

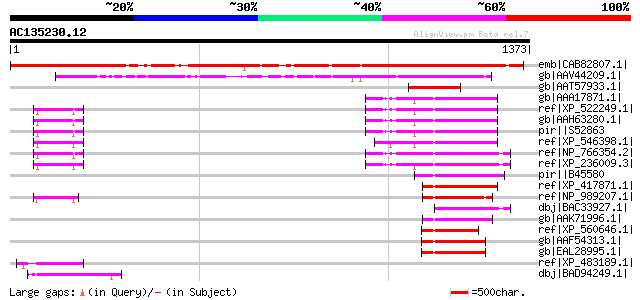
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135230.12 + phase: 0
(1373 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAB82807.1| DNA-binding protein-like [Arabidopsis thaliana] ... 1149 0.0
gb|AAV44209.1| unknow protein [Oryza sativa (japonica cultivar-g... 698 0.0
gb|AAT57933.1| putative DNA-binding protein [Gossypium hirsutum]... 262 5e-68
gb|AAA17871.1| R kappa B 176 5e-42
ref|XP_522249.1| PREDICTED: similar to nuclear factor related to... 176 5e-42
gb|AAH63280.1| NFRKB protein [Homo sapiens] 176 5e-42
pir||S52863 DNA-binding protein R kappa B - human gi|695579|emb|... 176 5e-42
ref|XP_546398.1| PREDICTED: similar to nuclear factor related to... 174 2e-41
ref|NP_766354.2| nuclear factor related to kappa B binding prote... 173 4e-41
ref|XP_236009.3| PREDICTED: similar to Nuclear factor related to... 169 5e-40
pir||B45580 transcription factor R-kappa-B - mouse (fragment) 169 5e-40
ref|XP_417871.1| PREDICTED: similar to RIKEN cDNA A530090G11 [Ga... 169 8e-40
ref|NP_989207.1| hypothetical protein LOC394815 [Xenopus tropica... 165 9e-39
dbj|BAC33927.1| unnamed protein product [Mus musculus] 153 3e-35
gb|AAK71996.1| NFRKB-like protein [Apis mellifera] 147 3e-33
ref|XP_560646.1| ENSANGP00000025456 [Anopheles gambiae str. PEST... 138 1e-30
gb|AAF54313.1| CG11970-PA [Drosophila melanogaster] gi|5052658|g... 137 3e-30
gb|EAL28995.1| GA11303-PA [Drosophila pseudoobscura] 136 4e-30
ref|XP_483189.1| DNA-binding protein-like [Oryza sativa (japonic... 135 1e-29
dbj|BAD94249.1| hypothetical protein [Arabidopsis thaliana] gi|5... 131 1e-28
>emb|CAB82807.1| DNA-binding protein-like [Arabidopsis thaliana]
gi|15231253|ref|NP_190169.1| expressed protein
[Arabidopsis thaliana] gi|11357267|pir||T47523
DNA-binding protein-like - Arabidopsis thaliana
Length = 1298
Score = 1149 bits (2971), Expect = 0.0
Identities = 678/1388 (48%), Positives = 865/1388 (61%), Gaps = 120/1388 (8%)
Query: 1 MAIEKNSFKVSRVDTECEPMSKESMSSGDEEDVQRRNSGNESDEDDDEFDDADSGAGSDD 60
MAIEK++ KVSR D E S +SMSS +E + +S+++DD+FD+ DSGAGSDD
Sbjct: 1 MAIEKSNVKVSRFDLEYSHGSGDSMSSYEERRKNSVVNNVDSEDEDDDFDEDDSGAGSDD 60
Query: 61 FDLLELGETGAEFCQIGNQTCSIPLELYDLSGLEDILSVDVWNDCLSEEERFELAKYLPD 120
FDLLEL ETGAEFCQ+GN TCSIP ELYDL LEDILSVDVWN+CL+E+ERF L+ YLPD
Sbjct: 61 FDLLELAETGAEFCQVGNVTCSIPFELYDLPSLEDILSVDVWNECLTEKERFSLSSYLPD 120
Query: 121 MDQETFVQTLKELFTGCNFQFGSPVKKLFDMLKGGLCEPRVALYREGLNFVQKRQHYHLL 180
+DQ TF++TLKELF GCNF FGSPVKKLFDMLKGG CEPR LY EG + + +HYH L
Sbjct: 121 VDQLTFMRTLKELFEGCNFHFGSPVKKLFDMLKGGQCEPRNTLYLEGRSLFLRTKHYHSL 180
Query: 181 KKHQNTMVSNLCQMRDAWLNCRGYSIEERLRVLNIMTSQKSLMGEKMDDLEADSSE--ES 238
+K+ N MV NLCQ RDAW +C+GYSI+E+LRVLNI+ SQK+LM EK DD E DSSE E
Sbjct: 181 RKYHNDMVVNLCQTRDAWTSCKGYSIDEKLRVLNIVKSQKTLMREKKDDFEDDSSEKDEP 240
Query: 239 GEGMWSRKNKDKKNAQK-LGRFPFQGVGSGLDFHPREQSMVMEQEKYSKQNPKGILKLAG 297
+ W RK KD+K+ QK L R GV SGL+F PR Q +EQ+ Y K PK K
Sbjct: 241 FDKPWGRKGKDRKSTQKKLARHAGYGVDSGLEF-PRRQLAAVEQDLYGK--PKSKPKFPF 297
Query: 298 SKTHLAKDPTAHSSSVYHGLDMNPRLNGSAFAHPQHNISTGYDLGSIRRTRDQLWNGDNE 357
+KT + T Y+G MN N S+ ++ D D++
Sbjct: 298 AKTSVGPYATG-----YNGYGMNSAYNPSSLVRQRYGSEDNID-------------DDDQ 339
Query: 358 EEI----SYRDRNALRGSLMDMSSALRVGKRHDLLRGDEIEGGNLMGLSMSSKTDLRGYT 413
+ + S RDR + R GK+H R E + MG SS+ Y+
Sbjct: 340 DPLFGMGSRRDR--------EKPGYSRPGKKHKSSRDGEPISEHFMGPPYSSRQYHSNYS 391
Query: 414 RNPNQSSDMQLFAAKPPSKK-KGKYAENVQQFVGSRGSKLSHNVDSIHSPDPDDLFYNKR 472
++ ++++Q A K KG A+ RG H K
Sbjct: 392 KSSRYANNIQPHAFADQMKPVKGSLAD-------LRGDLYRHG---------------KN 429
Query: 473 PAQELGMSSLFKYEDWNPKSKKRKAERESPDLSYTAYRSSSPQVSNRLFSSDFRTKSSQE 532
+ + +D N KSKK K+ER+SPD S +YR+S Q++ R +SDF QE
Sbjct: 430 HGDGFSVDPRYISDDLNSKSKKLKSERDSPDTSLRSYRASMQQMNERFLNSDFGENHVQE 489
Query: 533 KIRGSFVQNGRKDMKPLRGSHMLARGEETESDSSEQWDDDDDNNPLLQSKFAYPIGKAAG 592
KIR + V N R + R S M ++TESDSS +DD+++ N L+++K + +G
Sbjct: 490 KIRVNVVPNARSGVAAFRDSRMFMGNDDTESDSSHGYDDEEERNRLMRNKSSVSVGGMNN 549
Query: 593 SLTKPLKSHLDPMKAKFSRTDMKATQS--------KKIGGFAEQGNMHGADNYLSKNAKK 644
S LKS D K+K + DM+ + K +G E G + + K+ +K
Sbjct: 550 SHFPILKSRQDIKKSKSRKKDMQENELLDGRSAYLKYLGVSGEHIYAPGTEKHSFKSKQK 609
Query: 645 SKIFNGSPVRNPAGKFMEENYPSVSDMLNGGHDDWRQLYKS-KNDQIRDEPVQR--FDMP 701
K+ + SP+ N + + E+ P S ++ ++ ++S +N Q R++ + R F P
Sbjct: 610 GKMRDRSPLENFSSRDFEDG-PITSLSEFQDRNNRKEFFRSNRNSQTREQMIDRTLFQRP 668
Query: 702 SSTSYAAEHKKKGRIGLDHSSMRSKYLHDYGNDEDDSLENRLLADENGVGQSRFWRKGQK 761
S+ + K+ +++D+S E R L + + R RK Q
Sbjct: 669 SAKPNLSGRKR------------------VFDEDDESHEMRTLVN----ARDRLSRKYQV 706
Query: 762 NVAHKDDRDERSEVPL-LGCNSAMKKRKMKFGAADFGERDEDANLLSSNPSKIDDLPAFS 820
+ + DE E L + CN+ KKRK + D R+++ +L ++
Sbjct: 707 SEDDGNSGDENLEARLFVSCNALSKKRKTRESLMDMERREDNGDLQLYPDIQLPVGDVTV 766
Query: 821 LKRKSKKKPGAEMVISEMENSELPLTHTVTADVEVETKPQKKPYILITPTVHTGFSFSIM 880
KRK KKK ++ ++E S++P ++ EVETKPQKKP++LITPTVHTGFSFSI+
Sbjct: 767 SKRKGKKKMEVDVGFLDLETSDIPKA----SEAEVETKPQKKPFVLITPTVHTGFSFSIV 822
Query: 881 HLLTAVRTAMISPPEVESLEAGKPVEQQNKAQEDSLNGVISSDKVDDKVAANVEPSDQK- 939
HLL+AVR AM S +SL+ K V +N E NG ++ +D N P
Sbjct: 823 HLLSAVRMAMTSLRPEDSLDVSKSVAVENAEHETGENGASVPEEAED----NKSPQQGNG 878
Query: 940 NVPSLTIQEIVNRVRSNPGDPCILETQEPLQDLVRGVLKIFSSKTAPLGAKGWKVLAVYE 999
N+PSLTIQEIV+ V+SNPGDPCILETQEPLQDL+RGVLKIFSSKT+PLGAKGWK L +E
Sbjct: 879 NLPSLTIQEIVSCVKSNPGDPCILETQEPLQDLIRGVLKIFSSKTSPLGAKGWKPLVTFE 938
Query: 1000 KSTRSWSWIGPVLHNSSDHDPIEEVTSPEAWGLPHKMLVKLVDSFANWLKCGQDTLKQIG 1059
+ST+ WSWIGPVL SD + +EEVTSPEAW LPHKMLVKLVDSFANWLK GQ+TL+QIG
Sbjct: 939 RSTKCWSWIGPVL-GPSDQETVEEVTSPEAWSLPHKMLVKLVDSFANWLKTGQETLQQIG 997
Query: 1060 SLPAPPLELMQINLDEKERFRDLRAQKSLNTISPSSEEVRAYFRKEELLRYSIPDRAFSY 1119
SLP PPL LMQ NLDEKERF+DLRAQKSL+TI+ SSEE RAYFRKEE LRYSIPDRAF Y
Sbjct: 998 SLPEPPLSLMQCNLDEKERFKDLRAQKSLSTITQSSEEARAYFRKEEFLRYSIPDRAFVY 1057
Query: 1120 TAADGKKSIVAPLRRCGGKPTSKARDHFMLKRDRPPHVTILCLVRDAAARLPGSIGTRAD 1179
TAADGKKSIVAPLRR GGKPTSKARDHFMLKR+RPPHVTILCLVRDAAARLPGSIGTRAD
Sbjct: 1058 TAADGKKSIVAPLRRGGGKPTSKARDHFMLKRERPPHVTILCLVRDAAARLPGSIGTRAD 1117
Query: 1180 VCTLIRDSQYIVEDVSDEKINQVVSGALDRLHYERDPCVLFDQERKLWVYLHREREEEDF 1239
VCTLIRDSQYIVEDVSD ++NQVVSGALDRLHYERDPCV FD ERKLWVYLHR+RE+EDF
Sbjct: 1118 VCTLIRDSQYIVEDVSDSQVNQVVSGALDRLHYERDPCVQFDSERKLWVYLHRDREQEDF 1177
Query: 1240 DDDGTSSTKKWKRQKKDVADQS-DQAPVTVACNGTGEQSGYDLCSDLNV-DPPCIEDDKE 1297
+DDGTSSTKKWKR KK+ A+Q+ +Q VTVA G EQ+ ++ S+ +P ++ D+
Sbjct: 1178 EDDGTSSTKKWKRPKKEAAEQTEEQEAVTVAFLGNEEQTETEMGSEPKTGEPTGLDGDQG 1237
Query: 1298 AVQLLTTDTRPNAEDQVVVNPVSEVGNSCEDNSMT-WE---ALDLNPTRE---LCQENST 1350
A L +T AE+Q + N+ + N T WE A+ NP + +CQENS
Sbjct: 1238 ATDQLCNETEQAAEEQ-------DGENTAQGNEPTIWEPDPAVVSNPVEDNTFICQENSV 1290
Query: 1351 NEDFGDES 1358
N+DF DE+
Sbjct: 1291 NDDFDDET 1298
>gb|AAV44209.1| unknow protein [Oryza sativa (japonica cultivar-group)]
Length = 1121
Score = 698 bits (1802), Expect = 0.0
Identities = 480/1195 (40%), Positives = 647/1195 (53%), Gaps = 159/1195 (13%)
Query: 121 MDQETFVQTLKELFTGCNFQFGSPVKKLFDMLKGGLCEPRVALYREGLNFVQKRQHYHLL 180
MDQETF +TL EL G NF FGSP+ LFD LKGGLC+PR+ALYR G F ++R+HY+ L
Sbjct: 1 MDQETFARTLAELLRGDNFHFGSPLAALFDRLKGGLCDPRIALYRRGTRFAERRKHYYRL 60
Query: 181 KKHQNTMVSNLCQMRDAWLNCRGYSIEERLRVLNIMTSQKSLMGEKMDDLEADSSEESGE 240
+ + N+MV L ++D W C+GY+I E+LR L+ M +Q+ ++ +A G
Sbjct: 61 QSYHNSMVRGLWDVKDCWKGCQGYNINEKLRALDAMKAQQQQQQQQQQQQKA----HLGL 116
Query: 241 GMWSRKNKDKKNAQKLGRFPFQGVGSGLDFHPREQSMVMEQEKYSKQNPKGILKLAGSKT 300
G + D ++ + G S + P +++++ + K K+ KG+L+L
Sbjct: 117 GGRAGSETDSESRE-------YGDPSLMRLKP-DKTVLKKSGKPEKERSKGLLRLGA--- 165
Query: 301 HLAKDPTAHSSSVYHGLDMNPRLNGSAFAHPQHNISTGYDLGSIRRTRD-QLWNGDNEEE 359
P G + + S + + + GYD G +RR + + G + EE
Sbjct: 166 -----PKGLGEEYIGGAGRDAAMALSELSRQDN--AYGYDSGVMRRGKPRRSQQGLHSEE 218
Query: 360 ISYRDRNALRGSLMDMSSALRVGKRHDLLRGDEIEGGNLMGLSMSSKTDLRGYTRNPNQS 419
+ D LR + + GK+ D GN + + + G N NQ
Sbjct: 219 LG--DDRDLR-MIRSHRPMPKPGKKELAASYDGNLYGNNYHENQNGSSYYYGRNANANQG 275
Query: 420 SDMQLFAAKP--PSKKKGKYAENVQQFVGSRGSKLSHNVDSIHSPDPDDLFYNKRPAQEL 477
+ +P + K KY++ + G S + D + P + +
Sbjct: 276 VTVAAAYDRPYFDTAKNAKYSDRDWMYGGQGMSSKALKGDEMDWPAGS--YAGSMNDWQR 333
Query: 478 GMSSLFKYEDWNPKSKKRKAERESPDLSYTAYRSSSPQVSNRLFSSDFRTKSSQEKIRG- 536
G S+ + +S+K +A L +Y+S Q+S+ F SD R K KI+G
Sbjct: 334 GQSA------GDYRSRKTQA---GHGLKVKSYKSIEKQISDANFGSDHRGK-IPGKIKGK 383
Query: 537 SFVQNGRKDMKPLRGSHMLARGEETESDSSEQWDDDDDNNPLLQSKFAYPIGKAAGSLTK 596
S Q R K R + + + EETESDSSE+++ D
Sbjct: 384 STSQYDRIGQKYSRSNAVYTQSEETESDSSEKFEGGGD---------------------- 421
Query: 597 PLKSHLDPMKAKFSRTDMKATQSKKIGGFAEQGNMHGADNYLSKNAKK-SKIFNGSPVRN 655
D+K Q H + + +AKK +K+ S V
Sbjct: 422 ---------------MDLK-----------RQPEHHSGSHRPAYSAKKLNKLPKASKVNY 455
Query: 656 PAGKFMEENYPSVSDMLNGGHDDWRQLYKSKNDQIRD----EPVQRFDMPSSTSYAAEHK 711
P E Y S G H ++ +++D +RD E Q +M + E K
Sbjct: 456 PTATEDFEPYQS-----KGTH----RVNVTESDYLRDVHVTETEQISEMMRPPAARGERK 506
Query: 712 KKGRIGLDHSSMRSKYLHDYGN----DEDDSLENRLLADENGVGQSRFWRKGQKNVAHKD 767
+K +D HD+GN D +++ + L + ENG + G V
Sbjct: 507 RKVMASVD--------THDHGNTELPDSNENADESLRSPENGERLA----SGSGCVDSNG 554
Query: 768 DRDERSEVPLLGCNSAMKKRKMKFGAADFGERDEDANLLSSNPSKIDDLPAFSLKRKSKK 827
D E+ ++PL C+S KK+K + A E EDA S P +++ + S K+K KK
Sbjct: 555 D-VEKKKMPLASCSSGSKKQKRRVEATSPAEHGEDA---PSAPKLVEN--SSSSKKKGKK 608
Query: 828 KPGAEMVISE--MENSELPLTHTVTADVEVETKPQKKPYILITPTVHTGFSFSIMHLLTA 885
KP A +++ + + P+ V V VE + KK Y+ ITPT+HTGFSFSI+HLLTA
Sbjct: 609 KPAAPEAVTDAVVVDEPAPVLPEVNV-VVVEPEKPKKKYVPITPTIHTGFSFSIVHLLTA 667
Query: 886 VRTAMISPPEVESLEAGKP----------------VEQQNKAQEDSLNGVISSDKV---- 925
VR AM +P E ++L A +P Q A E + G + D
Sbjct: 668 VRKAMATPTE-DTLSAKQPDGEESRKCFNNEEHCKTPQDPSATEQAQQGHEAVDASGPEK 726
Query: 926 -----DDKVAANVEPSDQKNVPSLTIQEIVNRVRSNPGDPCILETQEPLQDLVRGVLKIF 980
+ A+ E + N+P+ T+QEIV R+RSNP DP ILETQEPLQDLVRGVLKI
Sbjct: 727 AQQGHETADASAAEQTTPSNLPAFTVQEIVTRIRSNPSDPNILETQEPLQDLVRGVLKIL 786
Query: 981 SSKTAPLGAKGWKVLAVYEKSTRSWSWIGPVLHNSSDHDPIEEVTSPEAWGLPHKMLVKL 1040
SS+TAPLGAKGWK L Y+KS +SW W+GP+ SSD DP EE TSP+AWG+PHKMLVKL
Sbjct: 787 SSRTAPLGAKGWKALVSYDKSNKSWLWVGPLPSGSSDGDPNEE-TSPDAWGIPHKMLVKL 845
Query: 1041 VDSFANWLKCGQDTLKQIGSLPAPPLELMQINLDEKERFRDLRAQKSLNTISPSSEEVRA 1100
VD+FANWLK GQ+TLKQIGSLP PP NLD KERF+DLRAQKSLNTISPSSEE RA
Sbjct: 846 VDAFANWLKSGQETLKQIGSLPPPPAP-DPANLDLKERFKDLRAQKSLNTISPSSEEARA 904
Query: 1101 YFRKEELLRYSIPDRAFSYTAADGKKSIVAPLRRCGGKPTSKARDHFMLKRDRPPHVTIL 1160
YF++EE LRYSIPDRAF YTAADG+KSIVAPLRR GGKPT+KAR H ML DRPPHVTIL
Sbjct: 905 YFQREEFLRYSIPDRAFCYTAADGEKSIVAPLRRGGGKPTAKARGHPMLLPDRPPHVTIL 964
Query: 1161 CLVRDAAARLPGSIGTRADVCTLIRDSQYIVEDVSDEK--INQVVSGALDRLHYERDPCV 1218
CLVRDAAARLP GTRADVCTL++DSQY+ + S+++ +NQVVSGALDRLHYERDPCV
Sbjct: 965 CLVRDAAARLPARTGTRADVCTLLKDSQYLNHEESNKEAAVNQVVSGALDRLHYERDPCV 1024
Query: 1219 LFDQERKLWVYLHREREEEDFDDDGTSSTKKWKRQKKDVADQSDQAPVTVACNGT 1273
L+D ++KLW YLHR REEEDF+DDGTSSTKKWKR +K+ +D +D + +GT
Sbjct: 1025 LYDNDKKLWTYLHRGREEEDFEDDGTSSTKKWKRPRKE-SDPADPGNDDLEDDGT 1078
>gb|AAT57933.1| putative DNA-binding protein [Gossypium hirsutum]
gi|49234777|gb|AAT57932.1| putative DNA-binding protein
[Gossypium hirsutum]
Length = 139
Score = 262 bits (670), Expect = 5e-68
Identities = 129/137 (94%), Positives = 134/137 (97%)
Query: 1056 KQIGSLPAPPLELMQINLDEKERFRDLRAQKSLNTISPSSEEVRAYFRKEELLRYSIPDR 1115
+QIG PAPPLELMQ+NLDEKERFRDL+AQKSLNTIS SSEEVRAYFR+EELLRYSIPDR
Sbjct: 1 QQIGKSPAPPLELMQVNLDEKERFRDLKAQKSLNTISSSSEEVRAYFRREELLRYSIPDR 60
Query: 1116 AFSYTAADGKKSIVAPLRRCGGKPTSKARDHFMLKRDRPPHVTILCLVRDAAARLPGSIG 1175
AFSYTAADGKKSIVAPLRRCGGKPTSKARDHFMLKRDRPPHVTILCLVRDAAARLPGSIG
Sbjct: 61 AFSYTAADGKKSIVAPLRRCGGKPTSKARDHFMLKRDRPPHVTILCLVRDAAARLPGSIG 120
Query: 1176 TRADVCTLIRDSQYIVE 1192
TRADVCTLIRDSQY+VE
Sbjct: 121 TRADVCTLIRDSQYVVE 137
>gb|AAA17871.1| R kappa B
Length = 998
Score = 176 bits (446), Expect = 5e-42
Identities = 131/369 (35%), Positives = 192/369 (51%), Gaps = 40/369 (10%)
Query: 942 PSLTIQEIVNRVRSNPGDPCILETQEPLQDLVRGVLKIFSSKTAPLGAKGWKVLAVYEKS 1001
P L I EI + S + +LE+Q L L VL SS + L + W A
Sbjct: 81 PCLGINEISSSFFSLLLEILLLESQASLPMLEERVLDWQSSPASSLNS--WFSAAP---- 134
Query: 1002 TRSWSWIGPVLHNSSDHDPIEEVTSPEAWGLPHKM--LVKLVDSFANWLKCGQ--DTLKQ 1057
+W VL P + + E+ +P V+ + W GQ D K+
Sbjct: 135 ----NWAELVL-------PALQYLAGESRAVPSSFSPFVEFKEKTQQWKLLGQSQDNEKE 183
Query: 1058 IGSLPAPPLELM------QINLDEKERFRDLRAQKSLNTISPSSEEVRAYFRKEELLRYS 1111
+ +L LE Q N D + + ++ + PS+ E + F+++E RYS
Sbjct: 184 LAALFQLWLETKDQAFCKQENEDSSDATTPVPRVRTDYVVRPSTGEEKRVFQEQERYRYS 243
Query: 1112 IPDRAFSYTAADGKKSIVAPLRRCGGKPTS--KARDHFMLKRDRPPHVTILCLVRDAAAR 1169
P +AF++ G +S+V P++ K TS KAR+H +L+ DRP +VTIL LVRDAAAR
Sbjct: 244 QPHKAFTFRM-HGFESVVGPVKGVFDKETSLNKAREHSLLRSDRPAYVTILSLVRDAAAR 302
Query: 1170 LPGSIGTRADVCTLIRDSQYIVEDVSDEKINQVVSGALDRLHYERDPCVLFDQERKLWVY 1229
LP GTRA++C L++DSQ++ DV+ ++N VVSGALDRLHYE+DPCV +D RKLW+Y
Sbjct: 303 LPNGEGTRAEICELLKDSQFLAPDVTSTQVNTVVSGALDRLHYEKDPCVKYDIGRKLWIY 362
Query: 1230 LHREREEEDFD-----DDGTSSTKKWKRQK----KDVADQSDQAPVTVACNGTGEQSGYD 1280
LHR+R EE+F+ + +K +QK V S ++ + V +G EQS
Sbjct: 363 LHRDRSEEEFERIHQAQAAAAKARKALQQKPKPPSKVKSSSKESSIKVLSSGPSEQSQMS 422
Query: 1281 LCSDLNVDP 1289
L SD ++ P
Sbjct: 423 L-SDSSMPP 430
>ref|XP_522249.1| PREDICTED: similar to nuclear factor related to kappa B binding
protein [Pan troglodytes]
Length = 1356
Score = 176 bits (446), Expect = 5e-42
Identities = 131/369 (35%), Positives = 192/369 (51%), Gaps = 40/369 (10%)
Query: 942 PSLTIQEIVNRVRSNPGDPCILETQEPLQDLVRGVLKIFSSKTAPLGAKGWKVLAVYEKS 1001
P L I EI + S + +LE+Q L L VL SS + L + W A
Sbjct: 439 PCLGINEISSSFFSLLLEILLLESQASLPMLEERVLDWQSSPASSLNS--WFSAAP---- 492
Query: 1002 TRSWSWIGPVLHNSSDHDPIEEVTSPEAWGLPHKM--LVKLVDSFANWLKCGQ--DTLKQ 1057
+W VL P + + E+ +P V+ + W GQ D K+
Sbjct: 493 ----NWAELVL-------PALQYLAGESRAVPSSFSPFVEFKEKTQQWKLLGQSQDNEKE 541
Query: 1058 IGSLPAPPLELM------QINLDEKERFRDLRAQKSLNTISPSSEEVRAYFRKEELLRYS 1111
+ +L LE Q N D + + ++ + PS+ E + F+++E RYS
Sbjct: 542 LAALFQLWLETKDQAFCKQENEDSSDATTPVPRVRTDYVVRPSTGEEKRVFQEQERYRYS 601
Query: 1112 IPDRAFSYTAADGKKSIVAPLRRCGGKPTS--KARDHFMLKRDRPPHVTILCLVRDAAAR 1169
P +AF++ G +S+V P++ K TS KAR+H +L+ DRP +VTIL LVRDAAAR
Sbjct: 602 QPHKAFTFRM-HGFESVVGPVKGVFDKETSLNKAREHSLLRSDRPAYVTILSLVRDAAAR 660
Query: 1170 LPGSIGTRADVCTLIRDSQYIVEDVSDEKINQVVSGALDRLHYERDPCVLFDQERKLWVY 1229
LP GTRA++C L++DSQ++ DV+ ++N VVSGALDRLHYE+DPCV +D RKLW+Y
Sbjct: 661 LPNGEGTRAEICELLKDSQFLAPDVTSTQVNTVVSGALDRLHYEKDPCVKYDIGRKLWIY 720
Query: 1230 LHREREEEDFD-----DDGTSSTKKWKRQK----KDVADQSDQAPVTVACNGTGEQSGYD 1280
LHR+R EE+F+ + +K +QK V S ++ + V +G EQS
Sbjct: 721 LHRDRSEEEFERIHQAQAAAAKARKALQQKPKPPSKVKSSSKESSIKVLSSGPSEQSQMS 780
Query: 1281 LCSDLNVDP 1289
L SD ++ P
Sbjct: 781 L-SDSSMPP 788
Score = 57.4 bits (137), Expect = 3e-06
Identities = 45/153 (29%), Positives = 73/153 (47%), Gaps = 22/153 (14%)
Query: 62 DLLELGETG-------AEFCQIGNQTCSIPLELY-DLSGLEDILSVDVWNDCLSEEERFE 113
D LELG G E C +G S+P +L D D++S+ W + LS+ +R
Sbjct: 48 DPLELGPCGDGHGTRIMEDCLLGGTRVSLPEDLLEDPEIFFDVVSLSTWQEVLSDSQREH 107
Query: 114 LAKYLPDMDQETFVQT---LKELFTGCNFQFGSPVKKLFDMLKGGLCEPRVALYRE---- 166
L ++LP +++ Q + LF+G NF+FG+P+ + + G P V YR+
Sbjct: 108 LQQFLPQFPEDSAEQQNELILALFSGENFRFGNPLHIAQKLFRDGHFNPEVVKYRQLCFK 167
Query: 167 -----GLNFVQKRQHYHLLKKHQNTMVSNLCQM 194
LN ++Q++H L K S+L +M
Sbjct: 168 SQYKRYLN--SQQQYFHRLLKQILASRSDLLEM 198
>gb|AAH63280.1| NFRKB protein [Homo sapiens]
Length = 1299
Score = 176 bits (446), Expect = 5e-42
Identities = 131/369 (35%), Positives = 192/369 (51%), Gaps = 40/369 (10%)
Query: 942 PSLTIQEIVNRVRSNPGDPCILETQEPLQDLVRGVLKIFSSKTAPLGAKGWKVLAVYEKS 1001
P L I EI + S + +LE+Q L L VL SS + L + W A
Sbjct: 368 PCLGINEISSSFFSLLLEILLLESQASLPMLEERVLDWQSSPASSLNS--WFSAAP---- 421
Query: 1002 TRSWSWIGPVLHNSSDHDPIEEVTSPEAWGLPHKM--LVKLVDSFANWLKCGQ--DTLKQ 1057
+W VL P + + E+ +P V+ + W GQ D K+
Sbjct: 422 ----NWAELVL-------PALQYLAGESRAVPSSFSPFVEFKEKTQQWKLLGQSQDNEKE 470
Query: 1058 IGSLPAPPLELM------QINLDEKERFRDLRAQKSLNTISPSSEEVRAYFRKEELLRYS 1111
+ +L LE Q N D + + ++ + PS+ E + F+++E RYS
Sbjct: 471 LAALFQLWLETKDQAFCKQENEDSSDATTPVPRVRTDYVVRPSTGEEKRVFQEQERYRYS 530
Query: 1112 IPDRAFSYTAADGKKSIVAPLRRCGGKPTS--KARDHFMLKRDRPPHVTILCLVRDAAAR 1169
P +AF++ G +S+V P++ K TS KAR+H +L+ DRP +VTIL LVRDAAAR
Sbjct: 531 QPHKAFTFRM-HGFESVVGPVKGVFDKETSLNKAREHSLLRSDRPAYVTILSLVRDAAAR 589
Query: 1170 LPGSIGTRADVCTLIRDSQYIVEDVSDEKINQVVSGALDRLHYERDPCVLFDQERKLWVY 1229
LP GTRA++C L++DSQ++ DV+ ++N VVSGALDRLHYE+DPCV +D RKLW+Y
Sbjct: 590 LPNGEGTRAEICELLKDSQFLAPDVTSTQVNTVVSGALDRLHYEKDPCVKYDIGRKLWIY 649
Query: 1230 LHREREEEDFD-----DDGTSSTKKWKRQK----KDVADQSDQAPVTVACNGTGEQSGYD 1280
LHR+R EE+F+ + +K +QK V S ++ + V +G EQS
Sbjct: 650 LHRDRSEEEFERIHQAQAAAAKARKALQQKPKPPSKVKSSSKESSIKVLSSGPSEQSQMS 709
Query: 1281 LCSDLNVDP 1289
L SD ++ P
Sbjct: 710 L-SDSSMPP 717
Score = 57.4 bits (137), Expect = 3e-06
Identities = 45/153 (29%), Positives = 73/153 (47%), Gaps = 22/153 (14%)
Query: 62 DLLELGETG-------AEFCQIGNQTCSIPLELY-DLSGLEDILSVDVWNDCLSEEERFE 113
D LELG G E C +G S+P +L D D++S+ W + LS+ +R
Sbjct: 10 DPLELGPCGDGHGTRIMEDCLLGGTRVSLPEDLLEDPEIFFDVVSLSTWQEVLSDSQREH 69
Query: 114 LAKYLPDMDQETFVQT---LKELFTGCNFQFGSPVKKLFDMLKGGLCEPRVALYRE---- 166
L ++LP +++ Q + LF+G NF+FG+P+ + + G P V YR+
Sbjct: 70 LQQFLPQFPEDSAEQQNELILALFSGENFRFGNPLHIAQKLFRDGHFNPEVVKYRQLCFK 129
Query: 167 -----GLNFVQKRQHYHLLKKHQNTMVSNLCQM 194
LN ++Q++H L K S+L +M
Sbjct: 130 SQYKRYLN--SQQQYFHRLLKQILASRSDLLEM 160
>pir||S52863 DNA-binding protein R kappa B - human gi|695579|emb|CAA56846.1| R
kappa B [Homo sapiens] gi|23346420|ref|NP_006156.2|
nuclear factor related to kappaB binding protein [Homo
sapiens]
Length = 1324
Score = 176 bits (446), Expect = 5e-42
Identities = 131/369 (35%), Positives = 192/369 (51%), Gaps = 40/369 (10%)
Query: 942 PSLTIQEIVNRVRSNPGDPCILETQEPLQDLVRGVLKIFSSKTAPLGAKGWKVLAVYEKS 1001
P L I EI + S + +LE+Q L L VL SS + L + W A
Sbjct: 393 PCLGINEISSSFFSLLLEILLLESQASLPMLEERVLDWQSSPASSLNS--WFSAAP---- 446
Query: 1002 TRSWSWIGPVLHNSSDHDPIEEVTSPEAWGLPHKM--LVKLVDSFANWLKCGQ--DTLKQ 1057
+W VL P + + E+ +P V+ + W GQ D K+
Sbjct: 447 ----NWAELVL-------PALQYLAGESRAVPSSFSPFVEFKEKTQQWKLLGQSQDNEKE 495
Query: 1058 IGSLPAPPLELM------QINLDEKERFRDLRAQKSLNTISPSSEEVRAYFRKEELLRYS 1111
+ +L LE Q N D + + ++ + PS+ E + F+++E RYS
Sbjct: 496 LAALFQLWLETKDQAFCKQENEDSSDATTPVPRVRTDYVVRPSTGEEKRVFQEQERYRYS 555
Query: 1112 IPDRAFSYTAADGKKSIVAPLRRCGGKPTS--KARDHFMLKRDRPPHVTILCLVRDAAAR 1169
P +AF++ G +S+V P++ K TS KAR+H +L+ DRP +VTIL LVRDAAAR
Sbjct: 556 QPHKAFTFRM-HGFESVVGPVKGVFDKETSLNKAREHSLLRSDRPAYVTILSLVRDAAAR 614
Query: 1170 LPGSIGTRADVCTLIRDSQYIVEDVSDEKINQVVSGALDRLHYERDPCVLFDQERKLWVY 1229
LP GTRA++C L++DSQ++ DV+ ++N VVSGALDRLHYE+DPCV +D RKLW+Y
Sbjct: 615 LPNGEGTRAEICELLKDSQFLAPDVTSTQVNTVVSGALDRLHYEKDPCVKYDIGRKLWIY 674
Query: 1230 LHREREEEDFD-----DDGTSSTKKWKRQK----KDVADQSDQAPVTVACNGTGEQSGYD 1280
LHR+R EE+F+ + +K +QK V S ++ + V +G EQS
Sbjct: 675 LHRDRSEEEFERIHQAQAAAAKARKALQQKPKPPSKVKSSSKESSIKVLSSGPSEQSQMS 734
Query: 1281 LCSDLNVDP 1289
L SD ++ P
Sbjct: 735 L-SDSSMPP 742
Score = 57.4 bits (137), Expect = 3e-06
Identities = 45/153 (29%), Positives = 73/153 (47%), Gaps = 22/153 (14%)
Query: 62 DLLELGETG-------AEFCQIGNQTCSIPLELY-DLSGLEDILSVDVWNDCLSEEERFE 113
D LELG G E C +G S+P +L D D++S+ W + LS+ +R
Sbjct: 23 DPLELGPCGDGHGTRIMEDCLLGGTRVSLPEDLLEDPEIFFDVVSLSTWQEVLSDSQREH 82
Query: 114 LAKYLPDMDQETFVQT---LKELFTGCNFQFGSPVKKLFDMLKGGLCEPRVALYRE---- 166
L ++LP +++ Q + LF+G NF+FG+P+ + + G P V YR+
Sbjct: 83 LQQFLPQFPEDSAEQQNELILALFSGENFRFGNPLHIAQKLFRDGHFNPEVVKYRQLCFK 142
Query: 167 -----GLNFVQKRQHYHLLKKHQNTMVSNLCQM 194
LN ++Q++H L K S+L +M
Sbjct: 143 SQYKRYLN--SQQQYFHRLLKQILASRSDLLEM 173
>ref|XP_546398.1| PREDICTED: similar to nuclear factor related to kappa B binding
protein [Canis familiaris]
Length = 1435
Score = 174 bits (441), Expect = 2e-41
Identities = 126/360 (35%), Positives = 189/360 (52%), Gaps = 38/360 (10%)
Query: 966 QEPLQDL--VRGVLKI----FSSKTAPLGAKGWKVLAVYEKSTRSW---------SWIGP 1010
+EPL+DL G+ +I FS L +G L + E+ W SW
Sbjct: 496 EEPLEDLKPCLGINEISSSFFSLLLEILLLEGQASLPMLEERVLDWQSSPASSLNSWFSA 555
Query: 1011 VLHNSSDHDPIEEVTSPEAWGLPHKM--LVKLVDSFANWLKCGQ--DTLKQIGSLPAPPL 1066
+ + P + + E+ +P V+ + W GQ D K++ +L L
Sbjct: 556 APNWAELVLPALQYLAGESRAVPSSFSPFVEFKEKTQQWKLLGQSQDNEKELAALFQLWL 615
Query: 1067 ELM------QINLDEKERFRDLRAQKSLNTISPSSEEVRAYFRKEELLRYSIPDRAFSYT 1120
E Q N D + + ++ + PS+ E + F+++E RYS P +AF++
Sbjct: 616 ETKDQAFCKQENEDSSDATTPVPRVRTDYVVRPSTGEEKRVFQEQERYRYSQPHKAFTFR 675
Query: 1121 AADGKKSIVAPLRRCGGKPTS--KARDHFMLKRDRPPHVTILCLVRDAAARLPGSIGTRA 1178
G +S+V P++ K TS KAR+H +L+ DRP +VTIL LVRDAAARLP GTRA
Sbjct: 676 M-HGFESVVGPVKGVFDKETSLNKAREHSLLRSDRPAYVTILSLVRDAAARLPNGEGTRA 734
Query: 1179 DVCTLIRDSQYIVEDVSDEKINQVVSGALDRLHYERDPCVLFDQERKLWVYLHREREEED 1238
++C L++DSQ++ DV+ ++N VVSGALDRLHYE+DPCV +D RKLW+YLHR+R EE+
Sbjct: 735 EICELLKDSQFLAPDVTSTQVNTVVSGALDRLHYEKDPCVKYDIGRKLWIYLHRDRSEEE 794
Query: 1239 FD-----DDGTSSTKKWKRQK----KDVADQSDQAPVTVACNGTGEQSGYDLCSDLNVDP 1289
F+ + +K +QK V S ++ + V +G EQS L SD ++ P
Sbjct: 795 FERIHQAQAAAAKARKALQQKPKPPSKVKSGSKESSLKVLGSGPSEQSQMSL-SDSSMPP 853
Score = 60.8 bits (146), Expect = 3e-07
Identities = 44/151 (29%), Positives = 72/151 (47%), Gaps = 18/151 (11%)
Query: 62 DLLELGETG-------AEFCQIGNQTCSIPLELY-DLSGLEDILSVDVWNDCLSEEERFE 113
D LELG G E C +G S+P +L D D++S+ W + LS+ +R
Sbjct: 146 DPLELGPCGDGHGTRIMEDCLLGGTRVSLPEDLLEDPEIFFDVVSLSTWQEVLSDSQREH 205
Query: 114 LAKYLPDMDQETFV---QTLKELFTGCNFQFGSPVKKLFDMLKGGLCEPRVALYRE---- 166
L ++LP +++F Q + LF+G NF+FG+P+ + + G P V YR+
Sbjct: 206 LQQFLPHFPEDSFEQQNQLILALFSGENFRFGNPLHIAQKLFRDGHFNPEVVKYRQLCFK 265
Query: 167 ---GLNFVQKRQHYHLLKKHQNTMVSNLCQM 194
++Q++H L K S+L +M
Sbjct: 266 SQYKRYLTSQQQYFHRLLKQILASRSDLLEM 296
>ref|NP_766354.2| nuclear factor related to kappa B binding protein [Mus musculus]
gi|37537243|gb|AAH33595.1| Nuclear factor related to
kappa B binding protein [Mus musculus]
Length = 1296
Score = 173 bits (438), Expect = 4e-41
Identities = 137/403 (33%), Positives = 200/403 (48%), Gaps = 46/403 (11%)
Query: 942 PSLTIQEIVNRVRSNPGDPCILETQEPLQDLVRGVLKIFSSKTAPLGAKGWKVLAVYEKS 1001
P L I EI + S + +LE+Q L L VL SS + L + W A
Sbjct: 368 PCLGINEISSSFFSLLLEILLLESQASLPMLEDRVLDWQSSPASSLNS--WFSAAP---- 421
Query: 1002 TRSWSWIGPVLHNSSDHDPIEEVTSPEAWGLPHKM--LVKLVDSFANWLKCGQ--DTLKQ 1057
+W VL P + + E+ +P V+ + W GQ D K+
Sbjct: 422 ----NWAELVL-------PALQYLAGESRAVPSSFSPFVEFKEKTQQWKLLGQSQDNEKE 470
Query: 1058 IGSLPAPPLE-----LMQINLDEKERFRDLRAQKSLNTISPSSEEVRAYFRKEELLRYSI 1112
+ +L LE + N D + + ++ + PS+ E + F+++E RYS
Sbjct: 471 LAALFHLWLETKDQAFCKENEDSSDAMTPVPRVRTDYVVRPSTGEEKRVFQEQERYRYSQ 530
Query: 1113 PDRAFSYTAADGKKSIVAPLRRCGGKPTS--KARDHFMLKRDRPPHVTILCLVRDAAARL 1170
P +AF++ G +S+V P++ K TS KAR+H +L+ DRP +VTIL LVRDAAARL
Sbjct: 531 PHKAFTFRM-HGFESVVGPVKGVFDKETSLNKAREHSLLRSDRPAYVTILSLVRDAAARL 589
Query: 1171 PGSIGTRADVCTLIRDSQYIVEDVSDEKINQVVSGALDRLHYERDPCVLFDQERKLWVYL 1230
P GTRA++C L++DSQ++ DV+ ++N VVSGALDRLHYE+DPCV +D RKLW+YL
Sbjct: 590 PNGEGTRAEICELLKDSQFLAPDVTSTQVNTVVSGALDRLHYEKDPCVKYDIGRKLWIYL 649
Query: 1231 HREREEEDFD-----DDGTSSTKKWKRQK---KDVADQSDQAPVTVACNGTGEQSGYDLC 1282
HR+R EE+F+ + +K +QK S++ T +G EQS L
Sbjct: 650 HRDRSEEEFERIHQAQAAAAKARKALQQKPKPPSKVKSSNKEGSTKGLSGPSEQSQMSL- 708
Query: 1283 SDLNVDPPCIEDDKEAVQLLTTDTRPNAEDQVVVNPVSEVGNS 1325
SD ++ P + T T P + PVS V S
Sbjct: 709 SDSSMPPTPVTP--------VTPTTPALPTPISPPPVSAVNRS 743
Score = 55.8 bits (133), Expect = 1e-05
Identities = 45/153 (29%), Positives = 72/153 (46%), Gaps = 22/153 (14%)
Query: 62 DLLELGETG-------AEFCQIGNQTCSIPLELY-DLSGLEDILSVDVWNDCLSEEERFE 113
D LELG G E C +G S+P +L D D++S+ W + LS+ +R
Sbjct: 10 DPLELGPCGDGHSTGIMEDCLLGGTRVSLPEDLLEDPEIFFDVVSLSTWQEVLSDSQREH 69
Query: 114 LAKYLPDMDQETFVQT---LKELFTGCNFQFGSPVKKLFDMLKGGLCEPRVALYRE---- 166
L ++LP ++ Q + LF+G NF+FG+P+ + + G P V YR+
Sbjct: 70 LQQFLPRFPADSVEQQRELILALFSGENFRFGNPLHIAQKLFRDGHFNPEVVKYRQLCFK 129
Query: 167 -----GLNFVQKRQHYHLLKKHQNTMVSNLCQM 194
LN ++Q++H L K S+L +M
Sbjct: 130 SQYKRYLN--SQQQYFHRLLKQILASRSDLLEM 160
>ref|XP_236009.3| PREDICTED: similar to Nuclear factor related to kappa B binding
protein [Rattus norvegicus]
Length = 1297
Score = 169 bits (429), Expect = 5e-40
Identities = 136/404 (33%), Positives = 198/404 (48%), Gaps = 47/404 (11%)
Query: 942 PSLTIQEIVNRVRSNPGDPCILETQEPLQDLVRGVLKIFSSKTAPLGAKGWKVLAVYEKS 1001
P L I EI + S + +LE+Q L L VL SS + L + W A
Sbjct: 368 PCLGINEISSSFFSLLLEILLLESQASLPMLEDRVLDWQSSPASSLNS--WFSAAP---- 421
Query: 1002 TRSWSWIGPVLHNSSDHDPIEEVTSPEAWGLPHKM--LVKLVDSFANWLKCGQ--DTLKQ 1057
+W VL P + + E+ +P V+ + W GQ D K+
Sbjct: 422 ----NWAELVL-------PALQYLAGESRAVPSSFSPFVEFKEKTQQWKLLGQSQDNEKE 470
Query: 1058 IGSLPAPPLELM------QINLDEKERFRDLRAQKSLNTISPSSEEVRAYFRKEELLRYS 1111
+ +L LE Q N D + + ++ + PS+ E + F+++E RYS
Sbjct: 471 LAALFHLWLETKDQAFCKQENEDSSDAMTPVPRVRTDYVVRPSTGEEKRVFQEQERYRYS 530
Query: 1112 IPDRAFSYTAADGKKSIVAPLRRCGGKPTS--KARDHFMLKRDRPPHVTILCLVRDAAAR 1169
P +AF++ G +S+V P++ K TS KAR+H +L+ DRP +VTIL LVRDAAAR
Sbjct: 531 QPHKAFTFRM-HGFESVVGPVKGVFDKETSLNKAREHSLLRSDRPAYVTILSLVRDAAAR 589
Query: 1170 LPGSIGTRADVCTLIRDSQYIVEDVSDEKINQVVSGALDRLHYERDPCVLFDQERKLWVY 1229
LP GTRA++C L++DSQ++ DV+ ++N VVSGALDRLHYE+DPCV +D RKLW+Y
Sbjct: 590 LPNGEGTRAEICELLKDSQFLAPDVTSTQVNTVVSGALDRLHYEKDPCVKYDIGRKLWIY 649
Query: 1230 LHREREEEDFD-----DDGTSSTKKWKRQK---KDVADQSDQAPVTVACNGTGEQSGYDL 1281
LHR+R EE+F+ + +K +QK S++ +G E S L
Sbjct: 650 LHRDRSEEEFERIHQAQAAAAKARKALQQKPKPPSKVKSSNKEGSVKGLSGPSEPSQMSL 709
Query: 1282 CSDLNVDPPCIEDDKEAVQLLTTDTRPNAEDQVVVNPVSEVGNS 1325
SD ++ P + T T P + PVS V S
Sbjct: 710 -SDSSMPPTPVTP--------VTPTTPALPTPISPPPVSAVSRS 744
Score = 57.0 bits (136), Expect = 4e-06
Identities = 43/151 (28%), Positives = 70/151 (45%), Gaps = 18/151 (11%)
Query: 62 DLLELGETG-------AEFCQIGNQTCSIPLELY-DLSGLEDILSVDVWNDCLSEEERFE 113
D LELG G E C +G S+P +L D D++S+ W + LS+ +R
Sbjct: 10 DPLELGPCGDGHSTGIMEDCLLGGTRVSLPEDLLEDPEIFFDVVSLSTWQEVLSDSQREH 69
Query: 114 LAKYLPDMDQETFVQ---TLKELFTGCNFQFGSPVKKLFDMLKGGLCEPRVALYRE---- 166
L ++LP +T Q + LF+G NF+FG+P+ + + G P V YR+
Sbjct: 70 LQQFLPQFPTDTVEQQNELILALFSGENFRFGNPLHIAQKLFRDGHFNPEVVKYRQLCFK 129
Query: 167 ---GLNFVQKRQHYHLLKKHQNTMVSNLCQM 194
++Q++H L + S+L +M
Sbjct: 130 SQYKRYLSSQQQYFHRLLRQILASRSDLLEM 160
>pir||B45580 transcription factor R-kappa-B - mouse (fragment)
Length = 288
Score = 169 bits (429), Expect = 5e-40
Identities = 99/247 (40%), Positives = 143/247 (57%), Gaps = 12/247 (4%)
Query: 1072 NLDEKERFRDLRAQKSLNTISPSSEEVRAYFRKEELLRYSIPDRAFSYTAADGKKSIVAP 1131
N D + + ++ + PS+ E + F+++E RYS P +AF++ G +S+V P
Sbjct: 39 NEDSSDAMTPVPRVRTDYVVRPSTGEEKRVFQEQERYRYSQPHKAFTFRM-HGFESVVGP 97
Query: 1132 LRRCGGKPTS--KARDHFMLKRDRPPHVTILCLVRDAAARLPGSIGTRADVCTLIRDSQY 1189
++ K TS KAR+H +L+ DRP +VTIL LVRDAAARLP GTRA++C L++DSQ+
Sbjct: 98 VKGVFDKETSLNKAREHSLLRSDRPAYVTILSLVRDAAARLPNGEGTRAEICELLKDSQF 157
Query: 1190 IVEDVSDEKINQVVSGALDRLHYERDPCVLFDQERKLWVYLHREREEEDFD-----DDGT 1244
+ DV+ ++N VVSGALDRLHYE+DPCV +D RKLW+YLHR+R EE+F+
Sbjct: 158 LAPDVTSTQVNTVVSGALDRLHYEKDPCVKYDIGRKLWIYLHRDRSEEEFERIHQAQAAA 217
Query: 1245 SSTKKWKRQK---KDVADQSDQAPVTVACNGTGEQSGYDLCSDLNVDPPCIEDDKEAVQL 1301
+K +QK S++ T +G EQS L SD ++ P +
Sbjct: 218 RKARKALQQKPKPPSKVKSSNKEGSTKGLSGPSEQSQMSL-SDSSMPPTPVTPVTPTTPA 276
Query: 1302 LTTDTRP 1308
L T P
Sbjct: 277 LPTPISP 283
>ref|XP_417871.1| PREDICTED: similar to RIKEN cDNA A530090G11 [Gallus gallus]
Length = 1370
Score = 169 bits (427), Expect = 8e-40
Identities = 94/210 (44%), Positives = 134/210 (63%), Gaps = 13/210 (6%)
Query: 1091 ISPSSEEVRAYFRKEELLRYSIPDRAFSYTAADGKKSIVAPLRRCGGKPTS--KARDHFM 1148
+ PS+ E + F+++E RYS P +AF++ G +S+V P++ K TS KAR+H +
Sbjct: 584 VRPSTGEEKRVFQEQERYRYSQPHKAFTFRM-HGFESVVGPVKGVFDKETSLNKAREHSL 642
Query: 1149 LKRDRPPHVTILCLVRDAAARLPGSIGTRADVCTLIRDSQYIVEDVSDEKINQVVSGALD 1208
L+ DRP +VTIL LVRDAAARLP GTRA++C L++DSQ++ DV+ ++N VVSGALD
Sbjct: 643 LRSDRPAYVTILSLVRDAAARLPNGEGTRAEICELLKDSQFLAPDVTSAQVNTVVSGALD 702
Query: 1209 RLHYERDPCVLFDQERKLWVYLHREREEEDFD-----DDGTSSTKKWKRQK----KDVAD 1259
RLHYE+DPCV +D RKLW+YLHR+R EE+F+ + KK +QK V
Sbjct: 703 RLHYEKDPCVKYDIGRKLWIYLHRDRSEEEFERIHQAQAAAAKAKKALQQKPKPPAKVKS 762
Query: 1260 QSDQAPVTVACNGTGEQSGYDLCSDLNVDP 1289
+ ++ V + T E S L SD ++ P
Sbjct: 763 SNKESSVKALPSSTSEPSQLSL-SDSSMPP 791
Score = 49.3 bits (116), Expect = 0.001
Identities = 33/116 (28%), Positives = 58/116 (49%), Gaps = 11/116 (9%)
Query: 62 DLLELGETG-------AEFCQIGNQTCSIPLELY-DLSGLEDILSVDVWNDCLSEEERFE 113
D LELG G E C +G S+P +L D +++S+ W + L++ ++
Sbjct: 85 DPLELGPCGDGNGARIMEDCLLGTTRVSLPEDLLEDPEIFFEVVSLSTWQEVLTDAQQEH 144
Query: 114 LAKYLP---DMDQETFVQTLKELFTGCNFQFGSPVKKLFDMLKGGLCEPRVALYRE 166
L +LP + ++E + + LF+G NF+FG+P+ + + G P V YR+
Sbjct: 145 LKTFLPHFPENNREHQNKIISALFSGENFRFGNPLHIAQKLFRDGHFNPEVVKYRQ 200
>ref|NP_989207.1| hypothetical protein LOC394815 [Xenopus tropicalis]
gi|38648973|gb|AAH63348.1| Hypothetical protein MGC75871
[Xenopus tropicalis]
Length = 1265
Score = 165 bits (418), Expect = 9e-39
Identities = 90/189 (47%), Positives = 125/189 (65%), Gaps = 6/189 (3%)
Query: 1091 ISPSSEEVRAYFRKEELLRYSIPDRAFSYTAADGKKSIVAPLRRCGGKPTS--KARDHFM 1148
+ PSS E + F+++E RY P +AF++ G +S+V P++ K TS KAR+H +
Sbjct: 510 VRPSSGEEKHVFQEQERHRYIQPHKAFTFRM-HGFESVVGPVKGVFDKETSLNKAREHSL 568
Query: 1149 LKRDRPPHVTILCLVRDAAARLPGSIGTRADVCTLIRDSQYIVEDVSDEKINQVVSGALD 1208
L+ DRP +VTIL LVRDAAARLP GTRA++C L++DSQ++ DV+ ++N VVSGALD
Sbjct: 569 LRSDRPAYVTILSLVRDAAARLPNGEGTRAEICELLKDSQFLAPDVTSAQVNTVVSGALD 628
Query: 1209 RLHYERDPCVLFDQERKLWVYLHREREEEDFDDDGTSSTKKWKRQKKDVADQSDQAPVTV 1268
RLHYE+DPCV +D RKLW+YLHR+R EE+F+ + K +K A Q P T
Sbjct: 629 RLHYEKDPCVKYDIGRKLWIYLHRDRSEEEFERIHQAQAAAAKAKK---ALQQKPKPPTK 685
Query: 1269 ACNGTGEQS 1277
+G+ E S
Sbjct: 686 MKSGSKECS 694
Score = 52.8 bits (125), Expect = 8e-05
Identities = 41/139 (29%), Positives = 66/139 (46%), Gaps = 18/139 (12%)
Query: 62 DLLELGE------TGA-EFCQIGNQTCSIPLELYDLSGLE-DILSVDVWNDCLSEEERFE 113
D LELG TG E C +G S+P +L + L +++S D W + LS+ +R
Sbjct: 10 DPLELGPCVESNGTGIMEECMLGGTRVSLPEDLLEDPDLFFEVVSRDTWKNVLSDSQREH 69
Query: 114 LAKYLPDMDQETFVQT---LKELFTGCNFQFGSPVKKLFDMLKGGLCEPRVALYRE---- 166
L ++LP + Q ++ LF G NF+FG+P+ + + G P V YR+
Sbjct: 70 LKQFLPVFPDDNANQQEKIIQSLFRGDNFRFGNPLHIAQKLFRDGHFNPEVVKYRQLCLK 129
Query: 167 ---GLNFVQKRQHYHLLKK 182
++Q++H L K
Sbjct: 130 SQYKRYLTSQQQYFHKLLK 148
>dbj|BAC33927.1| unnamed protein product [Mus musculus]
Length = 758
Score = 153 bits (387), Expect = 3e-35
Identities = 91/212 (42%), Positives = 124/212 (57%), Gaps = 19/212 (8%)
Query: 1124 GKKSIVAPLRRCGGKPTS--KARDHFMLKRDRPPHVTILCLVRDAAARLPGSIGTRADVC 1181
G +S+V P++ K TS KAR+H +L+ DRP +VTIL LVRDAAARLP GTRA++C
Sbjct: 3 GFESVVGPVKGVFDKETSLNKAREHSLLRSDRPAYVTILSLVRDAAARLPNGEGTRAEIC 62
Query: 1182 TLIRDSQYIVEDVSDEKINQVVSGALDRLHYERDPCVLFDQERKLWVYLHREREEEDFD- 1240
L++DSQ++ DV+ ++N VVSGALDRLHYE+DPCV +D RKLW+YLHR+R EE+F+
Sbjct: 63 ELLKDSQFLAPDVTSTQVNTVVSGALDRLHYEKDPCVKYDIGRKLWIYLHRDRSEEEFER 122
Query: 1241 ----DDGTSSTKKWKRQK---KDVADQSDQAPVTVACNGTGEQSGYDLCSDLNVDPPCIE 1293
+ +K +QK S++ T +G EQS L SD ++ P +
Sbjct: 123 IHQAQAAAAKARKALQQKPKPPSKVKSSNKEGSTKGLSGPSEQSQMSL-SDSSMPPTPVT 181
Query: 1294 DDKEAVQLLTTDTRPNAEDQVVVNPVSEVGNS 1325
T T P + PVS V S
Sbjct: 182 P--------VTPTTPALPTPISPPPVSAVNRS 205
>gb|AAK71996.1| NFRKB-like protein [Apis mellifera]
Length = 1308
Score = 147 bits (370), Expect = 3e-33
Identities = 88/197 (44%), Positives = 114/197 (57%), Gaps = 11/197 (5%)
Query: 1091 ISPSSEEVRAYFRKEELLRYSIPDRAFSYTAADGKKSIVAPLRRCG----GKPTSKARDH 1146
+ PS+ E R FR++E +RY+ P +AF+Y G S+V P++ +KAR H
Sbjct: 587 VRPSTVEEREEFRRQERMRYAAPHKAFTYRM-HGYASVVGPVKGIYQHNVASGLTKARGH 645
Query: 1147 FMLKRDRPPHVTILCLVRDAAARLPGSIGTRADVCTLIRDSQYIVEDV-SDEK---INQV 1202
+L DRP VTIL LVRDA ARLP GTRAD+C L++DSQYI E SD+K ++ V
Sbjct: 646 SLLVADRPNFVTILALVRDATARLPNGEGTRADICQLLKDSQYIREQTESDDKEGYLHSV 705
Query: 1203 VSGALDRLHYERDPCVLFDQERKLWVYLHREREEEDFD--DDGTSSTKKWKRQKKDVADQ 1260
VSGALDRLHYE DPCV + RK W+YLHR R E +F+ K K+ ++
Sbjct: 706 VSGALDRLHYETDPCVRYYPRRKEWLYLHRARSESEFEMYHQQLQGVAKNKKNVGSMSRN 765
Query: 1261 SDQAPVTVACNGTGEQS 1277
PV N EQS
Sbjct: 766 KALLPVIKPANVNKEQS 782
>ref|XP_560646.1| ENSANGP00000025456 [Anopheles gambiae str. PEST]
gi|57916213|ref|XP_555922.1| ENSANGP00000026191
[Anopheles gambiae str. PEST] gi|55246597|gb|EAL42109.1|
ENSANGP00000025456 [Anopheles gambiae str. PEST]
gi|55238101|gb|EAL39779.1| ENSANGP00000026191 [Anopheles
gambiae str. PEST]
Length = 787
Score = 138 bits (348), Expect = 1e-30
Identities = 71/153 (46%), Positives = 99/153 (64%), Gaps = 3/153 (1%)
Query: 1090 TISPSSEEVRAYFRKEELLRYSIPDRAFSYTAADGKKSIVAPLRRCGGKPT--SKARDHF 1147
++ +++E FR++E RY P AF+Y S+V P++ + SKAR H
Sbjct: 622 SVRKATDEEIQRFREQEKCRYENPHMAFTYKQ-HSYDSVVGPVKGIYTQAPGISKARGHS 680
Query: 1148 MLKRDRPPHVTILCLVRDAAARLPGSIGTRADVCTLIRDSQYIVEDVSDEKINQVVSGAL 1207
ML +RP VTIL LVRDA ARLP GTRAD+C L++ SQYI +D+ + +VSGAL
Sbjct: 681 MLVANRPNFVTILTLVRDATARLPNGEGTRADICELLKSSQYISPTATDQILQTIVSGAL 740
Query: 1208 DRLHYERDPCVLFDQERKLWVYLHREREEEDFD 1240
DR+H E DPCV +D +RK+W+YLHR R E++F+
Sbjct: 741 DRMHTEHDPCVKYDAKRKIWIYLHRNRTEQEFE 773
>gb|AAF54313.1| CG11970-PA [Drosophila melanogaster] gi|5052658|gb|AAD38659.1|
BcDNA.LD23876 [Drosophila melanogaster]
gi|21355615|ref|NP_649851.1| CG11970-PA [Drosophila
melanogaster]
Length = 1658
Score = 137 bits (344), Expect = 3e-30
Identities = 71/173 (41%), Positives = 108/173 (62%), Gaps = 6/173 (3%)
Query: 1090 TISPSSEEVRAYFRKEELLRYSIPDRAFSYTAADGKKSIVAPLRRC--GGKPTSKARDHF 1147
T+ +++ F+ +E +R+ P R F+Y +G +S+V P++ G P +KAR H
Sbjct: 711 TVRSATQAEILEFQSQERIRFEYPHRPFTYRL-NGYESVVGPVKGIYTQGLPLNKARGHA 769
Query: 1148 MLKRDRPPHVTILCLVRDAAARLPGSIGTRADVCTLIRDSQYIVEDVSDEKINQVVSGAL 1207
++ DRPP VTIL LVRDA ARLP GTR+++ L++ SQYI + +D + +VSGAL
Sbjct: 770 VMVDDRPPFVTILTLVRDATARLPNGEGTRSEISELLKCSQYISQQAADNVLQTIVSGAL 829
Query: 1208 DRLHYERDPCVLFDQERKLWVYLHREREEEDFDD---DGTSSTKKWKRQKKDV 1257
DR+H DPCV +D RK+W+YLHR R E +F+ S K+ ++Q+K +
Sbjct: 830 DRMHGGPDPCVKYDARRKIWIYLHRNRSEAEFEKMHRHNQSLVKQKQQQRKQL 882
Score = 40.8 bits (94), Expect = 0.32
Identities = 44/189 (23%), Positives = 78/189 (40%), Gaps = 24/189 (12%)
Query: 21 SKESMSSGDEEDVQRRNSGNESDEDDDEFDDADSGAGSDDFDLLELGETGAEFCQIGNQT 80
S E G++ED+ +++G+ S+ +DD S SD+ +L +L TG + ++
Sbjct: 66 SLEDDPDGEDEDMSAQDAGSASETEDD-----TSTCQSDELELAQL--TGRDIFRLPRGL 118
Query: 81 CSIPLELYDLSGLEDILSVDVWNDCLSEEERFELAKYLPDMDQETFVQ---------TLK 131
C + + + S++ W L + L LPD + Q TL
Sbjct: 119 CE------NAAIFHEFFSLETWQQ-LPHHLQARLQPMLPDFSRLVSGQAQACAEQQRTLV 171
Query: 132 ELFTGCNFQFG-SPVKKLFDMLKGGLCEPRVALYREGLNFVQKRQHYHLLKKHQNTMVSN 190
+LF G +FG SP+ +L L+ G P V R + +R+ + + +
Sbjct: 172 QLFNGGLQRFGQSPLLQLQRQLEEGNLRPDVLQLRRSIQHSGRREFRFQQAERLSLLAKE 231
Query: 191 LCQMRDAWL 199
L R+ L
Sbjct: 232 LFMSREQLL 240
>gb|EAL28995.1| GA11303-PA [Drosophila pseudoobscura]
Length = 1517
Score = 136 bits (343), Expect = 4e-30
Identities = 72/174 (41%), Positives = 112/174 (63%), Gaps = 6/174 (3%)
Query: 1090 TISPSSEEVRAYFRKEELLRYSIPDRAFSYTAADGKKSIVAPLRRCGGK--PTSKARDHF 1147
T+ +S+ F+++E LR+ P R F+Y +G +S+V P++ + P +KAR H
Sbjct: 636 TVRTASQADILEFQRQERLRFEYPHRPFTYRL-NGYESVVGPVKGIYTQSLPLNKARGHA 694
Query: 1148 MLKRDRPPHVTILCLVRDAAARLPGSIGTRADVCTLIRDSQYIVEDVSDEKINQVVSGAL 1207
++ DRPP VTIL LVRDA ARLP GTR+++ L++ SQ++ ++ + +VSGAL
Sbjct: 695 VMVDDRPPFVTILTLVRDATARLPNGEGTRSEISELLKCSQFVNHQAAENVLQTIVSGAL 754
Query: 1208 DRLHYERDPCVLFDQERKLWVYLHREREEEDFD---DDGTSSTKKWKRQKKDVA 1258
DR+H DPCV +D +RK+W+YLHR R EE+F+ S K+ ++QKK++A
Sbjct: 755 DRMHGGIDPCVKYDAKRKIWIYLHRNRSEEEFERMHKLNMSLVKQKQQQKKNLA 808
>ref|XP_483189.1| DNA-binding protein-like [Oryza sativa (japonica cultivar-group)]
gi|42407749|dbj|BAD08895.1| DNA-binding protein-like
[Oryza sativa (japonica cultivar-group)]
gi|42407355|dbj|BAD08816.1| DNA-binding protein-like
[Oryza sativa (japonica cultivar-group)]
Length = 558
Score = 135 bits (340), Expect = 1e-29
Identities = 74/184 (40%), Positives = 99/184 (53%), Gaps = 25/184 (13%)
Query: 17 CEPMSKESMSSGDEEDVQ------RRNSGNESDEDDDEFDDADSGAGSDDFDLLELGETG 70
C P + + S GDE+ Q +R +ESD+ + E E
Sbjct: 9 CAPTEERAYSCGDEDITQEEKMLLQRFPIHESDDYEHE-------------------EVN 49
Query: 71 AEFCQIGNQTCSIPLELYDLSGLEDILSVDVWNDCLSEEERFELAKYLPDMDQETFVQTL 130
E + G+Q CS+P LYDL L DILS++ WN CL+E++RF LA YLPDMDQ F T+
Sbjct: 50 CELAKSGDQICSVPYGLYDLPELNDILSLETWNLCLTEDDRFRLAAYLPDMDQHDFFVTM 109
Query: 131 KELFTGCNFQFGSPVKKLFDMLKGGLCEPRVALYREGLNFVQKRQHYHLLKKHQNTMVSN 190
KELF+G + FGSPVK F L GG P V+ RE L ++R++YH LK H + M+
Sbjct: 110 KELFSGSDLFFGSPVKSFFHRLNGGFYSPEVSQARELLMIFERRRYYHFLKSHHDGMIFK 169
Query: 191 LCQM 194
M
Sbjct: 170 FASM 173
>dbj|BAD94249.1| hypothetical protein [Arabidopsis thaliana]
gi|51536560|gb|AAU05518.1| At1g02290 [Arabidopsis
thaliana] gi|15217706|ref|NP_171731.1| expressed protein
[Arabidopsis thaliana] gi|48958489|gb|AAT47797.1|
At1g02290 [Arabidopsis thaliana]
Length = 443
Score = 131 bits (330), Expect = 1e-28
Identities = 87/263 (33%), Positives = 132/263 (50%), Gaps = 24/263 (9%)
Query: 46 DDEFDDADSGAGSDDFDLLELGETGAEFCQIGNQTCSIPLELYDLSGLEDILSVDVWNDC 105
D E DD SDD+D+ ++ E + Q C+IP ELYDL L ILSV+ WN
Sbjct: 10 DSEDDD------SDDYDIAQVN---CELALVEGQLCNIPYELYDLPDLTGILSVETWNSL 60
Query: 106 LSEEERFELAKYLPDMDQETFVQTLKELFTGCNFQFGSPVKKLFDMLKGGLCEPRVALYR 165
L+EEERF L+ +LPDMD +TF T++EL G N FG+P K + L GGL P+VA ++
Sbjct: 61 LTEEERFFLSCFLPDMDPQTFSLTMQELLDGANLYFGNPEDKFYKNLLGGLFTPKVACFK 120
Query: 166 EGLNFVQKRQHYHLLKKHQNTMVSNLCQMRDAWLNCRGYSIEERLRVLNIMTSQKSLMGE 225
EG+ FV++R++Y+ LK + ++ +M+ W+ G + R+L I + +
Sbjct: 121 EGVMFVKRRKYYYSLKFYHEKLIRTFTEMQRVWVQ-YGNKLGNYSRLL-IWSGRTQTGNL 178
Query: 226 KMDDLEADSSEESGEGMWSRKNKD-----KKNAQKLGRFPFQGVGSGL--------DFHP 272
K+ DL S+E K + ++N K FP G
Sbjct: 179 KLLDLNRVPSKEMDSATCRFKTPNVVKPVERNRSKSLTFPRSGSSKNSLKIKITKEGVFR 238
Query: 273 REQSMVMEQEKYSKQNPKGILKL 295
+ S ++ + + PKG+LKL
Sbjct: 239 YQGSSLVSAGHHHQTLPKGVLKL 261
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.312 0.131 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,449,988,619
Number of Sequences: 2540612
Number of extensions: 110937254
Number of successful extensions: 323335
Number of sequences better than 10.0: 227
Number of HSP's better than 10.0 without gapping: 58
Number of HSP's successfully gapped in prelim test: 172
Number of HSP's that attempted gapping in prelim test: 321337
Number of HSP's gapped (non-prelim): 1326
length of query: 1373
length of database: 863,360,394
effective HSP length: 141
effective length of query: 1232
effective length of database: 505,134,102
effective search space: 622325213664
effective search space used: 622325213664
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 82 (36.2 bits)
Medicago: description of AC135230.12


