

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC130806.6 + phase: 0 /pseudo
(2359 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
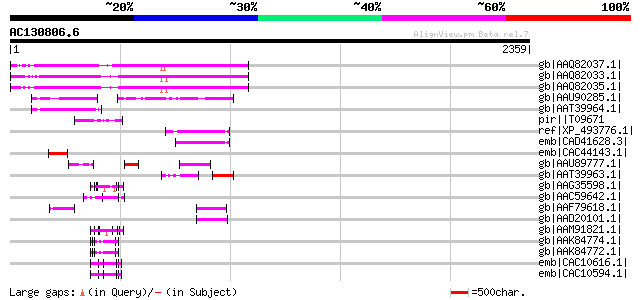
Score E
Sequences producing significant alignments: (bits) Value
gb|AAQ82037.1| gag/pol polyprotein [Pisum sativum] 589 e-166
gb|AAQ82033.1| gag/pol polyprotein [Pisum sativum] 580 e-163
gb|AAQ82035.1| gag/pol polyprotein [Pisum sativum] 579 e-163
gb|AAU90285.1| putative gag/pol polyprotein, 3'-partial [Solanum... 218 3e-54
gb|AAT39964.1| putative gag polyprotein [Solanum demissum] 177 3e-42
pir||T09671 RPE15 protein - alfalfa (fragment) gi|840619|gb|AAB5... 121 3e-25
ref|XP_493776.1| unnamed protein product [Oryza sativa (japonica... 117 6e-24
emb|CAD41628.3| OSJNBa0091D06.8 [Oryza sativa (japonica cultivar... 116 1e-23
emb|CAC44143.1| putative gag polyprotein [Cicer arietinum] 111 3e-22
gb|AAU89777.1| gag-pol polyprotein-like [Solanum tuberosum] 100 1e-18
gb|AAT39963.1| putative polyprotein [Solanum demissum] 87 5e-15
gb|AAG35598.1| secalin precursor [Secale cereale] 87 9e-15
gb|AAC59642.1| zona pellucida protein [Pseudopleuronectes americ... 85 3e-14
gb|AAF79618.1| F5M15.26 [Arabidopsis thaliana] gi|25402907|pir||... 81 5e-13
gb|AAD20101.1| putative retroelement pol polyprotein [Arabidopsi... 80 8e-13
gb|AAM91821.1| choriogenin Hminor [Oryzias latipes] 80 8e-13
gb|AAK84774.1| gamma-gliadin [Triticum aestivum] 80 8e-13
gb|AAK84772.1| gamma-gliadin [Triticum aestivum] 80 8e-13
emb|CAC10616.1| gamma-gliadin [Aegilops longissima] 80 1e-12
emb|CAC10594.1| gamma-gliadin [Aegilops bicornis] gi|10635141|em... 80 1e-12
>gb|AAQ82037.1| gag/pol polyprotein [Pisum sativum]
Length = 2262
Score = 589 bits (1519), Expect = e-166
Identities = 395/1139 (34%), Positives = 587/1139 (50%), Gaps = 143/1139 (12%)
Query: 3 EMAELIKTMAETQTQAQAQIQAQAQTQAQAVTEAQARSQAPPPPPPVRTQAEASSSWTLC 62
E+AE+ MA+ Q Q Q + +A R +A PPP +A
Sbjct: 7 ELAEMRANMAQFMHMMQGVAQGQEELRALV-----QRQEAVTPPPN-----QALPEGNPV 56
Query: 63 ADTPTRSAPQRSTPWFPPFTAGEIFRPITCEAQMPTHQYTAQVPLPAMRVTPATMTYSAP 122
DTP + P + + GE I + Q P AP
Sbjct: 57 HDTPAAAIPVNN------YAVGEELMGIRVDGQ------------PIAPDAANARVIHAP 98
Query: 123 VIHTIP--QTEEPIFHSGNAEAYEEV---SDLRE-KYDELRRDMKALHEKGKFGKTAYDL 176
+ IP +E +F + + E++ +D R+ K D L ++A+ + G ++
Sbjct: 99 ARNRIPIVDRQEDLFTMFSED--EDIPGRNDARDRKVDALAEKIRAMECQNSLGFDVTNM 156
Query: 177 CLVPSVQVPHKFKIPDFEKYKGSSCPEEHLKMYVRRMPAYAQDDQILIYYFQESLTGPAS 236
LV +++P+KFK P F+KY G+SCP H++ Y R++ AY D+++ +Y+FQ+SL+G +
Sbjct: 157 GLVEGLRIPYKFKAPSFDKYNGTSCPRTHVQAYYRKISAYTDDEKMWMYFFQDSLSGASL 216
Query: 237 KWYTNLDKTRVQTFRDLCEAFVEQYSYNVDMTPDRSDLQAMTQGDKETFKEYAQRWRDTA 296
WY L ++ +RDL EAF+ QY +N+DM P R+ LQ++ Q E+FKEYAQRWR+ A
Sbjct: 217 DWYMELKSDSIRCWRDLGEAFLRQYKHNMDMAPSRTQLQSLCQKSNESFKEYAQRWRELA 276
Query: 297 AQVSPRIEEKEMTKLFLKTLNHFYYKKMVGSTP-KSFAEMVGMGVQLEEGVREGRLVKDA 355
A+V P + E+E+T +F+ TL + +M GS P SF+++V G + E ++ G++
Sbjct: 277 ARVQPPMLERELTDMFIGTLQGVFMDRM-GSCPFGSFSDVVICGERTESLIKTGKI--QD 333
Query: 356 TPASGTKKTGNHFPRKKEQEVGMVTHGRPQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQ 415
+S +KK PR++E E V H R Q Q A P P QQ
Sbjct: 334 VGSSSSKKPFAGAPRRREGETNAVQHRRDQNRIEYRQAAAVTIPAPQPRQQ--------- 384
Query: 416 YPQIPQYPQIPQYPQFPQNPSPQNTQQQNFQQQPYQQQPYQQFPYQQYPQQNFQQQPYQQ 475
QQQ QQ PQQ QQ+PYQ
Sbjct: 385 -------------------------QQQRVQQ----------------PQQQQQQRPYQ- 402
Query: 476 RPQQPRPPRMPINPIPVTYAELLPGLLKKNLVQTRTAPPIPEKLPSWYRLDQTCDFHEGG 535
P+Q P R + +P++YAELLP LL+ LV+ T P P LP Y + CDFH G
Sbjct: 403 -PRQRMPDRR-FDSLPMSYAELLPELLRLGLVELCTMAP-PTVLPPGYDANVRCDFHSGA 459
Query: 536 RGHNIETCYAFKSIVQRLINDGKITFTDSAPNVQTNPLPNHGAATVNMIEDCQKTRPILN 595
GH+ E C A + VQ LI+ I F PNV NP+P HG VN IE + + N
Sbjct: 460 PGHHTEKCRALQHKVQDLIDANAINFA-PVPNVVNNPMPQHGGHRVNNIEGKEAEDLVDN 518
Query: 596 VQHIRTPLVPLHAKLCKVDLFEHDHDLCEICLMNSGGCQKVRNDIQGLLNRGELVVERRC 655
V ++T L+ + ++L ++ + C C + GC ++R IQG+++ G L R
Sbjct: 519 VDDVQTSLLVVKSRLLNEGVYSGCDEDCLGCAESENGCDQLRAGIQGMMDEGCLQFSRAV 578
Query: 656 DD---VCVIT-----PEGPLEVFYDSRKSTITPLVICLP------------GPLPYASEK 695
D V IT EG + + + TP+ I +P G + +
Sbjct: 579 KDRGTVSTITIYFKPSEGHGQRVVSAPATNGTPVTIPVPVTISAPTTIVASGRRAVENSR 638
Query: 696 AIPYKY-----------------NATMIEEG--REVPIPPLSSVDNIVEDSRVLRNGRVV 736
A+P+KY N T + G VP +VDN+ R+GR+
Sbjct: 639 AVPWKYDNAYRSNRRVESQTKPVNQTPVTIGLANRVPATVGPAVDNVGGPGGFTRSGRLF 698
Query: 737 PIVFPKKIDATINKEVRTKDAGIAKEVDQPNGAGTSAEFD--EILKLIKKSEYKVVDQLM 794
+ +A + + K A + +E Q S E D E +K+IKKS+YK+VDQL
Sbjct: 699 APQPLRDNNAEALAKAKGKQAVVEEEPVQKEAPEGSFEKDVEEFMKMIKKSDYKIVDQLN 758
Query: 795 QTPSKISIMSLLLNSEAHKDALMKVLEQAFVDYDVMVGQFGGIVGNITACNNLSFSDEEL 854
QTPSKISI+SLLL SEAH++AL+K+L A+V ++ V Q G++ N++ + + F++ +L
Sbjct: 759 QTPSKISILSLLLCSEAHRNALLKMLNLAYVPQEISVNQLEGVMANVSTRHGVGFTNLDL 818
Query: 855 PAEGRNHNRALHISVNCKTDALSNVLVDTGSSLNVLSKTTYAQLAYQDAPLRRSGVMVKA 914
P EGRNHN+ALHI++ CK LS+VLVDTGSSLNVL K ++ + L S ++V+A
Sbjct: 819 PPEGRNHNKALHITMECKGAVLSHVLVDTGSSLNVLPKQILKKIDVEGFVLTPSDLIVRA 878
Query: 915 FDGSRKDVLGEVVLPITVGPQVFQVNFQVMDIQASYSCLLGRPWIHEAGAVTSTLHQKLK 974
FD S++ V GEV LP+ +GP+VF + F VMDIQ +YSCLLGRPWIH AGAV+STLHQKLK
Sbjct: 879 FDRSKRSVCGEVTLPVKIGPEVFDIIFYVMDIQPAYSCLLGRPWIHAAGAVSSTLHQKLK 938
Query: 975 FVKNGKLVTVNGEEALLVSHLSSFSFIGAD-DVEGTPFQGF------TIEDKNAKRNEAS 1027
+V NG++VTV GEE +LVSHLSSF ++ D ++ T Q F + ++ S
Sbjct: 939 YVWNGQIVTVCGEEEILVSHLSSFKYVEVDGEIHETLCQVFETVALEKVAYAEQRKPGVS 998
Query: 1028 ISSLKDAQKVIQAGGSASWGKLIELPENKHREGLGFFPSTGLSTAKRGISTVRALSMQS 1086
I+S K A++V+ +G + WGK+++LP + + G+G+ P + G ST + + +
Sbjct: 999 ITSYKQAKEVVDSGKAEGWGKMVDLPVKEDKFGVGYEPLQAEQNGQAGPSTFTSAGLMN 1057
>gb|AAQ82033.1| gag/pol polyprotein [Pisum sativum]
Length = 1814
Score = 580 bits (1495), Expect = e-163
Identities = 383/1137 (33%), Positives = 581/1137 (50%), Gaps = 129/1137 (11%)
Query: 1 MDEMAELIKTMAETQTQAQAQIQAQAQTQAQAVTEAQARSQAPPPPPPVRTQAEASSSWT 60
MD++ + M Q +Q AQ Q + Q + A PP + +
Sbjct: 1 MDQVQAELAEMKANMAQFMNMMQGVAQGQEELRALVQRQEAAIPPVNHAPPEGGPVNGNN 60
Query: 61 LCADTPTRSAPQRSTPWFPPFTAGEIFRPITCEAQMPTHQYTAQVPLPAMRVTPATMTYS 120
+ A P + + G+ I Q P A R A
Sbjct: 61 VAAAVPINN-----------YAVGDELGGIRINGQ-PIAPDVANA-----RAVRAPARNP 103
Query: 121 APVIHTIPQTEEPIFH--SGNAEAYEEVSDLREKYDELRRDMKALHEKGKFGKTAYDLCL 178
AP++ +E +F S + + V + K D L ++A+ + G ++ L
Sbjct: 104 APIV----DRQEDMFSLLSEDEDVLGRVDERDRKVDALAEKIRAMECQNSLGFDVTNMGL 159
Query: 179 VPSVQVPHKFKIPDFEKYKGSSCPEEHLKMYVRRMPAYAQDDQILIYYFQESLTGPASKW 238
V +++P+KFK P F+KY G+SCP H++ Y R++ AY D+++ +Y+FQ+SL+G + W
Sbjct: 160 VEGLRIPYKFKAPSFDKYNGTSCPRTHVQAYYRKISAYTDDEKMWMYFFQDSLSGASLDW 219
Query: 239 YTNLDKTRVQTFRDLCEAFVEQYSYNVDMTPDRSDLQAMTQGDKETFKEYAQRWRDTAAQ 298
Y L + ++ ++DL EAF+ QY +N+DM P R+ LQ++ Q E+FKEYAQRWR+ AA+
Sbjct: 220 YMELKRDSIRCWKDLGEAFLRQYKHNMDMAPSRTQLQSLCQKSGESFKEYAQRWRELAAR 279
Query: 299 VSPRIEEKEMTKLFLKTLNHFYYKKMVGSTP-KSFAEMVGMGVQLEEGVREGRLVKDATP 357
V P + E+E+T +F+ TL + +M GS P SF+++V G + E ++ G+ ++DA
Sbjct: 280 VQPPMLERELTDMFIGTLQGVFMDRM-GSCPFVSFSDVVICGERTESLIKTGK-IQDAG- 336
Query: 358 ASGTKKTGNHFPRKKEQEVGMVTHGRPQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYP 417
+S +KK PR++E E V + R Q Q A P P QQ Q PQ
Sbjct: 337 SSSSKKPFAGAPRRREGETNAVQYRRDQNRSQRCQVAAVTIPAPQPRQQQQQRVQQPQ-- 394
Query: 418 QIPQYPQIPQYPQFPQNPSPQNTQQQNFQQQPYQQQPYQQFPYQQYPQQNFQQQPYQQRP 477
QQ QQ+P QP Q+ P +++
Sbjct: 395 ------------------------QQQQQQRP--YQPRQRMPDRRF-------------- 414
Query: 478 QQPRPPRMPINPIPVTYAELLPGLLKKNLVQTRTAPPIPEKLPSWYRLDQTCDFHEGGRG 537
+ +P++YAELLP LL+ +V+ RT P P LP Y + CDFH G G
Sbjct: 415 ----------DSLPMSYAELLPELLRLGMVELRTMAP-PTVLPPGYDANVRCDFHSGAPG 463
Query: 538 HNIETCYAFKSIVQRLINDGKITFTDSAPNVQTNPLPNHGAATVNMIEDCQKTRPILNVQ 597
H+ E C A + VQ LI+ I F PNV NP+P HG VN IE + ++NV
Sbjct: 464 HHTEKCRALQHKVQDLIDAKAINFA-PVPNVVNNPMPQHGGHRVNNIEGKEAEDLVVNVD 522
Query: 598 HIRTPLVPLHAKLCKVDLFEHDHDLCEICLMNSGGCQKVRNDIQGLLNRGELVVERRCDD 657
++T L+ + +L ++ + C C + GC ++R IQG+++ G L R D
Sbjct: 523 DVQTSLLVVKGRLLNGGVYSGCDEDCLGCAESENGCDQLRTGIQGMMDEGCLQFSRAVKD 582
Query: 658 ---VCVIT-----PEGPLEVFYDSRKSTITPLVICLP------------GPLPYASEKAI 697
V IT EG + + + TP+ I +P G + +A+
Sbjct: 583 RGTVSTITIYFKPSEGRGQRVVSAPATNGTPVTISVPVTISAPTTIAASGRRAVENSRAV 642
Query: 698 PYKYNATMIEEGR-------------------EVPIPPLSSVDNIVEDSRVLRNGRVVPI 738
P+KY+ R VP +VDN+ R+GR+
Sbjct: 643 PWKYDNAYRSNRRAESQTRPVNQAPVTIGLPNRVPATVGPAVDNVGGPGGFTRSGRLFAP 702
Query: 739 VFPKKIDATINKEVRTKDAGIAKEVDQPNGAGTSAEFD--EILKLIKKSEYKVVDQLMQT 796
+ +A + + K A + +E Q S E D E +K+IKKS+YK+VDQL QT
Sbjct: 703 QPLRDNNAEALAKAKGKQAVVEEEPVQKEAPEGSFEKDVEEFMKIIKKSDYKIVDQLNQT 762
Query: 797 PSKISIMSLLLNSEAHKDALMKVLEQAFVDYDVMVGQFGGIVGNITACNNLSFSDEELPA 856
PSKISI+SLLL SEAH++AL+K+L A+V ++ V Q G++ N++ + + F++ +L
Sbjct: 763 PSKISILSLLLCSEAHRNALLKMLNLAYVPQEISVNQLEGVMANVSTRHGVGFTNLDLTP 822
Query: 857 EGRNHNRALHISVNCKTDALSNVLVDTGSSLNVLSKTTYAQLAYQDAPLRRSGVMVKAFD 916
EGRNHN+ALHI++ CK LS+VLVDTGSSLNVL K ++ + L S ++V+AFD
Sbjct: 823 EGRNHNKALHITMECKGAVLSHVLVDTGSSLNVLPKQILKKIDVEGFVLTPSDLIVRAFD 882
Query: 917 GSRKDVLGEVVLPITVGPQVFQVNFQVMDIQASYSCLLGRPWIHEAGAVTSTLHQKLKFV 976
GS++ V GEV LP+ +GP+VF + F VMDIQ +YSCLLGRPWIH AGAV+STLHQKLK+V
Sbjct: 883 GSKRSVCGEVTLPVKIGPEVFDIIFYVMDIQPAYSCLLGRPWIHAAGAVSSTLHQKLKYV 942
Query: 977 KNGKLVTVNGEEALLVSHLSSFSFIGAD-DVEGTPFQGF------TIEDKNAKRNEASIS 1029
NG++VTV GEE +LVSHLSSF ++ D ++ T Q F + ++ ASI+
Sbjct: 943 WNGQIVTVCGEEEILVSHLSSFKYVEVDGEIHETLCQAFETVALEKVAYAEQRKPGASIT 1002
Query: 1030 SLKDAQKVIQAGGSASWGKLIELPENKHREGLGFFPSTGLSTAKRGISTVRALSMQS 1086
S K A++V+ +G + WGK++ LP + + G+G+ P + G ST + + +
Sbjct: 1003 SYKQAKEVVDSGKAEGWGKMVYLPVKEDKFGVGYEPLQAEQNGQAGPSTFTSAGLMN 1059
>gb|AAQ82035.1| gag/pol polyprotein [Pisum sativum]
Length = 1105
Score = 579 bits (1493), Expect = e-163
Identities = 380/1140 (33%), Positives = 582/1140 (50%), Gaps = 143/1140 (12%)
Query: 3 EMAELIKTMAETQTQAQAQIQAQAQTQAQAVTEAQAR---SQAPPPPPPVRTQAEASSSW 59
E+AE+ MA+ Q Q Q + +A + A + APP PV
Sbjct: 7 ELAEMKANMAQFMNMMQGVAQGQEELRAMVQRQEAAIPPVNHAPPEGGPVNDN------- 59
Query: 60 TLCADTPTRSAPQRSTPWFPPFTAGEIFRPITCEAQMPTHQYTAQVPLPAMRVTPATMTY 119
+ A P + + G+ I +Q P + T
Sbjct: 60 NVAAAVPINN-----------YDVGDELGGIRINSQ------------PIVPDAANARTI 96
Query: 120 SAPVIHTIP--QTEEPIFH--SGNAEAYEEVSDLREKYDELRRDMKALHEKGKFGKTAYD 175
AP + +P +E +F S + + V + K D L ++A+ + +
Sbjct: 97 RAPARNPVPIVDRQEDMFTLLSEDEDVLGRVDERDRKVDALAEKIRAMECQNSLSFDVTN 156
Query: 176 LCLVPSVQVPHKFKIPDFEKYKGSSCPEEHLKMYVRRMPAYAQDDQILIYYFQESLTGPA 235
+ LV +++P+KFK P F+KY +SCP H++ Y R++ AY D+++ +Y+FQ+SL+G +
Sbjct: 157 MGLVEGLRIPYKFKAPSFDKYNDTSCPRTHVQAYYRKISAYTDDEKMWMYFFQDSLSGAS 216
Query: 236 SKWYTNLDKTRVQTFRDLCEAFVEQYSYNVDMTPDRSDLQAMTQGDKETFKEYAQRWRDT 295
WY L + ++ +RDL EAF+ QY +N+DM P R+ LQ++ Q E+FKEYAQRWR+
Sbjct: 217 LDWYMELKRDSIRCWRDLGEAFLRQYKHNMDMAPSRTQLQSLCQKSGESFKEYAQRWREL 276
Query: 296 AAQVSPRIEEKEMTKLFLKTLNHFYYKKMVGSTP-KSFAEMVGMGVQLEEGVREGRLVKD 354
AA+V P + E+E+T +F+ TL + +M GS P SF+++V G + E ++ G++
Sbjct: 277 AARVQPPMLERELTDMFIGTLQGVFMDRM-GSCPFGSFSDVVICGERTESLIKTGKI--Q 333
Query: 355 ATPASGTKKTGNHFPRKKEQEVGMVTHGRPQQTYPAYQHIAAITPTSHPFQQTNNHPQIP 414
+S +KK PR++E E +V H R Q Q A P P QQ Q P
Sbjct: 334 DVGSSSSKKPFAGAPRRREGETNVVQHRRDQNRIEYRQAAAVTIPAPQPRQQQQQRVQQP 393
Query: 415 QYPQIPQYPQIPQYPQFPQNPSPQNTQQQNFQQQPYQQQPYQQFPYQQYPQQNFQQQPYQ 474
Q QQ QQ+P QP Q+ P +++
Sbjct: 394 Q--------------------------QQQQQQRP--YQPRQRMPDRRF----------- 414
Query: 475 QRPQQPRPPRMPINPIPVTYAELLPGLLKKNLVQTRTAPPIPEKLPSWYRLDQTCDFHEG 534
+ +P++YAELLP LL+ +V+ RT P P LP Y + CDFH G
Sbjct: 415 -------------DSLPMSYAELLPELLRLGMVELRTMAP-PTVLPPGYDANVRCDFHSG 460
Query: 535 GRGHNIETCYAFKSIVQRLINDGKITFTDSAPNVQTNPLPNHGAATVNMIEDCQKTRPIL 594
GH+ E C A + VQ LI+ I F PNV NP+P HG VN IE + ++
Sbjct: 461 APGHHTEKCRALQHKVQDLIDAKAINFA-PVPNVVNNPMPQHGGHRVNNIEGKEAEDLVV 519
Query: 595 NVQHIRTPLVPLHAKLCKVDLFEHDHDLCEICLMNSGGCQKVRNDIQGLLNRGELVVERR 654
NV ++T L+ + +L ++ + C C + GC ++R IQG+++ G L R
Sbjct: 520 NVDDVQTSLLVVKGRLLNGGVYSGCDEDCLGCAESENGCDQLRAGIQGMMDEGCLQFSRA 579
Query: 655 CDD---VCVIT-----PEGPLEVFYDSRKSTITPLVICLP------------GPLPYASE 694
D V IT EG + + + TP+ I +P G +
Sbjct: 580 VKDRGTVSTITIYFKPTEGRGQRVVSAPATNDTPVTISVPVTISAPTTIVASGRRAVENS 639
Query: 695 KAIPYKYNATMIEEGR-------------------EVPIPPLSSVDNIVEDSRVLRNGRV 735
+ +P+KY+ R VP +VDN+ R+GR+
Sbjct: 640 RVVPWKYDNAYRSNRRAESQTRPVNQAPVTIGLPNRVPATVGPAVDNVGGPGGFTRSGRL 699
Query: 736 VPIVFPKKIDATINKEVRTKDAGIAKE-VDQPNGAGT-SAEFDEILKLIKKSEYKVVDQL 793
+ +A + + K A + +E V + GT + +E +K+IKKS+YK+VDQL
Sbjct: 700 FAPQPLRDNNAEALAKAKGKQAVVEEEPVQKEAPEGTFEKDVEEFMKIIKKSDYKIVDQL 759
Query: 794 MQTPSKISIMSLLLNSEAHKDALMKVLEQAFVDYDVMVGQFGGIVGNITACNNLSFSDEE 853
QTPSKISI+SLL+ SEAH++AL+K+L A+V ++ V Q G++ N++ + + F++ +
Sbjct: 760 NQTPSKISILSLLMCSEAHRNALLKMLNLAYVPQEISVNQLEGVMANVSTRHGVGFTNLD 819
Query: 854 LPAEGRNHNRALHISVNCKTDALSNVLVDTGSSLNVLSKTTYAQLAYQDAPLRRSGVMVK 913
LP EGRNHN+ALHI++ CK LS+VLVDTGSSLNVL ++ + L S ++V+
Sbjct: 820 LPPEGRNHNKALHITMECKGAVLSHVLVDTGSSLNVLPNQILKKIDVEGFVLTPSDLIVR 879
Query: 914 AFDGSRKDVLGEVVLPITVGPQVFQVNFQVMDIQASYSCLLGRPWIHEAGAVTSTLHQKL 973
AFDGS++ V GEV LP+ +GP+VF + F VMDIQ +YSCLLGRPWIH AGAV+STLHQKL
Sbjct: 880 AFDGSKRSVCGEVTLPVKIGPEVFDIIFYVMDIQPAYSCLLGRPWIHAAGAVSSTLHQKL 939
Query: 974 KFVKNGKLVTVNGEEALLVSHLSSFSFIGAD-DVEGTPFQGF------TIEDKNAKRNEA 1026
K+V NG++VTV GEE +LVSHLSSF ++ D ++ T Q F + ++ A
Sbjct: 940 KYVWNGQIVTVCGEEEILVSHLSSFKYVEVDGEIHETLCQAFETVALEKVAYAEQRKPGA 999
Query: 1027 SISSLKDAQKVIQAGGSASWGKLIELPENKHREGLGFFPSTGLSTAKRGISTVRALSMQS 1086
SI+S K A++V+ +G + WGK+++LP + + G+G+ P + G ST + + +
Sbjct: 1000 SITSYKQAKEVVDSGKAEGWGKMVDLPVKEDKFGVGYEPLRAEQNGQAGPSTFTSAGLMN 1059
>gb|AAU90285.1| putative gag/pol polyprotein, 3'-partial [Solanum demissum]
Length = 1784
Score = 218 bits (554), Expect = 3e-54
Identities = 165/542 (30%), Positives = 265/542 (48%), Gaps = 79/542 (14%)
Query: 489 PIPVTYAELLPGLLKKNL---VQTRTAPPIPEKLPSWYRLDQTCDFHEGGRGHNIETCYA 545
PI +YA L L N+ ++ + + P P L Q C + GHNIE C+
Sbjct: 894 PIGESYASLFQKLRTLNVLSPIERKMSNPPPRNLD----YSQHCAYCSDAPGHNIERCWY 949
Query: 546 FKSIVQRLINDGKITF-TDSAPNVQTNPLPNHGAATVNMIEDCQKTRPILNVQHIRTPLV 604
K +Q LI+ +I + + PN+ NPLP H NM+E + N + + P
Sbjct: 950 LKKAIQDLIDTRRIIVESPNGPNINQNPLPRH--TETNMLE------MVNNHEEVAVPYK 1001
Query: 605 PLHAKLCKVDL-FEHDHDLCEICLMNSGGCQKVRNDIQGLLNRGELVVERRCDDVC---- 659
P+ KV+ E ++ ++ + G + + + L ++ +DV
Sbjct: 1002 PI----LKVETGMESSTNVIDLTKIMPSGAESTSEKLTPS-SAPILTIKGALEDVWASQR 1056
Query: 660 ---VITPEGPLEVFYDSRKSTITPLVICLPGPLPYASEKAIPYKYNATMIE-EGREVPIP 715
++ P GP + + + I P++I LP + KA+P+ Y T++ EG+E+
Sbjct: 1057 EARLVVPRGPDKPILIVQGAYIPPVIIRPVSQLPMTNPKAVPWNYEPTVVTYEGKEIN-- 1114
Query: 716 PLSSVDNIVEDSRVLRNGRVVPIVFPKKIDATINKEVRTKDAGIAKEVDQPNGAGTSAEF 775
+ + E + R+GR + E+R K+ +V P T E
Sbjct: 1115 -----EEVDEIGGMTRSGRCYALT-----------ELR-KNKNDQMQVKSPV---TEGEA 1154
Query: 776 DEILKLIKKSEYKVVDQLMQTPSKISIMSLLLNSEAHKDALMKVLEQAFVDYDVMVGQFG 835
+E L+ +K S+Y +V+QL +TP++IS++SLL++S+AH+ A++K+L +A V +V V Q
Sbjct: 1155 EEFLRKMKLSDYSIVEQLRKTPAQISLLSLLIHSDAHRKAVIKILNEAHVPNEVTVSQLE 1214
Query: 836 GIVGNITACNNLSFSDEELPAEGRNHNRALHISVNCKTDALSNVLVDTGSSLNVLSKTTY 895
I G I N ++FSD+ELP EG S N+ +T
Sbjct: 1215 KIAGRIFEVNRITFSDDELPVEG--------------------------SGANICPLSTL 1248
Query: 896 AQLAYQDAPLRRSGVMVKAFDGSRKDVLGEVVLPITVGPQVFQVNFQVMDIQASYSCLLG 955
+L +R + V V+AFDG++ D +GE+ L + +G F VNFQV++I ASY+ LLG
Sbjct: 1249 QKLNVNVERVRPNNVCVRAFDGTKTDAIGEIELILKIGHVDFTVNFQVLNINASYNLLLG 1308
Query: 956 RPWIHEAGAVTSTLHQKLKFVKNGKLVTVNGEEALLVSHLSSFSFIGA-DDVEGTPFQGF 1014
RPW+H AGAV STLHQ +KF + + V V+ E L V SS FI A ++ E +Q F
Sbjct: 1309 RPWVHRAGAVPSTLHQMVKFEYDRQEVIVHSEGDLSVYKDSSLPFIKANNENEALVYQAF 1368
Query: 1015 TI 1016
+
Sbjct: 1369 EV 1370
Score = 131 bits (329), Expect = 3e-28
Identities = 88/303 (29%), Positives = 146/303 (48%), Gaps = 43/303 (14%)
Query: 99 HQYTAQVPLPAMRVTPATMTYSAPVIHTIPQTEEPIFHSGNAEAYEEVSDLREKYDELRR 158
H YT + L P T YS+P I + E +EE++ + ++ R
Sbjct: 639 HGYTPEEALKIPSSYPQTHQYSSPF---------KIEKTVKNEEHEEMAKKMKSLEQSIR 689
Query: 159 DMKALHEKGKFGKTAYDLCLVPSVQVPHKFKIPDFEKYKGSSCPEEHLKMYVRRMPAYAQ 218
DM+ L G G + DLC+ P V +P G+ EE
Sbjct: 690 DMQGLG--GHKGISFSDLCMFPHVHLP-----------TGAEGKEE-------------- 722
Query: 219 DDQILIYYFQESLTGPASKWYTNLDKTRVQTFRDLCEAFVEQYSYNVDMTPDRSDLQAMT 278
+L+ YF ESL G AS+W+ + D T+ +L FV+Q+ YN+D+ PDRS L M
Sbjct: 723 ---LLMAYFGESLVGIASEWFIDQDIANWHTWDNLARCFVQQFQYNIDIVPDRSSLANMR 779
Query: 279 QGDKETFKEYAQRWRDTAAQVSPRIEEKEMTKLFLKTLNHFYYKKMVGSTPKSFAEMVGM 338
+ E F+EYA RWR+ AA+V ++E EM +FL+ Y+ ++ + K+F E++ +
Sbjct: 780 KKTTENFREYAVRWREQAARVKLSMKESEMIDVFLQAQEPDYFHYLLSAVGKTFVEVIKV 839
Query: 339 GVQLEEGVREGRLVKDATPASGTKK----TGNHFPRKKEQEVGMVTHGRPQQTYPAYQHI 394
G +E G++ G++V A + T+ +GN +K+ ++V V +G + P +
Sbjct: 840 GEMVENGIKSGKIVSQAALKATTQALQNGSGNIGGKKRREDVATVGNGAKDEFTPIGESY 899
Query: 395 AAI 397
A++
Sbjct: 900 ASL 902
>gb|AAT39964.1| putative gag polyprotein [Solanum demissum]
Length = 421
Score = 177 bits (450), Expect = 3e-42
Identities = 110/322 (34%), Positives = 165/322 (51%), Gaps = 28/322 (8%)
Query: 99 HQYTAQVPLPAMRVTPATMTYSAPVIHTIPQTEEPIFHSGNAEAYEEVSDLREKYDELRR 158
H YT + L P T YS+P I + E +EE++ + + R
Sbjct: 119 HGYTPEEALKIPSSYPQTHQYSSPF---------KIEKTVKNEEHEEMTKKMKSLEHSIR 169
Query: 159 DMKALHEKGKFGKTAYDLCLVPSVQVPHKFKIPDFEKYKGSSCPEEHLKMYVRRMPAYAQ 218
DM+ L G G + DLC+ P V +P FK P+FEKY G P HLK Y ++
Sbjct: 170 DMQGLG--GHKGISFSDLCMFPHVHLPTGFKTPNFEKYDGHGDPIAHLKRYYNQLRGAEG 227
Query: 219 DDQILIYYFQESLTGPASKWYTNLDKTRVQTFRDLCEAFVEQYSYNVDMTPDRSDLQAMT 278
+++L+ YF ESL G ASKW+ + D T+ DL FV+Q+ YN+D+ PDRS L M
Sbjct: 228 KEELLMAYFGESLVGIASKWFIDQDIANWHTWDDLARCFVQQFQYNIDIVPDRSSLANMR 287
Query: 279 QGDKETFKEYAQRWRDTAAQVSPRIEEKEMTKLFLKTLNHFYYKKMVGSTPKSFAEMVGM 338
+ E F+EYA RWR+ AA+V P ++E EM +FL+ Y+ ++ + K+F E++ +
Sbjct: 288 KKTTENFREYAVRWREQAARVKPPMKELEMIDVFLQAQEPDYFHYLLSAVGKTFTEVIKV 347
Query: 339 GVQLEEGVREGRLVKDATPASGTKK----TGNHFPRKKEQEVGMVTHGRPQQTYPAYQHI 394
G +E G++ G++V A + T+ +GN +K+ ++V V +TY H
Sbjct: 348 GEMVENGIKSGKIVSQAALKATTQALPNGSGNIGGKKRREDVATVVSA--PRTYAQDNH- 404
Query: 395 AAITPTSHPFQQTNNHPQIPQY 416
T H F PQIPQY
Sbjct: 405 -----TQHYFP-----PQIPQY 416
>pir||T09671 RPE15 protein - alfalfa (fragment) gi|840619|gb|AAB50037.1| RPE15
gene product
Length = 219
Score = 121 bits (304), Expect = 3e-25
Identities = 87/222 (39%), Positives = 113/222 (50%), Gaps = 40/222 (18%)
Query: 296 AAQVSPRIEEKEMTKLFLKTLNHFYYKKMVGSTPKSFAEMVGMGVQLEEGVREGRLV--K 353
AA+VSPR++E+E+T+ FLKTL + ++M+ S P +F++MV MG +LEE VREG +V K
Sbjct: 14 AARVSPRLDEEELTQTFLKTLKKEHKERMIVSAPNNFSDMVTMGTRLEEAVREGIIVLEK 73
Query: 354 DATPASGTKKTGN-HFPRKKEQEVGMVT--HGRPQQTYPAYQHIAAITPTSHPFQQTNNH 410
AS KK GN H RK+ ++VGMV+ H P A P + + Q + H
Sbjct: 74 GECSASAPKKYGNGHHKRKESKKVGMVSGNHDTPVN--------ATQLPLPYQYMQISQH 125
Query: 411 PQIPQYPQIPQYPQIPQYPQFPQNPSPQNTQQQNFQQQPYQQQPYQQFPYQQYPQQNFQQ 470
P P + Q QYP P P P P N Q Q QP QQ
Sbjct: 126 PFFPPFYQ--QYPLPPGQP-----PVPVNAMAQQVQHQPPAQQ----------------- 161
Query: 471 QPYQQRPQQPRPPRMPINPIPVTYAELLPGLLKKNLVQTRTA 512
QQ+ QQ P+ PIP+ YAELLP LL K TR A
Sbjct: 162 ---QQQQQQGPRPKTYFPPIPMRYAELLPTLLAKGHCVTRPA 200
>ref|XP_493776.1| unnamed protein product [Oryza sativa (japonica cultivar-group)]
gi|9927274|dbj|BAB08213.2| Similar to Arabidopsis
thaliana chromosome II BAC F26H6; putative retroelement
pol polyprotein (AC006920) [Oryza sativa (japonica
cultivar-group)]
Length = 2876
Score = 117 bits (292), Expect = 6e-24
Identities = 86/291 (29%), Positives = 142/291 (48%), Gaps = 18/291 (6%)
Query: 710 REVPIPPLSSVDNIVEDSRV-LRNGRVVPIVFPKKIDATINKEVRTKDAGIAKEVDQPNG 768
RE+ L VE+ + LR G+ +P K+ ++K AK+ P
Sbjct: 1237 REIFHAELDPEKTKVEEVNISLRGGKTLPDPHKSKVP-NVDKP--------AKKASPPGE 1287
Query: 769 AGTSAEFDEILKLIKKSEYKVVDQLMQTPSKISIMSLLLNSEAHKDALMKVLEQAFVDYD 828
A + E K +YKV+ L + P+ +S+ L+ ++AL+K L+ V Y+
Sbjct: 1288 APEAPETKTGSKEKPAVDYKVLAHLKRIPALLSVYDALMMVPDLREALIKALQAPEV-YE 1346
Query: 829 VMVGQFGGIVGNITACNNLSFSDEELPAEGRNHNRALHISVNCKTDALSNVLVDTGSSLN 888
V + + + N N ++F+DE+ +G +HNR L+I N + L +L+D GS++N
Sbjct: 1347 VDMAKHR-LYDNPLFVNEITFADEDNIIKGGDHNRPLYIEGNIGSAHLRRILIDLGSAVN 1405
Query: 889 VLSKTTYAQLAYQDAPLRRSGVMVKAFDGSRKDVLGEVVLPITVGPQVFQVNFQVMDIQA 948
+L + + + L V++ FD K LG + + I + F+V F V++
Sbjct: 1406 ILPVRSLTRAGFTTKDLEPIDVVICGFDNQGKPTLGAITIKIQMSTFSFKVRFFVIEANT 1465
Query: 949 SYSCLLGRPWIHEAGAVTSTLHQKLKFVKNGKLVTVNGEEALLVSHLSSFS 999
SYS LLGRPWIH+ V STLHQ LKF+ NG + + S+ S ++
Sbjct: 1466 SYSALLGRPWIHKYRVVPSTLHQCLKFLDG------NGVQQRITSNFSPYT 1510
>emb|CAD41628.3| OSJNBa0091D06.8 [Oryza sativa (japonica cultivar-group)]
gi|50926818|ref|XP_473330.1| OSJNBa0091D06.8 [Oryza
sativa (japonica cultivar-group)]
Length = 2657
Score = 116 bits (290), Expect = 1e-23
Identities = 75/247 (30%), Positives = 130/247 (52%), Gaps = 8/247 (3%)
Query: 753 RTKDAGIAKEVDQPNGAGTSAEFDEILKLIKKSEYKVVDQLMQTPSKISIMSLLLNSEAH 812
+ K + + K + + + G + E K +Y V+ L + P+ +S+ L+
Sbjct: 1354 KAKPSKVDKPIKEASPPGEAPETKARSKEKPDIDYNVLAHLKRIPALLSVYDALILVPDL 1413
Query: 813 KDALMKVLEQAFVDYDVMVGQFGGIVGNITACNNLSFSDEELPAEGRNHNRALHISVNCK 872
++AL+K L+ V Y+V + + + N N ++F+DE+ EG +HNR L+I N
Sbjct: 1414 REALIKALQTPEV-YEVDMAKHR-LFNNPLFVNEITFADEDNIIEGSDHNRPLYIEGNIG 1471
Query: 873 TDALSNVLVDTGSSLNVLSKTTYAQLAYQDAPLRRSGVMVKAFDGSRKDVLGEVVLPITV 932
L +L+D GS++N+L + + + L + +++ FD K LG + + I +
Sbjct: 1472 PAHLRRILIDPGSAVNILPVRSLTRAGFTTKDLEPTDMVICGFDNQGKSTLGAITVKIQM 1531
Query: 933 GPQVFQVNFQVMDIQASYSCLLGRPWIHEAGAVTSTLHQKLKFVKNGKLVTVNGEEALLV 992
F+V+F V++ SYS LLGRPWIH+ V STLHQ LKF+ +GK G + +V
Sbjct: 1532 STFSFKVHFFVIEANTSYSALLGRPWIHKYRVVPSTLHQCLKFL-DGK-----GAQQRIV 1585
Query: 993 SHLSSFS 999
+ SS++
Sbjct: 1586 GNSSSYT 1592
Score = 47.0 bits (110), Expect = 0.008
Identities = 31/136 (22%), Positives = 63/136 (45%), Gaps = 5/136 (3%)
Query: 182 VQVPHKFKIPDFEKYKGSSCPEEHLKMYVRRMPAYAQDDQILIYYFQESLTGPASKWYTN 241
V P ++ P F ++ + EHL + A + +L+ F SLTGPA WY+
Sbjct: 842 VAFPARWHPPKFRQFDRTGDAREHLAYFEAACGDTANNSSLLLRQFSGSLTGPAFHWYSR 901
Query: 242 LDKTRVQTFRDLCEAFVEQYSYNVDMTPDRS--DLQAMTQGDKETFKEYAQRWRDTAAQV 299
L ++++ + E + + V M D S +L + Q E +Y R+R++ ++
Sbjct: 902 LPTGSIRSWASMKEVLKKHF---VAMKKDFSIVELSQVRQRRDEAIDDYVIRFRNSFVRL 958
Query: 300 SPRIEEKEMTKLFLKT 315
+ + ++ ++ L +
Sbjct: 959 AREMHLEDAIEIALSS 974
>emb|CAC44143.1| putative gag polyprotein [Cicer arietinum]
Length = 88
Score = 111 bits (277), Expect = 3e-22
Identities = 43/88 (48%), Positives = 70/88 (78%)
Query: 176 LCLVPSVQVPHKFKIPDFEKYKGSSCPEEHLKMYVRRMPAYAQDDQILIYYFQESLTGPA 235
+CLVP V +P KFK+ +FEKYKG++CP+ HL MY R+M ++A +D++LI++FQ+SL+G +
Sbjct: 1 MCLVPDVTIPRKFKVLEFEKYKGATCPKNHLTMYGRKMASHAHNDKLLIHFFQDSLSGAS 60
Query: 236 SKWYTNLDKTRVQTFRDLCEAFVEQYSY 263
WY +L++ R+ +++DL +AF+ QY Y
Sbjct: 61 LSWYMHLERNRIHSWKDLADAFLRQYKY 88
>gb|AAU89777.1| gag-pol polyprotein-like [Solanum tuberosum]
Length = 1716
Score = 99.8 bits (247), Expect = 1e-18
Identities = 47/138 (34%), Positives = 81/138 (58%)
Query: 773 AEFDEILKLIKKSEYKVVDQLMQTPSKISIMSLLLNSEAHKDALMKVLEQAFVDYDVMVG 832
A+ +E + ++ +Y +V L +TP++IS+ +LL++S+ H+ ALMK L+ +V
Sbjct: 614 AKAEEFWRRMQPKDYSIVKYLEKTPAQISVWALLMSSQLHRQALMKALDDTYVPVGTNSD 673
Query: 833 QFGGIVGNITACNNLSFSDEELPAEGRNHNRALHISVNCKTDALSNVLVDTGSSLNVLSK 892
++ + + +SF DEELP EGR HN+ALH++ C+ ++ VLVD GS LN+
Sbjct: 674 NLAAMINQVIRGHRISFCDEELPFEGRMHNKALHVTTMCRDKIINRVLVDDGSGLNICPL 733
Query: 893 TTYAQLAYQDAPLRRSGV 910
+T L + L ++ V
Sbjct: 734 STLRHLKFDLGKLHQNQV 751
Score = 70.5 bits (171), Expect = 7e-10
Identities = 31/64 (48%), Positives = 39/64 (60%)
Query: 522 WYRLDQTCDFHEGGRGHNIETCYAFKSIVQRLINDGKITFTDSAPNVQTNPLPNHGAATV 581
+YR DQ C +H GH+ E C K +Q LIN ++ APNV +NPLPNHG T+
Sbjct: 457 FYRPDQRCAYHSNSVGHDTEDCINLKHKIQDLINQNVVSLQTVAPNVNSNPLPNHGGITI 516
Query: 582 NMIE 585
NMIE
Sbjct: 517 NMIE 520
Score = 61.2 bits (147), Expect = 4e-07
Identities = 36/117 (30%), Positives = 63/117 (53%), Gaps = 21/117 (17%)
Query: 266 DMTPDRSDLQAMTQGDKETFKEYAQRWRDTAAQVSPRIEEKEMTKLFLKTLNHFYYKKMV 325
++ PDR L+ M Q E+++E+A RWR AA+V P + EKE+ ++F++
Sbjct: 296 EIVPDRYSLEKMKQKSTESYREFAYRWRKEAARVRPPMSEKEIVEVFVRV---------- 345
Query: 326 GSTPKSFAEMVGMGVQLEEGVREGRLV-KDATPASGTKKTGNHFPRKKEQEVGMVTH 381
FAE+V +G +E+G+R G++V A+P S +KK +V +++
Sbjct: 346 ----AKFAEIVKIGETIEDGLRTGKIVYVAASPESSI------LLKKKRDDVSSISY 392
>gb|AAT39963.1| putative polyprotein [Solanum demissum]
Length = 1483
Score = 87.4 bits (215), Expect = 5e-15
Identities = 55/171 (32%), Positives = 92/171 (53%), Gaps = 23/171 (13%)
Query: 688 PLPYASEKAIPYKYNATMIE-EGREVPIPPLSSVDNIVEDSRVLRNGRVVPIVFPKKIDA 746
PLP + KA+P+KY T++ +G+E+ VD I +R R P ++
Sbjct: 65 PLPMTNPKAVPWKYEPTVVTYKGKEIN----EKVDEIEGMTRSRR------CYAPAELRK 114
Query: 747 TINKEVRTKDAGIAKEVDQPNGAGTSAEFDEILKLIKKSEYKVVDQLMQTPSKISIMSLL 806
N +++ K T E +E L+ +K S+Y +V+QL +TP++IS++SLL
Sbjct: 115 NNNDQMQVKSPV------------TEREAEEFLRKMKLSDYSIVEQLRKTPAQISLLSLL 162
Query: 807 LNSEAHKDALMKVLEQAFVDYDVMVGQFGGIVGNITACNNLSFSDEELPAE 857
++S+ H+ +MK+L +A V + + Q I G I N ++FSD+ELP E
Sbjct: 163 IHSDEHRKVVMKILNEAHVPNEGTMSQLEKIAGRIFEVNCITFSDDELPVE 213
Score = 86.7 bits (213), Expect = 9e-15
Identities = 47/98 (47%), Positives = 64/98 (64%), Gaps = 1/98 (1%)
Query: 920 KDVLGEVVLPITVGPQVFQVNFQVMDIQASYSCLLGRPWIHEAGAVTSTLHQKLKFVKNG 979
+D +GE+ L + +GP F VNFQV++I ASY+ LLGRPW+H AGAV STLHQ +KF +
Sbjct: 213 EDAIGEIELILKIGPVDFTVNFQVLNINASYNLLLGRPWVHRAGAVPSTLHQMVKFEYDR 272
Query: 980 KLVTVNGEEALLVSHLSSFSFIGA-DDVEGTPFQGFTI 1016
+ V V+ E L V SS FI A ++ E +Q F +
Sbjct: 273 QEVIVHCERDLSVYKDSSLPFIKANNENEALVYQAFEV 310
>gb|AAG35598.1| secalin precursor [Secale cereale]
Length = 455
Score = 86.7 bits (213), Expect = 9e-15
Identities = 59/125 (47%), Positives = 69/125 (55%), Gaps = 18/125 (14%)
Query: 383 RPQQTYPAYQHIAAITPTSHPF----QQTNNHPQIP----QYPQIPQ--YPQIPQ--YPQ 430
+PQQ+ P + PF QQ++ PQ P +PQ PQ YPQ PQ +PQ
Sbjct: 39 QPQQSSPQQPQQPFPQQSQQPFPQQPQQSSPQPQQPYPQQPFPQQPQQPYPQQPQQPFPQ 98
Query: 431 FPQNPSPQNTQQQNFQ--QQPYQQQPYQQFPYQQYPQQNFQQQPYQQRPQQPRP--PRMP 486
PQ P PQ QQQ Q QQP QQP QQFP Q PQQ F QQP QQ PQQP+ P+ P
Sbjct: 99 QPQQPYPQQPQQQFPQQPQQPVPQQPLQQFP--QQPQQPFPQQPLQQFPQQPQQPFPQQP 156
Query: 487 INPIP 491
P+P
Sbjct: 157 QQPVP 161
Score = 78.6 bits (192), Expect = 2e-12
Identities = 63/148 (42%), Positives = 74/148 (49%), Gaps = 30/148 (20%)
Query: 368 FPRKKEQEVGMVTHGRPQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQ--YPQIPQ--YP 423
FP++ +Q +PQQ+ P Q P P Q +PQ PQ +PQ PQ YP
Sbjct: 52 FPQQSQQPFPQ----QPQQSSPQPQQPYPQQP--FPQQPQQPYPQQPQQPFPQQPQQPYP 105
Query: 424 QIPQ--YPQFPQNPSPQNTQQQNFQQ--QPYQQQPYQQFPYQQYPQQNFQQQPYQ----- 474
Q PQ +PQ PQ P PQ QQ QQ QP+ QQP QQFP Q PQQ F QQP Q
Sbjct: 106 QQPQQQFPQQPQQPVPQQPLQQFPQQPQQPFPQQPLQQFPQQ--PQQPFPQQPQQPVPQQ 163
Query: 475 --------QRPQQPRP-PRMPINPIPVT 493
Q+PQQP P P+ P P T
Sbjct: 164 SQQPFPQTQQPQQPFPQPQQPQQLFPQT 191
Score = 74.7 bits (182), Expect = 4e-11
Identities = 56/135 (41%), Positives = 67/135 (49%), Gaps = 18/135 (13%)
Query: 399 PTSHPFQQTN-NHPQIPQYPQIPQYPQIPQYPQFPQNPSPQNTQ---QQNFQQQP---YQ 451
P PF Q + PQ PQ P PQ Q P +PQ PQ SPQ Q QQ F QQP Y
Sbjct: 32 PQQQPFPQPQQSSPQQPQQP-FPQQSQQP-FPQQPQQSSPQPQQPYPQQPFPQQPQQPYP 89
Query: 452 QQPYQQFP------YQQYPQQNFQQQPYQQRPQQP--RPPRMPINPIPVTYAELLPGLLK 503
QQP Q FP Y Q PQQ F QQP Q PQQP + P+ P P P + P +
Sbjct: 90 QQPQQPFPQQPQQPYPQQPQQQFPQQPQQPVPQQPLQQFPQQPQQPFPQQPLQQFPQQPQ 149
Query: 504 KNLVQTRTAPPIPEK 518
+ Q + P+P++
Sbjct: 150 QPFPQ-QPQQPVPQQ 163
Score = 68.2 bits (165), Expect = 3e-09
Identities = 47/119 (39%), Positives = 55/119 (45%), Gaps = 20/119 (16%)
Query: 383 RPQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQ--YPQI----PQYPQ------ 430
+PQQ +P + PF QT Q PQ PQ +PQ PQ PQ
Sbjct: 147 QPQQPFPQQPQQPVPQQSQQPFPQTQQPQQPFPQPQQPQQLFPQTQQSSPQQPQQVTSQP 206
Query: 431 ---FPQNPSPQNTQQQNFQQQPYQQQPYQQFPYQQYPQQNFQQQPYQQRPQQPRPPRMP 486
FPQ PQ + Q+ QQPY Q+P Q FP Q PQQ F P Q+PQQP P P
Sbjct: 207 QQPFPQAQPPQQSSPQS--QQPYPQEPQQLFPQSQQPQQPF---PQPQQPQQPFPQPQP 260
Score = 67.8 bits (164), Expect = 4e-09
Identities = 56/140 (40%), Positives = 62/140 (44%), Gaps = 21/140 (15%)
Query: 368 FPRKKEQEVGMVTHGRPQQTYPAYQHIAAITPTSHPFQQTNNHP-------QIPQYPQ-- 418
FP++ +Q PQQ +P PF Q P Q PQ PQ
Sbjct: 60 FPQQPQQSSPQPQQPYPQQPFPQQPQQPYPQQPQQPFPQQPQQPYPQQPQQQFPQQPQQP 119
Query: 419 IPQYP--QIPQYPQ--FPQNPSPQNTQQ--QNFQQQPYQ---QQPYQQFPYQQYPQQNF- 468
+PQ P Q PQ PQ FPQ P Q QQ Q F QQP Q QQ Q FP Q PQQ F
Sbjct: 120 VPQQPLQQFPQQPQQPFPQQPLQQFPQQPQQPFPQQPQQPVPQQSQQPFPQTQQPQQPFP 179
Query: 469 -QQQPYQQRPQ-QPRPPRMP 486
QQP Q PQ Q P+ P
Sbjct: 180 QPQQPQQLFPQTQQSSPQQP 199
Score = 66.6 bits (161), Expect = 1e-08
Identities = 45/101 (44%), Positives = 52/101 (50%), Gaps = 8/101 (7%)
Query: 394 IAAITPTSHPFQQTNNHPQIPQYPQIPQYPQIPQY-PQFPQNPSPQNTQQQNFQQQPYQQ 452
+A T + Q N Q+ Q PQ +PQ Q PQ PQ P PQ +QQ F QQP Q
Sbjct: 10 LAMATTIATANMQVNPSGQV-QCPQQQPFPQPQQSSPQQPQQPFPQQSQQP-FPQQPQQS 67
Query: 453 QPYQQFPYQQYPQQNFQQQPYQQRPQQPRP--PRMPINPIP 491
P P Q YPQQ F QQP Q PQQP+ P+ P P P
Sbjct: 68 SPQ---PQQPYPQQPFPQQPQQPYPQQPQQPFPQQPQQPYP 105
Score = 64.3 bits (155), Expect = 5e-08
Identities = 59/158 (37%), Positives = 69/158 (43%), Gaps = 41/158 (25%)
Query: 368 FPRKKEQEVGMVTHGRPQQTYPAYQHIAAITPTSHPFQQTNNHPQIP-------QYPQIP 420
FP++ +Q +PQQ +P Q P P QQ PQ P Q+PQ P
Sbjct: 96 FPQQPQQPYPQ----QPQQQFP--QQPQQPVP-QQPLQQFPQQPQQPFPQQPLQQFPQQP 148
Query: 421 Q--YPQIPQYP------------QFPQNPSPQNTQ--------QQNFQQQPYQ--QQPYQ 456
Q +PQ PQ P Q PQ P PQ Q QQ+ QQP Q QP Q
Sbjct: 149 QQPFPQQPQQPVPQQSQQPFPQTQQPQQPFPQPQQPQQLFPQTQQSSPQQPQQVTSQPQQ 208
Query: 457 QFPYQQYPQQNF--QQQPYQQRPQQPRP-PRMPINPIP 491
FP Q PQQ+ QQPY Q PQQ P + P P P
Sbjct: 209 PFPQAQPPQQSSPQSQQPYPQEPQQLFPQSQQPQQPFP 246
Score = 57.4 bits (137), Expect = 6e-06
Identities = 46/121 (38%), Positives = 56/121 (46%), Gaps = 10/121 (8%)
Query: 369 PRKKEQEVGMVTHGRPQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQ--YPQIP 426
P++ +Q PQQ P + P QQ++ Q P YPQ PQ +PQ
Sbjct: 181 PQQPQQLFPQTQQSSPQQPQQVTSQPQQPFPQAQPPQQSSPQSQQP-YPQEPQQLFPQ-S 238
Query: 427 QYPQFPQNPSPQNTQQQNFQQQPYQQQ----PYQQFPYQQYP-QQNFQQQPYQQRPQQPR 481
Q PQ P P PQ QQ Q QP QQ P Q FP Q P Q+ +Q P +PQQP
Sbjct: 239 QQPQQPF-PQPQQPQQPFPQPQPQTQQSIPQPQQPFPQPQQPFPQSQEQFPQVHQPQQPS 297
Query: 482 P 482
P
Sbjct: 298 P 298
Score = 48.9 bits (115), Expect = 0.002
Identities = 40/119 (33%), Positives = 48/119 (39%), Gaps = 9/119 (7%)
Query: 357 PASGTKKTGNHFPRKKEQEVGMVTHGRPQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQY 416
P T + FP+ + + PQ P Q + P S QQ PQ PQ
Sbjct: 199 PQQVTSQPQQPFPQAQPPQ-----QSSPQSQQPYPQEPQQLFPQSQQPQQPFPQPQQPQQ 253
Query: 417 PQIPQYPQIPQYPQFPQNPSPQNTQ--QQNFQQQPYQQQPYQQFPYQQYP--QQNFQQQ 471
P PQ Q PQ P PQ Q Q+ +Q P QP Q P QQ P Q + QQQ
Sbjct: 254 PFPQPQPQTQQSIPQPQQPFPQPQQPFPQSQEQFPQVHQPQQPSPQQQQPSIQLSLQQQ 312
Score = 37.7 bits (86), Expect = 4.8
Identities = 47/125 (37%), Positives = 54/125 (42%), Gaps = 31/125 (24%)
Query: 383 RPQQTYPAYQHIAAITPTSHP-FQQTNNHPQIPQYPQIPQYP------QIPQYPQFPQNP 435
+PQQ +P Q P P QQ+ PQ P +PQ PQ P Q PQ Q PQ P
Sbjct: 240 QPQQPFPQPQQPQQPFPQPQPQTQQSIPQPQQP-FPQ-PQQPFPQSQEQFPQVHQ-PQQP 296
Query: 436 SPQNTQQQNFQQQPYQQQ--PYQQFPYQQ--------------YPQ---QNFQQQPYQQR 476
SPQ QQQ Q QQQ P + QQ +PQ Q QQQ QQ
Sbjct: 297 SPQ--QQQPSIQLSLQQQLNPCKNVLLQQCSPVALVSSLRSKIFPQSECQVMQQQCCQQL 354
Query: 477 PQQPR 481
Q P+
Sbjct: 355 AQIPQ 359
>gb|AAC59642.1| zona pellucida protein [Pseudopleuronectes americanus]
gi|1079271|pir||A48048 egg envelope protein wf - winter
flounder
Length = 509
Score = 85.1 bits (209), Expect = 3e-14
Identities = 62/160 (38%), Positives = 79/160 (48%), Gaps = 21/160 (13%)
Query: 334 EMVGMGVQLEEGVREGRLVKDATPASGTKKTGNHFPRKKEQEVGMVTHGRPQQTYPAYQH 393
E++G + R GR++ ++TG++ P K Q QQ +
Sbjct: 25 EVLGSRRRSRSSERGGRII--------VQQTGHYHPAGKGQRY-------VQQRRRLHHD 69
Query: 394 IAAITPTSHPFQQTNNHPQIPQYPQIPQYPQIPQYPQFPQNPSPQNTQQQNFQQQPYQQQ 453
+ P + P QT P PQ PQ PQ PQ P+YPQ PQ PQ QQ + QQP Q Q
Sbjct: 70 FSPQNPGAEP-PQTPQQPTYPQQPQQPQQPQQPKYPQQPQ--QPQQPQQPKYPQQPQQPQ 126
Query: 454 PYQQFPYQQYPQQNFQ-QQP-YQQRPQQPRPPRMPINPIP 491
QQ Y Q PQQ Q QQP Y Q+PQQP+ P+ P NP P
Sbjct: 127 QPQQPKYPQQPQQPQQPQQPKYPQQPQQPKNPQ-PKNPQP 165
Score = 59.3 bits (142), Expect = 2e-06
Identities = 45/109 (41%), Positives = 51/109 (46%), Gaps = 16/109 (14%)
Query: 420 PQYP--QIPQYPQFPQNP----SPQNTQQQNFQQQPYQQQPYQQFPYQQYPQQNFQ-QQP 472
PQ P + PQ PQ P P PQ QQ + QQP Q Q QQ Y Q PQQ Q QQP
Sbjct: 72 PQNPGAEPPQTPQQPTYPQQPQQPQQPQQPKYPQQPQQPQQPQQPKYPQQPQQPQQPQQP 131
Query: 473 -YQQRPQQPRPPRMPINPIPVTYAELLPGLLKKNLVQTRTAPPIPEKLP 520
Y Q+PQQP+ P+ P P KN PP P+K P
Sbjct: 132 KYPQQPQQPQQPQQPKYPQQPQQ--------PKNPQPKNPQPPQPQKNP 172
>gb|AAF79618.1| F5M15.26 [Arabidopsis thaliana] gi|25402907|pir||H86337 protein
F5M15.26 [imported] - Arabidopsis thaliana
Length = 1838
Score = 80.9 bits (198), Expect = 5e-13
Identities = 44/138 (31%), Positives = 72/138 (51%)
Query: 847 LSFSDEELPAEGRNHNRALHISVNCKTDALSNVLVDTGSSLNVLSKTTYAQLAYQDAPLR 906
+SF+D +L HN L + + ++ VL+DTGSS++++ K + D ++
Sbjct: 600 ISFTDVDLEGLDTPHNDPLVVELIISDSRVTRVLIDTGSSVDLIFKDVLTAMNITDRQIK 659
Query: 907 RSGVMVKAFDGSRKDVLGEVVLPITVGPQVFQVNFQVMDIQASYSCLLGRPWIHEAGAVT 966
+ FDG +G + LPI VG + V F V+ A Y+ +LG PWIH+ A+
Sbjct: 660 PVSKPLAGFDGDFVMTIGTIKLPIFVGGLIAWVKFVVIGKPAVYNVILGTPWIHQMQAIP 719
Query: 967 STLHQKLKFVKNGKLVTV 984
ST HQ +KF + + T+
Sbjct: 720 STYHQCVKFPTHNGIFTL 737
Score = 52.8 bits (125), Expect = 1e-04
Identities = 31/117 (26%), Positives = 58/117 (49%), Gaps = 7/117 (5%)
Query: 181 SVQVPHKFKIPDFEKYKGSSCPEEHLKMY---VRRMPAYAQD-DQILIYYFQESLTGPAS 236
S++ K K+ E Y G + P+E L + + R + D F E LTGPA
Sbjct: 212 SIRGAQKIKL---ESYNGRNDPKEFLTSFNVAINRAELTIDNFDAGRCQIFIEHLTGPAH 268
Query: 237 KWYTNLDKTRVQTFRDLCEAFVEQYSYNVDMTPDRSDLQAMTQGDKETFKEYAQRWR 293
W++ L + +F L +F++ Y+ ++ +DL +++QG KE+ + + R++
Sbjct: 269 NWFSRLKPNSIDSFHQLTSSFLKHYAPLIENQTSNADLWSISQGAKESLRSFVDRFK 325
>gb|AAD20101.1| putative retroelement pol polyprotein [Arabidopsis thaliana]
gi|25411560|pir||E84516 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana
Length = 764
Score = 80.1 bits (196), Expect = 8e-13
Identities = 45/144 (31%), Positives = 75/144 (51%)
Query: 847 LSFSDEELPAEGRNHNRALHISVNCKTDALSNVLVDTGSSLNVLSKTTYAQLAYQDAPLR 906
L+F+ E+L HN L I ++ ++ +L+DTGSS+NV+ K ++ D ++
Sbjct: 330 LTFTSEDLFGVDLPHNDPLVIELHIGESEVTRILIDTGSSVNVVFKDVLQKMKVHDRHIK 389
Query: 907 RSGVMVKAFDGSRKDVLGEVVLPITVGPQVFQVNFQVMDIQASYSCLLGRPWIHEAGAVT 966
S + FDG+ G + LPI +G F V+D Y+ +LG PWIH+ A+
Sbjct: 390 PSVRPLTGFDGNTMMTNGTIKLPIYLGGAATWHKFVVVDKPTIYNIILGTPWIHDMQAIP 449
Query: 967 STLHQKLKFVKNGKLVTVNGEEAL 990
S+ HQ +K + + T+ G + L
Sbjct: 450 SSYHQCIKIPTSIGIETIRGNQNL 473
>gb|AAM91821.1| choriogenin Hminor [Oryzias latipes]
Length = 600
Score = 80.1 bits (196), Expect = 8e-13
Identities = 50/114 (43%), Positives = 59/114 (50%), Gaps = 8/114 (7%)
Query: 406 QTNNHPQIPQYPQIPQYPQIPQYPQFPQNPS-PQNTQQQNFQQQPYQQQPYQQFPYQQYP 464
Q ++PQ PQ PQ PQ PQ PQ+PQ PQ PS PQN QQ + +P QQP+Q QYP
Sbjct: 19 QPPHYPQYPQKPQNPQQPQQPQHPQQPQYPSKPQNPQQPQYPSKP--QQPHQ----PQYP 72
Query: 465 QQNFQQQPYQQRPQQPRPPRMPINPIPVTYAELLPGLLKKNLVQTRTAPPIPEK 518
QN QQ Y +PQQP P+ P NP Y + Q P P K
Sbjct: 73 -QNPQQPQYPSKPQQPHQPQYPQNPQQPQYPSKPQNPQQPQHPQNPQQPQYPSK 125
Score = 77.0 bits (188), Expect = 7e-12
Identities = 53/141 (37%), Positives = 65/141 (45%), Gaps = 21/141 (14%)
Query: 368 FPRKKEQEVGMVTHGRPQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQIPQ 427
+P+K + PQQ P Y P+ Q +P PQ P PQYPQ PQ
Sbjct: 26 YPQKPQNPQQPQQPQHPQQ--PQY-------PSKPQNPQQPQYPSKPQQPHQPQYPQNPQ 76
Query: 428 YPQFPQNPS-------PQNTQQQNFQQQPY--QQQPYQQFPYQ-QYPQ--QNFQQQPYQQ 475
PQ+P P PQN QQ + +P QQ + Q P Q QYP QN QQ + Q
Sbjct: 77 QPQYPSKPQQPHQPQYPQNPQQPQYPSKPQNPQQPQHPQNPQQPQYPSKPQNPQQPQHPQ 136
Query: 476 RPQQPRPPRMPINPIPVTYAE 496
PQQP+ P P NP P Y +
Sbjct: 137 NPQQPQYPSKPQNPQPPQYPQ 157
Score = 76.3 bits (186), Expect = 1e-11
Identities = 49/123 (39%), Positives = 59/123 (47%), Gaps = 11/123 (8%)
Query: 368 FPRKKEQEVGMVTHGRPQQTY-PAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQIP 426
+P K + +PQQ + P Y P + Q + PQ P PQ PQ PQ P
Sbjct: 47 YPSKPQNPQQPQYPSKPQQPHQPQY-------PQNPQQPQYPSKPQQPHQPQYPQNPQQP 99
Query: 427 QYPQFPQNPSPQNTQQQNFQQQPYQQQPYQQFPYQQYPQQNFQQQPYQQRPQQPRPPRMP 486
QYP PQNP Q QN QQ Y +P Q P Q QN QQ Y +PQ P+PP+ P
Sbjct: 100 QYPSKPQNPQ-QPQHPQNPQQPQYPSKP--QNPQQPQHPQNPQQPQYPSKPQNPQPPQYP 156
Query: 487 INP 489
NP
Sbjct: 157 QNP 159
Score = 75.1 bits (183), Expect = 3e-11
Identities = 47/107 (43%), Positives = 53/107 (48%), Gaps = 21/107 (19%)
Query: 404 FQQTNNHPQIPQYPQIPQYPQIPQYPQFPQNPS----------------PQNTQQQNFQQ 447
+ Q PQ PQ PQ PQ+PQ PQYP PQNP PQN QQ +
Sbjct: 23 YPQYPQKPQNPQQPQQPQHPQQPQYPSKPQNPQQPQYPSKPQQPHQPQYPQNPQQPQYPS 82
Query: 448 QPYQ--QQPYQQFPYQ-QYPQ--QNFQQQPYQQRPQQPRPPRMPINP 489
+P Q Q Y Q P Q QYP QN QQ + Q PQQP+ P P NP
Sbjct: 83 KPQQPHQPQYPQNPQQPQYPSKPQNPQQPQHPQNPQQPQYPSKPQNP 129
Score = 73.9 bits (180), Expect = 6e-11
Identities = 55/128 (42%), Positives = 66/128 (50%), Gaps = 9/128 (7%)
Query: 368 FPRKKEQEVGMVTHGRPQQT-YPAY-QHIAAITPTSHPFQ-QTNNHPQIPQYPQIPQYPQ 424
+P K +Q PQQ YP+ Q+ +P Q Q + PQ PQ PQ PQ PQ
Sbjct: 80 YPSKPQQPHQPQYPQNPQQPQYPSKPQNPQQPQHPQNPQQPQYPSKPQNPQQPQHPQNPQ 139
Query: 425 IPQYPQFPQNPSPQNTQQQNFQQQPYQQQPYQQFPYQ-QYP--QQNFQQQPYQQRPQQPR 481
PQYP PQNP P Q +QP+Q Q Y Q P Q QYP QN QQ Y Q P+QP+
Sbjct: 140 QPQYPSKPQNPQPPQYPQN--PKQPHQPQ-YPQNPKQPQYPSKHQNDQQPQYPQNPKQPQ 196
Query: 482 PPRMPINP 489
P+ P P
Sbjct: 197 QPQNPKQP 204
Score = 67.8 bits (164), Expect = 4e-09
Identities = 46/110 (41%), Positives = 54/110 (48%), Gaps = 3/110 (2%)
Query: 383 RPQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQIPQYPQFPQNP-SPQNTQ 441
+PQ Q P + Q +PQ PQYP PQ PQ PQYPQ P+ P PQ Q
Sbjct: 110 QPQHPQNPQQPQYPSKPQNPQQPQHPQNPQQPQYPSKPQNPQPPQYPQNPKQPHQPQYPQ 169
Query: 442 QQNFQQQPYQQQPYQQFPYQQYPQQNFQ-QQPYQ-QRPQQPRPPRMPINP 489
Q P + Q QQ Y Q P+Q Q Q P Q Q+PQQP+ P NP
Sbjct: 170 NPKQPQYPSKHQNDQQPQYPQNPKQPQQPQNPKQPQQPQQPQYPSKSQNP 219
Score = 50.8 bits (120), Expect = 5e-04
Identities = 35/93 (37%), Positives = 40/93 (42%), Gaps = 12/93 (12%)
Query: 384 PQQT-YPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQIP------QYPQFPQNP- 435
PQQ YP+ P + Q P PQYPQ P+ PQ P Q PQ+PQNP
Sbjct: 138 PQQPQYPSKPQ----NPQPPQYPQNPKQPHQPQYPQNPKQPQYPSKHQNDQQPQYPQNPK 193
Query: 436 SPQNTQQQNFQQQPYQQQPYQQFPYQQYPQQNF 468
PQ Q QQP Q Q + Q P F
Sbjct: 194 QPQQPQNPKQPQQPQQPQYPSKSQNPQTPPSTF 226
Score = 39.7 bits (91), Expect = 1.3
Identities = 19/51 (37%), Positives = 25/51 (48%)
Query: 404 FQQTNNHPQIPQYPQIPQYPQIPQYPQFPQNPSPQNTQQQNFQQQPYQQQP 454
+ Q PQ PQ P+ PQ PQ PQYP QNP + + + Y + P
Sbjct: 188 YPQNPKQPQQPQNPKQPQQPQQPQYPSKSQNPQTPPSTFHSCEVDEYYKIP 238
>gb|AAK84774.1| gamma-gliadin [Triticum aestivum]
Length = 337
Score = 80.1 bits (196), Expect = 8e-13
Identities = 52/111 (46%), Positives = 60/111 (53%), Gaps = 9/111 (8%)
Query: 383 RPQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQIPQYPQFPQNPSPQNTQ- 441
+PQQT+P A P QQT H Q+PQ PQ PQ P +PQ PQ PQ Q
Sbjct: 55 QPQQTFPHQPQQAFPQP-----QQTFPHQPQQQFPQ-PQQPQQP-FPQQPQQQFPQPQQP 107
Query: 442 QQNFQQQPYQQQPYQQFPYQQYPQQNFQQQPYQQRPQQPRP-PRMPINPIP 491
QQ F QQP QQ P Q P Q +PQ Q P+ Q+PQQP P P+ P P P
Sbjct: 108 QQPFPQQPQQQFPQPQQPQQPFPQPQQPQLPFPQQPQQPFPQPQQPQQPFP 158
Score = 69.7 bits (169), Expect = 1e-09
Identities = 53/123 (43%), Positives = 60/123 (48%), Gaps = 17/123 (13%)
Query: 377 GMVTHGRPQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQ--YPQIPQYPQFPQN 434
G V + QQ +P Q P S QQ PQ +P PQ +PQ Q FP
Sbjct: 27 GQVQWPQQQQPFPQPQQ-----PFSQQPQQIFPQPQ-QTFPHQPQQAFPQPQQ--TFPHQ 78
Query: 435 PSPQNTQQQNFQQQPYQQQPYQQFPYQQYPQQNFQQQPYQ-----QRPQQPRP-PRMPIN 488
P Q Q Q QQP+ QQP QQFP Q PQQ F QQP Q Q+PQQP P P+ P
Sbjct: 79 PQQQFPQPQQ-PQQPFPQQPQQQFPQPQQPQQPFPQQPQQQFPQPQQPQQPFPQPQQPQL 137
Query: 489 PIP 491
P P
Sbjct: 138 PFP 140
Score = 62.4 bits (150), Expect = 2e-07
Identities = 49/126 (38%), Positives = 57/126 (44%), Gaps = 18/126 (14%)
Query: 368 FPRKKEQEVGMVTHGRPQQTYPAY--QHIAAITPTSHPF----QQTNNHPQIPQYP-QIP 420
FP + +Q+ +PQQ +P Q PF QQ PQ PQ P P
Sbjct: 75 FPHQPQQQFPQPQ--QPQQPFPQQPQQQFPQPQQPQQPFPQQPQQQFPQPQQPQQPFPQP 132
Query: 421 QYPQIPQYPQFPQNPSPQNTQQQNFQQQPYQQQPYQQFPYQQYPQQNFQQ------QPYQ 474
Q PQ+P +PQ PQ P PQ Q Q Q P QQP Q P Q PQQ F Q QPY
Sbjct: 133 QQPQLP-FPQQPQQPFPQPQQPQ--QPFPQLQQPQQPLPQPQQPQQPFPQQQQPLIQPYL 189
Query: 475 QRPQQP 480
Q+ P
Sbjct: 190 QQQMNP 195
Score = 38.5 bits (88), Expect = 2.8
Identities = 27/71 (38%), Positives = 32/71 (45%), Gaps = 13/71 (18%)
Query: 434 NPSPQNTQQQNFQQQPYQQQPYQQFPYQQY--PQQNFQQQPYQ----------QRPQQPR 481
+PS Q Q Q P QQP+ Q P Q + PQQ F QP Q +PQQ
Sbjct: 24 DPSGQVQWPQQQQPFPQPQQPFSQQPQQIFPQPQQTFPHQPQQAFPQPQQTFPHQPQQQF 83
Query: 482 P-PRMPINPIP 491
P P+ P P P
Sbjct: 84 PQPQQPQQPFP 94
>gb|AAK84772.1| gamma-gliadin [Triticum aestivum]
Length = 241
Score = 80.1 bits (196), Expect = 8e-13
Identities = 52/111 (46%), Positives = 60/111 (53%), Gaps = 9/111 (8%)
Query: 383 RPQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQIPQYPQFPQNPSPQNTQ- 441
+PQQT+P A P QQT H Q+PQ PQ PQ P +PQ PQ PQ Q
Sbjct: 55 QPQQTFPHQPQQAFPQP-----QQTFPHQPQQQFPQ-PQQPQQP-FPQQPQQQFPQPQQP 107
Query: 442 QQNFQQQPYQQQPYQQFPYQQYPQQNFQQQPYQQRPQQPRP-PRMPINPIP 491
QQ F QQP QQ P Q P Q +PQ Q P+ Q+PQQP P P+ P P P
Sbjct: 108 QQPFPQQPQQQFPQPQQPQQPFPQPQQPQLPFPQQPQQPFPQPQQPQQPFP 158
Score = 69.7 bits (169), Expect = 1e-09
Identities = 53/123 (43%), Positives = 60/123 (48%), Gaps = 17/123 (13%)
Query: 377 GMVTHGRPQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQ--YPQIPQYPQFPQN 434
G V + QQ +P Q P S QQ PQ +P PQ +PQ Q FP
Sbjct: 27 GQVQWPQQQQPFPQPQQ-----PFSQQPQQIFPQPQ-QTFPHQPQQAFPQPQQ--TFPHQ 78
Query: 435 PSPQNTQQQNFQQQPYQQQPYQQFPYQQYPQQNFQQQPYQ-----QRPQQPRP-PRMPIN 488
P Q Q Q QQP+ QQP QQFP Q PQQ F QQP Q Q+PQQP P P+ P
Sbjct: 79 PQQQFPQPQQ-PQQPFPQQPQQQFPQPQQPQQPFPQQPQQQFPQPQQPQQPFPQPQQPQL 137
Query: 489 PIP 491
P P
Sbjct: 138 PFP 140
Score = 62.4 bits (150), Expect = 2e-07
Identities = 49/126 (38%), Positives = 57/126 (44%), Gaps = 18/126 (14%)
Query: 368 FPRKKEQEVGMVTHGRPQQTYPAY--QHIAAITPTSHPF----QQTNNHPQIPQYP-QIP 420
FP + +Q+ +PQQ +P Q PF QQ PQ PQ P P
Sbjct: 75 FPHQPQQQFPQPQ--QPQQPFPQQPQQQFPQPQQPQQPFPQQPQQQFPQPQQPQQPFPQP 132
Query: 421 QYPQIPQYPQFPQNPSPQNTQQQNFQQQPYQQQPYQQFPYQQYPQQNFQQ------QPYQ 474
Q PQ+P +PQ PQ P PQ Q Q Q P QQP Q P Q PQQ F Q QPY
Sbjct: 133 QQPQLP-FPQQPQQPFPQPQQPQ--QPFPQLQQPQQPLPQPQQPQQPFPQQQQPLIQPYL 189
Query: 475 QRPQQP 480
Q+ P
Sbjct: 190 QQQMNP 195
Score = 38.5 bits (88), Expect = 2.8
Identities = 27/71 (38%), Positives = 32/71 (45%), Gaps = 13/71 (18%)
Query: 434 NPSPQNTQQQNFQQQPYQQQPYQQFPYQQY--PQQNFQQQPYQ----------QRPQQPR 481
+PS Q Q Q P QQP+ Q P Q + PQQ F QP Q +PQQ
Sbjct: 24 DPSGQVQWPQQQQPFPQPQQPFSQQPQQIFPQPQQTFPHQPQQAFPQPQQTFPHQPQQQF 83
Query: 482 P-PRMPINPIP 491
P P+ P P P
Sbjct: 84 PQPQQPQQPFP 94
>emb|CAC10616.1| gamma-gliadin [Aegilops longissima]
Length = 300
Score = 79.7 bits (195), Expect = 1e-12
Identities = 58/141 (41%), Positives = 70/141 (49%), Gaps = 9/141 (6%)
Query: 369 PRKKEQEVGMVTHGRPQQTYPAYQHIAAITPTSHPFQQTNNHPQ-IPQYPQIPQYPQIPQ 427
P++ + T +PQQT+P +Q I P QQ Q PQ PQ P YPQ PQ
Sbjct: 24 PQQPFSQQPQQTFPQPQQTFP-HQPQQQIPQPQQPQQQFLQPQQPFPQQPQQP-YPQQPQ 81
Query: 428 YPQFPQNPSPQNTQQQNFQ-QQPYQQQPYQQFPYQQYPQQNFQQQPYQQRPQQPRP-PRM 485
P FPQ PQ Q+ Q QQPY QQP Q FP Q PQQ F P Q+PQQP P P+
Sbjct: 82 QP-FPQTQQPQQLFPQSQQPQQPYPQQPQQPFPQTQQPQQQF---PQSQQPQQPFPQPQQ 137
Query: 486 PINPIPVTYAELLPGLLKKNL 506
P P + L++ L
Sbjct: 138 PQQSFPQQQPPFIQPSLQQQL 158
Score = 55.1 bits (131), Expect = 3e-05
Identities = 33/63 (52%), Positives = 36/63 (56%), Gaps = 6/63 (9%)
Query: 425 IPQYP--QFPQNPSPQNTQQQNFQQQPYQQQPYQQFPYQQY--PQQNFQQQPYQQRPQQP 480
+PQ P Q PQ PQ QQ F QP QQ P Q P QQ+ PQQ F QQP Q PQQP
Sbjct: 23 LPQQPFSQQPQQTFPQ--PQQTFPHQPQQQIPQPQQPQQQFLQPQQPFPQQPQQPYPQQP 80
Query: 481 RPP 483
+ P
Sbjct: 81 QQP 83
Score = 51.2 bits (121), Expect = 4e-04
Identities = 46/106 (43%), Positives = 50/106 (46%), Gaps = 21/106 (19%)
Query: 405 QQTNNHP-QIPQYPQIPQYPQIPQYPQFPQNPSPQNTQQQNFQ-QQPYQQ--QPYQQF-- 458
Q T P +PQ P Q PQ +PQ PQ P QQQ Q QQP QQ QP Q F
Sbjct: 14 QWTQQQPVPLPQQP-FSQQPQ-QTFPQ-PQQTFPHQPQQQIPQPQQPQQQFLQPQQPFPQ 70
Query: 459 ----PYQQYPQQNFQQQ-------PYQQRPQQPRPPRMPINPIPVT 493
PY Q PQQ F Q P Q+PQQP P+ P P P T
Sbjct: 71 QPQQPYPQQPQQPFPQTQQPQQLFPQSQQPQQPY-PQQPQQPFPQT 115
>emb|CAC10594.1| gamma-gliadin [Aegilops bicornis] gi|10635141|emb|CAC10593.1|
gamma-gliadin [Aegilops bicornis]
Length = 300
Score = 79.7 bits (195), Expect = 1e-12
Identities = 58/141 (41%), Positives = 70/141 (49%), Gaps = 9/141 (6%)
Query: 369 PRKKEQEVGMVTHGRPQQTYPAYQHIAAITPTSHPFQQTNNHPQ-IPQYPQIPQYPQIPQ 427
P++ + T +PQQT+P +Q I P QQ Q PQ PQ P YPQ PQ
Sbjct: 24 PQQPFSQQPQQTFPQPQQTFP-HQPQQQIPQPQQPQQQFLQPQQPFPQQPQQP-YPQQPQ 81
Query: 428 YPQFPQNPSPQNTQQQNFQ-QQPYQQQPYQQFPYQQYPQQNFQQQPYQQRPQQPRP-PRM 485
P FPQ PQ Q+ Q QQPY QQP Q FP Q PQQ F P Q+PQQP P P+
Sbjct: 82 QP-FPQTQQPQQLFPQSQQPQQPYPQQPQQPFPQTQQPQQQF---PQSQQPQQPFPQPQQ 137
Query: 486 PINPIPVTYAELLPGLLKKNL 506
P P + L++ L
Sbjct: 138 PQQSFPQQQPPFIQPSLQQQL 158
Score = 55.1 bits (131), Expect = 3e-05
Identities = 33/63 (52%), Positives = 36/63 (56%), Gaps = 6/63 (9%)
Query: 425 IPQYP--QFPQNPSPQNTQQQNFQQQPYQQQPYQQFPYQQY--PQQNFQQQPYQQRPQQP 480
+PQ P Q PQ PQ QQ F QP QQ P Q P QQ+ PQQ F QQP Q PQQP
Sbjct: 23 LPQQPFSQQPQQTFPQ--PQQTFPHQPQQQIPQPQQPQQQFLQPQQPFPQQPQQPYPQQP 80
Query: 481 RPP 483
+ P
Sbjct: 81 QQP 83
Score = 51.2 bits (121), Expect = 4e-04
Identities = 46/106 (43%), Positives = 50/106 (46%), Gaps = 21/106 (19%)
Query: 405 QQTNNHP-QIPQYPQIPQYPQIPQYPQFPQNPSPQNTQQQNFQ-QQPYQQ--QPYQQF-- 458
Q T P +PQ P Q PQ +PQ PQ P QQQ Q QQP QQ QP Q F
Sbjct: 14 QWTQQQPVPLPQQP-FSQQPQ-QTFPQ-PQQTFPHQPQQQIPQPQQPQQQFLQPQQPFPQ 70
Query: 459 ----PYQQYPQQNFQQQ-------PYQQRPQQPRPPRMPINPIPVT 493
PY Q PQQ F Q P Q+PQQP P+ P P P T
Sbjct: 71 QPQQPYPQQPQQPFPQTQQPQQLFPQSQQPQQPY-PQQPQQPFPQT 115
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.343 0.149 0.504
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,709,692,042
Number of Sequences: 2540612
Number of extensions: 155012065
Number of successful extensions: 1250716
Number of sequences better than 10.0: 5580
Number of HSP's better than 10.0 without gapping: 2751
Number of HSP's successfully gapped in prelim test: 3141
Number of HSP's that attempted gapping in prelim test: 992499
Number of HSP's gapped (non-prelim): 72724
length of query: 2359
length of database: 863,360,394
effective HSP length: 145
effective length of query: 2214
effective length of database: 494,971,654
effective search space: 1095867241956
effective search space used: 1095867241956
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.5 bits)
S2: 84 (37.0 bits)
Medicago: description of AC130806.6


