

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133572.9 + phase: 0
(326 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
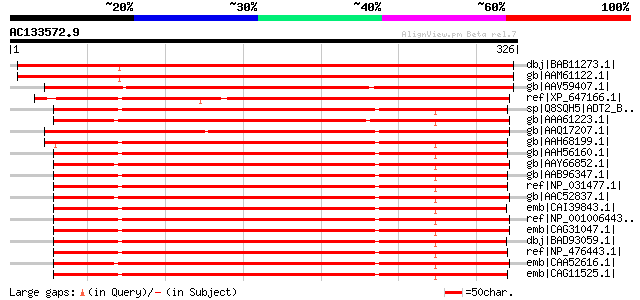
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAB11273.1| ADP/ATP translocase-like protein [Arabidopsis th... 503 e-141
gb|AAM61122.1| ADP/ATP translocase-like protein [Arabidopsis tha... 502 e-141
gb|AAV59407.1| putative ADP/ATP translocase [Oryza sativa (japon... 441 e-122
ref|XP_647166.1| hypothetical protein DDB0201558 [Dictyostelium ... 295 9e-79
sp|Q8SQH5|ADT2_BOVIN ADP,ATP carrier protein 2 (ADP/ATP transloc... 289 9e-77
gb|AAA61223.1| ADP/ADT translocator protein 287 3e-76
gb|AAQ17207.1| ADP/ATP translocase [Branchiostoma belcheri tsing... 286 4e-76
gb|AAH68199.1| SLC25A5 protein [Homo sapiens] 286 4e-76
gb|AAH56160.1| Solute carrier family 25, member 5 [Homo sapiens]... 286 8e-76
gb|AAY66852.1| ADP/ATP translocase [Ixodes scapularis] 286 8e-76
gb|AAB96347.1| ADP/ATP carrier protein (adenine nucleotide trans... 285 1e-75
ref|NP_031477.1| solute carrier family 25, member 5 [Mus musculu... 285 1e-75
gb|AAC52837.1| adenine nucleotide translocase-1 285 1e-75
emb|CAI39843.1| solute carrier family 25 (mitochondrial carrier;... 285 2e-75
ref|NP_001006443.1| similar to ADP/ATP translocase [Gallus gallus] 285 2e-75
emb|CAG31047.1| hypothetical protein [Gallus gallus] 285 2e-75
dbj|BAD93059.1| ADP,ATP carrier protein, liver isoform T2 varian... 285 2e-75
ref|NP_476443.1| solute carrier family 25, member 5 [Rattus norv... 284 2e-75
emb|CAA52616.1| adenine nucleotide carrier [Mus musculus] gi|200... 284 2e-75
emb|CAG11525.1| unnamed protein product [Tetraodon nigroviridis] 284 2e-75
>dbj|BAB11273.1| ADP/ATP translocase-like protein [Arabidopsis thaliana]
gi|15241218|ref|NP_200456.1| mitochondrial substrate
carrier family protein [Arabidopsis thaliana]
Length = 330
Score = 503 bits (1295), Expect = e-141
Identities = 246/323 (76%), Positives = 285/323 (88%), Gaps = 4/323 (1%)
Query: 6 EDPEEKALNLKRKRNSFSSSRFNNFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNL 65
ED E+ + N + + +FQ+DL+AGAVMGG VHTIVAPIERAKLLLQTQESN+
Sbjct: 6 EDEEDPSRNRRNQSPLSLPQTLKHFQKDLLAGAVMGGVVHTIVAPIERAKLLLQTQESNI 65
Query: 66 AIVA----SGRRKFKGMFDCIIRTVREEGVISLWRGNGSSVLRYYPSVALNFSLKDLYKS 121
AIV +G+R+FKGMFD I RTVREEGV+SLWRGNGSSVLRYYPSVALNFSLKDLY+S
Sbjct: 66 AIVGDEGHAGKRRFKGMFDFIFRTVREEGVLSLWRGNGSSVLRYYPSVALNFSLKDLYRS 125
Query: 122 ILRGGNSNPDNIFSGASANFVAGAAAGCTSLILVYPLDIAHTRLAADIGRTEVRQFRGIH 181
ILR +S ++IFSGA ANF+AG+AAGCT+LI+VYPLDIAHTRLAADIG+ E RQFRGIH
Sbjct: 126 ILRNSSSQENHIFSGALANFMAGSAAGCTALIVVYPLDIAHTRLAADIGKPEARQFRGIH 185
Query: 182 HFLATIFQKDGVRGIYRGLPASLHGMVIHRGLYFGGFDTIKEMLSEESKPELALWKRWMV 241
HFL+TI +KDGVRGIYRGLPASLHG++IHRGLYFGGFDT+KE+ SE++KPELALWKRW +
Sbjct: 186 HFLSTIHKKDGVRGIYRGLPASLHGVIIHRGLYFGGFDTVKEIFSEDTKPELALWKRWGL 245
Query: 242 AQAVTTSAGLVSYPLDTVRRRMMMQSGMEHPVYNSTLDCWRKIYRTEGLISFYRGAVSNV 301
AQAVTTSAGL SYPLDTVRRR+MMQSGMEHP+Y STLDCW+KIYR+EGL SFYRGA+SN+
Sbjct: 246 AQAVTTSAGLASYPLDTVRRRIMMQSGMEHPMYRSTLDCWKKIYRSEGLASFYRGALSNM 305
Query: 302 FRSTGAAAILVLYDEVKKFMNLG 324
FRSTG+AAILV YDEVK+F+N G
Sbjct: 306 FRSTGSAAILVFYDEVKRFLNWG 328
>gb|AAM61122.1| ADP/ATP translocase-like protein [Arabidopsis thaliana]
Length = 330
Score = 502 bits (1293), Expect = e-141
Identities = 246/323 (76%), Positives = 285/323 (88%), Gaps = 4/323 (1%)
Query: 6 EDPEEKALNLKRKRNSFSSSRFNNFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNL 65
ED E+ + N + + +FQ+DL+AGAVMGG VHTIVAPIERAKLLLQTQESN+
Sbjct: 6 EDEEDPSRNRRNQSPLSLPQTLKHFQKDLLAGAVMGGVVHTIVAPIERAKLLLQTQESNI 65
Query: 66 AIVA----SGRRKFKGMFDCIIRTVREEGVISLWRGNGSSVLRYYPSVALNFSLKDLYKS 121
AIV +G+R+FKGMFD I RTVREEGV+SLWRGNGSSVLRYYPSVALNFSLKDLY+S
Sbjct: 66 AIVGDEGHAGKRRFKGMFDFIFRTVREEGVLSLWRGNGSSVLRYYPSVALNFSLKDLYRS 125
Query: 122 ILRGGNSNPDNIFSGASANFVAGAAAGCTSLILVYPLDIAHTRLAADIGRTEVRQFRGIH 181
ILR +S ++IFSGA ANF+AG+AAGCT+LI+VYPLDIAHTRLAADIG+ E RQFRGIH
Sbjct: 126 ILRNSSSQENHIFSGALANFMAGSAAGCTALIVVYPLDIAHTRLAADIGKPESRQFRGIH 185
Query: 182 HFLATIFQKDGVRGIYRGLPASLHGMVIHRGLYFGGFDTIKEMLSEESKPELALWKRWMV 241
HFL+TI +KDGVRGIYRGLPASLHG++IHRGLYFGGFDT+KE+ SE++KPELALWKRW +
Sbjct: 186 HFLSTIHKKDGVRGIYRGLPASLHGVIIHRGLYFGGFDTVKEIFSEDTKPELALWKRWGL 245
Query: 242 AQAVTTSAGLVSYPLDTVRRRMMMQSGMEHPVYNSTLDCWRKIYRTEGLISFYRGAVSNV 301
AQAVTTSAGL SYPLDTVRRR+MMQSGMEHP+Y STLDCW+KIYR+EGL SFYRGA+SN+
Sbjct: 246 AQAVTTSAGLASYPLDTVRRRIMMQSGMEHPMYRSTLDCWKKIYRSEGLASFYRGALSNM 305
Query: 302 FRSTGAAAILVLYDEVKKFMNLG 324
FRSTG+AAILV YDEVK+F+N G
Sbjct: 306 FRSTGSAAILVFYDEVKRFLNWG 328
>gb|AAV59407.1| putative ADP/ATP translocase [Oryza sativa (japonica
cultivar-group)] gi|50932533|ref|XP_475794.1| putative
ADP/ATP translocase [Oryza sativa (japonica
cultivar-group)]
Length = 363
Score = 441 bits (1134), Expect = e-122
Identities = 215/303 (70%), Positives = 260/303 (84%), Gaps = 5/303 (1%)
Query: 23 SSSRFNNFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCI 82
+++R F+RDL+AGA MGGAVHT+VAPIERAKLLLQTQ+ N A++ RR F+G DC+
Sbjct: 5 AAARVWEFERDLVAGAAMGGAVHTVVAPIERAKLLLQTQDGNAALLGRARR-FRGFADCV 63
Query: 83 IRTVREEGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSG-ASANF 141
RTVR+EGV+SLWRGNG++V+RYYPSVALNFSLKDLY+SIL+ ++ DN FS A NF
Sbjct: 64 ARTVRDEGVLSLWRGNGTAVIRYYPSVALNFSLKDLYRSILKDAGTSADNKFSSIALTNF 123
Query: 142 VAGAAAGCTSLILVYPLDIAHTRLAADIGRTEVRQFRGIHHFLATIFQKDGVRGIYRGLP 201
+AGAAAGCT+L+L+YPLDIAHTRLAADIGRT+ RQFRGI HF+ TI+ K+G+RGIYRGLP
Sbjct: 124 IAGAAAGCTTLVLIYPLDIAHTRLAADIGRTDTRQFRGICHFVQTIYNKNGIRGIYRGLP 183
Query: 202 ASLHGMVIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRR 261
ASL GMV+HRGLYFGGFDT K+++ P LW+RW+ AQAVT+ AGL+SYPLDTVRR
Sbjct: 184 ASLQGMVVHRGLYFGGFDTAKDVMVPLDSP---LWQRWVTAQAVTSMAGLISYPLDTVRR 240
Query: 262 RMMMQSGMEHPVYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVKKFM 321
RMMMQSGM+ +Y+STLDCWRKIY+ EG+ SFYRGA+SN+FRSTGAAAILVLYDEVKKFM
Sbjct: 241 RMMMQSGMDVQMYSSTLDCWRKIYKVEGIKSFYRGALSNMFRSTGAAAILVLYDEVKKFM 300
Query: 322 NLG 324
+ G
Sbjct: 301 DRG 303
>ref|XP_647166.1| hypothetical protein DDB0201558 [Dictyostelium discoideum]
gi|3916729|gb|AAC79081.1| ADP/ATP translocase
[Dictyostelium discoideum] gi|3885438|gb|AAC77879.1|
ADP/ATP translocase [Dictyostelium discoideum]
gi|60475245|gb|EAL73180.1| hypothetical protein
DDB0201558 [Dictyostelium discoideum]
Length = 309
Score = 295 bits (756), Expect = 9e-79
Identities = 148/307 (48%), Positives = 206/307 (66%), Gaps = 12/307 (3%)
Query: 17 RKRNSFSSSRFNNFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFK 76
+K+N SS F +D + G GG TIVAPIER KLLLQ Q ++ I A +++K
Sbjct: 4 QKKNDVSS-----FVKDSLIGGTAGGVSKTIVAPIERVKLLLQVQSASTQIAAD--KQYK 56
Query: 77 GMFDCIIRTVREEGVISLWRGNGSSVLRYYPSVALNFSLKDLYKS--ILRGGNSNPDNIF 134
G+ DC +R +E+GVISLWRGN ++V+RY+P+ ALNF+ KD YK + NP F
Sbjct: 57 GIVDCFVRVSKEQGVISLWRGNLANVIRYFPTQALNFAFKDKYKKFFVRHTAKENPTKFF 116
Query: 135 SGASANFVAGAAAGCTSLILVYPLDIAHTRLAADIGRTEVRQFRGIHHFLATIFQKDGVR 194
G N ++G AAG TSL+ VYPLD A TRLAAD+G RQF G+ + +++I+++DG+
Sbjct: 117 IG---NLLSGGAAGATSLLFVYPLDFARTRLAADVGTGSARQFTGLGNCISSIYKRDGLI 173
Query: 195 GIYRGLPASLHGMVIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSY 254
G+YRG S+ G+ ++R +FGG+DT K +L E+ + + W W +AQ VTT AG+VSY
Sbjct: 174 GLYRGFGVSVGGIFVYRAAFFGGYDTAKGILLGENNKKASFWASWGIAQVVTTIAGVVSY 233
Query: 255 PLDTVRRRMMMQSGMEHPVYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLY 314
P DTVRRRMMMQ+G +Y+ST DCW KI EG +F++GA+SN R +G A +LV+Y
Sbjct: 234 PFDTVRRRMMMQAGRADILYSSTWDCWVKIATREGPTAFFKGALSNAIRGSGGALVLVIY 293
Query: 315 DEVKKFM 321
DE++K M
Sbjct: 294 DEIQKLM 300
>sp|Q8SQH5|ADT2_BOVIN ADP,ATP carrier protein 2 (ADP/ATP translocase 2) (Adenine
nucleotide translocator 2) (ANT 2) (Solute carrier
family 25, member 5) gi|32189334|ref|NP_777084.1| solute
carrier family 25 member 5 [Bos taurus]
gi|18642496|dbj|BAB84673.1| adenine nucleotide
translocator 2 [Bos taurus]
Length = 298
Score = 289 bits (739), Expect = 9e-77
Identities = 147/295 (49%), Positives = 196/295 (65%), Gaps = 7/295 (2%)
Query: 29 NFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVRE 88
+F +D +AG V T VAPIER KLLLQ Q ++ I A +++KG+ DC++R +E
Sbjct: 7 SFAKDFLAGGVAAAISKTAVAPIERVKLLLQVQHASKQITAD--KQYKGIIDCVVRIPKE 64
Query: 89 EGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAG 148
+GV+S WRGN ++V+RY+P+ ALNF+ KD YK I GG + + N +G AAG
Sbjct: 65 QGVLSFWRGNLANVIRYFPTQALNFAFKDKYKQIFLGGVDKRTQFWRYFAGNLASGGAAG 124
Query: 149 CTSLILVYPLDIAHTRLAADIGRTEV-RQFRGIHHFLATIFQKDGVRGIYRGLPASLHGM 207
TSL VYPLD A TRLAAD+G+ R+FRG+ L I++ DG+RG+Y+G S+ G+
Sbjct: 125 ATSLCFVYPLDFARTRLAADVGKAGAEREFRGLGDCLVKIYKSDGIRGLYQGFNVSVQGI 184
Query: 208 VIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQS 267
+I+R YFG +DT K ML + + + WM+AQ+VT AGL SYP DTVRRRMMMQS
Sbjct: 185 IIYRAAYFGIYDTAKGMLPDPKNTHIFI--SWMIAQSVTAVAGLTSYPFDTVRRRMMMQS 242
Query: 268 GMEHP--VYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVKKF 320
G + +Y TLDCWRKI R EG +F++GA SNV R G A +LVLYDE+KKF
Sbjct: 243 GRKGTDIMYTGTLDCWRKIARDEGAKAFFKGAWSNVLRGMGGAFVLVLYDEIKKF 297
Score = 33.9 bits (76), Expect = 6.7
Identities = 23/88 (26%), Positives = 45/88 (51%), Gaps = 7/88 (7%)
Query: 241 VAQAVTTSAGLVSYPLDTVRRRMMMQSGMEHPV----YNSTLDCWRKIYRTEGLISFYRG 296
VA A++ +A P++ V+ + +Q + Y +DC +I + +G++SF+RG
Sbjct: 17 VAAAISKTAVA---PIERVKLLLQVQHASKQITADKQYKGIIDCVVRIPKEQGVLSFWRG 73
Query: 297 AVSNVFRSTGAAAILVLYDEVKKFMNLG 324
++NV R A+ + + K + LG
Sbjct: 74 NLANVIRYFPTQALNFAFKDKYKQIFLG 101
>gb|AAA61223.1| ADP/ADT translocator protein
Length = 297
Score = 287 bits (735), Expect = 3e-76
Identities = 143/295 (48%), Positives = 198/295 (66%), Gaps = 6/295 (2%)
Query: 29 NFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVRE 88
+F +D +AGAV T VAPIER KLLLQ Q ++ I S +++KG+ DC++R +E
Sbjct: 7 SFLKDFLAGAVAAAVSKTAVAPIERVKLLLQVQHASKQI--SAEKQYKGIIDCVVRIPKE 64
Query: 89 EGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAG 148
+G +S WRGN ++V+RY+P+ ALNF+ KD YK + GG + + N +G AAG
Sbjct: 65 QGFLSFWRGNLANVIRYFPTQALNFAFKDKYKQLFLGGVDRHKQFWRYFAGNLASGGAAG 124
Query: 149 CTSLILVYPLDIAHTRLAADIGRTEVRQFRGIHHFLATIFQKDGVRGIYRGLPASLHGMV 208
TSL VYPLD A TRLAAD+GR R+F G+ + IF+ DG+RG+Y+G S+ G++
Sbjct: 125 ATSLCFVYPLDFARTRLAADVGRRAQREFHGLGDCIIKIFKSDGLRGLYQGFNVSVQGII 184
Query: 209 IHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQSG 268
I+R YFG +DT K ML + + ++ WM+AQ+VT AGL+SYP DTVRRRMMMQSG
Sbjct: 185 IYRAAYFGVYDTAKGMLPDPK--NVHIFVSWMIAQSVTAVAGLLSYPFDTVRRRMMMQSG 242
Query: 269 MEHP--VYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVKKFM 321
+ +Y T+DCWRKI + EG +F++GA SNV R G A +LVLYDE+KK++
Sbjct: 243 RKGADIMYTGTVDCWRKIAKDEGAKAFFKGAWSNVLRGMGGAFVLVLYDEIKKYV 297
Score = 34.3 bits (77), Expect = 5.1
Identities = 24/88 (27%), Positives = 44/88 (49%), Gaps = 7/88 (7%)
Query: 241 VAQAVTTSAGLVSYPLDTVRRRMMMQSGMEH----PVYNSTLDCWRKIYRTEGLISFYRG 296
VA AV+ +A P++ V+ + +Q + Y +DC +I + +G +SF+RG
Sbjct: 17 VAAAVSKTAVA---PIERVKLLLQVQHASKQISAEKQYKGIIDCVVRIPKEQGFLSFWRG 73
Query: 297 AVSNVFRSTGAAAILVLYDEVKKFMNLG 324
++NV R A+ + + K + LG
Sbjct: 74 NLANVIRYFPTQALNFAFKDKYKQLFLG 101
>gb|AAQ17207.1| ADP/ATP translocase [Branchiostoma belcheri tsingtaunese]
Length = 298
Score = 286 bits (733), Expect = 4e-76
Identities = 143/300 (47%), Positives = 200/300 (66%), Gaps = 4/300 (1%)
Query: 23 SSSRFNNFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCI 82
+S + +F +D +AG + T VAPIER KLLLQ Q+++ + A+G +K+ G+ DC+
Sbjct: 2 ASDQVVSFLKDFLAGGISAAIAKTAVAPIERVKLLLQVQDASKQMQAAGAKKYTGIIDCV 61
Query: 83 IRTVREEGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFV 142
R +E+G +S WRGN ++V+RY+P+ ALNF+ KD YK + GG + + + N
Sbjct: 62 TRIPKEQGFLSFWRGNLANVIRYFPTQALNFAFKDKYKQLFLGG-VDKNRFWRYFVGNLA 120
Query: 143 AGAAAGCTSLILVYPLDIAHTRLAADIGR-TEVRQFRGIHHFLATIFQKDGVRGIYRGLP 201
+G AAG TSL VYPLD A TRLAADIG+ R + G+ + L ++ DG+ G+YRG
Sbjct: 121 SGGAAGATSLCFVYPLDFARTRLAADIGKGAGERLYSGLGNCLMQTYRSDGLYGLYRGFS 180
Query: 202 ASLHGMVIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRR 261
S+ G++I+R YFG FDT K M+ + K + WM+AQ VTTSAG+VSYP DTVRR
Sbjct: 181 VSVQGIIIYRAAYFGCFDTAKGMMPDPKKTPFVV--SWMIAQVVTTSAGIVSYPFDTVRR 238
Query: 262 RMMMQSGMEHPVYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVKKFM 321
RMMMQSG++ +Y +TLDCWRKI + EG + ++GA SNV R G A +LVLYDE+KK +
Sbjct: 239 RMMMQSGLKELIYKNTLDCWRKIIKDEGGSALFKGAFSNVLRGMGGAFVLVLYDELKKII 298
Score = 33.9 bits (76), Expect = 6.7
Identities = 16/51 (31%), Positives = 28/51 (54%)
Query: 274 YNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVKKFMNLG 324
Y +DC +I + +G +SF+RG ++NV R A+ + + K + LG
Sbjct: 54 YTGIIDCVTRIPKEQGFLSFWRGNLANVIRYFPTQALNFAFKDKYKQLFLG 104
>gb|AAH68199.1| SLC25A5 protein [Homo sapiens]
Length = 323
Score = 286 bits (733), Expect = 4e-76
Identities = 149/307 (48%), Positives = 200/307 (64%), Gaps = 13/307 (4%)
Query: 23 SSSRFN------NFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFK 76
S+S FN +F +D +AG V T VAPIER KLLLQ Q ++ I A +++K
Sbjct: 20 SASSFNMTDAAVSFAKDFLAGGVAAAISKTAVAPIERVKLLLQVQHASKQITAD--KQYK 77
Query: 77 GMFDCIIRTVREEGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSG 136
G+ DC++R +E+GV+S WRGN ++V+RY+P+ ALNF+ KD YK I GG +
Sbjct: 78 GIIDCVVRIPKEQGVLSFWRGNLANVIRYFPTQALNFAFKDKYKQIFLGGVDKRTQFWLY 137
Query: 137 ASANFVAGAAAGCTSLILVYPLDIAHTRLAADIGRTEV-RQFRGIHHFLATIFQKDGVRG 195
+ N +G AAG TSL VYPLD A TRLAAD+G+ R+FRG+ L I++ DG++G
Sbjct: 138 FAGNLASGGAAGATSLCFVYPLDFARTRLAADVGKAGAEREFRGLGDCLVKIYKSDGIKG 197
Query: 196 IYRGLPASLHGMVIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYP 255
+Y+G S+ G++I+R YFG +DT K ML + + + WM+AQ VT AGL SYP
Sbjct: 198 LYQGFNVSVQGIIIYRAAYFGIYDTAKGMLPDPKNTHIVI--SWMIAQTVTAVAGLTSYP 255
Query: 256 LDTVRRRMMMQSGMEHP--VYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVL 313
DTVRRRMMMQSG + +Y TLDCWRKI R EG +F++GA SNV R G A +LVL
Sbjct: 256 FDTVRRRMMMQSGRKGTDIMYTGTLDCWRKIARDEGGKAFFKGAWSNVLRGMGGAFVLVL 315
Query: 314 YDEVKKF 320
YDE+KK+
Sbjct: 316 YDEIKKY 322
Score = 33.9 bits (76), Expect = 6.7
Identities = 23/88 (26%), Positives = 45/88 (51%), Gaps = 7/88 (7%)
Query: 241 VAQAVTTSAGLVSYPLDTVRRRMMMQSGMEHPV----YNSTLDCWRKIYRTEGLISFYRG 296
VA A++ +A P++ V+ + +Q + Y +DC +I + +G++SF+RG
Sbjct: 42 VAAAISKTAVA---PIERVKLLLQVQHASKQITADKQYKGIIDCVVRIPKEQGVLSFWRG 98
Query: 297 AVSNVFRSTGAAAILVLYDEVKKFMNLG 324
++NV R A+ + + K + LG
Sbjct: 99 NLANVIRYFPTQALNFAFKDKYKQIFLG 126
>gb|AAH56160.1| Solute carrier family 25, member 5 [Homo sapiens]
gi|113459|sp|P05141|ADT2_HUMAN ADP,ATP carrier protein,
fibroblast isoform (ADP/ATP translocase 2) (Adenine
nucleotide translocator 2) (ANT 2) (Solute carrier
family 25, member 5) gi|4502099|ref|NP_001143.1| solute
carrier family 25, member 5 [Homo sapiens]
gi|1381112|gb|AAB39266.1| ANT-2 gene product
gi|178661|gb|AAA51737.1| adenine nucleotide
translocator-2
Length = 298
Score = 286 bits (731), Expect = 8e-76
Identities = 145/295 (49%), Positives = 195/295 (65%), Gaps = 7/295 (2%)
Query: 29 NFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVRE 88
+F +D +AG V T VAPIER KLLLQ Q ++ I A +++KG+ DC++R +E
Sbjct: 7 SFAKDFLAGGVAAAISKTAVAPIERVKLLLQVQHASKQITAD--KQYKGIIDCVVRIPKE 64
Query: 89 EGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAG 148
+GV+S WRGN ++V+RY+P+ ALNF+ KD YK I GG + + N +G AAG
Sbjct: 65 QGVLSFWRGNLANVIRYFPTQALNFAFKDKYKQIFLGGVDKRTQFWRYFAGNLASGGAAG 124
Query: 149 CTSLILVYPLDIAHTRLAADIGRTEV-RQFRGIHHFLATIFQKDGVRGIYRGLPASLHGM 207
TSL VYPLD A TRLAAD+G+ R+FRG+ L I++ DG++G+Y+G S+ G+
Sbjct: 125 ATSLCFVYPLDFARTRLAADVGKAGAEREFRGLGDCLVKIYKSDGIKGLYQGFNVSVQGI 184
Query: 208 VIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQS 267
+I+R YFG +DT K ML + + + WM+AQ VT AGL SYP DTVRRRMMMQS
Sbjct: 185 IIYRAAYFGIYDTAKGMLPDPKNTHIVI--SWMIAQTVTAVAGLTSYPFDTVRRRMMMQS 242
Query: 268 GMEHP--VYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVKKF 320
G + +Y TLDCWRKI R EG +F++GA SNV R G A +LVLYDE+KK+
Sbjct: 243 GRKGTDIMYTGTLDCWRKIARDEGGKAFFKGAWSNVLRGMGGAFVLVLYDEIKKY 297
Score = 33.9 bits (76), Expect = 6.7
Identities = 23/88 (26%), Positives = 45/88 (51%), Gaps = 7/88 (7%)
Query: 241 VAQAVTTSAGLVSYPLDTVRRRMMMQSGMEHPV----YNSTLDCWRKIYRTEGLISFYRG 296
VA A++ +A P++ V+ + +Q + Y +DC +I + +G++SF+RG
Sbjct: 17 VAAAISKTAVA---PIERVKLLLQVQHASKQITADKQYKGIIDCVVRIPKEQGVLSFWRG 73
Query: 297 AVSNVFRSTGAAAILVLYDEVKKFMNLG 324
++NV R A+ + + K + LG
Sbjct: 74 NLANVIRYFPTQALNFAFKDKYKQIFLG 101
>gb|AAY66852.1| ADP/ATP translocase [Ixodes scapularis]
Length = 299
Score = 286 bits (731), Expect = 8e-76
Identities = 147/296 (49%), Positives = 194/296 (64%), Gaps = 7/296 (2%)
Query: 29 NFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVRE 88
+F +D +AG V T VAPIER KLLLQ Q ++ I S +++KGM DC +R RE
Sbjct: 7 SFAKDFIAGGVAAAISKTAVAPIERVKLLLQVQHASTQI--SEAQRYKGMVDCFVRIPRE 64
Query: 89 EGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAG 148
+G +S WRGN ++V+RY+P+ ALNF+ KD YK + GG + + N +G AAG
Sbjct: 65 QGFLSFWRGNLANVIRYFPTQALNFAFKDKYKQVFLGGVDKKTQFWRYFAGNLASGGAAG 124
Query: 149 CTSLILVYPLDIAHTRLAADIGR-TEVRQFRGIHHFLATIFQKDGVRGIYRGLPASLHGM 207
TSL VYPLD A TRLAADIG+ R+F G+ + L IF+ DG+ G+YRG S+ G+
Sbjct: 125 ATSLCFVYPLDFARTRLAADIGKGAGQREFSGLGNCLTKIFKSDGLMGLYRGFGVSVQGI 184
Query: 208 VIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQS 267
+I+R YFG FDT K ML + L + W++AQ VTT AG++SYP DTVRRRMMMQS
Sbjct: 185 IIYRAAYFGFFDTAKGMLPDPKNTPLVI--SWLIAQTVTTVAGIMSYPFDTVRRRMMMQS 242
Query: 268 GMEHP--VYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVKKFM 321
G +Y ST CW KIY+TEG +F++GA SNV R TG + +LVLYDE+K F+
Sbjct: 243 GRAKADLMYKSTAHCWGKIYKTEGGAAFFKGAFSNVLRGTGGSLVLVLYDEIKAFI 298
Score = 36.2 bits (82), Expect = 1.4
Identities = 25/88 (28%), Positives = 45/88 (50%), Gaps = 7/88 (7%)
Query: 241 VAQAVTTSAGLVSYPLDTVRRRMMMQSGM----EHPVYNSTLDCWRKIYRTEGLISFYRG 296
VA A++ +A P++ V+ + +Q E Y +DC+ +I R +G +SF+RG
Sbjct: 17 VAAAISKTAVA---PIERVKLLLQVQHASTQISEAQRYKGMVDCFVRIPREQGFLSFWRG 73
Query: 297 AVSNVFRSTGAAAILVLYDEVKKFMNLG 324
++NV R A+ + + K + LG
Sbjct: 74 NLANVIRYFPTQALNFAFKDKYKQVFLG 101
>gb|AAB96347.1| ADP/ATP carrier protein (adenine nucleotide translocator 2) [Homo
sapiens]
Length = 298
Score = 285 bits (730), Expect = 1e-75
Identities = 145/295 (49%), Positives = 195/295 (65%), Gaps = 7/295 (2%)
Query: 29 NFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVRE 88
+F +D +AG V T VAPIER KLLLQ Q ++ I A +++KG+ DC++R +E
Sbjct: 7 SFAKDFLAGGVAAAISKTAVAPIERVKLLLQVQHASKQITAD--KQYKGIIDCVVRIPKE 64
Query: 89 EGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAG 148
+GV+S WRGN ++V+RY+P+ ALNF+ KD YK I GG + + N +G AAG
Sbjct: 65 QGVLSFWRGNLANVIRYFPTQALNFAFKDKYKQIFLGGVDKRTQFWLYFAGNLASGGAAG 124
Query: 149 CTSLILVYPLDIAHTRLAADIGRTEV-RQFRGIHHFLATIFQKDGVRGIYRGLPASLHGM 207
TSL VYPLD A TRLAAD+G+ R+FRG+ L I++ DG++G+Y+G S+ G+
Sbjct: 125 ATSLCFVYPLDFARTRLAADVGKAGAEREFRGLGDCLVKIYKSDGIKGLYQGFNVSVQGI 184
Query: 208 VIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQS 267
+I+R YFG +DT K ML + + + WM+AQ VT AGL SYP DTVRRRMMMQS
Sbjct: 185 IIYRAAYFGIYDTAKGMLPDPKNTHIVI--SWMIAQTVTAVAGLTSYPFDTVRRRMMMQS 242
Query: 268 GMEHP--VYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVKKF 320
G + +Y TLDCWRKI R EG +F++GA SNV R G A +LVLYDE+KK+
Sbjct: 243 GRKGTDIMYTGTLDCWRKIARDEGGKAFFKGAWSNVLRGMGGAFVLVLYDEIKKY 297
Score = 33.9 bits (76), Expect = 6.7
Identities = 23/88 (26%), Positives = 45/88 (51%), Gaps = 7/88 (7%)
Query: 241 VAQAVTTSAGLVSYPLDTVRRRMMMQSGMEHPV----YNSTLDCWRKIYRTEGLISFYRG 296
VA A++ +A P++ V+ + +Q + Y +DC +I + +G++SF+RG
Sbjct: 17 VAAAISKTAVA---PIERVKLLLQVQHASKQITADKQYKGIIDCVVRIPKEQGVLSFWRG 73
Query: 297 AVSNVFRSTGAAAILVLYDEVKKFMNLG 324
++NV R A+ + + K + LG
Sbjct: 74 NLANVIRYFPTQALNFAFKDKYKQIFLG 101
>ref|NP_031477.1| solute carrier family 25, member 5 [Mus musculus]
gi|56270535|gb|AAH86756.1| Solute carrier family 25,
member 5 [Mus musculus] gi|1816495|emb|CAA50196.1|
adenine nucleotide translocase [Mus musculus]
gi|13435412|gb|AAH04570.1| Solute carrier family 25,
member 5 [Mus musculus] gi|1703188|sp|P51881|ADT2_MOUSE
ADP,ATP carrier protein 2 (ADP/ATP translocase 2)
(Adenine nucleotide translocator 2) (ANT 2) (Solute
carrier family 25, member 5) gi|902010|gb|AAC52838.1|
adenine nucleotide translocase-2
gi|7595833|gb|AAF64471.1| adenine nucleotide translocase
2 [Mus musculus] gi|26353806|dbj|BAC40533.1| unnamed
protein product [Mus musculus]
gi|12849700|dbj|BAB28445.1| unnamed protein product [Mus
musculus] gi|499132|gb|AAA19009.1| adenine nucleotide
translocase gi|12834153|dbj|BAB22804.1| unnamed protein
product [Mus musculus]
Length = 298
Score = 285 bits (729), Expect = 1e-75
Identities = 144/295 (48%), Positives = 196/295 (65%), Gaps = 7/295 (2%)
Query: 29 NFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVRE 88
+F +D +AG V T VAPIER KLLLQ Q ++ I A +++KG+ DC++R +E
Sbjct: 7 SFAKDFLAGGVAAAISKTAVAPIERVKLLLQVQHASKQITAD--KQYKGIIDCVVRIPKE 64
Query: 89 EGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAG 148
+GV+S WRGN ++V+RY+P+ ALNF+ KD YK I GG + + N +G AAG
Sbjct: 65 QGVLSFWRGNLANVIRYFPTQALNFAFKDKYKQIFLGGVDKRTQFWRYFAGNLASGGAAG 124
Query: 149 CTSLILVYPLDIAHTRLAADIGRTEV-RQFRGIHHFLATIFQKDGVRGIYRGLPASLHGM 207
TSL VYPLD A TRLAAD+G+ R+F+G+ L I++ DG++G+Y+G S+ G+
Sbjct: 125 ATSLCFVYPLDFARTRLAADVGKAGAEREFKGLGDCLVKIYKSDGIKGLYQGFNVSVQGI 184
Query: 208 VIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQS 267
+I+R YFG +DT K ML + + + WM+AQ+VT AGL SYP DTVRRRMMMQS
Sbjct: 185 IIYRAAYFGIYDTAKGMLPDPKNTHIFI--SWMIAQSVTAVAGLTSYPFDTVRRRMMMQS 242
Query: 268 GMEHP--VYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVKKF 320
G + +Y TLDCWRKI R EG +F++GA SNV R G A +LVLYDE+KK+
Sbjct: 243 GRKGTDIMYTGTLDCWRKIARDEGSKAFFKGAWSNVLRGMGGAFVLVLYDEIKKY 297
Score = 33.9 bits (76), Expect = 6.7
Identities = 23/88 (26%), Positives = 45/88 (51%), Gaps = 7/88 (7%)
Query: 241 VAQAVTTSAGLVSYPLDTVRRRMMMQSGMEHPV----YNSTLDCWRKIYRTEGLISFYRG 296
VA A++ +A P++ V+ + +Q + Y +DC +I + +G++SF+RG
Sbjct: 17 VAAAISKTAVA---PIERVKLLLQVQHASKQITADKQYKGIIDCVVRIPKEQGVLSFWRG 73
Query: 297 AVSNVFRSTGAAAILVLYDEVKKFMNLG 324
++NV R A+ + + K + LG
Sbjct: 74 NLANVIRYFPTQALNFAFKDKYKQIFLG 101
>gb|AAC52837.1| adenine nucleotide translocase-1
Length = 298
Score = 285 bits (729), Expect = 1e-75
Identities = 143/296 (48%), Positives = 198/296 (66%), Gaps = 7/296 (2%)
Query: 29 NFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVRE 88
+F +D +AG + T VAPIER KLLLQ Q ++ I S +++KG+ DC++R +E
Sbjct: 7 SFLKDFLAGGIAAAVSKTAVAPIERVKLLLQVQHASKQI--SAEKQYKGIIDCVVRIPKE 64
Query: 89 EGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAG 148
+G +S WRGN ++V+RY+P+ ALNF+ KD YK I GG + + N +G AAG
Sbjct: 65 QGFLSFWRGNLANVIRYFPTQALNFAFKDKYKQIFLGGVDRHKQFWRYFAGNLASGGAAG 124
Query: 149 CTSLILVYPLDIAHTRLAADIGR-TEVRQFRGIHHFLATIFQKDGVRGIYRGLPASLHGM 207
TSL VYPLD+A TRLAAD+G+ + R+F G+ L IF+ DG++G+Y+G S+ G+
Sbjct: 125 ATSLCFVYPLDLARTRLAADVGKGSSQREFNGLGDCLTKIFKSDGLKGLYQGFSVSVQGI 184
Query: 208 VIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQS 267
+I+R YFG +DT K ML + + + WM+AQ+VT AGLVSYP DTVRRRMMMQS
Sbjct: 185 IIYRAAYFGVYDTAKGMLPDPKNVHIIV--SWMIAQSVTAVAGLVSYPFDTVRRRMMMQS 242
Query: 268 GMEHP--VYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVKKFM 321
G + +Y TLDCWRKI + EG +F++GA SNV R G A +LVLYDE+KK++
Sbjct: 243 GRKGADIMYTGTLDCWRKIAKDEGANAFFKGAWSNVLRGMGGAFVLVLYDEIKKYV 298
Score = 33.5 bits (75), Expect = 8.8
Identities = 23/88 (26%), Positives = 44/88 (49%), Gaps = 7/88 (7%)
Query: 241 VAQAVTTSAGLVSYPLDTVRRRMMMQSGMEH----PVYNSTLDCWRKIYRTEGLISFYRG 296
+A AV+ +A P++ V+ + +Q + Y +DC +I + +G +SF+RG
Sbjct: 17 IAAAVSKTAVA---PIERVKLLLQVQHASKQISAEKQYKGIIDCVVRIPKEQGFLSFWRG 73
Query: 297 AVSNVFRSTGAAAILVLYDEVKKFMNLG 324
++NV R A+ + + K + LG
Sbjct: 74 NLANVIRYFPTQALNFAFKDKYKQIFLG 101
>emb|CAI39843.1| solute carrier family 25 (mitochondrial carrier; adenine nucleotide
translocator), member 6 [Homo sapiens]
gi|21594693|gb|AAH31912.1| Solute carrier family 25,
member A6 [Homo sapiens] gi|14286274|gb|AAH08935.1|
Solute carrier family 25, member A6 [Homo sapiens]
gi|14250567|gb|AAH08737.1| Solute carrier family 25,
member A6 [Homo sapiens] gi|14043791|gb|AAH07850.1|
Solute carrier family 25, member A6 [Homo sapiens]
gi|13938331|gb|AAH07295.1| Solute carrier family 25,
member A6 [Homo sapiens] gi|113463|sp|P12236|ADT3_HUMAN
ADP,ATP carrier protein, liver isoform T2 (ADP/ATP
translocase 3) (Adenine nucleotide translocator 3) (ANT
3) (Solute carrier family 25, member 6)
gi|9956039|gb|AAG01998.1| similar to bovine ADP/ATP
translocase T1 mRNA with GenBank Accession Number
M24102.1 [Homo sapiens] gi|48146917|emb|CAG33681.1|
SLC25A6 [Homo sapiens]
Length = 298
Score = 285 bits (728), Expect = 2e-75
Identities = 145/294 (49%), Positives = 196/294 (66%), Gaps = 7/294 (2%)
Query: 29 NFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVRE 88
+F +D +AG + T VAPIER KLLLQ Q ++ I A +++KG+ DCI+R +E
Sbjct: 7 SFAKDFLAGGIAAAISKTAVAPIERVKLLLQVQHASKQIAAD--KQYKGIVDCIVRIPKE 64
Query: 89 EGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAG 148
+GV+S WRGN ++V+RY+P+ ALNF+ KD YK I GG + + N +G AAG
Sbjct: 65 QGVLSFWRGNLANVIRYFPTQALNFAFKDKYKQIFLGGVDKHTQFWRYFAGNLASGGAAG 124
Query: 149 CTSLILVYPLDIAHTRLAADIGRTEV-RQFRGIHHFLATIFQKDGVRGIYRGLPASLHGM 207
TSL VYPLD A TRLAAD+G++ R+FRG+ L I + DG+RG+Y+G S+ G+
Sbjct: 125 ATSLCFVYPLDFARTRLAADVGKSGTEREFRGLGDCLVKITKSDGIRGLYQGFSVSVQGI 184
Query: 208 VIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQS 267
+I+R YFG +DT K ML + + + WM+AQ VT AG+VSYP DTVRRRMMMQS
Sbjct: 185 IIYRAAYFGVYDTAKGMLPDPKNTHIVV--SWMIAQTVTAVAGVVSYPFDTVRRRMMMQS 242
Query: 268 GMEHP--VYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVKK 319
G + +Y T+DCWRKI+R EG +F++GA SNV R G A +LVLYDE+KK
Sbjct: 243 GRKGADIMYTGTVDCWRKIFRDEGGKAFFKGAWSNVLRGMGGAFVLVLYDELKK 296
>ref|NP_001006443.1| similar to ADP/ATP translocase [Gallus gallus]
Length = 298
Score = 285 bits (728), Expect = 2e-75
Identities = 143/296 (48%), Positives = 198/296 (66%), Gaps = 7/296 (2%)
Query: 29 NFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVRE 88
+F +D +AG V T VAPIER KLLLQ Q ++ I A +++KG+ DC++R +E
Sbjct: 7 SFLKDFLAGGVAAAISKTAVAPIERVKLLLQVQHASKQITAE--KQYKGIIDCVVRIPKE 64
Query: 89 EGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAG 148
+G+IS WRGN ++V+RY+P+ ALNF+ KD YK I GG + + N +G AAG
Sbjct: 65 QGIISFWRGNLANVIRYFPTQALNFAFKDKYKQIFLGGVDRHKQFWRYFAGNLASGGAAG 124
Query: 149 CTSLILVYPLDIAHTRLAADIGR-TEVRQFRGIHHFLATIFQKDGVRGIYRGLPASLHGM 207
TSL VYPLD A TRLAAD+G+ R+F G+ + IF+ DG+RG+Y+G S+ G+
Sbjct: 125 ATSLCFVYPLDFARTRLAADVGKGVSEREFTGLGDCIVKIFKSDGLRGLYQGFNVSVQGI 184
Query: 208 VIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQS 267
+I+R YFG +DT K ML + + + WM+AQ+VT +AGLVSYP DTVRRRMMMQS
Sbjct: 185 IIYRAAYFGVYDTAKGMLPDPKNVHIIV--SWMIAQSVTAAAGLVSYPFDTVRRRMMMQS 242
Query: 268 GMEHP--VYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVKKFM 321
G + +Y T+DCW+KI + EG +F++GA SNV R G A +LVLYDE+KK++
Sbjct: 243 GRKGADIMYKGTIDCWKKIAKDEGSKAFFKGAWSNVLRGMGGAFVLVLYDEIKKYV 298
Score = 35.0 bits (79), Expect = 3.0
Identities = 24/88 (27%), Positives = 45/88 (50%), Gaps = 7/88 (7%)
Query: 241 VAQAVTTSAGLVSYPLDTVRRRMMMQSGMEHPV----YNSTLDCWRKIYRTEGLISFYRG 296
VA A++ +A P++ V+ + +Q + Y +DC +I + +G+ISF+RG
Sbjct: 17 VAAAISKTAVA---PIERVKLLLQVQHASKQITAEKQYKGIIDCVVRIPKEQGIISFWRG 73
Query: 297 AVSNVFRSTGAAAILVLYDEVKKFMNLG 324
++NV R A+ + + K + LG
Sbjct: 74 NLANVIRYFPTQALNFAFKDKYKQIFLG 101
>emb|CAG31047.1| hypothetical protein [Gallus gallus]
Length = 298
Score = 285 bits (728), Expect = 2e-75
Identities = 143/296 (48%), Positives = 198/296 (66%), Gaps = 7/296 (2%)
Query: 29 NFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVRE 88
+F +D +AG V T VAPIER KLLLQ Q ++ I A +++KG+ DC++R +E
Sbjct: 7 SFLKDFLAGGVAAAISKTAVAPIERVKLLLQVQHASKQITAE--KQYKGIIDCVVRIPKE 64
Query: 89 EGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAG 148
+G+IS WRGN ++V+RY+P+ ALNF+ KD YK I GG + + N +G AAG
Sbjct: 65 QGIISFWRGNLANVIRYFPTQALNFAFKDKYKQIFLGGVDRHKQFWRYFAGNLASGGAAG 124
Query: 149 CTSLILVYPLDIAHTRLAADIGR-TEVRQFRGIHHFLATIFQKDGVRGIYRGLPASLHGM 207
TSL VYPLD A TRLAAD+G+ R+F G+ + IF+ DG+RG+Y+G S+ G+
Sbjct: 125 ATSLCFVYPLDFARTRLAADVGKGVSEREFTGLGDCIVKIFKSDGLRGLYQGFNVSVQGI 184
Query: 208 VIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQS 267
+I+R YFG +DT K ML + + + WM+AQ+VT +AGLVSYP DTVRRRMMMQS
Sbjct: 185 IIYRAAYFGVYDTAKGMLPDPKNVHIIV--SWMIAQSVTAAAGLVSYPFDTVRRRMMMQS 242
Query: 268 GMEHP--VYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVKKFM 321
G + +Y T+DCW+KI + EG +F++GA SNV R G A +LVLYDE+KK++
Sbjct: 243 GRKGADIMYKGTIDCWKKIAKDEGSKAFFKGAWSNVLRGMGGAFVLVLYDEIKKYV 298
Score = 35.0 bits (79), Expect = 3.0
Identities = 24/88 (27%), Positives = 45/88 (50%), Gaps = 7/88 (7%)
Query: 241 VAQAVTTSAGLVSYPLDTVRRRMMMQSGMEHPV----YNSTLDCWRKIYRTEGLISFYRG 296
VA A++ +A P++ V+ + +Q + Y +DC +I + +G+ISF+RG
Sbjct: 17 VAAAISKTAVA---PIERVKLLLQVQHASKQITAEKQYKGIIDCVVRIPKEQGIISFWRG 73
Query: 297 AVSNVFRSTGAAAILVLYDEVKKFMNLG 324
++NV R A+ + + K + LG
Sbjct: 74 NLANVIRYFPTQALNFAFKDKYKQIFLG 101
>dbj|BAD93059.1| ADP,ATP carrier protein, liver isoform T2 variant [Homo sapiens]
Length = 323
Score = 285 bits (728), Expect = 2e-75
Identities = 145/294 (49%), Positives = 196/294 (66%), Gaps = 7/294 (2%)
Query: 29 NFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVRE 88
+F +D +AG + T VAPIER KLLLQ Q ++ I A +++KG+ DCI+R +E
Sbjct: 32 SFAKDFLAGGIAAAISKTAVAPIERVKLLLQVQHASKQIAAD--KQYKGIVDCIVRIPKE 89
Query: 89 EGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAG 148
+GV+S WRGN ++V+RY+P+ ALNF+ KD YK I GG + + N +G AAG
Sbjct: 90 QGVLSFWRGNLANVIRYFPTQALNFAFKDKYKQIFLGGVDKHTQFWRYFAGNLASGGAAG 149
Query: 149 CTSLILVYPLDIAHTRLAADIGRTEV-RQFRGIHHFLATIFQKDGVRGIYRGLPASLHGM 207
TSL VYPLD A TRLAAD+G++ R+FRG+ L I + DG+RG+Y+G S+ G+
Sbjct: 150 ATSLCFVYPLDFARTRLAADVGKSGTEREFRGLGDCLVKITKSDGIRGLYQGFSVSVQGI 209
Query: 208 VIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQS 267
+I+R YFG +DT K ML + + + WM+AQ VT AG+VSYP DTVRRRMMMQS
Sbjct: 210 IIYRAAYFGVYDTAKGMLPDPKNTHIVV--SWMIAQTVTAVAGVVSYPFDTVRRRMMMQS 267
Query: 268 GMEHP--VYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVKK 319
G + +Y T+DCWRKI+R EG +F++GA SNV R G A +LVLYDE+KK
Sbjct: 268 GRKGADIMYTGTVDCWRKIFRDEGGKAFFKGAWSNVLRGMGGAFVLVLYDELKK 321
>ref|NP_476443.1| solute carrier family 25, member 5 [Rattus norvegicus]
gi|37590229|gb|AAH59108.1| Solute carrier family 25,
member 5 [Rattus norvegicus]
gi|728810|sp|Q09073|ADT2_RAT ADP,ATP carrier protein 2
(ADP/ATP translocase 2) (Adenine nucleotide translocator
2) (ANT 2) (Solute carrier family 25, member 5)
gi|398595|dbj|BAA02238.1| adenine nucleotide
translocator [Rattus norvegicus]
Length = 298
Score = 284 bits (727), Expect = 2e-75
Identities = 144/295 (48%), Positives = 196/295 (65%), Gaps = 7/295 (2%)
Query: 29 NFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVRE 88
+F +D +AG V T VAPIER KLLLQ Q ++ I A +++KG+ DC++R +E
Sbjct: 7 SFAKDFLAGGVAAAISKTAVAPIERVKLLLQVQHASKQITAD--KQYKGIIDCVVRIPKE 64
Query: 89 EGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAG 148
+GV+S WRGN ++V+RY+P+ ALNF+ KD YK I GG + + N +G AAG
Sbjct: 65 QGVLSFWRGNLANVIRYFPTQALNFAFKDKYKQIFLGGVDKRTQFWRYFAGNLASGGAAG 124
Query: 149 CTSLILVYPLDIAHTRLAADIGRTEV-RQFRGIHHFLATIFQKDGVRGIYRGLPASLHGM 207
TSL VYPLD A TRLAAD+G+ R+F+G+ L I++ DG++G+Y+G S+ G+
Sbjct: 125 ATSLCFVYPLDFARTRLAADVGKAGAEREFKGLGDCLVKIYKSDGIKGLYQGFNVSVQGI 184
Query: 208 VIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQS 267
+I+R YFG +DT K ML + + + WM+AQ+VT AGL SYP DTVRRRMMMQS
Sbjct: 185 IIYRAAYFGIYDTAKGMLPDPKNTHIFI--SWMIAQSVTAVAGLTSYPFDTVRRRMMMQS 242
Query: 268 GMEHP--VYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVKKF 320
G + +Y TLDCWRKI R EG +F++GA SNV R G A +LVLYDE+KK+
Sbjct: 243 GRKGTDIMYTGTLDCWRKIARDEGGKAFFKGAWSNVLRGMGGAFVLVLYDEIKKY 297
Score = 33.9 bits (76), Expect = 6.7
Identities = 23/88 (26%), Positives = 45/88 (51%), Gaps = 7/88 (7%)
Query: 241 VAQAVTTSAGLVSYPLDTVRRRMMMQSGMEHPV----YNSTLDCWRKIYRTEGLISFYRG 296
VA A++ +A P++ V+ + +Q + Y +DC +I + +G++SF+RG
Sbjct: 17 VAAAISKTAVA---PIERVKLLLQVQHASKQITADKQYKGIIDCVVRIPKEQGVLSFWRG 73
Query: 297 AVSNVFRSTGAAAILVLYDEVKKFMNLG 324
++NV R A+ + + K + LG
Sbjct: 74 NLANVIRYFPTQALNFAFKDKYKQIFLG 101
>emb|CAA52616.1| adenine nucleotide carrier [Mus musculus]
gi|20070838|gb|AAH26925.1| Slc25a4 protein [Mus
musculus] gi|13277810|gb|AAH03791.1| Slc25a4 protein
[Mus musculus] gi|21903382|sp|P48962|ADT1_MOUSE ADP,ATP
carrier protein, heart/skeletal muscle isoform T1
(ADP/ATP translocase 1) (Adenine nucleotide translocator
1) (ANT 1) (Solute carrier family 25, member 4) (mANC1)
gi|7595831|gb|AAF64470.1| adenine nucleotide translocase
1 [Mus musculus]
Length = 298
Score = 284 bits (727), Expect = 2e-75
Identities = 143/296 (48%), Positives = 197/296 (66%), Gaps = 7/296 (2%)
Query: 29 NFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVRE 88
+F +D +AG + T VAPIER KLLLQ Q ++ I S +++KG+ DC++R +E
Sbjct: 7 SFLKDFLAGGIAAAVSKTAVAPIERVKLLLQVQHASKQI--SAEKQYKGIIDCVVRIPKE 64
Query: 89 EGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAG 148
+G +S WRGN ++V+RY+P+ ALNF+ KD YK I GG + + N +G AAG
Sbjct: 65 QGFLSFWRGNLANVIRYFPTQALNFAFKDKYKQIFLGGVDRHKQFWRYFAGNLASGGAAG 124
Query: 149 CTSLILVYPLDIAHTRLAADIGR-TEVRQFRGIHHFLATIFQKDGVRGIYRGLPASLHGM 207
TSL VYPLD A TRLAAD+G+ + R+F G+ L IF+ DG++G+Y+G S+ G+
Sbjct: 125 ATSLCFVYPLDFARTRLAADVGKGSSQREFNGLGDCLTKIFKSDGLKGLYQGFSVSVQGI 184
Query: 208 VIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQS 267
+I+R YFG +DT K ML + + + WM+AQ+VT AGLVSYP DTVRRRMMMQS
Sbjct: 185 IIYRAAYFGVYDTAKGMLPDPKNVHIIV--SWMIAQSVTAVAGLVSYPFDTVRRRMMMQS 242
Query: 268 GMEHP--VYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVKKFM 321
G + +Y TLDCWRKI + EG +F++GA SNV R G A +LVLYDE+KK++
Sbjct: 243 GRKGADIMYTGTLDCWRKIAKDEGANAFFKGAWSNVLRGMGGAFVLVLYDEIKKYV 298
Score = 33.5 bits (75), Expect = 8.8
Identities = 23/88 (26%), Positives = 44/88 (49%), Gaps = 7/88 (7%)
Query: 241 VAQAVTTSAGLVSYPLDTVRRRMMMQSGMEH----PVYNSTLDCWRKIYRTEGLISFYRG 296
+A AV+ +A P++ V+ + +Q + Y +DC +I + +G +SF+RG
Sbjct: 17 IAAAVSKTAVA---PIERVKLLLQVQHASKQISAEKQYKGIIDCVVRIPKEQGFLSFWRG 73
Query: 297 AVSNVFRSTGAAAILVLYDEVKKFMNLG 324
++NV R A+ + + K + LG
Sbjct: 74 NLANVIRYFPTQALNFAFKDKYKQIFLG 101
>emb|CAG11525.1| unnamed protein product [Tetraodon nigroviridis]
Length = 299
Score = 284 bits (727), Expect = 2e-75
Identities = 145/295 (49%), Positives = 194/295 (65%), Gaps = 7/295 (2%)
Query: 29 NFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVRE 88
+F +D +AG + T VAPIER KLLLQ Q ++ I A ++KG+ DC++R +E
Sbjct: 7 SFMKDFLAGGIAAAISKTAVAPIERVKLLLQVQHASKQITAE--TQYKGIIDCVVRIPKE 64
Query: 89 EGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAG 148
+G IS WRGN ++V+RY+P+ ALNF+ KD YK I GG + + N +G AAG
Sbjct: 65 QGFISFWRGNLANVIRYFPTQALNFAFKDKYKKIFLGGVDQKTQFWRYFAGNLASGGAAG 124
Query: 149 CTSLILVYPLDIAHTRLAADIGRTEV-RQFRGIHHFLATIFQKDGVRGIYRGLPASLHGM 207
TSL VYPLD A TRLAADIG+ R+F G+ + +A IF+ DG++G+Y G S+ G+
Sbjct: 125 ATSLCFVYPLDFARTRLAADIGKGPAEREFTGLGNCIAKIFKTDGIKGLYLGFNVSVQGI 184
Query: 208 VIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQS 267
+I+R YFG FDT K ML + + + WM+AQ VT AGL+SYP DTVRRRMMMQS
Sbjct: 185 IIYRAAYFGCFDTAKGMLPDPKNTHIIV--SWMIAQTVTAVAGLISYPFDTVRRRMMMQS 242
Query: 268 GMEHP--VYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVKKF 320
G + +Y T+DCWRKI + EG +F++GA SNV R G A +LVLYDE+KKF
Sbjct: 243 GRKGADIMYKGTIDCWRKILKDEGGKAFFKGAWSNVIRGMGGAFVLVLYDEIKKF 297
Score = 33.9 bits (76), Expect = 6.7
Identities = 20/74 (27%), Positives = 37/74 (49%), Gaps = 4/74 (5%)
Query: 255 PLDTVRRRMMMQSGMEHPV----YNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAI 310
P++ V+ + +Q + Y +DC +I + +G ISF+RG ++NV R A+
Sbjct: 28 PIERVKLLLQVQHASKQITAETQYKGIIDCVVRIPKEQGFISFWRGNLANVIRYFPTQAL 87
Query: 311 LVLYDEVKKFMNLG 324
+ + K + LG
Sbjct: 88 NFAFKDKYKKIFLG 101
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.323 0.137 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 523,338,167
Number of Sequences: 2540612
Number of extensions: 20882833
Number of successful extensions: 64191
Number of sequences better than 10.0: 2078
Number of HSP's better than 10.0 without gapping: 1326
Number of HSP's successfully gapped in prelim test: 752
Number of HSP's that attempted gapping in prelim test: 50348
Number of HSP's gapped (non-prelim): 5462
length of query: 326
length of database: 863,360,394
effective HSP length: 128
effective length of query: 198
effective length of database: 538,162,058
effective search space: 106556087484
effective search space used: 106556087484
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 75 (33.5 bits)
Medicago: description of AC133572.9


