

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136472.15 + phase: 0
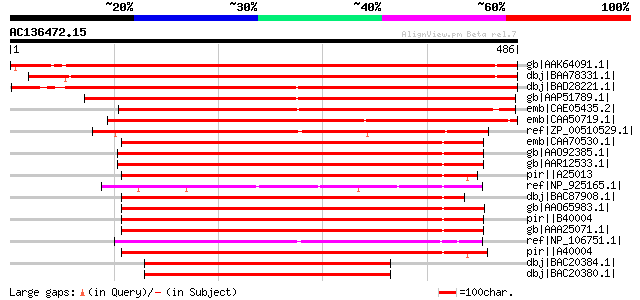
(486 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAK64091.1| putative histidine decarboxylase [Arabidopsis tha... 785 0.0
dbj|BAA78331.1| serine decarboxylase [Brassica napus] 772 0.0
dbj|BAD28221.1| putative serine decarboxylase [Oryza sativa (jap... 770 0.0
gb|AAP51789.1| putative histidine decarboxylase [Oryza sativa (j... 523 e-147
emb|CAE05435.2| OSJNBa0059H15.18 [Oryza sativa (japonica cultiva... 501 e-140
emb|CAA50719.1| histidine decarboxylase [Lycopersicon esculentum... 482 e-134
ref|ZP_00510529.1| Pyridoxal-dependent decarboxylase [Clostridiu... 291 3e-77
emb|CAA70530.1| pyridoxal-dependent histidine decarboxylase [Pse... 283 9e-75
gb|AAO92385.1| histidine decarboxylase [Listonella anguillarum] ... 283 1e-74
gb|AAR12533.1| histidine decarboxylase [Listonella anguillarum] ... 283 1e-74
pir||A25013 histidine decarboxylase (EC 4.1.1.22) - Morganella m... 282 2e-74
ref|NP_925165.1| histidine decarboxylase [Gloeobacter violaceus ... 280 9e-74
dbj|BAC87908.1| probable acinetobactin biosynthesis protein [Aci... 278 4e-73
gb|AAO65983.1| putative pyridoxal 5' phosphate-dependent histidi... 277 6e-73
pir||B40004 histidine decarboxylase (EC 4.1.1.22) - Klebsiella p... 276 8e-73
gb|AAA25071.1| histidine decarboxylase 274 4e-72
ref|NP_106751.1| histidine decarboxylase [Mesorhizobium loti MAF... 270 7e-71
pir||A40004 histidine decarboxylase (EC 4.1.1.22) - Enterobacter... 261 3e-68
dbj|BAC20384.1| histidine decarboxylase [Proteus vulgaris] 228 2e-58
dbj|BAC20380.1| histidine decarboxylase [Morganella morganii] 227 6e-58
>gb|AAK64091.1| putative histidine decarboxylase [Arabidopsis thaliana]
gi|13430642|gb|AAK25943.1| putative histidine
decarboxylase [Arabidopsis thaliana]
gi|17907848|dbj|BAB79456.1| histidine decarboxylase
[Arabidopsis thaliana] gi|7523682|gb|AAF63121.1|
Putative histidine decarboxylase [Arabidopsis thaliana]
gi|15218445|ref|NP_175036.1| serine decarboxylase
[Arabidopsis thaliana] gi|15011302|gb|AAK77493.1| serine
decarboxylase [Arabidopsis thaliana]
gi|25290978|pir||E96500 probable histidine decarboxylase
[imported] - Arabidopsis thaliana
gi|17907853|dbj|BAB79457.1| histidine decarboxylase
[Arabidopsis thaliana]
Length = 482
Score = 785 bits (2028), Expect = 0.0
Identities = 380/488 (77%), Positives = 430/488 (87%), Gaps = 8/488 (1%)
Query: 1 MVGS--ADVLVNGLSTNGAVELLPDDFDVSAIIKDPVPPVVAADNGIGKEEAKINGGKEK 58
MVGS +D ++ + +++L DDFD +A++ +P+PP V NGIG ++ GG +
Sbjct: 1 MVGSLESDQTLSMATLIEKLDILSDDFDPTAVVTEPLPPPVT--NGIGADKG---GGGGE 55
Query: 59 REIVLGRNIHTTCLEVTEPEADDEITGDRDAHMASVLARYRKSLTERTKYHLGYPYNLDF 118
RE+VLGRNIHTT L VTEPE +DE TGD++A+MASVLARYRK+L ERTK HLGYPYNLDF
Sbjct: 56 REMVLGRNIHTTSLAVTEPEVNDEFTGDKEAYMASVLARYRKTLVERTKNHLGYPYNLDF 115
Query: 119 DYGALSQLQHFSINNLGDPFIESNYGVHSRQFEVGVLDWFARLWELEKNEYWGYITNCGT 178
DYGAL QLQHFSINNLGDPFIESNYGVHSR FEVGVLDWFARLWE+E+++YWGYITNCGT
Sbjct: 116 DYGALGQLQHFSINNLGDPFIESNYGVHSRPFEVGVLDWFARLWEIERDDYWGYITNCGT 175
Query: 179 EGNLHGILVGREVLPDGILYASRESHYSIFKAARMYRMECEKVETLNSGEIDCDDFKAKL 238
EGNLHGILVGRE+ PDGILYASRESHYS+FKAARMYRMECEKV+TL SGEIDCDD + KL
Sbjct: 176 EGNLHGILVGREMFPDGILYASRESHYSVFKAARMYRMECEKVDTLMSGEIDCDDLRKKL 235
Query: 239 LRHQDKPAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKR 298
L ++DKPAI+NVNIGTTVKGAVDDLDLVI+ LEE GFS DRFYIH DGALFGLMMPFVKR
Sbjct: 236 LANKDKPAILNVNIGTTVKGAVDDLDLVIKTLEECGFSHDRFYIHCDGALFGLMMPFVKR 295
Query: 299 APKVTFKKPIGSVSVSGHKFVGCPMPCGVQITRLEHINALSRNVEYLASRDATIMGSRNG 358
APKVTF KPIGSVSVSGHKFVGCPMPCGVQITR+EHI LS NVEYLASRDATIMGSRNG
Sbjct: 296 APKVTFNKPIGSVSVSGHKFVGCPMPCGVQITRMEHIKVLSSNVEYLASRDATIMGSRNG 355
Query: 359 HAPIFLWYTLNRKGYRGFQKEVQKCLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDE 418
HAP+FLWYTLNRKGY+GFQKEVQKCLRNAHY KDRL EAGI AMLNELSSTVVFERP DE
Sbjct: 356 HAPLFLWYTLNRKGYKGFQKEVQKCLRNAHYLKDRLREAGISAMLNELSSTVVFERPKDE 415
Query: 419 EFIRKWQLACKGNIAHVVVMPNVTIEKLDDFLNELVQKRATWFEDGTFQPYCIASDVGEN 478
EF+R+WQLAC+G+IAHVVVMP+VTIEKLD+FL +LV+ R W+EDG+ QP C+AS+VG N
Sbjct: 416 EFVRRWQLACQGDIAHVVVMPSVTIEKLDNFLKDLVKHRLIWYEDGS-QPPCLASEVGTN 474
Query: 479 SCLCAQHK 486
+C+C HK
Sbjct: 475 NCICPAHK 482
>dbj|BAA78331.1| serine decarboxylase [Brassica napus]
Length = 490
Score = 772 bits (1994), Expect = 0.0
Identities = 370/475 (77%), Positives = 416/475 (86%), Gaps = 9/475 (1%)
Query: 19 ELLPDDFDVSAIIKDPVPPVVAADNGIGKEEAKI-------NGGKEKREIVLGRNIHTTC 71
++L + FD +A+ +P+P V G +EE + NGG E RE+VLGRN+HTT
Sbjct: 18 DILSEGFDPTAVAPEPLPLPVTNGTGADQEEDNLKKTKVVTNGGGE-REMVLGRNVHTTS 76
Query: 72 LEVTEPEADDEITGDRDAHMASVLARYRKSLTERTKYHLGYPYNLDFDYGALSQLQHFSI 131
L VTEPE++DE TGD++A+MASVLARYRK+L ERTKYHLGYPYNLDFDYGAL QLQHFSI
Sbjct: 77 LAVTEPESNDEFTGDKEAYMASVLARYRKTLVERTKYHLGYPYNLDFDYGALGQLQHFSI 136
Query: 132 NNLGDPFIESNYGVHSRQFEVGVLDWFARLWELEKNEYWGYITNCGTEGNLHGILVGREV 191
NNLGDPFIESNYGVHSR FEVGVLDWFARLWE+E+++YWGYITNCGTEGNLHGILVGREV
Sbjct: 137 NNLGDPFIESNYGVHSRPFEVGVLDWFARLWEIERDDYWGYITNCGTEGNLHGILVGREV 196
Query: 192 LPDGILYASRESHYSIFKAARMYRMECEKVETLNSGEIDCDDFKAKLLRHQDKPAIINVN 251
PDGILYAS ESHYS+FKAARMYRMECEKV+TL SGEIDCDDF+ KLL ++DKPAI+NVN
Sbjct: 197 FPDGILYASSESHYSVFKAARMYRMECEKVDTLISGEIDCDDFRRKLLANKDKPAILNVN 256
Query: 252 IGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAPKVTFKKPIGSV 311
IGTTVKGAVDDLDLVI+ LEE GFS DRFYIH DGALFGLMMPFVKRAPKVTF KPIGSV
Sbjct: 257 IGTTVKGAVDDLDLVIKTLEECGFSHDRFYIHCDGALFGLMMPFVKRAPKVTFNKPIGSV 316
Query: 312 SVSGHKFVGCPMPCGVQITRLEHINALSRNVEYLASRDATIMGSRNGHAPIFLWYTLNRK 371
SVSGHKFVGCPMPCGVQITR++HI LS NVEYLASRDATIMGSRNGHAP+FLWYTLNRK
Sbjct: 317 SVSGHKFVGCPMPCGVQITRMKHIKVLSNNVEYLASRDATIMGSRNGHAPLFLWYTLNRK 376
Query: 372 GYRGFQKEVQKCLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEFIRKWQLACKGN 431
GY+GFQKEVQKCLRNAHY KDRL EAGI AMLNELSSTVVFERP +EEF+R+WQLAC+G+
Sbjct: 377 GYKGFQKEVQKCLRNAHYLKDRLREAGISAMLNELSSTVVFERPKEEEFVRRWQLACQGD 436
Query: 432 IAHVVVMPNVTIEKLDDFLNELVQKRATWFEDGTFQPYCIASDVGENSCLCAQHK 486
IAHVVVMP+VT+EKLD FL +LV+ R W+EDG+ QP C+ DVG N+C+C HK
Sbjct: 437 IAHVVVMPSVTVEKLDHFLKDLVEHRLVWYEDGS-QPPCLVKDVGINNCICPAHK 490
>dbj|BAD28221.1| putative serine decarboxylase [Oryza sativa (japonica
cultivar-group)] gi|50251290|dbj|BAD28070.1| putative
serine decarboxylase [Oryza sativa (japonica
cultivar-group)]
Length = 482
Score = 770 bits (1989), Expect = 0.0
Identities = 378/485 (77%), Positives = 417/485 (85%), Gaps = 15/485 (3%)
Query: 2 VGSADVLVNGLSTNGAVELLPDDFDVSAIIKDPVPPVVAADNGIGKEEAKINGGKEKREI 61
+G A V VNG+ E + +V + PP A + G G +REI
Sbjct: 12 LGGAAVAVNGVGKGMRPEAVAVAMEVES------PPRPAEEEG--------EGSPTRREI 57
Query: 62 VLGRNIHTTCLEVTEPEADDEITGDRDAHMASVLARYRKSLTERTKYHLGYPYNLDFDYG 121
VLGRN+HT V EP+ADDE TG+R+A MASVLA YR++L ERTK+HLGYPYNLDFDYG
Sbjct: 58 VLGRNVHTASFAVKEPDADDEETGEREAAMASVLALYRRNLVERTKHHLGYPYNLDFDYG 117
Query: 122 ALSQLQHFSINNLGDPFIESNYGVHSRQFEVGVLDWFARLWELEKNEYWGYITNCGTEGN 181
AL QLQHFSINNLGDPFIESNYGVHSRQFEVGVLDWFAR+WELEKNEYWGYITNCGTEGN
Sbjct: 118 ALGQLQHFSINNLGDPFIESNYGVHSRQFEVGVLDWFARIWELEKNEYWGYITNCGTEGN 177
Query: 182 LHGILVGREVLPDGILYASRESHYSIFKAARMYRMECEKVETLNSGEIDCDDFKAKLLRH 241
LHGILVGREV PDGILYASRESHYS+FKAARMYRM+C KV+TL SGEIDC+DF+ KLL +
Sbjct: 178 LHGILVGREVFPDGILYASRESHYSVFKAARMYRMDCVKVDTLISGEIDCEDFQRKLLLN 237
Query: 242 QDKPAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAPK 301
+DKPAIINVNIGTTVKGAVDDLDLVI+ LEE GF +DRFYIH DGALFGLM+PFVK+APK
Sbjct: 238 RDKPAIINVNIGTTVKGAVDDLDLVIKTLEEGGF-KDRFYIHCDGALFGLMIPFVKKAPK 296
Query: 302 VTFKKPIGSVSVSGHKFVGCPMPCGVQITRLEHINALSRNVEYLASRDATIMGSRNGHAP 361
V+FKKPIGSVSVSGHKFVGCPMPCGVQITRLEHIN LS NVEYLASRDATIMGSRNGHAP
Sbjct: 297 VSFKKPIGSVSVSGHKFVGCPMPCGVQITRLEHINRLSSNVEYLASRDATIMGSRNGHAP 356
Query: 362 IFLWYTLNRKGYRGFQKEVQKCLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEFI 421
IFLWYTLNRKGYRGFQKEVQKCLRNAHY KDRL EAGIGAMLNELSSTVVFERP DEEF+
Sbjct: 357 IFLWYTLNRKGYRGFQKEVQKCLRNAHYLKDRLKEAGIGAMLNELSSTVVFERPKDEEFV 416
Query: 422 RKWQLACKGNIAHVVVMPNVTIEKLDDFLNELVQKRATWFEDGTFQPYCIASDVGENSCL 481
R+WQLAC+GNIAHVVVMP+VTI+KLD FLNEL +KRATW++DG+ QP C+A DVGE +CL
Sbjct: 417 RRWQLACEGNIAHVVVMPSVTIDKLDYFLNELTEKRATWYQDGSCQPPCLAKDVGEENCL 476
Query: 482 CAQHK 486
C+ HK
Sbjct: 477 CSIHK 481
>gb|AAP51789.1| putative histidine decarboxylase [Oryza sativa (japonica
cultivar-group)] gi|37530400|ref|NP_919502.1| putative
histidine decarboxylase [Oryza sativa (japonica
cultivar-group)] gi|18542940|gb|AAL75763.1| Putative
histidine decarboxylase [Oryza sativa]
gi|18542909|gb|AAG12476.2| Putative histidine
decarboxylase [Oryza sativa]
Length = 467
Score = 523 bits (1346), Expect = e-147
Identities = 241/414 (58%), Positives = 316/414 (76%), Gaps = 1/414 (0%)
Query: 72 LEVTEPEADDEITGDRDAHMASVLARYRKSLTERTKYHLGYPYNLDFDYGALSQLQHFSI 131
LE+ EP AD+ ++ A ++ ++A Y + L R+ YHLGYP N D+D+ L+ +FS+
Sbjct: 46 LEIEEPPADEMEAAEKKAGISRLMAGYVQHLQHRSAYHLGYPLNFDYDFSPLAPFLNFSL 105
Query: 132 NNLGDPFIESNYGVHSRQFEVGVLDWFARLWELEKNEYWGYITNCGTEGNLHGILVGREV 191
NN GDPF + N VHSRQFEV VL+WFA W+++++++WGYIT+ GTEGNL+G+LVGRE+
Sbjct: 106 NNAGDPFAKVNNSVHSRQFEVAVLNWFANFWDVQRDQFWGYITSGGTEGNLYGLLVGREL 165
Query: 192 LPDGILYASRESHYSIFKAARMYRMECEKVETLNSGEIDCDDFKAKLLRHQDKPAIINVN 251
PDGILYAS +SHYS+FKAA+MYR++C ++ T SGE++ D K+KL + + PAIIN N
Sbjct: 166 FPDGILYASNDSHYSVFKAAKMYRVKCIRIATTVSGEMNYADLKSKLQHNTNSPAIINAN 225
Query: 252 IGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAPKVTFKKPIGSV 311
IGTT KGAVDD+D +I LE+ GF Q+R+YIH D AL G+M PF+K+APKV+FKKPIGS+
Sbjct: 226 IGTTFKGAVDDIDQIISTLEKCGF-QNRYYIHCDSALSGMMTPFMKQAPKVSFKKPIGSI 284
Query: 312 SVSGHKFVGCPMPCGVQITRLEHINALSRNVEYLASRDATIMGSRNGHAPIFLWYTLNRK 371
SVSGHKF+GCPMPCGV ITRLEH LS ++EY+ASRD+TI GSRNGHAPIFLWYTL++K
Sbjct: 285 SVSGHKFLGCPMPCGVVITRLEHAEVLSTDIEYIASRDSTITGSRNGHAPIFLWYTLSKK 344
Query: 372 GYRGFQKEVQKCLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEFIRKWQLACKGN 431
GY+G KEV C+ NA Y + L + GI A N LS+ VVFERP DE + +WQLAC+GN
Sbjct: 345 GYKGLLKEVHICMGNARYLEVLLKQVGISASCNTLSNIVVFERPKDERIVCRWQLACEGN 404
Query: 432 IAHVVVMPNVTIEKLDDFLNELVQKRATWFEDGTFQPYCIASDVGENSCLCAQH 485
+AH+VVMPNVT EKL F+ EL +KR W++D F C+A D+G+ +C C H
Sbjct: 405 LAHIVVMPNVTFEKLTVFVEELAEKRKDWYQDKGFDIPCLAVDIGKENCYCNLH 458
>emb|CAE05435.2| OSJNBa0059H15.18 [Oryza sativa (japonica cultivar-group)]
gi|50921683|ref|XP_471202.1| OSJNBa0059H15.18 [Oryza
sativa (japonica cultivar-group)]
gi|38347364|emb|CAE04954.2| OSJNBa0070D17.5 [Oryza
sativa (japonica cultivar-group)]
Length = 446
Score = 501 bits (1290), Expect = e-140
Identities = 232/382 (60%), Positives = 297/382 (77%), Gaps = 7/382 (1%)
Query: 105 RTKYHLGYPYNLDFDYGALSQLQHFSINNLGDPFIESNYGVHSRQFEVGVLDWFARLWEL 164
RT+ + GYP N +FD+G + + + +NN GDPF+E NYG+HS++FE+ VLDWFARLWEL
Sbjct: 63 RTRRNAGYPINFEFDFGPVIEFLNMRLNNAGDPFMECNYGIHSKKFEIAVLDWFARLWEL 122
Query: 165 EKNEYWGYITNCGTEGNLHGILVGREVLPDGILYASRESHYSIFKAARMYRMECEKVETL 224
K++YWGY+T+ GTEGN+HG+LVGRE+ P+GI+Y S +SHYSIFKAA+MYR++C K++TL
Sbjct: 123 PKDQYWGYVTSGGTEGNMHGLLVGRELFPEGIIYTSCDSHYSIFKAAKMYRVQCIKIDTL 182
Query: 225 NSGEIDCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHV 284
SGE+D DF+ KLL++ PAI+NVNIGTT+KGAVDDLD V+ LE GF+ +RFYIH
Sbjct: 183 FSGEMDYADFRRKLLQNTRSPAIVNVNIGTTMKGAVDDLDEVVMILENCGFA-NRFYIHC 241
Query: 285 DGALFGLMMPFVKRAPKVTFKKPIGSVSVSGHKFVGCPMPCGVQITRLEHIN-ALSRNVE 343
D AL GLMMPF+K+APK+TFKKPIGS+ +SGHKF+GCP+PCGV ITRL IN +S N+E
Sbjct: 242 DSALVGLMMPFIKQAPKLTFKKPIGSICISGHKFIGCPIPCGVLITRLMDINHVMSTNIE 301
Query: 344 YLASRDATIMGSRNGHAPIFLWYTLNRKGYRGFQKEVQKCLRNAHYFKDRLIEAGIGAML 403
Y++S D TI GSRNGHAPIFLWY L R GY G K V+ CL+NA Y RL E G+ L
Sbjct: 302 YISSNDTTIAGSRNGHAPIFLWYALKRIGYNGLCKTVENCLKNAQYLALRLREMGVSVFL 361
Query: 404 NELSSTVVFERPHDEEFIRKWQLACKGNIAHVVVMPNVTIEKLDDFLNELVQKRATWFED 463
N LS TVVFERP+DE F+RKWQLAC+G IAHVVVMPNV++E+++ FL E + R +D
Sbjct: 362 NALSITVVFERPNDETFVRKWQLACQGKIAHVVVMPNVSLERINMFLEEFTKSRIALHQD 421
Query: 464 GTFQPYCIASDVGENSCLCAQH 485
C+A DV + +CLC+ H
Sbjct: 422 -----KCVAGDVSQENCLCSLH 438
>emb|CAA50719.1| histidine decarboxylase [Lycopersicon esculentum]
gi|481829|pir||S39554 histidine decarboxylase (EC
4.1.1.22) - tomato gi|1706319|sp|P54772|DCHS_LYCES
Histidine decarboxylase (HDC) (TOM92)
Length = 413
Score = 482 bits (1240), Expect = e-134
Identities = 229/396 (57%), Positives = 301/396 (75%), Gaps = 5/396 (1%)
Query: 94 VLARYRKSLTERTKYHLGYPYNLDFDYGA-LSQLQHFSINNLGDPFIESNYGVHSRQFEV 152
+L +Y ++L+ER KYH+GYP N+ +++ A L+ L F +NN GDPF + HS+ FEV
Sbjct: 17 ILTQYLETLSERKKYHIGYPINMCYEHHATLAPLLQFHLNNCGDPFTQHPTDFHSKDFEV 76
Query: 153 GVLDWFARLWELEKNEYWGYITNCGTEGNLHGILVGR-EVLPDGILYASRESHYSIFKAA 211
VLDWFA+LWE+EK+EYWGYIT+ GTEGNLHG +GR E+LP+G LYAS++SHYSIFKAA
Sbjct: 77 AVLDWFAQLWEIEKDEYWGYITSGGTEGNLHGFWLGRRELLPNGYLYASKDSHYSIFKAA 136
Query: 212 RMYRMECEKVETLNSGEIDCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDDLDLVIQKLE 271
RMYRME + + TL +GEID +D ++KLL +++KPAIIN+NIGTT KGA+DDLD VIQ LE
Sbjct: 137 RMYRMELQTINTLVNGEIDYEDLQSKLLVNKNKPAIININIGTTFKGAIDDLDFVIQTLE 196
Query: 272 EAGFSQDRFYIHVDGALFGLMMPFVKRAPKVTFKKPIGSVSVSGHKFVGCPMPCGVQITR 331
G+S D +YIH D AL GL++PF+K A K+TFKKPIGS+S+SGHKF+GCPM CGVQITR
Sbjct: 197 NCGYSNDNYYIHCDRALCGLILPFIKHAKKITFKKPIGSISISGHKFLGCPMSCGVQITR 256
Query: 332 LEHINALSRNVEYLASRDATIMGSRNGHAPIFLWYTLNRKGYRGFQKEVQKCLRNAHYFK 391
+++ LS+ +EY+ S DATI GSRNG PIFLWY L++KG+ Q++ C+ NA Y K
Sbjct: 257 RSYVSTLSK-IEYINSADATISGSRNGFTPIFLWYCLSKKGHARLQQDSITCIENARYLK 315
Query: 392 DRLIEAGIGAMLNELSSTVVFERPHDEEFIRKWQLACKGNIAHVVVMPNVTIEKLDDFLN 451
DRL+EAGI MLN+ S TVVFERP D +FIR+W L C +AHVV+MP +T E +D F
Sbjct: 316 DRLLEAGISVMLNDFSITVVFERPCDHKFIRRWNLCCLRGMAHVVIMPGITRETIDSFFK 375
Query: 452 ELVQKR-ATWFEDGTFQPYCIASDVGENSCLCAQHK 486
+L+Q+R W++D P C+A D+ N C+C+ K
Sbjct: 376 DLMQERNYKWYQDVKALPPCLADDLALN-CMCSNKK 410
>ref|ZP_00510529.1| Pyridoxal-dependent decarboxylase [Clostridium thermocellum ATCC
27405] gi|67849223|gb|EAM44842.1| Pyridoxal-dependent
decarboxylase [Clostridium thermocellum ATCC 27405]
Length = 398
Score = 291 bits (745), Expect = 3e-77
Identities = 148/386 (38%), Positives = 238/386 (61%), Gaps = 9/386 (2%)
Query: 80 DDEITGDRDAHMASVLARYRK---SLTERTKYHLGYPYNLDFDYGALSQLQHFSINNLGD 136
D +T D ++ S+L K L E+ +++LGYP+NL+ +Y + + NNLGD
Sbjct: 14 DTSVTADLKSNSESLLEEIDKIYEDLYEKHQHYLGYPFNLNLEYAEFGKFLNLQPNNLGD 73
Query: 137 PFIESNYGVHSRQFEVGVLDWFARLWELEKNEYWGYITNCGTEGNLHGILVGREVLPDGI 196
F S + +++ E VL +FA +++L E WGYI + GTEGNL G+LV RE PDGI
Sbjct: 74 AFYSSTVNIDTKKQEREVLKFFADVYKLPWEEAWGYIGHGGTEGNLCGMLVARERYPDGI 133
Query: 197 LYASRESHYSIFKAARMYRMECEKVETLNSGEIDCDDFKAKLLRHQDKPAIINVNIGTTV 256
Y S SHYSI K A + E + + +GE D + ++L++ +KP ++ +GTT+
Sbjct: 134 FYFSEASHYSIKKNAWILGKPGEVIPSQPNGEFDYNALIERILKNGNKPVLLVATLGTTM 193
Query: 257 KGAVDDLDLVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAPKVTFKK-PIGSVSVSG 315
GA+D++ +++ ++ + ++IH DGALFG M+PF++ P++ F+ PI S+++SG
Sbjct: 194 TGAIDNVQIIVDLFKKHNIKE--YHIHYDGALFGGMIPFMENGPELNFETLPIDSIAISG 251
Query: 316 HKFVGCPMPCGVQITRLEHINALSRN--VEYLASRDATIMGSRNGHAPIFLWYTLNRKGY 373
HKFVGCPMP G+ +TR ++I + N V Y+ ++D TI G RNG + + LWY +NRKG
Sbjct: 252 HKFVGCPMPAGIFLTRKKYIQKILENSDVSYVGTKDTTISGCRNGLSALLLWYQINRKGV 311
Query: 374 RGFQKEVQKCLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEFIRKWQLACKGNIA 433
GF+++V++C+ Y K RL G +N S+T+V ++P+D W LAC+G+ A
Sbjct: 312 EGFKQDVRQCMEVTAYAKARLDSIGWNNFVNPWSNTIVIDKPND-AICNYWSLACEGDKA 370
Query: 434 HVVVMPNVTIEKLDDFLNELVQKRAT 459
H+++M +VT E +D F+ L+ + T
Sbjct: 371 HIIIMQHVTKEHIDLFIEHLLNSKYT 396
>emb|CAA70530.1| pyridoxal-dependent histidine decarboxylase [Pseudomonas
fluorescens] gi|27151483|sp|P95477|DCHS_PSEFL Histidine
decarboxylase (HDC)
Length = 405
Score = 283 bits (724), Expect = 9e-75
Identities = 142/349 (40%), Positives = 216/349 (61%), Gaps = 3/349 (0%)
Query: 108 YHLGYPYNLDFDYGALSQLQHFSINNL-GDPFIESNYGVHSRQFEVGVLDWFARLWELEK 166
+++GYP + DFDY L + FSINNL G SNY ++S FE V+ +FA L+ +
Sbjct: 24 FNIGYPESADFDYSQLHRFLQFSINNLLGTGNEYSNYLLNSFDFEKDVMTYFAELFNIAL 83
Query: 167 NEYWGYITNCGTEGNLHGILVGREVLPDGILYASRESHYSIFKAARMYRMECEKVETLNS 226
+ WGY+TN GTEGN+ G +GRE+ PDG LY S+++HYS+ K ++ R++C VE+L +
Sbjct: 84 EDSWGYVTNGGTEGNMFGCYLGRELFPDGTLYYSKDTHYSVAKIVKLLRIKCRAVESLPN 143
Query: 227 GEIDCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDG 286
GEID DD AK+ Q++ II NIGTT++GA+D++ + Q+L++AG ++ +Y+H D
Sbjct: 144 GEIDYDDLMAKITADQERHPIIFANIGTTMRGALDNIVTIQQRLQQAGIARHDYYLHADA 203
Query: 287 ALFGLMMPFVKRAPKVTFKKPIGSVSVSGHKFVGCPMPCGVQITRLEHINALSRNVEYLA 346
AL G+++PFV +F I S+ VSGHK +G P+PCG+ + + ++ +S V+Y+
Sbjct: 204 ALSGMILPFVDHPQPFSFADGIDSICVSGHKMIGSPIPCGIVVAKRNNVARISVEVDYIR 263
Query: 347 SRDATIMGSRNGHAPIFLWYTLNRKGYRGFQKEVQKCLRNAHYFKDRLIEAGIGAMLNEL 406
+ D TI GSRNGH P+ +W L + ++ ++ L A Y DR +GI A NE
Sbjct: 264 AHDKTISGSRNGHTPLMMWAALRSYSWAEWRHRIKHSLDTAQYAVDRFQASGIDAWRNEN 323
Query: 407 SSTVVFERPHDEEFIRKWQLACKGNIAHVVVMP-NVTIEKLDDFLNELV 454
S TVVF P E K+ LA GN AH++ P + +D ++E+V
Sbjct: 324 SITVVFPCP-SERIATKYCLATSGNSAHLITTPHHHDCSMIDALIDEVV 371
>gb|AAO92385.1| histidine decarboxylase [Listonella anguillarum]
gi|1073745|pir||S49218 histidine decarboxylase (EC
4.1.1.22) - Vibrio anguillarum
gi|27151484|sp|Q56581|DCHS_VIBAN Histidine decarboxylase
(HDC)
Length = 386
Score = 283 bits (723), Expect = 1e-74
Identities = 140/352 (39%), Positives = 219/352 (61%), Gaps = 2/352 (0%)
Query: 104 ERTKYHLGYPYNLDFDYGALSQLQHFSINNLGDPFIESNYGVHSRQFEVGVLDWFARLWE 163
E +++GYP + FDY L + FSINN GD ESNY ++S +FE V+ +F++L++
Sbjct: 20 ENQYFNVGYPESAAFDYSILEKFMKFSINNCGDWREESNYKLNSFEFEKEVMRFFSQLFK 79
Query: 164 LEKNEYWGYITNCGTEGNLHGILVGREVLPDGILYASRESHYSIFKAARMYRMECEKVET 223
+ N+ WGYI+N GTEGN+ + RE+ P +Y S E+HYS+ K R+ + K+ +
Sbjct: 80 IPYNDSWGYISNGGTEGNMFSCYLAREIFPTAYIYYSEETHYSVDKIVRLLNIPARKIRS 139
Query: 224 LNSGEIDCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIH 283
L SGEID + ++ + + K II NIGTT++GA D++ + Q L G ++ +YIH
Sbjct: 140 LPSGEIDYQNLVDQIQKDKQKNPIIFANIGTTMRGATDNIQRIQQDLASIGLERNDYYIH 199
Query: 284 VDGALFGLMMPFVKRAPKVTFKKPIGSVSVSGHKFVGCPMPCGVQITRLEHINALSRNVE 343
D AL G++MPFV++ +F+ I S+SVSGHK +G P+PCG+ + + ++ +S V+
Sbjct: 200 ADAALSGMIMPFVEQPHPYSFEDGIDSISVSGHKMIGSPIPCGIVLAKRHMVDQISVEVD 259
Query: 344 YLASRDATIMGSRNGHAPIFLWYTLNRKGYRGFQKEVQKCLRNAHYFKDRLIEAGIGAML 403
Y++SRD TI GSRNGH+ +F+W + + +Q +V +CL A Y R E GI A
Sbjct: 260 YISSRDQTISGSRNGHSALFMWTAIKSHSFVDWQGKVNQCLNMAEYTVQRFQEVGINAWR 319
Query: 404 NELSSTVVFERPHDEEFIRKWQLACKGNIAHVVVMPNVT-IEKLDDFLNELV 454
N+ S+TVVF P E RK LA G++AH++ MP++ +KLD + +++
Sbjct: 320 NKNSNTVVFPCP-SEPVWRKHSLANSGSVAHIITMPHLDGPDKLDPLIEDVI 370
>gb|AAR12533.1| histidine decarboxylase [Listonella anguillarum]
gi|38638327|ref|NP_943559.1| histidine decarboxylase
[Listonella anguillarum]
Length = 400
Score = 283 bits (723), Expect = 1e-74
Identities = 140/352 (39%), Positives = 219/352 (61%), Gaps = 2/352 (0%)
Query: 104 ERTKYHLGYPYNLDFDYGALSQLQHFSINNLGDPFIESNYGVHSRQFEVGVLDWFARLWE 163
E +++GYP + FDY L + FSINN GD ESNY ++S +FE V+ +F++L++
Sbjct: 34 ENQYFNVGYPESAAFDYSILEKFMKFSINNCGDWREESNYKLNSFEFEKEVMRFFSQLFK 93
Query: 164 LEKNEYWGYITNCGTEGNLHGILVGREVLPDGILYASRESHYSIFKAARMYRMECEKVET 223
+ N+ WGYI+N GTEGN+ + RE+ P +Y S E+HYS+ K R+ + K+ +
Sbjct: 94 IPYNDSWGYISNGGTEGNMFSCYLAREIFPTAYIYYSEETHYSVDKIVRLLNIPARKIRS 153
Query: 224 LNSGEIDCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIH 283
L SGEID + ++ + + K II NIGTT++GA D++ + Q L G ++ +YIH
Sbjct: 154 LPSGEIDYQNLVDQIQKDKQKNPIIFANIGTTMRGATDNIQRIQQDLASIGLERNDYYIH 213
Query: 284 VDGALFGLMMPFVKRAPKVTFKKPIGSVSVSGHKFVGCPMPCGVQITRLEHINALSRNVE 343
D AL G++MPFV++ +F+ I S+SVSGHK +G P+PCG+ + + ++ +S V+
Sbjct: 214 ADAALSGMIMPFVEQPHPYSFEDGIDSISVSGHKMIGSPIPCGIVLAKRHMVDQISVEVD 273
Query: 344 YLASRDATIMGSRNGHAPIFLWYTLNRKGYRGFQKEVQKCLRNAHYFKDRLIEAGIGAML 403
Y++SRD TI GSRNGH+ +F+W + + +Q +V +CL A Y R E GI A
Sbjct: 274 YISSRDQTISGSRNGHSALFMWTAIKSHSFVDWQGKVNQCLNMAEYTVQRFQEVGINAWR 333
Query: 404 NELSSTVVFERPHDEEFIRKWQLACKGNIAHVVVMPNVT-IEKLDDFLNELV 454
N+ S+TVVF P E RK LA G++AH++ MP++ +KLD + +++
Sbjct: 334 NKNSNTVVFPCP-SEPVWRKHSLANSGSVAHIITMPHLDGPDKLDPLIEDVI 384
>pir||A25013 histidine decarboxylase (EC 4.1.1.22) - Morganella morganii
gi|118325|sp|P05034|DCHS_MORMO Histidine decarboxylase
(HDC) gi|149859|gb|AAA25321.1| Histidine decarboxylase
Length = 378
Score = 282 bits (721), Expect = 2e-74
Identities = 140/345 (40%), Positives = 218/345 (62%), Gaps = 5/345 (1%)
Query: 108 YHLGYPYNLDFDYGALSQLQHFSINNLGDPFIESNYGVHSRQFEVGVLDWFARLWELEKN 167
+++GYP + DFDY L + FSINN GD NY ++S FE V+++FA L+++
Sbjct: 24 FNIGYPESADFDYTNLERFLRFSINNCGDWGEYCNYLLNSFDFEKEVMEYFADLFKIPFE 83
Query: 168 EYWGYITNCGTEGNLHGILVGREVLPDGILYASRESHYSIFKAARMYRMECEKVETLNSG 227
+ WGY+TN GTEGN+ G +GRE+ PDG LY S+++HYS+ K ++ R++ + VE+ +G
Sbjct: 84 QSWGYVTNGGTEGNMFGCYLGREIFPDGTLYYSKDTHYSVAKIVKLLRIKSQVVESQPNG 143
Query: 228 EIDCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDGA 287
EID DD K+ ++ II NIGTTV+GA+DD+ + ++L+ AG ++ +Y+H D A
Sbjct: 144 EIDYDDLMKKIADDKEAHPIIFANIGTTVRGAIDDIAEIQKRLKAAGIKREDYYLHADAA 203
Query: 288 LFGLMMPFVKRAPKVTFKKPIGSVSVSGHKFVGCPMPCGVQITRLEHINALSRNVEYLAS 347
L G+++PFV A TF I S+ VSGHK +G P+PCG+ + + E+++ +S ++Y+++
Sbjct: 204 LSGMILPFVDDAQPFTFADGIDSIGVSGHKMIGSPIPCGIVVAKKENVDRISVEIDYISA 263
Query: 348 RDATIMGSRNGHAPIFLWYTLNRKGYRGFQKEVQKCLRNAHYFKDRLIEAGIGAMLNELS 407
D TI GSRNGH P+ LW + +++ + + L A Y DR+ +AGI A N+ S
Sbjct: 264 HDKTITGSRNGHTPLMLWEAIRSHSTEEWKRRITRSLDMAQYAVDRMQKAGINAWRNKNS 323
Query: 408 STVVFERPHDEEFIRKWQLACKGNIAHVVV----MPNVTIEKLDD 448
TVVF P E R+ LA G++AH++ + V I+KL D
Sbjct: 324 ITVVFPCP-SERVWREHCLATSGDVAHLITTAHHLDTVQIDKLID 367
>ref|NP_925165.1| histidine decarboxylase [Gloeobacter violaceus PCC 7421]
gi|35212786|dbj|BAC90160.1| histidine decarboxylase
[Gloeobacter violaceus PCC 7421]
Length = 382
Score = 280 bits (715), Expect = 9e-74
Identities = 157/375 (41%), Positives = 226/375 (59%), Gaps = 15/375 (4%)
Query: 89 AHMASVLARYRKSLTERTKYHLGYPYNLDFDYGA----LSQLQHFSINNLGDPFIESNYG 144
A +A L Y SL + HLGYP+ L +D+ Q Q +++ N+GDPF Y
Sbjct: 8 ASVADELVTYGLSLDIHKRNHLGYPFCLKYDHAEQLAETIQDQRYTLINIGDPFSSPIYQ 67
Query: 145 VHSRQFEVGVLDWFARLWELEKNE--YWGYITNCGTEGNLHGILVGREVLPDGILYASRE 202
+ S ++E VL +FA L+ L++ +WGYI +CGTEGNL+G+L+GR P+GILY S
Sbjct: 68 ISSLEYERQVLGFFAELFGLDRQPRPWWGYIGSCGTEGNLYGLLLGRLAQPEGILYFSEA 127
Query: 203 SHYSIFKAARMYRMECEKVETLNSGEIDCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDD 262
+HYS+ KAARM+RM KV + SGE+D + +P IIN+ +GTT GAVD+
Sbjct: 128 AHYSVGKAARMFRMPYRKVRSQASGEMDYAHLAE--IVESGQPVIINLTLGTTFTGAVDE 185
Query: 263 LDLVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAPKVTFKKPIGSVSVSGHKFVGCP 322
+ ++ L G D+ YIHVD AL G++ ++ R ++F PIGS+++SGHKF+GCP
Sbjct: 186 IGRTVEALTGRGIGLDQVYIHVDAALGGMIACYI-RPELISFDWPIGSLAISGHKFIGCP 244
Query: 323 MPCGVQITRLE----HINALSRNVEYLASRDATIMGSRNGHAPIFLWYTLNRKGYRGFQK 378
PCGV +T E + +S VEY+ S D TIMGSRNGH P++LW + R+ F
Sbjct: 245 HPCGVVLTYKETADRFSSEISAEVEYIGSTDLTIMGSRNGHTPLYLWAEIQRR-KSTFHL 303
Query: 379 EVQKCLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEFIRKWQLACKGNIAHVVVM 438
E + + A + +L + G+ A+LN LSSTVVF RP + I K+QLA + + AH V+M
Sbjct: 304 EAEAIVDKARFLHQKLSDQGLPALLNPLSSTVVFPRP-PQPVIAKYQLAVQVDQAHAVIM 362
Query: 439 PNVTIEKLDDFLNEL 453
+ E L++F L
Sbjct: 363 QQHSYELLEEFAGVL 377
>dbj|BAC87908.1| probable acinetobactin biosynthesis protein [Acinetobacter
baumannii]
Length = 383
Score = 278 bits (710), Expect = 4e-73
Identities = 134/329 (40%), Positives = 211/329 (63%), Gaps = 1/329 (0%)
Query: 108 YHLGYPYNLDFDYGALSQLQHFSINNLGDPFIESNYGVHSRQFEVGVLDWFARLWELEKN 167
+++GYP + DFDY AL + FSINN GD SNY ++S +FE V+ +FA ++++
Sbjct: 24 FNIGYPESADFDYSALFRFFKFSINNCGDWKDYSNYALNSFEFEKDVMAYFAEIFQIPFE 83
Query: 168 EYWGYITNCGTEGNLHGILVGREVLPDGILYASRESHYSIFKAARMYRMECEKVETLNSG 227
E WGY+TN GTEGN+ G + RE+ PD LY+S+++HYS+ K A++ +M+ +E+L++G
Sbjct: 84 ESWGYVTNGGTEGNMFGCYLARELFPDSTLYSSKDTHYSVRKIAKLLQMKSCVIESLDNG 143
Query: 228 EIDCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDGA 287
EID DD K+ +++ II NIGTT+ GA+DD++++ ++L + G + +YIH D A
Sbjct: 144 EIDYDDLIHKIKTNKESHPIIFANIGTTMTGAIDDIEMIQERLAQIGIMRRDYYIHADAA 203
Query: 288 LFGLMMPFVKRAPKVTFKKPIGSVSVSGHKFVGCPMPCGVQITRLEHINALSRNVEYLAS 347
L G+++PFV +F I S+ VSGHK +G P+PCG+ + + +++ +S +V+Y+++
Sbjct: 204 LSGMILPFVDHPQAFSFAHGIDSICVSGHKMIGSPIPCGIVVAKRQNVECISVDVDYIST 263
Query: 348 RDATIMGSRNGHAPIFLWYTLNRKGYRGFQKEVQKCLRNAHYFKDRLIEAGIGAMLNELS 407
RD TI GSRNGH + +W + + ++ +Q CL+ A Y DR GI A N S
Sbjct: 264 RDQTISGSRNGHTVLLMWAAIRSQTNLQRRQRIQHCLKMAQYAVDRFQVVGIPAWRNPNS 323
Query: 408 STVVFERPHDEEFIRKWQLACKGNIAHVV 436
TVVF P E +K LA GN+AH++
Sbjct: 324 ITVVFPCP-SEHIWKKHYLATSGNMAHLI 351
>gb|AAO65983.1| putative pyridoxal 5' phosphate-dependent histidine decarboxylase
[Photobacterium phosphoreum]
Length = 380
Score = 277 bits (708), Expect = 6e-73
Identities = 136/349 (38%), Positives = 216/349 (60%), Gaps = 2/349 (0%)
Query: 108 YHLGYPYNLDFDYGALSQLQHFSINNLGDPFIESNYGVHSRQFEVGVLDWFARLWELEKN 167
+++GYP + DFDY L + FSINN GD NY ++S FE V+++FA L+++
Sbjct: 24 FNIGYPESADFDYTILERFMRFSINNCGDWAEYCNYLLNSFDFEKEVMEYFADLFKIPFE 83
Query: 168 EYWGYITNCGTEGNLHGILVGREVLPDGILYASRESHYSIFKAARMYRMECEKVETLNSG 227
+ WGY+TN GTE N+ G +GRE+ PDG LY S+++HYS+ K ++ R++ + VE+L +G
Sbjct: 84 DSWGYVTNGGTESNMFGCYLGRELFPDGTLYYSKDTHYSVAKIVKLLRIKSQLVESLPNG 143
Query: 228 EIDCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDGA 287
EID DD AK+ + +K II NIGTTV+GA+DD+ + + E G ++ +YIH D A
Sbjct: 144 EIDYDDLIAKIKQDDEKHPIIFANIGTTVRGAIDDISKIQAMIGELGIKREDYYIHADAA 203
Query: 288 LFGLMMPFVKRAPKVTFKKPIGSVSVSGHKFVGCPMPCGVQITRLEHINALSRNVEYLAS 347
L G+++PFV F I S+ VSGHK +G P+PCG+ + + +++A+S ++Y+++
Sbjct: 204 LSGMILPFVDEPQGFNFADGIDSIGVSGHKMIGSPIPCGIVVAKKRNVDAISVEIDYISA 263
Query: 348 RDATIMGSRNGHAPIFLWYTLNRKGYRGFQKEVQKCLRNAHYFKDRLIEAGIGAMLNELS 407
D TI GSRNGH P+ +W + + F++ + + L A + RL AGI A N+ S
Sbjct: 264 HDKTITGSRNGHTPLMMWCAVKSHTHEDFKRRINRSLDLAQHAVQRLQSAGINAWCNKNS 323
Query: 408 STVVFERPHDEEFIRKWQLACKGNIAHVVVMP-NVTIEKLDDFLNELVQ 455
TVVF P E +K LA G AH++ ++ K+D ++++++
Sbjct: 324 ITVVFPCP-SEAVWKKHCLATSGGQAHLITTAHHLDASKVDALIDDVIK 371
>pir||B40004 histidine decarboxylase (EC 4.1.1.22) - Klebsiella planticola
gi|27151767|sp|P28578|DCHS_KLEPL Histidine decarboxylase
(HDC)
Length = 378
Score = 276 bits (707), Expect = 8e-73
Identities = 133/348 (38%), Positives = 217/348 (62%), Gaps = 2/348 (0%)
Query: 108 YHLGYPYNLDFDYGALSQLQHFSINNLGDPFIESNYGVHSRQFEVGVLDWFARLWELEKN 167
+++GYP + DFDY L + FSINN GD NY ++S FE V+++FA+L+++
Sbjct: 24 FNIGYPESADFDYTILERFMRFSINNCGDWGEYCNYLLNSFDFEKEVMEYFAQLFKIPFE 83
Query: 168 EYWGYITNCGTEGNLHGILVGREVLPDGILYASRESHYSIFKAARMYRMECEKVETLNSG 227
E WGY+TN GTEGN+ G +GRE+ P+G LY S+++HYS+ K ++ R++ VE+ +G
Sbjct: 84 ESWGYVTNGGTEGNMFGCYLGREIFPNGTLYYSKDTHYSVAKIVKLLRIKSTLVESQPNG 143
Query: 228 EIDCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDGA 287
E+D D K+ R +K II NIGTTV+GA+D++ ++ Q + E G + +Y+H D A
Sbjct: 144 EMDYADLIKKIKRDNEKHPIIFANIGTTVRGAIDNIAIIQQSISELGIERKDYYLHADAA 203
Query: 288 LFGLMMPFVKRAPKVTFKKPIGSVSVSGHKFVGCPMPCGVQITRLEHINALSRNVEYLAS 347
L G+++PFV F I S+ VSGHK +G P+PCG+ + + ++++ +S ++Y+++
Sbjct: 204 LSGMILPFVDNPQPFNFADGIDSIGVSGHKMIGSPIPCGIVVAKKKNVDRISVEIDYISA 263
Query: 348 RDATIMGSRNGHAPIFLWYTLNRKGYRGFQKEVQKCLRNAHYFKDRLIEAGIGAMLNELS 407
D TI GSRNGH P+ +W + + +++ +++ L A Y DR AGI A N+ S
Sbjct: 264 HDKTISGSRNGHTPLMMWEAIRSHSWEEWRRRIERSLNMAQYAVDRFQSAGIDAWRNKNS 323
Query: 408 STVVFERPHDEEFIRKWQLACKGNIAHVVVMP-NVTIEKLDDFLNELV 454
TVVF P E +K LA G+IAH++ ++ K+D +++++
Sbjct: 324 ITVVFPCP-SEAVWKKHCLATSGDIAHLIATAHHLDSSKIDALIDDVI 370
>gb|AAA25071.1| histidine decarboxylase
Length = 378
Score = 274 bits (701), Expect = 4e-72
Identities = 132/348 (37%), Positives = 216/348 (61%), Gaps = 2/348 (0%)
Query: 108 YHLGYPYNLDFDYGALSQLQHFSINNLGDPFIESNYGVHSRQFEVGVLDWFARLWELEKN 167
+++GYP + DFDY L + FSINN GD NY ++S FE V+++FA+L+++
Sbjct: 24 FNIGYPESADFDYTILERFMRFSINNCGDWGEYCNYLLNSFDFEKEVMEYFAQLFKIPFE 83
Query: 168 EYWGYITNCGTEGNLHGILVGREVLPDGILYASRESHYSIFKAARMYRMECEKVETLNSG 227
E WGY+TN GTEGN+ G +GRE+ P+G LY S+++HYS+ K ++ R++ VE+ +G
Sbjct: 84 ESWGYVTNGGTEGNMFGCYLGREIFPNGTLYYSKDTHYSVAKIVKLLRIKSTLVESQPNG 143
Query: 228 EIDCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDGA 287
E+D D K+ +K II NIGTTV+GA+D++ ++ Q + E G + +Y+H D A
Sbjct: 144 EMDYADLIKKIKADNEKHPIIFANIGTTVRGAIDNIAIIQQSISELGIERKDYYLHADAA 203
Query: 288 LFGLMMPFVKRAPKVTFKKPIGSVSVSGHKFVGCPMPCGVQITRLEHINALSRNVEYLAS 347
L G+++PFV F I S+ VSGHK +G P+PCG+ + + ++++ +S ++Y+++
Sbjct: 204 LSGMILPFVDNPQPFNFADGIDSIGVSGHKMIGSPIPCGIVVAKKKNVDRISVEIDYISA 263
Query: 348 RDATIMGSRNGHAPIFLWYTLNRKGYRGFQKEVQKCLRNAHYFKDRLIEAGIGAMLNELS 407
D TI GSRNGH P+ +W + + +++ +++ L A Y DR AGI A N+ S
Sbjct: 264 HDKTISGSRNGHTPLMMWEAIRSHSWEEWRRRIERSLNMAQYAVDRFQSAGIDAWRNKNS 323
Query: 408 STVVFERPHDEEFIRKWQLACKGNIAHVVVMP-NVTIEKLDDFLNELV 454
TVVF P E +K LA G+IAH++ ++ K+D +++++
Sbjct: 324 ITVVFPCP-SEAVWKKHCLATSGDIAHLIATAHHLDSSKIDALIDDVI 370
>ref|NP_106751.1| histidine decarboxylase [Mesorhizobium loti MAFF303099]
gi|27151489|sp|Q98A07|DCHS_RHILO Histidine decarboxylase
(HDC) gi|14025938|dbj|BAB52537.1| histidine
decarboxylase [Mesorhizobium loti MAFF303099]
Length = 369
Score = 270 bits (690), Expect = 7e-71
Identities = 138/353 (39%), Positives = 204/353 (57%), Gaps = 4/353 (1%)
Query: 101 SLTERTKYHLGYPYNLDFDYGALSQLQHFSINNLGDPFIESNYGVHSRQFEVGVLDWFAR 160
S+ E LGYP+ DFDY L + + NNLGDPF Y V+S FE V+D+FAR
Sbjct: 16 SMQEANGCFLGYPFAKDFDYEPLWRFMSLTGNNLGDPFEPGTYRVNSHAFECDVVDFFAR 75
Query: 161 LWELEKNEYWGYITNCGTEGNLHGILVGREVLPDGILYASRESHYSIFKAARMYRMECEK 220
L+ E WGY+TN GTEGN++G+ + RE+ P+ + Y S+++HYS+ K R+ R+E
Sbjct: 76 LFRACSCEVWGYVTNGGTEGNIYGLYLARELYPNAVAYFSQDTHYSVSKGVRLLRLEHSV 135
Query: 221 VETLNSGEIDCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRF 280
V + ++GEI+ DD K R++ +PA++ NIGTT+K DD + L + G S
Sbjct: 136 VRSQSNGEINYDDLAQKATRYRTRPAVVVANIGTTMKEGKDDTLKIRAVLHDVGIS--AI 193
Query: 281 YIHVDGALFGLMMPFVKRAPKVTFKKPIGSVSVSGHKFVGCPMPCGVQITRLEHINALSR 340
Y+H D AL G P + P F S+++SGHKF+G PMPCGV ++ H+ + R
Sbjct: 194 YVHSDAALCGPYAPLLNPKPAFDFADGADSITLSGHKFLGAPMPCGVVLSHKLHVQRVMR 253
Query: 341 NVEYLASRDATIMGSRNGHAPIFLWYTLNRKGYRGFQKEVQKCLRNAHYFKDRLIEAGIG 400
N++Y+ S D T+ GSRN PI LWY + G G ++ Q+C R A Y D L G+
Sbjct: 254 NIDYIGSSDTTLSGSRNAFTPIILWYAIRSLGIEGIKQTFQQCERLAAYTADELNVRGVS 313
Query: 401 AMLNELSSTVVFERPHDEEFIRKWQLACKGNIAHVVVMPNVTIEKLDDFLNEL 453
A N + TVV P ++ KWQ+A + +++H+VV P T ++ D + +
Sbjct: 314 AWRNPNALTVVLP-PVEDSIKTKWQIATQ-DVSHLVVTPGTTKQQADALIETI 364
>pir||A40004 histidine decarboxylase (EC 4.1.1.22) - Enterobacter aerogenes
gi|118319|sp|P28577|DCHS_ENTAE Histidine decarboxylase
(HDC) gi|435594|gb|AAA24802.1| histidine decarboxylase
Length = 378
Score = 261 bits (668), Expect = 3e-68
Identities = 133/356 (37%), Positives = 217/356 (60%), Gaps = 6/356 (1%)
Query: 108 YHLGYPYNLDFDYGALSQLQHFSINNLGDPFIESNYGVHSRQFEVGVLDWFARLWELEKN 167
+++GYP + DFDY L + FSINN GD NY ++S FE V+++F+ ++++
Sbjct: 24 FNIGYPESADFDYTMLERFLRFSINNCGDWGEYCNYLLNSFDFEKEVMEYFSGIFKIPFA 83
Query: 168 EYWGYITNCGTEGNLHGILVGREVLPDGILYASRESHYSIFKAARMYRMECEKVETLNSG 227
E WGY+TN GTE N+ G +GRE+ P+G LY S+++HYS+ K ++ R++ + VE+ G
Sbjct: 84 ESWGYVTNGGTESNMFGCYLGRELFPEGTLYYSKDTHYSVAKIVKLLRIKSQLVESQPDG 143
Query: 228 EIDCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDGA 287
E+D DD K+ ++ II NIGTTV+GAVD++ + +++ G ++ +Y+H D A
Sbjct: 144 EMDYDDLINKIRTSGERHPIIFANIGTTVRGAVDNIAEIQKRIAALGIPREDYYLHADAA 203
Query: 288 LFGLMMPFVKRAPKVTFKKPIGSVSVSGHKFVGCPMPCGVQITRLEHINALSRNVEYLAS 347
L G+++PFV+ TF I S+ VSGHK +G P+PCG+ + + +++ +S ++Y+++
Sbjct: 204 LSGMILPFVEDPQPFTFADGIDSIGVSGHKMIGSPIPCGIVVAKKANVDRISVEIDYISA 263
Query: 348 RDATIMGSRNGHAPIFLWYTLNRKGYRGFQKEVQKCLRNAHYFKDRLIEAGIGAMLNELS 407
D TI GSRNGH P+ +W + + + + L A Y DR AGI A+ ++ S
Sbjct: 264 HDKTISGSRNGHTPLMMWAAVRSHTDAEWHRRIGHSLNMAKYAVDRFKAAGIDALCHKNS 323
Query: 408 STVVFERPHDEEFIRKWQLACKGNIAHVVV----MPNVTIEKL-DDFLNELVQKRA 458
TVVF +P E +K LA GN+AH++ + + I+ L DD + +L Q+ A
Sbjct: 324 ITVVFPKP-SEWVWKKHCLATSGNVAHLITTAHHLDSSRIDALIDDVIADLAQRAA 378
>dbj|BAC20384.1| histidine decarboxylase [Proteus vulgaris]
Length = 236
Score = 228 bits (582), Expect = 2e-58
Identities = 103/236 (43%), Positives = 161/236 (67%)
Query: 130 SINNLGDPFIESNYGVHSRQFEVGVLDWFARLWELEKNEYWGYITNCGTEGNLHGILVGR 189
SINN GD NY ++S FE V+ +FA+L+++ + WGY+TN GTEGN+ G +GR
Sbjct: 1 SINNCGDWGEYCNYLLNSFDFEKEVMAYFAQLFKIPFEKSWGYVTNGGTEGNMFGCYLGR 60
Query: 190 EVLPDGILYASRESHYSIFKAARMYRMECEKVETLNSGEIDCDDFKAKLLRHQDKPAIIN 249
E+ PDG LY S+++HYS+ K ++ R++ VE+L +GE+D DD K+ +++ II
Sbjct: 61 EIFPDGTLYYSKDTHYSVAKIVKLLRIKSRLVESLPNGEVDYDDLIKKITMDKEQHPIIF 120
Query: 250 VNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAPKVTFKKPIG 309
NIGTTV+GA+DD+ L+ Q+L++AGF ++ +Y+H D AL G+++PFV TF I
Sbjct: 121 ANIGTTVRGAIDDIALIQQRLKDAGFKREDYYLHADAALSGMILPFVDDPQGFTFADGID 180
Query: 310 SVSVSGHKFVGCPMPCGVQITRLEHINALSRNVEYLASRDATIMGSRNGHAPIFLW 365
S+ VSGHK +G P+PCG+ + + +++ +S ++Y++ D TI GSRNGH P+ +W
Sbjct: 181 SIGVSGHKMIGSPIPCGIVVAKKANVDRISVEIDYISGHDKTITGSRNGHTPLMMW 236
>dbj|BAC20380.1| histidine decarboxylase [Morganella morganii]
Length = 236
Score = 227 bits (579), Expect = 6e-58
Identities = 103/236 (43%), Positives = 161/236 (67%)
Query: 130 SINNLGDPFIESNYGVHSRQFEVGVLDWFARLWELEKNEYWGYITNCGTEGNLHGILVGR 189
SINN GD NY ++S FE V+++FA L+++ + WGY+TN GTEGN+ G +GR
Sbjct: 1 SINNCGDWGEYCNYLLNSFDFEKEVMEYFADLFKIPFEQSWGYVTNGGTEGNMFGCYLGR 60
Query: 190 EVLPDGILYASRESHYSIFKAARMYRMECEKVETLNSGEIDCDDFKAKLLRHQDKPAIIN 249
E+ PDG LY S+++HYS+ K ++ R++ + VE+L +GEID DD K+ ++ II
Sbjct: 61 EIFPDGTLYYSKDTHYSVAKIVKLLRIKSQVVESLPNGEIDYDDLMKKIADDKEAHPIIF 120
Query: 250 VNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAPKVTFKKPIG 309
NIGTTV+GA+DD+ + ++L+ AG ++ +Y+H D AL G+++PFV A TF I
Sbjct: 121 ANIGTTVRGAIDDIAEIQKRLKAAGIKREDYYLHADAALSGMILPFVDDAQPFTFADGID 180
Query: 310 SVSVSGHKFVGCPMPCGVQITRLEHINALSRNVEYLASRDATIMGSRNGHAPIFLW 365
S+ VSGHK +G P+PCG+ + + E+++ +S ++Y+++ D TI GSRNGH P+ +W
Sbjct: 181 SIGVSGHKMIGSPIPCGIVVAKKENVDRISVEIDYISAHDKTITGSRNGHTPLMMW 236
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.320 0.139 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 848,543,717
Number of Sequences: 2540612
Number of extensions: 36423823
Number of successful extensions: 74935
Number of sequences better than 10.0: 570
Number of HSP's better than 10.0 without gapping: 44
Number of HSP's successfully gapped in prelim test: 526
Number of HSP's that attempted gapping in prelim test: 74489
Number of HSP's gapped (non-prelim): 610
length of query: 486
length of database: 863,360,394
effective HSP length: 132
effective length of query: 354
effective length of database: 527,999,610
effective search space: 186911861940
effective search space used: 186911861940
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 77 (34.3 bits)
Medicago: description of AC136472.15


