

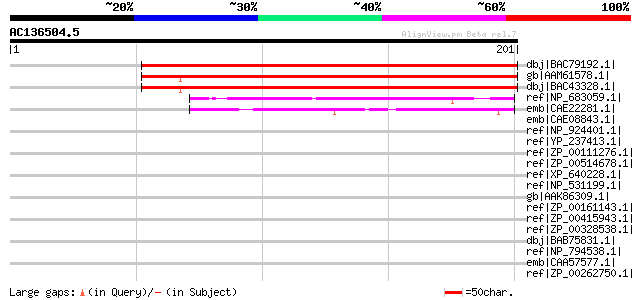
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136504.5 + phase: 0
(201 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAC79192.1| unknown protein [Oryza sativa (japonica cultivar... 222 5e-57
gb|AAM61578.1| unknown [Arabidopsis thaliana] gi|10177124|dbj|BA... 214 1e-54
dbj|BAC43328.1| unknown protein [Arabidopsis thaliana] 211 1e-53
ref|NP_683059.1| hypothetical protein tlr2269 [Thermosynechococc... 47 2e-04
emb|CAE22281.1| Universal stress protein (Usp) [Prochlorococcus ... 47 3e-04
emb|CAE08843.1| conserved hypothetical protein [Synechococcus sp... 45 0.001
ref|NP_924401.1| hypothetical protein gll1455 [Gloeobacter viola... 42 0.008
ref|YP_237413.1| Protein of unknown function UPF0004:tRNA-i(6)A3... 40 0.029
ref|ZP_00111276.1| COG0589: Universal stress protein UspA and re... 39 0.066
ref|ZP_00514678.1| conserved hypothetical protein [Crocosphaera ... 39 0.086
ref|XP_640228.1| hypothetical protein DDB0204950 [Dictyostelium ... 38 0.15
ref|NP_531199.1| hypothetical protein Atu0496 [Agrobacterium tum... 37 0.25
gb|AAK86309.1| AGR_C_878p [Agrobacterium tumefaciens str. C58] g... 37 0.25
ref|ZP_00161143.1| COG0589: Universal stress protein UspA and re... 37 0.33
ref|ZP_00415943.1| Protein of unknown function UPF0004:tRNA-i(6)... 36 0.56
ref|ZP_00328538.1| COG0589: Universal stress protein UspA and re... 36 0.56
dbj|BAB75831.1| alr4132 [Nostoc sp. PCC 7120] gi|17231624|ref|NP... 36 0.56
ref|NP_794538.1| tRNA-i(6)A37 modification enzyme MiaB [Pseudomo... 36 0.73
emb|CAA57577.1| hypothetical protein [Pseudomonas aeruginosa] 35 1.2
ref|ZP_00262750.1| COG0621: 2-methylthioadenine synthetase [Pseu... 35 1.2
>dbj|BAC79192.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|52076079|dbj|BAD46592.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 194
Score = 222 bits (565), Expect = 5e-57
Identities = 105/149 (70%), Positives = 128/149 (85%)
Query: 53 VKAKPQEPEVAIATDSFTQFKHLLLPITDRKPYLSEGTKQAIATTIALANKYGADITVVV 112
V+A+ E A+A D+F Q KH+LLP+TDR PYLSEGT+QA AT+ +LA KYGA+ITVVV
Sbjct: 46 VRAEVNESGSALAADAFAQVKHVLLPVTDRNPYLSEGTRQAAATSASLAKKYGANITVVV 105
Query: 113 IDEQQKESLPEHETQLSSIRWHLSEGGLKDYKLLERLGDGSKPTAIIGDVADELNLDLVV 172
ID++ KE PEH+TQ+SSIRWHLSEGG ++ L+ERLG+G KPTAIIG+VADEL LDLVV
Sbjct: 106 IDDKPKEEFPEHDTQMSSIRWHLSEGGFTEFGLMERLGEGKKPTAIIGEVADELELDLVV 165
Query: 173 ISMEAIHTKHIDANLLAEFIPCPVMLLPL 201
+SMEAIH+KH+D NLLAEFIPCPV+LLPL
Sbjct: 166 LSMEAIHSKHVDGNLLAEFIPCPVLLLPL 194
>gb|AAM61578.1| unknown [Arabidopsis thaliana] gi|10177124|dbj|BAB10414.1| unnamed
protein product [Arabidopsis thaliana]
gi|18425032|ref|NP_569028.1| expressed protein
[Arabidopsis thaliana]
Length = 210
Score = 214 bits (545), Expect = 1e-54
Identities = 106/153 (69%), Positives = 131/153 (85%), Gaps = 4/153 (2%)
Query: 53 VKAKPQEPEVAIAT----DSFTQFKHLLLPITDRKPYLSEGTKQAIATTIALANKYGADI 108
VKA+ +E E + + D+F+ KHLLLP+ DR PYLSEGT+QA ATT +LA KYGADI
Sbjct: 58 VKAQAKEAEASPSPVGVGDAFSNVKHLLLPVIDRNPYLSEGTRQAAATTTSLAKKYGADI 117
Query: 109 TVVVIDEQQKESLPEHETQLSSIRWHLSEGGLKDYKLLERLGDGSKPTAIIGDVADELNL 168
TVVVIDE+++ES EHETQ+S+IRWHLSEGG +++KLLERLG+G K TAIIG+VADEL +
Sbjct: 118 TVVVIDEEKRESSSEHETQVSNIRWHLSEGGFEEFKLLERLGEGKKATAIIGEVADELKM 177
Query: 169 DLVVISMEAIHTKHIDANLLAEFIPCPVMLLPL 201
+LVV+SMEAIH+K+IDANLLAEFIPCPV+LLPL
Sbjct: 178 ELVVMSMEAIHSKYIDANLLAEFIPCPVLLLPL 210
>dbj|BAC43328.1| unknown protein [Arabidopsis thaliana]
Length = 210
Score = 211 bits (537), Expect = 1e-53
Identities = 105/153 (68%), Positives = 129/153 (83%), Gaps = 4/153 (2%)
Query: 53 VKAKPQEPEVAIAT----DSFTQFKHLLLPITDRKPYLSEGTKQAIATTIALANKYGADI 108
VKA+ +E E + + D+F+ KHLLLP+ DR PYLS GT+QA ATT LA KYGADI
Sbjct: 58 VKAQAKEAEASPSPVGVGDAFSNVKHLLLPVIDRNPYLSGGTRQAAATTTGLAKKYGADI 117
Query: 109 TVVVIDEQQKESLPEHETQLSSIRWHLSEGGLKDYKLLERLGDGSKPTAIIGDVADELNL 168
TVVVIDE+++ES EHETQ+S+IRWHLSEGG +++KLLERLG+G K TAIIG+VADEL +
Sbjct: 118 TVVVIDEEKRESSSEHETQVSNIRWHLSEGGFEEFKLLERLGEGKKATAIIGEVADELKM 177
Query: 169 DLVVISMEAIHTKHIDANLLAEFIPCPVMLLPL 201
+LVV+SMEAIH+K+IDANLLAEFIPCPV+LLPL
Sbjct: 178 ELVVMSMEAIHSKYIDANLLAEFIPCPVLLLPL 210
>ref|NP_683059.1| hypothetical protein tlr2269 [Thermosynechococcus elongatus BP-1]
gi|22295996|dbj|BAC09821.1| tlr2269 [Thermosynechococcus
elongatus BP-1]
Length = 129
Score = 47.4 bits (111), Expect = 2e-04
Identities = 36/140 (25%), Positives = 67/140 (47%), Gaps = 23/140 (16%)
Query: 72 FKHLLLPITDRKPYLSEGTKQAIATTIALANKYGADITVVVIDEQQKESLPEHETQLSSI 131
FK +L PI DR S T++A+ T + + + + + ++ ++E + PE E ++
Sbjct: 2 FKTILFPI-DR----SRDTQEAMPTVVEMVKLFQSQLFLLSVEESPEPD-PELEAAIAEF 55
Query: 132 RWHLSEGGLKDYKLLERLGDGSKPTAIIGDVADELNLDLVVIS-----------MEAIHT 180
+ E + + E L KP +I DVADE+N L+++ E++ T
Sbjct: 56 LNRVKEAFAQQAIVAETLLRRGKPAFVICDVADEINASLIIMGCRGTALTPEGFQESVST 115
Query: 181 KHIDANLLAEFIPCPVMLLP 200
+ I+ PCPV+++P
Sbjct: 116 RVIN------LAPCPVLVVP 129
>emb|CAE22281.1| Universal stress protein (Usp) [Prochlorococcus marinus str. MIT
9313] gi|33864371|ref|NP_895931.1| Universal stress
protein (Usp) [Prochlorococcus marinus str. MIT 9313]
Length = 127
Score = 47.0 bits (110), Expect = 3e-04
Identities = 34/135 (25%), Positives = 67/135 (49%), Gaps = 15/135 (11%)
Query: 72 FKHLLLPITDRKPYLSEGTKQAIATTIALANKYGADITVVVIDEQQKESLPEHETQ---L 128
F+ +L PI + + +K + LA + + + ++ + + ++ + + E L
Sbjct: 2 FETVLFPIDQSREAMETASK-----ALELARSHNSRLIILSVVQAERPEMHDPEAVAVLL 56
Query: 129 SSIRWHLSEGGLKDYKLLERLGDGSKPTAIIGDVADELNLDLVVISMEAIHTKHIDANLL 188
R + + G+ ++LER G KP +I DVADELN+D++V+ ++ +
Sbjct: 57 KRAREQVKQAGIA-CEVLEREG---KPAFVICDVADELNVDVIVMGTRGVNLDGDSESTA 112
Query: 189 AEFI---PCPVMLLP 200
A I PCPV+++P
Sbjct: 113 ARVIQLAPCPVLVVP 127
>emb|CAE08843.1| conserved hypothetical protein [Synechococcus sp. WH 8102]
gi|33866858|ref|NP_898417.1| hypothetical protein
SYNW2328 [Synechococcus sp. WH 8102]
Length = 119
Score = 45.1 bits (105), Expect = 0.001
Identities = 22/57 (38%), Positives = 34/57 (59%), Gaps = 3/57 (5%)
Query: 147 ERLGDGSKPTAIIGDVADELNLDLVVISMEAIHTKHIDANLLA---EFIPCPVMLLP 200
+RL G +P +I DVADELN+D++V+ + D + A E PCPV+++P
Sbjct: 63 QRLERGGEPLTVILDVADELNVDVIVMGTGGVSLNSNDGSTAARVIELAPCPVLVVP 119
>ref|NP_924401.1| hypothetical protein gll1455 [Gloeobacter violaceus PCC 7421]
gi|35212020|dbj|BAC89396.1| gll1455 [Gloeobacter
violaceus PCC 7421]
Length = 142
Score = 42.4 bits (98), Expect = 0.008
Identities = 33/141 (23%), Positives = 65/141 (45%), Gaps = 18/141 (12%)
Query: 72 FKHLLLPITDRKPYLSEGTKQAIATTIALANKYGADITVVVIDEQQKESLPEHETQLSSI 131
F+ +LLP+ S +++A+ ++LA +Y + + ++ + + +E +PE E L+ +
Sbjct: 8 FELILLPLDS-----SPESERALQVALSLARQYSSKLLLLSVVDLPEE-MPEREAHLAEV 61
Query: 132 RWHLSEGGLKDYKLLERLG-------DGSKPTAIIGDVADELNLDLVVISMEAI-----H 179
R L+ G D K +I DVADE+ L+V+ +
Sbjct: 62 RAKAQALAQTVRDRLQGEGYPTDARIDAGKAAFVICDVADEVGAGLIVMGSRGLGLIEEG 121
Query: 180 TKHIDANLLAEFIPCPVMLLP 200
H ++ + PCPV+++P
Sbjct: 122 KHHSTSSRVINLSPCPVLVVP 142
>ref|YP_237413.1| Protein of unknown function UPF0004:tRNA-i(6)A37 modification
enzyme MiaB [Pseudomonas syringae pv. syringae B728a]
gi|63258279|gb|AAY39375.1| Protein of unknown function
UPF0004:tRNA-i(6)A37 modification enzyme MiaB
[Pseudomonas syringae pv. syringae B728a]
Length = 442
Score = 40.4 bits (93), Expect = 0.029
Identities = 37/138 (26%), Positives = 60/138 (42%), Gaps = 11/138 (7%)
Query: 41 PSTLHTLPSGIIVKAKPQEPEVAIATDSFTQFKHLLLPITDRKPYLSEGTKQAIATTIAL 100
P TLH LP I + P+V ++ +F HL P D A + +
Sbjct: 105 PQTLHRLPEMIDAARVTRLPQVDVSFPEIEKFDHLPEPRVDGP--------SAYVSVMEG 156
Query: 101 ANKYGADITVVVIDEQQKESLPEHETQLSSIRWHLSEGGLKDYKLLERLGDGSKPTAIIG 160
+KY T V+ + E + + S HL+E G+++ LL + +G + T G
Sbjct: 157 CSKY---CTFCVVPYTRGEEVSRPFDDVLSEVIHLAENGVREVTLLGQNVNGYRGTTHDG 213
Query: 161 DVADELNLDLVVISMEAI 178
VAD +L VV +++ I
Sbjct: 214 RVADLADLIRVVAAVDGI 231
>ref|ZP_00111276.1| COG0589: Universal stress protein UspA and related
nucleotide-binding proteins [Nostoc punctiforme PCC
73102]
Length = 141
Score = 39.3 bits (90), Expect = 0.066
Identities = 35/146 (23%), Positives = 63/146 (42%), Gaps = 26/146 (17%)
Query: 72 FKHLLLPITDRKPYLSEGTKQAIATTIALANKYGADITVVVIDEQQKESLPEHETQ---- 127
FK +L PI S T++A + KY + + ++ + E+ P+ +
Sbjct: 5 FKTVLFPIDQ-----SRETREAADVVTNVVKKYSSRLVLLSVVEEPPPDAPDAPSADPMV 59
Query: 128 --------LSSIRWHLSEGGLKDYKLLERLGDGSKPTAIIGDVADELNLDLVVISMEAIH 179
L + + S G++ ++LER G KP I DVADE+ DL+++ +
Sbjct: 60 SPEVVAKLLENAQSLFSGQGIQS-EILERQG---KPAFTICDVADEIGADLIIMGCRGLG 115
Query: 180 TKHIDAN-----LLAEFIPCPVMLLP 200
A+ + PCPV+++P
Sbjct: 116 LTEEGADDSVTTRVINLSPCPVLIVP 141
>ref|ZP_00514678.1| conserved hypothetical protein [Crocosphaera watsonii WH 8501]
gi|67857276|gb|EAM52516.1| conserved hypothetical
protein [Crocosphaera watsonii WH 8501]
Length = 78
Score = 38.9 bits (89), Expect = 0.086
Identities = 23/71 (32%), Positives = 42/71 (58%), Gaps = 9/71 (12%)
Query: 135 LSEGGLKDYKLLERLGDGSKPTAIIGDVADELNLDLVV-----ISMEAIHTKHIDANLLA 189
++E G+ + +++E+ G KP II DVADE N++L+V + + A + NL+
Sbjct: 12 VAEQGI-EAEVIEKEG---KPRFIICDVADETNVELIVMGSRGLGLTAEGSSESVTNLVI 67
Query: 190 EFIPCPVMLLP 200
PCP++++P
Sbjct: 68 NLAPCPILIIP 78
>ref|XP_640228.1| hypothetical protein DDB0204950 [Dictyostelium discoideum]
gi|60468212|gb|EAL66222.1| hypothetical protein
DDB0204950 [Dictyostelium discoideum]
Length = 1775
Score = 38.1 bits (87), Expect = 0.15
Identities = 22/82 (26%), Positives = 34/82 (40%), Gaps = 7/82 (8%)
Query: 9 PLQLVKPSLISPKPSLFFNNPPISITSHFFNPPSTLHTLPSGIIVKAKPQEPEVAIATDS 68
P V P + +P PP+S H FNP + ++++ KP EP V +
Sbjct: 736 PSSPVLPKIEKQQPPTVVTQPPVSTYKHTFNPVPQQKPVEPEVVIQ-KPVEPVVVVQKTP 794
Query: 69 FTQ------FKHLLLPITDRKP 84
Q +KH P+ +KP
Sbjct: 795 VVQQPPVSTYKHTFNPVPQQKP 816
>ref|NP_531199.1| hypothetical protein Atu0496 [Agrobacterium tumefaciens str. C58]
gi|17738844|gb|AAL41515.1| conserved hypothetical
protein [Agrobacterium tumefaciens str. C58]
gi|25522774|pir||AE2637 conserved hypothetical protein
Atu0496 [imported] - Agrobacterium tumefaciens (strain
C58, Dupont)
Length = 145
Score = 37.4 bits (85), Expect = 0.25
Identities = 35/120 (29%), Positives = 51/120 (42%), Gaps = 25/120 (20%)
Query: 72 FKHLLLPITDRKPYLSEGTKQAIATTIALANKYGADITVVVID----------------- 114
FKH+L+P TD P + AI ALA + GA +TVV +
Sbjct: 2 FKHILIP-TDGSPL----AQIAIDQGFALAREAGAKVTVVTVSEPFHVIASDVEDIAAIA 56
Query: 115 EQQKESLPEHETQLSSIRWHLSEGGLKDYKLLERLGDGSKPTAIIGDVADELNLDLVVIS 174
E++ E E L + H + GL LL R G +P I ++AD DL+ ++
Sbjct: 57 EEEFHRCEEAEHLLRDTQAHAAAMGLDCEALLARAG---RPDEAIIEIADRTGCDLIAMA 113
>gb|AAK86309.1| AGR_C_878p [Agrobacterium tumefaciens str. C58]
gi|25519868|pir||D97419 VGN1536C protein (AE005067)
[imported] - Agrobacterium tumefaciens (strain C58,
Cereon) gi|15887843|ref|NP_353524.1| hypothetical
protein AGR_C_878 [Agrobacterium tumefaciens str. C58]
Length = 160
Score = 37.4 bits (85), Expect = 0.25
Identities = 35/120 (29%), Positives = 51/120 (42%), Gaps = 25/120 (20%)
Query: 72 FKHLLLPITDRKPYLSEGTKQAIATTIALANKYGADITVVVID----------------- 114
FKH+L+P TD P + AI ALA + GA +TVV +
Sbjct: 17 FKHILIP-TDGSPL----AQIAIDQGFALAREAGAKVTVVTVSEPFHVIASDVEDIAAIA 71
Query: 115 EQQKESLPEHETQLSSIRWHLSEGGLKDYKLLERLGDGSKPTAIIGDVADELNLDLVVIS 174
E++ E E L + H + GL LL R G +P I ++AD DL+ ++
Sbjct: 72 EEEFHRCEEAEHLLRDTQAHAAAMGLDCEALLARAG---RPDEAIIEIADRTGCDLIAMA 128
>ref|ZP_00161143.1| COG0589: Universal stress protein UspA and related
nucleotide-binding proteins [Anabaena variabilis ATCC
29413]
Length = 135
Score = 37.0 bits (84), Expect = 0.33
Identities = 34/143 (23%), Positives = 62/143 (42%), Gaps = 23/143 (16%)
Query: 72 FKHLLLPITDRKPYLSEGTKQAIATTIALANKYGADITVVVI------DEQQKESLPEHE 125
FK +L PI S ++A + KYG+ + ++ + DE + + E
Sbjct: 2 FKTVLFPIDQ-----SREAREAADVVTNVVQKYGSRLILLSVVEESSPDESSADPMVSPE 56
Query: 126 TQLSSIRWH---LSEGGLKDYKLLERLGDGSKPTAIIGDVADELNLDLVVISMEAIHTKH 182
+R S+ G+ +++E+ G KP I DVADE++ DL+++ +
Sbjct: 57 AVAKLLRDAQALFSQQGISS-EIVEKQG---KPAFTICDVADEISADLIIMGCRGLGLTE 112
Query: 183 IDA-----NLLAEFIPCPVMLLP 200
A + PCPV+++P
Sbjct: 113 EGATDSVTTRVINLSPCPVLIVP 135
>ref|ZP_00415943.1| Protein of unknown function UPF0004:tRNA-i(6)A37 modification
enzyme MiaB [Azotobacter vinelandii AvOP]
gi|67088331|gb|EAM07797.1| Protein of unknown function
UPF0004:tRNA-i(6)A37 modification enzyme MiaB
[Azotobacter vinelandii AvOP]
Length = 515
Score = 36.2 bits (82), Expect = 0.56
Identities = 34/138 (24%), Positives = 57/138 (40%), Gaps = 11/138 (7%)
Query: 41 PSTLHTLPSGIIVKAKPQEPEVAIATDSFTQFKHLLLPITDRKPYLSEGTKQAIATTIAL 100
P TLH LP I ++P+V ++ +F DR P A + +
Sbjct: 178 PQTLHRLPEMIDAARSTRQPQVDVSFPEIEKF--------DRLPEPRVDGPSAFVSVMEG 229
Query: 101 ANKYGADITVVVIDEQQKESLPEHETQLSSIRWHLSEGGLKDYKLLERLGDGSKPTAIIG 160
+KY VV ++ S P + I HL+E G+++ LL + +G + T G
Sbjct: 230 CSKY-CSYCVVPYTRGEEVSRPPEDVLAEVI--HLAENGVREVTLLGQNVNGYRATTADG 286
Query: 161 DVADELNLDLVVISMEAI 178
D L +V +++ I
Sbjct: 287 RPVDFAELLHLVAAVDGI 304
>ref|ZP_00328538.1| COG0589: Universal stress protein UspA and related
nucleotide-binding proteins [Trichodesmium erythraeum
IMS101]
Length = 136
Score = 36.2 bits (82), Expect = 0.56
Identities = 36/144 (25%), Positives = 59/144 (40%), Gaps = 24/144 (16%)
Query: 72 FKHLLLPITDRKPYLSEGTKQAIATTIALANKYGADITVV----VIDEQQKESLPEHETQ 127
FK +L P+ LS ++A I + Y + + +V V+ E QK PE +
Sbjct: 2 FKTVLFPVD-----LSREAREAGDKVINIVKTYQSRLVLVSVVEVVSEGQKPFHPEMTSS 56
Query: 128 ------LSSIRWHLSEGGLKDYKLLERLGDGSKPTAIIGDVADELNLDLVVISMEAI--- 178
L + ++ G++ E L P I DVADE+ DL+VI
Sbjct: 57 ETVNQLLEETKSIFTQQGIEP----ETLQKEGHPAFTICDVADEIEADLIVIGCRGTGLS 112
Query: 179 --HTKHIDANLLAEFIPCPVMLLP 200
+N + PCP++++P
Sbjct: 113 DGGNNDSISNRIINLSPCPILVVP 136
>dbj|BAB75831.1| alr4132 [Nostoc sp. PCC 7120] gi|17231624|ref|NP_488172.1|
hypothetical protein alr4132 [Nostoc sp. PCC 7120]
gi|25303073|pir||AE2322 hypothetical protein alr4132
[imported] - Nostoc sp. (strain PCC 7120)
Length = 135
Score = 36.2 bits (82), Expect = 0.56
Identities = 34/142 (23%), Positives = 58/142 (39%), Gaps = 21/142 (14%)
Query: 72 FKHLLLPITDRKPYLSEGTKQAIATTIALANKYGADITVVVIDEQQKESLPEHETQLSSI 131
FK +L PI S ++A + KYG+ + ++ + E ES P+ + +
Sbjct: 2 FKTVLFPIDQ-----SREAREAADVVTNVVQKYGSRLILLSVVE---ESSPDESSADPMV 53
Query: 132 RWHLSEGGLKDYKLL--------ERLGDGSKPTAIIGDVADELNLDLVVISMEAIHTKHI 183
L+D + L E + KP I DVADE+ DL+++ +
Sbjct: 54 SPEAVAKLLRDAQALFLQQGIPSEIVEKQGKPAFTICDVADEIGADLIIMGCRGLGLTEE 113
Query: 184 DA-----NLLAEFIPCPVMLLP 200
A + PCPV+++P
Sbjct: 114 GATDSVTTRVINLSPCPVLIVP 135
>ref|NP_794538.1| tRNA-i(6)A37 modification enzyme MiaB [Pseudomonas syringae pv.
tomato str. DC3000] gi|28855172|gb|AAO58233.1|
tRNA-i(6)A37 modification enzyme MiaB [Pseudomonas
syringae pv. tomato str. DC3000]
Length = 442
Score = 35.8 bits (81), Expect = 0.73
Identities = 35/138 (25%), Positives = 58/138 (41%), Gaps = 11/138 (7%)
Query: 41 PSTLHTLPSGIIVKAKPQEPEVAIATDSFTQFKHLLLPITDRKPYLSEGTKQAIATTIAL 100
P TLH LP I + P+V ++ +F HL P D A + +
Sbjct: 105 PQTLHRLPEMIDAARITRLPQVDVSFPEIEKFDHLPEPRVD--------GPSAYVSVMEG 156
Query: 101 ANKYGADITVVVIDEQQKESLPEHETQLSSIRWHLSEGGLKDYKLLERLGDGSKPTAIIG 160
+KY T V+ + E + + + HL+E G+++ LL + +G + T G
Sbjct: 157 CSKY---CTFCVVPYTRGEEVSRPFDDVLTEVTHLAEHGVREVTLLGQNVNGYRGTTHDG 213
Query: 161 DVADELNLDLVVISMEAI 178
VAD +L V ++ I
Sbjct: 214 RVADLADLIRAVAVVDGI 231
>emb|CAA57577.1| hypothetical protein [Pseudomonas aeruginosa]
Length = 446
Score = 35.0 bits (79), Expect = 1.2
Identities = 35/138 (25%), Positives = 58/138 (41%), Gaps = 11/138 (7%)
Query: 41 PSTLHTLPSGIIVKAKPQEPEVAIATDSFTQFKHLLLPITDRKPYLSEGTKQAIATTIAL 100
P TLH LP I ++P+V ++ +F DR P A + +
Sbjct: 105 PQTLHRLPEMIDAARSTRKPQVDVSFPEIEKF--------DRLPEPRVDGPTAFVSVMEG 156
Query: 101 ANKYGADITVVVIDEQQKESLPEHETQLSSIRWHLSEGGLKDYKLLERLGDGSKPTAIIG 160
+KY VV ++ S P + I HL+E G+++ LL + +G + G
Sbjct: 157 CSKY-CSFCVVPYTRGEEVSRPFDDVIAEVI--HLAENGVREVTLLGQNVNGFRGLTHDG 213
Query: 161 DVADELNLDLVVISMEAI 178
+AD L VV +++ I
Sbjct: 214 RLADFAELLRVVAAVDGI 231
>ref|ZP_00262750.1| COG0621: 2-methylthioadenine synthetase [Pseudomonas fluorescens
PfO-1]
Length = 442
Score = 35.0 bits (79), Expect = 1.2
Identities = 36/138 (26%), Positives = 58/138 (41%), Gaps = 11/138 (7%)
Query: 41 PSTLHTLPSGIIVKAKPQEPEVAIATDSFTQFKHLLLPITDRKPYLSEGTKQAIATTIAL 100
P TLH LP I + P+V ++ +F HL P D A + +
Sbjct: 105 PQTLHRLPEMIDAARISKLPQVDVSFPEIEKFDHLPEPRID--------GPSAYVSVMEG 156
Query: 101 ANKYGADITVVVIDEQQKESLPEHETQLSSIRWHLSEGGLKDYKLLERLGDGSKPTAIIG 160
+KY VV ++ S P + I HL+E G+++ LL + +G + G
Sbjct: 157 CSKY-CTFCVVPYTRGEEVSRPFDDVIAEII--HLAENGVREVTLLGQNVNGYRGLTHDG 213
Query: 161 DVADELNLDLVVISMEAI 178
+AD L VV +++ I
Sbjct: 214 RLADLAELIRVVAAVDGI 231
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.318 0.137 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 355,964,169
Number of Sequences: 2540612
Number of extensions: 15027236
Number of successful extensions: 40433
Number of sequences better than 10.0: 66
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 58
Number of HSP's that attempted gapping in prelim test: 40401
Number of HSP's gapped (non-prelim): 77
length of query: 201
length of database: 863,360,394
effective HSP length: 122
effective length of query: 79
effective length of database: 553,405,730
effective search space: 43719052670
effective search space used: 43719052670
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 72 (32.3 bits)
Medicago: description of AC136504.5


