

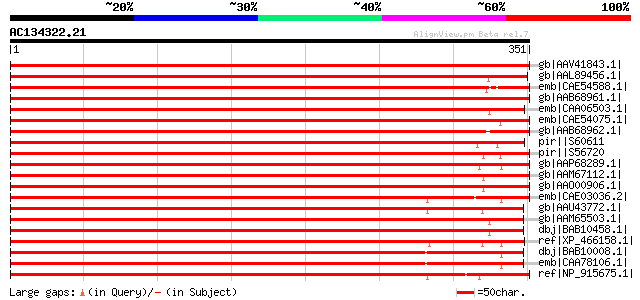
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134322.21 - phase: 0
(351 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAV41843.1| stress kinase [Medicago truncatula] gi|55140609|g... 713 0.0
gb|AAL89456.1| osmotic stress-activated protein kinase [Nicotian... 639 0.0
emb|CAE54588.1| serin/threonine protein kinase [Fagus sylvatica] 623 e-177
gb|AAB68961.1| protein kinase 3 [Glycine max] gi|1362051|pir||S5... 619 e-176
emb|CAA06503.1| protein kinase [Craterostigma plantagineum] gi|7... 614 e-174
emb|CAE54075.1| serine/threonine-protein kinase [Fagus sylvatica] 610 e-173
gb|AAB68962.1| protein kinase [Glycine max] 607 e-172
pir||S60611 probable serine/threonine-specific protein kinase (E... 607 e-172
pir||S56720 probable serine/threonine-specific protein kinase (E... 603 e-171
gb|AAP68289.1| At1g10940 [Arabidopsis thaliana] gi|21536490|gb|A... 602 e-171
gb|AAM67112.1| putative serine/threonine-protein kinase [Arabido... 595 e-169
gb|AAO00906.1| putative serine/threonine-protein kinase [Arabido... 594 e-168
emb|CAE03036.2| OSJNBa0084A10.11 [Oryza sativa (japonica cultiva... 589 e-167
gb|AAU43772.1| putative salt-inducible protein kinase [Zea mays] 582 e-165
gb|AAM65503.1| serine/threonine-protein kinase [Arabidopsis thal... 580 e-164
dbj|BAB10458.1| serine/threonine-protein kinase [Arabidopsis tha... 579 e-164
ref|XP_466158.1| putative osmotic stress-activated protein kinas... 576 e-163
dbj|BAB10008.1| serine/threonine-protein kinase [Arabidopsis tha... 572 e-162
emb|CAA78106.1| protein kinase [Arabidopsis thaliana] 569 e-161
ref|NP_915675.1| putative protein kinase SPK-3 [Oryza sativa (ja... 560 e-158
>gb|AAV41843.1| stress kinase [Medicago truncatula] gi|55140609|gb|AAV41842.1|
stress kinase [Medicago truncatula]
Length = 351
Score = 713 bits (1841), Expect = 0.0
Identities = 351/351 (100%), Positives = 351/351 (100%)
Query: 1 MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNI 60
MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNI
Sbjct: 1 MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNI 60
Query: 61 IRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQIC 120
IRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQIC
Sbjct: 61 IRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQIC 120
Query: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL 180
HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL
Sbjct: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL 180
Query: 181 ADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRI 240
ADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRI
Sbjct: 181 ADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRI 240
Query: 241 FVASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENPTYSLQSIEDIMKIVEEAK 300
FVASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENPTYSLQSIEDIMKIVEEAK
Sbjct: 241 FVASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENPTYSLQSIEDIMKIVEEAK 300
Query: 301 NPPQASRSVGGFGWGGEEDDDEINEAEAELEEDEYEKRVKEAQESGEIHVN 351
NPPQASRSVGGFGWGGEEDDDEINEAEAELEEDEYEKRVKEAQESGEIHVN
Sbjct: 301 NPPQASRSVGGFGWGGEEDDDEINEAEAELEEDEYEKRVKEAQESGEIHVN 351
>gb|AAL89456.1| osmotic stress-activated protein kinase [Nicotiana tabacum]
Length = 356
Score = 639 bits (1648), Expect = 0.0
Identities = 311/355 (87%), Positives = 334/355 (93%), Gaps = 5/355 (1%)
Query: 1 MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNI 60
M+KYE+VKDIGSGNFGVARLMRHK+TK+LVAMKYIERGHKIDENVAREIINHRSLRHPNI
Sbjct: 1 MDKYELVKDIGSGNFGVARLMRHKETKELVAMKYIERGHKIDENVAREIINHRSLRHPNI 60
Query: 61 IRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQIC 120
IRFKEV++TPTHL IVMEYAAGGELF+RIC+AGRFSEDEARYFFQQLISGV YCH+MQIC
Sbjct: 61 IRFKEVLVTPTHLAIVMEYAAGGELFERICNAGRFSEDEARYFFQQLISGVHYCHNMQIC 120
Query: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL 180
HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL
Sbjct: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL 180
Query: 181 ADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRI 240
ADVWSCGVTLYVMLVGAYPFEDQ+DPKNFRKTI RIMAVQYKIPDYVHISQ+CRHLLSRI
Sbjct: 181 ADVWSCGVTLYVMLVGAYPFEDQEDPKNFRKTIQRIMAVQYKIPDYVHISQDCRHLLSRI 240
Query: 241 FVASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENPTYSLQSIEDIMKIVEEAK 300
FVA+PARRITIKEIKSHPWFLKNLPRELTE AQA YYR+ENPT+SLQS+E+IMKIVEEAK
Sbjct: 241 FVANPARRITIKEIKSHPWFLKNLPRELTEAAQAAYYRRENPTFSLQSVEEIMKIVEEAK 300
Query: 301 NPPQASRSVGGFGWGGEEDDDE-----INEAEAELEEDEYEKRVKEAQESGEIHV 350
P ASRSV GFGWGGEE+++E E E E EEDEYEK+VK+A ESGE+ +
Sbjct: 301 TPAPASRSVSGFGWGGEEEEEEKEGDVEEEEEDEEEEDEYEKQVKQAHESGEVRL 355
>emb|CAE54588.1| serin/threonine protein kinase [Fagus sylvatica]
Length = 353
Score = 623 bits (1607), Expect = e-177
Identities = 307/355 (86%), Positives = 332/355 (93%), Gaps = 6/355 (1%)
Query: 1 MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNI 60
MEKYE+VKDIGSGNFGVARLMR+K++K+LVAMKYIERG KIDENVAREIINHRSLRHPNI
Sbjct: 1 MEKYELVKDIGSGNFGVARLMRNKESKELVAMKYIERGRKIDENVAREIINHRSLRHPNI 60
Query: 61 IRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQIC 120
IRFKEVVLTPTHL IVMEYAAGGELF+RIC+AGRFSEDEARYFFQQLISGV YCHSMQIC
Sbjct: 61 IRFKEVVLTPTHLAIVMEYAAGGELFERICAAGRFSEDEARYFFQQLISGVPYCHSMQIC 120
Query: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL 180
HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL
Sbjct: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL 180
Query: 181 ADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRI 240
ADVWSCGVTLYVMLVGAYPFED +DPKNFRK INRIMAVQYKIPDYVHISQEC+HLLSRI
Sbjct: 181 ADVWSCGVTLYVMLVGAYPFEDPEDPKNFRKPINRIMAVQYKIPDYVHISQECKHLLSRI 240
Query: 241 FVASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENPTYSLQSIEDIMKIVEEAK 300
F A+P++RITIKEIK+HPWFLKNLPRELTE AQ +YY+KENP++SLQS +I KIVE+AK
Sbjct: 241 FAANPSKRITIKEIKNHPWFLKNLPRELTEAAQIMYYKKENPSFSLQSDREITKIVEDAK 300
Query: 301 NPPQASRSVGGFGWGGEEDDD----EINEAEAELEEDEYEKRVKEAQESGEIHVN 351
PP SRS+GGFGWGGEED D E+N AE E EEDEYEK+VKEAQESG++ V+
Sbjct: 301 IPPPTSRSIGGFGWGGEEDGDGKEEEVN-AEDE-EEDEYEKKVKEAQESGDVQVS 353
>gb|AAB68961.1| protein kinase 3 [Glycine max] gi|1362051|pir||S56716 protein
kinase SPK-3 (EC 2.7.1.-) - soybean
Length = 351
Score = 619 bits (1596), Expect = e-176
Identities = 298/351 (84%), Positives = 326/351 (91%)
Query: 1 MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNI 60
M+KYE VKD+G+GNFGVARLMR+K+TK+LVAMKYIERG KIDENVAREIINHRSLRHPNI
Sbjct: 1 MDKYEAVKDLGAGNFGVARLMRNKETKELVAMKYIERGQKIDENVAREIINHRSLRHPNI 60
Query: 61 IRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQIC 120
IRFKEVVLTPTHL IVMEYAAGGELF+RIC+AGRFSEDEARYFFQQLISGV YCH+MQIC
Sbjct: 61 IRFKEVVLTPTHLAIVMEYAAGGELFERICNAGRFSEDEARYFFQQLISGVHYCHAMQIC 120
Query: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL 180
HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL
Sbjct: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL 180
Query: 181 ADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRI 240
ADVWSCGVTLYVMLVGAYPFEDQDDP+NFRKTI RIMAVQYKIPDYVHISQ+CRHLLSRI
Sbjct: 181 ADVWSCGVTLYVMLVGAYPFEDQDDPRNFRKTIQRIMAVQYKIPDYVHISQDCRHLLSRI 240
Query: 241 FVASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENPTYSLQSIEDIMKIVEEAK 300
FVA+P RRI++KEIKSHPWFLKNLPRELTE AQAVYY++ NP++S+QS+E+IMKIV EA+
Sbjct: 241 FVANPLRRISLKEIKSHPWFLKNLPRELTESAQAVYYQRGNPSFSIQSVEEIMKIVGEAR 300
Query: 301 NPPQASRSVGGFGWGGEEDDDEINEAEAELEEDEYEKRVKEAQESGEIHVN 351
+PP SR V GFGW GEED+ E + E E EEDEY+KRVKE SGE ++
Sbjct: 301 DPPPVSRPVKGFGWDGEEDEGEEDVEEEEDEEDEYDKRVKEVHASGEFQIS 351
>emb|CAA06503.1| protein kinase [Craterostigma plantagineum] gi|7434329|pir||T09738
protein kinase PK1 (EC 2.7.1.-) - Craterostigma
plantagineum
Length = 355
Score = 614 bits (1584), Expect = e-174
Identities = 303/351 (86%), Positives = 325/351 (92%), Gaps = 3/351 (0%)
Query: 1 MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNI 60
MEKYE+VKDIGSGNFGVARLMR+K+TK+LVAMKYIERGHKIDENVAREIINHRSLRHPNI
Sbjct: 1 MEKYELVKDIGSGNFGVARLMRNKETKELVAMKYIERGHKIDENVAREIINHRSLRHPNI 60
Query: 61 IRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQIC 120
IRFKEVVLTPTHL IVMEYAAGGELF+RIC+AGRFSEDEARYFFQQLI GV YCH++QIC
Sbjct: 61 IRFKEVVLTPTHLAIVMEYAAGGELFERICNAGRFSEDEARYFFQQLICGVHYCHALQIC 120
Query: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL 180
HRDLKLENTLLDGSPAPRLK CDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL
Sbjct: 121 HRDLKLENTLLDGSPAPRLKSCDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL 180
Query: 181 ADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRI 240
ADVWSCGVTLYVMLVGAYPFEDQ+DP FRKTI IMAVQYKIPDYV ISQ CRHLLSRI
Sbjct: 181 ADVWSCGVTLYVMLVGAYPFEDQEDPFFFRKTILWIMAVQYKIPDYVRISQSCRHLLSRI 240
Query: 241 FVASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENPTYSLQSIEDIMKIVEEAK 300
FVA+P RRITIKEIKSH WFLKNLPRELTE+AQA Y+R++NPTYSLQS E+IMKIVEEAK
Sbjct: 241 FVANPLRRITIKEIKSHLWFLKNLPRELTEVAQAAYFRRDNPTYSLQSEEEIMKIVEEAK 300
Query: 301 NPPQASRSVGGFGWGGEEDDDEI---NEAEAELEEDEYEKRVKEAQESGEI 348
PPQASRS+GGFGWG EED++ + E E+E EEDEYEK+VK A SGE+
Sbjct: 301 VPPQASRSIGGFGWGTEEDEENVEEEEEGESEGEEDEYEKQVKAAHASGEV 351
>emb|CAE54075.1| serine/threonine-protein kinase [Fagus sylvatica]
Length = 359
Score = 610 bits (1574), Expect = e-173
Identities = 289/359 (80%), Positives = 332/359 (91%), Gaps = 8/359 (2%)
Query: 1 MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNI 60
M+KYEVVKD+G+GNFGVARL+RHK+TK+LVAMKYIERG KIDENVAREIINHRSLRHPNI
Sbjct: 1 MDKYEVVKDLGAGNFGVARLLRHKETKELVAMKYIERGLKIDENVAREIINHRSLRHPNI 60
Query: 61 IRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQIC 120
IRFKEVVLTPTHL IVMEYAAGGELF+RIC+AG+FSEDEARYFFQQLISGV+YCHSMQIC
Sbjct: 61 IRFKEVVLTPTHLAIVMEYAAGGELFERICTAGKFSEDEARYFFQQLISGVNYCHSMQIC 120
Query: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL 180
HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL
Sbjct: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL 180
Query: 181 ADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRI 240
ADVWSCGVTLYVMLVGAYPFEDQDDP+NFRKTI +IMAVQYK+PDYVHISQ+C+HL SR+
Sbjct: 181 ADVWSCGVTLYVMLVGAYPFEDQDDPRNFRKTIQKIMAVQYKVPDYVHISQDCKHLFSRL 240
Query: 241 FVASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENPTYSLQSIEDIMKIVEEAK 300
FVA+P+RRIT+KEIK+HPWFLKNLPRELTE +QA+YY+++NP++SLQS+++IMKIV EA+
Sbjct: 241 FVANPSRRITLKEIKNHPWFLKNLPRELTESSQAIYYQRDNPSFSLQSVDEIMKIVGEAR 300
Query: 301 NPPQASRSVGGFGWGGEEDDDEINEAEAEL--------EEDEYEKRVKEAQESGEIHVN 351
NPP +S+ V GF WG +E+++ + +AE+ EEDEY+KRVKE SGE H++
Sbjct: 301 NPPPSSKIVRGFSWGPDENEEGNEDIDAEVEEEEEEEDEEDEYDKRVKEVHASGEFHIS 359
>gb|AAB68962.1| protein kinase [Glycine max]
Length = 349
Score = 607 bits (1566), Expect = e-172
Identities = 293/351 (83%), Positives = 324/351 (91%), Gaps = 2/351 (0%)
Query: 1 MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNI 60
M+KYE VKD+G+GNFGVARLMR+K TK+LVAMKYIERG KIDENVAREI+NHRSLRHPNI
Sbjct: 1 MDKYEAVKDLGAGNFGVARLMRNKVTKELVAMKYIERGPKIDENVAREIMNHRSLRHPNI 60
Query: 61 IRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQIC 120
IR+KEVVLTPTHL IVMEYAAGGELF+RICSAGRFSEDEARYFFQQLISGV +CH+MQIC
Sbjct: 61 IRYKEVVLTPTHLAIVMEYAAGGELFERICSAGRFSEDEARYFFQQLISGVHFCHTMQIC 120
Query: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL 180
HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL
Sbjct: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL 180
Query: 181 ADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRI 240
ADVWSC VTLYVMLVGAYPFEDQDDP+NFRKTI RIMAVQYKIPDYVHISQ+CRHLLSRI
Sbjct: 181 ADVWSCAVTLYVMLVGAYPFEDQDDPRNFRKTIQRIMAVQYKIPDYVHISQDCRHLLSRI 240
Query: 241 FVASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENPTYSLQSIEDIMKIVEEAK 300
FVA+P RRITIKEIK+HPWFL+NLPRELTE AQA+YY++++P + LQS+++IMKIV EA+
Sbjct: 241 FVANPLRRITIKEIKNHPWFLRNLPRELTESAQAIYYQRDSPNFHLQSVDEIMKIVGEAR 300
Query: 301 NPPQASRSVGGFGWGGEEDDDEINEAEAELEEDEYEKRVKEAQESGEIHVN 351
NPP SR++ GFGW GEED DE E E E +EDEY+KRVKE SGE ++
Sbjct: 301 NPPPVSRALKGFGWEGEEDLDE--EVEEEEDEDEYDKRVKEVHASGEFQIS 349
>pir||S60611 probable serine/threonine-specific protein kinase (EC 2.7.1.-) BSK2
- rape gi|289374|gb|AAA33004.1| serine/threonine protein
kinase gi|1097354|prf||2113401B protein kinase
Length = 354
Score = 607 bits (1564), Expect = e-172
Identities = 295/354 (83%), Positives = 326/354 (91%), Gaps = 6/354 (1%)
Query: 1 MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNI 60
MEKYE+VKDIG+GNFGVARLM+ K++K+LVAMKYIERG KIDENVAREIINHRSLRHPNI
Sbjct: 1 MEKYELVKDIGAGNFGVARLMKVKNSKELVAMKYIERGPKIDENVAREIINHRSLRHPNI 60
Query: 61 IRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQIC 120
IRFKEVVLTPTHL I MEYAAGGELF+RICSAGRFSEDEARYFFQQLISGVSYCH+MQIC
Sbjct: 61 IRFKEVVLTPTHLAIAMEYAAGGELFERICSAGRFSEDEARYFFQQLISGVSYCHAMQIC 120
Query: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL 180
HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGK+
Sbjct: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKM 180
Query: 181 ADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRI 240
ADVWSCGVTLYVMLVGAYPFEDQ+DPKNFRKTI +IMAVQYKIPDYVHISQ+C+HLLSRI
Sbjct: 181 ADVWSCGVTLYVMLVGAYPFEDQEDPKNFRKTIQKIMAVQYKIPDYVHISQDCKHLLSRI 240
Query: 241 FVASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENPTYSLQSIEDIMKIVEEAK 300
FVA+ +RITI EIK HPWFLKNLPRELTE AQA Y++KENPT+S Q+ E+IMKIV++AK
Sbjct: 241 FVANSLKRITIAEIKKHPWFLKNLPRELTETAQAAYFKKENPTFSPQTAEEIMKIVDDAK 300
Query: 301 NPPQASRSVGGFGWG--GEEDDDEINEAEA----ELEEDEYEKRVKEAQESGEI 348
PP SRS+GGFGWG G+E+++E++E E E EEDEY+K VKEA SGE+
Sbjct: 301 TPPPVSRSIGGFGWGGKGDEEEEEVDEEEVVEEEEDEEDEYDKTVKEAHASGEV 354
>pir||S56720 probable serine/threonine-specific protein kinase (EC 2.7.1.-) BSK1
- rape gi|289372|gb|AAA33003.1| serine/threonine protein
kinase gi|1097353|prf||2113401A protein kinase
Length = 359
Score = 603 bits (1556), Expect = e-171
Identities = 293/359 (81%), Positives = 323/359 (89%), Gaps = 8/359 (2%)
Query: 1 MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNI 60
MEKYE+VKDIG+GNFGVARLM+ KD+K+LVAMKYIERG KIDENVAREI NHRSLRHPNI
Sbjct: 1 MEKYELVKDIGAGNFGVARLMKVKDSKELVAMKYIERGPKIDENVAREIYNHRSLRHPNI 60
Query: 61 IRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQIC 120
IRFKEVVLTPTHL I MEYAAGGELF+RIC AGRFSEDEARYFFQQLISGVSYCH+MQIC
Sbjct: 61 IRFKEVVLTPTHLAIAMEYAAGGELFERICGAGRFSEDEARYFFQQLISGVSYCHAMQIC 120
Query: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL 180
HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGK+
Sbjct: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKM 180
Query: 181 ADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRI 240
ADVWSCGVTLYVMLVGAYPFEDQ+DPKNFRKTI +IMAVQYKIPDYVHISQ+C+HLLSRI
Sbjct: 181 ADVWSCGVTLYVMLVGAYPFEDQEDPKNFRKTIQKIMAVQYKIPDYVHISQDCKHLLSRI 240
Query: 241 FVASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENPTYSLQSIEDIMKIVEEAK 300
FVA+ +RITI EIK HPWF KNLPRELTE AQA Y++KENPT+S Q+ E+IMKIV++AK
Sbjct: 241 FVANSLKRITIAEIKKHPWFTKNLPRELTETAQAAYFKKENPTFSAQTAEEIMKIVDDAK 300
Query: 301 NPPQASRSVGGFGWGGEED-----DDEINEAEAEL---EEDEYEKRVKEAQESGEIHVN 351
PP SRS+GGFGWGGE D ++E++E E E EEDEY+K VKE SGE+ ++
Sbjct: 301 TPPPVSRSIGGFGWGGEGDLEGKEEEEVDEEEVEEEEDEEDEYDKTVKEVHASGEVRIS 359
>gb|AAP68289.1| At1g10940 [Arabidopsis thaliana] gi|21536490|gb|AAM60822.1| Ser/Thr
kinase [Arabidopsis thaliana] gi|20260462|gb|AAM13129.1|
Ser/Thr kinase [Arabidopsis thaliana]
gi|15220251|ref|NP_172563.1| serine/threonine protein
kinase, putative [Arabidopsis thaliana]
gi|1168529|sp|P43291|ASK1_ARATH Serine/threonine-protein
kinase ASK1 gi|1931648|gb|AAB65483.1| Ser/Thr kinase;
69816-71936 [Arabidopsis thaliana]
gi|166882|gb|AAA02840.1| serine/threonine kinase
Length = 363
Score = 602 bits (1551), Expect = e-171
Identities = 293/363 (80%), Positives = 326/363 (89%), Gaps = 12/363 (3%)
Query: 1 MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNI 60
M+KYE+VKDIG+GNFGVARLM+ K++K+LVAMKYIERG KIDENVAREIINHRSLRHPNI
Sbjct: 1 MDKYELVKDIGAGNFGVARLMKVKNSKELVAMKYIERGPKIDENVAREIINHRSLRHPNI 60
Query: 61 IRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQIC 120
IRFKEVVLTPTHL I MEYAAGGELF+RICSAGRFSEDEARYFFQQLISGVSYCH+MQIC
Sbjct: 61 IRFKEVVLTPTHLAIAMEYAAGGELFERICSAGRFSEDEARYFFQQLISGVSYCHAMQIC 120
Query: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL 180
HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGK+
Sbjct: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKM 180
Query: 181 ADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRI 240
ADVWSCGVTLYVMLVGAYPFEDQ+DPKNFRKTI +IMAVQYKIPDYVHISQ+C++LLSRI
Sbjct: 181 ADVWSCGVTLYVMLVGAYPFEDQEDPKNFRKTIQKIMAVQYKIPDYVHISQDCKNLLSRI 240
Query: 241 FVASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENPTYSLQSIEDIMKIVEEAK 300
FVA+ +RITI EIK H WFLKNLPRELTE AQA Y++KENPT+SLQ++E+IMKIV +AK
Sbjct: 241 FVANSLKRITIAEIKKHSWFLKNLPRELTETAQAAYFKKENPTFSLQTVEEIMKIVADAK 300
Query: 301 NPPQASRSVGGFGWGG-------EEDDDEINEAEAELE-----EDEYEKRVKEAQESGEI 348
PP SRS+GGFGWGG EED +++ E E E+E EDEY+K VKE SGE+
Sbjct: 301 TPPPVSRSIGGFGWGGNGDADGKEEDAEDVEEEEEEVEEEEDDEDEYDKTVKEVHASGEV 360
Query: 349 HVN 351
++
Sbjct: 361 RIS 363
>gb|AAM67112.1| putative serine/threonine-protein kinase [Arabidopsis thaliana]
Length = 361
Score = 595 bits (1533), Expect = e-169
Identities = 292/361 (80%), Positives = 324/361 (88%), Gaps = 10/361 (2%)
Query: 1 MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNI 60
M+KYE+VKDIG+GNFGVARLMR K++K+LVAMKYIERG KIDENVAREIINHRSLRHPNI
Sbjct: 1 MDKYELVKDIGAGNFGVARLMRVKNSKELVAMKYIERGPKIDENVAREIINHRSLRHPNI 60
Query: 61 IRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQIC 120
IRFKEVVLTPTH+ I MEYAAGGELF+RICSAGRFSEDEARYFFQQLISGVSYCH+MQIC
Sbjct: 61 IRFKEVVLTPTHIAIAMEYAAGGELFERICSAGRFSEDEARYFFQQLISGVSYCHAMQIC 120
Query: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL 180
HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHS PKSTVGTPAYIAPEVLSR EYDGK+
Sbjct: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSMPKSTVGTPAYIAPEVLSRGEYDGKM 180
Query: 181 ADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRI 240
ADVWSCGVTLYVMLVGAYPFEDQ+DPKNF+KTI RIMAV+YKIPDYVHISQ+C+HLLSRI
Sbjct: 181 ADVWSCGVTLYVMLVGAYPFEDQEDPKNFKKTIQRIMAVKYKIPDYVHISQDCKHLLSRI 240
Query: 241 FVASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENPTYSLQSIEDIMKIVEEAK 300
FV + +RITI +IK HPWFLKNLPRELTE+AQA Y+RKENPT+SLQS+E+IMKIVEEAK
Sbjct: 241 FVTNSNKRITIADIKKHPWFLKNLPRELTEIAQAAYFRKENPTFSLQSVEEIMKIVEEAK 300
Query: 301 NPPQASRSVGGFGWGGEED--------DDEINEAEAEL-EEDEYEKRVKEAQES-GEIHV 350
P + SRS+G FGWGG ED ++E+ E E E EEDEY+K VK+ S GE+ V
Sbjct: 301 TPARVSRSIGAFGWGGGEDAEGKEEDAEEEVEEVEEEEDEEDEYDKTVKQVHASMGEVRV 360
Query: 351 N 351
+
Sbjct: 361 S 361
>gb|AAO00906.1| putative serine/threonine-protein kinase [Arabidopsis thaliana]
gi|18252229|gb|AAL61947.1| putative
serine/threonine-protein kinase [Arabidopsis thaliana]
gi|15219836|ref|NP_176290.1| serine/threonine protein
kinase, putative [Arabidopsis thaliana]
gi|30696495|ref|NP_849834.1| serine/threonine protein
kinase, putative [Arabidopsis thaliana]
gi|12323341|gb|AAG51649.1| putative
serine/threonine-protein kinase; 36981-34483
[Arabidopsis thaliana] gi|25287615|pir||H96634
hypothetical protein T7P1.8 [imported] - Arabidopsis
thaliana
Length = 361
Score = 594 bits (1532), Expect = e-168
Identities = 292/361 (80%), Positives = 324/361 (88%), Gaps = 10/361 (2%)
Query: 1 MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNI 60
M+KYE+VKDIG+GNFGVARLMR K++K+LVAMKYIERG KIDENVAREIINHRSLRHPNI
Sbjct: 1 MDKYELVKDIGAGNFGVARLMRVKNSKELVAMKYIERGPKIDENVAREIINHRSLRHPNI 60
Query: 61 IRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQIC 120
IRFKEVVLTPTH+ I MEYAAGGELF+RICSAGRFSEDEARYFFQQLISGVSYCH+MQIC
Sbjct: 61 IRFKEVVLTPTHIAIAMEYAAGGELFERICSAGRFSEDEARYFFQQLISGVSYCHAMQIC 120
Query: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL 180
HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHS PKSTVGTPAYIAPEVLSR EYDGK+
Sbjct: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSMPKSTVGTPAYIAPEVLSRGEYDGKM 180
Query: 181 ADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRI 240
ADVWSCGVTLYVMLVGAYPFEDQ+DPKNF+KTI RIMAV+YKIPDYVHISQ+C+HLLSRI
Sbjct: 181 ADVWSCGVTLYVMLVGAYPFEDQEDPKNFKKTIQRIMAVKYKIPDYVHISQDCKHLLSRI 240
Query: 241 FVASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENPTYSLQSIEDIMKIVEEAK 300
FV + +RITI +IK HPWFLKNLPRELTE+AQA Y+RKENPT+SLQS+E+IMKIVEEAK
Sbjct: 241 FVTNSNKRITIGDIKKHPWFLKNLPRELTEIAQAAYFRKENPTFSLQSVEEIMKIVEEAK 300
Query: 301 NPPQASRSVGGFGWGGEED--------DDEINEAEAEL-EEDEYEKRVKEAQES-GEIHV 350
P + SRS+G FGWGG ED ++E+ E E E EEDEY+K VK+ S GE+ V
Sbjct: 301 TPARVSRSIGAFGWGGGEDAEGKEEDAEEEVEEVEEEEDEEDEYDKTVKQVHASMGEVRV 360
Query: 351 N 351
+
Sbjct: 361 S 361
>emb|CAE03036.2| OSJNBa0084A10.11 [Oryza sativa (japonica cultivar-group)]
gi|50924380|ref|XP_472550.1| OSJNBa0084A10.11 [Oryza
sativa (japonica cultivar-group)]
gi|46917342|dbj|BAD18003.1| serine/threonine protein
kinase SAPK7 [Oryza sativa (japonica cultivar-group)]
Length = 359
Score = 589 bits (1519), Expect = e-167
Identities = 288/360 (80%), Positives = 324/360 (90%), Gaps = 10/360 (2%)
Query: 1 MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNI 60
ME+YE++KDIG+GNFGVARLMR+K+TK+LVAMKYI RG KIDENVAREIINHRSLRHPNI
Sbjct: 1 MERYELLKDIGAGNFGVARLMRNKETKELVAMKYIPRGLKIDENVAREIINHRSLRHPNI 60
Query: 61 IRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQIC 120
IRFKEVV+TPTHL IVMEYAAGGELFDRIC+AGRFSEDEARYFFQQLI GVSYCH MQIC
Sbjct: 61 IRFKEVVVTPTHLAIVMEYAAGGELFDRICNAGRFSEDEARYFFQQLICGVSYCHFMQIC 120
Query: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL 180
HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHS+PKSTVGTPAYIAPEVLSRREYDGK
Sbjct: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSKPKSTVGTPAYIAPEVLSRREYDGKT 180
Query: 181 ADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRI 240
ADVWSCGVTLYVMLVGAYPFED DDPKNFRKTI RIM++QYKIP+YVH+SQ+CR LLSRI
Sbjct: 181 ADVWSCGVTLYVMLVGAYPFEDPDDPKNFRKTIGRIMSIQYKIPEYVHVSQDCRQLLSRI 240
Query: 241 FVASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKEN--PTYSLQSIEDIMKIVEE 298
FVA+PA+RITI+EI++HPWFLKNLPRELTE AQA+YY+K+N PTYS+QS+E+IMKIVEE
Sbjct: 241 FVANPAKRITIREIRNHPWFLKNLPRELTEAAQAMYYKKDNSAPTYSVQSVEEIMKIVEE 300
Query: 299 AKNPPQASRSVGGFGWGGEEDDDEINEAEAELE-------EDEYEKRVKEAQESGEIHVN 351
A+ PP++S V GFGW EED+ E N + E E EDEY+K+VK+ SGE ++
Sbjct: 301 ARTPPRSSTPVAGFGW-QEEDEQEDNSKKPEEEQEEEEDAEDEYDKQVKQVHASGEFQLS 359
>gb|AAU43772.1| putative salt-inducible protein kinase [Zea mays]
Length = 364
Score = 582 bits (1499), Expect = e-165
Identities = 286/358 (79%), Positives = 318/358 (87%), Gaps = 11/358 (3%)
Query: 1 MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNI 60
MEKYE++KDIGSGNFGVARLMR+KDTK+LVAMKYI RG KIDENVAREIINHRSLRH NI
Sbjct: 1 MEKYELLKDIGSGNFGVARLMRNKDTKELVAMKYIPRGLKIDENVAREIINHRSLRHHNI 60
Query: 61 IRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQIC 120
IRFKEVVLTPTHL IVMEYAAGGELFDRICSAGRFSEDEARYFFQQLI GVSYCH MQIC
Sbjct: 61 IRFKEVVLTPTHLAIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLICGVSYCHLMQIC 120
Query: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL 180
HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHS+PKSTVGTPAYIAPEVLSRREYDGK+
Sbjct: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSKPKSTVGTPAYIAPEVLSRREYDGKM 180
Query: 181 ADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRI 240
ADVWSCGVTLYVMLVGAYPFED DDPKNFRKTI RI+++QY+IP+YVHISQ+CR LL+RI
Sbjct: 181 ADVWSCGVTLYVMLVGAYPFEDPDDPKNFRKTIGRIVSIQYQIPEYVHISQDCRQLLARI 240
Query: 241 FVASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKEN--PTYSLQSIEDIMKIVEE 298
FVA+PA+RITI+EI++HPWFL+NLPRELTE AQA YY+K+N PT+S Q++E+IMKIVEE
Sbjct: 241 FVANPAKRITIREIRNHPWFLRNLPRELTEAAQAKYYKKDNSAPTFSDQTVEEIMKIVEE 300
Query: 299 AKNPPQASRSVGGFGWGGEE---------DDDEINEAEAELEEDEYEKRVKEAQESGE 347
A+ PPQ+S V GFGW EE DD++ E E EDEY+K+VK SG+
Sbjct: 301 ARTPPQSSTPVAGFGWAEEEEQEDGKRSDDDEQYGEDEDYDGEDEYDKQVKHVHASGD 358
>gb|AAM65503.1| serine/threonine-protein kinase [Arabidopsis thaliana]
Length = 358
Score = 580 bits (1495), Expect = e-164
Identities = 284/354 (80%), Positives = 312/354 (87%), Gaps = 7/354 (1%)
Query: 1 MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNI 60
M+KYEVVKD+G+GNFGVARL+RHK+TK+LVAMKYIERG KIDENVAREIINHRSLRHPNI
Sbjct: 1 MDKYEVVKDLGAGNFGVARLLRHKETKELVAMKYIERGRKIDENVAREIINHRSLRHPNI 60
Query: 61 IRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQIC 120
IRFKEV+LTPTHL IVMEYA+GGELF+RIC+AGRFSE EARYFFQQLI GV YCHS+QIC
Sbjct: 61 IRFKEVILTPTHLAIVMEYASGGELFERICNAGRFSEAEARYFFQQLICGVDYCHSLQIC 120
Query: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL 180
HRDLKLENTLLDGSPAP LKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGK
Sbjct: 121 HRDLKLENTLLDGSPAPLLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKH 180
Query: 181 ADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRI 240
ADVWSCGVTLYVMLVG YPFED DDP+NFRKTI RIMAVQYKIPDYVHISQECRHLLSRI
Sbjct: 181 ADVWSCGVTLYVMLVGGYPFEDPDDPRNFRKTIQRIMAVQYKIPDYVHISQECRHLLSRI 240
Query: 241 FVASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENPTYSLQSIEDIMKIVEEAK 300
FV + A+RIT+KEIK HPW+LKNLP+ELTE AQA YY++E P++SLQS+EDIMKIV EA+
Sbjct: 241 FVTNSAKRITLKEIKKHPWYLKNLPKELTEPAQAAYYKRETPSFSLQSVEDIMKIVGEAR 300
Query: 301 NPPQASRSVGGFGWGGEEDDDEI-------NEAEAELEEDEYEKRVKEAQESGE 347
NP +S +V GF E+ +DE+ E E E EEDEYEK VKEA E
Sbjct: 301 NPAPSSNAVKGFDDDEEDVEDEVEEEEEEEEEEEEEEEEDEYEKHVKEAHSCQE 354
>dbj|BAB10458.1| serine/threonine-protein kinase [Arabidopsis thaliana]
gi|15242858|ref|NP_201170.1| serine/threonine protein
kinase, putative [Arabidopsis thaliana]
Length = 360
Score = 579 bits (1493), Expect = e-164
Identities = 284/356 (79%), Positives = 312/356 (86%), Gaps = 9/356 (2%)
Query: 1 MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNI 60
M+KYEVVKD+G+GNFGVARL+RHK+TK+LVAMKYIERG KIDENVAREIINHRSLRHPNI
Sbjct: 1 MDKYEVVKDLGAGNFGVARLLRHKETKELVAMKYIERGRKIDENVAREIINHRSLRHPNI 60
Query: 61 IRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQIC 120
IRFKEV+LTPTHL IVMEYA+GGELF+RIC+AGRFSE EARYFFQQLI GV YCHS+QIC
Sbjct: 61 IRFKEVILTPTHLAIVMEYASGGELFERICNAGRFSEAEARYFFQQLICGVDYCHSLQIC 120
Query: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL 180
HRDLKLENTLLDGSPAP LKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGK
Sbjct: 121 HRDLKLENTLLDGSPAPLLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKH 180
Query: 181 ADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRI 240
ADVWSCGVTLYVMLVG YPFED DDP+NFRKTI RIMAVQYKIPDYVHISQECRHLLSRI
Sbjct: 181 ADVWSCGVTLYVMLVGGYPFEDPDDPRNFRKTIQRIMAVQYKIPDYVHISQECRHLLSRI 240
Query: 241 FVASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENPTYSLQSIEDIMKIVEEAK 300
FV + A+RIT+KEIK HPW+LKNLP+ELTE AQA YY++E P++SLQS+EDIMKIV EA+
Sbjct: 241 FVTNSAKRITLKEIKKHPWYLKNLPKELTEPAQAAYYKRETPSFSLQSVEDIMKIVGEAR 300
Query: 301 NPPQASRSVGGFGWGGEEDDDEI---------NEAEAELEEDEYEKRVKEAQESGE 347
NP +S +V GF E+ +DE+ E E E EEDEYEK VKEA E
Sbjct: 301 NPAPSSNAVKGFDDDEEDVEDEVEEEEEEEEEEEEEEEEEEDEYEKHVKEAHSCQE 356
>ref|XP_466158.1| putative osmotic stress-activated protein kinase [Oryza sativa
(japonica cultivar-group)] gi|50725759|dbj|BAD33270.1|
putative osmotic stress-activated protein kinase [Oryza
sativa (japonica cultivar-group)]
gi|46917340|dbj|BAD18002.1| serine/threonine protein
kinase SAPK6 [Oryza sativa (japonica cultivar-group)]
gi|46389873|dbj|BAD15474.1| putative osmotic
stress-activated protein kinase [Oryza sativa (japonica
cultivar-group)]
Length = 365
Score = 576 bits (1484), Expect = e-163
Identities = 286/362 (79%), Positives = 320/362 (88%), Gaps = 14/362 (3%)
Query: 1 MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNI 60
MEKYE++KDIGSGNFGVARLMR+++TK+LVAMKYI RG KIDENVAREIINHRSLRHPNI
Sbjct: 1 MEKYELLKDIGSGNFGVARLMRNRETKELVAMKYIPRGLKIDENVAREIINHRSLRHPNI 60
Query: 61 IRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQIC 120
IRFKEVVLTPTHL IVMEYAAGGELFDRICSAGRFSEDE+RYFFQQLI GVSYCH MQIC
Sbjct: 61 IRFKEVVLTPTHLAIVMEYAAGGELFDRICSAGRFSEDESRYFFQQLICGVSYCHFMQIC 120
Query: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL 180
HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHS+PKSTVGTPAYIAPEVLSRREYDGK+
Sbjct: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSKPKSTVGTPAYIAPEVLSRREYDGKM 180
Query: 181 ADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRI 240
ADVWSCGVTLYVMLVGAYPFED DDPKNFRKTI RI+++QYKIP+YVHISQ+CR LLSRI
Sbjct: 181 ADVWSCGVTLYVMLVGAYPFEDPDDPKNFRKTIGRIVSIQYKIPEYVHISQDCRQLLSRI 240
Query: 241 FVASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENP--TYSLQSIEDIMKIVEE 298
FVA+PA+RITI+EI++HPWF+KNLPRELTE AQA YY+K+N T+S Q++++IMKIV+E
Sbjct: 241 FVANPAKRITIREIRNHPWFMKNLPRELTEAAQAKYYKKDNSARTFSDQTVDEIMKIVQE 300
Query: 299 AKNPPQASRSVGGFGWGGEE--------DDDEINEAEAELE----EDEYEKRVKEAQESG 346
AK PP +S V GFGW EE DDDE + E E E EDEY K+VK+A S
Sbjct: 301 AKTPPPSSTPVAGFGWTEEEEQEDGKNPDDDEGDRDEEEGEEGDSEDEYTKQVKQAHASC 360
Query: 347 EI 348
++
Sbjct: 361 DL 362
>dbj|BAB10008.1| serine/threonine-protein kinase [Arabidopsis thaliana]
gi|15242317|ref|NP_196476.1| serine/threonine protein
kinase (ASK2) [Arabidopsis thaliana]
gi|26454616|sp|P43292|ASK2_ARATH
Serine/threonine-protein kinase ASK2
Length = 353
Score = 572 bits (1474), Expect = e-162
Identities = 280/350 (80%), Positives = 314/350 (89%), Gaps = 4/350 (1%)
Query: 1 MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNI 60
M+KY+VVKD+G+GNFGVARL+RHKDTK+LVAMKYIERG KIDENVAREIINHRSL+HPNI
Sbjct: 1 MDKYDVVKDLGAGNFGVARLLRHKDTKELVAMKYIERGRKIDENVAREIINHRSLKHPNI 60
Query: 61 IRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQIC 120
IRFKEV+LTPTHL IVMEYA+GGELFDRIC+AGRFSE EARYFFQQLI GV YCHS+QIC
Sbjct: 61 IRFKEVILTPTHLAIVMEYASGGELFDRICTAGRFSEAEARYFFQQLICGVDYCHSLQIC 120
Query: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL 180
HRDLKLENTLLDGSPAP LKICDFGYSKSS+LHSRPKSTVGTPAYIAPEVLSRREYDGK
Sbjct: 121 HRDLKLENTLLDGSPAPLLKICDFGYSKSSILHSRPKSTVGTPAYIAPEVLSRREYDGKH 180
Query: 181 ADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRI 240
ADVWSCGVTLYVMLVGAYPFED +DPKNFRKTI RIMAVQYKIPDYVHISQEC+HLLSRI
Sbjct: 181 ADVWSCGVTLYVMLVGAYPFEDPNDPKNFRKTIQRIMAVQYKIPDYVHISQECKHLLSRI 240
Query: 241 FVASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENPTYSLQSIEDIMKIVEEAK 300
FV + A+RIT+KEIK+HPW+LKNLP+EL E AQA YY+++ ++SLQS+EDIMKIV EA+
Sbjct: 241 FVTNSAKRITLKEIKNHPWYLKNLPKELLESAQAAYYKRDT-SFSLQSVEDIMKIVGEAR 299
Query: 301 NPPQASRSVGGFGWGGEEDDDEINEAEAELE---EDEYEKRVKEAQESGE 347
NP ++ +V G G +E+++E EAE E E EDEYEK VKEAQ E
Sbjct: 300 NPAPSTSAVKSSGSGADEEEEEDVEAEVEEEEDDEDEYEKHVKEAQSCQE 349
>emb|CAA78106.1| protein kinase [Arabidopsis thaliana]
Length = 353
Score = 569 bits (1467), Expect = e-161
Identities = 278/350 (79%), Positives = 313/350 (89%), Gaps = 4/350 (1%)
Query: 1 MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNI 60
M+KY+VVKD+G+GNFGVARL+RHKDTK+LVAMKYIERG KIDENVAREIINHRS +HPNI
Sbjct: 1 MDKYDVVKDLGAGNFGVARLLRHKDTKELVAMKYIERGRKIDENVAREIINHRSFKHPNI 60
Query: 61 IRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQIC 120
IRFKEV+LTPTHL IVMEYA+GGELFDRIC+AGRFSE EARYFFQQLI GV YCHS+QIC
Sbjct: 61 IRFKEVILTPTHLAIVMEYASGGELFDRICTAGRFSEAEARYFFQQLICGVDYCHSLQIC 120
Query: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL 180
HRDLKLENTLLDG+PAP LKICDFGYSKSS+LHSRPKSTVGTPAYIAPEVLSRREYDGK
Sbjct: 121 HRDLKLENTLLDGAPAPLLKICDFGYSKSSILHSRPKSTVGTPAYIAPEVLSRREYDGKH 180
Query: 181 ADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRI 240
ADVWSCGVTLYVMLVGAYPFED +DPKNFRKTI RIMAVQYKIPDYVHISQEC+HLLSRI
Sbjct: 181 ADVWSCGVTLYVMLVGAYPFEDPNDPKNFRKTIQRIMAVQYKIPDYVHISQECKHLLSRI 240
Query: 241 FVASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENPTYSLQSIEDIMKIVEEAK 300
FV + A+RIT+KEIK+HPW+LKNLP+EL E AQA YY+++ ++SLQS+EDIMKIV EA+
Sbjct: 241 FVTNSAKRITLKEIKNHPWYLKNLPKELLESAQAAYYKRDT-SFSLQSVEDIMKIVGEAR 299
Query: 301 NPPQASRSVGGFGWGGEEDDDEINEAEAELE---EDEYEKRVKEAQESGE 347
NP ++ +V G G +E+++E EAE E E EDEYEK VKEAQ E
Sbjct: 300 NPAPSTSAVKSSGSGADEEEEEDVEAEVEEEEDDEDEYEKHVKEAQSCQE 349
>ref|NP_915675.1| putative protein kinase SPK-3 [Oryza sativa (japonica
cultivar-group)] gi|20160874|dbj|BAB89813.1|
serine/threonine protein kinase SAPK4 [Oryza sativa
(japonica cultivar-group)] gi|15408695|dbj|BAB64101.1|
serine/threonine protein kinase SAPK4 [Oryza sativa
(japonica cultivar-group)] gi|46917336|dbj|BAD18000.1|
serine/threonine protein kinase SAPK4 [Oryza sativa
(japonica cultivar-group)]
Length = 360
Score = 560 bits (1443), Expect = e-158
Identities = 277/357 (77%), Positives = 315/357 (87%), Gaps = 7/357 (1%)
Query: 1 MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNI 60
MEKYE V+DIGSGNFGVARLMR+++T++LVA+K IERGH+IDENV REIINHRSLRHPNI
Sbjct: 1 MEKYEAVRDIGSGNFGVARLMRNRETRELVAVKCIERGHRIDENVYREIINHRSLRHPNI 60
Query: 61 IRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQIC 120
IRFKEV+LTPTHL IVME+AAGGELFDRIC GRFSEDEARYFFQQLI GVSYCH MQIC
Sbjct: 61 IRFKEVILTPTHLMIVMEFAAGGELFDRICDRGRFSEDEARYFFQQLICGVSYCHHMQIC 120
Query: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL 180
HRDLKLEN LLDGSPAPRLKICDFGYSKSS+LHSRPKS VGTPAYIAPEVLSRREYDGKL
Sbjct: 121 HRDLKLENVLLDGSPAPRLKICDFGYSKSSVLHSRPKSAVGTPAYIAPEVLSRREYDGKL 180
Query: 181 ADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRI 240
ADVWSCGVTLYVMLVGAYPFEDQDDPKN RKTI RIM+VQYKIPDYVHIS EC+ L++RI
Sbjct: 181 ADVWSCGVTLYVMLVGAYPFEDQDDPKNIRKTIQRIMSVQYKIPDYVHISAECKQLIARI 240
Query: 241 FVASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKEN--PTYSLQSIEDIMKIVEE 298
FV +P RRIT+KEIKSHPWFLKNLPRELTE AQA+YYR++N P++S Q+ E+IMKIV+E
Sbjct: 241 FVNNPLRRITMKEIKSHPWFLKNLPRELTETAQAMYYRRDNSVPSFSDQTSEEIMKIVQE 300
Query: 299 AKNPPQASRSVGGFGWGG----EEDDDEINEAEAELEEDEYEKRVKEAQESGEIHVN 351
A+ P++SR+ G + G E++++E E E EEDEY+KRVKE SGE+ ++
Sbjct: 301 ARTMPKSSRT-GYWSDAGSDEEEKEEEERPEENEEEEEDEYDKRVKEVHASGELRMS 356
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.318 0.136 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 606,349,485
Number of Sequences: 2540612
Number of extensions: 25683270
Number of successful extensions: 171550
Number of sequences better than 10.0: 19751
Number of HSP's better than 10.0 without gapping: 8862
Number of HSP's successfully gapped in prelim test: 10892
Number of HSP's that attempted gapping in prelim test: 124606
Number of HSP's gapped (non-prelim): 24650
length of query: 351
length of database: 863,360,394
effective HSP length: 129
effective length of query: 222
effective length of database: 535,621,446
effective search space: 118907961012
effective search space used: 118907961012
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 75 (33.5 bits)
Medicago: description of AC134322.21


