

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136287.2 + phase: 0
(365 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
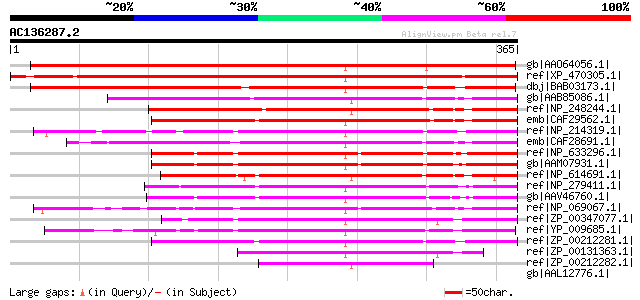
Score E
Sequences producing significant alignments: (bits) Value
gb|AAO64056.1| unknown protein [Arabidopsis thaliana] gi|2775438... 465 e-130
ref|XP_470305.1| unknown protein [Oryza sativa (japonica cultiva... 445 e-123
dbj|BAB03173.1| unnamed protein product [Arabidopsis thaliana] 441 e-122
gb|AAB85086.1| conserved protein [Methanothermobacter thermautot... 216 7e-55
ref|NP_248244.1| hypothetical protein MJ1249 [Methanocaldococcus... 209 1e-52
emb|CAF29562.1| Conserved hypothetical protein [Methanococcus ma... 205 2e-51
ref|NP_214319.1| hypothetical protein aq_1922 [Aquifex aeolicus ... 201 2e-50
emb|CAF28691.1| hypothetical protein [uncultured crenarchaeote] 201 4e-50
ref|NP_633296.1| 3-dehydroquinate synthase [Methanosarcina mazei... 198 2e-49
gb|AAM07931.1| conserved hypothetical protein [Methanosarcina ac... 196 9e-49
ref|NP_614691.1| Predicted alternative 3-dehydroquinate synthase... 196 1e-48
ref|NP_279411.1| hypothetical protein VNG0310C [Halobacterium sp... 187 4e-46
gb|AAV46760.1| 3-dehydroquinate synthase [Haloarcula marismortui... 186 7e-46
ref|NP_069067.1| hypothetical protein AF0229 [Archaeoglobus fulg... 182 1e-44
ref|ZP_00347077.1| COG1465: Predicted alternative 3-dehydroquina... 167 6e-40
ref|YP_009685.1| predicted 3-dehydroquinate synthase [Desulfovib... 153 9e-36
ref|ZP_00212281.1| COG1465: Predicted alternative 3-dehydroquina... 136 1e-30
ref|ZP_00131363.1| COG1465: Predicted alternative 3-dehydroquina... 128 3e-28
ref|ZP_00212282.1| COG1465: Predicted alternative 3-dehydroquina... 55 4e-06
gb|AAL12776.1| LktB [Mannheimia haemolytica] 42 0.022
>gb|AAO64056.1| unknown protein [Arabidopsis thaliana] gi|27754381|gb|AAO22639.1|
unknown protein [Arabidopsis thaliana]
gi|30689333|ref|NP_189518.2| expressed protein
[Arabidopsis thaliana]
Length = 422
Score = 465 bits (1197), Expect = e-130
Identities = 237/353 (67%), Positives = 289/353 (81%), Gaps = 4/353 (1%)
Query: 16 KSKKVWIWTKNKQVMTAAVERGWNTFIFPSNLPHLANDWSSIAVICPLFASEKEILDAQN 75
K+KKVWIWT K+VMT AVERGWNTFIF S+ L+N+WSSIA++ LF EK+++D
Sbjct: 69 KAKKVWIWTMCKEVMTVAVERGWNTFIFSSDNRKLSNEWSSIALMDTLFIEEKKVIDGTG 128
Query: 76 KKVATIFDVSTPEELERLRPEDEQAENIVVNLLDWQVIPAENIIAAFQNSQKTVFAISDN 135
VA++F+VSTPEEL L E+EQ ENIV++ LDW+ IPAEN++AA Q S+KTVFA+S+
Sbjct: 129 NVVASVFEVSTPEELRSLNIENEQIENIVLDFLDWKSIPAENLVAALQGSEKTVFAVSNT 188
Query: 136 ASEAQTFLEALEHGLDGIVLKVEDVEPMLELKEYFDRRTEESNVLNLTKATVTQIQVAGM 195
SEA+ FLEALEHGL GI+LK EDV+ +L+LKEYFD+R EES+ L+LT+AT+T++Q+ GM
Sbjct: 189 PSEAKLFLEALEHGLGGIILKSEDVKAVLDLKEYFDKRNEESDTLSLTEATITRVQMVGM 248
Query: 196 GDRVCVDLCSLMRPGEGLLVGSFARGLFLVHSECLESKLHCKQTF--SGECVHAYIAVPG 253
GDRVCVDLCSLMRPGEGLLVGSFARGLFLVHSECLES + F + VHAY+AVPG
Sbjct: 249 GDRVCVDLCSLMRPGEGLLVGSFARGLFLVHSECLESNYIESRPFRVNAGPVHAYVAVPG 308
Query: 254 GRTSYLSELKSGKEVIVVDQQGRQRIAIVGRVKIESRPLILVEAK--TDSDNQTISILLQ 311
G+T YLSEL++G+EVIVVDQ+G+QR A+VGRVKIE RPLI+VEAK T + SI+LQ
Sbjct: 309 GKTCYLSELRTGREVIVVDQKGKQRTAVVGRVKIEKRPLIVVEAKLSTKEEETVYSIILQ 368
Query: 312 NAETVALVCPVQGNKLLKTAFPVTSLKVGDEVLLRVQGGARHTGIEIQEFIVE 364
NAETVALV P Q N +TA PVTSLK GD+VL+R+QGGARHTGIEIQEFIVE
Sbjct: 369 NAETVALVTPHQVNSSGRTAVPVTSLKPGDQVLIRLQGGARHTGIEIQEFIVE 421
>ref|XP_470305.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|19071650|gb|AAL84317.1| unknown protein [Oryza sativa
(japonica cultivar-group)]
Length = 390
Score = 445 bits (1144), Expect = e-123
Identities = 235/368 (63%), Positives = 287/368 (77%), Gaps = 11/368 (2%)
Query: 1 MSSSSWTPLEDSGNKKSKKVWIWTKNKQVMTAAVERGWNTFIFPSNLPHLANDWSSIAVI 60
M +SS +P E K VW+WT N+QVMTAAVERGW+TF+F S L DWSS A I
Sbjct: 31 MCASSASPSESK-----KTVWVWTTNRQVMTAAVERGWSTFLFGSK--DLGKDWSSTARI 83
Query: 61 CPLFASEKEILDAQNKKVATIFDVSTPEELERLRPEDEQAENIVVNLLD-WQVIPAENII 119
PLF EI D + +K+A I +VS+P ELE ++P++ + ENIV++ WQVIPAENI+
Sbjct: 84 NPLFIDGLEIFDEKKQKIAVISEVSSPGELELIQPDNVEVENIVIDFRGGWQVIPAENIV 143
Query: 120 AAFQNSQKTVFAISDNASEAQTFLEALEHGLDGIVLKVEDVEPMLELKEYFDRRTEESNV 179
AAFQ + TV A+S N++EAQ FLEALE GLDG++LKVED++ +++LK+YFDRR E +
Sbjct: 144 AAFQGCRGTVLAVSTNSTEAQVFLEALEQGLDGVILKVEDMDDIIKLKDYFDRRNEAKSQ 203
Query: 180 LNLTKATVTQIQVAGMGDRVCVDLCSLMRPGEGLLVGSFARGLFLVHSECLESKLHCKQT 239
L LTKATV++++V GMGDRVCVDLCS+MRPGEGLLVGS+ARG+FLVHSECLE+ +
Sbjct: 204 LMLTKATVSKVEVVGMGDRVCVDLCSMMRPGEGLLVGSYARGMFLVHSECLETNYIASRP 263
Query: 240 F--SGECVHAYIAVPGGRTSYLSELKSGKEVIVVDQQGRQRIAIVGRVKIESRPLILVEA 297
F + VHAY+AVPGGRTSYLSEL+SG+EVIVVDQ G R AIVGRVKIESRPLILVEA
Sbjct: 264 FRVNAGPVHAYVAVPGGRTSYLSELQSGREVIVVDQNGLWRTAIVGRVKIESRPLILVEA 323
Query: 298 KTDSDNQTISILLQNAETVALVCPVQGNKLLKTAFPVTSLKVGDEVLLRVQGGARHTGIE 357
K + + T SI LQNAETVAL+ P +G+ +TA PVTSLKVGDEVL+R QGGARHTGIE
Sbjct: 324 KENGGDDTYSIFLQNAETVALITPEKGSS-GRTAIPVTSLKVGDEVLVRKQGGARHTGIE 382
Query: 358 IQEFIVEK 365
IQEFIVEK
Sbjct: 383 IQEFIVEK 390
>dbj|BAB03173.1| unnamed protein product [Arabidopsis thaliana]
Length = 434
Score = 441 bits (1133), Expect = e-122
Identities = 230/351 (65%), Positives = 281/351 (79%), Gaps = 15/351 (4%)
Query: 16 KSKKVWIWTKNKQVMTAAVERGWNTFIFPSNLPHLANDWSSIAVICPLFASEKEILDAQN 75
K+KKVWIWT K+VMT AVERGWNTFIF S+ L+N+WSSIA++ LF EK+++D
Sbjct: 96 KAKKVWIWTMCKEVMTVAVERGWNTFIFSSDNRKLSNEWSSIALMDTLFIEEKKVIDGTG 155
Query: 76 KKVATIFDVSTPEELERLRPEDEQAENIVVNLLDWQVIPAENIIAAFQNSQKTVFAISDN 135
VA++F+VSTPEEL L E+EQ ENIV++ LDW+ IPAEN++AA Q S+KTVFA+S+
Sbjct: 156 NVVASVFEVSTPEELRSLNIENEQIENIVLDFLDWKSIPAENLVAALQGSEKTVFAVSNT 215
Query: 136 ASEAQTFLEALEHGLDGIVLKVEDVEPMLELKEYFDRRTEESNVLNLTKATVTQIQVAGM 195
SEA+ FLEALEHGL GI+LK EDV+ +L+LK R EES+ L+LT+AT+T++Q+ GM
Sbjct: 216 PSEAKLFLEALEHGLGGIILKSEDVKAVLDLK-----RNEESDTLSLTEATITRVQMVGM 270
Query: 196 GDRVCVDLCSLMRPGEGLLVGSFARGLFLVHSECLESKLHCKQTF--SGECVHAYIAVPG 253
GDRVCVDLCSLMRPGEGLLVGSFARGLFLVHSECLES + F + VHAY+AVPG
Sbjct: 271 GDRVCVDLCSLMRPGEGLLVGSFARGLFLVHSECLESNYIESRPFRVNAGPVHAYVAVPG 330
Query: 254 GRTSYLSELKSGKEVIVVDQQGRQRIAIVGRVKIESRPLILVEAKTDSDNQTISILLQNA 313
G+T YLSEL++G+EVIVVDQ+G+QR A+VGRVKIE RPLI+VEAK + SI+LQNA
Sbjct: 331 GKTCYLSELRTGREVIVVDQKGKQRTAVVGRVKIEKRPLIVVEAK--EEETVYSIILQNA 388
Query: 314 ETVALVCPVQGNKLLKTAFPVTSLKVGDEVLLRVQGGARHTGIEIQEFIVE 364
ETVALV P +TA PVTSLK GD+VL+R+QGGARHTGIEIQEFIVE
Sbjct: 389 ETVALVTP------RRTAVPVTSLKPGDQVLIRLQGGARHTGIEIQEFIVE 433
>gb|AAB85086.1| conserved protein [Methanothermobacter thermautotrophicus str.
Delta H] gi|15678608|ref|NP_275723.1| hypothetical
protein MTH580 [Methanothermobacter thermautotrophicus
str. Delta H] gi|7451237|pir||H69176 conserved
hypothetical protein MTH580 - Methanobacterium
thermoautotrophicum (strain Delta H)
gi|32130310|sp|O26680|Y580_METTH Hypothetical UPF0245
protein MTH580
Length = 378
Score = 216 bits (551), Expect = 7e-55
Identities = 128/297 (43%), Positives = 178/297 (59%), Gaps = 12/297 (4%)
Query: 71 LDAQNKKVATIFDVSTPEELERLRPEDEQAENIVVNLLDWQVIPAENIIAAFQNSQKTVF 130
L ++VA ++ + E R + +++ DW++IP ENIIA Q +
Sbjct: 92 LSESGRQVAAYVEIRSKAHEELARRLGRVVDYLILVGEDWKIIPLENIIADLQEEDVKLI 151
Query: 131 AISDNASEAQTFLEALEHGLDGIVLKVEDVEPMLELKEYFDRRTEESNVLNLTKATVTQI 190
A + EA+ LE LEHG DG++++ D+ + ++ + ES L AT+T+I
Sbjct: 152 AAVADVDEARVALETLEHGTDGVLIEPADISQIKDIAALLENI--ESETYELKPATITRI 209
Query: 191 QVAGMGDRVCVDLCSLMRPGEGLLVGSFARGLFLVHSECLESKLHCKQTFSGEC--VHAY 248
+ G GDRVCVD CS+M GEG+LVGS+++GLFLVHSE LES+ + F V AY
Sbjct: 210 EPIGSGDRVCVDTCSIMGIGEGMLVGSYSQGLFLVHSESLESEYVASRPFRVNAGPVQAY 269
Query: 249 IAVPGGRTSYLSELKSGKEVIVVDQQGRQRIAIVGRVKIESRPLILVEAKTDSDNQTISI 308
+ VPGGRT YLSEL++G EVI+VD+ GR R AIVGRVKIE RPL+LVEA + + +
Sbjct: 270 VMVPGGRTRYLSELETGDEVIIVDRDGRSRSAIVGRVKIEKRPLMLVEA--EYEGMKVRT 327
Query: 309 LLQNAETVALVCPVQGNKLLKTAFPVTSLKVGDEVLLRVQGGARHTGIEIQEFIVEK 365
LLQNAET+ LV +G + V+ L GD VL+ ARH G+ I+E I+EK
Sbjct: 328 LLQNAETIRLVND-KGEPV-----SVSELGEGDRVLVYFDESARHFGMAIKETIIEK 378
>ref|NP_248244.1| hypothetical protein MJ1249 [Methanocaldococcus jannaschii DSM
2661] gi|1591882|gb|AAB99252.1| conserved hypothetical
protein [Methanocaldococcus jannaschii DSM 2661]
gi|2128748|pir||H64455 conserved hypothetical protein
MJ1249 - Methanococcus jannaschii
gi|32130336|sp|Q58646|YC49_METJA Hypothetical UPF0245
protein MJ1249
Length = 361
Score = 209 bits (532), Expect = 1e-52
Identities = 118/267 (44%), Positives = 171/267 (63%), Gaps = 12/267 (4%)
Query: 101 ENIVVNLLDWQVIPAENIIAAFQNSQKTVFAISDNASEAQTFLEALEHGLDGIVLKVEDV 160
+NI++ DW +IP EN+IA + + A ++ EA+ E LE G DG++L +++
Sbjct: 105 DNIILEGRDWTIIPLENLIADLFHRDVKIVASVNSVDEAKVAYEILEKGTDGVLLNPKNL 164
Query: 161 EPMLELKEYFDRRTEESNVLNLTKATVTQIQVAGMGDRVCVDLCSLMRPGEGLLVGSFAR 220
E + EL + + +E L++ ATVT+++ G GDRVC+D CSLM+ GEG+L+GS++R
Sbjct: 165 EDIKELSKLIEEMNKEKVALDV--ATVTKVEPIGSGDRVCIDTCSLMKIGEGMLIGSYSR 222
Query: 221 GLFLVHSECLESKLHCKQTFSGEC--VHAYIAVPGGRTSYLSELKSGKEVIVVDQQGRQR 278
LFLVHSE +E+ + F VHAYI PG +T YLSELK+G +V++VD+ G R
Sbjct: 223 ALFLVHSETVENPYVATRPFRVNAGPVHAYILCPGNKTKYLSELKAGDKVLIVDKDGNTR 282
Query: 279 IAIVGRVKIESRPLILVEAKTDSDNQTISILLQNAETVALVCPVQGNKLLKTAFPVTSLK 338
AIVGRVKIE RPL+L+EA+ D I +LQNAET+ LV +G + V LK
Sbjct: 283 EAIVGRVKIERRPLVLIEAEYKGD--IIRTILQNAETIRLV-NEKGEPI-----SVVDLK 334
Query: 339 VGDEVLLRVQGGARHTGIEIQEFIVEK 365
GD+VL++ + ARH G+ I+E I+EK
Sbjct: 335 PGDKVLIKPEEYARHFGMAIKETIIEK 361
>emb|CAF29562.1| Conserved hypothetical protein [Methanococcus maripaludis S2]
gi|45357569|ref|NP_987126.1| hypothetical protein
MMP0006 [Methanococcus maripaludis S2]
Length = 361
Score = 205 bits (522), Expect = 2e-51
Identities = 115/265 (43%), Positives = 168/265 (63%), Gaps = 12/265 (4%)
Query: 103 IVVNLLDWQVIPAENIIAAFQNSQKTVFAISDNASEAQTFLEALEHGLDGIVLKVEDVEP 162
+V+ DW +IP ENIIA + + + ++ N +A+ E LE G+DG+VL +D+
Sbjct: 107 VVLEGSDWTIIPLENIIADLFSEEIKIVSVVTNVKDAEAAYEILEKGVDGVVLIPKDINE 166
Query: 163 MLELKEYFDRRTEESNVLNLTKATVTQIQVAGMGDRVCVDLCSLMRPGEGLLVGSFARGL 222
+ + + +R ES + L ATVT+I+ G GDRVC+D CS+M GEG+L+GS++RG+
Sbjct: 167 VKDFSKLIERMNSES--VKLDYATVTKIEPVGSGDRVCIDTCSMMEMGEGMLIGSYSRGM 224
Query: 223 FLVHSECLESKLHCKQTF--SGECVHAYIAVPGGRTSYLSELKSGKEVIVVDQQGRQRIA 280
FLVHSE +E+ + F + VHAYI P +T YLS+LK+G +V+VV++ G R A
Sbjct: 225 FLVHSETVENPYVATRPFRVNAGPVHAYILCPENKTKYLSDLKAGDKVLVVNKNGETREA 284
Query: 281 IVGRVKIESRPLILVEAKTDSDNQTISILLQNAETVALVCPVQGNKLLKTAFPVTSLKVG 340
I+GRVKIE RPL LVEA+ + +N + +LQNAET+ LV G + V LKVG
Sbjct: 285 IIGRVKIEKRPLFLVEAEYNGEN--LRTILQNAETIRLV-GEDGKPV-----SVVDLKVG 336
Query: 341 DEVLLRVQGGARHTGIEIQEFIVEK 365
+VL++ ARH G+ I+E I+EK
Sbjct: 337 TKVLIKPDENARHFGMAIKETIIEK 361
>ref|NP_214319.1| hypothetical protein aq_1922 [Aquifex aeolicus VF5]
gi|2984189|gb|AAC07721.1| hypothetical protein [Aquifex
aeolicus VF5] gi|7451238|pir||C70465 conserved
hypothetical protein aq_1922 - Aquifex aeolicus
gi|32130364|sp|O67751|YJ22_AQUAE Hypothetical UPF0245
protein AQ_1922
Length = 331
Score = 201 bits (512), Expect = 2e-50
Identities = 126/353 (35%), Positives = 194/353 (54%), Gaps = 28/353 (7%)
Query: 18 KKVWIWTK--NKQVMTAAVERGWNTFIFPSN-LPHLANDWSSIAVICPLFASEKEILDAQ 74
K+ W+W + ++++++ A+E G N + P I VI P + ++ +
Sbjct: 2 KEFWVWVEPFDRKIVSVALEAGANAVVIPEKGRVEEVKKVGRITVIAP----DGDLKLGE 57
Query: 75 NKKVATIFDVSTPEELERLRPEDEQAENIVVNLLDWQVIPAENIIAAFQNSQKTVFAISD 134
+ I E + P + ++V DW VIP EN+IA ++ ++A+
Sbjct: 58 DVVYVLIKGKEDEERAAKYPPNVK----VIVETTDWTVIPLENLIA----QREELYAVVK 109
Query: 135 NASEAQTFLEALEHGLDGIVLKVEDVEPMLELKEYFDRRTEESNVLNLTKATVTQIQVAG 194
NA EA+ L+ LE G+ G+VLK D+ E+K+ +E L L +T+I G
Sbjct: 110 NAQEAEVALQTLEKGVKGVVLKSRDIN---EIKKVGQIVSEHEEKLELVTIKITKILPLG 166
Query: 195 MGDRVCVDLCSLMRPGEGLLVGSFARGLFLVHSECLESKLHCKQTF--SGECVHAYIAVP 252
+GDRVCVD SL+ GEG+LVG+ + G+FLVH+E E+ + F + VH YI VP
Sbjct: 167 LGDRVCVDTISLLHRGEGMLVGNSSGGMFLVHAETEENPYVAARPFRVNAGAVHMYIRVP 226
Query: 253 GGRTSYLSELKSGKEVIVVDQQGRQRIAIVGRVKIESRPLILVEAKTDSDNQTISILLQN 312
RT YL ELK+G +V+V D +GR R+ VGR K+E RP++L+E + +N+ +S +LQN
Sbjct: 227 NNRTKYLCELKAGDKVMVYDYKGRGRVTYVGRAKVERRPMLLIEGR--YENKKLSCILQN 284
Query: 313 AETVALVCPVQGNKLLKTAFPVTSLKVGDEVLLRVQGGARHTGIEIQEFIVEK 365
AET+ L P T V+ LK GDEVL V+ RH G++++E I+EK
Sbjct: 285 AETIRLTKPD------GTPISVSELKEGDEVLGYVEEAGRHFGMKVEETIIEK 331
>emb|CAF28691.1| hypothetical protein [uncultured crenarchaeote]
Length = 339
Score = 201 bits (510), Expect = 4e-50
Identities = 128/329 (38%), Positives = 193/329 (57%), Gaps = 28/329 (8%)
Query: 42 IFPSNLPHLANDWSSIAVICPLFASEKEILDAQNKKVATIFDVSTPEELERL-RPEDEQA 100
IFPS L V+C S ++I + K V V T E+L+ + E A
Sbjct: 34 IFPSENADL--------VVCDSIESLRKI-KRKKKSVGYYKKVVTNEDLDEISEASKEGA 84
Query: 101 ENIVVNLLDWQVIPAENIIAAFQNSQKTVFAISDNASEAQTFLEALEHGLDGIVLKVEDV 160
+ ++V+ DW++IP EN+IA Q S +FA + N+ E +T LE G+DG++L +D
Sbjct: 85 QFVIVDATDWKIIPLENLIAKLQTSGTKIFANAMNSDEVRTMFSVLELGVDGVILTTDD- 143
Query: 161 EPMLELKEYFDRRTEESNVL-NLTKATVTQIQVAGMGDRVCVDLCSLMRPGEGLLVGSFA 219
LKE + ++ NVL +L A V +++ G+G+RVCVD S+M GEG+LVG+ +
Sbjct: 144 -----LKEVENSKSNLENVLFSLRNARVVEVKDVGVGERVCVDTASIMVYGEGMLVGNKS 198
Query: 220 RGLFLVHSECLESKLHCKQTF--SGECVHAYIAVPGGRTSYLSELKSGKEVIVVDQQGRQ 277
LFL+H+E + S + F + VH Y +P G T YLSEL+SG EV++V+ +G
Sbjct: 199 NFLFLIHNESIGSSFTSPRPFRVNAGAVHCYTILPNGTTKYLSELESGVEVLIVNNEGVG 258
Query: 278 RIAIVGRVKIESRPLILVEAKTDSDNQTISILLQNAETVALVCPVQGNKLLKTAFPVTSL 337
R + VGR KIE+RPL L+ A +SD + S+++QNAET+ LV + KL+ PVT L
Sbjct: 259 RRSTVGRSKIETRPLRLIRA--ESDGEFGSVIVQNAETIRLVS--KDKKLI----PVTEL 310
Query: 338 KVGDEVLLRVQ-GGARHTGIEIQEFIVEK 365
K+GDE++ + RH G+++ EFI+EK
Sbjct: 311 KIGDEIVTYAKPTSGRHFGMQVNEFILEK 339
>ref|NP_633296.1| 3-dehydroquinate synthase [Methanosarcina mazei Go1]
gi|20905734|gb|AAM30968.1| 3-dehydroquinate synthase
[Methanosarcina mazei Goe1]
gi|32130354|sp|Q8PXE8|YC72_METMA Hypothetical UPF0245
protein MM1272
Length = 380
Score = 198 bits (503), Expect = 2e-49
Identities = 120/267 (44%), Positives = 174/267 (64%), Gaps = 17/267 (6%)
Query: 103 IVVNLLDWQVIPAENIIAAFQNSQ-KTVFAISDNASEAQTFLEALEHGLDGIVLKVEDVE 161
++V DW+VIP EN+IA Q + K +F + +A EA+ + LE G DG++L + +
Sbjct: 127 LLVTGTDWKVIPLENLIADLQREKVKIIFGVK-SAEEARLAFQTLETGADGVLLDSGNPQ 185
Query: 162 PMLELKEYFDRRTE-ESNVLNLTKATVTQIQVAGMGDRVCVDLCSLMRPGEGLLVGSFAR 220
E+K+ E ES L A VT+++ GMGDRVCVD C+LM+ GEG+L+GS A
Sbjct: 186 ---EIKDTIKAARELESESTELESAVVTRVEPLGMGDRVCVDTCNLMQRGEGMLIGSQAS 242
Query: 221 GLFLVHSECLESKLHCKQTF--SGECVHAYIAVPGGRTSYLSELKSGKEVIVVDQQGRQR 278
G+FLV+SE +S + F + VH+YI + G +T YLSEL++G V ++D +G+QR
Sbjct: 243 GMFLVNSESDDSPYVAARPFRVNAGAVHSYIKI-GDKTRYLSELQAGDSVTIIDSRGKQR 301
Query: 279 IAIVGRVKIESRPLILVEAKTDSDNQTISILLQNAETVALVCPVQGNKLLKTAFPVTSLK 338
IVGR+KIESRPL+L+EAK + ++T++ +LQNAET+ LV +G T V LK
Sbjct: 302 EGIVGRIKIESRPLMLIEAK--AGDRTLTAILQNAETIKLV--GKGG----TPISVAKLK 353
Query: 339 VGDEVLLRVQGGARHTGIEIQEFIVEK 365
GDEVL+R++ GARH G +I+E I+EK
Sbjct: 354 KGDEVLVRLEEGARHFGKKIEETIIEK 380
>gb|AAM07931.1| conserved hypothetical protein [Methanosarcina acetivorans str.
C2A] gi|20093376|ref|NP_619451.1| hypothetical protein
MA4592 [Methanosarcina acetivorans C2A]
gi|32130355|sp|Q8THC5|Y9J2_METAC Hypothetical UPF0245
protein MA4592
Length = 380
Score = 196 bits (498), Expect = 9e-49
Identities = 121/267 (45%), Positives = 174/267 (64%), Gaps = 17/267 (6%)
Query: 103 IVVNLLDWQVIPAENIIAAFQNSQ-KTVFAISDNASEAQTFLEALEHGLDGIVLKVEDVE 161
++V DW+VIP EN+IA Q+ + K +F + +A EA+ + LE G DG++L + +
Sbjct: 127 LLVTGTDWKVIPLENLIADLQHQKVKIIFGVK-SAEEARLAFQTLEAGADGVLLDSGNPQ 185
Query: 162 PMLELKEYFDRRTE-ESNVLNLTKATVTQIQVAGMGDRVCVDLCSLMRPGEGLLVGSFAR 220
E+K+ E ES L A VT+++ GMGDRVCVD C+LM+ GEG+L+GS A
Sbjct: 186 ---EIKDTIKAARELESESAELEAAVVTRVEPLGMGDRVCVDTCNLMQRGEGMLIGSQAS 242
Query: 221 GLFLVHSECLESKLHCKQTF--SGECVHAYIAVPGGRTSYLSELKSGKEVIVVDQQGRQR 278
G+FLV+SE +S + F + VH+YI + G +T YLSEL++G V +VD +G+QR
Sbjct: 243 GMFLVNSESDDSPYVAARPFRVNAGAVHSYIKI-GEKTRYLSELRAGDPVTIVDSKGKQR 301
Query: 279 IAIVGRVKIESRPLILVEAKTDSDNQTISILLQNAETVALVCPVQGNKLLKTAFPVTSLK 338
IVGRVKIESRPL+L+EAK + ++T++ +LQNAET+ LV G T V L+
Sbjct: 302 EGIVGRVKIESRPLMLIEAK--ARDRTLTAILQNAETIKLV----GKD--GTPISVAKLE 353
Query: 339 VGDEVLLRVQGGARHTGIEIQEFIVEK 365
GDEVL+R++ GARH G +I+E I+EK
Sbjct: 354 KGDEVLVRLEEGARHFGKKIEETIIEK 380
>ref|NP_614691.1| Predicted alternative 3-dehydroquinate synthase [Methanopyrus
kandleri AV19] gi|19888068|gb|AAM02621.1| Predicted
alternative 3-dehydroquinate synthase [Methanopyrus
kandleri AV19] gi|32130356|sp|Q8TVI1|YE08_METKA
Hypothetical UPF0245 protein MK1408
Length = 402
Score = 196 bits (497), Expect = 1e-48
Identities = 119/265 (44%), Positives = 163/265 (60%), Gaps = 19/265 (7%)
Query: 109 DWQVIPAENIIAAFQNSQKTVFAISDNASEAQTFLEALEHGLDGIVLKVEDVEPMLELK- 167
DW++IP EN+IA Q + + A + +A EA+ E LE G DG++L E ++P E+K
Sbjct: 149 DWKIIPLENLIAELQGEKTQLIAGARDAEEARIAFETLEVGSDGVLLDAERIDPS-EIKK 207
Query: 168 --EYFDRRTEESNVLNLTKATVTQIQVAGMGDRVCVDLCSLMRPGEGLLVGSFARGLFLV 225
E +R E L V +++ G GDRVCVD CSLM GEG+LVGS +RG+FL+
Sbjct: 208 TAEIAERAAAER--FELVAVEVKEVKPIGKGDRVCVDTCSLMSEGEGMLVGSTSRGMFLI 265
Query: 226 HSECLESKLHCKQTFSGEC--VHAYIAVPGGRTSYLSELKSGKEVIVVDQQGRQRIAIVG 283
HSE LE+ + F VHAYI VPGG+T YL+EL+ G EV++VD +GR R A+VG
Sbjct: 266 HSESLENPYVEPRPFRVNAGPVHAYIRVPGGKTKYLAELRPGDEVLIVDTEGRTRAAVVG 325
Query: 284 RVKIESRPLILVEAKTDSDNQTISILLQNAETVALVCPVQGNKLLKTAFPVTSLKVGDEV 343
R+KIE RPL+L+ A + + I ++QNAET+ LV G + V LK GD+V
Sbjct: 326 RLKIERRPLMLIRA--EYEGVEIQTIVQNAETIHLV-REDGEPV-----SVVDLKPGDKV 377
Query: 344 LLRV---QGGARHTGIEIQEFIVEK 365
L V +G RH G+E++E IVEK
Sbjct: 378 LAYVETEEGKGRHFGMEVEETIVEK 402
>ref|NP_279411.1| hypothetical protein VNG0310C [Halobacterium sp. NRC-1]
gi|10579939|gb|AAG18891.1| Vng0310c [Halobacterium sp.
NRC-1] gi|32130333|sp|Q9HSB6|Y310_HALSA Hypothetical
UPF0245 protein Vng0310c
Length = 387
Score = 187 bits (475), Expect = 4e-46
Identities = 118/270 (43%), Positives = 159/270 (58%), Gaps = 13/270 (4%)
Query: 98 EQAENIVVNLLDWQVIPAENIIAAFQNSQKTVFAISDNASEAQTFLEALEHGLDGIVLKV 157
E A++ +V DW +IP EN+IA + T+ A ++A+EA+T E L+ G D ++L
Sbjct: 129 EVADHTIVVGDDWTIIPLENLIARI-GEETTLVAGVESAAEAETAFETLDIGADAVLLDS 187
Query: 158 EDVEPMLELKEYFDRRTEESNVLNLTKATVTQIQVAGMGDRVCVDLCSLMRPGEGLLVGS 217
+D + + D E L L+ AT+T I+ AG DRVCVD SL+ EG+LVGS
Sbjct: 188 DDPDEIRRTVSVRDAADREH--LALSTATITTIEEAGSADRVCVDTGSLLADDEGMLVGS 245
Query: 218 FARGLFLVHSECLESKLHCKQTF--SGECVHAYIAVPGGRTSYLSELKSGKEVIVVDQQG 275
+RGLF VH+E +S + F + VHAY+ P G T YL+EL SG EV VVD G
Sbjct: 246 MSRGLFFVHAETAQSPYVAARPFRVNAGAVHAYVRTPDGGTKYLAELGSGDEVQVVDGDG 305
Query: 276 RQRIAIVGRVKIESRPLILVEAKTDSDNQTISILLQNAETVALVCPVQGNKLLKTAFPVT 335
R R A+VGR KIE RP+ VEA+TD D I LLQNAET+ + P +TA VT
Sbjct: 306 RTRSAVVGRAKIEKRPMFRVEAETD-DGDRIETLLQNAETIKVATPNG-----RTA--VT 357
Query: 336 SLKVGDEVLLRVQGGARHTGIEIQEFIVEK 365
L VGD++ + +Q G RH G I E I+E+
Sbjct: 358 DLSVGDDLHVFLQDGGRHFGEAIDERIIEQ 387
>gb|AAV46760.1| 3-dehydroquinate synthase [Haloarcula marismortui ATCC 43049]
gi|55378616|ref|YP_136466.1| 3-dehydroquinate synthase
[Haloarcula marismortui ATCC 43049]
Length = 388
Score = 186 bits (473), Expect = 7e-46
Identities = 119/269 (44%), Positives = 159/269 (58%), Gaps = 13/269 (4%)
Query: 99 QAENIVVNLLDWQVIPAENIIAAFQNSQKTVFAISDNASEAQTFLEALEHGLDGIVLKVE 158
+A+ +V +WQ+IP EN+IA + + A +A+T E LE G DG++L +
Sbjct: 131 EADFTIVIGENWQIIPLENLIARVGEETDLIAGVR-TAEDARTAYETLELGADGVLLDTD 189
Query: 159 DVEPMLELKEYFDRRTEESNVLNLTKATVTQIQVAGMGDRVCVDLCSLMRPGEGLLVGSF 218
+V+ + + E D ES L+L A VT I+ G DRVC+D SLM EG+LVGS
Sbjct: 190 EVDEIRKTVEVRDEMGRES--LDLEYAEVTAIEQTGSADRVCIDTGSLMEHDEGMLVGSM 247
Query: 219 ARGLFLVHSECLESKLHCKQTF--SGECVHAYIAVPGGRTSYLSELKSGKEVIVVDQQGR 276
ARGLF VH+E ES + F + VHAY+ P G T YLSEL+SG EV +VD GR
Sbjct: 248 ARGLFFVHAETAESPYVASRPFRVNAGAVHAYVRTPDGGTKYLSELQSGDEVQIVDANGR 307
Query: 277 QRIAIVGRVKIESRPLILVEAKTDSDNQTISILLQNAETVALVCPVQGNKLLKTAFPVTS 336
R AIVGR KIE RP+ V+A+T+ D+ I LLQNAET+ V G +TA VT
Sbjct: 308 TREAIVGRAKIEKRPMFRVQAETE-DSDRIETLLQNAETIK-VHTQDG----RTA--VTD 359
Query: 337 LKVGDEVLLRVQGGARHTGIEIQEFIVEK 365
L+ GDE+L+ + A H G I+E I+EK
Sbjct: 360 LEPGDEILIHHEDTATHFGERIEESIIEK 388
>ref|NP_069067.1| hypothetical protein AF0229 [Archaeoglobus fulgidus DSM 4304]
gi|2650412|gb|AAB91003.1| conserved hypothetical protein
[Archaeoglobus fulgidus DSM 4304] gi|7451239|pir||E69278
conserved hypothetical protein AF0229 - Archaeoglobus
fulgidus gi|32130311|sp|O30010|Y229_ARCFU Hypothetical
UPF0245 protein AF0229
Length = 323
Score = 182 bits (463), Expect = 1e-44
Identities = 130/356 (36%), Positives = 197/356 (54%), Gaps = 42/356 (11%)
Query: 18 KKVWI------WTKNKQVMTAAVERGWNTFIFPSNLPHLANDWSSIAVICPLFASEKEIL 71
K++W+ W + K+++ A+E G++ + + A + ++ I
Sbjct: 2 KEIWLLAESESWDEAKEMLKDAIEIGFDGALVRRDFLERAEKLGRMKIV--------PIE 53
Query: 72 DAQNKKVATIFDVSTPEELERLRPEDEQAENIVVNLLDWQVIPAENIIAAFQNSQKTVFA 131
DA + +S+ E+ ER Q E +V+ DW+VIP ENI+A + S K + A
Sbjct: 54 DA-------VVKISSAEDQERAL----QREVVVLKFEDWKVIPLENIVA-MKKSGKVIAA 101
Query: 132 ISDNASEAQTFLEALEHGLDGIVLKVEDVEPMLELKEYFDRRTEESNVLNLTKATVTQIQ 191
+ D +A+ L LE G DGI + + L+++++ EE + L +A V +I+
Sbjct: 102 V-DTIEDAKLALTTLERGADGIAVSGDRET----LRKFYEVVKEEGERVELVRARVKEIR 156
Query: 192 VAGMGDRVCVDLCSLMRPGEGLLVGSFARGLFLVHSECLESKLHCKQTF--SGECVHAYI 249
G+G+RVC+D +LM PGEG+LVG+ A +FLV SE ES+ + F + V+AY+
Sbjct: 157 PLGVGERVCIDTVTLMTPGEGMLVGNQASFMFLVASESEESEYVASRPFRVNAGSVNAYL 216
Query: 250 AVPGGRTSYLSELKSGKEVIVVDQQGRQRIAIVGRVKIESRPLILVEAKTDSDNQTISIL 309
V G +T YL+ELK+G EV VV G R + VGRVKIE RPLIL+ A+ D S++
Sbjct: 217 KV-GDKTRYLAELKAGDEVEVVKFDGAVRKSYVGRVKIERRPLILIRAEVDGVEG--SVI 273
Query: 310 LQNAETVALVCPVQGNKLLKTAFPVTSLKVGDEVLLRVQGGARHTGIEIQEFIVEK 365
LQNAET+ LV P G + V LK GDE+L+ + ARH G+E+ EFIVE+
Sbjct: 274 LQNAETIKLVAP-DGKHV-----SVAELKPGDEILVWLGKKARHFGVEVDEFIVER 323
>ref|ZP_00347077.1| COG1465: Predicted alternative 3-dehydroquinate synthase
[Desulfovibrio desulfuricans G20]
Length = 328
Score = 167 bits (422), Expect = 6e-40
Identities = 105/261 (40%), Positives = 144/261 (54%), Gaps = 18/261 (6%)
Query: 110 WQVIPAENIIAAFQNSQKTVFAISDNASEAQTFLEALEHGLDGIVLKVEDVEPMLELKEY 169
W++IP EN++A N V A + EA+ LE G+ + + VE ELK+
Sbjct: 81 WEIIPVENLLAVSDN----VVAEVASLEEARLAAGILERGVSAVAVSARGVE---ELKDI 133
Query: 170 FDRRTEESNVLNLTKATVTQIQVAGMGDRVCVDLCSLMRPGEGLLVGSFARGLFLVHSEC 229
L L ATVT + AG+G RVCVD SLMR G+G+LVG+ + FLV++E
Sbjct: 134 VKELKLSQGTLELVPATVTAVSAAGLGHRVCVDTMSLMRTGQGMLVGNSSAFTFLVNAET 193
Query: 230 LESKLHCKQTF--SGECVHAYIAVPGGRTSYLSELKSGKEVIVVDQQGRQRIAIVGRVKI 287
++ + F + VHAY +PG RT+YL EL +G EV+VVD G IA VGRVK+
Sbjct: 194 EHNEYVAARPFRVNAGAVHAYAVMPGDRTTYLEELSAGTEVLVVDHTGATGIATVGRVKV 253
Query: 288 ESRPLILVEAKTDS-DNQTI--SILLQNAETVALVCPVQGNKLLKTAFPVTSLKVGDEVL 344
E RP++LV A+ D Q ++ LQNAET+ LV + T V +LK GDE+L
Sbjct: 254 EVRPMLLVRARVQGPDGQCTEGAVFLQNAETIRLV------RTDGTPVSVVALKEGDEIL 307
Query: 345 LRVQGGARHTGIEIQEFIVEK 365
+R RH G+ I E I E+
Sbjct: 308 VRTDAAGRHFGMRISEEIREE 328
>ref|YP_009685.1| predicted 3-dehydroquinate synthase [Desulfovibrio vulgaris subsp.
vulgaris str. Hildenborough] gi|46448289|gb|AAS94944.1|
predicted 3-dehydroquinate synthase [Desulfovibrio
vulgaris subsp. vulgaris str. Hildenborough]
Length = 323
Score = 153 bits (386), Expect = 9e-36
Identities = 116/346 (33%), Positives = 173/346 (49%), Gaps = 42/346 (12%)
Query: 26 NKQVMTAAVERGWNTFIFPSNLPHLANDWSSIAVICPLFASEKEILDAQNKKVATIFDVS 85
+K+ +T A+E G + I P + V+C + V T+
Sbjct: 12 SKEAVTLALESGVDGIIVPEKHVAPVQALARCKVLC-------------DDAVCTL---- 54
Query: 86 TPEELERLRPEDEQAENI-----VVNLLDWQVIPAENIIAAFQNSQKTVFAISDNASEAQ 140
EL E+E A + VV W++IP EN++A ++ + TV S EA+
Sbjct: 55 ---ELTAKADEEEAASRLAAGEDVVLARGWEIIPVENLLA--RSDRVTVEVAS--LDEAR 107
Query: 141 TFLEALEHGLDGIVLKVEDVEPMLELKEYFDRRTEESNVLNLTKATVTQIQVAGMGDRVC 200
LE G+ +V+ + V ELK ++L A VT ++ G+G RVC
Sbjct: 108 LAAGILERGVSTVVVLPQAVT---ELKAIVQELKLSQGRIDLAPAVVTAVRPVGLGHRVC 164
Query: 201 VDLCSLMRPGEGLLVGSFARGLFLVHSECLESKLHCKQTF--SGECVHAYIAVPGGRTSY 258
VD SL+R G+G+LVG+ + FLV++E ++ + F + VHAY +PG RT+Y
Sbjct: 165 VDTMSLLRTGQGMLVGNSSAFTFLVNAETEHNEYVAARPFRINAGAVHAYAVMPGDRTTY 224
Query: 259 LSELKSGKEVIVVDQQGRQRIAIVGRVKIESRPLILVEAKTDSDNQTISILLQNAETVAL 318
L EL+SG EV++V G IA VGR+K+E RP++LVEA+ + ++ LQNAET+ L
Sbjct: 225 LEELRSGSEVLIVGADGATTIATVGRLKVEVRPMLLVEARV--GDTVGAVFLQNAETIRL 282
Query: 319 VCPVQGNKLLKTAFPVTSLKVGDEVLLRVQGGARHTGIEIQEFIVE 364
V + T V +L+ GDEVL R RH G+ I E I E
Sbjct: 283 V------RADGTPVSVVALREGDEVLCRTDVAGRHFGMRITEDIRE 322
>ref|ZP_00212281.1| COG1465: Predicted alternative 3-dehydroquinate synthase
[Burkholderia cepacia R18194]
Length = 366
Score = 136 bits (342), Expect = 1e-30
Identities = 85/265 (32%), Positives = 139/265 (52%), Gaps = 12/265 (4%)
Query: 103 IVVNLLDWQVIPAENIIAAFQNSQKTVFAISDNASEAQTFLEALEHGLDGIVLKVEDVEP 162
+V L D IP E ++AA + + + + +A+ L L+ G +G+++ V
Sbjct: 112 LVEFLQDPSKIPLEILLAAADQASGELITVVADLEDAEVTLNVLQRGPEGVLVSPTQVGQ 171
Query: 163 MLELKEYFDRRTEESNVLNLTKATVTQIQVAGMGDRVCVDLCSLMRPGEGLLVGSFARGL 222
+L E+ L L + + + G G+RVCVD CS EG+LVGS++ G+
Sbjct: 172 ATQLVSICSDTVED---LQLEEIEIVGLTHLGPGERVCVDTCSRFEQDEGILVGSYSTGM 228
Query: 223 FLVHSECLESKLHCKQTF--SGECVHAYIAVPGGRTSYLSELKSGKEVIVVDQQGRQRIA 280
L+ SE + F + +H+Y+ P RT YL+EL+SG E++ V+ QG+ R
Sbjct: 229 ILISSETHPLPYMPTRPFRVNAAALHSYVVAPDNRTRYLAELESGAEILAVNVQGKARRV 288
Query: 281 IVGRVKIESRPLILVEAKTDSDNQTISILLQNAETVALVCPVQGNKLLKTAFPVTSLKVG 340
+VGR K+E+RPL+L+EAK ++ N+ I+I+ Q+ V ++ P + +T LK G
Sbjct: 289 VVGRAKVETRPLLLIEAK-NAANRAINIIAQDDWHVRVLGP------KGSVHNITELKRG 341
Query: 341 DEVLLRVQGGARHTGIEIQEFIVEK 365
D +L RH G I EF+ E+
Sbjct: 342 DRILGYTLDAQRHVGYPITEFLHEQ 366
>ref|ZP_00131363.1| COG1465: Predicted alternative 3-dehydroquinate synthase
[Desulfovibrio desulfuricans G20]
Length = 185
Score = 128 bits (321), Expect = 3e-28
Identities = 79/182 (43%), Positives = 105/182 (57%), Gaps = 11/182 (6%)
Query: 165 ELKEYFDRRTEESNVLNLTKATVTQIQVAGMGDRVCVDLCSLMRPGEGLLVGSFARGLFL 224
ELK+ L L ATVT + AG+G RVCVD SLMR G+G+LVG+ + FL
Sbjct: 10 ELKDIVKELKLSQGTLELVPATVTAVSAAGLGHRVCVDTMSLMRTGQGMLVGNSSAFTFL 69
Query: 225 VHSECLESKLHCKQTF--SGECVHAYIAVPGGRTSYLSELKSGKEVIVVDQQGRQRIAIV 282
V++E ++ + F + VHAY +PG RT+YL EL +G EV+VVD G IA V
Sbjct: 70 VNAETEHNEYVAARPFRVNAGAVHAYAVMPGDRTTYLEELSAGTEVLVVDHTGATGIATV 129
Query: 283 GRVKIESRPLILVEAKTDS-DNQTI--SILLQNAETVALVCPVQGNKLLKTAFPVTSLKV 339
GRVK+E RP++LV A+ D Q ++ LQNAET+ LV + T V +LK
Sbjct: 130 GRVKVEVRPMLLVRARVQGPDGQCTEGAVFLQNAETIRLV------RTDGTPVSVVALKE 183
Query: 340 GD 341
GD
Sbjct: 184 GD 185
>ref|ZP_00212282.1| COG1465: Predicted alternative 3-dehydroquinate synthase
[Burkholderia cepacia R18194]
Length = 354
Score = 54.7 bits (130), Expect = 4e-06
Identities = 37/128 (28%), Positives = 61/128 (46%), Gaps = 2/128 (1%)
Query: 180 LNLTKATVTQIQVAGMGDRVCVDLCSLMRPGEGLLVGSFARGLFLVHSECLESKLHCKQT 239
L L V +++ G+G +D CS + E +L G+ A + LV + T
Sbjct: 169 LKLKTFEVQRVEPLGVGLHAVIDGCSQLDGEECVLAGASATSMLLVVAAGAAHPPGRPLT 228
Query: 240 FSGEC--VHAYIAVPGGRTSYLSELKSGKEVIVVDQQGRQRIAIVGRVKIESRPLILVEA 297
F + +++ G R LS L+SG+ ++ V G R +VGRV+IES PL+ +
Sbjct: 229 FGIDAGSAESFVFCGGDRARQLSLLRSGERILTVGAAGDTRAIVVGRVRIESAPLVALHT 288
Query: 298 KTDSDNQT 305
++ S T
Sbjct: 289 RSASGANT 296
>gb|AAL12776.1| LktB [Mannheimia haemolytica]
Length = 708
Score = 42.4 bits (98), Expect = 0.022
Identities = 31/108 (28%), Positives = 53/108 (48%), Gaps = 11/108 (10%)
Query: 248 YIAVPGGRTSYLSELKSGKEVIVVDQQG------RQRIAIVGRVKIESRPLILVEAKTDS 301
Y A G ++SEL+ G IV +Q RQRIAI + + LI EA +
Sbjct: 578 YAAKLAGAHDFISELREGYNTIVGEQGAGLSGGQRQRIAIARALVNNPKILIFDEATSAL 637
Query: 302 DNQTISILLQNAETVALVCPVQGNKLLKTAFPVTSLKVGDEVLLRVQG 349
D Q+ I++QN + + QG ++ A ++++K D +++ +G
Sbjct: 638 DYQSEHIIMQNMQKIC-----QGRTVILIAHRLSTVKNADRIIVMEKG 680
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.317 0.133 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 588,333,713
Number of Sequences: 2540612
Number of extensions: 23240298
Number of successful extensions: 65694
Number of sequences better than 10.0: 79
Number of HSP's better than 10.0 without gapping: 23
Number of HSP's successfully gapped in prelim test: 56
Number of HSP's that attempted gapping in prelim test: 65582
Number of HSP's gapped (non-prelim): 86
length of query: 365
length of database: 863,360,394
effective HSP length: 129
effective length of query: 236
effective length of database: 535,621,446
effective search space: 126406661256
effective search space used: 126406661256
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 76 (33.9 bits)
Medicago: description of AC136287.2


