

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135795.1 - phase: 0
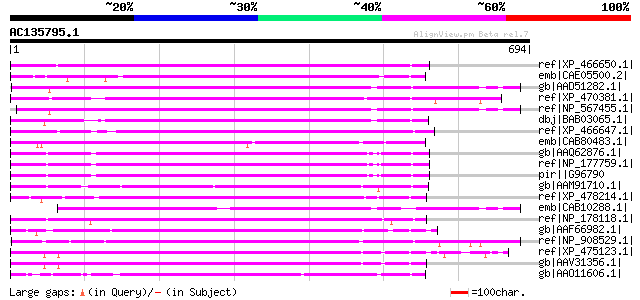
(694 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|XP_466650.1| putative far-red impaired response protein [Ory... 447 e-124
emb|CAE05500.2| OSJNBa0022H21.20 [Oryza sativa (japonica cultiva... 408 e-112
gb|AAD51282.1| far-red impaired response protein [Arabidopsis th... 398 e-109
ref|XP_470381.1| putative far-red impaired response protein [Ory... 391 e-107
ref|NP_567455.1| far-red impaired response protein (FAR1) / far-... 383 e-104
dbj|BAB03065.1| far-red impaired response protein; Mutator-like ... 376 e-102
ref|XP_466647.1| putative far-red impaired response protein [Ory... 375 e-102
emb|CAB80483.1| hypothetical protein [Arabidopsis thaliana] gi|4... 371 e-101
gb|AAQ62876.1| At1g76320 [Arabidopsis thaliana] gi|62320160|dbj|... 368 e-100
ref|NP_177759.1| far-red impaired responsive protein, putative [... 368 e-100
pir||G96790 hypothetical protein F15M4.18 [imported] - Arabidops... 368 e-100
gb|AAM91710.1| unknown protein [Arabidopsis thaliana] gi|1942405... 356 2e-96
ref|XP_478214.1| putative far-red impaired response protein [Ory... 348 4e-94
emb|CAB10288.1| hypothetical protein [Arabidopsis thaliana] gi|7... 332 3e-89
ref|NP_178118.1| far-red impaired responsive protein, putative [... 327 1e-87
gb|AAF66982.1| transposase [Zea mays] 319 2e-85
ref|NP_908529.1| P0702D12.15 [Oryza sativa (japonica cultivar-gr... 315 4e-84
ref|XP_475123.1| putative Mutator-like transposase [Oryza sativa... 313 2e-83
gb|AAV31356.1| hypothetical protein [Oryza sativa (japonica cult... 312 3e-83
gb|AAO11606.1| At2g27110/T20P8.16 [Arabidopsis thaliana] gi|2019... 308 3e-82
>ref|XP_466650.1| putative far-red impaired response protein [Oryza sativa (japonica
cultivar-group)] gi|47497945|dbj|BAD20150.1| putative
far-red impaired response protein [Oryza sativa
(japonica cultivar-group)] gi|47496830|dbj|BAD19590.1|
putative far-red impaired response protein [Oryza sativa
(japonica cultivar-group)]
Length = 817
Score = 447 bits (1151), Expect = e-124
Identities = 230/564 (40%), Positives = 338/564 (59%), Gaps = 7/564 (1%)
Query: 1 MTFSSEEEVIKYYKSYARCMGFGTVKINSKNAKDGK-KYFTLGCTRARSYVSNSKNLLKP 59
M F++ EV ++Y+ YAR +GFG S +++G Y L C + K +
Sbjct: 43 MVFNNHTEVNRFYRRYARRVGFGVSVRRSSFSQEGTCLYLELMCCKGGRPRYEPKFRKRA 102
Query: 60 NPTIRAQCMARVNMSMSLDGKITITKVALEHNHGLSPTKSRYFRCNKNLDPHTKRRLDIN 119
+ T C A++ + + D + + L+HNH +SP +R+ K L KRRL +
Sbjct: 103 SST--TNCPAKIRVKLWGDKLLHVELAILDHNHPVSPAMARFLNSYKQLSGPAKRRLRMG 160
Query: 120 DQAGINVSRNFRSMVVEANGYDNLTFGEKDCRNYIDKARRLRLGTGDAEAIQNYFVRMQK 179
+ V + V + + L FGE +++++ R L+ GD+EA++ +F RMQ
Sbjct: 161 GPGAMPVEEPSKMPVDKLGALEELLFGESKHHSFVERGR-LKFQPGDSEALRLFFTRMQA 219
Query: 180 KNSQFYYVMDVDDKSRLRNVFWVDARCRAAYEYFGEVITFDTTYLTNKYDMPFAPFVGVN 239
KN+ F+ V+D+DD+ +RNVFWVDAR R+ YE++ +V+T DT+Y+ KYDMP A F+GVN
Sbjct: 220 KNANFFNVIDLDDEGCVRNVFWVDARSRSMYEFYNDVVTLDTSYVVGKYDMPLATFIGVN 279
Query: 240 HHGQSVLLGCALLSNEDTKTFSWLFKTWLECMHGRAPNAIITDQDRAMKKAIEDVFPKAR 299
HHGQSVLLGC LLS+E +T+SWLFK W+ CM+G P AIIT R ++ A+ +V P
Sbjct: 280 HHGQSVLLGCGLLSDETAETYSWLFKAWIACMYGNLPKAIITGHCRGIQSAVAEVIPGVH 339
Query: 300 HRWCLWHLMKKVPEKLGRHSHYESIKLLLHDAVYDSSSISDFMEKWKKMIECYELHDNEW 359
HR CL+H+M+K E+LG S Y +I AVYDS +I +F W +I L N+W
Sbjct: 340 HRICLFHIMRKATERLGGLSEYAAISKAFQKAVYDSLTIDEFEGNWNALITYNGLQGNDW 399
Query: 360 LKGLFDERYRWVPVYVRDTFWAGMSTTQRSESMNSFFDGYVSSKTTLKQFVEQYDNALKD 419
L+ +++ RY WVPV+++DTFWAGMS TQR+E++ FFDGYV KTTLK F+ +Y+ AL+
Sbjct: 400 LRSIYECRYSWVPVFLKDTFWAGMSATQRNENIIPFFDGYVDLKTTLKHFLGKYEMALQS 459
Query: 420 KIEKESMADFVSFNTTIACISLFGFESQFQKAFTNAKFKEFQIEIASMIYCNAFLEGMEN 479
K EKE+ ADF +F+ +S F E Q K +T+ FK+FQ EI +++YC+ ++
Sbjct: 460 KYEKEAQADFETFHKQRPPVSKFYMEEQLSKVYTHNIFKKFQDEIEAIMYCHVSFINVDG 519
Query: 480 LNSTFCVIESKKVYD--RFKDTRFRVIFNEKDFEIQCACCLFEFKGILCRHILCVLQLTG 537
L STF V E + D R F V N ++ C C F+F GILCRH L VL+
Sbjct: 520 LISTFDVKEWIFLEDGKRTMSKIFTVTNNTDKNDLTCICGGFQFNGILCRHSLSVLKFQ- 578
Query: 538 KTESVPSCYILSRWRKDIKRRHTL 561
+ +P Y+L RW+KD ++ H +
Sbjct: 579 QVREIPPHYVLDRWKKDFRQLHVM 602
>emb|CAE05500.2| OSJNBa0022H21.20 [Oryza sativa (japonica cultivar-group)]
gi|50925121|ref|XP_472870.1| OSJNBa0022H21.20 [Oryza
sativa (japonica cultivar-group)]
Length = 803
Score = 408 bits (1049), Expect = e-112
Identities = 223/572 (38%), Positives = 326/572 (56%), Gaps = 32/572 (5%)
Query: 1 MTFSSEEEVIKYYKSYARCMGFGTVKINSKNAKDGKKY-FTLGCTRARSYVSNSKNLLKP 59
MTF + + ++Y Y GFG K S + DG KY T C + ++ K LK
Sbjct: 44 MTFETADLAYRFYLEYGYRAGFGVSKRTSHSV-DGVKYRATFVCYKGG--IARIKPGLKA 100
Query: 60 NPTIRAQCMARVNMSM---SLDGKITITKVALEHNHGLSPTKSRYFRCNKNLDPHTKRRL 116
+ A+ + M + + + + + V LEHNH +P R+ C K+L +
Sbjct: 101 RRRLVAKTGCKAMMVVKYNTTENQWEVVFVELEHNHPCNPEMVRFMMCFKDLPDWQREHR 160
Query: 117 DINDQAGINV----------SRNFRSMVVEANGYDNLTFGEKDCRNYIDKARRLRLGTGD 166
N + +N ++N MV + N + KA +LR GD
Sbjct: 161 PFNAKTRLNPKIHSGRGRPPNQNKDFMVKKTFSQSNYSI------EAAGKAGKLRFAEGD 214
Query: 167 AEAIQNYFVRMQKKNSQFYYVMDVDDKSRLRNVFWVDARCRAAYEYFGEVITFDTTYLTN 226
EA+ +F +MQ +NS F+Y D+DD+ RL+NV WVDAR RAAY++F +V+ FDT YLT
Sbjct: 215 VEALLVFFDKMQAQNSNFFYNWDMDDEGRLKNVCWVDARSRAAYQHFCDVVCFDTVYLTY 274
Query: 227 KYDMPFAPFVGVNHHGQSVLLGCALLSNEDTKTFSWLFKTWLECMHGRAPNAIITDQDRA 286
++ +P F+G+NHHGQ VLLGC LL +E +TFSWLFK WL+CM+ ++P AIIT R
Sbjct: 275 QFVIPLVAFLGINHHGQFVLLGCGLLGDESPETFSWLFKKWLKCMNDKSPEAIITTHSRP 334
Query: 287 MKKAIEDVFPKARHRWCLWHLMKKVPEKLGRHSHYESIKLLLHDAVYDSSSISDFMEKWK 346
+ KA+ +VFP RHR+ LWH+MK++PE GR E+I L + V+D+ + +DF +W
Sbjct: 335 VVKAVAEVFPNTRHRYNLWHIMKELPEMSGRVEDKEAISLRMKKVVFDTITSTDFEREWV 394
Query: 347 KMIECYELHDNEWLKGLFDERYRWVPVYVRDTFWAGMSTTQRSESMNSFFDGYVSSKTTL 406
+M+ Y LHDN WL LF+ER +WVP YV+DTFWAG+ST +RSE + +FFDGY++ +TT+
Sbjct: 395 EMVNQYNLHDNHWLTTLFEERAKWVPAYVKDTFWAGISTVRRSERLEAFFDGYITPETTI 454
Query: 407 KQFVEQYDNALKDKIEKESMADFVSFNTTIACISLFGFESQFQKAFTNAKFKEFQIEIAS 466
K F+EQ+D A+K + ++E+ DF SF +S FE QF +T F++FQ +
Sbjct: 455 KTFIEQFDTAMKLRSDREAYDDFRSFQQRPQVLSGLLFEEQFANVYTINMFQKFQDHLKQ 514
Query: 467 M--IYCNAFLEGMENLNSTFCVIESKKVYDRFKDTRFRVIFNEKDFEIQCACCLFEFKGI 524
+ + C + T VI ++ YD ++V++N + E+ C C F+FKGI
Sbjct: 515 LMNVTCTEVSRNGSIVTYTVTVIGKERKYD------YKVMYNSAEKEVWCICRSFQFKGI 568
Query: 525 LCRHILCVLQLTGKTESVPSCYILSRWRKDIK 556
LC H L VL+ +P YIL RWRKD K
Sbjct: 569 LCSHALAVLR-QELVMLIPYKYILDRWRKDYK 599
>gb|AAD51282.1| far-red impaired response protein [Arabidopsis thaliana]
Length = 827
Score = 398 bits (1022), Expect = e-109
Identities = 228/688 (33%), Positives = 371/688 (53%), Gaps = 28/688 (4%)
Query: 3 FSSEEEVIKYYKSYARCMGFGTVKINSKNAKDGKKYFTLGCTRARSYVS----NSKNLLK 58
F + E +Y+ YA+ MGF T NS+ +K K + +R V+ +S + +
Sbjct: 57 FDTHEAAYIFYQEYAKSMGFTTSIKNSRRSKKTKDFIDAKFACSRYGVTPESESSGSSSR 116
Query: 59 PNPTIRAQCMARVNMSMSLDGKITITKVALEHNHGLSPTKSRYFRCNKNLDPHTKRRLDI 118
+ + C A +++ DGK I + +HNH L P + +FR +N+ K +DI
Sbjct: 117 RSTVKKTDCKASMHVKRRPDGKWIIHEFVKDHNHELLPALAYHFRIQRNVKLAEKNNIDI 176
Query: 119 NDQAGINVSRNFRSMVVEANGYDNL-TFGEKDCRNYIDKARRLRLGTGDAEAIQNYFVRM 177
+ + M ++ GY N+ + + D + +DK R L L GD++ + YF R+
Sbjct: 177 LHAVSERTKKMYVEMSRQSGGYKNIGSLLQTDVSSQVDKGRYLALEEGDSQVLLEYFKRI 236
Query: 178 QKKNSQFYYVMDVDDKSRLRNVFWVDARCRAAYEYFGEVITFDTTYLTNKYDMPFAPFVG 237
+K+N +F+Y +D+++ RLRN+FW DA+ R Y F +V++FDTTY+ +P A F+G
Sbjct: 237 KKENPKFFYAIDLNEDQRLRNLFWADAKSRDDYLSFNDVVSFDTTYVKFNDKLPLALFIG 296
Query: 238 VNHHGQSVLLGCALLSNEDTKTFSWLFKTWLECMHGRAPNAIITDQDRAMKKAIEDVFPK 297
VNHH Q +LLGCAL+++E +TF WL KTWL M GRAP I+TDQD+ + A+ ++ P
Sbjct: 297 VNHHSQPMLLGCALVADESMETFVWLIKTWLRAMGGRAPKVILTDQDKFLMSAVSELLPN 356
Query: 298 ARHRWCLWHLMKKVPEKLGR-HSHYESIKLLLHDAVYDSSSISDFMEKWKKMIECYELHD 356
RH + LWH+++K+PE +E+ L + ++ S + +F +W KM+ + L +
Sbjct: 357 TRHCFALWHVLEKIPEYFSHVMKRHENFLLKFNKCIFRSWTDDEFDMRWWKMVSQFGLEN 416
Query: 357 NEWLKGLFDERYRWVPVYVRDTFWAGMSTTQRSESMNSFFDGYVSSKTTLKQFVEQYDNA 416
+EWL L + R +WVP ++ D F AGMST+QRSES+NSFFD Y+ K TLK+F+ QY
Sbjct: 417 DEWLLWLHEHRQKWVPTFMSDVFLAGMSTSQRSESVNSFFDKYIHKKITLKEFLRQYGVI 476
Query: 417 LKDKIEKESMADFVSFNTTIACISLFGFESQFQKAFTNAKFKEFQIEIASMIYCNAFLEG 476
L+++ E+ES+ADF + + A S +E Q +T+ FK+FQ+E+ ++ C+ E
Sbjct: 477 LQNRYEEESVADFDTCHKQPALKSPSPWEKQMATTYTHTIFKKFQVEVLGVVACHPRKEK 536
Query: 477 MENLNSTFCVIESKKVYDRFKDTRFRVIFNEKDFEIQCACCLFEFKGILCRHILCVLQLT 536
+ +TF +V D KD F V +++ E+ C C +FE+KG LCRH L +LQ+
Sbjct: 537 EDENMATF------RVQDCEKDDDFLVTWSKTKSELCCFCRMFEYKGFLCRHALMILQMC 590
Query: 537 GKTESVPSCYILSRWRKDIKRRHTLIKCGFDGNVELQLVGKACDAFYEVVSVGIHTEDDL 596
G S+P YIL RW KD K + +Q C E+ G +E++
Sbjct: 591 G-FASIPPQYILKRWTKDAKSGVLAGEGADQIQTRVQRYNDLCSRATELSEEGCVSEENY 649
Query: 597 -LKVMNRINELKIELNCEGSSSVIIEEDSLVQNQMAKILDPATTRSKGRPPSKRKASKLD 655
+ + + LK ++ + + I E +S + N T + + + KA+K
Sbjct: 650 NIALRTLVETLKNCVDMNNARNNITESNSQLNN--------GTHEEENQVMAGVKATK-- 699
Query: 656 QIVQKKVLRKKTQNRNNKSNNYLSQEEV 683
KK + +K + + S SQ+ +
Sbjct: 700 ----KKTVYRKRKGQQEASQMLESQQSL 723
>ref|XP_470381.1| putative far-red impaired response protein [Oryza sativa (japonica
cultivar-group)] gi|41469648|gb|AAS07371.1| putative
far-red impaired response protein [Oryza sativa
(japonica cultivar-group)]
Length = 786
Score = 391 bits (1005), Expect = e-107
Identities = 223/668 (33%), Positives = 342/668 (50%), Gaps = 35/668 (5%)
Query: 1 MTFSSEEEVIKYYKSYARCMGFGTVKINSKNAKDGK-KYFTLGCTRARSYVSNSKNLLKP 59
M F S EEV+ +YK YA GFG S K G + L C + ++ +
Sbjct: 32 MVFKSYEEVLNFYKRYALRTGFGVCVKKSSFTKAGLCRRLVLVCNKWGDGKEDA--CYQA 89
Query: 60 NPTIRAQCMARVNMSMSLDGKITITKVALEHNHGLSPTKSRYFRCNKNLDPHTKRRLDIN 119
PT + C A V + DG + + V LEHNH L+P+ +R+ RC K L
Sbjct: 90 RPTAKTNCQATVVARLWSDGLLHLMDVNLEHNHALNPSAARFLRCYKTLP---------- 139
Query: 120 DQAGINVSRNFRSMVVEANGYDNLTFGEKDCRNYIDKARRLRLGTGDAEAIQNYFVRMQK 179
S + +VV A + T G+ + + D R L++G D AI +F MQ
Sbjct: 140 -------SGMSKDLVVRAARGECSTSGDIEVPIFDDWGR-LKIGEDDVAAINGFFADMQA 191
Query: 180 KNSQFYYVMDVDDKSRLRNVFWVDARCRAAYEYFGEVITFDTTYLTNKYDMPFAPFVGVN 239
K + F+Y+MD + LR+VFW D+R RAAY+YF + + DTT L N++D P F+GVN
Sbjct: 192 KQANFFYLMDFYGEGHLRSVFWADSRSRAAYQYFNDAVWIDTTCLRNRFDTPLVLFLGVN 251
Query: 240 HHGQSVLLGCALLSNEDTKTFSWLFKTWLECMHGRAPNAIITDQDRAMKKAIEDVFPKAR 299
HHG+ VLLGC L S+E T++F WL K+WL CM G PNAI+TD A+K A+ +VFP AR
Sbjct: 252 HHGELVLLGCGLFSDESTESFLWLLKSWLTCMKGWPPNAIVTDDCAAIKAAVREVFPNAR 311
Query: 300 HRWCLWHLMKKVPEKLGRHSHYESIKLLLHDAVYDSSSISDFMEKWKKMIECYELHDNEW 359
HR WH+++ + EKLG + +E +K L +YDS +F +W +I + L DNEW
Sbjct: 312 HRISDWHVLRSISEKLGESAQFEGMKTELETVIYDSLKDDEFEARWNNLISRFGLQDNEW 371
Query: 360 LKGLFDERYRWVPVYVRDTFWAGMSTTQRSESMNSFFDGYVSSKTTLKQFVEQYDNALKD 419
+ L++ R+ WVP +++DTFWAG+ST ES N+FF+ ++ +T L F+ Y N L++
Sbjct: 372 ITFLYENRHFWVPAFLKDTFWAGLSTVNHHESPNAFFEDSINPETKLVTFLSSYVNLLQN 431
Query: 420 KIEKESMADFVSFNTTIACISLFGFESQFQKAFTNAKFKEFQIEIASMIYCNAFLEGMEN 479
K + E D S + + +S F E Q + +T F + Q E+ + + C L ++
Sbjct: 432 KYKMEEDDDLESLSRSRVLVSKFPMEEQLSRLYTFKMFTKLQNELNATMNCEVQL---DD 488
Query: 480 LNSTFCVIESKKVYDRFKDTRFRVIFNEKDFEIQCACCLFEFKGILCRHILCVLQLTGKT 539
S+ VI+ + + ++ V+ + ++C C LF+F GI+CRH L VL+
Sbjct: 489 STSSIVVIDLAESSGEMVNKKYEVVHCMETDRMECNCGLFQFSGIVCRHTLSVLKCQ-HV 547
Query: 540 ESVPSCYILSRWRKDIKRRHTLIKCGFD----GNVE-LQLVGKACDAFYEVVSVGIHTED 594
+P CY+L+RWR D K+ H L D ++E V C E+ +
Sbjct: 548 FDIPPCYVLNRWRNDFKQLHALDNPWKDLVTSNHIERYDYVSLQCLRLVEIGASSDEKHQ 607
Query: 595 DLLKVMNRINELKIELN-CEGSSSVIIEEDSLVQN----QMAKILDPATTRSKGRPPSKR 649
LK++ I ++ N C + + + Q + +GRPP K
Sbjct: 608 HALKLIRDIRRTLLDDNLCRELEQKLTPSERAINGDSHMQAGSSEGGPAKKRRGRPPKKS 667
Query: 650 KASKLDQI 657
K + ++ +
Sbjct: 668 KDTNVESV 675
>ref|NP_567455.1| far-red impaired response protein (FAR1) / far-red impaired
responsive protein (FAR1) [Arabidopsis thaliana]
Length = 768
Score = 383 bits (983), Expect = e-104
Identities = 222/681 (32%), Positives = 364/681 (52%), Gaps = 28/681 (4%)
Query: 10 IKYYKSYARCMGFGTVKINSKNAKDGKKYFTLGCTRARSYVS----NSKNLLKPNPTIRA 65
I + ++ F T NS+ +K K + +R V+ +S + + + +
Sbjct: 5 ISFIRNMQNLWVFTTSIKNSRRSKKTKDFIDAKFACSRYGVTPESESSGSSSRRSTVKKT 64
Query: 66 QCMARVNMSMSLDGKITITKVALEHNHGLSPTKSRYFRCNKNLDPHTKRRLDINDQAGIN 125
C A +++ DGK I + +HNH L P + +FR +N+ K +DI
Sbjct: 65 DCKASMHVKRRPDGKWIIHEFVKDHNHELLPALAYHFRIQRNVKLAEKNNIDILHAVSER 124
Query: 126 VSRNFRSMVVEANGYDNL-TFGEKDCRNYIDKARRLRLGTGDAEAIQNYFVRMQKKNSQF 184
+ + M ++ GY N+ + + D + +DK R L L GD++ + YF R++K+N +F
Sbjct: 125 TKKMYVEMSRQSGGYKNIGSLLQTDVSSQVDKGRYLALEEGDSQVLLEYFKRIKKENPKF 184
Query: 185 YYVMDVDDKSRLRNVFWVDARCRAAYEYFGEVITFDTTYLTNKYDMPFAPFVGVNHHGQS 244
+Y +D+++ RLRN+FW DA+ R Y F +V++FDTTY+ +P A F+GVNHH Q
Sbjct: 185 FYAIDLNEDQRLRNLFWADAKSRDDYLSFNDVVSFDTTYVKFNDKLPLALFIGVNHHSQP 244
Query: 245 VLLGCALLSNEDTKTFSWLFKTWLECMHGRAPNAIITDQDRAMKKAIEDVFPKARHRWCL 304
+LLGCAL+++E +TF WL KTWL M GRAP I+TDQD+ + A+ ++ P RH + L
Sbjct: 245 MLLGCALVADESMETFVWLIKTWLRAMGGRAPKVILTDQDKFLMSAVSELLPNTRHCFAL 304
Query: 305 WHLMKKVPEKLGR-HSHYESIKLLLHDAVYDSSSISDFMEKWKKMIECYELHDNEWLKGL 363
WH+++K+PE +E+ L + ++ S + +F +W KM+ + L ++EWL L
Sbjct: 305 WHVLEKIPEYFSHVMKRHENFLLKFNKCIFRSWTDDEFDMRWWKMVSQFGLENDEWLLWL 364
Query: 364 FDERYRWVPVYVRDTFWAGMSTTQRSESMNSFFDGYVSSKTTLKQFVEQYDNALKDKIEK 423
+ R +WVP ++ D F AGMST+QRSES+NSFFD Y+ K TLK+F+ QY L+++ E+
Sbjct: 365 HEHRQKWVPTFMSDVFLAGMSTSQRSESVNSFFDKYIHKKITLKEFLRQYGVILQNRYEE 424
Query: 424 ESMADFVSFNTTIACISLFGFESQFQKAFTNAKFKEFQIEIASMIYCNAFLEGMENLNST 483
ES+ADF + + A S +E Q +T+ FK+FQ+E+ ++ C+ E + +T
Sbjct: 425 ESVADFDTCHKQPALKSPSPWEKQMATTYTHTIFKKFQVEVLGVVACHPRKEKEDENMAT 484
Query: 484 FCVIESKKVYDRFKDTRFRVIFNEKDFEIQCACCLFEFKGILCRHILCVLQLTGKTESVP 543
F +V D KD F V +++ E+ C C +FE+KG LCRH L +LQ+ G S+P
Sbjct: 485 F------RVQDCEKDDDFLVTWSKTKSELCCFCRMFEYKGFLCRHALMILQMCG-FASIP 537
Query: 544 SCYILSRWRKDIKRRHTLIKCGFDGNVELQLVGKACDAFYEVVSVGIHTEDDL-LKVMNR 602
YIL RW KD K + +Q C E+ G +E++ + +
Sbjct: 538 PQYILKRWTKDAKSGVLAGEGADQIQTRVQRYNDLCSRATELSEEGCVSEENYNIALRTL 597
Query: 603 INELKIELNCEGSSSVIIEEDSLVQNQMAKILDPATTRSKGRPPSKRKASKLDQIVQKKV 662
+ LK ++ + + I E +S + N T + + + KA+K KK
Sbjct: 598 VETLKNCVDMNNARNNITESNSQLNN--------GTHEEENQVMAGVKATK------KKT 643
Query: 663 LRKKTQNRNNKSNNYLSQEEV 683
+ +K + + S SQ+ +
Sbjct: 644 VYRKRKGQQEASQMLESQQSL 664
>dbj|BAB03065.1| far-red impaired response protein; Mutator-like transposase-like
protein; phytochrome A signaling protein-like
[Arabidopsis thaliana] gi|22331250|ref|NP_188856.2|
far-red impaired responsive protein, putative
[Arabidopsis thaliana]
Length = 839
Score = 376 bits (965), Expect = e-102
Identities = 206/577 (35%), Positives = 310/577 (53%), Gaps = 47/577 (8%)
Query: 1 MTFSSEEEVIKYYKSYARCMGFGTVKINSKNAKDGKKYFT--LGCTR------------- 45
M F S E +Y+ Y+R MGF T NS+ +K +++ C+R
Sbjct: 74 MEFESHGEAYSFYQEYSRAMGFNTAIQNSRRSKTTREFIDAKFACSRYGTKREYDKSFNR 133
Query: 46 --ARSYVSNSKNLLKPNPTIRAQCMARVNMSMSLDGKITITKVALEHNHGLSPTKSRYFR 103
AR + +N+ + C A +++ DGK I EHNH L P ++
Sbjct: 134 PRARQSKQDPENMAGRRTCAKTDCKASMHVKRRPDGKWVIHSFVREHNHELLPAQA---- 189
Query: 104 CNKNLDPHTKRRLDINDQAGINVSRNFRSMVVEANGYDNLTFGEKDCRNYIDKARRLRLG 163
+++Q + + +M + Y + + D ++ +K R L +
Sbjct: 190 --------------VSEQT----RKIYAAMAKQFAEYKTVISLKSDSKSSFEKGRTLSVE 231
Query: 164 TGDAEAIQNYFVRMQKKNSQFYYVMDVDDKSRLRNVFWVDARCRAAYEYFGEVITFDTTY 223
TGD + + ++ RMQ NS F+Y +D+ D R++NVFWVDA+ R Y F +V++ DTTY
Sbjct: 232 TGDFKILLDFLSRMQSLNSNFFYAVDLGDDQRVKNVFWVDAKSRHNYGSFCDVVSLDTTY 291
Query: 224 LTNKYDMPFAPFVGVNHHGQSVLLGCALLSNEDTKTFSWLFKTWLECMHGRAPNAIITDQ 283
+ NKY MP A FVGVN H Q ++LGCAL+S+E T+SWL +TWL + G+AP +IT+
Sbjct: 292 VRNKYKMPLAIFVGVNQHYQYMVLGCALISDESAATYSWLMETWLRAIGGQAPKVLITEL 351
Query: 284 DRAMKKAIEDVFPKARHRWCLWHLMKKVPEKLGR-HSHYESIKLLLHDAVYDSSSISDFM 342
D M + ++FP RH LWH++ KV E LG+ +++ +Y S DF
Sbjct: 352 DVVMNSIVPEIFPNTRHCLFLWHVLMKVSENLGQVVKQHDNFMPKFEKCIYKSGKDEDFA 411
Query: 343 EKWKKMIECYELHDNEWLKGLFDERYRWVPVYVRDTFWAGMSTTQRSESMNSFFDGYVSS 402
KW K + + L D++W+ L+++R +W P Y+ D AGMST+QR++S+N+FFD Y+
Sbjct: 412 RKWYKNLARFGLKDDQWMISLYEDRKKWAPTYMTDVLLAGMSTSQRADSINAFFDKYMHK 471
Query: 403 KTTLKQFVEQYDNALKDKIEKESMADFVSFNTTIACISLFGFESQFQKAFTNAKFKEFQI 462
KT++++FV+ YD L+D+ E+E+ AD +N A S FE + +T A FK+FQI
Sbjct: 472 KTSVQEFVKVYDTVLQDRCEEEAKADSEMWNKQPAMKSPSPFEKSVSEVYTPAVFKKFQI 531
Query: 463 EIASMIYCNAFLEGMENLNSTFCVIESKKVYDRFKDTRFRVIFNEKDFEIQCACCLFEFK 522
E+ I C+ E + STF +V D + F V +N+ E+ C C LFE+K
Sbjct: 532 EVLGAIACSPREENRDATCSTF------RVQDFENNQDFMVTWNQTKAEVSCICRLFEYK 585
Query: 523 GILCRHILCVLQLTGKTESVPSCYILSRWRKDIKRRH 559
G LCRH L VLQ S+PS YIL RW KD K RH
Sbjct: 586 GYLCRHTLNVLQCC-HLSSIPSQYILKRWTKDAKSRH 621
>ref|XP_466647.1| putative far-red impaired response protein [Oryza sativa (japonica
cultivar-group)] gi|47497942|dbj|BAD20147.1| putative
far-red impaired response protein [Oryza sativa
(japonica cultivar-group)] gi|47496827|dbj|BAD19587.1|
putative far-red impaired response protein [Oryza sativa
(japonica cultivar-group)]
Length = 853
Score = 375 bits (962), Expect = e-102
Identities = 212/572 (37%), Positives = 323/572 (56%), Gaps = 26/572 (4%)
Query: 1 MTFSSEEEVIKYYKSYARCMGFGTVKINSKNAKDGK-KYFTLGCTRARSYVSNSKNLLKP 59
+ F + ++ +KYYK YA GF + + S K G + LGC+RA +N+ L +
Sbjct: 84 LRFKTYDDALKYYKQYAADSGFSAIILKSSYLKSGVCRRLVLGCSRAGRGRANACYLSRE 143
Query: 60 NPTIRAQCMARVNMSMSLDGKITITKVALEHNHGLSPTKSRYFRCNKNLDPHTKRRLDIN 119
+ I C AR+++ + D + I L+HNH + + + C K L D
Sbjct: 144 STKIN--CPARISLKLRQDRWLHIDDAKLDHNHPPNQSSVSHMNCYKKLT-------DAK 194
Query: 120 DQAGINVSRNFRSMVVEANGYDNLTFGEKDCRNYIDKARRLRLGTGDAEAIQNYFVRMQK 179
++ + S+ R N+ G+K+ ++ + R L+ G GD E I +F MQ
Sbjct: 195 NEETASRSKGRR----------NVPIGDKEQGSFTEIGR-LKFGEGDDEYIHKFFGSMQN 243
Query: 180 KNSQFYYVMDVDDKSRLRNVFWVDARCRAAYEYFG-EVITFDTTYLTNKYDMPFAPFVGV 238
KN F+Y++D+D + RLRN+FW DAR +AA++Y+G +VI FDT+YLT KYD+P F GV
Sbjct: 244 KNPNFFYLVDLDKQGRLRNLFWSDARSQAAHDYYGRDVIYFDTSYLTEKYDLPLVFFTGV 303
Query: 239 NHHGQSVLLGCALLSNEDTKTFSWLFKTWLECMHGRAPNAIITDQDRAMKKAIEDVFPKA 298
N+HGQ VL G LLS+ ++ WLF+ + CM G P+AIIT+ A+ A+ DV P+
Sbjct: 304 NNHGQPVLFGTGLLSDLGVDSYVWLFRAFFACMKGCYPDAIITEHYNAILDAVRDVLPQV 363
Query: 299 RHRWCLWHLMKKVPEKLGRHSHYESIKLLLHDAVYDSSSISDFMEKWKKMIECYELHDNE 358
+HR CL+ +MK V E L H+ +++IK L Y S S+F WKK+I + L +NE
Sbjct: 364 KHRLCLYRIMKDVAENLKAHAEFKTIKKALKKVTYGSLKASEFEADWKKIILEHGLGENE 423
Query: 359 WLKGLFDERYRWVPVYVRDTFWAGMSTTQRSESMNSFFDGYVSSKTTLKQFVEQYDNALK 418
L L++ R W P Y++ FWAGMS +QR ES+ S++DG+V KT+LKQF +Y+ L+
Sbjct: 424 CLSSLYEHRQLWAPAYLKGQFWAGMSVSQRGESIVSYYDGFVYPKTSLKQFFSKYEIILE 483
Query: 419 DKIEKESMADFVSFNTTIACISLFGFESQFQKAFTNAKFKEFQIEIASMIYCNAFLEGME 478
+K +KE AD S + T ++ F E Q K +T FK+FQ E+ + +YC+ ++
Sbjct: 484 NKYKKELQADEESSHRTPLTVTKFYMEEQLAKEYTINMFKKFQDELKATMYCDGMPTKVD 543
Query: 479 NLNSTFCVIESKKVYD-RFKDTR-FRVIFNEKDF-EIQCACCLFEFKGILCRHILCVLQL 535
TF V E + D + K+ R + V F +++ +C C F+F GILCRHIL V +L
Sbjct: 544 GQFVTFVVKECSYMEDGKEKEGRNYEVYFCKQELVNCECECGFFQFTGILCRHILSVFKL 603
Query: 536 TGKTESVPSCYILSRWRKDIKRRHTLIKCGFD 567
E +P ++L RW++D K+ H C D
Sbjct: 604 QEMFE-IPIRFVLDRWKRDYKKLHADALCKND 634
>emb|CAB80483.1| hypothetical protein [Arabidopsis thaliana]
gi|4467124|emb|CAB37558.1| hypothetical protein
[Arabidopsis thaliana] gi|22531237|gb|AAM97122.1|
unknown protein [Arabidopsis thaliana]
gi|15233732|ref|NP_195531.1| far-red impaired responsive
protein, putative [Arabidopsis thaliana]
gi|7485859|pir||T05645 hypothetical protein F20D10.300 -
Arabidopsis thaliana
Length = 788
Score = 371 bits (952), Expect = e-101
Identities = 201/568 (35%), Positives = 307/568 (53%), Gaps = 22/568 (3%)
Query: 1 MTFSSEEEVIKYYKSYARCMGFGT-VKINSKNAKDG---KKYFTL---GCTRARSYVSNS 53
+ F SEE +Y SYAR +GF T V + ++ +DG ++ F G +
Sbjct: 77 LEFESEEAAKAFYNSYARRIGFSTRVSSSRRSRRDGAIIQRQFVCAKEGFRNMNEKRTKD 136
Query: 54 KNLLKPNPTIRAQCMARVNMSMSLDGKITITKVALEHNHGLSPTKSRY-FRCNKNLDPHT 112
+ + +P R C A +++ M GK ++ +HNH L P + R ++ +
Sbjct: 137 REIKRPRTITRVGCKASLSVKMQDSGKWLVSGFVKDHNHELVPPDQVHCLRSHRQISGPA 196
Query: 113 KRRLDINDQAGINVSRNFRSMVVEANGYDNLTFGEKDCRNYIDKARRLRLGTGDAEAIQN 172
K +D AG+ R +++ E G + F E DCRNY+ R+ + G+ + + +
Sbjct: 197 KTLIDTLQAAGMGPRRIMSALIKEYGGISKVGFTEVDCRNYMRNNRQKSI-EGEIQLLLD 255
Query: 173 YFVRMQKKNSQFYYVMDVDDKSRLRNVFWVDARCRAAYEYFGEVITFDTTYLTNKYDMPF 232
Y +M N F+Y + + + NVFW D + + +FG+ +TFDTTY +N+Y +PF
Sbjct: 256 YLRQMNADNPNFFYSVQGSEDQSVGNVFWADPKAIMDFTHFGDTVTFDTTYRSNRYRLPF 315
Query: 233 APFVGVNHHGQSVLLGCALLSNEDTKTFSWLFKTWLECMHGRAPNAIITDQDRAMKKAIE 292
APF GVNHHGQ +L GCA + NE +F WLF TWL M P +I TD D ++ AI
Sbjct: 316 APFTGVNHHGQPILFGCAFIINETEASFVWLFNTWLAAMSAHPPVSITTDHDAVIRAAIM 375
Query: 293 DVFPKARHRWCLWHLMKKVPEKLG----RHSHYESIKLLLHDAVYDSSSISDFMEKWKKM 348
VFP ARHR+C WH++KK EKL +H +ES H V + S+ DF W +
Sbjct: 376 HVFPGARHRFCKWHILKKCQEKLSHVFLKHPSFESD---FHKCVNLTESVEDFERCWFSL 432
Query: 349 IECYELHDNEWLKGLFDERYRWVPVYVRDTFWAGMSTTQRSESMNSFFDGYVSSKTTLKQ 408
++ YEL D+EWL+ ++ +R +WVPVY+RDTF+A MS T RS+S+NS+FDGY+++ T L Q
Sbjct: 433 LDKYELRDHEWLQAIYSDRRQWVPVYLRDTFFADMSLTHRSDSINSYFDGYINASTNLSQ 492
Query: 409 FVEQYDNALKDKIEKESMADFVSFNTTIACISLFGFESQFQKAFTNAKFKEFQIEIASMI 468
F + Y+ AL+ ++EKE AD+ + N+ + E Q + +T F FQ E+ +
Sbjct: 493 FFKLYEKALESRLEKEVKADYDTMNSPPVLKTPSPMEKQASELYTRKLFMRFQEELVGTL 552
Query: 469 YCNAFLEGMENLNSTFCVIESKKVYDRFKDTRFRVIFNEKDFEIQCACCLFEFKGILCRH 528
F+ + + + K Y F V FN + C+C +FEF GI+CRH
Sbjct: 553 ---TFMASKADDDGDLVTYQVAK-YGEAHKAHF-VKFNVLEMRANCSCQMFEFSGIICRH 607
Query: 529 ILCVLQLTGKTESVPSCYILSRWRKDIK 556
IL V ++T ++P YIL RW ++ K
Sbjct: 608 ILAVFRVTNLL-TLPPYYILKRWTRNAK 634
>gb|AAQ62876.1| At1g76320 [Arabidopsis thaliana] gi|62320160|dbj|BAD94369.1|
putative phytochrome A signaling protein [Arabidopsis
thaliana]
Length = 732
Score = 368 bits (945), Expect = e-100
Identities = 199/564 (35%), Positives = 320/564 (56%), Gaps = 18/564 (3%)
Query: 1 MTFSSEEEVIKYYKSYARCMGFGTVKINSKNAKDGKKYFT--LGCTRARSYVSNSKNLLK 58
M F + E+ +YK YA+ +GFGT K++S+ ++ K++ C R S S + +
Sbjct: 1 MEFETHEDAYLFYKDYAKSVGFGTAKLSSRRSRASKEFIDAKFSCIRYGSK-QQSDDAIN 59
Query: 59 PNPTIRAQCMARVNMSMSLDGKITITKVALEHNHGLSPTKSRYFRCNKNLDPHTKRRLDI 118
P + + C A +++ DGK + EHNH L P ++ YFR ++N + +
Sbjct: 60 PRASPKIGCKASMHVKRRPDGKWYVYSFVKEHNHDLLPEQAHYFRSHRNTE-----LVKS 114
Query: 119 NDQAGINVSRNFRSMVVEANGYDNLTFGEKDCRNYIDKARRLRLGTGDAEAIQNYFVRMQ 178
ND + + Y +L F + RN DK RRL L TGDAE + + +RMQ
Sbjct: 115 NDSRLRRKKNTPLTDCKHLSAYHDLDFIDGYMRNQHDKGRRLVLDTGDAEILLEFLMRMQ 174
Query: 179 KKNSQFYYVMDVDDKSRLRNVFWVDARCRAAYEYFGEVITFDTTYLTNKYDMPFAPFVGV 238
++N +F++ +D + LRNVFWVDA+ Y+ F +V++F+T+Y +KY +P FVGV
Sbjct: 175 EENPKFFFAVDFSEDHLLRNVFWVDAKGIEDYKSFSDVVSFETSYFVSKYKVPLVLFVGV 234
Query: 239 NHHGQSVLLGCALLSNEDTKTFSWLFKTWLECMHGRAPNAIITDQDRAMKKAIEDVFPKA 298
NHH Q VLLGC LL+++ T+ WL ++WL M G+ P ++TDQ+ A+K AI V P+
Sbjct: 235 NHHVQPVLLGCGLLADDTVYTYVWLMQSWLVAMGGQKPKVMLTDQNNAIKAAIAAVLPET 294
Query: 299 RHRWCLWHLMKKVPEKLGRHSHY-ESIKLLLHDAVYDSSSISDFMEKWKKMIECYELHDN 357
RH +CLWH++ ++P L S + ++ L +Y S S +F +W K+I+ + L D
Sbjct: 295 RHCYCLWHVLDQLPRNLDYWSMWQDTFMKKLFKCIYRSWSEEEFDRRWLKLIDKFHLRDV 354
Query: 358 EWLKGLFDERYRWVPVYVRDTFWAGMSTTQRSESMNSFFDGYVSSKTTLKQFVEQYDNAL 417
W++ L++ER W P ++R +AG+S RSES+NS FD YV +T+LK+F+E Y L
Sbjct: 355 PWMRSLYEERKFWAPTFMRGITFAGLSMRCRSESVNSLFDRYVHPETSLKEFLEGYGLML 414
Query: 418 KDKIEKESMADFVSFNTTIACISLFGFESQFQKAFTNAKFKEFQIEIASMIYCNAFLEGM 477
+D+ E+E+ ADF +++ S FE Q +++ F+ FQ+E+ C+ E
Sbjct: 415 EDRYEEEAKADFDAWHEAPELKSPSPFEKQMLLVYSHEIFRRFQLEVLGAAACHLTKESE 474
Query: 478 ENLNSTFCVIESKKVYDRFKDTRFRVIFNEKDFEIQCACCLFEFKGILCRHILCVLQLTG 537
E +T+ V K +D + ++ V ++E +I C+C FE+KG LCRH + VLQ++G
Sbjct: 475 E--GTTYSV----KDFD--DEQKYLVDWDEFKSDIYCSCRSFEYKGYLCRHAIVVLQMSG 526
Query: 538 KTESVPSCYILSRWRKDIKRRHTL 561
++P Y+L RW + RH +
Sbjct: 527 -VFTIPINYVLQRWTNAARNRHQI 549
>ref|NP_177759.1| far-red impaired responsive protein, putative [Arabidopsis
thaliana] gi|6573712|gb|AAF17632.1| T23E18.25
[Arabidopsis thaliana]
Length = 732
Score = 368 bits (945), Expect = e-100
Identities = 199/564 (35%), Positives = 320/564 (56%), Gaps = 18/564 (3%)
Query: 1 MTFSSEEEVIKYYKSYARCMGFGTVKINSKNAKDGKKYFT--LGCTRARSYVSNSKNLLK 58
M F + E+ +YK YA+ +GFGT K++S+ ++ K++ C R S S + +
Sbjct: 1 MEFETHEDAYLFYKDYAKSVGFGTAKLSSRRSRASKEFIDAKFSCIRYGSK-QQSDDAIN 59
Query: 59 PNPTIRAQCMARVNMSMSLDGKITITKVALEHNHGLSPTKSRYFRCNKNLDPHTKRRLDI 118
P + + C A +++ DGK + EHNH L P ++ YFR ++N + +
Sbjct: 60 PRASPKIGCKASMHVKRRPDGKWYVYSFVKEHNHDLLPEQAHYFRSHRNTE-----LVKS 114
Query: 119 NDQAGINVSRNFRSMVVEANGYDNLTFGEKDCRNYIDKARRLRLGTGDAEAIQNYFVRMQ 178
ND + + Y +L F + RN DK RRL L TGDAE + + +RMQ
Sbjct: 115 NDSRLRRKKNTPLTDCKHLSAYHDLDFIDGYMRNQHDKGRRLVLDTGDAEILLEFLMRMQ 174
Query: 179 KKNSQFYYVMDVDDKSRLRNVFWVDARCRAAYEYFGEVITFDTTYLTNKYDMPFAPFVGV 238
++N +F++ +D + LRNVFWVDA+ Y+ F +V++F+T+Y +KY +P FVGV
Sbjct: 175 EENPKFFFAVDFSEDHLLRNVFWVDAKGIEDYKSFSDVVSFETSYFVSKYKVPLVLFVGV 234
Query: 239 NHHGQSVLLGCALLSNEDTKTFSWLFKTWLECMHGRAPNAIITDQDRAMKKAIEDVFPKA 298
NHH Q VLLGC LL+++ T+ WL ++WL M G+ P ++TDQ+ A+K AI V P+
Sbjct: 235 NHHVQPVLLGCGLLADDTVYTYVWLMQSWLVAMGGQKPKVMLTDQNNAIKAAIAAVLPET 294
Query: 299 RHRWCLWHLMKKVPEKLGRHSHY-ESIKLLLHDAVYDSSSISDFMEKWKKMIECYELHDN 357
RH +CLWH++ ++P L S + ++ L +Y S S +F +W K+I+ + L D
Sbjct: 295 RHCYCLWHVLDQLPRNLDYWSMWQDTFMKKLFKCIYRSWSEEEFDRRWLKLIDKFHLRDV 354
Query: 358 EWLKGLFDERYRWVPVYVRDTFWAGMSTTQRSESMNSFFDGYVSSKTTLKQFVEQYDNAL 417
W++ L++ER W P ++R +AG+S RSES+NS FD YV +T+LK+F+E Y L
Sbjct: 355 PWMRSLYEERKFWAPTFMRGITFAGLSMRCRSESVNSLFDRYVHPETSLKEFLEGYGLML 414
Query: 418 KDKIEKESMADFVSFNTTIACISLFGFESQFQKAFTNAKFKEFQIEIASMIYCNAFLEGM 477
+D+ E+E+ ADF +++ S FE Q +++ F+ FQ+E+ C+ E
Sbjct: 415 EDRYEEEAKADFDAWHEAPELKSPSPFEKQMLLVYSHEIFRRFQLEVLGAAACHLTKESE 474
Query: 478 ENLNSTFCVIESKKVYDRFKDTRFRVIFNEKDFEIQCACCLFEFKGILCRHILCVLQLTG 537
E +T+ V K +D + ++ V ++E +I C+C FE+KG LCRH + VLQ++G
Sbjct: 475 E--GTTYSV----KDFD--DEQKYLVDWDEFKSDIYCSCRSFEYKGYLCRHAIVVLQMSG 526
Query: 538 KTESVPSCYILSRWRKDIKRRHTL 561
++P Y+L RW + RH +
Sbjct: 527 -VFTIPINYVLQRWTNAARNRHQI 549
>pir||G96790 hypothetical protein F15M4.18 [imported] - Arabidopsis thaliana
gi|6554486|gb|AAF16668.1| putative phytochrome A
signaling protein; 74057-72045 [Arabidopsis thaliana]
Length = 670
Score = 368 bits (945), Expect = e-100
Identities = 199/564 (35%), Positives = 320/564 (56%), Gaps = 18/564 (3%)
Query: 1 MTFSSEEEVIKYYKSYARCMGFGTVKINSKNAKDGKKYFT--LGCTRARSYVSNSKNLLK 58
M F + E+ +YK YA+ +GFGT K++S+ ++ K++ C R S S + +
Sbjct: 1 MEFETHEDAYLFYKDYAKSVGFGTAKLSSRRSRASKEFIDAKFSCIRYGSK-QQSDDAIN 59
Query: 59 PNPTIRAQCMARVNMSMSLDGKITITKVALEHNHGLSPTKSRYFRCNKNLDPHTKRRLDI 118
P + + C A +++ DGK + EHNH L P ++ YFR ++N + +
Sbjct: 60 PRASPKIGCKASMHVKRRPDGKWYVYSFVKEHNHDLLPEQAHYFRSHRNTE-----LVKS 114
Query: 119 NDQAGINVSRNFRSMVVEANGYDNLTFGEKDCRNYIDKARRLRLGTGDAEAIQNYFVRMQ 178
ND + + Y +L F + RN DK RRL L TGDAE + + +RMQ
Sbjct: 115 NDSRLRRKKNTPLTDCKHLSAYHDLDFIDGYMRNQHDKGRRLVLDTGDAEILLEFLMRMQ 174
Query: 179 KKNSQFYYVMDVDDKSRLRNVFWVDARCRAAYEYFGEVITFDTTYLTNKYDMPFAPFVGV 238
++N +F++ +D + LRNVFWVDA+ Y+ F +V++F+T+Y +KY +P FVGV
Sbjct: 175 EENPKFFFAVDFSEDHLLRNVFWVDAKGIEDYKSFSDVVSFETSYFVSKYKVPLVLFVGV 234
Query: 239 NHHGQSVLLGCALLSNEDTKTFSWLFKTWLECMHGRAPNAIITDQDRAMKKAIEDVFPKA 298
NHH Q VLLGC LL+++ T+ WL ++WL M G+ P ++TDQ+ A+K AI V P+
Sbjct: 235 NHHVQPVLLGCGLLADDTVYTYVWLMQSWLVAMGGQKPKVMLTDQNNAIKAAIAAVLPET 294
Query: 299 RHRWCLWHLMKKVPEKLGRHSHY-ESIKLLLHDAVYDSSSISDFMEKWKKMIECYELHDN 357
RH +CLWH++ ++P L S + ++ L +Y S S +F +W K+I+ + L D
Sbjct: 295 RHCYCLWHVLDQLPRNLDYWSMWQDTFMKKLFKCIYRSWSEEEFDRRWLKLIDKFHLRDV 354
Query: 358 EWLKGLFDERYRWVPVYVRDTFWAGMSTTQRSESMNSFFDGYVSSKTTLKQFVEQYDNAL 417
W++ L++ER W P ++R +AG+S RSES+NS FD YV +T+LK+F+E Y L
Sbjct: 355 PWMRSLYEERKFWAPTFMRGITFAGLSMRCRSESVNSLFDRYVHPETSLKEFLEGYGLML 414
Query: 418 KDKIEKESMADFVSFNTTIACISLFGFESQFQKAFTNAKFKEFQIEIASMIYCNAFLEGM 477
+D+ E+E+ ADF +++ S FE Q +++ F+ FQ+E+ C+ E
Sbjct: 415 EDRYEEEAKADFDAWHEAPELKSPSPFEKQMLLVYSHEIFRRFQLEVLGAAACHLTKESE 474
Query: 478 ENLNSTFCVIESKKVYDRFKDTRFRVIFNEKDFEIQCACCLFEFKGILCRHILCVLQLTG 537
E +T+ V K +D + ++ V ++E +I C+C FE+KG LCRH + VLQ++G
Sbjct: 475 E--GTTYSV----KDFD--DEQKYLVDWDEFKSDIYCSCRSFEYKGYLCRHAIVVLQMSG 526
Query: 538 KTESVPSCYILSRWRKDIKRRHTL 561
++P Y+L RW + RH +
Sbjct: 527 -VFTIPINYVLQRWTNAARNRHQI 549
>gb|AAM91710.1| unknown protein [Arabidopsis thaliana] gi|19424057|gb|AAL87259.1|
unknown protein [Arabidopsis thaliana]
gi|15219020|ref|NP_175661.1| far-red impaired responsive
protein, putative [Arabidopsis thaliana]
gi|5903066|gb|AAD55625.1| F6D8.26 [Arabidopsis thaliana]
gi|25405564|pir||G96565 F6D8.26 [imported] - Arabidopsis
thaliana
Length = 703
Score = 356 bits (913), Expect = 2e-96
Identities = 196/567 (34%), Positives = 312/567 (54%), Gaps = 22/567 (3%)
Query: 1 MTFSSEEEVIKYYKSYARCMGFGT-VKINSKNAKDGKKYFTLGCTRARSYVSNSKNLLKP 59
M F S ++ YY YA +GF VK + + +KY + C ++ + ++ +
Sbjct: 89 MEFESYDDAYNYYNCYASEVGFRVRVKNSWFKRRSKEKYGAVLCCSSQGF-KRINDVNRV 147
Query: 60 NPTIRAQCMARVNMSMSLDGKITITKVALEHNHGLSPTKSRYFRCNKNLDPHTKRRLDIN 119
R C A + M + + +V L+HNH L C KR+ +
Sbjct: 148 RKETRTGCPAMIRMRQVDSKRWRVVEVTLDHNHLLG--------CKLYKSVKRKRKCVSS 199
Query: 120 DQAGINVSRNFRSMVVE--ANGYDNLTFGEKDCRNYIDKARRLRLGTGDAEAIQNYFVRM 177
+ + +R+ VV+ +N N T +K +N L L GD+ AI NYF RM
Sbjct: 200 PVSDAKTIKLYRACVVDNGSNVNPNSTLNKK-FQNSTGSPDLLNLKRGDSAAIYNYFCRM 258
Query: 178 QKKNSQFYYVMDVDDKSRLRNVFWVDARCRAAYEYFGEVITFDTTYLTNKYDMPFAPFVG 237
Q N F+Y+MDV+D+ +LRNVFW DA + + YFG+VI D++Y++ K+++P F G
Sbjct: 259 QLTNPNFFYLMDVNDEGQLRNVFWADAFSKVSCSYFGDVIFIDSSYISGKFEIPLVTFTG 318
Query: 238 VNHHGQSVLLGCALLSNEDTKTFSWLFKTWLECMHGRAPNAIITDQDRAMKKAIEDVFPK 297
VNHHG++ LL C L+ E +++ WL K WL M R+P I+TD+ + ++ AI VFP+
Sbjct: 319 VNHHGKTTLLSCGFLAGETMESYHWLLKVWLSVMK-RSPQTIVTDRCKPLEAAISQVFPR 377
Query: 298 ARHRWCLWHLMKKVPEKLGRHSHYESIKLLLHDAVYDSSSISDFMEKWKKMIECYELHDN 357
+ R+ L H+M+K+PEKLG +Y++++ AVY++ + +F W M+ + + +N
Sbjct: 378 SHQRFSLTHIMRKIPEKLGGLHNYDAVRKAFTKAVYETLKVVEFEAAWGFMVHNFGVIEN 437
Query: 358 EWLKGLFDERYRWVPVYVRDTFWAGMSTTQRSESMNSFFDGYVSSKTTLKQFVEQYDNAL 417
EWL+ L++ER +W PVY++DTF+AG++ E++ FF+ YV +T LK+F+++Y+ AL
Sbjct: 438 EWLRSLYEERAKWAPVYLKDTFFAGIAAAHPGETLKPFFERYVHKQTPLKEFLDKYELAL 497
Query: 418 KDKIEKESMADFVSFNTTIACI-SLFGFESQFQKAFTNAKFKEFQIEIASMIYCNAFLEG 476
+ K +E+++D S A + + FE+Q + +T FK+FQIE+ M C F
Sbjct: 498 QKKHREETLSDIESQTLNTAELKTKCSFETQLSRIYTRDMFKKFQIEVEEMYSC--FSTT 555
Query: 477 MENLNSTFCVIESKK----VYDRFKDTRFRVIFNEKDFEIQCACCLFEFKGILCRHILCV 532
+++ F + K+ R + F V++N E++C C F F G LCRH LCV
Sbjct: 556 QVHVDGPFVIFLVKERVRGESSRREIRDFEVLYNRSVGEVRCICSCFNFYGYLCRHALCV 615
Query: 533 LQLTGKTESVPSCYILSRWRKDIKRRH 559
L G E +P YIL RWRKD KR H
Sbjct: 616 LNFNG-VEEIPLRYILPRWRKDYKRLH 641
>ref|XP_478214.1| putative far-red impaired response protein [Oryza sativa (japonica
cultivar-group)] gi|22093808|dbj|BAC07095.1| putative
far-red impaired response protein [Oryza sativa
(japonica cultivar-group)] gi|50509975|dbj|BAD30438.1|
putative far-red impaired response protein [Oryza sativa
(japonica cultivar-group)]
Length = 809
Score = 348 bits (892), Expect = 4e-94
Identities = 193/565 (34%), Positives = 311/565 (54%), Gaps = 22/565 (3%)
Query: 1 MTFSSEEEVIKYYKSYARCMGFGTVKINSKNAKDGKKYFTL------GCTRARSYVSNSK 54
M F SEE+ ++Y SY GFG + S N DG +Y + G +R RS + S
Sbjct: 48 MVFESEEDAFQFYVSYGCRSGFGITR-RSNNTFDGFRYRSTFICSKGGQSRLRSGATRSA 106
Query: 55 NLLKPNPTIRAQCMARVNMSMSLDGKITITKVALEHNHGLSPTKSRYFRCNKNLDPHTKR 114
+A+ + + + + + LEHNH L P+ ++ + KN P +
Sbjct: 107 RKRGTKTGCKAKMIVK---DAHFQNRWEVIVLELEHNHPLDPSLLKFKKQLKN-SPFLQ- 161
Query: 115 RLDINDQAGINVSRNFRSMVVEANGYDN--LTFGEKDCRNYIDKARRLRLGTGDAEAIQN 172
N + + + + + G D+ + + R ID+ R+L+L GD +A+ +
Sbjct: 162 ----NPPLMLEAPDSSSAAALSSRGGDSGIPLSTQIEFRTKIDRNRKLKLAEGDLDALLS 217
Query: 173 YFVRMQKKNSQFYYVMDVDDKSRLRNVFWVDARCRAAYEYFGEVITFDTTYLTNKYDMPF 232
+ +MQ +N F+Y +D++++ +LRNVFW DA+ R++Y YFG+V+ + +++Y++ F
Sbjct: 218 FLNKMQDQNPYFFYSLDMNEQGQLRNVFWADAKSRSSYNYFGDVVAINVRNFSDQYEIQF 277
Query: 233 APFVGVNHHGQSVLLGCALLSNEDTKTFSWLFKTWLECMHGRAPNAIITDQDRAMKKAIE 292
FVG NHH Q VLLGC LL+ + WLF TW+ CM+ P ++IT+ + A++
Sbjct: 278 VSFVGTNHHAQQVLLGCGLLAGRSLGAYVWLFDTWVRCMNSTPPPSVITNYCHDVAIAVK 337
Query: 293 DVFPKARHRWCLWHLMKKVPEKLGRHSHYESIKLLLHDAVYDSSSISDFMEKWKKMIECY 352
VFP AR+R+CL ++ ++PEKL + I +DS ++ DF + W++M+E +
Sbjct: 338 KVFPNARYRFCLLDILNELPEKLEETEKKDEIVSAFSSLAFDSITMPDFDKDWQEMVEQF 397
Query: 353 ELHDNEWLKGLFDERYRWVPVYVRDTFWAGMSTTQRSESMNSFFDGYVSSKTTLKQFVEQ 412
L NEWL LF+ R +W PVYV+D+FWAGMS T+RS+S + +FDG++ T++K FVEQ
Sbjct: 398 HLEGNEWLSKLFEVRTQWAPVYVKDSFWAGMSITERSDSASDYFDGWLMPDTSVKMFVEQ 457
Query: 413 YDNALKDKIEKESMADFVSFNTTIACISLFGFESQFQKAFTNAKFKEFQIEIASMIYCNA 472
Y++A+K K+EKE+ D S ++ E Q K +T F++F EI +CN
Sbjct: 458 YESAVKVKLEKENYEDLRSSQMRPPVMTGVSVEEQAAKMYTLEIFQKFLDEIGHSFHCNY 517
Query: 473 FLEGMENLNSTFCVIESKKVYDRFKDTRFRVIFNEKDFEIQCACCLFEFKGILCRHILCV 532
+ ++ S I S + D K ++V ++ + +I C C LF+FKGILCRH L V
Sbjct: 518 SI--LDRNESVVTYIVSDHI-DETKKVDYKVAYDNVEDDILCLCRLFQFKGILCRHALTV 574
Query: 533 LQLTGKTESVPSCYILSRWRKDIKR 557
L+ +P YI+ RW KD K+
Sbjct: 575 LR-QEFVPMIPPKYIIHRWCKDCKQ 598
>emb|CAB10288.1| hypothetical protein [Arabidopsis thaliana]
gi|7268255|emb|CAB78551.1| hypothetical protein
[Arabidopsis thaliana] gi|7485146|pir||F71414
hypothetical protein - Arabidopsis thaliana
Length = 1705
Score = 332 bits (850), Expect = 3e-89
Identities = 198/623 (31%), Positives = 320/623 (50%), Gaps = 57/623 (9%)
Query: 64 RAQCMARVNMSMSLDGKITITKVALEHNHGLSPTKSRYFRCNKNLDPHTKRRLDINDQAG 123
+ C A +++ DGK I + +HNH L P + +FR +N+ K +DI
Sbjct: 102 KTDCKASMHVKRRPDGKWIIHEFVKDHNHELLPALAYHFRIQRNVKLAEKNNIDILHAVS 161
Query: 124 INVSRNFRSMVVEANGYDNL-TFGEKDCRNYIDKARRLRLGTGDAEAIQNYFVRMQKKNS 182
+ + M ++ GY N+ + + D + +DK R L L GD++ + YF R++K+N
Sbjct: 162 ERTKKMYVEMSRQSGGYKNIGSLLQTDVSSQVDKGRYLALEEGDSQVLLEYFKRIKKENP 221
Query: 183 QFYYVMDVDDKSRLRNVFWVDARCRAAYEYFGEVITFDTTYLTNKYDMPFAPFVGVNHHG 242
+F+Y +D+++ RLRN+FW DA+ R Y F +V++FDTTY+ +P A F+GVNHH
Sbjct: 222 KFFYAIDLNEDQRLRNLFWADAKSRDDYLSFNDVVSFDTTYVKFNDKLPLALFIGVNHHS 281
Query: 243 QSVLLGCALLSNEDTKTFSWLFKTWLECMHGRAPNAIITDQDRAMKKAIEDVFPKARHRW 302
Q +LLGCAL+++E +TF WL KTWL M GRAP + P RH +
Sbjct: 282 QPMLLGCALVADESMETFVWLIKTWLRAMGGRAPK----------------LLPNTRHCF 325
Query: 303 CLWHLMKKVPEKLGR-HSHYESIKLLLHDAVYDSSSISDFMEKWKKMIECYELHDNEWLK 361
LWH+++K+PE +E+ L + ++ S + +F +W KM+ + L ++EWL
Sbjct: 326 ALWHVLEKIPEYFSHVMKRHENFLLKFNKCIFRSWTDDEFDMRWWKMVSQFGLENDEWLL 385
Query: 362 GLFDERYRWVPVYVRDTFWAGMSTTQRSESMNSFFDGYVSSKTTLKQFVEQYDNALKDKI 421
L + R +WVP ++ D F AGMST+QRSES+NSFFD Y+ K TLK+F+ QY L+++
Sbjct: 386 WLHEHRQKWVPTFMSDVFLAGMSTSQRSESVNSFFDKYIHKKITLKEFLRQYGVILQNRY 445
Query: 422 EKESMADFVSFNTTIACISLFGFESQFQKAFTNAKFKEFQIEIASMIYCNAFLEGMENLN 481
E+ES+ADF + + A S +E Q +T+ FK+FQ+E+ ++ C+ E +
Sbjct: 446 EEESVADFDTCHKQPALKSPSPWEKQMATTYTHTIFKKFQVEVLGVVACHPRKEKEDENM 505
Query: 482 STFCVIESKKVYDRFKDTRFRVIFNEKDFEIQCACCLFEFKGILCRHILCVLQLTGKTES 541
+TF +V D KD F V +++ E+ C C +FE+KGI
Sbjct: 506 ATF------RVQDCEKDDDFLVTWSKTKSELCCFCRMFEYKGI----------------- 542
Query: 542 VPSCYILSRWRKDIKRRHTLIKCGFDGNVELQLVGKACDAFYEVVSVGIHTEDDL-LKVM 600
P YIL RW KD K + +Q C E+ G +E++ + +
Sbjct: 543 -PPQYILKRWTKDAKSGVLAGEGADQIQTRVQRYNDLCSRATELSEEGCVSEENYNIALR 601
Query: 601 NRINELKIELNCEGSSSVIIEEDSLVQNQMAKILDPATTRSKGRPPSKRKASKLDQIVQK 660
+ LK ++ + + I E +S + N T + + + KA+K K
Sbjct: 602 TLVETLKNCVDMNNARNNITESNSQLNN--------GTHEEENQVMAGVKATK------K 647
Query: 661 KVLRKKTQNRNNKSNNYLSQEEV 683
K + +K + + S SQ+ +
Sbjct: 648 KTVYRKRKGQQEASQMLESQQSL 670
>ref|NP_178118.1| far-red impaired responsive protein, putative [Arabidopsis
thaliana] gi|5902364|gb|AAD55466.1| Hypothetical protein
[Arabidopsis thaliana] gi|12324580|gb|AAG52241.1|
hypothetical protein; 6424-4334 [Arabidopsis thaliana]
gi|25406623|pir||E96831 hypothetical protein F18B13.10
[imported] - Arabidopsis thaliana
Length = 696
Score = 327 bits (837), Expect = 1e-87
Identities = 187/570 (32%), Positives = 303/570 (52%), Gaps = 19/570 (3%)
Query: 1 MTFSSEEEVIKYYKSYARCMGFGT-VKINSKNAKDGKKYFTLGCTRARSYVSNSKNLLKP 59
M F S ++ +Y SYAR +GF VK + +K + C + + K+
Sbjct: 70 MEFESYDDAYSFYNSYARELGFAIRVKSSWTKRNSKEKRGAVLCCNCQGF-KLLKDAHSR 128
Query: 60 NPTIRAQCMARVNMSMSLDGKITITKVALEHNHGLSPTKSRYFRCNK----NLDPHTKRR 115
R C A + + + + + +V L+HNH P ++ + +K + P TK
Sbjct: 129 RKETRTGCQAMIRLRLIHFDRWKVDQVKLDHNHSFDPQRAHNSKSHKKSSSSASPATKTN 188
Query: 116 LDINDQAGINVSRNFRSMVVEANGY--DNLTFGEKD--CRNYIDKARRLRLGTGDAEAIQ 171
+ + + +R++ ++ +L+ GE ++ +RRL L G A+Q
Sbjct: 189 PEPPPHVQVRTIKLYRTLALDTPPALGTSLSSGETSDLSLDHFQSSRRLEL-RGGFRALQ 247
Query: 172 NYFVRMQKKNSQFYYVMDVDDKSRLRNVFWVDARCRAAYEYFGEVITFDTTYLTNKYDMP 231
++F ++Q + F Y+MD+ D LRNVFW+DAR RAAY +FG+V+ FDTT L+N Y++P
Sbjct: 248 DFFFQIQLSSPNFLYLMDLADDGSLRNVFWIDARARAAYSHFGDVLLFDTTCLSNAYELP 307
Query: 232 FAPFVGVNHHGQSVLLGCALLSNEDTKTFSWLFKTWLECMHGRAPNAIITDQDRAMKKAI 291
FVG+NHHG ++LLGC LL+++ +T+ WLF+ WL CM GR P IT+Q +AM+ A+
Sbjct: 308 LVAFVGINHHGDTILLGCGLLADQSFETYVWLFRAWLTCMLGRPPQIFITEQCKAMRTAV 367
Query: 292 EDVFPKARHRWCLWHLMKKVPEKLGRHSHYESIKLLLHDAVYDSSSISDFMEKWKKMIEC 351
+VFP+A HR L H++ + + + + + + L+ VY + +F W++MI
Sbjct: 368 SEVFPRAHHRLSLTHVLHNICQSVVQLQDSDLFPMALNRVVYGCLKVEEFETAWEEMIIR 427
Query: 352 YELHDNEWLKGLFDERYRWVPVYVRDTFWAGMSTTQRSESMNSF-FDGYVSSKTTLKQFV 410
+ + +NE ++ +F +R W PVY++DTF AG T F F GYV T+L++F+
Sbjct: 428 FGMTNNETIRDMFQDRELWAPVYLKDTFLAGALTFPLGNVAAPFIFSGYVHENTSLREFL 487
Query: 411 EQYDNALKDKIEKESMADFVSFNTTIACISLFGFESQFQKAFTNAKFKEFQIEIASMIYC 470
E Y++ L K +E++ D S + +ESQ K FT F+ FQ E+++M C
Sbjct: 488 EGYESFLDKKYTREALCDSESLKLIPKLKTTHPYESQMAKVFTMEIFRRFQDEVSAMSSC 547
Query: 471 NAFLEGMENLNSTFCVIESKKVYDRFKDTRFRVIFNEK---DFEIQCACCLFEFKGILCR 527
+ N +++ V++ ++ D+ +D F VI+ C C F F G CR
Sbjct: 548 FGVTQVHSNGSASSYVVKERE-GDKVRD--FEVIYETSAAAQVRCFCVCGGFSFNGYQCR 604
Query: 528 HILCVLQLTGKTESVPSCYILSRWRKDIKR 557
H+L +L G E VP YIL RWRKD+KR
Sbjct: 605 HVLLLLSHNGLQE-VPPQYILQRWRKDVKR 633
>gb|AAF66982.1| transposase [Zea mays]
Length = 709
Score = 319 bits (817), Expect = 2e-85
Identities = 201/586 (34%), Positives = 309/586 (52%), Gaps = 38/586 (6%)
Query: 1 MTFSSEEEVIKYYKSYARCMGFGTVKINSKNAKD----------GKKYFTLGCTRARSYV 50
M+F S +E+ +YK+YA GF +V+I ++ K+ ++ FT C A++
Sbjct: 29 MSFDSLDELEGFYKTYAHECGF-SVRIGAQGKKNDVVEHKRFVCSREGFTRRCAEAKNQ- 86
Query: 51 SNSKNLLKPNPTIRAQCMARVNMSMSLDGKITITKVALEHNHGL-SPTKSRYFRCNKNLD 109
K + R C ARV + + D + I EHNHGL SP K + R N+ +
Sbjct: 87 -------KKHFETRCGCNARVYVRLGQDKRYYIASFVEEHNHGLVSPDKIPFLRSNRTIC 139
Query: 110 PHTKRRLDINDQAGINVSRNFRSMVVEANGYDNLTFGEKDCRNYIDKARRLRLGTGDAEA 169
K L +A I S+ +R + V ++G+DN+ ++D +NY + R ++ DA+
Sbjct: 140 QRAKTTLFTCHKASIGTSQAYRLLQV-SDGFDNIGCMKRDLQNYY-RGLREKIKNADAQL 197
Query: 170 IQNYFVRMQKKNSQFYYVMDVDDKSRLRNVFWVDARCRAAYEYFGEVITFDTTYLTNKYD 229
R ++ NS F+Y VD+ +L + W DA CR +Y +FG++++ D TY TN+Y+
Sbjct: 198 FVAQMERKKEANSAFFYDFAVDEHGKLVYICWADATCRKSYTHFGDLVSVDATYSTNQYN 257
Query: 230 MPFAPFVGVNHHGQSVLLGCALLSNEDTKTFSWLFKTWLECMHGRAPNAIITDQDRAMKK 289
M FAPF GVNHH Q V G A L+NE +++ WLF+T+L M G+AP IITD+D ++K
Sbjct: 258 MRFAPFTGVNHHMQRVFFGAAFLANEKIESYEWLFRTFLVAMGGKAPRLIITDEDASIKS 317
Query: 290 AIEDVFPKARHRWCLWHLMKKVPEKLGR-HSHYESIKLLLHDAVYDSSSISDFMEKWKKM 348
AI P HR C+WH+M+KV EK+G SH + L+ V+ S + +F +W +
Sbjct: 318 AIRTTLPDTIHRLCMWHIMEKVSEKVGHPTSHDKEFWDALNTCVWGSETPEEFEMRWNAL 377
Query: 349 IECYELHDNEWLKGLFDERYRWVPVYVRDTFWAG-MSTTQRSESMNSFFDGYVSSKTTLK 407
++ Y L NEWL + R W+P + DT AG + TT RSES NSFF+ ++ K
Sbjct: 378 MDAYGLESNEWLANRYKIRESWIPAFFMDTPLAGVLRTTSRSESANSFFNRFIHRKLCFV 437
Query: 408 QFVEQYDNALKDKIEKESMADFVSFNTTIACISLFGFESQFQKAFTNAKFKEFQIE-IAS 466
+F ++D AL+ + +E AD +S ++T + + E Q +T+ FK FQ E IA+
Sbjct: 438 EFWLRFDTALERQRHEELKADHISIHSTPVLRTPWVVEKQASILYTHKVFKIFQEEVIAA 497
Query: 467 MIYCNAFLEGMENLNSTFCVIESKKVYDRFKDTRFRVIFNEKDFEIQCACCLFEFKGILC 526
+C+ L + F V+ + DR IF +C+C LFE GI C
Sbjct: 498 RDHCSV-LGTTQQDAVKFVVVSDGSMRDRVVQWCTSNIFG------RCSCKLFEKIGIPC 550
Query: 527 RHILCVLQLTGKTESVPSCYILSRWRKDIKRRHTLIKCGFDGNVEL 572
HI+ ++ K +PS YIL RW KR +C +D + L
Sbjct: 551 CHIILAMR-GEKLYELPSSYILKRWETRCKR-----ECVYDDDGNL 590
>ref|NP_908529.1| P0702D12.15 [Oryza sativa (japonica cultivar-group)]
Length = 832
Score = 315 bits (806), Expect = 4e-84
Identities = 210/730 (28%), Positives = 358/730 (48%), Gaps = 60/730 (8%)
Query: 3 FSSEEEVIKYYKSYARCMGFGT-VKINSKNAKDGKKYFTLGCTRARSYVSNSKNLLKPNP 61
F +E + +Y YA GFG + + KN K+ + + C S+ + KP+
Sbjct: 77 FKTERDAYNFYNVYAVSKGFGIRLDKDRKNTKNQRTMRQICC----SHQGRNPKTKKPSV 132
Query: 62 TIRAQCMARVNMSMSLDGKITITKVALEHNHGLSPTK--SRYFRCNKNLDPHTKRRLDIN 119
I M ++N S + G ++TKV HNH + + ++ ++ + +D T+ ++
Sbjct: 133 RIGCPAMMKINRSRAGSGW-SVTKVVSAHNHPMKKSVGVTKNYQSHNQIDEGTRGIIEEM 191
Query: 120 DQAGINVSRNFRSMVVEANGYDNLTFGEKDCRNYIDKARRLRLGTGDAEAIQNYFVRMQK 179
+ ++++ N M+ +G ++ + + + A R + D + + +QK
Sbjct: 192 VDSSMSLT-NMYGMLSGMHGGPSMVPFTRKAMDRLAYAIRRDESSDDMQKTLDVLKDLQK 250
Query: 180 KNSQFYYVMDVDDKSRLRNVFWVDARCRAAYEYFGEVITFDTTYLTNKYDMPFAPFVGVN 239
++ F+Y + VD+ R++N+FW A R +E+FG+VITFDTTY TNKY+MPFAPFVGVN
Sbjct: 251 RSKNFFYSIQVDEACRVKNIFWSHAVSRLNFEHFGDVITFDTTYKTNKYNMPFAPFVGVN 310
Query: 240 HHGQSVLLGCALLSNEDTKTFSWLFKTWLECMHGRAPNAIITDQDRAMKKAIEDVFPKAR 299
+H QS GCALL E ++F+WLF T+ ECM+G+ P I+TD +M AI VFP
Sbjct: 311 NHFQSTFFGCALLREETEESFTWLFNTFKECMNGKVPIGILTDNCPSMAAAIRTVFPNTI 370
Query: 300 HRWCLWHLMKKVPEKLGR-HSHYESIKLLLHDAVYDSSSISDFMEKWKKMIECYELHDNE 358
HR C WH++KK E +G +S + K H + + + +F+ W K+I Y L +
Sbjct: 371 HRVCKWHVLKKAKEFMGNIYSKRHTFKKAFHKVLTQTLTEEEFVAAWHKLIRDYNLEKSV 430
Query: 359 WLKGLFDERYRWVPVYVRDTFWAGMSTTQRSESMNSFFDGYVSSKTTLKQFVEQYDNALK 418
+L+ ++D R +W VY F+AGM+TTQRSES N F +VS +++ FV++YD
Sbjct: 431 YLRHIWDIRRKWAFVYFSHRFFAGMTTTQRSESANHVFKMFVSPSSSMNGFVKRYDRFFN 490
Query: 419 DKIEKESMADFVSFNTTIACISLFGFESQFQKAFTNAKFKEFQIEIASMIYCNAFLEGME 478
+K++KE +F + N + + E + +T A F+ F E+ + +++
Sbjct: 491 EKLQKEDSEEFQTSNDKVEIKTRSPIEIHASQVYTRAVFQLFSEELTDSL---SYMVKPG 547
Query: 479 NLNSTFCVIESKKVYDRFKDTRFRVIFNEKDFEIQCACCLFEFKGILCRHILCVLQLTGK 538
ST V+ + F ++V + + E C C +FE KGILC HIL VL G
Sbjct: 548 EDESTVQVVRMNS-QESFLRKEYQVSCDLEREEFSCVCKMFEHKGILCSHILRVLVQYGL 606
Query: 539 TESVPSCYILSRWRKDIKRRHTLIKCGFDGNVELQ--------LVGKACDAFYEVVSVGI 590
+ +P YIL RW KD + G+ +V+ ++ + ++ + +
Sbjct: 607 SR-IPERYILKRWTKDARDTIPPHLHGYKDDVDASQSRSYRHVMLNRKTVEVAKIANKDV 665
Query: 591 HTEDDLLKVMNR-INELKIELNCEG---------------SSSVIIEEDSLVQ------- 627
T ++VMN+ + ++K +L+ + S S ++ ED
Sbjct: 666 QTFKMAMRVMNKLLEDMKNQLSLDDGDNSREAPKRAARHKSRSTVLTEDGNEDVGGGDED 725
Query: 628 ------------NQMAKILDPATTRSKGRPPSKRKAS--KLDQIVQKKVLRKKTQNRNNK 673
N A IL P RS+GRP R S ++ + ++K + + +N N+
Sbjct: 726 SEYDVPSEDEGGNLTADILPPLKKRSRGRPKVNRYKSGGEVASLKRRKEVGIEKKNTTNE 785
Query: 674 SNNYLSQEEV 683
+ + +V
Sbjct: 786 CESVEEENQV 795
>ref|XP_475123.1| putative Mutator-like transposase [Oryza sativa (japonica
cultivar-group)] gi|46063440|gb|AAS79743.1| putative
Mutator-like transposase [Oryza sativa (japonica
cultivar-group)]
Length = 1510
Score = 313 bits (801), Expect = 2e-83
Identities = 221/689 (32%), Positives = 341/689 (49%), Gaps = 54/689 (7%)
Query: 1 MTFSSEEEVIKYYKSYARCMGFGTVKINSKNAKDGKKYFTLGCTR-------ARSYVSNS 53
M F + +V K+YKSYA GF K + + C+R + S
Sbjct: 711 MIFDTLTDVEKFYKSYAHEAGFSVRVGQHKKQNEEILFKRYYCSREGYIKERVKDVSDES 770
Query: 54 KNLLKPNPTI---RAQCMARVNMSMSLDGKITITKVALEHNHG-LSPTKSRYFRCNKNLD 109
+ P + R C A + + + D K I+ + EH+HG +SP K R N+ +
Sbjct: 771 GKKKRKTPYMMETRCGCEAHIVVKLGSDKKYRISSMIGEHSHGFVSPDKRHLLRSNRTVS 830
Query: 110 PHTKRRLDINDQAGINVSRNFRSMVVEANGYDNLTFGEKDCRNYIDKARRLRLGTGDAEA 169
K L +A I S+ FR + V G+ N+ ++ +NY R ++ DA+
Sbjct: 831 ERAKSTLFSCHKASIGTSQAFRLLQVSDGGFQNVGCTLRNLQNYYHDLR-CKIKDADAQM 889
Query: 170 IQNYFVRMQKKNSQFYYVMDVDDKSRLRNVFWVDARCRAAYEYFGEVITFDTTYLTNKYD 229
R ++ N F+Y VD + RL VFW DA CR Y FG+V++ D+TY TN+Y+
Sbjct: 890 FVGQLERKKEVNPAFFYEFMVDKQGRLVRVFWADAICRKNYSVFGDVLSVDSTYSTNQYN 949
Query: 230 MPFAPFVGVNHHGQSVLLGCALLSNEDTKTFSWLFKTWLECMHGRAPNAIITDQDRAMKK 289
M F PF GVNHH QSV LG + L++E ++F WLF+T+L+ G AP IITD+D +MK
Sbjct: 950 MKFVPFTGVNHHLQSVFLGASFLADEKIESFVWLFQTFLKATGGVAPRLIITDEDASMKA 1009
Query: 290 AIEDVFPKARHRWCLWHLMKKVPEKLGRHSHYE-SIKLLLHDAVYDSSSISDFMEKWKKM 348
AI + P HR C+WH+M+KVPEK+G + LH V+ S DF +W +
Sbjct: 1010 AIAQILPNTVHRLCMWHIMEKVPEKVGPSIREDGEFWDRLHKCVWGSEDSDDFESEWNSI 1069
Query: 349 IECYELHDNEWLKGLFDERYRWVPVYVRDTFWAG-MSTTQRSESMNSFFDGYVSSKTTLK 407
+ Y L NEW FD R W+ Y D AG +STT RSES NSFF+ ++ K T
Sbjct: 1070 MAKYGLIGNEWFSTKFDIRQSWILAYFMDIPLAGILSTTSRSESANSFFNRFIHRKLTFV 1129
Query: 408 QFVEQYDNALKDKIEKESMADFVSFNTTIACISLFGFESQFQKAFTNAKFKEFQIE-IAS 466
+F ++D AL+ + ++E AD +S +T ++ + E Q +T+ F +FQ + I +
Sbjct: 1130 EFWLRFDTALECQRQEELKADNISLHTNPKLMTPWDMEKQCSGIYTHEVFSKFQEQLIVA 1189
Query: 467 MIYCNAFLEGMENLNSTFCVIESKKVYDRFKDTRFRVIFNEKDFEIQCACCLFEFKGILC 526
+C ++G+ + ++ ++++ + V N+ + C+C L+E GI C
Sbjct: 1190 RDHC--IIQGISK-SGDMKIVTISSLFEKER----VVQMNKSNMFGTCSCKLYESYGIPC 1242
Query: 527 RHILCVLQLTGKTESVPSCYILSRWRKDIKRRHTLIKCGFDGNVELQLVGKACD----AF 582
RHI+ VL+ K +PS YI+ RW K KR + FD L L KA D A
Sbjct: 1243 RHIIQVLR-GEKQNEIPSIYIMKRWEKICKR-----EMFFDDEGNL-LDEKAKDPMEVAM 1295
Query: 583 YEVVSVGIHTEDDLLKVMNRINELKIELNCEGSSSVIIEEDSLVQNQMAKILD-----PA 637
+ +S + DL++ + +I ++++ ++D +KI D P
Sbjct: 1296 RKKISDSRNNLSDLVEPLQKII----------PAAIVNKQDEFESFLGSKIPDEVEIHPP 1345
Query: 638 TTRSKGRPPSKRKASKLDQIVQKKVLRKK 666
+SKG P + K SK +KK RK+
Sbjct: 1346 NIKSKG-PCKRIKKSK-----EKKAPRKR 1368
>gb|AAV31356.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 896
Score = 312 bits (799), Expect = 3e-83
Identities = 193/571 (33%), Positives = 294/571 (50%), Gaps = 23/571 (4%)
Query: 1 MTFSSEEEVIKYYKSYARCMGFGTVKINSKNAKDGKKYFTLGCTR-------ARSYVSNS 53
M F + +V K+YKSYA GF K + + C+R + S
Sbjct: 102 MIFDTLTDVEKFYKSYAHEAGFSVRVGQHKKQNEEILFKRYYCSREGYIKERVKDVSDES 161
Query: 54 KNLLKPNPTI---RAQCMARVNMSMSLDGKITITKVALEHNHG-LSPTKSRYFRCNKNLD 109
+ P + R C A + + + D K I+ + EH+HG +SP K R N+ +
Sbjct: 162 GKKKRKTPYMMETRCGCEAHIVVKLGSDKKYRISSMIGEHSHGFVSPDKRHLLRSNRTVS 221
Query: 110 PHTKRRLDINDQAGINVSRNFRSMVVEANGYDNLTFGEKDCRNYIDKARRLRLGTGDAEA 169
K L +A I S+ FR + V G+ N+ ++ +NY R ++ DA+
Sbjct: 222 ERAKSTLFSCHKASIGTSQAFRLLQVSDGGFQNVGCTLRNLQNYYHDLR-CKIKDADAQM 280
Query: 170 IQNYFVRMQKKNSQFYYVMDVDDKSRLRNVFWVDARCRAAYEYFGEVITFDTTYLTNKYD 229
R ++ N F+Y VD + RL VFW DA CR Y FG+V++ D+TY TN+Y+
Sbjct: 281 FVGQLERKKEVNPAFFYEFMVDKQGRLVRVFWADAICRKNYSVFGDVLSVDSTYSTNQYN 340
Query: 230 MPFAPFVGVNHHGQSVLLGCALLSNEDTKTFSWLFKTWLECMHGRAPNAIITDQDRAMKK 289
M F PF GVNHH QSV LG + L++E ++F WLF+T+L+ G AP IITD+D +MK
Sbjct: 341 MKFVPFTGVNHHLQSVFLGASFLADEKIESFVWLFQTFLKATGGVAPRLIITDEDASMKA 400
Query: 290 AIEDVFPKARHRWCLWHLMKKVPEKLGRHSHYE-SIKLLLHDAVYDSSSISDFMEKWKKM 348
AI + P HR C+WH+M+KVPEK+G + LH V+ S DF +W +
Sbjct: 401 AIAQILPNTVHRLCMWHIMEKVPEKVGPSIREDGEFWDRLHKCVWGSEDSDDFESEWNSI 460
Query: 349 IECYELHDNEWLKGLFDERYRWVPVYVRDTFWAG-MSTTQRSESMNSFFDGYVSSKTTLK 407
+ Y L NEW FD R W+ Y D AG +STT RSES NSFF+ ++ K T
Sbjct: 461 MAKYGLIGNEWFSTKFDIRQSWILAYFMDIPLAGILSTTSRSESANSFFNRFIHRKLTFV 520
Query: 408 QFVEQYDNALKDKIEKESMADFVSFNTTIACISLFGFESQFQKAFTNAKFKEFQIE-IAS 466
+F ++D AL+ + ++E AD +S +T ++ + E Q +T+ F +FQ + I +
Sbjct: 521 EFWLRFDTALECQRQEELKADNISLHTNPKLMTPWDMEKQCSGIYTHEVFSKFQEQLIVA 580
Query: 467 MIYCNAFLEGMENLNSTFCVIESKKVYDRFKDTRFRVIFNEKDFEIQCACCLFEFKGILC 526
+C ++G+ + ++ ++++ + V N+ + C+C L+E GI C
Sbjct: 581 RDHC--IIQGISK-SGDMKIVTISSLFEKER----VVQMNKSNMFGTCSCKLYESYGIPC 633
Query: 527 RHILCVLQLTGKTESVPSCYILSRWRKDIKR 557
RHI+ VL+ K +PS YI+ RW K KR
Sbjct: 634 RHIIQVLR-GEKQNEIPSIYIMKRWEKICKR 663
>gb|AAO11606.1| At2g27110/T20P8.16 [Arabidopsis thaliana]
gi|20197414|gb|AAC77869.2| Mutator-like transposase
[Arabidopsis thaliana] gi|15982769|gb|AAL09732.1|
At2g27110/T20P8.16 [Arabidopsis thaliana]
gi|18401324|ref|NP_565636.1| far-red impaired responsive
protein, putative [Arabidopsis thaliana]
gi|30683396|ref|NP_850098.1| far-red impaired responsive
protein, putative [Arabidopsis thaliana]
Length = 851
Score = 308 bits (790), Expect = 3e-82
Identities = 191/559 (34%), Positives = 287/559 (51%), Gaps = 35/559 (6%)
Query: 1 MTFSSEEEVIKYYKSYARCMGFGTVKINSKNAKDGKKYFTLGCTRARSYVSNSKNLLKPN 60
M F+SE+E +Y Y+R +GF T K+ + T G R +V +S + +
Sbjct: 53 MEFNSEKEAKSFYDEYSRQLGF-TSKLLPR---------TDGSVSVREFVCSSSSK-RSK 101
Query: 61 PTIRAQCMARVNMSMSLDGKITITKVALEHNHGLSPTKSRYFRCNKNLDP--HTKRRLDI 118
+ C A V + + K +TK EH HGL+ S C L P H
Sbjct: 102 RRLSESCDAMVRIELQGHEKWVVTKFVKEHTHGLA--SSNMLHC---LRPRRHFANSEKS 156
Query: 119 NDQAGINVSRNFRSMVVEANGYDNLTFGEKDCRNYIDKARRLRLGTGDAEAIQNYFVRMQ 178
+ Q G+NV + ++AN + RN R DA + YF RMQ
Sbjct: 157 SYQEGVNVPSGMMYVSMDANS--------RGARNASMATNTKRTIGRDAHNLLEYFKRMQ 208
Query: 179 KKNSQFYYVMDVDDKSRLRNVFWVDARCRAAYEYFGEVITFDTTYLTNKYDMPFAPFVGV 238
+N F+Y + +D+ +++ NVFW D+R R AY +FG+ +T DT Y N++ +PFAPF GV
Sbjct: 209 AENPGFFYAVQLDEDNQMSNVFWADSRSRVAYTHFGDTVTLDTRYRCNQFRVPFAPFTGV 268
Query: 239 NHHGQSVLLGCALLSNEDTKTFSWLFKTWLECMHGRAPNAIITDQDRAMKKAIEDVFPKA 298
NHHGQ++L GCAL+ +E +F WLFKT+L M + P +++TDQDRA++ A VFP A
Sbjct: 269 NHHGQAILFGCALILDESDTSFIWLFKTFLTAMRDQPPVSLVTDQDRAIQIAAGQVFPGA 328
Query: 299 RHRWCLWHLMKKVPEKLGRHS-HYESIKLLLHDAVYDSSSISDFMEKWKKMIECYELHDN 357
RH W ++++ EKL Y S ++ L++ + + +I +F W +I+ Y+L +
Sbjct: 329 RHCINKWDVLREGQEKLAHVCLAYPSFQVELYNCINFTETIEEFESSWSSVIDKYDLGRH 388
Query: 358 EWLKGLFDERYRWVPVYVRDTFWAGMSTTQRSESMNSFFDGYVSSKTTLKQFVEQYDNAL 417
EWL L++ R +WVPVY RD+F+A + +Q SFFDGYV+ +TTL F Y+ A+
Sbjct: 389 EWLNSLYNARAQWVPVYFRDSFFAAVFPSQGYS--GSFFDGYVNQQTTLPMFFRLYERAM 446
Query: 418 KDKIEKESMADFVSFNTTIACISLFGFESQFQKAFTNAKFKEFQIEIASMIYCNAFLEGM 477
+ E E AD + NT + E+Q FT F +FQ E+ A
Sbjct: 447 ESWFEMEIEADLDTVNTPPVLKTPSPMENQAANLFTRKIFGKFQEELVETFAHTANRIED 506
Query: 478 ENLNSTFCVIESKKVYDRFKDTRFRVIFNEKDFEIQCACCLFEFKGILCRHILCVLQLTG 537
+ STF V + + + V F + C+C +FE GILCRH+L V +T
Sbjct: 507 DGTTSTFRVANFEN-----DNKAYIVTFCYPEMRANCSCQMFEHSGILCRHVLTVFTVT- 560
Query: 538 KTESVPSCYILSRWRKDIK 556
++P YIL RW ++ K
Sbjct: 561 NILTLPPHYILRRWTRNAK 579
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.322 0.135 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,146,017,139
Number of Sequences: 2540612
Number of extensions: 47652151
Number of successful extensions: 121176
Number of sequences better than 10.0: 491
Number of HSP's better than 10.0 without gapping: 302
Number of HSP's successfully gapped in prelim test: 189
Number of HSP's that attempted gapping in prelim test: 120134
Number of HSP's gapped (non-prelim): 731
length of query: 694
length of database: 863,360,394
effective HSP length: 135
effective length of query: 559
effective length of database: 520,377,774
effective search space: 290891175666
effective search space used: 290891175666
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 79 (35.0 bits)
Medicago: description of AC135795.1


