

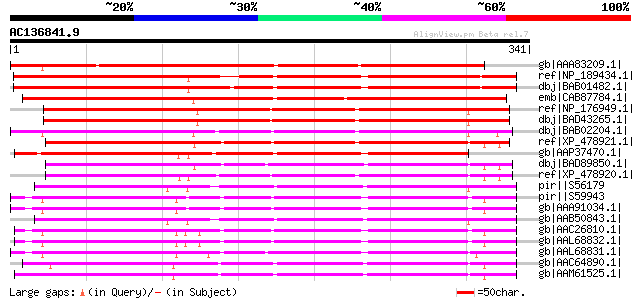
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136841.9 + phase: 0
(341 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAA83209.1| coil protein [Medicago sativa] gi|7488730|pir||T0... 363 5e-99
ref|NP_189434.1| early nodule-specific protein, putative [Arabid... 292 9e-78
dbj|BAB01482.1| unnamed protein product [Arabidopsis thaliana] 290 3e-77
emb|CAB87784.1| early nodule-specific protein-like [Arabidopsis ... 279 8e-74
ref|NP_176949.1| GDSL-motif lipase/hydrolase family protein [Ara... 261 2e-68
dbj|BAD43265.1| ENOD8-like protein [Arabidopsis thaliana] 261 2e-68
dbj|BAB02204.1| nodulin-like protein protein [Arabidopsis thalia... 256 9e-67
ref|XP_478921.1| putative early nodulin 8 precursor [Oryza sativ... 249 8e-65
gb|AAP37470.1| ENSP-like protein [Hevea brasiliensis] gi|5131578... 249 1e-64
dbj|BAD89850.1| hypothetical protein [Zea mays] 247 3e-64
ref|XP_478920.1| putative early nodulin 8 precursor [Oryza sativ... 238 2e-61
pir||S56179 secreted glycoprotein EP4, 47K, precursor - carrot ... 221 2e-56
pir||S59943 early nodulin 8 precursor - alfalfa gi|304037|gb|AAB... 221 3e-56
gb|AAA91034.1| nodulin 221 3e-56
gb|AAB50843.1| iEP4 [Daucus carota] gi|4204870|gb|AAD11468.1| iE... 220 5e-56
gb|AAC26810.1| early nodule-specific protein [Medicago truncatul... 216 1e-54
gb|AAL68832.1| Enod8.1 [Medicago truncatula] 215 1e-54
gb|AAL68831.1| Enod8.2 [Medicago truncatula] 213 9e-54
gb|AAC64890.1| Similar to nodulins and lipase homolog F14J9.5 gi... 209 7e-53
gb|AAM61525.1| early nodule-specific protein, putative [Arabidop... 209 1e-52
>gb|AAA83209.1| coil protein [Medicago sativa] gi|7488730|pir||T09416 coil protein
PO22, microspore/pollen-specific - alfalfa
Length = 340
Score = 363 bits (931), Expect = 5e-99
Identities = 189/318 (59%), Positives = 233/318 (72%), Gaps = 10/318 (3%)
Query: 1 MNTMTLIYILCFFNLCVACP-----SKKCVYPAIYNFGDSNSDTGAGYATMAAVEHPNGI 55
MN+M LI++L NL V C S +CVYPAIYNFGDSNSDTG YA + PNGI
Sbjct: 1 MNSMRLIHVLWCLNLYVTCTFIQVSSHECVYPAIYNFGDSNSDTGTAYAIFKRNQPPNGI 60
Query: 56 SFFGSISGRCCDGRLILDFISEELELPYLSSYLNSVGSNYRHGANFAVASAPIRPIIAGL 115
SF G+ISGR DGRLI+D+I+EEL+ PYLS+YLNSVGSNYR+GANFA A I P
Sbjct: 61 SF-GNISGRASDGRLIIDYITEELKAPYLSAYLNSVGSNYRYGANFASGGASICPGSGWS 119
Query: 116 TY-LGFQVSQFILFKSHTKILFDQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQK 174
+ LG QV+QF FKS T+ILF+ TEP L+SG+PR EDFSKA+YTIDIG ND+ G +
Sbjct: 120 PFDLGLQVTQFRQFKSQTRILFNNETEPSLKSGLPRPEDFSKALYTIDIGLNDLASGFLR 179
Query: 175 PNSSEEEVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGN 234
SEE+V+RS P+IL F+QAV++LYNE ARVFWIHN GP+ C+P Y+ +K +KGN
Sbjct: 180 --FSEEQVQRSFPEILGNFSQAVKQLYNEGARVFWIHNVGPVGCLPLNYYSNQNK-KKGN 236
Query: 235 LDANGCVKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVN 294
LDAN CV+ N++ QE N +LKDQV QLR+ AKFTYVD+Y KY LISNA++QGFV+
Sbjct: 237 LDANVCVESENKITQELNNKLKDQVSQLRKELVQAKFTYVDMYKAKYELISNAKSQGFVS 296
Query: 295 PLEFCCGSYQGNEIHYCG 312
++FCCGSY G+ CG
Sbjct: 297 LIDFCCGSYTGDYSVNCG 314
>ref|NP_189434.1| early nodule-specific protein, putative [Arabidopsis thaliana]
Length = 361
Score = 292 bits (748), Expect = 9e-78
Identities = 157/334 (47%), Positives = 216/334 (64%), Gaps = 22/334 (6%)
Query: 3 TMTLIYILCFFNLCVACPSKKCVYPAIYNFGDSNSDTGAGYATMAAVEHPNGISFFGSIS 62
T+ +I +L F ++ S C +PA++NFGDSNSDTGA A + V PNG++FFG +
Sbjct: 7 TLAIIVLLLGFTEKLSALSSSCNFPAVFNFGDSNSDTGAISAAIGEVPPPNGVAFFGRSA 66
Query: 63 GRCCDGRLILDFISEELELPYLSSYLNSVGSNYRHGANFAVASAPIRPIIAGLT--YLGF 120
GR DGRLI+DFI+E L LPYL+ YL+SVG+NYRHGANFA + IRP +A + +LG
Sbjct: 67 GRHSDGRLIIDFITENLTLPYLTPYLDSVGANYRHGANFATGGSCIRPTLACFSPFHLGT 126
Query: 121 QVSQFILFKSHTKILFDQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQKPNSSEE 180
QVSQFI FK+ T L++Q T DFSKA+YT+DIGQND+ G Q N +EE
Sbjct: 127 QVSQFIHFKTRTLSLYNQ------------TNDFSKALYTLDIGQNDLAIGFQ--NMTEE 172
Query: 181 EVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGC 240
+++ +IP I+ FT A++ LY E AR F IHNTGP C+PY +P D GC
Sbjct: 173 QLKATIPLIIENFTIALKLLYKEGARFFSIHNTGPTGCLPYLLKAFPAIPR----DPYGC 228
Query: 241 VKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVNPLEFCC 300
+KP N +A E+N+QLK+++ QL++ P + FTYVDVY+ KY LI+ A+ GF++P ++CC
Sbjct: 229 LKPLNNVAIEFNKQLKNKITQLKKELPSSFFTYVDVYSAKYNLITKAKALGFIDPFDYCC 288
Query: 301 GSYQGNEIHYCGKKSIKNGT-VYGIACDDSSTYI 333
G + CGK NGT +Y +C + +I
Sbjct: 289 VGAIGRGMG-CGKTIFLNGTELYSSSCQNRKNFI 321
>dbj|BAB01482.1| unnamed protein product [Arabidopsis thaliana]
Length = 371
Score = 290 bits (743), Expect = 3e-77
Identities = 157/334 (47%), Positives = 217/334 (64%), Gaps = 12/334 (3%)
Query: 3 TMTLIYILCFFNLCVACPSKKCVYPAIYNFGDSNSDTGAGYATMAAVEHPNGISFFGSIS 62
T+ +I +L F ++ S C +PA++NFGDSNSDTGA A + V PNG++FFG +
Sbjct: 7 TLAIIVLLLGFTEKLSALSSSCNFPAVFNFGDSNSDTGAISAAIGEVPPPNGVAFFGRSA 66
Query: 63 GRCCDGRLILDFISEELELPYLSSYLNSVGSNYRHGANFAVASAPIRPIIAGLT--YLGF 120
GR DGRLI+DFI+E L LPYL+ YL+SVG+NYRHGANFA + IRP +A + +LG
Sbjct: 67 GRHSDGRLIIDFITENLTLPYLTPYLDSVGANYRHGANFATGGSCIRPTLACFSPFHLGT 126
Query: 121 QVSQFILFKSHTKILFDQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQKPNSSEE 180
QVSQFI FK+ T L++Q R + T FSKA+YT+DIGQND+ G Q N +EE
Sbjct: 127 QVSQFIHFKTRTLSLYNQTNGKFNR--LSHTNYFSKALYTLDIGQNDLAIGFQ--NMTEE 182
Query: 181 EVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGC 240
+++ +IP I+ FT A++ LY E AR F IHNTGP C+PY +P D GC
Sbjct: 183 QLKATIPLIIENFTIALKLLYKEGARFFSIHNTGPTGCLPYLLKAFPAIPR----DPYGC 238
Query: 241 VKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVNPLEFCC 300
+KP N +A E+N+QLK+++ QL++ P + FTYVDVY+ KY LI+ A+ GF++P ++CC
Sbjct: 239 LKPLNNVAIEFNKQLKNKITQLKKELPSSFFTYVDVYSAKYNLITKAKALGFIDPFDYCC 298
Query: 301 GSYQGNEIHYCGKKSIKNGT-VYGIACDDSSTYI 333
G + CGK NGT +Y +C + +I
Sbjct: 299 VGAIGRGMG-CGKTIFLNGTELYSSSCQNRKNFI 331
>emb|CAB87784.1| early nodule-specific protein-like [Arabidopsis thaliana]
gi|26451820|dbj|BAC43003.1| putative early
nodule-specific protein [Arabidopsis thaliana]
gi|28950957|gb|AAO63402.1| At5g14450 [Arabidopsis
thaliana] gi|15241404|ref|NP_196949.1| GDSL-motif
lipase/hydrolase family protein [Arabidopsis thaliana]
gi|11357276|pir||T48618 early nodule-specific
protein-like - Arabidopsis thaliana
Length = 389
Score = 279 bits (714), Expect = 8e-74
Identities = 145/327 (44%), Positives = 199/327 (60%), Gaps = 12/327 (3%)
Query: 9 ILCFFNLCVACPSKK-CVYPAIYNFGDSNSDTGAGYATMAAVEHPNGISFFGSISGRCCD 67
+LC F + + + C +PAIYNFGDSNSDTG A + P G FF +GR D
Sbjct: 21 LLCLFAVTTSVSVQPTCTFPAIYNFGDSNSDTGGISAAFEPIRDPYGQGFFHRPTGRDSD 80
Query: 68 GRLILDFISEELELPYLSSYLNSVGSNYRHGANFAVASAPIRPIIAGLTYLG-------F 120
GRL +DFI+E L LPYLS+YLNS+GSN+RHGANFA + IR + G
Sbjct: 81 GRLTIDFIAERLGLPYLSAYLNSLGSNFRHGANFATGGSTIRRQNETIFQYGISPFSLDM 140
Query: 121 QVSQFILFKSHTKILFDQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQKPNSSEE 180
Q++QF FK+ + +LF Q R +PR E+F+KA+YT DIGQND+ G + S +
Sbjct: 141 QIAQFDQFKARSALLFTQIKSRYDREKLPRQEEFAKALYTFDIGQNDLSVGFR--TMSVD 198
Query: 181 EVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGC 240
+++ +IPDI++ AV+ +Y + R FW+HNTGP C+P FY G LD +GC
Sbjct: 199 QLKATIPDIVNHLASAVRNIYQQGGRTFWVHNTGPFGCLP-VNMFYMGTPAPGYLDKSGC 257
Query: 241 VKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVNPLEFCC 300
VK NE+A E+NR+LK+ V LR+ A TYVDVYT KY ++SN + GF NPL+ CC
Sbjct: 258 VKAQNEMAMEFNRKLKETVINLRKELTQAAITYVDVYTAKYEMMSNPKKLGFANPLKVCC 317
Query: 301 GSYQGNEIHYCGKK-SIKNGTVYGIAC 326
G ++ + +CG K + N +YG +C
Sbjct: 318 GYHEKYDHIWCGNKGKVNNTEIYGGSC 344
>ref|NP_176949.1| GDSL-motif lipase/hydrolase family protein [Arabidopsis thaliana]
gi|11072007|gb|AAG28886.1| F12A21.4 [Arabidopsis
thaliana]
Length = 372
Score = 261 bits (667), Expect = 2e-68
Identities = 143/316 (45%), Positives = 195/316 (61%), Gaps = 13/316 (4%)
Query: 23 KCVYPAIYNFGDSNSDTGAGYATMAAVEHPNGISFFGSISGRCCDGRLILDFISEELELP 82
+C +PAI+NFGDSNSDTG A P+G SFFGS +GR CDGRL++DFI+E L LP
Sbjct: 25 QCHFPAIFNFGDSNSDTGGLSAAFGQAGPPHGSSFFGSPAGRYCDGRLVIDFIAESLGLP 84
Query: 83 YLSSYLNSVGSNYRHGANFAVASAPIRPIIAGLTYLGFQV----SQFILFKS-HTKILFD 137
YLS++L+SVGSN+ HGANFA A +PIR + + L GF QF+ F + H +
Sbjct: 85 YLSAFLDSVGSNFSHGANFATAGSPIRALNSTLRQSGFSPFSLDVQFVQFYNFHNRSQTV 144
Query: 138 QRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQKPNSSEEEVRRSIPDILSQFTQAV 197
+ ++ +P ++ FSKA+YT DIGQND+ G N + E+V +P+I+SQF A+
Sbjct: 145 RSRGGVYKTMLPESDSFSKALYTFDIGQNDLTAG-YFANKTVEQVETEVPEIISQFMNAI 203
Query: 198 QKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGCVKPHNELAQEYNRQLKD 257
+ +Y + R FWIHNTGPI C+ Y +P N+ + D++GCV P N LAQ++N LK
Sbjct: 204 KNIYGQGGRYFWIHNTGPIGCLAYVIERFP--NKASDFDSHGCVSPLNHLAQQFNHALKQ 261
Query: 258 QVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVNPLEFCC---GSYQGNEIHYCG-K 313
V +LR A TYVDVY++K+ L +A+ GF L CC G Y N+ CG K
Sbjct: 262 AVIELRSSLSEAAITYVDVYSLKHELFVHAQGHGFKGSLVSCCGHGGKYNYNKGIGCGMK 321
Query: 314 KSIKNGTVY-GIACDD 328
K +K VY G CD+
Sbjct: 322 KIVKGKEVYIGKPCDE 337
>dbj|BAD43265.1| ENOD8-like protein [Arabidopsis thaliana]
Length = 364
Score = 261 bits (667), Expect = 2e-68
Identities = 143/316 (45%), Positives = 195/316 (61%), Gaps = 13/316 (4%)
Query: 23 KCVYPAIYNFGDSNSDTGAGYATMAAVEHPNGISFFGSISGRCCDGRLILDFISEELELP 82
+C +PAI+NFGDSNSDTG A P+G SFFGS +GR CDGRL++DFI+E L LP
Sbjct: 17 QCHFPAIFNFGDSNSDTGGLSAAFGQAGPPHGSSFFGSPAGRYCDGRLVIDFIAESLGLP 76
Query: 83 YLSSYLNSVGSNYRHGANFAVASAPIRPIIAGLTYLGFQV----SQFILFKS-HTKILFD 137
YLS++L+SVGSN+ HGANFA A +PIR + + L GF QF+ F + H +
Sbjct: 77 YLSAFLDSVGSNFSHGANFATAGSPIRALNSTLRQSGFSPFSLDVQFVQFYNFHNRSQTV 136
Query: 138 QRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQKPNSSEEEVRRSIPDILSQFTQAV 197
+ ++ +P ++ FSKA+YT DIGQND+ G N + E+V +P+I+SQF A+
Sbjct: 137 RSRGGVYKTMLPESDSFSKALYTFDIGQNDLTAG-YFANKTVEQVETEVPEIISQFMNAI 195
Query: 198 QKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGCVKPHNELAQEYNRQLKD 257
+ +Y + R FWIHNTGPI C+ Y +P N+ + D++GCV P N LAQ++N LK
Sbjct: 196 KNIYGQGGRYFWIHNTGPIGCLAYVIERFP--NKASDFDSHGCVSPLNHLAQQFNHALKQ 253
Query: 258 QVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVNPLEFCC---GSYQGNEIHYCG-K 313
V +LR A TYVDVY++K+ L +A+ GF L CC G Y N+ CG K
Sbjct: 254 AVIELRSSLSEAAITYVDVYSLKHELFVHAQGHGFKGSLVSCCGHGGKYNYNKGIGCGMK 313
Query: 314 KSIKNGTVY-GIACDD 328
K +K VY G CD+
Sbjct: 314 KIVKGKEVYIGKPCDE 329
>dbj|BAB02204.1| nodulin-like protein protein [Arabidopsis thaliana]
gi|20259934|gb|AAM13314.1| unknown protein [Arabidopsis
thaliana] gi|17064918|gb|AAL32613.1| Unknown protein
[Arabidopsis thaliana] gi|15231558|ref|NP_189274.1|
GDSL-motif lipase/hydrolase family protein [Arabidopsis
thaliana]
Length = 380
Score = 256 bits (653), Expect = 9e-67
Identities = 143/345 (41%), Positives = 207/345 (59%), Gaps = 20/345 (5%)
Query: 1 MNTMTLIYILCFFNLCVACP---SKKCVYPAIYNFGDSNSDTGAGYATMAAVEHPNGISF 57
M T L+ C+ P S C +PAI+NFGDSNSDTG A+ +PNG +F
Sbjct: 1 METNLLLVKCVLLASCLIHPRACSPSCNFPAIFNFGDSNSDTGGLSASFGQAPYPNGQTF 60
Query: 58 FGSISGRCCDGRLILDFISEELELPYLSSYLNSVGSNYRHGANFAVASAPIRPIIAGLTY 117
F S SGR DGRLI+DFI+EEL LPYL+++L+S+GSN+ HGANFA A + +RP A +
Sbjct: 61 FHSPSGRFSDGRLIIDFIAEELGLPYLNAFLDSIGSNFSHGANFATAGSTVRPPNATIAQ 120
Query: 118 LG-------FQVSQFILFKSHTKILFDQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGY 170
G Q+ QF F + ++++ + + +P+ E FS+A+YT DIGQND+
Sbjct: 121 SGVSPISLDVQLVQFSDFITRSQLI--RNRGGVFKKLLPKKEYFSQALYTFDIGQNDLTA 178
Query: 171 GLQKPNSSEEEVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKN 230
GL K N + ++++ IPD+ Q + ++K+Y++ R FWIHNT P+ C+PY +P
Sbjct: 179 GL-KLNMTSDQIKAYIPDVHDQLSNVIRKVYSKGGRRFWIHNTAPLGCLPYVLDRFP--V 235
Query: 231 EKGNLDANGCVKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQ 290
+D +GC P NE+A+ YN +LK +V +LR+ A FTYVD+Y++K TLI+ A+
Sbjct: 236 PASQIDNHGCAIPRNEIARYYNSELKRRVIELRKELSEAAFTYVDIYSIKLTLITQAKKL 295
Query: 291 GFVNPLEFCC---GSYQGNEIHYCGKKSIKNG--TVYGIACDDSS 330
GF PL CC G Y N++ CG K + G V +C+D S
Sbjct: 296 GFRYPLVACCGHGGKYNFNKLIKCGAKVMIKGKEIVLAKSCNDVS 340
>ref|XP_478921.1| putative early nodulin 8 precursor [Oryza sativa (japonica
cultivar-group)] gi|33147016|dbj|BAC80100.1| putative
early nodulin 8 precursor [Oryza sativa (japonica
cultivar-group)]
Length = 391
Score = 249 bits (636), Expect = 8e-65
Identities = 144/319 (45%), Positives = 192/319 (60%), Gaps = 19/319 (5%)
Query: 24 CVYPAIYNFGDSNSDTGAGYATMAAVEHPNGISFFGSISGRCCDGRLILDFISEELELPY 83
C +PA++NFGDSNSDTG AT A PNG +FFG GR CDGRL++DFI+E L LPY
Sbjct: 32 CRFPAVFNFGDSNSDTGGLSATFGAAPPPNGRTFFGMPVGRYCDGRLVIDFIAESLGLPY 91
Query: 84 LSSYLNSVGSNYRHGANFAVASAPIRPIIAGLTYLGF-------QVSQFILFKSHTKILF 136
LS+YLNS+GSN+ GANFA A + IR L GF Q +F F + ++ ++
Sbjct: 92 LSAYLNSIGSNFTQGANFATAGSSIRRQNTSLFLSGFSPISLDVQSWEFEQFINRSQFVY 151
Query: 137 DQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQKPNSSEEEVRRSIPDILSQFTQA 196
+ + R +P+ E FS+A+YT DIGQNDI G N + E+V IPD++ + T
Sbjct: 152 NNK-GGIYRELLPKAEYFSQALYTFDIGQNDITTGF-FINMTSEQVIAYIPDLMERLTNI 209
Query: 197 VQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGCVKPHNELAQEYNRQLK 256
+Q +Y R FWIHNTGPI C+PY P + D +GC +NE+AQ +N++LK
Sbjct: 210 IQNVYGLGGRYFWIHNTGPIGCLPYAMVHRP--DLAVVKDGSGCSVAYNEVAQLFNQRLK 267
Query: 257 DQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVNPLEFCCGSYQGNEIHY-----C 311
+ V LR+ A FTYVDVY+ KY LIS+A+ G +P+ CCG Y G ++ C
Sbjct: 268 ETVGHLRKTHADAAFTYVDVYSAKYKLISDAKKLGMDDPMLTCCG-YGGGRYNFDDRVGC 326
Query: 312 GKKSIKNGT--VYGIACDD 328
G K NGT V G +CDD
Sbjct: 327 GGKVKVNGTWVVAGKSCDD 345
>gb|AAP37470.1| ENSP-like protein [Hevea brasiliensis]
gi|51315784|sp|Q7Y1X1|EST_HEVBR Esterase precursor
(Early nodule-specific protein homolog) (Latex allergen
Hev b 13)
Length = 391
Score = 249 bits (635), Expect = 1e-64
Identities = 137/306 (44%), Positives = 188/306 (60%), Gaps = 18/306 (5%)
Query: 4 MTLIYILCFFNLCVACPSKKCVYPAIYNFGDSNSDTGAGYATMAAVEHPNGISFFGSISG 63
+TL ++LC +L A S+ C +PAI+NFGDSNSDTG A + P G +FF +G
Sbjct: 12 ITLSFLLCMLSLAYA--SETCDFPAIFNFGDSNSDTGGKAAAFYPLNPPYGETFFHRSTG 69
Query: 64 RCCDGRLILDFISEELELPYLSSYLNSVGSNYRHGANFAVASAPIR------PIIAGLT- 116
R DGRLI+DFI+E LPYLS YL+S+GSN++HGA+FA A + I+ P G +
Sbjct: 70 RYSDGRLIIDFIAESFNLPYLSPYLSSLGSNFKHGADFATAGSTIKLPTTIIPAHGGFSP 129
Query: 117 -YLGFQVSQFILFKSHTKILFDQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQKP 175
YL Q SQF F ++ F + T VP F KA+YT DIGQND+ G
Sbjct: 130 FYLDVQYSQFRQFIPRSQ--FIRETGGIFAELVPEEYYFEKALYTFDIGQNDLTEGFL-- 185
Query: 176 NSSEEEVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNL 235
N + EEV ++PD+++ F+ V+K+Y+ AR FWIHNTGPI C+ + ++P +
Sbjct: 186 NLTVEEVNATVPDLVNSFSANVKKIYDLGARTFWIHNTGPIGCLSFILTYFPWAEK---- 241
Query: 236 DANGCVKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVNP 295
D+ GC K +NE+AQ +N +LK+ V QLR+ PLA F +VD+Y+VKY+L S GF P
Sbjct: 242 DSAGCAKAYNEVAQHFNHKLKEIVAQLRKDLPLATFVHVDIYSVKYSLFSEPEKHGFEFP 301
Query: 296 LEFCCG 301
L CCG
Sbjct: 302 LITCCG 307
>dbj|BAD89850.1| hypothetical protein [Zea mays]
Length = 394
Score = 247 bits (631), Expect = 3e-64
Identities = 141/321 (43%), Positives = 190/321 (58%), Gaps = 19/321 (5%)
Query: 24 CVYPAIYNFGDSNSDTGAGYATMAAVEHPNGISFFGSISGRCCDGRLILDFISEELELPY 83
C +PA++NFGDSNSDTG + A PNG +FFG +GR CDGRL++DFI+E L L +
Sbjct: 36 CHFPAVFNFGDSNSDTGGLSSLFGAAPPPNGRTFFGMPAGRYCDGRLVIDFIAESLGLTH 95
Query: 84 LSSYLNSVGSNYRHGANFAVASAPIRPIIAGLTYLGF-------QVSQFILFKSHTKILF 136
LS+YLNS+GSN+ GANFA A + IR L GF Q +F F + +++++
Sbjct: 96 LSAYLNSIGSNFTQGANFATAGSSIRRQNTSLFLSGFSPISLDVQFWEFEQFINRSQLVY 155
Query: 137 DQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQKPNSSEEEVRRSIPDILSQFTQA 196
+ + R +PR E FS+A+YT DIGQNDI N++ EEV IPD++ + T
Sbjct: 156 NNK-GGIYREILPRAEYFSQALYTFDIGQNDI-TSSYFVNNTTEEVEAIIPDLMERLTSI 213
Query: 197 VQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGCVKPHNELAQEYNRQLK 256
+Q +Y+ R FWIHNTGP+ C+PY P D GC +N++ Q +N +LK
Sbjct: 214 IQSVYSRGGRYFWIHNTGPLGCLPYALLHRPDLATPA--DGTGCSVTYNKVPQLFNLRLK 271
Query: 257 DQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVNPLEFCCGSYQGNEIHY-----C 311
+ V LR+ P A FTYVDVYT KY LIS A+ GF +PL CCG Y G + C
Sbjct: 272 ETVASLRKTHPDAAFTYVDVYTAKYKLISQAKKLGFDDPLLTCCG-YGGGRYNLDLSVGC 330
Query: 312 GKKSIKNGT--VYGIACDDSS 330
G K NGT V G +C++ S
Sbjct: 331 GGKKQVNGTSVVVGKSCENPS 351
>ref|XP_478920.1| putative early nodulin 8 precursor [Oryza sativa (japonica
cultivar-group)] gi|33147015|dbj|BAC80099.1| putative
early nodulin 8 precursor [Oryza sativa (japonica
cultivar-group)]
Length = 405
Score = 238 bits (607), Expect = 2e-61
Identities = 138/321 (42%), Positives = 193/321 (59%), Gaps = 19/321 (5%)
Query: 24 CVYPAIYNFGDSNSDTGAGYATMAAVEHPNGISFFGSISGRCCDGRLILDFISEELELPY 83
C +PAI+NFGDSNSDTG A +A V P G ++FG +GR DGRL +DF+++ L + Y
Sbjct: 45 CDFPAIFNFGDSNSDTGGLSALIAVVPPPFGRTYFGMPAGRFSDGRLTIDFMAQSLGIRY 104
Query: 84 LSSYLNSVGSNYRHGANFAVASAPIRP-----IIAGLT--YLGFQVSQFILFKSHTKILF 136
LS+YL+SVGSN+ GANFA A+A IRP ++G++ L Q SQF F + ++ ++
Sbjct: 105 LSAYLDSVGSNFSQGANFATAAASIRPANGSIFVSGISPISLDVQTSQFEQFINRSQFVY 164
Query: 137 DQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQKPNSSEEEVRRSIPDILSQFTQA 196
R +P+ E FS+A+YT DIGQND+ G N S E+V +PD++ +F+ A
Sbjct: 165 -SNIGGIYREILPKAEYFSRALYTFDIGQNDLTMG-YFDNMSTEQVEAYVPDLMERFSAA 222
Query: 197 VQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGCVKPHNELAQEYNRQLK 256
+QK+Y+ R FW+HNT P+ C+ Y P D GC +N A+ +N +L+
Sbjct: 223 IQKVYSLGGRYFWVHNTAPLGCLTYAVVLLP--KLAAPRDDAGCSVAYNAAARFFNARLR 280
Query: 257 DQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVNPLEFCCGSYQGNEIHY-----C 311
+ V +LR P A TYVDVY+ KY LIS A+ GF +PL CCG Y G E ++ C
Sbjct: 281 ETVDRLRAALPDAALTYVDVYSAKYRLISQAKQLGFGDPLLVCCG-YGGGEYNFDRDIRC 339
Query: 312 GKKSIKNGT--VYGIACDDSS 330
G K NGT + G +CDD S
Sbjct: 340 GGKVEVNGTSVLAGKSCDDPS 360
>pir||S56179 secreted glycoprotein EP4, 47K, precursor - carrot (fragment)
gi|886223|gb|AAA98926.1| secreted glycoprotein
Length = 383
Score = 221 bits (563), Expect = 2e-56
Identities = 128/333 (38%), Positives = 186/333 (55%), Gaps = 24/333 (7%)
Query: 17 VACPSKKCVYPAIYNFGDSNSDTGAGYATMAAVEHPNGISFFGSISGRCCDGRLILDFIS 76
V+ S+ C +PAI+NFGD+NSDTGA A G S+F +GR DGRL++DF++
Sbjct: 13 VSASSQTCDFPAIFNFGDANSDTGAFAAWFFGNPPFFGQSYFNGSAGRVSDGRLLIDFMA 72
Query: 77 EELELPYLSSYLNSVGSNYRHGANFA----VASAPIRPIIAGL--------TYLGFQVSQ 124
+L LP+L Y++S+G+N+ HGANFA + P II G+ L QV+Q
Sbjct: 73 TDLGLPFLHPYMDSLGANFSHGANFANILSTIALPTSNIIPGVRPPRGLNPVNLDIQVAQ 132
Query: 125 FILFKSHTKILFDQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQKPNSSEEEVRR 184
F F + + Q + +P+ + FS+A+YT+DIGQ DI N +++E++
Sbjct: 133 FAQFVNRS-----QTQGEAFDNFMPKQDYFSQALYTLDIGQIDITQEFLT-NKTDDEIKA 186
Query: 185 SIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGCVKPH 244
+P ++S + +Q LY+ R FWIHN GP C+P + P +++ +D+ GC K +
Sbjct: 187 VVPGLISSLSSNIQILYSLGGRSFWIHNLGPNGCLPILWTLAPVPDDQ--IDSAGCAKRY 244
Query: 245 NELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVNPLEFCC---G 301
N+L Q +N +LK V QLR PLA TYVDVYT KY+L GF +PLE CC G
Sbjct: 245 NDLTQYFNSELKKGVDQLRTDLPLAAVTYVDVYTAKYSLYQEPAKYGFTHPLETCCGFGG 304
Query: 302 SYQGNEIHYCGKKSIKNGTVYGIA-CDDSSTYI 333
Y E CG NGT + C++ + YI
Sbjct: 305 RYNYGEFSLCGSTITVNGTQLAVGPCENPAEYI 337
>pir||S59943 early nodulin 8 precursor - alfalfa gi|304037|gb|AAB41547.1| early
nodulin [Medicago sativa]
Length = 381
Score = 221 bits (562), Expect = 3e-56
Identities = 137/348 (39%), Positives = 197/348 (56%), Gaps = 29/348 (8%)
Query: 1 MNTMTLIYILCFFNLCVACP---SKKCVYPAIYNFGDSNSDTGAGYATMAAVEHPNGISF 57
M +TLI ++ LC P S C +PAI++FG SN DTG A A P G ++
Sbjct: 10 MPLVTLIVLV----LCTTPPIFASTHCDFPAIFSFGASNVDTGGLAAAFRAPPSPYGQTY 65
Query: 58 FGSISGRCCDGRLILDFISEELELPYLSSYLNSVGSNYRHGANFAVASAPI--------R 109
F +GR DGR+I+DFI++ LPY S YLNS+GSN+ HGANFA A + I +
Sbjct: 66 FNRSTGRFSDGRIIIDFIAQSFRLPYPSPYLNSLGSNFTHGANFATAGSTINIPTSILPK 125
Query: 110 PIIAGLTYLGFQVSQFILFKSHTKILFDQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIG 169
I++ + L Q QF F S TK++ DQ + VP+ + FSKA+Y DIGQND+
Sbjct: 126 GILSPFS-LQIQYIQFKDFISKTKLIRDQ--GGVFATLVPKEDYFSKALYVFDIGQNDLT 182
Query: 170 YGLQKPNSSEEEVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHK 229
G N + ++V ++PD+++ + + ++ +YN AR FWIH+TGP C P +P
Sbjct: 183 IGF-FGNKTIQQVNATVPDLVNNYIENIKNIYNLGARSFWIHSTGPKGCAPVILANFPSA 241
Query: 230 NEKGNLDANGCVKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARN 289
+ D+ GC K +NE++Q +N +LK+ + QLR PLA TYVD+Y+ KY+L +N +
Sbjct: 242 IK----DSYGCAKQYNEVSQYFNLKLKEALAQLRSDLPLAAITYVDIYSPKYSLFTNPKK 297
Query: 290 QGFVNPLEFCCGSYQGNEIHY---CGKKSIKNGT-VYGIACDDSSTYI 333
GF P CCG G E + CG NGT + +C + ST I
Sbjct: 298 YGFELPYVACCG--YGGEYNIGAGCGATINVNGTKIVAGSCKNPSTRI 343
>gb|AAA91034.1| nodulin
Length = 381
Score = 221 bits (562), Expect = 3e-56
Identities = 137/348 (39%), Positives = 197/348 (56%), Gaps = 29/348 (8%)
Query: 1 MNTMTLIYILCFFNLCVACP---SKKCVYPAIYNFGDSNSDTGAGYATMAAVEHPNGISF 57
M +TLI ++ LC P S C +PAI++FG SN DTG A A P G ++
Sbjct: 10 MPLVTLIVLV----LCTTPPIFASTHCDFPAIFSFGASNVDTGGLAAAFRAPPSPYGQTY 65
Query: 58 FGSISGRCCDGRLILDFISEELELPYLSSYLNSVGSNYRHGANFAVASAPI--------R 109
F +GR DGR+I+DFI++ LPY S YLNS+GSN+ HGANFA A + I +
Sbjct: 66 FNRSTGRFSDGRIIIDFIAQSFRLPYPSPYLNSLGSNFTHGANFATAGSTINIPTSILPK 125
Query: 110 PIIAGLTYLGFQVSQFILFKSHTKILFDQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIG 169
I++ + L Q QF F S TK++ DQ + VP+ + FSKA+Y DIGQND+
Sbjct: 126 GILSPFS-LQIQYIQFKDFISKTKLIRDQ--GGVFATLVPKEDYFSKALYVFDIGQNDLT 182
Query: 170 YGLQKPNSSEEEVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHK 229
G N + ++V ++PD+++ + + ++ +YN AR FWIH+TGP C P +P
Sbjct: 183 IGF-FGNKTIQQVNATVPDLVNNYIENIKNIYNLGARSFWIHSTGPKGCAPVILANFPSA 241
Query: 230 NEKGNLDANGCVKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARN 289
+ D+ GC K +NE++Q +N +LK+ + QLR PLA TYVD+Y+ KY+L +N +
Sbjct: 242 IK----DSYGCAKQYNEVSQYFNLKLKEALAQLRSDLPLAAITYVDIYSPKYSLFTNPKK 297
Query: 290 QGFVNPLEFCCGSYQGNEIHY---CGKKSIKNGT-VYGIACDDSSTYI 333
GF P CCG G E + CG NGT + +C + ST I
Sbjct: 298 YGFELPYVACCG--YGGEYNIGAGCGATINVNGTKIVAGSCKNPSTRI 343
>gb|AAB50843.1| iEP4 [Daucus carota] gi|4204870|gb|AAD11468.1| iEP4 [Daucus carota]
Length = 391
Score = 220 bits (560), Expect = 5e-56
Identities = 129/333 (38%), Positives = 185/333 (54%), Gaps = 24/333 (7%)
Query: 17 VACPSKKCVYPAIYNFGDSNSDTGAGYATMAAVEHPNGISFFGSISGRCCDGRLILDFIS 76
V+ S+ C +PAI+NFGD+NSDTGA A G S+F +GR DGRL++DF++
Sbjct: 21 VSASSQTCDFPAIFNFGDANSDTGAFAAWFFGNPPFFGQSYFNGSAGRVSDGRLLIDFMA 80
Query: 77 EELELPYLSSYLNSVGSNYRHGANFA----VASAPIRPIIAGL--------TYLGFQVSQ 124
+L LP+L Y++S+G+N+ HGANFA + P II G+ L QV+Q
Sbjct: 81 TDLGLPFLHPYMDSLGANFSHGANFANILSTIALPTSNIIPGVRPPRGLNPVNLDIQVAQ 140
Query: 125 FILFKSHTKILFDQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQKPNSSEEEVRR 184
F F + + Q + +P+ + FS+A+YT+DIGQ DI N +++E++
Sbjct: 141 FAQFVNRS-----QTQGEAFDNFMPKQDYFSQALYTLDIGQIDITQEFLT-NKTDDEIKA 194
Query: 185 SIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGCVKPH 244
+P ++S + +Q LY+ R FWIHN GP C+P P +++ LD+ GC K +
Sbjct: 195 VVPGLISSLSSNIQILYSLGGRSFWIHNLGPNGCLPILLTLAPVPDDQ--LDSAGCAKRY 252
Query: 245 NELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVNPLEFCC---G 301
N+L Q +N +LK V QLR PLA TYVDVYT KY+L GF +PLE CC G
Sbjct: 253 NDLTQYFNSELKKGVDQLRTDLPLAAVTYVDVYTAKYSLYQEPAKYGFTHPLETCCGFGG 312
Query: 302 SYQGNEIHYCGKKSIKNGTVYGIA-CDDSSTYI 333
Y E CG NGT + C++ + YI
Sbjct: 313 RYNYGEFSLCGSTITVNGTQLTVGPCENPAEYI 345
>gb|AAC26810.1| early nodule-specific protein [Medicago truncatula]
gi|25457244|pir||T52338 early nodule-specific protein
ENOD8 [imported] - barrel medic
Length = 381
Score = 216 bits (549), Expect = 1e-54
Identities = 135/344 (39%), Positives = 193/344 (55%), Gaps = 27/344 (7%)
Query: 4 MTLIYILCFFNLCVACP---SKKCVYPAIYNFGDSNSDTGAGYATMAAVEHPNGISFFGS 60
+TLI ++ LC+ P +K C +PAI++FG SN DTG A A P G ++F
Sbjct: 13 VTLIVLV----LCITPPIFATKNCDFPAIFSFGASNVDTGGLAAAFRAPPSPYGETYFHR 68
Query: 61 ISGRCCDGRLILDFISEELELPYLSSYLNSVGSNYRHGANFAVASAPI---RPIIAG--L 115
+GR DGR+ILDFI+ LPYLS YLNS+GSN+ HGANFA + I + I+ L
Sbjct: 69 STGRFSDGRIILDFIARSFRLPYLSPYLNSLGSNFTHGANFASGGSTINIPKSILPNGKL 128
Query: 116 TYLGFQVS--QFILFKSHTKILFDQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQ 173
+ Q+ QF F S TK++ DQ + +P+ + FSKA+Y DIGQND+ G
Sbjct: 129 SPFSLQIQYIQFKEFISKTKLIRDQ--GGVFATLIPKEDYFSKALYIFDIGQNDLTIGF- 185
Query: 174 KPNSSEEEVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKG 233
N + ++V ++PDI++ + + ++ +YN AR FWIH TGP C P +P +
Sbjct: 186 FGNKTIQQVNATVPDIVNNYIENIKNIYNLGARSFWIHGTGPKGCAPVILANFPSAIK-- 243
Query: 234 NLDANGCVKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFV 293
D+ GC K +NE++Q +N +LK+ + +LR A TYVD+YT KY+L +N GF
Sbjct: 244 --DSYGCAKQYNEVSQYFNFKLKEALAELRSNLSSAAITYVDIYTPKYSLFTNPEKYGFE 301
Query: 294 NPLEFCCGSYQGNEIHY---CGKKSIKNGT-VYGIACDDSSTYI 333
P CCG G E + CG NGT + +C + ST I
Sbjct: 302 LPFVACCG--YGGEYNIGVGCGASININGTKIVAGSCKNPSTRI 343
>gb|AAL68832.1| Enod8.1 [Medicago truncatula]
Length = 381
Score = 215 bits (548), Expect = 1e-54
Identities = 135/344 (39%), Positives = 193/344 (55%), Gaps = 27/344 (7%)
Query: 4 MTLIYILCFFNLCVACP---SKKCVYPAIYNFGDSNSDTGAGYATMAAVEHPNGISFFGS 60
+TLI ++ LC+ P +K C +PAI++FG SN DTG A A P G ++F
Sbjct: 13 VTLIVLV----LCITPPIFATKNCDFPAIFSFGASNVDTGGLAAAFRAPPSPYGETYFHR 68
Query: 61 ISGRCCDGRLILDFISEELELPYLSSYLNSVGSNYRHGANFAVASAPI---RPIIAG--L 115
+GR DGR+ILDFI+ LPYLS YLNS+GSN+ HGANFA + I + I+ L
Sbjct: 69 STGRFSDGRIILDFIARSFRLPYLSPYLNSLGSNFTHGANFASGGSTINIPKSILPNGKL 128
Query: 116 TYLGFQVS--QFILFKSHTKILFDQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQ 173
+ Q+ QF F S TK++ DQ + +P+ + FSKA+Y DIGQND+ G
Sbjct: 129 SPFSLQIQYIQFKEFISKTKLIRDQ--GGVFATLIPKEDYFSKALYIFDIGQNDLTIGF- 185
Query: 174 KPNSSEEEVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKG 233
N + ++V ++PDI++ + + ++ +YN AR FWIH TGP C P +P +
Sbjct: 186 FGNKTIQQVNATVPDIVNNYIKNIKNIYNLGARSFWIHGTGPKGCAPVILANFPSAIK-- 243
Query: 234 NLDANGCVKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFV 293
D+ GC K +NE++Q +N +LK+ + +LR A TYVD+YT KY+L +N GF
Sbjct: 244 --DSYGCAKQYNEVSQYFNFKLKEALAELRSNLSSAAITYVDIYTPKYSLFTNPEKYGFE 301
Query: 294 NPLEFCCGSYQGNEIHY---CGKKSIKNGT-VYGIACDDSSTYI 333
P CCG G E + CG NGT + +C + ST I
Sbjct: 302 LPFVACCG--YGGEYNIGVGCGASININGTKIVAGSCKNPSTRI 343
>gb|AAL68831.1| Enod8.2 [Medicago truncatula]
Length = 385
Score = 213 bits (541), Expect = 9e-54
Identities = 137/347 (39%), Positives = 196/347 (56%), Gaps = 26/347 (7%)
Query: 1 MNTMTLIYILCFFNLCVACP----SKKCVYPAIYNFGDSNSDTGAGYATMAAVEHPNGIS 56
M+ ++LI ++ LC+ P ++ C +PAI++FG SN DTG A A P G +
Sbjct: 10 MSLVSLIVLI----LCIITPPIFATRNCDFPAIFSFGASNVDTGGLAAAFQAPPSPYGET 65
Query: 57 FFGSISGRCCDGRLILDFISEELELPYLSSYLNSVGSNYRHGANFAVASAPI---RPIIA 113
+F +GR DGR+ILDFI++ LPYLS YLNS+GSN+ HGANFA + I II
Sbjct: 66 YFHRSTGRFSDGRIILDFIAQSFGLPYLSPYLNSLGSNFTHGANFATGGSTINIPNSIIP 125
Query: 114 GLTYLGFQVS-QFILFK---SHTKILFDQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIG 169
+ F + Q+I FK S T ++ DQ + +P+ + FSKA+YT DIGQND+
Sbjct: 126 NGIFSPFSLQIQYIQFKDFISKTNLIRDQ--GGVFATLIPKEDYFSKALYTFDIGQNDL- 182
Query: 170 YGLQKPNSSEEEVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHK 229
G N + ++V ++PDI++ F ++ +YN AR FWIH+T P C P +P
Sbjct: 183 IGGYFGNKTIKQVNATVPDIVNNFIVNIKNIYNLGARSFWIHSTVPSGCTPTILANFPSA 242
Query: 230 NEKGNLDANGCVKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARN 289
+ D+ GC K +NE++Q +N +LK + QLR PLA TYVD+Y+ Y+L N +
Sbjct: 243 IK----DSYGCAKQYNEVSQYFNLKLKKALAQLRVDLPLAAITYVDIYSPNYSLFQNPKK 298
Query: 290 QGFVNPLEFCCGSYQG--NEIHYCGKKSIKNGT-VYGIACDDSSTYI 333
GF P CCG Y G N CG+ NGT + +C + ST I
Sbjct: 299 YGFELPHVACCG-YGGKYNIRVGCGETLNINGTKIEAGSCKNPSTRI 344
>gb|AAC64890.1| Similar to nodulins and lipase homolog F14J9.5 gi|3482914 from
Arabidopsis thaliana BAC gb|AC003970. Alternate first
exon from 72258 to 72509 gi|25405771|pir||A96590
hypothetical protein T22H22.20 [imported] - Arabidopsis
thaliana
Length = 383
Score = 209 bits (533), Expect = 7e-53
Identities = 133/339 (39%), Positives = 187/339 (54%), Gaps = 20/339 (5%)
Query: 9 ILCFFNLCVACPSKKCV---YPAIYNFGDSNSDTGAGY-ATMAAVEHPNGISFFGSISGR 64
I+ F + + PS + YPAI NFGDSNSDTG A + V P G ++F SGR
Sbjct: 10 IVLFILISLFLPSSFSIILKYPAIINFGDSNSDTGNLISAGIENVNPPYGQTYFNLPSGR 69
Query: 65 CCDGRLILDFISEELELPYLSSYLNSVG-SNYRHGANFAVASA---PIRPIIAGLTYLGF 120
CDGRLI+DF+ +E++LP+L+ YL+S+G N++ G NFA A + P P
Sbjct: 70 YCDGRLIVDFLLDEMDLPFLNPYLDSLGLPNFKKGCNFAAAGSTILPANPTSVSPFSFDL 129
Query: 121 QVSQFILFKSHTKILFDQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQKPNSSEE 180
Q+SQFI FKS L +T +P + +SK +Y IDIGQNDI + + +
Sbjct: 130 QISQFIRFKSRAIELLS-KTGRKYEKYLPPIDYYSKGLYMIDIGQNDIAGAFY--SKTLD 186
Query: 181 EVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGC 240
+V SIP IL F +++LY E R WIHNTGP+ C+ + + K LD GC
Sbjct: 187 QVLASIPSILETFEAGLKRLYEEGGRNIWIHNTGPLGCLAQNIAKFGTDSTK--LDEFGC 244
Query: 241 VKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVNPLEFCC 300
V HN+ A+ +N QL + + +P A TYVD++++K LI+N GF PL CC
Sbjct: 245 VSSHNQAAKLFNLQLHAMSNKFQAQYPDANVTYVDIFSIKSNLIANYSRFGFEKPLMACC 304
Query: 301 GSYQGNEIHY-----CGKKSIKNG-TVYGIACDDSSTYI 333
G G ++Y CG+ + +G +V AC+DSS YI
Sbjct: 305 G-VGGAPLNYDSRITCGQTKVLDGISVTAKACNDSSEYI 342
>gb|AAM61525.1| early nodule-specific protein, putative [Arabidopsis thaliana]
Length = 377
Score = 209 bits (532), Expect = 1e-52
Identities = 130/342 (38%), Positives = 189/342 (55%), Gaps = 18/342 (5%)
Query: 4 MTLIYILCFFNLCVACPSK-KCVYPAIYNFGDSNSDTGAGYATMAA-VEHPNGISFFGSI 61
M L Y++ FF + + YP+ +NFGDSNSDTG A + ++ PNG + F +
Sbjct: 1 MKLFYVILFFISSLQISNSIDFNYPSAFNFGDSNSDTGDLVAGLGIRLDLPNGQNSFKTS 60
Query: 62 SGRCCDGRLILDFISEELELPYLSSYLNSVG-SNYRHGANFAVASA---PIRPIIAGLTY 117
S R CDGRL++DF+ +E++LP+L+ YL+S+G N++ G NFA A + P P
Sbjct: 61 SQRFCDGRLVIDFLMDEMDLPFLNPYLDSLGLPNFKKGCNFAAAGSTILPANPTSVSPFS 120
Query: 118 LGFQVSQFILFKSHTKILFDQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQKPNS 177
Q+SQFI FKS L +T +P + +SK +Y IDIGQNDI +
Sbjct: 121 FDLQISQFIRFKSRAIELLS-KTGRKYEKYLPPIDYYSKGLYMIDIGQNDIAGAFY--SK 177
Query: 178 SEEEVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDA 237
+ ++V SIP IL F +++LY E R WIHNTGP+ C+ + + K LD
Sbjct: 178 TLDQVLASIPSILETFEAGLKRLYEEGGRNIWIHNTGPLGCLAQNIAKFGTDSTK--LDE 235
Query: 238 NGCVKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVNPLE 297
GCV HN+ A+ +N QL + + +P A TYVD++++K LI+N GF PL
Sbjct: 236 FGCVSSHNQAAKLFNLQLHAMSNKFQAQYPDANVTYVDIFSIKSNLIANYSRFGFEKPLM 295
Query: 298 FCCGSYQGNEIHY-----CGKKSIKNG-TVYGIACDDSSTYI 333
CCG G ++Y CG+ + +G +V AC+DSS YI
Sbjct: 296 ACCG-VGGAPLNYDSRITCGQTKVLDGISVTAKACNDSSEYI 336
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.322 0.139 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 606,364,029
Number of Sequences: 2540612
Number of extensions: 26342783
Number of successful extensions: 56101
Number of sequences better than 10.0: 375
Number of HSP's better than 10.0 without gapping: 254
Number of HSP's successfully gapped in prelim test: 121
Number of HSP's that attempted gapping in prelim test: 54971
Number of HSP's gapped (non-prelim): 436
length of query: 341
length of database: 863,360,394
effective HSP length: 128
effective length of query: 213
effective length of database: 538,162,058
effective search space: 114628518354
effective search space used: 114628518354
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 75 (33.5 bits)
Medicago: description of AC136841.9


