

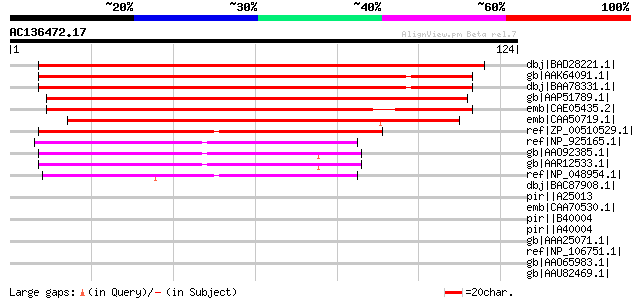
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136472.17 - phase: 0
(124 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAD28221.1| putative serine decarboxylase [Oryza sativa (jap... 181 5e-45
gb|AAK64091.1| putative histidine decarboxylase [Arabidopsis tha... 166 9e-41
dbj|BAA78331.1| serine decarboxylase [Brassica napus] 162 2e-39
gb|AAP51789.1| putative histidine decarboxylase [Oryza sativa (j... 114 4e-25
emb|CAE05435.2| OSJNBa0059H15.18 [Oryza sativa (japonica cultiva... 113 9e-25
emb|CAA50719.1| histidine decarboxylase [Lycopersicon esculentum... 105 3e-22
ref|ZP_00510529.1| Pyridoxal-dependent decarboxylase [Clostridiu... 62 2e-09
ref|NP_925165.1| histidine decarboxylase [Gloeobacter violaceus ... 53 1e-06
gb|AAO92385.1| histidine decarboxylase [Listonella anguillarum] ... 53 2e-06
gb|AAR12533.1| histidine decarboxylase [Listonella anguillarum] ... 53 2e-06
ref|NP_048954.1| similar to tomato histidine decarboxylase, corr... 52 3e-06
dbj|BAC87908.1| probable acinetobactin biosynthesis protein [Aci... 43 0.002
pir||A25013 histidine decarboxylase (EC 4.1.1.22) - Morganella m... 42 0.003
emb|CAA70530.1| pyridoxal-dependent histidine decarboxylase [Pse... 42 0.003
pir||B40004 histidine decarboxylase (EC 4.1.1.22) - Klebsiella p... 42 0.003
pir||A40004 histidine decarboxylase (EC 4.1.1.22) - Enterobacter... 42 0.003
gb|AAA25071.1| histidine decarboxylase 42 0.003
ref|NP_106751.1| histidine decarboxylase [Mesorhizobium loti MAF... 40 0.010
gb|AAO65983.1| putative pyridoxal 5' phosphate-dependent histidi... 38 0.064
gb|AAU82469.1| group II decarboxylase [uncultured archaeon GZfos... 33 1.2
>dbj|BAD28221.1| putative serine decarboxylase [Oryza sativa (japonica
cultivar-group)] gi|50251290|dbj|BAD28070.1| putative
serine decarboxylase [Oryza sativa (japonica
cultivar-group)]
Length = 482
Score = 181 bits (458), Expect = 5e-45
Identities = 84/109 (77%), Positives = 97/109 (88%)
Query: 8 FEKEVQKCLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEFIRKWQLACKGNIAHV 67
F+KEVQKCLRNAHY KDRL EAGIGAMLNELSSTVVFERP DEEF+R+WQLAC+GNIAHV
Sbjct: 371 FQKEVQKCLRNAHYLKDRLKEAGIGAMLNELSSTVVFERPKDEEFVRRWQLACEGNIAHV 430
Query: 68 VVMPNVTIEKLDDFLNELVQKRATWFEYGTFQPYCIASDVGENSCLAYV 116
VVMP+VTI+KLD FLNEL +KRATW++ G+ QP C+A DVGE +CL +
Sbjct: 431 VVMPSVTIDKLDYFLNELTEKRATWYQDGSCQPPCLAKDVGEENCLCSI 479
>gb|AAK64091.1| putative histidine decarboxylase [Arabidopsis thaliana]
gi|13430642|gb|AAK25943.1| putative histidine
decarboxylase [Arabidopsis thaliana]
gi|17907848|dbj|BAB79456.1| histidine decarboxylase
[Arabidopsis thaliana] gi|7523682|gb|AAF63121.1|
Putative histidine decarboxylase [Arabidopsis thaliana]
gi|15218445|ref|NP_175036.1| serine decarboxylase
[Arabidopsis thaliana] gi|15011302|gb|AAK77493.1| serine
decarboxylase [Arabidopsis thaliana]
gi|25290978|pir||E96500 probable histidine decarboxylase
[imported] - Arabidopsis thaliana
gi|17907853|dbj|BAB79457.1| histidine decarboxylase
[Arabidopsis thaliana]
Length = 482
Score = 166 bits (421), Expect = 9e-41
Identities = 79/106 (74%), Positives = 94/106 (88%), Gaps = 1/106 (0%)
Query: 8 FEKEVQKCLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEFIRKWQLACKGNIAHV 67
F+KEVQKCLRNAHY KDRL EAGI AMLNELSSTVVFERP DEEF+R+WQLAC+G+IAHV
Sbjct: 373 FQKEVQKCLRNAHYLKDRLREAGISAMLNELSSTVVFERPKDEEFVRRWQLACQGDIAHV 432
Query: 68 VVMPNVTIEKLDDFLNELVQKRATWFEYGTFQPYCIASDVGENSCL 113
VVMP+VTIEKLD+FL +LV+ R W+E G+ QP C+AS+VG N+C+
Sbjct: 433 VVMPSVTIEKLDNFLKDLVKHRLIWYEDGS-QPPCLASEVGTNNCI 477
>dbj|BAA78331.1| serine decarboxylase [Brassica napus]
Length = 490
Score = 162 bits (410), Expect = 2e-39
Identities = 76/106 (71%), Positives = 91/106 (85%), Gaps = 1/106 (0%)
Query: 8 FEKEVQKCLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEFIRKWQLACKGNIAHV 67
F+KEVQKCLRNAHY KDRL EAGI AMLNELSSTVVFERP +EEF+R+WQLAC+G+IAHV
Sbjct: 381 FQKEVQKCLRNAHYLKDRLREAGISAMLNELSSTVVFERPKEEEFVRRWQLACQGDIAHV 440
Query: 68 VVMPNVTIEKLDDFLNELVQKRATWFEYGTFQPYCIASDVGENSCL 113
VVMP+VT+EKLD FL +LV+ R W+E G+ QP C+ DVG N+C+
Sbjct: 441 VVMPSVTVEKLDHFLKDLVEHRLVWYEDGS-QPPCLVKDVGINNCI 485
>gb|AAP51789.1| putative histidine decarboxylase [Oryza sativa (japonica
cultivar-group)] gi|37530400|ref|NP_919502.1| putative
histidine decarboxylase [Oryza sativa (japonica
cultivar-group)] gi|18542940|gb|AAL75763.1| Putative
histidine decarboxylase [Oryza sativa]
gi|18542909|gb|AAG12476.2| Putative histidine
decarboxylase [Oryza sativa]
Length = 467
Score = 114 bits (286), Expect = 4e-25
Identities = 53/103 (51%), Positives = 70/103 (67%)
Query: 10 KEVQKCLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEFIRKWQLACKGNIAHVVV 69
KEV C+ NA Y + L + GI A N LS+ VVFERP DE + +WQLAC+GN+AH+VV
Sbjct: 351 KEVHICMGNARYLEVLLKQVGISASCNTLSNIVVFERPKDERIVCRWQLACEGNLAHIVV 410
Query: 70 MPNVTIEKLDDFLNELVQKRATWFEYGTFQPYCIASDVGENSC 112
MPNVT EKL F+ EL +KR W++ F C+A D+G+ +C
Sbjct: 411 MPNVTFEKLTVFVEELAEKRKDWYQDKGFDIPCLAVDIGKENC 453
>emb|CAE05435.2| OSJNBa0059H15.18 [Oryza sativa (japonica cultivar-group)]
gi|50921683|ref|XP_471202.1| OSJNBa0059H15.18 [Oryza
sativa (japonica cultivar-group)]
gi|38347364|emb|CAE04954.2| OSJNBa0070D17.5 [Oryza
sativa (japonica cultivar-group)]
Length = 446
Score = 113 bits (283), Expect = 9e-25
Identities = 55/104 (52%), Positives = 70/104 (66%), Gaps = 5/104 (4%)
Query: 10 KEVQKCLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEFIRKWQLACKGNIAHVVV 69
K V+ CL+NA Y RL E G+ LN LS TVVFERP+DE F+RKWQLAC+G IAHVVV
Sbjct: 336 KTVENCLKNAQYLALRLREMGVSVFLNALSITVVFERPNDETFVRKWQLACQGKIAHVVV 395
Query: 70 MPNVTIEKLDDFLNELVQKRATWFEYGTFQPYCIASDVGENSCL 113
MPNV++E+++ FL E + R Q C+A DV + +CL
Sbjct: 396 MPNVSLERINMFLEEFTKSR-----IALHQDKCVAGDVSQENCL 434
>emb|CAA50719.1| histidine decarboxylase [Lycopersicon esculentum]
gi|481829|pir||S39554 histidine decarboxylase (EC
4.1.1.22) - tomato gi|1706319|sp|P54772|DCHS_LYCES
Histidine decarboxylase (HDC) (TOM92)
Length = 413
Score = 105 bits (261), Expect = 3e-22
Identities = 49/97 (50%), Positives = 65/97 (66%), Gaps = 1/97 (1%)
Query: 15 CLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEFIRKWQLACKGNIAHVVVMPNVT 74
C+ NA Y KDRL+EAGI MLN+ S TVVFERP D +FIR+W L C +AHVV+MP +T
Sbjct: 307 CIENARYLKDRLLEAGISVMLNDFSITVVFERPCDHKFIRRWNLCCLRGMAHVVIMPGIT 366
Query: 75 IEKLDDFLNELVQKR-ATWFEYGTFQPYCIASDVGEN 110
E +D F +L+Q+R W++ P C+A D+ N
Sbjct: 367 RETIDSFFKDLMQERNYKWYQDVKALPPCLADDLALN 403
>ref|ZP_00510529.1| Pyridoxal-dependent decarboxylase [Clostridium thermocellum ATCC
27405] gi|67849223|gb|EAM44842.1| Pyridoxal-dependent
decarboxylase [Clostridium thermocellum ATCC 27405]
Length = 398
Score = 62.4 bits (150), Expect = 2e-09
Identities = 29/84 (34%), Positives = 51/84 (60%), Gaps = 1/84 (1%)
Query: 8 FEKEVQKCLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEFIRKWQLACKGNIAHV 67
F+++V++C+ Y K RL G +N S+T+V ++P+D W LAC+G+ AH+
Sbjct: 314 FKQDVRQCMEVTAYAKARLDSIGWNNFVNPWSNTIVIDKPNDA-ICNYWSLACEGDKAHI 372
Query: 68 VVMPNVTIEKLDDFLNELVQKRAT 91
++M +VT E +D F+ L+ + T
Sbjct: 373 IIMQHVTKEHIDLFIEHLLNSKYT 396
>ref|NP_925165.1| histidine decarboxylase [Gloeobacter violaceus PCC 7421]
gi|35212786|dbj|BAC90160.1| histidine decarboxylase
[Gloeobacter violaceus PCC 7421]
Length = 382
Score = 53.1 bits (126), Expect = 1e-06
Identities = 30/79 (37%), Positives = 46/79 (57%), Gaps = 1/79 (1%)
Query: 7 SFEKEVQKCLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEFIRKWQLACKGNIAH 66
+F E + + A + +L + G+ A+LN LSSTVVF RP + I K+QLA + + AH
Sbjct: 300 TFHLEAEAIVDKARFLHQKLSDQGLPALLNPLSSTVVFPRP-PQPVIAKYQLAVQVDQAH 358
Query: 67 VVVMPNVTIEKLDDFLNEL 85
V+M + E L++F L
Sbjct: 359 AVIMQQHSYELLEEFAGVL 377
>gb|AAO92385.1| histidine decarboxylase [Listonella anguillarum]
gi|1073745|pir||S49218 histidine decarboxylase (EC
4.1.1.22) - Vibrio anguillarum
gi|27151484|sp|Q56581|DCHS_VIBAN Histidine decarboxylase
(HDC)
Length = 386
Score = 52.8 bits (125), Expect = 2e-06
Identities = 30/80 (37%), Positives = 47/80 (58%), Gaps = 2/80 (2%)
Query: 8 FEKEVQKCLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEFIRKWQLACKGNIAHV 67
++ +V +CL A Y R E GI A N+ S+TVVF P E RK LA G++AH+
Sbjct: 292 WQGKVNQCLNMAEYTVQRFQEVGINAWRNKNSNTVVFPCP-SEPVWRKHSLANSGSVAHI 350
Query: 68 VVMPNVT-IEKLDDFLNELV 86
+ MP++ +KLD + +++
Sbjct: 351 ITMPHLDGPDKLDPLIEDVI 370
>gb|AAR12533.1| histidine decarboxylase [Listonella anguillarum]
gi|38638327|ref|NP_943559.1| histidine decarboxylase
[Listonella anguillarum]
Length = 400
Score = 52.8 bits (125), Expect = 2e-06
Identities = 30/80 (37%), Positives = 47/80 (58%), Gaps = 2/80 (2%)
Query: 8 FEKEVQKCLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEFIRKWQLACKGNIAHV 67
++ +V +CL A Y R E GI A N+ S+TVVF P E RK LA G++AH+
Sbjct: 306 WQGKVNQCLNMAEYTVQRFQEVGINAWRNKNSNTVVFPCP-SEPVWRKHSLANSGSVAHI 364
Query: 68 VVMPNVT-IEKLDDFLNELV 86
+ MP++ +KLD + +++
Sbjct: 365 ITMPHLDGPDKLDPLIEDVI 384
>ref|NP_048954.1| similar to tomato histidine decarboxylase, corresponds to
Swiss-Prot Accession Number P54772 [Paramecium bursaria
Chlorella virus 1] gi|2447079|gb|AAC96937.1| similar to
tomato histidine decarboxylase, corresponds to
Swiss-Prot Accession Number P54772 [Paramecium bursaria
Chlorella virus 1] gi|7461245|pir||T18100 histidine
decarboxylase homolog A598L - Chlorella virus PBCV-1
Length = 363
Score = 52.4 bits (124), Expect = 3e-06
Identities = 31/79 (39%), Positives = 46/79 (57%), Gaps = 3/79 (3%)
Query: 9 EKEVQKCLRNAHYFKDRLIEAGIGAM--LNELSSTVVFERPHDEEFIRKWQLACKGNIAH 66
EKEV+ CL Y +RL EA M N+ S VVF++P DE I KW LA +H
Sbjct: 279 EKEVEDCLERTEYLFERLREAVPECMPWKNDRSVIVVFKKPSDE-IIFKWSLATVKGRSH 337
Query: 67 VVVMPNVTIEKLDDFLNEL 85
+V+ +VT + +D F++++
Sbjct: 338 AIVLNHVTTDIIDAFVHDM 356
>dbj|BAC87908.1| probable acinetobactin biosynthesis protein [Acinetobacter
baumannii]
Length = 383
Score = 42.7 bits (99), Expect = 0.002
Identities = 25/59 (42%), Positives = 32/59 (53%), Gaps = 1/59 (1%)
Query: 10 KEVQKCLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEFIRKWQLACKGNIAHVV 68
+ +Q CL+ A Y DR GI A N S TVVF P E +K LA GN+AH++
Sbjct: 294 QRIQHCLKMAQYAVDRFQVVGIPAWRNPNSITVVFPCP-SEHIWKKHYLATSGNMAHLI 351
>pir||A25013 histidine decarboxylase (EC 4.1.1.22) - Morganella morganii
gi|118325|sp|P05034|DCHS_MORMO Histidine decarboxylase
(HDC) gi|149859|gb|AAA25321.1| Histidine decarboxylase
Length = 378
Score = 42.4 bits (98), Expect = 0.003
Identities = 28/77 (36%), Positives = 43/77 (55%), Gaps = 5/77 (6%)
Query: 8 FEKEVQKCLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEFIRKWQLACKGNIAHV 67
+++ + + L A Y DR+ +AGI A N+ S TVVF P E R+ LA G++AH+
Sbjct: 292 WKRRITRSLDMAQYAVDRMQKAGINAWRNKNSITVVFPCP-SERVWREHCLATSGDVAHL 350
Query: 68 VV----MPNVTIEKLDD 80
+ + V I+KL D
Sbjct: 351 ITTAHHLDTVQIDKLID 367
>emb|CAA70530.1| pyridoxal-dependent histidine decarboxylase [Pseudomonas
fluorescens] gi|27151483|sp|P95477|DCHS_PSEFL Histidine
decarboxylase (HDC)
Length = 405
Score = 42.4 bits (98), Expect = 0.003
Identities = 31/89 (34%), Positives = 45/89 (49%), Gaps = 5/89 (5%)
Query: 2 AMLPYSFEK---EVQKCLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEFIRKWQL 58
A+ YS+ + ++ L A Y DR +GI A NE S TVVF P E K+ L
Sbjct: 284 ALRSYSWAEWRHRIKHSLDTAQYAVDRFQASGIDAWRNENSITVVFPCP-SERIATKYCL 342
Query: 59 ACKGNIAHVVVMP-NVTIEKLDDFLNELV 86
A GN AH++ P + +D ++E+V
Sbjct: 343 ATSGNSAHLITTPHHHDCSMIDALIDEVV 371
>pir||B40004 histidine decarboxylase (EC 4.1.1.22) - Klebsiella planticola
gi|27151767|sp|P28578|DCHS_KLEPL Histidine decarboxylase
(HDC)
Length = 378
Score = 42.0 bits (97), Expect = 0.003
Identities = 26/80 (32%), Positives = 44/80 (54%), Gaps = 2/80 (2%)
Query: 8 FEKEVQKCLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEFIRKWQLACKGNIAHV 67
+ + +++ L A Y DR AGI A N+ S TVVF P E +K LA G+IAH+
Sbjct: 292 WRRRIERSLNMAQYAVDRFQSAGIDAWRNKNSITVVFPCP-SEAVWKKHCLATSGDIAHL 350
Query: 68 VVMP-NVTIEKLDDFLNELV 86
+ ++ K+D +++++
Sbjct: 351 IATAHHLDSSKIDALIDDVI 370
>pir||A40004 histidine decarboxylase (EC 4.1.1.22) - Enterobacter aerogenes
gi|118319|sp|P28577|DCHS_ENTAE Histidine decarboxylase
(HDC) gi|435594|gb|AAA24802.1| histidine decarboxylase
Length = 378
Score = 42.0 bits (97), Expect = 0.003
Identities = 30/88 (34%), Positives = 47/88 (53%), Gaps = 6/88 (6%)
Query: 8 FEKEVQKCLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEFIRKWQLACKGNIAHV 67
+ + + L A Y DR AGI A+ ++ S TVVF +P E +K LA GN+AH+
Sbjct: 292 WHRRIGHSLNMAKYAVDRFKAAGIDALCHKNSITVVFPKP-SEWVWKKHCLATSGNVAHL 350
Query: 68 VV----MPNVTIEKL-DDFLNELVQKRA 90
+ + + I+ L DD + +L Q+ A
Sbjct: 351 ITTAHHLDSSRIDALIDDVIADLAQRAA 378
>gb|AAA25071.1| histidine decarboxylase
Length = 378
Score = 42.0 bits (97), Expect = 0.003
Identities = 26/80 (32%), Positives = 44/80 (54%), Gaps = 2/80 (2%)
Query: 8 FEKEVQKCLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEFIRKWQLACKGNIAHV 67
+ + +++ L A Y DR AGI A N+ S TVVF P E +K LA G+IAH+
Sbjct: 292 WRRRIERSLNMAQYAVDRFQSAGIDAWRNKNSITVVFPCP-SEAVWKKHCLATSGDIAHL 350
Query: 68 VVMP-NVTIEKLDDFLNELV 86
+ ++ K+D +++++
Sbjct: 351 IATAHHLDSSKIDALIDDVI 370
>ref|NP_106751.1| histidine decarboxylase [Mesorhizobium loti MAFF303099]
gi|27151489|sp|Q98A07|DCHS_RHILO Histidine decarboxylase
(HDC) gi|14025938|dbj|BAB52537.1| histidine
decarboxylase [Mesorhizobium loti MAFF303099]
Length = 369
Score = 40.4 bits (93), Expect = 0.010
Identities = 24/73 (32%), Positives = 39/73 (52%), Gaps = 2/73 (2%)
Query: 13 QKCLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEFIRKWQLACKGNIAHVVVMPN 72
Q+C R A Y D L G+ A N + TVV P ++ KWQ+A + +++H+VV P
Sbjct: 294 QQCERLAAYTADELNVRGVSAWRNPNALTVVLP-PVEDSIKTKWQIATQ-DVSHLVVTPG 351
Query: 73 VTIEKLDDFLNEL 85
T ++ D + +
Sbjct: 352 TTKQQADALIETI 364
>gb|AAO65983.1| putative pyridoxal 5' phosphate-dependent histidine decarboxylase
[Photobacterium phosphoreum]
Length = 380
Score = 37.7 bits (86), Expect = 0.064
Identities = 25/81 (30%), Positives = 43/81 (52%), Gaps = 2/81 (2%)
Query: 8 FEKEVQKCLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEFIRKWQLACKGNIAHV 67
F++ + + L A + RL AGI A N+ S TVVF P E +K LA G AH+
Sbjct: 292 FKRRINRSLDLAQHAVQRLQSAGINAWCNKNSITVVFPCP-SEAVWKKHCLATSGGQAHL 350
Query: 68 VVMP-NVTIEKLDDFLNELVQ 87
+ ++ K+D ++++++
Sbjct: 351 ITTAHHLDASKVDALIDDVIK 371
>gb|AAU82469.1| group II decarboxylase [uncultured archaeon GZfos17F1]
Length = 374
Score = 33.5 bits (75), Expect = 1.2
Identities = 20/86 (23%), Positives = 47/86 (54%), Gaps = 9/86 (10%)
Query: 8 FEKEVQKCLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEFIRK------WQLACK 61
+E+ VQ+C+ + R E+GI +++ +++ +V + P D + +R W ++
Sbjct: 280 YERIVQRCMELTYELVARASESGIEPLIDPVTNVLVLDVP-DADSVRSALKKRGWDVSIT 338
Query: 62 GN--IAHVVVMPNVTIEKLDDFLNEL 85
+ +V+MP+++ E L+ F ++L
Sbjct: 339 RDPRALRLVIMPHISSENLNLFADDL 364
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.324 0.138 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 207,844,233
Number of Sequences: 2540612
Number of extensions: 7309334
Number of successful extensions: 19788
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 18
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 19757
Number of HSP's gapped (non-prelim): 26
length of query: 124
length of database: 863,360,394
effective HSP length: 100
effective length of query: 24
effective length of database: 609,299,194
effective search space: 14623180656
effective search space used: 14623180656
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 68 (30.8 bits)
Medicago: description of AC136472.17


