

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135796.9 - phase: 0 /pseudo
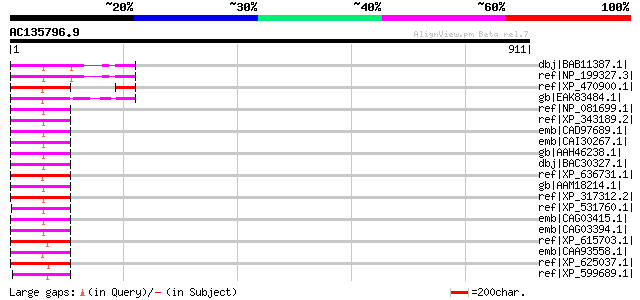
(911 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAB11387.1| DNA-directed RNA polymerase subunit [Arabidopsis... 160 2e-37
ref|NP_199327.3| DNA-directed RNA polymerase, putative [Arabidop... 160 2e-37
ref|XP_470900.1| putative RNA polymerase III [Oryza sativa (japo... 133 2e-29
gb|EAK83484.1| hypothetical protein UM02446.1 [Ustilago maydis 5... 100 4e-19
ref|NP_081699.1| RNA polymerase III subunit RPC2 [Mus musculus] ... 94 2e-17
ref|XP_343189.2| PREDICTED: similar to RNA polymerase III subuni... 94 2e-17
emb|CAD97689.1| hypothetical protein [Homo sapiens] gi|42476127|... 94 2e-17
emb|CAI30267.1| hypothetical protein [Pongo pygmaeus] 94 2e-17
gb|AAH46238.1| Polymerase (RNA) III (DNA directed) polypeptide B... 94 2e-17
dbj|BAC30327.1| unnamed protein product [Mus musculus] 94 2e-17
ref|XP_636731.1| RNA polymerase III, second largest subunit [Dic... 94 2e-17
gb|AAM18214.1| RNA polymerase III subunit RPC2 [Homo sapiens] 93 5e-17
ref|XP_317312.2| ENSANGP00000006850 [Anopheles gambiae str. PEST... 91 2e-16
ref|XP_531760.1| PREDICTED: similar to DNA-directed RNA polymera... 91 2e-16
emb|CAG03415.1| unnamed protein product [Tetraodon nigroviridis] 91 2e-16
emb|CAG03394.1| unnamed protein product [Tetraodon nigroviridis] 91 2e-16
ref|XP_615703.1| PREDICTED: similar to DNA-directed RNA polymera... 87 3e-15
emb|CAA93558.1| SPAC4G9.08c [Schizosaccharomyces pombe] gi|19114... 86 4e-15
ref|XP_625037.1| PREDICTED: similar to DNA-directed RNA polymera... 83 5e-14
ref|XP_599689.1| PREDICTED: similar to hypothetical protein, par... 82 6e-14
>dbj|BAB11387.1| DNA-directed RNA polymerase subunit [Arabidopsis thaliana]
Length = 1194
Score = 160 bits (404), Expect = 2e-37
Identities = 96/243 (39%), Positives = 139/243 (56%), Gaps = 60/243 (24%)
Query: 1 MVVMKAMGMEHDQEVVQLIGRDPRYSFLLMPSIEECTKIGIFTQAQALDYVDSKAER--- 57
++V+KAMGME DQE+VQ++GRDPR+S L+PSIEEC G+ TQ QALDY+++K ++
Sbjct: 267 IIVLKAMGMESDQEIVQMVGRDPRFSASLLPSIEECVSEGVNTQKQALDYLEAKVKKISY 326
Query: 58 --------QAFNILRETFLANVPVHGYNFRPKCIYVAVMIRRIMDAILNKDAMDDKSF-- 107
+A +ILR+ FLA+VPV NFR KC YV VM+RR+++A+LNKDAMDDK +
Sbjct: 327 GTPPEKDGRALSILRDLFLAHVPVPDNNFRQKCFYVGVMLRRMIEAMLNKDAMDDKDYVG 386
Query: 108 -------KKIIGLCWKQTIGVIWPSYLSSI*GSF*VDDN*N*VCGGQIVG*ARQS*EV*L 160
++I L ++ + + ++ D+
Sbjct: 387 NKRLELSGQLISLLFEDLFKTMLSEAIKNV-------DH--------------------- 418
Query: 161 SYFQPDLSISGFQFT*FLWMCL*QLINRD---CITPGLERTLSTGNFDVKRFKMHKSGVT 217
+ S F F+ Q +N+D I+ GLERTLSTGNFD+KRF+MH+ G+T
Sbjct: 419 -ILNKPIRASRFDFS--------QCLNKDSRYSISLGLERTLSTGNFDIKRFRMHRKGMT 469
Query: 218 QVI 220
QV+
Sbjct: 470 QVL 472
>ref|NP_199327.3| DNA-directed RNA polymerase, putative [Arabidopsis thaliana]
Length = 1150
Score = 160 bits (404), Expect = 2e-37
Identities = 96/243 (39%), Positives = 139/243 (56%), Gaps = 60/243 (24%)
Query: 1 MVVMKAMGMEHDQEVVQLIGRDPRYSFLLMPSIEECTKIGIFTQAQALDYVDSKAER--- 57
++V+KAMGME DQE+VQ++GRDPR+S L+PSIEEC G+ TQ QALDY+++K ++
Sbjct: 246 IIVLKAMGMESDQEIVQMVGRDPRFSASLLPSIEECVSEGVNTQKQALDYLEAKVKKISY 305
Query: 58 --------QAFNILRETFLANVPVHGYNFRPKCIYVAVMIRRIMDAILNKDAMDDKSF-- 107
+A +ILR+ FLA+VPV NFR KC YV VM+RR+++A+LNKDAMDDK +
Sbjct: 306 GTPPEKDGRALSILRDLFLAHVPVPDNNFRQKCFYVGVMLRRMIEAMLNKDAMDDKDYVG 365
Query: 108 -------KKIIGLCWKQTIGVIWPSYLSSI*GSF*VDDN*N*VCGGQIVG*ARQS*EV*L 160
++I L ++ + + ++ D+
Sbjct: 366 NKRLELSGQLISLLFEDLFKTMLSEAIKNV-------DH--------------------- 397
Query: 161 SYFQPDLSISGFQFT*FLWMCL*QLINRD---CITPGLERTLSTGNFDVKRFKMHKSGVT 217
+ S F F+ Q +N+D I+ GLERTLSTGNFD+KRF+MH+ G+T
Sbjct: 398 -ILNKPIRASRFDFS--------QCLNKDSRYSISLGLERTLSTGNFDIKRFRMHRKGMT 448
Query: 218 QVI 220
QV+
Sbjct: 449 QVL 451
>ref|XP_470900.1| putative RNA polymerase III [Oryza sativa (japonica
cultivar-group)] gi|29788820|gb|AAP03366.1| putative RNA
polymerase III [Oryza sativa (japonica cultivar-group)]
Length = 1174
Score = 133 bits (335), Expect = 2e-29
Identities = 68/116 (58%), Positives = 83/116 (70%), Gaps = 9/116 (7%)
Query: 1 MVVMKAMGMEHDQEVVQLIGRDPRYSFLLMPSIEECTKIGIFTQAQALDYVDSKA----- 55
+VVMKAMGME DQEV Q++GRDPRY LL PSI+EC I+TQ QAL Y+D K
Sbjct: 268 IVVMKAMGMESDQEVAQMVGRDPRYGDLLYPSIQECAFERIYTQKQALQYMDDKVMYPGA 327
Query: 56 ----ERQAFNILRETFLANVPVHGYNFRPKCIYVAVMIRRIMDAILNKDAMDDKSF 107
E ++ +ILR+ F+A+VPV NFRPKCIY AVM+RR+MDAILN D DDK +
Sbjct: 328 GNQKEGRSKSILRDVFVAHVPVESGNFRPKCIYTAVMLRRMMDAILNADTFDDKDY 383
Score = 50.4 bits (119), Expect = 3e-04
Identities = 21/35 (60%), Positives = 29/35 (82%)
Query: 186 INRDCITPGLERTLSTGNFDVKRFKMHKSGVTQVI 220
I + IT GLER +STGN+D+KRF+MH+ GV+QV+
Sbjct: 433 IKENIITHGLERAISTGNWDIKRFRMHRKGVSQVL 467
>gb|EAK83484.1| hypothetical protein UM02446.1 [Ustilago maydis 521]
gi|49071544|ref|XP_400061.1| hypothetical protein
UM02446.1 [Ustilago maydis 521]
Length = 1169
Score = 99.8 bits (247), Expect = 4e-19
Identities = 68/233 (29%), Positives = 118/233 (50%), Gaps = 40/233 (17%)
Query: 2 VVMKAMGMEHDQEVVQLI-GRDPRYSFLLMPSIEECTKIGIFTQAQALDYVDSKAE---- 56
V KAMG++ D+E++QL+ G D + L ++EE ++GI+T+ QALD++ ++A+
Sbjct: 262 VAFKAMGIQADREILQLVAGHDDIHKELFAINLEEAARLGIYTRKQALDFIGARAKASRK 321
Query: 57 ---------RQAFNILRETFLANVPVHGYNFRPKCIYVAVMIRRIMDAILNKDAMDDKSF 107
+A ++L +A+VPV NFRPK IY+A M+RR++ A+ ++ +DD+ +
Sbjct: 322 PLSMRRPLSEEALDVLATVIMAHVPVERLNFRPKAIYIASMVRRVLMAMQDEKKVDDRDY 381
Query: 108 KKIIGLCWKQTIGVIWPSYLSSI*GSF*VDDN*N*VCGGQIVG*ARQS*EV*LSYFQPDL 167
+G + G + + F D N + +P+
Sbjct: 382 ---VGNKRLELAGQLLSLLFEDLFKKFNSDLKMN----------------IDKVLKKPNR 422
Query: 168 SISGFQFT*FLWMCL*QLINRDCITPGLERTLSTGNFDVKRFKMHKSGVTQVI 220
++ F F + N D IT G R +STGN+ +KRFKM ++G+T V+
Sbjct: 423 TVEFDAFNQFHF-------NGDYITSGFVRAISTGNWSLKRFKMERAGITHVL 468
>ref|NP_081699.1| RNA polymerase III subunit RPC2 [Mus musculus]
gi|45829714|gb|AAH68143.1| RNA polymerase III subunit
RPC2 [Mus musculus] gi|51704303|sp|P59470|RPC2_MOUSE
DNA-directed RNA polymerase III subunit 127.6 kDa
polypeptide (RNA polymerase III subunit 2) (RPC2)
Length = 1133
Score = 94.4 bits (233), Expect = 2e-17
Identities = 51/121 (42%), Positives = 71/121 (58%), Gaps = 15/121 (12%)
Query: 1 MVVMKAMGMEHDQEVVQLIGRDPRYSFLLMPSIEECTKIGIFTQAQALDYVDSKAERQ-- 58
+++ KAMG+E DQE+VQ+IG + PS+EEC K IFTQ QAL Y+ +K RQ
Sbjct: 237 VIIFKAMGVESDQEIVQMIGTEEHVMAAFGPSLEECQKAQIFTQMQALKYIGNKVRRQRM 296
Query: 59 ------------AFNILRETFLANVPVHGYNFRPKCIYVAVMIRRIMDAILNKDAMDDKS 106
A +L T L +VPV +NFR KCIY AVM+RR++ A + +DD+
Sbjct: 297 WGGGPKKTKIEEARELLASTILTHVPVKEFNFRAKCIYTAVMVRRVILA-QGDNKVDDRD 355
Query: 107 F 107
+
Sbjct: 356 Y 356
Score = 41.6 bits (96), Expect = 0.12
Identities = 18/35 (51%), Positives = 26/35 (73%)
Query: 186 INRDCITPGLERTLSTGNFDVKRFKMHKSGVTQVI 220
+ +D IT G+ +STGN+ +KRFKM + GVTQV+
Sbjct: 406 MRQDQITNGMVNAISTGNWSLKRFKMDRQGVTQVL 440
>ref|XP_343189.2| PREDICTED: similar to RNA polymerase III subunit RPC2 [Rattus
norvegicus]
Length = 1213
Score = 94.4 bits (233), Expect = 2e-17
Identities = 51/121 (42%), Positives = 71/121 (58%), Gaps = 15/121 (12%)
Query: 1 MVVMKAMGMEHDQEVVQLIGRDPRYSFLLMPSIEECTKIGIFTQAQALDYVDSKAERQ-- 58
+++ KAMG+E DQE+VQ+IG + PS+EEC K IFTQ QAL Y+ +K RQ
Sbjct: 317 VIIFKAMGVESDQEIVQMIGTEEHVMAAFGPSLEECQKAQIFTQMQALKYIGNKVRRQRM 376
Query: 59 ------------AFNILRETFLANVPVHGYNFRPKCIYVAVMIRRIMDAILNKDAMDDKS 106
A +L T L +VPV +NFR KCIY AVM+RR++ A + +DD+
Sbjct: 377 WGGGPKKTKIEEARELLASTILTHVPVKEFNFRAKCIYTAVMVRRVILA-QGDNKVDDRD 435
Query: 107 F 107
+
Sbjct: 436 Y 436
Score = 41.6 bits (96), Expect = 0.12
Identities = 18/35 (51%), Positives = 26/35 (73%)
Query: 186 INRDCITPGLERTLSTGNFDVKRFKMHKSGVTQVI 220
+ +D IT G+ +STGN+ +KRFKM + GVTQV+
Sbjct: 486 MRQDQITNGMVNAISTGNWSLKRFKMDRQGVTQVL 520
>emb|CAD97689.1| hypothetical protein [Homo sapiens] gi|42476127|ref|NP_060552.3|
polymerase (RNA) III (DNA directed) polypeptide B [Homo
sapiens]
Length = 1133
Score = 94.4 bits (233), Expect = 2e-17
Identities = 51/121 (42%), Positives = 71/121 (58%), Gaps = 15/121 (12%)
Query: 1 MVVMKAMGMEHDQEVVQLIGRDPRYSFLLMPSIEECTKIGIFTQAQALDYVDSKAERQ-- 58
+++ KAMG+E DQE+VQ+IG + PS+EEC K IFTQ QAL Y+ +K RQ
Sbjct: 237 VIIFKAMGVESDQEIVQMIGTEEHVMAAFGPSLEECQKAQIFTQMQALKYIGNKVRRQRM 296
Query: 59 ------------AFNILRETFLANVPVHGYNFRPKCIYVAVMIRRIMDAILNKDAMDDKS 106
A +L T L +VPV +NFR KCIY AVM+RR++ A + +DD+
Sbjct: 297 WGGGPKKTKIEEARELLASTILTHVPVKEFNFRAKCIYTAVMVRRVILA-QGDNKVDDRD 355
Query: 107 F 107
+
Sbjct: 356 Y 356
Score = 41.6 bits (96), Expect = 0.12
Identities = 18/35 (51%), Positives = 26/35 (73%)
Query: 186 INRDCITPGLERTLSTGNFDVKRFKMHKSGVTQVI 220
+ +D IT G+ +STGN+ +KRFKM + GVTQV+
Sbjct: 406 MRQDQITNGMVNAISTGNWSLKRFKMDRQGVTQVL 440
>emb|CAI30267.1| hypothetical protein [Pongo pygmaeus]
Length = 1116
Score = 94.4 bits (233), Expect = 2e-17
Identities = 51/121 (42%), Positives = 71/121 (58%), Gaps = 15/121 (12%)
Query: 1 MVVMKAMGMEHDQEVVQLIGRDPRYSFLLMPSIEECTKIGIFTQAQALDYVDSKAERQ-- 58
+++ KAMG+E DQE+VQ+IG + PS+EEC K IFTQ QAL Y+ +K RQ
Sbjct: 237 VIIFKAMGVESDQEIVQMIGTEEHVMAAFGPSLEECQKAQIFTQMQALKYIGNKVRRQRM 296
Query: 59 ------------AFNILRETFLANVPVHGYNFRPKCIYVAVMIRRIMDAILNKDAMDDKS 106
A +L T L +VPV +NFR KCIY AVM+RR++ A + +DD+
Sbjct: 297 WGGGPKKTKIEEARELLASTILTHVPVKEFNFRAKCIYTAVMVRRVILA-QGDNKVDDRD 355
Query: 107 F 107
+
Sbjct: 356 Y 356
>gb|AAH46238.1| Polymerase (RNA) III (DNA directed) polypeptide B [Homo sapiens]
gi|29428029|sp|Q9NW08|RPC2_HUMAN DNA-directed RNA
polymerase III subunit 127.6 kDa polypeptide (RNA
polymerase III subunit 2) (RPC2)
Length = 1133
Score = 94.4 bits (233), Expect = 2e-17
Identities = 51/121 (42%), Positives = 71/121 (58%), Gaps = 15/121 (12%)
Query: 1 MVVMKAMGMEHDQEVVQLIGRDPRYSFLLMPSIEECTKIGIFTQAQALDYVDSKAERQ-- 58
+++ KAMG+E DQE+VQ+IG + PS+EEC K IFTQ QAL Y+ +K RQ
Sbjct: 237 VIIFKAMGVESDQEIVQMIGTEEHVMAAFGPSLEECQKAQIFTQMQALKYIGNKVRRQRM 296
Query: 59 ------------AFNILRETFLANVPVHGYNFRPKCIYVAVMIRRIMDAILNKDAMDDKS 106
A +L T L +VPV +NFR KCIY AVM+RR++ A + +DD+
Sbjct: 297 WGGGPKKTKIEEARELLASTILTHVPVKEFNFRAKCIYTAVMVRRVILA-QGDNKVDDRD 355
Query: 107 F 107
+
Sbjct: 356 Y 356
Score = 41.6 bits (96), Expect = 0.12
Identities = 18/35 (51%), Positives = 26/35 (73%)
Query: 186 INRDCITPGLERTLSTGNFDVKRFKMHKSGVTQVI 220
+ +D IT G+ +STGN+ +KRFKM + GVTQV+
Sbjct: 406 MRQDQITNGMVNAISTGNWSLKRFKMDRQGVTQVL 440
>dbj|BAC30327.1| unnamed protein product [Mus musculus]
Length = 551
Score = 94.4 bits (233), Expect = 2e-17
Identities = 51/121 (42%), Positives = 71/121 (58%), Gaps = 15/121 (12%)
Query: 1 MVVMKAMGMEHDQEVVQLIGRDPRYSFLLMPSIEECTKIGIFTQAQALDYVDSKAERQ-- 58
+++ KAMG+E DQE+VQ+IG + PS+EEC K IFTQ QAL Y+ +K RQ
Sbjct: 237 VIIFKAMGVESDQEIVQMIGTEEHVMAAFGPSLEECQKAQIFTQMQALKYIGNKVRRQRM 296
Query: 59 ------------AFNILRETFLANVPVHGYNFRPKCIYVAVMIRRIMDAILNKDAMDDKS 106
A +L T L +VPV +NFR KCIY AVM+RR++ A + +DD+
Sbjct: 297 WGGGPKKTKIEEARELLASTILTHVPVKEFNFRAKCIYTAVMVRRVILA-QGDNKVDDRD 355
Query: 107 F 107
+
Sbjct: 356 Y 356
Score = 41.6 bits (96), Expect = 0.12
Identities = 18/35 (51%), Positives = 26/35 (73%)
Query: 186 INRDCITPGLERTLSTGNFDVKRFKMHKSGVTQVI 220
+ +D IT G+ +STGN+ +KRFKM + GVTQV+
Sbjct: 406 MRQDQITNGMVNAISTGNWSLKRFKMDRQGVTQVL 440
>ref|XP_636731.1| RNA polymerase III, second largest subunit [Dictyostelium
discoideum] gi|60465151|gb|EAL63250.1| RNA polymerase
III, second largest subunit [Dictyostelium discoideum]
Length = 1608
Score = 94.0 bits (232), Expect = 2e-17
Identities = 51/121 (42%), Positives = 73/121 (60%), Gaps = 15/121 (12%)
Query: 2 VVMKAMGMEHDQEVVQLIGRDPRYSFLLMPSIEECTKIGIFTQAQALDYVDSKAE----- 56
+V+K MG+E DQE+ QL+G D + + PS+EEC K G+ T AQALDY+ S+ +
Sbjct: 251 IVLKGMGVETDQEMAQLVGSDDVFLNAITPSLEECQKCGVHTAAQALDYLGSRIKVFRRP 310
Query: 57 ----------RQAFNILRETFLANVPVHGYNFRPKCIYVAVMIRRIMDAILNKDAMDDKS 106
+A +IL L +VPV YNFR K IY+++MIRRI+ A +K +DDK
Sbjct: 311 YGVQNKKTKSEEARDILAGVVLNHVPVRRYNFRLKVIYLSLMIRRIIMASKDKSCLDDKD 370
Query: 107 F 107
+
Sbjct: 371 Y 371
>gb|AAM18214.1| RNA polymerase III subunit RPC2 [Homo sapiens]
Length = 1133
Score = 92.8 bits (229), Expect = 5e-17
Identities = 51/121 (42%), Positives = 70/121 (57%), Gaps = 15/121 (12%)
Query: 1 MVVMKAMGMEHDQEVVQLIGRDPRYSFLLMPSIEECTKIGIFTQAQALDYVDSKAERQ-- 58
+++ KAMG+E DQE+VQ+IG PS+EEC K IFTQ QAL Y+ +K RQ
Sbjct: 237 VIIFKAMGVESDQEIVQMIGTAEHVMAAFGPSLEECQKAQIFTQMQALKYIGNKVRRQRM 296
Query: 59 ------------AFNILRETFLANVPVHGYNFRPKCIYVAVMIRRIMDAILNKDAMDDKS 106
A +L T L +VPV +NFR KCIY AVM+RR++ A + +DD+
Sbjct: 297 WGGGPKKTKIEEARELLASTILTHVPVKEFNFRAKCIYTAVMVRRVILA-QGDNKVDDRD 355
Query: 107 F 107
+
Sbjct: 356 Y 356
Score = 41.6 bits (96), Expect = 0.12
Identities = 18/35 (51%), Positives = 26/35 (73%)
Query: 186 INRDCITPGLERTLSTGNFDVKRFKMHKSGVTQVI 220
+ +D IT G+ +STGN+ +KRFKM + GVTQV+
Sbjct: 406 MRQDQITNGMVNAISTGNWSLKRFKMDRQGVTQVL 440
>ref|XP_317312.2| ENSANGP00000006850 [Anopheles gambiae str. PEST]
gi|55237532|gb|EAA12689.2| ENSANGP00000006850 [Anopheles
gambiae str. PEST]
Length = 1112
Score = 90.9 bits (224), Expect = 2e-16
Identities = 50/121 (41%), Positives = 76/121 (62%), Gaps = 15/121 (12%)
Query: 2 VVMKAMGMEHDQEVVQLIGRDPRYSFLLMPSIEECTKIGIFTQAQALDYVDSK--AER-- 57
+++KAMG+ DQE++QL+G DP PS+ E +FTQ +AL+Y+ SK A+R
Sbjct: 216 IILKAMGIASDQEIMQLVGIDPETQKRFAPSLLEAANHKVFTQQRALEYMGSKLIAKRFT 275
Query: 58 -----------QAFNILRETFLANVPVHGYNFRPKCIYVAVMIRRIMDAILNKDAMDDKS 106
+A ++L T LA+VPV +NF+ K IYVA+MIRR+M A L++ A+DD+
Sbjct: 276 TAATKYKTTADEARDLLATTILAHVPVPSFNFQVKAIYVALMIRRVMAAELDRSAVDDRD 335
Query: 107 F 107
+
Sbjct: 336 Y 336
Score = 44.3 bits (103), Expect = 0.019
Identities = 19/30 (63%), Positives = 25/30 (83%)
Query: 191 ITPGLERTLSTGNFDVKRFKMHKSGVTQVI 220
IT GLE +STGN+ +KRFKM ++GVTQV+
Sbjct: 391 ITAGLETAISTGNWTIKRFKMERAGVTQVL 420
>ref|XP_531760.1| PREDICTED: similar to DNA-directed RNA polymerase III subunit 127.6
kDa polypeptide (RNA polymerase III subunit 2) (RPC2)
[Canis familiaris]
Length = 1422
Score = 90.9 bits (224), Expect = 2e-16
Identities = 50/118 (42%), Positives = 69/118 (58%), Gaps = 15/118 (12%)
Query: 4 MKAMGMEHDQEVVQLIGRDPRYSFLLMPSIEECTKIGIFTQAQALDYVDSKAERQ----- 58
++AMG+E DQE+VQ+IG + PS+EEC K IFTQ QAL Y+ +K RQ
Sbjct: 450 LEAMGVESDQEIVQMIGTEEHVMAAFGPSLEECQKAQIFTQMQALKYIGNKVRRQRMWGG 509
Query: 59 ---------AFNILRETFLANVPVHGYNFRPKCIYVAVMIRRIMDAILNKDAMDDKSF 107
A +L T L +VPV +NFR KCIY AVM+RR++ A + +DD+ +
Sbjct: 510 GPKKTKIEEARELLASTILTHVPVKEFNFRAKCIYTAVMVRRVILA-QGDNKVDDRDY 566
Score = 41.6 bits (96), Expect = 0.12
Identities = 18/35 (51%), Positives = 26/35 (73%)
Query: 186 INRDCITPGLERTLSTGNFDVKRFKMHKSGVTQVI 220
+ +D IT G+ +STGN+ +KRFKM + GVTQV+
Sbjct: 616 MRQDQITNGMVNAISTGNWSLKRFKMDRQGVTQVL 650
>emb|CAG03415.1| unnamed protein product [Tetraodon nigroviridis]
Length = 1171
Score = 90.5 bits (223), Expect = 2e-16
Identities = 49/119 (41%), Positives = 69/119 (57%), Gaps = 14/119 (11%)
Query: 2 VVMKAMGMEHDQEVVQLIGRDPRYSFLLMPSIEECTKIGIFTQAQALDYVDSKAERQ--- 58
++ K MG+E DQE+VQ+IG + PS+EEC K IFTQ QAL Y+ +K RQ
Sbjct: 267 IIFKGMGVESDQEIVQMIGTEENVMASFAPSLEECQKAQIFTQTQALRYLGNKVRRQRMW 326
Query: 59 ----------AFNILRETFLANVPVHGYNFRPKCIYVAVMIRRIMDAILNKDAMDDKSF 107
A +L L +VPV +NFR KCIY+AVM+RR++ A + +DD+ +
Sbjct: 327 GGPKKTKMEEARELLASLILTHVPVKEFNFRAKCIYLAVMVRRVILA-QGDNKVDDRDY 384
Score = 41.6 bits (96), Expect = 0.12
Identities = 18/35 (51%), Positives = 26/35 (73%)
Query: 186 INRDCITPGLERTLSTGNFDVKRFKMHKSGVTQVI 220
+ +D IT G+ +STGN+ +KRFKM + GVTQV+
Sbjct: 434 MRQDQITNGMVNAISTGNWSLKRFKMDRQGVTQVL 468
>emb|CAG03394.1| unnamed protein product [Tetraodon nigroviridis]
Length = 271
Score = 90.5 bits (223), Expect = 2e-16
Identities = 49/119 (41%), Positives = 69/119 (57%), Gaps = 14/119 (11%)
Query: 2 VVMKAMGMEHDQEVVQLIGRDPRYSFLLMPSIEECTKIGIFTQAQALDYVDSKAERQ--- 58
++ K MG+E DQE+VQ+IG + PS+EEC K IFTQ QAL Y+ +K RQ
Sbjct: 138 IIFKGMGVESDQEIVQMIGTEENVMASFAPSLEECQKAQIFTQTQALRYLGNKVRRQRMW 197
Query: 59 ----------AFNILRETFLANVPVHGYNFRPKCIYVAVMIRRIMDAILNKDAMDDKSF 107
A +L L +VPV +NFR KCIY+AVM+RR++ A + +DD+ +
Sbjct: 198 GGPKKTKMEEARELLASLILTHVPVKEFNFRAKCIYLAVMVRRVILA-QGDNKVDDRDY 255
>ref|XP_615703.1| PREDICTED: similar to DNA-directed RNA polymerase III subunit 127.6
kDa polypeptide (RNA polymerase III subunit 2) (RPC2),
partial [Bos taurus]
Length = 384
Score = 87.0 bits (214), Expect = 3e-15
Identities = 48/113 (42%), Positives = 69/113 (60%), Gaps = 7/113 (6%)
Query: 1 MVVMKAMGMEHDQEVVQLIGRDPRYSFLLMPSIEECTKIGIFTQAQALDYVDSKAERQAF 60
+++ KAMG+E DQE+VQ+IG + PS+EEC K IFTQ QAL Y+ +K RQ
Sbjct: 102 VIIFKAMGVESDQEIVQMIGTEEHVMAAFGPSLEECQKAQIFTQMQALKYIGNKVRRQRM 161
Query: 61 NILRE-----TFLANVP-VHGYNFRPKCIYVAVMIRRIMDAILNKDAMDDKSF 107
+ R+ + AN V +NFR KCIY AVM+RR++ A + +DD+ +
Sbjct: 162 TLKRKEVEAASLQANPKMVKEFNFRAKCIYTAVMVRRVILA-QGDNKVDDRDY 213
>emb|CAA93558.1| SPAC4G9.08c [Schizosaccharomyces pombe]
gi|19114602|ref|NP_593690.1| hypothetical protein
SPAC4G9.08c [Schizosaccharomyces pombe 972h-]
gi|1710663|sp|Q10233|RPC2_SCHPO Putative DNA-directed
RNA polymerase III 130 kDa polypeptide (RNA polymerase
III subunit 2)
Length = 1165
Score = 86.3 bits (212), Expect = 4e-15
Identities = 42/121 (34%), Positives = 72/121 (58%), Gaps = 14/121 (11%)
Query: 1 MVVMKAMGMEHDQEVVQLI-GRDPRYSFLLMPSIEECTKIGIFTQAQALDYVDSKAE--- 56
+VV+KAMG++ DQE+ +L+ G + Y L PSIEEC K+ I+T QAL+Y+ ++ +
Sbjct: 262 VVVLKAMGLQSDQEIFELVAGAEASYQDLFAPSIEECAKLNIYTAQQALEYIGARVKVNR 321
Query: 57 ----------RQAFNILRETFLANVPVHGYNFRPKCIYVAVMIRRIMDAILNKDAMDDKS 106
+A +L LA++ V FRPK +Y+ +M RR++ A+++ +DD+
Sbjct: 322 RAGANRLPPHEEALEVLAAVVLAHINVFNLEFRPKAVYIGIMARRVLMAMVDPLQVDDRD 381
Query: 107 F 107
+
Sbjct: 382 Y 382
Score = 43.9 bits (102), Expect = 0.024
Identities = 19/35 (54%), Positives = 27/35 (76%)
Query: 186 INRDCITPGLERTLSTGNFDVKRFKMHKSGVTQVI 220
++ D IT G+ R LSTGN+ +KRFKM ++GVT V+
Sbjct: 435 VHSDHITQGMVRALSTGNWSLKRFKMERAGVTHVL 469
>ref|XP_625037.1| PREDICTED: similar to DNA-directed RNA polymerase III subunit 127.6
kDa polypeptide (RNA polymerase III subunit 2) (RPC2)
[Apis mellifera]
Length = 1134
Score = 82.8 bits (203), Expect = 5e-14
Identities = 39/118 (33%), Positives = 72/118 (60%), Gaps = 12/118 (10%)
Query: 2 VVMKAMGMEHDQEVVQLIGRDPRYSFLLMPSIEECTKIGIFTQAQALDYVDSKAERQAFN 61
++ KAMG+ DQE++QLIG + + PS+EEC + +F Q QAL ++ +K +++ F+
Sbjct: 240 IIFKAMGIVSDQEIMQLIGTEEEFMKKFAPSLEECHVLNVFAQNQALRFLSNKRKQKRFS 299
Query: 62 ILRET------------FLANVPVHGYNFRPKCIYVAVMIRRIMDAILNKDAMDDKSF 107
+++ + L++VPV +NF+ K Y+A+MIR++M A + +DD+ +
Sbjct: 300 VIKSSITDEMKDALATNILSHVPVIDFNFKMKATYIALMIRKVMKAQSDGKLVDDRDY 357
Score = 40.4 bits (93), Expect = 0.27
Identities = 18/35 (51%), Positives = 26/35 (73%)
Query: 186 INRDCITPGLERTLSTGNFDVKRFKMHKSGVTQVI 220
+ +D IT GL +S+GN+ +KRFKM + GVTQV+
Sbjct: 407 MRQDQITNGLAFAISSGNWTIKRFKMERHGVTQVL 441
>ref|XP_599689.1| PREDICTED: similar to hypothetical protein, partial [Bos taurus]
Length = 278
Score = 82.4 bits (202), Expect = 6e-14
Identities = 47/108 (43%), Positives = 65/108 (59%), Gaps = 7/108 (6%)
Query: 6 AMGMEHDQEVVQLIGRDPRYSFLLMPSIEECTKIGIFTQAQALDYVDSKAERQAFNILRE 65
AMG+E DQE+VQ+IG + PS+EEC K IFTQ QAL Y+ +K RQ + R+
Sbjct: 1 AMGVESDQEIVQMIGTEEHVMAAFGPSLEECQKAQIFTQMQALKYIGNKVRRQRMTLKRK 60
Query: 66 -----TFLANVP-VHGYNFRPKCIYVAVMIRRIMDAILNKDAMDDKSF 107
+ AN V +NFR KCIY AVM+RR++ A + +DD+ +
Sbjct: 61 EVEAASLQANPKMVKEFNFRAKCIYTAVMVRRVILA-QGDNKVDDRDY 107
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.365 0.160 0.668
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,341,102,903
Number of Sequences: 2540612
Number of extensions: 47212353
Number of successful extensions: 211387
Number of sequences better than 10.0: 83
Number of HSP's better than 10.0 without gapping: 57
Number of HSP's successfully gapped in prelim test: 26
Number of HSP's that attempted gapping in prelim test: 211181
Number of HSP's gapped (non-prelim): 162
length of query: 911
length of database: 863,360,394
effective HSP length: 137
effective length of query: 774
effective length of database: 515,296,550
effective search space: 398839529700
effective search space used: 398839529700
T: 11
A: 40
X1: 14 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 36 (21.6 bits)
S2: 80 (35.4 bits)
Medicago: description of AC135796.9


