

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135801.7 - phase: 0
(189 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
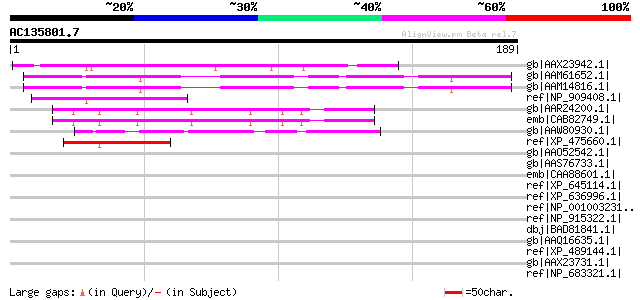
Score E
Sequences producing significant alignments: (bits) Value
gb|AAX23942.1| hypothetical protein At5g57400 [Arabidopsis thali... 79 6e-14
gb|AAM61652.1| unknown [Arabidopsis thaliana] 59 5e-08
gb|AAM14816.1| Expressed protein [Arabidopsis thaliana] gi|18405... 59 5e-08
ref|NP_909408.1| unknown protein [Oryza sativa (japonica cultiva... 53 4e-06
gb|AAR24200.1| At5g01790 [Arabidopsis thaliana] gi|40824035|gb|A... 49 7e-05
emb|CAB82749.1| putative protein [Arabidopsis thaliana] gi|15241... 49 7e-05
gb|AAW80930.1| unknown [Astragalus membranaceus] 47 2e-04
ref|XP_475660.1| unknown protein [Oryza sativa (japonica cultiva... 45 8e-04
gb|AAO52542.1| similar to PH (pleckstrin homology) domain [Caeno... 44 0.003
gb|AAS76733.1| At3g56260 [Arabidopsis thaliana] gi|44681332|gb|A... 43 0.005
emb|CAA88601.1| Hypothetical protein F59B10.2 [Caenorhabditis el... 41 0.015
ref|XP_645114.1| hypothetical protein DDB0168851 [Dictyostelium ... 39 0.057
ref|XP_636996.1| hypothetical protein DDB0187577 [Dictyostelium ... 39 0.057
ref|NP_001003231.1| pinin [Canis familiaris] gi|1684845|gb|AAB48... 39 0.074
ref|NP_915322.1| B1088C09.23 [Oryza sativa (japonica cultivar-gr... 39 0.074
dbj|BAD81841.1| unknown protein [Oryza sativa (japonica cultivar... 39 0.074
gb|AAQ16635.1| vitellogenin [Rivulus marmoratus] 39 0.074
ref|XP_489144.1| PREDICTED: hypothetical protein XP_489144 [Mus ... 39 0.074
gb|AAX23731.1| hypothetical protein At1g06930 [Arabidopsis thali... 38 0.13
ref|NP_683321.1| hydroxyproline-rich glycoprotein family protein... 38 0.17
>gb|AAX23942.1| hypothetical protein At5g57400 [Arabidopsis thaliana]
Length = 231
Score = 79.0 bits (193), Expect = 6e-14
Identities = 62/164 (37%), Positives = 88/164 (52%), Gaps = 25/164 (15%)
Query: 2 DLFEIESNKILSRRSSVGCSSRISYY---RS-GEGVPFKWEMQPGIAKESSPKEVLPPLT 57
DL + ES +SR SSVG SSR+ YY RS EGVPFKWEMQPG P+E + P+T
Sbjct: 55 DLCKRESE--ISRSSSVGVSSRLFYYYHHRSLNEGVPFKWEMQPGTPINPPPEENVRPIT 112
Query: 58 PPPKNLSLGLPKPSILEL---KKPASTMSKLKFWKKRVVKIK--SKKPQEVCFHED---- 108
PPP LSLG P+PS+ + K + + +K W+ + ++ + S+ + ++D
Sbjct: 113 PPPAFLSLGFPEPSVSVVGNSNKHSLFPANIKLWRWKNLRKRYFSRWSSQSMLNKDNNNA 172
Query: 109 -------FDILSRLDCSSDSESMVSPRGSSFSSSSSSMSLMKST 145
D L R + +D S S SS SSSS L+K++
Sbjct: 173 RDGNSNSCDDLERFEGFNDYRSSSS---SSSSSSSKDRRLIKAS 213
>gb|AAM61652.1| unknown [Arabidopsis thaliana]
Length = 190
Score = 59.3 bits (142), Expect = 5e-08
Identities = 62/190 (32%), Positives = 78/190 (40%), Gaps = 35/190 (18%)
Query: 6 IESNKILSRRSSVGCSSRISYYRSGEGVPFKWEMQPGIAKES--SPKEVLPPLTPPPKNL 63
I S I+ S V SSRI YY G VPF WE +PG K + S LPPLTPPP
Sbjct: 10 ISSKIIVKESSQVNSSSRIYYY-GGASVPFLWETRPGTPKHALFSESLRLPPLTPPPSYY 68
Query: 64 SLGLPKPSILELKKPASTMSKLKFWKKRVVKIKSKKPQEVCFHEDFDILSRLDCSSDSES 123
S +S+ S K K R +K + H +SR S S S
Sbjct: 69 S--------------SSSSSGNKLSKARTIKQTRFVKTLLSRH-----VSRPSFSWSSAS 109
Query: 124 MVSPRGSSFSSSSSSMSLMKSTRSSLHSVCSSCSEGNTKQ------VSRKPSTLGCFPMH 177
S SS+SSSS + R C SCS K+ VS P++ C+
Sbjct: 110 --SSSSSSYSSSSPPSKVEHRPRK-----CYSCSRSYVKEDDEEEIVSSSPTSTLCYKRG 162
Query: 178 ISRVLVSITR 187
S + S+ R
Sbjct: 163 FSSSMGSMKR 172
>gb|AAM14816.1| Expressed protein [Arabidopsis thaliana]
gi|18405403|ref|NP_565934.1| expressed protein
[Arabidopsis thaliana]
Length = 193
Score = 59.3 bits (142), Expect = 5e-08
Identities = 62/190 (32%), Positives = 78/190 (40%), Gaps = 35/190 (18%)
Query: 6 IESNKILSRRSSVGCSSRISYYRSGEGVPFKWEMQPGIAKES--SPKEVLPPLTPPPKNL 63
I S I+ S V SSRI YY G VPF WE +PG K + S LPPLTPPP
Sbjct: 13 ISSKIIVKESSQVNSSSRIYYY-GGASVPFLWETRPGTPKHALFSESLRLPPLTPPPSYY 71
Query: 64 SLGLPKPSILELKKPASTMSKLKFWKKRVVKIKSKKPQEVCFHEDFDILSRLDCSSDSES 123
S +S+ S K K R +K + H +SR S S S
Sbjct: 72 S--------------SSSSSGNKLSKARTIKQTRFVKTLLSRH-----VSRPSFSWSSAS 112
Query: 124 MVSPRGSSFSSSSSSMSLMKSTRSSLHSVCSSCSEGNTKQ------VSRKPSTLGCFPMH 177
S SS+SSSS + R C SCS K+ VS P++ C+
Sbjct: 113 --SSSSSSYSSSSPPSKVEHRPRK-----CYSCSRSYVKEDDEEEIVSSSPTSTLCYKRG 165
Query: 178 ISRVLVSITR 187
S + S+ R
Sbjct: 166 FSSSMGSMKR 175
>ref|NP_909408.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|13486661|dbj|BAB39898.1| contains EST
AU081362(R10239)~unknown protein [Oryza sativa
(japonica cultivar-group)] gi|15528762|dbj|BAB64804.1|
unknown protein [Oryza sativa (japonica
cultivar-group)]
Length = 246
Score = 53.1 bits (126), Expect = 4e-06
Identities = 28/60 (46%), Positives = 33/60 (54%), Gaps = 2/60 (3%)
Query: 9 NKILSRRSSVGCSSRISYY--RSGEGVPFKWEMQPGIAKESSPKEVLPPLTPPPKNLSLG 66
+K+LSR S+ S YY S VPF WE QPG K + VLPPLTPPP + G
Sbjct: 28 SKLLSRESAAAAPSFRVYYGVASAGSVPFLWESQPGTPKNAMSDAVLPPLTPPPSYYTAG 87
>gb|AAR24200.1| At5g01790 [Arabidopsis thaliana] gi|40824035|gb|AAR92330.1|
At5g01790 [Arabidopsis thaliana]
Length = 208
Score = 48.9 bits (115), Expect = 7e-05
Identities = 45/136 (33%), Positives = 60/136 (44%), Gaps = 21/136 (15%)
Query: 17 SVGCSS-RISYYRSGEG-VPFKWEMQPGIAKE-SSPKEVLPPLTPPPKNLSLG---LPKP 70
S C S RI YY G VPF+WE PG K SS LPPLTPPP + S + +
Sbjct: 57 SAACPSFRIYYYDGAAGAVPFEWESHPGTPKHPSSELPTLPPLTPPPSHFSFSGDQIRRR 116
Query: 71 SILELKKPASTMSKLKFW-----KKRVVKIKSKKP----QEVCFHE-DFDILSRLDCSSD 120
S KK + + FW + K+ + P + V E ++D+
Sbjct: 117 SKKSTKKILALIPTRLFWLSGDHNGKAKKLSASSPPSLSERVLIDENEYDLF-----KFQ 171
Query: 121 SESMVSPRGSSFSSSS 136
+E V R SSF SS+
Sbjct: 172 TERNVMRRFSSFDSSA 187
>emb|CAB82749.1| putative protein [Arabidopsis thaliana]
gi|15241064|ref|NP_195799.1| expressed protein
[Arabidopsis thaliana] gi|11283441|pir||T48200
hypothetical protein T20L15.60 - Arabidopsis thaliana
Length = 188
Score = 48.9 bits (115), Expect = 7e-05
Identities = 45/136 (33%), Positives = 60/136 (44%), Gaps = 21/136 (15%)
Query: 17 SVGCSS-RISYYRSGEG-VPFKWEMQPGIAKE-SSPKEVLPPLTPPPKNLSLG---LPKP 70
S C S RI YY G VPF+WE PG K SS LPPLTPPP + S + +
Sbjct: 37 SAACPSFRIYYYDGAAGAVPFEWESHPGTPKHPSSELPTLPPLTPPPSHFSFSGDQIRRR 96
Query: 71 SILELKKPASTMSKLKFW-----KKRVVKIKSKKP----QEVCFHE-DFDILSRLDCSSD 120
S KK + + FW + K+ + P + V E ++D+
Sbjct: 97 SKKSTKKILALIPTRLFWLSGDHNGKAKKLSASSPPSLSERVLIDENEYDLF-----KFQ 151
Query: 121 SESMVSPRGSSFSSSS 136
+E V R SSF SS+
Sbjct: 152 TERNVMRRFSSFDSSA 167
>gb|AAW80930.1| unknown [Astragalus membranaceus]
Length = 139
Score = 47.4 bits (111), Expect = 2e-04
Identities = 41/114 (35%), Positives = 53/114 (45%), Gaps = 14/114 (12%)
Query: 25 SYYRSGEGVPFKWEMQPGIAKESSPKEVLPPLTPPPKNLSLGLPKPSILELKKPASTMSK 84
S+ RSG VPF+WE+QPG+ PK PP +P P N L P PS A + S
Sbjct: 13 SFKRSGS-VPFQWEVQPGV-----PKSYAPPPSPSPTNTKL-RPPPSSPSHGLTAPSRSD 65
Query: 85 LKFWKKRVVKIKSKKPQEVCFHEDFDILSRLDCSSDSESMVSPRGSSFSSSSSS 138
+ R K ++P CF S L SS ++ + G S S SSSS
Sbjct: 66 IGILSPR----KPRRPSRGCFPA---AASFLGSSSKRKADLDFGGGSDSRSSSS 112
>ref|XP_475660.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|50080297|gb|AAT69632.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 227
Score = 45.4 bits (106), Expect = 8e-04
Identities = 23/41 (56%), Positives = 25/41 (60%), Gaps = 1/41 (2%)
Query: 21 SSRISYYRSGEG-VPFKWEMQPGIAKESSPKEVLPPLTPPP 60
S R+ Y + G VPF WE QPG K S VLPPLTPPP
Sbjct: 38 SFRVYYGVASAGSVPFLWESQPGTPKSSPSTAVLPPLTPPP 78
>gb|AAO52542.1| similar to PH (pleckstrin homology) domain [Caenorhabditis elegans]
[Dictyostelium discoideum] gi|66821193|ref|XP_644103.1|
hypothetical protein DDB0217532 [Dictyostelium
discoideum] gi|60472384|gb|EAL70337.1| hypothetical
protein DDB0217532 [Dictyostelium discoideum]
Length = 1165
Score = 43.5 bits (101), Expect = 0.003
Identities = 45/146 (30%), Positives = 62/146 (41%), Gaps = 16/146 (10%)
Query: 49 PKEVLPPL----TPPPKNLSLGLPKPSILELKKPASTMSK--LKFWKKRVVKIKSKKPQE 102
P+ LPP+ TPP +L PS+ + P S + L K+V + S+ P
Sbjct: 841 PQHPLPPIPQSHTPPQHSL------PSVPQSHTPPSPSQQHPLIMQTKKVTRSTSQPPLP 894
Query: 103 VCFHEDFDILSRLDCSSDSESMVSPRGSSFSSSSSSMSLMKSTRSSLHSVCSSCSEGNTK 162
H + SS S S S SS SSSSSS S S+ SS S SS S ++
Sbjct: 895 EQTHSSTTLQPSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSS 954
Query: 163 QVSRKPSTLGCFPMHISRVLVSITRR 188
S S P+ S ++ +RR
Sbjct: 955 SSSSSISN----PLGKSAPTITPSRR 976
>gb|AAS76733.1| At3g56260 [Arabidopsis thaliana] gi|44681332|gb|AAS47606.1|
At3g56260 [Arabidopsis thaliana]
Length = 197
Score = 42.7 bits (99), Expect = 0.005
Identities = 41/129 (31%), Positives = 54/129 (41%), Gaps = 25/129 (19%)
Query: 23 RISYYRSGEGVPFKWEMQPGIAKE--SSPKEVLPPLTPPPKNLSLGLPKPSILELKKPAS 80
R SYY G +PF WE +PG K S + PLTPPP S G IL + S
Sbjct: 15 RSSYY-GGASIPFIWESRPGTPKNHLCSDSSLPLPLTPPPSYYSSG-----ILSTPRTQS 68
Query: 81 TMSKLKFWKKRVVKIKSKKPQEVCFHEDFDILSRLDCSSDSESMVSPRGSSFSSSSSSMS 140
K++SK + + F + S L S+ + S+SS+SSS S
Sbjct: 69 -------------KVRSKLSKFL----SFSMFSDLRRSNHGSKKTASSSFSWSSTSSSSS 111
Query: 141 LMKSTRSSL 149
S SL
Sbjct: 112 FSSSPPHSL 120
>emb|CAA88601.1| Hypothetical protein F59B10.2 [Caenorhabditis elegans]
gi|2496975|sp|Q09950|YSR2_CAEEL Hypothetical protein
F59B10.2 in chromosome II gi|17534561|ref|NP_496263.1|
putative nuclear protein, with 2 coiled coil-4 domains
(2K798) [Caenorhabditis elegans]
Length = 482
Score = 41.2 bits (95), Expect = 0.015
Identities = 43/144 (29%), Positives = 62/144 (42%), Gaps = 14/144 (9%)
Query: 34 PFKWEMQPGIAKESSPK----EVLPPLTP-PPKNLSLGLPKPSILELK--KPASTMSKLK 86
PFK +Q G S EV + P PP+ + P ++E K KPA +K
Sbjct: 143 PFKKAIQSGSESSDSDSIIFDEVFEEVLPSPPRKPAPARTAPIVVEKKIEKPA-----VK 197
Query: 87 FWKKRVVKIKSKKPQEVCFHEDFDILSRLDCSSDSESMVSPRGSSFSSSSSSMSLMKSTR 146
K R K K+ P E F D S + S+ SES S S S S S S + S++
Sbjct: 198 EQKARKKKEKTPTPTESSFESSSDSSSTSESSTSSES--SSSASESESESKSESQVSSSK 255
Query: 147 SSLHSVCSSCSEGNTKQVSRKPST 170
+S SS + G+ + + S+
Sbjct: 256 TSTSKASSSKAYGSDFESEKSSSS 279
>ref|XP_645114.1| hypothetical protein DDB0168851 [Dictyostelium discoideum]
gi|42733633|gb|AAL92651.3| hypothetical protein
[Dictyostelium discoideum] gi|60473228|gb|EAL71175.1|
hypothetical protein DDB0168851 [Dictyostelium
discoideum]
Length = 1126
Score = 39.3 bits (90), Expect = 0.057
Identities = 34/98 (34%), Positives = 45/98 (45%), Gaps = 10/98 (10%)
Query: 68 PKPSILELKKPASTMSKLKFWKKRVVKIKSKKPQEVCFHEDFDILSRLDCSSDSESMVSP 127
P+P I+ELKK +S MS F+ K + + + S SS S S S
Sbjct: 345 PQPPIVELKKTSSWMSS--FFTKVLPSLSTNTSSSSS--------SSSSSSSSSSSSSSS 394
Query: 128 RGSSFSSSSSSMSLMKSTRSSLHSVCSSCSEGNTKQVS 165
SS SSSSSS S S+ SS + +S S T +S
Sbjct: 395 SSSSSSSSSSSSSSSSSSSSSNIATMTSASTTTTSTIS 432
>ref|XP_636996.1| hypothetical protein DDB0187577 [Dictyostelium discoideum]
gi|60465411|gb|EAL63496.1| hypothetical protein
DDB0187577 [Dictyostelium discoideum]
Length = 742
Score = 39.3 bits (90), Expect = 0.057
Identities = 30/78 (38%), Positives = 41/78 (52%), Gaps = 8/78 (10%)
Query: 98 KKPQEVCFHEDFDILSRLDCSSDSESMVSPRGSSFSSS--SSSMSLMKSTRSSL-----H 150
K + VC +F+ ++ L SS S S S SS SSS SSS SL+ S+ SSL
Sbjct: 153 KSVERVCL-SNFNFITSLSSSSPSSSSSSSSSSSSSSSSSSSSSSLLLSSLSSLSSSITS 211
Query: 151 SVCSSCSEGNTKQVSRKP 168
S+ SSCS + ++ P
Sbjct: 212 SITSSCSSSTSTSLTTSP 229
>ref|NP_001003231.1| pinin [Canis familiaris] gi|1684845|gb|AAB48303.1| pinin [Canis
familiaris]
Length = 773
Score = 38.9 bits (89), Expect = 0.074
Identities = 39/149 (26%), Positives = 65/149 (43%), Gaps = 11/149 (7%)
Query: 33 VPFKWEMQPGIAKESSPKEVLPPL-TPPPKNLSLGLPKPSILELKKPASTMSKLKFWKKR 91
+P + QP + +S P+ VL P P+ L L + + + +++ + + K +
Sbjct: 530 LPLPLQPQPQVQAQSQPQAVLQPQPVSQPETLPLAVLQAPVQVIQEQGHLLPERKEFPVE 589
Query: 92 VVKIKSKKPQEVCF-HEDFDILSRL---------DCSSDSESMVSPRGSSFSSSSSSMSL 141
VK+ + V H D ++ + +S S S S SS SSS+SS S
Sbjct: 590 SVKLTEVTVEPVLIVHSDSKTKTKTRSRSRGRARNKTSKSRSRSSSSSSSSSSSTSSSSG 649
Query: 142 MKSTRSSLHSVCSSCSEGNTKQVSRKPST 170
S+ S S SS S +T SR+ S+
Sbjct: 650 GSSSSGSSSSRSSSSSSSSTSGSSRRDSS 678
>ref|NP_915322.1| B1088C09.23 [Oryza sativa (japonica cultivar-group)]
Length = 467
Score = 38.9 bits (89), Expect = 0.074
Identities = 21/50 (42%), Positives = 27/50 (54%), Gaps = 7/50 (14%)
Query: 16 SSVGCSSRISYYRSGEGVPFKWEMQPGIAKESS-----PKEVLPPLTPPP 60
+SV C S R+ VPF WE PG+ K SS +E++PP PPP
Sbjct: 126 ASVRCRREASPLRTQ--VPFSWESSPGVPKRSSACMHMAQEIMPPPKPPP 173
>dbj|BAD81841.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|56202100|dbj|BAD73629.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 386
Score = 38.9 bits (89), Expect = 0.074
Identities = 21/50 (42%), Positives = 27/50 (54%), Gaps = 7/50 (14%)
Query: 16 SSVGCSSRISYYRSGEGVPFKWEMQPGIAKESS-----PKEVLPPLTPPP 60
+SV C S R+ VPF WE PG+ K SS +E++PP PPP
Sbjct: 45 ASVRCRREASPLRTQ--VPFSWESSPGVPKRSSACMHMAQEIMPPPKPPP 92
>gb|AAQ16635.1| vitellogenin [Rivulus marmoratus]
Length = 1711
Score = 38.9 bits (89), Expect = 0.074
Identities = 41/126 (32%), Positives = 54/126 (42%), Gaps = 1/126 (0%)
Query: 45 KESSPKEVLPPLTPPPKNLSLGLPKPSILELKKPASTMSKLKFWKKRVVKIKSKKPQEVC 104
KE + V P L K LS L S +S+ S+ F V S +
Sbjct: 1060 KEDIIESVEPVLMKLNKILSSRLNSSSSSSSSSRSSSSSR-SFKTSSVSSSSSSRSNRKI 1118
Query: 105 FHEDFDILSRLDCSSDSESMVSPRGSSFSSSSSSMSLMKSTRSSLHSVCSSCSEGNTKQV 164
+ + SS S S S R SS SSSSSS S +S+ SS S SS S ++K+
Sbjct: 1119 INAAAISTNSSSSSSSSSSSSSRRSSSSSSSSSSSSSRRSSSSSSRSSSSSSSSRSSKRS 1178
Query: 165 SRKPST 170
S K S+
Sbjct: 1179 SSKLSS 1184
Score = 32.0 bits (71), Expect = 9.1
Identities = 23/54 (42%), Positives = 26/54 (47%)
Query: 118 SSDSESMVSPRGSSFSSSSSSMSLMKSTRSSLHSVCSSCSEGNTKQVSRKPSTL 171
SS S S S SSSSSS S S RSS S SS S +++ R S L
Sbjct: 1129 SSSSSSSSSSSSRRSSSSSSSSSSSSSRRSSSSSSRSSSSSSSSRSSKRSSSKL 1182
>ref|XP_489144.1| PREDICTED: hypothetical protein XP_489144 [Mus musculus]
Length = 485
Score = 38.9 bits (89), Expect = 0.074
Identities = 25/65 (38%), Positives = 35/65 (53%), Gaps = 2/65 (3%)
Query: 108 DFDILSRLDCSSDSESMVSPRGSSFSSSSSSMSLMKSTRSSLHSVCSSCSEGNTKQVSRK 167
D+ SR CSS S S +S R S S+SS + S ++ S+ S +S S ++ SRK
Sbjct: 3 DYKFSSRRRCSSSSRSSLSSRSSDTSTSSDTSSDTSTSTST--STSTSHSNNSSSNSSRK 60
Query: 168 PSTLG 172
PS G
Sbjct: 61 PSNKG 65
Score = 32.7 bits (73), Expect = 5.3
Identities = 24/61 (39%), Positives = 31/61 (50%)
Query: 113 SRLDCSSDSESMVSPRGSSFSSSSSSMSLMKSTRSSLHSVCSSCSEGNTKQVSRKPSTLG 172
S SS+S+S SPR S+ SSSS + S+ SS S SS S + + S S G
Sbjct: 330 SNTTSSSNSQSNSSPRSSNTDSSSSPGNSHTSSSSSSSSSSSSSSSSSPQLPSPIESFPG 389
Query: 173 C 173
C
Sbjct: 390 C 390
>gb|AAX23731.1| hypothetical protein At1g06930 [Arabidopsis thaliana]
gi|15222240|ref|NP_172175.1| expressed protein
[Arabidopsis thaliana]
Length = 169
Score = 38.1 bits (87), Expect = 0.13
Identities = 39/145 (26%), Positives = 49/145 (32%), Gaps = 14/145 (9%)
Query: 26 YYRSGEGVPFKWEMQPG----IAKESSPK----------EVLPPLTPPPKNLSLGLPKPS 71
Y S VPFKWE QPG ++K SS V PLTPPP
Sbjct: 25 YGGSSAAVPFKWESQPGTPRRLSKRSSSSGFNSDSDFNFPVSAPLTPPPSYFYASPSSTK 84
Query: 72 ILELKKPASTMSKLKFWKKRVVKIKSKKPQEVCFHEDFDILSRLDCSSDSESMVSPRGSS 131
+ KK + S L + V + L D S+ SM GSS
Sbjct: 85 HVSPKKTNTLFSSLLSKNRSVPSSPASSSSSSSSSVPSSPLRTSDLSNRRRSMWFESGSS 144
Query: 132 FSSSSSSMSLMKSTRSSLHSVCSSC 156
S+S S + + SC
Sbjct: 145 LDYGSNSAKSSGCYASLVKVLLRSC 169
>ref|NP_683321.1| hydroxyproline-rich glycoprotein family protein [Arabidopsis
thaliana] gi|5263322|gb|AAD41424.1| F8K7.12 [Arabidopsis
thaliana] gi|45752758|gb|AAS76277.1| At1g21695
[Arabidopsis thaliana] gi|25402937|pir||C86350 protein
F8K7.12 [imported] - Arabidopsis thaliana
Length = 217
Score = 37.7 bits (86), Expect = 0.17
Identities = 33/113 (29%), Positives = 46/113 (40%), Gaps = 10/113 (8%)
Query: 25 SYYRSGEGVPFKWEMQPGIAKESSPKEVLPPLTPPPKNLSLGLPKPSILELKKPASTMSK 84
S+ R G VPFKWE++PG+ K + L PPP N L S P+ + S
Sbjct: 7 SFKRPG-AVPFKWEIRPGVPKTRPGDDDPALLQPPPPNKLPPLKLKSTPPSNSPSPSSSS 65
Query: 85 LKFWKKRVVKIKSKKPQEVCFHEDFDILSRLDCSSDSESMVSPRGSSFSSSSS 137
F + R + P +L SDS+S S + + SSS
Sbjct: 66 SFFSESRSRPVSPFAPPP---------SFKLKSPSDSDSNCSASPTPYFRSSS 109
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.313 0.128 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 313,835,872
Number of Sequences: 2540612
Number of extensions: 12790726
Number of successful extensions: 68046
Number of sequences better than 10.0: 346
Number of HSP's better than 10.0 without gapping: 174
Number of HSP's successfully gapped in prelim test: 176
Number of HSP's that attempted gapping in prelim test: 61109
Number of HSP's gapped (non-prelim): 2595
length of query: 189
length of database: 863,360,394
effective HSP length: 121
effective length of query: 68
effective length of database: 555,946,342
effective search space: 37804351256
effective search space used: 37804351256
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 71 (32.0 bits)
Medicago: description of AC135801.7


