

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133139.2 + phase: 0 /pseudo
(155 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
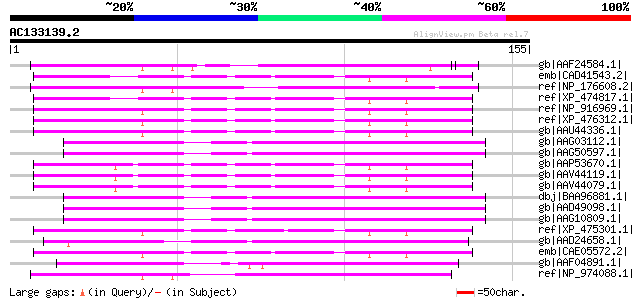
Score E
Sequences producing significant alignments: (bits) Value
gb|AAF24584.1| F22C12.1 [Arabidopsis thaliana] 61 8e-09
emb|CAD41543.2| OSJNBb0091E11.13 [Oryza sativa (japonica cultiva... 60 2e-08
ref|NP_176608.2| zinc finger protein-related [Arabidopsis thalia... 59 5e-08
ref|XP_474817.1| OSJNBa0014F04.28 [Oryza sativa (japonica cultiv... 58 7e-08
ref|NP_916969.1| P0445E10.20 [Oryza sativa (japonica cultivar-gr... 56 3e-07
ref|XP_476312.1| putative mutator-like transposase [Oryza sativa... 56 3e-07
gb|AAU44336.1| putative polyprotein [Oryza sativa (japonica cult... 55 6e-07
gb|AAG03112.1| F5A9.11 [Arabidopsis thaliana] 55 7e-07
gb|AAG50597.1| hypothetical protein [Arabidopsis thaliana] gi|25... 55 7e-07
gb|AAP53670.1| putative transposable element [Oryza sativa (japo... 55 7e-07
gb|AAV44119.1| putative polyprotein [Oryza sativa (japonica cult... 55 7e-07
gb|AAV44079.1| putative polyprotein [Oryza sativa (japonica cult... 55 7e-07
dbj|BAA96881.1| mutator-like transposase [Arabidopsis thaliana] 55 7e-07
gb|AAD49098.1| contains similarity to maize transposon MuDR (GB:... 55 7e-07
gb|AAG10809.1| Similar to mutator transposase [Arabidopsis thali... 55 7e-07
ref|XP_475301.1| unknown protein [Oryza sativa (japonica cultiva... 54 1e-06
gb|AAD24658.1| Mutator-like transposase [Arabidopsis thaliana] g... 54 2e-06
emb|CAE05572.2| OSJNBb0013O03.13 [Oryza sativa (japonica cultiva... 54 2e-06
gb|AAF04891.1| Mutator-like transposase [Arabidopsis thaliana] g... 52 5e-06
ref|NP_974088.1| SWIM zinc finger family protein [Arabidopsis th... 52 5e-06
>gb|AAF24584.1| F22C12.1 [Arabidopsis thaliana]
Length = 3290
Score = 61.2 bits (147), Expect = 8e-09
Identities = 42/132 (31%), Positives = 62/132 (46%), Gaps = 16/132 (12%)
Query: 7 LRLWVTPQPDLGLISDRHESIKSVVRRLDSNW-----HHVFCIRHI-TQLFEDFQEHRDE 60
+R VT + D+ LIS H I V+ S W +H+FC+ I TQ FQ+ D
Sbjct: 1503 IREKVTQRKDICLISRPHPDILDVINEPGSQWQEPWAYHMFCLDDICTQFHYVFQD--DY 1560
Query: 61 EKSKKYGLNNINFLPSGYSMTQPSITHYRNQIRDWSDDVVKWVDDNPKKQWLQAHDQGRR 120
K+ Y +G + + Y N+I + + KW+D P+ QW QAHD GRR
Sbjct: 1561 LKNLVY--------EAGSTSEKEEFDSYMNEIEKKNSEARKWLDQFPQYQWAQAHDSGRR 1612
Query: 121 WGHITTNISECF 132
+ +T + F
Sbjct: 1613 YRVMTIDAENLF 1624
Score = 58.5 bits (140), Expect = 5e-08
Identities = 42/140 (30%), Positives = 64/140 (45%), Gaps = 17/140 (12%)
Query: 7 LRLWVTPQPDLGLISDRHESIKSVVRRLDSNW-----HHVFCIRHI-TQLFEDFQEHRDE 60
+R VT + DL LIS I +VV S W HH FC+ H+ +Q F+++ E
Sbjct: 3000 IREKVTQRKDLCLISSPLRDIVAVVNEPGSLWQEPWAHHKFCLNHLRSQFLGVFRDYNLE 3059
Query: 61 EKSKKYGLNNINFLPSGYSMTQPSITHYRNQIRDWSDDVVKWVDDNPKKQWLQAHDQGRR 120
++ G N + Y N I++ + + KW+D P+ +W AHD G R
Sbjct: 3060 SLVEQAGSTN----------QKEEFDSYMNDIKEKNPEAWKWLDQIPRHKWALAHDSGLR 3109
Query: 121 WGHITTNISECFNNVLKGGP 140
+G I + E V +G P
Sbjct: 3110 YGIIEID-REALFAVCRGFP 3128
Score = 57.0 bits (136), Expect = 1e-07
Identities = 39/136 (28%), Positives = 63/136 (45%), Gaps = 19/136 (13%)
Query: 7 LRLWVTPQPDLGLISDRHESIKSVVRRLDSNW-----HHVFCIRHITQLFED-FQEHRDE 60
+R VT + L LIS + +V+ S W ++ FC+RH+ F F+++ E
Sbjct: 675 IRERVTQRKGLCLISSPDPDLLAVINESGSQWQEPWAYNRFCLRHLLSQFSGIFRDYYLE 734
Query: 61 EKSKKYGLNNINFLPSGYSMTQPSITHYRNQIRDWSDDVVKWVDDNPKKQWLQAHDQGRR 120
+ K+ +G + + Y I + + KW+D P+ QW AHD GRR
Sbjct: 735 DLVKR----------AGSTSQKEEFDSYMKDIEKKNSEARKWLDQFPQNQWALAHDNGRR 784
Query: 121 WGHI---TTNISECFN 133
+G + TT + E FN
Sbjct: 785 YGIMEIETTTLFEDFN 800
Score = 52.0 bits (123), Expect = 5e-06
Identities = 36/135 (26%), Positives = 56/135 (40%), Gaps = 22/135 (16%)
Query: 7 LRLWVTPQPDLGLISDRHESIKSVVRRLDSNW-----HHVFCIRHI----TQLFEDFQEH 57
+R VT + L LIS H I +VV S W +H F + H +++F F
Sbjct: 2277 IREKVTQRKGLCLISSPHPDIIAVVNESGSQWQEPWAYHRFSLNHFYSQFSRVFPSF--- 2333
Query: 58 RDEEKSKKYGLNNINFLPSGYSMTQPSITHYRNQIRDWSDDVVKWVDDNPKKQWLQAHDQ 117
+G + + Y N I++ + + KW+D P+ +W AHD
Sbjct: 2334 ----------CLGARIRRAGSTSQKDEFVSYMNDIKEKNPEARKWLDQFPQNRWALAHDN 2383
Query: 118 GRRWGHITTNISECF 132
GRR+G + N F
Sbjct: 2384 GRRYGIMEINTKALF 2398
>emb|CAD41543.2| OSJNBb0091E11.13 [Oryza sativa (japonica cultivar-group)]
gi|50925901|ref|XP_473014.1| OSJNBb0091E11.13 [Oryza
sativa (japonica cultivar-group)]
Length = 1107
Score = 59.7 bits (143), Expect = 2e-08
Identities = 44/134 (32%), Positives = 74/134 (54%), Gaps = 19/134 (14%)
Query: 8 RLWVTPQPDLGLISDRHESIKSVVRRLDSNWHHVFCIRHITQLFEDFQEHRDEEKSKKYG 67
R+ V P ++ +ISDRH + SV HH +C+RH + F + D+ ++K+
Sbjct: 609 RVVVGPHREVCIISDRHAGLPSV--------HHRWCMRHFSANFH--KAGADKHQTKE-- 656
Query: 68 LNNINFLPSGYSMTQPSITHYRNQIRDWSDDVVKWVDDN--PKKQWLQAHDQ-GRRWGHI 124
L I + + + + + R I + S KW++D K +W +A+D+ GRRWG++
Sbjct: 657 LLRICQIDEKW-IFERDVEALRQHIPEGSR---KWLEDELLDKDKWSRAYDRNGRRWGYM 712
Query: 125 TTNISECFNNVLKG 138
TTN++E FN+VL G
Sbjct: 713 TTNMAEQFNSVLVG 726
>ref|NP_176608.2| zinc finger protein-related [Arabidopsis thaliana]
gi|12323472|gb|AAG51711.1| hypothetical protein;
95918-93759 [Arabidopsis thaliana]
Length = 719
Score = 58.5 bits (140), Expect = 5e-08
Identities = 42/140 (30%), Positives = 64/140 (45%), Gaps = 17/140 (12%)
Query: 7 LRLWVTPQPDLGLISDRHESIKSVVRRLDSNW-----HHVFCIRHI-TQLFEDFQEHRDE 60
+R VT + DL LIS I +VV S W HH FC+ H+ +Q F+++ E
Sbjct: 429 IREKVTQRKDLCLISSPLRDIVAVVNEPGSLWQEPWAHHKFCLNHLRSQFLGVFRDYNLE 488
Query: 61 EKSKKYGLNNINFLPSGYSMTQPSITHYRNQIRDWSDDVVKWVDDNPKKQWLQAHDQGRR 120
++ G N + Y N I++ + + KW+D P+ +W AHD G R
Sbjct: 489 SLVEQAGSTN----------QKEEFDSYMNDIKEKNPEAWKWLDQIPRHKWALAHDSGLR 538
Query: 121 WGHITTNISECFNNVLKGGP 140
+G I + E V +G P
Sbjct: 539 YGIIEID-REALFAVCRGFP 557
>ref|XP_474817.1| OSJNBa0014F04.28 [Oryza sativa (japonica cultivar-group)]
gi|32488049|emb|CAE02862.1| OSJNBa0014F04.28 [Oryza
sativa (japonica cultivar-group)]
gi|38345180|emb|CAE03336.2| OSJNBb0005B05.3 [Oryza
sativa (japonica cultivar-group)]
Length = 1575
Score = 58.2 bits (139), Expect = 7e-08
Identities = 43/134 (32%), Positives = 74/134 (55%), Gaps = 19/134 (14%)
Query: 8 RLWVTPQPDLGLISDRHESIKSVVRRLDSNWHHVFCIRHITQLFEDFQEHRDEEKSKKYG 67
R+ V P ++ +ISDRH + SV HH +C+RH + F + D+ ++K+
Sbjct: 562 RVVVGPHREVCIISDRHAGLPSV--------HHRWCMRHFSANFH--KAGADKHQTKE-- 609
Query: 68 LNNINFLPSGYSMTQPSITHYRNQIRDWSDDVVKWVDDN--PKKQWLQAHDQ-GRRWGHI 124
L I + + + + + R +I + KW++D K +W +A+D+ GRRWG++
Sbjct: 610 LLRICQIDEKW-IFERDVEALRQRIPEGPR---KWLEDELLDKDKWSRAYDRNGRRWGYM 665
Query: 125 TTNISECFNNVLKG 138
TTN++E FN+VL G
Sbjct: 666 TTNMAEQFNSVLVG 679
>ref|NP_916969.1| P0445E10.20 [Oryza sativa (japonica cultivar-group)]
Length = 1130
Score = 56.2 bits (134), Expect = 3e-07
Identities = 43/137 (31%), Positives = 74/137 (53%), Gaps = 14/137 (10%)
Query: 8 RLWVTPQPDLGLISDRHESIKSVVRRLDSNW---HHVFCIRHITQLFEDFQEHRDEEKSK 64
R+ V P ++ +ISDRH I + + HH +C+RH + F + D+ ++K
Sbjct: 428 RVVVGPHREVCIISDRHAGIMNAMTTPVPGLPPVHHRWCMRHFSANFH--KAGADKHQTK 485
Query: 65 KYGLNNINFLPSGYSMTQPSITHYRNQIRDWSDDVVKWVDDN--PKKQWLQAHDQ-GRRW 121
+ L I + + + + + R +I + KW++D K +W +A+D+ GRRW
Sbjct: 486 E--LLRICQIDEKW-IFERDVEALRQRIPEGPR---KWLEDELLDKDKWSRAYDRNGRRW 539
Query: 122 GHITTNISECFNNVLKG 138
G++TTN++E FNNVL G
Sbjct: 540 GYMTTNMAEQFNNVLVG 556
>ref|XP_476312.1| putative mutator-like transposase [Oryza sativa (japonica
cultivar-group)]
Length = 1316
Score = 55.8 bits (133), Expect = 3e-07
Identities = 43/137 (31%), Positives = 75/137 (54%), Gaps = 14/137 (10%)
Query: 8 RLWVTPQPDLGLISDRHESIKSVVRRLDSNW---HHVFCIRHITQLFEDFQEHRDEEKSK 64
R+ V P ++ +ISDRH SI + + HH +C+RH + F + D+ ++K
Sbjct: 403 RVVVGPHREVCIISDRHASIMNAMTTPVPGLPPVHHRWCMRHFSANFH--KAGADKHQTK 460
Query: 65 KYGLNNINFLPSGYSMTQPSITHYRNQIRDWSDDVVKWVDDN--PKKQWLQAHDQ-GRRW 121
+ L I + + + + + R +I + KW++D K +W +A+D+ GRRW
Sbjct: 461 E--LLRICQIDEKW-IFERDVEALRQRIPEGPR---KWLEDELLDKDKWSRAYDRNGRRW 514
Query: 122 GHITTNISECFNNVLKG 138
G++TTN++E FN+VL G
Sbjct: 515 GYMTTNMAEQFNSVLVG 531
>gb|AAU44336.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
Length = 1378
Score = 55.1 bits (131), Expect = 6e-07
Identities = 42/137 (30%), Positives = 74/137 (53%), Gaps = 14/137 (10%)
Query: 8 RLWVTPQPDLGLISDRHESIKSVVRRLDSNW---HHVFCIRHITQLFEDFQEHRDEEKSK 64
R+ V P ++ +ISDRH I + + HH +C+RH + F + D+ ++K
Sbjct: 404 RVMVGPHREVCIISDRHAGIMNAMTTPVPGLPPVHHRWCMRHFSANFH--KAGADKHQTK 461
Query: 65 KYGLNNINFLPSGYSMTQPSITHYRNQIRDWSDDVVKWVDDN--PKKQWLQAHDQ-GRRW 121
+ L I + + + + + R +I + KW++D K +W +A+D+ GRRW
Sbjct: 462 E--LLRICQIDEKW-IFERDVEALRQRIPEGPR---KWLEDELVDKDKWSRAYDRNGRRW 515
Query: 122 GHITTNISECFNNVLKG 138
G++TTN++E FN+VL G
Sbjct: 516 GYMTTNMAEQFNSVLVG 532
>gb|AAG03112.1| F5A9.11 [Arabidopsis thaliana]
Length = 843
Score = 54.7 bits (130), Expect = 7e-07
Identities = 37/126 (29%), Positives = 59/126 (46%), Gaps = 9/126 (7%)
Query: 17 LGLISDRHESIKSVVRRLDSNWHHVFCIRHITQLFEDFQEHRDEEKSKKYGLNNINFLPS 76
L ++SDRHESIK V+++ HH CI H+ + + + K GL + +
Sbjct: 354 LTILSDRHESIKVGVKKVFPQAHHGACIIHLCRNIQ--------ARFKNRGLTQL-VKNA 404
Query: 77 GYSMTQPSITHYRNQIRDWSDDVVKWVDDNPKKQWLQAHDQGRRWGHITTNISECFNNVL 136
GY T NQI + +K++ D W + + G+R+ +T+NI+E N L
Sbjct: 405 GYEFTSGKFKTLYNQINAINPLCIKYLHDVGMAHWTRLYFPGQRFNLMTSNIAETLNKAL 464
Query: 137 KGGPQS 142
G S
Sbjct: 465 FKGRSS 470
>gb|AAG50597.1| hypothetical protein [Arabidopsis thaliana] gi|25350250|pir||B86471
hypothetical protein T32G9.38 [imported] - Arabidopsis
thaliana
Length = 873
Score = 54.7 bits (130), Expect = 7e-07
Identities = 37/126 (29%), Positives = 59/126 (46%), Gaps = 9/126 (7%)
Query: 17 LGLISDRHESIKSVVRRLDSNWHHVFCIRHITQLFEDFQEHRDEEKSKKYGLNNINFLPS 76
L ++SDRHESIK V+++ HH CI H+ + + + K GL + +
Sbjct: 586 LTILSDRHESIKVGVKKVFPQAHHGACIIHLCRNIQ--------ARFKNRGLTQL-VKNA 636
Query: 77 GYSMTQPSITHYRNQIRDWSDDVVKWVDDNPKKQWLQAHDQGRRWGHITTNISECFNNVL 136
GY T NQI + +K++ D W + + G+R+ +T+NI+E N L
Sbjct: 637 GYEFTSGKFKTLYNQINAINPLCIKYLHDVGMAHWTRLYFPGQRFNLMTSNIAETLNKAL 696
Query: 137 KGGPQS 142
G S
Sbjct: 697 FKGRSS 702
>gb|AAP53670.1| putative transposable element [Oryza sativa (japonica
cultivar-group)] gi|37534162|ref|NP_921383.1| putative
transposable element [Oryza sativa (japonica
cultivar-group)] gi|21671916|gb|AAM74278.1| Putative
transposable element [Oryza sativa (japonica
cultivar-group)]
Length = 1592
Score = 54.7 bits (130), Expect = 7e-07
Identities = 45/138 (32%), Positives = 76/138 (54%), Gaps = 16/138 (11%)
Query: 8 RLWVTPQPDLGLISDRHESIKSV----VRRLDSNWHHVFCIRHITQLFEDFQEHRDEEKS 63
R+ V P ++ +ISDRH I + V L S HH +C+RH + F + D+ ++
Sbjct: 594 RVVVGPHREVCIISDRHAGIMNAMMTPVPGLPSV-HHRWCMRHFSANFH--KAGADKHQT 650
Query: 64 KKYGLNNINFLPSGYSMTQPSITHYRNQIRDWSDDVVKWVDDN--PKKQWLQAHDQ-GRR 120
K+ L I + + + + + R +I + KW++D K +W +A+D+ GRR
Sbjct: 651 KE--LLRICQIDEKW-IFERDVEALRQRIPEGPR---KWLEDELLDKDKWSRAYDRNGRR 704
Query: 121 WGHITTNISECFNNVLKG 138
WG++TTN++E FN+VL G
Sbjct: 705 WGYMTTNMAEQFNSVLVG 722
>gb|AAV44119.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
Length = 1525
Score = 54.7 bits (130), Expect = 7e-07
Identities = 45/138 (32%), Positives = 76/138 (54%), Gaps = 16/138 (11%)
Query: 8 RLWVTPQPDLGLISDRHESIKSV----VRRLDSNWHHVFCIRHITQLFEDFQEHRDEEKS 63
R+ V P ++ +ISDRH I + V L S HH +C+RH + F + D+ ++
Sbjct: 594 RVVVGPHREVCIISDRHAGIMNAMMTPVPGLPSV-HHRWCMRHFSANFH--KAGADKHQT 650
Query: 64 KKYGLNNINFLPSGYSMTQPSITHYRNQIRDWSDDVVKWVDDN--PKKQWLQAHDQ-GRR 120
K+ L I + + + + + R +I + KW++D K +W +A+D+ GRR
Sbjct: 651 KE--LLRICQIDEKW-IFERDVEALRQRIPEGPR---KWLEDELLDKDKWSRAYDRNGRR 704
Query: 121 WGHITTNISECFNNVLKG 138
WG++TTN++E FN+VL G
Sbjct: 705 WGYMTTNMAEQFNSVLVG 722
>gb|AAV44079.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
Length = 1567
Score = 54.7 bits (130), Expect = 7e-07
Identities = 45/138 (32%), Positives = 76/138 (54%), Gaps = 16/138 (11%)
Query: 8 RLWVTPQPDLGLISDRHESIKSV----VRRLDSNWHHVFCIRHITQLFEDFQEHRDEEKS 63
R+ V P ++ +ISDRH I + V L S HH +C+RH + F + D+ ++
Sbjct: 609 RVVVGPHREVCIISDRHAGIMNAMMTPVPGLPSV-HHRWCMRHFSANFH--KAGADKHQT 665
Query: 64 KKYGLNNINFLPSGYSMTQPSITHYRNQIRDWSDDVVKWVDDN--PKKQWLQAHDQ-GRR 120
K+ L I + + + + + R +I + KW++D K +W +A+D+ GRR
Sbjct: 666 KE--LLRICQIDEKW-IFERDVEALRQRIPEGPR---KWLEDELLDKDKWSRAYDRNGRR 719
Query: 121 WGHITTNISECFNNVLKG 138
WG++TTN++E FN+VL G
Sbjct: 720 WGYMTTNMAEQFNSVLVG 737
>dbj|BAA96881.1| mutator-like transposase [Arabidopsis thaliana]
Length = 875
Score = 54.7 bits (130), Expect = 7e-07
Identities = 37/126 (29%), Positives = 59/126 (46%), Gaps = 9/126 (7%)
Query: 17 LGLISDRHESIKSVVRRLDSNWHHVFCIRHITQLFEDFQEHRDEEKSKKYGLNNINFLPS 76
L ++SDRHESIK V+++ HH CI H+ + + + K GL + +
Sbjct: 588 LTILSDRHESIKVGVKKVFPQAHHGACIIHLCRNIQ--------ARFKNRGLTQL-VKNA 638
Query: 77 GYSMTQPSITHYRNQIRDWSDDVVKWVDDNPKKQWLQAHDQGRRWGHITTNISECFNNVL 136
GY T NQI + +K++ D W + + G+R+ +T+NI+E N L
Sbjct: 639 GYEFTSGKFKTLYNQINAINPLCIKYLHDVGMAHWTRLYFPGQRFNLMTSNIAETLNKAL 698
Query: 137 KGGPQS 142
G S
Sbjct: 699 FKGRSS 704
>gb|AAD49098.1| contains similarity to maize transposon MuDR (GB:M76978)
[Arabidopsis thaliana]
Length = 872
Score = 54.7 bits (130), Expect = 7e-07
Identities = 37/126 (29%), Positives = 59/126 (46%), Gaps = 9/126 (7%)
Query: 17 LGLISDRHESIKSVVRRLDSNWHHVFCIRHITQLFEDFQEHRDEEKSKKYGLNNINFLPS 76
L ++SDRHESIK V+++ HH CI H+ + + + K GL + +
Sbjct: 588 LTILSDRHESIKVGVKKVFPQAHHGACIIHLCRNIQ--------ARFKNRGLTQL-VKNA 638
Query: 77 GYSMTQPSITHYRNQIRDWSDDVVKWVDDNPKKQWLQAHDQGRRWGHITTNISECFNNVL 136
GY T NQI + +K++ D W + + G+R+ +T+NI+E N L
Sbjct: 639 GYEFTSGKFKTLYNQINAINPLCIKYLHDVGMAHWTRLYFPGQRFNLMTSNIAETLNKAL 698
Query: 137 KGGPQS 142
G S
Sbjct: 699 FKGRSS 704
>gb|AAG10809.1| Similar to mutator transposase [Arabidopsis thaliana]
Length = 884
Score = 54.7 bits (130), Expect = 7e-07
Identities = 37/126 (29%), Positives = 59/126 (46%), Gaps = 9/126 (7%)
Query: 17 LGLISDRHESIKSVVRRLDSNWHHVFCIRHITQLFEDFQEHRDEEKSKKYGLNNINFLPS 76
L ++SDRHESIK V+++ HH CI H+ + + + K GL + +
Sbjct: 570 LTILSDRHESIKVGVKKVFPQAHHGACIIHLCRNIQ--------ARFKNRGLTQL-VKNA 620
Query: 77 GYSMTQPSITHYRNQIRDWSDDVVKWVDDNPKKQWLQAHDQGRRWGHITTNISECFNNVL 136
GY T NQI + +K++ D W + + G+R+ +T+NI+E N L
Sbjct: 621 GYEFTSGKFKTLYNQINAINPLCIKYLHDVGMAHWTRLYFPGQRFNLMTSNIAETLNKAL 680
Query: 137 KGGPQS 142
G S
Sbjct: 681 FKGRSS 686
>ref|XP_475301.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|49328188|gb|AAT58884.1| unknown protein [Oryza sativa
(japonica cultivar-group)]
Length = 1564
Score = 53.9 bits (128), Expect = 1e-06
Identities = 42/137 (30%), Positives = 73/137 (52%), Gaps = 14/137 (10%)
Query: 8 RLWVTPQPDLGLISDRHESIKSVVRRLDSNW---HHVFCIRHITQLFEDFQEHRDEEKSK 64
R+ V P ++ +ISDRH I + + HH +C+RH + F + D+ ++K
Sbjct: 666 RVVVGPHREVCIISDRHAGIMNAMTTPVPGLPLVHHRWCMRHFSANFH--KAGADKHQTK 723
Query: 65 KYGLNNINFLPSGYSMTQPSITHYRNQIRDWSDDVVKWVDDN--PKKQWLQAHDQ-GRRW 121
+ L I + + + + R +I + KW++D K +W +A+D+ GRRW
Sbjct: 724 E--LLRICQIDEKWIFKR-DVEALRQRIPEGPR---KWLEDELLDKDKWSRAYDRNGRRW 777
Query: 122 GHITTNISECFNNVLKG 138
G++TTN++E FN+VL G
Sbjct: 778 GYMTTNMAEQFNSVLVG 794
>gb|AAD24658.1| Mutator-like transposase [Arabidopsis thaliana]
gi|25350245|pir||B84469 Mutator-like transposase
[imported] - Arabidopsis thaliana
Length = 942
Score = 53.5 bits (127), Expect = 2e-06
Identities = 35/128 (27%), Positives = 59/128 (45%), Gaps = 10/128 (7%)
Query: 11 VTPQPD-LGLISDRHESIKSVVRRLDSNWHHVFCIRHITQLFEDFQEHRDEEKSKKYGLN 69
+ P D L ++SD+H SI V + HH CI H E +YG++
Sbjct: 628 IVPDTDNLMIVSDKHSSIYKGVSVVYPKAHHGACIVH--------PEQNISVSYARYGVS 679
Query: 70 NINFLPSGYSMTQPSITHYRNQIRDWSDDVVKWVDDNPKKQWLQAHDQGRRWGHITTNIS 129
+ F + + Y ++R WS K+++D + W +A+ + R+ +T+N S
Sbjct: 680 GL-FFSAAKAYRVRDFEKYFEELRGWSPGCAKYLEDVGFEHWTRAYCKEERYNIMTSNNS 738
Query: 130 ECFNNVLK 137
E NNVL+
Sbjct: 739 EAMNNVLR 746
>emb|CAE05572.2| OSJNBb0013O03.13 [Oryza sativa (japonica cultivar-group)]
Length = 1611
Score = 53.5 bits (127), Expect = 2e-06
Identities = 42/137 (30%), Positives = 74/137 (53%), Gaps = 14/137 (10%)
Query: 8 RLWVTPQPDLGLISDRHESIKSVVRRLDSNW---HHVFCIRHITQLFEDFQEHRDEEKSK 64
R+ V P ++ +ISDRH I + + HH +C+RH + F + D+ ++K
Sbjct: 561 RVVVGPHWEVCIISDRHAGIMNAMTTPVPGLPPVHHRWCMRHFSANFH--KAGADKHQTK 618
Query: 65 KYGLNNINFLPSGYSMTQPSITHYRNQIRDWSDDVVKWVDDN--PKKQWLQAHDQ-GRRW 121
+ L I + + + + + R +I + KW++D K +W +A+D+ GRRW
Sbjct: 619 E--LLRICQIDEKW-IFERDVEAVRQRIPEGPR---KWLEDELLDKDKWSRAYDRNGRRW 672
Query: 122 GHITTNISECFNNVLKG 138
G++TTN++E FN+VL G
Sbjct: 673 GYMTTNMAEQFNSVLVG 689
>gb|AAF04891.1| Mutator-like transposase [Arabidopsis thaliana]
gi|20465953|gb|AAM20162.1| putative mutator transposase
[Arabidopsis thaliana] gi|18389286|gb|AAL67086.1|
putative Mutator transposase [Arabidopsis thaliana]
Length = 757
Score = 52.0 bits (123), Expect = 5e-06
Identities = 35/124 (28%), Positives = 60/124 (48%), Gaps = 17/124 (13%)
Query: 15 PDLGLISDRHESIKSVVRRLDSNWHHVFCIRHITQLFEDFQEHRDEEKSKKYGLNN---I 71
P L ++SDR + I V + H FC+RH+++ F K++ NN +
Sbjct: 439 PRLTILSDRQKGIVEGVEQNFPTAFHGFCMRHLSESFR-----------KEF--NNTLLV 485
Query: 72 NFL-PSGYSMTQPSITHYRNQIRDWSDDVVKWVDDNPKKQWLQAHDQGRRWGHITTNISE 130
N+L + ++T +I + S D W+ P + W A+ +G+R+GH+T NI E
Sbjct: 486 NYLWEAAQALTVIEFEAKILEIEEISQDAAYWIRRIPPRLWATAYFEGQRFGHLTANIVE 545
Query: 131 CFNN 134
N+
Sbjct: 546 SLNS 549
>ref|NP_974088.1| SWIM zinc finger family protein [Arabidopsis thaliana]
Length = 750
Score = 52.0 bits (123), Expect = 5e-06
Identities = 36/135 (26%), Positives = 56/135 (40%), Gaps = 22/135 (16%)
Query: 7 LRLWVTPQPDLGLISDRHESIKSVVRRLDSNW-----HHVFCIRHI----TQLFEDFQEH 57
+R VT + L LIS H I +VV S W +H F + H +++F F
Sbjct: 436 IREKVTQRKGLCLISSPHPDIIAVVNESGSQWQEPWAYHRFSLNHFYSQFSRVFPSF--- 492
Query: 58 RDEEKSKKYGLNNINFLPSGYSMTQPSITHYRNQIRDWSDDVVKWVDDNPKKQWLQAHDQ 117
+G + + Y N I++ + + KW+D P+ +W AHD
Sbjct: 493 ----------CLGARIRRAGSTSQKDEFVSYMNDIKEKNPEARKWLDQFPQNRWALAHDN 542
Query: 118 GRRWGHITTNISECF 132
GRR+G + N F
Sbjct: 543 GRRYGIMEINTKALF 557
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.318 0.135 0.441
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 310,927,313
Number of Sequences: 2540612
Number of extensions: 12830406
Number of successful extensions: 31858
Number of sequences better than 10.0: 247
Number of HSP's better than 10.0 without gapping: 60
Number of HSP's successfully gapped in prelim test: 187
Number of HSP's that attempted gapping in prelim test: 31535
Number of HSP's gapped (non-prelim): 314
length of query: 155
length of database: 863,360,394
effective HSP length: 117
effective length of query: 38
effective length of database: 566,108,790
effective search space: 21512134020
effective search space used: 21512134020
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 69 (31.2 bits)
Medicago: description of AC133139.2


