

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135160.10 + phase: 0
(373 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
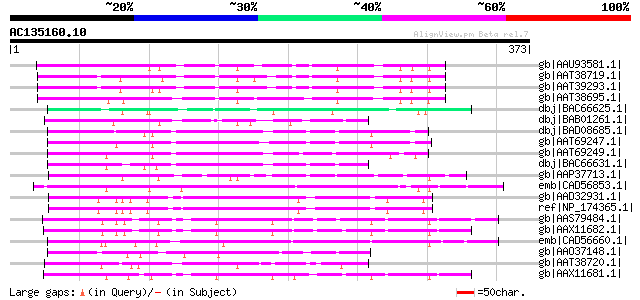
Score E
Sequences producing significant alignments: (bits) Value
gb|AAU93581.1| putative F-box protein [Solanum demissum] 100 7e-20
gb|AAT38719.1| putative F-Box protein [Solanum demissum] 100 1e-19
gb|AAT39293.1| putative F-box-like protein [Solanum demissum] 100 1e-19
gb|AAT38695.1| putative F-box protein [Solanum demissum] 94 5e-18
dbj|BAC66625.1| F-box [Prunus mume] 92 3e-17
dbj|BAB01261.1| unnamed protein product [Arabidopsis thaliana] g... 88 4e-16
dbj|BAD08685.1| S haplotype-specific F-box protein 5 [Prunus avium] 86 2e-15
gb|AAT69247.1| F-box protein 1 [Prunus armeniaca] 85 3e-15
gb|AAT69249.1| F-box protein 4 [Prunus armeniaca] 84 7e-15
dbj|BAC66631.1| F-box [Prunus mume] 84 9e-15
gb|AAP37713.1| At3g23880 [Arabidopsis thaliana] gi|9294656|dbj|B... 83 2e-14
emb|CAD56853.1| S locus F-box (SLF)-S5A protein [Antirrhinum his... 83 2e-14
gb|AAD32931.1| T17H7.6 [Arabidopsis thaliana] 83 2e-14
ref|NP_174365.1| F-box family protein [Arabidopsis thaliana] 83 2e-14
gb|AAS79484.1| S1-locus linked F-box protein [Petunia integrifol... 83 2e-14
gb|AAX11682.1| non-S F-box protein 1 [Petunia axillaris subsp. a... 82 2e-14
emb|CAD56660.1| S locus F-box (SLF)-S4D protein [Antirrhinum his... 81 5e-14
gb|AAO37148.1| hypothetical protein [Arabidopsis thaliana] gi|61... 80 1e-13
gb|AAT38720.1| putative F-Box protein [Solanum demissum] 79 3e-13
gb|AAX11681.1| S19-locus linked F-box protein [Petunia axillaris... 78 4e-13
>gb|AAU93581.1| putative F-box protein [Solanum demissum]
Length = 383
Score = 100 bits (249), Expect = 7e-20
Identities = 93/323 (28%), Positives = 143/323 (43%), Gaps = 51/323 (15%)
Query: 20 SIERMKLGEELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATRRNRLLILQNA-PNM 78
SI + L +EL EILLRLP KSLS+F CV KSW +I SP F + +L
Sbjct: 5 SIIPLLLPDELITEILLRLPIKSLSKFMCVSKSWLQLISSPAFVKKHIKLTANDKGYIYH 64
Query: 79 KFIFCDGGNDQKSIPIKSLFP-----QDVARIE-----------IYGSCDGVFCLKGISS 122
+ IF + ND K P+ LF +++ I+ I GS +G+ C+ +
Sbjct: 65 RLIFRNTNNDFKFCPLPPLFTNQQLIEEILHIDSPIERTTLSTHIVGSVNGLICVAHV-- 122
Query: 123 CITRHDQLILWNPTTKEVHLIPRAPSLGNHYSDESLYGFG--AVNDDFKVVKLNISNSNR 180
R + +WNP + +P++ S N SD GFG DD+KVV ++
Sbjct: 123 ---RQREAYIWNPAITKSKELPKSTS--NLCSDGIKCGFGYDESRDDYKVVFIDYP---- 173
Query: 181 MAKINSLLKADIYDLSTKSWTPLVSHPPITMVTRIQPSRYNTLVNGVYYWITSS--DGSD 238
+ N +IY L T SWT L H + + + + + VNG YW +S+ +
Sbjct: 174 -IRHNHRTVVNIYSLRTNSWTTL--HDQLQGIFLL--NLHGRFVNGKLYWTSSTCINNYK 228
Query: 239 AARILCFDFRDNQFRKLEAPKLGHYIPFFCDDVFEIKGYLG--YVVQYRCRI--VWLEIW 294
I FD D + LE P G D+ + G +G + Y C++ ++W
Sbjct: 229 VCNITSFDLADGTWGSLELPSCGK------DNSYINVGVVGSDLSLLYTCQLGAATSDVW 282
Query: 295 TLEQNG----WAKKYNIDTKMSI 313
++ +G W K + I +I
Sbjct: 283 IMKHSGVNVSWTKLFTIKYPQNI 305
>gb|AAT38719.1| putative F-Box protein [Solanum demissum]
Length = 327
Score = 99.8 bits (247), Expect = 1e-19
Identities = 95/326 (29%), Positives = 142/326 (43%), Gaps = 58/326 (17%)
Query: 21 IERMKLGEELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATRRNRLLILQNAPNM-- 78
I + L +EL EILL+LP KSL +F CV KSW +I SP F +N + + +
Sbjct: 7 IPPLLLPDELITEILLKLPIKSLLKFMCVSKSWLQLISSPAFV--KNHIKLTADDKGYIY 64
Query: 79 -KFIFCDGGNDQKSIPIKSLFPQDVARIEIY----------------GSCDGVFCLKGIS 121
+ IF + +D K P+ LF Q E+Y GS +G+ C +
Sbjct: 65 HRLIFRNTNDDFKFCPLPPLFTQQQLIKELYHIDSPIERTTLSTHIVGSVNGLICAAHV- 123
Query: 122 SCITRHDQLILWNPTTKEVHLIPRAPSLGNHYSDESLYGFG--AVNDDFKVVKLN--ISN 177
R + +WNPT + +P++ S N SD GFG DD+KVV ++ I
Sbjct: 124 ----RQREAYIWNPTITKSKELPKSRS--NLCSDGIKCGFGYDESRDDYKVVFIDYPIHR 177
Query: 178 SNRMAKINSLLKADIYDLSTKSWTPLVSHPPITMVTRIQPSRYNTLVNGVYYWITSS--D 235
N +N IY L TKSWT L H + + + + VNG YW +SS +
Sbjct: 178 HNHRTVVN------IYSLRTKSWTTL--HDQLQGFFLL--NLHGRFVNGKLYWTSSSCIN 227
Query: 236 GSDAARILCFDFRDNQFRKLEAPKLGHYIPFFCDDVFEIKGYLG--YVVQYRCR--IVWL 291
I FD D + +LE P G D+ + G +G + Y C+
Sbjct: 228 NYKVCNITSFDLADGTWERLELPSCGK------DNSYINVGVVGSDLSLLYTCQRGAATS 281
Query: 292 EIWTLEQNG----WAKKYNIDTKMSI 313
++W ++ +G W K + I +I
Sbjct: 282 DVWIMKHSGVNVSWTKLFTIKYPQNI 307
>gb|AAT39293.1| putative F-box-like protein [Solanum demissum]
Length = 384
Score = 99.8 bits (247), Expect = 1e-19
Identities = 97/325 (29%), Positives = 146/325 (44%), Gaps = 57/325 (17%)
Query: 21 IERMKLGEELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATRRNRLLILQNAPNM-- 78
I + L +EL EILLRLP KSLS+F CV KSW +I SP F +N + + N
Sbjct: 7 IPLLLLPDELITEILLRLPIKSLSKFMCVSKSWLQLISSPAFV--KNHIKLTANGKGYIY 64
Query: 79 -KFIFCDGGNDQKSIPIKSLFPQD---------VARIE-------IYGSCDGVFCLKGIS 121
+ IF + +D K P+ SLF + V+ IE I GS +G+ C +
Sbjct: 65 HRLIFRNTNDDFKFCPLPSLFTKQQLIEELFDIVSPIERTTLSTHIVGSVNGLICAAHV- 123
Query: 122 SCITRHDQLILWNPTTKEVHLIPRAPSLGNHYSDESLYGFG--AVNDDFKVVKLNI-SNS 178
R + +WNPT + +P++ S N SD GFG +DD+KVV +N S+
Sbjct: 124 ----RQREAYIWNPTITKSKELPKSRS--NLCSDGIKCGFGYDESHDDYKVVFINYPSHH 177
Query: 179 NRMAKINSLLKADIYDLSTKSWTPLVSHPPITMVTRIQPSRYNTLVNGVYYWITSS--DG 236
N + +N IY L T SWT L H + + + + + V YW +S+ +
Sbjct: 178 NHRSVVN------IYSLRTNSWTTL--HDQLQGIFLL--NLHCRFVKEKLYWTSSTCINN 227
Query: 237 SDAARILCFDFRDNQFRKLEAPKLGHYIPFFCDDVFEIKGYLG--YVVQYRCR--IVWLE 292
I FD D + LE P G D+ + G +G + Y C+ +
Sbjct: 228 YKVCNITSFDLADGTWESLELPSCGK------DNSYINVGVVGSDLSLLYTCQRGAANSD 281
Query: 293 IWTLEQNG----WAKKYNIDTKMSI 313
+W ++ +G W K + I +I
Sbjct: 282 VWIMKHSGVNVSWTKLFTIKYPQNI 306
>gb|AAT38695.1| putative F-box protein [Solanum demissum]
Length = 427
Score = 94.4 bits (233), Expect = 5e-18
Identities = 91/323 (28%), Positives = 134/323 (41%), Gaps = 51/323 (15%)
Query: 21 IERMKLGEELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATRRNRL----------- 69
I + L +EL EILL+LP KSLS+F CV KSW +I SP F +L
Sbjct: 48 IPLLLLPDELITEILLKLPVKSLSKFMCVSKSWLQLISSPTFVKNHIKLTADDKGYIHHR 107
Query: 70 LILQNAP-NMKF-----IFCDGGNDQKSIPIKSLFPQDVARIEIYGSCDGVFCLKGISSC 123
LI +N N KF +F + ++ I S + I GS +G+ C+
Sbjct: 108 LIFRNIDGNFKFCSLPPLFTKQQHTEELFHIDSPIERSTLSTHIVGSVNGLICV------ 161
Query: 124 ITRHDQLILWNPTTKEVHLIPRAPSLGNHYSDESLYGFG--AVNDDFKVVKLNISNSNRM 181
+ + +WNPT + +P+ S N S YGFG DD+KVV ++ N
Sbjct: 162 VHGQKEAYIWNPTITKSKELPKFTS--NMCSSSIKYGFGYDESRDDYKVVFIHYP-YNHS 218
Query: 182 AKINSLLKADIYDLSTKSWTPLVSHPPITMVTRIQPSRYNTLVNGVYYWITSS--DGSDA 239
+ N IY L SWT +V + Y VNG YW +S+ +
Sbjct: 219 SSSNMTTVVHIYSLRNNSWTTFRDQLQCFLV-----NHYGRFVNGKLYWTSSTCINKYKV 273
Query: 240 ARILCFDFRDNQFRKLEAPKLGHYIPFFCDDVFEIK-GYLG--YVVQYRCR--IVWLEIW 294
I FD D + L+ P G D F+I G +G + Y C+ ++W
Sbjct: 274 CNITSFDLADGTWGSLDLPICG-------KDNFDINLGVVGSDLSLLYTCQRGAATSDVW 326
Query: 295 TLEQN----GWAKKYNIDTKMSI 313
++ + W K + I +I
Sbjct: 327 IMKHSAVNVSWTKLFTIKYPQNI 349
>dbj|BAC66625.1| F-box [Prunus mume]
Length = 399
Score = 91.7 bits (226), Expect = 3e-17
Identities = 92/381 (24%), Positives = 149/381 (38%), Gaps = 96/381 (25%)
Query: 28 EELEIE-ILLRLPTKSLSRFKCVQKSWNNIIKSPYFATRRNRLLILQNAPNMK------- 79
EE+ + IL RLP+KSL RFKCV+KSW +I +P F L N+ + K
Sbjct: 3 EEMALRHILPRLPSKSLMRFKCVRKSWYTVINNPTFVENH-----LSNSMHNKLSTCILV 57
Query: 80 --FIFCDGGNDQKSIPIKSL---------------------FP---------QDVARIEI 107
F+ D +D+K + L FP +DV I
Sbjct: 58 SRFVQSDTNSDEKELAFSFLHLRNDYDDAEHNLNFVVEDIKFPLSSGRFIGLEDVESPSI 117
Query: 108 YGSCDGVFCLKGISSCITRHDQLILWNPTTKEVHLIPRAPSLGNHYSDESLYGFGAVNDD 167
G C+G+ CL S D L+L NP KE+ L+P++ L + + +G+ + D
Sbjct: 118 LGHCNGIVCLSPCS------DNLVLCNPAIKEIKLLPKS-GLPDWWGCAVGFGYDPKSKD 170
Query: 168 FKVVKLNISNSNRMAKINSLL---KADIYDLSTKSWTPLVSHPPITMVTRIQPSRYNTLV 224
+KV ++ ++ A+I+ L+ + +IY LST SW + ++ T T P +
Sbjct: 171 YKVSRI----ASYQAEIDGLIPPPRVEIYTLSTDSWREINNNSLETDSTCFFPDYFQIYF 226
Query: 225 NGVYYW---------ITSSDGSDAARILCFDFRDNQFRKLEAPKLGHYIPFFCDDVFEIK 275
G+ YW + D ++ FD D F L P + +E
Sbjct: 227 QGICYWVGYEQPKQSVEYEDEEQKPMVIFFDTGDEIFHNLLFPDSFYMYEEGSSYAYE-- 284
Query: 276 GYLGYVVQYRCRIVWLE-------------------IWTLE-----QNGWAKKYNIDTKM 311
+ Y++ RI+ + +W L+ + W K N + M
Sbjct: 285 --MSYLMYCDLRIILWDGSIALFGFNRFSVCPDSYGVWVLDDFDGAKGSWTKHLNFEPLM 342
Query: 312 SIFHIYGLWNDGAEILVGEFG 332
I + W ++V E G
Sbjct: 343 GIKRVLEFWRSDEILMVTEDG 363
>dbj|BAB01261.1| unnamed protein product [Arabidopsis thaliana]
gi|15233315|ref|NP_188242.1| F-box family protein
[Arabidopsis thaliana]
Length = 360
Score = 88.2 bits (217), Expect = 4e-16
Identities = 73/246 (29%), Positives = 115/246 (46%), Gaps = 41/246 (16%)
Query: 26 LGEELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATRRNRLLILQ--NAPNMKFIFC 83
L EEL IEIL+RL K L+RF+CV K+W ++I P F + + + + F
Sbjct: 5 LPEELAIEILVRLSMKDLARFRCVCKTWRDLINDPGFTETYRDMSPAKFVSFYDKNFYML 64
Query: 84 DGGNDQKSIPIKSLFPQDVARIE---IYGSCDGVFCLKGISSCITRHDQLILWNPTTKEV 140
D I K FP D + I+ CDG C+ ++ L++WNP +K+
Sbjct: 65 DVEGKHPVITNKLDFPLDQSMIDESTCVLHCDGTLCV------TLKNHTLMVWNPFSKQF 118
Query: 141 HLIPRAPSLGNHYSDESLYGFG--AVNDDFKVV----KLNISNSNRMAKINSLLKADIYD 194
++P P + Y D ++ GFG V+DD+KVV +L++S A +++
Sbjct: 119 KIVPN-PGI---YQDSNILGFGYDPVHDDYKVVTFIDRLDVST------------AHVFE 162
Query: 195 LSTKSW--TPLVSHPPITMVTRIQPSRYNTLVNGVYYWITSSDGSDAARILCFDFRDNQF 252
T SW + +S+P R T ++ YWI +D ILCF+ +++
Sbjct: 163 FRTGSWGESLRISYPDWH-----YRDRRGTFLDQYLYWIAYRSSADRF-ILCFNLSTHEY 216
Query: 253 RKLEAP 258
RKL P
Sbjct: 217 RKLPLP 222
>dbj|BAD08685.1| S haplotype-specific F-box protein 5 [Prunus avium]
Length = 377
Score = 85.5 bits (210), Expect = 2e-15
Identities = 78/305 (25%), Positives = 134/305 (43%), Gaps = 49/305 (16%)
Query: 28 EELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATRRNRLLILQNAPNMKFIFCDGGN 87
EE+ I+IL+RLP KSL RF C KSW++++ S F + I ++A ++ + N
Sbjct: 8 EEILIDILVRLPAKSLVRFLCTCKSWSDLVGSSSFVSTHLHRNITKHA-HVHLLCLHHPN 66
Query: 88 DQKSIPIK----------SLFPQD--------------VARIEIYGSCDGVFCLKGISSC 123
++ + SLFP + IYGS +G+ C IS
Sbjct: 67 VRRQVNPDDPYVTQEFQWSLFPNETFEECSKLSHPLGSTEHYGIYGSSNGLVC---ISDE 123
Query: 124 ITRHDQ-LILWNPTTKEVHLIPRAPSLGNHYSDESL-YGFGAVNDDFKVVKLNISNSNRM 181
I D +++WNP+ ++ P + ++ ++ +L +GF +D+KVV++ +N + +
Sbjct: 124 ILNFDSPILMWNPSVRKFRTAPTSTNINLKFAYVALQFGFHHAVNDYKVVRMMRTNKDAL 183
Query: 182 AKINSLLKADIYDLSTKSWTPLVSHPPITMVTRIQPSRYNTLVNGVYYWITSSDGSDAAR 241
A ++Y L T SW + + PP T T NGV Y I
Sbjct: 184 A-------VEVYSLRTDSWKMIEAIPPWLKCT--WQHHRGTFFNGVAYHIIQK--GPIFS 232
Query: 242 ILCFDFRDNQFRKLEAP-----KLGHYIPFFCDDVFEIKGYLGYVVQYRCRIVWLEIWTL 296
I+ FD +F + AP G I + D + + G+ G + +I ++W L
Sbjct: 233 IMSFDSGSEEFEEFIAPDAVCSSWGLCIDVYNDQICLLSGFYGCEDEGLDKI---DLWVL 289
Query: 297 EQNGW 301
++ W
Sbjct: 290 QEKQW 294
>gb|AAT69247.1| F-box protein 1 [Prunus armeniaca]
Length = 377
Score = 85.1 bits (209), Expect = 3e-15
Identities = 83/306 (27%), Positives = 131/306 (42%), Gaps = 47/306 (15%)
Query: 28 EELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATRRNRLLILQNA---------PNM 78
+E+ ++IL+RLP KSL RF C KSW+ +I S F + + ++A PN
Sbjct: 8 KEILVDILVRLPAKSLVRFLCTCKSWSGLIGSSSFVSIHLNRNVTEHAHVYLLCLLHPNF 67
Query: 79 KFIFCDGGNDQKSIPIKSLFPQD--------------VARIEIYGSCDGVFCLKGISSCI 124
+ + K SLF + IYGS +G+ C IS I
Sbjct: 68 ERLADPDDPYVKQEFSWSLFSNETFEESSKITHPLGSTEHYGIYGSSNGLVC---ISDEI 124
Query: 125 TRHDQLI-LWNPTTKEVHLIPRAPSLGNHYSDESL-YGFGAVNDDFKVVKLNISNSNRMA 182
D I +WNP+ K+ P + ++ +S SL +GF +D+K V++ +N N
Sbjct: 125 LNFDSPIHIWNPSVKKFKTPPMSTNINIKFSLVSLQFGFHPRVNDYKAVRMMRTNKN--- 181
Query: 183 KINSLLKADIYDLSTKSWTPLVSHPPITMVTRIQPSRYNTLVNGVYYWITSSDGSDAARI 242
+L ++Y LST SW + + PP T T NGV Y I I
Sbjct: 182 ----VLAVEVYSLSTDSWKMVEAIPPWLKCT--WQHHKGTFFNGVAYHIIQK--GPLFSI 233
Query: 243 LCFDFRDNQFRKLEAP-----KLGHYIPFFCDDVFEIKGYLGYVVQYRCRIVWLEIWTLE 297
+ FD +F + AP LG I + + + + G+ G + +I ++W L+
Sbjct: 234 MSFDSGSEEFEEFIAPDAICRSLGLCIDVYKEHICLLFGFYGCEEEGMDKI---DLWVLQ 290
Query: 298 QNGWAK 303
+ W K
Sbjct: 291 EKRWKK 296
>gb|AAT69249.1| F-box protein 4 [Prunus armeniaca]
Length = 373
Score = 84.0 bits (206), Expect = 7e-15
Identities = 80/303 (26%), Positives = 132/303 (43%), Gaps = 46/303 (15%)
Query: 28 EELEIEILLRLPTKSLSRFKCVQKSWNNII-KSPYFATRRNR--------LLILQNAPNM 78
+E+ ++IL+RLP KSL RF C KSW+++I S + +T NR L+ + PN
Sbjct: 8 KEILVDILVRLPAKSLVRFMCTCKSWSDLIGSSSFVSTHLNRNVNKHAHVYLLCLHHPNF 67
Query: 79 KFIFCDGGNDQKSIPIKSLFPQD--------------VARIEIYGSCDGVFCLKGISSCI 124
+ + + SLFP + IYGS +G+ C IS I
Sbjct: 68 ECVVDPDDPYLEEEVQWSLFPNETFEECSKLNHPLGSTEHYGIYGSSNGLVC---ISDEI 124
Query: 125 TRHDQLI-LWNPTTKEVHLIPRAPSLGNHYSDESLYGFGAVNDDFKVVKLNISNSNRMAK 183
D I +WNP+ +++ +P + ++ +GF +D+K V++ +N N +A
Sbjct: 125 LNFDSPIHIWNPSVRKLRTLPISTNIIKFSHVALQFGFHPGVNDYKAVRMMRTNKNALA- 183
Query: 184 INSLLKADIYDLSTKSWTPLVSHPPITMVTRIQPSRYNTLVNGVYYWITSSDGSDAARIL 243
++Y L T SW + + PP + T NGV Y I + I+
Sbjct: 184 ------IEVYSLRTDSWKMIEAIPP--WLKCAWQHYKGTFFNGVAYHII--EKGPIFSIM 233
Query: 244 CFDFRDNQFRKLEAPKLGHYIPFFCDDVF--EIKGYLGYVVQYRCRIVW---LEIWTLEQ 298
FD +F + AP C DV+ +I G+ YRC + +++W L++
Sbjct: 234 SFDSGSEEFEEFIAPDAICRPSEVCIDVYKEQICLLFGF---YRCEEMGMDKIDLWVLQE 290
Query: 299 NGW 301
W
Sbjct: 291 KRW 293
>dbj|BAC66631.1| F-box [Prunus mume]
Length = 374
Score = 83.6 bits (205), Expect = 9e-15
Identities = 72/261 (27%), Positives = 121/261 (45%), Gaps = 49/261 (18%)
Query: 28 EELEIEILLRLPTKSLSRFKCVQKSWNNII-KSPYFATRRNR--------LLILQNAPNM 78
+E+ I+IL+RLP KSL RF C KSW+++I S + +T +R L+ N PN+
Sbjct: 8 KEIVIDILVRLPVKSLVRFLCTCKSWSDLIGSSSFVSTHLHRNVTKHAHVYLLCLNHPNV 67
Query: 79 KFIFCDGGNDQKSIPIK-----SLFPQDVAR--------------IEIYGSCDGVFCLKG 119
+++ +D+ +K S+FP ++ IYGS +G+ C
Sbjct: 68 EYL-----DDRDDPYVKQEFQWSIFPNEIFEECSKLTHPLRSTEDYMIYGSSNGLVC--- 119
Query: 120 ISSCITRHDQ-LILWNPTTKEVHLIPRAPSLGNHYSDESL-YGFGAVNDDFKVVKLNISN 177
+S I D +++WNP+ K+ P + ++ +S +L +GF +D+K V++ +N
Sbjct: 120 VSDEILNFDSPILIWNPSVKKFRTPPMSININIKFSYVALQFGFHPGVNDYKAVRMMRTN 179
Query: 178 SNRMAKINSLLKADIYDLSTKSWTPLVSHPPITMVTRIQPSRYNTLVNGVYYWITSSDGS 237
N +A ++Y L T SW + + PP T T NGV Y +
Sbjct: 180 KNALA-------IEVYSLGTDSWKMIEAIPPWLKCT--WQHLKGTFFNGVAYHVIQK--G 228
Query: 238 DAARILCFDFRDNQFRKLEAP 258
I+ FD +F + AP
Sbjct: 229 PIFSIISFDSGSEEFEEFIAP 249
>gb|AAP37713.1| At3g23880 [Arabidopsis thaliana] gi|9294656|dbj|BAB03005.1| unnamed
protein product [Arabidopsis thaliana]
gi|26449693|dbj|BAC41970.1| unknown protein [Arabidopsis
thaliana] gi|15229536|ref|NP_189030.1| F-box family
protein [Arabidopsis thaliana]
Length = 364
Score = 82.8 bits (203), Expect = 2e-14
Identities = 91/328 (27%), Positives = 134/328 (40%), Gaps = 51/328 (15%)
Query: 29 ELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATRRNRLLILQNAPNMK----FIFCD 84
E+ EILLRLP KSL+RFKCV SW ++I FA + +L A +
Sbjct: 17 EMMEEILLRLPVKSLTRFKCVCSSWRSLISETLFALKHALILETSKATTSTKSPYGVITT 76
Query: 85 GGNDQKSIPIKSLFPQDVARI-------------EIYGSCDGVFCLKGISSCITRHDQLI 131
KS I SL+ + ++ G+C G+ C + L
Sbjct: 77 SRYHLKSCCIHSLYNASTVYVSEHDGELLGRDYYQVVGTCHGLVCFH-----VDYDKSLY 131
Query: 132 LWNPTTKEVHLIPRAPSLGNHYSDES---LYGFG--AVNDDFKVVKLNISNSNRMAKINS 186
LWNPT K L R S SD+ YGFG DD+KVV L KI
Sbjct: 132 LWNPTIK---LQQRLSSSDLETSDDECVVTYGFGYDESEDDYKVVAL--LQQRHQVKI-- 184
Query: 187 LLKADIYDLSTKSWTPLVSHPPITMVTRIQPSRYNTLVNGVYYWITSSDGSDAARILCFD 246
+ IY K W S P +V SR +NG W +S S + I+ +D
Sbjct: 185 --ETKIYSTRQKLWRSNTSFPSGVVVA--DKSRSGIYINGTLNWAATS-SSSSWTIISYD 239
Query: 247 FRDNQFRKLEAPK-LGHYIPFFCDDVFEIKGYLGYVVQYRCRIVWLEIWTLEQNG----W 301
++F++L P G F + +++G L V C+ ++W +++ G W
Sbjct: 240 MSRDEFKELPGPVCCGR--GCFTMTLGDLRGCLSMVCY--CKGANADVWVMKEFGEVYSW 295
Query: 302 AKKYNIDTKMSIFHIYGLW-NDGAEILV 328
+K +I + LW +DG +L+
Sbjct: 296 SKLLSIPGLTDF--VRPLWISDGLVVLL 321
>emb|CAD56853.1| S locus F-box (SLF)-S5A protein [Antirrhinum hispanicum]
Length = 404
Score = 82.8 bits (203), Expect = 2e-14
Identities = 97/368 (26%), Positives = 159/368 (42%), Gaps = 38/368 (10%)
Query: 18 KLSIERMKLGEELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATRRNR-----LLIL 72
K+S ++++ +++ I+IL++L ++L RF+C+ KSW +IKS F R+R LL+
Sbjct: 13 KMSDDQLQ-SKDVFIQILVQLSVRALMRFRCISKSWCALIKSSTFHLLRDRKYDNVLLVR 71
Query: 73 QNAPNMKFIFCDGGNDQKSIPIKSLFP-------QDVARIEIYGSCDGVFCLKGISS--- 122
+ P + C D S+ +K + P +D+ + C L G S
Sbjct: 72 RYLPPPEDEDCFSFYDLNSLEVKQVLPNLSIPLLKDLRFKYDHPYCPEAAYLLGPDSGLI 131
Query: 123 CITRHDQLILWNPTTKEVHLIPRAPSL-GNHYSDESL-YGFGAV-NDDFKVVKLNISNSN 179
CI L NP +E +P P + +S+E GFG +DFK+V +
Sbjct: 132 CIACIGNYYLCNPALREFKQLPPCPFVCPKGFSNEIFAEGFGCTCTNDFKIVLIRRVTLY 191
Query: 180 RMAKINSLLKADIYDLSTKSWTPLVSHPPITMVTRIQPSRYNTLVNGVYYWITSSDG-SD 238
+ + +Y +T W I++ + L NGV +W +S G S
Sbjct: 192 DDYDPDLYIMVHLYTSNTNLWRTFAG-DVISVKNLCNYACSELLFNGVCHWNANSTGFSS 250
Query: 239 AARILCFDFRDNQFRKLE-APKLGHYIPFFCDDVFEIKGYLGYVVQYRCRIVWLE----- 292
IL F+ R F +LE P + +C + I LG +V+Y WLE
Sbjct: 251 PDTILTFNIRTEVFGQLEFIPDWEEEVYGYCVSLIAIDNCLG-MVRYE---GWLEEPQLI 306
Query: 293 -IWTLEQNG----WAKKYNIDTKMSIFHIYGLWNDGAEILVGEFGQRQLTSCDHHGNVLR 347
IW + + G W K + I + + WN+ +LV E + QL +C H N R
Sbjct: 307 DIWVMNEYGVGESWTKSFVIG-PYEVICPFMFWNNDEWLLV-ESTEGQLVACALHTNEAR 364
Query: 348 QFQLDTLE 355
+ Q+ +E
Sbjct: 365 RLQICGVE 372
>gb|AAD32931.1| T17H7.6 [Arabidopsis thaliana]
Length = 502
Score = 82.8 bits (203), Expect = 2e-14
Identities = 87/306 (28%), Positives = 139/306 (44%), Gaps = 43/306 (14%)
Query: 29 ELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYF--------ATRRNRLLILQNA--PNM 78
+L +EIL RLP KSL +FKCV K W++II + F +TR ++ N P+
Sbjct: 12 DLTVEILTRLPAKSLMKFKCVSKLWSSIIHNQSFIDSFYSISSTRPRFIVAFSNGSFPSD 71
Query: 79 K----FIFCDG--GNDQKSIPIKSL---FPQDVARIEIYGSCDGVFCLKGISSCITRHDQ 129
K FIF G++ S I +L P + C V G +C + + +
Sbjct: 72 KEKRLFIFSSSHEGHESSSSVITNLDTTIPSLTVSNNLASRCISV---NGFIAC-SLYTR 127
Query: 130 LILWNPTTKEVHLIPRAPSLGNHYSDESLYGFGAVNDDFKVVKLNIS-NSNRMAKINSLL 188
+ NP+T++V ++P PS + G+ V+D FK + L S N+ + + L+
Sbjct: 128 FTICNPSTRQVIVLPILPSGRAPDMRSTCIGYDPVDDQFKALALISSCIPNKDSTVEHLV 187
Query: 189 KADIYDLSTKSWTPLVSH---PPITMVTRIQPSRYNTLVNG-VYYWITSSDGSDAARILC 244
D SW + + PP + VT +NG VYY + S A I+C
Sbjct: 188 LTLKGDKKNYSWRQIQGNNNIPPYSPVT------MRVCINGVVYYGAWTPRQSMNAVIVC 241
Query: 245 FDFRDNQFRKLEAPKLGHYIPFFCDD--VFEIKGYLGYVVQYR-CRIVWLEIWTL---EQ 298
FD R + ++ PK + +C+D + E KG L +V+ R R ++W L E+
Sbjct: 242 FDVRSEKITFIKTPK---DVVRWCNDSILMEYKGKLASIVRNRYSRFDTFDLWVLEDIEK 298
Query: 299 NGWAKK 304
W+K+
Sbjct: 299 QEWSKQ 304
>ref|NP_174365.1| F-box family protein [Arabidopsis thaliana]
Length = 399
Score = 82.8 bits (203), Expect = 2e-14
Identities = 87/306 (28%), Positives = 139/306 (44%), Gaps = 43/306 (14%)
Query: 29 ELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYF--------ATRRNRLLILQNA--PNM 78
+L +EIL RLP KSL +FKCV K W++II + F +TR ++ N P+
Sbjct: 12 DLTVEILTRLPAKSLMKFKCVSKLWSSIIHNQSFIDSFYSISSTRPRFIVAFSNGSFPSD 71
Query: 79 K----FIFCDG--GNDQKSIPIKSL---FPQDVARIEIYGSCDGVFCLKGISSCITRHDQ 129
K FIF G++ S I +L P + C V G +C + + +
Sbjct: 72 KEKRLFIFSSSHEGHESSSSVITNLDTTIPSLTVSNNLASRCISV---NGFIAC-SLYTR 127
Query: 130 LILWNPTTKEVHLIPRAPSLGNHYSDESLYGFGAVNDDFKVVKLNIS-NSNRMAKINSLL 188
+ NP+T++V ++P PS + G+ V+D FK + L S N+ + + L+
Sbjct: 128 FTICNPSTRQVIVLPILPSGRAPDMRSTCIGYDPVDDQFKALALISSCIPNKDSTVEHLV 187
Query: 189 KADIYDLSTKSWTPLVSH---PPITMVTRIQPSRYNTLVNG-VYYWITSSDGSDAARILC 244
D SW + + PP + VT +NG VYY + S A I+C
Sbjct: 188 LTLKGDKKNYSWRQIQGNNNIPPYSPVT------MRVCINGVVYYGAWTPRQSMNAVIVC 241
Query: 245 FDFRDNQFRKLEAPKLGHYIPFFCDD--VFEIKGYLGYVVQYR-CRIVWLEIWTL---EQ 298
FD R + ++ PK + +C+D + E KG L +V+ R R ++W L E+
Sbjct: 242 FDVRSEKITFIKTPK---DVVRWCNDSILMEYKGKLASIVRNRYSRFDTFDLWVLEDIEK 298
Query: 299 NGWAKK 304
W+K+
Sbjct: 299 QEWSKQ 304
>gb|AAS79484.1| S1-locus linked F-box protein [Petunia integrifolia subsp. inflata]
Length = 389
Score = 82.8 bits (203), Expect = 2e-14
Identities = 98/371 (26%), Positives = 156/371 (41%), Gaps = 63/371 (16%)
Query: 24 MKLGEELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATR--------RNRLLILQNA 75
MKL E+L +LL P KSL RFKC+ K+W+ +I+S F R ++ +I + +
Sbjct: 7 MKLPEDLVFLVLLTFPVKSLLRFKCISKAWSILIQSTTFINRHVNRKTNTKDEFIIFKRS 66
Query: 76 -----PNMKFI--FCDGGNDQKSIPIKSLFPQ-DVARI---------EIYGSCDGVFCLK 118
K I F G +D + LFP +V+ + + G CDG+ L
Sbjct: 67 IKDEQEGFKDILSFFSGHDD----VLNPLFPDVEVSYMTSKCNCTFNPLIGPCDGLIAL- 121
Query: 119 GISSCITRHDQLILWNPTTKEVHLIPRAP---SLGNHYSDESL-YGFGAVNDDFKVVKLN 174
S IT IL NP T+ L+P +P G H S E + G +++ +KVV+++
Sbjct: 122 -TDSIIT-----ILLNPATRNFRLLPPSPFGCPKGYHRSVEGVGLGLDTISNYYKVVRIS 175
Query: 175 ISNSNRMAKINSL--LKADIYDLSTKSWTPLVSHPPITMVTRIQPSRYNTLVNGVYYWIT 232
K D+ DL T SW L H + ++ + S L + +W
Sbjct: 176 EVYCEEAGGYPGPKDSKIDVCDLGTDSWREL-DHVQLPLIYWVPCS--GMLYKEMVHWFA 232
Query: 233 SSDGSDAARILCFDFRDNQFRKLEAP--------KLGHYIPFFCDDVFEIKGYLGYVVQY 284
++D ++ ILCFD FR +E P +L + + C+ F + GY +
Sbjct: 233 TTD--ESMVILCFDMSTEMFRNMEMPDSCSPITHELYYGLVILCES-FTLIGYSNPISSI 289
Query: 285 RCRIVWLEIWTLEQNG----WAKKYNIDTKMSIFHIYGLWNDGAEILVGEFGQRQLTSCD 340
+ IW + + G W KY I +SI +W +L G +L S D
Sbjct: 290 DPVKDKMHIWVMMEYGVSESWIMKYTI-KPLSIESPLAVWKKNILLLQSRSG--RLISYD 346
Query: 341 HHGNVLRQFQL 351
+ ++ L
Sbjct: 347 LNSGEAKELNL 357
>gb|AAX11682.1| non-S F-box protein 1 [Petunia axillaris subsp. axillaris]
Length = 389
Score = 82.4 bits (202), Expect = 2e-14
Identities = 96/351 (27%), Positives = 149/351 (42%), Gaps = 61/351 (17%)
Query: 25 KLGEELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATR--------RNRLLILQNA- 75
KL E+L ILL P KSL RFKC+ KSW+ +I+S F R ++ ++ + A
Sbjct: 8 KLPEDLVFLILLTFPVKSLMRFKCISKSWSFLIQSTGFINRHVNRKTNTKDEFILFKRAI 67
Query: 76 --PNMKFI----FCDGGNDQKSIPIKSLFPQ-DVARI---------EIYGSCDGVFCLKG 119
+FI F G +D + LFP DV+ + + G CDG+ L
Sbjct: 68 KDEEEEFINILSFFSGYDD----VLNPLFPDIDVSYMTSNCNCTFNPLIGPCDGLIAL-- 121
Query: 120 ISSCITRHDQLILWNPTTKEVHLIPRAP---SLGNHYSDESL-YGFGAVNDDFKVVKLNI 175
+ IT IL NP T+ L+P +P G H S E + +G +++ +KVV+++
Sbjct: 122 TDTIIT-----ILLNPATRNFRLLPPSPFACPKGYHRSIEGVGFGLDTISNYYKVVRISE 176
Query: 176 SNSNRMAKINSL--LKADIYDLSTKSWTPLVSHPPITMVTRIQPSRYNTLVNGVYYWITS 233
K D+ DL T SW L H + ++ + S L + +W +
Sbjct: 177 VYCEEADGYPGPKDSKIDVCDLVTDSWREL-DHIQLPLIYWVPCS--GMLYMEMVHWFAT 233
Query: 234 SDGSDAARILCFDFRDNQFRKLEAP--------KLGHYIPFFCDDVFEIKGYLGYVVQYR 285
+D S ILCFD FR ++ P +L + + CD F + GY +
Sbjct: 234 TDISMV--ILCFDMSTEVFRNMKMPDTCTRITHELYYGLVILCDS-FTLIGYSNPIGSID 290
Query: 286 CRIVWLEIWTLEQNG----WAKKYNIDTKMSIFHIYGLWNDGAEILVGEFG 332
+ IW + + G W KY I +SI +W + +L G
Sbjct: 291 SARDKMHIWVMMEYGVSESWIMKYTI-KPLSIESPLAVWKNNILLLQSRSG 340
>emb|CAD56660.1| S locus F-box (SLF)-S4D protein [Antirrhinum hispanicum]
Length = 374
Score = 81.3 bits (199), Expect = 5e-14
Identities = 93/354 (26%), Positives = 148/354 (41%), Gaps = 41/354 (11%)
Query: 28 EELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFAT------RRN------RLLILQNA 75
E++ EIL+ LP KSL R KC K + +IKS F T RRN R ++ +
Sbjct: 9 EDILKEILVWLPVKSLIRLKCASKHLDMLIKSQAFITSHMIKQRRNDGMLLVRRILPPST 68
Query: 76 PNMKFIFCDGGNDQ-----KSIPIKSLFPQDVAR-----IEIYGSCDGVFCLKGISSCIT 125
N F F D + + +PI L D A +++ G C+G+ CIT
Sbjct: 69 YNDVFSFHDVNSPELEEVLPKLPITLLSNPDEASFNPNIVDVLGPCNGIV-------CIT 121
Query: 126 RHDQLILWNPTTKEVHLIPRAPSLGNH--YSDESLYGFGAV-NDDFKVVKLNISNSNRMA 182
+ +IL NP +E +P AP YS + GFG+ ++FKV+ +N + R+
Sbjct: 122 GQEDIILCNPALREFRKLPSAPISCRPPCYSIRTGGGFGSTCTNNFKVILMNTLYTARVD 181
Query: 183 KINSLLKADIYDLSTKSWTPLVSHPPITMVTRIQPSRYNTLVNGVYYWITSSDGSDAAR- 241
++ + +Y+ + SW ++ I M G +W + G
Sbjct: 182 GRDAQHRIHLYNSNNDSWRE-INDFAIVMPVVFSYQCSELFFKGACHWNGRTSGETTPDV 240
Query: 242 ILCFDFRDNQFRKLEAPKLGHYIPFFCDDVFEIKGYLGYVVQYRCRIVWLEIWTLEQNG- 300
IL FD F + E P G + F I V+ +E+W +++ G
Sbjct: 241 ILTFDVSTEVFGQFEHPS-GFKLCTGLQHNFMILNECFASVRSEVVRCLIEVWVMKEYGI 299
Query: 301 ---WAKKYNIDTKMSIFHIYGLWNDGAEILVGEFGQRQLTSCDHHGNVLRQFQL 351
W KK+ I I + W + E+L G+ G QL S H N ++ F++
Sbjct: 300 KQSWTKKFVIGPH-EIGCPFLFWRNNEELL-GDNGDGQLASFVLHTNEIKTFEV 351
>gb|AAO37148.1| hypothetical protein [Arabidopsis thaliana]
gi|61742586|gb|AAX55114.1| hypothetical protein
At2g16810 [Arabidopsis thaliana] gi|25411735|pir||E84544
hypothetical protein At2g16810 [imported] - Arabidopsis
thaliana gi|42569080|ref|NP_565395.2| F-box family
protein (FBX8) [Arabidopsis thaliana]
Length = 295
Score = 79.7 bits (195), Expect = 1e-13
Identities = 69/250 (27%), Positives = 121/250 (47%), Gaps = 33/250 (13%)
Query: 28 EELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATRRNRLLILQNAPNMKFIFC--DG 85
++L IEIL RLP KS+ RFKCV K W++++ S YF NR LI+ + P C D
Sbjct: 43 QDLLIEILTRLPPKSVMRFKCVSKFWSSLLSSRYFC---NRFLIVPSQPQPSLYMCLLDR 99
Query: 86 GNDQKSI---------PIKSLFPQDVARIEIYGSCDGVFCLKGISSCI--TRHDQLILWN 134
N KS+ P +F QD+ ++ G F L+ + I TR+ + ++N
Sbjct: 100 YNYSKSLILSSAPSTSPYSFVFDQDLTIRKM-----GGFFLRILRGFIFFTRNLKARIYN 154
Query: 135 PTTKEVHLIPRAPS----LGNHYSDESLYGFGAVNDDFKVV-KLNISNSNRMAKINSLLK 189
PTT+++ ++P G Y+ VND +K++ ++ ++ N + + S L
Sbjct: 155 PTTRQLVILPTIKESDIIAGPPYNILYFICHDPVNDRYKLLCTVSYASDNDLQNLKSELW 214
Query: 190 ADIYDLSTKSWTPLVSHPPITMVTRIQPSRYNTLVNGVYYWITSSDGSDAARILCFDFRD 249
+ + + SW + + P + PS + +NGV Y++ +D ++ FD R
Sbjct: 215 IFVLE-AGGSWKRVANEFPHHV-----PSHLDLNMNGVLYFLAWTD-PHTCMLVSFDVRS 267
Query: 250 NQFRKLEAPK 259
+F ++ P+
Sbjct: 268 EEFNTMQVPR 277
>gb|AAT38720.1| putative F-Box protein [Solanum demissum]
Length = 372
Score = 78.6 bits (192), Expect = 3e-13
Identities = 74/255 (29%), Positives = 113/255 (44%), Gaps = 37/255 (14%)
Query: 26 LGEELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFAT------------RRNRLLILQ 73
L E+ IEILL++P KSL +F CV K+W +I S F +R++ +
Sbjct: 9 LPHEIIIEILLKVPPKSLLKFMCVSKTWLELISSAKFIKTHLELIANDKEYSHHRIIFQE 68
Query: 74 NAPNMKFIFCDGG--NDQKS---IPIKSLFPQDVARIEIYGSCDGVFCLKGISSCITRHD 128
+A N K + C N ++S I S I GS +G+ CL ++ +
Sbjct: 69 SACNFK-VCCLPSMLNKERSTELFDIGSPMENPTIYTWIVGSVNGLICL------YSKIE 121
Query: 129 QLILWNPTTKEVHLIPR-APSLGNHYSDESLYGFG--AVNDDFKVVKLNISNSNRMAKIN 185
+ +LWNP K+ +P L N S YGFG DD+KVV + + +
Sbjct: 122 ETVLWNPAVKKSKKLPTLGAKLRNGCSYYLKYGFGYDETRDDYKVVVIQCIYED-SGSCD 180
Query: 186 SLLKADIYDLSTKSWTPLVSHPPITMVTRIQPSRYNTLVNGVYYWITSSDGS--DAARIL 243
S++ +IY L SW + +V P ++ VNG YW S+D + I+
Sbjct: 181 SVV--NIYSLKADSWRTINKFQGNFLVN--SPGKF---VNGKIYWALSADVDTFNMCNII 233
Query: 244 CFDFRDNQFRKLEAP 258
D D +R+LE P
Sbjct: 234 SLDLADETWRRLELP 248
>gb|AAX11681.1| S19-locus linked F-box protein [Petunia axillaris subsp. axillaris]
Length = 389
Score = 78.2 bits (191), Expect = 4e-13
Identities = 96/352 (27%), Positives = 144/352 (40%), Gaps = 63/352 (17%)
Query: 25 KLGEELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATRR-NR--------LLILQNA 75
KL E+L ILL P KSL RFKC+ K+W+ +I+S F R NR +L ++
Sbjct: 8 KLPEDLVFLILLTFPVKSLLRFKCISKAWSILIQSTTFINRHINRKTNTKAEFILFKRSI 67
Query: 76 PNMKFIFCD-----GGNDQKSIPIKSLFPQ----------DVARIEIYGSCDGVFCLKGI 120
+ + F + GND P LFP D + G CDG+ L
Sbjct: 68 KDEEEEFINILSFFSGNDDVLNP---LFPDIDVSYMTSKCDCTFTPLIGPCDGLIAL--T 122
Query: 121 SSCITRHDQLILWNPTTKEVHLIPRAP---SLGNHYSDESL-YGFGAVNDDFKVVKLNIS 176
+ IT I+ NP T+ ++P +P G H S E + +GF ++ +KVV+++
Sbjct: 123 DTIIT-----IVLNPATRNFRVLPPSPFGCPKGYHRSVEGVGFGFDTISYYYKVVRISEV 177
Query: 177 NSNRMAKINSL--LKADIYDLSTKSWTPL--VSHPPITMVTRIQPSRYNTLVNGVYYWIT 232
K D+ DLST SW L V P I V L + +W
Sbjct: 178 YCEEADGYPGPKDSKIDVCDLSTDSWRELDHVQLPSIYWVPCA-----GMLYKEMVHWFA 232
Query: 233 SSDGSDAARILCFDFRDNQFRKLEAP--------KLGHYIPFFCDDVFEIKGYLGYVVQY 284
++D S ILCFD F ++ P +L + + C+ F + GY +
Sbjct: 233 TTDTSMV--ILCFDMSTEMFHDMKMPDTCSRITHELYYGLVILCES-FTLIGYSNPISSI 289
Query: 285 RCRIVWLEIWTLEQNG----WAKKYNIDTKMSIFHIYGLWNDGAEILVGEFG 332
+ IW + + G W KY I +SI +W + +L G
Sbjct: 290 DPVEDKMHIWVMMEYGVSESWIMKYTI-RPLSIESPLAVWKNHILLLQSRSG 340
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.324 0.140 0.444
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 659,569,595
Number of Sequences: 2540612
Number of extensions: 27443424
Number of successful extensions: 49397
Number of sequences better than 10.0: 447
Number of HSP's better than 10.0 without gapping: 286
Number of HSP's successfully gapped in prelim test: 161
Number of HSP's that attempted gapping in prelim test: 48749
Number of HSP's gapped (non-prelim): 558
length of query: 373
length of database: 863,360,394
effective HSP length: 129
effective length of query: 244
effective length of database: 535,621,446
effective search space: 130691632824
effective search space used: 130691632824
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 76 (33.9 bits)
Medicago: description of AC135160.10


