 221
S S+ AS S S+S + S+S ST + A+ A S
Sbjct: 846 SASTSASASASTSASASASTSASE---SASTSASASASTSASESASTSASASASTSASAS 902
Query: 222 SGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVVSVSP 281
+ A S S + + ++ ASTS SA+T +S S + +
Sbjct: 903 ASTSASASASTSASASASTSASASASTSASASASTSASASTSASESASTSASASASTSAS 962
Query: 282 LADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHKNSGF 341
+ + S A A +++ SA+A + + S
Sbjct: 963 ASASTSASASASTSASESASTSASASASTSASASASTSASASASTSASASASTSASASAS 1022
Query: 342 MSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENTSQGNN 401
+S E+ SA+ + + +A+V+ + +A+A+ +++A+ A A + +S E+ S +
Sbjct: 1023 ISASESASTSASASASTSASVSASTSASASASTSASESASTSASASASTSASESASTSAS 1082
Query: 402 TFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAENMDAIL 461
+++ + ASI + S + + A +ASA+T +E+
Sbjct: 1083 A-----SASTSASASASISASESASTSASASASTSASASASTSASASASTSASESASTST 1137
Query: 462 KAA 464
A+
Sbjct: 1138 SAS 1140
Score = 57.8 bits (138), Expect = 2e-06
Identities = 85/438 (19%), Positives = 159/438 (35%), Gaps = 2/438 (0%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S +A TS S + +S AS+ A+ + + +S S ++ S
Sbjct: 2538 SASASASTSASASASTSASASASTSASVSASTSASESASTSASASASTSASASASTSASA 2597
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
+S A S + A S S STS + A S + + +T
Sbjct: 2598 SASTSASESASTSASASASTSASASASTSASESASTSASASASTSASASASTSASASAST 2657
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIK-DVTPGPPASSSGLQSTFVGTDSQLDANNVTVP 216
SAS S+ T AS S S+S + AS S S + A+
Sbjct: 2658 SASASASASTSASASASTSASASASTSASASASISASESASTSASASASASTSASASAST 2717
Query: 217 PAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSV 276
A S+ A SE +S + + ++ ASTS S + +S S
Sbjct: 2718 SASASASTSASASASISASESASTSASESASTSTSASASTSASESASTSASASASTSAS- 2776
Query: 277 VSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKH 336
S S A + S+ + + + ASA ++A+ + S
Sbjct: 2777 ASASTSASASASTSASASTSASESASTSASASASTSASASASTSASASASTSASASASTS 2836
Query: 337 KNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENT 396
++ + +++A A A+ +A A A+ +A+ AS +A + ++ A S+ +
Sbjct: 2837 ASASASTSASVSASTSASASASTSASASASTSASESASTSASASTSASESASTSASASAS 2896
Query: 397 SQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAEN 456
+ + + +++ + S A+ S + A+ A +ASA+ + +
Sbjct: 2897 TSASASASTSASASASTSASESASTSASASASTSASASASTSASESASTSASASASTSAS 2956
Query: 457 MDAILKAAELAAEAVSQA 474
A A+ A+ + S++
Sbjct: 2957 ASASTSASASASTSASES 2974
Score = 57.8 bits (138), Expect = 2e-06
Identities = 89/436 (20%), Positives = 165/436 (37%), Gaps = 19/436 (4%)
Query: 42 AARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSA 101
A+ ++ ++ S+ + S+ ++S +T A+ +S S T ++ S +SA
Sbjct: 543 ASASTSASASASISASESASTSASESASTSTSASAS-----TSASESASTSASASASTSA 597
Query: 102 LARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSA 161
A S + A T S S STS + A S + + +T SA
Sbjct: 598 SASASTSASASASTSASASTSASE----SASTSASASASTSASASASTSASASASTSASA 653
Query: 162 SPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQS 221
S S+ AS S S+S + S+S ST + A+ A S
Sbjct: 654 SASTSASVSASTSASASASTS---ASASASTSASESASTSASASASTSASASASTSASAS 710
Query: 222 SGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVVSVSP 281
+ A S +S + + ++ ASTS SA+ +S S S S
Sbjct: 711 ASTSASASASTSASASASTSASESASTSASASASTSASASASTSASASASTSASA-SAST 769
Query: 282 LADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHKNSGF 341
A + E S + + + ASA ++ + + S ++
Sbjct: 770 SASASASISASESASTSASASASTSASASASTSASASASTSASESASTSASASASTSASA 829
Query: 342 MSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENTSQGNN 401
+ A ASA+ + +A+A+ + +A+A+ + +++A+ A A + +S + S+ +
Sbjct: 830 SASTSAS-ASASTSASASASTSASASASTSASASASTSASESASTSASASASTSASESAS 888
Query: 402 TFLPEGTSNLGQATP-----ASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAEN 456
T S A+ AS A+ S + A+ A +ASA+T +E+
Sbjct: 889 TSASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSASASTSASES 948
Query: 457 MDAILKAAELAAEAVS 472
A+ + + S
Sbjct: 949 ASTSASASASTSASAS 964
Score = 57.8 bits (138), Expect = 2e-06
Identities = 91/437 (20%), Positives = 157/437 (35%), Gaps = 28/437 (6%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S +A TS S + +S AS+ A+ + + +S S ++ S
Sbjct: 2906 SASASASTSASESASTSASASASTSASASASTSASESASTSASASASTSASASASTSASA 2965
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
+S A S + A +S S STS + A S + + +T
Sbjct: 2966 SASTSASESASTSASASASTSASESASTSASASASTSASASASTSASGSASTSTSASAST 3025
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIKD-VTPGPPASSSGLQSTFVGTDSQLDANNVTVP 216
SAS S+ AS+ S S+S + + AS+S S T + A+
Sbjct: 3026 SASASASTSASASASISASESASTSASESASTSTSASASTSASESASTSASASASTSASA 3085
Query: 217 PAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSV 276
A S+ A S S + + ++ AS S SA+ +S S
Sbjct: 3086 SASTSASASASTSASASTSASESASTSASASASTSASASASTSASASASTSASASASTSA 3145
Query: 277 -VSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDK 335
VS S A + S A A E+++ SA+A
Sbjct: 3146 SVSASTSAS-------------ASASTSASASASTSASESASTSASA------------S 3180
Query: 336 HKNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYEN 395
S S + ASA+ + +A+A+ + +A+A+ + + +A+ A A + +S +
Sbjct: 3181 ASTSASASASTSASASASTSASASASTSASASASTSASESASTSASASASTSASASASTS 3240
Query: 396 TSQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAE 455
S +T S A+ ASI + S + + A +ASA+T +E
Sbjct: 3241 ASASASTSASASASTSASAS-ASISASESASTSASASASTSASASASTSASASASTSASE 3299
Query: 456 NMDAILKAAELAAEAVS 472
+ A+ + + S
Sbjct: 3300 SASTSASASASTSASAS 3316
Score = 57.8 bits (138), Expect = 2e-06
Identities = 89/436 (20%), Positives = 165/436 (37%), Gaps = 19/436 (4%)
Query: 42 AARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSA 101
A+ ++ ++ S+ + S+ ++S +T A+ +S S T ++ S +SA
Sbjct: 3029 ASASTSASASASISASESASTSASESASTSTSASAS-----TSASESASTSASASASTSA 3083
Query: 102 LARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSA 161
A S + A T S S STS + A S + + +T SA
Sbjct: 3084 SASASTSASASASTSASASTSASE----SASTSASASASTSASASASTSASASASTSASA 3139
Query: 162 SPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQS 221
S S+ AS S S+S + S+S ST + A+ A S
Sbjct: 3140 SASTSASVSASTSASASASTS---ASASASTSASESASTSASASASTSASASASTSASAS 3196
Query: 222 SGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVVSVSP 281
+ A S +S + + ++ ASTS SA+ +S S S S
Sbjct: 3197 ASTSASASASTSASASASTSASESASTSASASASTSASASASTSASASASTSASA-SAST 3255
Query: 282 LADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHKNSGF 341
A + E S + + + ASA ++ + + S ++
Sbjct: 3256 SASASASISASESASTSASASASTSASASASTSASASASTSASESASTSASASASTSASA 3315
Query: 342 MSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENTSQGNN 401
+ A ASA+ + +A+A+ + +A+A+ + +++A+ A A + +S + S+ +
Sbjct: 3316 SASTSAS-ASASTSASASASTSASASASTSASASASTSASESASTSASASASTSASESAS 3374
Query: 402 TFLPEGTSNLGQATP-----ASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAEN 456
T S A+ AS A+ S + A+ A +ASA+T +E+
Sbjct: 3375 TSASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSASASTSASES 3434
Query: 457 MDAILKAAELAAEAVS 472
A+ + + S
Sbjct: 3435 ASTSASASASTSASAS 3450
Score = 57.4 bits (137), Expect = 3e-06
Identities = 86/438 (19%), Positives = 156/438 (34%), Gaps = 8/438 (1%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S +A TS S + +S AS+ A+ + + +S S+ ++ S
Sbjct: 4298 SASASASTSASESASTSASASASTSASASASTSASASASTSASASASTSASVSASTSASE 4357
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
+S A S + A +S S STS A S + + +T
Sbjct: 4358 SASTSASASASTSASASASTSASESASTSASASASTSASESASTSASASASTSASASAST 4417
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPP 217
SAS S+ AS S S+S + S+S ST + A+
Sbjct: 4418 SASASASTSASASASTSASASASTS---ASASASTSASASASTSASASASTSASASASTS 4474
Query: 218 AQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVV 277
A S+ A S S + + + ASTS SA+ +S SV
Sbjct: 4475 ASASASTSASASASTSASASASTSASASASISASESASTSASASASTSASASASTSASVS 4534
Query: 278 SVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHK 337
+ + + + I + ES A AS ++A+ + S
Sbjct: 4535 ASTSASASASTSASASASISASESASTSASA-----SASTSASASASTSASASASTSASA 4589
Query: 338 NSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENTS 397
++ + A +++A A +A+A A +A+A+ S +A + + A S+ ++
Sbjct: 4590 SASISASESASTSASASASTSASASASTSASASASTSASASASTSASASASTSASASAST 4649
Query: 398 QGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAENM 457
+ + +++ + AS A+ S + A+ A +ASA+ + +
Sbjct: 4650 SASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSASA 4709
Query: 458 DAILKAAELAAEAVSQAG 475
A + A + SQ G
Sbjct: 4710 SASTSVSNSANHSNSQVG 4727
Score = 57.4 bits (137), Expect = 3e-06
Identities = 81/434 (18%), Positives = 155/434 (35%), Gaps = 1/434 (0%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S +A TS S + +S AS+ A+ + + +S S ++ S
Sbjct: 2204 SASASASTSASASASTSASESASTSASASASTSASASASTSASASASTSASASASTSASA 2263
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
+S A S + A S S STS A S + + +T
Sbjct: 2264 SASISASESASTSASASASTSASVSASTSASASASTSASESASTSASASASTSASESAST 2323
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIK-DVTPGPPASSSGLQSTFVGTDSQLDANNVTVP 216
SAS S+ AS+ S S+S + AS+S S T + A+ T
Sbjct: 2324 SASASASTSASASASISASESASTSASASASTSASASASTSASASASTSASESASTSTSA 2383
Query: 217 PAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSV 276
A S+ A S S + + ++ AS S S + S S
Sbjct: 2384 SASTSASESASTSASASASTSASASASTSASASASTSASASASTSASASTSASESASTSA 2443
Query: 277 VSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKH 336
+ + + + + S A A +++ SA+ + +
Sbjct: 2444 SASASTSASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSASASA 2503
Query: 337 KNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENT 396
S S + ASA+ + +A+A+ + + +A+ + +++A+ A A + +S +
Sbjct: 2504 STSASASASTSASASASTSASASASTSASESASTSASASASTSASASASTSASASASTSA 2563
Query: 397 SQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAEN 456
S +T E S A+ ++ + ++ S +A E+ A+++ + A
Sbjct: 2564 SVSASTSASESASTSASASASTSASASASTSASASASTSASESASTSASASASTSASASA 2623
Query: 457 MDAILKAAELAAEA 470
+ ++A +A A
Sbjct: 2624 STSASESASTSASA 2637
Score = 57.4 bits (137), Expect = 3e-06
Identities = 89/436 (20%), Positives = 161/436 (36%), Gaps = 17/436 (3%)
Query: 42 AARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSA 101
A+ ++ ++ +S S+ + S +T A+ S+ S T ++ S +SA
Sbjct: 3345 ASASASTSASESASTSASASASTSASESASTSASASASTSASA---SASTSASASASTSA 3401
Query: 102 LARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSA 161
A S + A T S S STS A S + + +T SA
Sbjct: 3402 SASASTSASASASTSA----SASASTSASASTSASESASTSASASASTSASASASTSASA 3457
Query: 162 SPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQS 221
S S+ + AS S S+S + S+S ST + A+ A +S
Sbjct: 3458 SASTSASESASTSASASASTS---ASASASTSASASASTSASASASTSASASASISASES 3514
Query: 222 SGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVVSVSP 281
+ A S S + + + ASTS SA+ +S S S S
Sbjct: 3515 ASTSASASASTSASVSASTSASASASTSASESASTSASASASTSASESASTSASA-SAST 3573
Query: 282 LADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHKNSGF 341
A + E S + + + ASA ++ + + S ++
Sbjct: 3574 SASASASISASESASTSASASASTSASASASTSASASASTSASESASTSTSASASTSASE 3633
Query: 342 MSDIEAKL-----ASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENT 396
+ A ASA+ + +A+A+ + +A+A+A+ +++A+ A A + +S +
Sbjct: 3634 SASTSASASASTSASASASTSASASASTSASASASTSASASTSASESASTSASASASTSA 3693
Query: 397 SQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAEN 456
S +T S A+ AS A+ S + + A +ASA+T +E+
Sbjct: 3694 SASASTSASASASTSASAS-ASTSASASASTSASASASTSASASASTSASASASTSASES 3752
Query: 457 MDAILKAAELAAEAVS 472
A+ + + S
Sbjct: 3753 ASTSASASASTSASAS 3768
Score = 57.4 bits (137), Expect = 3e-06
Identities = 86/437 (19%), Positives = 163/437 (36%), Gaps = 12/437 (2%)
Query: 42 AARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSA 101
+A TS S + +S AS+ A+ + + +S S ++ S +S
Sbjct: 2192 SASTSASASASTSASASASTSASASASTSASESASTSASASASTSASASASTSASASAST 2251
Query: 102 LARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSA 161
A S + A + +S S STS A S + + +T SA
Sbjct: 2252 SASASASTSASASASISASESASTSASASASTSASVSASTSASASASTSASESASTSASA 2311
Query: 162 SPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQS 221
S S+ + AS S S+S + S+S ST + A+ A S
Sbjct: 2312 SASTSASESASTSASASASTS---ASASASISASESASTSASASASTSASASASTSASAS 2368
Query: 222 SGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVVSVSP 281
+ A + S +S + + ++ ASTS SA+ +S S + +
Sbjct: 2369 ASTSASESASTSTSASASTSASESASTSASASASTSASASASTSASASASTSASASASTS 2428
Query: 282 LADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHKNSGF 341
+ ++ + S A A ASA ++A+ + S ++
Sbjct: 2429 ASASTSASESASTSASASASTSASASASTSAS-ASASTSASASASTSASASASTSASASA 2487
Query: 342 MSDIEAKLASAAVAIAAAAAVAKAA-AAAANVASNAAFQAKLMADEALISSGYENTSQGN 400
+ A +++A A A+ +A A A+ +A+A+ +++A+ A A E+ +S + S
Sbjct: 2488 STSASASASTSASASASTSASASASTSASASASTSASASASTSASESASTSA--SASAST 2545
Query: 401 NTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEA-----ASAATKRAE 455
+ TS A+ ++ + + + S +A + A ASA+T +E
Sbjct: 2546 SASASASTSASASASTSASVSASTSASESASTSASASASTSASASASTSASASASTSASE 2605
Query: 456 NMDAILKAAELAAEAVS 472
+ A+ + + S
Sbjct: 2606 SASTSASASASTSASAS 2622
Score = 57.4 bits (137), Expect = 3e-06
Identities = 89/436 (20%), Positives = 161/436 (36%), Gaps = 17/436 (3%)
Query: 42 AARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSA 101
A+ ++ ++ +S S+ + S +T A+ S+ S T ++ S +SA
Sbjct: 859 ASASASTSASESASTSASASASTSASESASTSASASASTSASA---SASTSASASASTSA 915
Query: 102 LARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSA 161
A S + A T S S STS A S + + +T SA
Sbjct: 916 SASASTSASASASTSA----SASASTSASASTSASESASTSASASASTSASASASTSASA 971
Query: 162 SPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQS 221
S S+ + AS S S+S + S+S ST + A+ A +S
Sbjct: 972 SASTSASESASTSASASASTS---ASASASTSASASASTSASASASTSASASASISASES 1028
Query: 222 SGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVVSVSP 281
+ A S S + + + ASTS SA+ +S S S S
Sbjct: 1029 ASTSASASASTSASVSASTSASASASTSASESASTSASASASTSASESASTSASA-SAST 1087
Query: 282 LADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHKNSGF 341
A + E S + + + ASA ++ + + S ++
Sbjct: 1088 SASASASISASESASTSASASASTSASASASTSASASASTSASESASTSTSASASTSASE 1147
Query: 342 MSDIEAKL-----ASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENT 396
+ A ASA+ + +A+A+ + +A+A+A+ +++A+ A A + +S +
Sbjct: 1148 SASTSASASASTSASASASTSASASASTSASASASTSASASTSASESASTSASASASTSA 1207
Query: 397 SQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAEN 456
S +T S A+ AS A+ S + + A +ASA+T +E+
Sbjct: 1208 SASASTSASASASTSASAS-ASTSASASASTSASASASTSASASASTSASASASTSASES 1266
Query: 457 MDAILKAAELAAEAVS 472
A+ + + S
Sbjct: 1267 ASTSASASASTSASAS 1282
Score = 57.0 bits (136), Expect = 3e-06
Identities = 89/449 (19%), Positives = 163/449 (35%), Gaps = 12/449 (2%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S +A TS S + +S AS+ A+ + + +S S ++ S
Sbjct: 3304 SASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSASESASTSASA 3363
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
+S A S + A S S STS + A S + + +T
Sbjct: 3364 SASTSASESASTSASASASTSASASASTSASASASTSASASASTSASASASTSASASAST 3423
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIK-----DVTPGPPASSSGLQSTFVGTDSQLDANN 212
SAS S+ S S S+S + AS S S + A+
Sbjct: 3424 SASASTSASESASTSASASASTSASASASTSASASASTSASESASTSASASASTSASASA 3483
Query: 213 VTVPPAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSV 272
T A S+ A S S + + ++ S SVSA+T +S
Sbjct: 3484 STSASASASTSASASASTSASASASISASESASTSASASASTSASVSASTSASASASTSA 3543
Query: 273 EKSVVSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQ 332
+S + + + ++ + S A + A E+++ SA+A + +
Sbjct: 3544 SESASTSASASASTSASESASTSASASASTSASASASISASESASTSASASASTSASASA 3603
Query: 333 LDKHKNSGFMSDIE--AKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALIS 390
S S E + SA+ + +A+ + + +A+A+A+ +++A+ A + +
Sbjct: 3604 STSASASASTSASESASTSTSASASTSASESASTSASASASTSASASASTSASASASTSA 3663
Query: 391 SGYENTSQGNNTFLPEGTSNLGQAT-----PASILKGANGPNSPGSFIVAAKEAIRRRVE 445
S +TS +T E S A+ AS A+ S + A+ A
Sbjct: 3664 SASASTSASASTSASESASTSASASASTSASASASTSASASASTSASASASTSASASAST 3723
Query: 446 AASAATKRAENMDAILKAAELAAEAVSQA 474
+ASA+ + + A A+ A+ + S++
Sbjct: 3724 SASASASTSASASASTSASASASTSASES 3752
Score = 57.0 bits (136), Expect = 3e-06
Identities = 85/435 (19%), Positives = 157/435 (35%), Gaps = 24/435 (5%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S +A TS S + +S AS A+ + + +S S ++ S+
Sbjct: 3916 SASASASTSASASASTSASASASTSASASASISASESASTSASASASTSASASASTSASV 3975
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
+S A S + A + +S S STS + A S + + +
Sbjct: 3976 SASTSASASASTSASASASISASESASTSASASASTSASASASTSAS--ASASTSASASA 4033
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPP 217
+SAS S+ T AS S S+S + S+S ST + A+
Sbjct: 4034 SISASESASTSASASASTSASASASTS-ASASASTSASASASTSASASASTSASASASTS 4092
Query: 218 AQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVV 277
A S+ A S +S + + ++ ASTS SA+ + +S S
Sbjct: 4093 ASASASTSASASASTSASASASTSASESASTSASASASTSASASASISASESASTSASAS 4152
Query: 278 SVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHK 337
+ + + + + ES A + + S +A
Sbjct: 4153 ASTSASASASTSASASASTSASESASTSTSASASTSASESASTSA--------------- 4197
Query: 338 NSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENTS 397
S + ASA+ + +A+A+ + +A+A+A+ +++A+ A A + +S + S
Sbjct: 4198 -----SASASTSASASASTSASASASTSASASASTSASASTSASESASTSASASASTSAS 4252
Query: 398 QGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAENM 457
+T S A+ AS A+ S + + A +ASA+T +E+
Sbjct: 4253 ASASTSASASASTSASAS-ASTSASASASTSASASASTSASASASTSASASASTSASESA 4311
Query: 458 DAILKAAELAAEAVS 472
A+ + + S
Sbjct: 4312 STSASASASTSASAS 4326
Score = 57.0 bits (136), Expect = 3e-06
Identities = 89/449 (19%), Positives = 163/449 (35%), Gaps = 12/449 (2%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S +A TS S + +S AS+ A+ + + +S S ++ S
Sbjct: 818 SASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSASESASTSASA 877
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
+S A S + A S S STS + A S + + +T
Sbjct: 878 SASTSASESASTSASASASTSASASASTSASASASTSASASASTSASASASTSASASAST 937
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIK-----DVTPGPPASSSGLQSTFVGTDSQLDANN 212
SAS S+ S S S+S + AS S S + A+
Sbjct: 938 SASASTSASESASTSASASASTSASASASTSASASASTSASESASTSASASASTSASASA 997
Query: 213 VTVPPAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSV 272
T A S+ A S S + + ++ S SVSA+T +S
Sbjct: 998 STSASASASTSASASASTSASASASISASESASTSASASASTSASVSASTSASASASTSA 1057
Query: 273 EKSVVSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQ 332
+S + + + ++ + S A + A E+++ SA+A + +
Sbjct: 1058 SESASTSASASASTSASESASTSASASASTSASASASISASESASTSASASASTSASASA 1117
Query: 333 LDKHKNSGFMSDIE--AKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALIS 390
S S E + SA+ + +A+ + + +A+A+A+ +++A+ A + +
Sbjct: 1118 STSASASASTSASESASTSTSASASTSASESASTSASASASTSASASASTSASASASTSA 1177
Query: 391 SGYENTSQGNNTFLPEGTSNLGQAT-----PASILKGANGPNSPGSFIVAAKEAIRRRVE 445
S +TS +T E S A+ AS A+ S + A+ A
Sbjct: 1178 SASASTSASASTSASESASTSASASASTSASASASTSASASASTSASASASTSASASAST 1237
Query: 446 AASAATKRAENMDAILKAAELAAEAVSQA 474
+ASA+ + + A A+ A+ + S++
Sbjct: 1238 SASASASTSASASASTSASASASTSASES 1266
Score = 56.2 bits (134), Expect = 6e-06
Identities = 93/447 (20%), Positives = 164/447 (35%), Gaps = 20/447 (4%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S +A S ST + +S AS+ A+ + + +S S T ++ S
Sbjct: 2622 SASTSASESASTSASASASTSASASASTSASASASTSASASASASTSASASASTSASASA 2681
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
+SA A S+ A T S S STS + A S + + +T
Sbjct: 2682 STSASASASISASESASTSASASASASTSASASASTSASASASTSASASASISASESAST 2741
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIK---DVTPGPPASSSGLQSTFVGTDSQLDANNVT 214
S S S+ T AS S S+S + AS+S S + A+
Sbjct: 2742 SASESASTSTSASASTSASESASTSASASASTSASASASTSASASASTSASASTSASESA 2801
Query: 215 VPPAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEK 274
A S+ A S S + + ++ ASTS S + +S
Sbjct: 2802 STSASASASTSASASASTSASASASTSASASASTSASASASTSASVSASTSASASASTSA 2861
Query: 275 SVVSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLD 334
S S S A + + S+ + + + ASA ++A+ + S
Sbjct: 2862 SA-SASTSASESASTSASASTSASESASTSASASASTSASASASTSASASASTSASESAS 2920
Query: 335 KHKNSGFMSDIEAKLASAAV------AIAAAAAVAKAAA---AAANVASNAAFQAKLMAD 385
++ + A +++A A A+A+ A A+A A+A+ +++A+ A A
Sbjct: 2921 TSASASASTSASASASTSASESASTSASASASTSASASASTSASASASTSASESASTSAS 2980
Query: 386 EALISSGYENTSQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVE 445
+ +S E+ S +++ + AS A+G S + A+ A
Sbjct: 2981 ASASTSASESAS-------TSASASASTSASASASTSASGSASTSTSASASTSASASAST 3033
Query: 446 AASAATKRAENMDAILKAAELAAEAVS 472
+ASA+ + + A A+E A+ + S
Sbjct: 3034 SASASASISASESASTSASESASTSTS 3060
Score = 56.2 bits (134), Expect = 6e-06
Identities = 83/440 (18%), Positives = 159/440 (35%), Gaps = 7/440 (1%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S +A TS S + +S AS+ A+ + + +S S ++ S
Sbjct: 2062 SASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSASESASTSASA 2121
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
+S A S + A S S STS + A S + + +T
Sbjct: 2122 SASTSASESASTSASASASTSASASASTSASASASTSASASASTSASASASTSASASAST 2181
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIK-----DVTPGPPASSSGLQSTFVGTDSQLDANN 212
SAS S+ S S S+S + AS S S + A+
Sbjct: 2182 SASASTSASESASTSASASASTSASASASTSASASASTSASESASTSASASASTSASASA 2241
Query: 213 VTVPPAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSV 272
T A S+ A S S + + ++ S SVSA+T +S
Sbjct: 2242 STSASASASTSASASASTSASASASISASESASTSASASASTSASVSASTSASASASTSA 2301
Query: 273 EKSVVSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQ 332
+S + + + ++ + S A + A E+++ SA+A + +
Sbjct: 2302 SESASTSASASASTSASESASTSASASASTSASASASISASESASTSASASASTSASASA 2361
Query: 333 LDKHKNSGFMSDIE--AKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALIS 390
S S E + SA+ + +A+ + + +A+A+A+ +++A+ A + +
Sbjct: 2362 STSASASASTSASESASTSTSASASTSASESASTSASASASTSASASASTSASASASTSA 2421
Query: 391 SGYENTSQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAA 450
S +TS +T E S A+ ++ + ++ S +A + A+++
Sbjct: 2422 SASASTSASASTSASESASTSASASASTSASASASTSASASASTSASASASTSASASAST 2481
Query: 451 TKRAENMDAILKAAELAAEA 470
+ A + +A +A A
Sbjct: 2482 SASASASTSASASASTSASA 2501
Score = 55.1 bits (131), Expect = 1e-05
Identities = 92/439 (20%), Positives = 159/439 (35%), Gaps = 16/439 (3%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S +A TS S + +S AS+ A+ + + +S S T ++ S
Sbjct: 1694 SASASASTSASESASTSASASASTSASASASTSASESAS------TSASASASTSASASA 1747
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
+SA A S + A S S STS + A + S + + +T
Sbjct: 1748 STSASASASTSASASASASTSASASASTSASASASTSASASASISASESASTSASESAST 1807
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPP 217
SAS S+ + AS S S+S + S+S ST + + T
Sbjct: 1808 STSASASTSASESASTSASASASTS---ASASASTSASASASTSASASTSASESASTSAS 1864
Query: 218 AQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSV- 276
A S+ A S S + + ++ S SVSA+T +S S
Sbjct: 1865 ASASTSASASASTSASASASTSASASASTSASASASTSASVSASTSASASASTSASASAS 1924
Query: 277 --VSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLD 334
S S + T S + + + SA ++A+ + S
Sbjct: 1925 TSASESASTSASASASTSASASASTSASASASTSASASASTSASASASTSASESASTSAS 1984
Query: 335 KHKNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVAS-NAAFQAKLMADEALISSGY 393
++ + +++A A+A+A A+A+A+ AS +A+ A A + +S
Sbjct: 1985 ASASTSASASASTSASASASTSASASASTSASASASISASESASTSASASASTSASASAS 2044
Query: 394 ENTSQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKR 453
+ S +T E S + AS A+ S + A+ A +ASA+
Sbjct: 2045 TSASASASTSASESAST---SASASASTSASASASTSASASASTSASASASTSASASAST 2101
Query: 454 AENMDAILKAAELAAEAVS 472
+ + A A+E A+ + S
Sbjct: 2102 SASASASTSASESASTSAS 2120
Score = 54.7 bits (130), Expect = 2e-05
Identities = 89/436 (20%), Positives = 161/436 (36%), Gaps = 13/436 (2%)
Query: 42 AARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSA 101
A+ ++ ++ S S+ ++S +T A+ I S S T ++ S +SA
Sbjct: 3997 ASESASTSASASASTSASASASTSASASASTSASASASISASE---SASTSASASASTSA 4053
Query: 102 LARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSA 161
A S + A T S S STS + A S + + +T SA
Sbjct: 4054 SASASTSASASASTSASASASTSAS--ASASTSASASASTSASASASTSASASASTSASA 4111
Query: 162 SPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQS 221
S S+ + AS S S+S + S+S ST + A+ A S
Sbjct: 4112 SASTSASESASTSASASASTS---ASASASISASESASTSASASASTSASASASTSASAS 4168
Query: 222 SGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSV---VS 278
+ A + S +S + + ++ ASTS SA+ +S S S
Sbjct: 4169 ASTSASESASTSTSASASTSASESASTSASASASTSASASASTSASASASTSASASASTS 4228
Query: 279 VSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHKN 338
S ++ T S + + + SA ++A+ + S +
Sbjct: 4229 ASASTSASESASTSASASASTSASASASTSASASASTSASASASTSASASASTSASASAS 4288
Query: 339 SGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVAS-NAAFQAKLMADEALISSGYENTS 397
+ + +++A A+ +A A+A+A+ AS +A+ A A + +S + S
Sbjct: 4289 TSASASASTSASASASTSASESASTSASASASTSASASASTSASASASTSASASASTSAS 4348
Query: 398 QGNNTFLPEGTSNLGQATPA-SILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAEN 456
+T E S A+ + S A+ S + A+ A E+AS + + +
Sbjct: 4349 VSASTSASESASTSASASASTSASASASTSASESASTSASASASTSASESASTSASASAS 4408
Query: 457 MDAILKAAELAAEAVS 472
A A+ A+ + S
Sbjct: 4409 TSASASASTSASASAS 4424
Score = 53.5 bits (127), Expect = 4e-05
Identities = 91/449 (20%), Positives = 165/449 (36%), Gaps = 12/449 (2%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S +A TS S + +S AS A+ + + +S S ++ S
Sbjct: 1064 SASASASTSASESASTSASASASTSASASASISASESASTSASASASTSASASASTSASA 1123
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
+S A S + A +S S STS + A S + + +T
Sbjct: 1124 SASTSASESASTSTSASASTSASESASTSASASASTSASASASTSASASASTSASASAST 1183
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASS---SGLQSTFVGTDSQLDANNVT 214
SAS S+ S S S+S T ++S S ST + A+
Sbjct: 1184 SASASTSASESASTSASASASTSASASASTSASASASTSASASASTSASASASTSASASA 1243
Query: 215 VPPAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEK 274
A S+ A SE S + + ++ ASTS SA+ +S
Sbjct: 1244 STSASASASTSASASASTSASESASTSASASASTSASASASTSASASASTSASASASTSA 1303
Query: 275 SVVSVSPLADQPKNDQTVEKRILSDESLM-KVKEARVHAEEASALSAAAVNHSLELWNQL 333
SV + + ++ + + S E+ + ASA ++A+ + S
Sbjct: 1304 SVSASTSASESASTSASASASTSASASASTSASESASTSASASASTSASESASTSASASA 1363
Query: 334 DKHKNSGFMSDIEAKLASAAVAIAA----AAAVAKAAAAAANVAS-NAAFQAKLMADEAL 388
++ + A +++A A A+ A+A A+A+A+ AS +A+ A A +
Sbjct: 1364 STSASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSA 1423
Query: 389 ISSGYENTSQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAAS 448
+S + S +T +++ + AS A+ S + A+ A +AS
Sbjct: 1424 SASASTSASASAST---SASASASTSASASASTSASASASISASESASTSASASASTSAS 1480
Query: 449 AATKRAENMDAILKAAELAAEAVSQAGKI 477
A+ + ++ A A+ A+ + S + I
Sbjct: 1481 ASASTSASVSASTSASASASTSASASASI 1509
Score = 53.5 bits (127), Expect = 4e-05
Identities = 91/449 (20%), Positives = 165/449 (36%), Gaps = 12/449 (2%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S +A TS S + +S AS A+ + + +S S ++ S
Sbjct: 3550 SASASASTSASESASTSASASASTSASASASISASESASTSASASASTSASASASTSASA 3609
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
+S A S + A +S S STS + A S + + +T
Sbjct: 3610 SASTSASESASTSTSASASTSASESASTSASASASTSASASASTSASASASTSASASAST 3669
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASS---SGLQSTFVGTDSQLDANNVT 214
SAS S+ S S S+S T ++S S ST + A+
Sbjct: 3670 SASASTSASESASTSASASASTSASASASTSASASASTSASASASTSASASASTSASASA 3729
Query: 215 VPPAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEK 274
A S+ A SE S + + ++ ASTS SA+ +S
Sbjct: 3730 STSASASASTSASASASTSASESASTSASASASTSASASASTSASASASTSASASASTSA 3789
Query: 275 SVVSVSPLADQPKNDQTVEKRILSDESLM-KVKEARVHAEEASALSAAAVNHSLELWNQL 333
SV + + ++ + + S E+ + ASA ++A+ + S
Sbjct: 3790 SVSASTSASESASTSASASASTSASASASTSASESASTSASASASTSASESASTSASASA 3849
Query: 334 DKHKNSGFMSDIEAKLASAAVAIAA----AAAVAKAAAAAANVAS-NAAFQAKLMADEAL 388
++ + A +++A A A+ A+A A+A+A+ AS +A+ A A +
Sbjct: 3850 STSASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSA 3909
Query: 389 ISSGYENTSQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAAS 448
+S + S +T +++ + AS A+ S + A+ A +AS
Sbjct: 3910 SASASTSASASAST---SASASASTSASASASTSASASASISASESASTSASASASTSAS 3966
Query: 449 AATKRAENMDAILKAAELAAEAVSQAGKI 477
A+ + ++ A A+ A+ + S + I
Sbjct: 3967 ASASTSASVSASTSASASASTSASASASI 3995
Score = 53.5 bits (127), Expect = 4e-05
Identities = 91/449 (20%), Positives = 165/449 (36%), Gaps = 12/449 (2%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S +A TS S + +S AS A+ + + +S S ++ S
Sbjct: 4108 SASASASTSASESASTSASASASTSASASASISASESASTSASASASTSASASASTSASA 4167
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
+S A S + A +S S STS + A S + + +T
Sbjct: 4168 SASTSASESASTSTSASASTSASESASTSASASASTSASASASTSASASASTSASASAST 4227
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASS---SGLQSTFVGTDSQLDANNVT 214
SAS S+ S S S+S T ++S S ST + A+
Sbjct: 4228 SASASTSASESASTSASASASTSASASASTSASASASTSASASASTSASASASTSASASA 4287
Query: 215 VPPAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEK 274
A S+ A SE S + + ++ ASTS SA+ +S
Sbjct: 4288 STSASASASTSASASASTSASESASTSASASASTSASASASTSASASASTSASASASTSA 4347
Query: 275 SVVSVSPLADQPKNDQTVEKRILSDESLM-KVKEARVHAEEASALSAAAVNHSLELWNQL 333
SV + + ++ + + S E+ + ASA ++A+ + S
Sbjct: 4348 SVSASTSASESASTSASASASTSASASASTSASESASTSASASASTSASESASTSASASA 4407
Query: 334 DKHKNSGFMSDIEAKLASAAVAIAA----AAAVAKAAAAAANVAS-NAAFQAKLMADEAL 388
++ + A +++A A A+ A+A A+A+A+ AS +A+ A A +
Sbjct: 4408 STSASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSA 4467
Query: 389 ISSGYENTSQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAAS 448
+S + S +T +++ + AS A+ S + A+ A +AS
Sbjct: 4468 SASASTSASASAST---SASASASTSASASASTSASASASISASESASTSASASASTSAS 4524
Query: 449 AATKRAENMDAILKAAELAAEAVSQAGKI 477
A+ + ++ A A+ A+ + S + I
Sbjct: 4525 ASASTSASVSASTSASASASTSASASASI 4553
Score = 52.4 bits (124), Expect = 8e-05
Identities = 89/447 (19%), Positives = 166/447 (36%), Gaps = 29/447 (6%)
Query: 42 AARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSA 101
A+ ++ ++ +S S+ + S +T A+ S+ S T ++ S +SA
Sbjct: 2103 ASASASTSASESASTSASASASTSASESASTSASASASTSASA---SASTSASASASTSA 2159
Query: 102 LARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSA 161
A S + A T S S STS A S + + +T SA
Sbjct: 2160 SASASTSASASASTSA----SASASTSASASTSASESASTSASASASTSASASASTSASA 2215
Query: 162 SPSSDTIKLASVKGSLPPSSSIK-----DVTPGPPASSSGLQSTFVGTDSQLDANNVTVP 216
S S+ + AS S S+S + S+S ST + + A+
Sbjct: 2216 SASTSASESASTSASASASTSASASASTSASASASTSASASASTSASASASISASESAST 2275
Query: 217 PAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSV 276
A S+ A S +S + + ++ ASTS S + +S S
Sbjct: 2276 SASASASTSASVSASTSASASASTSASESASTSASASASTSASESASTSASASASTSASA 2335
Query: 277 ---VSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQL 333
+S S A + S + + +E AS ++A+ + S
Sbjct: 2336 SASISASESASTSASASASTSASASASTSASASASTSASESASTSTSASASTSAS----- 2390
Query: 334 DKHKNSGFMSDIEAKLASAAVAIAAAAAVAK-AAAAAANVASNAAFQAKLMADEALISSG 392
+++ + A +++A A +A+A A +A+A+A+ +++A+ A A + +S
Sbjct: 2391 ---ESASTSASASASTSASASASTSASASASTSASASASTSASASTSASESASTSASASA 2447
Query: 393 YENTSQGNNTFLPEGTSNLGQAT-----PASILKGANGPNSPGSFIVAAKEAIRRRVEAA 447
+ S +T S A+ AS A+ S + A+ A +A
Sbjct: 2448 STSASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSA 2507
Query: 448 SAATKRAENMDAILKAAELAAEAVSQA 474
SA+ + + A A+ A+ + S++
Sbjct: 2508 SASASTSASASASTSASASASTSASES 2534
Score = 52.0 bits (123), Expect = 1e-04
Identities = 92/436 (21%), Positives = 163/436 (37%), Gaps = 24/436 (5%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S +A TS S + +S AS+ A+ + + +S S T ++ S
Sbjct: 650 SASASASTSASVSASTSASASASTSASASASTSASESAS------TSASASASTSASASA 703
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
+SA A S + A T S S STS + A S + + +T
Sbjct: 704 STSASASASTSASASASTSASASASTSASE--SASTSASASASTSASASASTSASASAST 761
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPP 217
SAS S+ AS+ S S+S + S+S ST + A+
Sbjct: 762 SASASASTSASASASISASESASTS---ASASASTSASASASTSASASASTSASESASTS 818
Query: 218 AQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVV 277
A S+ A S S + + ++ ASTS SA+ S+ E +
Sbjct: 819 ASASASTSASASASTSASASASTSASASASTSASASASTSASASAST-----SASESAST 873
Query: 278 SVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHK 337
S S A ++ S + + + AS ++A+ + S
Sbjct: 874 SASASASTSASESASTSASASASTSASASASTSASASASTSASASASTSASASASTSASA 933
Query: 338 NSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVAS-NAAFQAKLMADEALISSGYENT 396
++ + + +A A+A+A A+A+A+ AS +A+ A A + +S +
Sbjct: 934 SASTSASASTSASESASTSASASASTSASASASTSASASASTSASESASTSASASASTSA 993
Query: 397 SQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAEN 456
S +T +++ +T AS A+ S + I A++ A +AS + + +
Sbjct: 994 SASAST-----SASASASTSAS--ASASTSASASASISASESASTSASASASTSASVSAS 1046
Query: 457 MDAILKAAELAAEAVS 472
A A+ A+E+ S
Sbjct: 1047 TSASASASTSASESAS 1062
Score = 52.0 bits (123), Expect = 1e-04
Identities = 92/436 (21%), Positives = 163/436 (37%), Gaps = 24/436 (5%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S +A TS S + +S AS+ A+ + + +S S T ++ S
Sbjct: 3136 SASASASTSASVSASTSASASASTSASASASTSASESAS------TSASASASTSASASA 3189
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
+SA A S + A T S S STS + A S + + +T
Sbjct: 3190 STSASASASTSASASASTSASASASTSASE--SASTSASASASTSASASASTSASASAST 3247
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPP 217
SAS S+ AS+ S S+S + S+S ST + A+
Sbjct: 3248 SASASASTSASASASISASESASTS---ASASASTSASASASTSASASASTSASESASTS 3304
Query: 218 AQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVV 277
A S+ A S S + + ++ ASTS SA+ S+ E +
Sbjct: 3305 ASASASTSASASASTSASASASTSASASASTSASASASTSASASAST-----SASESAST 3359
Query: 278 SVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHK 337
S S A ++ S + + + AS ++A+ + S
Sbjct: 3360 SASASASTSASESASTSASASASTSASASASTSASASASTSASASASTSASASASTSASA 3419
Query: 338 NSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVAS-NAAFQAKLMADEALISSGYENT 396
++ + + +A A+A+A A+A+A+ AS +A+ A A + +S +
Sbjct: 3420 SASTSASASTSASESASTSASASASTSASASASTSASASASTSASESASTSASASASTSA 3479
Query: 397 SQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAEN 456
S +T +++ +T AS A+ S + I A++ A +AS + + +
Sbjct: 3480 SASAST-----SASASASTSAS--ASASTSASASASISASESASTSASASASTSASVSAS 3532
Query: 457 MDAILKAAELAAEAVS 472
A A+ A+E+ S
Sbjct: 3533 TSASASASTSASESAS 3548
Score = 52.0 bits (123), Expect = 1e-04
Identities = 92/436 (21%), Positives = 163/436 (37%), Gaps = 24/436 (5%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S +A TS S + +S AS+ A+ + + +S S T ++ S
Sbjct: 1894 SASASASTSASVSASTSASASASTSASASASTSASESAS------TSASASASTSASASA 1947
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
+SA A S + A T S S STS + A S + + +T
Sbjct: 1948 STSASASASTSASASASTSASASASTSASE--SASTSASASASTSASASASTSASASAST 2005
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPP 217
SAS S+ AS+ S S+S + S+S ST + A+
Sbjct: 2006 SASASASTSASASASISASESASTS---ASASASTSASASASTSASASASTSASESASTS 2062
Query: 218 AQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVV 277
A S+ A S S + + ++ ASTS SA+ S+ E +
Sbjct: 2063 ASASASTSASASASTSASASASTSASASASTSASASASTSASASAST-----SASESAST 2117
Query: 278 SVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHK 337
S S A ++ S + + + AS ++A+ + S
Sbjct: 2118 SASASASTSASESASTSASASASTSASASASTSASASASTSASASASTSASASASTSASA 2177
Query: 338 NSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVAS-NAAFQAKLMADEALISSGYENT 396
++ + + +A A+A+A A+A+A+ AS +A+ A A + +S +
Sbjct: 2178 SASTSASASTSASESASTSASASASTSASASASTSASASASTSASESASTSASASASTSA 2237
Query: 397 SQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAEN 456
S +T +++ +T AS A+ S + I A++ A +AS + + +
Sbjct: 2238 SASAST-----SASASASTSAS--ASASTSASASASISASESASTSASASASTSASVSAS 2290
Query: 457 MDAILKAAELAAEAVS 472
A A+ A+E+ S
Sbjct: 2291 TSASASASTSASESAS 2306
Score = 51.6 bits (122), Expect = 1e-04
Identities = 80/438 (18%), Positives = 150/438 (33%), Gaps = 6/438 (1%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S +A TS S + +S AS A+ + + +S S ++ S
Sbjct: 4004 SASASASTSASASASTSASASASTSASASASISASESASTSASASASTSASASASTSASA 4063
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
+S A S + A S S STS + A S + + +T
Sbjct: 4064 SASTSASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSASESAST 4123
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPP 217
SAS S+ AS+ S S+S + S+S ST + A+
Sbjct: 4124 SASASASTSASASASISASESASTS---ASASASTSASASASTSASASASTSASESASTS 4180
Query: 218 AQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSV- 276
S+ A + S S + + ++ ASTS SA+ +S +S
Sbjct: 4181 TSASASTSASESASTSASASASTSASASASTSASASASTSASASASTSASASTSASESAS 4240
Query: 277 --VSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLD 334
S S + T S + + + SA ++A+ + S
Sbjct: 4241 TSASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSAS 4300
Query: 335 KHKNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYE 394
++ +++A A+A+A A+A+A+ AS +A + ++ S
Sbjct: 4301 ASASTSASESASTSASASASTSASASASTSASASASTSASASASTSASVSASTSASESAS 4360
Query: 395 NTSQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRA 454
++ + + +++ + AS A+ S + A +ASA+T +
Sbjct: 4361 TSASASASTSASASASTSASESASTSASASASTSASESASTSASASASTSASASASTSAS 4420
Query: 455 ENMDAILKAAELAAEAVS 472
+ A+ + + S
Sbjct: 4421 ASASTSASASASTSASAS 4438
Score = 51.2 bits (121), Expect = 2e-04
Identities = 86/441 (19%), Positives = 162/441 (36%), Gaps = 19/441 (4%)
Query: 42 AARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSA 101
A+ ++ ++ S+ + S+ ++S + A+ +S S T +++S +SA
Sbjct: 3251 ASASTSASASASISASESASTSASASASTSASASAS-----TSASASASTSASESASTSA 3305
Query: 102 LARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSA 161
A S + A T S S STS + A S + + +T SA
Sbjct: 3306 SASASTSASASASTSASASASTSAS--ASASTSASASASTSASASASTSASESASTSASA 3363
Query: 162 SPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQS 221
S S+ + AS S S+S + AS+S S + A+ T A S
Sbjct: 3364 SASTSASESASTSASASASTSAS-ASASTSASASASTSASASASTSASASASTSASASAS 3422
Query: 222 SGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVS-AATPVGNVPMSSVEKSVVSVS 280
+ A + S S + + ++ AS S S +A+ + S+ + S S
Sbjct: 3423 TSASASTSASESASTSASASASTSASASASTSASASASTSASESASTSASASASTSASAS 3482
Query: 281 PLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHKNSG 340
+ T S + + + SA ++A+ + S+ ++
Sbjct: 3483 ASTSASASASTSASASASTSASASASISASESASTSASASASTSASVSASTSASASASTS 3542
Query: 341 FMSDIEAKLASAAVAIAAAAAVAKAAAAAANVAS---------NAAFQAKLMADEALISS 391
+++A A+ +A A+A+A+ AS +A+ A A + +S
Sbjct: 3543 ASESASTSASASASTSASESASTSASASASTSASASASISASESASTSASASASTSASAS 3602
Query: 392 GYENTSQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAAT 451
+ S +T E S A+ AS + S + + A +ASA+T
Sbjct: 3603 ASTSASASASTSASESASTSTSAS-ASTSASESASTSASASASTSASASASTSASASAST 3661
Query: 452 KRAENMDAILKAAELAAEAVS 472
+ + A+ A+E+ S
Sbjct: 3662 SASASASTSASASTSASESAS 3682
Score = 51.2 bits (121), Expect = 2e-04
Identities = 86/441 (19%), Positives = 162/441 (36%), Gaps = 19/441 (4%)
Query: 42 AARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSA 101
A+ ++ ++ S+ + S+ ++S + A+ +S S T +++S +SA
Sbjct: 2009 ASASTSASASASISASESASTSASASASTSASASAS-----TSASASASTSASESASTSA 2063
Query: 102 LARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSA 161
A S + A T S S STS + A S + + +T SA
Sbjct: 2064 SASASTSASASASTSASASASTSAS--ASASTSASASASTSASASASTSASESASTSASA 2121
Query: 162 SPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQS 221
S S+ + AS S S+S + AS+S S + A+ T A S
Sbjct: 2122 SASTSASESASTSASASASTSAS-ASASTSASASASTSASASASTSASASASTSASASAS 2180
Query: 222 SGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVS-AATPVGNVPMSSVEKSVVSVS 280
+ A + S S + + ++ AS S S +A+ + S+ + S S
Sbjct: 2181 TSASASTSASESASTSASASASTSASASASTSASASASTSASESASTSASASASTSASAS 2240
Query: 281 PLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHKNSG 340
+ T S + + + SA ++A+ + S+ ++
Sbjct: 2241 ASTSASASASTSASASASTSASASASISASESASTSASASASTSASVSASTSASASASTS 2300
Query: 341 FMSDIEAKLASAAVAIAAAAAVAKAAAAAANVAS---------NAAFQAKLMADEALISS 391
+++A A+ +A A+A+A+ AS +A+ A A + +S
Sbjct: 2301 ASESASTSASASASTSASESASTSASASASTSASASASISASESASTSASASASTSASAS 2360
Query: 392 GYENTSQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAAT 451
+ S +T E S A+ AS + S + + A +ASA+T
Sbjct: 2361 ASTSASASASTSASESASTSTSAS-ASTSASESASTSASASASTSASASASTSASASAST 2419
Query: 452 KRAENMDAILKAAELAAEAVS 472
+ + A+ A+E+ S
Sbjct: 2420 SASASASTSASASTSASESAS 2440
Score = 51.2 bits (121), Expect = 2e-04
Identities = 86/441 (19%), Positives = 162/441 (36%), Gaps = 19/441 (4%)
Query: 42 AARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSA 101
A+ ++ ++ S+ + S+ ++S + A+ +S S T +++S +SA
Sbjct: 765 ASASTSASASASISASESASTSASASASTSASASAS-----TSASASASTSASESASTSA 819
Query: 102 LARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSA 161
A S + A T S S STS + A S + + +T SA
Sbjct: 820 SASASTSASASASTSASASASTSAS--ASASTSASASASTSASASASTSASESASTSASA 877
Query: 162 SPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQS 221
S S+ + AS S S+S + AS+S S + A+ T A S
Sbjct: 878 SASTSASESASTSASASASTSAS-ASASTSASASASTSASASASTSASASASTSASASAS 936
Query: 222 SGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVS-AATPVGNVPMSSVEKSVVSVS 280
+ A + S S + + ++ AS S S +A+ + S+ + S S
Sbjct: 937 TSASASTSASESASTSASASASTSASASASTSASASASTSASESASTSASASASTSASAS 996
Query: 281 PLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHKNSG 340
+ T S + + + SA ++A+ + S+ ++
Sbjct: 997 ASTSASASASTSASASASTSASASASISASESASTSASASASTSASVSASTSASASASTS 1056
Query: 341 FMSDIEAKLASAAVAIAAAAAVAKAAAAAANVAS---------NAAFQAKLMADEALISS 391
+++A A+ +A A+A+A+ AS +A+ A A + +S
Sbjct: 1057 ASESASTSASASASTSASESASTSASASASTSASASASISASESASTSASASASTSASAS 1116
Query: 392 GYENTSQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAAT 451
+ S +T E S A+ AS + S + + A +ASA+T
Sbjct: 1117 ASTSASASASTSASESASTSTSAS-ASTSASESASTSASASASTSASASASTSASASAST 1175
Query: 452 KRAENMDAILKAAELAAEAVS 472
+ + A+ A+E+ S
Sbjct: 1176 SASASASTSASASTSASESAS 1196
Score = 50.4 bits (119), Expect = 3e-04
Identities = 80/447 (17%), Positives = 151/447 (32%), Gaps = 16/447 (3%)
Query: 42 AARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSA 101
+A TS S + +S AS+ A+ + + +S S ++ S +S
Sbjct: 948 SASTSASASASTSASASASTSASASASTSASESASTSASASASTSASASASTSASASAST 1007
Query: 102 LARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSA 161
A S + A + +S S STS A S + + +T SA
Sbjct: 1008 SASASASTSASASASISASESASTSASASASTSASVSASTSASASASTSASESASTSASA 1067
Query: 162 SPSSDTIKLASVKGSLPPSSSIK-----DVTPGPPASSSGLQSTFVGTDSQLDANNVTVP 216
S S+ + AS S S+S + S+S ST + A+
Sbjct: 1068 SASTSASESASTSASASASTSASASASISASESASTSASASASTSASASASTSASASAST 1127
Query: 217 PAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSV 276
A +S+ SE S + + ++ ASTS SA+ +S S
Sbjct: 1128 SASESASTSTSASASTSASESASTSASASASTSASASASTSASASASTSASASASTSASA 1187
Query: 277 -----------VSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNH 325
S S + T S + + + SA ++A+ +
Sbjct: 1188 STSASESASTSASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSA 1247
Query: 326 SLELWNQLDKHKNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMAD 385
S ++ +++A A+A+A A+A+A+ AS +A + ++
Sbjct: 1248 SASASTSASASASTSASESASTSASASASTSASASASTSASASASTSASASASTSASVSA 1307
Query: 386 EALISSGYENTSQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVE 445
S ++ + + +++ + AS A+ S + A
Sbjct: 1308 STSASESASTSASASASTSASASASTSASESASTSASASASTSASESASTSASASASTSA 1367
Query: 446 AASAATKRAENMDAILKAAELAAEAVS 472
+ASA+T + + A+ + + S
Sbjct: 1368 SASASTSASASASTSASASASTSASAS 1394
Score = 50.4 bits (119), Expect = 3e-04
Identities = 80/447 (17%), Positives = 151/447 (32%), Gaps = 16/447 (3%)
Query: 42 AARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSA 101
+A TS S + +S AS+ A+ + + +S S ++ S +S
Sbjct: 3434 SASTSASASASTSASASASTSASASASTSASESASTSASASASTSASASASTSASASAST 3493
Query: 102 LARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSA 161
A S + A + +S S STS A S + + +T SA
Sbjct: 3494 SASASASTSASASASISASESASTSASASASTSASVSASTSASASASTSASESASTSASA 3553
Query: 162 SPSSDTIKLASVKGSLPPSSSIK-----DVTPGPPASSSGLQSTFVGTDSQLDANNVTVP 216
S S+ + AS S S+S + S+S ST + A+
Sbjct: 3554 SASTSASESASTSASASASTSASASASISASESASTSASASASTSASASASTSASASAST 3613
Query: 217 PAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSV 276
A +S+ SE S + + ++ ASTS SA+ +S S
Sbjct: 3614 SASESASTSTSASASTSASESASTSASASASTSASASASTSASASASTSASASASTSASA 3673
Query: 277 -----------VSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNH 325
S S + T S + + + SA ++A+ +
Sbjct: 3674 STSASESASTSASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSA 3733
Query: 326 SLELWNQLDKHKNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMAD 385
S ++ +++A A+A+A A+A+A+ AS +A + ++
Sbjct: 3734 SASASTSASASASTSASESASTSASASASTSASASASTSASASASTSASASASTSASVSA 3793
Query: 386 EALISSGYENTSQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVE 445
S ++ + + +++ + AS A+ S + A
Sbjct: 3794 STSASESASTSASASASTSASASASTSASESASTSASASASTSASESASTSASASASTSA 3853
Query: 446 AASAATKRAENMDAILKAAELAAEAVS 472
+ASA+T + + A+ + + S
Sbjct: 3854 SASASTSASASASTSASASASTSASAS 3880
Score = 50.1 bits (118), Expect = 4e-04
Identities = 78/376 (20%), Positives = 139/376 (36%), Gaps = 13/376 (3%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S +A TS S + +S AS+ A+ + + +S S ++ S
Sbjct: 4370 SASASASTSASESASTSASASASTSASESASTSASASASTSASASASTSASASASTSASA 4429
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
+S A S + A S S STS + A S + + +T
Sbjct: 4430 SASTSASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSASASAST 4489
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPP 217
SAS S+ AS+ S S+S + S+S ST + A+
Sbjct: 4490 SASASASTSASASASISASESASTS---ASASASTSASASASTSASVSASTSASASASTS 4546
Query: 218 AQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSV- 276
A S+ A + S S + + ++ ASTS SA+ + +S S
Sbjct: 4547 ASASASISASESASTSASASASTSASASASTSASASASTSASASASISASESASTSASAS 4606
Query: 277 VSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKH 336
S S A + S + + + ASA ++A+ + S
Sbjct: 4607 ASTSASASASTSASASASTSASASASTSASASASTSASASASTSASASASTSASASASTS 4666
Query: 337 KNSGFMSDIEAKL---ASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGY 393
++ + A ASA+ + +A+A+ + +A+A+A+ +++A+ A ++ +S
Sbjct: 4667 ASASASTSASASASTSASASASTSASASASTSASASASTSASAS------ASTSVSNSAN 4720
Query: 394 ENTSQGNNTFLPEGTS 409
+ SQ NT G S
Sbjct: 4721 HSNSQVGNTSGSTGKS 4736
Score = 49.3 bits (116), Expect = 7e-04
Identities = 83/447 (18%), Positives = 157/447 (34%), Gaps = 15/447 (3%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S +A S S + +S AS+ A+ + + +S S ++ S+
Sbjct: 1732 STSASASASTSASASASTSASASASTSASASASASTSASASASTSASASASTSASASASI 1791
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
+S A S + + T S S S S + A S + + +
Sbjct: 1792 SASESASTSASESASTSTSASASTSASESASTSASASASTSASASASTSASASASTSASA 1851
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPP 217
SAS S+ T AS S S+S + S+S ST + A+
Sbjct: 1852 STSASESASTSASASASTSASASASTS-ASASASTSASASASTSASASASTSASVSASTS 1910
Query: 218 AQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVV 277
A S+ A SE S + + ++ ASTS SA+ +S S
Sbjct: 1911 ASASASTSASASASTSASESASTSASASASTSASASASTSASASASTSASASASTSASAS 1970
Query: 278 SVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAE-EASALSAAAVNHSLELWNQLDKH 336
+ + ++ + + S A ASA ++A+ + S+
Sbjct: 1971 ASTSASESASTSASASASTSASASASTSASASASTSASASASTSASASASISASESASTS 2030
Query: 337 KNSGFMSDIEAKLASAAVAIAAAAAV----AKAAAAAANVAS---------NAAFQAKLM 383
++ + A +++A A A+ +A A+A+A+ AS +A+ A
Sbjct: 2031 ASASASTSASASASTSASASASTSASESASTSASASASTSASASASTSASASASTSASAS 2090
Query: 384 ADEALISSGYENTSQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRR 443
A + +S + S +T E S A+ ++ + ++ S +A +
Sbjct: 2091 ASTSASASASTSASASASTSASESASTSASASASTSASESASTSASASASTSASASASTS 2150
Query: 444 VEAASAATKRAENMDAILKAAELAAEA 470
A+++ + A + +A +A A
Sbjct: 2151 ASASASTSASASASTSASASASTSASA 2177
>gb|AAK31375.1| ppg3 [Leishmania major]
Length = 1325
Score = 66.6 bits (161), Expect = 4e-09
Identities = 90/438 (20%), Positives = 173/438 (38%), Gaps = 11/438 (2%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S A S S+ S SS +SS + P+ ++ P SS S P+ S+ S
Sbjct: 681 SSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSS--SAPSASSSSA 738
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
SS+ + S S + P +PS + S+S S AP + + +
Sbjct: 739 PSSSSSAPSA---SSSSAPSSSSSAPSASSSSAPSSS-SSSAPSASSSSAPSSSSSAPSA 794
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPP 217
S++PSS + S S PSSS + ++ S S + S +++ + P
Sbjct: 795 SSSSAPSSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPS 854
Query: 218 AQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVV 277
A SS P + S S A +S A +S S++ P + SS S
Sbjct: 855 ASSSSAPSSSSSAPSA-SSSSAPSSSSSAPSASSSSAPSSSSSSAP--SASSSSAPSSSS 911
Query: 278 SVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHK 337
S +P A + S S + A AS+ SA + + S +
Sbjct: 912 SSAPSASSSSAPSSSSSSAPSASSSSAPSSSSSSAPSASSSSAPSSSSSAPSASSSSAPS 971
Query: 338 NSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENTS 397
+S + A ++ + +A +A + +A ++++ A +A+ + + + S +++
Sbjct: 972 SSSSAPSASSSSAPSSSSSSAPSASSSSAPSSSSSAPSAS-SSSAPSSSSSAPSASSSSA 1030
Query: 398 QGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAENM 457
+++ P +S+ ++ +S A+ ++P S +A A ++S++ A +
Sbjct: 1031 PSSSSSAPSASSSSAPSSSSS-APSASSSSAPSSSSSSAPSASSSSAPSSSSSAPSASSS 1089
Query: 458 DAILKAAELAAEAVSQAG 475
A ++ + + S G
Sbjct: 1090 SAPSSSSSFLSGSGSDGG 1107
Score = 63.2 bits (152), Expect = 5e-08
Identities = 99/471 (21%), Positives = 181/471 (38%), Gaps = 28/471 (5%)
Query: 11 CGRMSGVSAWKGSVVKNLILIPQKHLCSQDLAARTSDSTVKQSVLQGKGISSPLGRASSK 70
CG + A K V ++++ Q L + D A + +S SS +SS
Sbjct: 626 CGCLPSTWADKNGV--SMMITAQPELLASDCATENACKPETESSSSAPSASSSSASSSSS 683
Query: 71 ATPTIANPLIPLSSPLWSLPTLSADSLQSSALARGSVVDYSQALTPLHPYQSPSPRNFLG 130
+ P+ ++ P SS S P+ S+ S SS+ + S S + P +PS +
Sbjct: 684 SAPSASSSSAPSSSS--SAPSASSSSAPSSSSSAPSA---SSSSAPSSSSSAPSASSSSA 738
Query: 131 HSTSWISQAPLRGPWIGSPTPAPDNNTHLSASPSSDTIKLASVKGSLPPSSSIKDVTPGP 190
S+S + + S + AP ++ + S SS + AS S P SSS
Sbjct: 739 PSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSSAPSAS-SSSAPSSSSSAPSASSS 797
Query: 191 PASSSGLQSTFVGTDSQLDANNVTVP-------PAQQSSGPKAKKRKKDVLSEDHGQKLL 243
A SS S + S +++ + P P+ SS P A S
Sbjct: 798 SAPSSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPSASSSSAP-SSSSSAPSAS 856
Query: 244 QSLTPAVASRASTSVSAATPVGNVPMSSVEKSVVSVSPLADQPKNDQTVEKRILSDESLM 303
S P+ +S A ++ S++ P + S S S + P + S +
Sbjct: 857 SSSAPSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSSAPSASSSSAPSSSSSSAPS 916
Query: 304 KVKEARVHAEEASALSAAAVNHSLELWNQLDKHKNSGFMSDIEAKLASAAVAIAAAAAVA 363
+ + +SA SA++ + + +S S +S+A + ++++A +
Sbjct: 917 ASSSSAPSSSSSSAPSASSSSAPSSSSSSAPSASSSSAPSS-----SSSAPSASSSSAPS 971
Query: 364 KAAAAAANVASNAAFQAKLMADEALISSGYENTSQGNNTFLPEGTSNLGQATPASILKGA 423
+++A + +S+A + A A SS ++S + S+ A AS +
Sbjct: 972 SSSSAPSASSSSAPSSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPSAS---SS 1028
Query: 424 NGPNSPGSFIVAAKEAIRRRVEAASAATKRAENMDAILKAAELAAEAVSQA 474
+ P+S S A A ++S++ A + A ++ A A S +
Sbjct: 1029 SAPSSSSS----APSASSSSAPSSSSSAPSASSSSAPSSSSSSAPSASSSS 1075
>emb|CAG44357.1| putative cell wall-anchored protein [Staphylococcus aureus subsp.
aureus MSSA476] gi|49487433|ref|YP_044654.1| putative
cell wall-anchored protein [Staphylococcus aureus subsp.
aureus MSSA476]
Length = 2275
Score = 65.1 bits (157), Expect = 1e-08
Identities = 200/1074 (18%), Positives = 398/1074 (36%), Gaps = 119/1074 (11%)
Query: 38 SQDLAARTSDSTVKQ-SVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADS 96
S ++ TS ST + S+ + S + A S + T I LS S T ++S
Sbjct: 1152 STSMSDSTSLSTSESDSISESTSTSDSISEAISASESTF----ISLSE---SNSTSDSES 1204
Query: 97 LQSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNN 156
+SA S+ + + T S S L STS S + + +
Sbjct: 1205 QSASAFLSESLSESTSESTSESVSSSTSESTSLSDSTS--ESGSTSTSLSNSTSGSASIS 1262
Query: 157 THLSASPSSDTIKLASVKGSLP--PSSSIKDVTP--------GPPASSSGLQSTFVGTDS 206
T S S S+ T K SV SL S+S+ D T + S L ++ +DS
Sbjct: 1263 TSTSISESTSTFKSESVSTSLSMSTSTSLSDSTSLSTSLSDSTSDSKSDSLSTSMSTSDS 1322
Query: 207 QLDANNVTVPPAQQSSGPKAKKRKKDVLSEDHGQ-------KLLQSLTPAVASRASTSVS 259
+ + ++ + SG ++ S + + QS + + ++ STS+S
Sbjct: 1323 ISTSKSDSISTSTSLSGSTSESESDSTSSSESKSDSTSMSISMSQSTSGSTSTSTSTSLS 1382
Query: 260 AATPVGNVPMSSVEKSVVSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALS 319
+T +S+ +S V S A Q ++ T SD ++ ++ S +
Sbjct: 1383 DSTSTSLSLSASMNQSGVD-SNSASQSASNSTSTSTSESDSQSTSSYTSQSTSQSESTST 1441
Query: 320 AAAVNHSLEL---WNQLDKHKNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNA 376
+ +++ S + +Q S +S E++ S +++ +A+ + +++A+ + + +++
Sbjct: 1442 STSLSDSTSISKSTSQSGSVSTSASLSGSESESDSQSISTSASESTSESASTSLSDSTST 1501
Query: 377 AFQAKLMADEALISSGYENTSQGNNTFLPEGTS-------NLGQATPASILKGANGPNSP 429
+ +L +S + S ++T L + TS + Q+T AS+ + S
Sbjct: 1502 SNSGSASTSTSLSNSASASESDSSSTSLSDSTSASMQSSESDSQSTSASLSDSLSTSTSN 1561
Query: 430 GSFIVAAKEAIRRRVEAASAA--TKRAENMDAILKAAELAAEAVSQAGKIVTMGDPLPLI 487
+A+ E+ S + T +++ L ++ + + S +G T
Sbjct: 1562 RMSTIASLSTSVSTSESGSTSESTSESDSTSTSLSDSQSTSRSTSASGSASTSTSTSDS- 1620
Query: 488 ELIEAGPEGCWKASRESSREVGLLKDMTRDLVNIDMVRD-IPETSHAQNRDILSSEISAS 546
A + S S+ + L + + + + D + +++ S +S S
Sbjct: 1621 RSTSASTSTSMRTSTSDSQSMSLSTSTSTSMSDSTSLSDSVSDSTSDSTSASTSGSMSVS 1680
Query: 547 IMINEKNTRGQQA-----------RTVSDLVKPVDMVLGSESETQDPSFTVRNGSENLEE 595
I +++ + A +++S+ V + V S SE+ S + GS ++ +
Sbjct: 1681 ISLSDSTSTSTSASEVMSASISDSQSMSESVNDSESVSESNSESDSKSMS---GSTSVSD 1737
Query: 596 NTFKEGSL---VEVFKDEEGHKAAWFMGNILSLKDGKVYVCYTSLVAVEGPLKEWVSLEC 652
+ GSL + K E +++ G+ S+ D V +S ++V L+ S+
Sbjct: 1738 S----GSLSVSTSLRKSESVSESSSLSGS-QSMSDS-VSTSDSSSLSVSTSLRSSESVS- 1790
Query: 653 EGDKPPRIRTARPLTSLQHEGTRKRRRAAMGDYAWSVGDRVDAWIQESWREGVITEKNKK 712
E D ++ TS G+ + G + S + I S +
Sbjct: 1791 ESDSLSDSKSTSGSTSTSTSGSLSTSTSLSGSESVSESTSLSDSISMS------DSTSTS 1844
Query: 713 DETTLTVHIPASGETSVLRAWNLRPSLIWKDGQWLDFSKVGANDSSTHKGDTPHEK--RP 770
D +L+ I SG TS+ + +L S Q S G+ +ST D+
Sbjct: 1845 DSDSLSGSISLSGSTSLSTSDSLSDSKSLSSSQ----SMSGSESTSTSVSDSQSSSTSNS 1900
Query: 771 KLGSNAVEVKGKDKMSKNIDAAESANPDEMRSLNLTEN--------------------EI 810
+ S ++ D MS + ++ S + SL+ +++
Sbjct: 1901 QFDSMSISASESDSMSTSDSSSISGSNSTSTSLSTSDSMSGSVSVSTSTSLSDSISGSTS 1960
Query: 811 VFNIGKSSTNESKQDPQRQVRSGLQKEGSKVIFGVPKPGKKRKFMEVSKHYVAHGSSKVN 870
+ + +ST+ S D Q +S + + S+ S +
Sbjct: 1961 LSDSSSTSTSTSLSDSMSQSQSTSTSASGSLSTSISTSMSMSASTSSSQSTSVSTSLSTS 2020
Query: 871 DK-NDSVKIANFSMPQGSELRGWRNSSKNDSKEKLGADSKPKTKFGKPPGVLGRVNPP-- 927
D +DS I+ E +S+ E L T G +
Sbjct: 2021 DSISDSTSISISGSQSTVESESTSDSTSISDSESLSTSDSDSTSTSTSDSTSGSTSTSIS 2080
Query: 928 --------RNTSVSNTEMNKDSSNHTKNASQSESRVERAPYSTTDGATQVPIVFSSQATS 979
+TSVS++ +S + + + SQS S + Y++T V S++ S
Sbjct: 2081 ESLSTSGSGSTSVSDSTSMSESDSTSVSMSQSMS---GSTYNSTS-------VSDSESVS 2130
Query: 980 TNTLPTKRTFTSRASKGKLAPASDKLRKGGGGKALNDKPTTSTSEPDALEPRRS 1033
T+T + T S ++ L+ + + + + +TSTSE +++ P S
Sbjct: 2131 TSTSTSLSTSDSTSTSESLSTSMSGSQSISDSTSTSMSGSTSTSESNSMHPSDS 2184
Score = 60.1 bits (144), Expect = 4e-07
Identities = 180/987 (18%), Positives = 373/987 (37%), Gaps = 68/987 (6%)
Query: 88 SLPTLSADSLQSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIG 147
S+ T ADS +S GS+V + A T S S S S +
Sbjct: 768 SVSTSKADSQSASTSTSGSIVVSTSASTSKSTSVSLSDSVSASKSLSTSESNSVSS---- 823
Query: 148 SPTPAPDNNTHLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQ 207
S + + N+ +S+S S K S+ S+ SSS + ++S L+++ +DS
Sbjct: 824 STSTSLVNSQSVSSSMSDSASKSTSLSDSISNSSSTEKSESLSTSTSDSLRTSTSLSDS- 882
Query: 208 LDANNVTVPPAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNV 267
S+ K K S SL+ + ++ STS S +
Sbjct: 883 ----------LSMSTSGSLSKSKSLSTSTSESSSTSASLSDSTSNAISTSESLSESASTS 932
Query: 268 PMSSVEKSVVSVSPLADQPKNDQTVEKRI-LSDESLMKVKEARVHAEEASALSAAAVNHS 326
S+ S+ + + + Q+ + SD M E+ S ++ +V+ S
Sbjct: 933 DSISISNSIANSQSASTSKSDSQSTSISLSTSDSKSMSTSESL----SDSTSTSGSVSGS 988
Query: 327 LEL-WNQLDKHKNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMAD 385
L + +Q S MS E S + + + +A+ +K+ + +++++++ +
Sbjct: 989 LSIAASQSVSTSTSDSMSTSEIVSDSISTSGSLSASDSKSMSVSSSMSTSQSGSTSESLS 1048
Query: 386 EALISSGYENTSQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVE 445
++ +S ++ S +T G+++ +T AS+ + + ++ GS + +++
Sbjct: 1049 DSQSTSDSDSKSLSLST-SQSGSTSTSTSTSASV-RTSESQSTSGSMSASQSDSM----- 1101
Query: 446 AASAATKRAENMDAILKAAELAAEAVSQAGKIVTMGDPLPLIELIEAGPEGCWKASRESS 505
S +T +++ A+ ++E++SQ+ T G L + E + S S
Sbjct: 1102 --SISTSFSDSTSDSKSASTASSESISQSASTSTSGSVSTSTSLSTSNSE---RTSTSMS 1156
Query: 506 REVGLLKDMTRDLVNIDMVRDIPETSHAQNRDILSSEISASIMINEKN-TRGQQARTVSD 564
L + D + + TS + + I +SE S I ++E N T ++++ S
Sbjct: 1157 DSTSLSTSES------DSISESTSTSDSISEAISASE-STFISLSESNSTSDSESQSASA 1209
Query: 565 LVKPVDMVLGSESETQDPSFTVRNGSENLEENTFKEGSLVEVFKDEEGHKAAWFMGNILS 624
+ SES ++ S + + S +L ++T + GS + A+ +S
Sbjct: 1210 FLSESLSESTSESTSESVSSST-SESTSLSDSTSESGSTSTSLSNSTSGSASISTSTSIS 1268
Query: 625 LKDG--KVYVCYTSL-VAVEGPLKEWVSLECE-GDKPPRIRTARPLTSLQHEGTRKRRRA 680
K TSL ++ L + SL D ++ TS+ + ++
Sbjct: 1269 ESTSTFKSESVSTSLSMSTSTSLSDSTSLSTSLSDSTSDSKSDSLSTSMSTSDSISTSKS 1328
Query: 681 AMGDYAWSVGDRVDAWIQESWREGVITEKNKKDETTLTVHI--PASGETSVLRAWNL--R 736
+ S + ES + + ++K D T++++ + SG TS + +L
Sbjct: 1329 D----SISTSTSLSGSTSESESDSTSSSESKSDSTSMSISMSQSTSGSTSTSTSTSLSDS 1384
Query: 737 PSLIWKDGQWLDFSKVGANDSSTHKGDTPHEKRPKLGSNAVEVKGKDKMSKNIDAAESAN 796
S ++ S V +N +S ++ + S + S++ + S +
Sbjct: 1385 TSTSLSLSASMNQSGVDSNSASQSASNSTSTSTSESDSQSTSSYTSQSTSQSESTSTSTS 1444
Query: 797 PDEMRSL--NLTENEIVFNIGKSSTNESKQDPQRQVRSGLQKEGSKVIFGVPKPGKKRKF 854
+ S+ + +++ V S +ES+ D Q S + +
Sbjct: 1445 LSDSTSISKSTSQSGSVSTSASLSGSESESDSQSISTSASESTSESASTSLSDSTSTSNS 1504
Query: 855 MEVSKHYVAHGSSKVNDKNDSVKIANFSMPQGSELRGWRNSSKNDSKEKLGADSKP-KTK 913
S S+ ++ + S S SS++DS+ + S T
Sbjct: 1505 GSASTSTSLSNSASASESDSS------STSLSDSTSASMQSSESDSQSTSASLSDSLSTS 1558
Query: 914 FGKPPGVLGRVNPPRNTSVSNTEMNKDSSNHTKNASQSESRVERAPYSTTDGATQVPIVF 973
+ ++ +TS S + S + + + S S+S+ S + A+
Sbjct: 1559 TSNRMSTIASLSTSVSTSESGSTSESTSESDSTSTSLSDSQSTSRSTSASGSASTSTSTS 1618
Query: 974 SSQATSTNTLPTKRTFTSRASKGKLAPASDKLRKGGGGKALNDKPTTSTSEPDALEPRRS 1033
S++TS +T + RT TS + L+ ++ +L+D + STS+ + S
Sbjct: 1619 DSRSTSASTSTSMRTSTSDSQSMSLSTSTS--TSMSDSTSLSDSVSDSTSDSTSASTSGS 1676
Query: 1034 NRRIQPTSRLLEGLQSSLMVSKIPSVS 1060
+ + L + +S S++ S S
Sbjct: 1677 ---MSVSISLSDSTSTSTSASEVMSAS 1700
Score = 53.5 bits (127), Expect = 4e-05
Identities = 190/1042 (18%), Positives = 380/1042 (36%), Gaps = 102/1042 (9%)
Query: 45 TSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLS---SPLWSLPTLSADSLQSSA 101
TSDS K +S ++ S +T T + + S S S+ +DS+ S
Sbjct: 1053 TSDS-------DSKSLSLSTSQSGSTSTSTSTSASVRTSESQSTSGSMSASQSDSMSIST 1105
Query: 102 LARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSA 161
S D A T S S S S + S + + +T +S
Sbjct: 1106 SFSDSTSDSKSASTASSESISQSASTSTSGSVSTSTSL--------STSNSERTSTSMSD 1157
Query: 162 SPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQS 221
S S T + S+ S S SI + S +STF+ L +N T QS
Sbjct: 1158 STSLSTSESDSISESTSTSDSISEAI-------SASESTFIS----LSESNSTSDSESQS 1206
Query: 222 SGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVVSVSP 281
+ + + SE + + S + + + STS S +T + +S+ S+S
Sbjct: 1207 ASAFLSESLSESTSESTSESVSSSTSESTSLSDSTSESGST---STSLSNSTSGSASIST 1263
Query: 282 LADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHKNSGF 341
++ T + +S M + ++++LS + + + + ++ D S
Sbjct: 1264 STSISESTSTFKSESVSTSLSMSTSTS---LSDSTSLSTSLSDSTSD--SKSDSLSTSMS 1318
Query: 342 MSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENTSQGNN 401
SD + S +++ + + + + + + + + +S+ + ++ S +TS +
Sbjct: 1319 TSDSISTSKSDSISTSTSLSGSTSESESDSTSSSESKSDSTSMSISMSQSTSGSTSTSTS 1378
Query: 402 TFLPEGTS-------NLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRA 454
T L + TS ++ Q+ S + NS + + + S + +
Sbjct: 1379 TSLSDSTSTSLSLSASMNQSGVDSNSASQSASNSTSTSTSESDSQSTSSYTSQSTSQSES 1438
Query: 455 ENMDAILKAAELAAEAVSQAGKIVTMGDPLPLIELIEAGPEGCWKASRESSREVGLLKDM 514
+ L + +++ SQ+G + T +G E S S+ +
Sbjct: 1439 TSTSTSLSDSTSISKSTSQSGSVSTSASL--------SGSE-----SESDSQSISTSASE 1485
Query: 515 TRDLVNIDMVRDIPETSHAQNRDILSSEISASIMINEKNTRGQQARTVSDLVKPVDMVLG 574
+ + D TS++ + S+ +S S +E ++ + ++SD +
Sbjct: 1486 STSESASTSLSDSTSTSNSGSAST-STSLSNSASASESDS---SSTSLSDSTSA--SMQS 1539
Query: 575 SESETQDPSFTVRNGSENLEENTFKEGSLVEVFKDEEGHKAAWFMGNILSLKDGKVYVCY 634
SES++Q S ++ S++L +T S + + S D
Sbjct: 1540 SESDSQSTSASL---SDSLSTSTSNRMSTIASLSTSVSTSESGSTSESTSESDSTSTSLS 1596
Query: 635 TSLVAVEGPLKEWVSLECEGDKPPRIRTARPLTSLQHEGTRKRRRAAMGDYAWSVGDRVD 694
S + R +A TS++ + + + + S+ D
Sbjct: 1597 DSQSTSRSTSASGSASTSTSTSDSRSTSASTSTSMRTSTSDSQSMSLSTSTSTSMSDSTS 1656
Query: 695 AWIQESWREGVITEKNKKDETTLTVHIPASGETSVLRAWNLRPSLIWKDGQWLDFSKVGA 754
+ +S + + +++V I S TS + + S D Q + S
Sbjct: 1657 --LSDSVSDSTSDSTSASTSGSMSVSISLSDSTSTSTSASEVMSASISDSQSMSES---V 1711
Query: 755 NDS-STHKGDTPHEKRPKLGSNAVEVKGKDKMSKNIDAAESANPDEMRSLNLTENEIVFN 813
NDS S + ++ + + GS +V G +S ++ +ES + S + + ++ V
Sbjct: 1712 NDSESVSESNSESDSKSMSGSTSVSDSGSLSVSTSLRKSESVSESSSLSGSQSMSDSVST 1771
Query: 814 IGKSSTNESKQ-DPQRQVRSGLQKEGSKVIFGVPKPGKKRKFMEVSKHYVAHGSSKVNDK 872
SS + S V SK G + + S+
Sbjct: 1772 SDSSSLSVSTSLRSSESVSESDSLSDSKSTSGSTSTSTSGSLSTSTSLSGSESVSESTSL 1831
Query: 873 NDSVKIANFSMPQGSE-------LRGWRNSSKNDSKEKLGADSKPKTKFGKPPGVLGRVN 925
+DS+ +++ + S+ L G + S +DS +DSK + G
Sbjct: 1832 SDSISMSDSTSTSDSDSLSGSISLSGSTSLSTSDSL----SDSKSLSSSQSMSG------ 1881
Query: 926 PPRNTSVSNTEMNKDSSNHTKNASQSESRVERAPYSTTDGATQVPIVFSSQATSTNTLPT 985
+TS S ++ S+++++ S S S E ST+D ++ + S +TST+ L T
Sbjct: 1882 -SESTSTSVSDSQSSSTSNSQFDSMSISASESDSMSTSDSSS----ISGSNSTSTS-LST 1935
Query: 986 KRTFTSRASKGKLAPASDKLRKGGGGKALNDKPTTSTSE--PDALEPRRSNRRIQPTSRL 1043
+ + S SD + G +L+D +TSTS D++ +S + L
Sbjct: 1936 SDSMSGSVSVSTSTSLSDSI---SGSTSLSDSSSTSTSTSLSDSMSQSQST-STSASGSL 1991
Query: 1044 LEGLQSSLMVSKIPSVSHNRNI 1065
+ +S+ +S S S + ++
Sbjct: 1992 STSISTSMSMSASTSSSQSTSV 2013
>dbj|BAB96440.1| MW2575 [Staphylococcus aureus subsp. aureus MW2]
gi|21284304|ref|NP_647392.1| hypothetical protein MW2575
[Staphylococcus aureus subsp. aureus MW2]
Length = 2275
Score = 65.1 bits (157), Expect = 1e-08
Identities = 200/1074 (18%), Positives = 398/1074 (36%), Gaps = 119/1074 (11%)
Query: 38 SQDLAARTSDSTVKQ-SVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADS 96
S ++ TS ST + S+ + S + A S + T I LS S T ++S
Sbjct: 1152 STSMSDSTSLSTSESDSISESTSTSDSISEAISASESTF----ISLSE---SNSTSDSES 1204
Query: 97 LQSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNN 156
+SA S+ + + T S S L STS S + + +
Sbjct: 1205 QSASAFLSESLSESTSESTSESVSSSTSESTSLSDSTS--ESGSTSTSLSNSTSGSASIS 1262
Query: 157 THLSASPSSDTIKLASVKGSLP--PSSSIKDVTP--------GPPASSSGLQSTFVGTDS 206
T S S S+ T K SV SL S+S+ D T + S L ++ +DS
Sbjct: 1263 TSTSISESTSTFKSESVSTSLSMSTSTSLSDSTSLSTSLSDSTSDSKSDSLSTSMSTSDS 1322
Query: 207 QLDANNVTVPPAQQSSGPKAKKRKKDVLSEDHGQ-------KLLQSLTPAVASRASTSVS 259
+ + ++ + SG ++ S + + QS + + ++ STS+S
Sbjct: 1323 ISTSKSDSISTSTSLSGSTSESESDSTSSSESKSDSTSMSISMSQSTSGSTSTSTSTSLS 1382
Query: 260 AATPVGNVPMSSVEKSVVSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALS 319
+T +S+ +S V S A Q ++ T SD ++ ++ S +
Sbjct: 1383 DSTSTSLSLSASMNQSGVD-SNSASQSASNSTSTSTSESDSQSTSSYTSQSTSQSESTST 1441
Query: 320 AAAVNHSLEL---WNQLDKHKNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNA 376
+ +++ S + +Q S +S E++ S +++ +A+ + +++A+ + + +++
Sbjct: 1442 STSLSDSTSISKSTSQSGSVSTSASLSGSESESDSQSISTSASESTSESASTSLSDSTST 1501
Query: 377 AFQAKLMADEALISSGYENTSQGNNTFLPEGTS-------NLGQATPASILKGANGPNSP 429
+ +L +S + S ++T L + TS + Q+T AS+ + S
Sbjct: 1502 SNSGSASTSTSLSNSASASESDSSSTSLSDSTSASMQSSESDSQSTSASLSDSLSTSTSN 1561
Query: 430 GSFIVAAKEAIRRRVEAASAA--TKRAENMDAILKAAELAAEAVSQAGKIVTMGDPLPLI 487
+A+ E+ S + T +++ L ++ + + S +G T
Sbjct: 1562 RMSTIASLSTSVSTSESGSTSESTSESDSTSTSLSDSQSTSRSTSASGSASTSTSTSDS- 1620
Query: 488 ELIEAGPEGCWKASRESSREVGLLKDMTRDLVNIDMVRD-IPETSHAQNRDILSSEISAS 546
A + S S+ + L + + + + D + +++ S +S S
Sbjct: 1621 RSTSASTSTSMRTSTSDSQSMSLSTSTSTSMSDSTSLSDSVSDSTSDSTSASTSGSMSVS 1680
Query: 547 IMINEKNTRGQQA-----------RTVSDLVKPVDMVLGSESETQDPSFTVRNGSENLEE 595
I +++ + A +++S+ V + V S SE+ S + GS ++ +
Sbjct: 1681 ISLSDSTSTSTSASEVMSASISDSQSMSESVNDSESVSESNSESDSKSMS---GSTSVSD 1737
Query: 596 NTFKEGSL---VEVFKDEEGHKAAWFMGNILSLKDGKVYVCYTSLVAVEGPLKEWVSLEC 652
+ GSL + K E +++ G+ S+ D V +S ++V L+ S+
Sbjct: 1738 S----GSLSVSTSLRKSESVSESSSLSGS-QSMSDS-VSTSDSSSLSVSTSLRSSESVS- 1790
Query: 653 EGDKPPRIRTARPLTSLQHEGTRKRRRAAMGDYAWSVGDRVDAWIQESWREGVITEKNKK 712
E D ++ TS G+ + G + S + I S +
Sbjct: 1791 ESDSLSDSKSTSGSTSTSTSGSLSTSTSLSGSESVSESTSLSDSISMS------DSTSTS 1844
Query: 713 DETTLTVHIPASGETSVLRAWNLRPSLIWKDGQWLDFSKVGANDSSTHKGDTPHEK--RP 770
D +L+ I SG TS+ + +L S Q S G+ +ST D+
Sbjct: 1845 DSDSLSGSISLSGSTSLSTSDSLSDSKSLSSSQ----SMSGSESTSTSVSDSQSSSTSNS 1900
Query: 771 KLGSNAVEVKGKDKMSKNIDAAESANPDEMRSLNLTEN--------------------EI 810
+ S ++ D MS + ++ S + SL+ +++
Sbjct: 1901 QFDSMSISASESDSMSTSDSSSISGSNSTSTSLSTSDSMSGSVSVSTSTSLSDSISGSTS 1960
Query: 811 VFNIGKSSTNESKQDPQRQVRSGLQKEGSKVIFGVPKPGKKRKFMEVSKHYVAHGSSKVN 870
+ + +ST+ S D Q +S + + S+ S +
Sbjct: 1961 LSDSSSTSTSTSLSDSMSQSQSTSTSASGSLSTSISTSMSMSASTSSSQSTSVSTSLSTS 2020
Query: 871 DK-NDSVKIANFSMPQGSELRGWRNSSKNDSKEKLGADSKPKTKFGKPPGVLGRVNPP-- 927
D +DS I+ E +S+ E L T G +
Sbjct: 2021 DSISDSTSISISGSQSTVESESTSDSTSISDSESLSTSDSDSTSTSTSDSTSGSTSTSIS 2080
Query: 928 --------RNTSVSNTEMNKDSSNHTKNASQSESRVERAPYSTTDGATQVPIVFSSQATS 979
+TSVS++ +S + + + SQS S + Y++T V S++ S
Sbjct: 2081 ESLSTSGSGSTSVSDSTSMSESDSTSVSMSQSMS---GSTYNSTS-------VSDSESVS 2130
Query: 980 TNTLPTKRTFTSRASKGKLAPASDKLRKGGGGKALNDKPTTSTSEPDALEPRRS 1033
T+T + T S ++ L+ + + + + +TSTSE +++ P S
Sbjct: 2131 TSTSTSLSTSDSTSTSESLSTSMSGSQSISDSTSTSMSGSTSTSESNSMHPSDS 2184
Score = 59.3 bits (142), Expect = 7e-07
Identities = 180/987 (18%), Positives = 376/987 (37%), Gaps = 68/987 (6%)
Query: 88 SLPTLSADSLQSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIG 147
S+ T ADS +S GS+V + A T S S S S +
Sbjct: 768 SVSTSKADSQSASTSTSGSIVVSTSASTSKSTSVSLSDSVSASKSLSTSESNSVSS---- 823
Query: 148 SPTPAPDNNTHLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQ 207
S + + N+ +S+S S K S+ S+ SSS + ++S L+++ +DS
Sbjct: 824 STSTSLVNSQSVSSSMSDSASKSTSLSDSISNSSSTEKSESLSTSTSDSLRTSTSLSDSL 883
Query: 208 LDANNVTVPPAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNV 267
+ +SG +K + S SL+ + ++ STS S +
Sbjct: 884 ----------SMSTSGSLSKSQSLST-STSESSSTSASLSDSTSNAISTSESLSESASTS 932
Query: 268 PMSSVEKSVVSVSPLADQPKNDQTVEKRI-LSDESLMKVKEARVHAEEASALSAAAVNHS 326
S+ S+ + + + Q+ + SD M E+ S ++ +V+ S
Sbjct: 933 DSISISNSIANSQSASTSKSDSQSTSISLSTSDSKSMSTSESL----SDSTSTSGSVSGS 988
Query: 327 LEL-WNQLDKHKNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMAD 385
L + +Q S MS E S + + + +A+ +K+ + +++++++ +
Sbjct: 989 LSIAASQSVSTSTSDSMSTSEIVSDSISTSGSLSASDSKSMSVSSSMSTSQSGSTSESLS 1048
Query: 386 EALISSGYENTSQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVE 445
++ +S ++ S +T G+++ +T AS+ + + ++ GS + +++
Sbjct: 1049 DSQSTSDSDSKSLSLST-SQSGSTSTSTSTSASV-RTSESQSTSGSMSASQSDSM----- 1101
Query: 446 AASAATKRAENMDAILKAAELAAEAVSQAGKIVTMGDPLPLIELIEAGPEGCWKASRESS 505
S +T +++ A+ ++E++SQ+ T G L + E + S S
Sbjct: 1102 --SISTSFSDSTSDSKSASTASSESISQSASTSTSGSVSTSTSLSTSNSE---RTSTSMS 1156
Query: 506 REVGLLKDMTRDLVNIDMVRDIPETSHAQNRDILSSEISASIMINEKN-TRGQQARTVSD 564
L + D + + TS + + I +SE S I ++E N T ++++ S
Sbjct: 1157 DSTSLSTSES------DSISESTSTSDSISEAISASE-STFISLSESNSTSDSESQSASA 1209
Query: 565 LVKPVDMVLGSESETQDPSFTVRNGSENLEENTFKEGSLVEVFKDEEGHKAAWFMGNILS 624
+ SES ++ S + + S +L ++T + GS + A+ +S
Sbjct: 1210 FLSESLSESTSESTSESVSSST-SESTSLSDSTSESGSTSTSLSNSTSGSASISTSTSIS 1268
Query: 625 LKDG--KVYVCYTSL-VAVEGPLKEWVSLECE-GDKPPRIRTARPLTSLQHEGTRKRRRA 680
K TSL ++ L + SL D ++ TS+ + ++
Sbjct: 1269 ESTSTFKSESVSTSLSMSTSTSLSDSTSLSTSLSDSTSDSKSDSLSTSMSTSDSISTSKS 1328
Query: 681 AMGDYAWSVGDRVDAWIQESWREGVITEKNKKDETTLTVHI--PASGETSVLRAWNL--R 736
+ S + ES + + ++K D T++++ + SG TS + +L
Sbjct: 1329 D----SISTSTSLSGSTSESESDSTSSSESKSDSTSMSISMSQSTSGSTSTSTSTSLSDS 1384
Query: 737 PSLIWKDGQWLDFSKVGANDSSTHKGDTPHEKRPKLGSNAVEVKGKDKMSKNIDAAESAN 796
S ++ S V +N +S ++ + S + S++ + S +
Sbjct: 1385 TSTSLSLSASMNQSGVDSNSASQSASNSTSTSTSESDSQSTSSYTSQSTSQSESTSTSTS 1444
Query: 797 PDEMRSL--NLTENEIVFNIGKSSTNESKQDPQRQVRSGLQKEGSKVIFGVPKPGKKRKF 854
+ S+ + +++ V S +ES+ D Q S + +
Sbjct: 1445 LSDSTSISKSTSQSGSVSTSASLSGSESESDSQSISTSASESTSESASTSLSDSTSTSNS 1504
Query: 855 MEVSKHYVAHGSSKVNDKNDSVKIANFSMPQGSELRGWRNSSKNDSKEKLGADSKP-KTK 913
S S+ ++ + S S SS++DS+ + S T
Sbjct: 1505 GSASTSTSLSNSASASESDSS------STSLSDSTSASMQSSESDSQSTSASLSDSLSTS 1558
Query: 914 FGKPPGVLGRVNPPRNTSVSNTEMNKDSSNHTKNASQSESRVERAPYSTTDGATQVPIVF 973
+ ++ +TS S + S + + + S S+S+ S + A+
Sbjct: 1559 TSNRMSTIASLSTSVSTSESGSTSESTSESDSTSTSLSDSQSTSRSTSASGSASTSTSTS 1618
Query: 974 SSQATSTNTLPTKRTFTSRASKGKLAPASDKLRKGGGGKALNDKPTTSTSEPDALEPRRS 1033
S++TS +T + RT TS + L+ ++ +L+D + STS+ + S
Sbjct: 1619 DSRSTSASTSTSMRTSTSDSQSMSLSTSTS--TSMSDSTSLSDSVSDSTSDSTSASTSGS 1676
Query: 1034 NRRIQPTSRLLEGLQSSLMVSKIPSVS 1060
+ + L + +S S++ S S
Sbjct: 1677 ---MSVSISLSDSTSTSTSASEVMSAS 1700
Score = 53.5 bits (127), Expect = 4e-05
Identities = 190/1042 (18%), Positives = 380/1042 (36%), Gaps = 102/1042 (9%)
Query: 45 TSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLS---SPLWSLPTLSADSLQSSA 101
TSDS K +S ++ S +T T + + S S S+ +DS+ S
Sbjct: 1053 TSDS-------DSKSLSLSTSQSGSTSTSTSTSASVRTSESQSTSGSMSASQSDSMSIST 1105
Query: 102 LARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSA 161
S D A T S S S S + S + + +T +S
Sbjct: 1106 SFSDSTSDSKSASTASSESISQSASTSTSGSVSTSTSL--------STSNSERTSTSMSD 1157
Query: 162 SPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQS 221
S S T + S+ S S SI + S +STF+ L +N T QS
Sbjct: 1158 STSLSTSESDSISESTSTSDSISEAI-------SASESTFIS----LSESNSTSDSESQS 1206
Query: 222 SGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVVSVSP 281
+ + + SE + + S + + + STS S +T + +S+ S+S
Sbjct: 1207 ASAFLSESLSESTSESTSESVSSSTSESTSLSDSTSESGST---STSLSNSTSGSASIST 1263
Query: 282 LADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHKNSGF 341
++ T + +S M + ++++LS + + + + ++ D S
Sbjct: 1264 STSISESTSTFKSESVSTSLSMSTSTS---LSDSTSLSTSLSDSTSD--SKSDSLSTSMS 1318
Query: 342 MSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENTSQGNN 401
SD + S +++ + + + + + + + + +S+ + ++ S +TS +
Sbjct: 1319 TSDSISTSKSDSISTSTSLSGSTSESESDSTSSSESKSDSTSMSISMSQSTSGSTSTSTS 1378
Query: 402 TFLPEGTS-------NLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRA 454
T L + TS ++ Q+ S + NS + + + S + +
Sbjct: 1379 TSLSDSTSTSLSLSASMNQSGVDSNSASQSASNSTSTSTSESDSQSTSSYTSQSTSQSES 1438
Query: 455 ENMDAILKAAELAAEAVSQAGKIVTMGDPLPLIELIEAGPEGCWKASRESSREVGLLKDM 514
+ L + +++ SQ+G + T +G E S S+ +
Sbjct: 1439 TSTSTSLSDSTSISKSTSQSGSVSTSASL--------SGSE-----SESDSQSISTSASE 1485
Query: 515 TRDLVNIDMVRDIPETSHAQNRDILSSEISASIMINEKNTRGQQARTVSDLVKPVDMVLG 574
+ + D TS++ + S+ +S S +E ++ + ++SD +
Sbjct: 1486 STSESASTSLSDSTSTSNSGSAST-STSLSNSASASESDS---SSTSLSDSTSA--SMQS 1539
Query: 575 SESETQDPSFTVRNGSENLEENTFKEGSLVEVFKDEEGHKAAWFMGNILSLKDGKVYVCY 634
SES++Q S ++ S++L +T S + + S D
Sbjct: 1540 SESDSQSTSASL---SDSLSTSTSNRMSTIASLSTSVSTSESGSTSESTSESDSTSTSLS 1596
Query: 635 TSLVAVEGPLKEWVSLECEGDKPPRIRTARPLTSLQHEGTRKRRRAAMGDYAWSVGDRVD 694
S + R +A TS++ + + + + S+ D
Sbjct: 1597 DSQSTSRSTSASGSASTSTSTSDSRSTSASTSTSMRTSTSDSQSMSLSTSTSTSMSDSTS 1656
Query: 695 AWIQESWREGVITEKNKKDETTLTVHIPASGETSVLRAWNLRPSLIWKDGQWLDFSKVGA 754
+ +S + + +++V I S TS + + S D Q + S
Sbjct: 1657 --LSDSVSDSTSDSTSASTSGSMSVSISLSDSTSTSTSASEVMSASISDSQSMSES---V 1711
Query: 755 NDS-STHKGDTPHEKRPKLGSNAVEVKGKDKMSKNIDAAESANPDEMRSLNLTENEIVFN 813
NDS S + ++ + + GS +V G +S ++ +ES + S + + ++ V
Sbjct: 1712 NDSESVSESNSESDSKSMSGSTSVSDSGSLSVSTSLRKSESVSESSSLSGSQSMSDSVST 1771
Query: 814 IGKSSTNESKQ-DPQRQVRSGLQKEGSKVIFGVPKPGKKRKFMEVSKHYVAHGSSKVNDK 872
SS + S V SK G + + S+
Sbjct: 1772 SDSSSLSVSTSLRSSESVSESDSLSDSKSTSGSTSTSTSGSLSTSTSLSGSESVSESTSL 1831
Query: 873 NDSVKIANFSMPQGSE-------LRGWRNSSKNDSKEKLGADSKPKTKFGKPPGVLGRVN 925
+DS+ +++ + S+ L G + S +DS +DSK + G
Sbjct: 1832 SDSISMSDSTSTSDSDSLSGSISLSGSTSLSTSDSL----SDSKSLSSSQSMSG------ 1881
Query: 926 PPRNTSVSNTEMNKDSSNHTKNASQSESRVERAPYSTTDGATQVPIVFSSQATSTNTLPT 985
+TS S ++ S+++++ S S S E ST+D ++ + S +TST+ L T
Sbjct: 1882 -SESTSTSVSDSQSSSTSNSQFDSMSISASESDSMSTSDSSS----ISGSNSTSTS-LST 1935
Query: 986 KRTFTSRASKGKLAPASDKLRKGGGGKALNDKPTTSTSE--PDALEPRRSNRRIQPTSRL 1043
+ + S SD + G +L+D +TSTS D++ +S + L
Sbjct: 1936 SDSMSGSVSVSTSTSLSDSI---SGSTSLSDSSSTSTSTSLSDSMSQSQST-STSASGSL 1991
Query: 1044 LEGLQSSLMVSKIPSVSHNRNI 1065
+ +S+ +S S S + ++
Sbjct: 1992 STSISTSMSMSASTSSSQSTSV 2013
>gb|EAL20680.1| hypothetical protein CNBE0450 [Cryptococcus neoformans var.
neoformans B-3501A]
Length = 566
Score = 64.3 bits (155), Expect = 2e-08
Identities = 72/330 (21%), Positives = 130/330 (38%), Gaps = 13/330 (3%)
Query: 57 GKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSALARG-SVVDYSQALT 115
G G + P AS +A P+ A P + S P SA Q+S L S D +Q+ +
Sbjct: 30 GNGTTEPSAEASQEAAPSSAAPAQSSDAQQDSSPAASASPTQTSELQPSPSSDDAAQSTS 89
Query: 116 PLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSASPSSDTIKLASVKG 175
P SP+P + +TS S P P +PT P ++ AS + T +
Sbjct: 90 P-----SPTPTSDAQTTTSEASAEPQ--PSSAAPTSDPAPSSEAPASSAQPTSEAQPTTS 142
Query: 176 SLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQSSGPKAKKRKKDVLS 235
PPSS+ + + P S S +T + S+ + + T P+Q +S + +
Sbjct: 143 EAPPSSAQESQSASPSPSPSPSVTT-TASPSESPSASPTPSPSQSASPSATESASPSEQA 201
Query: 236 EDHGQKLLQSLTPAVASRASTS----VSAATPVGNVPMSSVEKSVVSVSPLADQPKNDQT 291
G + S T V+S+A+ S S+ATP + V + S A Q DQ+
Sbjct: 202 SSQGVTTIYSTTQPVSSQAAQSSAAQASSATPSTTEVTTVVVGGSTTASSTAQQSSADQS 261
Query: 292 VEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHKNSGFMSDIEAKLAS 351
L+ ++ + SA A + + S +++ + ++ +
Sbjct: 262 SRAGTLTSTIVISDTSSSQSTAAESAFVATTTDSAGHSITTTPASYTSSYVTTSDGEVYT 321
Query: 352 AAVAIAAAAAVAKAAAAAANVASNAAFQAK 381
+ + +++++ ++N F K
Sbjct: 322 VTQIVHNPTGALDSGSSSSDSSTNTFFSNK 351
>dbj|BAB58816.1| serine-threoinine rich antigen [Staphylococcus aureus subsp. aureus
Mu50] gi|15928240|ref|NP_375773.1| hypothetical protein
SA2447 [Staphylococcus aureus subsp. aureus N315]
gi|13702612|dbj|BAB43752.1| SA2447 [Staphylococcus aureus
subsp. aureus N315] gi|25507441|pir||F90073 hypothetical
protein SA2447 [imported] - Staphylococcus aureus (strain
N315) gi|15925644|ref|NP_373178.1| serine-threoinine rich
antigen [Staphylococcus aureus subsp. aureus Mu50]
Length = 2271
Score = 63.9 bits (154), Expect = 3e-08
Identities = 184/1042 (17%), Positives = 389/1042 (36%), Gaps = 92/1042 (8%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
SQ ++ +DS + G + S ++SK+T + + S L + + S S
Sbjct: 766 SQSVSTSKADSQSASTSTSGSIVVST-SASTSKSTSVSLSDSVSASKSLSTSESNSVSSS 824
Query: 98 QSSALARGSVVDYSQA-----LTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPA 152
S++L V S + T L S S S S + LR S + +
Sbjct: 825 TSTSLVNSQSVSSSMSGSVSKSTSLSDSISNSNSTEKSESLSTSTSDSLRTSTSLSDSLS 884
Query: 153 PDNNTHLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANN 212
+ LS S S T S+ GS S+S+ D T ++S+ L + +DS +N+
Sbjct: 885 MSTSGSLSKSQSLST----SISGSSSTSASLSDSTSNAISTSTSLSESASTSDSISISNS 940
Query: 213 VTVPPAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSV 272
+ +Q +S K+ + + K + S + +++ STS S + + SV
Sbjct: 941 IA--NSQSASTSKSDSQSTSISLSTSDSKSM-STSESLSDSTSTSGSVSGSLSIAASQSV 997
Query: 273 EKSVVSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQ 332
S +D + V I + SL A ++ ++S ++ + + +
Sbjct: 998 STST------SDSMSTSEIVSDSISTSGSLS--------ASDSKSMSVSSSMSTSQSGST 1043
Query: 333 LDKHKNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMA---DEALI 389
+ +S SD ++K S + + + + + + + +A+ + + + + A D I
Sbjct: 1044 SESLSDSQSTSDSDSKSLSLSTSQSGSTSTSTSTSASVRTSESQSTSGSMSASQSDSMSI 1103
Query: 390 SSGYENTSQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRV-EAAS 448
S+ + +++ + + + ++ Q+ S + ++ S + E V ++ S
Sbjct: 1104 STSFSDSTSDSKSASTASSESISQSASTST---SGSVSTSTSLSTSNSERTSTSVSDSTS 1160
Query: 449 AATKRAENMDAILKAAELAAEAVSQAGKIVTMGDPLPLIELIEAGPEGCWKASRESSREV 508
+T ++++ ++ +EA+S + I L E+ ++ S+
Sbjct: 1161 LSTSESDSISESTSTSDSISEAISASESTS--------ISLSESNSTSDSESQSASAFLS 1212
Query: 509 GLLKDMTRDLVNIDMVRDIPETSHAQNRDILSSEISASIMINEKNTRGQQARTVSDLVKP 568
L + T + + + E++ + S S S+ +T G + + S +
Sbjct: 1213 ESLSESTSESTSESVSSSTSESTSLSDSTSESGSTSTSL---SNSTSGSASISTSTSISE 1269
Query: 569 VDMVLGSESETQDPSFTVRNGSENLEENTFKEGSLVEVFKDEEGHKAAWFMGNILSLKDG 628
SES + S + S +L +T SL + D + + M S+
Sbjct: 1270 STSTFKSESVSTSLSMST---STSLSNSTSLSTSLSDSTSDSKSDSLSTSMSTSDSISTS 1326
Query: 629 KVYVCYTSLVAVEGPLKEWVSLECEGDKPPRIRTARPLTSLQH-EGTRKRRRAAMGDYAW 687
K TS ++ G E S + T+ ++ Q G+ + +
Sbjct: 1327 KSDSISTS-TSLSGSTSESESDSTSSSESKSDSTSMSISMSQSTSGSTSTSTSTSLSDST 1385
Query: 688 SVGDRVDAWIQESWREGVITEKNKKDETTLTVHIPASGETSVLRAWNLRPSLIWKDGQWL 747
S + A + +S + ++ + T+ + S TS + + S L
Sbjct: 1386 STSLSLSASMNQSGVDSNSASQSASNSTSTSTSESDSQSTSTYTSQSTSQSESTSTSTSL 1445
Query: 748 DFSKVGANDSSTHKGDTPHEKRPKLGSNAVEVKGKDKMS--KNIDAAESANPDEMRSLNL 805
S + S++ G T S + + G + S ++I + S + E S +L
Sbjct: 1446 SDS-TSISKSTSQSGST---------STSASLSGSESESDSQSISTSASESTSESASTSL 1495
Query: 806 TENEIVFNIGKSSTNESKQDPQRQVRSGLQKEGSKVIFGVPKPGKKRKFMEVSKHYVAHG 865
+++ N G +ST+ S + S + S
Sbjct: 1496 SDSTSTSNSGSASTSTSLSNSASASESDSSSTSLSDSTSASMQSSESDSQSTSASLSDSL 1555
Query: 866 SSKVNDKNDSVKIANFSMPQGSELRGWRNSSKNDSKEKLGADSKPKTKFGKPPGVLGRVN 925
S+ +++ ++ + S+ ++S++DS +DS+ +
Sbjct: 1556 STSTSNRMSTIASLSTSVSTSESGSTSESTSESDSTSTSLSDSQSTS------------- 1602
Query: 926 PPRNTSVSNTEMNKDSSNHTKNASQSESRVERAPYSTTDGATQVPIVFSSQATSTNTLPT 985
R+TS S + S++ +++ S S S R ST+D + S +TST+T +
Sbjct: 1603 --RSTSASGSASTSTSTSDSRSTSASTSTSMRT--STSDSQSM------SLSTSTSTSMS 1652
Query: 986 KRTFTSRASKGKLAPASDKLRKGGGGK--ALNDKPTTSTSEPDALEPRRSNRRIQPTSRL 1043
T S + + ++ G +L+D +TSTS + + + I + +
Sbjct: 1653 DSTSLSDSVSDSTSDSTSASTSGSMSVSISLSDSTSTSTSASEVM-----SASISDSQSM 1707
Query: 1044 LEGLQSSLMVSKIPSVSHNRNI 1065
E + S VS+ S S ++++
Sbjct: 1708 SESVNDSESVSESNSESDSKSM 1729
Score = 58.2 bits (139), Expect = 2e-06
Identities = 170/924 (18%), Positives = 324/924 (34%), Gaps = 79/924 (8%)
Query: 38 SQDLAARTSDS-TVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADS 96
S L+ TS+S + S + K S+ + + S++T + +S SL ++ S
Sbjct: 1334 STSLSGSTSESESDSTSSSESKSDSTSMSISMSQSTSGSTS-----TSTSTSLSDSTSTS 1388
Query: 97 LQSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTS-WISQAPLRGPWIGSPTPAPDN 155
L SA S VD + A S S STS + SQ+ + + T D+
Sbjct: 1389 LSLSASMNQSGVDSNSASQSASNSTSTSTSESDSQSTSTYTSQSTSQSESTSTSTSLSDS 1448
Query: 156 NT-HLSASPSSDTIKLASVKGSLPPS---------------SSIKDVTPGPPASSSGLQS 199
+ S S S T AS+ GS S S+ ++ S+SG S
Sbjct: 1449 TSISKSTSQSGSTSTSASLSGSESESDSQSISTSASESTSESASTSLSDSTSTSNSGSAS 1508
Query: 200 TFVGTDSQLDANNVTVPPAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVS 259
T + A+ S A + + S+ L SL+ + ++R ST S
Sbjct: 1509 TSTSLSNSASASESDSSSTSLSDSTSASMQSSESDSQSTSASLSDSLSTSTSNRMSTIAS 1568
Query: 260 AATPVGNVPMSSVEKSVVSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALS 319
+T V S +S S S +D R S + + SA +
Sbjct: 1569 LSTSVSTSESGSTSES-TSESDSTSTSLSDSQSTSRSTSASGSASTSTSTSDSRSTSAST 1627
Query: 320 AAAVNHSLELWNQLD-KHKNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAF 378
+ ++ S + S MSD + S + + + + + + + + + +++ + +
Sbjct: 1628 STSMRTSTSDSQSMSLSTSTSTSMSDSTSLSDSVSDSTSDSTSASTSGSMSVSISLSDST 1687
Query: 379 QAKLMADEALISSGYENTSQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKE 438
A E + +S ++ S + E S + + + G+ + GS V+
Sbjct: 1688 STSTSASEVMSASISDSQSMSESVNDSESVSESNSESDSKSMSGSTSVSDSGSLSVSTSL 1747
Query: 439 AIRRRVEAASAATKRAENMDAI--LKAAELAAEAVSQAGKIVTMGDPLPLIELIEAGPEG 496
V +S+ + D++ ++ L+ ++ + V+ D L +
Sbjct: 1748 RKSESVSESSSLSGSQSMSDSVSTSDSSSLSVSTSLRSSESVSESDSLSDSKSTSGSTST 1807
Query: 497 CWKASRESSREVGLLKDMTRDLVNIDMVRDIPETSHAQNRDILSSEISASIMINEKNTRG 556
S +S + + ++ D + + +++ + D LS IS S G
Sbjct: 1808 STSGSLSTSTSLSGSESVSESTSLSDSI-SMSDSTSTSDSDSLSGSISLS---------G 1857
Query: 557 QQARTVSDLVKPVDMVLGSESETQDPSFTVRNGSENLEENTFKEGSLVEVFKDEEGHKAA 616
+ + SD + + S+S + S + + + + + E +
Sbjct: 1858 STSLSTSDSLSDSKSLSSSQSMSGSESTSTSVSDSQSSSTSNSQFDSMSISASESDSMST 1917
Query: 617 WFMGNI---------LSLKDGKVYVCYTSLVAVEGPLKEWVSLECEGDKPPRIRTARPLT 667
NI LS D + + V+ L + +S T+ L+
Sbjct: 1918 SDSSNISGSNSTSTSLSTSDS---MSGSVSVSTSTSLSDSISGSTSVSDSSSTSTSTSLS 1974
Query: 668 SLQHEGTRKRRRAAMGDYAWSVGD--RVDAWIQESWREGVITEKNKKDETTLTVHIPASG 725
+ ++ +A G + S+ + A S V T + D + + I SG
Sbjct: 1975 DSMSQ-SQSTSTSASGSLSTSISTSMSMSASTSSSQSTSVSTSLSTSDSISDSTSISISG 2033
Query: 726 ETSVLRAWNLRPSLIWKDGQWLDFSKVGANDSSTH---KGDTP---HEKRPKLGSNAVEV 779
S + + + S D + L S + +ST G T E GS + V
Sbjct: 2034 SQSTVESESTSDSTSISDSESLSTSDSDSTSTSTSDSTSGSTSTSISESLSTSGSGSTSV 2093
Query: 780 KGKDKMSKNIDAAESANPDEMRSLNLTENEIVFNIGKSSTNESKQDPQRQVRS-GLQKEG 838
MS++ + S + D+ S +++++E V +ST+ S D S G
Sbjct: 2094 SDSTSMSESDSTSVSMSQDKSDSTSISDSESVST--STSTSLSTSDSTSTSESLSTSMSG 2151
Query: 839 SKVIFGVPKPGKKRKFMEVSKHYVAHGSSKVNDKN-----DSVKIANFSMPQGSELRGWR 893
S+ I S GS+ ++ N DS+ + + S L
Sbjct: 2152 SQSI-------------SDSTSTSMSGSTSTSESNSMHPSDSMSMHHTHSTSTSRLSSEA 2198
Query: 894 NSSKNDSKEKLGADSKPKTKFGKP 917
+S ++S+ L A S+ G P
Sbjct: 2199 TTSTSESQSTLSATSEVTKHNGTP 2222
>gb|AAL58470.1| serine-threonine rich antigen [Staphylococcus aureus]
Length = 2283
Score = 63.5 bits (153), Expect = 4e-08
Identities = 201/1051 (19%), Positives = 391/1051 (37%), Gaps = 115/1051 (10%)
Query: 42 AARTSDSTVKQSVLQGKG----ISSPLGRASSKATPTIANPLIPLS---SPLWSLPTLSA 94
A+ S ++ QSV S+ L ++S+ T T + LS S S T ++
Sbjct: 1117 ASTASSESISQSVSTSTSGSVSTSTSLSTSNSERTSTSMSDSTSLSTSESDSTSDSTSTS 1176
Query: 95 DSLQSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPD 154
DS+ + S +S S FL S S + GS +
Sbjct: 1177 DSISEAISGSESTSISLSESNSTGDSESKSASAFLSESLSESTSESTSESLSGSTS---- 1232
Query: 155 NNTHLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTD-------SQ 207
++T LS S S S+ S S+SI T G AS+S ++S V T S
Sbjct: 1233 DSTSLSDSNSESGSTSTSLSNSTSGSTSISTSTSGS-ASTSTVKSESVSTSLSTSTSTSL 1291
Query: 208 LDANNVTVPPAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNV 267
D+ +++ + +SG K+ + + D ++++R S S+SA+T +
Sbjct: 1292 SDSTSLSTSLSDSTSGSKSNSLSASMSTSD-----------SISTRKSESLSASTSLSGS 1340
Query: 268 PMSSVEKSVVSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSL 327
S S S + +D ++ + I S + +LSA+ +
Sbjct: 1341 TSESESGSTSSSASQSDSTSMSLSMSQSISGSTSTSTSTSLSDSTSTSLSLSASMNQSGV 1400
Query: 328 ELWNQLDKHKNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQ------AK 381
+ + S +S E+ S + + + + +++ + + +++ + +
Sbjct: 1401 DSNSASQSASTSTSISTSESDSQSTSTYTSQSTSQSESTSTSTSISDSTSISKSTSQSGS 1460
Query: 382 LMADEALISSGYENTSQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIR 441
+L S E+ SQ +T E TS ++ S+ + NS A
Sbjct: 1461 TSTSASLSGSESESDSQSVSTSASESTS---ESASTSLSDSTSTSNSTSESTSNAISTSA 1517
Query: 442 RRVEAASAATKRAENMDAILKAAELAAEAVSQA-GKIVTMGDPLPLIELIEAGPEGCWKA 500
E+ S++T +++ A ++++E +++ S + + + + ++ ++
Sbjct: 1518 SASESDSSSTSLSDSTSASMQSSESDSQSTSTSLSNSQSTSTSIRMSTIVS-------ES 1570
Query: 501 SRESSREVGLLKDMTRDLVNIDM-VRDIPETSHAQNRDILSSEISASIMINEKNTRGQQA 559
ES+ E G + T + + + D TS + S+ SAS + ++R A
Sbjct: 1571 VSESTSESGSTSESTSESDSTSTSLSDSQSTSRST-----SASGSASTSTSTSDSRSTSA 1625
Query: 560 RTVSDLVKPVDMVLGSESETQDPSFTVRNGSENLEENTFKEGSLVEVFKDEEGHKAAWFM 619
T + + S ++Q S + S ++ ++T S+ + D + M
Sbjct: 1626 PTSTSMRT-------STLDSQSMSLSTST-STSVSDSTSLSDSVSDSTSDSTSTSTSGSM 1677
Query: 620 GNILSLKDGKVYVCYTSLVAVEGPLKEWVSLECEGDKPPRIRTARPLTSLQHEGTRKRRR 679
+SL D TS A E V D + S+ + +
Sbjct: 1678 SASISLSDSTS----TSTSASE------VMSASISDSQSMSESVNDSESVSESNSESDSK 1727
Query: 680 AAMGDYAWSVGDRVDAWIQESWR--EGVITEKNKKDETTLTVHIPASGETSVLRAWNLRP 737
+ G + SV D + S R E V + +++ + S +S+ + +LR
Sbjct: 1728 SMSG--STSVSDSGSLSVSTSLRKSESVSESSSLSGSQSMSDSVSTSDSSSLSVSMSLRS 1785
Query: 738 SLIWKDGQWLDFSKVGANDSSTHKGDTPHEKRPKLGSNAVEVKGKDKMSKNIDAAESANP 797
S + L SK + +ST GS + + G + +S++ ++S +
Sbjct: 1786 SESVSESDSLSDSKSTSGSTSTSTS----------GSLSTSLSGSESVSESTSLSDSISM 1835
Query: 798 DEMRSLNLTENEIVFNIGKSSTNESKQDPQRQVRSGLQKEGSKVIFGVPKPGKKRKFMEV 857
+ S + +++ ST+ S D +S + G + V
Sbjct: 1836 SDSTSTSDSDSLSGSISLSGSTSLSTSDSLSDSKSLSSSQSMS--------GSESTSTSV 1887
Query: 858 SKHYVAHGSSKVNDKNDSVKIANFSMPQGSELRGWRNSSKNDSKEKLGADSKPKTKFGKP 917
S + SS N + DS+ I+ + S +DS G++S T
Sbjct: 1888 SD---SQSSSASNSQFDSMSISASESD---------SVSTSDSSSISGSNST-STSLSTS 1934
Query: 918 PGVLGRVNPPRNTSVSNTEMNK----DSSNHTKNASQSESRVERAPYSTTDGATQVPIVF 973
+ G V+ +TS+S++ DSS+ + + S S+S + ST+ + +
Sbjct: 1935 DSMSGSVSVSTSTSLSDSISGSISVSDSSSTSTSESLSDSMAQSQSTSTSASGSLSTSIS 1994
Query: 974 SSQATSTNTLPTKRTFTSRA-SKGKLAPASDKLRKGGGGKALNDKPT---TSTSEPDALE 1029
+S + S +T ++ T S + S S + G A+ + T TS S+ ++L
Sbjct: 1995 TSMSMSASTSTSQSTSVSTSLSTSDSISDSTSISISGSQSAVESESTSDSTSISDSESLS 2054
Query: 1030 PRRSNRRIQPTSRLLEGLQSSLMVSKIPSVS 1060
S+ TS G +S VS+ S S
Sbjct: 2055 TSDSDSTSTSTSVSTSG-STSTSVSESLSTS 2084
Score = 54.7 bits (130), Expect = 2e-05
Identities = 181/995 (18%), Positives = 373/995 (37%), Gaps = 94/995 (9%)
Query: 88 SLPTLSADSLQSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIG 147
S+ T ADS +S GS+V + A T S S S S +
Sbjct: 768 SVSTSKADSQSASTSTSGSIVVSTSASTSKSTSVSLSDSVSASKSLSTSESNSVSS---- 823
Query: 148 SPTPAPDNNTHLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQ 207
S + + N+ +S+S S K S+ S+ SSS + ++S L+++ +DS
Sbjct: 824 STSTSLVNSQSVSSSMSDSASKSTSLSDSISNSSSTEKSESLSTSTSDSLRTSTSLSDSL 883
Query: 208 LDANNVTVPPAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNV 267
+ +SG +K + + D QS++ + ++ ST+ S +
Sbjct: 884 ----------SMSTSGSLSKSQSLSTSTSD-SASTSQSVSDSTSNSISTAESLSESASTS 932
Query: 268 PMSSVEKSVVSVSPLADQPKNDQTVEKRI-LSDESLMKVKEARVHAEEASALSAAAVNHS 326
S+ S+ + + + Q+ + SD M E+ S ++ +V+ S
Sbjct: 933 DSISISNSIANSQSASTSKSDSQSTSISLSTSDSKSMSTSESL----SDSTSTSDSVSGS 988
Query: 327 LELWNQLDKHKNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADE 386
L + S SD + + +I+ + +++ + + + +V+S+ + E
Sbjct: 989 LSV---AGSQSVSTSTSDSMSTSEIVSDSISTSGSLSASDSKSMSVSSSMSTSQSGSTSE 1045
Query: 387 ALISSGYENTSQGNNTFLPEGTSNLG----QATPASILKGANGPNSPGSFIVAAKEAIRR 442
+L S ++TS ++ L TS G + +S ++ + ++ GS +
Sbjct: 1046 SLSDS--QSTSDSDSKSLSLSTSQSGSTSTSTSTSSSVRTSESQSTSGSMSTS------- 1096
Query: 443 RVEAASAATKRAENMDAILKAAELAAEAVSQAGKIVTMGDPLPLIELIEAGPEGCWKASR 502
+ ++ S +T ++ A+ ++E++SQ+ T G L + E + S
Sbjct: 1097 QSDSTSISTSFSDPTSDSKSASTASSESISQSVSTSTSGSVSTSTSLSTSNSE---RTST 1153
Query: 503 ESSREVGLLKDMTRDLVNIDMVRDIPETSHAQNRDILSSEISASIMINEKNTRG-QQART 561
S L + D D TS + + I SE S SI ++E N+ G ++++
Sbjct: 1154 SMSDSTSLSTSES------DSTSDSTSTSDSISEAISGSE-STSISLSESNSTGDSESKS 1206
Query: 562 VSDLVKPVDMVLGSESETQDPSFTVRNGSENLEENTFKEGSLVEVFKDEEGHKA-AWFMG 620
S + SES ++ S + + + + N+ + + G + +
Sbjct: 1207 ASAFLSESLSESTSESTSESLSGSTSDSTSLSDSNSESGSTSTSLSNSTSGSTSISTSTS 1266
Query: 621 NILSLKDGKVYVCYTSL-VAVEGPLKEWVSLECE-GDKPPRIRTARPLTSLQHEGTRKRR 678
S K TSL + L + SL D ++ S+ + R
Sbjct: 1267 GSASTSTVKSESVSTSLSTSTSTSLSDSTSLSTSLSDSTSGSKSNSLSASMSTSDSISTR 1326
Query: 679 RAAMGDYAWSVGDRVDAWIQESWREGVITEKNKKDETTLTVHI--PASGETSVLRAWNLR 736
++ + S + ES + ++ D T++++ + SG TS + +L
Sbjct: 1327 KSE----SLSASTSLSGSTSESESGSTSSSASQSDSTSMSLSMSQSISGSTSTSTSTSLS 1382
Query: 737 PSLIWKDGQWLDFSKVGANDSSTHKGDTPHEKRPKLGSNAVEVKGKDKMSKNIDAAESAN 796
S ++ G + +S + + S ++ D S + ++S +
Sbjct: 1383 DSTSTSLSLSASMNQSGVDSNSASQSAST--------STSISTSESDSQSTSTYTSQSTS 1434
Query: 797 PDEMRSLNLTENEIVFNIGKSSTNESKQDPQRQVRSGLQKEGSKVIFGVPKPGKKRKFME 856
E S + + ++ +I KS++ + + S+ + E
Sbjct: 1435 QSESTSTSTSISDST-SISKSTSQSGSTSTSASLSGSESESDSQSV----STSASESTSE 1489
Query: 857 VSKHYVAHGSSKVNDKNDSVKIANFSMPQGSELRGWRNSSKNDSKEKLGADSKPKTKFGK 916
+ ++ +S N ++S A + SE ++S +DS S+ ++
Sbjct: 1490 SASTSLSDSTSTSNSTSESTSNAISTSASASESDS-SSTSLSDSTSASMQSSESDSQ--- 1545
Query: 917 PPGVLGRVNPPRNTSVSNTEMNKDSSNH----TKNASQSESRVERAPYSTTDGATQVPIV 972
+TS+SN++ S +++ S+S S ST++ + +
Sbjct: 1546 ----------STSTSLSNSQSTSTSIRMSTIVSESVSESTSESGSTSESTSESDSTSTSL 1595
Query: 973 FSSQATSTNTLPTKRTFTSRA---SKGKLAPASDKLRKG---GGGKALNDKPTTSTSEPD 1026
SQ+TS +T + TS + S+ AP S +R +L+ +TS S+
Sbjct: 1596 SDSQSTSRSTSASGSASTSTSTSDSRSTSAPTSTSMRTSTLDSQSMSLSTSTSTSVSDST 1655
Query: 1027 ALEPRRSNRRIQPTSRLLEG-LQSSLMVSKIPSVS 1060
+L S+ TS G + +S+ +S S S
Sbjct: 1656 SLSDSVSDSTSDSTSTSTSGSMSASISLSDSTSTS 1690
Score = 48.9 bits (115), Expect = 0.001
Identities = 154/892 (17%), Positives = 308/892 (34%), Gaps = 51/892 (5%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
SQ ++ TS ST S+ S L AS + +N +S S+ T +DS
Sbjct: 1366 SQSISGSTSTST-STSLSDSTSTSLSLS-ASMNQSGVDSNSASQSASTSTSISTSESDSQ 1423
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
+S S T S S S S + A L G S + + +
Sbjct: 1424 STSTYTSQSTSQSESTSTSTSISDSTSISKSTSQSGSTSTSASLSG----SESESDSQSV 1479
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPP 217
SAS S+ S+ S S+S + T ++S+ + S+ D+++ ++
Sbjct: 1480 STSASESTSESASTSLSDSTSTSNSTSESTSNAISTSA--------SASESDSSSTSL-- 1529
Query: 218 AQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVV 277
S A + + S+ L S + + + R ST VS + S +S
Sbjct: 1530 ---SDSTSASMQSSESDSQSTSTSLSNSQSTSTSIRMSTIVSESVSESTSESGSTSESTS 1586
Query: 278 SVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHK 337
+ + Q+ + + S ++ S + +L+ +
Sbjct: 1587 ESDSTSTSLSDSQSTSRSTSASGSASTSTSTSDSRSTSAPTSTSMRTSTLDSQSMSLSTS 1646
Query: 338 NSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENTS 397
S +SD + S + + + + + + + + +A+++ + + A E + +S ++ S
Sbjct: 1647 TSTSVSDSTSLSDSVSDSTSDSTSTSTSGSMSASISLSDSTSTSTSASEVMSASISDSQS 1706
Query: 398 QGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAENM 457
+ E S + + + G+ + GS V+ V +S+ +
Sbjct: 1707 MSESVNDSESVSESNSESDSKSMSGSTSVSDSGSLSVSTSLRKSESVSESSSLSGSQSMS 1766
Query: 458 DAI--LKAAELAAEAVSQAGKIVTMGDPLPLIELIEAGPEGCWKASRESSREVGLL-KDM 514
D++ ++ L+ ++ + V+ D L G S S L +
Sbjct: 1767 DSVSTSDSSSLSVSMSLRSSESVSESDSLS----DSKSTSGSTSTSTSGSLSTSLSGSES 1822
Query: 515 TRDLVNIDMVRDIPETSHAQNRDILSSEISASIMINEKNTRGQQARTVSDLVKPVDMVLG 574
+ ++ + +++ + D LS IS S + + + S + + G
Sbjct: 1823 VSESTSLSDSISMSDSTSTSDSDSLSGSISLS---GSTSLSTSDSLSDSKSLSSSQSMSG 1879
Query: 575 SESETQDPSFTVRNGSENLEENTF----KEGSLVEVFKDEEGHKAAWFMGNILSLKDGKV 630
SES + S + + + N + ++ E V D + LS D
Sbjct: 1880 SESTSTSVSDSQSSSASNSQFDSMSISASESDSVST-SDSSSISGSNSTSTSLSTSDS-- 1936
Query: 631 YVCYTSLVAVEGPLKEWVSLECEGDKPPRIRTARPLTSLQHEGTRKRRRAAMGDYAWSVG 690
+ + V+ L + +S T+ L+ + ++ +A G + S+
Sbjct: 1937 -MSGSVSVSTSTSLSDSISGSISVSDSSSTSTSESLSDSMAQ-SQSTSTSASGSLSTSIS 1994
Query: 691 D--RVDAWIQESWREGVITEKNKKDETTLTVHIPASGETSVLRAWNLRPSLIWKDGQWLD 748
+ A S V T + D + + I SG S + + + S D + L
Sbjct: 1995 TSMSMSASTSTSQSTSVSTSLSTSDSISDSTSISISGSQSAVESESTSDSTSISDSESLS 2054
Query: 749 FSKVGANDSSTH---KGDTP---HEKRPKLGSNAVEVKGKDKMSKNIDAAESANPDEMRS 802
S + +ST G T E GS + V MS++ + S + D+ S
Sbjct: 2055 TSDSDSTSTSTSVSTSGSTSTSVSESLSTSGSGSTSVSDSTSMSESDSTSASMSQDKSDS 2114
Query: 803 LNLTENEIVFNIGKSSTNESKQDPQRQVRSGLQKEGSKVIFGVPKPGKKRKFMEVSKHYV 862
+++ +E V +S + S + S + M S
Sbjct: 2115 TSISNSESVSTSTSTSLSTSDSTSTSESLSTSMSGSQSISDSTSTSMSNSTSMSNSTSTS 2174
Query: 863 AHGSSKVNDKN-----DSVKIANFSMPQGSELRGWRNSSKNDSKEKLGADSK 909
GS+ ++ N DS+ + + S +S +DS+ L A S+
Sbjct: 2175 MSGSTSTSESNSMHPSDSMSMHHTHSTSTSISTSEATTSTSDSQSTLSATSE 2226
Score = 39.7 bits (91), Expect = 0.56
Identities = 79/383 (20%), Positives = 142/383 (36%), Gaps = 47/383 (12%)
Query: 710 NKKDETTLTVHIPASGETSVLRAWNLRPSLIWKDGQWLDFSKVGANDSSTHKGDTPHEKR 769
+K D + ++ + S S+ + +L S D S G+ ST D+
Sbjct: 950 SKSDSQSTSISLSTSDSKSMSTSESLSDSTSTSDSVSGSLSVAGSQSVSTSTSDSMSTS- 1008
Query: 770 PKLGSNAVEVKGK----DKMSKNIDAAESANPDEMRSLNLTENEIVFNIGKSSTNESKQD 825
++ S+++ G D S ++ ++ S + S +L++++ +S ++SK
Sbjct: 1009 -EIVSDSISTSGSLSASDSKSMSVSSSMSTSQSGSTSESLSDSQ------STSDSDSKSL 1061
Query: 826 PQRQVRSGLQKEGSKVIFGVPKPGKKRKFMEVSKHYVAHGSSKVNDKNDSVKIANFSMPQ 885
+SG + V S+ GS + + + +FS P
Sbjct: 1062 SLSTSQSGSTSTSTSTSSSV----------RTSESQSTSGSMSTSQSDSTSISTSFSDPT 1111
Query: 886 GSELRGWRNSSKNDSKEKLGADSKPKTKFGKPPGVLGRVNPPRNTSVSNTEMNKDSSNHT 945
S+ S E + T G V+ + S SN+E S + +
Sbjct: 1112 SDS-----KSASTASSESISQSVSTSTS--------GSVSTSTSLSTSNSERTSTSMSDS 1158
Query: 946 KNASQSESRVERAPYSTTDGATQVPIVFSSQATSTNTLPTKRTFTSRASKGKLAPASDKL 1005
+ S SES ST+D ++ + S++TS + + T S SK A S+ L
Sbjct: 1159 TSLSTSESDSTSDSTSTSDSISEA--ISGSESTSISLSESNSTGDSE-SKSASAFLSESL 1215
Query: 1006 RKGGGGKALNDKPTTSTSEPDALEPRRSNRRIQPTSRLLEGLQSSLMVSKIPSVSHNRNI 1065
+ ++ ++ + STS+ +L S TS L S +S S S + +
Sbjct: 1216 SE-STSESTSESLSGSTSDSTSLSDSNSESGSTSTS-LSNSTSGSTSISTSTSGSASTST 1273
Query: 1066 PKGEHPQVLLLLVEFSLSFSIQT 1088
K E V SLS S T
Sbjct: 1274 VKSES-------VSTSLSTSTST 1289
Score = 37.7 bits (86), Expect = 2.1
Identities = 68/376 (18%), Positives = 147/376 (39%), Gaps = 26/376 (6%)
Query: 707 TEKNKKDETTLTVHIPASGETSVLRAWNLRPSLIWKDGQWLDFSKVGANDSSTHKGDTPH 766
T ++ D T+ + + SG SV + ++ S + S++ ++ ST +
Sbjct: 971 TSESLSDSTSTSDSV--SGSLSVAGSQSVSTST----SDSMSTSEIVSDSISTSGSLSAS 1024
Query: 767 EKRPKLGSNAVEVKGKDKMSKNIDAAESANPDEMRSLNLTENEIVFNIGKSSTNESKQDP 826
+ + S+++ S+++ ++S + + +SL+L+ ++ +ST+ S +
Sbjct: 1025 DSKSMSVSSSMSTSQSGSTSESLSDSQSTSDSDSKSLSLSTSQSGSTSTSTSTSSSVRTS 1084
Query: 827 QRQVRSGL----QKEGSKVIFGVPKPGKKRKFMEVSKHYVAHGSSKVNDKNDSVKIANFS 882
+ Q SG Q + + + P K + S V+ + S
Sbjct: 1085 ESQSTSGSMSTSQSDSTSISTSFSDPTSDSKSASTASSESI--SQSVSTSTSGSVSTSTS 1142
Query: 883 MPQGSELRGWRNSSKNDSKEKLGADSKPKTKFGKPPGVLGRVNPPRNTSVSNTEMNKDSS 942
+ + R + S + S +DS + + ++ +TS+S +E N
Sbjct: 1143 LSTSNSERTSTSMSDSTSLSTSESDSTSDST-STSDSISEAISGSESTSISLSESNSTGD 1201
Query: 943 NHTKNASQ----------SESRVERAPYSTTDGATQVPIVFSSQATSTNTLPTKRTFTSR 992
+ +K+AS SES E ST+D + S +TST+ + TS
Sbjct: 1202 SESKSASAFLSESLSESTSESTSESLSGSTSDSTSLSDSNSESGSTSTSLSNSTSGSTSI 1261
Query: 993 ASKGKLAPASDKLRKGGGGKALNDKPTTSTSEPDALEPRRSNRRIQPTSRLLEGLQS--- 1049
++ + ++ ++ +L+ +TS S+ +L S+ S L S
Sbjct: 1262 STSTSGSASTSTVKSESVSTSLSTSTSTSLSDSTSLSTSLSDSTSGSKSNSLSASMSTSD 1321
Query: 1050 SLMVSKIPSVSHNRNI 1065
S+ K S+S + ++
Sbjct: 1322 SISTRKSESLSASTSL 1337
>ref|YP_187464.1| LPXTG cell wall surface anchor family protein [Staphylococcus aureus
subsp. aureus COL] gi|57286580|gb|AAW38674.1| LPXTG cell
wall surface anchor family protein [Staphylococcus aureus
subsp. aureus COL]
Length = 2261
Score = 63.2 bits (152), Expect = 5e-08
Identities = 193/1076 (17%), Positives = 392/1076 (35%), Gaps = 113/1076 (10%)
Query: 42 AARTSDSTVKQSVLQGKG----ISSPLGRASSKATPTIANPLIPLS---SPLWSLPTLSA 94
A+ S ++ QS S+ L ++S+ T T + LS S S T ++
Sbjct: 1117 ASTASSESISQSASTSTSGSVSTSTSLSTSNSERTSTSMSDSTSLSTSESDSISESTSTS 1176
Query: 95 DSLQSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPD 154
DS+ + A S +S S FL S S + S + +
Sbjct: 1177 DSISEAISASESTFISLSESNSTSDSESQSASAFLSESLSESTSESTSESVSSSTSESTS 1236
Query: 155 NNTHLSASPSSDTIKLASVKGS--LPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANN 212
+ S S S+ T S GS + S+SI + T + S + + S D+ +
Sbjct: 1237 LSDSTSESGSTSTSLSNSTSGSTSISTSTSISESTSTFKSESVSTSLSMSTSTSLSDSTS 1296
Query: 213 VTVPPAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPV-GNVPMSS 271
++ + +S K+ D LS S + ++++ S S+S +T + G+ S
Sbjct: 1297 LSTSLSDSTSDSKS-----DSLSTS------MSTSDSISTSKSDSISTSTSLSGSTSESK 1345
Query: 272 VEKSVVSVSPLADQPKNDQTVEKRILSDE---SLMKVKEARVHAEEASALSAAAVNHSLE 328
+ + +S+S + T LSD SL ++++ S +A N +
Sbjct: 1346 SDSTSMSISMSQSTSGSTSTSTSTSLSDSTSTSLSLSASMNQSGVDSNSASQSASNSTST 1405
Query: 329 LWNQLDKHKNSGFMSDI--EAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADE 386
++ D S + S +++ S + +++ + +++K+ + + +V+++A+ +
Sbjct: 1406 STSESDSQSTSSYTSQSTSQSESTSTSTSLSDSTSISKSTSQSGSVSTSASLSGSESESD 1465
Query: 387 A--LISSGYENTSQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRV 444
+ + +S E+TS+ +T L + TS + ++ +N ++ S + + +
Sbjct: 1466 SQSISTSASESTSESASTSLSDSTSTSNSGSASTSTSLSNSASASESDLSSTSLS----- 1520
Query: 445 EAASAATKRAENMDAILKAAELAAEAVSQAGKIVTMGDPLPLIELIEAGPEGCWKASRES 504
++ SA+ + +E+ A+ + + S + ++ T+ + E+G +++ ES
Sbjct: 1521 DSTSASMQSSESDSQSTSASLSDSLSTSTSNRMSTIASLSTSVSTSESGSTS--ESTSES 1578
Query: 505 SREVGLLKDMTRDLVNIDMVRDIPETSHAQNRDILSSEISASIMINEKNTRGQQART--- 561
L D + ++ + S+ S S+ + +++ T
Sbjct: 1579 DSTSTSLSDSQSTSRSTSASGSASTSTSTSDSRSTSASTSTSMRTSTSDSQSMSLSTSTS 1638
Query: 562 --VSDLVKPVDMVLGSESET---------------QDPSFTVRNGSENLEENTFKEGSLV 604
+SD D V S S++ D + T + SE + + S+
Sbjct: 1639 TSMSDSTSLSDSVSDSTSDSTSASTSGSMSVSISLSDSTSTSTSASEVMSASISDSQSMS 1698
Query: 605 EVFKDEEG------HKAAWFMGNILSLKDGKVYVCYTSLVAVEGPLKEWVSLECEGDKPP 658
E D E + M S+ D TSL E + E SL C
Sbjct: 1699 ESVNDSESVSESNSESDSKSMSGSTSVSDSGSLSVSTSLRKSES-VSESSSLSCSQSMSD 1757
Query: 659 RIRTARPLTSLQHEGTRKRRRAAMGDYAWSVGDRVDAWIQESWREGVITEKNKKDETTLT 718
+ T+ + R + D S+ D S T + T+L
Sbjct: 1758 SVSTSDSSSLSVSTSLRSSESVSESD---SLSDSKST----SGSTSTSTSGSLSTSTSL- 1809
Query: 719 VHIPASGETSVLRAWNLRPSLIWKDGQWLDFSKVGANDSSTHKGDTPHEKRPKLGSNAVE 778
SG SV + +L S+ D +ST D+ GS ++
Sbjct: 1810 -----SGSESVSESTSLSDSISMSDS------------TSTSDSDSLSGSISLSGSTSLS 1852
Query: 779 VKGKDKMSKNIDAAESANPDEMRSLNLTENEIVFNIGK--------SSTNESKQDPQRQV 830
SK++ +++S + E S ++++++ +S ++S
Sbjct: 1853 TSDSLSDSKSLSSSQSMSGSESTSTSVSDSQSSSTSNSQFDSMSISASESDSMSTSDSSS 1912
Query: 831 RSGLQKEGSKVIFGVPKPGKKRKFMEVSKHYVAHGSSKVNDKNDSVKIANFSMPQGSELR 890
SG + + G S GS+ V+D + S + L
Sbjct: 1913 ISGSNSTSTSLSTSDSMSGSVSVSTSTSLSDSISGSTSVSDSS--------STSTSTSLS 1964
Query: 891 GWRNSSKNDSKEKLGADSKP-KTKFGKPPGVLGRVNPPRNTSVSNTEMNKDSSNHTKNAS 949
+ S++ S G+ S T + +TS+S ++ DS++ + + S
Sbjct: 1965 DSMSQSQSTSTSASGSLSTSISTSMSMSASTSSSQSTSVSTSLSTSDSISDSTSISISGS 2024
Query: 950 QSESRVERAPYSTTDGATQVPIVFSSQATSTNTLPTKRTFTSRASKGKLAPASDKLRKGG 1009
QS E ST+ ++ S +TST+T + TS + L+ + G
Sbjct: 2025 QSTVESESTSDSTSISDSESLSTSDSDSTSTSTSDSTSGSTSTSISESLSTS------GS 2078
Query: 1010 GGKALNDKPTTSTSEPDAL---EPRRSNRRIQPTSRLLEGLQSSLMVSKIPSVSHN 1062
G +++D + S S ++ + + + I + + +SL S S S +
Sbjct: 2079 GSTSVSDSTSMSESNSSSVSMSQDKSDSTSISDSESVSTSTSTSLSTSDSTSTSES 2134
Score = 58.2 bits (139), Expect = 2e-06
Identities = 166/915 (18%), Positives = 319/915 (34%), Gaps = 57/915 (6%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S ++ SDS + L G S S + + + +S SL ++ SL
Sbjct: 1320 SDSISTSKSDSISTSTSLSGSTSESKSDSTSMSISMSQSTSGSTSTSTSTSLSDSTSTSL 1379
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTS-WISQAPLRGPWIGSPTPAPDNN 156
SA S VD + A S S STS + SQ+ + + T D+
Sbjct: 1380 SLSASMNQSGVDSNSASQSASNSTSTSTSESDSQSTSSYTSQSTSQSESTSTSTSLSDST 1439
Query: 157 T-HLSASPSSDTIKLASVKGSLPPS---------------SSIKDVTPGPPASSSGLQST 200
+ S S S AS+ GS S S+ ++ S+SG ST
Sbjct: 1440 SISKSTSQSGSVSTSASLSGSESESDSQSISTSASESTSESASTSLSDSTSTSNSGSAST 1499
Query: 201 FVGTDSQLDANNVTVPPAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSA 260
+ A+ + S A + + S+ L SL+ + ++R ST S
Sbjct: 1500 STSLSNSASASESDLSSTSLSDSTSASMQSSESDSQSTSASLSDSLSTSTSNRMSTIASL 1559
Query: 261 ATPVGNVPMSSVEKSVVSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSA 320
+T V S +S S S +D R S + + SA ++
Sbjct: 1560 STSVSTSESGSTSES-TSESDSTSTSLSDSQSTSRSTSASGSASTSTSTSDSRSTSASTS 1618
Query: 321 AAVNHSLELWNQLD-KHKNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQ 379
++ S + S MSD + S + + + + + + + + + +++ + +
Sbjct: 1619 TSMRTSTSDSQSMSLSTSTSTSMSDSTSLSDSVSDSTSDSTSASTSGSMSVSISLSDSTS 1678
Query: 380 AKLMADEALISSGYENTSQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEA 439
A E + +S ++ S + E S + + + G+ + GS V+
Sbjct: 1679 TSTSASEVMSASISDSQSMSESVNDSESVSESNSESDSKSMSGSTSVSDSGSLSVSTSLR 1738
Query: 440 IRRRVEAASAATKRAENMDAI--LKAAELAAEAVSQAGKIVTMGDPLPLIELIEAGPEGC 497
V +S+ + D++ ++ L+ ++ + V+ D L +
Sbjct: 1739 KSESVSESSSLSCSQSMSDSVSTSDSSSLSVSTSLRSSESVSESDSLSDSKSTSGSTSTS 1798
Query: 498 WKASRESSREVGLLKDMTRDLVNIDMVRDIPETSHAQNRDILSSEISASIMINEKNTRGQ 557
S +S + + ++ D + + +++ + D LS IS S +
Sbjct: 1799 TSGSLSTSTSLSGSESVSESTSLSDSI-SMSDSTSTSDSDSLSGSISLS---GSTSLSTS 1854
Query: 558 QARTVSDLVKPVDMVLGSESETQDPSFTVRNGSENLEENTFK-EGSLVEVFKDEEGHKAA 616
+ + S + + GSES + S + + + N + ++ S + + +
Sbjct: 1855 DSLSDSKSLSSSQSMSGSESTSTSVSDSQSSSTSNSQFDSMSISASESDSMSTSDSSSIS 1914
Query: 617 WFMGNILSLKDGKVYVCYTSLVAVEGPLKEWVSLECEGDKPPRIRTARPLTSLQHEGTRK 676
SL S V+ L + +S T+ L+ + ++
Sbjct: 1915 GSNSTSTSLSTSDSMSGSVS-VSTSTSLSDSISGSTSVSDSSSTSTSTSLSDSMSQ-SQS 1972
Query: 677 RRRAAMGDYAWSVGD--RVDAWIQESWREGVITEKNKKDETTLTVHIPASGETSVLRAWN 734
+A G + S+ + A S V T + D + + I SG S + + +
Sbjct: 1973 TSTSASGSLSTSISTSMSMSASTSSSQSTSVSTSLSTSDSISDSTSISISGSQSTVESES 2032
Query: 735 LRPSLIWKDGQWLDFSKVGANDSSTH---KGDTP---HEKRPKLGSNAVEVKGKDKMSKN 788
S D + L S + +ST G T E GS + V MS++
Sbjct: 2033 TSDSTSISDSESLSTSDSDSTSTSTSDSTSGSTSTSISESLSTSGSGSTSVSDSTSMSES 2092
Query: 789 IDAAESANPDEMRSLNLTENEIVFNIGKSSTNESKQDPQRQVRS-GLQKEGSKVIFGVPK 847
++ S + D+ S +++++E V +ST+ S D S GS+ I
Sbjct: 2093 NSSSVSMSQDKSDSTSISDSESVST--STSTSLSTSDSTSTSESLSTSMSGSQSI----- 2145
Query: 848 PGKKRKFMEVSKHYVAHGSSKVNDKN-----DSVKIANFSMPQGSELRGWRNSSKNDSKE 902
S GS+ ++ N DS+ + + S L +S ++S+
Sbjct: 2146 --------SDSTSTSMSGSTSTSESNSMHPSDSMSMHHTHSTSTSRLSSEATTSTSESQS 2197
Query: 903 KLGADSKPKTKFGKP 917
L A S+ G P
Sbjct: 2198 TLSATSEVTKHNGTP 2212
Score = 57.8 bits (138), Expect = 2e-06
Identities = 185/997 (18%), Positives = 381/997 (37%), Gaps = 98/997 (9%)
Query: 88 SLPTLSADSLQSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIG 147
S+ T ADS +S GS+V + A T S S S S +
Sbjct: 768 SVSTSKADSQSASTSTSGSIVVSTSASTSKSTSVSLSDSVSASKSLSTSESNSVSS---- 823
Query: 148 SPTPAPDNNTHLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQ 207
S + + N+ +S+S S K S+ S+ SSS + ++S L+++ +DS
Sbjct: 824 STSTSLVNSQSVSSSMSDSASKSTSLSDSISNSSSTEKSESLSTSTSDSLRTSTSLSDSL 883
Query: 208 LDANNVTVPPAQQ-SSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGN 266
+ + ++ +Q S+ LS+ + S + + ++ S S+S + + N
Sbjct: 884 SMSTSGSLSKSQSLSTSISGSSSTSASLSDSTSNAISTSTSLSESASTSDSISISNSIAN 943
Query: 267 VPMSSVEKS---VVSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAV 323
+S KS S+S K+ T E LSD + + +LS AA
Sbjct: 944 SQSASTSKSDSQSTSISLSTSDSKSMSTSES--LSDST-------STSGSVSGSLSIAA- 993
Query: 324 NHSLELWNQLDKHKNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLM 383
+Q S MS E S + + + +A+ +K+ + +++++++ +
Sbjct: 994 -------SQSVSTSTSDSMSTSEIVSDSISTSGSLSASDSKSMSVSSSMSTSQSGSTSES 1046
Query: 384 ADEALISSGYENTSQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRR 443
++ +S ++ S +T G+++ +T AS+ + + ++ GS + +++
Sbjct: 1047 LSDSQSTSDSDSKSLSQST-SQSGSTSTSTSTSASV-RTSESQSTSGSMSASQSDSM--- 1101
Query: 444 VEAASAATKRAENMDAILKAAELAAEAVSQAGKIVTMGDPLPLIELIEAGPEGCWKASRE 503
S +T +++ A+ ++E++SQ+ T G L + E + S
Sbjct: 1102 ----SISTSFSDSTSDSKSASTASSESISQSASTSTSGSVSTSTSLSTSNSE---RTSTS 1154
Query: 504 SSREVGLLKDMTRDLVNIDMVRDIPETSHAQNRDILSSEISASIMINEKN-TRGQQARTV 562
S L + D + + TS + + I +SE S I ++E N T ++++
Sbjct: 1155 MSDSTSLSTSES------DSISESTSTSDSISEAISASE-STFISLSESNSTSDSESQSA 1207
Query: 563 SDLVKPVDMVLGSESETQDPSFTVRNGSENLEENTFKEGSLVEVFKDEEGHKAAWFMGNI 622
S + SES ++ T + SE++ +T + SL + E + + N
Sbjct: 1208 SAFL--------SESLSES---TSESTSESVSSSTSESTSLSD--STSESGSTSTSLSN- 1253
Query: 623 LSLKDGKVYVCYTSLVAVEGPLK-EWVSLECEGDKPPRIRTARPLTSLQHEGTRKRRRAA 681
S TS+ K E VS + + L++ + T + +
Sbjct: 1254 -STSGSTSISTSTSISESTSTFKSESVSTSLSMSTSTSLSDSTSLSTSLSDSTSDSKSDS 1312
Query: 682 MGDYAWSVGDRVDA----WIQESWREGVITEKNKKDETTLTVHI--PASGETSVLRAWNL 735
+ + S D + I S T ++K D T++++ + SG TS + +L
Sbjct: 1313 LST-SMSTSDSISTSKSDSISTSTSLSGSTSESKSDSTSMSISMSQSTSGSTSTSTSTSL 1371
Query: 736 RPSLIWKDGQWLDFSKVGANDSSTHKGDTPHEKRPKLGSNAVEVKGKDKMSKNIDAAESA 795
S ++ G + +S + + S + D S + ++S
Sbjct: 1372 SDSTSTSLSLSASMNQSGVDSNSASQSAS--------NSTSTSTSESDSQSTSSYTSQST 1423
Query: 796 NPDEMRSLNLTENEIVFNIGKSSTNESKQDPQRQVRSGLQKEGSKVIFGVPKPGKKRKFM 855
+ E S + + ++ +I KS++ + + S+ I
Sbjct: 1424 SQSESTSTSTSLSDST-SISKSTSQSGSVSTSASLSGSESESDSQSI----STSASESTS 1478
Query: 856 EVSKHYVAHGSSKVN--DKNDSVKIANFSMPQGSELRGW---------RNSSKNDSKEKL 904
E + ++ +S N + S ++N + S+L SS++DS+
Sbjct: 1479 ESASTSLSDSTSTSNSGSASTSTSLSNSASASESDLSSTSLSDSTSASMQSSESDSQSTS 1538
Query: 905 GADSKP-KTKFGKPPGVLGRVNPPRNTSVSNTEMNKDSSNHTKNASQSESRVERAPYSTT 963
+ S T + ++ +TS S + S + + + S S+S+ S +
Sbjct: 1539 ASLSDSLSTSTSNRMSTIASLSTSVSTSESGSTSESTSESDSTSTSLSDSQSTSRSTSAS 1598
Query: 964 DGATQVPIVFSSQATSTNTLPTKRTFTSRASKGKLAPASDKLRKGGGGKALNDKPTTSTS 1023
A+ S++TS +T + RT TS + L+ ++ +L+D + STS
Sbjct: 1599 GSASTSTSTSDSRSTSASTSTSMRTSTSDSQSMSLSTSTS--TSMSDSTSLSDSVSDSTS 1656
Query: 1024 EPDALEPRRSNRRIQPTSRLLEGLQSSLMVSKIPSVS 1060
+ + S + + L + +S S++ S S
Sbjct: 1657 DSTSASTSGS---MSVSISLSDSTSTSTSASEVMSAS 1690
>gb|AAW43455.1| hypothetical protein CNE00530 [Cryptococcus neoformans var.
neoformans JEC21] gi|58267212|ref|XP_570762.1|
hypothetical protein CNE00530 [Cryptococcus neoformans
var. neoformans JEC21]
Length = 566
Score = 62.0 bits (149), Expect = 1e-07
Identities = 71/330 (21%), Positives = 128/330 (38%), Gaps = 13/330 (3%)
Query: 57 GKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSALARG-SVVDYSQALT 115
G G + P AS +A P+ A P + S P SA Q+S L S D +Q+ +
Sbjct: 30 GNGTTEPSAEASQEAAPSSAAPAQSSDAQQDSSPAASASPTQTSELQPSPSSDDAAQSTS 89
Query: 116 PLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSASPSSDTIKLASVKG 175
P SP+P + +TS S P P +PT P ++ AS + T +
Sbjct: 90 P-----SPTPTSDAQTTTSEASAEPQ--PSSAAPTSDPAPSSEAPASSAQPTSEAQPTTS 142
Query: 176 SLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQSSGPKAKKRKKDVLS 235
PPSS+ + + P S S +T + S+ + + T P+Q +S + +
Sbjct: 143 EAPPSSAQESQSASPSPSPSPSVTT-TASPSESPSASPTPSPSQSASPSATESASPSEQA 201
Query: 236 EDHGQKLLQSLTPAVASRASTS----VSAATPVGNVPMSSVEKSVVSVSPLADQPKNDQT 291
G + S T V+S+A+ S S+ATP + V + S A Q D +
Sbjct: 202 SSQGVTTIYSTTQPVSSQAAQSSAAQASSATPSTTEVTTVVVGGSTTASSTAQQSSADNS 261
Query: 292 VEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHKNSGFMSDIEAKLAS 351
L+ ++ + SA A + + S +++ + ++ +
Sbjct: 262 SRAGTLTSTIVISDTSSSQSTAAESAFVATTTDSAGHSITTTPASYTSSYVTTSDGEVYT 321
Query: 352 AAVAIAAAAAVAKAAAAAANVASNAAFQAK 381
+ +++++ ++N F K
Sbjct: 322 VTQIVHNPTGALDTGSSSSDSSTNTFFSNK 351
>gb|AAF34780.1| srpA [Streptococcus cristatus]
Length = 3381
Score = 61.2 bits (147), Expect = 2e-07
Identities = 88/456 (19%), Positives = 192/456 (41%), Gaps = 21/456 (4%)
Query: 38 SQDLAARTSDS----TVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLS 93
SQ +A S++ ++ S Q IS+ ++S + A+ +S+ + ++S
Sbjct: 2335 SQSASASASNNQNSASISVSASQSASISASQSASASISASISASQSASVSASQSASASVS 2394
Query: 94 ADSLQSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAP 153
A S++++ S + S S S S + A + S + +
Sbjct: 2395 ASQSTSASVSASQSASASASNNQNSASISVSASQSASISASQSASASISASISASQSASV 2454
Query: 154 DNNTHLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNV 213
+ SAS S+ ASV S S+S + S S QS + A+
Sbjct: 2455 SASQSASASVSASQSTSASVSASQSASASASNNQNSASISVSASQSASISASQSASASIS 2514
Query: 214 TVPPAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVE 273
A QS+ A + +S +S + ++++ S S+SA+ V +SV
Sbjct: 2515 ASISASQSASVSASQSASASVSASQSTSASESASTSMSNSVSASISASESVSTSMSNSVS 2574
Query: 274 KSVVSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAA-AVNHSLELWNQ 332
S+ + + N +V I + +S V A ++++SA+ ++++S+
Sbjct: 2575 ASISASVSASTSMSN--SVSASISASQSASTSMSNSVSASISASVSASTSMSNSVSASIS 2632
Query: 333 LDKHKNSGFMSDIEAKLA---SAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALI 389
++ + + A ++ SA+ +++ + + + +A+ +A+ + + + A + A E+
Sbjct: 2633 ASVSASTSMSNSVSASISASVSASTSMSNSVSASISASVSASTSMSNSVSASISASESAS 2692
Query: 390 SSGYENTSQGNNTFLPEGTS---NLGQATPASILKGANGPNSPGSFIVAAKEA---IRRR 443
+S + S + + TS ++ + AS+ + NS + I A++ A +
Sbjct: 2693 TSMSNSVSASISASVSASTSMSNSVSASISASVSASTSMSNSVSASISASESASTSMSNS 2752
Query: 444 VEAA-----SAATKRAENMDAILKAAELAAEAVSQA 474
V A+ SA+T + ++ A + A+E A+ ++S +
Sbjct: 2753 VSASISASVSASTSMSNSVSASISASESASTSMSNS 2788
Score = 57.4 bits (137), Expect = 3e-06
Identities = 92/523 (17%), Positives = 206/523 (38%), Gaps = 28/523 (5%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S ++A S S + IS +++S + A+ I S ++SA
Sbjct: 1777 SASVSASQSASASASNNQNSASISVSASQSASISASQSASASISASISASQSASVSASQS 1836
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
S++++ S + + + + +N S S A + S + + +
Sbjct: 1837 ASASVSASQSTSASVSASQSASASASNNQNSASISVSASQSASISASQSTSASVSASQSA 1896
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPP 217
+SAS S ASV S S+S+ + AS S QS + ++ +++V
Sbjct: 1897 SVSASQS------ASVSASQSASASVS-ASQSTSASVSASQSASASASNNQNSASISVSA 1949
Query: 218 AQQSSGPKAKKRKKDV-----LSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSV 272
+ +S ++ + S+ QS + +V++ STS S + +S
Sbjct: 1950 SHSASISASQSTSASISASISASQSASVSASQSASASVSASQSTSASVSASQSVSASASN 2009
Query: 273 EKSVVSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQ 332
++ S+S A + + S + + ++ + SA ++ + + S
Sbjct: 2010 NQNSASISVSASHSASISASQSTSASISASISASQSASVSASQSASASVSASQSTSASVS 2069
Query: 333 LDKHKNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSG 392
+ ++ ++ + SA+++ + +A+V+ + +A+A+V+++ + A + A +++ +S
Sbjct: 2070 ASQSASASASNNQNSASISASISASQSASVSASQSASASVSASQSTSASVSASQSVSASA 2129
Query: 393 YENTSQGNNTFLPEGTSNL--GQATPASILKGANGPNS-------PGSFIVAAKEAIRRR 443
N + + + ++++ Q+T ASI + S S V+A ++
Sbjct: 2130 SNNQNSASISVSASHSASISASQSTSASISASISASQSASVSASQSASASVSASQSTSAS 2189
Query: 444 VEAASAATKRAENMDAILKAAELAAEAVSQAGKIVTMGDPLPLIELIEAGPEGCWKASRE 503
V A+ +A+ A N +A ++ A A + + I A AS+
Sbjct: 2190 VSASQSASASASNNQ---NSASISVSASQSASISASQSASASISASISASQSASVSASQS 2246
Query: 504 SSREVGLLKDMTRDLVNIDMVRDIPETSHAQNRDILSSEISAS 546
+S V + + + S + N++ S +SAS
Sbjct: 2247 ASASVSASQSTSASV----SASQSASASASNNQNSASISVSAS 2285
Score = 57.4 bits (137), Expect = 3e-06
Identities = 92/523 (17%), Positives = 206/523 (38%), Gaps = 28/523 (5%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S ++A S S + IS +++S + A+ I S ++SA
Sbjct: 1225 SASVSASQSASASASNNQNSASISVSASQSASISASQSASASISASISASQSASVSASQS 1284
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
S++++ S + + + + +N S S A + S + + +
Sbjct: 1285 ASASVSASQSTSASVSASQSASASASNNQNSASISVSASQSASISASQSTSASVSASQSA 1344
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPP 217
+SAS S ASV S S+S+ + AS S QS + ++ +++V
Sbjct: 1345 SVSASQS------ASVSASQSASASVS-ASQSTSASVSASQSASASASNNQNSASISVSA 1397
Query: 218 AQQSSGPKAKKRKKDV-----LSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSV 272
+ +S ++ + S+ QS + +V++ STS S + +S
Sbjct: 1398 SHSASISASQSTSASISASISASQSASVSASQSASASVSASQSTSASVSASQSVSASASN 1457
Query: 273 EKSVVSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQ 332
++ S+S A + + S + + ++ + SA ++ + + S
Sbjct: 1458 NQNSASISVSASHSASISASQSTSASISASISASQSASVSASQSASASVSASQSTSASVS 1517
Query: 333 LDKHKNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSG 392
+ ++ ++ + SA+++ + +A+V+ + +A+A+V+++ + A + A +++ +S
Sbjct: 1518 ASQSASASASNNQNSASISASISASQSASVSASQSASASVSASQSTSASVSASQSVSASA 1577
Query: 393 YENTSQGNNTFLPEGTSNL--GQATPASILKGANGPNS-------PGSFIVAAKEAIRRR 443
N + + + ++++ Q+T ASI + S S V+A ++
Sbjct: 1578 SNNQNSASISVSASHSASISASQSTSASISASISASQSASVSASQSASASVSASQSTSAS 1637
Query: 444 VEAASAATKRAENMDAILKAAELAAEAVSQAGKIVTMGDPLPLIELIEAGPEGCWKASRE 503
V A+ +A+ A N +A ++ A A + + I A AS+
Sbjct: 1638 VSASQSASASASNNQ---NSASISVSASQSASISASQSASASISASISASQSASVSASQS 1694
Query: 504 SSREVGLLKDMTRDLVNIDMVRDIPETSHAQNRDILSSEISAS 546
+S V + + + S + N++ S +SAS
Sbjct: 1695 ASASVSASQSTSASV----SASQSASASASNNQNSASISVSAS 1733
Score = 54.7 bits (130), Expect = 2e-05
Identities = 111/550 (20%), Positives = 217/550 (39%), Gaps = 50/550 (9%)
Query: 38 SQDLAARTSDS-TVKQSVLQGKGISSPLGRASSKATPTI-ANPLIPLSSPLWSLPTLSAD 95
SQ +A S S + SV + S+ + A+ ++ A+ +S+ + ++SA
Sbjct: 1002 SQSASASVSASQSTSASVSASQSASASASNNQNSASISVSASHSASISASQSASASISA- 1060
Query: 96 SLQSSALARGSVVDYSQALTPLHPYQSPSP-----RNFLGHSTSWISQAPLRGPWIGSPT 150
S+ +S A SV + QS S +N S S A + S +
Sbjct: 1061 SISASQSASASVSASQSTSASVSASQSASASASNNQNSASISVSASQSASISASQSASAS 1120
Query: 151 PAPDNNTHLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDA 210
+ + SAS S+ ASV S S+S+ ++S+ S + + A
Sbjct: 1121 ISASISASQSASVSASQSASASVSASQSTSASVSASQSASASASNNQNSASISVSASQSA 1180
Query: 211 NNVTVPPAQQSSGPKAKKRKKDV-LSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPM 269
+ + +Q +S + + V S+ QS + +V++ STS S +
Sbjct: 1181 S---ISASQSTSASVSASQSASVSASQSASVSASQSASASVSASQSTSASVSASQSASAS 1237
Query: 270 SSVEKSVVSVSPLADQPKNDQTVEKRILS-DESLMKVKEARVHAEE-------ASALSAA 321
+S ++ S+S A Q + + S S+ + A V A + AS ++A
Sbjct: 1238 ASNNQNSASISVSASQSASISASQSASASISASISASQSASVSASQSASASVSASQSTSA 1297
Query: 322 AVNHSLELWNQLDKHKNSGFMSDIEAKLA----------------SAAVAIAAAAAVAKA 365
+V+ S ++NS +S ++ A SA+V+ + +A+V+ +
Sbjct: 1298 SVSASQSASASASNNQNSASISVSASQSASISASQSTSASVSASQSASVSASQSASVSAS 1357
Query: 366 AAAAANVASNAAFQAKLMADEALISSGYENTSQGNNTFLPEGTSNL--GQATPASILKGA 423
+A+A+V+++ + A + A ++ +S N + + + ++++ Q+T ASI
Sbjct: 1358 QSASASVSASQSTSASVSASQSASASASNNQNSASISVSASHSASISASQSTSASISASI 1417
Query: 424 NGPNS-------PGSFIVAAKEAIRRRVEAASAATKRAENMDAILKAAELAAEAVSQAGK 476
+ S S V+A ++ V A+ + + A N +A ++ A A
Sbjct: 1418 SASQSASVSASQSASASVSASQSTSASVSASQSVSASASNNQ---NSASISVSASHSASI 1474
Query: 477 IVTMGDPLPLIELIEAGPEGCWKASRESSREVGLLKDMTRDLVNIDMVRDIPETSHAQNR 536
+ + I A AS+ +S V + + + S+ QN
Sbjct: 1475 SASQSTSASISASISASQSASVSASQSASASVSASQSTSASVSASQSAS--ASASNNQNS 1532
Query: 537 DILSSEISAS 546
+S+ ISAS
Sbjct: 1533 ASISASISAS 1542
Score = 54.3 bits (129), Expect = 2e-05
Identities = 88/457 (19%), Positives = 188/457 (40%), Gaps = 37/457 (8%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S ++A S S + IS +++S + A+ I S ++SA
Sbjct: 2187 SASVSASQSASASASNNQNSASISVSASQSASISASQSASASISASISASQSASVSASQS 2246
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
S++++ S + + + + +N S S A + S + + +
Sbjct: 2247 ASASVSASQSTSASVSASQSASASASNNQNSASISVSASHSASISASQSASASISASISA 2306
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPP 217
SAS S+ ASV S S+S+ S QS + ++ +++V
Sbjct: 2307 SQSASVSASQSASASVSASQSTSASV-----------SASQSASASASNNQNSASISVSA 2355
Query: 218 AQQSSGPKAKKRKKDV-----LSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSV 272
+Q +S ++ + S+ QS + +V++ STS S + +S
Sbjct: 2356 SQSASISASQSASASISASISASQSASVSASQSASASVSASQSTSASVSASQSASASASN 2415
Query: 273 EKSVVSVSPLADQPKNDQTVEKRILS-DESLMKVKEARVHAEE-------ASALSAAAVN 324
++ S+S A Q + + S S+ + A V A + AS ++A+V+
Sbjct: 2416 NQNSASISVSASQSASISASQSASASISASISASQSASVSASQSASASVSASQSTSASVS 2475
Query: 325 HSLELWNQLDKHKNSGFMSDIEAKLASAAVAIAAAAAVAK--AAAAAANVASNAAFQAKL 382
S ++NS +S ++ AS + + +A+A+++ +A+ +A+V+++ + A +
Sbjct: 2476 ASQSASASASNNQNSASISVSASQSASISASQSASASISASISASQSASVSASQSASASV 2535
Query: 383 MADEALISSGYENTSQGNNTFLPEGTS-----NLGQATPASILKGANGPNSPGSFIVAAK 437
A ++ +S +TS N+ S ++ + ASI + S + + A+
Sbjct: 2536 SASQSTSASESASTSMSNSVSASISASESVSTSMSNSVSASISASVSASTSMSNSVSASI 2595
Query: 438 EAIRRRVEAASAATKRAENMDAILKAAELAAEAVSQA 474
A + SA+T + ++ A + A+ A+ ++S +
Sbjct: 2596 SA------SQSASTSMSNSVSASISASVSASTSMSNS 2626
Score = 53.9 bits (128), Expect = 3e-05
Identities = 105/542 (19%), Positives = 217/542 (39%), Gaps = 50/542 (9%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
SQ ++A S++ S+ S+ + ++S++T + I S + SA +
Sbjct: 1570 SQSVSASASNNQNSASISVSASHSASI--SASQSTSASISASISASQSASVSASQSASAS 1627
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
S++ + + V SQ+ + + + +N S S A + S + + +
Sbjct: 1628 VSASQSTSASVSASQSASA----SASNNQNSASISVSASQSASISASQSASASISASISA 1683
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPP 217
SAS S+ ASV S S+S+ ++S+ S + + +++ ++
Sbjct: 1684 SQSASVSASQSASASVSASQSTSASVSASQSASASASNNQNSASISVSA---SHSASISA 1740
Query: 218 AQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVV 277
+Q +S A S+ QS + +V++ STS S + +S ++
Sbjct: 1741 SQSAS---ASISASISASQSASVSASQSASASVSASQSTSASVSASQSASASASNNQNSA 1797
Query: 278 SVSPLADQPKNDQTVEKRILS-DESLMKVKEARVHAEE-------ASALSAAAVNHSLEL 329
S+S A Q + + S S+ + A V A + AS ++A+V+ S
Sbjct: 1798 SISVSASQSASISASQSASASISASISASQSASVSASQSASASVSASQSTSASVSASQSA 1857
Query: 330 WNQLDKHKNSGFMSDIEAKLA----------------SAAVAIAAAAAVAKAAAAAANVA 373
++NS +S ++ A SA+V+ + +A+V+ + +A+A+V+
Sbjct: 1858 SASASNNQNSASISVSASQSASISASQSTSASVSASQSASVSASQSASVSASQSASASVS 1917
Query: 374 SNAAFQAKLMADEALISSGYENTSQGNNTFLPEGTSNL--GQATPASILKGANGPNS--- 428
++ + A + A ++ +S N + + + ++++ Q+T ASI + S
Sbjct: 1918 ASQSTSASVSASQSASASASNNQNSASISVSASHSASISASQSTSASISASISASQSASV 1977
Query: 429 ----PGSFIVAAKEAIRRRVEAASAATKRAENMDAILKAAELAAEAVSQAGKIVTMGDPL 484
S V+A ++ V A+ + + A N +A ++ A A +
Sbjct: 1978 SASQSASASVSASQSTSASVSASQSVSASASNNQ---NSASISVSASHSASISASQSTSA 2034
Query: 485 PLIELIEAGPEGCWKASRESSREVGLLKDMTRDLVNIDMVRDIPETSHAQNRDILSSEIS 544
+ I A AS+ +S V + + + S+ QN +S+ IS
Sbjct: 2035 SISASISASQSASVSASQSASASVSASQSTSASVSASQSAS--ASASNNQNSASISASIS 2092
Query: 545 AS 546
AS
Sbjct: 2093 AS 2094
Score = 53.9 bits (128), Expect = 3e-05
Identities = 89/449 (19%), Positives = 191/449 (41%), Gaps = 28/449 (6%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
SQ ++A S++ S+ S+ + ++S++T + I S + SA +
Sbjct: 2122 SQSVSASASNNQNSASISVSASHSASI--SASQSTSASISASISASQSASVSASQSASAS 2179
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
S++ + + V SQ+ + + + +N S S A + S + + +
Sbjct: 2180 VSASQSTSASVSASQSASA----SASNNQNSASISVSASQSASISASQSASASISASISA 2235
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPP 217
SAS S+ ASV S S+S+ ++S+ S + + +++ ++
Sbjct: 2236 SQSASVSASQSASASVSASQSTSASVSASQSASASASNNQNSASISVSA---SHSASISA 2292
Query: 218 AQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVV 277
+Q +S A S+ QS + +V++ STS S + +S ++
Sbjct: 2293 SQSAS---ASISASISASQSASVSASQSASASVSASQSTSASVSASQSASASASNNQNSA 2349
Query: 278 SVSPLADQPKNDQTVEKRILS-DESLMKVKEARVHAEE-------ASALSAAAVNHSLEL 329
S+S A Q + + S S+ + A V A + AS ++A+V+ S
Sbjct: 2350 SISVSASQSASISASQSASASISASISASQSASVSASQSASASVSASQSTSASVSASQSA 2409
Query: 330 WNQLDKHKNSGFMSDIEAKLA------SAAVAIAAAAAVAKAAAAAANVASNAAFQAKLM 383
++NS +S ++ A SA+ +I+A+ + +++A+ +A+ +++A+ A
Sbjct: 2410 SASASNNQNSASISVSASQSASISASQSASASISASISASQSASVSASQSASASVSASQS 2469
Query: 384 ADEALISSGYENTSQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRR 443
++ +S + S NN + ++ + ASI + S + I A++ A
Sbjct: 2470 TSASVSASQSASASASNNQ--NSASISVSASQSASISASQSASASISASISASQSASVSA 2527
Query: 444 VEAASAATKRAENMDAILKAAELAAEAVS 472
++ASA+ +++ A A+ + +VS
Sbjct: 2528 SQSASASVSASQSTSASESASTSMSNSVS 2556
Score = 53.5 bits (127), Expect = 4e-05
Identities = 94/457 (20%), Positives = 190/457 (41%), Gaps = 26/457 (5%)
Query: 42 AARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLS---SPLWSLPTLSADSLQ 98
A++++ ++V S +S+ + S + A+ + +S S S ++ S+
Sbjct: 783 ASQSASASVSASQSASTSVSASHSASISSSQSASASISVSISASHSASVSASQSTSASVS 842
Query: 99 SSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSW---ISQAPLRGPWIGSPTPAPDN 155
SS A SV A + QS S S S +SQ+ T A +
Sbjct: 843 SSQSASASVSASQSASASVSVSQSASASVSASQSASASVSVSQSASASVSASQSTSASVS 902
Query: 156 NTH-LSASPSSDTIKLASVKGSLPPSSSIK-------DVTPGPPASSSGLQSTFVGTD-S 206
+ SAS S+ ASV S S+S+ V+ AS S QS V S
Sbjct: 903 ASQSASASVSASQSASASVSSSHSASASVSVSQSASASVSASHSASVSASQSASVSVSAS 962
Query: 207 QLDANNVTVPPAQQSSGPKAKKRKKDV---LSEDHGQKLLQSLTPAVASRASTSVSAATP 263
Q + +V+ ++ S ++ S+ QS + +V++ STS S +
Sbjct: 963 QSASTSVSASHSESISASQSASASISASISASQSASVSASQSASASVSASQSTSASVSAS 1022
Query: 264 VGNVPMSSVEKSVVSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAV 323
+S ++ S+S A + + S + + ++ + AS ++A+V
Sbjct: 1023 QSASASASNNQNSASISVSASHSASISASQSASASISASISASQSASASVSASQSTSASV 1082
Query: 324 NHSLELWNQLDKHKNSGFMSDIEAKLA------SAAVAIAAAAAVAKAAAAAANVASNAA 377
+ S ++NS +S ++ A SA+ +I+A+ + +++A+ +A+ +++A+
Sbjct: 1083 SASQSASASASNNQNSASISVSASQSASISASQSASASISASISASQSASVSASQSASAS 1142
Query: 378 FQAKLMADEALISSGYENTSQGNNTFLPEGTSNLGQAT--PASILKGANGPNSPGSFIVA 435
A ++ +S + S NN + + Q+ AS A+ S + + A
Sbjct: 1143 VSASQSTSASVSASQSASASASNNQNSASISVSASQSASISASQSTSASVSASQSASVSA 1202
Query: 436 AKEAIRRRVEAASAATKRAENMDAILKAAELAAEAVS 472
++ A ++ASA+ +++ A + A++ A+ + S
Sbjct: 1203 SQSASVSASQSASASVSASQSTSASVSASQSASASAS 1239
Score = 53.1 bits (126), Expect = 5e-05
Identities = 84/470 (17%), Positives = 185/470 (38%), Gaps = 33/470 (7%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S ++A S S + IS +++S + A+ I S ++SA
Sbjct: 1635 SASVSASQSASASASNNQNSASISVSASQSASISASQSASASISASISASQSASVSASQS 1694
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
S++++ S + + + + +N S S A + S + + +
Sbjct: 1695 ASASVSASQSTSASVSASQSASASASNNQNSASISVSASHSASISASQSASASISASISA 1754
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPP 217
SAS S+ ASV S S+S+ ++S+ S + + A+
Sbjct: 1755 SQSASVSASQSASASVSASQSTSASVSASQSASASASNNQNSASISVSASQSASISASQS 1814
Query: 218 AQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGN------VPMSS 271
A S + +S S + + ++ S S SA+ N + +S+
Sbjct: 1815 ASASISASISASQSASVSASQSASASVSASQSTSASVSASQSASASASNNQNSASISVSA 1874
Query: 272 VEKSVVSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWN 331
+ + +S S + S + + ++ + AS ++A+V+ S
Sbjct: 1875 SQSASISASQSTSASVSASQSASVSASQSASVSASQSASASVSASQSTSASVSASQSASA 1934
Query: 332 QLDKHKNSGFMSDIEAKLA------------SAAVAIAAAAAVAKAAAAAANVASNAAFQ 379
++NS +S + A SA+++ + +A+V+ + +A+A+V+++ +
Sbjct: 1935 SASNNQNSASISVSASHSASISASQSTSASISASISASQSASVSASQSASASVSASQSTS 1994
Query: 380 AKLMADEALISSGYENTSQGNNTFLPEGTSNL--GQATPASILKGANGPNS-------PG 430
A + A +++ +S N + + + ++++ Q+T ASI + S
Sbjct: 1995 ASVSASQSVSASASNNQNSASISVSASHSASISASQSTSASISASISASQSASVSASQSA 2054
Query: 431 SFIVAAKEAIRRRVEAASAATKRAEN------MDAILKAAELAAEAVSQA 474
S V+A ++ V A+ +A+ A N + A + A++ A+ + SQ+
Sbjct: 2055 SASVSASQSTSASVSASQSASASASNNQNSASISASISASQSASVSASQS 2104
Score = 52.0 bits (123), Expect = 1e-04
Identities = 79/432 (18%), Positives = 172/432 (39%), Gaps = 46/432 (10%)
Query: 42 AARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSA 101
A++++ +++ S+ + S +++S + + +S+ + SA + Q+SA
Sbjct: 2028 ASQSTSASISASISASQSASVSASQSASASVSASQSTSASVSAS--QSASASASNNQNSA 2085
Query: 102 LARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSA 161
S+ A + QS S STS A A +N SA
Sbjct: 2086 SISASISASQSA--SVSASQSASASVSASQSTSASVSA-------SQSVSASASNNQNSA 2136
Query: 162 SPSSDTIKLASVKGSLPPSSSIK-DVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQ 220
S S AS+ S S+SI ++ AS S QS + + + +V +Q
Sbjct: 2137 SISVSASHSASISASQSTSASISASISASQSASVSASQSASASVSAS-QSTSASVSASQS 2195
Query: 221 SSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVVSVS 280
+S + + +S Q S + + ++ S S+SA+ S SV +
Sbjct: 2196 ASASASNNQNSASISVSASQSASISASQSASASISASISASQSASVSASQSASASVSASQ 2255
Query: 281 PLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHKNSG 340
+ Q+ ++++ + + H+ SA +A+ +
Sbjct: 2256 STSASVSASQSASASASNNQNSASISVSASHSASISASQSASAS---------------- 2299
Query: 341 FMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENTSQGN 400
SA+++ + +A+V+ + +A+A+V+++ + A + A ++ +S N +
Sbjct: 2300 ---------ISASISASQSASVSASQSASASVSASQSTSASVSASQSASASASNNQN--- 2347
Query: 401 NTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAENMDAI 460
+ ++ + ASI + S + I A++ A ++ASA+ +++ A
Sbjct: 2348 -----SASISVSASQSASISASQSASASISASISASQSASVSASQSASASVSASQSTSAS 2402
Query: 461 LKAAELAAEAVS 472
+ A++ A+ + S
Sbjct: 2403 VSASQSASASAS 2414
Score = 52.0 bits (123), Expect = 1e-04
Identities = 79/432 (18%), Positives = 172/432 (39%), Gaps = 46/432 (10%)
Query: 42 AARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSA 101
A++++ +++ S+ + S +++S + + +S+ + SA + Q+SA
Sbjct: 1476 ASQSTSASISASISASQSASVSASQSASASVSASQSTSASVSAS--QSASASASNNQNSA 1533
Query: 102 LARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSA 161
S+ A + QS S STS A A +N SA
Sbjct: 1534 SISASISASQSA--SVSASQSASASVSASQSTSASVSA-------SQSVSASASNNQNSA 1584
Query: 162 SPSSDTIKLASVKGSLPPSSSIK-DVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQ 220
S S AS+ S S+SI ++ AS S QS + + + +V +Q
Sbjct: 1585 SISVSASHSASISASQSTSASISASISASQSASVSASQSASASVSAS-QSTSASVSASQS 1643
Query: 221 SSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVVSVS 280
+S + + +S Q S + + ++ S S+SA+ S SV +
Sbjct: 1644 ASASASNNQNSASISVSASQSASISASQSASASISASISASQSASVSASQSASASVSASQ 1703
Query: 281 PLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHKNSG 340
+ Q+ ++++ + + H+ SA +A+ +
Sbjct: 1704 STSASVSASQSASASASNNQNSASISVSASHSASISASQSASAS---------------- 1747
Query: 341 FMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENTSQGN 400
SA+++ + +A+V+ + +A+A+V+++ + A + A ++ +S N +
Sbjct: 1748 ---------ISASISASQSASVSASQSASASVSASQSTSASVSASQSASASASNNQN--- 1795
Query: 401 NTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAENMDAI 460
+ ++ + ASI + S + I A++ A ++ASA+ +++ A
Sbjct: 1796 -----SASISVSASQSASISASQSASASISASISASQSASVSASQSASASVSASQSTSAS 1850
Query: 461 LKAAELAAEAVS 472
+ A++ A+ + S
Sbjct: 1851 VSASQSASASAS 1862
Score = 52.0 bits (123), Expect = 1e-04
Identities = 89/457 (19%), Positives = 185/457 (40%), Gaps = 44/457 (9%)
Query: 42 AARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSA 101
A++++ ++V S +S AS A+ + +N S + + + S + QS++
Sbjct: 2172 ASQSASASVSASQSTSASVS-----ASQSASASASNNQNSASISVSASQSASISASQSAS 2226
Query: 102 LARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSA 161
+ + + SQ+ + + QS S STS A A +N SA
Sbjct: 2227 ASISASISASQSAS-VSASQSASASVSASQSTSASVSA-------SQSASASASNNQNSA 2278
Query: 162 SPSSDTIKLASVKGSLPPSSSIK-DVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQ 220
S S AS+ S S+SI ++ AS S QS + + + +V +Q
Sbjct: 2279 SISVSASHSASISASQSASASISASISASQSASVSASQSASASVSAS-QSTSASVSASQS 2337
Query: 221 SSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSV---- 276
+S + + +S Q S + + ++ S S+SA+ S SV
Sbjct: 2338 ASASASNNQNSASISVSASQSASISASQSASASISASISASQSASVSASQSASASVSASQ 2397
Query: 277 -VSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDK 335
S S A Q + + + S+ + A + A ++++ S +A + + +
Sbjct: 2398 STSASVSASQSASASASNNQNSASISVSASQSASISASQSASASISASISASQSASVSAS 2457
Query: 336 HKNSGFMSDIEAKLA-----------------SAAVAIAAAAAVAKAAAAAANVASNAAF 378
S +S ++ A SA+++++A+ + + +A+ +A+ + +A+
Sbjct: 2458 QSASASVSASQSTSASVSASQSASASASNNQNSASISVSASQSASISASQSASASISASI 2517
Query: 379 QAKLMADEALISSGYENTSQGNNTFLPEGTS-NLGQATPASILKGANGPNSPGSFIVAAK 437
A A + S + S +T E S ++ + ASI + S + + A+
Sbjct: 2518 SASQSASVSASQSASASVSASQSTSASESASTSMSNSVSASISASESVSTSMSNSVSASI 2577
Query: 438 EAIRRRVEAASAATKRAENMDAILKAAELAAEAVSQA 474
A + SA+T + ++ A + A++ A+ ++S +
Sbjct: 2578 SA------SVSASTSMSNSVSASISASQSASTSMSNS 2608
Score = 51.2 bits (121), Expect = 2e-04
Identities = 84/454 (18%), Positives = 187/454 (40%), Gaps = 30/454 (6%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S ++A S S + IS ++S + A+ I S ++SA
Sbjct: 2258 SASVSASQSASASASNNQNSASISVSASHSASISASQSASASISASISASQSASVSASQS 2317
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
S++++ S + + + + +N S S A + S + + +
Sbjct: 2318 ASASVSASQSTSASVSASQSASASASNNQNSASISVSASQSASISASQSASASISASISA 2377
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPP 217
SAS S+ ASV S S+S+ S QS + ++ +++V
Sbjct: 2378 SQSASVSASQSASASVSASQSTSASV-----------SASQSASASASNNQNSASISVSA 2426
Query: 218 AQQSSGPKAKKRKKDV-----LSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSV 272
+Q +S ++ + S+ QS + +V++ STS S + +S
Sbjct: 2427 SQSASISASQSASASISASISASQSASVSASQSASASVSASQSTSASVSASQSASASASN 2486
Query: 273 EKSVVSVSPLADQPKNDQTVEKRILS-DESLMKVKEARVHAEEASALSAAAVNHSLELWN 331
++ S+S A Q + + S S+ + A V A ++++ S +A + +
Sbjct: 2487 NQNSASISVSASQSASISASQSASASISASISASQSASVSASQSASASVSASQSTSASES 2546
Query: 332 QLDKHKNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISS 391
NS S ++ S +++ + +A+++ + +A+ +++++ + A + A ++ +S
Sbjct: 2547 ASTSMSNSVSASISASESVSTSMSNSVSASISASVSASTSMSNSVS--ASISASQSASTS 2604
Query: 392 GYENTSQGNNTFLPEGTS---NLGQATPASILKGANGPNSPGSFI---VAAKEAIRRRVE 445
+ S + + TS ++ + AS+ + NS + I V+A ++ V
Sbjct: 2605 MSNSVSASISASVSASTSMSNSVSASISASVSASTSMSNSVSASISASVSASTSMSNSVS 2664
Query: 446 AA-----SAATKRAENMDAILKAAELAAEAVSQA 474
A+ SA+T + ++ A + A+E A+ ++S +
Sbjct: 2665 ASISASVSASTSMSNSVSASISASESASTSMSNS 2698
Score = 51.2 bits (121), Expect = 2e-04
Identities = 90/463 (19%), Positives = 193/463 (41%), Gaps = 37/463 (7%)
Query: 42 AARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLS---SPLWSLPTLSADSLQ 98
A++++ ++V S +S+ ++S + A+ + S S S ++ S+
Sbjct: 853 ASQSASASVSVSQSASASVSASQSASASVSVSQSASASVSASQSTSASVSASQSASASVS 912
Query: 99 SSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTS----WISQAPLRGPWIGSPTPAPD 154
+S A SV A + QS S HS S + + S + +
Sbjct: 913 ASQSASASVSSSHSASASVSVSQSASASVSASHSASVSASQSASVSVSASQSASTSVSAS 972
Query: 155 NNTHLSASPSSDTIKLASVKGSLPPSSSIKD-------VTPGPPASSSGLQSTFVGTDSQ 207
++ +SAS S+ AS+ S S S + AS S QS +
Sbjct: 973 HSESISASQSASASISASISASQSASVSASQSASASVSASQSTSASVSASQSASASASNN 1032
Query: 208 LDANNVTVPPAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNV 267
++ +++V + +S ++ + + QS + +V++ STS S +
Sbjct: 1033 QNSASISVSASHSASISASQSASASISASISAS---QSASASVSASQSTSASVSASQSAS 1089
Query: 268 PMSSVEKSVVSVSPLADQPKNDQTVEKRILS-DESLMKVKEARVHAEEASALS------- 319
+S ++ S+S A Q + + S S+ + A V A ++++ S
Sbjct: 1090 ASASNNQNSASISVSASQSASISASQSASASISASISASQSASVSASQSASASVSASQST 1149
Query: 320 AAAVNHSLELWNQLDKHKNSGFMSDIEAKLAS--------AAVAIAAAAAV--AKAAAAA 369
+A+V+ S ++NS +S ++ AS A+V+ + +A+V +++A+ +
Sbjct: 1150 SASVSASQSASASASNNQNSASISVSASQSASISASQSTSASVSASQSASVSASQSASVS 1209
Query: 370 ANVASNAAFQAKLMADEALISSGYENTSQGNNTFLPEGTSNLGQATPASILKGANGPNSP 429
A+ +++A+ A ++ +S + S NN + ++ + ASI + S
Sbjct: 1210 ASQSASASVSASQSTSASVSASQSASASASNNQ--NSASISVSASQSASISASQSASASI 1267
Query: 430 GSFIVAAKEAIRRRVEAASAATKRAENMDAILKAAELAAEAVS 472
+ I A++ A ++ASA+ +++ A + A++ A+ + S
Sbjct: 1268 SASISASQSASVSASQSASASVSASQSTSASVSASQSASASAS 1310
Score = 44.3 bits (103), Expect = 0.023
Identities = 99/461 (21%), Positives = 191/461 (40%), Gaps = 50/461 (10%)
Query: 42 AARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSA 101
A++++ ++V S +S AS A+ + +N S + + + S + QS++
Sbjct: 2456 ASQSASASVSASQSTSASVS-----ASQSASASASNNQNSASISVSASQSASISASQSAS 2510
Query: 102 LARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSA 161
+ + + SQ+ + + QS S STS A S + + + +SA
Sbjct: 2511 ASISASISASQSAS-VSASQSASASVSASQSTSASESA--------STSMSNSVSASISA 2561
Query: 162 SPSSDTIKLASVKGSLPPS-SSIKDVTPGPPASSSGLQSTFVGTDSQLDAN-NVTVPPAQ 219
S S T SV S+ S S+ ++ AS S QS + + A+ + +V +
Sbjct: 2562 SESVSTSMSNSVSASISASVSASTSMSNSVSASISASQSASTSMSNSVSASISASVSAST 2621
Query: 220 QSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVV-- 277
S + V + S++ +V++ S SVSA+T + N +S+ SV
Sbjct: 2622 SMSNSVSASISASVSAST-------SMSNSVSASISASVSASTSMSNSVSASISASVSAS 2674
Query: 278 -----SVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQ 332
SVS ++ T +S V + + SA +A+V+ S + N
Sbjct: 2675 TSMSNSVSASISASESASTSMSNSVSASISASVSASTSMSNSVSASISASVSASTSMSNS 2734
Query: 333 LD-----KHKNSGFMSDIEAKLASAAVAIAAA-----AAVAKAAAAAANVASNAAFQAKL 382
+ S MS+ + SA+V+ + + +A A+ +A+ SN+ A +
Sbjct: 2735 VSASISASESASTSMSNSVSASISASVSASTSMSNSVSASISASESASTSMSNSV-SASI 2793
Query: 383 MADEALISSGYENTSQGNNTFLPEGTS-NLGQATPASILKGANGPNSPGSFIVAAKE--- 438
A E+ +S + S + S + Q+T AS + NS + I A++
Sbjct: 2794 SASESASTSMSNSVSASISASQSASASVSASQSTSASESASTSMSNSVSASISASQSVST 2853
Query: 439 AIRRRVEAA-----SAATKRAENMDAILKAAELAAEAVSQA 474
++ V A+ SA+T + ++ A + A+ A+ ++S +
Sbjct: 2854 SMSNSVSASISASVSASTSMSNSVSASISASASASTSLSNS 2894
>gb|AAP37755.1| At1g68580 [Arabidopsis thaliana] gi|22135896|gb|AAM91530.1| unknown
protein [Arabidopsis thaliana]
gi|30697711|ref|NP_177025.2| agenet domain-containing
protein / bromo-adjacent homology (BAH)
domain-containing protein [Arabidopsis thaliana]
Length = 648
Score = 60.8 bits (146), Expect = 2e-07
Identities = 44/170 (25%), Positives = 76/170 (43%), Gaps = 13/170 (7%)
Query: 585 TVRNGSENLEENTFKEGSLVEVFKDEEGHKAAWFMGNILSLKDGKVYVCYTSLVAVEG-- 642
++ G E+ + K+GSL+EV ++ G + WF +L KV V Y + +
Sbjct: 350 SIFKGDEDGSSHHIKKGSLIEVLSEDSGIRGCWFKALVLKKHKDKVKVQYQDIQDADDES 409
Query: 643 -PLKEWV--SLECEGDKPPRIR-TARPLTSLQHEGTRKRRRAAMGDYAWSVGDRVDAWIQ 698
L+EW+ S GD +R R + + +++ +G VG VD W
Sbjct: 410 KKLEEWILTSRVAAGDHLGDLRIKGRKVVRPMLKPSKENDVCVIG-----VGMPVDVWWC 464
Query: 699 ESWREGVITEKNKKDETTLTVHIPASGETSVLRAWNLRPSLIWKDGQWLD 748
+ W EG++ + + E V++P + S +LR S W D +WL+
Sbjct: 465 DGWWEGIVVQ--EVSEEKFEVYLPGEKKMSAFHRNDLRQSREWLDDEWLN 512
>gb|AAD49985.1| Contains PF|01426 BAH (bromo-adjacent homology) domain. ESTs
gb|N96349, gb|T42710, gb|H77084, gb|AA395147 and
gb|AA605500 come from this gene. [Arabidopsis thaliana]
gi|25404756|pir||B96710 hypothetical protein F24J5.18
[imported] - Arabidopsis thaliana
Length = 625
Score = 60.8 bits (146), Expect = 2e-07
Identities = 44/170 (25%), Positives = 76/170 (43%), Gaps = 13/170 (7%)
Query: 585 TVRNGSENLEENTFKEGSLVEVFKDEEGHKAAWFMGNILSLKDGKVYVCYTSLVAVEG-- 642
++ G E+ + K+GSL+EV ++ G + WF +L KV V Y + +
Sbjct: 327 SIFKGDEDGSSHHIKKGSLIEVLSEDSGIRGCWFKALVLKKHKDKVKVQYQDIQDADDES 386
Query: 643 -PLKEWV--SLECEGDKPPRIR-TARPLTSLQHEGTRKRRRAAMGDYAWSVGDRVDAWIQ 698
L+EW+ S GD +R R + + +++ +G VG VD W
Sbjct: 387 KKLEEWILTSRVAAGDHLGDLRIKGRKVVRPMLKPSKENDVCVIG-----VGMPVDVWWC 441
Query: 699 ESWREGVITEKNKKDETTLTVHIPASGETSVLRAWNLRPSLIWKDGQWLD 748
+ W EG++ + + E V++P + S +LR S W D +WL+
Sbjct: 442 DGWWEGIVVQ--EVSEEKFEVYLPGEKKMSAFHRNDLRQSREWLDDEWLN 489
>gb|AAN46759.1| At5g55600/MDF20_4 [Arabidopsis thaliana] gi|18087548|gb|AAL58906.1|
AT5g55600/MDF20_4 [Arabidopsis thaliana]
gi|22327858|ref|NP_200371.2| agenet domain-containing
protein / bromo-adjacent homology (BAH)
domain-containing protein [Arabidopsis thaliana]
Length = 663
Score = 59.3 bits (142), Expect = 7e-07
Identities = 61/287 (21%), Positives = 105/287 (36%), Gaps = 36/287 (12%)
Query: 599 KEGSLVEVFKDEEGHKAAWFMGNILSLKDGKVYVCYTSLVAVEG--PLKEWVSLECEGDK 656
K + +E + G + WF +L + +V + Y + +G L+EWV
Sbjct: 382 KTDAKIEFLCQDSGIRGCWFRCTVLDVSRKQVKLQYDDIEDEDGYGNLEEWV-------- 433
Query: 657 PPRIRTARPLTSLQHEGTRKRRRAAMGD-----YAWSVGDRVDAWIQESWREGVITEKNK 711
P ++A P R R A D + ++G+ VDAW + W EGV+ K
Sbjct: 434 -PAFKSAMPDKLGIRLSNRPTIRPAPRDVKTAYFDLTIGEAVDAWWNDGWWEGVVIATGK 492
Query: 712 KDETTLTVHIPASGETSVLRAWNLRPSLIWKDGQWLDFSKVGANDSSTHKGDTPHEKRPK 771
D L ++IP + ++R S W W+D + + K
Sbjct: 493 PDTEDLKIYIPGENLCLTVLRKDIRISRDWVGDSWVDIDPKPEILAIVSSDSSSESKLSM 552
Query: 772 LGSNAVEVKGKDKMSKNIDAAESANPDEMRSLNLTENEIVFNIGKSSTNESKQDPQRQVR 831
L + + + K K NI E A P+ ++ N +G+ + +
Sbjct: 553 LSTLSKDTKAKPSAMPNI--VEEAEPEGEKAYNSL-------LGEQNKEHKDVVVKEDDE 603
Query: 832 SGLQKEGSKVIFGVPKPGKKRKFMEVSKHYVAHGSSKVNDKNDSVKI 878
+ L KE K + +K YV H ++ + K D V +
Sbjct: 604 NKLSKEEDKEVGS-----------NETKTYVNHENTVEDHKEDDVTL 639
>emb|CAD79181.1| hypothetical protein-signal peptide prediction [Rhodopirellula
baltica SH 1] gi|32477034|ref|NP_870028.1| hypothetical
protein-signal peptide prediction [Rhodopirellula
baltica SH 1]
Length = 669
Score = 59.3 bits (142), Expect = 7e-07
Identities = 96/447 (21%), Positives = 178/447 (39%), Gaps = 33/447 (7%)
Query: 3 SAYGGTGTCGRMSGVSAWKGSVVKNLILIPQKHLCSQDLAARTSDSTVKQSVLQGKGISS 62
S+ G+ + S S+ S + H S ++ +SDS+ S SS
Sbjct: 105 SSSSGSSSSSSKSSSSSDSSSNSSSSDSSDSSHSSSASSSSDSSDSSTSSSSSVSSSQSS 164
Query: 63 PLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSALARGSVVDYSQALTPLHPYQS 122
+S +T T + P + SS + S+ S SS+ ++ S S +
Sbjct: 165 DSSDSSDSSTST-SLPSVSSSSSSDGSSSSSSGSSDSSSNSQSSDSSNSDSSDSSDSTSL 223
Query: 123 PSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSASPSSDTIKLASVKGSLPPSSS 182
PS G S+S S A S + + D++ S S SSD+I L S+ GS + S
Sbjct: 224 PS---ISGSSSSDGSSASSDSSDSSSNSQSSDSSNSES-SDSSDSISLPSISGSSSSAGS 279
Query: 183 IKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQSSGPKAKKRKKDVLSEDHGQKL 242
+ +SS+ S ++S + +V++P SS + + S D G
Sbjct: 280 SSASSDSSNSSSNSQSSDSSTSESNDSSASVSLPSVSGSSSSASDSSSSESQSSDSG--- 336
Query: 243 LQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVVSVSPLADQPKN---DQTVEKRILSD 299
+ + +S + +S SA+TP + SS +S S S ++ + T E R S+
Sbjct: 337 ----SSSDSSTSQSSESASTPSVSSSQSSTSESASSTSESSNSESSQSESSTSESRSRSE 392
Query: 300 ESLMKVKEARVHAEEASALSAAAVNHSLELWNQ--------LDKHKNSGFMSDIEAKLAS 351
+ ++ +S + + + E +Q D +S S I+ S
Sbjct: 393 SDSESLPSYSWNSSTSSDSDSESSDSESESQSQSDSTSQSVSDSGSDSQSESSIDRSSDS 452
Query: 352 AAVAIAAAAAVAKAAAAAANVASNA----------AFQAKLMADEALISSGYENTSQGNN 401
+ + +A+A+ + +A+A+ + + +A + + A E+ SG ++ S+ +
Sbjct: 453 QSYSASASASYSDSASASGSQSGSASGSGSNSGSDSGSGSVSASESDSGSGSDSDSRSTS 512
Query: 402 TFLPEGTSNLGQATPASILKGANGPNS 428
T E S +T A +G +S
Sbjct: 513 TSDSESDSRSDDSTSAPTSSSGSGSDS 539
Score = 43.1 bits (100), Expect = 0.051
Identities = 98/455 (21%), Positives = 187/455 (40%), Gaps = 38/455 (8%)
Query: 28 LILIPQKHLCSQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLW 87
L +I QK S +A +S S+ S SSP +SS ++ + ++ SS
Sbjct: 2 LFIILQK---SPPVAGSSSSSSSDSS--SSSSSSSP---SSSSSSGSSSSSSGSSSSSSM 53
Query: 88 SLPTLSADSLQSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIG 147
S + S+ S SS+ S S + + S S + S+S S + G
Sbjct: 54 SGSSSSSMSSSSSSSLSSSSSSSSGSSSSSSGSSSSSSGSPSSSSSSGSSSSSSSGSSSS 113
Query: 148 S--PTPAPDNNTHLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTD 205
S + + D++++ S+S SSD +S S SS D + +S S QS+ +D
Sbjct: 114 SSKSSSSSDSSSNSSSSDSSD----SSHSSSASSSSDSSDSSTSSSSSVSSSQSS-DSSD 168
Query: 206 SQLDANNVTVPPAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVG 265
S + + ++P SS S+ + + +S +S S S + G
Sbjct: 169 SSDSSTSTSLPSVSSSSSSDGSSSSSSGSSDSSSNSQSSDSSNSDSSDSSDSTSLPSISG 228
Query: 266 NVPMSSVEKSVVSVSPLADQPKNDQTVE-KRILSDESLMKVKEARVHAEEASALSAAAVN 324
+ SS + S S S +D N Q+ + S +S + + +SA S++A +
Sbjct: 229 S---SSSDGSSAS-SDSSDSSSNSQSSDSSNSESSDSSDSISLPSISGSSSSAGSSSASS 284
Query: 325 HSLELWNQLDKHKNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMA 384
S + + S S E+ +SA+V++ + + + +A+ +++ S ++
Sbjct: 285 DS----SNSSSNSQSSDSSTSESNDSSASVSLPSVSGSSSSASDSSSSESQSSDSG---- 336
Query: 385 DEALISSGYENTSQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRV 444
SS +TSQ + + S+ +T S + NS S ++ R R
Sbjct: 337 -----SSSDSSTSQSSESASTPSVSSSQSSTSESASSTSESSNSESSQSESSTSESRSRS 391
Query: 445 EAASAAT-----KRAENMDAILKAAELAAEAVSQA 474
E+ S + + + D+ ++++ +E+ SQ+
Sbjct: 392 ESDSESLPSYSWNSSTSSDSDSESSDSESESQSQS 426
Score = 39.7 bits (91), Expect = 0.56
Identities = 92/503 (18%), Positives = 201/503 (39%), Gaps = 34/503 (6%)
Query: 62 SPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSALARGSVVDYSQALTPLHPYQ 121
SP SS ++ + ++ SSP S + S S SS + S + S + +
Sbjct: 9 SPPVAGSSSSSSSDSSSSSSSSSP--SSSSSSGSSSSSSGSSSSSSMSGSSSSS-----M 61
Query: 122 SPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSASPSSDTIKLASVKGSLPPSS 181
S S + L S+S S GS + + +++ S SPSS + +S S SS
Sbjct: 62 SSSSSSSLSSSSSSSS---------GSSSSSSGSSSSSSGSPSSSSSSGSSSSSSSGSSS 112
Query: 182 SIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQSSGPKAKKRKKDVLSEDHGQK 241
S + +SS+ S + A++ + +S + + S D
Sbjct: 113 SSSKSSSSSDSSSNSSSSDSSDSSHSSSASSSSDSSDSSTSSSSSVSSSQSSDSSDSSDS 172
Query: 242 LLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVVSVSPLADQPKNDQTVEKRILSDES 301
+ P+V+S +S+ S+++ G+ SS +S S S +D + + +S S
Sbjct: 173 STSTSLPSVSSSSSSDGSSSSSSGSSDSSSNSQS--SDSSNSDSSDSSDSTSLPSISGSS 230
Query: 302 LMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHKNSGFMSDIEAKLASAAVAIAAAAA 361
A + ++S+ S ++ + + E + D +S+A + +A++
Sbjct: 231 SSDGSSASSDSSDSSSNSQSSDSSNSESSDSSDSISLPSISGS-----SSSAGSSSASSD 285
Query: 362 VAKAAAAAANVASNAAFQAKLMADEALISSGYENTSQGNNTFLPEGTSNLGQATPASILK 421
+ +++ + + S+ + A +L S ++S +++ +S+ G ++ +S +
Sbjct: 286 SSNSSSNSQSSDSSTSESNDSSASVSLPSVSGSSSSASDSSSSESQSSDSGSSSDSSTSQ 345
Query: 422 GANGPNSPGSFIVAAKEAIRRRVEAASAATKRAENMDAILKAAELAAEAVSQAGKIVTMG 481
+ ++P V++ ++ ++++ + +E+ + +E + + S +
Sbjct: 346 SSESASTPS---VSSSQSSTSESASSTSESSNSESSQSESSTSESRSRSESDS------- 395
Query: 482 DPLPLIELIEAGPEGCWKASRESSREVGLLKDMTRDLVNIDMVRDIPETSHAQNRDILSS 541
+ LP + S +S E D T V+ E+S ++ D S
Sbjct: 396 ESLPSYSWNSSTSSDSDSESSDSESESQSQSDSTSQSVSDSGSDSQSESSIDRSSDSQSY 455
Query: 542 EISASIMINEK-NTRGQQARTVS 563
SAS ++ + G Q+ + S
Sbjct: 456 SASASASYSDSASASGSQSGSAS 478
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.308 0.125 0.347
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,827,743,130
Number of Sequences: 2540612
Number of extensions: 78856902
Number of successful extensions: 293023
Number of sequences better than 10.0: 1607
Number of HSP's better than 10.0 without gapping: 55
Number of HSP's successfully gapped in prelim test: 1647
Number of HSP's that attempted gapping in prelim test: 283695
Number of HSP's gapped (non-prelim): 6190
length of query: 1091
length of database: 863,360,394
effective HSP length: 139
effective length of query: 952
effective length of database: 510,215,326
effective search space: 485724990352
effective search space used: 485724990352
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 81 (35.8 bits)
Medicago: description of AC137077.8


