

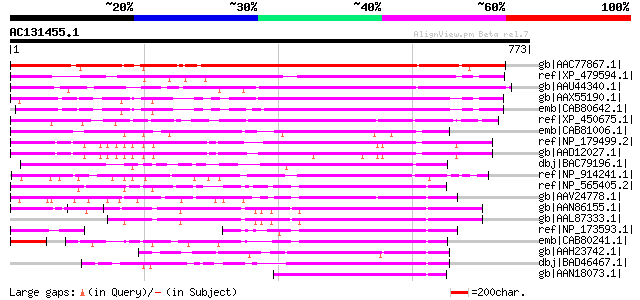
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC131455.1 + phase: 0
(773 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAC77867.1| hypothetical protein [Arabidopsis thaliana] gi|46... 808 0.0
ref|XP_479594.1| hypothetical protein [Oryza sativa (japonica cu... 394 e-108
gb|AAU44340.1| hypothetical protein [Oryza sativa (japonica cult... 359 2e-97
gb|AAX55190.1| hypothetical protein At4g39790 [Arabidopsis thali... 351 5e-95
emb|CAB80642.1| putative protein [Arabidopsis thaliana] gi|30804... 341 6e-92
ref|XP_450675.1| leucine zipper protein-like [Oryza sativa (japo... 286 2e-75
emb|CAB81006.1| putative protein [Arabidopsis thaliana] gi|49384... 268 7e-70
ref|NP_179499.2| expressed protein [Arabidopsis thaliana] 258 5e-67
gb|AAD12027.1| hypothetical protein [Arabidopsis thaliana] gi|74... 246 3e-63
dbj|BAC79196.1| bzip-like transcription factor-like protein [Ory... 207 8e-52
ref|NP_914241.1| bzip-like transcription factor-like [Oryza sati... 200 2e-49
ref|NP_565405.2| expressed protein [Arabidopsis thaliana] 191 6e-47
gb|AAV24778.1| hypothetical protein [Oryza sativa (japonica cult... 185 6e-45
gb|AAN86155.1| unknown protein [Arabidopsis thaliana] gi|1522397... 181 1e-43
gb|AAL87333.1| unknown protein [Arabidopsis thaliana] 179 4e-43
ref|NP_173593.1| expressed protein [Arabidopsis thaliana] gi|526... 177 2e-42
emb|CAB80241.1| putative protein [Arabidopsis thaliana] gi|30804... 168 5e-40
gb|AAH23742.1| Unknown (protein for MGC:38531) [Mus musculus] gi... 165 6e-39
dbj|BAD46467.1| bzip-related transcription factor -like [Oryza s... 147 1e-33
gb|AAN18073.1| At4g35240/F23E12_200 [Arabidopsis thaliana] gi|15... 144 1e-32
>gb|AAC77867.1| hypothetical protein [Arabidopsis thaliana]
gi|46518491|gb|AAS99727.1| At2g27090 [Arabidopsis
thaliana] gi|25407857|pir||F84668 hypothetical protein
At2g27090 [imported] - Arabidopsis thaliana
gi|15225846|ref|NP_180277.1| expressed protein
[Arabidopsis thaliana]
Length = 743
Score = 808 bits (2086), Expect = 0.0
Identities = 443/758 (58%), Positives = 542/758 (71%), Gaps = 44/758 (5%)
Query: 1 MGASSSKMEEDKALQLCRERKKFVRQALDGRCSLAAAHISYVQSLRIVGTALSKFTEPEP 60
MGAS+S+++EDKALQLCRERKKFV+QALDGRC LAAAH+SYVQSL+ GTAL KF+E E
Sbjct: 1 MGASTSRIDEDKALQLCRERKKFVQQALDGRCLLAAAHVSYVQSLKSTGTALRKFSETEV 60
Query: 61 HSESSLYTN--ATPEA-LALTDKAMSQSSPSMSQQIDAGETHSFYTT----PSPPSSSKF 113
ESSLYT+ ATPE LAL +K++S S S +HS + T PSPPS+S F
Sbjct: 61 PVESSLYTSTSATPEQPLALIEKSVSHLSYSPPPA-----SHSHHDTYSPPPSPPSTSPF 115
Query: 114 HANHMKFGSFSSKKVEEKPPVPVIATVTSSGIAQNGS--RMSETEAFEDSSVPDGTPQWD 171
NHMKF FSSKKVEEKPPV +IATVTSS I + S +M T E SS+P P WD
Sbjct: 116 QVNHMKFKGFSSKKVEEKPPVSIIATVTSSSIPPSRSIEKMESTPFEESSSMPPEAP-WD 174
Query: 172 FFGLFNPVDHQFSFQEPKGMPHHIGNA-------EEGGIPDLEDDEEKASSCGSEGSRDS 224
+FGL +P+D+QFS H+GN EE G P+ EDD E S E SRDS
Sbjct: 175 YFGLSHPIDNQFSSS-------HVGNGHVSRSVKEEDGTPEEEDDGEDFSFQEREESRDS 227
Query: 225 EDEFDEEPTAETLVQRFENLNRANNHVQENVLPVKGDSVSEVELVSEKGNFPNSSPSKKL 284
+D+ +EPT++TLV+ FEN NR + LP + + V +EK P SP
Sbjct: 228 DDDEFDEPTSDTLVRSFENFNRVRR--DHSTLPQR-EGVESDSSDAEKSKTPELSP---- 280
Query: 285 PMAALLPPEANKSTEKENHSENKVTPKNFFSSMKDIEVLFNRASDSGKEVPRMLEANKFH 344
P+ L+ NK+ K +H+ENK+ P++F SSMK+IE+LF +AS++GKEVPRMLEANK H
Sbjct: 281 PVTPLVATPVNKTPNKGDHTENKLPPRDFLSSMKEIELLFVKASETGKEVPRMLEANKLH 340
Query: 345 FRPIFQGKKDGSLASFICKACFSCGEDPSQVPEEPAQNSVKYLTWHRTASSRSSSSKNPL 404
FRPI K+ GS AS + K C SCGEDP VPEEPAQNSVKYLTWHRT SSRSSSS+NPL
Sbjct: 341 FRPIVPSKESGSGASSLFKTCLSCGEDPKDVPEEPAQNSVKYLTWHRTESSRSSSSRNPL 400
Query: 405 DANSRENVEDLTNNLFDNSCMNAGSHASTLDRLHAWERKLYDEVKASEIVRKEYDAKCKI 464
+ ++VE+L +NLF+N CM AGSHASTLDRL+AWERKLYDEVK S+ VR+EYD KC+I
Sbjct: 401 GGMNSDDVEELNSNLFENICMIAGSHASTLDRLYAWERKLYDEVKGSQTVRREYDEKCRI 460
Query: 465 LRNLESKAEKTSTVDKTRAAVKDLHSRIRVAIHRIDSISKRIEELRDKELQPQLEELIEG 524
LR LES+ + + +DKTRA VKDLHSRIRVAIHRIDSIS+RIEELRD ELQPQLEELIEG
Sbjct: 461 LRELESEGKGSQRIDKTRAVVKDLHSRIRVAIHRIDSISRRIEELRDNELQPQLEELIEG 520
Query: 525 LNRMWEVMHECHKLQFQIMSASYNNSHARITMHSELRRQITAYLENELQFLSSSFTKWIE 584
L+RMWEVM ECHK+QFQ++ A Y + ++ M SEL RQ+T++LE+EL L+SSFTKWI
Sbjct: 521 LSRMWEVMLECHKVQFQLIKACYRGGNIKLNMQSELHRQVTSHLEDELCALASSFTKWIT 580
Query: 585 AQKSYLQAINGWIHKCVPLQQKSVKRKRRPQSELLIQYGPPIYATCDVWLKKLGTLPVKD 644
QKSY+QAIN W+ KCV L Q+S KRKRR L YGPPIYATC +WL+KL LP K+
Sbjct: 581 GQKSYIQAINEWLVKCVALPQRS-KRKRRAPQPSLRNYGPPIYATCGIWLEKLEVLPTKE 639
Query: 645 VVDSIKSLAADTARFLPYQDKNQGKEPHSHIGGESADGL----LRDDISEDWISGFDRFR 700
V SIK+LA+D ARFLP Q+KN+ K+ H GE+ + L L+D+ ED GFDRFR
Sbjct: 640 VSGSIKALASDVARFLPRQEKNRTKK---HRSGENKNDLTAHMLQDETLEDCGPGFDRFR 696
Query: 701 ANLIRFLGQLNSLSGSSVKMYRELRQAIQEAKNNYHRL 738
+L F+GQLN + SSVKMY EL++ I AKNNY +L
Sbjct: 697 TSLEGFVGQLNQFAESSVKMYEELKEGIHGAKNNYEQL 734
>ref|XP_479594.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|50509145|dbj|BAD30285.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
gi|22324428|dbj|BAC10345.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 909
Score = 394 bits (1012), Expect = e-108
Identities = 265/779 (34%), Positives = 390/779 (50%), Gaps = 131/779 (16%)
Query: 1 MGASSSKMEEDKALQLCRERKKFVRQALDGRCSLAAAHISYVQSLRIVGTALSKFTEPEP 60
MG + SK+E+D+AL LC+ERK+FVR+A+DGRC+LAAAH Y++SLR G L K E E
Sbjct: 218 MGVAPSKIEDDRALVLCQERKRFVREAIDGRCALAAAHCDYIRSLRDTGFLLRKCFEHEA 277
Query: 61 HSESSLYTNATPEALALTDKAMSQSSPSMSQQIDAGETHSFYTTPSPPSSSKFHANHMKF 120
ES P+ S S F A+HMK
Sbjct: 278 SEES---------------------------------------IPNNKSPSSFQASHMKA 298
Query: 121 GSFSSKKVEEKPPVPVIATVTSSGIAQNGSRMSETEAFEDSSVPDGTPQWDFFGLFNPVD 180
S + EK PV T+ S+ S P GT D F +P D
Sbjct: 299 AMNSIRTYLEKVATPVTVTMVSA----------------SSQDPTGTSPLDHFDQIHPGD 342
Query: 181 HQFSFQEPKGMPHHIGNAE------EGGIPDLEDDEEKASSCGSEGSRDSEDEF--DEEP 232
+QFS +E + + E GIP+LE++ E+ S +G +S+D+F +EE
Sbjct: 343 NQFSPKEKDRSGQCLDKVDDPRPFLEEGIPELEEEGERTPSNEEDGFAESKDDFANEEEN 402
Query: 233 TAET-------LVQRFENLNRANNHVQENVLPVKG------DSVSEVELVSEKGNFPNSS 279
+E+ ++ F ++ +N+ +N K SV+ ++ N +
Sbjct: 403 FSESNDAFLSPSIETFVPVSNSNDVSDKNSSTDKAPEHHGHGSVASKDIALPNTGCQNDN 462
Query: 280 PSKKLPMAAL----------------LPPE--ANKSTEKENHSENKVTPKNFFSSMKDIE 321
P + M + +PP A KE + ++ K ++ M +IE
Sbjct: 463 PQNERRMTDIHTNENYSNSAVSPVNVVPPSGAAFPMVSKEPYPYLSISVKGLYTGMVEIE 522
Query: 322 VLFNRASDSGKEVPRMLEANKFHFRPIF-QGKKDGSLASFICKACFSCGEDPSQVPEEPA 380
LF+RA DSGKEV R+L+ +K FR + Q GS +S F+C + +PE P+
Sbjct: 523 RLFSRACDSGKEVTRVLDEDKLQFRALLPQETARGSASSSFLSTLFACCREDVPLPETPS 582
Query: 381 QNSVKYLTWHRTASSRSSSSKNPLDANSRENVEDLTNNLFDNSCMNAGSHASTLDRLHAW 440
Q VKYLTWHR+ SS+ S S+NP A + H STLD+L+AW
Sbjct: 583 QAEVKYLTWHRSVSSQLSLSRNPPGAIT-------------------VMHTSTLDKLYAW 623
Query: 441 ERKLYDEVKASEIVRKEYDAKCKILRNLESKAEKTSTVDKTRAAVKDLHSRIRVAIHRID 500
E KLYDEVK + + + YD KCK LR+ ES+ + VD TRA VKDLHSRI VAI +ID
Sbjct: 624 EEKLYDEVKVNSAICRRYDEKCKQLRDQESRGKNQILVDFTRATVKDLHSRILVAIQKID 683
Query: 501 SISKRIEELRDKELQPQLEELIEGLNRMWEVMHECHKLQFQIMSASYNNSHARITMHSEL 560
ISK IE++RDKELQPQL+ELI L RMWE M ECH LQ IM + +++ SE
Sbjct: 684 FISKNIEDIRDKELQPQLDELIRSLTRMWETMLECHHLQLAIMKLVSSKRSVKLSFQSES 743
Query: 561 RRQITAYLENELQFLSSSFTKWIEAQKSYLQAINGWIHKCV-PLQQKSVKRKRRPQSELL 619
Q L +L L S F W+ + K YL ++N W+HKC+ PL+++ RK+ L
Sbjct: 744 ECQDALLLSAKLIKLCSDFQNWVASHKVYLSSLNLWLHKCMKPLKKRKGSRKQNVVDVSL 803
Query: 620 IQYG-PPIYATCDVWLKKLGTLPVKDVVDSIKSLAADTARFLPYQDKNQGKEPHSHIGGE 678
+ PI+ TC++W+K + L ++V +I++L AD R P+Q++ + GE
Sbjct: 804 TECAVAPIFTTCEIWIKLIDDLLTNELVKAIENLVADVGRSFPHQEQ--------VLNGE 855
Query: 679 SADGLLRDDISEDWISGFDRFRANLIRFLGQLNSLSGSSVKMYRELRQAIQEAKNNYHR 737
+ +LR++ D +++L+ FL +L + S S++ Y +L++ I EAK+ + R
Sbjct: 856 TGGEILRNNAPAD-------VQSSLMAFLEKLEAFSAVSLQKYIDLQKNIDEAKDRFSR 907
>gb|AAU44340.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 722
Score = 359 bits (921), Expect = 2e-97
Identities = 260/777 (33%), Positives = 391/777 (49%), Gaps = 86/777 (11%)
Query: 1 MGASSSKMEEDKALQLCRERKKFVRQALDGRCSLAAAHISYVQSLRIVGTALSKFTEPEP 60
MG+ SK + AL LC+ER + +++A+D R +L+A+H+SY QSLR VGTAL ++ E E
Sbjct: 1 MGSLPSKATGEDALVLCKERMRHIKRAIDSRDALSASHLSYTQSLRSVGTALRRYAESEI 60
Query: 61 HSESSLYTNATPEALALTDKAMSQSS---PSMSQQIDAGETHSFYTTPSPPSSSKFHANH 117
+ESSL +++ DK+ S SS PS SQ +++ + + P S+K H
Sbjct: 61 STESSL-------SISEADKSPSHSSMASPSPSQALESTGSPVHRGSQLTPPSTKIHY-- 111
Query: 118 MKFGSFSSKKVEEKPPVPVIATVTSSGIAQNGSRMSETEAFEDSSVPDGTPQWDFFGLFN 177
K P+ + +++ S +S P+ WDFF
Sbjct: 112 --------MKAAGTKPLTITIDPSAADFVGQESPVSTFVPPPPPLPPELCTSWDFFD--- 160
Query: 178 PVDHQFSFQEPKGMPHHIGNAEEGGIPDLEDDEEKASSCGSEGSRDSEDEFDEEPTAETL 237
N G ++ + +G RDS + +E +
Sbjct: 161 ------------------SNYASGSATSNNENGVTLNFSRLKGLRDSRE-------SEAV 195
Query: 238 VQRFENLNRANNHVQENVLPVKGDSVS-EVELVSEKGNFPNSSPSKKLPMAALLPPEANK 296
R E NR++ E + GD+ + + E ++K S S ++ A + +
Sbjct: 196 SLREEATNRSDGMHPE----LPGDNAAPKQEAQAKKSGMSKPSGSVEVTTEAATSGQVGE 251
Query: 297 STEKENHSENKVTP------------KNFFSSMKDIEVLFNRASDSGKEVPRMLEANKFH 344
E+++ + T K+F SSMKDIE+ F RA+++G EV RMLE K
Sbjct: 252 KVEEDDMEKELCTEAEDPSEFITHRAKDFVSSMKDIEIRFMRAAEAGNEVSRMLETKKIR 311
Query: 345 FR-----PIFQGKKDGSLASFICKACFSCGEDPSQVPEEPAQNSVKYLTWHRTASSRSSS 399
P GK + + CF+ + + +E AQ+ K +TW R+ SS SSS
Sbjct: 312 LDICAKIPGSPGKPPTARFVSALRVCFN---RENILNQETAQHVSKVVTWKRSVSSLSSS 368
Query: 400 SKNPLDANS-RENVEDLTNNLFDNSCMNAGSHASTLDRLHAWERKLYDEVKASEIVRKEY 458
SK+PL A ++V D ++ + M +GSH+STLDRLHAWERKL+DE+KASE VRK Y
Sbjct: 369 SKSPLTAAMITDDVGDSNSDFVEQFAMVSGSHSSTLDRLHAWERKLHDEIKASEHVRKTY 428
Query: 459 DAKCKILRNLESKAEKTSTVDKTRAAVKDLHSRIRVAIHRIDSISKRIEELRDKELQPQL 518
D KC +LR+ ++ +DKTRA VKDLHSR+ VAI +D+ISKRIE++RD+ELQPQL
Sbjct: 429 DEKCNLLRHQFARGLNAQLIDKTRAIVKDLHSRVSVAIQAVDAISKRIEKIRDEELQPQL 488
Query: 519 EELIEGLNRMWEVMHECHKLQFQIMSASYNNSHARITMHSELRRQITAYLENELQFLSSS 578
ELI+GL RMW+ M ECH QF +S +Y+ A E + +L NEL SSS
Sbjct: 489 VELIQGLIRMWKTMLECHHKQFITISLAYHVKSATTVQQGEHHHRAATHLWNELDCFSSS 548
Query: 579 FTKWIEAQKSYLQAINGWIHKCV--PLQQKSVKRKRRPQSELLIQYGPPIYATCDVWLKK 636
F W+ A KSY++++N W+ KCV P Q + +RKR+ PPI+ C WL
Sbjct: 549 FKIWVTAHKSYVESLNAWLQKCVLQPAQDR-WRRKRKVSFPPRHALSPPIFVLCRDWLTM 607
Query: 637 L--GTLPVKDVVDSIKSLAADTARFLPYQDKNQGK-EPHSHIGGESAD-GLLRDDISE-- 690
+ +LP ++ SIK + +Q +Q K SH+ ES + G+L ++ E
Sbjct: 608 MESQSLPTDELCKSIKEVVQLLRGSFDHQADHQNKMTTESHLRNESQECGMLENNEQEVS 667
Query: 691 DWISGFDRFRANLIRFLGQLNSLSGSSVKMYRELRQAIQEAKNNYHRLNSQSQNGHL 747
+ + ++ L L +L S +S+K Y EL+Q + A+++Y S N HL
Sbjct: 668 GSVEAVEGLQSKLTTVLDRLTKFSEASLKHYEELKQNYEMARDDYKMGRS---NAHL 721
>gb|AAX55190.1| hypothetical protein At4g39790 [Arabidopsis thaliana]
gi|52354435|gb|AAU44538.1| hypothetical protein
AT4G39790 [Arabidopsis thaliana]
Length = 657
Score = 351 bits (901), Expect = 5e-95
Identities = 263/753 (34%), Positives = 374/753 (48%), Gaps = 125/753 (16%)
Query: 1 MGASSSKMEEDKA---LQLCRERKKFVRQALDGRCSLAAAHISYVQSLRIVGTALSKFTE 57
MG S+SK +K L LC+ERK+FV+QA+D RC+LAAAH+SY++SLR +G L ++ E
Sbjct: 9 MGCSNSKASMNKKNEPLHLCKERKRFVKQAMDSRCALAAAHVSYIRSLRNIGACLRQYAE 68
Query: 58 PEPHSESSLYTNATPEALALTDKAMSQSSPSMSQQIDAGETHSFYTTPSPPSSSKFHANH 117
E ESS AT +K+ S +S +D+ +H+ + P+P F+ ++
Sbjct: 69 AETAHESSPSLTATE-----PEKSPSHNSSYPDDSVDSPLSHN--SNPNPNPKPLFNLSY 121
Query: 118 MKFGSFSSKKVEEKPPVPVIATVTSSGIAQNGSRMSET-EAFEDSSVP----DGTPQWDF 172
MK + SS T T + ++ + T AF P T WD+
Sbjct: 122 MKTETASS-----------TVTFTINPLSDGDDDLEVTMPAFSPPPPPRPRRPETSSWDY 170
Query: 173 FGLFNPVDHQFSFQEPKGMPHHIGNAEEGGIPDLEDDE-----EKASSCGSEGSRDSEDE 227
F + D F F +G +E+ I D E D EK +S G+ SE
Sbjct: 171 FDTCDDFD-SFRF---------VGLSEQTEI-DSECDAAVIGLEKITSQGNVAKSGSETL 219
Query: 228 FDEEPTAETLVQRFENLNRANNHVQENVLPVKGDSVSEVELVSEKGNFPNSSPSKKLPMA 287
D + Q E+ N PS+ +
Sbjct: 220 QDSSFKTKQRKQSCED------------------------------NDEREDPSEFITHR 249
Query: 288 ALLPPEANKSTEKENHSENKVTPKNFFSSMKDIEVLFNRASDSGKEVPRMLEANKFHFRP 347
A K+ S K FF RAS+SG+EV RMLE NK
Sbjct: 250 A-----------KDFVSSMKDIEHKFF-----------RASESGREVSRMLEVNKIRVGF 287
Query: 348 IFQGKKDGSLA--SFICKACFSCGEDPSQVPEEPAQNSV-KYLTWHRTASSRSSSSKNPL 404
K S+A + + +AC G+ S V +EP + V K + W RT+SSRSS+S+NPL
Sbjct: 288 ADMTGKGNSIAFLAALKRACCR-GKSYSPVSQEPLSHQVTKVIVWKRTSSSRSSTSRNPL 346
Query: 405 DANSRENVEDLTNNLF-DNSCMNAGSHASTLDRLHAWERKLYDEVKASEIVRKEYDAKCK 463
S+E+ +D + + F + CM +GSH+S+LDRL+AWERKLYDEVKASE++RKEYD KC+
Sbjct: 347 IQTSKEDHDDESGSDFIEEFCMISGSHSSSLDRLYAWERKLYDEVKASEMIRKEYDRKCE 406
Query: 464 ILRNLESKAEKTSTVDKTRAAVKDLHSRIRVAIHRIDSISKRIEELRDKELQPQLEELIE 523
LRN +K ++DKTRAA KDLHSRIRVAI ++SISKRIE +RD EL PQL E ++
Sbjct: 407 QLRNQFAKDHSAKSMDKTRAAAKDLHSRIRVAIQSVESISKRIERIRDDELHPQLLEFLQ 466
Query: 524 GLNRMWEVMHECHKLQFQIMSASYNNSHARITMH-SELRRQITAYLENELQFLSSSFTKW 582
GL RMW+ M ECH Q+ +S +Y+ H+ T H S L+R+I A L E + SF
Sbjct: 467 GLIRMWKAMLECHHTQYITISLAYHCRHSSKTAHESVLKRRILAELLEETECFGLSFVDL 526
Query: 583 IEAQKSYLQAINGWIHKCVPLQQKSVKRKRRPQSELLIQYGPPIYATCDVWLKKLGTLPV 642
+ + SY++A+NGW+H CV L Q+ R RRP S + PPI+ C W + TLP
Sbjct: 527 VHSMASYVEALNGWLHNCVLLPQERSTRNRRPWSPRRV-LAPPIFVLCRDWSAGIKTLPS 585
Query: 643 KDVVDSIKSLAADTARFLPYQDKNQGKEPHSHIGGESADGLLRDDISEDWISGFDRFRAN 702
++ SIK + D +G E LL D+S ++
Sbjct: 586 DELSGSIKGFSLDM----------------EMLGEEKGGSLLVSDLSS--------VHSS 621
Query: 703 LIRFLGQLNSLSGSSVKMYRELRQAIQEAKNNY 735
L + L +L S +S+KMY +++ + A+ Y
Sbjct: 622 LAKLLERLKKFSEASLKMYEDVKVKSEAARVAY 654
>emb|CAB80642.1| putative protein [Arabidopsis thaliana] gi|3080448|emb|CAA18765.1|
putative protein [Arabidopsis thaliana]
gi|15236022|ref|NP_195689.1| expressed protein
[Arabidopsis thaliana] gi|7487214|pir||T05016
hypothetical protein T19P19.180 - Arabidopsis thaliana
Length = 631
Score = 341 bits (874), Expect = 6e-92
Identities = 255/742 (34%), Positives = 365/742 (48%), Gaps = 131/742 (17%)
Query: 9 EEDKALQLCRERKKFVRQALDGRCSLAAAHISYVQSLRIVGTALSKFTEPEPHSESSLYT 68
++++ L LC+ERK+FV+QA+D RC+LAAAH+SY++SLR +G L ++ E E ESS
Sbjct: 3 KKNEPLHLCKERKRFVKQAMDSRCALAAAHVSYIRSLRNIGACLRQYAEAETAHESSPSL 62
Query: 69 NATPEALALTDKAMSQSSPSMSQQIDAGETHSFYTTPSPPSSSKFHANHMKFGSFSSKKV 128
AT +K+ S +S +D+ +H+ + P+P F+ ++MK + SS
Sbjct: 63 TATE-----PEKSPSHNSSYPDDSVDSPLSHN--SNPNPNPKPLFNLSYMKTETASS--- 112
Query: 129 EEKPPVPVIATVTSSGIAQNGSRMSET-EAFEDSSVP----DGTPQWDFFGLFNPVDHQF 183
T T + ++ + T AF P T WD+F + D F
Sbjct: 113 --------TVTFTINPLSDGDDDLEVTMPAFSPPPPPRPRRPETSSWDYFDTCDDFD-SF 163
Query: 184 SFQEPKGMPHHIGNAEEGGIPDLEDDE-----EKASSCGSEGSRDSEDEFDEEPTAETLV 238
F +G +E+ I D E D EK +S G+ SE D +
Sbjct: 164 RF---------VGLSEQTEI-DSECDAAVIGLEKITSQGNVAKSGSETLQDSSFKTKQRK 213
Query: 239 QRFENLNRANNHVQENVLPVKGDSVSEVELVSEKGNFPNSSPSKKLPMAALLPPEANKST 298
Q E+ N PS+ + A
Sbjct: 214 QSCED------------------------------NDEREDPSEFITHRA---------- 233
Query: 299 EKENHSENKVTPKNFFSSMKDIEVLFNRASDSGKEVPRMLEANKFH--FRPIFQGKKDGS 356
K+ S K FF RAS+SG+EV RMLE NK F + +GK
Sbjct: 234 -KDFVSSMKDIEHKFF-----------RASESGREVSRMLEVNKIRVGFADMTEGK---- 277
Query: 357 LASFICKACFSCGEDPSQVPEEPAQNSV-KYLTWHRTASSRSSSSKNPLDANSRENVEDL 415
+ C E V EP + V K + W RT+SSRSS+S+NPL S+E+ +D
Sbjct: 278 ----VIHQFLKCSELNFNV--EPLSHQVTKVIVWKRTSSSRSSTSRNPLIQTSKEDHDDE 331
Query: 416 TNNLF-DNSCMNAGSHASTLDRLHAWERKLYDEVKASEIVRKEYDAKCKILRNLESKAEK 474
+ + F + CM +GSH+S+LDRL+AWERKLYDEVKASE++RKEYD KC+ LRN +K
Sbjct: 332 SGSDFIEEFCMISGSHSSSLDRLYAWERKLYDEVKASEMIRKEYDRKCEQLRNQFAKDHS 391
Query: 475 TSTVDKTRAAVKDLHSRIRVAIHRIDSISKRIEELRDKELQPQLEELIEGLNRMWEVMHE 534
++DKTRAA KDLHSRIRVAI ++SISKRIE +RD EL PQL E ++GL RMW+ M E
Sbjct: 392 AKSMDKTRAAAKDLHSRIRVAIQSVESISKRIERIRDDELHPQLLEFLQGLIRMWKAMLE 451
Query: 535 CHKLQFQIMSASYNNSHARITMH-SELRRQITAYLENELQFLSSSFTKWIEAQKSYLQAI 593
CH Q+ +S +Y+ H+ T H S L+R+I A L E + SF + + SY++A+
Sbjct: 452 CHHTQYITISLAYHCRHSSKTAHESVLKRRILAELLEETECFGLSFVDLVHSMASYVEAL 511
Query: 594 NGWIHKCVPLQQKSVKRKRRPQSELLIQYGPPIYATCDVWLKKLGTLPVKDVVDSIKSLA 653
NGW+H CV L Q+ R RRP S + PPI+ C W + TLP ++ SIK +
Sbjct: 512 NGWLHNCVLLPQERSTRNRRPWSPRRV-LAPPIFVLCRDWSAGIKTLPSDELSGSIKGFS 570
Query: 654 ADTARFLPYQDKNQGKEPHSHIGGESADGLLRDDISEDWISGFDRFRANLIRFLGQLNSL 713
D +G E LL D+S ++L + L +L
Sbjct: 571 LDM----------------EMLGEEKGGSLLVSDLSS--------VHSSLAKLLERLKKF 606
Query: 714 SGSSVKMYRELRQAIQEAKNNY 735
S +S+KMY +++ + A+ Y
Sbjct: 607 SEASLKMYEDVKVKSEAARVAY 628
>ref|XP_450675.1| leucine zipper protein-like [Oryza sativa (japonica
cultivar-group)] gi|49388784|dbj|BAD25979.1| leucine
zipper protein-like [Oryza sativa (japonica
cultivar-group)] gi|49387676|dbj|BAD25922.1| leucine
zipper protein-like [Oryza sativa (japonica
cultivar-group)]
Length = 735
Score = 286 bits (731), Expect = 2e-75
Identities = 231/746 (30%), Positives = 352/746 (46%), Gaps = 86/746 (11%)
Query: 1 MGASSSKMEEDKALQLCRERKKFVRQALDGRCSLAAAHISYVQSLRIVGTALSKFTEPEP 60
MG+S SK ++D AL +C++R + + QA+D R +L+AA ++Y QSLR +G AL +F E
Sbjct: 1 MGSSVSKQDDDTALLICKDRLRHIEQAIDARYALSAAQLAYEQSLRSLGIALRQFVEAHK 60
Query: 61 ------HSESSLYT--NATPEALALTDKAMSQSSPSMSQQIDAGETHSFYTTPS------ 106
S SS Y +++P + + S++S S++ I+ + S S
Sbjct: 61 DDDDIERSPSSSYAIVSSSPPHRSDVNHMKSEASTSVTVTINTSQASSVQKEQSVTAFLP 120
Query: 107 PPSSSKFHANHMKFGSFSSKKVEEKPPVPVIATVTSSGIAQNGSRMSET---EAFEDSSV 163
PP +F ++ F TV S +A + S S+T ED S
Sbjct: 121 PPLQLEFCSSWDFFDP----------------TVVSENVASDASVNSQTFELRTLEDLSN 164
Query: 164 PDGTPQWDFFGLFNPVDHQFSFQEPKGMPHHIGNAEEGGIPDLEDDEEKASSCGSEGSRD 223
P+ G + + QE G P + +PDL ++ G+
Sbjct: 165 PNEMGLASSIGNTSEI---VEVQEVFGAPGWKQVHKNDNLPDLH--HSNSNEIQMSGTHL 219
Query: 224 SEDEFDEEPTAETLVQRFENLNRANNHVQENVLPVKGDSVSEVELVSEKGNFPNSSPSKK 283
D EE + Q N +N V +N+ + N N + K
Sbjct: 220 PNDSSLEEELEQVQTQAIGGQN--SNDVSDNI--------------KSEANHINVNAPKN 263
Query: 284 LPMAALLPPEANKSTEKENHSENKVTPKNFFSSMKDIEVLFNRASDSGKEVPRMLEANKF 343
A+ +++ S K+F S +KD+E F+RA+ S EV RMLE K
Sbjct: 264 EEAKAIFITDSDSS-------------KDFLSCVKDLERQFSRAAVSCHEVSRMLETKKI 310
Query: 344 HFRPIFQGKKDGSLASFICKACFSCGEDPSQVPEEPAQNSV-KYLTWHRTASSRSSSSKN 402
Q K G + + + F G + ++ V K +TW+R+ SSRSS+SKN
Sbjct: 311 RLSISSQTK--GKSSDVLFRPTFLIGCKAGTAASDGSEKRVTKAITWNRSLSSRSSASKN 368
Query: 403 PLD-ANSRENVEDLTNNLFDNSCMNAGSHASTLDRLHAWERKLYDEVKASEIVRKEYDAK 461
PL A + ++ ++ + CM +GSHAS+LDRL+AWE KLY+E+K +E ++K YD K
Sbjct: 369 PLTPAQMDDEFSEICSDFVEEFCMISGSHASSLDRLYAWEMKLYNELKGTESLKKIYDKK 428
Query: 462 CKILRNLESKAEKTSTVDKTRAAVKDLHSRIRVAIHRIDSISKRIEELRDKELQPQLEEL 521
C LR+ + VDKTR VKDL+SR++V + SISK IE+LRD+ELQPQL EL
Sbjct: 429 CVQLRHQFERDASARQVDKTRVIVKDLYSRLKVETEVLYSISKIIEKLRDEELQPQLLEL 488
Query: 522 IEGLNRMWEVMHECHKLQFQIMSASYNNSHARITMHSELRRQITAYLENELQFLSSSFTK 581
++GL RMW +MHE H++Q I+S+S + + + E +Q L NE+ F SS T
Sbjct: 489 LKGLTRMWAMMHEIHRVQQTIVSSS-DIVYVLRSPRGEPYKQPLVNLVNEMGFFYSSLTN 547
Query: 582 WIEAQKSYLQAINGWIHKCVPLQQKSVKRKRRPQSELLIQYGPPIYATCDVWLKKLGTLP 641
WI A K Y+ ++ W+ KCV LQ R RR PP++ D W + +LP
Sbjct: 548 WIAAYKCYVDGLHSWLQKCV-LQPYDHTRGRRLTLSPRRHLAPPMFVLLDDWSSAIASLP 606
Query: 642 VKDVVDSIKSLAADTARFLPYQDKNQGKEPHSHIGGESADGLLRDDISEDWISGFDRFRA 701
++ + SIK++ +D + KN H G + G + + FDR
Sbjct: 607 GEETLGSIKNIMSDLKKMF----KN-----HQAEGNKPETGSKLATLQAGLATMFDR--- 654
Query: 702 NLIRFLGQLNSLSGSSVKMYRELRQA 727
L +F ++SLS S R+A
Sbjct: 655 -LSKFSTAMSSLSESVKNSTEAAREA 679
>emb|CAB81006.1| putative protein [Arabidopsis thaliana] gi|4938489|emb|CAB43848.1|
putative protein [Arabidopsis thaliana]
gi|15810479|gb|AAL07127.1| unknown protein [Arabidopsis
thaliana] gi|15234640|ref|NP_194742.1| expressed protein
[Arabidopsis thaliana] gi|25055036|gb|AAN71978.1|
unknown protein [Arabidopsis thaliana]
gi|7486512|pir||T08989 hypothetical protein F6G3.160 -
Arabidopsis thaliana
Length = 725
Score = 268 bits (684), Expect = 7e-70
Identities = 210/687 (30%), Positives = 332/687 (47%), Gaps = 91/687 (13%)
Query: 1 MGASSSKMEEDKALQLCRERKKFVRQALDGRCSLAAAHISYVQSLRIVGTALSKFTEPEP 60
MG S SK ++D+A+Q+C++RK+F++QA++ R A+ HI+Y+QSLR V AL ++ E +
Sbjct: 1 MGCSHSKFDDDEAVQICKDRKRFIKQAVEHRTGFASGHIAYIQSLRKVSDALREYIEGDE 60
Query: 61 HSESSLYTNATP-EALALTDKAMSQSSPSMSQQIDAGETHSFYTTPSPPSSSKFHANHMK 119
E L T TP + ++ + + S PS Q +A SK + N
Sbjct: 61 PHEFMLDTCVTPVKRMSSSGGFIEISPPSKMVQSEA--------------ESKLNVNSYL 106
Query: 120 FGSFSSK-KVEEKPP-VPVIATVTSSGIAQN-GSRMS-ETEAFEDSSVPDGTPQ---WDF 172
S S +VEEKPP P V + G G M+ + ++P +PQ WDF
Sbjct: 107 MASGSRPVRVEEKPPRSPETFQVETYGADSFFGMNMNMNSPGLGSHNIPPPSPQNSQWDF 166
Query: 173 F-GLFNPVD-HQFSFQEPKGMPHHIGNA----EEGGIPDLEDDEEKASSCGSEGSRDSED 226
F F+ +D + +S+ GM + EE GIPDLE+DE ED
Sbjct: 167 FWNPFSALDQYGYSYDNQNGMDDDMRRLRRVREEEGIPDLEEDEYVKF----------ED 216
Query: 227 EFDEEPTAET---LVQRFENLNRANNHVQENVLPVKGDSVSEVELVSEKGNFPNSSPSKK 283
+ + T + + + + + N ++ ++ +S E+ + S
Sbjct: 217 HHNMKATEDFNGGKMYQEDKVEHVNEEFTDSGCKIENESDKNCNGTQERRSLEVSRGGTT 276
Query: 284 LPMAALLPPEANKSTEKENHSENKVTPKNFFSSMKDIEVLFNRASDSGKEVPRMLEANKF 343
+ + + T N+ P + +KD+E F +GKEV +LEA++
Sbjct: 277 GHVVGVTSDDGKGETPGFTVYLNR-RPTSMAEVIKDLEDQFAIICTAGKEVSGLLEASRV 335
Query: 344 HFRPIFQGKKDGSLASFICKACFSCGEDPSQVPEEPAQNSVKYLTWHRTASSRSSSSKNP 403
+ + ++ A F G SSRSSSS
Sbjct: 336 QYT---SSNELSAMTMLNPVALFRSG-----------------------GSSRSSSSSRF 369
Query: 404 L---DANSRENVEDLTNNLFDNSCMNAGSHASTLDRLHAWERKLYDEVKASEIVRKEYDA 460
L SR + + ++ + SCM +GSH STLDRL+AWE+KLYDEVK+ + +R Y+
Sbjct: 370 LISSSGGSRASEFESSSEFSEESCMLSGSHQSTLDRLYAWEKKLYDEVKSGDRIRIAYEK 429
Query: 461 KCKILRNLESKAEKTSTVDKTRAAVKDLHSRIRVAIHRIDSISKRIEELRDKELQPQLEE 520
KC +LRN + K +S VDKTRA ++DLH++I+V+IH I+SIS+RIE LRD+EL PQL E
Sbjct: 430 KCLVLRNQDVKGADSSAVDKTRATIRDLHTQIKVSIHSIESISERIETLRDQELLPQLLE 489
Query: 521 LIEGLNRMWEVMHECHKLQFQ-------IMSASYNNSHARITMHS--ELRRQITA----Y 567
L++GL +MW+VM ECH++Q + +++ + +N H + S E+ Q A +
Sbjct: 490 LVQGLAQMWKVMAECHQIQKRTLDEAKLLLATTPSNRHKKQQQTSLPEINSQRLARSALH 549
Query: 568 LENELQFLSSSFTKWIEAQKSYLQAINGWIHKCVPLQQKSVKRKRRPQSELLIQYGPPIY 627
L +L+ + F WI +Q+SY+ ++ GW+ +C P+ L PIY
Sbjct: 550 LVVQLRNWRACFQAWITSQRSYILSLTGWLLRCFRCDPD-------PEKVTLTSCPHPIY 602
Query: 628 ATCDVWLKKLGTLPVKDVVDSIKSLAA 654
C W + L L K V+D + A+
Sbjct: 603 EVCIQWSRLLNGLNEKPVLDKLDFFAS 629
>ref|NP_179499.2| expressed protein [Arabidopsis thaliana]
Length = 814
Score = 258 bits (659), Expect = 5e-67
Identities = 227/818 (27%), Positives = 371/818 (44%), Gaps = 138/818 (16%)
Query: 1 MGASSSKMEEDKALQLCRERKKFVRQALDGRCSLAAAHISYVQSLRIVGTALSKFTEPEP 60
MG S SK+++++A+Q+C++RK+F++QA++ R A+ HI+Y+ SLR V AL F
Sbjct: 1 MGCSHSKLDDEEAVQICKDRKRFIKQAIEHRTKFASGHIAYIHSLRKVSDALHDFILQGD 60
Query: 61 HSESSLYTNATPEALALTDKAMSQSSPSMSQQIDAGETHSFYTTPSPPSS------SKFH 114
++ + T ++ K M Q S+ ++GE S + PP S
Sbjct: 61 NNNEFVPT-LCQDSFVTPVKRMPQRRRRSSRS-NSGEFISISPSSIPPKMIQGRPRSNVK 118
Query: 115 ANHMKFGSFSSKKVEEKPP----VPVIATVTSS----------GIAQNGSRMSETEA--- 157
AN++ +VE++ P V + +SS G+ N + + T +
Sbjct: 119 ANYLMANRSRPVRVEQRSPETFRVETFSPPSSSNQYGESDGFFGMNMNMNTSASTSSSFW 178
Query: 158 ---------FEDSSVPDGTPQ---WDFF-GLFNPVDH-------QFSFQEPKGMPHHIGN 197
++P +PQ WDFF F+ +D+ + S G+ I
Sbjct: 179 NPLSSPEQRLSSHNIPPPSPQNSQWDFFWNPFSSLDYYGYNSYDRGSVDSRSGIDDEIRG 238
Query: 198 A----EEGGIPDLEDDEEK-----------------------------ASSCGSEGSRDS 224
EE GIPDLE+D+E S C E +
Sbjct: 239 LRRVREEEGIPDLEEDDEPHKPEPVVNVRFQNHNPNPKATEENRGKVDKSCCNEEVKVED 298
Query: 225 EDEFDEEPTAETLVQRFENLNRANNHVQENVL--PVKGDSVSEVELVSEK-GNFPNSSPS 281
DE ++E E + ++ N E + P + +EV E GN
Sbjct: 299 VDEDEDEDGDEDDDEFTDSGCETENEGDEKCVAPPTQEQRKAEVCRGGEATGNVVGVGKV 358
Query: 282 KKLPMAALLPPEANKSTEKENHSENKVTPKNFFSSMKDIEVLFNRASDSGKEVPRMLEAN 341
+++ + +A K+T + + P + +KD+E F D+ KEV +LEA
Sbjct: 359 QEMKNVVGVRDDA-KTTGFTVYVNRR--PTSMAEVIKDLEDQFTTICDAAKEVSGLLEAG 415
Query: 342 KFHFRPIFQGKKDGSLASFICKACFSCGEDPSQVPEEPAQNSVKYLTWHRTASSRSSSSK 401
+ + F + A+ + + R+ SSRSSSS+
Sbjct: 416 RAQYTSSFN--------------------------DHSARKMLNPVALFRSGSSRSSSSR 449
Query: 402 NPLDAN--SRENVEDLTNNLFDNSCMNAGSHASTLDRLHAWERKLYDEVKASEIVRKEYD 459
+ ++ SRE+ + +++ D SCM +GSH +TLDRL AWE+KLYDEV++ E VR+ Y+
Sbjct: 450 FLITSSGGSRESGSESRSDVSDESCMISGSHQTTLDRLFAWEKKLYDEVRSGERVRRAYE 509
Query: 460 AKCKILRNLESKAEKTSTVDKTRAAVKDLHSRIRVAIHRIDSISKRIEELRDKELQPQLE 519
KC LRN + K + VDKTRA ++DL ++I+V+IH I+SISKRIE LRD+EL PQL
Sbjct: 510 KKCMQLRNQDVKGDDPLAVDKTRATIRDLDTQIKVSIHSIESISKRIETLRDQELLPQLL 569
Query: 520 ELIEGLNRMWEVMHECHKLQFQIMSAS--------YNNSHAR-------ITMHSELRRQI 564
EL+EGL RMW+VM E H++Q + + + + H + ++S+ Q
Sbjct: 570 ELVEGLTRMWKVMAESHQIQKRTLDEAKLLLAGTPVSKRHKKRQPPIMPEAINSQRLAQS 629
Query: 565 TAYLENELQFLSSSFTKWIEAQKSYLQAINGWIHKCVPLQQKSVKRKRRPQSELLIQYGP 624
LE +L+ + F WI +Q+SY++A++GW+ +C P+ L
Sbjct: 630 ALNLEAQLRNWRTCFEFWITSQRSYMKALSGWLLRCFRCDPD-------PEKVRLSSCLH 682
Query: 625 PIYATCDVWLKKLGTLPVKDVVDSIKSLAADTARFLPYQ---DKN-QGKEPHSHIGGESA 680
PIY C W + L +L K V+D ++ A+ Q D N G + G ES
Sbjct: 683 PIYRVCIQWSRLLNSLNEKPVLDKLEFFASGMGSIYARQVREDPNWSGSGSRRYSGSESM 742
Query: 681 DGLLRDDISEDWISGFDRFRANLIRFLGQLNSLSGSSV 718
+ ++ D ED + ++ ++ L S++ SS+
Sbjct: 743 ELVVADKGEEDVVMTAEKLAEVAVKVLCHGMSVAVSSL 780
>gb|AAD12027.1| hypothetical protein [Arabidopsis thaliana] gi|7487277|pir||T00529
hypothetical protein At2g19090 [imported] - Arabidopsis
thaliana
Length = 844
Score = 246 bits (627), Expect = 3e-63
Identities = 230/837 (27%), Positives = 374/837 (44%), Gaps = 146/837 (17%)
Query: 1 MGASSSKMEEDKALQLCRERKKFVRQALDGRCSLAAAHISYVQSLRIVGTALSKFTEPEP 60
MG S SK+++++A+Q+C++RK+F++QA++ R A+ HI+Y+ SLR V AL F
Sbjct: 1 MGCSHSKLDDEEAVQICKDRKRFIKQAIEHRTKFASGHIAYIHSLRKVSDALHDFILQGD 60
Query: 61 HSESSLYTNATPEALALTDKAMSQSSPSMSQQIDAGETHSFYTTPSPPSS------SKFH 114
++ + T ++ K M Q S+ ++GE S + PP S
Sbjct: 61 NNNEFVPT-LCQDSFVTPVKRMPQRRRRSSRS-NSGEFISISPSSIPPKMIQGRPRSNVK 118
Query: 115 ANHMKFGSFSSKKVEEKPP----VPVIATVTSS----------GIAQNGSRMSETEA--- 157
AN++ +VE++ P V + +SS G+ N + + T +
Sbjct: 119 ANYLMANRSRPVRVEQRSPETFRVETFSPPSSSNQYGESDGFFGMNMNMNTSASTSSSFW 178
Query: 158 ---------FEDSSVPDGTPQ---WDFF-GLFNPVDH-------QFSFQEPKGMPHHIGN 197
++P +PQ WDFF F+ +D+ + S G+ I
Sbjct: 179 NPLSSPEQRLSSHNIPPPSPQNSQWDFFWNPFSSLDYYGYNSYDRGSVDSRSGIDDEIRG 238
Query: 198 A----EEGGIPDLEDDEEK-----------------------------ASSCGSEGSRDS 224
EE GIPDLE+D+E S C E +
Sbjct: 239 LRRVREEEGIPDLEEDDEPHKPEPVVNVRFQNHNPNPKATEENRGKVDKSCCNEEVKVED 298
Query: 225 EDEFDEEPTAETLVQRFENLNRANNHVQENVL--PVKGDSVSEVELVSEK-GNFPNSSPS 281
DE ++E E + ++ N E + P + +EV E GN
Sbjct: 299 VDEDEDEDGDEDDDEFTDSGCETENEGDEKCVAPPTQEQRKAEVCRGGEATGNVVGVGKV 358
Query: 282 KKLPMAALLPPEANKSTEKENHSENKVTPKNFFSSMKDIEVLFNRASDSGKEVPRMLEAN 341
+++ + +A K+T + + P + +KD+E F D+ KEV +LEA
Sbjct: 359 QEMKNVVGVRDDA-KTTGFTVYVNRR--PTSMAEVIKDLEDQFTTICDAAKEVSGLLEAG 415
Query: 342 KFHFRPIFQGKKDGSLASFICKACFSCGEDPSQVPEEPAQNSVKYLTWHRTASSRSSSSK 401
+ + F GE + V ++ + + R+ SSRSSSS+
Sbjct: 416 RAQYTSSFNDHS---------------GEFMNNVAIFLSRKMLNPVALFRSGSSRSSSSR 460
Query: 402 NPLDAN--SRENVEDLTNNLFDNSCMNAGSHASTLDRLHAWERKLYDEVKAS-------- 451
+ ++ SRE+ + +++ D SCM +GSH +TLDRL AWE+KLYDEV+ S
Sbjct: 461 FLITSSGGSRESGSESRSDVSDESCMISGSHQTTLDRLFAWEKKLYDEVRVSNKRYKQLS 520
Query: 452 -EIVRKEYDAKCKILRNLESKAEKTSTVDKTRAAVKDLHSRIRVAIHRIDSISKRIEELR 510
E VR+ Y+ KC LRN + K + VDKTRA ++DL ++I+V+IH I+SISKRIE LR
Sbjct: 521 GERVRRAYEKKCMQLRNQDVKGDDPLAVDKTRATIRDLDTQIKVSIHSIESISKRIETLR 580
Query: 511 DKELQPQLEELIEG----------LNRMWEVMHECHKLQFQIMSAS--------YNNSHA 552
D+EL PQL EL+EG L RMW+VM E H++Q + + + + H
Sbjct: 581 DQELLPQLLELVEGLTNGNHDHNRLTRMWKVMAESHQIQKRTLDEAKLLLAGTPVSKRHK 640
Query: 553 R-------ITMHSELRRQITAYLENELQFLSSSFTKWIEAQKSYLQAINGWIHKCVPLQQ 605
+ ++S+ Q LE +L+ + F WI +Q+SY++A++GW+ +C
Sbjct: 641 KRQPPIMPEAINSQRLAQSALNLEAQLRNWRTCFEFWITSQRSYMKALSGWLLRCFRCDP 700
Query: 606 KSVKRKRRPQSELLIQYGPPIYATCDVWLKKLGTLPVKDVVDSIKSLAADTARFLPYQ-- 663
P+ L PIY C W + L +L K V+D ++ A+ Q
Sbjct: 701 D-------PEKVRLSSCLHPIYRVCIQWSRLLNSLNEKPVLDKLEFFASGMGSIYARQVR 753
Query: 664 -DKN-QGKEPHSHIGGESADGLLRDDISEDWISGFDRFRANLIRFLGQLNSLSGSSV 718
D N G + G ES + ++ D ED + ++ ++ L S++ SS+
Sbjct: 754 EDPNWSGSGSRRYSGSESMELVVADKGEEDVVMTAEKLAEVAVKVLCHGMSVAVSSL 810
>dbj|BAC79196.1| bzip-like transcription factor-like protein [Oryza sativa (japonica
cultivar-group)] gi|52076085|dbj|BAD46598.1| bzip-like
transcription factor-like [Oryza sativa (japonica
cultivar-group)]
Length = 722
Score = 207 bits (528), Expect = 8e-52
Identities = 177/650 (27%), Positives = 283/650 (43%), Gaps = 121/650 (18%)
Query: 16 LCRERKKFVRQALDGRCSLAAAHISYVQSLRIVGTALSKFTEPEPHSESSLYTNATPEAL 75
+CR+RK+ ++ A D R +LA AH +Y +LR V A+ F L T TP
Sbjct: 24 VCRDRKRLIKAAADRRFALAGAHAAYAAALRSVADAVDVFVARHTAPAPILITLPTPTGS 83
Query: 76 ALTDKAMSQSSPSMSQQIDAGETHSFYTTPSPPSSSKFHANHMKFGSFSSKKVEEKPPVP 135
A + + +++ GE ++ + + PP P
Sbjct: 84 PPASPAPAPAPAALASVAQGGEEEEGKAEVDDGGGARTPDLGCPYYYAPPETATATPPPP 143
Query: 136 VIATVTSSGIAQNGSRMSETEAFEDSSVPDGTPQWDFFGLFNPVDH----QFSFQEPKGM 191
A G WDFF F + S +E + +
Sbjct: 144 PPAASAVGG-------------------------WDFFNPFYGTEEVAAAAISDEEMRAV 178
Query: 192 PHHIGNAEEGGIPDLEDDEEKASSCGSEGSRDSEDEFDEEPTAETLVQRFENLNRANNHV 251
E GIP+LE+ EE+ EG++ + + P A
Sbjct: 179 ------REREGIPELEEAEEE----DDEGAKSAAAANAKTPKA----------------A 212
Query: 252 QENVLPVKGDSVSEVELVSEKGNFPNSSPSKKLPMAALLPPEANKSTEKENHSENKVTPK 311
+ ++ K + +V V+ N L +A P +
Sbjct: 213 ETSLGVTKQEEAKDVCEVASN----NGGRGGGLEVAVSQPG------------------R 250
Query: 312 NFFSSMKDIEVLFNRASDSGKEVPRMLEANKFHFRPIFQGKKDGSLASFICKACFSCGED 371
+++K+IE LF RA+++GKEV MLEA
Sbjct: 251 ELLAALKEIEELFARAAEAGKEVTAMLEA------------------------------- 279
Query: 372 PSQVPE--EPAQNSVKYLTWHRTASSRSSSSKNPLDANSRE----NVEDLTNNLFDN-SC 424
S+VPE E + + +TWHR+ SS SSS ++ L A+S + + +++FD+
Sbjct: 280 ASRVPELKENSSKIIHAITWHRSPSSVSSSYRSELGASSNSLSWTDKSETKSDIFDDYGG 339
Query: 425 MNAGSHASTLDRLHAWERKLYDEVKASEIVRKEYDAKCKILRNLESKAEKTSTVDKTRAA 484
M +GSH+ TL RL+AWE+KLY+EVKA + +R+ Y+ KC LRN ++K + +KTR
Sbjct: 340 MKSGSHSQTLGRLYAWEKKLYEEVKAIDQIRQTYEKKCVQLRNQDAKGSELRCAEKTRTT 399
Query: 485 VKDLHSRIRVAIHRIDSISKRIEELRDKELQPQLEELIEGLNRMWEVMHECHKLQFQIM- 543
V+DL++RI V++ +SIS RI++LRD+ELQPQL EL++GL R W++M + H+ Q QIM
Sbjct: 400 VRDLYTRIWVSLRAAESISDRIQKLRDEELQPQLVELLQGLTRTWKIMVDSHETQRQIMF 459
Query: 544 SASYNNSHARITMHSELRRQITAYLENELQFLSSSFTKWIEAQKSYLQAINGWIHKCVPL 603
+ A ++ +R T LE EL+ S F ++ AQK+Y++A++GW+ K +
Sbjct: 460 EVNSFTCPAYGKFCNDAQRHATLKLEAELRNWRSCFMIYVSAQKAYIEALDGWLSKFI-- 517
Query: 604 QQKSVKRKRRPQSELL--IQYGPPIYATCDVWLKKLGTLPVKDVVDSIKS 651
R R S + PP+ C W L K V ++++
Sbjct: 518 -LTDTIRYSRGISSIAPDRSSAPPLVVICHDWYTTLSKFQNKRVAFTMRN 566
>ref|NP_914241.1| bzip-like transcription factor-like [Oryza sativa (japonica
cultivar-group)] gi|20146230|dbj|BAB89012.1| bzip
transcription factor-like [Oryza sativa (japonica
cultivar-group)]
Length = 834
Score = 200 bits (508), Expect = 2e-49
Identities = 212/789 (26%), Positives = 338/789 (41%), Gaps = 133/789 (16%)
Query: 3 ASSSKMEEDKALQ-----LCRERKKFVRQALDGRCSLAAAHISYVQSLRIVGTALSKFTE 57
ASSS +E + Q LCRER +R A D R +LA+AH +Y +SL VG AL +F
Sbjct: 19 ASSSSLERPRRAQADPAALCRERAALIRAAADRRYALASAHAAYFRSLAAVGGALRRFAA 78
Query: 58 PE-----PHSESSLYTNATP------EALALTDKAMSQSSPSMSQQIDAGETHSF----- 101
P S SS P +A A ++ SPS S + +H+
Sbjct: 79 AALAPGTPPSGSSPVLTLPPSPAKPVDASAAAARSSLPPSPSSSSTVSP-LSHTLSDEDL 137
Query: 102 -------YTTPSPPSSSKFHANHMKFGSFSSKKVEEKPPVPVIATVTSSGIAQNGSR--- 151
+ T + SS+++H ++M+ P VP +G A G
Sbjct: 138 DAHGAAKHATAAAASSTRYHYHYMR----------NSPTVPTTVYEDPNGEASYGGYGGY 187
Query: 152 --------MSETEAFE--DSSVPDGTPQ---WDFFGLFNPVDH---QFSFQEPKGMPHHI 195
E A E +++ P T + WDFF F D + + +P +
Sbjct: 188 GYTYSYGPYGEVVAEERPETATPPPTAEVAAWDFFDPFTSYDQFIEDYKGHDGGNLPSNS 247
Query: 196 GNAEE----GGIPDLEDDEE-KASSCGSEGSRDSEDEF-DEEPTAETLVQRFENLNRANN 249
N E GIP+LED+ E +A+ ++ S+ S D+ + + +++
Sbjct: 248 PNYSELRRVEGIPELEDEAELEAAEAKAKASKPSTSVVADQGGKGKRPIS--SDVSSKGE 305
Query: 250 HVQENVLPVKGDSVS-EVELVSEKG------NFPNSSPSKKLPMAALLPPEA-------- 294
+L KG + E E S KG N ++S K + + P A
Sbjct: 306 ASDGKLLQRKGSGGNGEPENASLKGSGSGDNNGSSTSKKKGIAFDGIEQPIAAAQGEGGS 365
Query: 295 NKSTEKENHSENKVTP-----KNFFSSMKDIEVLFNRASDSGKEVPRMLEANKFHFRPIF 349
KS + S +P ++ +M +I+ F+ A + G+EV ++LE K
Sbjct: 366 GKSVQSTAVSSESFSPLHQGNRSVMEAMDEIKERFDEALNCGEEVSKLLEVGKV------ 419
Query: 350 QGKKDGSLASFICKACFSCGEDPSQVPEEPAQNSVKYLTWHRTASSRSSSSKNPLDANSR 409
S + + S DP + + K RT S ++S+S NP A R
Sbjct: 420 ---PPQSSTPRVLRYLSSRVMDPLSLTVPSSSCLPKPRRKSRTLSGKASTSSNPSVAGRR 476
Query: 410 ENVEDLTNNLFDNSCMNAGSHASTLDRLHAWERKLYDEVKASEIVRKEYDAKCKILRNLE 469
+ AGS +STL++L AWE+KLY E+K E +R Y+ K + L++L+
Sbjct: 477 NS---------------AGSLSSTLEKLCAWEKKLYQEIKDEEKLRILYEKKYRRLKSLD 521
Query: 470 SKAEKTSTVDKTRAAVKDLHSRIRVAIHRIDSISKRIEELRDKELQPQLEELIEGLNRMW 529
+ ++T+D TR +V++L SRI + I ++ S +I+ +RD+EL PQL +LI GL RMW
Sbjct: 522 ERGLDSTTIDATRLSVRNLQSRITINIRTANAFSSKIQNIRDEELYPQLVDLIIGLRRMW 581
Query: 530 EVMHECHKLQFQIMSASYNNSHARITM-HSELRRQITAYLENELQFLSSSFTKWIEAQKS 588
+ + CH+ Q + S + +T+ S T LE EL F KWI +Q+S
Sbjct: 582 KAVLLCHEKQLSAIQDSKMHLIKAVTISQSNAAAVATVELERELAKWYRCFNKWISSQRS 641
Query: 589 YLQAINGWIHKCVPLQQKSVKRKRRPQSELLIQYG----PPIYATCDVWLKKLGTLPVKD 644
Y +A+NGW+ K L + V+ + P G PP++ + WL+ + + +
Sbjct: 642 YAEALNGWLRKW--LTEPEVQEENTPDGAPPFSPGKLGAPPVFVISNDWLQVIEMVSKNE 699
Query: 645 VVDSIKSLAADTARFLPYQDKNQGKEPHSHIGGESADGLLRDDISEDWISGFDRFRANLI 704
V+ +I ++ + K Q KE H AD RD +++ R L
Sbjct: 700 VLKTIDQF----SKLVHEYKKTQEKE---HRQKRKADHASRD---------YNKRRKVLQ 743
Query: 705 RFLGQLNSL 713
R LG SL
Sbjct: 744 RELGLSTSL 752
>ref|NP_565405.2| expressed protein [Arabidopsis thaliana]
Length = 733
Score = 191 bits (486), Expect = 6e-47
Identities = 179/691 (25%), Positives = 288/691 (40%), Gaps = 111/691 (16%)
Query: 1 MGASSSKMEEDKALQLCRERKKFVRQALDGRCSLAAAHISYVQSLRIVGTALSKF-TEPE 59
MG S+SK+++ A+ LCR+R F+ A+ R +L+ AH+SY QSL+ + +L +F
Sbjct: 1 MGCSTSKLDDLPAVALCRDRCSFLEAAIHQRYALSEAHVSYTQSLKAISHSLHQFINHHH 60
Query: 60 PHSESSLYTNATPEALALTDKAMSQSSPSMSQQIDAGETHS------FYTTPSPPSSSKF 113
+++S A P+ + + S D HS S P S
Sbjct: 61 RYNDSDSPKKAKPKMDSGSGHLDFDSDSDSDDDDDIDSLHSSPLHHHLEDDDSNPKSY-L 119
Query: 114 HANHMKFGSFSSKKV-EEKPPVPVIATVTSSGIAQNGSRMSETEAFEDSSVPDGTPQ--- 169
H N+MK V E++P P S + + S +P P
Sbjct: 120 HMNYMKNSYMPPSLVYEQRPSSPQRVHFGESSSSSTSEYNPYLNSNYGSKLPPPPPSPPR 179
Query: 170 ---WDFFGLFNPVDHQFSFQEPKGMPHHIGNAEEGGIPDLEDDE---------------- 210
WDF +P D ++ P + +E G+PDLE+D+
Sbjct: 180 EKVWDFL---DPFDTYYTPYTPSRDTREL--RDELGVPDLEEDDVVVKEVHGKQKFVAAV 234
Query: 211 --------EKASSCGSEGSRDSEDEFDEEPTAETLVQRFENLNRANNHVQEN--VLPVKG 260
AS+ G G + P+ + E+ H+ E V G
Sbjct: 235 SVEEPLGNSGASTSGGGGGGGKASLYQTRPSVSVEKEEMEH----EVHIVEKKIVEDSGG 290
Query: 261 DSVSEVELVSEKGNFPNSSPSKKLPMAALLPPEANKSTEKENHSENKVTPKNFFSSMKDI 320
D V + + +G + +P A K+I
Sbjct: 291 DEVRKSKAAVARGG---GGVRRGVPEVA-----------------------------KEI 318
Query: 321 EVLFNRASDSGKEVPRMLEANKFHFRPIFQGKKDGSLASFICKACFSCGEDPSQVPEEPA 380
E F RA++SG E+ MLE K + G+K+ S K + PS V +
Sbjct: 319 EAQFLRAAESGNEIAVMLEVGKHPY-----GRKNVS-----SKKLYEGTPSPSVVSSAQS 368
Query: 381 QNSVKYLTWHRTASSRSSSSKNPLDANSRENVEDLTNNLFDNSCMNAGSHASTLDRLHAW 440
S K + SSS P A+ + + NL +STL +LH W
Sbjct: 369 STSKK------AKAEASSSVTAPTYADIEAELALKSRNL-----------SSTLHKLHLW 411
Query: 441 ERKLYDEVKASEIVRKEYDAKCKILRNLESKAEKTSTVDKTRAAVKDLHSRIRVAIHRID 500
E+KLYDEVKA E +R ++ K + L+ ++ + + VD TR V+ L ++IR+AI +D
Sbjct: 412 EKKLYDEVKAEEKMRVNHEKKLRKLKRMDERGAENQKVDSTRKLVRSLSTKIRIAIQVVD 471
Query: 501 SISKRIEELRDKELQPQLEELIEGLNRMWEVMHECHKLQFQIMSASYNNSHARITMH-SE 559
IS I ++RD+EL QL ELI+GL++MW+ M ECHK Q + + + R + +
Sbjct: 472 KISVTINKIRDEELWLQLNELIQGLSKMWKSMLECHKSQCEAIKEARGLGPIRASKNFGG 531
Query: 560 LRRQITAYLENELQFLSSSFTKWIEAQKSYLQAINGWIHKCVPLQQKSVKRKRRPQSELL 619
++T L EL F+ W+ AQK +++ +N W+ KC+ + + P S
Sbjct: 532 EHLEVTRTLGYELINWIVGFSSWVSAQKGFVRELNSWLMKCLFYEPEETPDGIVPFSPGR 591
Query: 620 IQYGPPIYATCDVWLKKLGTLPVKDVVDSIK 650
I P I+ C+ W + L + K+V+++I+
Sbjct: 592 IG-APMIFVICNQWEQALDRISEKEVIEAIR 621
>gb|AAV24778.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 872
Score = 185 bits (469), Expect = 6e-45
Identities = 200/797 (25%), Positives = 325/797 (40%), Gaps = 171/797 (21%)
Query: 1 MGASSSKMEEDKA---LQLCRERKKFVRQALDGRCSLAAAHISYVQSLRIVGTALSKF-- 55
MG SS+++ A LCRER+ +R A + R +LAAAH +Y ++L V AL++F
Sbjct: 1 MGCRSSRLDAADASPVAALCRERRDLLRAAAERRAALAAAHAAYFRALPRVADALARFAE 60
Query: 56 -----------------TEPEPH-----SESSLYTNATPEALALTDKAMSQSSPSMSQQI 93
+EP+ H S S+L+T++ L S S P+ +
Sbjct: 61 QHHAATPPGSPVLTLPPSEPDEHKKRSASSSTLHTDSGHSHLHFHTDGGSDSEPNSADDD 120
Query: 94 DA-------------------GETHSFYTTPSPPSSSKFHAN------------------ 116
A E P P +SS+
Sbjct: 121 CACGAAAAAAGHGVRGEISPPAEELQERRIPEPGASSRLQMPWEYAPYDPYPSSFPNVTF 180
Query: 117 ----HMKFGSFSSKKVEEKPPV--PVIATVTSSG-------------IAQNGSRMSETEA 157
+MK S + V ++P A V+ +G + +G R++E A
Sbjct: 181 PNYYYMKASSTPANTVYQEPYGYGNFAANVSYNGYDYGYSNPMYGIPVPPDGERLAEDRA 240
Query: 158 FEDSS-----------VPDGTPQWDFFGLFNPVDHQFSFQEPK-------GMPHHIGNAE 199
E ++ +P+ +P WDFF F+ D+ + K P+ E
Sbjct: 241 REAAAAAAPAPPPPMPMPEVSP-WDFFNPFDSYDYNQQLPQYKDANGSFTSSPNSSEVRE 299
Query: 200 EGGIPDLEDDEEKASSCGSEGSRDSEDEFDEEPTAETLVQRFENLN-RANNHVQEN---- 254
GIP+LE++ E+ S S +R + E TA T + + +N +A E+
Sbjct: 300 REGIPELEEETEQESMRESIKARKAV-----ESTASTRIDNADAVNAKAKTASMEHKECE 354
Query: 255 --------VLPVKGDSVSEVELVSEKGNFPNSSPSKKLPMAALLPPEANKSTEKENHSE- 305
VL +SV E GN + A P K E HS
Sbjct: 355 IESVGSASVLDSGEESVCSCECDHADGN---AGAGAATAAPAGDDPRMVKKVASEEHSSM 411
Query: 306 ---NKVTPKNFFSS-----MKDIEVLFNRASDSGKEVPRMLEANKFHFRP------IFQG 351
V PKNF + + +I+ FN G +V R+LE +RP +
Sbjct: 412 VVAEDVLPKNFGTRDVADVVNEIKEQFNSVVACGDDVARILEVGSMRYRPRRRIVRLVFS 471
Query: 352 KKDGSLASFICKACFSCGEDPSQVPEEPAQNSVKYLTWHRTASSRSSSSKNPLDANSREN 411
+ G+ A S V E P +N + ++ S+S +N + +
Sbjct: 472 RLMGAFALLF-----------SSVSEPPVENLEQ--------TALSASGRNHNSSQRIGS 512
Query: 412 VEDLTNNLFDNSCMNAGSHASTLDRLHAWERKLYDEVKASEIVRKEYDAKCKILRNLESK 471
D+ N +S +DRL+ WE++L+ E+ E +R YD + K L+ L+
Sbjct: 513 ASDIEFNTL----------SSVMDRLYVWEKRLHKEIMEEEKLRITYDKQWKRLKELDDN 562
Query: 472 AEKTSTVDKTRAAVKDLHSRIRVAIHRIDSISKRIEELRDKELQPQLEELIEGLNRMWEV 531
+ +D TRA++ L +RI + I IS+RI LRD EL P L +LI+GL RMW+
Sbjct: 563 GAEPYKIDSTRASISTLLTRINITIRSAKVISRRIHILRDDELHPHLVKLIQGLVRMWKF 622
Query: 532 MHECHKLQFQIMSASYNNSHARITMHS-ELRRQITAYLENELQFLSSSFTKWIEAQKSYL 590
+ ECH+ QF + + SH I + E +IT LE EL S F+ WI +QK+Y+
Sbjct: 623 ILECHRKQFHAILET--KSHVLIPKNGPERNSKITLELEMELLNWCSCFSNWILSQKAYI 680
Query: 591 QAINGWIHKCVPLQQKSVKRKRRPQSELLIQYGPPIYATCDVWLKKLGTLPVKDVVDSIK 650
+ +NGW+ K +P +++ P S + P ++ T + W + + +P V+ +++
Sbjct: 681 ETLNGWLVKWLPEEKEETPDGIAPFSPGRLG-APAVFITANDWCQSMKRIPEGTVIGAME 739
Query: 651 SLAADTARFLPYQDKNQ 667
+ A + QD+ Q
Sbjct: 740 AFAVNVHMLRERQDEEQ 756
>gb|AAN86155.1| unknown protein [Arabidopsis thaliana] gi|15223973|ref|NP_177874.1|
expressed protein [Arabidopsis thaliana]
gi|12323381|gb|AAG51662.1| unknown protein; 32274-35458
[Arabidopsis thaliana] gi|25406508|pir||D96804 unknown
protein T5M16.9 [imported] - Arabidopsis thaliana
Length = 879
Score = 181 bits (458), Expect = 1e-43
Identities = 167/658 (25%), Positives = 283/658 (42%), Gaps = 69/658 (10%)
Query: 87 PSMSQQIDAGETHSFYTTPSPPSSSKF---HANHMKFGSFSSKKVEEKPPVPVIATVTSS 143
P+ + + G +Y S P S F NH +VEE P + S
Sbjct: 187 PNYTGENPYGNRGMYYMKKSAPQSRPFIFQPENH---------RVEENAQWP-----SDS 232
Query: 144 GIAQNGSRMSETEAFEDSSVPDGTPQWDFFGLFNPVDHQFSFQEPKGMPHHIGNAEEGGI 203
G G + S P WDF +F+ D+ + G + +G A
Sbjct: 233 GFRNTGVQRRSPSPLPPPSPPT-VSTWDFLNVFDTYDYSNARSRASGY-YPMGMASISSS 290
Query: 204 PDLEDDEEKASSCGSEGSRDSEDEFDEEPTAETLVQ-RFENLNRANNHVQE---NVLPVK 259
PD ++ E+ EG + E+ ++E + + + L + H E NV P +
Sbjct: 291 PDSKEVRER------EGIPELEEVTEQEVIKQVYRRPKRPGLEKVKEHRDEHKHNVFPER 344
Query: 260 GDSVSEVELVSEKGNFPNSSPSKKLPMAALLPPEAN--KSTEKENHSENKVTPKNFFSSM 317
+ EV + + S + + + E + E ++ S + ++ S
Sbjct: 345 NINKREVPMPEQVTESSLDSETISSFSGSDVESEFHYVNGGEGKSSSISSISHGAGTKSS 404
Query: 318 KDIEVLFNRASDSGKEVPRMLEANKFHFRPIFQGKKDGSLASFICKA------------- 364
+++E + R E+ + F + K SL+S A
Sbjct: 405 REVEEQYGRKKGVSFELEETTSTSSFDV----ESSKISSLSSLSIHATRDLREVVKEIKS 460
Query: 365 ----CFSCGEDPS------QVPEEPAQNSVKYLT----WHRTASSRSSSSKNPLDANSRE 410
SCG++ + ++P + N VK + + S+RSS S+ L
Sbjct: 461 EFEIASSCGKEVALLLEVGKLPYQHKNNGVKVILSRIMYLVAPSTRSSHSQPRLSIRLTS 520
Query: 411 NVEDLTNNLFDNSCMNAGSH---ASTLDRLHAWERKLYDEVKASEIVRKEYDAKCKILRN 467
+ + ++ +N G + +STL++L+AWE+KLY EVK E +R Y+ KC+ L+
Sbjct: 521 RTRKMAKS-YNGQDVNGGFNGNLSSTLEKLYAWEKKLYKEVKDEEKLRAIYEEKCRRLKK 579
Query: 468 LESKAEKTSTVDKTRAAVKDLHSRIRVAIHRIDSISKRIEELRDKELQPQLEELIEGLNR 527
++S ++ +D TRAA++ L ++I V I +DSIS RI +LRD+ELQPQL +LI GL R
Sbjct: 580 MDSHGAESIKIDATRAAIRKLLTKIDVCIRSVDSISSRIHKLRDEELQPQLIQLIHGLIR 639
Query: 528 MWEVMHECHKLQFQ-IMSASYNNSHARITMHSELRRQITAYLENELQFLSSSFTKWIEAQ 586
MW M CH+ QFQ I + + A T+ ++ LE EL+ SF W+ Q
Sbjct: 640 MWRSMLRCHQKQFQAIRESKVRSLKANTTLQNDSGSTAILDLEIELREWCISFNNWVNTQ 699
Query: 587 KSYLQAINGWIHKCVPLQQKSVKRKRRPQSELLIQYGPPIYATCDVWLKKLGTLPVKDVV 646
KSY+Q ++GW+ KC+ + ++ P S I PPI+ C W + + + ++V
Sbjct: 700 KSYVQFLSGWLTKCLHYEPEATDDGIAPFSPSQIG-APPIFIICKDWQEAMCRISGENVT 758
Query: 647 DSIKSLAADTARFLPYQDKNQG-KEPHSHIGGESADGLLRDDISEDWISGFDRFRANL 703
++++ A+ Q++ Q K ES ++ SE IS D + +L
Sbjct: 759 NAMQGFASSLHELWEKQEEEQRVKAQSEQRDAESERSVVSKGRSESGISALDDLKVDL 816
Score = 59.3 bits (142), Expect = 5e-07
Identities = 40/140 (28%), Positives = 71/140 (50%), Gaps = 4/140 (2%)
Query: 1 MGASSSKMEEDKALQLCRERKKFVRQALDGRCSLAAAHISYVQSLRIVGTALSKFTEPEP 60
MG SK++ + LCRERK+ ++ A R +LA AH++Y QSL VG A+ +F + E
Sbjct: 1 MGCGGSKVDNQPIVILCRERKELLKAASYHRSALAVAHLTYFQSLSDVGEAIQRFVDDEV 60
Query: 61 HSESSLYTNATPEALALTDKAMSQSSPSMSQQIDAGETHSF-YTTPSPPSSSKFHANHMK 119
+ S ++++P++ LT + P+ ++I T S ++ + H+
Sbjct: 61 VAGFS--SSSSPDSPVLT-LPSDEGKPTKHKRISPSSTTSISHSVIEEEDTDDDSHLHLS 117
Query: 120 FGSFSSKKVEEKPPVPVIAT 139
GS S +V K + + +T
Sbjct: 118 SGSESESEVGSKDHIQITST 137
>gb|AAL87333.1| unknown protein [Arabidopsis thaliana]
Length = 666
Score = 179 bits (453), Expect = 4e-43
Identities = 154/600 (25%), Positives = 269/600 (44%), Gaps = 55/600 (9%)
Query: 146 AQNGSRMSETEAFEDSSVPDGTPQ----WDFFGLFNPVDHQFSFQEPKGMPHHIGNAEEG 201
+ +G R + + S +P +P WDF +F+ D+ + G + +G A
Sbjct: 17 SDSGFRNTGVQRRSPSPLPPPSPPTVSTWDFLNVFDTYDYSNARSRASGY-YPMGMASIS 75
Query: 202 GIPDLEDDEEKASSCGSEGSRDSEDEFDEEPTAETLVQ-RFENLNRANNHVQE---NVLP 257
PD ++ E+ EG + E+ ++E + + + L + H E NV P
Sbjct: 76 SSPDSKEVRER------EGIPELEEVTEQEVIKQVYRRPKRPGLEKVKEHRDEHKHNVFP 129
Query: 258 VKGDSVSEVELVSEKGNFPNSSPSKKLPMAALLPPEAN--KSTEKENHSENKVTPKNFFS 315
+ + EV + + S + + + E + E ++ S + ++
Sbjct: 130 ERNINKREVPMPEQVTESSLDSETISSFSGSDVESEFHYVNGGEGKSSSISSISHGAGTK 189
Query: 316 SMKDIEVLFNRASDSGKEVPRMLEANKFHFRPIFQGKKDGSLASFICKA----------- 364
S +++E + R E+ + F + K SL+S A
Sbjct: 190 SSREVEEQYGRKKGVSFELEETTSTSSFDV----ESSKISSLSSLSIHATRDLREVVKEI 245
Query: 365 ------CFSCGEDPS------QVPEEPAQNSVKYLT----WHRTASSRSSSSKNPLDANS 408
SCG++ + ++P + N VK + + S+RSS S+ L
Sbjct: 246 KSEFEIASSCGKEVALLLEVGKLPYQHKNNGVKVILSRIMYLVAPSTRSSHSQPRLSIRL 305
Query: 409 RENVEDLTNNLFDNSCMNAGSH---ASTLDRLHAWERKLYDEVKASEIVRKEYDAKCKIL 465
+ + ++ +N G + +STL++L+AWE+KLY EVK E +R Y+ KC+ L
Sbjct: 306 TSRTRKMAKS-YNGQDVNGGFNGNLSSTLEKLYAWEKKLYKEVKDEEKLRAIYEEKCRRL 364
Query: 466 RNLESKAEKTSTVDKTRAAVKDLHSRIRVAIHRIDSISKRIEELRDKELQPQLEELIEGL 525
+ ++S ++ +D TRAA++ L ++I V I +DSIS RI +LRD+ELQPQL +LI GL
Sbjct: 365 KKMDSHGAESIKIDATRAAIRKLLTKIDVCIRSVDSISSRIHKLRDEELQPQLIQLIHGL 424
Query: 526 NRMWEVMHECHKLQFQ-IMSASYNNSHARITMHSELRRQITAYLENELQFLSSSFTKWIE 584
RMW M CH+ QFQ I + + A T+ ++ LE EL+ SF W+
Sbjct: 425 IRMWRSMLRCHQKQFQAIRESKVRSLKANTTLQNDSGSTAILDLEIELREWCISFNNWVN 484
Query: 585 AQKSYLQAINGWIHKCVPLQQKSVKRKRRPQSELLIQYGPPIYATCDVWLKKLGTLPVKD 644
QKSY+Q ++GW+ KC+ + ++ P S I PPI+ C W + + + ++
Sbjct: 485 TQKSYVQFLSGWLTKCLHYEPEATDDGIAPFSPSQIG-APPIFIICKDWQEAMCRISGEN 543
Query: 645 VVDSIKSLAADTARFLPYQDKNQG-KEPHSHIGGESADGLLRDDISEDWISGFDRFRANL 703
V ++++ A+ Q++ Q K ES ++ SE IS D + +L
Sbjct: 544 VTNAMQGFASSLHELWEKQEEEQRVKAQSEQRDAESERSVVSKGRSESGISALDDLKVDL 603
>ref|NP_173593.1| expressed protein [Arabidopsis thaliana] gi|5263327|gb|AAD41429.1|
EST gb|T20649 comes from this gene. [Arabidopsis
thaliana] gi|25518309|pir||A86351 hypothetical protein
F8K7.18 - Arabidopsis thaliana
Length = 953
Score = 177 bits (448), Expect = 2e-42
Identities = 118/362 (32%), Positives = 183/362 (49%), Gaps = 48/362 (13%)
Query: 317 MKDIEVLFNRASDSGKEVPRMLEANKFHFRPIFQGKKDGSLASFICKACFSCGEDPSQVP 376
+K+I+ F AS GKEV +LE +K + Q K G K FS
Sbjct: 503 VKEIKSEFEVASSHGKEVAVLLEVSKLPY----QQKSSG------LKVIFS--------- 543
Query: 377 EEPAQNSVKYLTWHRTASSRSSSS----------KNPLDANSRENVEDLTNNLFDNSCMN 426
+ YL T SSRS K N ++ E LT NL
Sbjct: 544 ------RIMYLVAPSTVSSRSQPQPSIRLTSRILKIAKSYNGQDVREGLTGNL------- 590
Query: 427 AGSHASTLDRLHAWERKLYDEVKASEIVRKEYDAKCKILRNLESKAEKTSTVDKTRAAVK 486
++TL++L+AWE+KLY EVK E +R Y+ KC+ L+ L+S ++S +D TRAA++
Sbjct: 591 ----SATLEQLYAWEKKLYKEVKDEEKLRVVYEEKCRTLKKLDSLGAESSKIDTTRAAIR 646
Query: 487 DLHSRIRVAIHRIDSISKRIEELRDKELQPQLEELIEGLNRMWEVMHECHKLQFQ-IMSA 545
L +++ V I +DSIS RI +LRD+ELQPQL +LI GL RMW M +CH+ QFQ IM +
Sbjct: 647 KLLTKLDVCIRSVDSISSRIHKLRDEELQPQLTQLIHGLIRMWRSMLKCHQKQFQAIMES 706
Query: 546 SYNNSHARITMHSELRRQITAYLENELQFLSSSFTKWIEAQKSYLQAINGWIHKCVPLQQ 605
+ A + + + LE EL+ SF W+ QKSY++++NGW+ +C+ +
Sbjct: 707 KVRSLRANTGLQRDSGLKAILDLEMELREWCISFNDWVNTQKSYVESLNGWLSRCLHYEP 766
Query: 606 KSVKRKRRPQSELLIQYGPPIYATCDVWLKKLGTLPVKDVVDSIKSLAADTARFLPYQDK 665
+S + P S + P ++ C W + + + ++V ++++ A+ QD+
Sbjct: 767 ESTEDGIAPFSPSRVG-APQVFVICKDWQEAMARISGENVSNAMQGFASSLHELWERQDE 825
Query: 666 NQ 667
Q
Sbjct: 826 EQ 827
Score = 63.5 bits (153), Expect = 2e-08
Identities = 38/111 (34%), Positives = 56/111 (50%), Gaps = 13/111 (11%)
Query: 1 MGASSSKMEEDKALQLCRERKKFVRQALDGRCSLAAAHISYVQSLRIVGTALSKFTEPEP 60
MG SK+++ + LCRERK+ ++ A RC+LAAAH+SY QSL VG ++ +F + E
Sbjct: 1 MGCGGSKVDDQPLVILCRERKQLIKAASHHRCALAAAHLSYFQSLCDVGDSIKRFVDEE- 59
Query: 61 HSESSLYTNATPEALALTDKAMSQSSPSMSQQIDAGETHSFYTTPSPPSSS 111
L T + S SP ++ D G+ H + S S S
Sbjct: 60 ------------LVLVGTSSSSSPDSPVLTLPSDEGKPHKHKISSSSTSVS 98
>emb|CAB80241.1| putative protein [Arabidopsis thaliana] gi|3080426|emb|CAA18745.1|
putative protein [Arabidopsis thaliana]
gi|15236929|ref|NP_195250.1| expressed protein
[Arabidopsis thaliana] gi|7486021|pir||T06133
hypothetical protein F23E12.200 - Arabidopsis thaliana
Length = 828
Score = 168 bits (426), Expect = 5e-40
Identities = 156/592 (26%), Positives = 249/592 (41%), Gaps = 118/592 (19%)
Query: 84 QSSPSMSQQIDAGETHSFYTTPSPPSSSKFHANHMKFG---------SFSSKKVEEKPPV 134
+ P+ Q++ GE+ S Y P PP +S F ++ G S S+ KPP
Sbjct: 210 EQRPTSPQRVYIGESSSSY--PYPPQNSYFGYSNPVPGPGPGYYGSSSASTTAAATKPPP 267
Query: 135 PVIATVTSSGIAQNGSRMSETEAFEDSSVPDGTPQWDFFGLFNPVDHQFSFQEPKGMPHH 194
P + S+G WDF NP D + P
Sbjct: 268 PPPSPPRSNG-------------------------WDFL---NPFDTYYPPYTPSRDSRE 299
Query: 195 IGNAEEGGIPDLEDDE---EKASSCGSEGSRDSEDEFDEEPTAETLVQRFENLNRANNHV 251
+ EE GIPDLEDD+ E + + P A +++
Sbjct: 300 L--REEEGIPDLEDDDSHYEVVKEVYGKPKFAAGGGHQPNPAAVHMMRE---------ES 348
Query: 252 QENVLPVKGDSVS---EVELVSEKGNFPNSSPSKKLPMAALLPPEANKSTEKENHSENKV 308
L G S S +V S + P+ S K+ + E ++E S
Sbjct: 349 PSPPLDKSGASTSGGGDVGDASAYQSRPSVSVEKEGMEYEVHVVEKKVVEDEERRSNATA 408
Query: 309 T--------PKNFFSSMKDIEVLFNRASDSGKEVPRMLEANKFHFRPIFQGKKDGSLASF 360
T P+ K+IE F +A++SG E+ ++LE K + G+K
Sbjct: 409 TRGGGGGGGPRAVPEVAKEIENQFVKAAESGSEIAKLLEVGKHPY-----GRK------- 456
Query: 361 ICKACFSCGEDPSQVPEEPAQNSVKYLTWHRTASSRSSSSKNPLDANSRENVEDLTNNLF 420
H T+SS +++ P A+ E + + NL
Sbjct: 457 -----------------------------HGTSSSAAAAVVPPTYADIEEELASRSRNL- 486
Query: 421 DNSCMNAGSHASTLDRLHAWERKLYDEVKASEIVRKEYDAKCKILRNLESKAEKTSTVDK 480
+STL +LH WE+KLY EVKA E +R ++ K + L+ L+ + + VDK
Sbjct: 487 ----------SSTLHKLHLWEKKLYHEVKAEEKLRLAHEKKLRKLKRLDQRGAEAIKVDK 536
Query: 481 TRAAVKDLHSRIRVAIHRIDSISKRIEELRDKELQPQLEELIEGLNRMWEVMHECHKLQF 540
TR V+D+ ++IR+AI +D IS I ++RD++L PQL LI+GL RMW+ M ECH+ Q
Sbjct: 537 TRKLVRDMSTKIRIAIQVVDKISVTINKIRDEDLWPQLNALIQGLTRMWKTMLECHQSQC 596
Query: 541 QIMSASYNNSHARITMH-SELRRQITAYLENELQFLSSSFTKWIEAQKSYLQAINGWIHK 599
Q + + R + + + T+ L +EL F+ W+ AQK Y++ +N W+ K
Sbjct: 597 QAIREAQGLGPIRASKKLGDEHLEATSLLGHELINWILGFSSWVSAQKGYVKELNKWLMK 656
Query: 600 CVPLQQKSVKRKRRPQSELLIQYGPPIYATCDVWLKKLGTLPVKDVVDSIKS 651
C+ + + P S I PPI+ C+ W + L + K+V+++++S
Sbjct: 657 CLLYEPEETPDGIVPFSPGRIG-APPIFVICNQWSQALDRISEKEVIEAMRS 707
Score = 50.4 bits (119), Expect = 2e-04
Identities = 24/55 (43%), Positives = 37/55 (66%)
Query: 1 MGASSSKMEEDKALQLCRERKKFVRQALDGRCSLAAAHISYVQSLRIVGTALSKF 55
MG +SSK+++ A+ LCRER F+ A+ R +LA +H++Y SLR +G +L F
Sbjct: 1 MGCTSSKLDDLPAVALCRERCAFLEAAIHQRYALAESHVAYTHSLREIGHSLHLF 55
>gb|AAH23742.1| Unknown (protein for MGC:38531) [Mus musculus]
gi|23271726|gb|AAH23708.1| Unknown (protein for
MGC:38398) [Mus musculus]
Length = 642
Score = 165 bits (417), Expect = 6e-39
Identities = 139/473 (29%), Positives = 209/473 (43%), Gaps = 55/473 (11%)
Query: 193 HHIGNAEEGGIPDLEDDEEKASSCGSEGSRDSEDEFDEEPTAETLVQRFENLNRANNHVQ 252
H +A IP ++EE+ D E+E +EE + ++ + V+
Sbjct: 88 HSSNSAPPPQIPATSEEEEE----------DDEEEEEEERESGVCDHFYDEAEGGFSSVE 137
Query: 253 ENVLPVKGDSVSEVELVSEKGNFPNSSPSKKLPMAALLPPEANKSTEKENHSENKVTPKN 312
E K +F N KK+ + N+ + E N +
Sbjct: 138 EEC----------------KWDFFNLLDEKKVKEEREFKGKLNEIGKDEKMVNNN--GRE 179
Query: 313 FFSSMKDIEVLFNRASDSGKEVPRMLEANKFHFRPIFQGKKDG-------SLASFICKAC 365
++KD+E F RA +SG EV +MLE K P F+ KD S S CK+
Sbjct: 180 LLEALKDVEDHFLRACESGMEVSKMLEVEKIPSHPTFEDIKDNASRKRSVSSRSSSCKSL 239
Query: 366 FSCGEDPSQVPEEPAQNSVKYLTWHRTASSRSSSSKNPLDANSRENVEDLTNNLFDNSCM 425
S + TW + + R S D +S E+ + L +S M
Sbjct: 240 LSSSSRSTSS------------TWAESGADRRSLLSCSSDTSSTWT-ENKSEALDGHSGM 286
Query: 426 NAGSHASTLDRLHAWERKLYDEVKASEIVRKEYDAKCKILRNLESKAEKTSTVDKTRAAV 485
+GSH TL RL+AWE+KLY EVKA + RK Y KC L+ + + K + V
Sbjct: 287 VSGSHFFTLGRLYAWEKKLYHEVKAGQQTRKTYKQKCSQLQQKDVDGDDLCP-SKASSEV 345
Query: 486 KDLHSRIRVAIHRIDSISKRIEELRDKELQPQLEELIEGLNRMWEVMHECHKLQFQIMSA 545
DL+ + V++ R +SISK+IE+LRD+ELQPQL EL+ GL R W M E H +Q +IM
Sbjct: 346 TDLYHVMLVSLQRAESISKQIEKLRDEELQPQLFELLHGLLRTWNSMLETHDIQRKIMRE 405
Query: 546 SYNNS---HARITMHSELRRQITAYLENELQFLSSSFTKWIEAQKSYLQAINGWIHKCVP 602
+ S H + S+ R T LE +Q F+ ++ AQK+Y +++ W+ K +
Sbjct: 406 INSFSCPEHGKFC--SDSHRLATIQLEAVIQDWHYCFSDYVSAQKTYAKSLLSWLSKFID 463
Query: 603 LQQKSVKRKRRPQSELLIQYGPPIYATCDVWLKKLGTLPVKDVVDSIKSLAAD 655
+ + + GP + TC W + LP K V ++KSLA D
Sbjct: 464 AEAEYHYTSSFSLPPIRFN-GPTLIVTCQRWSTSMERLPDKSVKYAMKSLAKD 515
Score = 43.9 bits (102), Expect = 0.020
Identities = 34/131 (25%), Positives = 56/131 (41%), Gaps = 8/131 (6%)
Query: 9 EEDKALQLCRERKKFVRQALDGRCSLAAAHISYVQSLRIVGTALSKFTEPEPHSESSLY- 67
E + + LCRERK+ V++AL R A +H Y QSL + +A+S F S S +
Sbjct: 26 ETNDVVSLCRERKRLVKKALKHRQVFADSHFRYTQSLSSLSSAISVFVARYSSSPSHFFI 85
Query: 68 ------TNATPEALALTDKAMSQSSPSMSQQIDAGETHSFYTTPSPP-SSSKFHANHMKF 120
+ P+ A +++ ++ ++G FY SS + F
Sbjct: 86 TFHSSNSAPPPQIPATSEEEEEDDEEEEEEERESGVCDHFYDEAEGGFSSVEEECKWDFF 145
Query: 121 GSFSSKKVEEK 131
KKV+E+
Sbjct: 146 NLLDEKKVKEE 156
>dbj|BAD46467.1| bzip-related transcription factor -like [Oryza sativa (japonica
cultivar-group)]
Length = 912
Score = 147 bits (372), Expect = 1e-33
Identities = 141/568 (24%), Positives = 234/568 (40%), Gaps = 103/568 (18%)
Query: 107 PPSSSKFHANHMKFGSFSSKKVEEKPPVPVIATVTSSGIAQNGSRMSETEAFEDSSVPDG 166
PP S + + G + PP P TS G + S P
Sbjct: 308 PPQGSSSYNQYAYGGYYGGAS----PPPPADIPSTSRGEVTPPAPPS----------PPR 353
Query: 167 TPQWDFFGLFNPVDHQFSFQEPKGMPHHIGNA-----EEGGIPDLED-----------DE 210
WDF F + + + + EE GIPDLED DE
Sbjct: 354 VSTWDFLNPFETYESYYEQPTAAQASYTPSRSSKDVREEEGIPDLEDEDMEVVKEAYGDE 413
Query: 211 EKASSCGSEGSRDSEDEFDEEPTAETLVQ--RFENLNRANNHVQENVLPVKGDSVSEVEL 268
+ A++ S + +++E T + L + + + ++ + +V V+ V E
Sbjct: 414 KHAANGYSGKGKMAKEEGGRSSTGDELPHESKLSEASSSGSNQEHDVHVVEKSVVGE--- 470
Query: 269 VSEKGNFPNSSPSKKLPMAALLPPEANKSTEKENHSENKVTPKNFFSSMKDIEVLFNRAS 328
S P + + A LPP ++ T ++ + +I F RAS
Sbjct: 471 -----QVQRSEPRQHV---AGLPPIGSEKTYFDDAEV-----------VLEIRTQFERAS 511
Query: 329 DSGKEVPRMLEANKFHFRPIFQGKKDGSLASFICKACFSCGEDPSQVPEEPAQNSVKYLT 388
S EV +MLE K + P G F A CG +P
Sbjct: 512 KSAIEVSKMLEVGKMPYYPKSSG--------FKVSAMMICG-----IP------------ 546
Query: 389 WHRTASSRSSSSKNPLDANSRENVEDLTNNLFDNSCMNAGSHASTLDRLHAWERKLYDEV 448
+E+ ++ M G+ +STL +L+ WE+KL +EV
Sbjct: 547 ----------------------TMEEEFLRFEEDKAMGCGNLSSTLQKLYMWEKKLLEEV 584
Query: 449 KASEIVRKEYDAKCKILRNLESKAEKTSTVDKTRAAVKDLHSRIRVAIHRIDSISKRIEE 508
KA E +R YD + + L+ L+ K + ++ T +++ L ++I +AI +++IS +I +
Sbjct: 585 KAEEKMRALYDRQREELKILDEKGAEADKLEATERSIRKLSTKISIAIQVVNTISDKISK 644
Query: 509 LRDKELQPQLEELIEGLNRMWEVMHECHKLQFQIMSASYN-NSHARITMHSELRRQITAY 567
LRD+EL PQ ELI+GL RMW M ECH++Q +S + N +S E +
Sbjct: 645 LRDEELWPQTCELIQGLMRMWSTMLECHQIQLHAISHAKNIDSMINGAKFGEAHMDLIKR 704
Query: 568 LENELQFLSSSFTKWIEAQKSYLQAINGWIHKCVPLQQKSVKRKRRPQSELLIQYGPPIY 627
LE + +SF W+ AQKSY+ +N W+ K V + + P S + PPI+
Sbjct: 705 LELQHLDWIASFASWVNAQKSYVGTLNDWLRKGVTYEPEVTDDGVPPFSPGRLG-APPIF 763
Query: 628 ATCDVWLKKLGTLPVKDVVDSIKSLAAD 655
+ W +G + K+VV+++++ A++
Sbjct: 764 VIYNNWAVGVGRISEKEVVEAMQAFASN 791
Score = 41.6 bits (96), Expect = 0.100
Identities = 30/103 (29%), Positives = 47/103 (45%), Gaps = 17/103 (16%)
Query: 8 MEEDKALQLCRERKKFVRQALDGRCSLAAAHISYVQSLRIVGTALSKFTE-----PEPHS 62
MEE+ A++ CRER + + A+ R +LA H +Y +SL VG L F P P
Sbjct: 1 MEEEYAVRHCRERSELLALAIRHRYALADTHRAYAESLAAVGAVLHDFLRGVQSLPPPPL 60
Query: 63 ESSLYTNA------------TPEALALTDKAMSQSSPSMSQQI 93
E +L A P A+ + +Q P +++Q+
Sbjct: 61 EPTLRLPAHRKGDNLPTASPVPANPAIASSSAAQPLPPVAKQV 103
>gb|AAN18073.1| At4g35240/F23E12_200 [Arabidopsis thaliana]
gi|15724286|gb|AAL06536.1| AT4g35240/F23E12_200
[Arabidopsis thaliana]
Length = 399
Score = 144 bits (363), Expect = 1e-32
Identities = 83/259 (32%), Positives = 142/259 (54%), Gaps = 2/259 (0%)
Query: 393 ASSRSSSSKNPLDANSRENVEDLTNNLFDNSCMNAGSHASTLDRLHAWERKLYDEVKASE 452
+S++SS+SK S ++ + + + +STL +LH WE+KLYDEVKA E
Sbjct: 30 SSAQSSTSKKAKAEASSSVTAPTYADIEAELALKSRNLSSTLHKLHLWEKKLYDEVKAEE 89
Query: 453 IVRKEYDAKCKILRNLESKAEKTSTVDKTRAAVKDLHSRIRVAIHRIDSISKRIEELRDK 512
+R ++ K + L+ ++ + + VD TR V+ L ++IR+AI +D IS I ++RD+
Sbjct: 90 KMRVNHEKKLRKLKRMDERGAENQKVDSTRKLVRSLSTKIRIAIQVVDKISVTINKIRDE 149
Query: 513 ELQPQLEELIEGLNRMWEVMHECHKLQFQIMSASYNNSHARITMH-SELRRQITAYLENE 571
EL QL ELI+GL++MW+ M ECHK Q + + + R + + ++T L E
Sbjct: 150 ELWLQLNELIQGLSKMWKSMLECHKSQCEAIKEARGLGPIRASKNFGGEHLEVTRTLGYE 209
Query: 572 LQFLSSSFTKWIEAQKSYLQAINGWIHKCVPLQQKSVKRKRRPQSELLIQYGPPIYATCD 631
L F+ W+ AQK +++ +N W+ KC+ + + P S I P I+ C+
Sbjct: 210 LINWIVGFSSWVSAQKGFVRELNSWLMKCLFYEPEETPDGIVPFSPGRIG-APMIFVICN 268
Query: 632 VWLKKLGTLPVKDVVDSIK 650
W + L + K+V+++I+
Sbjct: 269 QWEQALDRISEKEVIEAIR 287
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.312 0.128 0.365
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,298,018,089
Number of Sequences: 2540612
Number of extensions: 55771937
Number of successful extensions: 184104
Number of sequences better than 10.0: 633
Number of HSP's better than 10.0 without gapping: 102
Number of HSP's successfully gapped in prelim test: 541
Number of HSP's that attempted gapping in prelim test: 182968
Number of HSP's gapped (non-prelim): 1368
length of query: 773
length of database: 863,360,394
effective HSP length: 136
effective length of query: 637
effective length of database: 517,837,162
effective search space: 329862272194
effective search space used: 329862272194
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 79 (35.0 bits)
Medicago: description of AC131455.1


