

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC130807.11 - phase: 0
(261 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
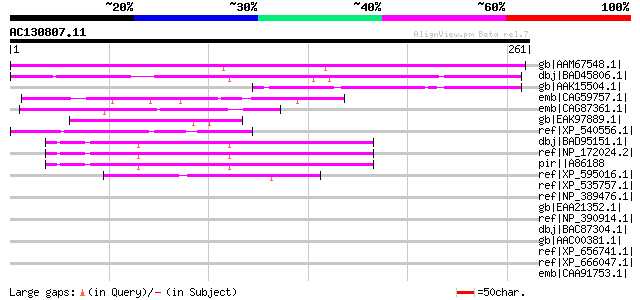
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM67548.1| unknown protein [Arabidopsis thaliana] gi|1738079... 199 8e-50
dbj|BAD45806.1| hypothetical protein [Oryza sativa (japonica cul... 135 1e-30
gb|AAK15504.1| unknown [Pennisetum ciliare] 84 3e-15
emb|CAG59757.1| unnamed protein product [Candida glabrata CBS138... 51 4e-05
emb|CAG87361.1| unnamed protein product [Debaryomyces hansenii C... 50 6e-05
gb|EAK97889.1| hypothetical protein CaO19.8591 [Candida albicans... 49 1e-04
ref|XP_540556.1| PREDICTED: similar to hypothetical protein 4931... 47 4e-04
dbj|BAD95151.1| hypothetical protein [Arabidopsis thaliana] 47 4e-04
ref|NP_172024.2| myosin-related [Arabidopsis thaliana] 47 4e-04
pir||A86188 hypothetical protein [imported] - Arabidopsis thalia... 47 4e-04
ref|XP_595016.1| PREDICTED: similar to hypothetical protein FLJ1... 47 5e-04
ref|XP_535757.1| PREDICTED: similar to Golgi autoantigen, golgin... 46 0.001
ref|NP_389476.1| chromosome segregation SMC protein homolg [Baci... 46 0.001
gb|EAA21352.1| hypothetical protein [Plasmodium yoelii yoelii] 46 0.001
ref|NP_390914.1| hypothetical protein BSU30360 [Bacillus subtili... 45 0.002
dbj|BAC87304.1| unnamed protein product [Homo sapiens] gi|566059... 45 0.002
gb|AAC00381.1| YttA [Bacillus subtilis] 45 0.002
ref|XP_656741.1| chromosome partition protein, putative [Entamoe... 45 0.002
ref|XP_666047.1| hypothetical protein Chro.80267 [Cryptosporidiu... 45 0.003
emb|CAA91753.1| Hypothetical protein C34E11.3 [Caenorhabditis el... 44 0.003
>gb|AAM67548.1| unknown protein [Arabidopsis thaliana] gi|17380796|gb|AAL36085.1|
unknown protein [Arabidopsis thaliana]
gi|9293961|dbj|BAB01864.1| unnamed protein product
[Arabidopsis thaliana] gi|42572493|ref|NP_974342.1|
expressed protein [Arabidopsis thaliana]
gi|22331190|ref|NP_188642.2| expressed protein
[Arabidopsis thaliana]
Length = 282
Score = 199 bits (505), Expect = 8e-50
Identities = 114/280 (40%), Positives = 169/280 (59%), Gaps = 21/280 (7%)
Query: 1 MEALYKKLYSKYTTLKTNKLSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQIN 60
MEALY KLY KYT L+ K SE ++++KEQE KFL FVSA+E++++HLR EN L +
Sbjct: 1 MEALYAKLYDKYTKLQKKKYSEYDEINKEQEEKFLTFVSASEELMEHLRGENQSSLEMVE 60
Query: 61 DLGNELTSVRVAKDNELVEHKRLLLEEKKKNEALFEEVEKLQKLLK-------EGTSGDL 113
L NE+ S+R +D++ +E ++LL+EE+ KN++L EEV KL++L++ E SG
Sbjct: 61 KLRNEIISIRSGRDDKFLECQKLLMEEELKNKSLSEEVVKLKELVQEEHPRNYEDQSGKK 120
Query: 114 SNRKVVNNTSNNSSIRMTRKRMRQEQDALDIEARCIPSENEGVD--------------TQ 159
RK + + + R R+ ++ D+ + I ++ +
Sbjct: 121 QKRKTPESARVTTRSMIKRSRLSEDLVETDMVSPDISKHHKAKEPLLVSQPQCCRTTYDG 180
Query: 160 ESDHQNWLVHALFEYTLDMKLSTDCQTGRLCLSAMHQSSGYSFSISWISRAPGEEAELLY 219
S + AL ++ L MKLST+ + R C+ A H ++G SFS+++I+ GEE+ELLY
Sbjct: 181 SSSSASCTFQALGKHLLGMKLSTNNKGKRACIVASHPTTGLSFSLTFINNPNGEESELLY 240
Query: 220 HVLSLGTLERLAPEWMREDIMFSPTMCPIFFERVTHVINL 259
SLGT +R+APEWMRE I FS +MCPIFFERV+ VI L
Sbjct: 241 KPASLGTFQRVAPEWMREVIKFSTSMCPIFFERVSRVIKL 280
>dbj|BAD45806.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 270
Score = 135 bits (340), Expect = 1e-30
Identities = 89/281 (31%), Positives = 142/281 (49%), Gaps = 38/281 (13%)
Query: 1 MEALYKKLYSKYTTLKTNKLSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQIN 60
ME L KLY +YT LK KL + E + +++ + A +D + L++EN++L+ ++
Sbjct: 1 MEQLNAKLYHRYTALKKRKLLD-EGLDQKRAADINELRQAMKDWVAELQSENERLVAKLT 59
Query: 61 DLGNELTSVRVAKDNELVEHKRLLLEEKKKNEALFEEVEKLQKLLKEGT--------SGD 112
K+ +LVE + LLL+E +K + L E+ KLQ LL E S D
Sbjct: 60 Q-----------KEQQLVEAQTLLLDETRKTKELNSEILKLQCLLAEKNDANHIATGSPD 108
Query: 113 LSNRKVVNNTSNNSSIRMTRKRMRQEQDALDIEARCIPS---ENEGVDTQ---------- 159
+ + N + S + T K +E++ IE +P + EG D
Sbjct: 109 TTAEMTIENQTPISPAKKTPKSNSRERNIRSIEKAIVPRNGFQEEGRDLDSCRRHMSISG 168
Query: 160 ---ESDHQNWLVHALFEYTLDMKLSTDCQTGRLCLSAMHQSSGYSFSISWISRAPGEEAE 216
E + H L E + MK S +T LS H++SGY+F+++W+ + G E
Sbjct: 169 SATEESSSTCMFHLLAESMVGMKFSVKNETEGFSLSVSHEASGYNFTLTWVDQPGGSEWS 228
Query: 217 LLYHVLSLGTLERLAPEWMREDIMFSPTMCPIFFERVTHVI 257
Y SLGTL+R+A WM+EDI FS TMCP+FF++++H++
Sbjct: 229 --YQYSSLGTLDRIAMGWMKEDIKFSSTMCPVFFKQISHIL 267
>gb|AAK15504.1| unknown [Pennisetum ciliare]
Length = 130
Score = 84.3 bits (207), Expect = 3e-15
Identities = 48/135 (35%), Positives = 76/135 (55%), Gaps = 9/135 (6%)
Query: 123 SNNSSIRMTRKRMRQEQDALDIEARCIPSENEGVDTQESDHQNWLVHALFEYTLDMKLST 182
S N+SI + + +E L+ R S D S + H + + + MK++
Sbjct: 2 SKNASI--PHRSLEEESQELECSRRYKCSSENETDEWPSAR---MFHLMLQSLVRMKVTV 56
Query: 183 DCQTGRLCLSAMHQSSGYSFSISWISRAPGEEAELLYHVLSLGTLERLAPEWMREDIMFS 242
D T +S H++SGYSF ++W+ PGE + Y + SLGTLER+A +WM++DI FS
Sbjct: 57 DDGTESFSVSVCHEASGYSFKLTWLEN-PGEWS---YKLSSLGTLERIAVDWMKQDITFS 112
Query: 243 PTMCPIFFERVTHVI 257
MC +FFER++++I
Sbjct: 113 MNMCRVFFERISNII 127
>emb|CAG59757.1| unnamed protein product [Candida glabrata CBS138]
gi|50288793|ref|XP_446826.1| unnamed protein product
[Candida glabrata]
Length = 584
Score = 50.8 bits (120), Expect = 4e-05
Identities = 50/178 (28%), Positives = 90/178 (50%), Gaps = 28/178 (15%)
Query: 7 KLYSKYTTLKTNKLSELEDVHKEQELKFLKFVSAAEDVIDHLRT--ENDKLLGQINDLGN 64
++ +KY + K +E + ++Q+ +A + I+ LR D+L+ QIN+L +
Sbjct: 111 EIENKYKEVIEQKAAEFSQIFEKQQ-------NAWREKIEELRNMPPEDELINQINNLKH 163
Query: 65 ELTSV------RVAK-DNELVEHKRLL----LEEKKKNEALFEEVEKLQKLLKEGTSGDL 113
L+S ++AK DN+L+ +K+ L E KKKN EE++ K L E T +L
Sbjct: 164 TLSSTEQNLEDKIAKNDNDLILYKQKLQVSFSEYKKKNSTSLEELQSQSKNL-ESTMAEL 222
Query: 114 SNRKVVNNTSNNSSIR-MTRKRMRQEQDALD--IEARCIPSENEGVDTQESDHQNWLV 168
N K T NS +R + ++ E+ +D ++ + +E E V+ + ++QN V
Sbjct: 223 QNIK----TEKNSQLRTLAEEQSELERIVMDKQLQVDKLNAEIEPVENKIKEYQNSFV 276
>emb|CAG87361.1| unnamed protein product [Debaryomyces hansenii CBS767]
gi|50421287|ref|XP_459190.1| unnamed protein product
[Debaryomyces hansenii]
Length = 734
Score = 50.1 bits (118), Expect = 6e-05
Identities = 42/138 (30%), Positives = 69/138 (49%), Gaps = 15/138 (10%)
Query: 6 KKLYSKYTTLKTNKLSELEDVHKEQELKFLKFVSAAEDVID------HLRTENDKLLGQI 59
+KL + N +E E + +E +K+ +++ D + LR N KL G++
Sbjct: 600 EKLIEELDKEWANTSTEYEKLCEENNTLSVKYQNSSTDAVQLSSKLIALRDANSKLEGKL 659
Query: 60 NDLGNELTSVRVAKDNELVEHKRLLLEEKKKNEALFEEV-EKLQKLLKEGTSGDLSNRKV 118
N G +L + KD EL+ H+ L + K +N+ + EKL KL+KE TS +
Sbjct: 660 NGAGIQLLPSLINKD-ELINHEVELNKLKSENQFMLRFFDEKLDKLIKERTS-------L 711
Query: 119 VNNTSNNSSIRMTRKRMR 136
+++TSN SS R T + R
Sbjct: 712 MDSTSNGSSSRPTNRVSR 729
>gb|EAK97889.1| hypothetical protein CaO19.8591 [Candida albicans SC5314]
gi|46438498|gb|EAK97828.1| hypothetical protein
CaO19.976 [Candida albicans SC5314]
Length = 681
Score = 48.9 bits (115), Expect = 1e-04
Identities = 36/95 (37%), Positives = 53/95 (54%), Gaps = 8/95 (8%)
Query: 31 ELKFLKF-VSAAEDVIDHLRTENDKLLGQINDLGNELTSVRVAKDNELVEHKRLLLEEKK 89
E K LK ++ ++D+I +L K L +I+ L N+L R+ K+N L E+ +L E K
Sbjct: 488 ENKLLKAQIAKSQDLIKNLNDLEKKYLNKIDLLSNQLVDFRIIKENSLAENSKLHDELKT 547
Query: 90 KN---EALFEEVE----KLQKLLKEGTSGDLSNRK 117
N +AL +EVE KL+ LKE T SN+K
Sbjct: 548 LNIAQDALHQEVERVNAKLESTLKEHTDLQESNKK 582
Score = 35.4 bits (80), Expect = 1.6
Identities = 34/126 (26%), Positives = 56/126 (43%), Gaps = 13/126 (10%)
Query: 45 IDHLRTENDKLLGQINDLGNELTSVRVAKDNELVEHKRLLLEEKKKN--EALFEEVEKLQ 102
+D L+T N L+ Q+ L K+N+ + K L LE K N E+ E+ +
Sbjct: 240 LDKLKTANQLLVQQVEQL---------TKENQSLIQKSLNLENKLHNLEESDLEDNTYYK 290
Query: 103 KLLKEGTSGDLSNRKVVNNTSNNSSIRMTRKRMRQEQDALDIEARCIPSENEGVDTQESD 162
K++K S K+ N+S++ + RQ+ D + I ENE + Q +
Sbjct: 291 KIVKNNQSLQEQIGKLTK--LNSSNVSRLNELERQQNDLKSVIEGEIIEENEKLKQQLQE 348
Query: 163 HQNWLV 168
+N LV
Sbjct: 349 SENNLV 354
>ref|XP_540556.1| PREDICTED: similar to hypothetical protein 4931417A20 [Canis
familiaris]
Length = 1167
Score = 47.4 bits (111), Expect = 4e-04
Identities = 37/122 (30%), Positives = 62/122 (50%), Gaps = 8/122 (6%)
Query: 1 MEALYKKLYSKYTTLKTNKLSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQIN 60
+EA K L TL+ + + E + KE E KFL + E + + ++L G+IN
Sbjct: 403 LEAELKVLKENNQTLERDNELQREKI-KENEEKFLNLQNEHEKALGTWKKHVEELNGEIN 461
Query: 61 DLGNELTSVRVAKDNELVEHKRLLLEEKKKNEALFEEVEKLQKLLKEGTSGDLSNRKVVN 120
++ NEL+S++ + ++L EH L ++KK FEE +K Q L + T + +
Sbjct: 462 EIKNELSSLK--ETHKLQEHYNELCDQKK-----FEEDKKFQNLPEVNTENSEMSMEKSE 514
Query: 121 NT 122
NT
Sbjct: 515 NT 516
>dbj|BAD95151.1| hypothetical protein [Arabidopsis thaliana]
Length = 699
Score = 47.4 bits (111), Expect = 4e-04
Identities = 44/175 (25%), Positives = 81/175 (46%), Gaps = 13/175 (7%)
Query: 19 KLSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQINDLG---NELTSVRVAKDN 75
KLS LE K Q+ K L+ ED+ L +E ++L QI+ L N++ + + N
Sbjct: 511 KLSVLE-AEKYQQAKELQIT--IEDLTKQLTSERERLRSQISSLEEEKNQVNEIYQSTKN 567
Query: 76 ELVEHKRLLLEEKKKNEALFEEVEKLQKLLKEGT-------SGDLSNRKVVNNTSNNSSI 128
ELV+ + L +K K++ + ++EKL L+ E + ++ ++ V + +S
Sbjct: 568 ELVKLQAQLQVDKSKSDDMVSQIEKLSALVAEKSVLESKFEQVEIHLKEEVEKVAELTSK 627
Query: 129 RMTRKRMRQEQDALDIEARCIPSENEGVDTQESDHQNWLVHALFEYTLDMKLSTD 183
K ++D L+ +A + E + T S+ + L H E +K S +
Sbjct: 628 LQEHKHKASDRDVLEEKAIQLHKELQASHTAISEQKEALSHKHSELEATLKKSQE 682
>ref|NP_172024.2| myosin-related [Arabidopsis thaliana]
Length = 828
Score = 47.4 bits (111), Expect = 4e-04
Identities = 44/175 (25%), Positives = 81/175 (46%), Gaps = 13/175 (7%)
Query: 19 KLSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQINDLG---NELTSVRVAKDN 75
KLS LE K Q+ K L+ ED+ L +E ++L QI+ L N++ + + N
Sbjct: 511 KLSVLE-AEKYQQAKELQIT--IEDLTKQLTSERERLRSQISSLEEEKNQVNEIYQSTKN 567
Query: 76 ELVEHKRLLLEEKKKNEALFEEVEKLQKLLKEGT-------SGDLSNRKVVNNTSNNSSI 128
ELV+ + L +K K++ + ++EKL L+ E + ++ ++ V + +S
Sbjct: 568 ELVKLQAQLQVDKSKSDDMVSQIEKLSALVAEKSVLESKFEQVEIHLKEEVEKVAELTSK 627
Query: 129 RMTRKRMRQEQDALDIEARCIPSENEGVDTQESDHQNWLVHALFEYTLDMKLSTD 183
K ++D L+ +A + E + T S+ + L H E +K S +
Sbjct: 628 LQEHKHKASDRDVLEEKAIQLHKELQASHTAISEQKEALSHKHSELEATLKKSQE 682
>pir||A86188 hypothetical protein [imported] - Arabidopsis thaliana
gi|2388564|gb|AAB71445.1| ESTs gb|AA042402,gb|ATTS1380
come from this gene. [Arabidopsis thaliana]
Length = 841
Score = 47.4 bits (111), Expect = 4e-04
Identities = 44/175 (25%), Positives = 81/175 (46%), Gaps = 13/175 (7%)
Query: 19 KLSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQINDLG---NELTSVRVAKDN 75
KLS LE K Q+ K L+ ED+ L +E ++L QI+ L N++ + + N
Sbjct: 524 KLSVLE-AEKYQQAKELQIT--IEDLTKQLTSERERLRSQISSLEEEKNQVNEIYQSTKN 580
Query: 76 ELVEHKRLLLEEKKKNEALFEEVEKLQKLLKEGT-------SGDLSNRKVVNNTSNNSSI 128
ELV+ + L +K K++ + ++EKL L+ E + ++ ++ V + +S
Sbjct: 581 ELVKLQAQLQVDKSKSDDMVSQIEKLSALVAEKSVLESKFEQVEIHLKEEVEKVAELTSK 640
Query: 129 RMTRKRMRQEQDALDIEARCIPSENEGVDTQESDHQNWLVHALFEYTLDMKLSTD 183
K ++D L+ +A + E + T S+ + L H E +K S +
Sbjct: 641 LQEHKHKASDRDVLEEKAIQLHKELQASHTAISEQKEALSHKHSELEATLKKSQE 695
>ref|XP_595016.1| PREDICTED: similar to hypothetical protein FLJ14640, partial [Bos
taurus]
Length = 203
Score = 47.0 bits (110), Expect = 5e-04
Identities = 35/117 (29%), Positives = 59/117 (49%), Gaps = 12/117 (10%)
Query: 48 LRTENDKLLGQINDLGNELTSVRVAKDNELVEHKRLLLEEKKKNEALFEEVEKLQKLLKE 107
L+ E +K ++ +L +L +++V K + L+E K L+ KN+AL E+EK QK+ +
Sbjct: 72 LQKEEEKENAEMEELMEKLAALQVQKKSLLLEKKNLMA----KNKALEAELEKAQKINRR 127
Query: 108 GTSG-DLSNRKVVNNTSNNSSIRM-------TRKRMRQEQDALDIEARCIPSENEGV 156
D+ ++V N S + + QE+D L A+C+ SEN GV
Sbjct: 128 SQKKIDVLKKQVEKAMENEMSAHQYLANLVGLAENVTQERDNLVFLAKCLESENHGV 184
>ref|XP_535757.1| PREDICTED: similar to Golgi autoantigen, golgin subfamily B member
1 (Giantin) (Macrogolgin) (Golgi complex-associated
protein, 372-kDa) (GCP372) [Canis familiaris]
Length = 2141
Score = 45.8 bits (107), Expect = 0.001
Identities = 41/165 (24%), Positives = 73/165 (43%), Gaps = 12/165 (7%)
Query: 6 KKLYSKYTTLKTNK------LSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQI 59
++LY K + + NK L E E +E + K KF + + I L END+L ++
Sbjct: 604 QELYGKLRSTEANKKEAEKQLQEAEQEMEEMKEKMRKFAKSKQQKILELEEENDRLRAEV 663
Query: 60 NDLG---NELTSVRVAKDNELVEHKRLLLEEKKKNEALFEEVEKLQKLLKEGTSGDLSNR 116
+ G N+ ++ ++ L E + E K FE + + L E DL +
Sbjct: 664 HSTGGTSNDGMEALLSSNSNLKEELERIKTEYKTLSQEFEGLMTEKNSLSEEVQ-DLKH- 721
Query: 117 KVVNNTSNNSSIRMTRKRMRQEQDALDIEARCIPSENEGVDTQES 161
+V +N S+ T K Q +D ++ + +P E+ D+Q +
Sbjct: 722 QVESNVIKQGSLEATEKHDNQ-RDVIEGATQSVPGESNEQDSQNT 765
Score = 34.3 bits (77), Expect = 3.6
Identities = 27/103 (26%), Positives = 49/103 (47%), Gaps = 4/103 (3%)
Query: 5 YKKLYSKYTTLKTNKLSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQINDLGN 64
Y++L ++ ++ K ++D E K + + +D L +EN KL ++
Sbjct: 1441 YQQLEERHLSVILEKDQLIQDAAAENN-KLKEEIRCLRSHMDDLNSENAKLDAELIQYRE 1499
Query: 65 ELTSVRVAKDNELVEHKRLLLEEKKKNEALFEEVEKLQKLLKE 107
+L V KD + K+LL + ++N+ L E KL+ LKE
Sbjct: 1500 DLNQVISRKD---CQQKQLLEAQLQQNKELKSECAKLEGKLKE 1539
Score = 33.1 bits (74), Expect = 8.0
Identities = 34/116 (29%), Positives = 52/116 (44%), Gaps = 6/116 (5%)
Query: 19 KLSELEDVHKEQELK-FLKFVSAAEDVIDHLRTENDKLLGQINDLGNE---LTSVRVAKD 74
KL E E+V E+ +L+ + ++ ID L E K L +E L S AKD
Sbjct: 783 KLDESENVSPRDEINNYLQQIDQLKERIDKLEEEKQKEREFSQTLESERSGLLSQISAKD 842
Query: 75 NELVEHKRLLLEEKKKNEALFEEVEKLQKLLK--EGTSGDLSNRKVVNNTSNNSSI 128
NEL + + + N+ + EE+ ++ KL + E DL R + N SI
Sbjct: 843 NELKVLQEEVTKINLSNQQIQEELARVTKLKETAEEEKDDLEERLMNQLAELNGSI 898
>ref|NP_389476.1| chromosome segregation SMC protein homolg [Bacillus subtilis subsp.
subtilis str. 168] gi|2633966|emb|CAB13467.1| chromosome
segregation SMC protein homolg [Bacillus subtilis subsp.
subtilis str. 168] gi|7446600|pir||G69708 chromosome
segregation SMC protein - Bacillus subtilis
gi|7531285|sp|P51834|SMC_BACSU Chromosome partition
protein smc
Length = 1186
Score = 45.8 bits (107), Expect = 0.001
Identities = 45/166 (27%), Positives = 87/166 (52%), Gaps = 23/166 (13%)
Query: 6 KKLYSKYTTLKTNKLSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQINDLGNE 65
+KL+ K++TLK E + KE+EL +SA E I+ R + L +N+L
Sbjct: 235 EKLHGKWSTLK-----EKVQMAKEEELAESSAISAKEAKIEDTRDKIQALDESVNELQQV 289
Query: 66 LTSVRVAKDNELVEHKRLLLEEKKKN-----EALFEEVEKLQK---LLKEGTSGDLSNRK 117
L + +++ E +E ++ +L+E+KKN E L E + + Q+ +LKE +LS ++
Sbjct: 290 L--LVTSEELEKLEGRKEVLKERKKNAVQNQEQLEEAIVQFQQKETVLKE----ELSKQE 343
Query: 118 VVNNTSNNSSIRMTRKRMRQEQDALDIEARCIPSENEGVDTQESDH 163
V T + ++ R +++++Q AL + + E ++ +SD+
Sbjct: 344 AVFETL-QAEVKQLRAQVKEKQQALSLHNENV---EEKIEQLKSDY 385
>gb|EAA21352.1| hypothetical protein [Plasmodium yoelii yoelii]
Length = 2095
Score = 45.8 bits (107), Expect = 0.001
Identities = 37/153 (24%), Positives = 73/153 (47%), Gaps = 9/153 (5%)
Query: 16 KTNKLSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQINDLGNELTSVRVAKDN 75
+T ++ E+E V K +E+K +K V E+V ++E K + ++ ++ +V +
Sbjct: 1046 ETEEVKEVEKVEKSEEIKEVKKVEKVEEVEKVEKSEEAKEVDEVKEVKK---VEKVEEVK 1102
Query: 76 ELVEHKRLLLEEKKKNEALFEEVEKLQKLLKEGTSGDLSNRKVVNNTSNNSSIRMTRKRM 135
E+VE K + EK +N +E+EK++++ K ++ K + N I+ K
Sbjct: 1103 EIVEIKEV---EKVENSEEVKEIEKVKEVEKGEEIKEMEKVKEIEEVEKNEEIKEVEKVE 1159
Query: 136 RQE---QDALDIEARCIPSENEGVDTQESDHQN 165
+ E +D L ENE + +E +H +
Sbjct: 1160 KGEEVKEDKLTEPQIKQAGENETKEIKEGNHND 1192
Score = 37.7 bits (86), Expect = 0.32
Identities = 24/109 (22%), Positives = 42/109 (38%), Gaps = 12/109 (11%)
Query: 24 EDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQINDLGNELTSVRVAKDNELVEHKRL 83
E + +E+E + +D ++H+ + DK +IN+L ++ + +N
Sbjct: 1998 EKMSEEKEFTLSSVIDNIKDPLEHIEFKEDKEKSEINELSGDIFVDNIESEN-------- 2049
Query: 84 LLEEKKKNEALFEEVEKLQKLLKEGTSGDLSNRKVVNNTSNNSSIRMTR 132
K EK + L+ EG S SN K N N R +
Sbjct: 2050 ----KNNENPQDTNFEKTETLISEGRSSYASNTKNYKNNKNKKKKRKNK 2094
Score = 35.8 bits (81), Expect = 1.2
Identities = 26/95 (27%), Positives = 50/95 (52%), Gaps = 13/95 (13%)
Query: 16 KTNKLSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQINDL-------GNELTS 68
K ++ E+E + K +E+K ++ V E+V + T+ + + + ++ G E+
Sbjct: 970 KNEEIKEVEKIEKGEEIKEMEKVKEVEEVETNEETKEIERVKETEEVKEVEVEKGGEVKE 1029
Query: 69 VRVAKDNELVEHKRLLLEEKKKNEALFEEVEKLQK 103
V V K EL E +E+ K+ E + +EVEK++K
Sbjct: 1030 VEVEKGGELKE-----IEKVKETEEV-KEVEKVEK 1058
>ref|NP_390914.1| hypothetical protein BSU30360 [Bacillus subtilis subsp. subtilis
str. 168] gi|2635520|emb|CAB15014.1| yttA [Bacillus
subtilis subsp. subtilis str. 168]
gi|7475416|pir||E70001 hypothetical protein yttA -
Bacillus subtilis
Length = 246
Score = 45.4 bits (106), Expect = 0.002
Identities = 42/132 (31%), Positives = 67/132 (49%), Gaps = 15/132 (11%)
Query: 2 EALYKKLYSKYTTLKTNKLSELEDVHKE--QELKFLKFVSAAEDVIDHLRTENDKLLGQI 59
E Y L ++Y L T + ELE +K E K LK +D ++ L+ EN
Sbjct: 66 EKKYSALNAEYKNL-TKEHEELEKEYKSVSSEAKKLKDNKEDQDKLEKLKNEN------- 117
Query: 60 NDLGNELTSVRVAKDNELVEHKRLLLEEKK----KNEALFEEVEKLQKLLKEGTSGDLSN 115
+DL S++ A+ EL E+++ L E+ K +NE L ++ KL+ LKE S S+
Sbjct: 118 SDLKKTQKSLK-AEIKELQENQKQLKEDAKTAKAENETLRQDKTKLENQLKETESQTASS 176
Query: 116 RKVVNNTSNNSS 127
+ ++SNN+S
Sbjct: 177 HEDTGSSSNNTS 188
>dbj|BAC87304.1| unnamed protein product [Homo sapiens]
gi|56605996|ref|NP_001008392.1| sarcoma antigen
NY-SAR-79 [Homo sapiens]
Length = 822
Score = 45.4 bits (106), Expect = 0.002
Identities = 53/196 (27%), Positives = 82/196 (41%), Gaps = 13/196 (6%)
Query: 1 MEALYKKLYSKYTTLKTNKLSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQIN 60
MEA K L TL+ + + E V KE E KFL + E + + ++L G+IN
Sbjct: 44 MEAELKVLKENNQTLERDNELQREKV-KENEEKFLNLQNEHEKALGTWKRHAEELNGEIN 102
Query: 61 DLGNELTSVRVAKDNELVEHKRLLLEEKKKNEALFEEVEKLQKLLK-EGTSGDLSNRKVV 119
+ NEL+S++ +L EH L N+ FEE +K Q + + + ++S K
Sbjct: 103 KIKNELSSLK-ETHIKLQEHYNKLC-----NQKTFEEDKKFQNVPEVNNENSEMSTEKSE 156
Query: 120 NNTSNNSSIRMTRKRMRQEQDALDIEARCIPSENEGVDTQE---SDHQNWLVHALFEYTL 176
N + + E D E R + EG +E D Q L F+ +
Sbjct: 157 NTIIQKYNTEQEIREENMENFCSDTEYREKEEKKEGSFIEEIIIDDLQ--LFEKSFKNEI 214
Query: 177 DMKLSTDCQTGRLCLS 192
D +S D + LS
Sbjct: 215 DTVVSQDENQSEISLS 230
>gb|AAC00381.1| YttA [Bacillus subtilis]
Length = 248
Score = 45.4 bits (106), Expect = 0.002
Identities = 42/132 (31%), Positives = 67/132 (49%), Gaps = 15/132 (11%)
Query: 2 EALYKKLYSKYTTLKTNKLSELEDVHKE--QELKFLKFVSAAEDVIDHLRTENDKLLGQI 59
E Y L ++Y L T + ELE +K E K LK +D ++ L+ EN
Sbjct: 68 EKKYSALNAEYKNL-TKEHEELEKEYKSVSSEAKKLKDNKEDQDKLEKLKNEN------- 119
Query: 60 NDLGNELTSVRVAKDNELVEHKRLLLEEKK----KNEALFEEVEKLQKLLKEGTSGDLSN 115
+DL S++ A+ EL E+++ L E+ K +NE L ++ KL+ LKE S S+
Sbjct: 120 SDLKKTQKSLK-AEIKELQENQKQLKEDAKTAKAENETLRQDKTKLENQLKETESQTASS 178
Query: 116 RKVVNNTSNNSS 127
+ ++SNN+S
Sbjct: 179 HEDTGSSSNNTS 190
>ref|XP_656741.1| chromosome partition protein, putative [Entamoeba histolytica
HM-1:IMSS] gi|56473964|gb|EAL51358.1| chromosome
partition protein, putative [Entamoeba histolytica
HM-1:IMSS]
Length = 605
Score = 45.1 bits (105), Expect = 0.002
Identities = 40/160 (25%), Positives = 68/160 (42%), Gaps = 13/160 (8%)
Query: 9 YSKYTTLKTNKLSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQINDLGNELTS 68
Y T NK+ ELE+V+ K K V+ E I+ + EN++ N+L L
Sbjct: 233 YQGKITEYRNKIGELEEVNG----KLTKKVNGMERKIEKMEKENEQNQANTNELIYNLKE 288
Query: 69 VRVAKDNELVEHKRLLLEEKKKNEALFEEVEKLQKLLKEGTSGDLSNRK---VVNNTSNN 125
KDN ++ K L +K E L E+ K++ KE + D+ K +
Sbjct: 289 DIKTKDNIIIGLKTDLNNTDEKIEGLKSEINKMKSAKKENKTDDIFEIKKEHQIQVEEMK 348
Query: 126 SSIRMTRKRMRQEQDALDIEARCIPSENEGVDTQESDHQN 165
I K M+++ D +CI E + +E++ ++
Sbjct: 349 KQIEERDKGMKEKND------KCIELMKENIQMKENNEKH 382
>ref|XP_666047.1| hypothetical protein Chro.80267 [Cryptosporidium hominis]
gi|54656956|gb|EAL35818.1| hypothetical protein
Chro.80267 [Cryptosporidium hominis]
Length = 893
Score = 44.7 bits (104), Expect = 0.003
Identities = 49/167 (29%), Positives = 81/167 (48%), Gaps = 37/167 (22%)
Query: 5 YKKLYSKYTTL-------KTNK------LSELEDVHKEQEL------KFLKFVSAAEDVI 45
Y KLY KYT L K N+ L EL+D++K+ ++ + K +SA E V+
Sbjct: 511 YNKLYQKYTALLSEYTASKKNEENLNASLVELQDINKKLDIEGRDMKEKYKTLSATERVL 570
Query: 46 DHLRTENDKLLGQINDLGNELTSVRVAKDNELVEHKRLLLEEKK---KNEAL--FEEVEK 100
+ L+ N +L+G+I+ L E ++ D +++E K EE KN+ L E +K
Sbjct: 571 EQLQESNAELIGEISALKEERDI--LSADYKILEKKYKTSEESMHEIKNQLLDFTLENQK 628
Query: 101 LQKLLKEGTSGDLSNRK---------VVNNTSNNSSIRMTRKRMRQE 138
+KL+K+ + N + +V N NS RM+ K + +E
Sbjct: 629 FKKLIKQTIIPLIPNAEYNSTIGHILMVMNAKENS--RMSTKVLEEE 673
>emb|CAA91753.1| Hypothetical protein C34E11.3 [Caenorhabditis elegans]
gi|3880366|emb|CAB02323.1| Hypothetical protein C34E11.3
[Caenorhabditis elegans] gi|3877587|emb|CAA91761.1|
Hypothetical protein C34E11.3 [Caenorhabditis elegans]
gi|17550806|ref|NP_509975.1| intracellular protein
transport like (XM453) [Caenorhabditis elegans]
gi|7497024|pir||T19722 hypothetical protein C34E11.3 -
Caenorhabditis elegans
Length = 1577
Score = 44.3 bits (103), Expect = 0.003
Identities = 44/149 (29%), Positives = 74/149 (49%), Gaps = 16/149 (10%)
Query: 6 KKLYSKYTTLKTNKLSELED---VHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQINDL 62
KKL ++ T L + ELE KE E+K+ K S E TE +K+ G+ N
Sbjct: 785 KKLRAE-TELLRKDMQELETDKKTVKEFEIKYKKLESIFE-------TEREKMNGERNRS 836
Query: 63 GNELTSVRVAKDNELVEHKRLLLEEKKKNEALFE-EVEKLQK---LLKEGTSGDLSNRKV 118
NEL +++ KD + EH + L +++KKN+A ++ E KL+K LLK+ + ++
Sbjct: 837 KNELAAMKKLKD-DAEEHLKKLSDDQKKNDAAWKIEKSKLEKDIALLKKQLPDEHEMKES 895
Query: 119 VNNTSNNSSIRMTRKRMRQEQDALDIEAR 147
N+ S + R + + L +E +
Sbjct: 896 TATPQNSISGESSPLRRQDSEKMLVLELK 924
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.315 0.131 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 420,824,510
Number of Sequences: 2540612
Number of extensions: 17160749
Number of successful extensions: 78414
Number of sequences better than 10.0: 1306
Number of HSP's better than 10.0 without gapping: 67
Number of HSP's successfully gapped in prelim test: 1266
Number of HSP's that attempted gapping in prelim test: 76489
Number of HSP's gapped (non-prelim): 2975
length of query: 261
length of database: 863,360,394
effective HSP length: 125
effective length of query: 136
effective length of database: 545,783,894
effective search space: 74226609584
effective search space used: 74226609584
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 74 (33.1 bits)
Medicago: description of AC130807.11


