

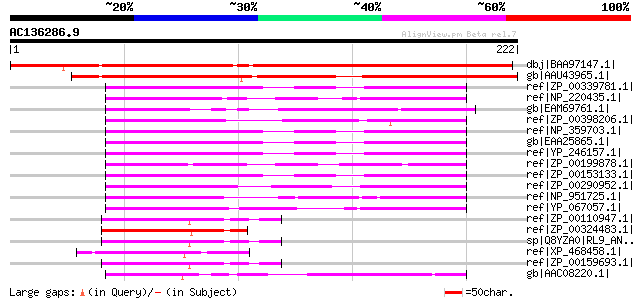
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136286.9 - phase: 0
(222 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAA97147.1| unnamed protein product [Arabidopsis thaliana] g... 237 2e-61
gb|AAU43965.1| unknown protein [Oryza sativa (japonica cultivar-... 219 4e-56
ref|ZP_00339781.1| COG0359: Ribosomal protein L9 [Rickettsia aka... 55 1e-06
ref|NP_220435.1| 50S RIBOSOMAL PROTEIN L9 (rplI) [Rickettsia pro... 53 5e-06
gb|EAM69761.1| Ribosomal protein L9 [Desulfuromonas acetoxidans ... 52 9e-06
ref|ZP_00398206.1| Ribosomal protein L9 [Anaeromyxobacter dehalo... 52 9e-06
ref|NP_359703.1| 50S ribosomal protein L9 [Rickettsia conorii st... 52 9e-06
gb|EAA25865.1| 50S ribosomal protein L9 [Rickettsia sibirica 246... 52 9e-06
ref|YP_246157.1| 50S ribosomal protein L9 [Rickettsia felis URRW... 52 9e-06
ref|ZP_00199878.1| COG0359: Ribosomal protein L9 [Rubrobacter xy... 52 1e-05
ref|ZP_00153133.1| COG0359: Ribosomal protein L9 [Rickettsia ric... 52 1e-05
ref|ZP_00290952.1| COG0359: Ribosomal protein L9 [Magnetococcus ... 50 4e-05
ref|NP_951725.1| ribosomal protein L9 [Geobacter sulfurreducens ... 50 4e-05
ref|YP_067057.1| 50S ribosomal protein L9 [Rickettsia typhi str.... 50 5e-05
ref|ZP_00110947.1| COG0359: Ribosomal protein L9 [Nostoc punctif... 49 8e-05
ref|ZP_00324483.1| COG0359: Ribosomal protein L9 [Trichodesmium ... 49 1e-04
sp|Q8YZA0|RL9_ANASP 50S ribosomal protein L9 gi|17129924|dbj|BAB... 49 1e-04
ref|XP_468458.1| putative 50S ribosomal protein L9, chloroplast ... 49 1e-04
ref|ZP_00159693.1| COG0359: Ribosomal protein L9 [Anabaena varia... 48 2e-04
gb|AAC08220.1| 50S ribosomal protein L9 [Porphyra purpurea] gi|1... 48 2e-04
>dbj|BAA97147.1| unnamed protein product [Arabidopsis thaliana]
gi|28393965|gb|AAO42390.1| unknown protein [Arabidopsis
thaliana] gi|27754507|gb|AAO22701.1| unknown protein
[Arabidopsis thaliana] gi|15237342|ref|NP_200119.1|
ribosomal protein L9 family protein [Arabidopsis
thaliana]
Length = 221
Score = 237 bits (604), Expect = 2e-61
Identities = 127/222 (57%), Positives = 164/222 (73%), Gaps = 5/222 (2%)
Query: 1 MGYLQFGRYSVRQIVRFKDVVN--EGVVVNPLMYASQGLRYNRKLQVILTTNIDKLGKAG 58
M Y+ R +R +V + E + +PL++A QG+RY RKL+VILTT I+KLGKAG
Sbjct: 1 MAYVGQSRNVIRHVVSRGTAYHKYENAIHHPLLFACQGVRY-RKLEVILTTGIEKLGKAG 59
Query: 59 ETVKVAPGFFRNHLMPKLLAVPNIDKFAYLLTEQRKIYQPTEEVKQEDVVVVTESKEDLM 118
ETVKVAPG+FRNHLMPKLLAVPNIDK+AYL+ EQRK+Y EEVK+E V VV ++ E
Sbjct: 60 ETVKVAPGYFRNHLMPKLLAVPNIDKYAYLIREQRKMYNH-EEVKEE-VKVVHKTSEVQT 117
Query: 119 KEYERAALILDKAKLVLRRLIDVKKAKARESKDEPLELQIPVSKNALVAEVARQLCVNIT 178
KEYE+AA L A LVLR+L+D +K K R SKD+ ++Q P++K +V+EVARQLCV I
Sbjct: 118 KEYEKAAKRLANANLVLRKLVDKEKFKNRSSKDDKPDVQTPITKEEIVSEVARQLCVKID 177
Query: 179 AENLHLPTPLSTIGEYEVPLRLPRSIPLPEGKLNWALKVKIR 220
+N+ L PL T GEYEVPL+ P++IPLP+G + W LKVK+R
Sbjct: 178 PDNVVLAAPLETFGEYEVPLKFPKTIPLPQGTVQWILKVKVR 219
>gb|AAU43965.1| unknown protein [Oryza sativa (japonica cultivar-group)]
Length = 206
Score = 219 bits (558), Expect = 4e-56
Identities = 120/200 (60%), Positives = 147/200 (73%), Gaps = 19/200 (9%)
Query: 28 NPLMYASQGLRYNRKLQVILTTNIDKLGKAGETVKVAPGFFRNHLMPKLLAVPNIDKFAY 87
NP++++ GLRY RKL+VILTT IDKLGKAGE VKVAPG FRNHLMPK+LAVPNIDKFA
Sbjct: 21 NPVLFSGHGLRY-RKLEVILTTTIDKLGKAGEVVKVAPGHFRNHLMPKMLAVPNIDKFAL 79
Query: 88 LLTEQRKIYQPTE-----EVKQEDVVVVTESKEDLMKEYERAALILDKAKLVLRRLIDVK 142
L+ EQRK+YQ E EV+QED + +E+ +KEY+ AA LD A LVLRR I V
Sbjct: 80 LIHEQRKLYQRQEEEVVKEVRQED--DDAKQQEEKLKEYQTAAKCLDNALLVLRRFISVG 137
Query: 143 KAKARESKDEPLELQIPVSKNALVAEVARQLCVNITAENLHLPTPLSTIGEYEVPLRLPR 202
EL+ PV+K+ +V+EVARQL +NI +NLHLP+PL+++GE+E+PLRLPR
Sbjct: 138 N-----------ELRSPVTKDEIVSEVARQLNINIHPDNLHLPSPLASLGEFELPLRLPR 186
Query: 203 SIPLPEGKLNWALKVKIRSK 222
IP PEGKL W L VKIR K
Sbjct: 187 DIPRPEGKLQWTLTVKIRRK 206
>ref|ZP_00339781.1| COG0359: Ribosomal protein L9 [Rickettsia akari str. Hartford]
Length = 171
Score = 55.1 bits (131), Expect = 1e-06
Identities = 40/158 (25%), Positives = 77/158 (48%), Gaps = 25/158 (15%)
Query: 43 LQVILTTNIDKLGKAGETVKVAPGFFRNHLMPKLLAVPNIDKFAYLLTEQRKIYQPTEEV 102
+++IL + KLGK G+ +KVA GF RN+L+P+ LA+ + L+ +Q+ ++ ++
Sbjct: 1 MEIILIKPVRKLGKIGDMLKVADGFGRNYLLPQKLAIRATEPNKELIVKQKHEFEAKDKQ 60
Query: 103 KQEDVVVVTESKEDLMKEYERAALILDKAKLVLRRLIDVKKAKARESKDEPLELQIPVSK 162
+E+V + ALI D+ + +R+ D K L V+
Sbjct: 61 IREEVEKIN-------------ALIKDQKLVFIRQASDDGK------------LFGSVTN 95
Query: 163 NALVAEVARQLCVNITAENLHLPTPLSTIGEYEVPLRL 200
+ ++++ + NI+ N+ L + + G Y V +RL
Sbjct: 96 KEIADKLSKNVSYNISHSNIILDKQIKSTGVYTVEIRL 133
>ref|NP_220435.1| 50S RIBOSOMAL PROTEIN L9 (rplI) [Rickettsia prowazekii str. Madrid
E] gi|3860611|emb|CAA14512.1| 50S RIBOSOMAL PROTEIN L9
(rplI) [Rickettsia prowazekii] gi|7440910|pir||A71712
ribosomal protein L9 - Rickettsia prowazekii
gi|6226013|sp|Q9ZEA4|RL9_RICPR 50S ribosomal protein L9
Length = 171
Score = 53.1 bits (126), Expect = 5e-06
Identities = 43/158 (27%), Positives = 78/158 (49%), Gaps = 25/158 (15%)
Query: 43 LQVILTTNIDKLGKAGETVKVAPGFFRNHLMPKLLAVPNIDKFAYLLTEQRKIYQPTEEV 102
++VIL + KLGK G+ +KVA GF RN+L+P+ LA+ L+ +Q+ Y+ E+
Sbjct: 1 MEVILIKPVRKLGKIGDILKVADGFGRNYLLPQKLAIRATKSNKELIVKQK--YEFEEKD 58
Query: 103 KQEDVVVVTESKEDLMKEYERAALILDKAKLVLRRLIDVKKAKARESKDEPLELQIPVSK 162
KQ + +E E+ I+ +LV R++ D+ +L V+
Sbjct: 59 KQ------------VREEVEKINTIIKDQQLVF----------IRQTSDD-CKLFGSVTN 95
Query: 163 NALVAEVARQLCVNITAENLHLPTPLSTIGEYEVPLRL 200
+ ++++ + NI+ N+ L + + G Y V +RL
Sbjct: 96 KDIADKLSKNISYNISHSNIILDKQIKSTGIYTVEIRL 133
>gb|EAM69761.1| Ribosomal protein L9 [Desulfuromonas acetoxidans DSM 684]
Length = 149
Score = 52.4 bits (124), Expect = 9e-06
Identities = 43/162 (26%), Positives = 77/162 (46%), Gaps = 26/162 (16%)
Query: 43 LQVILTTNIDKLGKAGETVKVAPGFFRNHLMPKLLAVPNIDKFAYLLTEQRKIYQPTEEV 102
+++ILT +ID LG+ GE V V PG+ RN+L+PK AV + +Q + +EV
Sbjct: 1 MEIILTESIDGLGEIGEIVNVKPGYARNYLLPKGFAV---------VADQGNV----KEV 47
Query: 103 KQEDVVVVTESKEDLMKEYERAALILDKAKLVLRRLIDVKKAKARESKDEPLELQIPVSK 162
+ + ++ ER AL + K L+ I+ + E +L V+
Sbjct: 48 EHQ------------KRQLERKALKIAKTSEALKAKIEAAACEFELRASEEGKLFGSVTS 95
Query: 163 NALVAEVARQLCVNITAENLHLPTPLSTIGEYEVPLRLPRSI 204
+ A++A + V I + + L + +GE V ++LP ++
Sbjct: 96 ADIAAKLA-EADVEIDRKKIQLDEAIKALGEQTVNVKLPGNV 136
>ref|ZP_00398206.1| Ribosomal protein L9 [Anaeromyxobacter dehalogenans 2CP-C]
gi|66779894|gb|EAL80969.1| Ribosomal protein L9
[Anaeromyxobacter dehalogenans 2CP-C]
Length = 149
Score = 52.4 bits (124), Expect = 9e-06
Identities = 43/159 (27%), Positives = 74/159 (46%), Gaps = 28/159 (17%)
Query: 43 LQVILTTNIDKLGKAGETVKVAPGFFRNHLMPKLLAVPNIDKFAYLLTEQRKIYQPTEEV 102
+++IL +++ LGK G+ V V PG+ RN+L+P+ LAV K L Q+ +
Sbjct: 1 MKLILREDVENLGKGGDLVDVKPGYGRNYLLPRGLAVSANPKNVKELEHQKAV------- 53
Query: 103 KQEDVVVVTESKEDLMKEYERAALILDKAKLVLRRLIDVKKAKARESKDEPLELQIPVSK 162
+AA + A+ V +RL + R+ ++ ++ S
Sbjct: 54 -----------------AAAKAAKLKASAQAVAKRLSETPVTLKRKVGEQD---KLYGSV 93
Query: 163 NAL-VAEVARQLCVNITAENLHLPTPLSTIGEYEVPLRL 200
AL VAE V + ++ L P+ T+GE+EVP++L
Sbjct: 94 TALDVAEALAARGVQLDRRSIVLDEPIKTLGEFEVPVKL 132
>ref|NP_359703.1| 50S ribosomal protein L9 [Rickettsia conorii str. Malish 7]
gi|15619103|gb|AAL02604.1| 50S ribosomal protein L9
[Rickettsia conorii str. Malish 7]
gi|25295284|pir||B97708 50S ribosomal protein L9
[imported] - Rickettsia conorii (strain Malish 7)
gi|20139337|sp|Q92JK1|RL9_RICCN 50S ribosomal protein L9
Length = 171
Score = 52.4 bits (124), Expect = 9e-06
Identities = 39/158 (24%), Positives = 76/158 (47%), Gaps = 25/158 (15%)
Query: 43 LQVILTTNIDKLGKAGETVKVAPGFFRNHLMPKLLAVPNIDKFAYLLTEQRKIYQPTEEV 102
+++IL + KLGK G+ +KVA GF RN+L+P+ LA+ + L+ +Q+ ++ ++
Sbjct: 1 MEIILIKPVRKLGKIGDILKVADGFGRNYLLPQKLAIRATEPNKELIVKQKHEFEAKDKQ 60
Query: 103 KQEDVVVVTESKEDLMKEYERAALILDKAKLVLRRLIDVKKAKARESKDEPLELQIPVSK 162
+E+V + ALI D+ + +R+ + K L V+
Sbjct: 61 IREEVEKIN-------------ALIKDQQLVFIRQTSNDGK------------LFGSVTN 95
Query: 163 NALVAEVARQLCVNITAENLHLPTPLSTIGEYEVPLRL 200
+ +++ + NI+ N+ L + + G Y V +RL
Sbjct: 96 KEIADKLSENISYNISHSNIILDKQIKSTGIYTVEIRL 133
>gb|EAA25865.1| 50S ribosomal protein L9 [Rickettsia sibirica 246]
gi|34580976|ref|ZP_00142456.1| 50S ribosomal protein L9
[Rickettsia sibirica 246]
Length = 171
Score = 52.4 bits (124), Expect = 9e-06
Identities = 39/158 (24%), Positives = 76/158 (47%), Gaps = 25/158 (15%)
Query: 43 LQVILTTNIDKLGKAGETVKVAPGFFRNHLMPKLLAVPNIDKFAYLLTEQRKIYQPTEEV 102
+++IL + KLGK G+ +KVA GF RN+L+P+ LA+ + L+ +Q+ ++ ++
Sbjct: 1 MEIILIKPVRKLGKIGDILKVADGFGRNYLLPQKLAIRATEPNKELIVKQKHEFEAKDKQ 60
Query: 103 KQEDVVVVTESKEDLMKEYERAALILDKAKLVLRRLIDVKKAKARESKDEPLELQIPVSK 162
+E+V + ALI D+ + +R+ + K L V+
Sbjct: 61 IREEVEKIN-------------ALIKDQQLVFIRQTSNDGK------------LFGSVTN 95
Query: 163 NALVAEVARQLCVNITAENLHLPTPLSTIGEYEVPLRL 200
+ +++ + NI+ N+ L + + G Y V +RL
Sbjct: 96 KEIADKLSENISYNISHSNIILDKQIKSTGIYTVEIRL 133
>ref|YP_246157.1| 50S ribosomal protein L9 [Rickettsia felis URRWXCal2]
gi|67004066|gb|AAY60992.1| 50S ribosomal protein L9
[Rickettsia felis URRWXCal2]
Length = 171
Score = 52.4 bits (124), Expect = 9e-06
Identities = 40/158 (25%), Positives = 74/158 (46%), Gaps = 25/158 (15%)
Query: 43 LQVILTTNIDKLGKAGETVKVAPGFFRNHLMPKLLAVPNIDKFAYLLTEQRKIYQPTEEV 102
+++IL + KLGK G+ +KVA GF RN+L+P+ LA+ + L+ +Q+ ++ ++
Sbjct: 1 MEIILIKPVRKLGKIGDMLKVADGFGRNYLLPQKLAIRATEPNKELIVKQKHEFEAKDKQ 60
Query: 103 KQEDVVVVTESKEDLMKEYERAALILDKAKLVLRRLIDVKKAKARESKDEPLELQIPVSK 162
+E+V + ALI D+ + +R+ D K L V+
Sbjct: 61 IREEVEKIN-------------ALIKDQKLVFIRQTSDDGK------------LFGSVTN 95
Query: 163 NALVAEVARQLCVNITAENLHLPTPLSTIGEYEVPLRL 200
+ +++ + NI N+ L + G Y V +RL
Sbjct: 96 KEIADKLSENVSYNIAHSNIILDKQIKFTGIYTVEIRL 133
>ref|ZP_00199878.1| COG0359: Ribosomal protein L9 [Rubrobacter xylanophilus DSM 9941]
Length = 149
Score = 52.0 bits (123), Expect = 1e-05
Identities = 40/158 (25%), Positives = 80/158 (50%), Gaps = 25/158 (15%)
Query: 43 LQVILTTNIDKLGKAGETVKVAPGFFRNHLMPKLLAVPNIDKFAYLLTEQRKIYQPTEEV 102
+QVILT +++K+G+ G+ V V+ G+ RN+L+P+ LA + A L +R++ + E
Sbjct: 1 MQVILTQDVEKVGRRGDIVDVSRGYVRNYLVPRGLA--EVATPAKLEEARRRMEEAAERE 58
Query: 103 KQEDVVVVTESKEDLMKEYERAALILDKAKLVLRRLIDVKKAKARESKDEPLELQIPVSK 162
++ L + E A L+K+ + + +AR +DE L + +
Sbjct: 59 RR------------LAERAEEIAETLNKSVITI---------EARTGEDERLFGSVTAAN 97
Query: 163 NALVAEVARQLCVNITAENLHLPTPLSTIGEYEVPLRL 200
A E AR +++ + L P+ ++G ++VP+++
Sbjct: 98 IAEAIEKARG--IHLDRRKIRLEEPIRSLGTHQVPVQV 133
>ref|ZP_00153133.1| COG0359: Ribosomal protein L9 [Rickettsia rickettsii]
Length = 171
Score = 52.0 bits (123), Expect = 1e-05
Identities = 39/158 (24%), Positives = 76/158 (47%), Gaps = 25/158 (15%)
Query: 43 LQVILTTNIDKLGKAGETVKVAPGFFRNHLMPKLLAVPNIDKFAYLLTEQRKIYQPTEEV 102
+++IL + KLGK G+ +KVA GF RN+L+P+ LA+ + L+ +Q+ ++ ++
Sbjct: 1 MEIILIKPVRKLGKIGDILKVADGFGRNYLLPQKLAIRATEPNKELIVKQKHEFEAKDKQ 60
Query: 103 KQEDVVVVTESKEDLMKEYERAALILDKAKLVLRRLIDVKKAKARESKDEPLELQIPVSK 162
+E+V + ALI D+ + +R+ + K L V+
Sbjct: 61 IREEVEKIN-------------ALIKDQQLVFIRQTSNDGK------------LFGSVTN 95
Query: 163 NALVAEVARQLCVNITAENLHLPTPLSTIGEYEVPLRL 200
+ +++ + NI+ N+ L + + G Y V +RL
Sbjct: 96 KEIANKLSENISYNISHSNVILDKQIKSTGIYTVEIRL 133
>ref|ZP_00290952.1| COG0359: Ribosomal protein L9 [Magnetococcus sp. MC-1]
Length = 149
Score = 50.4 bits (119), Expect = 4e-05
Identities = 39/158 (24%), Positives = 72/158 (44%), Gaps = 26/158 (16%)
Query: 43 LQVILTTNIDKLGKAGETVKVAPGFFRNHLMPKLLAVPNIDKFAYLLTEQRKIYQPTEEV 102
++VIL I K+G GE VKV G+ RN L+P+ A+P + + QR Y+ +
Sbjct: 1 MEVILLEKIGKIGNLGEVVKVRSGYGRNFLIPQGKALPATSENKSVFEAQRADYEARQ-- 58
Query: 103 KQEDVVVVTESKEDLMKEYERAALILDKAKLVLRRLIDVKKAKARESKDEPLELQIPVSK 162
+++ + E A +D+ +V++R ++
Sbjct: 59 ------------AEILADAETLAAKVDEVSVVMKRPAGA------------MDKLFGSVT 94
Query: 163 NALVAEVARQLCVNITAENLHLPTPLSTIGEYEVPLRL 200
+A +A + L +NI + + TP+ T+GE++V +RL
Sbjct: 95 SADIAVFYKDLGLNIPRNIIDVLTPIRTLGEHQVRVRL 132
>ref|NP_951725.1| ribosomal protein L9 [Geobacter sulfurreducens PCA]
gi|39982538|gb|AAR33998.1| ribosomal protein L9
[Geobacter sulfurreducens PCA]
Length = 148
Score = 50.4 bits (119), Expect = 4e-05
Identities = 39/158 (24%), Positives = 80/158 (49%), Gaps = 26/158 (16%)
Query: 43 LQVILTTNIDKLGKAGETVKVAPGFFRNHLMPKLLAVPNIDKFAYLLTEQRKIYQPTEEV 102
++VIL N++ LG G+ VKVAPG+ RN+L+P+ A+ +K A L ++
Sbjct: 1 MKVILKENVENLGHIGDIVKVAPGYARNYLIPRNFAIEATEKNAKALEHAKR-------- 52
Query: 103 KQEDVVVVTESKEDLMKEYERAALILDKAKLVLRRLIDVKKAKARESKDEPLELQIPVSK 162
EY+R +L++A+L++ ++ + + + ++ +E +L V+
Sbjct: 53 ---------------QLEYKRNK-VLEQARLLVAKIEGLSLSISHQAGEEG-KLFGSVT- 94
Query: 163 NALVAEVARQLCVNITAENLHLPTPLSTIGEYEVPLRL 200
N +AE+ + V I + + L P+ +GE+ +++
Sbjct: 95 NMELAELLKAQGVEIDRKKIVLAEPIKHVGEFTAVVKV 132
>ref|YP_067057.1| 50S ribosomal protein L9 [Rickettsia typhi str. Wilmington]
gi|51459612|gb|AAU03575.1| 50S ribosomal protein L9
[Rickettsia typhi str. Wilmington]
Length = 171
Score = 50.1 bits (118), Expect = 5e-05
Identities = 38/158 (24%), Positives = 74/158 (46%), Gaps = 25/158 (15%)
Query: 43 LQVILTTNIDKLGKAGETVKVAPGFFRNHLMPKLLAVPNIDKFAYLLTEQRKIYQPTEEV 102
+++IL + KLGK G+ +KVA GF RN+L+P+ LA+ L+ +Q+ + E
Sbjct: 1 MEIILIKPVRKLGKIGDILKVANGFGRNYLLPQKLAIRATKSNKELIVKQKHEF----EA 56
Query: 103 KQEDVVVVTESKEDLMKEYERAALILDKAKLVLRRLIDVKKAKARESKDEPLELQIPVSK 162
K + + E L+K+ + + R++ D+ +L V+
Sbjct: 57 KDKQIRAEVEKINTLIKDQQLVFI--------------------RQTSDD-CKLFGSVTN 95
Query: 163 NALVAEVARQLCVNITAENLHLPTPLSTIGEYEVPLRL 200
+ ++++ + NI+ N+ L + + G Y V +RL
Sbjct: 96 KDIADKLSKNISYNISHSNVILDKQIKSTGIYTVEIRL 133
>ref|ZP_00110947.1| COG0359: Ribosomal protein L9 [Nostoc punctiforme PCC 73102]
Length = 152
Score = 49.3 bits (116), Expect = 8e-05
Identities = 32/82 (39%), Positives = 49/82 (59%), Gaps = 9/82 (10%)
Query: 41 RKLQVILTTNIDKLGKAGETVKVAPGFFRNHLMPKLL---AVPNIDKFAYLLTEQRKIYQ 97
+++Q++L +I KLGK+G+ V+VAPG+ RN+L+P+ L A P I K EQ + Q
Sbjct: 3 KRVQLVLNQDISKLGKSGDLVEVAPGYARNYLIPQKLATNATPGILKQVERRREQER--Q 60
Query: 98 PTEEVKQEDVVVVTESKEDLMK 119
E++Q+ E KE L K
Sbjct: 61 RQLELRQQ----ALEQKESLEK 78
>ref|ZP_00324483.1| COG0359: Ribosomal protein L9 [Trichodesmium erythraeum IMS101]
Length = 152
Score = 48.9 bits (115), Expect = 1e-04
Identities = 28/67 (41%), Positives = 42/67 (61%), Gaps = 5/67 (7%)
Query: 41 RKLQVILTTNIDKLGKAGETVKVAPGFFRNHLMPKLLA---VPNIDKFAYLLTEQRKIYQ 97
+KLQV+L +++KLGK+G+ V VAPG+ RN+L+P +A P + K EQ + Q
Sbjct: 3 KKLQVVLNKDVNKLGKSGDVVDVAPGYARNYLIPNKIAFRTTPGVLKQVERRQEQER--Q 60
Query: 98 PTEEVKQ 104
E+KQ
Sbjct: 61 RQIELKQ 67
>sp|Q8YZA0|RL9_ANASP 50S ribosomal protein L9 gi|17129924|dbj|BAB72537.1| 50S ribosomal
protein L9 [Nostoc sp. PCC 7120]
gi|17228075|ref|NP_484623.1| 50S ribosomal protein L9
[Nostoc sp. PCC 7120]
Length = 152
Score = 48.9 bits (115), Expect = 1e-04
Identities = 31/82 (37%), Positives = 49/82 (58%), Gaps = 9/82 (10%)
Query: 41 RKLQVILTTNIDKLGKAGETVKVAPGFFRNHLMPKLL---AVPNIDKFAYLLTEQRKIYQ 97
+++Q++LT ++ KLG++G+ V VAPG+ RN+L+P+ L A P I K EQ + Q
Sbjct: 3 KRVQLVLTKDVSKLGRSGDLVDVAPGYARNYLIPQSLATHATPGILKQVERRREQER--Q 60
Query: 98 PTEEVKQEDVVVVTESKEDLMK 119
E++Q+ E KE L K
Sbjct: 61 RQLELRQQ----ALEQKESLEK 78
>ref|XP_468458.1| putative 50S ribosomal protein L9, chloroplast [Oryza sativa
(japonica cultivar-group)] gi|51979514|ref|XP_507552.1|
PREDICTED OJ1119_A01.21-1 gene product [Oryza sativa
(japonica cultivar-group)] gi|51964554|ref|XP_507062.1|
PREDICTED OJ1119_A01.21-1 gene product [Oryza sativa
(japonica cultivar-group)] gi|48716281|dbj|BAD22896.1|
putative 50S ribosomal protein L9, chloroplast [Oryza
sativa (japonica cultivar-group)]
Length = 187
Score = 48.5 bits (114), Expect = 1e-04
Identities = 31/79 (39%), Positives = 47/79 (59%), Gaps = 6/79 (7%)
Query: 30 LMYASQGLRYNRKLQVILTTNIDKLGKAGETVKVAPGFFRNHLMPK---LLAVPNIDKFA 86
L+ +QG + + QVILT +I ++GK G+T+KV GF+RN L+PK L P + K
Sbjct: 28 LVVVAQG-KVKKYRQVILTDDIAEVGKKGDTLKVRAGFYRNFLLPKGKATLLTPEVLK-- 84
Query: 87 YLLTEQRKIYQPTEEVKQE 105
+ EQ +I + VK+E
Sbjct: 85 EMQVEQERIDAEKKRVKEE 103
>ref|ZP_00159693.1| COG0359: Ribosomal protein L9 [Anabaena variabilis ATCC 29413]
Length = 152
Score = 48.1 bits (113), Expect = 2e-04
Identities = 31/82 (37%), Positives = 49/82 (58%), Gaps = 9/82 (10%)
Query: 41 RKLQVILTTNIDKLGKAGETVKVAPGFFRNHLMPKLL---AVPNIDKFAYLLTEQRKIYQ 97
+++Q++LT ++ KLG++G+ V VAPG+ RN+L+P+ L A P I K EQ + Q
Sbjct: 3 KRVQLVLTKDVSKLGRSGDLVDVAPGYARNYLIPQSLATHATPGILKQVERRREQER--Q 60
Query: 98 PTEEVKQEDVVVVTESKEDLMK 119
E++Q+ E KE L K
Sbjct: 61 RQLELRQQ----ALEQKEALEK 78
>gb|AAC08220.1| 50S ribosomal protein L9 [Porphyra purpurea]
gi|11465800|ref|NP_053944.1| ribosomal protein L9
[Porphyra purpurea] gi|1710478|sp|P51334|RK9_PORPU
Chloroplast 50S ribosomal protein L9
gi|2148020|pir||S73255 ribosomal protein L9, chloroplast
- red alga (Porphyra purpurea) chloroplast
Length = 155
Score = 47.8 bits (112), Expect = 2e-04
Identities = 38/159 (23%), Positives = 76/159 (46%), Gaps = 27/159 (16%)
Query: 43 LQVILTTNIDKLGKAGETVKVAPGFFRNHLMP-KLLAVPNIDKFAYLLTEQRKIYQPTEE 101
+QV+L NI KLGK+ + VKVA G+ RN L+P K+ +V + + Q+K+Y
Sbjct: 6 IQVVLKENIQKLGKSNDVVKVATGYARNFLIPNKMASVATVG-----MLNQQKLY---AA 57
Query: 102 VKQEDVVVVTESKEDLMKEYERAALILDKAKLVLRRLIDVKKAKARESKDEPLELQIPVS 161
+K++ ++ E+ A+ + L +++K + + + V+
Sbjct: 58 IKEKKIIFAKEN-----------------ARKTQQLLEEIQKFSISKKTGDGETIFGSVT 100
Query: 162 KNALVAEVARQLCVNITAENLHLPTPLSTIGEYEVPLRL 200
+ + + V+I +N+ +P + TIG Y + ++L
Sbjct: 101 EKEISQIIKNTTNVDIDKQNILIP-EIKTIGLYNIEIKL 138
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.318 0.137 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 351,535,868
Number of Sequences: 2540612
Number of extensions: 14077585
Number of successful extensions: 42613
Number of sequences better than 10.0: 256
Number of HSP's better than 10.0 without gapping: 186
Number of HSP's successfully gapped in prelim test: 70
Number of HSP's that attempted gapping in prelim test: 42378
Number of HSP's gapped (non-prelim): 277
length of query: 222
length of database: 863,360,394
effective HSP length: 123
effective length of query: 99
effective length of database: 550,865,118
effective search space: 54535646682
effective search space used: 54535646682
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 73 (32.7 bits)
Medicago: description of AC136286.9


