

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135959.8 + phase: 0 /pseudo/partial
(1454 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
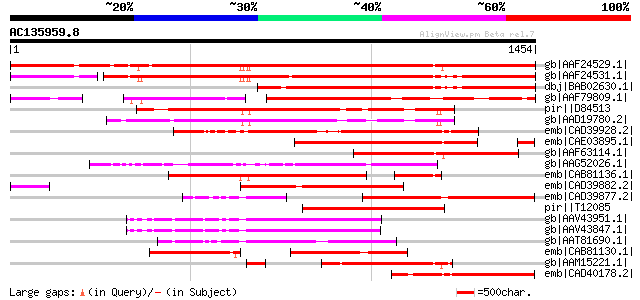
gb|AAF24529.1| F7F22.15 [Arabidopsis thaliana] 1486 0.0
gb|AAF24531.1| F7F22.17 [Arabidopsis thaliana] 1300 0.0
dbj|BAB02630.1| retroelement pol polyprotein-like [Arabidopsis t... 1024 0.0
gb|AAF79809.1| T32E20.9 [Arabidopsis thaliana] 910 0.0
pir||D84513 probable retroelement pol polyprotein [imported] - A... 853 0.0
gb|AAD19780.2| hypothetical protein [Arabidopsis thaliana] 814 0.0
emb|CAD39928.2| OSJNBa0091C12.6 [Oryza sativa (japonica cultivar... 735 0.0
emb|CAE03895.1| OSJNBb0026I12.3 [Oryza sativa (japonica cultivar... 692 0.0
gb|AAF63114.1| Hypothetical protein [Arabidopsis thaliana] gi|25... 650 0.0
gb|AAG52026.1| polyprotein, putative; 77260-80472 [Arabidopsis t... 646 0.0
emb|CAB81136.1| putative athila transposon protein [Arabidopsis ... 630 e-178
emb|CAD39882.2| OSJNBb0067G11.5 [Oryza sativa (japonica cultivar... 596 e-168
emb|CAD39877.2| OSJNBb0058J09.16 [Oryza sativa (japonica cultiva... 588 e-166
pir||T12085 reverse transcriptase homolog - fava bean (fragment)... 583 e-164
gb|AAV43951.1| putative polyprotein [Oryza sativa (japonica cult... 547 e-153
gb|AAV43847.1| putative polyprotein [Oryza sativa (japonica cult... 542 e-152
gb|AAT81690.1| putative reverse transcriptase [Oryza sativa (jap... 533 e-149
emb|CAB81130.1| AT4g07600 [Arabidopsis thaliana] gi|5724765|gb|A... 474 e-132
gb|AAM15221.1| putative retroelement pol polyprotein [Arabidopsi... 466 e-129
emb|CAD40178.2| OSJNBa0061A09.17 [Oryza sativa (japonica cultiva... 439 e-121
>gb|AAF24529.1| F7F22.15 [Arabidopsis thaliana]
Length = 1862
Score = 1486 bits (3848), Expect = 0.0
Identities = 789/1548 (50%), Positives = 1035/1548 (65%), Gaps = 124/1548 (8%)
Query: 1 NQFSGSPSEDPNLHISNFWRLSVTCKDD---QETVRLHLFPFSLKNKASNWFNSLKPGSI 57
N+F G P EDP H+ F RL K + ++ +L LFPFSL +KA W +L SI
Sbjct: 170 NKFHGLPMEDPLDHLDEFDRLCNLTKINGVSEDGFKLRLFPFSLGDKAHIWEKNLPHDSI 229
Query: 58 TSWDQLRREFLCRFFPPSKTAQLRGKLYQFTQMNGESLFDVWERFKEILRRCPHHGLEKW 117
T+WD ++ FL +FF ++TA+LR ++ F+Q GES + WERFK +C HHG K
Sbjct: 230 TTWDDCKKAFLSKFFSNARTARLRNEISGFSQKTGESFCEAWERFKGYTNQCSHHGFTKA 289
Query: 118 LIIHIFYNGLTSSTKLTIDAAAGGALMNKDFTTAYALIENMALNLFQWTEEKETVDPSPS 177
++ Y G+ ++ +D A+ G NKD + L+EN+A + + E + +
Sbjct: 290 SLLSTLYRGVLPRIRMLLDTASNGNFQNKDVEEGWELVENLAQSNGNYNENCDRTVRGTA 349
Query: 178 EKEAGINETSSIDYLSTKLNALSQRLDRMSTPTFSPPCETCGLSSHSDTDCNLTSVEQLN 237
+ + D ++ AL+ +LDR+ + D +E+++
Sbjct: 350 DSD---------DKHRKEIKALNDKLDRILLSQHKHVHFLVDDEQYEVQDGEGNQLEEVS 400
Query: 238 FVQNGQRVVQK-SHIELLMKNLFFKQSE----QLQELKDQTRLLNNSLATLTTKIDSIYS 292
++ N Q + ++ + NL ++ + Q Q Q + N
Sbjct: 401 YINNNQGGYKGYNNFKTNNPNLSYRSTNVANPQDQVYPPQQQQSQNK---------PFVP 451
Query: 293 HNKISETQLSQVVRKAGHPNK--MNAVTLRSGKPL---EDPIRRTETDHSEKEIREPPS- 346
+N+ ++ Q SQ+ KA K +A+TL SGK L E+P +T T+ SE + E S
Sbjct: 452 YNQATK-QTSQLPGKAVQNPKEYAHAITLHSGKALPTREEP--KTVTEDSEDQDGEDLSL 508
Query: 347 RETRAEKE-----------------EPRVEGNTTPPFKPKIPFPQRFAKSKLDEQFKKFM 389
++ +A+K +P ++ T P +P IP A + + K+
Sbjct: 509 KKDQADKPLDLSLEQPLDLSLQQSLDPPLDSFTRPTTRPVIPAASPTAPKPVAVKNKE-- 566
Query: 390 EMMNKIYIDVPFTEVLTQMPTYAKFLKEILSKKRKIEESETVNLTEECSAIIQNKL-PPK 448
K+ + +P + L +P KFLK+++ ++ + E V L+ ECSAIIQ K+ P K
Sbjct: 567 ----KVELRIPLVDALALIPDSHKFLKDLIVERIQ-EVQGMVVLSHECSAIIQKKIIPKK 621
Query: 449 LKDPGSFSIPCVIGSEVVKKAMCDLGASVSLMPLSLYERLGIGELKSTRMTLQLADRSVK 508
L DPGSF++PC +G + +CDLGASVSLMPLS+ +RLG + KS ++L LADRSV+
Sbjct: 622 LSDPGSFTLPCSLGPLAFNRCLCDLGASVSLMPLSVAKRLGFTQYKSCNISLILADRSVR 681
Query: 509 YPAGIIEDVPVKVGEVYIPADFVVMEMEEDNQVPILLGRPFLATAGAIIDVKKGKLAFNV 568
P G++E++P+++G V IP DFVV+EM+E+ + P++LGRPFLATAGA+IDVKKGK+ N+
Sbjct: 682 IPHGLLENLPIRIGAVEIPTDFVVLEMDEEPKDPLILGRPFLATAGAMIDVKKGKIDLNL 741
Query: 569 GKE-TVEFELAKLMKGPSIKDSCCMIDIIDHCVKECSLASTTHDGLEVCLVNNAGTK-LE 626
GK+ + F++ MK P+I+ I+ +D E D L L + L
Sbjct: 742 GKDFRMTFDVKDAMKKPTIEGQLFWIEEMDQLADELLEELAEEDHLNSALTKSGEDGFLH 801
Query: 627 GEAKAYEVLLD------RTPPIKDL----------SVEELIKEEPTL------------- 657
E Y+ LLD + P ++L S E + +P L
Sbjct: 802 LETLGYQKLLDSHKAMEESEPFEELNGPATEVMVMSEEGSTRVQPALSRTYSSNHSTLST 861
Query: 658 -QPKE-------------APKVELKTLPSNLRYEFLGPNSTYPVIVNASLDEVETEKLLY 703
+P+E APKV+LK LP LRY FLGPNSTYPVI+NA L+ E LL
Sbjct: 862 DEPREPIISTSDDWSELKAPKVDLKPLPKGLRYAFLGPNSTYPVIINAELNSDEVNLLLS 921
Query: 704 VLKKYPKAIGYTIDDIKGINPSLCMHRILLEDDYKPSIEHQRRLNPNMKEVVKKEVLKLL 763
L+KY +AIGY++ DIKGI+PSLC HRI LE++ SIE RRLNPN+KEVVKKE+LKLL
Sbjct: 922 ELRKYRRAIGYSLSDIKGISPSLCNHRIHLENESYSSIEPHRRLNPNLKEVVKKEILKLL 981
Query: 764 DAGVIYPISDSKWVSPVQVVPKKGGLTVIKNDKNESIATRTVTGWRMCIDYRKLNKATRK 823
DAGVIYPISDS WVSPV VPKK G+ V+KN+K+E I TRT+TG RMCIDYRKLN A+RK
Sbjct: 982 DAGVIYPISDSTWVSPVHCVPKKDGMIVVKNEKDELIPTRTITGHRMCIDYRKLNAASRK 1041
Query: 824 DHFPLPFIDQMLERLAKHSHFCYLDGYSGFFQIPIHPNDQEKTTFTCPFGTFAYRRMPFG 883
DHFPLPFIDQMLERLA H ++C+LDGYSGFFQIPIHPNDQEKTTFTCP+GTFAY+RMPFG
Sbjct: 1042 DHFPLPFIDQMLERLANHPYYCFLDGYSGFFQIPIHPNDQEKTTFTCPYGTFAYKRMPFG 1101
Query: 884 LCNAPATFQRCMMSIFSDFVEKIMEVFMDDFSVHGSNFDDCLTNLEKVLERCEQVNLVLN 943
LCNAPATFQRCM SIFSD +++++EVFMDDFSV+G +F CL NL +VL RCE+ NLVLN
Sbjct: 1102 LCNAPATFQRCMTSIFSDLIKEMVEVFMDDFSVYGPSFSSCLLNLGRVLTRCEETNLVLN 1161
Query: 944 WEKCHFMVREGIVLGHLVSDRGIEVDRAKVEIIEKMLPPTSVKEIRSFLGHAGFYRRFIK 1003
WEKCHFMV+EGIVLGH +S++GIEVD+ KVE++ ++ PP +VK+IRSFLGHAGFYRRFIK
Sbjct: 1162 WEKCHFMVKEGIVLGHKISEKGIEVDKGKVEVMMQLQPPKTVKDIRSFLGHAGFYRRFIK 1221
Query: 1004 DFSSITKPLTNLLLKDADFTFDDSCLQAFCRLKEALITAPIIQPPDWNLPFEIMCDASDY 1063
DFS I +PLT LL K+ +F FDD CL++F +K+AL++AP+++ P+W+ PFEIMCDASDY
Sbjct: 1222 DFSKIARPLTRLLCKETEFKFDDDCLKSFQTIKDALVSAPVVRAPNWDYPFEIMCDASDY 1281
Query: 1064 AVGVVLGQRKDKKMHAIYYASKTLDGAQVNYATTEKELLAVVYAIDKFRQYLVGSKIIVY 1123
AVG VLGQ+ DKK+H IYYAS+TLD AQ YATTEKELLAVV+A +KFR YLVGSK+ VY
Sbjct: 1282 AVGAVLGQKIDKKLHVIYYASRTLDDAQGRYATTEKELLAVVFAFEKFRSYLVGSKVTVY 1341
Query: 1124 TDHSAIKYLLNKKDAKPRLIRWILLLQEFDLEIKDKKGVENVVADHLSRLREANKDELPL 1183
TDH+A+++L KKD KPRL+RWILLLQEFD+EI DKKG+EN ADHLSR+R ++ L +
Sbjct: 1342 TDHAALRHLYAKKDTKPRLLRWILLLQEFDMEIVDKKGIENGAADHLSRMR--IEEPLLI 1399
Query: 1184 DDSFPDDQLFLL-----------------AQTDAPWYADFVNFLAAGVLPPELNYQQKKK 1226
DDS P++QL ++ + ++PWYAD VN+LA GV PP L ++KK
Sbjct: 1400 DDSMPEEQLMVVEFFGKSYSGKEFHQLNAVEGESPWYADHVNYLACGVEPPNLTSYERKK 1459
Query: 1227 FFNDLKHYYWDEPYLFRRGSDGIFRRCIPESEVSSILTHCHSSSYGGHASTQKTSFKILH 1286
FF D+ HYYWDEPYL+ D I+RRC+ E EV IL HCH S+YGGH +T KT KIL
Sbjct: 1460 FFRDIHHYYWDEPYLYTLCKDKIYRRCVSEDEVEGILLHCHGSAYGGHFATFKTVSKILQ 1519
Query: 1287 SGFWWPSLFKDVHLFISKCDKCQRTGSITKRNEMPLNNILEVEIFDVWGVDFMGPFPSSF 1346
+GFWWP++FKD F+SKCD CQR G+I++RNEMP N ILEVEIFDVWG+DFMGPFPSS+
Sbjct: 1520 AGFWWPTMFKDAQEFVSKCDSCQRKGNISRRNEMPQNPILEVEIFDVWGIDFMGPFPSSY 1579
Query: 1347 GNQYILVAVDYVSKWVEAIASPTNDAQVVIKMFKKVIFPRFGVPRVVISDGGSHFISKHF 1406
GN+YILVAVDYVSKWVEAIASPTNDA+VV+K+FK +IFPRFGVPRVVISDGG HFI+K F
Sbjct: 1580 GNKYILVAVDYVSKWVEAIASPTNDAKVVLKLFKTIIFPRFGVPRVVISDGGKHFINKVF 1639
Query: 1407 EKLLQKLGVRHKVATPYHPQTSGQVEVSNRQIKAILEKTVSTSRTDWS 1454
E LL+K GV+HKVATPY+PQTSGQVE+SNR+IK ILEKTV +R DWS
Sbjct: 1640 ENLLKKHGVKHKVATPYNPQTSGQVEISNREIKTILEKTVGITRKDWS 1687
>gb|AAF24531.1| F7F22.17 [Arabidopsis thaliana]
Length = 1799
Score = 1300 bits (3363), Expect = 0.0
Identities = 696/1294 (53%), Positives = 885/1294 (67%), Gaps = 143/1294 (11%)
Query: 261 KQSEQLQELKDQTRLLNNSLATLTTKIDSIYSHNKISET--QLSQVVRKAGHPNK--MNA 316
K SE +L LN + TL TK+ + H+ S Q SQ+ KA K +A
Sbjct: 374 KISELHNKLDCSYNDLNVKMETLDTKVRYLEGHSTSSSATKQTSQLPGKAVQNPKEYAHA 433
Query: 317 VTLRSGKPL---EDPIRRTETDHSEKEIREPPSRET-RAEKE-----------------E 355
+TLRSGK L E+P +T T+ SE + E S E +A+K +
Sbjct: 434 ITLRSGKALPTREEP--KTVTEDSEDQDGEDLSLEKDQADKPLDLSLEQPLDLSLQQSLD 491
Query: 356 PRVEGNTTP-----------------------------PFKPKIPFPQRFAKSKLDEQFK 386
P ++ T P P+K ++PFP R K+ D+
Sbjct: 492 PPLDSFTRPTTRPVIPAASPTAPKPVAVKNKEKVFVPPPYKSQLPFPGRHKKALADKYRA 551
Query: 387 KFMEMMNKIYIDVPFTEVLTQMPTYAKFLKEILSKKRKIEESETVNLTEECSAIIQNKL- 445
F + + ++ + +P + L +P KFLK+++ ++ + E V L+ ECSAIIQ K+
Sbjct: 552 MFAKNIKEVELRIPLVDALALIPDSHKFLKDLIVERIQ-EVQGMVVLSHECSAIIQKKII 610
Query: 446 PPKLKDPGSFSIPCVIGSEVVKKAMCDLGASVSLMPLSLYERLGIGELKSTRMTLQLADR 505
P KL DPGSF++PC +G + +CDLGASVSLMPLS+ +RLG + KS ++L LADR
Sbjct: 611 PKKLSDPGSFTLPCSLGPLAFNRCLCDLGASVSLMPLSVAKRLGFTQYKSCNISLILADR 670
Query: 506 SVKYPAGIIEDVPVKVGEVYIPADFVVMEMEEDNQVPILLGRPFLATAGAIIDVKKGKLA 565
SV+ P G++E++P+++G V IP DFVV+EM+E+ + P++LGRPFLATAGA+IDVKKGK+
Sbjct: 671 SVRIPHGLLENLPIRIGAVEIPTDFVVLEMDEEPKDPLILGRPFLATAGAMIDVKKGKID 730
Query: 566 FNVGKE-TVEFELAKLMKGPSIKDSCCMIDIIDHCVKECSLASTTHDGLEVCLVNNAGTK 624
N+GK+ + F++ MK P+I+ I+ +D E D L L +
Sbjct: 731 LNLGKDFRMTFDVKDAMKKPTIEGQLFWIEEMDQLADELLEELAEEDHLNSALTKSGEDG 790
Query: 625 -LEGEAKAYEVLLD------RTPPIKDL----------SVEELIKEEPTL---------- 657
L E Y+ LLD + P ++L S E + +P L
Sbjct: 791 FLHLETLGYQKLLDSHKAMEESEPFEELNGPATEVMVMSEEGSTRVQPALSRTYSSNHST 850
Query: 658 ----QPKE-------------APKVELKTLPSNLRYEFLGPNSTYPVIVNASLDEVETEK 700
+P+E APKV+LK LP LRY FLGPNSTYPVI+NA L+ E
Sbjct: 851 LSTDEPREPIIPTSDDWSELKAPKVDLKPLPKGLRYAFLGPNSTYPVIINAELNSDEVNL 910
Query: 701 LLYVLKKYPKAIGYTIDDIKGINPSLCMHRILLEDDYKPSIEHQRRLNPNMKEVVKKEVL 760
LL LKKY +AIGY++ DIKGI+PSLC HRI LE++ SIE QRRLNPN+KEVVKKE+L
Sbjct: 911 LLSELKKYRRAIGYSLSDIKGISPSLCNHRIHLENESYSSIEPQRRLNPNLKEVVKKEIL 970
Query: 761 KLLDAGVIYPISDSKWVSPVQVVPKKGGLTVIKNDKNESIATRTVTGWRMCIDYRKLNKA 820
KLLDAGVIYPISD + VPKK G+TV+KN+K+E I TRT+TG RMCIDYRKLN A
Sbjct: 971 KLLDAGVIYPISD------MHCVPKKDGMTVVKNEKDELIPTRTITGHRMCIDYRKLNAA 1024
Query: 821 TRKDHFPLPFIDQMLERLAKHSHFCYLDGYSGFFQIPIHPNDQEKTTFTCPFGTFAYRRM 880
+RKDHFPLPFIDQMLERLA H ++C+LDGY+GFFQIPIHPNDQEKTTFTCP+GTFAY+RM
Sbjct: 1025 SRKDHFPLPFIDQMLERLANHPYYCFLDGYNGFFQIPIHPNDQEKTTFTCPYGTFAYKRM 1084
Query: 881 PFGLCNAPATFQRCMMSIFSDFVEKIMEVFMDDFSVHGSNFDDCLTNLEKVLERCEQVNL 940
PFGLCNAPATFQRCM SIFSD +E+++EVFMDDFSV+G +F CL NL +VL RCE+ NL
Sbjct: 1085 PFGLCNAPATFQRCMTSIFSDLIEEMVEVFMDDFSVYGPSFSSCLLNLGRVLTRCEETNL 1144
Query: 941 VLNWEKCHFMVREGIVLGHLVSDRGIEVDRAKVEIIEKMLPPTSVKEIRSFLGHAGFYRR 1000
VLNWEKCHFMV+EGIVLGH +S++GIEVD+ KVE++ ++ PP +VK+IRSFLGHAGFYRR
Sbjct: 1145 VLNWEKCHFMVKEGIVLGHKISEKGIEVDKGKVEVMMQLQPPKTVKDIRSFLGHAGFYRR 1204
Query: 1001 FIKDFSSITKPLTNLLLKDADFTFDDSCLQAFCRLKEALITAPIIQPPDWNLPFEIMCDA 1060
FIKDFS I +PLT LL K+ +F FDD CL++F +K+AL++AP+++ P+W+ PFEIMCDA
Sbjct: 1205 FIKDFSKIARPLTRLLCKETEFKFDDDCLKSFQTIKDALVSAPVVRAPNWDYPFEIMCDA 1264
Query: 1061 SDYAVGVVLGQRKDKKMHAIYYASKTLDGAQVNYATTEKELLAVVYAIDKFRQYLVGSKI 1120
SDYAVG VLGQ+ DKK+H IYYAS+TLD AQ YATTEKELL VV+A +KFR YLVGSK+
Sbjct: 1265 SDYAVGAVLGQKIDKKLHVIYYASRTLDDAQGRYATTEKELLVVVFAFEKFRSYLVGSKV 1324
Query: 1121 IVYTDHSAIKYLLNKKDAKPRLIRWILLLQEFDLEIKDKKGVENVVADHLSRLREANKDE 1180
VYTDH+A+++L KKD KPRL+RWILLLQEFD+EI DKKG+EN ADHLSR+R ++
Sbjct: 1325 TVYTDHAALRHLYAKKDTKPRLLRWILLLQEFDMEIVDKKGIENGAADHLSRMR--IEEP 1382
Query: 1181 LPLDDSFPDDQLFLLAQTDAPWYADFVNFLAAGVLPPELNYQQKKKFFNDLKHYYWDEPY 1240
L +DDS P++QL ++ +F +G K F+ L + P+
Sbjct: 1383 LLIDDSMPEEQLMVV---------EFFGKSYSG------------KEFHQLNAVEGESPW 1421
Query: 1241 LFRRGSDGIFRRCIPESEVSSILTHCHSSSYGGHASTQKTSFKILHSGFWWPSLFKDVHL 1300
RC+ E EV IL HCH S+YGGH +T KT KIL +GFWWP++FKD
Sbjct: 1422 -----------RCVSEDEVEGILLHCHGSAYGGHFATFKTVSKILQAGFWWPTMFKDAQE 1470
Query: 1301 FISKCDKCQRTGSITKRNEMPLNNILEVEIFDVWGVDFMGPFPSSFGNQYILVAVDYVSK 1360
F+SKCD CQR G+I++RNEMP N ILEVEIFDVWG+DFMGPFPSS+GN+YILVAVDYVSK
Sbjct: 1471 FVSKCDSCQRKGNISRRNEMPQNPILEVEIFDVWGIDFMGPFPSSYGNKYILVAVDYVSK 1530
Query: 1361 WVEAIASPTNDAQVVIKMFKKVIFPRFGVPRVVISDGGSHFISKHFEKLLQKLGVRHKVA 1420
WVEAIASPTNDA+VV+K+FK +IFPRFGVPRVVISDGG HFI+K FE LL+K GV+HKVA
Sbjct: 1531 WVEAIASPTNDAKVVLKLFKTIIFPRFGVPRVVISDGGKHFINKVFENLLKKHGVKHKVA 1590
Query: 1421 TPYHPQTSGQVEVSNRQIKAILEKTVSTSRTDWS 1454
TPY+PQTSGQVE+SNR+IK ILEKTV +R DWS
Sbjct: 1591 TPYNPQTSGQVEISNREIKTILEKTVGITRKDWS 1624
Score = 132 bits (332), Expect = 9e-29
Identities = 75/246 (30%), Positives = 124/246 (49%), Gaps = 12/246 (4%)
Query: 1 NQFSGSPSEDPNLHISNFWRLSVTCKDD---QETVRLHLFPFSLKNKASNWFNSLKPGSI 57
N+F G P EDP H+ F RL K + ++ +L LFPFSL +KA W +L SI
Sbjct: 35 NKFHGLPMEDPLDHLDEFDRLCNLTKINGVSEDGFKLRLFPFSLGDKAHIWEKNLPHDSI 94
Query: 58 TSWDQLRREFLCRFFPPSKTAQLRGKLYQFTQMNGESLFDVWERFKEILRRCPHHGLEKW 117
T+WD ++ FL +FF ++TA+LR ++ F+Q GES + WERFK +CPHHG K
Sbjct: 95 TTWDDCKKVFLSKFFSNARTARLRNEISGFSQKTGESFCEAWERFKGYTNQCPHHGFTKA 154
Query: 118 LIIHIFYNGLTSSTKLTIDAAAGGALMNKDFTTAYALIENMALNLFQWTEEKETVDPSPS 177
++ Y G+ ++ +D A+ G NKD + L+EN+A + + E+ + +
Sbjct: 155 SLLSTLYRGVLPRIRMLLDTASNGNFQNKDVEEGWELVENLAQSDGNYNEDCDRTVRGTA 214
Query: 178 EKEAGINETSSIDYLSTKLNALSQRLDRMSTPTFSPPCETCGLSSHSDTDCNLTSVEQLN 237
+ + D ++ AL+ +LDR+ + D +E+++
Sbjct: 215 DSD---------DKHRKEIKALNDKLDRILLSQHKHVHFLVDDEQYEVQDGEGNQLEEVS 265
Query: 238 FVQNGQ 243
++ N Q
Sbjct: 266 YINNNQ 271
>dbj|BAB02630.1| retroelement pol polyprotein-like [Arabidopsis thaliana]
Length = 897
Score = 1024 bits (2648), Expect = 0.0
Identities = 497/768 (64%), Positives = 603/768 (77%), Gaps = 46/768 (5%)
Query: 687 VIVNASLDEVETEKLLYVLKKYPKAIGYTIDDIKGINPSLCMHRILLEDDYKPSIEHQRR 746
+I+NA L+ E L L+KY +AIGY++ DIKGI+PSLC HRI LE++ SIE QRR
Sbjct: 1 MIINAELNSDEVNLQLSELRKYRRAIGYSLSDIKGISPSLCNHRIHLENESYSSIEPQRR 60
Query: 747 LNPNMKEVVKKEVLKLLDAGVIYPISDSKWVSPVQVVPKKGGLTVIKNDKNESIATRTVT 806
LNPN DAGVIYPISDS WVS V VPKKGG+TV+KN+K+E I TRT+T
Sbjct: 61 LNPNF------------DAGVIYPISDSTWVSLVYCVPKKGGMTVVKNEKDELIPTRTIT 108
Query: 807 GWRMCIDYRKLNKATRKDHFPLPFIDQMLERLAKHSHFCYLDGYSGFFQIPIHPNDQEKT 866
G RMCIDYRKLN A+RKDHFPLPFIDQMLERLA H ++C+LDGYSGFFQIPIHPNDQEKT
Sbjct: 109 GHRMCIDYRKLNAASRKDHFPLPFIDQMLERLANHPYYCFLDGYSGFFQIPIHPNDQEKT 168
Query: 867 TFTCPFGTFAYRRMPFGLCNAPATFQRCMMSIFSDFVEKIMEVFMDDFSVHGSNFDDCLT 926
TFTCP+GTFAY+RMPFGLCNAPATFQRCM SIFSD +E+++EVFMDDFS +G +F CL
Sbjct: 169 TFTCPYGTFAYKRMPFGLCNAPATFQRCMTSIFSDLIEEMVEVFMDDFSGYGPSFSSCLL 228
Query: 927 NLEKVLERCEQVNLVLNWEKCHFMVREGIVLGHLVSDRGIEVDRAKVEIIEKMLPPTSVK 986
NL +VL RCE+ NLVLNWEKCHFMV+EGIVLGH +S++GIEVD+ KVE++ ++ PP +VK
Sbjct: 229 NLGRVLTRCEETNLVLNWEKCHFMVKEGIVLGHKISEKGIEVDKGKVEVMMQLQPPKTVK 288
Query: 987 EIRSFLGHAGFYRRFIKDFSSITKPLTNLLLKDADFTFDDSCLQAFCRLKEALITAPIIQ 1046
EIRSFLGHAGFYRRFIKDFS I +PLT LL K+ +F FD+ CL++F +K+AL++AP+++
Sbjct: 289 EIRSFLGHAGFYRRFIKDFSKIVRPLTRLLCKETEFEFDEDCLKSFQTIKDALVSAPVVR 348
Query: 1047 PPDWNLPFEIMCDASDYAVGVVLGQRKDKKMHAIYYASKTLDGAQVNYATTEKELLAVVY 1106
P+W+ PFEIMCDASDY VG VLGQ+ DKK+H IYYAS+TLD AQ YATTEKELLAVV+
Sbjct: 349 APNWDYPFEIMCDASDYVVGAVLGQKIDKKLHVIYYASRTLDDAQGRYATTEKELLAVVF 408
Query: 1107 AIDKFRQYLVGSKIIVYTDHSAIKYLLNKKDAKPRLIRWILLLQEFDLEIKDKKGVENVV 1166
A +KFR YLVGSK+ VYTDH+A+++L KKD KPRL+RWILLLQEFD+EI DKKG+EN
Sbjct: 409 AFEKFRSYLVGSKVTVYTDHAALRHLYAKKDTKPRLLRWILLLQEFDMEIVDKKGIENGA 468
Query: 1167 ADHLSRLREANKDELPLDDSFPDDQLFLLAQTDAPWYADFVNFLAAGVLPPELNYQQKKK 1226
ADHL R+R ++ LP+DDS P++QL ++ +F +G K
Sbjct: 469 ADHLLRMR--IEEPLPIDDSMPEEQLMVV---------EFFGKSYSG------------K 505
Query: 1227 FFNDLKHYYWDEPYLFRRGSDGIFRRCIPESEVSSILTHCHSSSYGGHASTQKTSFKILH 1286
F+ L + P+ RC+ E EV IL HCH S+YGGH +T KT KIL
Sbjct: 506 EFHQLNDVEGESPW-----------RCVSEDEVEGILLHCHCSAYGGHFATFKTVSKILQ 554
Query: 1287 SGFWWPSLFKDVHLFISKCDKCQRTGSITKRNEMPLNNILEVEIFDVWGVDFMGPFPSSF 1346
+GFWWP++FKD F+SKCD CQR +I++ NEMP N I+EVEIFDVWG+DFMG FPSS+
Sbjct: 555 AGFWWPTMFKDAQEFVSKCDSCQRKDNISRINEMPQNPIVEVEIFDVWGIDFMGLFPSSY 614
Query: 1347 GNQYILVAVDYVSKWVEAIASPTNDAQVVIKMFKKVIFPRFGVPRVVISDGGSHFISKHF 1406
GN+YILVA+DYVSKWVEAIA PTNDA+VV+K+FK +IFPRFGVPRVVISDGG HFI+K F
Sbjct: 615 GNKYILVAIDYVSKWVEAIAIPTNDAKVVLKLFKTIIFPRFGVPRVVISDGGKHFINKVF 674
Query: 1407 EKLLQKLGVRHKVATPYHPQTSGQVEVSNRQIKAILEKTVSTSRTDWS 1454
E LL+K GV+HKVATPYHPQTSGQVE+S+R+IK ILEKTV +R DWS
Sbjct: 675 ENLLKKHGVKHKVATPYHPQTSGQVEISDREIKTILEKTVGITRKDWS 722
>gb|AAF79809.1| T32E20.9 [Arabidopsis thaliana]
Length = 1586
Score = 910 bits (2351), Expect = 0.0
Identities = 449/745 (60%), Positives = 540/745 (72%), Gaps = 112/745 (15%)
Query: 710 KAIGYTIDDIKGINPSLCMHRILLEDDYKPSIEHQRRLNPNMKEVVKKEVLKLLDAGVIY 769
KAIGY++DDIKGI+P+LC HRI LE++ SIE QRRLNPN+KEVVKKE+LKLLDAGVIY
Sbjct: 779 KAIGYSLDDIKGISPTLCTHRIHLENESYSSIEPQRRLNPNLKEVVKKEILKLLDAGVIY 838
Query: 770 PISDSKWVSPVQVVPKKGGLTVIKNDKNESIATRTVTGWRMCIDYRKLNKATRKDHFPLP 829
PISDS WVSPV VPKKGG+TV+KN K+E I TRT+TG RMCI+YRKLN A+RK+HFPLP
Sbjct: 839 PISDSTWVSPVHCVPKKGGMTVVKNSKDELIPTRTITGHRMCIEYRKLNVASRKEHFPLP 898
Query: 830 FIDQMLERLAKHSHFCYLDGYSGFFQIPIHPNDQEKTTFTCPFGTFAYRRMPFGLCNAPA 889
FID MLERLA H ++C+LD YSGFFQIPIHPNDQ KTTFTCP+GTFAY+RMPFGLCNAPA
Sbjct: 899 FIDHMLERLANHPYYCFLDSYSGFFQIPIHPNDQGKTTFTCPYGTFAYKRMPFGLCNAPA 958
Query: 890 TFQRCMMSIFSDFVEKIMEVFMDDFSVHGSNFDDCLTNLEKVLERCEQVNLVLNWEKCHF 949
TFQRCM SIFSD +E+++EVFMDDFSV+GS+F CL NL +VL+RCE+ NLVLNWEKCHF
Sbjct: 959 TFQRCMTSIFSDLIEEMVEVFMDDFSVYGSSFSSCLLNLCRVLKRCEETNLVLNWEKCHF 1018
Query: 950 MVREGIVLGHLVSDRGIEVDRAKVEIIEKMLPPTSVKEIRSFLGHAGFYRRFIKDFSSIT 1009
MVREGIVLG +S+ GIEVD+AK++++ ++ PP +VK+IRSFLGHAGFYR FIKDFS +
Sbjct: 1019 MVREGIVLGRKISEEGIEVDKAKIDVMMQLQPPKTVKDIRSFLGHAGFYRIFIKDFSKLA 1078
Query: 1010 KPLTNLLLKDADFTFDDSCLQAFCRLKEALITAPIIQPPDWNLPFEIMCDASDYAVGVVL 1069
+PLT LL K+ +F FDD CL AF +KEALITAPI+Q P+W+ PFEI+
Sbjct: 1079 RPLTRLLCKETEFAFDDECLTAFKLIKEALITAPIVQAPNWDFPFEII------------ 1126
Query: 1070 GQRKDKKMHAIYYASKTLDGAQVNYATTEKELLAVVYAIDKFRQYLVGSKIIVYTDHSAI 1129
T+D AQV YATTEKELLAVV+A +KFR YLVGSK+ +YTDH+A+
Sbjct: 1127 ----------------TMDDAQVRYATTEKELLAVVFAFEKFRSYLVGSKVTIYTDHAAL 1170
Query: 1130 KYLLNKKDAKPRLIRWILLLQEFDLEIKDKKGVENVVADHLSRLREANKDELPLDDSFPD 1189
+++ KKD KPRL+RWILLLQEFD+EI DKKG+EN
Sbjct: 1171 RHIYAKKDTKPRLLRWILLLQEFDMEIVDKKGIEN------------------------- 1205
Query: 1190 DQLFLLAQTDAPWYADFVNFLAAGVLPPELNYQQKKKFFNDLKHYYWDEPYLFRRGSDGI 1249
PWYAD VN+L +G PP L+ +KKKFF D+ H+YWDEPYL+ D I
Sbjct: 1206 --------EKLPWYADHVNYLVSGEEPPNLSSYEKKKFFKDINHFYWDEPYLYTLCKDKI 1257
Query: 1250 FRRCIPESEVSSILTHCHSSSYGGHASTQKTSFKILHSGFWWPSLFKDVHLFISKCDKCQ 1309
+R C+ E E+ IL HCH +YGGH +T KT KIL +
Sbjct: 1258 YRTCVSEDEIEGILLHCHGFAYGGHFATFKTMSKILQA---------------------- 1295
Query: 1310 RTGSITKRNEMPLNNILEVEIFDVWGVDFMGPFPSSFGNQYILVAVDYVSKWVEAIASPT 1369
EVE FDVWG+DFMGPFPSS+GN+YILVA+DYVSKWVEAIAS T
Sbjct: 1296 -----------------EVENFDVWGIDFMGPFPSSYGNKYILVAIDYVSKWVEAIASHT 1338
Query: 1370 NDAQVVIKMFKKVIFPRFGVPRVVISDGGSHFISKHFEKLLQKLGVRHKVATPYHPQTSG 1429
NDA+VV+K+FK +IFPRFGVPR+VISDGG HFI+K FE LL+K GV+HK
Sbjct: 1339 NDARVVLKLFKTIIFPRFGVPRIVISDGGKHFINKGFENLLKKHGVKHK----------- 1387
Query: 1430 QVEVSNRQIKAILEKTVSTSRTDWS 1454
VE+SNR+IKAILEKTV ++R DWS
Sbjct: 1388 -VEISNREIKAILEKTVGSTRKDWS 1411
Score = 211 bits (537), Expect = 1e-52
Identities = 137/371 (36%), Positives = 210/371 (55%), Gaps = 38/371 (10%)
Query: 315 NAVTLRSGKPL---EDPIRRTET-------------DHSEKEIREP----PSRETRAEKE 354
+A+TLRSGK L E P + TE + +EK I EP P+R A
Sbjct: 465 HAITLRSGKELPTKESPNQNTEDSLDQDGEDFCQNGNSAEKAIEEPILHQPTRPL-APAA 523
Query: 355 EPRVEGNTT----------PPFKPKIPFPQRFAKSKLDEQFKKFMEMMNKIYIDVPFTEV 404
P VE PP+KP +PFP RF K + + + + + + +P +
Sbjct: 524 SPLVEKPAAAKTKENVFIPPPYKPPLPFPGRFKKVMIQKYKALLEKQLKNLEVTMPLVDC 583
Query: 405 LTQMPTYAKFLKEILSKKRKIEESETVNLTEECSAIIQNKL-PPKLKDPGSFSIPCVIGS 463
L +P K++K++++++ K E V L+ ECSAIIQ K+ P KL DPGSF++PC +G
Sbjct: 584 LALIPDSNKYVKDMITERIK-EVQGMVVLSHECSAIIQQKIIPKKLGDPGSFTLPCALGP 642
Query: 464 EVVKKAMCDLGASVSLMPLSLYERLGIGELKSTRMTLQLADRSVKYPAGIIEDVPVKVGE 523
K +CDLGASVSLMPL + ++LG + K ++L LADRSV+ G++ED+PV +G
Sbjct: 643 LAFSKCLCDLGASVSLMPLPVAKKLGFNKYKPCNISLILADRSVRISHGLLEDLPVMIGV 702
Query: 524 VYIPADFVVMEMEEDNQVPILLGRPFLATAGAIIDVKKGKLAFNVGKE-TVEFELAKLMK 582
V +P DFVV+EM+E+ + P++LGRPFLA A AIIDVKKGK+ N+G++ + F++ MK
Sbjct: 703 VEVPTDFVVLEMDEEPKDPLILGRPFLARARAIIDVKKGKIDLNLGRDLKMTFDITNTMK 762
Query: 583 GPSIKDSCCMIDIIDHCVKECSLASTTHDGLEVCLVNNAGTKLEGEA-KAYEVLLDRTPP 641
P+I+ + I+ +D +K + G+ L + LE E+ + E P
Sbjct: 763 KPTIEGNIFWIEEMD--MKAIGYSLDDIKGISPTLCTHR-IHLENESYSSIEPQRRLNPN 819
Query: 642 IKDLSVEELIK 652
+K++ +E++K
Sbjct: 820 LKEVVKKEILK 830
Score = 119 bits (299), Expect = 6e-25
Identities = 69/206 (33%), Positives = 107/206 (51%), Gaps = 23/206 (11%)
Query: 1 NQFSGSPSEDPNLHISNFWRLSVTCKDD---QETVRLHLFPFSLKNKASNWFNSLKPGSI 57
N+F G P EDP H+ F RL K + ++ +L LFPFSL +KA W SL GSI
Sbjct: 84 NKFHGLPMEDPLDHLDEFDRLCSLTKINRVSEDGFKLRLFPFSLGDKAHQWEKSLPQGSI 143
Query: 58 TSWDQLRREFLCRFFPPSKTAQLRGKLYQFTQMNGESLFDVWERFKEILRRCPHHGLEKW 117
TSW+ ++ FL +FF S+TA+LR + FTQ N E+ ++ WERFK +CPHH
Sbjct: 144 TSWNDCKKAFLAKFFSNSRTARLRNDISGFTQTNNETFYEAWERFKGYQTQCPHH----- 198
Query: 118 LIIHIFYNGLTSSTKLTIDAAAGGALMNKDFTTAYALIENMALNLFQWTEEKE-TVDPSP 176
++ +D A+ G +NKD + ++EN+A + + E+ + ++ S
Sbjct: 199 --------------EMLLDTASNGNFLNKDVEDGWEVVENLAQSDGNYNEDYDRSIRTSS 244
Query: 177 SEKEAGINETSSIDYLSTKLNALSQR 202
E E +++ KL + Q+
Sbjct: 245 DSDEKHRREMKAMNDKLDKLLLMQQK 270
>pir||D84513 probable retroelement pol polyprotein [imported] - Arabidopsis
thaliana
Length = 841
Score = 853 bits (2205), Expect = 0.0
Identities = 460/919 (50%), Positives = 589/919 (64%), Gaps = 128/919 (13%)
Query: 350 RAEKEEPRVEGNTTPPFKPKIPFPQRFAKSKLDEQFKKFMEMMNKIYIDVPFTEVLTQMP 409
R + E + + PP+ PK+PFP R K + ++++ F E+M ++ P
Sbjct: 4 RKQTLEEKAKKTVLPPYVPKLPFPGRQRKIQREKEYSLFDEIMRQLQRYSP--------- 54
Query: 410 TYAKFLKEILSKKRKIEESETVNLTEECSAIIQNKLPPKLKDPGSFSIPCVIGSEVVKKA 469
ECSAI+QN +P K +DPGSF +P IG +
Sbjct: 55 --------------------------ECSAILQNVIPVKREDPGSFVLPSRIGEYTFDRC 88
Query: 470 MCDLGASVSLMPLSLYERLGIGELKSTRMTLQLADRSVKYPAGIIEDVPVKVGEVYIPAD 529
+CDLGA VSLMP S+ +RLG T+M+L L DRS+ +P G+ EDV V+VG YIP D
Sbjct: 89 LCDLGAGVSLMPFSVAKRLGDTNFTPTKMSLVLGDRSISFPVGVAEDVQVRVGNFYIPTD 148
Query: 530 FVVMEMEEDNQVPILLGRPFLATAGAIIDVKKGKLAFNVGKETVEFELAKLMKGPSIKDS 589
FV++E++E+ + ++LGRPFL A+IDV+K K+ +G EF + ++M P+ +
Sbjct: 149 FVIIELDEEPRHRLILGRPFLNIVAALIDVRKSKINLRIGDIVQEFNMERIMSKPTTECQ 208
Query: 590 CCMIDIIDHCVKECSLASTTHDGLEVCLVNNAGT-KLEGEAKA-YEVLLDRTPPIK---- 643
+DI+D V E T D L+ L GEA + +LD + P+
Sbjct: 209 TFWVDIMDELVNELLAELNTEDPLQTVLTKEESEFGYLGEATTRFARILDSSSPMTKVVA 268
Query: 644 --DLSVEELIKEEPTLQPKE--------APKVELKTLPSNLRYEFLGPNSTYPVIVNASL 693
+L E+ K PK+ APK+ELK LP+ LR FLGPNSTY VI+N L
Sbjct: 269 FAELGDNEVEKALVVSSPKDCDDWSELNAPKMELKPLPARLRVAFLGPNSTYLVIINPEL 328
Query: 694 DEVETEKLLYVLKKYPKAIGYTIDDIKGINPSLCMHRILLEDDYKPSIEHQRRLNPNMKE 753
+ VE+ LL L+KY KA+GY++DDI GI+P+LCMH I LE + S+EHQRRLN N+++
Sbjct: 329 NNVESALLLCELRKYRKALGYSLDDITGISPTLCMHMIHLEGESITSVEHQRRLNSNLRD 388
Query: 754 VVKKEVLKLLDAGVIYPISDSKWVSPVQVVPKKGGLTVIKNDKNESIATRTVTGWRMCID 813
VVKKE++KLLDAG+IYPISDS WVSPV VVPKKGG+ VIKN+KNE I TRTVTG RMCID
Sbjct: 389 VVKKEIMKLLDAGIIYPISDSTWVSPVHVVPKKGGVIVIKNEKNELIPTRTVTGHRMCID 448
Query: 814 YRKLNKATRKDHFPLPFIDQMLERLAKHSHFCYLDGYSGFFQIPIHPNDQEKTTFTCPFG 873
YRKLN ATRKD+FPL FIDQMLERL+ ++C+LDGY GFFQI IHP+DQEKTTFTCP+G
Sbjct: 449 YRKLNSATRKDNFPLSFIDQMLERLSNQPYYCFLDGYLGFFQILIHPDDQEKTTFTCPYG 508
Query: 874 TFAYRRMPFGLCNAPATFQRCMMSIFSDFVEKIMEVFMDDFSVHGSNFDDCLTNLEKVLE 933
TFAYRRMPFGLCNAPATFQ CM IFSD +E MEVF+DDF VL+
Sbjct: 509 TFAYRRMPFGLCNAPATFQHCMKYIFSDMIEDFMEVFIDDF---------------LVLQ 553
Query: 934 RCEQVNLVLNWEKCHFMVREGIVLGHLVSDRGIEVDRAKVEIIEKMLPPTSVKEIRSFLG 993
RCE +LVLNWEK HFMVR+GIVLGH +S++G+EVDRAK+EI+ FLG
Sbjct: 554 RCEDKHLVLNWEKSHFMVRDGIVLGHKISEKGVEVDRAKIEIMR-------------FLG 600
Query: 994 HAGFYRRFIKDFSSITKPLTNLLLKDADFTFDDSCLQAFCRLKEALITAPIIQPPDWNLP 1053
HAGFYRRFIKDFS I +P T LL K+ FDD+CLQAF +KE+L++A I+QPP+W LP
Sbjct: 601 HAGFYRRFIKDFSKIARPCTQLLCKEQKSEFDDTCLQAFNIVKESLVSALIVQPPEWELP 660
Query: 1054 FEIMCDASDYAVGVVLGQRKDKKMHAIYYASKTLDGAQVNYATTEKELLAVVYAIDKFRQ 1113
FE++CDASDY VG VLGQRKDKK+HAIYYAS+TLDGAQV
Sbjct: 661 FEVICDASDYVVGAVLGQRKDKKLHAIYYASRTLDGAQV--------------------- 699
Query: 1114 YLVGSKIIVYTDHSAIKYLLNKKDAKPRLIRWILLLQEFDLEIKDKKGVENVVADHLSRL 1173
IV+T H+A++YLL+KKDAKPRL+RWILLLQ+FDLEIKDKKG+EN VAD+LSRL
Sbjct: 700 -------IVHTVHAALRYLLSKKDAKPRLLRWILLLQDFDLEIKDKKGIENEVADNLSRL 752
Query: 1174 REAN----KDELP------LDDSFPDD---------QLFLL--AQTDAPWYADFVNFLAA 1212
R +D LP ++D + D+ +L L +++ PWYADF N+L+
Sbjct: 753 RVQEEVLMRDILPGENLASIEDCYMDEVGRLRVSTLELMTLHTGESNLPWYADFANYLSC 812
Query: 1213 GVLPPELNYQQKKKFFNDL 1231
V PP+ KKK ++
Sbjct: 813 EVPPPDFTGYFKKKLLKEV 831
>gb|AAD19780.2| hypothetical protein [Arabidopsis thaliana]
Length = 1048
Score = 814 bits (2103), Expect = 0.0
Identities = 459/1005 (45%), Positives = 607/1005 (59%), Gaps = 144/1005 (14%)
Query: 268 ELKDQTRLLNNSLATLTTKIDSIYSHNKISETQLSQVVRKAGHPNKMNAVTLRSGKPLED 327
E+ + LL L+ + S N Q K GH +SG+ L +
Sbjct: 2 EISQECLLLYQILSLARVGNQNSRSENSGGRAPKHQNPEKGGH---------KSGREL-N 51
Query: 328 PIRRTETDHSEKEIREPPSRETRAEKE----EPRVEGNTTPPFKPKIPFPQRFAKSKLDE 383
PI + E + + E KE E + + PP+ PK+PFP R K + ++
Sbjct: 52 PILKKEKAKEKSKAASDDLEEDNTGKESDTLEEKAKKTVLPPYVPKLPFPGRQRKIQREK 111
Query: 384 QFKKFMEMMNKIYIDVPFTEVLTQMPTYAKFLKEILSKKRKIEESETVNLTEECSAIIQN 443
++ F E+M ++ + +PF +++ + Y K LK+IL+ KR +EE + ++ ECSAI+QN
Sbjct: 112 EYSLFDEIMRQLQVKLPFLDLVQNVSIYRKHLKDILTNKRTLEEGHVL-ISHECSAILQN 170
Query: 444 KLPPKLKDPGSFSIPCVIGSEVVKKAMCDLGASVSLMPLSLYERLGIGELKSTRMTLQLA 503
FS +IG + +CDLGA VSLMP S+ +RLG T+M+L L
Sbjct: 171 V--------ALFSR--MIGEYTFDRCLCDLGAGVSLMPFSVAKRLGDTNFTPTKMSLVLG 220
Query: 504 DRSVKYPAGIIEDVPVKVGEVYIPADFVVMEMEEDNQVPILLGRPFLATAGAIIDVKKGK 563
DRS+ +P G+ EDV V+VG YIP DFV++E++E+ + ++LGRPFL A+IDV+K K
Sbjct: 221 DRSISFPVGVAEDVQVRVGNFYIPTDFVIIELDEEPRHRLILGRPFLNIVAALIDVRKSK 280
Query: 564 LAFNVGKETVEFELAKLMKGPSIKDSCCMIDIIDHCVKECSLASTTHDGLEVCLVNNAGT 623
+ +G EF + ++M P+ + +DI+D V E T D L+ L
Sbjct: 281 INLRIGDIVQEFNMERIMSKPTTECQTFWVDIMDELVNELLAELNTEDPLQTVLTKEESE 340
Query: 624 -KLEGEAKA-YEVLLDRTPPIK------DLSVEELIKEEPTLQPKE--------APKVEL 667
GEA + +LD + P+ +L E+ K PK+ APK+EL
Sbjct: 341 FGYLGEATTRFARILDSSSPMTKVVAFAELGDNEVEKALVVSSPKDCDDWSELNAPKMEL 400
Query: 668 KTLPSNLRYEFLGPNSTYPVIVNASLDEVETEKLLYVLKKYPKAIGYTIDDIKGINPSLC 727
K LP+ LR FLGPNSTY VI+N L+ VE+ LL L+KY KA+GY++DDI GI+P+LC
Sbjct: 401 KPLPARLRVAFLGPNSTYLVIINPELNNVESALLLCELRKYRKALGYSLDDITGISPTLC 460
Query: 728 MHRILLEDDYKPSIEHQRRLNPNMKEVVKKEVLKLLDAGVIYPISDSKWVSPVQVVPKKG 787
MH I LE + S+EHQRRLN N+++VVKKE++KLLDAG+IYPISDS WVSPV VVPKKG
Sbjct: 461 MHMIHLEGESITSVEHQRRLNSNLRDVVKKEIMKLLDAGIIYPISDSTWVSPVHVVPKKG 520
Query: 788 GLTVIKNDKNESIATRTVTGWRMCIDYRKLNKATRKDHFPLPFIDQMLERLAKHSHFCYL 847
G+ VIKN+KNE I TRTVTG RMCIDYRKLN ATRKD+FPL FIDQMLERL+ ++C+L
Sbjct: 521 GVIVIKNEKNELIPTRTVTGHRMCIDYRKLNSATRKDNFPLSFIDQMLERLSNQPYYCFL 580
Query: 848 DGYSGFFQIPIHPNDQEKTTFTCPFGTFAYRRMPFGLCNAPATFQRCMMSIFSDFVEKIM 907
DGY GFFQI IHP+DQEKTTFTCP+GTFAYRRMPFGLCNAPATFQ CM IFSD +E M
Sbjct: 581 DGYLGFFQILIHPDDQEKTTFTCPYGTFAYRRMPFGLCNAPATFQHCMKYIFSDMIEDFM 640
Query: 908 EVFMDDFSVHGSNFDDCLTNLEKVLERCEQVNLVLNWEKCHFMVREGIVLGHLVSDRGIE 967
ERCE +LVLNWEK HFMVR+GIVLGH +S++G+E
Sbjct: 641 -------------------------ERCEDKHLVLNWEKSHFMVRDGIVLGHKISEKGVE 675
Query: 968 VDRAKVEIIEKMLPPTSVKEIRSFLGHAGFYRRFIKDFSSITKPLTNLLLKDADFTFDDS 1027
VDRAK+E++ M PP SVK+ ++ FDD+
Sbjct: 676 VDRAKIEVMMSMQPPNSVKD-----------------------------HEEQKSEFDDT 706
Query: 1028 CLQAFCRLKEALITAPIIQPPDWNLPFEIMCDASDYAVGVVLGQRKDKKMHAIYYASKTL 1087
CLQAF +KE+L++A I+QPP+W LPFE++CDASDY VG VLGQRKDKK+HAIYYAS+TL
Sbjct: 707 CLQAFNIVKESLVSALIVQPPEWELPFEVICDASDYVVGAVLGQRKDKKLHAIYYASRTL 766
Query: 1088 DGAQVNYATTEKELLAVVYAIDKFRQYLVGSKIIVYTDHSAIKYLLNKKDAKPRLIRWIL 1147
DGAQV IV+T H+A++YLL+KKDAKPRL+RWIL
Sbjct: 767 DGAQV----------------------------IVHTVHAALRYLLSKKDAKPRLLRWIL 798
Query: 1148 LLQEFDLEIKDKKGVENVVADHLSRLREAN----KDELP------LDDSFPDD------- 1190
LLQ+FDLEIKDKKG+EN VAD+LSRLR +D LP ++D + D+
Sbjct: 799 LLQDFDLEIKDKKGIENEVADNLSRLRVQEEVLMRDILPGENLASIEDCYMDEVGRLRVS 858
Query: 1191 --QLFLL--AQTDAPWYADFVNFLAAGVLPPELNYQQKKKFFNDL 1231
+L L +++ PWYADF N+L+ V PP+ KKK ++
Sbjct: 859 TLELMTLHTGESNLPWYADFANYLSCEVPPPDFTGYFKKKLLKEV 903
>emb|CAD39928.2| OSJNBa0091C12.6 [Oryza sativa (japonica cultivar-group)]
gi|50921841|ref|XP_471281.1| OSJNBa0091C12.6 [Oryza
sativa (japonica cultivar-group)]
Length = 781
Score = 735 bits (1897), Expect = 0.0
Identities = 410/846 (48%), Positives = 536/846 (62%), Gaps = 90/846 (10%)
Query: 454 SFSIPCVIGSEVVKKAMCDLGASVSLMPLSL-YERLGIGELKSTRMTLQLADRSVKYPAG 512
S +I C IG V+ A+ + ++M + ++ LG L+ T T +++ S G
Sbjct: 22 SSTIMCHIGGNSVR-ALYNPSVGANIMSATFAFDCLGDRSLEPTVRTFRISTNSTTEGLG 80
Query: 513 IIEDVPVKVGEVYIPADFVVMEMEEDNQVPILLGRPFLATAGAIIDVKK-GKLAFNVGKE 571
II VP + V + DF V E+ + + IL+G P ++DV + G L +G+
Sbjct: 81 IISGVPTRHNSVEVILDFHVFEIYDFD---ILIGHPI----EKLLDVPETGVLNLKIGR- 132
Query: 572 TVEFELAKLMKGPSIKDSCCMIDIIDHCVKECSLASTTHDGLEVCLVNNAGTKLEGEAKA 631
+ + L S+ +S + + I E +A + + LE + EGE
Sbjct: 133 -IATSVPMLQSTNSLMESLPVPEPI-----EGVMAISPFETLESIFDESIEEFNEGE--- 183
Query: 632 YEVLLDRTPPIKDLSVEELIKEEPTLQPKEAPKVELKTLPSNLRYEFLGPNSTYPVIVNA 691
D T DL +L P AP +ELK LPS+L Y FL ++ VI++
Sbjct: 184 -----DETGETMDLPKTDL--------PSRAP-IELKPLPSSLSYAFLNSDAESLVIISD 229
Query: 692 SLDEVETEKLLYVLKKYPKAIGYTIDDIKGINPSLCMHRILLEDDYKPSIEHQRRLNPNM 751
L E E +L+ +L+K+ A GY++ D+KGI+P+LC HRILLE PS E QRRLN M
Sbjct: 230 KLSEREMARLIAILEKHSAAFGYSLQDLKGISPTLCTHRILLEPSSTPSREPQRRLNNAM 289
Query: 752 KEVVKKEVLKLLDAGVIYPISDSKWVSPVQVVPKKGGLTVIKNDKNESIATRTVTGWRMC 811
+EV+KKEVLKLL G+IYP+ S+WVSPVQVVPKKGG+TV++N NE I RTVTGWRMC
Sbjct: 290 REVIKKEVLKLLHTGIIYPVPYSEWVSPVQVVPKKGGMTVVENSNNELIPQRTVTGWRMC 349
Query: 812 IDYRKLNKATRKDHFPLPFIDQMLERLAKHSHFCYLDGYSGFFQIPIHPNDQEKTTFTCP 871
IDYRKLNKAT+KDHF LPFID+MLERLA HS FC+LDGYS
Sbjct: 350 IDYRKLNKATKKDHFSLPFIDEMLERLANHSFFCFLDGYS-------------------- 389
Query: 872 FGTFAYRRMPFGLCNAPATFQRCMMSIFSDFVEKIMEVFMDDFSVHGSNFDDCLTNLEKV 931
+Y ++P ++ DD ++G F+ CL NL+KV
Sbjct: 390 ----SYHQIP---------------------------IYPDD-QIYGKTFNHCLENLDKV 417
Query: 932 LERCEQVNLVLNWEKCHFMVREGIVLGHLVSDRGIEVDRAKVEIIEKMLPPTSVKEIRSF 991
L+RC++ +LVLNWEK HFMVREGIVLGH +S+RG+EVDRAK+E+I ++ PP + +RSF
Sbjct: 418 LQRCQEKDLVLNWEKYHFMVREGIVLGHQISERGVEVDRAKIEVIRQLPPPVNDMGVRSF 477
Query: 992 LGHAGFYRRFIKDFSSITKPLTNLLLKDADFTFDDSCLQAFCRLKEALITAPIIQPPDWN 1051
LGHAGFYRRFIKDFS I +PLT LL D FDD C+++F LKE+LI+APIIQP DW+
Sbjct: 478 LGHAGFYRRFIKDFSKIARPLTALLANDVPVDFDDECMKSFKILKESLISAPIIQPSDWS 537
Query: 1052 LPFEIMCDASDYAVGVVLGQRKDKKMHAIYYASKTLDGAQVNYATTEKELLAVVYAIDKF 1111
L FEIMCDASD+AVG VLGQ KD+K HAI YASKTL GAQ+NYATTEKELLAVV+AI+KF
Sbjct: 538 LLFEIMCDASDFAVGAVLGQTKDRKHHAITYASKTLTGAQLNYATTEKELLAVVFAIEKF 597
Query: 1112 RQYLVGSKIIVYTDHSAIKYLLNKKDAKPRLIRWILLLQEFDLEIKDKKGVENVVADHLS 1171
R YLVG+K++VYTDH+A+KYLL KKDAKPR+IRWILLLQEFD+EI DKKGVEN VADHLS
Sbjct: 598 RSYLVGAKVVVYTDHAALKYLLTKKDAKPRVIRWILLLQEFDIEIMDKKGVENYVADHLS 657
Query: 1172 RLREANKDELPLDDSFPDDQLFLLAQTDAPWYADFVNFLAAGVLPPELNYQQKKKFFNDL 1231
R++ N +L ++D DD + +D PWYA VNF+A G +PP + KK+ +
Sbjct: 658 RMQITNMQKLLINDYLRDDMRLKVIDSD-PWYATIVNFMATGHVPP---VENKKRLIYES 713
Query: 1232 KHYYWDEPYLFRRGSDGIFRRCIPESEVSSILTHCHSSSYGGHASTQKTSFKILHSGFWW 1291
+ + WD PYLFR SDG+ R C+ E I+ CH++ YGGH +T KI SGF+W
Sbjct: 714 RRHLWDAPYLFRVCSDGLLRICVSTDEGIMIIEKCHAAPYGGHYGAFRTHAKIWQSGFFW 773
Query: 1292 PSLFKD 1297
PS++ D
Sbjct: 774 PSMYDD 779
>emb|CAE03895.1| OSJNBb0026I12.3 [Oryza sativa (japonica cultivar-group)]
gi|50921891|ref|XP_471306.1| OSJNBb0026I12.3 [Oryza
sativa (japonica cultivar-group)]
Length = 589
Score = 692 bits (1786), Expect = 0.0
Identities = 336/507 (66%), Positives = 410/507 (80%), Gaps = 5/507 (0%)
Query: 789 LTVIKNDKNESIATRTVTGWRMCIDYRKLNKATRKDHFPLPFIDQMLERLAKHSHFCYLD 848
+TV+ N NE I RTVTGWRMCIDY+KLNKAT+KD+FPLP ID+MLERLA HS FC+LD
Sbjct: 1 MTVVANVNNELIPQRTVTGWRMCIDYQKLNKATKKDNFPLPVIDEMLERLANHSLFCFLD 60
Query: 849 GYSGFFQIPIHPNDQEKTTFTCPFGTFAYRRMPFGLCNAPATFQRCMMSIFSDFVEKIME 908
GYSG+ QIPIHP DQ KTTFTCP+GT+AYRRM FGLCNAPA+FQRCMMSIFSD +E IME
Sbjct: 61 GYSGYHQIPIHPKDQCKTTFTCPYGTYAYRRMSFGLCNAPASFQRCMMSIFSDMIEAIME 120
Query: 909 VFMDDFSVHGSNFDDCLTNLEKVLERCEQVNLVLNWEKCHFMVREGIVLGHLVSDRGIEV 968
VFMDDFS++G FD CL NL+KV +RC++ +LVLNWEKCHFMV EGIVLGH VS+RGIEV
Sbjct: 121 VFMDDFSLYGKTFDHCLQNLDKVFQRCQEKDLVLNWEKCHFMVHEGIVLGHRVSERGIEV 180
Query: 969 DRAKVEIIEKMLPPTSVKEIRSFLGHAGFYRRFIKDFSSITKPLTNLLLKDADFTFDDSC 1028
DRAK+E+I+++ PP ++K IR+FLGHAGF+RRFIK+FS+I +PLTNLL KDA F FDD C
Sbjct: 181 DRAKIEVIDQLPPPVNIKGIRNFLGHAGFHRRFIKEFSTIARPLTNLLAKDAPFEFDDVC 240
Query: 1029 LQAFCRLKEALITAPIIQPPDWNLPFEIMCDASDYAVGVVLGQRKDKKMHAIYYASKTLD 1088
L++F LK+AL++A IIQPPD LPFEIMCDASD+ VG VLGQ KDKK+HAI YAS+TL
Sbjct: 241 LKSFKTLKKALVSALIIQPPDGMLPFEIMCDASDFIVGAVLGQTKDKKLHAICYASETLA 300
Query: 1089 GAQVNYATTEKELLAVVYAIDKFRQY-LVGSKIIVYTDHSAIKYLLNKKDAKPRLIRWIL 1147
GAQ NYATTEKELLAVV+AIDKFR+ LVG+K+IVYT+H+A+KYLL KKDAKPRL+RWI
Sbjct: 301 GAQSNYATTEKELLAVVFAIDKFRRSDLVGAKVIVYTEHAALKYLLTKKDAKPRLLRWIP 360
Query: 1148 LLQEFDLEIKDKKGVENVVADHLSRLREANKDELPLDDSFPDDQLFLLAQTDAPWYADFV 1207
LLQEFDLEIKDKKGVEN VADHLSRL+ N E P++ DD L + +D PWYA+ V
Sbjct: 361 LLQEFDLEIKDKKGVENSVADHLSRLQLTNMQEPPINYFLQDDMLMAVRNSD-PWYANIV 419
Query: 1208 NFLAAGVLPPELNYQQKKKFFNDLKHYYWDEPYLFRRGSDGIFRRCIPESEVSSILTHCH 1267
N++ + +P N Q+K K+ + + WDEPYL+R S+G+ RRC+P+ E I+ CH
Sbjct: 420 NYMVSKYVPQGEN-QRKLKY--ESHCHIWDEPYLYRVCSNGLLRRCVPKEEGFKIIERCH 476
Query: 1268 SSSYGGHASTQKTSFKILHSGFWWPSL 1294
++ Y GH +T KI S F+WP++
Sbjct: 477 AAPYRGHYGAFRTQAKIWQSRFFWPTM 503
Score = 66.2 bits (160), Expect = 8e-09
Identities = 28/48 (58%), Positives = 37/48 (76%)
Query: 1406 FEKLLQKLGVRHKVATPYHPQTSGQVEVSNRQIKAILEKTVSTSRTDW 1453
F +LL+K+ +H +ATPYHPQT+GQ E+SN+QIK IL+KTV T W
Sbjct: 505 FRELLRKMNAKHNIATPYHPQTNGQAEMSNKQIKNILQKTVHEMGTGW 552
>gb|AAF63114.1| Hypothetical protein [Arabidopsis thaliana] gi|25405147|pir||B96502
hypothetical protein F28H19.8 [imported] - Arabidopsis
thaliana
Length = 640
Score = 650 bits (1678), Expect = 0.0
Identities = 305/472 (64%), Positives = 378/472 (79%), Gaps = 18/472 (3%)
Query: 953 EGIVLGHLVSDRGIEVDRAKVEIIEKMLPPTSVKEIRSFLGHAGFYRRFIKDFSSITKPL 1012
EGIVLGH +S +GIEVD+AK++++ ++ P +VK+IRSFLGHAGFYRRFIKDFS I +PL
Sbjct: 170 EGIVLGHKISGKGIEVDKAKIDVMVRLQPLKTVKDIRSFLGHAGFYRRFIKDFSKIARPL 229
Query: 1013 TNLLLKDADFTFDDSCLQAFCRLKEALITAPIIQPPDWNLPFEIMCDASDYAVGVVLGQR 1072
T LL K+A+F FD+ CL+AF +KEAL+ API+Q P+WN PFEIMCDASDYAVG VLGQR
Sbjct: 230 TRLLCKEAEFDFDEDCLKAFHSIKEALVLAPIVQAPNWNHPFEIMCDASDYAVGAVLGQR 289
Query: 1073 KDKKMHAIYYASKTLDGAQVNYATTEKELLAVVYAIDKFRQYLVGSKIIVYTDHSAIKYL 1132
DKK+H IYYAS+TLD Q YATTEKELL VV+A +KFR YLVGSK+ VYTDH+A+K++
Sbjct: 290 IDKKLHVIYYASRTLDDTQSRYATTEKELLVVVFAFEKFRSYLVGSKVTVYTDHAALKHI 349
Query: 1133 LNKKDAKPRLIRWILLLQEFDLEIKDKKGVENVVADHLSRLREANKDELPLDDSFPDDQL 1192
KKD KPRL+RWIL LQEFD+EI DK+G EN VADHLSR+R + +P+DDS P++QL
Sbjct: 350 YAKKDTKPRLLRWILFLQEFDMEIVDKRGSENGVADHLSRMRMT--EAVPIDDSMPEEQL 407
Query: 1193 FLLAQT---------------DAPWYADFVNFLAAGVLPPELNYQQKKKFFNDLKHYYWD 1237
+ + PWY D VN+L AG++P EL+ QKKKFF D+ HYYWD
Sbjct: 408 LAVGTSFDKFKIETMCATRSGKLPWYVDLVNYLTAGIVPSELSSYQKKKFFRDINHYYWD 467
Query: 1238 EPYLFRRGSDGIFRRCIPESEVSSILTHCHSSSYGGHASTQKTSFKILHSGFWWPSLFKD 1297
EP L+++G DG+FRRCI E EV +L CH+S YGGH +T KT K+L +G WWPS+FKD
Sbjct: 468 EPVLYKKGVDGLFRRCIAEEEVQGVLELCHNSPYGGHFATFKTVQKVLQAGLWWPSMFKD 527
Query: 1298 VHLFISKCDKCQRTGSITKRNEMPLNNILEVEIFDVWGVDFMGPF-PSSFGNQYILVAVD 1356
H I++CD+CQR G+I++RNEMP N ILEVE+FDVWG+DFMGPF P+S+GN+YILVAVD
Sbjct: 528 AHEHITRCDRCQRVGNISRRNEMPQNPILEVEVFDVWGIDFMGPFEPASYGNKYILVAVD 587
Query: 1357 YVSKWVEAIASPTNDAQVVIKMFKKVIFPRFGVPRVVISDGGSHFISKHFEK 1408
YVSKWVEAIASPTND++VV+K+FK +IFPRFG+PRVVISDGGSHFI+K FEK
Sbjct: 588 YVSKWVEAIASPTNDSKVVLKLFKTIIFPRFGIPRVVISDGGSHFINKVFEK 639
>gb|AAG52026.1| polyprotein, putative; 77260-80472 [Arabidopsis thaliana]
gi|25405064|pir||B96492 probable polyprotein, 77260-80472
[imported] - Arabidopsis thaliana
Length = 884
Score = 646 bits (1667), Expect = 0.0
Identities = 396/970 (40%), Positives = 553/970 (56%), Gaps = 131/970 (13%)
Query: 221 SSHSDTDCNLTSVEQLNFVQNGQRVVQKSHIELLMKNLFFKQSEQLQELKDQTRLLNNSL 280
S+ S ++ Q+ +V + + ++ F +QS + ++ T + N
Sbjct: 28 SNISTKSVDIARKTQVQYVAHVDSLATSEKMDQQRYMSFNQQSTHQKPVQQTTSFIQNHA 87
Query: 281 ATLTTKIDSIYSHNKISETQLSQVVRKAGHPNKMNAVTLRSGKPLEDPIRRTETDHSEKE 340
++ T+ K E+ L Q+++ G + ++G IRR E + S E
Sbjct: 88 SS--TQAHPPPDTTKTIESMLEQILK--GQEKQTTDFDHKTGCI----IRRCEWEDSNIE 139
Query: 341 IREPPSRETRAEKEEPRVEGNTTPPFKPKIPFPQRFAKSKLDEQFKKFMEMMNKIYIDVP 400
+ + ++ R T KPK+PFP+ KSK + + MM+K+ +++P
Sbjct: 140 -----QLDQAIDYKQRRSAAKVT---KPKVPFPKSPRKSKQELDDARCKVMMDKLIVEMP 191
Query: 401 FTEVLTQMPTYAKFLKEILSKKRKIEESETVNLTEECSAIIQNKLPPKLKDPGSFSIPCV 460
+ + P +F+K I++K + E+ + ++ + S IIQNK+P KL D
Sbjct: 192 LIDAVKSSPMIRQFVKRIVTKDM-LTETVVMTMSTQVSDIIQNKIPQKLPDR-------- 242
Query: 461 IGSEVVKKAMCDLGASVSLMPLSLYERLGIGELKSTRMTLQLADRSVKYPAGIIEDVPVK 520
+L+ T++TL LADRSV+ GII DVPV+
Sbjct: 243 -------------------------------DLEPTQITLVLADRSVRRSDGIICDVPVQ 271
Query: 521 VGEVYIPADFVVMEMEEDNQVPILLGRPFLATAGAIIDVKKGKLAFNVGKETVEFELAKL 580
VG YIP D VV+ E++ + P++LGRPFL T GAIIDV+KG + NVG T++F++ K+
Sbjct: 272 VGTSYIPTDLVVLSYEKEPKDPLILGRPFLVTTGAIIDVRKGIIGLNVGDLTMQFDINKV 331
Query: 581 MKGPSIKDSCCMIDIIDHCVKECSLASTTHDGLEVCLVNNAGTKLEGEAKAYEVLLDRTP 640
+K P+I +D I E + T L+ L+ + ++ K Y LLDR
Sbjct: 332 VKKPTIDGKTFYLDTISFLADEFLMKMTLAVPLKHALIPS----IDKVPKGYGKLLDRIE 387
Query: 641 PIKDLSVEELIKEEPTLQPKEAPKVELKTLPSNLRYEFLGPNSTYPVIVNASLDEVETEK 700
V +L+ +E E +G +S I + SL+
Sbjct: 388 -----HVMQLVAQE----------------------ELIGTSSQAATIGDWSLE------ 414
Query: 701 LLYVLKKYPKAIGYTIDDIKGINPSLCMHRILLEDDYKPSIEHQRRLNPNMKEVVKKEVL 760
K PK D+K + P L + L E+ I + + ++ K L
Sbjct: 415 ------KAPKV------DLKPLPPGL-RYAFLGENSTYHVIVNASLNKVELTLLLSK--L 459
Query: 761 KLLDAGVIYPISDSKWVSPVQVVPKKGGLTVIKNDKNESIATRTVTGWRMCIDYRK---- 816
+ + Y + D +SP + + +K++ S+ R + +K
Sbjct: 460 RKYRKALGYSLDDITGISPDLCMHR----IHLKDESKPSVEHRRRLNPNLKDAVKKEIMK 515
Query: 817 -LNKATRKDHFPLPFIDQMLERLAKHSHFCYLDGYSGFFQIPIHPNDQEKTTFTCPFGTF 875
LN ATRK+HFPL FIDQ+LERL+ H ++C LDGYSGFFQIPIHP+DQEKT FTCP+GTF
Sbjct: 516 KLNAATRKNHFPLQFIDQLLERLSNHKYYCVLDGYSGFFQIPIHPDDQEKTMFTCPYGTF 575
Query: 876 AYRRMPFGLCNAPATFQRCMMSIFSDFVEKIMEVFMDDFSVHGSNFDDCLTNLEKVLERC 935
AY RMPFGLCNAPA F+RCMMSIF+D +E +EVFMDDFSV+GS+F+ CL NL KVL RC
Sbjct: 576 AYSRMPFGLCNAPAIFERCMMSIFTDMIENFIEVFMDDFSVYGSSFEACLENLRKVLARC 635
Query: 936 EQVNLVLNWEKCHFMVREGIVLGHLVSDRGIEVDRAKVEIIEKMLPPTSVKEIRSFLGHA 995
E+ NLVLNWEKCHFMV+EGIVLGH VS GIEV++AK+E++ + SV
Sbjct: 636 EEKNLVLNWEKCHFMVQEGIVLGHKVSGAGIEVNKAKIEVMTSLQALDSVN--------- 686
Query: 996 GFYRRFIKDFSSITKPLTNLLLKDADFTFDDSCLQAFCRLKEALITAPIIQPPDWNLPFE 1055
RF+KDFS I +PLT LL KD F F+ C AF ++K AL++API+QP DWNLPFE
Sbjct: 687 ---LRFVKDFSKIARPLTALLCKDVKFDFNSECHDAFNQIKNALVSAPIVQPLDWNLPFE 743
Query: 1056 IMCDASDYAVGVVLGQRKDKKMHAIYYASKTLDGAQVNYATTEKELLAVVYAIDKFRQYL 1115
IMCDA DYAVG VLGQRK K+HAI+YAS+TLD AQ N ATTEKELLAVV+A DKFR YL
Sbjct: 744 IMCDAGDYAVGAVLGQRKGNKLHAIHYASRTLDDAQRNNATTEKELLAVVFAFDKFRSYL 803
Query: 1116 VGSKIIVYTDHSAIKYLLNKKDAKPRLIRWILLLQEFDLEIKDKKGVENVVADHLSRLRE 1175
VGSK+IV+TDHSA+KYL+ KKDAKPRL+RWILLLQEFD+E++D+KG+EN VADHLSR++
Sbjct: 804 VGSKVIVHTDHSALKYLMQKKDAKPRLLRWILLLQEFDIEVRDRKGIENGVADHLSRIKV 863
Query: 1176 ANKDELPLDD 1185
+D++P++D
Sbjct: 864 --EDDIPIND 871
>emb|CAB81136.1| putative athila transposon protein [Arabidopsis thaliana]
gi|25407395|pir||D85075 probable athila transposon
protein [imported] - Arabidopsis thaliana
Length = 724
Score = 630 bits (1624), Expect = e-178
Identities = 328/596 (55%), Positives = 418/596 (70%), Gaps = 46/596 (7%)
Query: 439 AIIQNKL-PPKLKDPGSFSIPCVIGSEVVKKAMCDLGASVSLMPLSLYERLGIGELKSTR 497
AI Q K+ P KL DPGSF++PC +G K+ +CDLGA VSLMPLS+ +RLG + KS
Sbjct: 3 AITQKKIVPKKLIDPGSFTLPCSLGPLAFKRCLCDLGALVSLMPLSVAKRLGFTQYKSCN 62
Query: 498 MTLQLADRSVKYPAGIIEDVPVKVGEVYIPADFVVMEMEEDNQVPILLGRPFLATAGAII 557
++L LADRSV+ P + E++P+++G V IP DFVV+EM+E+ + P++LGRPFLATAGA+
Sbjct: 63 ISLILADRSVRIPHSLFENLPIRIGAVDIPTDFVVLEMDEEPKDPLILGRPFLATAGAMN 122
Query: 558 DVKKGKLAFNVGKET-VEFELAKLMKGPSIKDSCCMIDIIDHCVKECSLASTTHDGLEVC 616
DVKKGK+ N+GK + F++ MK P+IK I+ ID E D L
Sbjct: 123 DVKKGKIDLNLGKYCRMTFDVKDAMKKPTIKGQLFWIEEIDQLADELLEERAEEDHLYSA 182
Query: 617 LVNNAGTK-LEGEAKAYEVLLD------RTPPIKDLSVEE----LIKEEPTLQPK----- 660
L L E Y+ LLD + P ++L+ E ++ EE + Q +
Sbjct: 183 LTKRGEDGFLHLETLGYQKLLDSHKAMEESEPFEELNGPETEVMVMSEEGSTQVQPAHSR 242
Query: 661 ----------------------------EAPKVELKTLPSNLRYEFLGPNSTYPVIVNAS 692
+APKV+LK+LP LRY F GPNSTYPVI+N
Sbjct: 243 TYSTNNSTSTISNSGELIIPTSDDWSELKAPKVDLKSLPKGLRYVFFGPNSTYPVIINVE 302
Query: 693 LDEVETEKLLYVLKKYPKAIGYTIDDIKGINPSLCMHRILLEDDYKPSIEHQRRLNPNMK 752
L++ E LL L+KY +AIGY++ DIK I+PSLC HRI LE++ SIE QRRLN N+K
Sbjct: 303 LNDNEVNLLLSELRKYKRAIGYSLSDIKRISPSLCNHRIHLENESYSSIEPQRRLNLNLK 362
Query: 753 EVVKKEVLKLLDAGVIYPISDSKWVSPVQVVPKKGGLTVIKNDKNESIATRTVTGWRMCI 812
EVVKKE+LKLLDAGVIYPISDS WV PV VPKKGG+TV+KN+K+E I TRT+TG R+CI
Sbjct: 363 EVVKKEILKLLDAGVIYPISDSTWVFPVHCVPKKGGMTVVKNEKDELIPTRTITGHRVCI 422
Query: 813 DYRKLNKATRKDHFPLPFIDQMLERLAKHSHFCYLDGYSGFFQIPIHPNDQEKTTFTCPF 872
DYRKLN A+RKDHFPLPF +QMLE LA H + C+LDGYSGFFQIPIHPNDQEKTTFTCP+
Sbjct: 423 DYRKLNAASRKDHFPLPFTNQMLEGLANHLYNCFLDGYSGFFQIPIHPNDQEKTTFTCPY 482
Query: 873 GTFAYRRMPFGLCNAPATFQRCMMSIFSDFVEKIMEVFMDDFSVHGSNFDDCLTNLEKVL 932
GTFAY+RMPFGLCNAP TFQRCM SIFSD +EK++EVFMDDFSV+G +F CL NL +VL
Sbjct: 483 GTFAYKRMPFGLCNAPTTFQRCMTSIFSDLIEKMVEVFMDDFSVYGPSFSSCLLNLGRVL 542
Query: 933 ERCEQVNLVLNWEKCHFMVREGIVLGHLVSDRGIEVDRAKVEIIEKMLPPTSVKEI 988
+ E+ NLVLNWEKC+FMV+EGIVLGH +S++GIEVD+ K++++ ++ PP +VK+I
Sbjct: 543 TKWEETNLVLNWEKCYFMVKEGIVLGHKISEKGIEVDKEKIKVMMQLQPPKTVKDI 598
Score = 147 bits (371), Expect = 3e-33
Identities = 74/131 (56%), Positives = 100/131 (75%), Gaps = 2/131 (1%)
Query: 1065 VGVVLGQRKDKKMHAIYYASKTLDGAQVNYATTEKELLAVVYAIDKFRQYLVGSKIIVYT 1124
+ V++ + K + IYYAS+TLD AQ YATTEKELL VV+A KFR YLV SK+ VYT
Sbjct: 583 IKVMMQLQPPKTVKDIYYASRTLDEAQGRYATTEKELLVVVFAFKKFRSYLVESKVTVYT 642
Query: 1125 DHSAIKYLLNKKDAKPRLIRWILLLQEFDLEIKDKKGVENVVADHLSRLREANKDELPLD 1184
DH+A++++ KKD KPRL+R ILLLQEFD+EI +KKG+EN ADHLSR+R ++ + +D
Sbjct: 643 DHAALRHMYAKKDTKPRLLRGILLLQEFDMEIVEKKGIENGAADHLSRMR--IEEPILID 700
Query: 1185 DSFPDDQLFLL 1195
DS P++QL ++
Sbjct: 701 DSMPEEQLMVV 711
>emb|CAD39882.2| OSJNBb0067G11.5 [Oryza sativa (japonica cultivar-group)]
gi|50922247|ref|XP_471484.1| OSJNBb0067G11.5 [Oryza
sativa (japonica cultivar-group)]
Length = 791
Score = 596 bits (1537), Expect = e-168
Identities = 294/458 (64%), Positives = 354/458 (77%), Gaps = 19/458 (4%)
Query: 639 TPPIKDL-SVEELIKEEPTLQPK-----EAPKVELKTLPSNLRYEFLGPNSTYPVIVNAS 692
T P++++ S EE+ P P+ E P + LRY+FL + PVI++
Sbjct: 347 TDPLQEIESAEEVTAVPPHESPEALLEDEVPDFIKEEADPGLRYDFLHNDREAPVIISDK 406
Query: 693 LDEVETEKLLYVLKKYPKAIGYTIDDIKGINPSLCMHRILLEDDYKPSIEHQRRLNPNMK 752
L E ET+ LL VL+K+ +GY++ D++GINP+LC HRIL++ + PS E QR LN M+
Sbjct: 407 LSEDETQHLLTVLEKHRSILGYSLQDLRGINPALCTHRILIDPESTPSREPQRWLNNAMR 466
Query: 753 EVVKKEVLKLLDAGVIYPISDSKWVSPVQVVPKKGGLTVIKNDKNESIATRTVTGWRMCI 812
EVVKKEVLKLL AG+IYPI S+WVSPVQVVPKKGG+TV+ WRMCI
Sbjct: 467 EVVKKEVLKLLHAGIIYPIPYSEWVSPVQVVPKKGGMTVVYR-------------WRMCI 513
Query: 813 DYRKLNKATRKDHFPLPFIDQMLERLAKHSHFCYLDGYSGFFQIPIHPNDQEKTTFTCPF 872
DYRKLNKAT+KDHFPLPFID+MLERLA HS FC+LDGYSG+ QIPIHP DQ KTTFTCP+
Sbjct: 514 DYRKLNKATKKDHFPLPFIDEMLERLANHSFFCFLDGYSGYHQIPIHPEDQSKTTFTCPY 573
Query: 873 GTFAYRRMPFGLCNAPATFQRCMMSIFSDFVEKIMEVFMDDFSVHGSNFDDCLTNLEKVL 932
GT+AYRRM FGLCN PA+FQRCMMSIFSD +E IMEVFMDDFSV+G CL NL+KVL
Sbjct: 574 GTYAYRRMSFGLCNTPASFQRCMMSIFSDMIEDIMEVFMDDFSVYGKTLGHCLQNLDKVL 633
Query: 933 ERCEQVNLVLNWEKCHFMVREGIVLGHLVSDRGIEVDRAKVEIIEKMLPPTSVKEIRSFL 992
+RC++ +LVLN EKCHFMVREGIVLGH VS+RG+EVDRAK+++I+++ PP ++K I SF
Sbjct: 634 QRCQEKDLVLNREKCHFMVREGIVLGHRVSERGVEVDRAKIDVIDQLPPPMNIKGIHSFF 693
Query: 993 GHAGFYRRFIKDFSSITKPLTNLLLKDADFTFDDSCLQAFCRLKEALITAPIIQPPDWNL 1052
GHAGFYRRFIKDFS+I +PLTNLL KDA FDD CL++F LK+AL++APIIQPPDW L
Sbjct: 694 GHAGFYRRFIKDFSTIARPLTNLLAKDAPIEFDDVCLKSFEILKKALVSAPIIQPPDWTL 753
Query: 1053 PFEIMCDASDYAVGVVLGQRKDKKMHAIYYASKTLDGA 1090
PFEIMCDASD+AVG VLGQ KDKK HAI YASKTL GA
Sbjct: 754 PFEIMCDASDFAVGAVLGQTKDKKHHAICYASKTLTGA 791
Score = 71.2 bits (173), Expect = 2e-10
Identities = 40/113 (35%), Positives = 59/113 (51%), Gaps = 3/113 (2%)
Query: 1 NQFSGSPSEDPNLHISNFWRLSVTCKDD---QETVRLHLFPFSLKNKASNWFNSLKPGSI 57
N FSG E+P H+ +F ++ K QETV LFPFSL+ +A W+ S
Sbjct: 43 NPFSGFDLENPYHHLRDFEQVCSCLKIRGMRQETVWWKLFPFSLQERAKQWYTSTVGCVN 102
Query: 58 TSWDQLRREFLCRFFPPSKTAQLRGKLYQFTQMNGESLFDVWERFKEILRRCP 110
SW++LR F FFP ++ LR ++ F Q+ ES+ W RF +++ P
Sbjct: 103 GSWEKLRDRFCLTFFPVTRITALRVEILNFKQIENESIGAAWSRFTNLVQSGP 155
>emb|CAD39877.2| OSJNBb0058J09.16 [Oryza sativa (japonica cultivar-group)]
gi|50922333|ref|XP_471527.1| OSJNBb0058J09.16 [Oryza
sativa (japonica cultivar-group)]
Length = 1045
Score = 588 bits (1515), Expect = e-166
Identities = 293/476 (61%), Positives = 351/476 (73%), Gaps = 22/476 (4%)
Query: 978 KMLPPTSVKEIRSFLGHAGFYRRFIKDFSSITKPLTNLLLKDADFTFDDSCLQAFCRLKE 1037
++ PP ++K I SFLGHAGFYRRFIKDFS+I +PLTNLL KDA F F+D+CL++F LK+
Sbjct: 500 RLPPPVNIKGICSFLGHAGFYRRFIKDFSTIARPLTNLLAKDAPFEFNDACLRSFKILKK 559
Query: 1038 ALITAPIIQPPDWNLPFEIMCDASDYAVGVVLGQRKDKKMHAIYYASKTLDGAQVNYATT 1097
AL++APIIQPPDW LPFEIMCDASD+AVG VLGQ KDKK HAI YASKTL GAQ+NYATT
Sbjct: 560 ALVSAPIIQPPDWTLPFEIMCDASDFAVGAVLGQTKDKKHHAICYASKTLTGAQLNYATT 619
Query: 1098 EKELLAVVYAIDKFRQYLVGSKIIVYTDHSAIKYLLNKKDAKPRLIRWILLLQEFDLEIK 1157
EKELL VV+AIDKFR YLVG+K+I+YTDH+A+KYLL KKDAKPRL+RWILLLQEFD+EIK
Sbjct: 620 EKELLTVVFAIDKFRSYLVGAKVIIYTDHAALKYLLTKKDAKPRLLRWILLLQEFDIEIK 679
Query: 1158 DKKGVENVVADHLSRLREANKDELPLDDSFPDDQLFLLAQTDAPWYADFVNFLAAGVLPP 1217
DKKGVEN VADHLSRL N E P++D DD L +PP
Sbjct: 680 DKKGVENYVADHLSRLHITNMQEQPINDLLRDDMLMT-------------------CIPP 720
Query: 1218 ELNYQQKKKFFNDLKHYYWDEPYLFRRGSDGIFRRCIPESEVSSILTHCHSSSYGGHAST 1277
N Q+K K+ + + WDEPYL+R SDG+ RRC+P E I+ CH+S YGGH
Sbjct: 721 GEN-QRKLKY--ESHRHIWDEPYLYRVCSDGLLRRCVPTDEGLKIIERCHASPYGGHYGA 777
Query: 1278 QKTSFKILHSGFWWPSLFKDVHLFISKCDKCQRTGSITKRNEMPLNNILEVEIFDVWGVD 1337
+T KI SGF+WP++++D FI +C CQR G IT R+ MPL L+VEIFDVWG+D
Sbjct: 778 FRTQAKIWQSGFFWPTMYEDSKEFIRRCTSCQRQGGITARDAMPLTYNLQVEIFDVWGID 837
Query: 1338 FMGPFPSSFGNQYILVAVDYVSKWVEAIASPTNDAQVVIKMFKKVIFPRFGVPRVVISDG 1397
FMGPFP S +YILVAVDYVSKWVEA+ DA+ KMF + IFPRFG R+VISD
Sbjct: 838 FMGPFPKSRNCEYILVAVDYVSKWVEAMPCSAADARHAKKMFTETIFPRFGTRRMVISDR 897
Query: 1398 GSHFISKHFEKLLQKLGVRHKVATPYHPQTSGQVEVSNRQIKAILEKTVSTSRTDW 1453
G HFI K F LL+++ +H VATPYHPQTSGQ E SN+QIK IL+KTV+ T+W
Sbjct: 898 GFHFIDKTFRDLLREMEAKHNVATPYHPQTSGQAETSNKQIKNILQKTVNKMGTEW 953
Score = 114 bits (285), Expect = 2e-23
Identities = 91/296 (30%), Positives = 148/296 (49%), Gaps = 46/296 (15%)
Query: 480 MPLSLYERLGIGELKST--RMTLQLADRSVKYPAGIIEDVPVKVGEVYIPADFVVMEMEE 537
+PLS + +G L+ T +T ++D ++ I++DV V E DF V E+++
Sbjct: 210 VPLSPPNPMELGFLRETVRELTSIMSDEWLRE---IVQDVLVYFEEREAILDFHVFEIQD 266
Query: 538 DN---QVPI---LLGRPFLATAGAIIDVKKGKLAFNVGKETVEFELAKLMKGPSIKDSCC 591
+ ++PI L+ P L + K+ + ++ F A++ ++ D
Sbjct: 267 FDILIRLPIEQLLINTPHLGSL---------KITLGENEFSIPFSRARI----TLTDPLP 313
Query: 592 MIDIIDHCVKECSLASTTHDGLEVCLVNNAGTKLEGEAKAYEVLLDRTPPIKDLSVEELI 651
I++++ A H+ E L + ++ EA E L
Sbjct: 314 EIELVEEVT-----AVPPHESPEALLEDEVPDFIKEEADPGETL---------------- 352
Query: 652 KEEPTLQPKEAPKVELKTLPSNLRYEFLGPNSTYPVIVNASLDEVETEKLLYVLKKYPKA 711
+ P ++P P +ELK LP LRY FL + PVI++ L + ET++L VL+K+
Sbjct: 353 -DLPIMEPPPQPPLELKPLPLGLRYAFLHNDREAPVIISDKLSKDETQRLPTVLEKHRSV 411
Query: 712 IGYTIDDIKGINPSLCMHRILLEDDYKPSIEHQRRLNPNMKEVVKKEVLKLLDAGV 767
+GY++ D++GIN +LC HRI ++ PS E QRRLN M+EVVKKEVLKLL AG+
Sbjct: 412 LGYSLQDLRGINLALCTHRIPIDPKSTPSREPQRRLNNEMQEVVKKEVLKLLHAGI 467
Score = 49.3 bits (116), Expect = 0.001
Identities = 29/132 (21%), Positives = 63/132 (46%), Gaps = 6/132 (4%)
Query: 75 SKTAQLRGKLYQFTQMNGESLFDVWERFKEILRRCPHHGLEKWLIIHIFYNGLTSSTKLT 134
++ R ++ F Q+ ES+ W RF +++ P L +++++ F+ GL +
Sbjct: 39 TRITAFRVEILSFKQIKNESIGATWSRFTNLVQSGPTLSLPEYVLLQYFHTGLDKESAFY 98
Query: 135 IDAAAGGALMNKDFTTAYALIENMALNLFQWTEEKETVDPSPSEKEAGINETSSIDYL-- 192
+D GG+ M+K + +++ + N T+ E P P + I E +I+ L
Sbjct: 99 LDITVGGSFMHKTPSEGRTILDRILENTSFMTQPNE---PQPKASLSKIVEPLTIEPLTE 155
Query: 193 -STKLNALSQRL 203
ST +++ +++
Sbjct: 156 PSTSASSIDEKV 167
>pir||T12085 reverse transcriptase homolog - fava bean (fragment)
gi|2522228|dbj|BAA22787.1| reverse transcriptase-like
protein [Vicia faba]
Length = 407
Score = 583 bits (1504), Expect = e-164
Identities = 274/396 (69%), Positives = 329/396 (82%), Gaps = 4/396 (1%)
Query: 810 MCIDYRKLNKATRKDHFPLPFIDQMLERLAKHSHFCYLDGYSGFFQIPIHPNDQEKTTFT 869
+CIDYR+LN ATRKDHFPLPFIDQMLERLA H ++C+LDGYSG+ QI + P DQEKTTFT
Sbjct: 1 VCIDYRRLNLATRKDHFPLPFIDQMLERLADHEYYCFLDGYSGYNQIAVVPEDQEKTTFT 60
Query: 870 CPFGTFAYRRMPFGLCNAPATFQRCMMSIFSDFVEKIMEVFMDDFSVHGSNFDDCLTNLE 929
CPFG F+YRR+PFGLCNAPATFQRCM SIF+D +EK MEVFMDDFSV G +FD+CL+NL
Sbjct: 61 CPFGIFSYRRIPFGLCNAPATFQRCMQSIFADMLEKYMEVFMDDFSVFGKSFDNCLSNLA 120
Query: 930 KVLERCEQVNLVLNWEKCHFMVREGIVLGHLVSDRGIEVDRAKVEIIEKMLPPTSVKEIR 989
VLERC++ NL+LNWEKCHFMVREGIVLGH +S +GIEVD+AK+E+I K+ PPT+ K IR
Sbjct: 121 LVLERCQESNLILNWEKCHFMVREGIVLGHKISYKGIEVDQAKIEVISKLHPPTNEKGIR 180
Query: 990 SFLGHAGFYRRFIKDFSSITKPLTNLLLKDADFTFDDSCLQAFCRLKEALITAPIIQPPD 1049
SFLGH GFYRRFI+DFS I KPLT LL+KD DF FD C+ AF LK L++API+ PD
Sbjct: 181 SFLGHVGFYRRFIRDFSKIEKPLTTLLVKDKDFVFDKECVVAFETLKGKLVSAPIVVAPD 240
Query: 1050 WNLPFEIMCDASDYAVGVVLGQRKDKKMHAIYYASKTLDGAQVNYATTEKELLAVVYAID 1109
W LPFEIMCDASD AVG VLGQR++K++H IYYAS L+ AQ+NYATTEKELL +VYA D
Sbjct: 241 WYLPFEIMCDASDIAVGAVLGQRREKRLHVIYYASHVLNPAQMNYATTEKELLVMVYAFD 300
Query: 1110 KFRQYLVGSKIIVYTDHSAIKYLLNKKDAKPRLIRWILLLQEFDLEIKDKKGVENVVADH 1169
KFRQY++GSK+IVYTDH+A+KYL +K+D+KPRL+RWILLLQEFD+EI+DK+G EN VADH
Sbjct: 301 KFRQYMLGSKVIVYTDHAALKYLFSKQDSKPRLLRWILLLQEFDVEIRDKRGCENTVADH 360
Query: 1170 LSRLR--EANKDELPLDDSFPDDQLFLLAQTDAPWY 1203
LSR+ E K + P+ D F D+ +L PW+
Sbjct: 361 LSRMSHIEETKVKQPIKDEFADEH--ILVVIGVPWF 394
>gb|AAV43951.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
Length = 963
Score = 547 bits (1409), Expect = e-153
Identities = 317/708 (44%), Positives = 427/708 (59%), Gaps = 58/708 (8%)
Query: 323 KPLEDPIRRTETDHSEKEIREPPSRETRAEKEEPRVEGNTTPPFKPKIPFPQRFAKSKLD 382
+PL +P T ++++ E PS E EE + T FK D
Sbjct: 309 EPLAEP--STFASSIDEKVPEQPS----VENEEIQTPDCATILFK-----------DGFD 351
Query: 383 EQFKKFMEMMNKIYIDVPFTEVLTQMPTYAKFLKEILSKKRKIEESETVNLTEECSAIIQ 442
E + + +K P + P +FL+E + + I E + E S +I+
Sbjct: 352 EDYGNTLNYFSK---RKPLVPLPPPDPMELRFLRETIRELTTIMSDEWLREAELSSEVIR 408
Query: 443 NKLPPKLKDPGSFSIPCVI-GSEVVKKAMCDLGASVSLMPLSLYERLGIGELKSTRMTLQ 501
P++ IPC + G++V +GA++ + + L + T +
Sbjct: 409 INTAPRI-------IPCHLEGNDVSILYSPTVGANLVSGSFA-FAYLSDKAVTPTNKFFK 460
Query: 502 LADRSVKYPAGIIEDVPVKVGEVYIPADFVVMEMEEDNQVPILLGRPFLATAGAIIDVKK 561
++ GI++DVPV + DF V E+++ IL+G P +
Sbjct: 461 HPSGNIIKRFGIMQDVPVCFEDKEAILDFHVFEIQD---FDILIGLPIEQLLINTPRLDS 517
Query: 562 GKLAFNVGKETVEFELAKLMKGPSIKDSCCMIDIIDHCVKECSLASTTHDGLEVCLVNNA 621
K+ + ++ F A+++ + D I+ + + A H+ E L +
Sbjct: 518 LKITLGETEFSIPFSRARIV----LTDPLPEIESTEEVI-----AVPPHESPEALLEDEV 568
Query: 622 GTKLEGEAKAYEVLLDRTPPIKDLSVEELIKEEPTLQPKEAPKVELKTLPSNLRYEFLGP 681
++ EA E L + P ++P P +ELK LP +L Y FL
Sbjct: 569 PDFIKEEADPGETL-----------------DLPIMEPPPWPPLELKPLPPSLHYAFLHH 611
Query: 682 NSTYPVIVNASLDEVETEKLLYVLKKYPKAIGYTIDDIKGINPSLCMHRILLEDDYKPSI 741
+ PVI++ L E ET++LL VL+K+ +GY++ D+KGINP LC+HRI ++ + PS
Sbjct: 612 DREAPVIISDKLFEDETQRLLAVLEKHCSVLGYSLQDLKGINPVLCIHRIPIDPESTPSR 671
Query: 742 EHQRRLNPNMKEVVKKEVLKLLDAGVIYPISDSKWVSPVQVVPKKGGLTVIKNDKNESIA 801
E QRRLN M+EVVKKEVLKLL AG+IYP+ S+WVSPVQVVPKKGG+ V+ N +NE I
Sbjct: 672 EPQRRLNNAMREVVKKEVLKLLHAGIIYPVPYSEWVSPVQVVPKKGGIMVVANAQNELIP 731
Query: 802 TRTVTGWRMCIDYRKLNKATRKDHFPLPFIDQMLERLAKHSHFCYLDGYSGFFQIPIHPN 861
RTVT WRMCIDYRKLNKAT+KDHFPLPFID+MLERLA HS FC+LDGYSG+ QIPIHP
Sbjct: 732 QRTVTEWRMCIDYRKLNKATKKDHFPLPFIDEMLERLANHSFFCFLDGYSGYHQIPIHPE 791
Query: 862 DQEKTTFTCPFGTFAYRRMPFGLCNAPATFQRCMMSIFSDFVEKIMEVFMDDFSVHGSNF 921
DQ KTTFTCP+GT+AYRRM FGLCNAPA+FQRCMMSIFSD +E IMEVFMDDF V+G F
Sbjct: 792 DQSKTTFTCPYGTYAYRRMSFGLCNAPASFQRCMMSIFSDMIEYIMEVFMDDFLVYGKTF 851
Query: 922 DDCLTNLEKVLERCEQVNLVLNWEKCHFMVREGIVLGHLVSDRGIEVDRAKVEIIEKMLP 981
L NL+KVL+RC++ +LVLNWEKCHFMVREGIVLGH VS+RGIEVDRAK+++I+++ P
Sbjct: 852 GHYLQNLDKVLQRCQEKDLVLNWEKCHFMVREGIVLGHRVSERGIEVDRAKIDVIDQLPP 911
Query: 982 PTSVKEIRSFLGHAGFYRRFIKDFSSITKPLTNLLLKDADFTFDDSCL 1029
P ++K IRSFLGHA FYRRFI+D+S+I +PLTNLL KDA F FDD+CL
Sbjct: 912 PVNIKGIRSFLGHASFYRRFIEDYSTIVRPLTNLLAKDAPFEFDDACL 959
>gb|AAV43847.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
Length = 1005
Score = 542 bits (1396), Expect = e-152
Identities = 315/706 (44%), Positives = 425/706 (59%), Gaps = 58/706 (8%)
Query: 323 KPLEDPIRRTETDHSEKEIREPPSRETRAEKEEPRVEGNTTPPFKPKIPFPQRFAKSKLD 382
+PL +P T ++++ E PS E EE + T FK D
Sbjct: 314 EPLAEP--STFASSIDEKVPEQPS----VENEEIQTPDCATILFK-----------DGFD 356
Query: 383 EQFKKFMEMMNKIYIDVPFTEVLTQMPTYAKFLKEILSKKRKIEESETVNLTEECSAIIQ 442
E + + +K P + P +FL+E + + I E + E S +I+
Sbjct: 357 EDYGNTLNYFSK---RKPLVPLPPPDPMELRFLRETIRELTTIMSDEWLREAELSSEVIR 413
Query: 443 NKLPPKLKDPGSFSIPCVI-GSEVVKKAMCDLGASVSLMPLSLYERLGIGELKSTRMTLQ 501
P++ IPC + G++V +GA++ + + L + T +
Sbjct: 414 INTAPRI-------IPCHLEGNDVSILYSPTVGANLVSGSFA-FAYLSDKAVTPTNKFFK 465
Query: 502 LADRSVKYPAGIIEDVPVKVGEVYIPADFVVMEMEEDNQVPILLGRPFLATAGAIIDVKK 561
++ GI++DVPV + DF V E+++ IL+G P +
Sbjct: 466 HPSGNIIKRFGIMQDVPVCFEDKEAILDFHVFEIQD---FDILIGLPIEQLLINTPRLDS 522
Query: 562 GKLAFNVGKETVEFELAKLMKGPSIKDSCCMIDIIDHCVKECSLASTTHDGLEVCLVNNA 621
K+ + ++ F A+++ + D I+ + + A H+ E L +
Sbjct: 523 LKITLGETEFSIPFSRARIV----LTDPLPEIESTEEVI-----AVPPHESPEALLEDEV 573
Query: 622 GTKLEGEAKAYEVLLDRTPPIKDLSVEELIKEEPTLQPKEAPKVELKTLPSNLRYEFLGP 681
++ EA E L + P ++P P +ELK LP +L Y FL
Sbjct: 574 PDFIKEEADPGETL-----------------DLPIMEPPPWPPLELKPLPPSLHYAFLHH 616
Query: 682 NSTYPVIVNASLDEVETEKLLYVLKKYPKAIGYTIDDIKGINPSLCMHRILLEDDYKPSI 741
+ PVI++ L E ET++LL VL+K+ +GY++ D+KGINP LC+HRI ++ + PS
Sbjct: 617 DREAPVIISDKLFEDETQRLLAVLEKHCSVLGYSLQDLKGINPVLCIHRIPIDPESTPSR 676
Query: 742 EHQRRLNPNMKEVVKKEVLKLLDAGVIYPISDSKWVSPVQVVPKKGGLTVIKNDKNESIA 801
E QRRLN M+EVVKKEVLKLL AG+IYP+ S+WVSPVQVVPKKGG+ V+ N +NE I
Sbjct: 677 EPQRRLNNAMREVVKKEVLKLLHAGIIYPVPYSEWVSPVQVVPKKGGIMVVANAQNELIP 736
Query: 802 TRTVTGWRMCIDYRKLNKATRKDHFPLPFIDQMLERLAKHSHFCYLDGYSGFFQIPIHPN 861
RTVT WRMCIDYRKLNKAT+KDHFPLPFID+MLERLA HS FC+LDGYSG+ QIPIHP
Sbjct: 737 QRTVTEWRMCIDYRKLNKATKKDHFPLPFIDEMLERLANHSFFCFLDGYSGYHQIPIHPE 796
Query: 862 DQEKTTFTCPFGTFAYRRMPFGLCNAPATFQRCMMSIFSDFVEKIMEVFMDDFSVHGSNF 921
DQ KTTFTCP+GT+AYRRM FGLCNAPA+FQRCMMSIFSD +E IMEVFMDDF V+G F
Sbjct: 797 DQSKTTFTCPYGTYAYRRMSFGLCNAPASFQRCMMSIFSDMIEYIMEVFMDDFLVYGKTF 856
Query: 922 DDCLTNLEKVLERCEQVNLVLNWEKCHFMVREGIVLGHLVSDRGIEVDRAKVEIIEKMLP 981
L NL+KVL+RC++ +LVLNWEKCHFMVREGIVLGH VS+RGIEVDRAK+++I+++ P
Sbjct: 857 GHYLQNLDKVLQRCQEKDLVLNWEKCHFMVREGIVLGHRVSERGIEVDRAKIDVIDQLPP 916
Query: 982 PTSVKEIRSFLGHAGFYRRFIKDFSSITKPLTNLLLKDADFTFDDS 1027
P ++K IRSFLGHA FYRRFI+D+S+I +PLTNLL KDA F FDD+
Sbjct: 917 PVNIKGIRSFLGHASFYRRFIEDYSTIVRPLTNLLAKDAPFEFDDA 962
>gb|AAT81690.1| putative reverse transcriptase [Oryza sativa (japonica
cultivar-group)]
Length = 1017
Score = 533 bits (1374), Expect = e-149
Identities = 311/667 (46%), Positives = 404/667 (59%), Gaps = 76/667 (11%)
Query: 409 PTYAKFLKEILSKKRKIEESETVNLTEECSAIIQNKLPPKLKDPGSFSIPCVI-GSEVVK 467
P FL+E + + I E E S +I+ P++ IPC + G++V
Sbjct: 422 PMELGFLRETVRELTSIMSDEWQREAELSSEVIRINTAPRI-------IPCHLEGNDVGI 474
Query: 468 KAMCDLGASVSLMPLSLYERLGIGELKSTRMTLQLADRSVKYPAGIIEDVPVKVGEVYIP 527
+GA++ + + + L + T + +R++ GI++DVPV +
Sbjct: 475 LYSPTIGANLISVSFA-FAYLSDKAVTPTNKFFKHPNRNIIEGFGIVQDVPVFFEDREAI 533
Query: 528 ADFVVMEMEEDNQVPILLGRPFLATAGAIIDVKKGKLAFNVGKETVEFELAKLMKGPSIK 587
DF V E+++ IL+G P I + G L +G+ EF SI
Sbjct: 534 LDFHVFEIQD---FDILIGLPIEQL--LINTSRLGSLKITLGEN--EF---------SIP 577
Query: 588 DSCCMIDIIDHCVKECSLASTT----HDGLEVCLVNNAGTKLEGEAKAYEVLLDRTPPIK 643
S I + D + S T H+ + L + ++ EA E L
Sbjct: 578 FSRARITLTDPLPETESAEEVTAVPPHESTKALLEDEVPDFIKEEADPSETL-------- 629
Query: 644 DLSVEELIKEEPTLQPKEAPKVELKTLPSNLRYEFLGPNSTYPVIVNASLDEVETEKLLY 703
+ P ++P P +ELK LP LRY FL + PVI++ L E ET++LL
Sbjct: 630 ---------DLPIMEPPPRPPLELKPLPPGLRYAFLHNDREAPVIISDKLSEDETQRLLT 680
Query: 704 VLKKYPKAIGYTIDDIKGINPSLCMHRILLEDDYKPSIEHQRRLNPNMKEVVKKEVLKLL 763
VL+K+ +GY++ D++GINP+LC HRI ++ + PS E QRRLN M+EVVKKEVLKLL
Sbjct: 681 VLEKHRSILGYSLQDLRGINPALCTHRIPIDLESTPSREPQRRLNNAMREVVKKEVLKLL 740
Query: 764 DAGVIYPISDSKWVSPVQVVPKKGGLTVIKNDKNESIATRTVTGWRMCIDYRKLNKATRK 823
AG+IYP+ S+WVS VQV+PKKGG+TV+ N +NE I RTVTGWRMCIDYRKLNKAT+K
Sbjct: 741 HAGIIYPVPYSEWVSKVQVMPKKGGMTVVANSQNELIPQRTVTGWRMCIDYRKLNKATKK 800
Query: 824 DHFPLPFIDQMLERLAKHSHFCYLDGYSGFFQIPIHPNDQEKTTFTCPFGTFAYRRMPFG 883
DHF LPF D+MLE LA HS FC+LDG M FG
Sbjct: 801 DHFLLPFNDEMLEWLANHSFFCFLDG------------------------------MSFG 830
Query: 884 LCNAPATFQRCMMSIFSDFVEKIMEVFMDDFSVHGSNFDDCLTNLEKVLERCEQVNLVLN 943
LCNAPA FQRCMMSIFSD +E IMEVFMDDFSV+G F CL NL+KVL+RC++ +LVLN
Sbjct: 831 LCNAPAPFQRCMMSIFSDMIEDIMEVFMDDFSVYGKTFGHCLQNLDKVLQRCQEKDLVLN 890
Query: 944 WEKCHFMVREGIVLGHLVSDRGIEVDRAKVEIIEKMLPPTSVKEIRSFLGHAGFYRRFIK 1003
WEKCHFMVREGI+LGH VS+RG EVDRAK+++I+++ P ++K I SFLGHAGFYRRFIK
Sbjct: 891 WEKCHFMVREGIILGHRVSERGSEVDRAKIDVIDQLPPLVNIKGIHSFLGHAGFYRRFIK 950
Query: 1004 DFSSITKPLTNLLLKDADFTFDDSCLQAFCRLKEALITAPIIQPPDWNLPFEIMCDASDY 1063
DFS I +PLTNLL KDA F FDD+CL++F LK+AL+ APIIQPPDW LPFEIMCDASD+
Sbjct: 951 DFSIIARPLTNLLAKDAPFEFDDACLKSFEILKKALVFAPIIQPPDWTLPFEIMCDASDF 1010
Query: 1064 AVGVVLG 1070
AV VLG
Sbjct: 1011 AVAAVLG 1017
Score = 38.9 bits (89), Expect = 1.3
Identities = 29/146 (19%), Positives = 62/146 (41%), Gaps = 10/146 (6%)
Query: 89 QMNGESLFDVWERFKEILRRCPHHGLEKWLIIHIFYNGLTSSTKLTIDAAAGGALMNKDF 148
++ ES+ W RF +++ P L +++++ F+ GL + +D A G+ M+K
Sbjct: 258 RIENESIGAAWSRFTNLVQSGPTLSLPEYVLLQHFHTGLDKESAFYLDITARGSFMHKTP 317
Query: 149 TTAYALIENMALNLFQWTEEKETVDPSPSEKEAGINETSSIDYL---STKLNALSQRLDR 205
+ +++ + N T+ E P + I E +I+ ST +++ +++
Sbjct: 318 SEGKTILDRILENTSFMTQSNE---PQLEASVSKIEEPLTIEPFAEPSTSASSIDEKVPE 374
Query: 206 M----STPTFSPPCETCGLSSHSDTD 227
+ +P C T D D
Sbjct: 375 QPSVKNEEIQTPDCATIMFRDGFDVD 400
>emb|CAB81130.1| AT4g07600 [Arabidopsis thaliana] gi|5724765|gb|AAD48069.1| contains
similarity to Pfam family PF00078 -943 Reverse
transcriptase (RNA-dependent DNA polymerase); score 65.8,
E=9.4e-16, N=1; may be a pseudogene [Arabidopsis
thaliana] gi|25407387|pir||F85074 hypothetical protein
AT4g07600 [imported] - Arabidopsis thaliana
Length = 630
Score = 474 bits (1221), Expect = e-132
Identities = 221/325 (68%), Positives = 262/325 (80%), Gaps = 18/325 (5%)
Query: 777 VSPVQVVPKKGGLTVIKNDKNESIATRTVTGWRMCIDYRKLNKATRKDHFPLPFIDQMLE 836
VSPVQ +PKKGG+TVIKN+K+E I RT+TG RMCIDYR LN A+R DHFPLPF DQMLE
Sbjct: 324 VSPVQCIPKKGGITVIKNEKDELIPIRTITGLRMCIDYRNLNAASRNDHFPLPFTDQMLE 383
Query: 837 RLAKHSHFCYLDGYSGFFQIPIHPNDQEKTTFTCPFGTFAYRRMPFGLCNAPATFQRCMM 896
RLA H ++C+LDGYSGFFQIPIHPND EKTTFTCP+GTFAY RMPFGLCNAPATFQRCM
Sbjct: 384 RLANHPYYCFLDGYSGFFQIPIHPNDHEKTTFTCPYGTFAYERMPFGLCNAPATFQRCMT 443
Query: 897 SIFSDFVEKIMEVFMDDFSVHGSNFDDCLTNLEKVLERCEQVNLVLNWEKCHFMVREGIV 956
SIFSD +E+++EVFMDDFSV+G +F CL NL +VL RCE+ NLVLNWEKCHFMV+EGI+
Sbjct: 444 SIFSDIIEEMVEVFMDDFSVYGPSFSSCLLNLGRVLTRCEETNLVLNWEKCHFMVKEGIM 503
Query: 957 LGHLVSDRGIEVDRAKVEIIEKMLPPTSVKEIRSFLGHAGFYRRFIKDFSSITKPLTNLL 1016
LGH +S++GIEVD+ FLGHAGFYRRFIKDFS I +PLT LL
Sbjct: 504 LGHKISEKGIEVDKG------------------CFLGHAGFYRRFIKDFSKIARPLTRLL 545
Query: 1017 LKDADFTFDDSCLQAFCRLKEALITAPIIQPPDWNLPFEIMCDASDYAVGVVLGQRKDKK 1076
K+ +F FD+ C ++F +KEAL++AP+++ +W+ PFEIMCDA DYAVG VLGQ+ DKK
Sbjct: 546 CKETEFEFDEDCPKSFHTIKEALVSAPVVRATNWDYPFEIMCDALDYAVGAVLGQKIDKK 605
Query: 1077 MHAIYYASKTLDGAQVNYATTEKEL 1101
+H IYYAS+TLD AQ YATTEKEL
Sbjct: 606 LHVIYYASRTLDEAQGRYATTEKEL 630
Score = 177 bits (448), Expect = 3e-42
Identities = 103/260 (39%), Positives = 161/260 (61%), Gaps = 18/260 (6%)
Query: 388 FMEMMNKIYIDVPFTEVLTQMPTYAKFLKEILSKKRKIEESETVNLTEECSAIIQNKL-P 446
F + + ++ + +P + L + KFLK+++ ++ + E V L+ ECSAIIQ K+ P
Sbjct: 2 FAKNIKEVELRIPLVDALALILDTHKFLKDLIVERIQ-EVQGMVVLSHECSAIIQKKIVP 60
Query: 447 PKLKDPGSFSIPCVIGSEVVKKAMCDLGASVSLMPLSLYERLGIGELKSTRMTLQLADRS 506
KL DPGSF++PC +G+ + +CDLGA VS MPLS+ +RLG + KS ++L LADRS
Sbjct: 61 KKLSDPGSFTLPCFLGTVAFNRCLCDLGALVSPMPLSIAKRLGFTQYKSCNISLILADRS 120
Query: 507 VKYPAGIIEDVPVKVGEVYIPADFVVMEMEEDNQVPILLGRPFLATAGAIIDVKKGKLAF 566
V+ G+++++P+++G I DFV++EM+E+ + P++L RPFLATAGA+IDVKKGK+
Sbjct: 121 VRISHGLLKNLPIRIGAAEISTDFVILEMDEEPKDPLILRRPFLATAGAMIDVKKGKIDL 180
Query: 567 NVGKE-TVEFELAKLMKGPSIKDSCCMIDIIDHCVKEC--SLASTTHDGLEVCLVNNAGT 623
N+GK+ + F++ MK P+I+ I+ +D E LA H +N+A T
Sbjct: 181 NLGKDFRMTFDIKDAMKKPNIEGQLFWIEEMDQLADELLEELAQEDH-------LNSALT 233
Query: 624 K------LEGEAKAYEVLLD 637
K L + Y+ LLD
Sbjct: 234 KTGQDGFLHLKVLGYQKLLD 253
>gb|AAM15221.1| putative retroelement pol polyprotein [Arabidopsis thaliana]
gi|25411366|pir||D84493 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana
Length = 622
Score = 466 bits (1200), Expect = e-129
Identities = 230/382 (60%), Positives = 288/382 (75%), Gaps = 27/382 (7%)
Query: 863 QEKTTFTCPFGTFAYRRMPFGLCNAPATFQRCMMSIFSDFVEKIMEVFMDDFSVHGSNFD 922
++KTTFTCP GTFAY+RM FGLCNAP TFQR M SIFSDF+E+IMEVFMDDFS F
Sbjct: 163 KKKTTFTCPNGTFAYKRMSFGLCNAPGTFQRSMTSIFSDFIEEIMEVFMDDFS----GFS 218
Query: 923 DCLTNLEKVLERCEQVNLVLNWEKCHFMVREGIVLGHLVSDRGIEVDRAKVEIIEKMLPP 982
CL NL +VLERCE+ NLVLNWEKCHFMV EGIVLGH +S +GIEVD+AK++++ ++ PP
Sbjct: 219 SCLLNLCRVLERCEETNLVLNWEKCHFMVHEGIVLGHKISGKGIEVDKAKIDVMIQLQPP 278
Query: 983 TSVKEIRSFLGHAGFYRRFIKDFSSITKPLTNLLLKDADFTFDDSCLQAFCRLKEALITA 1042
+VK+IRSFLGHA FYRRFIKDFS I +PLT LL K+ +F FD+ CL+AF +KE L++A
Sbjct: 279 KTVKDIRSFLGHAEFYRRFIKDFSKIARPLTRLLCKETEFNFDEDCLKAFHLIKETLVSA 338
Query: 1043 PIIQPPDWNLPFEIMCDASDYAVGVVLGQRKDKKMHAIYYASKTLDGAQVNYATTEKELL 1102
PI Q P+W+ PFEIMCDA DYAVG VL Q+ D K+H IYYAS+T+D AQ YAT EKELL
Sbjct: 339 PIFQAPNWDHPFEIMCDAFDYAVGAVLDQKIDDKLHVIYYASRTMDEAQTRYATIEKELL 398
Query: 1103 AVVYAIDKFRQYLVGSKIIVYTDHSAIKYLLNKKDAKPRLIRWILLLQEFDLEIKDKKGV 1162
AVV+A +K R YLVG K+ VY DH+A++++ KK+ K RL+RWILLLQEFD+EI DKKGV
Sbjct: 399 AVVFAFEKIRSYLVGFKVKVYIDHAALRHIYAKKETKSRLLRWILLLQEFDMEIIDKKGV 458
Query: 1163 ENVVADHLSRLREANKDELPLDDSFPDDQLFL-------------------LAQTDAPWY 1203
EN VADHLSR+R +D +P+DD+ P++QL + + PWY
Sbjct: 459 ENGVADHLSRMR--IEDSVPIDDTMPEEQLMFYDLVNKSFDTKDMPEEADAVEEEKLPWY 516
Query: 1204 ADFVNFLAAGVLPPELNYQQKK 1225
AD VN+L +G + E++ +Q K
Sbjct: 517 ADLVNYLTSGQV--EVSNRQMK 536
Score = 61.2 bits (147), Expect = 2e-07
Identities = 28/52 (53%), Positives = 36/52 (68%)
Query: 657 LQPKEAPKVELKTLPSNLRYEFLGPNSTYPVIVNASLDEVETEKLLYVLKKY 708
L +APK++LK LP LRY FLGPN TYPVI+N L + + +LL L+KY
Sbjct: 95 LSELKAPKLDLKPLPKGLRYAFLGPNDTYPVIINDGLSDEQVNQLLNELRKY 146
Score = 42.7 bits (99), Expect = 0.090
Identities = 20/28 (71%), Positives = 22/28 (78%)
Query: 1427 TSGQVEVSNRQIKAILEKTVSTSRTDWS 1454
TSGQVEVSNRQ+K IL K V S+ DWS
Sbjct: 524 TSGQVEVSNRQMKDILTKVVGVSKRDWS 551
>emb|CAD40178.2| OSJNBa0061A09.17 [Oryza sativa (japonica cultivar-group)]
gi|50921885|ref|XP_471303.1| OSJNBa0061A09.17 [Oryza
sativa (japonica cultivar-group)]
Length = 430
Score = 439 bits (1130), Expect = e-121
Identities = 216/397 (54%), Positives = 274/397 (68%), Gaps = 15/397 (3%)
Query: 1057 MCDASDYAVGVVLGQRKDKKMHAIYYASKTLDGAQVNYATTEKELLAVVYAIDKFRQYLV 1116
MCDAS++AVG VLGQ KD+K HAI YASKTL GAQ+NY+TTEKELLAVV
Sbjct: 1 MCDASNFAVGAVLGQTKDRKQHAIAYASKTLTGAQLNYSTTEKELLAVV----------- 49
Query: 1117 GSKIIVYTDHSAIKYLLNKKDAKPRLIRWILLLQEFDLEIKDKKGVENVVADHLSRLREA 1176
G+K++VYTDH+A+KYLL KKDAKPRLIRWIL+LQEFD+EI +KKG+EN VADHLSR++
Sbjct: 50 GAKVVVYTDHAALKYLLTKKDAKPRLIRWILILQEFDIEINNKKGIENSVADHLSRMQIT 109
Query: 1177 NKDELPLDDSFPDDQLFLLAQTDAPWYADFVNFLAAGVLPPELNYQQKKKFFNDLKHYYW 1236
N EL ++D DD L + +D PWYA VNF+ A +PP N KK+ + + W
Sbjct: 110 NMQELSINDYLRDDMLLKVTDSD-PWYAPIVNFMVAWHVPPGEN---KKRLSYESRKRLW 165
Query: 1237 DEPYLFRRGSDGIFRRCIPESEVSSILTHCHSSSYGGHASTQKTSFKILHSGFWWPSLFK 1296
D PYL+R SD + RRC+P E I+ C + YG H +T KI SGF+WP+++
Sbjct: 166 DAPYLYRVCSDSLLRRCVPADEGMEIIEKCRVAPYGSHYGAFRTHAKIWQSGFFWPTMYG 225
Query: 1297 DVHLFISKCDKCQRTGSITKRNEMPLNNILEVEIFDVWGVDFMGPFPSSFGNQYILVAVD 1356
D FI +C CQ+ G IT ++ MPL L+VE+FDVWG+DFMGPFP + +YILVAVD
Sbjct: 226 DTKEFIRRCTSCQKHGGITAQDAMPLTYNLQVELFDVWGIDFMGPFPKFYDCEYILVAVD 285
Query: 1357 YVSKWVEAIASPTNDAQVVIKMFKKVIFPRFGVPRVVISDGGSHFISKHFEKLLQKLGVR 1416
YVSKWVEA+ + + +MF ++IFP FG PR VISDGGSHFI K F+ LQ+LG +
Sbjct: 286 YVSKWVEAMPCRAANTKHAHRMFDEIIFPCFGTPRTVISDGGSHFIDKTFQNFLQELGAQ 345
Query: 1417 HKVATPYHPQTSGQVEVSNRQIKAILEKTVSTSRTDW 1453
H + TPYHP+T GQ E N+QI IL+KTV+ W
Sbjct: 346 HNIVTPYHPKTGGQAETRNKQIMNILQKTVNEMGKAW 382
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.137 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,490,898,573
Number of Sequences: 2540612
Number of extensions: 108762963
Number of successful extensions: 364433
Number of sequences better than 10.0: 37230
Number of HSP's better than 10.0 without gapping: 4775
Number of HSP's successfully gapped in prelim test: 32455
Number of HSP's that attempted gapping in prelim test: 311541
Number of HSP's gapped (non-prelim): 45450
length of query: 1454
length of database: 863,360,394
effective HSP length: 141
effective length of query: 1313
effective length of database: 505,134,102
effective search space: 663241075926
effective search space used: 663241075926
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 82 (36.2 bits)
Medicago: description of AC135959.8


