

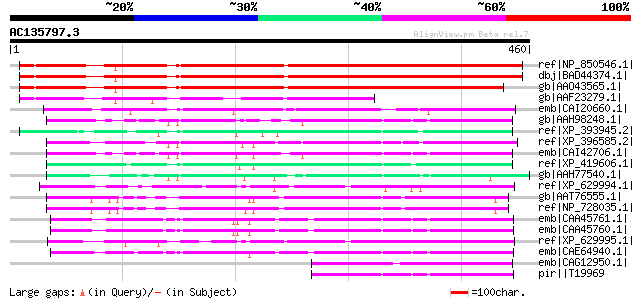
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135797.3 - phase: 0 /pseudo
(460 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_850546.1| expressed protein [Arabidopsis thaliana] gi|519... 412 e-113
dbj|BAD44374.1| unnamed protein product [Arabidopsis thaliana] g... 411 e-113
gb|AAO43565.1| At3g09470/F11F8_4 [Arabidopsis thaliana] gi|15146... 392 e-107
gb|AAF23279.1| hypothetical protein [Arabidopsis thaliana] 218 2e-55
emb|CAI20660.1| novel protein similar to vertebrate unc-93 homol... 114 8e-24
gb|AAH98248.1| Unknown (protein for MGC:119394) [Homo sapiens] 112 3e-23
ref|XP_393945.2| PREDICTED: similar to ENSANGP00000010215 [Apis ... 112 3e-23
ref|XP_396585.2| PREDICTED: similar to CG4928-PB, isoform B [Api... 110 7e-23
emb|CAI42706.1| unc-93 homolog A (C. elegans) [Homo sapiens] 108 3e-22
ref|XP_419606.1| PREDICTED: similar to unc-93 homolog A; unc93 (... 107 6e-22
gb|AAH77540.1| MGC83353 protein [Xenopus laevis] gi|67462040|sp|... 107 8e-22
ref|XP_629994.1| hypothetical protein DDB0184030 [Dictyostelium ... 105 3e-21
gb|AAT76555.1| CG4928 [Drosophila lutescens] 104 5e-21
ref|NP_728035.1| CG4928-PA, isoform A [Drosophila melanogaster] ... 104 7e-21
emb|CAA45761.1| unc-93 [Caenorhabditis elegans] gi|58081858|emb|... 97 1e-18
emb|CAA45760.1| unc-93 [Caenorhabditis elegans] gi|61855544|emb|... 97 1e-18
ref|XP_629995.1| hypothetical protein DDB0184029 [Dictyostelium ... 97 1e-18
emb|CAE64940.1| Hypothetical protein CBG09771 [Caenorhabditis br... 96 3e-18
emb|CAG12950.1| unnamed protein product [Tetraodon nigroviridis] 95 4e-18
pir||T19969 hypothetical protein C46F11.1 - Caenorhabditis elegans 94 9e-18
>ref|NP_850546.1| expressed protein [Arabidopsis thaliana]
gi|51971813|dbj|BAD44571.1| unnamed protein product
[Arabidopsis thaliana]
Length = 437
Score = 412 bits (1059), Expect = e-113
Identities = 237/453 (52%), Positives = 298/453 (65%), Gaps = 34/453 (7%)
Query: 9 DEETPLVVATDESPIQFQQHKSHTRDVHILSIAFLLIFLAYGAAQNLQSTLNTASGELIV 68
DEE PL+ A+ E + + K +TRDVHILSI+FLLIFLAYGAAQNL++T+N G + +
Sbjct: 6 DEEAPLISASGEDR-KVRAGKCYTRDVHILSISFLLIFLAYGAAQNLETTVNKDLGTISL 64
Query: 69 CIEFSGGGFGYDFAWDIIFVFYIF------FCVCFFGGADSRIQECFDCWNFGLLVIPGC 122
I++V ++F V G ++ + W F +
Sbjct: 65 ---------------GILYVSFMFCSMVASLVVRLMGSKNALVLGTTGYWLFVAANLKPS 109
Query: 123 KFEAQLVYVGSGFCLSWVLRFYIMGWAACQGTYLTSTARSHAIDNNFHEGAVIGDFNGEF 182
F + GF S + W QGTYLTS ARSHA D+ HEG+VIG FNGEF
Sbjct: 110 WFTMVPASLYLGFAASII-------WVG-QGTYLTSIARSHATDHGLHEGSVIGVFNGEF 161
Query: 183 WGVYTLHQFIGNLITFALLSDGQEGSTNGTTLLFVVFLSVMTFGAILTCFLHK-RGDYSK 241
W ++ HQ GNLIT ALL DG+EGST+GTTLL +VFL MT G IL F+ K G+ K
Sbjct: 162 WAMFACHQLFGNLITLALLKDGKEGSTSGTTLLMLVFLFSMTLGTILMFFIRKIDGEDGK 221
Query: 242 GGYKHLDAGTGQSKSLKSLCRSLTGALSDVKMLLIIPLIAYSGLQHAFVWAEFTKYVVTP 301
G + + G SL SL R + L D++MLLI+PL+AYSGLQ AFVWAEFTK +VTP
Sbjct: 222 GP---VGSPVGLVDSLASLPRMIITPLLDIRMLLIVPLLAYSGLQQAFVWAEFTKEIVTP 278
Query: 302 EIGVSGVGIAMAVYGAFDGICSLVAGRLTFGLTSITSIVSFGAFVQAVVLILLLLDFSMS 361
IGVSGVG AMAVYGA D +CS+ AGR T GL+SIT IVS GA QA V + LLL + +
Sbjct: 279 AIGVSGVGGAMAVYGALDAVCSMTAGRFTSGLSSITFIVSGGAVAQASVFLWLLLGYRQT 338
Query: 362 SGFIGTLYILFLAALLGIGDGVLMTQLNALLGMLFKHDMEGAFAQLKIWQSATIAMVFFL 421
SG +GT Y L +AA+LGIGDG+L TQ++ALL +LFKHD EGAFAQLK+WQSA IA+VFFL
Sbjct: 339 SGVLGTAYPLIMAAILGIGDGILNTQISALLALLFKHDTEGAFAQLKVWQSAAIAIVFFL 398
Query: 422 APYISFQAVIMVMLTLLCLSFCSFLWLALKVGN 454
+PYIS QA+++VML ++C+S SFL+LALKV N
Sbjct: 399 SPYISLQAMLIVMLVMVCVSLFSFLFLALKVEN 431
>dbj|BAD44374.1| unnamed protein product [Arabidopsis thaliana]
gi|51971130|dbj|BAD44257.1| unnamed protein product
[Arabidopsis thaliana]
Length = 437
Score = 411 bits (1056), Expect = e-113
Identities = 236/453 (52%), Positives = 298/453 (65%), Gaps = 34/453 (7%)
Query: 9 DEETPLVVATDESPIQFQQHKSHTRDVHILSIAFLLIFLAYGAAQNLQSTLNTASGELIV 68
DEE PL+ A+ E + + K +TRDVHILSI+FLLIFLAYGAAQNL++T+N G + +
Sbjct: 6 DEEAPLISASGEDR-KVRAGKCYTRDVHILSISFLLIFLAYGAAQNLETTVNKDLGTISL 64
Query: 69 CIEFSGGGFGYDFAWDIIFVFYIF------FCVCFFGGADSRIQECFDCWNFGLLVIPGC 122
I++V ++F V G ++ + W F +
Sbjct: 65 ---------------GILYVSFMFCSMVASLVVRLMGSKNALVLGTTGYWLFVAANLKPS 109
Query: 123 KFEAQLVYVGSGFCLSWVLRFYIMGWAACQGTYLTSTARSHAIDNNFHEGAVIGDFNGEF 182
F + GF S + W QGTYLTS ARSHA D+ HEG+VIG FNGEF
Sbjct: 110 WFTMVPASLYLGFAASII-------WVG-QGTYLTSIARSHATDHGLHEGSVIGVFNGEF 161
Query: 183 WGVYTLHQFIGNLITFALLSDGQEGSTNGTTLLFVVFLSVMTFGAILTCFLHK-RGDYSK 241
W ++ HQ GNLIT ALL DG+EGST+GTTLL +VFL MT G IL F+ K G+ K
Sbjct: 162 WAMFACHQLFGNLITLALLKDGKEGSTSGTTLLMLVFLFSMTLGTILMFFIRKIDGEDGK 221
Query: 242 GGYKHLDAGTGQSKSLKSLCRSLTGALSDVKMLLIIPLIAYSGLQHAFVWAEFTKYVVTP 301
G + + G SL SL R + L D++MLLI+PL+AYSGLQ AFVWAEFTK +VTP
Sbjct: 222 GP---VGSPVGLVDSLASLPRMIITPLLDIRMLLIVPLLAYSGLQQAFVWAEFTKEIVTP 278
Query: 302 EIGVSGVGIAMAVYGAFDGICSLVAGRLTFGLTSITSIVSFGAFVQAVVLILLLLDFSMS 361
IGVSGVG AMAVYGA D +CS+ AGR T GL+SIT IVS GA QA V + LLL + +
Sbjct: 279 AIGVSGVGGAMAVYGALDAVCSMTAGRFTSGLSSITFIVSGGAVAQASVFLWLLLGYRQT 338
Query: 362 SGFIGTLYILFLAALLGIGDGVLMTQLNALLGMLFKHDMEGAFAQLKIWQSATIAMVFFL 421
SG +GT Y L +AA+LGIGDG+L TQ++ALL +LFKHD EGAFAQL++WQSA IA+VFFL
Sbjct: 339 SGVLGTAYPLIMAAILGIGDGILNTQISALLALLFKHDTEGAFAQLRVWQSAAIAIVFFL 398
Query: 422 APYISFQAVIMVMLTLLCLSFCSFLWLALKVGN 454
+PYIS QA+++VML ++C+S SFL+LALKV N
Sbjct: 399 SPYISLQAMLIVMLVMVCVSLFSFLFLALKVEN 431
>gb|AAO43565.1| At3g09470/F11F8_4 [Arabidopsis thaliana] gi|15146218|gb|AAK83592.1|
AT3g09470/F11F8_4 [Arabidopsis thaliana]
gi|67462074|sp|Q94AA1|UN93L_ARATH UNC93-like protein
gi|30680871|ref|NP_187558.2| expressed protein
[Arabidopsis thaliana]
Length = 464
Score = 392 bits (1006), Expect = e-107
Identities = 226/436 (51%), Positives = 284/436 (64%), Gaps = 34/436 (7%)
Query: 9 DEETPLVVATDESPIQFQQHKSHTRDVHILSIAFLLIFLAYGAAQNLQSTLNTASGELIV 68
DEE PL+ A+ E + + K +TRDVHILSI+FLLIFLAYGAAQNL++T+N G + +
Sbjct: 6 DEEAPLISASGEDR-KVRAGKCYTRDVHILSISFLLIFLAYGAAQNLETTVNKDLGTISL 64
Query: 69 CIEFSGGGFGYDFAWDIIFVFYIF------FCVCFFGGADSRIQECFDCWNFGLLVIPGC 122
I++V ++F V G ++ + W F +
Sbjct: 65 ---------------GILYVSFMFCSMVASLVVRLMGSKNALVLGTTGYWLFVAANLKPS 109
Query: 123 KFEAQLVYVGSGFCLSWVLRFYIMGWAACQGTYLTSTARSHAIDNNFHEGAVIGDFNGEF 182
F + GF S + W QGTYLTS ARSHA D+ HEG+VIG FNGEF
Sbjct: 110 WFTMVPASLYLGFAASII-------WVG-QGTYLTSIARSHATDHGLHEGSVIGVFNGEF 161
Query: 183 WGVYTLHQFIGNLITFALLSDGQEGSTNGTTLLFVVFLSVMTFGAILTCFLHK-RGDYSK 241
W ++ HQ GNLIT ALL DG+EGST+GTTLL +VFL MT G IL F+ K G+ K
Sbjct: 162 WAMFACHQLFGNLITLALLKDGKEGSTSGTTLLMLVFLFSMTLGTILMFFIRKIDGEDGK 221
Query: 242 GGYKHLDAGTGQSKSLKSLCRSLTGALSDVKMLLIIPLIAYSGLQHAFVWAEFTKYVVTP 301
G + + G SL SL R + L D++MLLI+PL+AYSGLQ AFVWAEFTK +VTP
Sbjct: 222 GP---VGSPVGLVDSLASLPRMIITPLLDIRMLLIVPLLAYSGLQQAFVWAEFTKEIVTP 278
Query: 302 EIGVSGVGIAMAVYGAFDGICSLVAGRLTFGLTSITSIVSFGAFVQAVVLILLLLDFSMS 361
IGVSGVG AMAVYGA D +CS+ AGR T GL+SIT IVS GA QA V + LLL + +
Sbjct: 279 AIGVSGVGGAMAVYGALDAVCSMTAGRFTSGLSSITFIVSGGAVAQASVFLWLLLGYRQT 338
Query: 362 SGFIGTLYILFLAALLGIGDGVLMTQLNALLGMLFKHDMEGAFAQLKIWQSATIAMVFFL 421
SG +GT Y L +AA+LGIGDG+L TQ++ALL +LFKHD EGAFAQLK+WQSA IA+VFFL
Sbjct: 339 SGVLGTAYPLIMAAILGIGDGILNTQISALLALLFKHDTEGAFAQLKVWQSAAIAIVFFL 398
Query: 422 APYISFQAVIMVMLTL 437
+PYIS QA+++VML +
Sbjct: 399 SPYISLQAMLIVMLVM 414
>gb|AAF23279.1| hypothetical protein [Arabidopsis thaliana]
Length = 285
Score = 218 bits (556), Expect = 2e-55
Identities = 143/324 (44%), Positives = 182/324 (56%), Gaps = 53/324 (16%)
Query: 9 DEETPLVVATDESPIQFQQHKSHTRDVHILSIAFLLIFLAYGAAQNLQSTLNTASGELIV 68
DEE PL+ A+ E + + K +TRDVHILSI+FLLIFLAYGAAQNL++T+N G +
Sbjct: 6 DEEAPLISASGEDR-KVRAGKCYTRDVHILSISFLLIFLAYGAAQNLETTVNKDLGTI-- 62
Query: 69 CIEFSGGGFGYDFAWDIIFVFYIF------FCVCFFGGADSRIQECFDCWNFGLLVIPGC 122
+ I++V ++F V G ++ + W F +
Sbjct: 63 -------------SLGILYVSFMFCSMVASLVVRLMGSKNALVLGTTGYWLFVAANLKPS 109
Query: 123 KFE--AQLVYVGSGFCLSWVLRFYIMGWAACQGTYLTSTARSHAIDNNFHEGAVIGDFNG 180
F +Y+G + WV QGTYLTS ARSHA D+ HEG+VIG FNG
Sbjct: 110 WFTMVPASLYLGFAASIIWV----------GQGTYLTSIARSHATDHGLHEGSVIGVFNG 159
Query: 181 EFWGVYTLHQFIGNLITFALLSDGQEGSTNGTTLLFVVFLSVMTFGAILTCFLHK-RGDY 239
EFW ++ H QEGST+GTTLL +VFL MT G IL F+ K G+
Sbjct: 160 EFWAMFACH---------------QEGSTSGTTLLMLVFLFSMTLGTILMFFIRKIDGED 204
Query: 240 SKGGYKHLDAGTGQSKSLKSLCRSLTGALSDVKMLLIIPLIAYSGLQHAFVWAEFTKYVV 299
KG + + G SL SL R + L D++MLLI+PL+AYSGLQ AFVWAEFTK +V
Sbjct: 205 GKG---PVGSPVGLVDSLASLPRMIITPLLDIRMLLIVPLLAYSGLQQAFVWAEFTKEIV 261
Query: 300 TPEIGVSGVGIAMAVYGAFDGICS 323
TP IGVSGVG AMAVYGA D + S
Sbjct: 262 TPAIGVSGVGGAMAVYGALDAVVS 285
>emb|CAI20660.1| novel protein similar to vertebrate unc-93 homolog A (C. elegans)
(UNC93A) [Danio rerio] gi|67462027|sp|Q5SPF7|UN93A_BRARE
UNC93 homolog A
Length = 465
Score = 114 bits (284), Expect = 8e-24
Identities = 112/458 (24%), Positives = 186/458 (40%), Gaps = 74/458 (16%)
Query: 31 HTRDVHILSIAFLLIFLAYGAAQNLQSTLNTASGELIVCIEFSGGGFGYDFAWDIIFVFY 90
+T++V ++S FLL+F AYG Q+LQS+LN G ++ + +A I+ +
Sbjct: 4 NTKNVLVVSFGFLLLFTAYGGLQSLQSSLNAEEGMGVISLSVI-------YAAIILSSMF 56
Query: 91 IFFCVCFFGGADSRI---QECFDCWNFGLLVIPGCKFEAQLVYVGSGFCLSWVLRFYIMG 147
+ + G I C+ ++FG L + +G G W
Sbjct: 57 LPPIMIKNLGCKWTIVVSMGCYVAYSFGNLAPGWASLMSTSAILGMGGSPLWS------- 109
Query: 148 WAACQGTYLTSTARSHAIDNNFHEGAVIGDFNGEFWGVYTLHQFIGNLIT---------- 197
A C TYLT + +N +I + G F+ ++ GNL++
Sbjct: 110 -AKC--TYLTISGNRQGQKHNKKGQDLINQYFGIFFFIFQSSGVWGNLMSSLIFGQDQNI 166
Query: 198 -----------------FALLSDGQEGSTNGTTLLFVVFLSVMTFGAILTCFLHKRGDYS 240
F ++ + S + L ++ V I D
Sbjct: 167 VPKENLEFCGVSTCLDNFTVIGNSTRPSKHLVDTLLGCYIGVGLLAIIFVAVFLDNIDRD 226
Query: 241 KGGYKHLDAGTGQSKSLKSLCRSLTGALSDVKMLLIIPLIAYSGLQHAFVWAEFTKYVVT 300
+ K + G +KS + L D ++LL+IPL YSG + +F+ E+TK VT
Sbjct: 227 EA--KEFRSTKG-NKSFWDTFLATFKLLRDPRLLLLIPLTMYSGFEQSFLSGEYTKNYVT 283
Query: 301 PEIGVSGVGIAMAVYGAFDGICSLVAGRLTFGLTSITSIVSFGAFVQAVVLILLLLDFSM 360
+G+ VG M + A + +CS GRL + + L L ++
Sbjct: 284 CALGIHNVGFVMICFAASNSLCSFAFGRL-------------AQYTGRIALFCLAAAINL 330
Query: 361 SSGFIGTLY---------ILFL-AALLGIGDGVLMTQLNALLGMLFKHDMEGAFAQLKIW 410
S F+G LY I F+ AL G+ D V TQ NAL G+LF + E AFA ++W
Sbjct: 331 GS-FLGLLYWKPHPDQLAIFFVFPALWGMADAVWQTQTNALYGILFAKNKEAAFANYRMW 389
Query: 411 QSATIAMVFFLAPYISFQAVIMVMLTLLCLSFCSFLWL 448
+S + F + +I I + L +L L+ ++L++
Sbjct: 390 ESLGFVIAFAYSTFICLSTKIYIALAVLALTMVTYLYV 427
>gb|AAH98248.1| Unknown (protein for MGC:119394) [Homo sapiens]
Length = 415
Score = 112 bits (279), Expect = 3e-23
Identities = 109/424 (25%), Positives = 190/424 (44%), Gaps = 57/424 (13%)
Query: 33 RDVHILSIAFLLIFLAYGAAQNLQSTLNTASGELIVCIEFSGGGFGYDFAWDIIFVFYIF 92
R+V ++S FLL+F AYG Q+LQS+L + G + + GG ++ ++
Sbjct: 6 RNVLVVSFGFLLLFTAYGGLQSLQSSLYSEEGLGVTALSTLYGGM-------LLSSMFL- 57
Query: 93 FCVCFFGGADSRIQECFDCWNFGLLVIPGCKFEAQLVYVGSGFCLSW---VLRFYIMG-- 147
+ E C G +++ C + A V F SW + ++G
Sbjct: 58 ---------PPLLIERLGCK--GTIILSMCGYVAFSV---GNFFASWYTLIPTSILLGLG 103
Query: 148 ----WAACQGTYLTSTARSHAIDNNFHEGAVIGDFNGEFWGVYTLHQFIGNLITFALLSD 203
W+A Q TYLT T +HA ++ + G F+ ++ GNLI+ +
Sbjct: 104 AAPLWSA-QCTYLTITGNTHAEKAGKRGKDMVNQYFGIFFLIFQSSGVWGNLISSLVF-- 160
Query: 204 GQEGSTNGTTLLFVVFLSVMTFGAILTCFLHKRGDYSKGGYKHLDAGTGQSKSLK--SLC 261
GQ S G+ +L V+ ++ FL D + G+ KS+ S
Sbjct: 161 GQTPS-QGSGVLAVLMIAA---------FLQPIRDVQR-------ESEGEKKSVPFWSTL 203
Query: 262 RSLTGALSDVKMLLIIPLIAYSGLQHAFVWAEFTKYVVTPEIGVSGVGIAMAVYGAFDGI 321
S D ++ L+I L YSGLQ F+ +E+T+ VT +G+ VG M + A D +
Sbjct: 204 LSTFKLYRDKRLCLLILLPLYSGLQQGFLSSEYTRSYVTCTLGIQFVGYVMICFSATDAL 263
Query: 322 CSLVAGRLTFGLTSITSIVSFGAFVQAVVLILLLLDFSMSSGFIGTLYILFLAALLGIGD 381
CS++ G+++ T + GA +I LLL + + + ++ + L G+ D
Sbjct: 264 CSVLYGKVS-QYTGRAVLYVLGAVTHVSCMIALLL-WRPRADHLAVFFV--FSGLWGVAD 319
Query: 382 GVLMTQLNALLGMLFKHDMEGAFAQLKIWQSATIAMVFFLAPYISFQAVIMVMLTLLCLS 441
V TQ NAL G+LF+ E AFA ++W++ + F + ++ + ++L +L L+
Sbjct: 320 AVWQTQNNALYGVLFEKSKEAAFANYRLWEALGFVIAFGYSMFLCVHVKLYILLGVLSLT 379
Query: 442 FCSF 445
++
Sbjct: 380 MVAY 383
>ref|XP_393945.2| PREDICTED: similar to ENSANGP00000010215 [Apis mellifera]
Length = 495
Score = 112 bits (279), Expect = 3e-23
Identities = 120/491 (24%), Positives = 197/491 (39%), Gaps = 85/491 (17%)
Query: 9 DEETPLVVATDESPIQFQQHKSHTRDVHILSIAFLLIFLAYGAAQNLQSTLNTASGELIV 68
DE + V T ++ ++ + TR+V ++ +AF++ F A+ A NLQS++N A G L
Sbjct: 2 DEAQFVEVRTKKTGFRYSERWRITRNVLVIGLAFMVNFTAFMGATNLQSSIN-ADGSL-- 58
Query: 69 CIEFSGGGFGYDFAWDIIFVFYIFFCVCFFGGADSRIQECFDCWNFGLLVIPGCKFEAQL 128
G F + + IF + W L ++ F A
Sbjct: 59 ------GTFTLASIYGSLIFSNIFLPTLIISWLGCK-------WTISLSILAYVPFMAAQ 105
Query: 129 VY------------VGSGFCLSWVLRFYIMGWAACQGTYLTSTARSHAIDNNFHEGAVIG 176
Y VG G W A C TYLT A ++A ++ ++
Sbjct: 106 FYPKFYTMIPAGLLVGIGAAPLWC--------AKC--TYLTVVAEAYATLSDVATEILVT 155
Query: 177 DFNGEFWGVYTLHQFIGNLITFA---------LLSDGQEGSTNGTTLLFVVFLSV----- 222
F G F+ Y + Q GNLI+ A LLS G + + TL V V
Sbjct: 156 RFFGLFFMFYQMAQVWGNLISSAGAYILATGTLLSYGIDAAPTNITLNTSVVAEVCGAKF 215
Query: 223 ----------------------MTFGAILTCFLHKR-----GDYSKGGYKHLDAGTGQSK 255
+ G L C + G S Y G+ K
Sbjct: 216 CGATSADNDGSNLERPSEKRIYLISGIYLCCMIMASLIVAFGVDSLTRYNKNRTGSATGK 275
Query: 256 SLKSLCRSLTGALSDVKMLLIIPLIAYSGLQHAFVWAEFTKYVVTPEIGVSGVGIAMAVY 315
S L L + +LI+P+I + G + AF++A++ V+ G++ +G M +
Sbjct: 276 SGLKLLVVTLKLLKEKNQILILPIIMFIGAEQAFLFADYNASFVSCAWGINNIGYVMICF 335
Query: 316 GAFDGICSLVAGRLTFGLTSITSIVSFGAFVQAVVLILLLLDFSMSSGFIGTLYILFL-A 374
G + I +L AG + LT +++F FV VLI LL M +I FL +
Sbjct: 336 GVTNAIAALGAGSI-MKLTGRRPLMAFAFFVHMGVLIFLLHWKPMPE----QNFIFFLIS 390
Query: 375 ALLGIGDGVLMTQLNALLGMLFKHDMEGAFAQLKIWQSATIAMVFFLAPYISFQAVIMVM 434
L G+ D + + Q+NAL G+LF E AF+ ++W+S + + +PY+ + + ++
Sbjct: 391 GLWGLCDAMWLVQINALSGLLFPGKEEAAFSNFRLWESTGSVITYVYSPYLCTEIKLYIL 450
Query: 435 LTLLCLSFCSF 445
+ +LC +
Sbjct: 451 IGILCFGMIGY 461
>ref|XP_396585.2| PREDICTED: similar to CG4928-PB, isoform B [Apis mellifera]
Length = 564
Score = 110 bits (276), Expect = 7e-23
Identities = 112/462 (24%), Positives = 197/462 (42%), Gaps = 75/462 (16%)
Query: 33 RDVHILSIAFLLIFLAYGAAQNLQSTLNTASGELIVCIEFSGGGFGYDFAWDIIFVFYIF 92
+++ +S+AF++ F A+ NLQS++N + G V + I+ +
Sbjct: 89 KNISTVSVAFMVQFTAFQGTANLQSSINASDGLGTVSLS-------------AIYAALVL 135
Query: 93 FCVCFFGGADSRIQECFDCWNFGLLVIPGCKFEAQLVYVGSGFCLSW---VLRFYIMG-- 147
C+ R+ + L P Y+GS F + V ++G
Sbjct: 136 SCIFVPTFVIKRLTVKWTLCISMLCYAP---------YIGSQFYPKFYTLVPAGVLLGLG 186
Query: 148 ----WAACQGTYLTSTARSHAIDNNFHEGAVIGDFNGEFWGVYTLHQFIGNLITFALLSD 203
WAA Q TYLT +A + A++ F G F+ + + GNLI+ +LS
Sbjct: 187 AAPMWAA-QATYLTQVGGVYAKLTDQPVDAIVVRFFGFFFLAWQTAELWGNLISSLVLSG 245
Query: 204 GQ--EGSTNGTTL--------------------------------LFVVFLSVMTFGAIL 229
G+ GS N TT + ++LS + I+
Sbjct: 246 GEFGSGSENSTTNSNKIKHCGANFCVLGNGGHETLERPPESEIYEISAIYLSCVIVAVII 305
Query: 230 TCFLHKRGDYSKGGYKHLDAGTGQSKSLKSLCRSLTGALSDVKMLLIIPLIAYSGLQHAF 289
S+ G K + + ++ L + L L+IP+ + G++ AF
Sbjct: 306 VALFVD--PLSRYGEKQRKVDSQELSGIQ-LLSATAYQLKKPYQQLLIPITIWIGMEQAF 362
Query: 290 VWAEFTKYVVTPEIGVSGVGIAMAVYGAFDGICSLVAGRLTFGLTSITSIVSFGAFVQAV 349
+ A+FT+ ++ +GV VG M +G + ICSLV G L ++ GA V V
Sbjct: 363 IGADFTQAYISCALGVHRVGYVMICFGVVNAICSLVFGSL-MKFVGRQPLMVLGAIVH-V 420
Query: 350 VLILLLLDFSMSSGFIGTLYILF-LAALLGIGDGVLMTQLNALLGMLFKHDMEGAFAQLK 408
LI++LL + + Y+ + ++ L G+GD V TQ+N L G LF+ + E AF+ +
Sbjct: 421 SLIVVLLHWKPNPD---NPYVFYSVSGLWGVGDAVWQTQVNGLYGTLFRRNKEAAFSNYR 477
Query: 409 IWQSATIAMVFFLAPYISFQAVIMVMLTLLCLSFCSFLWLAL 450
+W+SA + + + ++ + + VMLT+L + C ++ + L
Sbjct: 478 LWESAGFVIAYAYSTHLCARMKLYVMLTVLLVGTCGYIVVEL 519
>emb|CAI42706.1| unc-93 homolog A (C. elegans) [Homo sapiens]
Length = 452
Score = 108 bits (271), Expect = 3e-22
Identities = 111/449 (24%), Positives = 189/449 (41%), Gaps = 70/449 (15%)
Query: 33 RDVHILSIAFLLIFLAYGAAQNLQSTLNTASGELIVCIEFSGGGFGYDFAWDIIFVFYIF 92
R+V ++S FLL+F AYG Q+LQS+L + G + + GG ++ ++
Sbjct: 6 RNVLVVSFGFLLLFTAYGGLQSLQSSLYSEEGLGVTALSTLYGGM-------LLSSMFL- 57
Query: 93 FCVCFFGGADSRIQECFDCWNFGLLVIPGCKFEAQLVYVGSGFCLSW---VLRFYIMG-- 147
+ E C G +++ C + A V F SW + ++G
Sbjct: 58 ---------PPLLIERLGCK--GTIILSMCGYVAFSV---GNFFASWYTLIPTSILLGLG 103
Query: 148 ----WAACQGTYLTSTARSHAIDNNFHEGAVIGDFNGEFWGVYTLHQFIGNLITFA---- 199
W+A Q TYLT T +HA ++ + G F+ ++ GNLI+
Sbjct: 104 AAPLWSA-QCTYLTITGNTHAEKAGKRGKDMVNQYFGIFFLIFQSSGVWGNLISSLVFGQ 162
Query: 200 -----------LLSDGQEGSTNGTTL----------LFVVFLSVMTFGAILTCFLHKRGD 238
L S G TT L L + T ++ FL D
Sbjct: 163 TPSQETLPEEQLTSCGASDCLMATTTTNSTQRPSQQLVYTLLGIYTAVLMIAAFLQPIRD 222
Query: 239 YSKGGYKHLDAGTGQSKSLK--SLCRSLTGALSDVKMLLIIPLIAYSGLQHAFVWAEFTK 296
+ G+ KS+ S S D ++ L+I L YSGLQ F+ +E+T+
Sbjct: 223 VQR-------ESEGEKKSVPFWSTLLSTFKLYRDKRLCLLILLPLYSGLQQGFLSSEYTR 275
Query: 297 YVVTPEIGVSGVGIAMAVYGAFDGICSLVAGRLTFGLTSITSIVSFGAFVQAVVLILLLL 356
VT +G+ VG M + A D +CS++ G+++ T + GA +I LLL
Sbjct: 276 SYVTCTLGIQFVGYVMICFSATDALCSVLYGKVS-QYTGRAVLYVLGAVTHVSCMIALLL 334
Query: 357 DFSMSSGFIGTLYILFLAALLGIGDGVLMTQLNALLGMLFKHDMEGAFAQLKIWQSATIA 416
+ + + ++ + L G+ D V TQ NAL G+LF+ E AFA ++W++
Sbjct: 335 -WRPRADHLAVFFV--FSGLWGVADAVWQTQNNALYGVLFEKSKEAAFANYRLWEALGFV 391
Query: 417 MVFFLAPYISFQAVIMVMLTLLCLSFCSF 445
+ F + ++ + ++L +L L+ ++
Sbjct: 392 IAFGYSMFLCVHVKLYILLGVLSLTMVAY 420
>ref|XP_419606.1| PREDICTED: similar to unc-93 homolog A; unc93 (C.elegans) homolog A
[Gallus gallus]
Length = 458
Score = 107 bits (268), Expect = 6e-22
Identities = 106/439 (24%), Positives = 173/439 (39%), Gaps = 44/439 (10%)
Query: 33 RDVHILSIAFLLIFLAYGAAQNLQSTLNTASGELIVCIEFSGGGFGYDFAWDIIFVFYIF 92
++V +++ FLL+F AYG Q+LQS+LN+ G + + + +
Sbjct: 6 KNVLVIAFGFLLLFTAYGGLQSLQSSLNSEEGLGVASLSVLYAALIVSSMFLPPILIKKL 65
Query: 93 FCVCFFGGADSRIQECFDCWNFGLLVIPGCKFEAQLVYVGSGFCLSWVLRFYIMGWAACQ 152
C G+ C+ ++ G V +G G W A C
Sbjct: 66 GCKWTIAGSMC----CYIAFSLGNFYASWYTLIPTSVILGLGGAPLWS--------AKC- 112
Query: 153 GTYLTSTARSHAIDNNFHEGAVIGDFNGEFWGVYTLHQFIGNLITFALLS---------- 202
TYLT S+A +I + G F+ ++ GNLI+ +LS
Sbjct: 113 -TYLTIAGNSYAEKAGKIGKDIINQYFGVFFLIFQSSGIWGNLISSLILSQSSNKVEISE 171
Query: 203 ---------DGQEGSTNGTTL------LFVVFLSVMTFGAILTCFLHKRG-DYSKGGYKH 246
D +TN T L L L V T +L L D K
Sbjct: 172 EDLECCGAYDCVSNTTNTTALAEPSKSLVYTLLGVYTASGVLAVLLIIIFLDQIKSDQAE 231
Query: 247 LDAGTGQSKSLKSLCRSLTGALSDVKMLLIIPLIAYSGLQHAFVWAEFTKYVVTPEIGVS 306
+ ++ S S + L D + L+IPL YSG + F+ ++TK VT +G+
Sbjct: 232 TEKEIQKTPSFWSTFLATFQHLKDKRQCLLIPLTMYSGFEQGFLAGDYTKTYVTCALGIH 291
Query: 307 GVGIAMAVYGAFDGICSLVAGRLTFGLTSITSIVSFGAFVQAVVLILLLLDFSMSSGFIG 366
VG M + A + +CSL+ G+++ T + + +I LLL F
Sbjct: 292 YVGYVMICFSAVNSLCSLLFGKIS-QFTGRKFLFALATVTNTSCIIALLL---WKPHFNQ 347
Query: 367 TLYILFLAALLGIGDGVLMTQLNALLGMLFKHDMEGAFAQLKIWQSATIAMVFFLAPYIS 426
+ AL G+ D + TQ N L G+LF+ E AFA ++W+S + F + +
Sbjct: 348 LVVFFIFPALWGLSDAIWQTQTNGLYGVLFEKHTEAAFANYRLWESCGFVIAFGYSTTLR 407
Query: 427 FQAVIMVMLTLLCLSFCSF 445
+ ++L +L LS ++
Sbjct: 408 VYIKLYILLAVLTLSMVTY 426
>gb|AAH77540.1| MGC83353 protein [Xenopus laevis] gi|67462040|sp|Q6DDL7|UN93A_XENLA
UNC93 homolog A
Length = 460
Score = 107 bits (267), Expect = 8e-22
Identities = 118/474 (24%), Positives = 193/474 (39%), Gaps = 81/474 (17%)
Query: 33 RDVHILSIAFLLIFLAYGAAQNLQSTLNTASGELIVCIEFSGGGFGYDFAWDIIFVFYIF 92
+++ I+S FLL+F A+G Q+LQS+LN+ G + + V Y
Sbjct: 5 KNILIVSFGFLLLFTAFGGLQSLQSSLNSDEGLGVASLS----------------VIYAA 48
Query: 93 FCVCFFGGADSRIQECFDCWNFGLLVIPGCKFEAQLVYVGSGFCLSW---VLRFYIMG-- 147
V I++ W +V C + + Y F SW + I+G
Sbjct: 49 LIVSSVFVPPIVIKKIGCKWT---IVASMCCY---ITYSLGNFYASWYTLIPTSLILGFG 102
Query: 148 ----WAACQGTYLTSTARSHAIDNNFHEGAVIGDFNGEFWGVYTLHQFIGNLITFALLSD 203
WAA + TYLT + +A ++ + G F+ ++ GNLI+ +
Sbjct: 103 GAPLWAA-KCTYLTESGNRYAEKKGKLAKDIVNQYFGLFFLIFQSSGVWGNLISSLIFGQ 161
Query: 204 GQE---------------------------GSTNGTTLLFVVFLSVMTFGAILTCFL--- 233
G+T T L L V T +L L
Sbjct: 162 NYPAGSNDSFTDYSQCGANDCPGTNFGNGTGTTKPTKSLIYTLLGVYTGSGVLAVILIAV 221
Query: 234 -----HKRGDYSKGGYKHLDAGTGQSKSLKSLCRSLTGALSDVKMLLIIPLIAYSGLQHA 288
+ R D K G K +S S K L + L D + L+IPL YSG +
Sbjct: 222 FLDTINLRTDQLKPGTKE------ESFSKKIL--ATVRHLKDKRQCLLIPLTMYSGFEQG 273
Query: 289 FVWAEFTKYVVTPEIGVSGVGIAMAVYGAFDGICSLVAGRLTFGLTSITSIVSFGAFVQA 348
F+ ++TK VT +G+ VG M + A + +CSL+ G+L+ T + A A
Sbjct: 274 FLSGDYTKSYVTCSLGIHFVGYVMICFAATNAVCSLLFGQLS-KYTGRICLFILAAVSNA 332
Query: 349 VVLILLLLDFSMSSGFIGTLYILFLAALLGIGDGVLMTQLNALLGMLFKHDMEGAFAQLK 408
+I LLL + F ++ +F A+ G+ D + TQ NAL G+LF E AFA +
Sbjct: 333 ACVIALLLWEPYPNDF--AVFFIF-PAIWGMADAIWQTQTNALYGVLFDEHKEAAFANYR 389
Query: 409 IWQSATIAMVFFLAPY--ISFQAVIMVMLTLLCLSFCSFLWLALKVGNASSPST 460
+W+S + + + + +S + I++ + L+ + F F+ V + + ST
Sbjct: 390 LWESLGFVIAYGYSTFLCVSVKLYILLAVLLIAIVFYGFVEYLEHVEHKKAAST 443
>ref|XP_629994.1| hypothetical protein DDB0184030 [Dictyostelium discoideum]
gi|60463369|gb|EAL61557.1| hypothetical protein
DDB0184030 [Dictyostelium discoideum]
Length = 447
Score = 105 bits (262), Expect = 3e-21
Identities = 113/447 (25%), Positives = 196/447 (43%), Gaps = 65/447 (14%)
Query: 27 QHKSHTR-DVHILSIAFLLIFLAYGAAQNLQSTLNTASGELIVCIEFSGGGFGYDFAWDI 85
Q+K++ + ++ IL + F ++F A+ Q LQ+T+N G + + +S
Sbjct: 32 QNKNNNKINIIILGLCFCILFSAFSPTQILQTTINQNLGYYTLAVLYSSLS--------- 82
Query: 86 IFVFYIFFCVCFFGGADSRIQECFDCWNFGLLVIPGCKFEAQLVYVGSGFCLSWVLRFYI 145
I F F V FG S I G L + +Y+GS ++ +
Sbjct: 83 ISNFISPFIVSKFGEKISLI--------IGTL--------SYAIYIGSNIYVTQPSLYIS 126
Query: 146 MGWAACQGTYLTSTARSHAIDNNFHEGAVIGDFNGEFWGVYTLHQFIGNLITFALLSDGQ 205
G L + S I + E IG G F+ ++ Q IGN+ + L++ +
Sbjct: 127 SILVGFGGAILWNAQGSLIIKYSTEE--TIGANTGLFFALFQTDQIIGNIGSATLIN--K 182
Query: 206 EGSTNGTTLLFVVFLSVMTFGAILTCFL---------------HKRGDYSKGGYKHLDAG 250
G +N ++LF +F+ + I FL + + + +KG ++ D
Sbjct: 183 AGLSN--SILFTIFMGISLMPIIGFLFLKCPITPKIKKTIKSINIQDEETKGNQENHD-- 238
Query: 251 TGQSKSLKSLCRSLTGALSDVKMLLIIPLIAYSGLQHAFVWAEFTKYVVTPEIGVSGVGI 310
S+K++ +S+ D + L+IP + YSG+ F + F IGV VG
Sbjct: 239 -DDDLSIKNIFKSIIILFKDKPIQLLIPSLLYSGISQTFFFGVFPSL-----IGVEWVGY 292
Query: 311 AMAVYGAFDGICSLVAGRLTF--GLTSITSIVSFGAFVQAVVLILL----LLDFSMSS-- 362
M+V+G FD + S + G+L+F G + I + + + V++IL+ ++ FS+++
Sbjct: 293 VMSVFGFFDALSSFILGKLSFKIGRKILILISTISSIIGTVLIILVNQSKIIYFSINNNN 352
Query: 363 --GFIGTLYILFLAALLGIGDGVLMTQLNALLGMLFKHDMEGAFAQLKIWQSATIAMVFF 420
G L +ALLG D TQL +LLG+L+ + E A K QS A+ F
Sbjct: 353 NYGEYKILCYFIGSALLGFSDAGFNTQLYSLLGVLYPTNGEAAVGVFKFVQSTATAVAFI 412
Query: 421 LAPYISFQAVIMVMLTLLCLSFCSFLW 447
PY S + V+ L+ +S F++
Sbjct: 413 YGPYASLFENVFVLDCLVIISCVFFIF 439
>gb|AAT76555.1| CG4928 [Drosophila lutescens]
Length = 515
Score = 104 bits (260), Expect = 5e-21
Identities = 109/457 (23%), Positives = 192/457 (41%), Gaps = 76/457 (16%)
Query: 33 RDVHILSIAFLLIFLAYGAAQNLQSTLNTASGELIVCIE--FSGGGFGYDFAWDIIF--- 87
+++ I+SIAF++ F A+ NLQS++N G V + ++ F +I
Sbjct: 35 KNISIISIAFMVQFTAFQGTANLQSSINAQDGLGTVSLSAIYAALVVSCIFLPTLIIRKL 94
Query: 88 -VFYIFFC--VCFFGGADSRIQECFDCWNFGLLVIPGCKFEAQLVYVGSGFCLSWVLRFY 144
V + C +C+ A + F F LV G + VG G W
Sbjct: 95 TVKWTLVCSMLCY---APYIAFQLFP--RFYTLVPAG-------ILVGMGAAPMW----- 137
Query: 145 IMGWAACQGTYLTSTARSHAIDNNFHEGAVIGDFNGEFWGVYTLHQFIGNLITFALLSDG 204
A + TYLT + +A A+I F G F+ + + GNLI+ +LS G
Sbjct: 138 -----ASKATYLTQVGQVYAKITEQAVDAIIVRFFGFFFLAWQSAELWGNLISSLVLSSG 192
Query: 205 QEGSTNG--TTL---------------------------------LFVVFLS-VMTFGAI 228
G T+ TT+ + +++LS ++ I
Sbjct: 193 AHGGTSSSNTTISDEDLQYCGANFCTTGSGGHGNLERPPEDEIFEISMIYLSCIVAAVCI 252
Query: 229 LTCFLHKRGDYSKGGYKHLDAGTGQSKSLKSLCRSLTGALSDVKMLLIIPLIAYSGLQHA 288
+ FL Y G K + + S L + + + L+IP+ + G++ A
Sbjct: 253 IAFFLDPLKRY---GEKRKGSNSAAELSGLQLLSATFRQMKKPNLQLLIPITVFIGMEQA 309
Query: 289 FVWAEFTKYVVTPEIGVSGVGIAMAVYGAFDGICSLVAGRLTFGLTSITSIVSFGAFVQA 348
F+ A+FT+ V +GV+ +G M +G + +CS++ G + T I+ GA V
Sbjct: 310 FIGADFTQAYVACALGVNKIGFVMICFGVVNALCSILFGSV-MKYIGRTPIIVLGAVVH- 367
Query: 349 VVLILLLLDFSMSSGFIGTLYILFLAALLGIGDGVLMTQLNALLGMLFKHDMEGAFAQLK 408
L+ ++ + ++ L G+GD V TQ+N L G+LF+ + E AF+ +
Sbjct: 368 --FTLITVELFWRPNPDNPIIFYAMSGLWGVGDAVWQTQINGLYGLLFRRNKEAAFSNYR 425
Query: 409 IWQSATIAMVFFLAPYISFQA---VIMVMLTLLCLSF 442
+W+SA + + A + Q +++ +LTL C+ +
Sbjct: 426 LWESAGFVIAYAYATTLCTQMKLYILLAVLTLGCIGY 462
>ref|NP_728035.1| CG4928-PA, isoform A [Drosophila melanogaster]
gi|18859663|ref|NP_573179.1| CG4928-PB, isoform B
[Drosophila melanogaster] gi|7293301|gb|AAF48682.1|
CG4928-PB, isoform B [Drosophila melanogaster]
gi|22832423|gb|AAN09429.1| CG4928-PA, isoform A
[Drosophila melanogaster]
gi|67462083|sp|Q9Y115|UN93L_DROME UNC93-like protein
gi|5052604|gb|AAD38632.1| BcDNA.GH10120 [Drosophila
melanogaster]
Length = 538
Score = 104 bits (259), Expect = 7e-21
Identities = 109/457 (23%), Positives = 193/457 (41%), Gaps = 76/457 (16%)
Query: 33 RDVHILSIAFLLIFLAYGAAQNLQSTLNTASGELIVCIE--FSGGGFGYDFAWDIIF--- 87
+++ I+SIAF++ F A+ NLQS++N G V + ++ F +I
Sbjct: 44 KNISIISIAFMVQFTAFQGTANLQSSINAKDGLGTVSLSAIYAALVVSCIFLPTLIIRKL 103
Query: 88 -VFYIFFC--VCFFGGADSRIQECFDCWNFGLLVIPGCKFEAQLVYVGSGFCLSWVLRFY 144
V + C +C+ A + F F LV G + VG G W
Sbjct: 104 TVKWTLVCSMLCY---APYIAFQLFP--RFYTLVPAG-------ILVGMGAAPMW----- 146
Query: 145 IMGWAACQGTYLTSTARSHAIDNNFHEGAVIGDFNGEFWGVYTLHQFIGNLITFALLSDG 204
A + TYLT + +A A+I F G F+ + + GNLI+ +LS G
Sbjct: 147 -----ASKATYLTQVGQVYAKITEQAVDAIIVRFFGFFFLAWQSAELWGNLISSLVLSSG 201
Query: 205 QEG--STNGTTL---------------------------------LFVVFLS-VMTFGAI 228
G S++ TT+ + +++LS ++ I
Sbjct: 202 AHGGGSSSNTTVSEEDLQFCGANFCTTGSGGHGNLERPPEDEIFEISMIYLSCIVAAVCI 261
Query: 229 LTCFLHKRGDYSKGGYKHLDAGTGQSKSLKSLCRSLTGALSDVKMLLIIPLIAYSGLQHA 288
+ FL Y G K + + S L + + + L+IP+ + G++ A
Sbjct: 262 IAFFLDPLKRY---GEKRKGSNSAAELSGLQLLSATFRQMKKPNLQLLIPITVFIGMEQA 318
Query: 289 FVWAEFTKYVVTPEIGVSGVGIAMAVYGAFDGICSLVAGRLTFGLTSITSIVSFGAFVQA 348
F+ A+FT+ V +GV+ +G M +G + +CS++ G + T I+ GA V
Sbjct: 319 FIGADFTQAYVACALGVNKIGFVMICFGVVNALCSILFGSV-MKYIGRTPIIVLGAVVH- 376
Query: 349 VVLILLLLDFSMSSGFIGTLYILFLAALLGIGDGVLMTQLNALLGMLFKHDMEGAFAQLK 408
L+ ++ + ++ L G+GD V TQ+N L G+LF+ + E AF+ +
Sbjct: 377 --FTLITVELFWRPNPDNPIIFYAMSGLWGVGDAVWQTQINGLYGLLFRRNKEAAFSNYR 434
Query: 409 IWQSATIAMVFFLAPYISFQA---VIMVMLTLLCLSF 442
+W+SA + + A + Q +++ +LTL C+ +
Sbjct: 435 LWESAGFVIAYAYATTLCTQMKLYILLAVLTLGCIGY 471
>emb|CAA45761.1| unc-93 [Caenorhabditis elegans] gi|58081858|emb|CAB03760.3|
Hypothetical protein C46F11.1b [Caenorhabditis elegans]
Length = 700
Score = 96.7 bits (239), Expect = 1e-18
Identities = 104/448 (23%), Positives = 194/448 (43%), Gaps = 66/448 (14%)
Query: 37 ILSIAFLLIFLAYGAAQNLQSTLNTASG-ELIVCIEFSGGGFGYDFAWDIIFVFYIFFCV 95
ILS+AFL +F A+ QNLQ+++N G + +V + S + + +F
Sbjct: 247 ILSVAFLFLFTAFNGLQNLQTSVNGDLGSDSLVALYLS------------LAISSLFVPS 294
Query: 96 CFFGGADSRIQECFDCWNFGLLVIPGCKFEAQLVYVGSGFCLSWVLRFYIMGWAACQGTY 155
++ + + L ++ + + S FC + I G A C Y
Sbjct: 295 FMINRLGCKLTFLIAIFVYFLYIVINLRPTYSSMIPASIFC--GIAASCIWG-AKC--AY 349
Query: 156 LTSTARSHAIDNNFHEGAVIGDFNGEFWGVYTLHQFIGNLIT---FAL------------ 200
+T +A N + VI F G F+ + Q +GN+++ F L
Sbjct: 350 ITEMGIRYASLNFESQTTVIVRFFGYFFMIVHCGQVVGNMVSSYIFTLSYSQALRGPEDS 409
Query: 201 ------------LSDGQEGSTNG---------TTLLFVVFLSVMTFGAILTCFLHKRGDY 239
LSD E + + + V+ G I++ FL+
Sbjct: 410 IYDSCGYQFPKNLSDLTELAESNLARPPQKVYVAVCLAYLACVIISGMIMSMFLNA---L 466
Query: 240 SKGGYKHLDAGTGQSKSLKSLCRSLTGALSDVKMLLIIPLIAYSGLQHAFVWAEFTKYVV 299
+K A S+ + + L ++K +L++PL ++GL+ AF+ +TK V
Sbjct: 467 AKDARNRKMAQKFNSEIFYLMLKHLI----NIKFMLLVPLTIFNGLEQAFLVGVYTKAFV 522
Query: 300 TPEIGVSGVGIAMAVYGAFDGICSLVAGRLTFGLTSITSIVSFGAFVQAVVLILLLLDFS 359
+G+ +G MA +G D +CSLV G L L + FGA V ++++ L++ +
Sbjct: 523 GCGLGIWQIGFVMACFGISDAVCSLVFGPL-IKLFGRMPLFVFGAVVNLLMIVTLMV-WP 580
Query: 360 MSSGFIGTLYILFLAALLGIGDGVLMTQLNAL-LGMLFKHDMEGAFAQLKIWQSATIAMV 418
+++ Y+ +AA+ G+ DGV TQ+N + ++ + ++ AF + + W+S IA+
Sbjct: 581 LNAADTQIFYV--VAAMWGMADGVWNTQINGFWVALVGRQSLQFAFTKYRFWESLGIAIG 638
Query: 419 FFLAPYISFQAVIMVMLTLLCLSFCSFL 446
F L +++ + +++ +L L C FL
Sbjct: 639 FALIRHVTVEIYLLITFFMLLLGMCGFL 666
>emb|CAA45760.1| unc-93 [Caenorhabditis elegans] gi|61855544|emb|CAI70399.1|
Hypothetical protein C46F11.1a [Caenorhabditis elegans]
gi|62511220|sp|Q93380|UNC93_CAEEL Putative potassium
channel regulatory protein unc-93 (Uncoordinated protein
93) gi|25144062|ref|NP_497738.2| UNCoordinated
locomotion UNC-93, putative protein, with at least 11
transmembrane domains, of eukaryotic origin (unc-93)
[Caenorhabditis elegans] gi|102475|pir||S23352 gene
unc-93 protein 1 - Caenorhabditis elegans
Length = 705
Score = 96.7 bits (239), Expect = 1e-18
Identities = 104/448 (23%), Positives = 194/448 (43%), Gaps = 66/448 (14%)
Query: 37 ILSIAFLLIFLAYGAAQNLQSTLNTASG-ELIVCIEFSGGGFGYDFAWDIIFVFYIFFCV 95
ILS+AFL +F A+ QNLQ+++N G + +V + S + + +F
Sbjct: 252 ILSVAFLFLFTAFNGLQNLQTSVNGDLGSDSLVALYLS------------LAISSLFVPS 299
Query: 96 CFFGGADSRIQECFDCWNFGLLVIPGCKFEAQLVYVGSGFCLSWVLRFYIMGWAACQGTY 155
++ + + L ++ + + S FC + I G A C Y
Sbjct: 300 FMINRLGCKLTFLIAIFVYFLYIVINLRPTYSSMIPASIFC--GIAASCIWG-AKC--AY 354
Query: 156 LTSTARSHAIDNNFHEGAVIGDFNGEFWGVYTLHQFIGNLIT---FAL------------ 200
+T +A N + VI F G F+ + Q +GN+++ F L
Sbjct: 355 ITEMGIRYASLNFESQTTVIVRFFGYFFMIVHCGQVVGNMVSSYIFTLSYSQALRGPEDS 414
Query: 201 ------------LSDGQEGSTNG---------TTLLFVVFLSVMTFGAILTCFLHKRGDY 239
LSD E + + + V+ G I++ FL+
Sbjct: 415 IYDSCGYQFPKNLSDLTELAESNLARPPQKVYVAVCLAYLACVIISGMIMSMFLNA---L 471
Query: 240 SKGGYKHLDAGTGQSKSLKSLCRSLTGALSDVKMLLIIPLIAYSGLQHAFVWAEFTKYVV 299
+K A S+ + + L ++K +L++PL ++GL+ AF+ +TK V
Sbjct: 472 AKDARNRKMAQKFNSEIFYLMLKHLI----NIKFMLLVPLTIFNGLEQAFLVGVYTKAFV 527
Query: 300 TPEIGVSGVGIAMAVYGAFDGICSLVAGRLTFGLTSITSIVSFGAFVQAVVLILLLLDFS 359
+G+ +G MA +G D +CSLV G L L + FGA V ++++ L++ +
Sbjct: 528 GCGLGIWQIGFVMACFGISDAVCSLVFGPL-IKLFGRMPLFVFGAVVNLLMIVTLMV-WP 585
Query: 360 MSSGFIGTLYILFLAALLGIGDGVLMTQLNAL-LGMLFKHDMEGAFAQLKIWQSATIAMV 418
+++ Y+ +AA+ G+ DGV TQ+N + ++ + ++ AF + + W+S IA+
Sbjct: 586 LNAADTQIFYV--VAAMWGMADGVWNTQINGFWVALVGRQSLQFAFTKYRFWESLGIAIG 643
Query: 419 FFLAPYISFQAVIMVMLTLLCLSFCSFL 446
F L +++ + +++ +L L C FL
Sbjct: 644 FALIRHVTVEIYLLITFFMLLLGMCGFL 671
>ref|XP_629995.1| hypothetical protein DDB0184029 [Dictyostelium discoideum]
gi|60463368|gb|EAL61556.1| hypothetical protein
DDB0184029 [Dictyostelium discoideum]
Length = 425
Score = 96.7 bits (239), Expect = 1e-18
Identities = 98/425 (23%), Positives = 172/425 (40%), Gaps = 60/425 (14%)
Query: 34 DVHILSIAFLLIFLAYGAAQNLQSTLNTASGELIVCIEFSGGGFGYDFAWDIIFVFYIFF 93
++ +L ++F ++F A+ Q LQ+T+N G D + V Y F
Sbjct: 35 NIIVLGLSFCILFSAFSPTQELQTTINKNLGS------------------DSLAVLYAFL 76
Query: 94 CVCFFGGA------DSRIQECFDCWNFGLLVIPGCKFEAQLVY-----VGSGFCLSWVLR 142
V F R+ + + K +Y +G G + W
Sbjct: 77 SVTNFISPFVILKLGERLSLIVGTLTYSAYIAANIKVTIPTLYGASVLLGIGGAILWT-- 134
Query: 143 FYIMGWAACQGTYLTSTARSHAIDNNFHEGAVIGDFNGEFWGVYTLHQFIGNLITFALLS 202
QG+++ + I N G F+ ++ ++Q IGNL T AL+
Sbjct: 135 --------AQGSFVIQCSTESTIGAN----------TGLFFALFQINQIIGNLGTAALID 176
Query: 203 DGQEGSTNGTTLLFVVFLSVMTFGAILTCFLHKRGDYSKGGYKHLDAGTGQSKSLKSLCR 262
+ G N +LF++FL V + L K + G + Q+ S++
Sbjct: 177 --KAGLQN--EVLFLIFLGVSLLSILGFAILGKPVKVNDKG--QIVEKENQTMSIRDRLL 230
Query: 263 SLTGALSDVKMLLIIPLIAYSGLQHAFVWAEFTKYVVTPEIGVSGVGIAMAVYGAFDGIC 322
S D + L++ + YSG+ +F + F + +G +G MAV+G D
Sbjct: 231 STIVLFKDRPIQLLVAPLLYSGISQSFFFGVFPQL-----LGKEWIGYVMAVFGFCDAAG 285
Query: 323 SLVAGRLTFGLTSITSIVSFGAFVQAVVLILLLLDFSMSSGFIGTLYILFLAALLGIGDG 382
S++ G+L+ +V F + ++ +LLD +S+ Y +AALLG+ D
Sbjct: 286 SMIMGKLSDIFGRKILVVFSTVFCISGTVLCILLDRLVSTQSERIPYYFIVAALLGLADA 345
Query: 383 VLMTQLNALLGMLFKHDMEGAFAQLKIWQSATIAMVFFLAPYISFQAVIMVMLTLLCLSF 442
TQL AL+G+++ E A K QS A F PY + ++++ +L+ LS
Sbjct: 346 GFNTQLYALIGVIYPKKGEAAAGVFKFVQSTATAAAFVYGPYATLFDNVIIVGSLVILSC 405
Query: 443 CSFLW 447
F++
Sbjct: 406 VLFMF 410
>emb|CAE64940.1| Hypothetical protein CBG09771 [Caenorhabditis briggsae]
Length = 716
Score = 95.5 bits (236), Expect = 3e-18
Identities = 104/448 (23%), Positives = 192/448 (42%), Gaps = 66/448 (14%)
Query: 37 ILSIAFLLIFLAYGAAQNLQSTLNTASG-ELIVCIEFSGGGFGYDFAWDIIFVFYIFFCV 95
ILS+A L +F A+ QNLQ+++N G + +V + S I +F F +
Sbjct: 247 ILSVALLFLFTAFNGLQNLQTSVNGDLGSDSLVALYLSLA---------ISSLFVPSFMI 297
Query: 96 CFFGGADSRIQECFDCWNFGLLVIPGCKFEAQLVYVGSGFCLSWVLRFYIMGWAACQGTY 155
G + + F + L ++ + + S FC + I G A C Y
Sbjct: 298 NRLGCKMTFLVAIFV---YFLYIVINLRPTYSSMIPASIFC--GIAASCIWG-AKC--AY 349
Query: 156 LTSTARSHAIDNNFHEGAVIGDFNGEFWGVYTLHQFIGNLITFALLSDGQEGSTNG---- 211
+T +A N + VI F G F+ + Q +GN+++ + + GS G
Sbjct: 350 ITEMGIRYASLNFESQTTVIVRFFGYFFMIVHCGQVVGNVVSSYIFTLSYSGSLRGPEDS 409
Query: 212 --------------------------------TTLLFVVFLSVMTFGAILTCFLHKRGDY 239
+ ++ G I++ FL+
Sbjct: 410 IYDSCGYQFPKNLSYLSELAESNLARPPQKVYVAVCLAYLACIIISGMIMSMFLNA---L 466
Query: 240 SKGGYKHLDAGTGQSKSLKSLCRSLTGALSDVKMLLIIPLIAYSGLQHAFVWAEFTKYVV 299
K A S+ + + + L ++K LL++PL ++GL+ AF+ +TK V
Sbjct: 467 VKDTRNRKMAQRFNSEIITLMVKHLI----NIKFLLLVPLTIFNGLEQAFLVGVYTKAFV 522
Query: 300 TPEIGVSGVGIAMAVYGAFDGICSLVAGRLTFGLTSITSIVSFGAFVQAVVLILLLLDFS 359
+G+ +G MA +G D ICSLV G L L + FGA V ++++ L++ +
Sbjct: 523 GCGLGIWQIGFVMACFGISDAICSLVFGPL-IKLFGRMPLFVFGAVVNLLMIVTLMV-WP 580
Query: 360 MSSGFIGTLYILFLAALLGIGDGVLMTQLNAL-LGMLFKHDMEGAFAQLKIWQSATIAMV 418
+++ Y+ +AA+ G+ DGV TQ+N + ++ + ++ AF + + W+S IA+
Sbjct: 581 LNAADTQIFYV--VAAMWGMADGVWNTQINGFWVALVGRQSLQFAFTKYRFWESLGIAIG 638
Query: 419 FFLAPYISFQAVIMVMLTLLCLSFCSFL 446
F L +++ + +++ +L L C F+
Sbjct: 639 FALIRHVTVEMYLLITFFVLLLGMCGFV 666
>emb|CAG12950.1| unnamed protein product [Tetraodon nigroviridis]
Length = 540
Score = 95.1 bits (235), Expect = 4e-18
Identities = 58/179 (32%), Positives = 94/179 (52%), Gaps = 6/179 (3%)
Query: 268 LSDVKMLLIIPLIAYSGLQHAFVWAEFTKYVVTPEIGVSGVGIAMAVYGAFDGICSLVAG 327
L D ++L +IPL YSG + +F+ E+TK VT +G+ VG M +GA + I S + G
Sbjct: 352 LKDWRLLALIPLTMYSGFEQSFLSGEYTKNYVTCALGIHYVGFVMMCFGASNSIFSFLFG 411
Query: 328 RLTFGLTSITSIVSFGAFVQAVVLILLLLDFSMSSGFIGTLYILFLA-ALLGIGDGVLMT 386
RL + I + AV + ++ F + LY+ FL AL G+ D V
Sbjct: 412 RLARYTGRLALIC-----LAAVTNLACIITFLLWKPDPQQLYLFFLLPALWGMADAVWQP 466
Query: 387 QLNALLGMLFKHDMEGAFAQLKIWQSATIAMVFFLAPYISFQAVIMVMLTLLCLSFCSF 445
Q+NAL G+LF D E AFA ++W+S + F + ++ + + +ML +L L+ ++
Sbjct: 467 QVNALYGILFPRDKEAAFANYRMWESLGFVIAFAYSTFLCLEYKVYIMLGVLLLTAVTY 525
>pir||T19969 hypothetical protein C46F11.1 - Caenorhabditis elegans
Length = 708
Score = 94.0 bits (232), Expect = 9e-18
Identities = 53/180 (29%), Positives = 102/180 (56%), Gaps = 5/180 (2%)
Query: 268 LSDVKMLLIIPLIAYSGLQHAFVWAEFTKYVVTPEIGVSGVGIAMAVYGAFDGICSLVAG 327
L ++K +L++PL ++GL+ AF+ +TK V +G+ +G MA +G D +CSLV G
Sbjct: 499 LINIKFMLLVPLTIFNGLEQAFLVGVYTKAFVGCGLGIWQIGFVMACFGISDAVCSLVFG 558
Query: 328 RLTFGLTSITSIVSFGAFVQAVVLILLLLDFSMSSGFIGTLYILFLAALLGIGDGVLMTQ 387
L L + FGA V ++++ L++ + +++ Y+ +AA+ G+ DGV TQ
Sbjct: 559 PL-IKLFGRMPLFVFGAVVNLLMIVTLMV-WPLNAADTQIFYV--VAAMWGMADGVWNTQ 614
Query: 388 LNAL-LGMLFKHDMEGAFAQLKIWQSATIAMVFFLAPYISFQAVIMVMLTLLCLSFCSFL 446
+N + ++ + ++ AF + + W+S IA+ F L +++ + +++ +L L C FL
Sbjct: 615 INGFWVALVGRQSLQFAFTKYRFWESLGIAIGFALIRHVTVEIYLLITFFMLLLGMCGFL 674
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.329 0.143 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 759,937,302
Number of Sequences: 2540612
Number of extensions: 31787761
Number of successful extensions: 96753
Number of sequences better than 10.0: 165
Number of HSP's better than 10.0 without gapping: 70
Number of HSP's successfully gapped in prelim test: 95
Number of HSP's that attempted gapping in prelim test: 96517
Number of HSP's gapped (non-prelim): 236
length of query: 460
length of database: 863,360,394
effective HSP length: 131
effective length of query: 329
effective length of database: 530,540,222
effective search space: 174547733038
effective search space used: 174547733038
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 77 (34.3 bits)
Medicago: description of AC135797.3


