

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135320.11 + phase: 0
(165 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
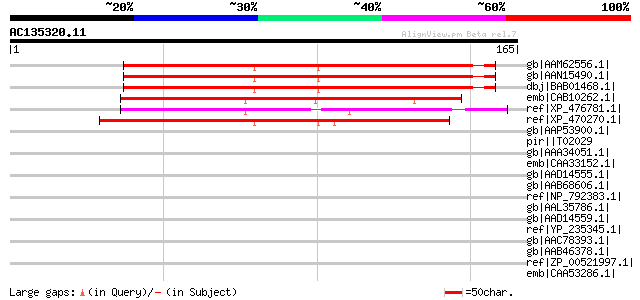
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM62556.1| unknown [Arabidopsis thaliana] 137 8e-32
gb|AAN15490.1| unknown protein [Arabidopsis thaliana] gi|1825291... 137 8e-32
dbj|BAB01468.1| unnamed protein product [Arabidopsis thaliana] 137 8e-32
emb|CAB10262.1| hypothetical protein [Arabidopsis thaliana] gi|7... 133 2e-30
ref|XP_476781.1| hypothetical protein [Oryza sativa (japonica cu... 112 3e-24
ref|XP_470270.1| Hypothetical protein [Oryza sativa (japonica cu... 100 2e-20
gb|AAP53900.1| putative DNA-binding protein [Oryza sativa (japon... 37 0.20
pir||T02029 DNA-binding protein pabf - common tobacco gi|555655|... 35 0.98
gb|AAA34051.1| DNA-binding protein 35 0.98
emb|CAA33152.1| unnamed protein product [Chlamydomonas reinhardt... 34 1.7
gb|AAD14555.1| structural polyprotein [Venezuelan equine encepha... 33 2.2
gb|AAB68606.1| structural protein precursor [Venezuelan equine e... 33 2.2
ref|NP_792383.1| peptide ABC transporter, ATP-binding protein [P... 33 3.7
gb|AAL35786.1| glycoprotein [71D1252 virus] 33 3.7
gb|AAD14559.1| structural polyprotein [Venezuelan equine encepha... 33 3.7
ref|YP_235345.1| Oligopeptide/dipeptide ABC transporter, ATP-bin... 32 4.9
gb|AAC78393.1| low molecular mass heat shock protein Oshsp18.0 [... 32 4.9
gb|AAB46378.1| LMW heat shock protein [Oryza sativa] gi|422000|p... 32 4.9
ref|ZP_00521997.1| Ankyrin [Solibacter usitatus Ellin6076] gi|67... 32 4.9
emb|CAA53286.1| heat shock protein 17.8 [Oryza sativa (japonica ... 32 4.9
>gb|AAM62556.1| unknown [Arabidopsis thaliana]
Length = 198
Score = 137 bits (346), Expect = 8e-32
Identities = 73/126 (57%), Positives = 90/126 (70%), Gaps = 8/126 (6%)
Query: 38 LRRLPHVFSRILQLPLRSDADVSIEEEPTCFRFVAETDSSL---GHVETHTLHIHPGVTK 94
LRRLPH+F+R+L+LPLRS+ADV++EE CFRFVAET G + + + IHPG+TK
Sbjct: 61 LRRLPHIFNRVLELPLRSEADVAVEERHDCFRFVAETVGLCNGDGEMRAYMVEIHPGITK 120
Query: 95 IVVRA--SHSLHFSLDDLHPDIWRFRLPESVVPELATAVFVDGELIVTVPKAFDDENTPE 152
IVVR S SL SLD+L D+WRFRLPES PEL T VDG+LIVTVPK ++E+
Sbjct: 121 IVVRTNGSSSLGLSLDELELDVWRFRLPESTRPELVTVASVDGDLIVTVPKNAEEEDDD- 179
Query: 153 PIGGGG 158
GGGG
Sbjct: 180 --GGGG 183
>gb|AAN15490.1| unknown protein [Arabidopsis thaliana] gi|18252913|gb|AAL62383.1|
unknown protein [Arabidopsis thaliana]
gi|18403425|ref|NP_566710.1| expressed protein
[Arabidopsis thaliana]
Length = 198
Score = 137 bits (346), Expect = 8e-32
Identities = 73/126 (57%), Positives = 90/126 (70%), Gaps = 8/126 (6%)
Query: 38 LRRLPHVFSRILQLPLRSDADVSIEEEPTCFRFVAETDSSL---GHVETHTLHIHPGVTK 94
LRRLPH+F+R+L+LPLRS+ADV++EE CFRFVAET G + + + IHPG+TK
Sbjct: 61 LRRLPHIFNRVLELPLRSEADVAVEERHDCFRFVAETVGLCNGDGEMRAYMVEIHPGITK 120
Query: 95 IVVRA--SHSLHFSLDDLHPDIWRFRLPESVVPELATAVFVDGELIVTVPKAFDDENTPE 152
IVVR S SL SLD+L D+WRFRLPES PEL T VDG+LIVTVPK ++E+
Sbjct: 121 IVVRTNGSSSLGLSLDELELDVWRFRLPESTRPELVTVACVDGDLIVTVPKNAEEEDDD- 179
Query: 153 PIGGGG 158
GGGG
Sbjct: 180 --GGGG 183
>dbj|BAB01468.1| unnamed protein product [Arabidopsis thaliana]
Length = 167
Score = 137 bits (346), Expect = 8e-32
Identities = 73/126 (57%), Positives = 90/126 (70%), Gaps = 8/126 (6%)
Query: 38 LRRLPHVFSRILQLPLRSDADVSIEEEPTCFRFVAETDSSL---GHVETHTLHIHPGVTK 94
LRRLPH+F+R+L+LPLRS+ADV++EE CFRFVAET G + + + IHPG+TK
Sbjct: 30 LRRLPHIFNRVLELPLRSEADVAVEERHDCFRFVAETVGLCNGDGEMRAYMVEIHPGITK 89
Query: 95 IVVRA--SHSLHFSLDDLHPDIWRFRLPESVVPELATAVFVDGELIVTVPKAFDDENTPE 152
IVVR S SL SLD+L D+WRFRLPES PEL T VDG+LIVTVPK ++E+
Sbjct: 90 IVVRTNGSSSLGLSLDELELDVWRFRLPESTRPELVTVACVDGDLIVTVPKNAEEEDDD- 148
Query: 153 PIGGGG 158
GGGG
Sbjct: 149 --GGGG 152
>emb|CAB10262.1| hypothetical protein [Arabidopsis thaliana]
gi|7268229|emb|CAB78525.1| hypothetical protein
[Arabidopsis thaliana] gi|48310385|gb|AAT41810.1|
At4g14830 [Arabidopsis thaliana]
gi|46931228|gb|AAT06418.1| At4g14830 [Arabidopsis
thaliana] gi|15233585|ref|NP_193219.1| expressed protein
[Arabidopsis thaliana] gi|7485090|pir||D71411
hypothetical protein - Arabidopsis thaliana
Length = 152
Score = 133 bits (335), Expect = 2e-30
Identities = 73/115 (63%), Positives = 85/115 (73%), Gaps = 4/115 (3%)
Query: 37 KLRRLPHVFSRILQLPLRSDADVSIEEEPTCFRFVAETD-SSLGHVETHTLHIHPGVTKI 95
KLRRLPH+FSR+L+LPL+SDADV++EE CFRFVAETD G V + + IHPGVTKI
Sbjct: 29 KLRRLPHIFSRVLELPLKSDADVAVEESHDCFRFVAETDGGGGGGVRAYMVEIHPGVTKI 88
Query: 96 VVR--ASHSLHFSLDDLHPDIWRFRLPESVVPELATA-VFVDGELIVTVPKAFDD 147
+VR S SL SLD+L D+WRFRLPES PEL T DGELIVTVPK D+
Sbjct: 89 LVRTNGSSSLGLSLDELELDVWRFRLPESTRPELVTVDCDGDGELIVTVPKIEDN 143
>ref|XP_476781.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|34394452|dbj|BAC83626.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 171
Score = 112 bits (281), Expect = 3e-24
Identities = 63/133 (47%), Positives = 79/133 (59%), Gaps = 14/133 (10%)
Query: 37 KLRRLPHVFSRILQLPLRSDADVSIEEEPTCFRFVAETD--SSLGHVETHTLHIHPGVTK 94
KLRRLPH+F+++L+LP +DADVS+EE+ RFVA D + G H + IHPGVTK
Sbjct: 32 KLRRLPHIFAKVLELPFAADADVSVEEDAAALRFVAAADGFTPSGGASAHAVEIHPGVTK 91
Query: 95 IVVRASHSLHFSLDD-----LHPDIWRFRLPESVVPELATAVFVDGELIVTVPKAFDDEN 149
+VVR L LD D WRFRLP +P +ATA + DGEL+VTVPK
Sbjct: 92 VVVR---DLSAGLDGDDGAVFELDRWRFRLPPCTLPAMATATYADGELVVTVPKG----A 144
Query: 150 TPEPIGGGGATLV 162
P+ G G A V
Sbjct: 145 APDDDGDGAAAAV 157
>ref|XP_470270.1| Hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|22773244|gb|AAN06850.1| Hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 176
Score = 99.8 bits (247), Expect = 2e-20
Identities = 53/118 (44%), Positives = 74/118 (61%), Gaps = 4/118 (3%)
Query: 30 GFSNNNNKLRRLPHVFSRILQLPLRSDADVSIEEEPTCFRFVAETDSSL-GHVETHTLHI 88
G LRRLPHV+S++L+LP +D DV++ E P F FV ++L G V T+ I
Sbjct: 13 GSCGGKKDLRRLPHVYSKVLELPFPADTDVAVFEGPDAFHFVVSAAAALAGEVRVRTVRI 72
Query: 89 HPGVTKIVVRA-SHSLH--FSLDDLHPDIWRFRLPESVVPELATAVFVDGELIVTVPK 143
HPGV ++VV+A +H DD+ D WR RLPE+ P +A A +V+G+L+VTVPK
Sbjct: 73 HPGVVRVVVQAGGGGVHDDGDDDDMELDKWRSRLPEASCPAMAVAGYVNGQLVVTVPK 130
>gb|AAP53900.1| putative DNA-binding protein [Oryza sativa (japonica
cultivar-group)] gi|37534622|ref|NP_921613.1| putative
DNA-binding protein [Oryza sativa (japonica
cultivar-group)]
Length = 1066
Score = 37.0 bits (84), Expect = 0.20
Identities = 29/97 (29%), Positives = 43/97 (43%), Gaps = 3/97 (3%)
Query: 37 KLRRLPHVFSRILQLPLRSDADVS--IEEEPTCFRFVAETDSSLGHVETHTLHIHP-GVT 93
KL+RLP VFSR+L+LP D +V F V+ H + + +T
Sbjct: 16 KLQRLPPVFSRVLELPFPRDTNVRKLFTTNADLFFVPHGVGGEPDVVKVHIVRLERWDMT 75
Query: 94 KIVVRASHSLHFSLDDLHPDIWRFRLPESVVPELATA 130
++VV +DL D WRF L E+ + + A
Sbjct: 76 RVVVHIGPGEPDLRNDLVYDKWRFPLAETSILSMVMA 112
>pir||T02029 DNA-binding protein pabf - common tobacco gi|555655|gb|AAA50196.1|
DNA-binding protein
Length = 546
Score = 34.7 bits (78), Expect = 0.98
Identities = 20/45 (44%), Positives = 22/45 (48%), Gaps = 4/45 (8%)
Query: 116 RFRLPESVVPELATAVFVDGELIVTVPKAFDDENTPEPIGGGGAT 160
R R P S P AT D V + AFD EN P +GGGG T
Sbjct: 392 RGRPPRSSGPPAATVGVTD----VPIAAAFDTENLPNAVGGGGVT 432
>gb|AAA34051.1| DNA-binding protein
Length = 380
Score = 34.7 bits (78), Expect = 0.98
Identities = 20/45 (44%), Positives = 22/45 (48%), Gaps = 4/45 (8%)
Query: 116 RFRLPESVVPELATAVFVDGELIVTVPKAFDDENTPEPIGGGGAT 160
R R P S P AT D V + AFD EN P +GGGG T
Sbjct: 226 RGRPPRSSGPPAATVGVTD----VPIAAAFDTENLPNAVGGGGVT 266
>emb|CAA33152.1| unnamed protein product [Chlamydomonas reinhardtii]
gi|81238|pir||S04939 heat shock 22K protein -
Chlamydomonas reinhardtii gi|123560|sp|P12811|HS2C_CHLRE
CHLOROPLAST HEAT SHOCK 22 KD PROTEIN
Length = 157
Score = 33.9 bits (76), Expect = 1.7
Identities = 31/108 (28%), Positives = 46/108 (41%), Gaps = 16/108 (14%)
Query: 57 ADVSIEEEPTCFRFVAETDSSLGHVETHTLHIHPGVTKIVVRASHSLHFSLDDLHPDIWR 116
A + I E PT F A+ +G + + + GV ++V L + + +WR
Sbjct: 47 APMDIIESPTAFELHADAPG-MGPDDVK-VELQEGV--LMVTGERKLSHTTKEAGGKVWR 102
Query: 117 -----------FRLPESVVPELATAVFVDGELIVTVPKAFDDENTPEP 153
F LPE+ P+ TA G L+VTVPK + PEP
Sbjct: 103 SERTAYSFSRAFSLPENANPDGITAAMDKGVLVVTVPKR-EPPAKPEP 149
>gb|AAD14555.1| structural polyprotein [Venezuelan equine encephalitis virus]
Length = 1254
Score = 33.5 bits (75), Expect = 2.2
Identities = 22/79 (27%), Positives = 38/79 (47%), Gaps = 9/79 (11%)
Query: 44 VFSRILQLPLRSDADVSIEE--EPTCFRFVAETDSSLG-------HVETHTLHIHPGVTK 94
+F+R+ + P S A+ S+ E T F +A S HV + T + + +
Sbjct: 1098 LFTRVSETPTLSSAECSLNECTYSTDFGGIATVKYSASKSGKCAVHVPSGTATLKESLVE 1157
Query: 95 IVVRASHSLHFSLDDLHPD 113
+V + S +LHFS +HP+
Sbjct: 1158 VVEQGSMTLHFSTASIHPE 1176
>gb|AAB68606.1| structural protein precursor [Venezuelan equine encephalitis virus]
Length = 1254
Score = 33.5 bits (75), Expect = 2.2
Identities = 22/79 (27%), Positives = 38/79 (47%), Gaps = 9/79 (11%)
Query: 44 VFSRILQLPLRSDADVSIEE--EPTCFRFVAETDSSLG-------HVETHTLHIHPGVTK 94
+F+R+ + P S A+ S+ E T F +A S HV + T + + +
Sbjct: 1098 LFTRVSETPTLSSAECSLNECTYSTDFGGIATVKYSASKSGKCAVHVPSGTATLKESLVE 1157
Query: 95 IVVRASHSLHFSLDDLHPD 113
+V + S +LHFS +HP+
Sbjct: 1158 VVEQGSMTLHFSTASIHPE 1176
>ref|NP_792383.1| peptide ABC transporter, ATP-binding protein [Pseudomonas syringae
pv. tomato str. DC3000] gi|28853009|gb|AAO56078.1|
peptide ABC transporter, ATP-binding protein
[Pseudomonas syringae pv. tomato str. DC3000]
Length = 333
Score = 32.7 bits (73), Expect = 3.7
Identities = 19/54 (35%), Positives = 28/54 (51%), Gaps = 6/54 (11%)
Query: 19 LSQAETLLSMSGFSNNNNKLRRLPHVFSR------ILQLPLRSDADVSIEEEPT 66
L +A +L G SN + +LR+ PH S I+ + L + DV I +EPT
Sbjct: 128 LQRAAQMLEKVGISNASERLRQYPHQLSGGLRQRVIIAMALMCEPDVIIADEPT 181
>gb|AAL35786.1| glycoprotein [71D1252 virus]
Length = 335
Score = 32.7 bits (73), Expect = 3.7
Identities = 22/79 (27%), Positives = 38/79 (47%), Gaps = 9/79 (11%)
Query: 44 VFSRILQLPLRSDADVSIEEEPTCFRF-----VAETDSSLG----HVETHTLHIHPGVTK 94
+F+R+ + P SDA S+ E F V + S G H+ + T + +
Sbjct: 196 LFTRVSETPTLSDAGCSLNECVYSSDFGGIATVKYSASKAGKCAVHIPSGTATLKESIVD 255
Query: 95 IVVRASHSLHFSLDDLHPD 113
+V + S +LHFS ++HP+
Sbjct: 256 VVEQGSLTLHFSTANIHPE 274
>gb|AAD14559.1| structural polyprotein [Venezuelan equine encephalitis virus]
Length = 1254
Score = 32.7 bits (73), Expect = 3.7
Identities = 22/79 (27%), Positives = 38/79 (47%), Gaps = 9/79 (11%)
Query: 44 VFSRILQLPLRSDADVSIEEEPTCFRF-----VAETDSSLG----HVETHTLHIHPGVTK 94
+F+R+ + P SDA S+ E F V + S G H+ + T + +
Sbjct: 1098 LFTRVSETPTLSDAGCSLNECVYSSDFGGIATVKYSASKAGKCAVHIPSGTATLKESIVD 1157
Query: 95 IVVRASHSLHFSLDDLHPD 113
+V + S +LHFS ++HP+
Sbjct: 1158 VVEQGSLTLHFSTANIHPE 1176
>ref|YP_235345.1| Oligopeptide/dipeptide ABC transporter, ATP-binding protein,
C-terminal [Pseudomonas syringae pv. syringae B728a]
gi|63256211|gb|AAY37307.1| Oligopeptide/dipeptide ABC
transporter, ATP-binding protein, C-terminal
[Pseudomonas syringae pv. syringae B728a]
Length = 333
Score = 32.3 bits (72), Expect = 4.9
Identities = 19/54 (35%), Positives = 27/54 (49%), Gaps = 6/54 (11%)
Query: 19 LSQAETLLSMSGFSNNNNKLRRLPHVFSR------ILQLPLRSDADVSIEEEPT 66
L +A +L G SN +LR+ PH S I+ + L + DV I +EPT
Sbjct: 128 LQRAAQMLEKVGISNAGERLRQYPHQLSGGLRQRVIIAMALMCEPDVIIADEPT 181
>gb|AAC78393.1| low molecular mass heat shock protein Oshsp18.0 [Oryza sativa]
gi|2119194|pir||JC4377 low-molecular-weight heat-shock
protein - rice
Length = 160
Score = 32.3 bits (72), Expect = 4.9
Identities = 16/28 (57%), Positives = 18/28 (64%)
Query: 116 RFRLPESVVPELATAVFVDGELIVTVPK 143
RFRLPE+ PE A +G L VTVPK
Sbjct: 118 RFRLPENTKPEQIKASMENGVLTVTVPK 145
>gb|AAB46378.1| LMW heat shock protein [Oryza sativa] gi|422000|pir||S24396 heat
shock protein, low molecular weight (clone pTS3) - rice
Length = 154
Score = 32.3 bits (72), Expect = 4.9
Identities = 16/28 (57%), Positives = 18/28 (64%)
Query: 116 RFRLPESVVPELATAVFVDGELIVTVPK 143
RFRLPE+ PE A +G L VTVPK
Sbjct: 112 RFRLPENTKPEQIKASMENGVLTVTVPK 139
>ref|ZP_00521997.1| Ankyrin [Solibacter usitatus Ellin6076] gi|67864040|gb|EAM59048.1|
Ankyrin [Solibacter usitatus Ellin6076]
Length = 493
Score = 32.3 bits (72), Expect = 4.9
Identities = 18/70 (25%), Positives = 31/70 (43%)
Query: 91 GVTKIVVRASHSLHFSLDDLHPDIWRFRLPESVVPELATAVFVDGELIVTVPKAFDDENT 150
G+ K+ + S +LH ++ + H ++ F L P + +I +V K +N
Sbjct: 225 GINKVGRQGSSALHLAVQNAHYELASFLLDAGADPNADGPGYTALHIIPSVRKPGGGDND 284
Query: 151 PEPIGGGGAT 160
P P G G T
Sbjct: 285 PAPDGSGSMT 294
>emb|CAA53286.1| heat shock protein 17.8 [Oryza sativa (japonica cultivar-group)]
Length = 160
Score = 32.3 bits (72), Expect = 4.9
Identities = 16/28 (57%), Positives = 18/28 (64%)
Query: 116 RFRLPESVVPELATAVFVDGELIVTVPK 143
RFRLPE+ PE A +G L VTVPK
Sbjct: 118 RFRLPENTKPEQIKASMENGVLTVTVPK 145
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.321 0.136 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 297,360,742
Number of Sequences: 2540612
Number of extensions: 11697892
Number of successful extensions: 25872
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 18
Number of HSP's successfully gapped in prelim test: 24
Number of HSP's that attempted gapping in prelim test: 25838
Number of HSP's gapped (non-prelim): 42
length of query: 165
length of database: 863,360,394
effective HSP length: 118
effective length of query: 47
effective length of database: 563,568,178
effective search space: 26487704366
effective search space used: 26487704366
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 70 (31.6 bits)
Medicago: description of AC135320.11


