

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136506.11 - phase: 0 /pseudo
(1043 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
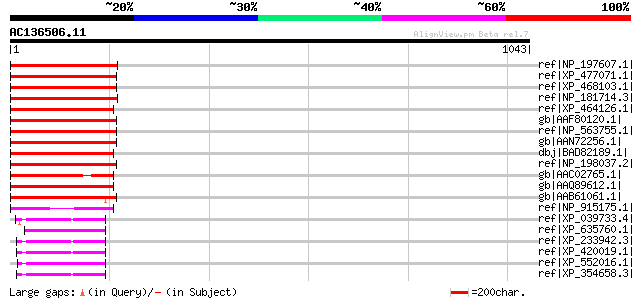
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_197607.1| expressed protein [Arabidopsis thaliana] 309 3e-82
ref|XP_477071.1| cyclin-like protein [Oryza sativa (japonica cul... 293 2e-77
ref|XP_468103.1| cyclin-like protein [Oryza sativa (japonica cul... 287 1e-75
ref|NP_181714.3| cyclin-related [Arabidopsis thaliana] 286 3e-75
ref|XP_464126.1| cyclin-like protein [Oryza sativa (japonica cul... 281 8e-74
gb|AAF80120.1| Contains similarity to an unknown protein T11A7.7... 277 2e-72
ref|NP_563755.1| expressed protein [Arabidopsis thaliana] 277 2e-72
gb|AAN72256.1| At1g05960/T21E18_20 [Arabidopsis thaliana] gi|141... 277 2e-72
dbj|BAD82189.1| cyclin-like [Oryza sativa (japonica cultivar-gro... 255 5e-66
ref|NP_198037.2| expressed protein [Arabidopsis thaliana] 251 7e-65
gb|AAC02765.1| hypothetical protein [Arabidopsis thaliana] gi|25... 251 9e-65
gb|AAQ89612.1| At5g26850 [Arabidopsis thaliana] gi|26449532|dbj|... 250 2e-64
gb|AAB61061.1| Hypothetical protein F2P16.24 [Arabidopsis thalia... 240 2e-61
ref|NP_915175.1| P0674H09.27 [Oryza sativa (japonica cultivar-gr... 162 5e-38
ref|XP_039733.4| PREDICTED: KIAA0953 protein [Homo sapiens] 75 1e-11
ref|XP_635760.1| hypothetical protein DDB0219543 [Dictyostelium ... 74 3e-11
ref|XP_233942.3| PREDICTED: similar to KIAA0953 protein [Rattus ... 74 3e-11
ref|XP_420019.1| PREDICTED: similar to KIAA0953 protein [Gallus ... 74 3e-11
ref|XP_552016.1| ENSANGP00000028460 [Anopheles gambiae str. PEST... 74 3e-11
ref|XP_354658.3| PREDICTED: similar to KIAA0953 protein [Mus mus... 74 3e-11
>ref|NP_197607.1| expressed protein [Arabidopsis thaliana]
Length = 980
Score = 309 bits (792), Expect = 3e-82
Identities = 145/216 (67%), Positives = 178/216 (82%)
Query: 1 MSVISRNIWPVCGSLCCFCPALRERSRHPIKRYKKLLADIFPRTPEEEPNDRKISKLCEY 60
M V+SR ++PVC SLCCFCPALR RSRHP+KRYK LLADIFPR+ +E+PNDRKI KLCEY
Sbjct: 1 MGVVSRTVFPVCESLCCFCPALRARSRHPVKRYKHLLADIFPRSQDEQPNDRKIGKLCEY 60
Query: 61 ASKNPLRVPKITSYLEQRCYKELRTENYQAVKVVICIYRKLLVSCRDQMPLFASSLLSII 120
A+KNPLR+PKIT+ LEQRCYKELR E + +VK+V+ IY+KLLVSC +QM LFASS L +I
Sbjct: 61 AAKNPLRIPKITTSLEQRCYKELRMEQFHSVKIVMSIYKKLLVSCNEQMLLFASSYLGLI 120
Query: 121 QILLDQSRQDEVQILGCQTLFDFVNNQRDGTYMFNLDSFILKLCHLAQQVGDDGKVEHLR 180
ILLDQ+R DE++ILGC+ L+DFV +Q +GTYMFNLD I K+C LA ++G++ +L
Sbjct: 121 HILLDQTRYDEMRILGCEALYDFVTSQAEGTYMFNLDGLIPKICPLAHELGEEDSTTNLC 180
Query: 181 ASGLQVLSSMVWFMGEFTHISVEFDNVSLASLVNSG 216
A+GLQ LSS+VWFMGEF+HISVEFDNV L N G
Sbjct: 181 AAGLQALSSLVWFMGEFSHISVEFDNVVSVVLENYG 216
>ref|XP_477071.1| cyclin-like protein [Oryza sativa (japonica cultivar-group)]
gi|34393302|dbj|BAC83231.1| cyclin-like protein [Oryza
sativa (japonica cultivar-group)]
Length = 1066
Score = 293 bits (750), Expect = 2e-77
Identities = 139/214 (64%), Positives = 170/214 (78%)
Query: 1 MSVISRNIWPVCGSLCCFCPALRERSRHPIKRYKKLLADIFPRTPEEEPNDRKISKLCEY 60
M VISR + P CGSLC FCP LR RSR P+KRYK +LA+IFP+T +EEPN+R+I KLCEY
Sbjct: 1 MGVISRKVLPACGSLCYFCPGLRARSRQPVKRYKSILAEIFPKTQDEEPNERRIGKLCEY 60
Query: 61 ASKNPLRVPKITSYLEQRCYKELRTENYQAVKVVICIYRKLLVSCRDQMPLFASSLLSII 120
S+NPLRVPKIT LEQR YKELR+E Y KVV+ IYR+LLVSC++QMPLFASSLLSI+
Sbjct: 61 CSRNPLRVPKITVSLEQRIYKELRSEQYGFAKVVMLIYRRLLVSCKEQMPLFASSLLSIV 120
Query: 121 QILLDQSRQDEVQILGCQTLFDFVNNQRDGTYMFNLDSFILKLCHLAQQVGDDGKVEHLR 180
LLDQ RQD+++I+GC+TLFDF NQ DGTY FNL+ + +LC L+Q+VG+D + LR
Sbjct: 121 HTLLDQKRQDDMRIIGCETLFDFAVNQVDGTYQFNLEGLVPRLCELSQEVGEDEQTIALR 180
Query: 181 ASGLQVLSSMVWFMGEFTHISVEFDNVSLASLVN 214
A+ LQ LS+M+WFMGE +HIS EFDNV L N
Sbjct: 181 AAALQALSAMIWFMGELSHISSEFDNVVQVVLEN 214
>ref|XP_468103.1| cyclin-like protein [Oryza sativa (japonica cultivar-group)]
gi|51964448|ref|XP_507009.1| PREDICTED OJ1293_A01.36
gene product [Oryza sativa (japonica cultivar-group)]
gi|47497474|dbj|BAD19529.1| cyclin-like protein [Oryza
sativa (japonica cultivar-group)]
Length = 997
Score = 287 bits (735), Expect = 1e-75
Identities = 135/214 (63%), Positives = 174/214 (81%)
Query: 1 MSVISRNIWPVCGSLCCFCPALRERSRHPIKRYKKLLADIFPRTPEEEPNDRKISKLCEY 60
M V+SR + P C LC CP+LR RSRHP+KRYKKLL++IFP++ +EEPNDRKI KLCEY
Sbjct: 3 MGVVSREVLPACERLCFLCPSLRTRSRHPVKRYKKLLSEIFPKSQDEEPNDRKIGKLCEY 62
Query: 61 ASKNPLRVPKITSYLEQRCYKELRTENYQAVKVVICIYRKLLVSCRDQMPLFASSLLSII 120
S+NPLRVPKIT YLEQ+ YKELR E++ +VKVV+ IYRK++ SC++Q+PLFA+SLL+I+
Sbjct: 63 ISRNPLRVPKITVYLEQKFYKELRVEHFGSVKVVMAIYRKVICSCQEQLPLFANSLLNIV 122
Query: 121 QILLDQSRQDEVQILGCQTLFDFVNNQRDGTYMFNLDSFILKLCHLAQQVGDDGKVEHLR 180
+ LL+Q+RQD+++ + C+TLF FVNNQ D TYMFNL+S I KLC LAQ++G+ K+ +
Sbjct: 123 EALLEQNRQDDLRTIACRTLFYFVNNQVDSTYMFNLESQIPKLCQLAQEMGEKEKISIVH 182
Query: 181 ASGLQVLSSMVWFMGEFTHISVEFDNVSLASLVN 214
A+GLQ LSSMVWFMGE +HIS E DNV A L N
Sbjct: 183 AAGLQALSSMVWFMGEHSHISAELDNVVSAVLEN 216
>ref|NP_181714.3| cyclin-related [Arabidopsis thaliana]
Length = 1025
Score = 286 bits (732), Expect = 3e-75
Identities = 133/214 (62%), Positives = 169/214 (78%)
Query: 3 VISRNIWPVCGSLCCFCPALRERSRHPIKRYKKLLADIFPRTPEEEPNDRKISKLCEYAS 62
VISR + PVCGSLC CPALR RSR P+KRYKKL+A+IFPR EE NDRKI KLCEYA+
Sbjct: 7 VISRQVLPVCGSLCILCPALRARSRQPVKRYKKLIAEIFPRNQEEGINDRKIGKLCEYAA 66
Query: 63 KNPLRVPKITSYLEQRCYKELRTENYQAVKVVICIYRKLLVSCRDQMPLFASSLLSIIQI 122
KN +R+PKI+ LE RCYKELR EN+ + K+ +CIYR+LLV+C++Q+PLF+S L +Q
Sbjct: 67 KNAVRMPKISDSLEHRCYKELRNENFHSAKIAMCIYRRLLVTCKEQIPLFSSGFLRTVQA 126
Query: 123 LLDQSRQDEVQILGCQTLFDFVNNQRDGTYMFNLDSFILKLCHLAQQVGDDGKVEHLRAS 182
LLDQ+RQDE+QI+GCQ+LF+FV NQ+DG+ +FNL+ F+ KLC L + GDD + LRA+
Sbjct: 127 LLDQTRQDEMQIVGCQSLFEFVINQKDGSSLFNLEGFLPKLCQLGLEGGDDDRSRSLRAA 186
Query: 183 GLQVLSSMVWFMGEFTHISVEFDNVSLASLVNSG 216
GLQ LS+M+W MGE++HI EFDNV A L N G
Sbjct: 187 GLQALSAMIWLMGEYSHIPSEFDNVVSAVLENYG 220
>ref|XP_464126.1| cyclin-like protein [Oryza sativa (japonica cultivar-group)]
gi|45736188|dbj|BAD13233.1| cyclin-like protein [Oryza
sativa (japonica cultivar-group)]
Length = 1035
Score = 281 bits (719), Expect = 8e-74
Identities = 135/205 (65%), Positives = 162/205 (78%)
Query: 3 VISRNIWPVCGSLCCFCPALRERSRHPIKRYKKLLADIFPRTPEEEPNDRKISKLCEYAS 62
V+SR + P CG LC FCP LR RSR P+KRYKK++ADIFP T +EEPN+R+I KLCEY +
Sbjct: 13 VVSRKVLPACGGLCYFCPGLRARSRQPVKRYKKIIADIFPATQDEEPNERRIGKLCEYVA 72
Query: 63 KNPLRVPKITSYLEQRCYKELRTENYQAVKVVICIYRKLLVSCRDQMPLFASSLLSIIQI 122
+N RVPKIT+YLEQRCYKELR E Y VKVV+ IYRKLLVSC+ QMPL ASS LSII
Sbjct: 73 RNHHRVPKITAYLEQRCYKELRNEQYGFVKVVVLIYRKLLVSCKKQMPLLASSALSIICT 132
Query: 123 LLDQSRQDEVQILGCQTLFDFVNNQRDGTYMFNLDSFILKLCHLAQQVGDDGKVEHLRAS 182
LLDQ+R+D+++I+GC+TLFDF +Q DGTY FNL+ + KLC LAQ V + K LRAS
Sbjct: 133 LLDQTRRDDMRIIGCETLFDFTVSQVDGTYQFNLEELVPKLCELAQIVKAEEKDNMLRAS 192
Query: 183 GLQVLSSMVWFMGEFTHISVEFDNV 207
LQ LS+M+WFMGEF+HIS FDNV
Sbjct: 193 TLQALSAMIWFMGEFSHISSAFDNV 217
>gb|AAF80120.1| Contains similarity to an unknown protein T11A7.7 gi|2335096 from
Arabidopsis thaliana BAC T11A7 gb|AC002339 and contains
a tropomyosin PF|00261 domain. ESTs gb|AI995205,
gb|N37925, gb|F13889, gb|AV523107, gb|AV535948,
gb|AV558461, gb|F13888 come from this gene
gi|25406886|pir||F86194 hypothetical protein [imported]
- Arabidopsis thaliana
Length = 1628
Score = 277 bits (708), Expect = 2e-72
Identities = 129/214 (60%), Positives = 170/214 (79%)
Query: 1 MSVISRNIWPVCGSLCCFCPALRERSRHPIKRYKKLLADIFPRTPEEEPNDRKISKLCEY 60
M V+SR + P CG+LC FCP+LR RSRHP+KRYKK+LA+IFPR E EPNDRKI KLCEY
Sbjct: 1 MGVMSRRVLPACGNLCFFCPSLRARSRHPVKRYKKMLAEIFPRNQEAEPNDRKIGKLCEY 60
Query: 61 ASKNPLRVPKITSYLEQRCYKELRTENYQAVKVVICIYRKLLVSCRDQMPLFASSLLSII 120
AS+NPLR+PKIT YLEQ+CYKELR N +VKVV+CIY+KLL SC++QMPLF+ SLLSI+
Sbjct: 61 ASRNPLRIPKITEYLEQKCYKELRNGNIGSVKVVLCIYKKLLSSCKEQMPLFSCSLLSIV 120
Query: 121 QILLDQSRQDEVQILGCQTLFDFVNNQRDGTYMFNLDSFILKLCHLAQQVGDDGKVEHLR 180
+ LL+Q++++EVQILGC TL DF++ Q ++MFNL+ I KLC LAQ++GDD + LR
Sbjct: 121 RTLLEQTKEEEVQILGCNTLVDFISLQTVNSHMFNLEGLIPKLCQLAQEMGDDERSLQLR 180
Query: 181 ASGLQVLSSMVWFMGEFTHISVEFDNVSLASLVN 214
++G+Q L+ MV F+GE + +S++ D + L N
Sbjct: 181 SAGMQALAFMVSFIGEHSQLSMDLDMIISVILEN 214
>ref|NP_563755.1| expressed protein [Arabidopsis thaliana]
Length = 982
Score = 277 bits (708), Expect = 2e-72
Identities = 129/214 (60%), Positives = 170/214 (79%)
Query: 1 MSVISRNIWPVCGSLCCFCPALRERSRHPIKRYKKLLADIFPRTPEEEPNDRKISKLCEY 60
M V+SR + P CG+LC FCP+LR RSRHP+KRYKK+LA+IFPR E EPNDRKI KLCEY
Sbjct: 1 MGVMSRRVLPACGNLCFFCPSLRARSRHPVKRYKKMLAEIFPRNQEAEPNDRKIGKLCEY 60
Query: 61 ASKNPLRVPKITSYLEQRCYKELRTENYQAVKVVICIYRKLLVSCRDQMPLFASSLLSII 120
AS+NPLR+PKIT YLEQ+CYKELR N +VKVV+CIY+KLL SC++QMPLF+ SLLSI+
Sbjct: 61 ASRNPLRIPKITEYLEQKCYKELRNGNIGSVKVVLCIYKKLLSSCKEQMPLFSCSLLSIV 120
Query: 121 QILLDQSRQDEVQILGCQTLFDFVNNQRDGTYMFNLDSFILKLCHLAQQVGDDGKVEHLR 180
+ LL+Q++++EVQILGC TL DF++ Q ++MFNL+ I KLC LAQ++GDD + LR
Sbjct: 121 RTLLEQTKEEEVQILGCNTLVDFISLQTVNSHMFNLEGLIPKLCQLAQEMGDDERSLQLR 180
Query: 181 ASGLQVLSSMVWFMGEFTHISVEFDNVSLASLVN 214
++G+Q L+ MV F+GE + +S++ D + L N
Sbjct: 181 SAGMQALAFMVSFIGEHSQLSMDLDMIISVILEN 214
>gb|AAN72256.1| At1g05960/T21E18_20 [Arabidopsis thaliana]
gi|14194169|gb|AAK56279.1| At1g05960/T21E18_20
[Arabidopsis thaliana]
Length = 731
Score = 277 bits (708), Expect = 2e-72
Identities = 129/214 (60%), Positives = 170/214 (79%)
Query: 1 MSVISRNIWPVCGSLCCFCPALRERSRHPIKRYKKLLADIFPRTPEEEPNDRKISKLCEY 60
M V+SR + P CG+LC FCP+LR RSRHP+KRYKK+LA+IFPR E EPNDRKI KLCEY
Sbjct: 1 MGVMSRRVLPACGNLCFFCPSLRARSRHPVKRYKKMLAEIFPRNQEAEPNDRKIGKLCEY 60
Query: 61 ASKNPLRVPKITSYLEQRCYKELRTENYQAVKVVICIYRKLLVSCRDQMPLFASSLLSII 120
AS+NPLR+PKIT YLEQ+CYKELR N +VKVV+CIY+KLL SC++QMPLF+ SLLSI+
Sbjct: 61 ASRNPLRIPKITEYLEQKCYKELRNGNIGSVKVVLCIYKKLLSSCKEQMPLFSCSLLSIV 120
Query: 121 QILLDQSRQDEVQILGCQTLFDFVNNQRDGTYMFNLDSFILKLCHLAQQVGDDGKVEHLR 180
+ LL+Q++++EVQILGC TL DF++ Q ++MFNL+ I KLC LAQ++GDD + LR
Sbjct: 121 RTLLEQTKEEEVQILGCNTLVDFISLQTVNSHMFNLEGLIPKLCQLAQEMGDDERSLQLR 180
Query: 181 ASGLQVLSSMVWFMGEFTHISVEFDNVSLASLVN 214
++G+Q L+ MV F+GE + +S++ D + L N
Sbjct: 181 SAGMQALAFMVSFIGEHSQLSMDLDMIISVILEN 214
>dbj|BAD82189.1| cyclin-like [Oryza sativa (japonica cultivar-group)]
gi|56784331|dbj|BAD82352.1| cyclin-like [Oryza sativa
(japonica cultivar-group)]
Length = 980
Score = 255 bits (652), Expect = 5e-66
Identities = 118/207 (57%), Positives = 155/207 (74%)
Query: 1 MSVISRNIWPVCGSLCCFCPALRERSRHPIKRYKKLLADIFPRTPEEEPNDRKISKLCEY 60
M V+SR + P C SLC FCP+LR RSR P+KRYKK++A+I+ P+ EPNDR+I KLC+Y
Sbjct: 1 MGVMSRRVLPACSSLCYFCPSLRARSRQPVKRYKKIIAEIYQLPPDGEPNDRRIGKLCDY 60
Query: 61 ASKNPLRVPKITSYLEQRCYKELRTENYQAVKVVICIYRKLLVSCRDQMPLFASSLLSII 120
S+NP R+PKIT YLE+RCYK+LR EN+ KVV CIYRKLL SC+D PL A+S LSII
Sbjct: 61 VSRNPTRIPKITEYLEERCYKDLRHENFTLAKVVPCIYRKLLCSCKDHTPLLATSTLSII 120
Query: 121 QILLDQSRQDEVQILGCQTLFDFVNNQRDGTYMFNLDSFILKLCHLAQQVGDDGKVEHLR 180
+ LLDQ D++++LGC L DF+N Q D T+MFNL+ I KLC ++Q++ +D K LR
Sbjct: 121 RTLLDQRMNDDLRVLGCLMLVDFLNGQVDSTHMFNLEGLIPKLCQISQELREDDKGFRLR 180
Query: 181 ASGLQVLSSMVWFMGEFTHISVEFDNV 207
+ LQ L+SMV +MG+ +HIS+E D V
Sbjct: 181 CAALQALASMVQYMGDHSHISMELDEV 207
>ref|NP_198037.2| expressed protein [Arabidopsis thaliana]
Length = 919
Score = 251 bits (642), Expect = 7e-65
Identities = 117/214 (54%), Positives = 159/214 (73%)
Query: 1 MSVISRNIWPVCGSLCCFCPALRERSRHPIKRYKKLLADIFPRTPEEEPNDRKISKLCEY 60
M ISRN++P C S+C CPALR RSR P+KRYKKLL +IFP++P+ PN+RKI KLCEY
Sbjct: 1 MGFISRNVFPACESMCICCPALRSRSRQPVKRYKKLLGEIFPKSPDGGPNERKIVKLCEY 60
Query: 61 ASKNPLRVPKITSYLEQRCYKELRTENYQAVKVVICIYRKLLVSCRDQMPLFASSLLSII 120
A+KNP+R+PKI +LE+RCYK+LR+E + + +V Y K+L C+DQM FA+SLL+++
Sbjct: 61 AAKNPIRIPKIAKFLEERCYKDLRSEQMKFINIVTEAYNKMLCHCKDQMAYFATSLLNVV 120
Query: 121 QILLDQSRQDEVQILGCQTLFDFVNNQRDGTYMFNLDSFILKLCHLAQQVGDDGKVEHLR 180
LLD S+QD ILGCQTL F+ +Q DGTY +++ F LK+C LA++ G++ + + LR
Sbjct: 121 TELLDNSKQDTPTILGCQTLTRFIYSQVDGTYTHSIEKFALKVCSLAREEGEEHQKQCLR 180
Query: 181 ASGLQVLSSMVWFMGEFTHISVEFDNVSLASLVN 214
ASGLQ LS+MVW+MGEF+HI D + A L N
Sbjct: 181 ASGLQCLSAMVWYMGEFSHIFATVDEIVHAILDN 214
>gb|AAC02765.1| hypothetical protein [Arabidopsis thaliana] gi|25408758|pir||F84846
hypothetical protein At2g41830 [imported] - Arabidopsis
thaliana
Length = 961
Score = 251 bits (641), Expect = 9e-65
Identities = 123/210 (58%), Positives = 154/210 (72%), Gaps = 18/210 (8%)
Query: 3 VISRNIWPVCGSLCCFCPALRERSRHPIKRYKKLLADIFPRTPEEEPNDRKISKLCEYAS 62
VISR + PVCGSLC CPALR RSR P+KRYKKL+A+IFPR EE NDRKI KLCEYA+
Sbjct: 7 VISRQVLPVCGSLCILCPALRARSRQPVKRYKKLIAEIFPRNQEEGINDRKIGKLCEYAA 66
Query: 63 KNPLRVPK---ITSYLEQRCYKELRTENYQAVKVVICIYRKLLVSCRDQMPLFASSLLSI 119
KN +R+PK I+ LE RCYKELR EN+ + K+ +CIYR+LLV+C++Q+PLF+S L
Sbjct: 67 KNAVRMPKFFQISDSLEHRCYKELRNENFHSAKIAMCIYRRLLVTCKEQIPLFSSGFLRT 126
Query: 120 IQILLDQSRQDEVQILGCQTLFDFVNNQRDGTYMFNLDSFILKLCHLAQQVGDDGKVEHL 179
+Q LLDQ+RQDE+QI+GCQ+LF+FV NQ LC L + GDD + L
Sbjct: 127 VQALLDQTRQDEMQIVGCQSLFEFVINQ---------------LCQLGLEGGDDDRSRSL 171
Query: 180 RASGLQVLSSMVWFMGEFTHISVEFDNVSL 209
RA+GLQ LS+M+W MGE++HI EFDNV L
Sbjct: 172 RAAGLQALSAMIWLMGEYSHIPSEFDNVLL 201
>gb|AAQ89612.1| At5g26850 [Arabidopsis thaliana] gi|26449532|dbj|BAC41892.1|
unknown protein [Arabidopsis thaliana]
Length = 970
Score = 250 bits (638), Expect = 2e-64
Identities = 114/207 (55%), Positives = 156/207 (75%)
Query: 1 MSVISRNIWPVCGSLCCFCPALRERSRHPIKRYKKLLADIFPRTPEEEPNDRKISKLCEY 60
M ISRN++P C S+C CPALR RSR P+KRYKKLL +IFP++P+ PN+RKI KLCEY
Sbjct: 1 MGFISRNVFPACESMCICCPALRSRSRQPVKRYKKLLGEIFPKSPDGGPNERKIVKLCEY 60
Query: 61 ASKNPLRVPKITSYLEQRCYKELRTENYQAVKVVICIYRKLLVSCRDQMPLFASSLLSII 120
A+KNP+R+PKI +LE+RCYK+LR+E + + +V Y K+L C+DQM FA+SLL+++
Sbjct: 61 AAKNPIRIPKIAKFLEERCYKDLRSEQMKFINIVTEAYNKMLCHCKDQMAYFATSLLNVV 120
Query: 121 QILLDQSRQDEVQILGCQTLFDFVNNQRDGTYMFNLDSFILKLCHLAQQVGDDGKVEHLR 180
LLD S+QD ILGCQTL F+ +Q DGTY +++ F LK+C LA++ G++ + + LR
Sbjct: 121 TELLDNSKQDTPTILGCQTLTRFIYSQVDGTYTHSIEKFALKVCSLAREEGEEHQKQCLR 180
Query: 181 ASGLQVLSSMVWFMGEFTHISVEFDNV 207
ASGLQ LS+MVW+MGEF+HI D +
Sbjct: 181 ASGLQCLSAMVWYMGEFSHIFATVDEI 207
>gb|AAB61061.1| Hypothetical protein F2P16.24 [Arabidopsis thaliana]
gi|7485250|pir||T01764 hypothetical protein
A_IG002P16.24 - Arabidopsis thaliana
Length = 907
Score = 240 bits (613), Expect = 2e-61
Identities = 117/232 (50%), Positives = 159/232 (68%), Gaps = 18/232 (7%)
Query: 1 MSVISRNIWPVCGSLCCFCPALRERSRHPIKRYKKLLADIFPRTPEEEPNDRKISKLCEY 60
M ISRN++P C S+C CPALR RSR P+KRYKKLL +IFP++P+ PN+RKI KLCEY
Sbjct: 1 MGFISRNVFPACESMCICCPALRSRSRQPVKRYKKLLGEIFPKSPDGGPNERKIVKLCEY 60
Query: 61 ASKNPLRVPKITSYLEQRCYKELRTENYQAVKVVICIYRKLLVSCRDQMPLFASSLLSII 120
A+KNP+R+PKI +LE+RCYK+LR+E + + +V Y K+L C+DQM FA+SLL+++
Sbjct: 61 AAKNPIRIPKIAKFLEERCYKDLRSEQMKFINIVTEAYNKMLCHCKDQMAYFATSLLNVV 120
Query: 121 QILLDQSRQDEVQILGCQTLFDFVNNQRDGTYMFNLDSFILKLCHLAQQVGDDGKVEHLR 180
LLD S+QD ILGCQTL F+ +Q DGTY +++ F LK+C LA++ G++ + + LR
Sbjct: 121 TELLDNSKQDTPTILGCQTLTRFIYSQVDGTYTHSIEKFALKVCSLAREEGEEHQKQCLR 180
Query: 181 ASGLQVLSSM------------------VWFMGEFTHISVEFDNVSLASLVN 214
ASGLQ LS+M VW+MGEF+HI D + A L N
Sbjct: 181 ASGLQCLSAMVLELVLLIFQIQSILLPKVWYMGEFSHIFATVDEIVHAILDN 232
>ref|NP_915175.1| P0674H09.27 [Oryza sativa (japonica cultivar-group)]
Length = 894
Score = 162 bits (410), Expect = 5e-38
Identities = 83/207 (40%), Positives = 115/207 (55%), Gaps = 46/207 (22%)
Query: 1 MSVISRNIWPVCGSLCCFCPALRERSRHPIKRYKKLLADIFPRTPEEEPNDRKISKLCEY 60
M V+SR + P C SLC FCP+LR RSR P+KRYKK++A+I+ P+ EPNDR+I KLC+Y
Sbjct: 1 MGVMSRRVLPACSSLCYFCPSLRARSRQPVKRYKKIIAEIYQLPPDGEPNDRRIGKLCDY 60
Query: 61 ASKNPLRVPKITSYLEQRCYKELRTENYQAVKVVICIYRKLLVSCRDQMPLFASSLLSII 120
S+NP R+PK S Q Y+
Sbjct: 61 VSRNPTRIPKEASVFMQGSYR--------------------------------------- 81
Query: 121 QILLDQSRQDEVQILGCQTLFDFVNNQRDGTYMFNLDSFILKLCHLAQQVGDDGKVEHLR 180
D++++LGC L DF+N Q D T+MFNL+ I KLC ++Q++ +D K LR
Sbjct: 82 -------MNDDLRVLGCLMLVDFLNGQVDSTHMFNLEGLIPKLCQISQELREDDKGFRLR 134
Query: 181 ASGLQVLSSMVWFMGEFTHISVEFDNV 207
+ LQ L+SMV +MG+ +HIS+E D V
Sbjct: 135 CAALQALASMVQYMGDHSHISMELDEV 161
>ref|XP_039733.4| PREDICTED: KIAA0953 protein [Homo sapiens]
Length = 1077
Score = 74.7 bits (182), Expect = 1e-11
Identities = 56/191 (29%), Positives = 95/191 (49%), Gaps = 19/191 (9%)
Query: 12 CGSLCCF--------CPALRERSRHPIKRYKKLLADIFPRTPEEEPNDRKISKLCEYASK 63
CGS+C C ALR R YK+L+ +IFP PE+ + KL YA
Sbjct: 253 CGSICGINKSGVCGCCGALRPR-------YKRLVDNIFPEDPEDGLVKTNMEKLTFYALS 305
Query: 64 NPLRVPKITSYLEQRCYKELRTENYQAVKVVICIYRKLLVSCRDQ-MPLFASSLLSIIQI 122
P ++ +I +YL +R +++ Y V + + +LL++C Q + LF S L ++
Sbjct: 306 APEKLDRIGAYLSERLIRDVGRHRYGYVCIAMEALDQLLMACHCQSINLFVESFLKMVAK 365
Query: 123 LLDQSRQDEVQILGCQTLFDFVNNQRD-GTYMFNLDSFILKLCHLAQQVGDDGKVE-HLR 180
LL +S + +QILG + F N + D +Y + D F+ + + DD +++ +R
Sbjct: 366 LL-ESEKPNLQILGTNSFVKFANIEEDTPSYHRSYDFFVSRFSEMCHSSHDDLEIKTKIR 424
Query: 181 ASGLQVLSSMV 191
SG++ L +V
Sbjct: 425 MSGIKGLQGVV 435
>ref|XP_635760.1| hypothetical protein DDB0219543 [Dictyostelium discoideum]
gi|60464145|gb|EAL62306.1| hypothetical protein
DDB0219543 [Dictyostelium discoideum]
Length = 946
Score = 73.9 bits (180), Expect = 3e-11
Identities = 45/163 (27%), Positives = 83/163 (50%), Gaps = 2/163 (1%)
Query: 31 KRYKKLLADIFPRTPEEEPNDRKISKLCEYASKNPLRVPKITSYLEQRCYKELRTENYQA 90
K+YK+++ IFP+ E ISKL + +P ++ K+ Y+E + K L +
Sbjct: 7 KKYKRIVKTIFPQVEGGEMQLGNISKLVYFCEMSPEQLQKVGPYIEAKAQKNLSKKRLVY 66
Query: 91 VKVVICIYRKLLVSCRDQMPLFASSLLSIIQILLDQSRQDEVQILGCQTLFDFVNNQRDG 150
V I I R+L++ CR + LF+ + +++ILL Q+ +QI +T F + Q D
Sbjct: 67 VSTCIIIIRELIIGCRKHLALFSGNATRLMKILLIQTEYPSLQIEATETFIRFASVQDDS 126
Query: 151 TYM-FNLDSFILKLCHLAQQVGDDGKVEH-LRASGLQVLSSMV 191
+ +D FI +A+ + +D + +R GL+ +++ V
Sbjct: 127 NQIPPEIDEFIKYFIIMAKNMDNDDTMRRTIRGEGLRGVTAYV 169
>ref|XP_233942.3| PREDICTED: similar to KIAA0953 protein [Rattus norvegicus]
Length = 817
Score = 73.6 bits (179), Expect = 3e-11
Identities = 53/180 (29%), Positives = 92/180 (50%), Gaps = 11/180 (6%)
Query: 15 LCCFCPALRERSRHPIKRYKKLLADIFPRTPEEEPNDRKISKLCEYASKNPLRVPKITSY 74
+C C ALR R YK+L+ +IFP PE+ + KL YA P ++ +I +Y
Sbjct: 4 VCGCCGALRPR-------YKRLVDNIFPEDPEDGLVKTNMEKLTFYALSAPEKLDRIGAY 56
Query: 75 LEQRCYKELRTENYQAVKVVICIYRKLLVSCRDQ-MPLFASSLLSIIQILLDQSRQDEVQ 133
L +R +++ Y V + + +LL++C Q + LF S L ++ LL +S + +Q
Sbjct: 57 LSERLIRDVGRHRYGYVCIAMEALDQLLMACHCQSINLFVESFLKMVAKLL-ESEKPNLQ 115
Query: 134 ILGCQTLFDFVNNQRD-GTYMFNLDSFILKLCHLAQQVGDDGKVE-HLRASGLQVLSSMV 191
ILG + F N + D +Y + D F+ + + DD +++ +R SG++ L +V
Sbjct: 116 ILGTNSFVKFANIEEDTPSYHRSYDFFVSRFSEMCHSSHDDLEIKTKIRMSGIKGLQGVV 175
>ref|XP_420019.1| PREDICTED: similar to KIAA0953 protein [Gallus gallus]
Length = 949
Score = 73.6 bits (179), Expect = 3e-11
Identities = 53/180 (29%), Positives = 91/180 (50%), Gaps = 11/180 (6%)
Query: 15 LCCFCPALRERSRHPIKRYKKLLADIFPRTPEEEPNDRKISKLCEYASKNPLRVPKITSY 74
+C C ALR R YK+L+ +IFP PE+ + KL YA P ++ +I +Y
Sbjct: 33 VCGCCGALRPR-------YKRLVDNIFPEDPEDGLVKTNMEKLTFYALSAPEKLDRIGAY 85
Query: 75 LEQRCYKELRTENYQAVKVVICIYRKLLVSCRDQ-MPLFASSLLSIIQILLDQSRQDEVQ 133
L +R +++ Y V + + +LL++C Q + LF S L ++ LL +S + +Q
Sbjct: 86 LSERLIRDVSRHRYGYVCIAMEALDQLLMACHCQSINLFVESFLKMVAKLL-ESEKPNLQ 144
Query: 134 ILGCQTLFDFVNNQRD-GTYMFNLDSFILKLCHLAQQVGDDGKVE-HLRASGLQVLSSMV 191
ILG + F N + D +Y + D F+ + + DD ++ +R SG++ L +V
Sbjct: 145 ILGTNSFVKFANIEEDTPSYHRSYDFFVSRFSEMCHSSHDDLEIRTKIRMSGIKGLQGVV 204
>ref|XP_552016.1| ENSANGP00000028460 [Anopheles gambiae str. PEST]
gi|55234481|gb|EAL38737.1| ENSANGP00000028460 [Anopheles
gambiae str. PEST]
Length = 793
Score = 73.6 bits (179), Expect = 3e-11
Identities = 52/179 (29%), Positives = 92/179 (51%), Gaps = 11/179 (6%)
Query: 16 CCFCPALRERSRHPIKRYKKLLADIFPRTPEEEPNDRKISKLCEYASKNPLRVPKITSYL 75
C C ALR R YK+L+ +IFP PE+ + KL Y+ ++P ++ +I YL
Sbjct: 5 CGCCSALRPR-------YKRLVDNIFPANPEDGLVKSNMEKLTFYSLRSPEKLDRIGEYL 57
Query: 76 EQRCYKELRTENYQAVKVVICIYRKLLVSCRDQ-MPLFASSLLSIIQILLDQSRQDEVQI 134
QR K++ + Y+ V++ + LL++C Q + LF S L ++Q LL+ + +QI
Sbjct: 58 YQRASKDIYRKRYKFVEIAMEAMDLLLMACHAQILNLFVESFLRMVQKLLEDT-NPTLQI 116
Query: 135 LGCQTLFDFVNNQRD-GTYMFNLDSFILKLCHLAQQVGDDGKV-EHLRASGLQVLSSMV 191
+ + F N + D +Y D FI K + DD ++ + +R +G++ L ++
Sbjct: 117 MATNSFVRFANIEEDTPSYHRRYDFFISKFSSMCYGNNDDMELRDSIRMAGIKGLQGVI 175
>ref|XP_354658.3| PREDICTED: similar to KIAA0953 protein [Mus musculus]
Length = 817
Score = 73.6 bits (179), Expect = 3e-11
Identities = 53/180 (29%), Positives = 92/180 (50%), Gaps = 11/180 (6%)
Query: 15 LCCFCPALRERSRHPIKRYKKLLADIFPRTPEEEPNDRKISKLCEYASKNPLRVPKITSY 74
+C C ALR R YK+L+ +IFP PE+ + KL YA P ++ +I +Y
Sbjct: 4 VCGCCGALRPR-------YKRLVDNIFPEDPEDGLVKTNMEKLTFYALSAPEKLDRIGAY 56
Query: 75 LEQRCYKELRTENYQAVKVVICIYRKLLVSCRDQ-MPLFASSLLSIIQILLDQSRQDEVQ 133
L +R +++ Y V + + +LL++C Q + LF S L ++ LL +S + +Q
Sbjct: 57 LSERLIRDVGRHRYGYVCIAMEALDQLLMACHCQSINLFVESFLKMVAKLL-ESEKPNLQ 115
Query: 134 ILGCQTLFDFVNNQRD-GTYMFNLDSFILKLCHLAQQVGDDGKVE-HLRASGLQVLSSMV 191
ILG + F N + D +Y + D F+ + + DD +++ +R SG++ L +V
Sbjct: 116 ILGTNSFVKFANIEEDTPSYHRSYDFFVSRFSEMCHSSHDDLEIKTKIRMSGIKGLQGVV 175
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.361 0.161 0.605
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,472,505,923
Number of Sequences: 2540612
Number of extensions: 54667720
Number of successful extensions: 191104
Number of sequences better than 10.0: 69
Number of HSP's better than 10.0 without gapping: 59
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 190989
Number of HSP's gapped (non-prelim): 82
length of query: 1043
length of database: 863,360,394
effective HSP length: 138
effective length of query: 905
effective length of database: 512,755,938
effective search space: 464044123890
effective search space used: 464044123890
T: 11
A: 40
X1: 14 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.9 bits)
S2: 81 (35.8 bits)
Medicago: description of AC136506.11


