

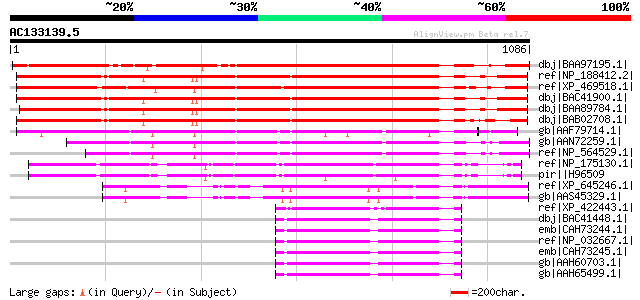
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133139.5 + phase: 0
(1086 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAA97195.1| IRE [Arabidopsis thaliana] gi|15241795|ref|NP_20... 1186 0.0
ref|NP_188412.2| protein kinase, putative [Arabidopsis thaliana] 962 0.0
ref|XP_469518.1| putative protein kinase [Oryza sativa] gi|13324... 958 0.0
dbj|BAC41900.1| putative protein kinase [Arabidopsis thaliana] 958 0.0
dbj|BAA89784.1| IRE homolog 1 [Arabidopsis thaliana] 957 0.0
dbj|BAB02708.1| IRE homolog; protein kinase-like protein [Arabid... 948 0.0
gb|AAF79714.1| T1N15.10 [Arabidopsis thaliana] 847 0.0
gb|AAN72259.1| At1g48490/T1N15_9 [Arabidopsis thaliana] gi|14326... 807 0.0
ref|NP_564529.1| protein kinase, putative [Arabidopsis thaliana] 754 0.0
ref|NP_175130.1| protein kinase family protein [Arabidopsis thal... 607 e-172
pir||H96509 protein F27F5.23 [imported] - Arabidopsis thaliana g... 590 e-166
ref|XP_645246.1| protein serine/threonine kinase [Dictyostelium ... 344 1e-92
gb|AAS45329.1| similar to cell wall biosynthesis kinase; Cbk1p [... 344 1e-92
ref|XP_422443.1| PREDICTED: similar to Mast2 protein [Gallus gal... 296 3e-78
dbj|BAC41448.1| mKIAA0807 protein [Mus musculus] 292 5e-77
emb|CAH73244.1| microtubule associated serine/threonine kinase 2... 291 6e-77
ref|NP_032667.1| microtubule associated serine/threonine kinase ... 291 6e-77
emb|CAH73245.1| microtubule associated serine/threonine kinase 2... 291 6e-77
gb|AAH60703.1| Mast2 protein [Mus musculus] 291 6e-77
gb|AAH65499.1| MAST205 protein [Homo sapiens] 291 6e-77
>dbj|BAA97195.1| IRE [Arabidopsis thaliana] gi|15241795|ref|NP_201037.1| incomplete
root hair elongation (IRE) / protein kinase, putative
[Arabidopsis thaliana] gi|6729346|dbj|BAA89783.1| IRE
[Arabidopsis thaliana]
Length = 1168
Score = 1186 bits (3067), Expect = 0.0
Identities = 658/1113 (59%), Positives = 792/1113 (71%), Gaps = 145/1113 (13%)
Query: 6 VQGNHAAYAKDFQSPRYQEILRLTSGKKRKNRPDIKSFSHELNSKGVRPFPVWKNRAFG- 64
V+ H AK+ QSPR+Q ILR+TSG+K+K DIKSFSHELNSKGVRPFPVW++RA G
Sbjct: 163 VESTHVGLAKETQSPRFQAILRVTSGRKKKAH-DIKSFSHELNSKGVRPFPVWRSRAVGH 221
Query: 65 -QEIMEEIRAKFEKLKEEVDSDLGGFAGDLVGTLEKIPGSHPEWKEGLEDLLVVARQCAK 123
+EIM IR KF+K KE+VD+DLG FAG LV TLE P S+ E + GLEDLLV ARQCA
Sbjct: 222 MEEIMAAIRTKFDKQKEDVDADLGVFAGYLVTTLESTPESNKELRVGLEDLLVEARQCAT 281
Query: 124 MTAAEFWINCETIVQKLDDKRQDIPVGILKQAHTRLLFILSRCTRLVQFQKESVKEQDHI 183
M A+EFW+ CE IVQKLDDKRQ++P+G LKQAH RLLFIL+RC RLVQF+KES ++HI
Sbjct: 282 MPASEFWLKCEGIVQKLDDKRQELPMGGLKQAHNRLLFILTRCNRLVQFRKESGYVEEHI 341
Query: 184 LGLHQLSDLGVYSEQIMKAEESCGFPPSDHEMAEKLIKKSHGKEQDKPITKQSQADQHAS 243
LG+HQLSDLGVY EQ+++ + EK I+K + K+ Q DQ+++
Sbjct: 342 LGMHQLSDLGVYPEQMVEISRQQDL------LREKEIQKINEKQN----LAGKQDDQNSN 391
Query: 244 VVIDNVEVTTA-STDSTPGSSYKMASWRKLPSAAEKNRVGQDAVK-------------DE 289
D VEV TA STDST S+++M+SW+KLPSAAEKNR + K DE
Sbjct: 392 SGADGVEVNTARSTDST-SSNFRMSSWKKLPSAAEKNRSLNNTPKAKGESKIQPKVYGDE 450
Query: 290 NAENWDTLSCHPDQHSQPSSRTRRPSWGYWGDQQNLLHDDSMICRICEVEIPILHVEEHS 349
NAEN + S QP+S R WG+W D Q + +D+SMICRICEVEIP++HVEEHS
Sbjct: 451 NAENLHSPS------GQPASADRSALWGFWADHQCVTYDNSMICRICEVEIPVVHVEEHS 504
Query: 350 RICTIADKCDLKGLTVNERLERVAETIEMLLDSLTPTSSLHEEFNELSLERNN------- 402
RICTIAD+CDLKG+ VN RLERVAE++E +L+S TP SS+ S +N
Sbjct: 505 RICTIADRCDLKGINVNLRLERVAESLEKILESWTPKSSVTPRAVADSARLSNSSRQEDL 564
Query: 403 --MSSRCSEDMLDLAP--DNTFVADDLNLSREISCEAHSLKPDHGAKISSPESLTPRSPL 458
+S RCS+DMLD P NTF D+LN+ E+S +G K SS SLTP SP
Sbjct: 565 DEISQRCSDDMLDCVPRSQNTFSLDELNILNEMSMT-------NGTKDSSAGSLTPPSPA 617
Query: 459 ITPRTSQIEMLLSVSGRRPISELESYDQINKLVEIARAVANANSCDESAFQDIVDCVEDL 518
TPR SQ+++LLS GR+ ISELE+Y QINKL++IAR+VAN N C S+ +++ +++L
Sbjct: 618 -TPRNSQVDLLLS--GRKTISELENYQQINKLLDIARSVANVNVCGYSSLDFMIEQLDEL 674
Query: 519 RCVIQNRKEDALIVDTFGRRIEKLLQEKYLTLCEQIHDERAESSNSMADEESSVDDDTIR 578
+ VIQ+RK DAL+V+TFGRRIEKLLQEKY+ LC I DE+ +SSN+M DEESS D+DT+R
Sbjct: 675 KYVIQDRKADALVVETFGRRIEKLLQEKYIELCGLIDDEKVDSSNAMPDEESSADEDTVR 734
Query: 579 SLRASPINGFSKDRTSIEDFEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKN 638
SLRASP+N +KDRTSIEDFEIIKPISRGAFGRVFLA+KR+TGDLFAIKVLKKADMIRKN
Sbjct: 735 SLRASPLNPRAKDRTSIEDFEIIKPISRGAFGRVFLAKKRATGDLFAIKVLKKADMIRKN 794
Query: 639 AVEGILAERDILISVRNPFVVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDMA 698
AVE ILAER+ILISVRNPFVVRF+YSFTC+ENLYLVMEYLNGGDL+S+LRNLGCLDEDMA
Sbjct: 795 AVESILAERNILISVRNPFVVRFFYSFTCRENLYLVMEYLNGGDLFSLLRNLGCLDEDMA 854
Query: 699 RVYIAEVVLALEYLHSQSIVHRDLKPDNLLIGQDGHIKLTDFGLSKVGLINSTEDLSAPA 758
R+YIAEVVLALEYLHS +I+HRDLKPDNLLI QDGHIKLTDFGLSKVGLINST+DLS +
Sbjct: 855 RIYIAEVVLALEYLHSVNIIHRDLKPDNLLINQDGHIKLTDFGLSKVGLINSTDDLSGES 914
Query: 759 SFTN-GFLVDDEPKPRHVSKREARQQQSIVGTPDYLAPEILLGMGHGTTADWWSVGVILY 817
S N GF +D K +H +++R++ ++VGTPDYLAPEILLGMGHG TADWWSVGVIL+
Sbjct: 915 SLGNSGFFAEDGSKAQHSQGKDSRKKHAVVGTPDYLAPEILLGMGHGKTADWWSVGVILF 974
Query: 818 ELLVGIPPFNADHAQQIFDNIINRDIQWPKHPEEISFEAYDLMNKLLIENPVQRLGVTGA 877
E+LVGIPPFNA+ QQIF+NIINRDI WP PEEIS+EA+DL+NKLL ENPVQRLG TGA
Sbjct: 975 EVLVGIPPFNAETPQQIFENIINRDIPWPNVPEEISYEAHDLINKLLTENPVQRLGATGA 1034
Query: 878 TEVKRHAFFKDVNWDTLARQKVVILILFEFDWFAPYLLISHTWDPTNFLQAMFIPSAEAH 937
EVK+H FFKD+NWDTLARQK AMF+PSAE
Sbjct: 1035 GEVKQHHFFKDINWDTLARQK-----------------------------AMFVPSAEPQ 1065
Query: 938 DTSYFMSRYIWNVEDDEHCAGGSDFYDHSETSSSGSGSDSLDEDAMFIPSAEAHDTSYFM 997
DTSYFMSRYIWN E DE+ GGSDF D ++T SS S
Sbjct: 1066 DTSYFMSRYIWNPE-DENVHGGSDFDDLTDTCSSSS------------------------ 1100
Query: 998 SRYIWNVEDDEHCAGGSDFYDHSETSSSGSGSDSLDEDGDECASLTEFGNSA-LGVQYSF 1056
+N +++ DGDEC SL EFGN L V+YSF
Sbjct: 1101 ----FNTQEE---------------------------DGDECGSLAEFGNGPNLAVKYSF 1129
Query: 1057 SNFSFKNISQLVSIN---MMHISKETPDDSNPS 1086
SNFSFKN+SQL SIN ++ +KE+ + SN S
Sbjct: 1130 SNFSFKNLSQLASINYDLVLKNAKESVEASNQS 1162
>ref|NP_188412.2| protein kinase, putative [Arabidopsis thaliana]
Length = 1296
Score = 962 bits (2486), Expect = 0.0
Identities = 549/1112 (49%), Positives = 696/1112 (62%), Gaps = 136/1112 (12%)
Query: 15 KDFQSPRYQEILRLTSGKKRKNRPDIKSFSHELNSKGVRPFPVWKNRAFG--QEIMEEIR 72
K+ +SPRYQ +LR+TS +++ DIKSFSHELNSKGVRPFP+WK R +E++ IR
Sbjct: 269 KESESPRYQALLRMTSAPRKRFPGDIKSFSHELNSKGVRPFPLWKPRRSNNVEEVLNLIR 328
Query: 73 AKFEKLKEEVDSDLGGFAGDLVGTLEKIPGSHPEWKEGLEDLLVVARQCAKMTAAEFWIN 132
AKFEK KEEV+SDL FA DLVG LEK SHPEW+E EDLL++AR CA T +FW+
Sbjct: 329 AKFEKAKEEVNSDLAVFAADLVGVLEKNAESHPEWEETFEDLLILARSCAMTTPGDFWLQ 388
Query: 133 CETIVQKLDDKRQDIPVGILKQAHTRLLFILSRCTRLVQFQKESVKEQDHILGLHQLSDL 192
CE IVQ LDD+RQ++P G+LKQ HTR+LFIL+RCTRL+QF KES E++ ++ L Q L
Sbjct: 389 CEGIVQDLDDRRQELPPGVLKQLHTRMLFILTRCTRLLQFHKESWGEEEQVVQLRQSRVL 448
Query: 193 GVYSEQIMKAEESCGFPPSDHEMAEKLIKKSHGKEQDKPITKQSQADQHASVVIDNVEVT 252
I K S G S KK++ +EQ K+ + +
Sbjct: 449 ----HSIEKIPPS-GAGRSYSAAKVPSTKKAYSQEQHGLDWKEDAVVRSVPPLAPPENYA 503
Query: 253 TASTDSTPGSSYKMASWRKLPSAAEK-------------------NRVGQDAVKDENAEN 293
++S P + +M+SW+KLPS A K N VG +D+ A
Sbjct: 504 IKESES-PANIDRMSSWKKLPSPALKTVKEAPASEEQNDSKVEPPNIVGSRQGRDDAAVA 562
Query: 294 WDTLSCHPDQHSQPSSRTRRPSWGYWGDQQNLLHDDSMICRICEVEIPILHVEEHSRICT 353
D H S SWGYWG+Q + + S++CRICE E+P HVE+HSR+CT
Sbjct: 563 ILNFPPAKDSHEHSSKHRHNISWGYWGEQPLISEESSIMCRICEEEVPTTHVEDHSRVCT 622
Query: 354 IADKCDLKGLTVNERLERVAETIEMLL------DSLTPTSS----------LHEEFNELS 397
+ADK D KGL+V+ERL VA T++ + DSL S L EE + LS
Sbjct: 623 LADKYDQKGLSVDERLMAVAGTLDKIAETFRHKDSLAAAESPDGMKVSNSHLTEESDVLS 682
Query: 398 LERNNMSSRCSEDMLDLAP--DNTFVADDLNLSREISCEAH-SLKPDHGAKISSPESLTP 454
++ S + SEDMLD P DN+ DDL +SC K D G SS S+TP
Sbjct: 683 PRLSDWSRKGSEDMLDCFPEADNSIFMDDLRGLPLMSCRTRFGPKSDQGMTTSSASSMTP 742
Query: 455 RSPLITPRTSQIEMLLSVSGRRPISELESYDQINKLVEIARAVANANSCDESAFQDIVDC 514
RSP+ TPR IE +L G+ + + Q+++L +IA+ A+A D+ + ++ C
Sbjct: 743 RSPIPTPRPDPIEQILG--GKGTFHDQDDIPQMSELADIAKCAADAIPGDDQSIPFLLSC 800
Query: 515 VEDLRCVIQNRKEDALIVDTFGRRIEKLLQEKYLTLCEQIHDERAESSNSMADEESSVDD 574
+EDLR VI RK DAL V+TFG RIEKL++EKY+ +CE + DE+ + +++ DE++ ++D
Sbjct: 801 LEDLRVVIDRRKFDALTVETFGTRIEKLIREKYVHMCELMDDEKVDLLSTVIDEDAPLED 860
Query: 575 DTIRSLRASPINGFSKDRTSIEDFEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADM 634
D +RSLR SP++ +DRTSI+DFEIIKPISRGAFGRVFLA+KR+TGDLFAIKVLKKADM
Sbjct: 861 DVVRSLRTSPVH--PRDRTSIDDFEIIKPISRGAFGRVFLAKKRTTGDLFAIKVLKKADM 918
Query: 635 IRKNAVEGILAERDILISVRNPFVVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLD 694
IRKNAVE ILAERDILI+VRNPFVVRF+YSFTC++NLYLVMEYLNGGDLYS+LRNLGCL+
Sbjct: 919 IRKNAVESILAERDILINVRNPFVVRFFYSFTCRDNLYLVMEYLNGGDLYSLLRNLGCLE 978
Query: 695 EDMARVYIAEVVLALEYLHSQSIVHRDLKPDNLLIGQDGHIKLTDFGLSKVGLINSTEDL 754
ED+ RVYIAEVVLALEYLHS+ +VHRDLKPDNLLI DGHIKLTDFGLSKVGLINST+DL
Sbjct: 979 EDIVRVYIAEVVLALEYLHSEGVVHRDLKPDNLLIAHDGHIKLTDFGLSKVGLINSTDDL 1038
Query: 755 SAPASFTNGFLVDDEPK-PRHVSKREARQQQSIVGTPDYLAPEILLGMGHGTTADWWSVG 813
+ PA L ++E + + E R+++S VGTPDYLAPEILLG GHG TADWWSVG
Sbjct: 1039 AGPAVSGTSLLDEEESRLAASEEQLERRKKRSAVGTPDYLAPEILLGTGHGATADWWSVG 1098
Query: 814 VILYELLVGIPPFNADHAQQIFDNIINRDIQWPKHPEEISFEAYDLMNKLLIENPVQRLG 873
+IL+EL+VGIPPFNA+H QQIFDNI+NR I WP PEE+S EA+D++++ L E+P QRLG
Sbjct: 1099 IILFELIVGIPPFNAEHPQQIFDNILNRKIPWPHVPEEMSAEAHDIIDRFLTEDPHQRLG 1158
Query: 874 VTGATEVKRHAFFKDVNWDTLARQKVVILILFEFDWFAPYLLISHTWDPTNFLQAMFIPS 933
GA EVK+H FFKD+NWDTLARQK A F+P+
Sbjct: 1159 ARGAAEVKQHIFFKDINWDTLARQK-----------------------------AAFVPA 1189
Query: 934 AE-AHDTSYFMSRYIWNVEDDEHCAGGSDFYDHSETSSSGSGSDSLDEDAMFIPSAEAHD 992
+E A DTSYF SRY WN D++ F PS E D
Sbjct: 1190 SESAIDTSYFRSRYSWNTSDEQ-----------------------------FFPSGEVPD 1220
Query: 993 TSYFMSRYIWNVEDDEHCAGGSDFYDHSETSSSGSGSDSLDEDG--DECASLTEFGNSAL 1050
S D ++S S + E+G +EC EF S +
Sbjct: 1221 YS-----------------------DADSMTNSSGCSSNHHEEGEAEECEGHAEF-ESGI 1256
Query: 1051 GVQYSFSNFSFKNISQLVSINMMHISKETPDD 1082
V YSFSNFSFKN+SQL SIN +SK D+
Sbjct: 1257 PVDYSFSNFSFKNLSQLASINYDLLSKGWKDE 1288
>ref|XP_469518.1| putative protein kinase [Oryza sativa] gi|13324795|gb|AAK18843.1|
putative protein kinase [Oryza sativa]
Length = 1274
Score = 958 bits (2477), Expect = 0.0
Identities = 546/1109 (49%), Positives = 705/1109 (63%), Gaps = 137/1109 (12%)
Query: 15 KDFQSPRYQEILRLTSGKKRKNRPDIKSFSHELNSKGVRPFPVWKNRAFG--QEIMEEIR 72
K+ +SPR++ I++ TS +++ DIKSFSHELNSKGVRPFP WK R +E+++ I+
Sbjct: 255 KESESPRFKAIMQATSAPRKRVPADIKSFSHELNSKGVRPFPFWKPRGIYNLKEVLKVIQ 314
Query: 73 AKFEKLKEEVDSDLGGFAGDLVGTLEKIPGSHPEWKEGLEDLLVVARQCAKMTAAEFWIN 132
+FEK KEEV+SDL FAGDLVG +EK SHPEWKE LEDLL++AR C MT EFW+
Sbjct: 315 VRFEKAKEEVNSDLAVFAGDLVGVMEKYADSHPEWKETLEDLLILARSCCVMTPGEFWLQ 374
Query: 133 CETIVQKLDDKRQDIPVGILKQAHTRLLFILSRCTRLVQFQKESVKEQDHILGLHQLSDL 192
CE IVQ LDD RQ++P+G+LK+ +TR+LFIL+RCTRL+QF KES +D ++ +
Sbjct: 375 CEGIVQDLDDHRQELPMGVLKKLYTRMLFILTRCTRLLQFHKESGFAEDEVV-------M 427
Query: 193 GVYSEQIMKAEESCGFPPSDHEM---AEKLIKKSHGKEQDKPITKQSQADQHASVVIDNV 249
+ I A+ P D ++ ++KS+ +EQ K+SQ + + +
Sbjct: 428 DQRDKIIQSADRQILAQPGDDTTTRGSKSDVRKSYSQEQHNLKWKRSQEIKPVKF-LSPL 486
Query: 250 EVTTASTDSTPGSSYKMASWRKLPSAAEKNRVGQDAVKDENA-ENWDTLSCHPDQ----- 303
+ T + + +++SW+ PS K +K+E+ + DT Q
Sbjct: 487 DTTDVKKEVESPTRERISSWKPFPSPVPKPPKDPTPIKEESPNKKTDTPPAVSSQAELNS 546
Query: 304 -------HSQPSSRTRRPSWGYWGDQQNLLHDDSMICRICEVEIPILHVEEHSRICTIAD 356
S P + SWG+W DQ N+ + S++CRICE +P +VE HS IC AD
Sbjct: 547 PVESTSHQSLPPKHQHKTSWGHWSDQPNISEEGSIMCRICEEYVPTHYVENHSAICASAD 606
Query: 357 KCDLKGLTVNERLERVAETIEMLLDSLTP----------------TSSLHEEFNELSLER 400
+CD KG++V+ERL RVAE +E L++S T SS++EE + S +
Sbjct: 607 RCDQKGVSVDERLIRVAEALEKLVESYTQKDLPNAVGSPDVAKVSNSSINEESDGPSPKL 666
Query: 401 NNMSSRCSEDMLDLAP--DNTFVADDLNLSREISCEAH-SLKPDHGAKISSPESLTPRSP 457
++ S R S DMLD D+T DD+ ++C+ K DHG SS S+TPRSP
Sbjct: 667 SDWSRRGSADMLDYLQEADSTISLDDIKNLPSMTCKTRFGPKSDHGMATSSAGSMTPRSP 726
Query: 458 LITPRTSQIEMLLSVSGRRPISELESYDQINKLVEIARAVANANSCDESAFQDIVDCVED 517
L TPR++ I+MLL+ GR I+E + QI +L +IAR +A +E A +V C+ED
Sbjct: 727 LTTPRSNHIDMLLA--GRSAINESDDLPQIVELADIARCIATTPLDEERALSLLVTCIED 784
Query: 518 LRCVIQNRKEDALIVDTFGRRIEKLLQEKYLTLCEQIHDERAESSNSMADEESSVDDDTI 577
L+ ++ RK +AL V TFG RIEKL +EKYL LC+ + ++ +S++++ DEE DD +
Sbjct: 785 LQEIVNRRKHEALTVQTFGTRIEKLHREKYLLLCDSVDMDKVDSASTVMDEE----DDVV 840
Query: 578 RSLRASPINGFSKDRTSIEDFEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRK 637
RSLRASP++ KDRTSI+DFEIIKPISRGAFGRVFLA+KR+TGDLFAIKVL+KADMIRK
Sbjct: 841 RSLRASPVHPV-KDRTSIDDFEIIKPISRGAFGRVFLAKKRTTGDLFAIKVLRKADMIRK 899
Query: 638 NAVEGILAERDILISVRNPFVVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDM 697
NAVE ILAERDILI+VRNPFVVRF+YSFT +ENLYLVMEYLNGGDLYS+LRNLGCLDED+
Sbjct: 900 NAVESILAERDILITVRNPFVVRFFYSFTSRENLYLVMEYLNGGDLYSLLRNLGCLDEDV 959
Query: 698 ARVYIAEVVLALEYLHSQSIVHRDLKPDNLLIGQDGHIKLTDFGLSKVGLINSTEDLSAP 757
AR+Y+AEVVLALEYLHS IVHRDLKPDNLLI DGHIKLTDFGLSKVGLINST+DLS P
Sbjct: 960 ARIYLAEVVLALEYLHSMHIVHRDLKPDNLLIAHDGHIKLTDFGLSKVGLINSTDDLSGP 1019
Query: 758 ASFTNGFLVDDEPKP---RHVSKREARQQQSIVGTPDYLAPEILLGMGHGTTADWWSVGV 814
A + DDEP+ + R RQ++S VGTPDYLAPEILLG GHGT+ADWWSVGV
Sbjct: 1020 AVSGSSLYGDDEPQMSEFEEMDHRARRQKRSAVGTPDYLAPEILLGTGHGTSADWWSVGV 1079
Query: 815 ILYELLVGIPPFNADHAQQIFDNIINRDIQWPKHPEEISFEAYDLMNKLLIENPVQRLGV 874
IL+EL+VGIPPFNA+H Q IFDNI+NR I WP PEE+S EA DL++KLL E+P QRLG
Sbjct: 1080 ILFELIVGIPPFNAEHPQTIFDNILNRKIPWPHVPEEMSSEAQDLIDKLLTEDPHQRLGA 1139
Query: 875 TGATEVKRHAFFKDVNWDTLARQKVVILILFEFDWFAPYLLISHTWDPTNFLQAMFIPSA 934
GA+EVK+H FFKD++WDTLARQK A F+PS+
Sbjct: 1140 NGASEVKQHQFFKDISWDTLARQK-----------------------------AAFVPSS 1170
Query: 935 E-AHDTSYFMSRYIWNVEDDEHCAGGSDFYDHSETSSSGSGSDSLDEDAMFIPSAEAHDT 993
+ A DTSYF SRY WN D+ + Y+ ++S +GS S S
Sbjct: 1171 DSAFDTSYFTSRYSWNPSDENI----YEAYEFEDSSDNGSLSGS---------------- 1210
Query: 994 SYFMSRYIWNVEDDEHCAGGSDFYDHSETSSSGSGSDSLDEDGDECASLTEFGNSALGVQ 1053
S + N +DD GDE + TEF +S+ V
Sbjct: 1211 ----SSCVSNHQDDM---------------------------GDESSGFTEFESSS-NVN 1238
Query: 1054 YSFSNFSFKNISQLVSINMMHISKETPDD 1082
YSFSNFSFKN+SQLVSIN ++K DD
Sbjct: 1239 YSFSNFSFKNLSQLVSINYDLLTKGLKDD 1267
>dbj|BAC41900.1| putative protein kinase [Arabidopsis thaliana]
Length = 1296
Score = 958 bits (2477), Expect = 0.0
Identities = 548/1112 (49%), Positives = 695/1112 (62%), Gaps = 136/1112 (12%)
Query: 15 KDFQSPRYQEILRLTSGKKRKNRPDIKSFSHELNSKGVRPFPVWKNRAFG--QEIMEEIR 72
K+ +SPRYQ +LR+TS +++ DIKSFSHELNSKGVRPFP+WK R +E++ IR
Sbjct: 269 KESESPRYQALLRMTSAPRKRFPGDIKSFSHELNSKGVRPFPLWKPRRSNNVEEVLNLIR 328
Query: 73 AKFEKLKEEVDSDLGGFAGDLVGTLEKIPGSHPEWKEGLEDLLVVARQCAKMTAAEFWIN 132
AKFEK KEEV+SDL FA DLVG LEK SHPEW+E EDLL++AR CA T +FW+
Sbjct: 329 AKFEKAKEEVNSDLAVFAADLVGVLEKNAESHPEWEETFEDLLILARSCAMTTPGDFWLQ 388
Query: 133 CETIVQKLDDKRQDIPVGILKQAHTRLLFILSRCTRLVQFQKESVKEQDHILGLHQLSDL 192
CE IVQ LDD+RQ++P G+LKQ HTR+LFIL+RCTRL+QF KES E++ ++ L Q L
Sbjct: 389 CEGIVQDLDDRRQELPPGVLKQLHTRMLFILTRCTRLLQFHKESWGEEEQVVQLRQSRVL 448
Query: 193 GVYSEQIMKAEESCGFPPSDHEMAEKLIKKSHGKEQDKPITKQSQADQHASVVIDNVEVT 252
I K S G S KK++ +EQ K+ + +
Sbjct: 449 ----HSIEKIPPS-GAGRSYSAAKVPSTKKAYSQEQHGLDWKEDAVVRSVPPLAPPENYA 503
Query: 253 TASTDSTPGSSYKMASWRKLPSAAEK-------------------NRVGQDAVKDENAEN 293
++S P + +M+SW+KLPS A K N VG +D+ A
Sbjct: 504 IKESES-PANIDRMSSWKKLPSPALKTVKEAPASEEQNDSKVEPPNIVGSRQGRDDAAVA 562
Query: 294 WDTLSCHPDQHSQPSSRTRRPSWGYWGDQQNLLHDDSMICRICEVEIPILHVEEHSRICT 353
D H S SWGYWG+Q + + S++CRICE E+P HVE+HSR+CT
Sbjct: 563 ILNFPPAKDSHEHSSKHRHNISWGYWGEQPLISEESSIMCRICEEEVPTTHVEDHSRVCT 622
Query: 354 IADKCDLKGLTVNERLERVAETIEMLL------DSLTPTSS----------LHEEFNELS 397
+ADK D KGL+V+ERL VA T++ + DSL S L EE + LS
Sbjct: 623 LADKYDQKGLSVDERLMAVAGTLDKIAETFRHKDSLAAAESPDGMKVSNSHLTEESDVLS 682
Query: 398 LERNNMSSRCSEDMLDLAP--DNTFVADDLNLSREISCEAH-SLKPDHGAKISSPESLTP 454
++ S + SEDMLD P DN+ DDL +SC K D G SS S+TP
Sbjct: 683 PRLSDWSRKGSEDMLDCFPEADNSIFMDDLRGLPLMSCRTRFGPKSDQGMTTSSASSMTP 742
Query: 455 RSPLITPRTSQIEMLLSVSGRRPISELESYDQINKLVEIARAVANANSCDESAFQDIVDC 514
RSP+ TPR IE +L G+ + + Q+++L +IA+ A+A D+ + ++ C
Sbjct: 743 RSPIPTPRPDPIEQILG--GKGTFHDQDDIPQMSELADIAKCAADAIPGDDQSIPFLLSC 800
Query: 515 VEDLRCVIQNRKEDALIVDTFGRRIEKLLQEKYLTLCEQIHDERAESSNSMADEESSVDD 574
+EDLR VI RK DAL V+TFG RIEKL++EKY+ +CE + DE+ + +++ DE++ ++D
Sbjct: 801 LEDLRVVIDRRKFDALTVETFGTRIEKLIREKYVHMCELMDDEKVDLLSTVIDEDAPLED 860
Query: 575 DTIRSLRASPINGFSKDRTSIEDFEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADM 634
D +RSLR SP++ +DRTSI+DFEIIKPISRGAFGRVFLA+KR+TGDLFAIKVLKKADM
Sbjct: 861 DVVRSLRTSPVH--PRDRTSIDDFEIIKPISRGAFGRVFLAKKRTTGDLFAIKVLKKADM 918
Query: 635 IRKNAVEGILAERDILISVRNPFVVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLD 694
IRKNAVE ILAERDILI+VRNPFVVRF+YSFTC++NLYLVMEYLNGGDLYS+LRNLGCL+
Sbjct: 919 IRKNAVESILAERDILINVRNPFVVRFFYSFTCRDNLYLVMEYLNGGDLYSLLRNLGCLE 978
Query: 695 EDMARVYIAEVVLALEYLHSQSIVHRDLKPDNLLIGQDGHIKLTDFGLSKVGLINSTEDL 754
ED+ RVYIAEVVLALEYLHS+ +VHRDLKPDNLLI DGHIKLTDFGLSKVGLINST+DL
Sbjct: 979 EDIVRVYIAEVVLALEYLHSEGVVHRDLKPDNLLIAHDGHIKLTDFGLSKVGLINSTDDL 1038
Query: 755 SAPASFTNGFLVDDEPK-PRHVSKREARQQQSIVGTPDYLAPEILLGMGHGTTADWWSVG 813
+ PA L ++E + + E R+++S VGTPDYLAPEILLG GHG TADWWSVG
Sbjct: 1039 AGPAVSGTSLLDEEESRLAASEEQLERRKKRSAVGTPDYLAPEILLGTGHGATADWWSVG 1098
Query: 814 VILYELLVGIPPFNADHAQQIFDNIINRDIQWPKHPEEISFEAYDLMNKLLIENPVQRLG 873
+IL+EL+VGIPPFNA+H QQIFDNI+NR I W PEE+S EA+D++++ L E+P QRLG
Sbjct: 1099 IILFELIVGIPPFNAEHPQQIFDNILNRKIPWHHVPEEMSAEAHDIIDRFLTEDPHQRLG 1158
Query: 874 VTGATEVKRHAFFKDVNWDTLARQKVVILILFEFDWFAPYLLISHTWDPTNFLQAMFIPS 933
GA EVK+H FFKD+NWDTLARQK A F+P+
Sbjct: 1159 ARGAAEVKQHIFFKDINWDTLARQK-----------------------------AAFVPA 1189
Query: 934 AE-AHDTSYFMSRYIWNVEDDEHCAGGSDFYDHSETSSSGSGSDSLDEDAMFIPSAEAHD 992
+E A DTSYF SRY WN D++ F PS E D
Sbjct: 1190 SESAIDTSYFRSRYSWNTSDEQ-----------------------------FFPSGEVPD 1220
Query: 993 TSYFMSRYIWNVEDDEHCAGGSDFYDHSETSSSGSGSDSLDEDG--DECASLTEFGNSAL 1050
S D ++S S + E+G +EC EF S +
Sbjct: 1221 YS-----------------------DADSMTNSSGCSSNHHEEGEAEECEGHAEF-ESGI 1256
Query: 1051 GVQYSFSNFSFKNISQLVSINMMHISKETPDD 1082
V YSFSNFSFKN+SQL SIN +SK D+
Sbjct: 1257 PVDYSFSNFSFKNLSQLASINYDLLSKGWKDE 1288
>dbj|BAA89784.1| IRE homolog 1 [Arabidopsis thaliana]
Length = 1023
Score = 957 bits (2475), Expect = 0.0
Identities = 547/1107 (49%), Positives = 692/1107 (62%), Gaps = 136/1107 (12%)
Query: 20 PRYQEILRLTSGKKRKNRPDIKSFSHELNSKGVRPFPVWKNRAFG--QEIMEEIRAKFEK 77
PRYQ +LR+TS +++ DIKSFSHELNSKGVRPFP+WK R +E++ IRAKFEK
Sbjct: 1 PRYQALLRMTSAPRKRFPGDIKSFSHELNSKGVRPFPLWKPRRSNNVEEVLNLIRAKFEK 60
Query: 78 LKEEVDSDLGGFAGDLVGTLEKIPGSHPEWKEGLEDLLVVARQCAKMTAAEFWINCETIV 137
KEEV+SDL FA DLVG LEK SHPEW+E EDLL++AR CA T +FW+ CE IV
Sbjct: 61 AKEEVNSDLAVFAADLVGVLEKNAESHPEWEETFEDLLILARSCAMTTPGDFWLQCEGIV 120
Query: 138 QKLDDKRQDIPVGILKQAHTRLLFILSRCTRLVQFQKESVKEQDHILGLHQLSDLGVYSE 197
Q LDD+RQ++P G+LKQ HTR+LFIL+RCTRL+QF KES E++ ++ L Q L
Sbjct: 121 QDLDDRRQELPPGVLKQLHTRMLFILTRCTRLLQFHKESWGEEEQVVQLRQSRVL----H 176
Query: 198 QIMKAEESCGFPPSDHEMAEKLIKKSHGKEQDKPITKQSQADQHASVVIDNVEVTTASTD 257
I K S G S KK++ +EQ K+ + + ++
Sbjct: 177 SIEKIPPS-GAGRSYSAAKVPSTKKAYSQEQHGLDWKEDAVVRSVPPLAPPENYAIKESE 235
Query: 258 STPGSSYKMASWRKLPSAAEK-------------------NRVGQDAVKDENAENWDTLS 298
S P + +M+SW+KLPS A K N VG +D+ A
Sbjct: 236 S-PANIDRMSSWKKLPSPALKTVKEAPASEEQNDSKVEPPNIVGSRQGRDDAAVAILNFP 294
Query: 299 CHPDQHSQPSSRTRRPSWGYWGDQQNLLHDDSMICRICEVEIPILHVEEHSRICTIADKC 358
D H S SWGYWG+Q + + S++CRICE E+P HVE+HSR+CT+ADK
Sbjct: 295 PAKDSHEHSSKHRHNISWGYWGEQPLISEESSIMCRICEEEVPTTHVEDHSRVCTLADKY 354
Query: 359 DLKGLTVNERLERVAETIEMLL------DSLTPTSS----------LHEEFNELSLERNN 402
D KGL+V+ERL VA T++ + DSL S L EE + LS ++
Sbjct: 355 DQKGLSVDERLMAVAGTLDKIAETFRHKDSLAAAESPDGMKVSNSHLTEESDVLSPRLSD 414
Query: 403 MSSRCSEDMLDLAP--DNTFVADDLNLSREISCEAH-SLKPDHGAKISSPESLTPRSPLI 459
S + SEDMLD P DN+ DDL +SC K D G SS S+TPRSP+
Sbjct: 415 WSRKGSEDMLDCFPEADNSIFMDDLRGLPLMSCRTRFGPKSDQGMTTSSASSMTPRSPIP 474
Query: 460 TPRTSQIEMLLSVSGRRPISELESYDQINKLVEIARAVANANSCDESAFQDIVDCVEDLR 519
TPR IE +L G+ + + Q+++L +IA+ A+A D+ + ++ C+EDLR
Sbjct: 475 TPRPDPIEQILG--GKGTFHDQDDIPQMSELADIAKCAADAIPGDDQSIPFLLSCLEDLR 532
Query: 520 CVIQNRKEDALIVDTFGRRIEKLLQEKYLTLCEQIHDERAESSNSMADEESSVDDDTIRS 579
VI RK DAL V+TFG RIEKL++EKY+ +CE + DE+ + +++ DE++ ++DD +RS
Sbjct: 533 VVIDRRKFDALTVETFGTRIEKLIREKYVHMCELMDDEKVDLLSTVIDEDAPLEDDVVRS 592
Query: 580 LRASPINGFSKDRTSIEDFEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNA 639
LR SP++ +DRTSI+DFEIIKPISRGAFGRVFLA+KR+TGDLFAIKVLKKADMIRKNA
Sbjct: 593 LRTSPVH--PRDRTSIDDFEIIKPISRGAFGRVFLAKKRTTGDLFAIKVLKKADMIRKNA 650
Query: 640 VEGILAERDILISVRNPFVVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDMAR 699
VE ILAERDILI+VRNPFVVRF+YSFTC++NLYLVMEYLNGGDLYS+LRNLGCL+ED+ R
Sbjct: 651 VESILAERDILINVRNPFVVRFFYSFTCRDNLYLVMEYLNGGDLYSLLRNLGCLEEDIVR 710
Query: 700 VYIAEVVLALEYLHSQSIVHRDLKPDNLLIGQDGHIKLTDFGLSKVGLINSTEDLSAPAS 759
VYIAEVVLALEYLHS+ +VHRDLKPDNLLI DGHIKLTDFGLSKVGLINST+DL+ PA
Sbjct: 711 VYIAEVVLALEYLHSEGVVHRDLKPDNLLIAHDGHIKLTDFGLSKVGLINSTDDLAGPAV 770
Query: 760 FTNGFLVDDEPK-PRHVSKREARQQQSIVGTPDYLAPEILLGMGHGTTADWWSVGVILYE 818
L ++E + + E R+++S VGTPDYLAPEILLG GHG TADWWSVG+IL+E
Sbjct: 771 SGTSLLDEEESRLAASEEQLERRKKRSAVGTPDYLAPEILLGTGHGATADWWSVGIILFE 830
Query: 819 LLVGIPPFNADHAQQIFDNIINRDIQWPKHPEEISFEAYDLMNKLLIENPVQRLGVTGAT 878
L+VGIPPFNA+H QQIFDNI+NR I WP PEE+S EA+D++++ L E+P QRLG GA
Sbjct: 831 LIVGIPPFNAEHPQQIFDNILNRKIPWPHVPEEMSAEAHDIIDRFLTEDPHQRLGARGAA 890
Query: 879 EVKRHAFFKDVNWDTLARQKVVILILFEFDWFAPYLLISHTWDPTNFLQAMFIPSAE-AH 937
EVK+H FFKD+NWDTLARQK A F+P++E A
Sbjct: 891 EVKQHIFFKDINWDTLARQK-----------------------------AAFVPASESAI 921
Query: 938 DTSYFMSRYIWNVEDDEHCAGGSDFYDHSETSSSGSGSDSLDEDAMFIPSAEAHDTSYFM 997
DTSYF SRY WN D++ F PS E D S
Sbjct: 922 DTSYFRSRYSWNTSDEQ-----------------------------FFPSGEVPDYS--- 949
Query: 998 SRYIWNVEDDEHCAGGSDFYDHSETSSSGSGSDSLDEDG--DECASLTEFGNSALGVQYS 1055
D ++S S + E+G +EC EF S + V YS
Sbjct: 950 --------------------DADSMTNSSGCSSNHHEEGEAEECEGHAEF-ESGIPVDYS 988
Query: 1056 FSNFSFKNISQLVSINMMHISKETPDD 1082
FSNFSFKN+SQL SIN +SK D+
Sbjct: 989 FSNFSFKNLSQLASINYDLLSKGWKDE 1015
>dbj|BAB02708.1| IRE homolog; protein kinase-like protein [Arabidopsis thaliana]
Length = 1398
Score = 948 bits (2450), Expect = 0.0
Identities = 545/1111 (49%), Positives = 693/1111 (62%), Gaps = 135/1111 (12%)
Query: 15 KDFQSPRYQEILRLTSGKKRKNRPDIKSFSHELNSKGVRPFPVWKNRAFG--QEIMEEIR 72
K+ +SPRYQ +LR+TS +++ DIKSFSHELNSKGVRPFP+WK R +E++ IR
Sbjct: 372 KESESPRYQALLRMTSAPRKRFPGDIKSFSHELNSKGVRPFPLWKPRRSNNVEEVLNLIR 431
Query: 73 AKFEKLKEEVDSDLGGFAGDLVGTLEKIPGSHPEWKEGLEDLLVVARQCAKMTAAEFWIN 132
AKFEK KEEV+SDL FA DLVG LEK SHPEW+E EDLL++AR CA T +FW+
Sbjct: 432 AKFEKAKEEVNSDLAVFAADLVGVLEKNAESHPEWEETFEDLLILARSCAMTTPGDFWLQ 491
Query: 133 CETIVQKLDDKRQDIPVGILKQAHTRLLFILSRCTRLVQFQKESVKEQDHILGLHQLSDL 192
CE IVQ LDD+RQ++P G+LKQ HTR+LFIL+RCTRL+QF KES E++ ++ L Q L
Sbjct: 492 CEGIVQDLDDRRQELPPGVLKQLHTRMLFILTRCTRLLQFHKESWGEEEQVVQLRQSRVL 551
Query: 193 GVYSEQIMKAEESCGFPPSDHEMAEKLIKKSHGKEQDKPITKQSQADQHASVVIDNVEVT 252
I K S G S KK++ +EQ K+ + +
Sbjct: 552 ----HSIEKIPPS-GAGRSYSAAKVPSTKKAYSQEQHGLDWKEDAVVRSVPPLAPPENYA 606
Query: 253 TASTDSTPGSSYKMASWRKLPSAAEK-------------------NRVGQDAVKDENAEN 293
++S P + +M+SW+KLPS A K N VG +D+ A
Sbjct: 607 IKESES-PANIDRMSSWKKLPSPALKTVKEAPASEEQNDSKVEPPNIVGSRQGRDDAAVA 665
Query: 294 WDTLSCHPDQHSQPSSRTRRPSWGYWGDQQNLLHDDSMICRICEVEIPILHVEEHSRICT 353
D H S SWGYWG+Q + + S++CRICE E+P HVE+HSR+CT
Sbjct: 666 ILNFPPAKDSHEHSSKHRHNISWGYWGEQPLISEESSIMCRICEEEVPTTHVEDHSRVCT 725
Query: 354 IADKCDLKGLTVNERLERVAETIEMLL------DSLTPTSS----------LHEEFNELS 397
+ADK D KGL+V+ERL VA T++ + DSL S L EE + LS
Sbjct: 726 LADKYDQKGLSVDERLMAVAGTLDKIAETFRHKDSLAAAESPDGMKVSNSHLTEESDVLS 785
Query: 398 LERNNMSSRCSEDMLDLAP--DNTFVADDLNLSREISCEAH-SLKPDHGAKISSPESLTP 454
++ S + SEDMLD P DN+ DDL +SC K D G SS S+TP
Sbjct: 786 PRLSDWSRKGSEDMLDCFPEADNSIFMDDLRGLPLMSCRTRFGPKSDQGMTTSSASSMTP 845
Query: 455 RSPLITPRTSQIEMLLSVSGRRPISELESYDQINKLVEIARAVANANSCDESAFQDIVDC 514
RSP+ TPR IE +L G+ + + Q+++L +IA+ A+A D+ + ++ C
Sbjct: 846 RSPIPTPRPDPIEQILG--GKGTFHDQDDIPQMSELADIAKCAADAIPGDDQSIPFLLSC 903
Query: 515 VEDLRCVIQNRKEDALIVDTFGRRIEKLLQEKYLTLCEQIHDERAESSNSMADEESSVDD 574
+EDLR VI RK DAL V+TFG RIEKL++EKY+ +CE + DE+ + +++ DE++ ++D
Sbjct: 904 LEDLRVVIDRRKFDALTVETFGTRIEKLIREKYVHMCELMDDEKVDLLSTVIDEDAPLED 963
Query: 575 DTIRSLRASPINGFSKDRTSIEDFEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADM 634
D +RSLR SP++ +DRTSI+DFEIIKPISRGAFGRVFLA+KR+TGDLFAIKVLKKADM
Sbjct: 964 DVVRSLRTSPVH--PRDRTSIDDFEIIKPISRGAFGRVFLAKKRTTGDLFAIKVLKKADM 1021
Query: 635 IRKNAVEGILAERDILISVRNPFVVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLD 694
IRKNAVE ILAERDILI+VRNPFVVRF+YSFTC++NLYLVMEYLNGGDLYS+LRNLGCL+
Sbjct: 1022 IRKNAVESILAERDILINVRNPFVVRFFYSFTCRDNLYLVMEYLNGGDLYSLLRNLGCLE 1081
Query: 695 EDMARVYIAEVVLALEYLHSQSIVHRDLKPDNLLIGQDGHIKLTDFGLSKVGLINSTEDL 754
ED+ RVYIAEVVLALEYLHS+ +VHRDLKPDNLLI DGHIKLTDFGLSKVGLINST+DL
Sbjct: 1082 EDIVRVYIAEVVLALEYLHSEGVVHRDLKPDNLLIAHDGHIKLTDFGLSKVGLINSTDDL 1141
Query: 755 SAPASFTNGFLVDDEPK-PRHVSKREARQQQSIVGTPDYLAPEILLGMGHGTTADWWSVG 813
+ PA L ++E + + E R+++S VGTPDYLAPEILLG GHG TADWWSVG
Sbjct: 1142 AGPAVSGTSLLDEEESRLAASEEQLERRKKRSAVGTPDYLAPEILLGTGHGATADWWSVG 1201
Query: 814 VILYELLVGIPPFNADHAQQIFDNIINRDIQWPKHPEEISFEAYDLMNKLLIENPVQRLG 873
+IL+EL+VGIPPFNA+H QQIFDNI+NR I WP PEE+S EA+D++++ L E+P QRLG
Sbjct: 1202 IILFELIVGIPPFNAEHPQQIFDNILNRKIPWPHVPEEMSAEAHDIIDRFLTEDPHQRLG 1261
Query: 874 VTGATEVKRHAFFKDVNWDTLARQKVVILILFEFDWFAPYLLISHTWDPTNFLQAMFIPS 933
GA EVK+H FFKD+NWDTLARQK A F+P+
Sbjct: 1262 ARGAAEVKQHIFFKDINWDTLARQK-----------------------------AAFVPA 1292
Query: 934 AE-AHDTSYFMSRYIWNVEDDEHCAGGSDFYDHSETSSSGSGSDSLDEDAMFIPSAEAHD 992
+E A DTSYF SRY WN D++ F+ S +F+
Sbjct: 1293 SESAIDTSYFRSRYSWNTSDEQ-------FFPSELVGKLLS--------LVFV------- 1330
Query: 993 TSYFMSRYIWNVEDD-EHCAGGSDFYDHSETSSSGSGSDSLDEDGDECASLTEFGNSALG 1051
Y +S NV E C G ++F S +
Sbjct: 1331 -GYAISLNWINVATQAEECEGHAEF------------------------------ESGIP 1359
Query: 1052 VQYSFSNFSFKNISQLVSINMMHISKETPDD 1082
V YSFSNFSFKN+SQL SIN +SK D+
Sbjct: 1360 VDYSFSNFSFKNLSQLASINYDLLSKGWKDE 1390
>gb|AAF79714.1| T1N15.10 [Arabidopsis thaliana]
Length = 1294
Score = 847 bits (2189), Expect = 0.0
Identities = 504/1059 (47%), Positives = 640/1059 (59%), Gaps = 140/1059 (13%)
Query: 15 KDFQSPRYQEILRLTSGKKRKNRPDIKSFSHELNSKGVRPFPVWKNRAFGQ--------- 65
K+ SPRYQ +LR+TS +++ DIKSFSHELNSKGVRPFP+WK R
Sbjct: 212 KESDSPRYQALLRMTSAPRKRFPGDIKSFSHELNSKGVRPFPLWKPRRLNNLEVVFEATF 271
Query: 66 -EIMEEIRAKFEKLKEEVDSDLGGFAGDLVGTLEKIPGSHPEWKEGLEDLLVVARQCAKM 124
+I+ IR KF+K KEEV+SDL F GDL+ +K SHPE +EDLLV+A+ CAK
Sbjct: 272 SDILNLIRTKFDKAKEEVNSDLFAFGGDLLDIYDKNKESHPELLVTIEDLLVLAKTCAKT 331
Query: 125 TAAEFWINCETIVQKLDDKRQDIPVGILKQAHTRLLFILSRCTRLVQFQKESVKEQDHIL 184
T+ EFW+ CE IVQ LDD+RQ++P G+LKQ HTR+LFIL+RCTRL+QF KES +++ +
Sbjct: 332 TSKEFWLQCEGIVQDLDDRRQELPPGVLKQLHTRMLFILTRCTRLLQFHKESWGQEEDAV 391
Query: 185 GLHQLSDLGVYSEQIMKAEESCGFPPSD-HEMAEKLIKKSHGKEQDKPITKQSQADQHAS 243
L Q L ++ E G S + + KK++ +EQ + + A
Sbjct: 392 QLRQSGVLHSADKRDPTGEVRDGKGSSTANALKVPSTKKAYSQEQRGLNWIEGFFVRPAP 451
Query: 244 VVIDNVEVTTASTDSTPGSSYKMASWRKLPSAAEKNRVGQDAVKDENAENWDT------- 296
+ E T+ +P + KM+SW++LPS A K K++N +
Sbjct: 452 LSSPYNE--TSKDSESPANIDKMSSWKRLPSPASKGVQEAAVSKEQNDRKVEPPQVVKKL 509
Query: 297 --------------LSCHPDQHSQPSSRTRRPSWGYWGDQQNLLHDDSMICRICEVEIPI 342
+S S SWGYWG Q + + S+ICRICE EIP
Sbjct: 510 VAISDDMAVAKLPEVSSAKASQEHMSKNRHNISWGYWGHQSCISEESSIICRICEEEIPT 569
Query: 343 LHVEEHSRICTIADKCDLKGLTVNERLERVAETIEMLLDSLTP----------------T 386
HVE+HSRIC +ADK D KG+ V+ERL VA T+E + D++
Sbjct: 570 THVEDHSRICALADKYDQKGVGVDERLMAVAVTLEKITDNVIQKDSLAAVESPEGMKISN 629
Query: 387 SSLHEEFNELSLERNNMSSRCSEDMLDLAP--DNTFVADDLNLSREISCEAH-SLKPDHG 443
+SL EE + LS + ++ S R SEDMLD P DN+ DD+ +SC K D G
Sbjct: 630 ASLTEELDVLSPKLSDWSRRGSEDMLDCFPETDNSVFMDDMGCLPSMSCRTRFGPKSDQG 689
Query: 444 AKISSPESLTPRSPLITPRTSQIEMLLSVSGRRPISELESYDQINKLVEIARAVANANSC 503
SS S+TPRSP+ TPR IE+LL G+ + + + Q+++L +IAR ANA
Sbjct: 690 MATSSAGSMTPRSPIPTPRPDPIELLLE--GKGTFHDQDDFPQMSELADIARCAANAIPV 747
Query: 504 DESAFQDIVDCVEDLRCVIQNRKEDALIVDTFGRRIEKLLQEKYLTLCEQIHDERAESSN 563
D+ + Q ++ C+EDLR VI RK L V+ + E LL EKYL LCE + DE+
Sbjct: 748 DDQSIQLLLSCLEDLRVVIDRRKTILLSVEASIAQAENLL-EKYLQLCELMDDEKG---- 802
Query: 564 SMADEESSVDDDTIRSLRASPINGFSKDRTSIEDFEIIKPISRGAFGRVFLAQKRSTGDL 623
++ DE++ ++DD +RSLR SP++ +DR SI+DFE++K ISRGAFG V LA+K +TGDL
Sbjct: 803 TIIDEDAPLEDDVVRSLRTSPVH--LRDRISIDDFEVMKSISRGAFGHVILARKNTTGDL 860
Query: 624 FAIKVLKKADMIRKNAVEGILAERDILISVRNPFVV-------------------RFYYS 664
FAIKVL+KADMIRKNAVE ILAERDILI+ RNPFVV RF+YS
Sbjct: 861 FAIKVLRKADMIRKNAVESILAERDILINARNPFVVSCSSFKHIKSSAEISYTNVRFFYS 920
Query: 665 FTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEV-------VLALEYLHSQSI 717
FTC ENLYLVMEYLNGGD YSMLR +GCLDE ARVYIAEV VLALEYLHS+ +
Sbjct: 921 FTCSENLYLVMEYLNGGDFYSMLRKIGCLDEANARVYIAEVCRYYSCQVLALEYLHSEGV 980
Query: 718 VHRDLKPDNLLIGQDGHIKLTDFGLSKVGLINSTEDLSAPASFTNGFLVDDEPK-PRHVS 776
VHRDLKPDNLLI DGH+KLTDFGLSKVGLIN+T+DLS P S LV+++PK P
Sbjct: 981 VHRDLKPDNLLIAHDGHVKLTDFGLSKVGLINNTDDLSGPVSSATSLLVEEKPKLPTLDH 1040
Query: 777 KREARQQQSIVGTPDYLAPEILLGMGHGTTADWWSVGVILYELLVGIPPFNADHAQQIFD 836
KR A VGTPDYLAPEILLG GHG TADWWSVG+ILYE LVGIPPFNADH QQIFD
Sbjct: 1041 KRSA------VGTPDYLAPEILLGTGHGATADWWSVGIILYEFLVGIPPFNADHPQQIFD 1094
Query: 837 NIINRDIQWPKHPEEISFEAYDLMNKLLIENPVQRLGVTGA----------TEVKRHAFF 886
NI+NR+IQWP PE++S EA DL+++LL E+P QRLG GA T+VK+H+FF
Sbjct: 1095 NILNRNIQWPPVPEDMSHEARDLIDRLLTEDPHQRLGARGAAEYSDETNIMTQVKQHSFF 1154
Query: 887 KDVNWDTLARQKVVILILFEFDWFAPYLLISHTWDPTNFLQAMFIPSAE-AHDTSYFMSR 945
KD++W+TLA+QK A F+P +E A DTSYF SR
Sbjct: 1155 KDIDWNTLAQQK-----------------------------AAFVPDSENAFDTSYFQSR 1185
Query: 946 YIWNVEDDEHCAGGSDFYDHSE----TSSSGSGSDSLDE 980
Y WN E C ++ D SE SSG S+ DE
Sbjct: 1186 YSWNY-SGERCFPTNENEDSSEGDSLCGSSGRLSNHHDE 1223
Score = 50.4 bits (119), Expect = 3e-04
Identities = 36/88 (40%), Positives = 45/88 (50%), Gaps = 7/88 (7%)
Query: 980 EDAMFIPSAE-AHDTSYFMSRYIWNVEDDEHCAGGSDFYDHSE----TSSSGSGSDSLDE 1034
+ A F+P +E A DTSYF SRY WN E C ++ D SE SSG S+ DE
Sbjct: 1165 QKAAFVPDSENAFDTSYFQSRYSWNY-SGERCFPTNENEDSSEGDSLCGSSGRLSNHHDE 1223
Query: 1035 DGDECASLTEFGNSALGVQYSFSNFSFK 1062
D EF +++ Y F NFSFK
Sbjct: 1224 GVDIPCGPAEF-ETSVSENYPFDNFSFK 1250
>gb|AAN72259.1| At1g48490/T1N15_9 [Arabidopsis thaliana] gi|14326578|gb|AAK60333.1|
At1g48490/T1N15_9 [Arabidopsis thaliana]
Length = 939
Score = 807 bits (2084), Expect = 0.0
Identities = 478/1011 (47%), Positives = 606/1011 (59%), Gaps = 140/1011 (13%)
Query: 119 RQCAKMTAAEFWINCETIVQKLDDKRQDIPVGILKQAHTRLLFILSRCTRLVQFQKESVK 178
++ AK T+ EFW+ CE IVQ LDD+RQ++P G+LKQ HTR+LFIL+RCTRL+QF KES
Sbjct: 22 QKLAKTTSKEFWLQCEGIVQDLDDRRQELPPGVLKQLHTRMLFILTRCTRLLQFHKESWG 81
Query: 179 EQDHILGLHQLSDLGVYSEQIMKAEESCGFPPSD-HEMAEKLIKKSHGKEQDKPITKQSQ 237
+++ + L Q L ++ E G S + + KK++ +EQ +
Sbjct: 82 QEEDAVQLRQSGVLHSADKRDPTGEVRDGKGSSTANALKVPSTKKAYSQEQRGLNWIEGF 141
Query: 238 ADQHASVVIDNVEVTTASTDSTPGSSYKMASWRKLPSAAEKNRVGQDAVKDENAENWDT- 296
+ A + E T+ +P + KM+SW++LPS A K K++N +
Sbjct: 142 FVRPAPLSSPYNE--TSKDSESPANIDKMSSWKRLPSPASKGVQEAAVSKEQNDRKVEPP 199
Query: 297 --------------------LSCHPDQHSQPSSRTRRPSWGYWGDQQNLLHDDSMICRIC 336
+S S SWGYWG Q + + S+ICRIC
Sbjct: 200 QVVKKLVAISDDMAVAKLPEVSSAKASQEHMSKNRHNISWGYWGHQSCISEESSIICRIC 259
Query: 337 EVEIPILHVEEHSRICTIADKCDLKGLTVNERLERVAETIEMLLDSLTP----------- 385
E EIP HVE+HSRIC +ADK D KG+ V+ERL VA T+E + D++
Sbjct: 260 EEEIPTTHVEDHSRICALADKYDQKGVGVDERLMAVAVTLEKITDNVIQKDSLAAVESPE 319
Query: 386 -----TSSLHEEFNELSLERNNMSSRCSEDMLDLAP--DNTFVADDLNLSREISCEAH-S 437
+SL EE + LS + ++ S R SEDMLD P DN+ DD+ +SC
Sbjct: 320 GMKISNASLTEELDVLSPKLSDWSRRGSEDMLDCFPETDNSVFMDDMGCLPSMSCRTRFG 379
Query: 438 LKPDHGAKISSPESLTPRSPLITPRTSQIEMLLSVSGRRPISELESYDQINKLVEIARAV 497
K D G SS S+TPRSP+ TPR IE+LL G+ + + + Q+++L +IAR
Sbjct: 380 PKSDQGMATSSAGSMTPRSPIPTPRPDPIELLLE--GKGTFHDQDDFPQMSELADIARCA 437
Query: 498 ANANSCDESAFQDIVDCVEDLRCVIQNRKEDALIVDTFGRRIEKLLQEKYLTLCEQIHDE 557
ANA D+ + Q ++ C+EDLR VI RK DALIV+TFG RIEKL+QEKYL LCE + DE
Sbjct: 438 ANAIPVDDQSIQLLLSCLEDLRVVIDRRKFDALIVETFGTRIEKLIQEKYLQLCELMDDE 497
Query: 558 RAESSNSMADEESSVDDDTIRSLRASPINGFSKDRTSIEDFEIIKPISRGAFGRVFLAQK 617
+ ++ DE++ ++DD +RSLR SP++ +DR SI+DFE++K ISRGAFG V LA+K
Sbjct: 498 KG----TIIDEDAPLEDDVVRSLRTSPVH--LRDRISIDDFEVMKSISRGAFGHVILARK 551
Query: 618 RSTGDLFAIKVLKKADMIRKNAVEGILAERDILISVRNPFVVRFYYSFTCKENLYLVMEY 677
+TGDLFAIKVL+KADMIRKNAVE ILAERDILI+ RNPFVVRF+YSFTC ENLYLVMEY
Sbjct: 552 NTTGDLFAIKVLRKADMIRKNAVESILAERDILINARNPFVVRFFYSFTCSENLYLVMEY 611
Query: 678 LNGGDLYSMLRNLGCLDEDMARVYIAEVVLALEYLHSQSIVHRDLKPDNLLIGQDGHIKL 737
LNGGD YSMLR +GCLDE ARVYIAEVVLALEYLHS+ +VHRDLKPDNLLI DGH+KL
Sbjct: 612 LNGGDFYSMLRKIGCLDEANARVYIAEVVLALEYLHSEGVVHRDLKPDNLLIAHDGHVKL 671
Query: 738 TDFGLSKVGLINSTEDLSAPASFTNGFLVDDEPK-PRHVSKREARQQQSIVGTPDYLAPE 796
TDFGLSKVGLIN+T+DLS P S LV+++PK P KR A VGTPDYLAPE
Sbjct: 672 TDFGLSKVGLINNTDDLSGPVSSATSLLVEEKPKLPTLDHKRSA------VGTPDYLAPE 725
Query: 797 ILLGMGHGTTADWWSVGVILYELLVGIPPFNADHAQQIFDNIINRDIQWPKHPEEISFEA 856
ILLG GHG TADWWSVG+ILYE LVGIPPFNADH QQIFDNI+NR+IQWP PE++S EA
Sbjct: 726 ILLGTGHGATADWWSVGIILYEFLVGIPPFNADHPQQIFDNILNRNIQWPPVPEDMSHEA 785
Query: 857 YDLMNKLLIENPVQRLGVTGATEVKRHAFFKDVNWDTLARQKVVILILFEFDWFAPYLLI 916
DL+++LL E+P QRLG GA EVK+H+FFKD++W+TLA+QK
Sbjct: 786 RDLIDRLLTEDPHQRLGARGAAEVKQHSFFKDIDWNTLAQQK------------------ 827
Query: 917 SHTWDPTNFLQAMFIPSAE-AHDTSYFMSRYIWNVEDDEHCAGGSDFYDHSETSSSGSGS 975
A F+P +E A DTSYF SRY WN E C
Sbjct: 828 -----------AAFVPDSENAFDTSYFQSRYSWNY-SGERC------------------- 856
Query: 976 DSLDEDAMFIPSAEAHDTSYFMSRYIWNVEDDEHCAGGSDFYDHSETSSSGSGSDSLDED 1035
P+ E D+S E D C SSG S+ DE
Sbjct: 857 ---------FPTNENKDSS----------EGDSLC------------GSSGRLSNHHDEG 885
Query: 1036 GDECASLTEFGNSALGVQYSFSNFSFKNISQLVSINMMHISKETPDDSNPS 1086
D EF +++ Y F NFSFKN+SQL IN +SK D++ PS
Sbjct: 886 VDIPCGPAEF-ETSVSENYPFDNFSFKNLSQLAYINYNLMSKGHKDETQPS 935
>ref|NP_564529.1| protein kinase, putative [Arabidopsis thaliana]
Length = 878
Score = 754 bits (1946), Expect = 0.0
Identities = 454/971 (46%), Positives = 574/971 (58%), Gaps = 140/971 (14%)
Query: 159 LLFILSRCTRLVQFQKESVKEQDHILGLHQLSDLGVYSEQIMKAEESCGFPPSD-HEMAE 217
+LFIL+RCTRL+QF KES +++ + L Q L ++ E G S + +
Sbjct: 1 MLFILTRCTRLLQFHKESWGQEEDAVQLRQSGVLHSADKRDPTGEVRDGKGSSTANALKV 60
Query: 218 KLIKKSHGKEQDKPITKQSQADQHASVVIDNVEVTTASTDSTPGSSYKMASWRKLPSAAE 277
KK++ +EQ + + A + E T+ +P + KM+SW++LPS A
Sbjct: 61 PSTKKAYSQEQRGLNWIEGFFVRPAPLSSPYNE--TSKDSESPANIDKMSSWKRLPSPAS 118
Query: 278 KNRVGQDAVKDENAENWDT---------------------LSCHPDQHSQPSSRTRRPSW 316
K K++N + +S S SW
Sbjct: 119 KGVQEAAVSKEQNDRKVEPPQVVKKLVAISDDMAVAKLPEVSSAKASQEHMSKNRHNISW 178
Query: 317 GYWGDQQNLLHDDSMICRICEVEIPILHVEEHSRICTIADKCDLKGLTVNERLERVAETI 376
GYWG Q + + S+ICRICE EIP HVE+HSRIC +ADK D KG+ V+ERL VA T+
Sbjct: 179 GYWGHQSCISEESSIICRICEEEIPTTHVEDHSRICALADKYDQKGVGVDERLMAVAVTL 238
Query: 377 EMLLDSLTP----------------TSSLHEEFNELSLERNNMSSRCSEDMLDLAP--DN 418
E + D++ +SL EE + LS + ++ S R SEDMLD P DN
Sbjct: 239 EKITDNVIQKDSLAAVESPEGMKISNASLTEELDVLSPKLSDWSRRGSEDMLDCFPETDN 298
Query: 419 TFVADDLNLSREISCEAH-SLKPDHGAKISSPESLTPRSPLITPRTSQIEMLLSVSGRRP 477
+ DD+ +SC K D G SS S+TPRSP+ TPR IE+LL G+
Sbjct: 299 SVFMDDMGCLPSMSCRTRFGPKSDQGMATSSAGSMTPRSPIPTPRPDPIELLLE--GKGT 356
Query: 478 ISELESYDQINKLVEIARAVANANSCDESAFQDIVDCVEDLRCVIQNRKEDALIVDTFGR 537
+ + + Q+++L +IAR ANA D+ + Q ++ C+EDLR VI RK DALIV+TFG
Sbjct: 357 FHDQDDFPQMSELADIARCAANAIPVDDQSIQLLLSCLEDLRVVIDRRKFDALIVETFGT 416
Query: 538 RIEKLLQEKYLTLCEQIHDERAESSNSMADEESSVDDDTIRSLRASPINGFSKDRTSIED 597
RIEKL+QEKYL LCE + DE+ ++ DE++ ++DD +RSLR SP++ +DR SI+D
Sbjct: 417 RIEKLIQEKYLQLCELMDDEKG----TIIDEDAPLEDDVVRSLRTSPVH--LRDRISIDD 470
Query: 598 FEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERDILISVRNPF 657
FE++K ISRGAFG V LA+K +TGDLFAIKVL+KADMIRKNAVE ILAERDILI+ RNPF
Sbjct: 471 FEVMKSISRGAFGHVILARKNTTGDLFAIKVLRKADMIRKNAVESILAERDILINARNPF 530
Query: 658 VVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALEYLHSQSI 717
VVRF+YSFTC ENLYLVMEYLNGGD YSMLR +GCLDE ARVYIAEVVLALEYLHS+ +
Sbjct: 531 VVRFFYSFTCSENLYLVMEYLNGGDFYSMLRKIGCLDEANARVYIAEVVLALEYLHSEGV 590
Query: 718 VHRDLKPDNLLIGQDGHIKLTDFGLSKVGLINSTEDLSAPASFTNGFLVDDEPK-PRHVS 776
VHRDLKPDNLLI DGH+KLTDFGLSKVGLIN+T+DLS P S LV+++PK P
Sbjct: 591 VHRDLKPDNLLIAHDGHVKLTDFGLSKVGLINNTDDLSGPVSSATSLLVEEKPKLPTLDH 650
Query: 777 KREARQQQSIVGTPDYLAPEILLGMGHGTTADWWSVGVILYELLVGIPPFNADHAQQIFD 836
KR A VGTPDYLAPEILLG GHG TADWWSVG+ILYE LVGIPPFNADH QQIFD
Sbjct: 651 KRSA------VGTPDYLAPEILLGTGHGATADWWSVGIILYEFLVGIPPFNADHPQQIFD 704
Query: 837 NIINRDIQWPKHPEEISFEAYDLMNKLLIENPVQRLGVTGATEVKRHAFFKDVNWDTLAR 896
NI+NR+IQWP PE++S EA DL+++LL E+P QRLG GA EVK+H+FFKD++W+TLA+
Sbjct: 705 NILNRNIQWPPVPEDMSHEARDLIDRLLTEDPHQRLGARGAAEVKQHSFFKDIDWNTLAQ 764
Query: 897 QKVVILILFEFDWFAPYLLISHTWDPTNFLQAMFIPSAE-AHDTSYFMSRYIWNVEDDEH 955
QK A F+P +E A DTSYF SRY WN E
Sbjct: 765 QK-----------------------------AAFVPDSENAFDTSYFQSRYSWNY-SGER 794
Query: 956 CAGGSDFYDHSETSSSGSGSDSLDEDAMFIPSAEAHDTSYFMSRYIWNVEDDEHCAGGSD 1015
C P+ E D+S E D C
Sbjct: 795 C----------------------------FPTNENEDSS----------EGDSLC----- 811
Query: 1016 FYDHSETSSSGSGSDSLDEDGDECASLTEFGNSALGVQYSFSNFSFKNISQLVSINMMHI 1075
SSG S+ DE D EF +++ Y F NFSFKN+SQL IN +
Sbjct: 812 -------GSSGRLSNHHDEGVDIPCGPAEF-ETSVSENYPFDNFSFKNLSQLAYINYNLM 863
Query: 1076 SKETPDDSNPS 1086
SK D++ PS
Sbjct: 864 SKGHKDETQPS 874
>ref|NP_175130.1| protein kinase family protein [Arabidopsis thaliana]
Length = 1067
Score = 607 bits (1566), Expect = e-172
Identities = 418/1058 (39%), Positives = 562/1058 (52%), Gaps = 173/1058 (16%)
Query: 40 IKSFSHELNSKGVRPFPVWKNRAFG--QEIMEEIRAKFEKLKEEVDSDLGGFAGDLVGTL 97
IKSFSHEL +G P + ++ +E++ + ++F+ KE VD L F D+ +
Sbjct: 134 IKSFSHELGPRGGVQTPYPRPHSYNDLKELLGSLHSRFDVAKETVDKKLDVFVRDVKEAM 193
Query: 98 EKIPGSHPEWKEGLEDLLVVARQCAKMTAAEFWINCETIVQKLDDKRQDIPVGILKQAHT 157
EK+ S PE +E E LL VAR C +MT+A+ CE+IVQ L KR+ G++K +
Sbjct: 194 EKMDPSCPEDREMAEQLLDVARACMEMTSAQLRATCESIVQDLTRKRKQCQAGLVKWLFS 253
Query: 158 RLLFILSRCTRLVQFQKESVKEQDHILGLHQLSDLGVYSEQIMKAEESCGFPP--SDHEM 215
+LLFIL+ CTR+V FQKE+ + E+I E G P D
Sbjct: 254 QLLFILTHCTRVVMFQKETEPIDES-----SFRKFKECLERIPALETDWGSTPRVDDSGS 308
Query: 216 AEKLIKKSHGKEQDKPITKQSQADQHA-SVVIDNVEVTTASTDSTPGSSYKMASWRKLPS 274
+++ ++ K K+S + A V+ N A+ + + + S P
Sbjct: 309 GYPEYQRNEAGQKFKRRDKESLESETALDYVVPNDHGNNAAREGYAAAKQEFPSHE--PQ 366
Query: 275 AAEKNRVGQDAVKDENAENWDTLSCHPDQHSQPSSRTRRPSWGYWGDQQNLLHDDSMICR 334
K + + DE D +S P + S D +ICR
Sbjct: 367 FDSKVVEQRFYLSDEYE---DKMSNEPGKELGGS--------------------DYVICR 403
Query: 335 ICEVEIPILHVEEHSRICTIADKCDLKGLTVNERLERVAETIEMLLDSLTPTSSLHEEFN 394
ICE E+P+ H+E HS IC ADKC++ + V+ERL ++ E +E ++DS + S
Sbjct: 404 ICEEEVPLFHLEPHSYICAYADKCEINCVDVDERLLKLEEILEQIIDSRSLNSFTQAGGL 463
Query: 395 ELSLERNN--MSSRCS---------------EDMLDLAPDNTFVADDLNLSREISCEAH- 436
E S+ R + S CS ED+ ++ D F+ D + + I ++H
Sbjct: 464 ENSVLRKSGVASEGCSPKINEWRNKGLEGMFEDLHEM--DTAFI--DESYTYPIHLKSHV 519
Query: 437 SLKPDHGAKISSPESLTPRSPLITPRTSQIEMLLSVSGRRPISELESYDQINKLVEIARA 496
K H A SS S+T S TPRTS + S R E E + L +IAR
Sbjct: 520 GAKFCHHATSSSTGSITSVSSTNTPRTSHFD---SYWLERHCPEQEDLRLMMDLSDIARC 576
Query: 497 VANANSCDESAFQDIVDCVEDLRCVIQNRKEDALIVDTFGRRIEKLLQEKYLTLCEQIHD 556
A+ + E + I+ C++D++ V++ K AL++DTFG RIEKLL EKYL E D
Sbjct: 577 GASTDFSKEGSCDYIMACMQDIQAVLKQGKLKALVIDTFGGRIEKLLCEKYLHARELTAD 636
Query: 557 ERAESSNSMADEESSVDDDTIRSLRASPINGFSKDRTSIEDFEIIKPISRGAFGRVFLAQ 616
+ +S+ + + S +D + A+P KDR SI+DFEIIKPISRGAFG+VFLA+
Sbjct: 637 K-----SSVGNIKES--EDVLEHASATP-QLLLKDRISIDDFEIIKPISRGAFGKVFLAR 688
Query: 617 KRSTGDLFAIKVLKKADMIRKNAVEGILAERDILISVRNPFVVRFYYSFTCKENLYLVME 676
KR+TGD FAIKVLKK DMIRKN +E IL ER+ILI+VR PF+VRF+YSFTC++NLYLVME
Sbjct: 689 KRTTGDFFAIKVLKKLDMIRKNDIERILQERNILITVRYPFLVRFFYSFTCRDNLYLVME 748
Query: 677 YLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALEYLHSQSIVHRDLKPDNLLIGQDGHIK 736
YLNGGDLYS+L+ +GCLDE++AR+YIAE+VLALEYLHS IVHRDLKPDNLLI +GHIK
Sbjct: 749 YLNGGDLYSLLQKVGCLDEEIARIYIAELVLALEYLHSLKIVHRDLKPDNLLIAYNGHIK 808
Query: 737 LTDFGLSKVGLINSTEDLSAPASFTNGFLVDDEPKPRHVSK--REARQQQSIVGTPDYLA 794
LTDFGLSK+GLIN+T DLS S V H K E R + S VGTPDYLA
Sbjct: 809 LTDFGLSKIGLINNTIDLSGHESD-----VSPRTNSHHFQKNQEEERIRHSAVGTPDYLA 863
Query: 795 PEILLGMGHGTTADWWSVGVILYELLVGIPPFNADHAQQIFDNIINRDIQWPKHPEEISF 854
PEILLG HG ADWWS G++L+ELL GIPPF A ++IFDNI+N + WP P E+S+
Sbjct: 864 PEILLGTEHGYAADWWSAGIVLFELLTGIPPFTASRPEKIFDNILNGKMPWPDVPGEMSY 923
Query: 855 EAYDLMNKLLIENPVQRLGVTGATEVKRHAFFKDVNWDTLARQKVVILILFEFDWFAPYL 914
EA DL+N+LL+ P +RLG GA EVK H FF+ V+W+ LA QK
Sbjct: 924 EAQDLINRLLVHEPEKRLGANGAAEVKSHPFFQGVDWENLALQK---------------- 967
Query: 915 LISHTWDPTNFLQAMFIPSAEA-HDTSYFMSRYIWNVEDDEHCAGGSDFYDHSETSSSGS 973
A F+P E+ +DTSYF+SR+ + C+ D ++SGS
Sbjct: 968 -------------AAFVPQPESINDTSYFVSRF-----SESSCS------DTETGNNSGS 1003
Query: 974 GSDSLDEDAMFIPSAEAHDTSYFMSRYIWNVEDDEHCAGGSDFYDHSETSSSGSGSDSLD 1033
DS DE LD
Sbjct: 1004 NPDSGDE---------------------------------------------------LD 1012
Query: 1034 EDGDECASLTEFGNSALGVQYSFSNFSFKNISQLVSIN 1071
E C +L +F + S NFSFKN+SQL SIN
Sbjct: 1013 E----CTNLEKFDSPP--YYLSLINFSFKNLSQLASIN 1044
>pir||H96509 protein F27F5.23 [imported] - Arabidopsis thaliana
gi|7767670|gb|AAF69167.1| F27F5.23 [Arabidopsis thaliana]
Length = 1092
Score = 590 bits (1520), Expect = e-166
Identities = 418/1083 (38%), Positives = 562/1083 (51%), Gaps = 198/1083 (18%)
Query: 40 IKSFSHELNSKGVRPFPVWKNRAFG--QEIMEEIRAKFEKLKEEVDSDLGGFAGDLVGTL 97
IKSFSHEL +G P + ++ +E++ + ++F+ KE VD L F D+ +
Sbjct: 134 IKSFSHELGPRGGVQTPYPRPHSYNDLKELLGSLHSRFDVAKETVDKKLDVFVRDVKEAM 193
Query: 98 EKIPGSHPEWKEGLEDLLVVARQCAKMTAAEFWINCETIVQKLDDKRQDIPVGILKQAHT 157
EK+ S PE +E E LL VAR C +MT+A+ CE+IVQ L KR+ G++K +
Sbjct: 194 EKMDPSCPEDREMAEQLLDVARACMEMTSAQLRATCESIVQDLTRKRKQCQAGLVKWLFS 253
Query: 158 RLLFILSRCTRLVQFQKESVKEQDHILGLHQLSDLGVYSEQIMKAEESCGFPP--SDHEM 215
+LLFIL+ CTR+V FQKE+ + E+I E G P D
Sbjct: 254 QLLFILTHCTRVVMFQKETEPIDES-----SFRKFKECLERIPALETDWGSTPRVDDSGS 308
Query: 216 AEKLIKKSHGKEQDKPITKQSQADQHA-SVVIDNVEVTTASTDSTPGSSYKMASWRKLPS 274
+++ ++ K K+S + A V+ N A+ + + + S P
Sbjct: 309 GYPEYQRNEAGQKFKRRDKESLESETALDYVVPNDHGNNAAREGYAAAKQEFPSHE--PQ 366
Query: 275 AAEKNRVGQDAVKDENAENWDTLSCHPDQHSQPSSRTRRPSWGYWGDQQNLLHDDSMICR 334
K + + DE D +S P + S D +ICR
Sbjct: 367 FDSKVVEQRFYLSDEYE---DKMSNEPGKELGGS--------------------DYVICR 403
Query: 335 ICEVEIPILHVEEHSRICTIADKCDLKGLTVNERLERVAETIEMLLDSLTPTSSLHEEFN 394
ICE E+P+ H+E HS IC ADKC++ + V+ERL ++ E +E ++DS + S
Sbjct: 404 ICEEEVPLFHLEPHSYICAYADKCEINCVDVDERLLKLEEILEQIIDSRSLNSFTQAGGL 463
Query: 395 ELSLERNN--MSSRCS---------------EDMLDLAPDNTFVADDLNLSREISCEAH- 436
E S+ R + S CS ED+ ++ D F+ D + + I ++H
Sbjct: 464 ENSVLRKSGVASEGCSPKINEWRNKGLEGMFEDLHEM--DTAFI--DESYTYPIHLKSHV 519
Query: 437 SLKPDHGAKISSPESLTPRSPLITPRTSQIEMLLSVSGRRPISELESYDQINKLVEIARA 496
K H A SS S+T S TPRTS + S R E E + L +IAR
Sbjct: 520 GAKFCHHATSSSTGSITSVSSTNTPRTSHFD---SYWLERHCPEQEDLRLMMDLSDIARC 576
Query: 497 VANANSCDESAFQDIVDCVEDLRCVIQNRKEDALIVDTFGRRIEKLLQEKYLTLCEQIHD 556
A+ + E + I+ C++D++ V++ K AL++DTFG RIEKLL EKYL E D
Sbjct: 577 GASTDFSKEGSCDYIMACMQDIQAVLKQGKLKALVIDTFGGRIEKLLCEKYLHARELTAD 636
Query: 557 ERAESSNSMADEESSVDDDTIRSLRASPINGFSKDRTSIEDFEIIKPISRGAFGRVFLAQ 616
+ +S+ + + S +D + A+P KDR SI+DFEIIKPISRGAFG+VFLA+
Sbjct: 637 K-----SSVGNIKES--EDVLEHASATP-QLLLKDRISIDDFEIIKPISRGAFGKVFLAR 688
Query: 617 KRSTGDLFAIKVLKKADMIRKNAVEGILAERDILISVRNPFV---------VRFYYSFTC 667
KR+TGD FAIKVLKK DMIRKN +E IL ER+ILI+VR PF+ VRF+YSFTC
Sbjct: 689 KRTTGDFFAIKVLKKLDMIRKNDIERILQERNILITVRYPFLAEHLMLLMQVRFFYSFTC 748
Query: 668 KENLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALEYLHSQSIVHRDLKPDNL 727
++NLYLVMEYLNGGDLYS+L+ +GCLDE++AR+YIAE+VLALEYLHS IVHRDLKPDNL
Sbjct: 749 RDNLYLVMEYLNGGDLYSLLQKVGCLDEEIARIYIAELVLALEYLHSLKIVHRDLKPDNL 808
Query: 728 LIGQDGHIKLTDFGLSKVGLINSTEDLSAPASFTNGFLVDDEPKPRHVSK--REARQQQS 785
LI +GHIKLTDFGLSK+GLIN+T DLS S V H K E R + S
Sbjct: 809 LIAYNGHIKLTDFGLSKIGLINNTIDLSGHESD-----VSPRTNSHHFQKNQEEERIRHS 863
Query: 786 IVGTPDYLAPEILLGMGHGT----------------TADWWSVGVILYELLVGIPPFNAD 829
VGTPDYLAPEILLG HG ADWWS G++L+ELL GIPPF A
Sbjct: 864 AVGTPDYLAPEILLGTEHGLDTTFKSGFHEAPVNCYAADWWSAGIVLFELLTGIPPFTAS 923
Query: 830 HAQQIFDNIINRDIQWPKHPEEISFEAYDLMNKLLIENPVQRLGVTGATEVKRHAFFKDV 889
++IFDNI+N + WP P E+S+EA DL+N+LL+ P +RLG GA EVK H FF+ V
Sbjct: 924 RPEKIFDNILNGKMPWPDVPGEMSYEAQDLINRLLVHEPEKRLGANGAAEVKSHPFFQGV 983
Query: 890 NWDTLARQKVVILILFEFDWFAPYLLISHTWDPTNFLQAMFIPSAEA-HDTSYFMSRYIW 948
+W+ LA QK A F+P E+ +DTSYF+SR+
Sbjct: 984 DWENLALQK-----------------------------AAFVPQPESINDTSYFVSRF-- 1012
Query: 949 NVEDDEHCAGGSDFYDHSETSSSGSGSDSLDEDAMFIPSAEAHDTSYFMSRYIWNVEDDE 1008
+ C+ D ++SGS DS DE
Sbjct: 1013 ---SESSCS------DTETGNNSGSNPDSGDE---------------------------- 1035
Query: 1009 HCAGGSDFYDHSETSSSGSGSDSLDEDGDECASLTEFGNSALGVQYSFSNFSFKNISQLV 1068
LDE C +L +F + S NFSFKN+SQL
Sbjct: 1036 -----------------------LDE----CTNLEKFDSPP--YYLSLINFSFKNLSQLA 1066
Query: 1069 SIN 1071
SIN
Sbjct: 1067 SIN 1069
>ref|XP_645246.1| protein serine/threonine kinase [Dictyostelium discoideum]
gi|60473347|gb|EAL71293.1| putative AGC family protein
kinase [Dictyostelium discoideum]
Length = 2102
Score = 344 bits (882), Expect = 1e-92
Identities = 284/985 (28%), Positives = 427/985 (42%), Gaps = 227/985 (23%)
Query: 194 VYSEQIMKAEESCGFPPSDHEMAEKLIKKSHGKEQ----DKPITKQSQADQH------AS 243
+ S + + S + P ++A +LI++ ++Q ++P+ Q Q Q ++
Sbjct: 1169 ISSRNDLISSNSSIYEPPLSDVASQLIEQQQEQQQQQHQEQPLQPQQQQQQQPQPQPIST 1228
Query: 244 VVIDNVEVTTASTDSTPGSSYK-MASWRKLPSAAEKNRVGQDAVKDENAENWDTLSCHPD 302
+V D ++S S + N +G + E + +P+
Sbjct: 1229 LVFDKANQRSSSPILEKKSILTDLLKENYQKETTNNNGIGNMIILSEYDKKLQKQQLYPN 1288
Query: 303 QHSQPSSRTRRPSWGYWGDQQNLLHDDSMICRICEVEIPILHVEEHSRICTIADKCDLKG 362
QH + R + S +CRICE + +H+ C + +K D K
Sbjct: 1289 QHVIITEEMRNKP------------ERSFVCRICEDTYTQSQLAKHTPFCALTNKHDFKN 1336
Query: 363 LTVN-ERLERVAETIEMLLDSLTPTSSLHEEFNELSLERNNMSSRCSEDMLDLAPDNTFV 421
+ + ERL +
Sbjct: 1337 HSSHDERLYSI------------------------------------------------- 1347
Query: 422 ADDLNLSREISCEAHSLKPDHGAKISSPESLTPRSPLITPRTSQIEMLLSVSGRRPISEL 481
LNL + I C++ + P++ S S +I+ Q+E L ++ P +
Sbjct: 1348 ---LNLVKGIICDSFA-SPNN----SEQYSYLIDDEIISQLGQQLEYLFNI----PYGPV 1395
Query: 482 ESYDQINKLVEIARAVANANSCDESAFQDIVDCVEDLRCVIQNRKEDALIVDTFGRRIEK 541
ES Q ++ +A+ + NS D + + FG+RI K
Sbjct: 1396 ESPKQCQDVINRVQAIIDENSTD-------------------------MALHVFGKRICK 1430
Query: 542 LLQEKYLTLCEQIHDERAESSNSMADEE-----------------SSVDDDTIRSLRASP 584
+++EK + + A + + ++ SS D + S +SP
Sbjct: 1431 IIEEKKALFVQYSKFQNAAQTTTSKGKKWSMWGIIPFIKDIIPSPSSKVDSSSSSQISSP 1490
Query: 585 I-----------------------NGFSKDRTSIEDFEIIKPISRGAFGRVFLAQKRSTG 621
I + SI DFEIIKPISRGAFGRV+LAQK+ TG
Sbjct: 1491 ILSSPPPPMKQPPPQVIVPPSSLTTSSNTTSISIADFEIIKPISRGAFGRVYLAQKKKTG 1550
Query: 622 DLFAIKVLKKADMIRKNAVEGILAERDILISVRNPFVVRFYYSFTCKENLYLVMEYLNGG 681
DL+AIKVLKK D IRKN V ++ ER+IL V+N FVV+ +Y+F + LYLVMEYL GG
Sbjct: 1551 DLYAIKVLKKLDTIRKNMVNHVIVERNILAMVQNEFVVKLFYAFQSTDKLYLVMEYLIGG 1610
Query: 682 DLYSMLRNLGCLDEDMARVYIAEVVLALEYLHSQSIVHRDLKPDNLLIGQDGHIKLTDFG 741
D S+LR LGC +E MA+ YIAE VL LEYLH +IVHRDLKPDN+LI GHIKLTDFG
Sbjct: 1611 DCASLLRALGCFEEHMAKHYIAETVLCLEYLHKSAIVHRDLKPDNMLIDGLGHIKLTDFG 1670
Query: 742 LSKVGLI-----------NSTEDLSAPASFTNGFLVDDE--------------------- 769
LSK+G+I N+ + S TN ++DD
Sbjct: 1671 LSKIGIIDDKKMEDSGNTNTNTHFNFSTSPTNTSMMDDSSTTGNPNGNGNTSLNSSQTNI 1730
Query: 770 ----PKPRHVSKREARQQ-QSIVGTPDYLAPEILLGMGHGTTADWWSVGVILYELLVGIP 824
P+ ++ K ++ + +VGTPDYL+PEILLG GHG T DWW++G+ILYE L G P
Sbjct: 1731 LSPYPQRKNTLKTPLKKPVKKVVGTPDYLSPEILLGTGHGQTVDWWALGIILYEFLTGSP 1790
Query: 825 PFNADHAQQIFDNIINRD--IQWPKHPEEISFEAYDLMNKLLIENPVQRLGVTGATEVKR 882
PFN D + IF +I++RD ++W PEEIS EA DL+ KLL +P +RLG GA EVK
Sbjct: 1791 PFNDDTPELIFQHILHRDREMEW---PEEISSEAKDLILKLLNPDPYKRLGANGAYEVKT 1847
Query: 883 HAFFKDVNWDTLARQKVVILILFEFDWFAPYLLISHTWDPTNFLQAMFIPSAE-AHDTSY 941
H FF +VNWDTL Q+ + +F+P E +DT Y
Sbjct: 1848 HPFFANVNWDTLIDQE---------------------------MDNIFLPKPENNYDTDY 1880
Query: 942 FMSRYIWNVEDDEHCAGGSDFYDHSETSSSGSGSDSLDEDAMFIPSAEAHDTSYFMSRYI 1001
F R ++ DDE DF +++ + P ++ + S
Sbjct: 1881 FWDRQ--SMYDDE---AEDDFLTINQSQPQHQSQHQSQPQSQPQPQSQNLGQNNNNSESN 1935
Query: 1002 WNVEDDEHCAGGSDFYDHSETSSSGSGSDSLDE--DGDECASLTEFGNSALGVQYSFSNF 1059
N + G + + +SS+ +GS +L+ + + N LG + N
Sbjct: 1936 NNNNNSNSNVSGQNINNSVSSSSNNNGSGNLNNICNNSNVNANNNNSNGNLGNNNNNKNN 1995
Query: 1060 SFKNISQLVSINMMHISKETPDDSN 1084
+ N + N ++ +++N
Sbjct: 1996 NINNDNDKNKNNNSNVQNNNNNNNN 2020
>gb|AAS45329.1| similar to cell wall biosynthesis kinase; Cbk1p [Saccharomyces
cerevisiae] [Dictyostelium discoideum]
Length = 2254
Score = 344 bits (882), Expect = 1e-92
Identities = 284/985 (28%), Positives = 427/985 (42%), Gaps = 227/985 (23%)
Query: 194 VYSEQIMKAEESCGFPPSDHEMAEKLIKKSHGKEQ----DKPITKQSQADQH------AS 243
+ S + + S + P ++A +LI++ ++Q ++P+ Q Q Q ++
Sbjct: 1321 ISSRNDLISSNSSIYEPPLSDVASQLIEQQQEQQQQQHQEQPLQPQQQQQQQPQPQPIST 1380
Query: 244 VVIDNVEVTTASTDSTPGSSYK-MASWRKLPSAAEKNRVGQDAVKDENAENWDTLSCHPD 302
+V D ++S S + N +G + E + +P+
Sbjct: 1381 LVFDKANQRSSSPILEKKSILTDLLKENYQKETTNNNGIGNMIILSEYDKKLQKQQLYPN 1440
Query: 303 QHSQPSSRTRRPSWGYWGDQQNLLHDDSMICRICEVEIPILHVEEHSRICTIADKCDLKG 362
QH + R + S +CRICE + +H+ C + +K D K
Sbjct: 1441 QHVIITEEMRNKP------------ERSFVCRICEDTYTQSQLAKHTPFCALTNKHDFKN 1488
Query: 363 LTVN-ERLERVAETIEMLLDSLTPTSSLHEEFNELSLERNNMSSRCSEDMLDLAPDNTFV 421
+ + ERL +
Sbjct: 1489 HSSHDERLYSI------------------------------------------------- 1499
Query: 422 ADDLNLSREISCEAHSLKPDHGAKISSPESLTPRSPLITPRTSQIEMLLSVSGRRPISEL 481
LNL + I C++ + P++ S S +I+ Q+E L ++ P +
Sbjct: 1500 ---LNLVKGIICDSFA-SPNN----SEQYSYLIDDEIISQLGQQLEYLFNI----PYGPV 1547
Query: 482 ESYDQINKLVEIARAVANANSCDESAFQDIVDCVEDLRCVIQNRKEDALIVDTFGRRIEK 541
ES Q ++ +A+ + NS D + + FG+RI K
Sbjct: 1548 ESPKQCQDVINRVQAIIDENSTD-------------------------MALHVFGKRICK 1582
Query: 542 LLQEKYLTLCEQIHDERAESSNSMADEE-----------------SSVDDDTIRSLRASP 584
+++EK + + A + + ++ SS D + S +SP
Sbjct: 1583 IIEEKKALFVQYSKFQNAAQTTTSKGKKWSMWGIIPFIKDIIPSPSSKVDSSSSSQISSP 1642
Query: 585 I-----------------------NGFSKDRTSIEDFEIIKPISRGAFGRVFLAQKRSTG 621
I + SI DFEIIKPISRGAFGRV+LAQK+ TG
Sbjct: 1643 ILSSPPPPMKQPPPQVIVPPSSLTTSSNTTSISIADFEIIKPISRGAFGRVYLAQKKKTG 1702
Query: 622 DLFAIKVLKKADMIRKNAVEGILAERDILISVRNPFVVRFYYSFTCKENLYLVMEYLNGG 681
DL+AIKVLKK D IRKN V ++ ER+IL V+N FVV+ +Y+F + LYLVMEYL GG
Sbjct: 1703 DLYAIKVLKKLDTIRKNMVNHVIVERNILAMVQNEFVVKLFYAFQSTDKLYLVMEYLIGG 1762
Query: 682 DLYSMLRNLGCLDEDMARVYIAEVVLALEYLHSQSIVHRDLKPDNLLIGQDGHIKLTDFG 741
D S+LR LGC +E MA+ YIAE VL LEYLH +IVHRDLKPDN+LI GHIKLTDFG
Sbjct: 1763 DCASLLRALGCFEEHMAKHYIAETVLCLEYLHKSAIVHRDLKPDNMLIDGLGHIKLTDFG 1822
Query: 742 LSKVGLI-----------NSTEDLSAPASFTNGFLVDDE--------------------- 769
LSK+G+I N+ + S TN ++DD
Sbjct: 1823 LSKIGIIDDKKMEDSGNTNTNTHFNFSTSPTNTSMMDDSSTTGNPNGNGNTSLNSSQTNI 1882
Query: 770 ----PKPRHVSKREARQQ-QSIVGTPDYLAPEILLGMGHGTTADWWSVGVILYELLVGIP 824
P+ ++ K ++ + +VGTPDYL+PEILLG GHG T DWW++G+ILYE L G P
Sbjct: 1883 LSPYPQRKNTLKTPLKKPVKKVVGTPDYLSPEILLGTGHGQTVDWWALGIILYEFLTGSP 1942
Query: 825 PFNADHAQQIFDNIINRD--IQWPKHPEEISFEAYDLMNKLLIENPVQRLGVTGATEVKR 882
PFN D + IF +I++RD ++W PEEIS EA DL+ KLL +P +RLG GA EVK
Sbjct: 1943 PFNDDTPELIFQHILHRDREMEW---PEEISSEAKDLILKLLNPDPYKRLGANGAYEVKT 1999
Query: 883 HAFFKDVNWDTLARQKVVILILFEFDWFAPYLLISHTWDPTNFLQAMFIPSAE-AHDTSY 941
H FF +VNWDTL Q+ + +F+P E +DT Y
Sbjct: 2000 HPFFANVNWDTLIDQE---------------------------MDNIFLPKPENNYDTDY 2032
Query: 942 FMSRYIWNVEDDEHCAGGSDFYDHSETSSSGSGSDSLDEDAMFIPSAEAHDTSYFMSRYI 1001
F R ++ DDE DF +++ + P ++ + S
Sbjct: 2033 FWDRQ--SMYDDE---AEDDFLTINQSQPQHQSQHQSQPQSQPQPQSQNLGQNNNNSESN 2087
Query: 1002 WNVEDDEHCAGGSDFYDHSETSSSGSGSDSLDE--DGDECASLTEFGNSALGVQYSFSNF 1059
N + G + + +SS+ +GS +L+ + + N LG + N
Sbjct: 2088 NNNNNSNSNVSGQNINNSVSSSSNNNGSGNLNNICNNSNVNANNNNSNGNLGNNNNNKNN 2147
Query: 1060 SFKNISQLVSINMMHISKETPDDSN 1084
+ N + N ++ +++N
Sbjct: 2148 NINNDNDKNKNNNSNVQNNNNNNNN 2172
>ref|XP_422443.1| PREDICTED: similar to Mast2 protein [Gallus gallus]
Length = 2424
Score = 296 bits (758), Expect = 3e-78
Identities = 168/390 (43%), Positives = 233/390 (59%), Gaps = 48/390 (12%)
Query: 557 ERAESSNSMADEESSVDDDTIRSLRASPINGFSKDRTSIEDFEIIKPISRGAFGRVFLAQ 616
E NS ++ DD++ S R + + SK S E+FE IK IS GA+G V+L +
Sbjct: 814 EEMAQLNSYDSPDTPETDDSVES-RGATVQ--SKKTPSEEEFETIKLISNGAYGAVYLVR 870
Query: 617 KRSTGDLFAIKVLKKADMIRKNAVEGILAERDILISVRNPFVVRFYYSFTCKENLYLVME 676
++T FA+K + K ++I +N ++ ERDIL NPFVV + SF K +L +VME
Sbjct: 871 HKTTRQRFAMKKINKQNLILRNQIQQAFVERDILTFAENPFVVSMFCSFETKRHLCMVME 930
Query: 677 YLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALEYLHSQSIVHRDLKPDNLLIGQDGHIK 736
Y+ GGD ++L+N+G L DMAR+Y AE VLALEYLH+ IVHRDLKPDNLLI GHIK
Sbjct: 931 YVEGGDCATLLKNIGALPVDMARMYFAETVLALEYLHNYGIVHRDLKPDNLLITSMGHIK 990
Query: 737 LTDFGLSKVGLINSTEDLSAPASFTNGFLVDDEPKPRHVSKREARQQQSIVGTPDYLAPE 796
LTDFGLSK+GL++ T +L G + D RE +Q + GTP+Y+APE
Sbjct: 991 LTDFGLSKIGLMSLTTNL------YEGHIEKD--------TREFLDKQ-VCGTPEYIAPE 1035
Query: 797 ILLGMGHGTTADWWSVGVILYELLVGIPPFNADHAQQIFDNIINRDIQWPKHPEEISFEA 856
++L G+G DWW++GVILYE LVG PF D +++F +I+ +I WP+ + + +A
Sbjct: 1036 VILRQGYGKPVDWWAMGVILYEFLVGCVPFFGDTPEELFGQVISDEIAWPEGDDALPPDA 1095
Query: 857 YDLMNKLLIENPVQRLGVTGATEVKRHAFFKDVNWDTLARQKVVILILFEFDWFAPYLLI 916
DL++KLL +NP++R+G A EVK+H FFKD++W+ L RQK
Sbjct: 1096 QDLISKLLRQNPLERMGTGSAFEVKQHRFFKDLDWNGLLRQK------------------ 1137
Query: 917 SHTWDPTNFLQAMFIPSAEAH-DTSYFMSR 945
A FIP E+ DTSYF +R
Sbjct: 1138 -----------AEFIPQLESEDDTSYFDTR 1156
>dbj|BAC41448.1| mKIAA0807 protein [Mus musculus]
Length = 1432
Score = 292 bits (747), Expect = 5e-77
Identities = 165/393 (41%), Positives = 231/393 (57%), Gaps = 53/393 (13%)
Query: 556 DERAESSNSMADEESSVDDDTIRSLRASPINGFSKDRTSIEDFEIIKPISRGAFGRVFLA 615
+E A+ S+ + + DD S+ ++ S+ S EDFE IK IS GA+G VFL
Sbjct: 114 EEMAQLSSYDSPDTPETDD----SVEGRGVSQPSQKTPSEEDFETIKLISNGAYGAVFLV 169
Query: 616 QKRSTGDLFAIKVLKKADMIRKNAVEGILAERDILISVRNPFVVRFYYSFTCKENLYLVM 675
+ +ST FA+K + K ++I +N ++ ERDIL NPFVV + SF K +L +VM
Sbjct: 170 RHKSTRQCFAMKKINKQNLILRNQIQQAFVERDILTFAENPFVVSMFCSFETKRHLCMVM 229
Query: 676 EYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALEYLHSQSIVHRDLKPDNLLIGQDGHI 735
EY+ GGD ++L+N+G L DM R+Y AE VLALEYLH+ IVHRDLKPDNLLI GHI
Sbjct: 230 EYVEGGDCATLLKNIGALPVDMVRLYFAETVLALEYLHNYGIVHRDLKPDNLLITSMGHI 289
Query: 736 KLTDFGLSKVGLINSTEDLSAPASFTNGFLVDDEPKPRHVSKREARQ--QQSIVGTPDYL 793
KLTDFGLSK+GL++ T +L H+ K +AR+ + + GTP+Y+
Sbjct: 290 KLTDFGLSKIGLMSLTTNL----------------YEGHIEK-DAREFLDKQVCGTPEYI 332
Query: 794 APEILLGMGHGTTADWWSVGVILYELLVGIPPFNADHAQQIFDNIINRDIQWPKHPEEIS 853
APE++L G+G DWW++G+ILYE LVG PF D +++F +I+ +I WP+ + +
Sbjct: 333 APEVILRQGYGKPVDWWAMGIILYEFLVGCVPFFGDTPEELFGQVISDEIVWPEGDDALP 392
Query: 854 FEAYDLMNKLLIENPVQRLGVTGATEVKRHAFFKDVNWDTLARQKVVILILFEFDWFAPY 913
+A DL +KLL +NP++RLG + A EVK+H FF ++W L RQK
Sbjct: 393 PDAQDLTSKLLHQNPLERLGTSSAYEVKQHPFFMGLDWTGLLRQK--------------- 437
Query: 914 LLISHTWDPTNFLQAMFIPSAEAH-DTSYFMSR 945
A FIP E+ DTSYF +R
Sbjct: 438 --------------AEFIPQLESEDDTSYFDTR 456
>emb|CAH73244.1| microtubule associated serine/threonine kinase 2 [Homo sapiens]
gi|55962055|emb|CAI16217.1| microtubule associated
serine/threonine kinase 2 [Homo sapiens]
gi|55961861|emb|CAI16562.1| microtubule associated
serine/threonine kinase 2 [Homo sapiens]
gi|56205428|emb|CAI21705.1| microtubule associated
serine/threonine kinase 2 [Homo sapiens]
gi|62287152|sp|Q6P0Q8|MAST2_HUMAN Microtubule-associated
serine/threonine-protein kinase 2
Length = 1798
Score = 291 bits (746), Expect = 6e-77
Identities = 167/393 (42%), Positives = 229/393 (57%), Gaps = 53/393 (13%)
Query: 556 DERAESSNSMADEESSVDDDTIRSLRASPINGFSKDRTSIEDFEIIKPISRGAFGRVFLA 615
+E A+ S+ + + DD S+ + SK S EDFE IK IS GA+G VFL
Sbjct: 474 EEMAQLSSCDSPDTPETDD----SIEGHGASLPSKKTPSEEDFETIKLISNGAYGAVFLV 529
Query: 616 QKRSTGDLFAIKVLKKADMIRKNAVEGILAERDILISVRNPFVVRFYYSFTCKENLYLVM 675
+ +ST FA+K + K ++I +N ++ ERDIL NPFVV + SF K +L +VM
Sbjct: 530 RHKSTRQRFAMKKINKQNLILRNQIQQAFVERDILTFAENPFVVSMFCSFDTKRHLCMVM 589
Query: 676 EYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALEYLHSQSIVHRDLKPDNLLIGQDGHI 735
EY+ GGD ++L+N+G L DM R+Y AE VLALEYLH+ IVHRDLKPDNLLI GHI
Sbjct: 590 EYVEGGDCATLLKNIGALPVDMVRLYFAETVLALEYLHNYGIVHRDLKPDNLLITSMGHI 649
Query: 736 KLTDFGLSKVGLINSTEDLSAPASFTNGFLVDDEPKPRHVSKREARQ--QQSIVGTPDYL 793
KLTDFGLSK+GL++ T +L H+ K +AR+ + + GTP+Y+
Sbjct: 650 KLTDFGLSKIGLMSLTTNL----------------YEGHIEK-DAREFLDKQVCGTPEYI 692
Query: 794 APEILLGMGHGTTADWWSVGVILYELLVGIPPFNADHAQQIFDNIINRDIQWPKHPEEIS 853
APE++L G+G DWW++G+ILYE LVG PF D +++F +I+ +I WP+ E +
Sbjct: 693 APEVILRQGYGKPVDWWAMGIILYEFLVGCVPFFGDTPEELFGQVISDEIVWPEGDEALP 752
Query: 854 FEAYDLMNKLLIENPVQRLGVTGATEVKRHAFFKDVNWDTLARQKVVILILFEFDWFAPY 913
+A DL +KLL +NP++RLG A EVK+H FF ++W L RQK
Sbjct: 753 PDAQDLTSKLLHQNPLERLGTGSAYEVKQHPFFTGLDWTGLLRQK--------------- 797
Query: 914 LLISHTWDPTNFLQAMFIPSAEAH-DTSYFMSR 945
A FIP E+ DTSYF +R
Sbjct: 798 --------------AEFIPQLESEDDTSYFDTR 816
>ref|NP_032667.1| microtubule associated serine/threonine kinase 2 [Mus musculus]
gi|62287151|sp|Q60592|MAST2_MOUSE Microtubule-associated
serine/threonine-protein kinase 2
gi|406058|gb|AAC04312.1| protein kinase [Mus musculus]
Length = 1734
Score = 291 bits (746), Expect = 6e-77
Identities = 165/393 (41%), Positives = 231/393 (57%), Gaps = 53/393 (13%)
Query: 556 DERAESSNSMADEESSVDDDTIRSLRASPINGFSKDRTSIEDFEIIKPISRGAFGRVFLA 615
+E A+ S+ + + DD S+ ++ S+ S EDFE IK IS GA+G VFL
Sbjct: 415 EEMAQLSSYDSPDTPETDD----SVEGRGVSQPSQKTPSEEDFETIKLISNGAYGAVFLV 470
Query: 616 QKRSTGDLFAIKVLKKADMIRKNAVEGILAERDILISVRNPFVVRFYYSFTCKENLYLVM 675
+ +ST FA+K + K ++I +N ++ ERDIL NPFVV + SF K +L +VM
Sbjct: 471 RHKSTRQRFAMKKINKQNLILRNQIQQAFVERDILTFAENPFVVSMFCSFETKRHLCMVM 530
Query: 676 EYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALEYLHSQSIVHRDLKPDNLLIGQDGHI 735
EY+ GGD ++L+N+G L DM R+Y AE VLALEYLH+ IVHRDLKPDNLLI GHI
Sbjct: 531 EYVEGGDCATLLKNIGALPVDMVRLYFAETVLALEYLHNYGIVHRDLKPDNLLITSMGHI 590
Query: 736 KLTDFGLSKVGLINSTEDLSAPASFTNGFLVDDEPKPRHVSKREARQ--QQSIVGTPDYL 793
KLTDFGLSK+GL++ T +L H+ K +AR+ + + GTP+Y+
Sbjct: 591 KLTDFGLSKIGLMSLTTNL----------------YEGHIEK-DAREFLDKQVCGTPEYI 633
Query: 794 APEILLGMGHGTTADWWSVGVILYELLVGIPPFNADHAQQIFDNIINRDIQWPKHPEEIS 853
APE++L G+G DWW++G+ILYE LVG PF D +++F +I+ +I WP+ + +
Sbjct: 634 APEVILRQGYGKPVDWWAMGIILYEFLVGCVPFFGDTPEELFGQVISDEIVWPEGDDALP 693
Query: 854 FEAYDLMNKLLIENPVQRLGVTGATEVKRHAFFKDVNWDTLARQKVVILILFEFDWFAPY 913
+A DL +KLL +NP++RLG + A EVK+H FF ++W L RQK
Sbjct: 694 PDAQDLTSKLLHQNPLERLGTSSAYEVKQHPFFMGLDWTGLLRQK--------------- 738
Query: 914 LLISHTWDPTNFLQAMFIPSAEAH-DTSYFMSR 945
A FIP E+ DTSYF +R
Sbjct: 739 --------------AEFIPQLESEDDTSYFDTR 757
>emb|CAH73245.1| microtubule associated serine/threonine kinase 2 [Homo sapiens]
gi|55961862|emb|CAI16563.1| microtubule associated
serine/threonine kinase 2 [Homo sapiens]
gi|56205429|emb|CAI21706.1| microtubule associated
serine/threonine kinase 2 [Homo sapiens]
Length = 792
Score = 291 bits (746), Expect = 6e-77
Identities = 167/393 (42%), Positives = 229/393 (57%), Gaps = 53/393 (13%)
Query: 556 DERAESSNSMADEESSVDDDTIRSLRASPINGFSKDRTSIEDFEIIKPISRGAFGRVFLA 615
+E A+ S+ + + DD S+ + SK S EDFE IK IS GA+G VFL
Sbjct: 359 EEMAQLSSCDSPDTPETDD----SIEGHGASLPSKKTPSEEDFETIKLISNGAYGAVFLV 414
Query: 616 QKRSTGDLFAIKVLKKADMIRKNAVEGILAERDILISVRNPFVVRFYYSFTCKENLYLVM 675
+ +ST FA+K + K ++I +N ++ ERDIL NPFVV + SF K +L +VM
Sbjct: 415 RHKSTRQRFAMKKINKQNLILRNQIQQAFVERDILTFAENPFVVSMFCSFDTKRHLCMVM 474
Query: 676 EYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALEYLHSQSIVHRDLKPDNLLIGQDGHI 735
EY+ GGD ++L+N+G L DM R+Y AE VLALEYLH+ IVHRDLKPDNLLI GHI
Sbjct: 475 EYVEGGDCATLLKNIGALPVDMVRLYFAETVLALEYLHNYGIVHRDLKPDNLLITSMGHI 534
Query: 736 KLTDFGLSKVGLINSTEDLSAPASFTNGFLVDDEPKPRHVSKREARQ--QQSIVGTPDYL 793
KLTDFGLSK+GL++ T +L H+ K +AR+ + + GTP+Y+
Sbjct: 535 KLTDFGLSKIGLMSLTTNL----------------YEGHIEK-DAREFLDKQVCGTPEYI 577
Query: 794 APEILLGMGHGTTADWWSVGVILYELLVGIPPFNADHAQQIFDNIINRDIQWPKHPEEIS 853
APE++L G+G DWW++G+ILYE LVG PF D +++F +I+ +I WP+ E +
Sbjct: 578 APEVILRQGYGKPVDWWAMGIILYEFLVGCVPFFGDTPEELFGQVISDEIVWPEGDEALP 637
Query: 854 FEAYDLMNKLLIENPVQRLGVTGATEVKRHAFFKDVNWDTLARQKVVILILFEFDWFAPY 913
+A DL +KLL +NP++RLG A EVK+H FF ++W L RQK
Sbjct: 638 PDAQDLTSKLLHQNPLERLGTGSAYEVKQHPFFTGLDWTGLLRQK--------------- 682
Query: 914 LLISHTWDPTNFLQAMFIPSAEAH-DTSYFMSR 945
A FIP E+ DTSYF +R
Sbjct: 683 --------------AEFIPQLESEDDTSYFDTR 701
>gb|AAH60703.1| Mast2 protein [Mus musculus]
Length = 1739
Score = 291 bits (746), Expect = 6e-77
Identities = 165/393 (41%), Positives = 231/393 (57%), Gaps = 53/393 (13%)
Query: 556 DERAESSNSMADEESSVDDDTIRSLRASPINGFSKDRTSIEDFEIIKPISRGAFGRVFLA 615
+E A+ S+ + + DD S+ ++ S+ S EDFE IK IS GA+G VFL
Sbjct: 421 EEMAQLSSYDSPDTPETDD----SVEGRGVSQPSQKTPSEEDFETIKLISNGAYGAVFLV 476
Query: 616 QKRSTGDLFAIKVLKKADMIRKNAVEGILAERDILISVRNPFVVRFYYSFTCKENLYLVM 675
+ +ST FA+K + K ++I +N ++ ERDIL NPFVV + SF K +L +VM
Sbjct: 477 RHKSTRQRFAMKKINKQNLILRNQIQQAFVERDILTFAENPFVVSMFCSFETKRHLCMVM 536
Query: 676 EYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALEYLHSQSIVHRDLKPDNLLIGQDGHI 735
EY+ GGD ++L+N+G L DM R+Y AE VLALEYLH+ IVHRDLKPDNLLI GHI
Sbjct: 537 EYVEGGDCATLLKNIGALPVDMVRLYFAETVLALEYLHNYGIVHRDLKPDNLLITSMGHI 596
Query: 736 KLTDFGLSKVGLINSTEDLSAPASFTNGFLVDDEPKPRHVSKREARQ--QQSIVGTPDYL 793
KLTDFGLSK+GL++ T +L H+ K +AR+ + + GTP+Y+
Sbjct: 597 KLTDFGLSKIGLMSLTTNL----------------YEGHIEK-DAREFLDKQVCGTPEYI 639
Query: 794 APEILLGMGHGTTADWWSVGVILYELLVGIPPFNADHAQQIFDNIINRDIQWPKHPEEIS 853
APE++L G+G DWW++G+ILYE LVG PF D +++F +I+ +I WP+ + +
Sbjct: 640 APEVILRQGYGKPVDWWAMGIILYEFLVGCVPFFGDTPEELFGQVISDEIVWPEGDDALP 699
Query: 854 FEAYDLMNKLLIENPVQRLGVTGATEVKRHAFFKDVNWDTLARQKVVILILFEFDWFAPY 913
+A DL +KLL +NP++RLG + A EVK+H FF ++W L RQK
Sbjct: 700 PDAQDLTSKLLHQNPLERLGTSSAYEVKQHPFFMGLDWTGLLRQK--------------- 744
Query: 914 LLISHTWDPTNFLQAMFIPSAEAH-DTSYFMSR 945
A FIP E+ DTSYF +R
Sbjct: 745 --------------AEFIPQLESEDDTSYFDTR 763
>gb|AAH65499.1| MAST205 protein [Homo sapiens]
Length = 1797
Score = 291 bits (746), Expect = 6e-77
Identities = 167/393 (42%), Positives = 229/393 (57%), Gaps = 53/393 (13%)
Query: 556 DERAESSNSMADEESSVDDDTIRSLRASPINGFSKDRTSIEDFEIIKPISRGAFGRVFLA 615
+E A+ S+ + + DD S+ + SK S EDFE IK IS GA+G VFL
Sbjct: 474 EEMAQLSSCDSPDTPETDD----SIEGHGASLPSKKTPSEEDFETIKLISNGAYGAVFLV 529
Query: 616 QKRSTGDLFAIKVLKKADMIRKNAVEGILAERDILISVRNPFVVRFYYSFTCKENLYLVM 675
+ +ST FA+K + K ++I +N ++ ERDIL NPFVV + SF K +L +VM
Sbjct: 530 RHKSTRQRFAMKKINKQNLILRNQIQQAFVERDILTFAENPFVVSMFCSFDTKRHLCMVM 589
Query: 676 EYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALEYLHSQSIVHRDLKPDNLLIGQDGHI 735
EY+ GGD ++L+N+G L DM R+Y AE VLALEYLH+ IVHRDLKPDNLLI GHI
Sbjct: 590 EYVEGGDCATLLKNIGALPVDMVRLYFAETVLALEYLHNYGIVHRDLKPDNLLITSMGHI 649
Query: 736 KLTDFGLSKVGLINSTEDLSAPASFTNGFLVDDEPKPRHVSKREARQ--QQSIVGTPDYL 793
KLTDFGLSK+GL++ T +L H+ K +AR+ + + GTP+Y+
Sbjct: 650 KLTDFGLSKIGLMSLTTNL----------------YEGHIEK-DAREFLDKQVCGTPEYI 692
Query: 794 APEILLGMGHGTTADWWSVGVILYELLVGIPPFNADHAQQIFDNIINRDIQWPKHPEEIS 853
APE++L G+G DWW++G+ILYE LVG PF D +++F +I+ +I WP+ E +
Sbjct: 693 APEVILRQGYGKPVDWWAMGIILYEFLVGCVPFFGDTPEELFGQVISDEIVWPEGDEALP 752
Query: 854 FEAYDLMNKLLIENPVQRLGVTGATEVKRHAFFKDVNWDTLARQKVVILILFEFDWFAPY 913
+A DL +KLL +NP++RLG A EVK+H FF ++W L RQK
Sbjct: 753 PDAQDLTSKLLHQNPLERLGTGSAYEVKQHPFFTGLDWTGLLRQK--------------- 797
Query: 914 LLISHTWDPTNFLQAMFIPSAEAH-DTSYFMSR 945
A FIP E+ DTSYF +R
Sbjct: 798 --------------AEFIPQLESEDDTSYFDTR 816
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.317 0.134 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,880,188,309
Number of Sequences: 2540612
Number of extensions: 81914733
Number of successful extensions: 298250
Number of sequences better than 10.0: 21096
Number of HSP's better than 10.0 without gapping: 16184
Number of HSP's successfully gapped in prelim test: 4919
Number of HSP's that attempted gapping in prelim test: 241103
Number of HSP's gapped (non-prelim): 37774
length of query: 1086
length of database: 863,360,394
effective HSP length: 139
effective length of query: 947
effective length of database: 510,215,326
effective search space: 483173913722
effective search space used: 483173913722
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 81 (35.8 bits)
Medicago: description of AC133139.5


