

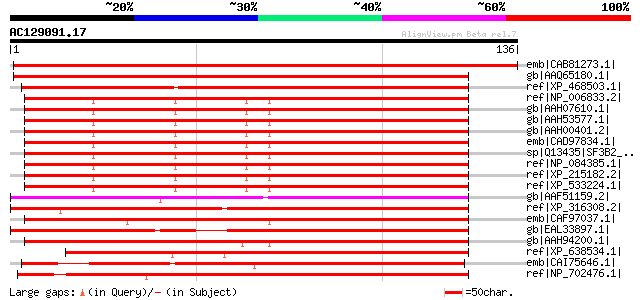
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC129091.17 + phase: 0 /pseudo
(136 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAB81273.1| spliceosome associated protein-like [Arabidopsis... 209 1e-53
gb|AAQ65180.1| At4g21660 [Arabidopsis thaliana] gi|30685521|ref|... 195 2e-49
ref|XP_468503.1| putative plicing factor 3B subunit 2 [Oryza sat... 163 1e-39
ref|NP_006833.2| splicing factor 3B subunit 2 [Homo sapiens] 114 4e-25
gb|AAH07610.1| SF3B2 protein [Homo sapiens] 114 4e-25
gb|AAH53577.1| SF3B2 protein [Homo sapiens] 114 4e-25
gb|AAH00401.2| SF3B2 protein [Homo sapiens] 114 4e-25
emb|CAD97834.1| hypothetical protein [Homo sapiens] 114 4e-25
sp|Q13435|SF3B2_HUMAN Splicing factor 3B subunit 2 (Spliceosome ... 114 4e-25
ref|NP_084385.1| splicing factor 3b, subunit 2 [Mus musculus] gi... 113 1e-24
ref|XP_215182.2| PREDICTED: similar to Splicing factor 3b, subun... 113 1e-24
ref|XP_533224.1| PREDICTED: similar to SF3B2 protein [Canis fami... 113 1e-24
gb|AAF51159.2| CG3605-PA [Drosophila melanogaster] gi|19920622|r... 110 7e-24
ref|XP_316308.2| ENSANGP00000020597 [Anopheles gambiae str. PEST... 106 1e-22
emb|CAF97037.1| unnamed protein product [Tetraodon nigroviridis] 105 2e-22
gb|EAL33897.1| GA17553-PA [Drosophila pseudoobscura] 105 3e-22
gb|AAH94200.1| Unknown (protein for MGC:115052) [Xenopus laevis] 103 1e-21
ref|XP_638534.1| hypothetical protein DDB0218611 [Dictyostelium ... 103 1e-21
emb|CAI75646.1| spliceosome-associated protein, putative [Theile... 100 6e-21
ref|NP_702476.1| hypothetical protein PF14_0587 [Plasmodium falc... 94 9e-19
>emb|CAB81273.1| spliceosome associated protein-like [Arabidopsis thaliana]
gi|4455274|emb|CAB36810.1| spliceosome associated
protein-like [Arabidopsis thaliana]
gi|7488345|pir||T05841 spliceosome-associated protein
homolog F17L22.120 - Arabidopsis thaliana
Length = 700
Score = 209 bits (532), Expect = 1e-53
Identities = 100/135 (74%), Positives = 115/135 (85%)
Query: 2 DEEFRKIFEKFSFTEVAASEETDKKDVAEETAATKKKANSDSDYEDEENDNEQKEKGVSN 61
++EF++IFEKF+F E ASEE KD +EE KKK NSDSD +D+E DN+ KEKG+SN
Sbjct: 91 NDEFKEIFEKFNFREPLASEEDGTKDESEEKEDVKKKVNSDSDSDDDEQDNQNKEKGISN 150
Query: 62 KKKKLQRRMKIAELKQVSSRPDVVEVWDATAADPKLLVFLKSYRNTVPVPRHWSQKRKFL 121
KKKKLQRRMKIAELKQVS+RPDVVEVWDAT+ADPKLLVFLKSYRNTVPVPRHWSQKRK+L
Sbjct: 151 KKKKLQRRMKIAELKQVSARPDVVEVWDATSADPKLLVFLKSYRNTVPVPRHWSQKRKYL 210
Query: 122 QGSGCGNHQSWTGRC 136
Q + C NH +WT RC
Sbjct: 211 QENICDNHGAWTCRC 225
>gb|AAQ65180.1| At4g21660 [Arabidopsis thaliana] gi|30685521|ref|NP_193897.2|
proline-rich spliceosome-associated (PSP) family protein
[Arabidopsis thaliana] gi|25082966|gb|AAN72024.1|
spliceosome associated protein - like [Arabidopsis
thaliana]
Length = 584
Score = 195 bits (495), Expect = 2e-49
Identities = 94/122 (77%), Positives = 107/122 (87%)
Query: 2 DEEFRKIFEKFSFTEVAASEETDKKDVAEETAATKKKANSDSDYEDEENDNEQKEKGVSN 61
++EF++IFEKF+F E ASEE KD +EE KKK NSDSD +D+E DN+ KEKG+SN
Sbjct: 95 NDEFKEIFEKFNFREPLASEEDGTKDESEEKEDVKKKVNSDSDSDDDEQDNQNKEKGISN 154
Query: 62 KKKKLQRRMKIAELKQVSSRPDVVEVWDATAADPKLLVFLKSYRNTVPVPRHWSQKRKFL 121
KKKKLQRRMKIAELKQVS+RPDVVEVWDAT+ADPKLLVFLKSYRNTVPVPRHWSQKRK+L
Sbjct: 155 KKKKLQRRMKIAELKQVSARPDVVEVWDATSADPKLLVFLKSYRNTVPVPRHWSQKRKYL 214
Query: 122 QG 123
QG
Sbjct: 215 QG 216
>ref|XP_468503.1| putative plicing factor 3B subunit 2 [Oryza sativa (japonica
cultivar-group)] gi|48716448|dbj|BAD23055.1| putative
splicing factor 3B subunit 2 [Oryza sativa (japonica
cultivar-group)]
Length = 582
Score = 163 bits (412), Expect = 1e-39
Identities = 80/120 (66%), Positives = 97/120 (80%), Gaps = 1/120 (0%)
Query: 4 EFRKIFEKFSFTEVAASEETDKKDVAEETAATKKKANSDSDYEDEENDNEQKEKGVSNKK 63
+F+ IF+KF+F + +A E D+K T A KK A SDSD +DE+ ++KE G+SNK+
Sbjct: 95 DFKSIFDKFTFKDSSADAEDDEKKDEAGTDAAKKAAGSDSD-DDEQGTQQKKEGGLSNKQ 153
Query: 64 KKLQRRMKIAELKQVSSRPDVVEVWDATAADPKLLVFLKSYRNTVPVPRHWSQKRKFLQG 123
KKLQRRMKIAELKQ+ +RPDVVEVWDATA+DP LLV+LKSYRNTVPVPRHW QKRKFLQG
Sbjct: 154 KKLQRRMKIAELKQICNRPDVVEVWDATASDPSLLVYLKSYRNTVPVPRHWCQKRKFLQG 213
>ref|NP_006833.2| splicing factor 3B subunit 2 [Homo sapiens]
Length = 895
Score = 114 bits (286), Expect = 4e-25
Identities = 66/128 (51%), Positives = 87/128 (67%), Gaps = 9/128 (7%)
Query: 5 FRKIFEKFSFTEVAASE---ETDKKDVAEETAATKKKANSDS--DYEDEENDNEQKEKGV 59
F++IFE F T+ E E +K D E +AA KKK + D +D+ +D+EQ++K
Sbjct: 386 FKRIFEAFKLTDDVKKEKEKEPEKLDKLENSAAPKKKGFEEEHKDSDDDSSDDEQEKKPE 445
Query: 60 SNK--KKKLQR--RMKIAELKQVSSRPDVVEVWDATAADPKLLVFLKSYRNTVPVPRHWS 115
+ K KKKL+R R +AELKQ+ +RPDVVE+ D TA DPKLLV LK+ RN+VPVPRHW
Sbjct: 446 APKLSKKKLRRMNRFTVAELKQLVARPDVVEMHDVTAQDPKLLVHLKATRNSVPVPRHWC 505
Query: 116 QKRKFLQG 123
KRK+LQG
Sbjct: 506 FKRKYLQG 513
>gb|AAH07610.1| SF3B2 protein [Homo sapiens]
Length = 636
Score = 114 bits (286), Expect = 4e-25
Identities = 66/128 (51%), Positives = 87/128 (67%), Gaps = 9/128 (7%)
Query: 5 FRKIFEKFSFTEVAASE---ETDKKDVAEETAATKKKANSDS--DYEDEENDNEQKEKGV 59
F++IFE F T+ E E +K D E +AA KKK + D +D+ +D+EQ++K
Sbjct: 127 FKRIFEAFKLTDDVKKEKEKEPEKLDKLENSAAPKKKGFEEEHKDSDDDSSDDEQEKKPE 186
Query: 60 SNK--KKKLQR--RMKIAELKQVSSRPDVVEVWDATAADPKLLVFLKSYRNTVPVPRHWS 115
+ K KKKL+R R +AELKQ+ +RPDVVE+ D TA DPKLLV LK+ RN+VPVPRHW
Sbjct: 187 APKLSKKKLRRMNRFTVAELKQLVARPDVVEMHDVTAQDPKLLVHLKATRNSVPVPRHWC 246
Query: 116 QKRKFLQG 123
KRK+LQG
Sbjct: 247 FKRKYLQG 254
>gb|AAH53577.1| SF3B2 protein [Homo sapiens]
Length = 877
Score = 114 bits (286), Expect = 4e-25
Identities = 66/128 (51%), Positives = 87/128 (67%), Gaps = 9/128 (7%)
Query: 5 FRKIFEKFSFTEVAASE---ETDKKDVAEETAATKKKANSDS--DYEDEENDNEQKEKGV 59
F++IFE F T+ E E +K D E +AA KKK + D +D+ +D+EQ++K
Sbjct: 386 FKRIFEAFKLTDDVKKEKEKEPEKLDKLENSAAPKKKGFEEEHKDSDDDSSDDEQEKKPE 445
Query: 60 SNK--KKKLQR--RMKIAELKQVSSRPDVVEVWDATAADPKLLVFLKSYRNTVPVPRHWS 115
+ K KKKL+R R +AELKQ+ +RPDVVE+ D TA DPKLLV LK+ RN+VPVPRHW
Sbjct: 446 APKLSKKKLRRMNRFTVAELKQLVARPDVVEMHDVTAQDPKLLVHLKATRNSVPVPRHWC 505
Query: 116 QKRKFLQG 123
KRK+LQG
Sbjct: 506 FKRKYLQG 513
>gb|AAH00401.2| SF3B2 protein [Homo sapiens]
Length = 894
Score = 114 bits (286), Expect = 4e-25
Identities = 66/128 (51%), Positives = 87/128 (67%), Gaps = 9/128 (7%)
Query: 5 FRKIFEKFSFTEVAASE---ETDKKDVAEETAATKKKANSDS--DYEDEENDNEQKEKGV 59
F++IFE F T+ E E +K D E +AA KKK + D +D+ +D+EQ++K
Sbjct: 385 FKRIFEAFKLTDDVKKEKEKEPEKLDKLENSAAPKKKGFEEEHKDSDDDSSDDEQEKKPE 444
Query: 60 SNK--KKKLQR--RMKIAELKQVSSRPDVVEVWDATAADPKLLVFLKSYRNTVPVPRHWS 115
+ K KKKL+R R +AELKQ+ +RPDVVE+ D TA DPKLLV LK+ RN+VPVPRHW
Sbjct: 445 APKLSKKKLRRMNRFTVAELKQLVARPDVVEMHDVTAQDPKLLVHLKATRNSVPVPRHWC 504
Query: 116 QKRKFLQG 123
KRK+LQG
Sbjct: 505 FKRKYLQG 512
>emb|CAD97834.1| hypothetical protein [Homo sapiens]
Length = 799
Score = 114 bits (286), Expect = 4e-25
Identities = 66/128 (51%), Positives = 87/128 (67%), Gaps = 9/128 (7%)
Query: 5 FRKIFEKFSFTEVAASE---ETDKKDVAEETAATKKKANSDS--DYEDEENDNEQKEKGV 59
F++IFE F T+ E E +K D E +AA KKK + D +D+ +D+EQ++K
Sbjct: 290 FKRIFEAFKLTDDVKKEKEKEPEKLDKLENSAAPKKKGFEEEHKDSDDDSSDDEQEKKPE 349
Query: 60 SNK--KKKLQR--RMKIAELKQVSSRPDVVEVWDATAADPKLLVFLKSYRNTVPVPRHWS 115
+ K KKKL+R R +AELKQ+ +RPDVVE+ D TA DPKLLV LK+ RN+VPVPRHW
Sbjct: 350 APKLSKKKLRRMNRFTVAELKQLVARPDVVEMHDVTAQDPKLLVHLKATRNSVPVPRHWC 409
Query: 116 QKRKFLQG 123
KRK+LQG
Sbjct: 410 FKRKYLQG 417
>sp|Q13435|SF3B2_HUMAN Splicing factor 3B subunit 2 (Spliceosome associated protein 145)
(SAP 145) (SF3b150) (Pre-mRNA splicing factor SF3b 145
kDa subunit) gi|1173905|gb|AAA97461.1| spliceosome
associated protein
Length = 872
Score = 114 bits (286), Expect = 4e-25
Identities = 66/128 (51%), Positives = 87/128 (67%), Gaps = 9/128 (7%)
Query: 5 FRKIFEKFSFTEVAASE---ETDKKDVAEETAATKKKANSDS--DYEDEENDNEQKEKGV 59
F++IFE F T+ E E +K D E +AA KKK + D +D+ +D+EQ++K
Sbjct: 363 FKRIFEAFKLTDDVKKEKEKEPEKLDKLENSAAPKKKGFEEEHKDSDDDSSDDEQEKKPE 422
Query: 60 SNK--KKKLQR--RMKIAELKQVSSRPDVVEVWDATAADPKLLVFLKSYRNTVPVPRHWS 115
+ K KKKL+R R +AELKQ+ +RPDVVE+ D TA DPKLLV LK+ RN+VPVPRHW
Sbjct: 423 APKLSKKKLRRMNRFTVAELKQLVARPDVVEMHDVTAQDPKLLVHLKATRNSVPVPRHWC 482
Query: 116 QKRKFLQG 123
KRK+LQG
Sbjct: 483 FKRKYLQG 490
>ref|NP_084385.1| splicing factor 3b, subunit 2 [Mus musculus]
gi|29144992|gb|AAH49118.1| Splicing factor 3b, subunit 2
[Mus musculus]
Length = 878
Score = 113 bits (282), Expect = 1e-24
Identities = 65/128 (50%), Positives = 86/128 (66%), Gaps = 9/128 (7%)
Query: 5 FRKIFEKFSFTEVAASE---ETDKKDVAEETAATKKKANSDS--DYEDEENDNEQKEKGV 59
F++IFE F T+ E E +K D E +A KKK + D +D+ +D+EQ++K
Sbjct: 369 FKRIFEAFKLTDDVKKEKEKEPEKLDKMESSAVPKKKGFEEEHKDSDDDSSDDEQEKKPE 428
Query: 60 SNK--KKKLQR--RMKIAELKQVSSRPDVVEVWDATAADPKLLVFLKSYRNTVPVPRHWS 115
+ K KKKL+R R +AELKQ+ +RPDVVE+ D TA DPKLLV LK+ RN+VPVPRHW
Sbjct: 429 APKLSKKKLRRMNRFTVAELKQLVARPDVVEMHDVTAQDPKLLVHLKATRNSVPVPRHWC 488
Query: 116 QKRKFLQG 123
KRK+LQG
Sbjct: 489 FKRKYLQG 496
>ref|XP_215182.2| PREDICTED: similar to Splicing factor 3b, subunit 2 [Rattus
norvegicus]
Length = 865
Score = 113 bits (282), Expect = 1e-24
Identities = 65/128 (50%), Positives = 86/128 (66%), Gaps = 9/128 (7%)
Query: 5 FRKIFEKFSFTEVAASE---ETDKKDVAEETAATKKKANSDS--DYEDEENDNEQKEKGV 59
F++IFE F T+ E E +K D E +A KKK + D +D+ +D+EQ++K
Sbjct: 369 FKRIFEAFKLTDDVKKEKEKEPEKLDKMENSAVPKKKGFEEEHKDSDDDSSDDEQEKKPE 428
Query: 60 SNK--KKKLQR--RMKIAELKQVSSRPDVVEVWDATAADPKLLVFLKSYRNTVPVPRHWS 115
+ K KKKL+R R +AELKQ+ +RPDVVE+ D TA DPKLLV LK+ RN+VPVPRHW
Sbjct: 429 APKLSKKKLRRMNRFTVAELKQLVARPDVVEMHDVTAQDPKLLVHLKATRNSVPVPRHWC 488
Query: 116 QKRKFLQG 123
KRK+LQG
Sbjct: 489 FKRKYLQG 496
>ref|XP_533224.1| PREDICTED: similar to SF3B2 protein [Canis familiaris]
Length = 992
Score = 113 bits (282), Expect = 1e-24
Identities = 65/128 (50%), Positives = 86/128 (66%), Gaps = 9/128 (7%)
Query: 5 FRKIFEKFSFTEVAASE---ETDKKDVAEETAATKKKANSDS--DYEDEENDNEQKEKGV 59
F++IFE F T+ E E +K D E + A KKK + D +D+ +D+EQ++K
Sbjct: 470 FKRIFEAFKLTDDVKKEKEKEPEKLDKLENSTAPKKKGFEEEHKDSDDDSSDDEQEKKPE 529
Query: 60 SNK--KKKLQR--RMKIAELKQVSSRPDVVEVWDATAADPKLLVFLKSYRNTVPVPRHWS 115
+ K KKKL+R R +AELKQ+ +RPDVVE+ D TA DPKLLV LK+ RN+VPVPRHW
Sbjct: 530 APKLSKKKLRRMNRFTVAELKQLVARPDVVEMHDVTAQDPKLLVHLKATRNSVPVPRHWC 589
Query: 116 QKRKFLQG 123
KRK+LQG
Sbjct: 590 FKRKYLQG 597
>gb|AAF51159.2| CG3605-PA [Drosophila melanogaster] gi|19920622|ref|NP_608739.1|
CG3605-PA [Drosophila melanogaster]
gi|16769632|gb|AAL29035.1| LD45152p [Drosophila
melanogaster]
Length = 749
Score = 110 bits (275), Expect = 7e-24
Identities = 64/129 (49%), Positives = 77/129 (59%), Gaps = 7/129 (5%)
Query: 1 MDEEFRKIFEKFSFTEVAASEETDKKDVAEETAATKKKA------NSDSDYEDEENDNEQ 54
M +F ++FE F E DK E+ KKA + D D +D+E E
Sbjct: 236 MYRQFYRVFEIFKLENKPKPVEKDKSSHDSESHPRDKKATDKQLEDEDDDGDDDEERKED 295
Query: 55 KEKGVSNKKKKLQRRMKIAELKQVSSRPDVVEVWDATAADPKLLVFLKSYRNTVPVPRHW 114
KEK K KKL R + +AELKQ+ SRPDVVE+ D TA DPKLLV LK+YRNTV VPRHW
Sbjct: 296 KEKLSKRKLKKLTR-LSVAELKQLVSRPDVVEMHDVTARDPKLLVQLKAYRNTVQVPRHW 354
Query: 115 SQKRKFLQG 123
KRK+LQG
Sbjct: 355 CFKRKYLQG 363
>ref|XP_316308.2| ENSANGP00000020597 [Anopheles gambiae str. PEST]
gi|55239045|gb|EAA10768.2| ENSANGP00000020597 [Anopheles
gambiae str. PEST]
Length = 715
Score = 106 bits (265), Expect = 1e-22
Identities = 60/128 (46%), Positives = 81/128 (62%), Gaps = 6/128 (4%)
Query: 1 MDEEFRKIFEKF-----SFTEVAASEETDKKDVAEETAATKKKANSDSDYEDEENDNEQK 55
M +F ++FE F S V ++EE +K+ A ET + D D + ++ + K
Sbjct: 200 MYRQFYRVFEIFKLDTKSKEAVKSTEEAEKEKAAAETKKVDDGLDDDDDDLGDLDEKDDK 259
Query: 56 EKGVSNKKKKLQRRMKIAELKQVSSRPDVVEVWDATAADPKLLVFLKSYRNTVPVPRHWS 115
EK +S +K K R+ +AELKQ+ +RPDVVE+ D TA DPKLLV LKS+RNTV VPRHW
Sbjct: 260 EK-ISKRKLKKLNRLTVAELKQLVTRPDVVEMHDVTARDPKLLVQLKSHRNTVQVPRHWC 318
Query: 116 QKRKFLQG 123
KRK+LQG
Sbjct: 319 FKRKYLQG 326
>emb|CAF97037.1| unnamed protein product [Tetraodon nigroviridis]
Length = 981
Score = 105 bits (263), Expect = 2e-22
Identities = 58/123 (47%), Positives = 78/123 (63%), Gaps = 4/123 (3%)
Query: 5 FRKIFEKFSFTEVAASEETDKKDVAE--ETAATKKKANSDSDYEDEENDNEQKEKGVSNK 62
F++IFE F T+ E+ + + E ET KK + +D ++D E +
Sbjct: 358 FKRIFEAFKLTDDVKKEKEKEPEKTEKQETTMVWKKGFVEEKKDDNDSDEEIRPDVPKMS 417
Query: 63 KKKLQR--RMKIAELKQVSSRPDVVEVWDATAADPKLLVFLKSYRNTVPVPRHWSQKRKF 120
KKKL+R R+ +AELKQ+ +RPDVVE+ D TA +PKLLV LK+ RNTVPVPRHW KRK+
Sbjct: 418 KKKLRRMNRLTVAELKQLVARPDVVEMHDVTAQEPKLLVHLKATRNTVPVPRHWCFKRKY 477
Query: 121 LQG 123
LQG
Sbjct: 478 LQG 480
>gb|EAL33897.1| GA17553-PA [Drosophila pseudoobscura]
Length = 716
Score = 105 bits (261), Expect = 3e-22
Identities = 61/123 (49%), Positives = 75/123 (60%), Gaps = 9/123 (7%)
Query: 1 MDEEFRKIFEKFSFTEVAASEETDKKDVAEETAATKKKANSDSDYEDEENDNEQKEKGVS 60
M +F ++FE F E DK + ET A KKA +D ED+E +S
Sbjct: 216 MYRQFYRVFELFKLENKPKPVEKDKLALDSETVAKAKKA-ADKLQEDKEK--------LS 266
Query: 61 NKKKKLQRRMKIAELKQVSSRPDVVEVWDATAADPKLLVFLKSYRNTVPVPRHWSQKRKF 120
+K K R+ +AELKQ+ SRPDVVE+ D TA DPKLLV LK+YRNTV VPRHW KRK+
Sbjct: 267 KRKLKKLTRLSVAELKQLVSRPDVVEMHDVTARDPKLLVQLKAYRNTVQVPRHWCFKRKY 326
Query: 121 LQG 123
LQG
Sbjct: 327 LQG 329
>gb|AAH94200.1| Unknown (protein for MGC:115052) [Xenopus laevis]
Length = 764
Score = 103 bits (256), Expect = 1e-21
Identities = 55/124 (44%), Positives = 78/124 (62%), Gaps = 5/124 (4%)
Query: 5 FRKIFEKFSFTEVAASEETDKKDVAEETAATKKKANSDSDYEDEENDNEQKEKGVSN--- 61
F++IF+ F T+ E+ + + E+ + + + D+++D++ EK +
Sbjct: 259 FKRIFQAFKLTDDVKREKEKEPEKVEKIEPAVPRKRFEEELMDDDSDSDADEKKLDAPKL 318
Query: 62 KKKKLQR--RMKIAELKQVSSRPDVVEVWDATAADPKLLVFLKSYRNTVPVPRHWSQKRK 119
KKKL+R R +AELKQ+ SRPDVVE+ D TA DPKLLV LK RN+VPVPRHW KRK
Sbjct: 319 SKKKLRRMNRFTVAELKQLVSRPDVVEMHDVTAPDPKLLVHLKGTRNSVPVPRHWCFKRK 378
Query: 120 FLQG 123
+LQG
Sbjct: 379 YLQG 382
>ref|XP_638534.1| hypothetical protein DDB0218611 [Dictyostelium discoideum]
gi|60467168|gb|EAL65204.1| hypothetical protein
DDB0218611 [Dictyostelium discoideum]
Length = 554
Score = 103 bits (256), Expect = 1e-21
Identities = 52/114 (45%), Positives = 75/114 (65%), Gaps = 6/114 (5%)
Query: 16 EVAASEETDKKDVAEETAATKKKANSD-----SDYEDEENDNEQKE-KGVSNKKKKLQRR 69
E EE ++K+ EE KK+ N D +D +D NDNE ++ K +SNK++K QR+
Sbjct: 120 EQEEQEEKERKEREEEEERKKKEDNEDDDDDNNDDDDNNNDNEDEDSKKLSNKERKRQRK 179
Query: 70 MKIAELKQVSSRPDVVEVWDATAADPKLLVFLKSYRNTVPVPRHWSQKRKFLQG 123
+ + LKQ+ RPDVVE+ D + +P L+ +KSYRNT+PVP HW QK+K+LQG
Sbjct: 180 LHLPILKQLVDRPDVVELHDVNSPNPGYLIAMKSYRNTIPVPAHWCQKKKYLQG 233
>emb|CAI75646.1| spliceosome-associated protein, putative [Theileria annulata]
Length = 706
Score = 100 bits (250), Expect = 6e-21
Identities = 50/120 (41%), Positives = 78/120 (64%), Gaps = 10/120 (8%)
Query: 4 EFRKIFEKFSFTEVAASEETDKKDVAEETAATKKKANSDSDYEDEENDNEQKEKGVSNKK 63
+F+ +FEKFS E + + V EE A +++ + ++D ++D ++ ++ +S KK
Sbjct: 94 DFKGVFEKFS--------ELENQSVEEEPAPVEQEPEEER-FDDSDSDEDESQRRMSAKK 144
Query: 64 K-KLQRRMKIAELKQVSSRPDVVEVWDATAADPKLLVFLKSYRNTVPVPRHWSQKRKFLQ 122
+L R + ELKQ + +P+VVE+WD TA+DPK LV+LK RNTVPVP HWS+K F+Q
Sbjct: 145 MMRLMNRPTLYELKQSAEKPEVVEIWDTTASDPKFLVWLKGQRNTVPVPSHWSEKTPFMQ 204
>ref|NP_702476.1| hypothetical protein PF14_0587 [Plasmodium falciparum 3D7]
gi|23497660|gb|AAN37200.1| hypothetical protein
[Plasmodium falciparum 3D7]
Length = 677
Score = 93.6 bits (231), Expect = 9e-19
Identities = 42/123 (34%), Positives = 79/123 (64%), Gaps = 5/123 (4%)
Query: 3 EEFRKIFEKFSFTEVAASEETDKKDVAEETAAT--KKKANSDSDYEDEENDNEQKEKGVS 60
E F +++KF ++ + + + K+ +E+ + + + D+D EDE ++ + +K +S
Sbjct: 80 ENFEDVYKKF---KLHSKDGIENKEFSEQIQKSYFEDSESDDNDEEDEHRNSMKYKKNIS 136
Query: 61 NKKKKLQRRMKIAELKQVSSRPDVVEVWDATAADPKLLVFLKSYRNTVPVPRHWSQKRKF 120
K KKL +R + +LK+ + +P++VEVWD T++DP V++K +N++PVP+ W QKRK+
Sbjct: 137 KKTKKLLKRPSVMKLKEFAPKPELVEVWDTTSSDPYFYVWIKCLKNSIPVPQQWCQKRKY 196
Query: 121 LQG 123
L G
Sbjct: 197 LHG 199
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.309 0.125 0.357
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 227,092,570
Number of Sequences: 2540612
Number of extensions: 8886406
Number of successful extensions: 124918
Number of sequences better than 10.0: 2850
Number of HSP's better than 10.0 without gapping: 1300
Number of HSP's successfully gapped in prelim test: 1618
Number of HSP's that attempted gapping in prelim test: 104514
Number of HSP's gapped (non-prelim): 13740
length of query: 136
length of database: 863,360,394
effective HSP length: 112
effective length of query: 24
effective length of database: 578,811,850
effective search space: 13891484400
effective search space used: 13891484400
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 67 (30.4 bits)
Medicago: description of AC129091.17


