

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136953.11 + phase: 0 /pseudo
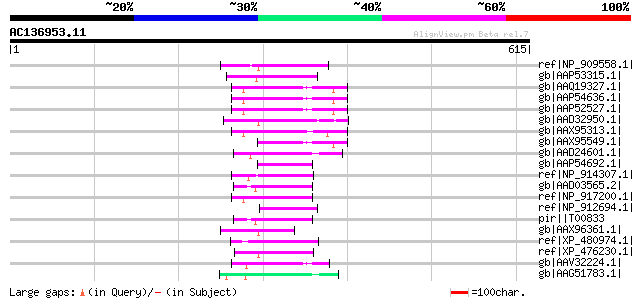
(615 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_909558.1| putative reverse transcriptase [Oryza sativa] g... 69 6e-10
gb|AAP53315.1| putative reverse transcriptase [Oryza sativa (jap... 67 1e-09
gb|AAQ19327.1| bZIP-like protein [Oryza sativa (japonica cultiva... 63 2e-08
gb|AAP54636.1| putative reverse transcriptase [Oryza sativa (jap... 63 2e-08
gb|AAP52527.1| putative reverse transcriptase. [Oryza sativa (ja... 63 2e-08
gb|AAD32950.1| putative non-LTR retroelement reverse transcripta... 61 1e-07
gb|AAX95313.1| hypothetical protein [Oryza sativa (japonica cult... 60 3e-07
gb|AAX95549.1| hypothetical protein [Oryza sativa (japonica cult... 59 6e-07
gb|AAD24601.1| putative non-LTR retroelement reverse transcripta... 57 1e-06
gb|AAP54692.1| putative reverse transcriptase [Oryza sativa (jap... 57 1e-06
ref|NP_914307.1| reverse transcriptase -like protein [Oryza sati... 57 1e-06
gb|AAD03565.2| putative non-LTR retroelement reverse transcripta... 57 2e-06
ref|NP_917200.1| P0707D10.17 [Oryza sativa (japonica cultivar-gr... 57 2e-06
ref|NP_912694.1| hypothetical protein [Oryza sativa (japonica cu... 57 2e-06
pir||T00833 RNA-directed DNA polymerase homolog T13L16.7 - Arabi... 57 2e-06
gb|AAX96361.1| hypothetical protein LOC_Os11g08530 [Oryza sativa... 57 2e-06
ref|XP_480974.1| cyst nematode resistance protein-like protein [... 56 3e-06
ref|XP_476230.1| hypothetical protein [Oryza sativa (japonica cu... 56 3e-06
gb|AAV32224.1| hypothetical protein [Oryza sativa (japonica cult... 56 3e-06
gb|AAG51783.1| reverse transcriptase, putative; 16838-20266 [Ara... 56 4e-06
>ref|NP_909558.1| putative reverse transcriptase [Oryza sativa]
gi|14018103|gb|AAK52166.1| putative reverse
transcriptase [Oryza sativa]
Length = 1185
Score = 68.6 bits (166), Expect = 6e-10
Identities = 40/135 (29%), Positives = 57/135 (41%), Gaps = 9/135 (6%)
Query: 251 IPFGNHDVPV*KFERDGAYSVKSAYTNILNHDVAVVHHRVPDN--------WTCIWSLKF 302
I G D R G +SV+SAY N R+PD W +W L+
Sbjct: 810 ISAGREDFVAWHHTRSGIFSVRSAYHCQWNAKFGS-KERLPDGASSSSRPVWDNLWKLEI 868
Query: 303 PSKVKIFLWRACRNCLPTRIRLQSKGVQCTDKCAVRDDFAEDSTHLFFMCNKSMICWQRS 362
P K+KIF WR +P + L K + C V ED H+ F C ++ + W++
Sbjct: 869 PPKIKIFAWRMLHGVIPCKGILADKHIAVQGGCPVCSSGVEDIKHMVFTCRRAKLIWKQL 928
Query: 363 GLWILLMAIFYIDLS 377
G+W + I D S
Sbjct: 929 GIWSRIQPILGFDRS 943
>gb|AAP53315.1| putative reverse transcriptase [Oryza sativa (japonica
cultivar-group)] gi|37533452|ref|NP_921028.1| putative
reverse transcriptase [Oryza sativa (japonica
cultivar-group)] gi|20279456|gb|AAM18736.1| putative
reverse transcriptase [Oryza sativa (japonica
cultivar-group)]
Length = 1509
Score = 67.4 bits (163), Expect = 1e-09
Identities = 38/118 (32%), Positives = 51/118 (43%), Gaps = 10/118 (8%)
Query: 257 DVPV*KFERDGAYSVKSAYTNILNHDVAVVHHRVP----------DNWTCIWSLKFPSKV 306
DV F+ G ++VKSAY + + P D W +W L P K+
Sbjct: 1200 DVLAWHFDARGCFTVKSAYKVQREMERRASRNGCPGVSNWESGDDDFWKKLWKLGVPGKI 1259
Query: 307 KIFLWRACRNCLPTRIRLQSKGVQCTDKCAVRDDFAEDSTHLFFMCNKSMICWQRSGL 364
K FLWR C N L R L +G+ +C + + ED+ HLFF C WQ L
Sbjct: 1260 KHFLWRMCHNTLALRANLHHRGMDVDTRCVMCGRYNEDAGHLFFKCKPVKKVWQALNL 1317
>gb|AAQ19327.1| bZIP-like protein [Oryza sativa (japonica cultivar-group)]
Length = 2367
Score = 63.2 bits (152), Expect = 2e-08
Identities = 40/148 (27%), Positives = 70/148 (47%), Gaps = 17/148 (11%)
Query: 264 ERDGAYSVKSAY---TNILNHDVAVVH--HRVPDNWTCIWSLKFPSKVKIFLWRACRNCL 318
+++G +SV+SAY ++N + + + + W IW K P KVKIF WR NCL
Sbjct: 1740 DKNGMFSVRSAYRLAAQLVNIEESSSSGTNNINKAWEMIWKCKVPQKVKIFAWRVASNCL 1799
Query: 319 PTRIRLQSKGVQCTDKCAVRDDFAEDSTHLFFMCNKSMICWQRSGLWILLMAIFYIDLSF 378
T + + + ++ +D C + D ED H +C C Q S LW + + +
Sbjct: 1800 ATMVNKKKRKLEQSDMCQICDRENEDDAHA--LCR----CIQASQLWSCMHKSGSVSVDI 1853
Query: 379 PTNV------FAILQHLDQQQKQVFSVT 400
+V F L+ + + ++ +F +T
Sbjct: 1854 KASVLGRFWLFDCLEKIPEYEQAMFLMT 1881
>gb|AAP54636.1| putative reverse transcriptase [Oryza sativa (japonica
cultivar-group)] gi|37536094|ref|NP_922349.1| putative
reverse transcriptase [Oryza sativa (japonica
cultivar-group)] gi|13786450|gb|AAK39575.1| putative
reverse transcriptase [Oryza sativa]
Length = 791
Score = 63.2 bits (152), Expect = 2e-08
Identities = 40/148 (27%), Positives = 70/148 (47%), Gaps = 17/148 (11%)
Query: 264 ERDGAYSVKSAY---TNILNHDVAVVH--HRVPDNWTCIWSLKFPSKVKIFLWRACRNCL 318
+++G +SV+SAY ++N + + + + W IW K P KVKIF WR NCL
Sbjct: 416 DKNGMFSVRSAYRLAAQLVNIEESSSSGTNNINKAWEMIWKCKVPQKVKIFAWRVASNCL 475
Query: 319 PTRIRLQSKGVQCTDKCAVRDDFAEDSTHLFFMCNKSMICWQRSGLWILLMAIFYIDLSF 378
T + + + ++ +D C + D ED H +C C Q S LW + + +
Sbjct: 476 ATMVNKKKRKLEQSDMCQICDRENEDDAHA--LCR----CIQASQLWSCMHKSGSVSVDI 529
Query: 379 PTNV------FAILQHLDQQQKQVFSVT 400
+V F L+ + + ++ +F +T
Sbjct: 530 KASVLGRFWLFDCLEKIPEYEQAMFLMT 557
>gb|AAP52527.1| putative reverse transcriptase. [Oryza sativa (japonica
cultivar-group)] gi|37531876|ref|NP_920240.1| putative
reverse transcriptase. [Oryza sativa (japonica
cultivar-group)]
Length = 493
Score = 63.2 bits (152), Expect = 2e-08
Identities = 40/148 (27%), Positives = 70/148 (47%), Gaps = 17/148 (11%)
Query: 264 ERDGAYSVKSAY---TNILNHDVAVVH--HRVPDNWTCIWSLKFPSKVKIFLWRACRNCL 318
+++G +SV+SAY ++N + + + + W IW K P KVKIF WR NCL
Sbjct: 118 DKNGMFSVRSAYRLAAQLVNIEESSSSGTNNINKAWEMIWKCKVPQKVKIFAWRVASNCL 177
Query: 319 PTRIRLQSKGVQCTDKCAVRDDFAEDSTHLFFMCNKSMICWQRSGLWILLMAIFYIDLSF 378
T + + + ++ +D C + D ED H +C C Q S LW + + +
Sbjct: 178 ATMVNKKKRKLEQSDMCQICDRENEDDAHA--LCR----CIQASQLWSCMHKSGSVSVDI 231
Query: 379 PTNV------FAILQHLDQQQKQVFSVT 400
+V F L+ + + ++ +F +T
Sbjct: 232 KASVLGRFWLFDCLEKIPEYEQAMFLMT 259
>gb|AAD32950.1| putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana] gi|25411805|pir||C84554 hypothetical protein
At2g17610 [imported] - Arabidopsis thaliana
Length = 773
Score = 60.8 bits (146), Expect = 1e-07
Identities = 47/156 (30%), Positives = 71/156 (45%), Gaps = 11/156 (7%)
Query: 254 GNHDVPV*KFERDGAYSVKSAYTNILNHDVAVVHHRVPDN--------WTCIWSLKFPSK 305
G +D + DG YSVKS Y ++L H +P +T IW P K
Sbjct: 451 GANDAITWIYTHDGNYSVKSGY-HLLRKLSQQQHASLPSPNEVSAQTVFTNIWKQNAPPK 509
Query: 306 VKIFLWRACRNCLPTRIRLQSKGVQCTDKCAVRDDFAEDSTHLFFMCNKSMICWQRSGLW 365
+K F WR+ N LPT L+ + + D C + +ED HL F C S W+++ +
Sbjct: 510 IKHFWWRSAHNALPTAGNLKRRRLITDDTCQRCGEASEDVNHLLFQCRVSKEIWEQAHI- 568
Query: 366 ILLMAIFYIDLSFPTNVFAILQHLDQQQKQVFSVTP 401
L + SF N+ +I Q L+Q ++ S+ P
Sbjct: 569 KLCPGDSLMSNSFNQNLESI-QKLNQSARKDVSLFP 603
>gb|AAX95313.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 1063
Score = 59.7 bits (143), Expect = 3e-07
Identities = 40/148 (27%), Positives = 66/148 (44%), Gaps = 17/148 (11%)
Query: 264 ERDGAYSVKSAY---TNILNHDVAVVHHRV--PDNWTCIWSLKFPSKVKIFLWRACRNCL 318
+R G +S++SAY T++ D + V W C+ K KVKI W+A N L
Sbjct: 688 DRLGRFSIRSAYHLATSLAESDSSSSSSGVILSKTWDCLRKCKVLQKVKIHAWKAASNYL 747
Query: 319 PTRIRLQSKGVQCTDKCAVRDDFAEDSTHLFFMCNKSMICWQRSGLWILLMAIFYID--- 375
PT + + ++ ++ C + ED+ H + C Q LWIL+ + Y+D
Sbjct: 748 PTMENKKKRNLEASEVCKICGIEKEDTIHALYRCP------QARQLWILMRDVIYLDPAV 801
Query: 376 ---LSFPTNVFAILQHLDQQQKQVFSVT 400
+ P V +L L + + F +T
Sbjct: 802 VGRCTGPNRVLDLLMLLPEDNRMFFLMT 829
>gb|AAX95549.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 472
Score = 58.5 bits (140), Expect = 6e-07
Identities = 33/113 (29%), Positives = 52/113 (45%), Gaps = 12/113 (10%)
Query: 294 WTCIWSLKFPSKVKIFLWRACRNCLPTRIRLQSKGVQCTDKCAVRDDFAEDSTHLFFMCN 353
W IW K P KVKIF WR NCL T + + + ++ +D C + D ED H +C
Sbjct: 132 WEMIWKCKVPQKVKIFAWRVASNCLATMVNKKKRKLEQSDMCQICDRENEDDAHA--LCR 189
Query: 354 KSMICWQRSGLWILLMAIFYIDLSFPTNV------FAILQHLDQQQKQVFSVT 400
C Q S LW + + + +V F L+ + + ++ +F +T
Sbjct: 190 ----CIQASQLWSCMHKSGSVSVDIKASVLGRFWLFDCLEKIPEYEQAMFLMT 238
>gb|AAD24601.1| putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana] gi|25411729|pir||H84542 hypothetical protein
At2g16680 [imported] - Arabidopsis thaliana
Length = 1319
Score = 57.4 bits (137), Expect = 1e-06
Identities = 37/135 (27%), Positives = 60/135 (44%), Gaps = 13/135 (9%)
Query: 266 DGAYSVKSAYTNILNHDV------AVVHHRVPDNWTCIWSLKFPSKVKIFLWRACRNCLP 319
+G Y+VKS Y A V+ V + +WSL+ K+K+F+W+A + L
Sbjct: 961 NGLYTVKSGYERSSRETFKNLFKEADVYPSVNPLFDKVWSLETVPKIKVFMWKALKGALA 1020
Query: 320 TRIRLQSKGVQCTDKCAVRDDFAEDSTHLFFMCNKSMICWQRSGLWILLMAIFYIDLSFP 379
RL+S+G++ D C + E HL F C + W L I F
Sbjct: 1021 VEDRLRSRGIRTADGCLFCKEEIETINHLLFQCPFARQVW-------ALSLIQAPATGFG 1073
Query: 380 TNVFAILQHLDQQQK 394
T++F+ + H+ Q +
Sbjct: 1074 TSIFSNINHVIQNSQ 1088
>gb|AAP54692.1| putative reverse transcriptase [Oryza sativa (japonica
cultivar-group)] gi|37536206|ref|NP_922405.1| putative
reverse transcriptase [Oryza sativa (japonica
cultivar-group)] gi|27311287|gb|AAO00713.1|
retrotransposon protein, putative, unclassified [Oryza
sativa (japonica cultivar-group)]
Length = 1557
Score = 57.4 bits (137), Expect = 1e-06
Identities = 24/66 (36%), Positives = 34/66 (51%)
Query: 294 WTCIWSLKFPSKVKIFLWRACRNCLPTRIRLQSKGVQCTDKCAVRDDFAEDSTHLFFMCN 353
W +W PSKVKIF WRA NCLPT + + ++ +D C + ED+ H C
Sbjct: 1410 WELLWKCNVPSKVKIFTWRATSNCLPTWDNKKKRNLEISDTCVICGMEKEDTMHALCRCP 1469
Query: 354 KSMICW 359
++ W
Sbjct: 1470 QAKHLW 1475
>ref|NP_914307.1| reverse transcriptase -like protein [Oryza sativa (japonica
cultivar-group)]
Length = 727
Score = 57.4 bits (137), Expect = 1e-06
Identities = 34/110 (30%), Positives = 49/110 (43%), Gaps = 13/110 (11%)
Query: 263 FERDGAYSVKSAYTNILNH------------DVAVVHHRVPDNWTCIWSLKFPSKVKIFL 310
F+ G YSVKSAY + + V+ R + W IWSL+ KVK F+
Sbjct: 448 FDTRGMYSVKSAYHVLEDQRERSAKKQEGGSSSGAVNQRNFE-WDKIWSLQCIPKVKQFI 506
Query: 311 WRACRNCLPTRIRLQSKGVQCTDKCAVRDDFAEDSTHLFFMCNKSMICWQ 360
WR N LP ++ ++ + C V F E+ H F C +CW+
Sbjct: 507 WRLAHNSLPLKLNIKRRVPDAETLCPVCKRFDENGGHCFLQCKPMKLCWR 556
>gb|AAD03565.2| putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana] gi|25411819|pir||H84557 hypothetical protein
At2g17910 [imported] - Arabidopsis thaliana
Length = 1344
Score = 56.6 bits (135), Expect = 2e-06
Identities = 32/104 (30%), Positives = 49/104 (46%), Gaps = 14/104 (13%)
Query: 266 DGAYSVKSAYTNILNHDVAVVHHR----------VPDNWTCIWSLKFPSKVKIFLWRACR 315
+G YSVK+ Y + VHHR V + IW+L K++IFLW+A
Sbjct: 987 NGLYSVKTGYEFLSKQ----VHHRLYQEAKVKPSVNSLFDKIWNLHTAPKIRIFLWKALH 1042
Query: 316 NCLPTRIRLQSKGVQCTDKCAVRDDFAEDSTHLFFMCNKSMICW 359
+P RL+++G++ D C + D E H+ F C + W
Sbjct: 1043 GAIPVEDRLRTRGIRSDDGCLMCDTENETINHILFECPLARQVW 1086
>ref|NP_917200.1| P0707D10.17 [Oryza sativa (japonica cultivar-group)]
Length = 892
Score = 56.6 bits (135), Expect = 2e-06
Identities = 32/101 (31%), Positives = 48/101 (46%), Gaps = 5/101 (4%)
Query: 264 ERDGAYSVKSAY---TNILNHDVAVVHH--RVPDNWTCIWSLKFPSKVKIFLWRACRNCL 318
++ G +SV+SAY N++N + + R W IW P KV+IF WRA NCL
Sbjct: 517 DKHGKFSVRSAYGLACNLVNMETSSSSSSARCKRMWNLIWQTNAPQKVRIFAWRAATNCL 576
Query: 319 PTRIRLQSKGVQCTDKCAVRDDFAEDSTHLFFMCNKSMICW 359
T + ++ +D C V ED+ H+ C + W
Sbjct: 577 ATMENKAKRKLEQSDICLVCGLEKEDTKHVLCRCPDARNLW 617
>ref|NP_912694.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|24060082|dbj|BAC21533.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 195
Score = 56.6 bits (135), Expect = 2e-06
Identities = 22/68 (32%), Positives = 38/68 (55%)
Query: 297 IWSLKFPSKVKIFLWRACRNCLPTRIRLQSKGVQCTDKCAVRDDFAEDSTHLFFMCNKSM 356
+W P KVKIF+WRA NCLPT + + ++ ++ C++ ED+ H + C +
Sbjct: 2 VWKCNVPQKVKIFVWRAVTNCLPTLENKKKRKLEVSNVCSICGVETEDTGHALYRCPHAC 61
Query: 357 ICWQRSGL 364
+ W+ G+
Sbjct: 62 LFWEAMGV 69
>pir||T00833 RNA-directed DNA polymerase homolog T13L16.7 - Arabidopsis thaliana
(fragment)
Length = 1365
Score = 56.6 bits (135), Expect = 2e-06
Identities = 32/104 (30%), Positives = 49/104 (46%), Gaps = 14/104 (13%)
Query: 266 DGAYSVKSAYTNILNHDVAVVHHR----------VPDNWTCIWSLKFPSKVKIFLWRACR 315
+G YSVK+ Y + VHHR V + IW+L K++IFLW+A
Sbjct: 1008 NGLYSVKTGYEFLSKQ----VHHRLYQEAKVKPSVNSLFDKIWNLHTAPKIRIFLWKALH 1063
Query: 316 NCLPTRIRLQSKGVQCTDKCAVRDDFAEDSTHLFFMCNKSMICW 359
+P RL+++G++ D C + D E H+ F C + W
Sbjct: 1064 GAIPVEDRLRTRGIRSDDGCLMCDTENETINHILFECPLARQVW 1107
>gb|AAX96361.1| hypothetical protein LOC_Os11g08530 [Oryza sativa (japonica
cultivar-group)] gi|62733013|gb|AAX95132.1| hypothetical
protein [Oryza sativa (japonica cultivar-group)]
Length = 371
Score = 56.6 bits (135), Expect = 2e-06
Identities = 31/98 (31%), Positives = 47/98 (47%), Gaps = 11/98 (11%)
Query: 251 IPFGNHDVPV*KFERDGAYSVKSAYTNILNHDVAVVHHRVPDN-----------WTCIWS 299
+ G D F+ G +SVKSAY + + + +V ++ W IWS
Sbjct: 42 VRLGFEDTAAWHFDPRGRFSVKSAYHTLEDQRERLSLRQVGESSSVSSGVAKLEWGRIWS 101
Query: 300 LKFPSKVKIFLWRACRNCLPTRIRLQSKGVQCTDKCAV 337
L +P KVK FLWR N LP R+ + +G++ +C V
Sbjct: 102 LHYPPKVKHFLWRLAHNSLPLRLNIDRRGLEVDTRCPV 139
>ref|XP_480974.1| cyst nematode resistance protein-like protein [Oryza sativa
(japonica cultivar-group)] gi|40253728|dbj|BAD05668.1|
cyst nematode resistance protein-like protein [Oryza
sativa (japonica cultivar-group)]
gi|40253549|dbj|BAD05496.1| cyst nematode resistance
protein-like protein [Oryza sativa (japonica
cultivar-group)]
Length = 184
Score = 56.2 bits (134), Expect = 3e-06
Identities = 34/105 (32%), Positives = 48/105 (45%), Gaps = 6/105 (5%)
Query: 262 KFERDGAYSVKSAYTNILNHDVAVVHHRVPDNWTCIWSLKFPSKVKIFLWRACRNCLPTR 321
K DG +SV SAY D+ + V + +W K PSKV+ FLW A ++ T
Sbjct: 21 KLTPDGFFSVSSAY------DLFFMAREVSLSGQLVWQTKAPSKVRFFLWLATKSRCLTA 74
Query: 322 IRLQSKGVQCTDKCAVRDDFAEDSTHLFFMCNKSMICWQRSGLWI 366
L +G D+C + ED HLF C+ + W+ WI
Sbjct: 75 DNLAKRGWPHQDQCVLCQRQQEDCLHLFVSCDYTKRVWRLLRDWI 119
>ref|XP_476230.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|46485866|gb|AAS98491.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 488
Score = 56.2 bits (134), Expect = 3e-06
Identities = 30/105 (28%), Positives = 47/105 (44%), Gaps = 11/105 (10%)
Query: 267 GAYSVKSAYTNILNHDVAVVHHRVPDN-----------WTCIWSLKFPSKVKIFLWRACR 315
G ++VKSAY + + + + ++ W +W L + KVK FLWR
Sbjct: 170 GIFTVKSAYHTLEDQREHLASRQTGESSSGVARTASLDWGKLWRLHYFPKVKHFLWRFAH 229
Query: 316 NCLPTRIRLQSKGVQCTDKCAVRDDFAEDSTHLFFMCNKSMICWQ 360
N LP R+ + +G++ +C V ED H FF C W+
Sbjct: 230 NTLPLRMNIDRRGMEIDTRCPVCHRLNEDGGHCFFKCKYVKAGWR 274
>gb|AAV32224.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|45267888|gb|AAS55787.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 1936
Score = 56.2 bits (134), Expect = 3e-06
Identities = 35/122 (28%), Positives = 57/122 (46%), Gaps = 12/122 (9%)
Query: 264 ERDGAYSVKSAYTNIL-----NHDVAVVHHRVPDNWTCIWSLKFPSKVKIFLWRACRNCL 318
++ G +SV+SAY L N+ + R+ +W IW P KV+IF WR N L
Sbjct: 1646 DKTGRFSVRSAYKLALQLADMNNCSSSSSSRLNKSWELIWKCNVPQKVRIFAWRVASNSL 1705
Query: 319 PTRIRLQSKGVQCTDKCAVRDDFAEDSTHLFFMCNKSMICWQRSGLWI-LLMAIFYIDLS 377
T + + ++ D C + D ED+ H +C C + LW+ L + + ++
Sbjct: 1706 ATMENKKKRNLERFDVCGICDREKEDAGHA--LCR----CVHANSLWVNLEKGKYVVSVN 1759
Query: 378 FP 379
FP
Sbjct: 1760 FP 1761
>gb|AAG51783.1| reverse transcriptase, putative; 16838-20266 [Arabidopsis thaliana]
gi|25405244|pir||E96519 probable reverse transcriptase,
16838-20266 [imported] - Arabidopsis thaliana
Length = 1142
Score = 55.8 bits (133), Expect = 4e-06
Identities = 41/148 (27%), Positives = 60/148 (39%), Gaps = 14/148 (9%)
Query: 249 SIIPFGN---HDVPV*KFERDGAYSVKSAYTNI---LNHDVAVVHHRVPDNWTCIWSLKF 302
S +P GN D F + G Y+VKS Y LN ++ + IW ++
Sbjct: 774 SALPIGNPNMEDTLGWHFTKAGNYTVKSGYHTARLDLNEGTTLIGPDLTTLKAYIWKVQC 833
Query: 303 PSKVKIFLWRACRNCLPTRIRLQSKGVQCTDKCAVRDDFAEDSTHLFFMCNKSMICWQRS 362
P K++ FLW+ C+P L+ +G+ C C E H F C+ + W
Sbjct: 834 PPKLRHFLWQILSGCVPVSENLRKRGILCDKGCVSCGASEESINHTLFQCHPARQIW--- 890
Query: 363 GLWILLMAIFYIDLSFPTN-VFAILQHL 389
L I FP+N +F L HL
Sbjct: 891 ----ALSQIPTAPGIFPSNSIFTNLDHL 914
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.364 0.161 0.620
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 848,324,108
Number of Sequences: 2540612
Number of extensions: 29047325
Number of successful extensions: 150310
Number of sequences better than 10.0: 143
Number of HSP's better than 10.0 without gapping: 92
Number of HSP's successfully gapped in prelim test: 51
Number of HSP's that attempted gapping in prelim test: 150115
Number of HSP's gapped (non-prelim): 182
length of query: 615
length of database: 863,360,394
effective HSP length: 134
effective length of query: 481
effective length of database: 522,918,386
effective search space: 251523743666
effective search space used: 251523743666
T: 11
A: 40
X1: 14 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 36 (21.5 bits)
S2: 78 (34.7 bits)
Medicago: description of AC136953.11


