

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC128638.4 - phase: 0
(561 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
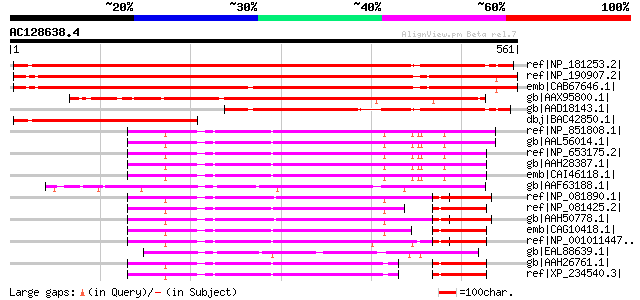
ref|NP_181253.2| transducin family protein / WD-40 repeat family... 843 0.0
ref|NP_190907.2| transducin family protein / WD-40 repeat family... 832 0.0
emb|CAB67646.1| putative protein [Arabidopsis thaliana] gi|11357... 811 0.0
gb|AAX95800.1| expressed protein [Oryza sativa (japonica cultiva... 473 e-132
gb|AAD18143.1| unknown protein [Arabidopsis thaliana] gi|2540853... 436 e-121
dbj|BAC42850.1| unknown protein [Arabidopsis thaliana] 285 2e-75
ref|NP_851808.1| WD repeat domain 20 isoform 1 [Homo sapiens] 239 2e-61
gb|AAL56014.1| DMR protein [Homo sapiens] 239 2e-61
ref|NP_653175.2| WD repeat domain 20 isoform 2 [Homo sapiens] 237 6e-61
gb|AAH28387.1| WD repeat domain 20, isoform 2 [Homo sapiens] gi|... 234 7e-60
emb|CAI46118.1| hypothetical protein [Homo sapiens] 233 2e-59
gb|AAF63188.1| CreC [Emericella nidulans] gi|25090015|sp|Q9P4R5|... 221 5e-56
ref|NP_081890.1| WD repeat domain 20 [Mus musculus] gi|28380207|... 214 6e-54
ref|NP_081425.2| WD repeat domain 20 [Mus musculus] 213 1e-53
gb|AAH50778.1| WD repeat domain 20 [Mus musculus] 213 1e-53
emb|CAG10418.1| unnamed protein product [Tetraodon nigroviridis] 211 4e-53
ref|NP_001011447.1| hypothetical protein LOC496935 [Xenopus trop... 211 5e-53
gb|EAL88639.1| catabolite repressor protein (CreC), putative [As... 202 3e-50
gb|AAH26761.1| 2310040A13Rik protein [Mus musculus] 196 2e-48
ref|XP_234540.3| PREDICTED: similar to 2310040A13Rik protein [Ra... 194 5e-48
>ref|NP_181253.2| transducin family protein / WD-40 repeat family protein
[Arabidopsis thaliana]
Length = 544
Score = 843 bits (2177), Expect = 0.0
Identities = 412/554 (74%), Positives = 468/554 (84%), Gaps = 15/554 (2%)
Query: 5 TTNGNMTAASSSSSSGPTTTTQSQQQGLKTYFKTPEGRYKLQFDKTHPSGLLQFNHGKTV 64
T+NG ++ SSSS+S Q G+KTYFKTPEG+YKL ++KTHPSGLL + HGKTV
Sbjct: 4 TSNGMISGPSSSSASA----ANPQSPGIKTYFKTPEGKYKLHYEKTHPSGLLHYTHGKTV 59
Query: 65 SMVTLAHLKEKPAPLTPTASSSSFSASSGVRSAAARLLGGSNGNRALSFVGGNGSSKSNG 124
+ VTLAHLK+KPAP TPT +SSS +ASSG RSA ARLLGG NGNRALSFVGGNG +K+ G
Sbjct: 60 TQVTLAHLKDKPAPSTPTGTSSSSTASSGFRSATARLLGGGNGNRALSFVGGNGGAKNVG 119
Query: 125 GASRIG-SIGSSSLSSSVANPNFDGKGSYLVFNAGDAILISDLNSQDKDPIKSIHFSNSN 183
+SRIG S +SS ++S N NFDGKG+YLVFN GDAI ISDLNSQDKDP+KSIH SNSN
Sbjct: 120 ASSRIGASFAASSSNTSATNTNFDGKGTYLVFNVGDAIFISDLNSQDKDPVKSIHISNSN 179
Query: 184 PVCHAFDQDAKDGHDLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCTC 243
P+CHAF DAKDGHDLLIGL SGDVY+VSLRQQLQDVGKK+V A HYNKDG +NNSRCT
Sbjct: 180 PMCHAFHPDAKDGHDLLIGLNSGDVYTVSLRQQLQDVGKKLVSAQHYNKDGSVNNSRCTS 239
Query: 244 ISWVPGGDGAFVVAHADGNLYVYEKNKDGAGESSFPILKDQTLFSVAHARYSKSNPIARW 303
I+WVPGGDG+FV AHADGNLYVYEKNK+GA +SSF ++D T FSV A+YSKSNP+ARW
Sbjct: 240 IAWVPGGDGSFVAAHADGNLYVYEKNKEGATDSSFSAIRDPTQFSVDKAKYSKSNPVARW 299
Query: 304 HICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKY 363
HI QG+INSI+FS DGAYLATVGRDGYLR+FD++ + LVCG KSYYG LLCCAWSMDGKY
Sbjct: 300 HIGQGAINSIAFSNDGAYLATVGRDGYLRIFDFSTQKLVCGVKSYYGALLCCAWSMDGKY 359
Query: 364 ILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVG 423
+LTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNS +GE + YRFGSVG
Sbjct: 360 LLTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSEGSGENVMYRFGSVG 419
Query: 424 QDTQLLLWELEMDEIVVPLRRGPPGGSPTFGSPTFSAGSQSSHWDNAVPLGTLQPAPSMR 483
QDTQLLLW+LEMDEIVVPLRR PPG GSPT+S GSQS+HWDN VP+GTLQPAP MR
Sbjct: 420 QDTQLLLWDLEMDEIVVPLRR-PPG-----GSPTYSTGSQSAHWDNIVPMGTLQPAPCMR 473
Query: 484 DVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTRPGVAESQPSNSETLLATS 543
DVPK+SP+VAHRVHTEPLS LIFTQES++TACREGH+KIWTRP ++QPS+S
Sbjct: 474 DVPKLSPVVAHRVHTEPLSGLIFTQESLITACREGHLKIWTRP---DTQPSSSSE-ATNP 529
Query: 544 LKEKPSLSSKISNS 557
KPSL+SK+ S
Sbjct: 530 TTSKPSLTSKVGGS 543
>ref|NP_190907.2| transducin family protein / WD-40 repeat family protein
[Arabidopsis thaliana]
Length = 558
Score = 832 bits (2148), Expect = 0.0
Identities = 416/566 (73%), Positives = 474/566 (83%), Gaps = 20/566 (3%)
Query: 5 TTNGNMTAASSSSSSGPTTTTQSQQQGLKTYFKTPEGRYKLQFDKTHPSGLLQFNHGKTV 64
T+NG ++A SSSSS P QS G+KTYFKTPEG+YKL ++KTH SGLL + HGKTV
Sbjct: 4 TSNGMISAPSSSSS--PAANPQSP--GIKTYFKTPEGKYKLHYEKTHSSGLLHYAHGKTV 59
Query: 65 SMVTLAHLKEKPAPLTPTASSSSFSASSGVRSAAARLLGGSNGNRALSFVGGNGSSKSNG 124
+ VT+A LKE+ AP TPT +SS +SASSG RSA ARLLG NG+RALSFVGGNG KS
Sbjct: 60 TQVTIAQLKERAAPSTPTGTSSGYSASSGFRSATARLLGTGNGSRALSFVGGNGGGKSVS 119
Query: 125 GASRI-GSIGSSSLSSSVANPNFDGKGSYLVFNAGDAILISDLNSQDKDPIKSIHFSNSN 183
+SRI GS +S+ ++S+ N NFDGKG+YLVFN DAI I DLNSQ+KDP+KSIHFSNSN
Sbjct: 120 TSSRISGSFAASNSNTSMTNTNFDGKGTYLVFNVSDAIFICDLNSQEKDPVKSIHFSNSN 179
Query: 184 PVCHAFDQDAKDGHDLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCTC 243
P+CHAFD DAKDGHDLLIGL SGDVY+VSLRQQLQDV KK+VGA HYNKDG +NNSRCT
Sbjct: 180 PMCHAFDPDAKDGHDLLIGLNSGDVYTVSLRQQLQDVSKKLVGALHYNKDGSVNNSRCTS 239
Query: 244 ISWVPGGDGAFVVAHADGNLYVYEKNKDGAGESSFPILKDQTLFSVAHARYSKSNPIARW 303
ISWVPGGDGAFVVAHADGNLYVYEKNKDGA ES+FP ++D T FSV A+YSKSNP+ARW
Sbjct: 240 ISWVPGGDGAFVVAHADGNLYVYEKNKDGATESTFPAIRDPTQFSVDKAKYSKSNPVARW 299
Query: 304 HICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKY 363
HICQGSINSI+FS DGA+LATVGRDGYLR+FD+ + LVCGGKSYYG LLCC+WSMDGKY
Sbjct: 300 HICQGSINSIAFSNDGAHLATVGRDGYLRIFDFLTQKLVCGGKSYYGALLCCSWSMDGKY 359
Query: 364 ILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVG 423
ILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDS WSSPNS+ +GE + YRFGSVG
Sbjct: 360 ILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSNWSSPNSDGSGEHVMYRFGSVG 419
Query: 424 QDTQLLLWELEMDEIVVPLRRGPPGGSPTFGSPTFSAGSQSSHWDNAVPLGTLQPAPSMR 483
QDTQLLLW+LEMDEIVVPLRR P GSPT+S GS S+HWDN +P+GTLQPAP R
Sbjct: 420 QDTQLLLWDLEMDEIVVPLRRAPG------GSPTYSTGS-SAHWDNVIPMGTLQPAPCKR 472
Query: 484 DVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTRPGVAESQPSNSE-----T 538
DVPK+SP++AHRVHTEPLS L+FTQESV+TACREGHIKIWTRP +E+Q ++SE
Sbjct: 473 DVPKLSPIIAHRVHTEPLSGLMFTQESVVTACREGHIKIWTRPSESETQTNSSEANPTTA 532
Query: 539 LLATSLKE--KPSLSSK-ISNSIYKQ 561
LL+TS + K SLSSK IS S +KQ
Sbjct: 533 LLSTSFTKDNKASLSSKIISGSTFKQ 558
>emb|CAB67646.1| putative protein [Arabidopsis thaliana] gi|11357803|pir||T45879
hypothetical protein F4P12.90 - Arabidopsis thaliana
Length = 556
Score = 811 bits (2095), Expect = 0.0
Identities = 411/568 (72%), Positives = 469/568 (82%), Gaps = 26/568 (4%)
Query: 5 TTNGNMTAASSSSSSGPTTTTQSQQQGLKTYFKTPEGRYKLQFDKTHPSGLLQFNHGKTV 64
T+NG ++A SSSSS P QS G+KTYFKTPEG+YKL ++KTH SGLL + HGKTV
Sbjct: 4 TSNGMISAPSSSSS--PAANPQSP--GIKTYFKTPEGKYKLHYEKTHSSGLLHYAHGKTV 59
Query: 65 SMVTLAHLKEKPAPLTPTASSSSFSASSGVRSAAARLLGGSNGNRALSFVGGNGSSKSNG 124
+ VT+A LKE+ AP TPT +SS +SASSG RSA ARLLG NG+RALSFVGGNG KS
Sbjct: 60 TQVTIAQLKERAAPSTPTGTSSGYSASSGFRSATARLLGTGNGSRALSFVGGNGGGKSVS 119
Query: 125 GASRI-GSIGSSSLSSSVANPNFDGKGSYLVFNAGDAILISDLNSQDKDPIKSIHFSNSN 183
+SRI GS +S+ ++S+ N NFDGKG+YLVFN DAI I DLNSQ+KDP+KSIHFSNSN
Sbjct: 120 TSSRISGSFAASNSNTSMTNTNFDGKGTYLVFNVSDAIFICDLNSQEKDPVKSIHFSNSN 179
Query: 184 PVCHAFDQDAKDGHDLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNN--SRC 241
P+CHAFD DAKDGHDLLIGL SGDVY+VSLRQQLQDV KK+VGA HYNKDG +NN RC
Sbjct: 180 PMCHAFDPDAKDGHDLLIGLNSGDVYTVSLRQQLQDVSKKLVGALHYNKDGSVNNRQDRC 239
Query: 242 TCISWVPGGDGAFVVAHADGNLYVYEKNKDGAGESSFPILKDQTLFSVAHARYSKSNPIA 301
T ISWVPGGDGAFVVAHADGNLY NKDGA ES+FP ++D T FSV A+YSKSNP+A
Sbjct: 240 TSISWVPGGDGAFVVAHADGNLY----NKDGATESTFPAIRDPTQFSVDKAKYSKSNPVA 295
Query: 302 RWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDG 361
RWHICQGSINSI+FS DGA+LATVGRDGYLR+FD+ + LVCGGKSYYG LLCC+WSMDG
Sbjct: 296 RWHICQGSINSIAFSNDGAHLATVGRDGYLRIFDFLTQKLVCGGKSYYGALLCCSWSMDG 355
Query: 362 KYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGS 421
KYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDS WSSPNS+ +GE + YRFGS
Sbjct: 356 KYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSNWSSPNSDGSGEHVMYRFGS 415
Query: 422 VGQDTQLLLWELEMDEIVVPLRRGPPGGSPTFGSPTFSAGSQSSHWDNAVPLGTLQPAPS 481
VGQDTQLLLW+LEMDEIVVPLRR P GSPT+S GS S+HWDN +P+GTLQPAP
Sbjct: 416 VGQDTQLLLWDLEMDEIVVPLRRAPG------GSPTYSTGS-SAHWDNVIPMGTLQPAPC 468
Query: 482 MRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTRPGVAESQPSNSE---- 537
RDVPK+SP++AHRVHTEPLS L+FTQESV+TACREGHIKIWTRP +E+Q ++SE
Sbjct: 469 KRDVPKLSPIIAHRVHTEPLSGLMFTQESVVTACREGHIKIWTRPSESETQTNSSEANPT 528
Query: 538 -TLLATSLKE--KPSLSSK-ISNSIYKQ 561
LL+TS + K SLSSK IS S +KQ
Sbjct: 529 TALLSTSFTKDNKASLSSKIISGSTFKQ 556
>gb|AAX95800.1| expressed protein [Oryza sativa (japonica cultivar-group)]
Length = 522
Score = 473 bits (1218), Expect = e-132
Identities = 256/498 (51%), Positives = 331/498 (66%), Gaps = 58/498 (11%)
Query: 67 VTLAHLKEKPAPLTPTASSSSFSASSGVRSAAARLLGGSNGNRALSFVGGNGSSKSNGGA 126
+T+A+LKE P+ TP+ S +SG+RS AARLLG G R LSFVG NG GG
Sbjct: 1 MTVAYLKEMPS--TPSIPS----CTSGMRSVAARLLGAGTGTRPLSFVGSNGV----GGR 50
Query: 127 SRIGSIGSSSLSSSVANPNFDGKGSYLVFNAGDAILISDLNSQDKDPIKSIHFSNSNPVC 186
S GS + S ++ N + D +G+Y++FN D++ I DLN + P+K I FS+ P+C
Sbjct: 51 SASGSCHAGQ-SRALVNYD-DDRGTYIIFNIADSLFIRDLNY--RRPVKRICFSDMKPLC 106
Query: 187 HAFDQDAKDGHDLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCTCISW 246
HAFD +AKDGHDL++G SG+VYS+SLRQQL++ G + + H+ NSRCT ++W
Sbjct: 107 HAFDSEAKDGHDLIVGFLSGEVYSMSLRQQLKEPGPEPIALQHFFN----TNSRCTGVAW 162
Query: 247 VPGGDGAFVVAHADGNLYVYEKNKDGAGESSFPILKDQTLFSVAHARYSKSNPIARWHIC 306
VPG +G FVV++ADGNL+VY+K+KD + +FP ++DQ+ +++A+ SKSNP+ARWHIC
Sbjct: 163 VPGHEGFFVVSNADGNLFVYDKSKDVNTDWTFPTVEDQSEMKISYAKSSKSNPVARWHIC 222
Query: 307 QGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILT 366
QG+IN+ISFS DG YLAT+GRDGYLRVFD+ KE L+ GGKSY+G LLCC+WS DGKY+L+
Sbjct: 223 QGAINAISFSPDGTYLATIGRDGYLRVFDFAKEQLIFGGKSYFGALLCCSWSTDGKYLLS 282
Query: 367 GGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWS---------------------- 404
GGEDDLVQVWSM DRK+VAWGEGH SWVS VAFDSYWS
Sbjct: 283 GGEDDLVQVWSMHDRKMVAWGEGHKSWVSAVAFDSYWSPPKPYERKQNSMHRFASPKSDE 342
Query: 405 ----------SPNSNDNGE--TITYRFGSVGQDTQLLLWELEMDEIVVPLRRGPP-GGSP 451
SP S++ E I YRF S+GQD QLLLW+L DE+ V L S
Sbjct: 343 AEEDPIYSFASPKSDETKENTNIMYRFASIGQDAQLLLWDLTKDELNVSLTHASSCSESS 402
Query: 452 TFGSPTFSAGSQSSHW---DNAVPLGTLQPAPSMRDVPKISPLVAHRVHTEPLSSLIFTQ 508
+ GS + S+ S SS D PLG L P+P +++VPK+SP VAH V EPL +L FT
Sbjct: 403 SSGSCSASSSSGSSSTEDRDKEFPLGFLHPSPRLQEVPKLSPEVAHLVGVEPLFTLEFTS 462
Query: 509 ESVLTACREGHIKIWTRP 526
ESV+T CR G +I TRP
Sbjct: 463 ESVITVCRRG--RITTRP 478
>gb|AAD18143.1| unknown protein [Arabidopsis thaliana] gi|25408536|pir||C84789
hypothetical protein At2g37160 [imported] - Arabidopsis
thaliana
Length = 444
Score = 436 bits (1121), Expect = e-121
Identities = 220/317 (69%), Positives = 251/317 (78%), Gaps = 36/317 (11%)
Query: 238 NSRCTCISWVPGGDGAFVVAHADGNLYVYEKNKDGAGESSFPILKDQTLFSVAHARYSKS 297
NSRCT I+WVPGGDG+FV AHADGNLY NK+GA +SSF ++D T FSV A+YSKS
Sbjct: 150 NSRCTSIAWVPGGDGSFVAAHADGNLY----NKEGATDSSFSAIRDPTQFSVDKAKYSKS 205
Query: 298 NPIARWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAW 357
NP+ARWHI QG+INSI+FS DGAYLATVGRDGYLR+FD++ + LVCG KSYYG LLCCAW
Sbjct: 206 NPVARWHIGQGAINSIAFSNDGAYLATVGRDGYLRIFDFSTQKLVCGVKSYYGALLCCAW 265
Query: 358 SMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITY 417
SMDGKY+LTGGEDDLVQVWSMEDRKVVAW EG +GE + Y
Sbjct: 266 SMDGKYLLTGGEDDLVQVWSMEDRKVVAW-EG---------------------SGENVMY 303
Query: 418 RFGSVGQDTQLLLWELEMDEIVVPLRRGPPGGSPTFGSPTFSAGSQSSHWDNAVPLGTLQ 477
RFGSVGQDTQLLLW+LEMDEIVVPLRR PPG GSPT+S GSQS+HWDN VP+GTLQ
Sbjct: 304 RFGSVGQDTQLLLWDLEMDEIVVPLRR-PPG-----GSPTYSTGSQSAHWDNIVPMGTLQ 357
Query: 478 PAPSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTRPGVAESQPSNSE 537
PAP MRDVPK+SP+VAHRVHTEPLS LIFTQES++TACREGH+KIWTRP ++QPS+S
Sbjct: 358 PAPCMRDVPKLSPVVAHRVHTEPLSGLIFTQESLITACREGHLKIWTRP---DTQPSSSS 414
Query: 538 TLLATSLKEKPSLSSKI 554
KPSL+SK+
Sbjct: 415 E-ATNPTTSKPSLTSKV 430
>dbj|BAC42850.1| unknown protein [Arabidopsis thaliana]
Length = 206
Score = 285 bits (730), Expect = 2e-75
Identities = 143/204 (70%), Positives = 165/204 (80%), Gaps = 5/204 (2%)
Query: 5 TTNGNMTAASSSSSSGPTTTTQSQQQGLKTYFKTPEGRYKLQFDKTHPSGLLQFNHGKTV 64
T+NG ++ SSSS+S Q G+KTYFKTPEG+YKL ++KTHPSGLL + HGKTV
Sbjct: 4 TSNGMISGPSSSSASA----ANPQSPGIKTYFKTPEGKYKLHYEKTHPSGLLHYTHGKTV 59
Query: 65 SMVTLAHLKEKPAPLTPTASSSSFSASSGVRSAAARLLGGSNGNRALSFVGGNGSSKSNG 124
+ VTLAHLK+KPAP TPT +SSS +ASSG RSA ARLLGG NGNRALSFVGGNG +K+ G
Sbjct: 60 TQVTLAHLKDKPAPSTPTGTSSSSTASSGFRSATARLLGGGNGNRALSFVGGNGGAKNVG 119
Query: 125 GASRIG-SIGSSSLSSSVANPNFDGKGSYLVFNAGDAILISDLNSQDKDPIKSIHFSNSN 183
+SRIG S +SS ++S N NFDGKG+YLVFN GDAI ISDLNSQDKDP+KSIH SNSN
Sbjct: 120 ASSRIGASFAASSSNTSATNTNFDGKGTYLVFNVGDAIFISDLNSQDKDPVKSIHISNSN 179
Query: 184 PVCHAFDQDAKDGHDLLIGLFSGD 207
P+CHAF DAKDGHDLLIGL SGD
Sbjct: 180 PMCHAFHPDAKDGHDLLIGLNSGD 203
>ref|NP_851808.1| WD repeat domain 20 isoform 1 [Homo sapiens]
Length = 581
Score = 239 bits (610), Expect = 2e-61
Identities = 179/540 (33%), Positives = 242/540 (44%), Gaps = 142/540 (26%)
Query: 131 SIGSSSLSSSVANPNFD-GKGSYLVFNAGDAILISDLNSQDK-----DPIKSIHFSNSNP 184
S GS+ + S N N G G L FN G + K PI + + P
Sbjct: 42 SQGSNPVRVSFVNLNDQSGNGDRLCFNVGRELYFYIYKGVRKAADLSKPIDKRIYKGTQP 101
Query: 185 VCHAFDQ--DAKDGHDLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCT 242
CH F+ + LL+G +G V QL D KK + +N++ +++ SR T
Sbjct: 102 TCHDFNHLTATAESVSLLVGFSAGQV-------QLIDPIKKET-SKLFNEERLIDKSRVT 153
Query: 243 CISWVPGGDGAFVVAHADGNLYVYE-KNKDGAGESSFPILKDQTLFSVAHARYSKS--NP 299
C+ WVPG + F+VAH+ GN+Y+Y ++ G + +LK F+V H SKS NP
Sbjct: 154 CVKWVPGSESLFLVAHSSGNMYLYNVEHTCGTTAPHYQLLKQGESFAV-HTCKSKSTRNP 212
Query: 300 IARWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSM 359
+ +W + +G++N +FS DG +LA V +DG+LRVF++ L KSY+GGLLC WS
Sbjct: 213 LLKWTVGEGALNEFAFSPDGKFLACVSQDGFLRVFNFDSVELHGTMKSYFGGLLCVCWSP 272
Query: 360 DGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGE------ 413
DGKYI+TGGEDDLV VWS D +V+A G GH SWVS VAFD Y +S D E
Sbjct: 273 DGKYIVTGGEDDLVTVWSFVDCRVIARGHGHKSWVSVVAFDPYTTSVEEGDPMEFSGSDE 332
Query: 414 -------------------------------TITYRFGSVGQDTQLLLWELEMDEIV--V 440
++TYRFGSVGQDTQL LW+L D +
Sbjct: 333 DFQDLLHFGRDRANSTQSRLSKRNSTDSRPVSVTYRFGSVGQDTQLCLWDLTEDILFPHQ 392
Query: 441 PLRR-----------GPPGGS-----PTFG---------------SPTFSAGSQSSHWDN 469
PL R PP GS T G S +AGS+SS D
Sbjct: 393 PLSRARTHTNVMNATSPPAGSNGNSVTTPGNSVPPPLPRSNSLPHSAVSNAGSKSSVMDG 452
Query: 470 AVPLGTLQPA-------------------------------------------------- 479
A+ G + A
Sbjct: 453 AIASGVSKFATLSLHDRKERHHEKDHKRNHSMGHISSKSSDKLNLVTKTKTDPAKTLGTP 512
Query: 480 --PSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTRPGVAESQPSNSE 537
P M DVP + PL+ ++ E L+ LIF ++ ++TAC+EG I W RPG S S S+
Sbjct: 513 LCPRMEDVPLLEPLICKKIAHERLTVLIFLEDCIVTACQEGFICTWGRPGKVGSLSSPSQ 572
>gb|AAL56014.1| DMR protein [Homo sapiens]
Length = 572
Score = 239 bits (609), Expect = 2e-61
Identities = 179/540 (33%), Positives = 242/540 (44%), Gaps = 142/540 (26%)
Query: 131 SIGSSSLSSSVANPNFD-GKGSYLVFNAGDAILISDLNSQDK-----DPIKSIHFSNSNP 184
S GS+ + S N N G G L FN G + K PI + + P
Sbjct: 33 SQGSNPVRVSFVNLNDQSGNGDRLCFNVGRELYFYIYKGVRKAADLSKPIDKRIYKGTQP 92
Query: 185 VCHAFDQ--DAKDGHDLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCT 242
CH F+ + LL+G +G V QL D KK + +N++ +++ SR T
Sbjct: 93 TCHDFNHLTATAESVSLLVGFSAGQV-------QLIDPIKKET-SKLFNEERLIDKSRVT 144
Query: 243 CISWVPGGDGAFVVAHADGNLYVYE-KNKDGAGESSFPILKDQTLFSVAHARYSKS--NP 299
C+ WVPG + F+VAH+ GN+Y+Y ++ G + +LK F+V H SKS NP
Sbjct: 145 CVKWVPGSESLFLVAHSSGNMYLYNVEHTCGTTAPHYQLLKHGESFAV-HTCKSKSTRNP 203
Query: 300 IARWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSM 359
+ +W + +G++N +FS DG +LA V +DG+LRVF++ L KSY+GGLLC WS
Sbjct: 204 LLKWTVGEGALNEFAFSPDGKFLACVSQDGFLRVFNFDSVELHGTMKSYFGGLLCVCWSP 263
Query: 360 DGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGE------ 413
DGKYI+TGGEDDLV VWS D +V+A G GH SWVS VAFD Y +S D E
Sbjct: 264 DGKYIVTGGEDDLVTVWSFVDCRVIAKGHGHKSWVSVVAFDPYTTSVEEGDPMEFSGSDE 323
Query: 414 -------------------------------TITYRFGSVGQDTQLLLWELEMDEIV--V 440
++TYRFGSVGQDTQL LW+L D +
Sbjct: 324 DFQDLLHFGRDRANSTQSRLSKRNSTDSRPVSVTYRFGSVGQDTQLCLWDLTEDILFPHQ 383
Query: 441 PLRR-----------GPPGGS-----PTFG---------------SPTFSAGSQSSHWDN 469
PL R PP GS T G S +AGS+SS D
Sbjct: 384 PLSRARTHTNVMNATSPPAGSNGNSVTTPGNSVPPPLPRSNSLPHSAVSNAGSKSSVMDG 443
Query: 470 AVPLGTLQPA-------------------------------------------------- 479
A+ G + A
Sbjct: 444 AIASGVSKFATLSLHDRKERHHEKDHKRNHSMGHISSKSSDKLNLVTKTKTDPAKTLGTP 503
Query: 480 --PSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTRPGVAESQPSNSE 537
P M DVP + PL+ ++ E L+ LIF ++ ++TAC+EG I W RPG S S S+
Sbjct: 504 LCPRMEDVPLLEPLICKKIAHERLTVLIFLEDCIVTACQEGFICTWGRPGKVGSLSSPSQ 563
>ref|NP_653175.2| WD repeat domain 20 isoform 2 [Homo sapiens]
Length = 569
Score = 237 bits (605), Expect = 6e-61
Identities = 176/530 (33%), Positives = 238/530 (44%), Gaps = 142/530 (26%)
Query: 131 SIGSSSLSSSVANPNFD-GKGSYLVFNAGDAILISDLNSQDK-----DPIKSIHFSNSNP 184
S GS+ + S N N G G L FN G + K PI + + P
Sbjct: 42 SQGSNPVRVSFVNLNDQSGNGDRLCFNVGRELYFYIYKGVRKAADLSKPIDKRIYKGTQP 101
Query: 185 VCHAFDQ--DAKDGHDLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCT 242
CH F+ + LL+G +G V QL D KK + +N++ +++ SR T
Sbjct: 102 TCHDFNHLTATAESVSLLVGFSAGQV-------QLIDPIKKET-SKLFNEERLIDKSRVT 153
Query: 243 CISWVPGGDGAFVVAHADGNLYVYE-KNKDGAGESSFPILKDQTLFSVAHARYSKS--NP 299
C+ WVPG + F+VAH+ GN+Y+Y ++ G + +LK F+V H SKS NP
Sbjct: 154 CVKWVPGSESLFLVAHSSGNMYLYNVEHTCGTTAPHYQLLKQGESFAV-HTCKSKSTRNP 212
Query: 300 IARWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSM 359
+ +W + +G++N +FS DG +LA V +DG+LRVF++ L KSY+GGLLC WS
Sbjct: 213 LLKWTVGEGALNEFAFSPDGKFLACVSQDGFLRVFNFDSVELHGTMKSYFGGLLCVCWSP 272
Query: 360 DGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGE------ 413
DGKYI+TGGEDDLV VWS D +V+A G GH SWVS VAFD Y +S D E
Sbjct: 273 DGKYIVTGGEDDLVTVWSFVDCRVIARGHGHKSWVSVVAFDPYTTSVEEGDPMEFSGSDE 332
Query: 414 -------------------------------TITYRFGSVGQDTQLLLWELEMDEIV--V 440
++TYRFGSVGQDTQL LW+L D +
Sbjct: 333 DFQDLLHFGRDRANSTQSRLSKRNSTDSRPVSVTYRFGSVGQDTQLCLWDLTEDILFPHQ 392
Query: 441 PLRR-----------GPPGGS-----PTFG---------------SPTFSAGSQSSHWDN 469
PL R PP GS T G S +AGS+SS D
Sbjct: 393 PLSRARTHTNVMNATSPPAGSNGNSVTTPGNSVPPPLPRSNSLPHSAVSNAGSKSSVMDG 452
Query: 470 AVPLGTLQPA-------------------------------------------------- 479
A+ G + A
Sbjct: 453 AIASGVSKFATLSLHDRKERHHEKDHKRNHSMGHISSKSSDKLNLVTKTKTDPAKTLGTP 512
Query: 480 --PSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTRPG 527
P M DVP + PL+ ++ E L+ LIF ++ ++TAC+EG I W RPG
Sbjct: 513 LCPRMEDVPLLEPLICKKIAHERLTVLIFLEDCIVTACQEGFICTWGRPG 562
>gb|AAH28387.1| WD repeat domain 20, isoform 2 [Homo sapiens]
gi|28380188|sp|Q8TBZ3|WDR20_HUMAN WD-repeat protein 20
(DMR protein)
Length = 569
Score = 234 bits (596), Expect = 7e-60
Identities = 175/530 (33%), Positives = 237/530 (44%), Gaps = 142/530 (26%)
Query: 131 SIGSSSLSSSVANPNFD-GKGSYLVFNAGDAILISDLNSQDK-----DPIKSIHFSNSNP 184
S GS+ + S N N G G L FN G + K PI + + P
Sbjct: 42 SQGSNPVRVSFVNLNDQSGNGDRLCFNVGRELYFYIYKGVRKAADLSKPIDKRIYKGTQP 101
Query: 185 VCHAFDQ--DAKDGHDLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCT 242
CH F+ + LL+G +G V QL D KK + +N++ +++ SR T
Sbjct: 102 TCHDFNHLTATAESVSLLVGFSAGQV-------QLIDPIKKET-SKLFNEERLIDKSRVT 153
Query: 243 CISWVPGGDGAFVVAHADGNLYVYE-KNKDGAGESSFPILKDQTLFSVAHARYSKS--NP 299
C+ WV G + F+VAH+ GN+Y+Y ++ G + +LK F+V H SKS NP
Sbjct: 154 CVKWVHGSESLFLVAHSSGNMYLYNVEHTCGTTAPHYQLLKQGESFAV-HTCKSKSTRNP 212
Query: 300 IARWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSM 359
+ +W + +G++N +FS DG +LA V +DG+LRVF++ L KSY+GGLLC WS
Sbjct: 213 LLKWTVGEGALNEFAFSPDGKFLACVSQDGFLRVFNFDSVELHGTMKSYFGGLLCVCWSP 272
Query: 360 DGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGE------ 413
DGKYI+TGGEDDLV VWS D +V+A G GH SWVS VAFD Y +S D E
Sbjct: 273 DGKYIVTGGEDDLVTVWSFVDCRVIARGHGHKSWVSVVAFDPYTTSVEEGDPMEFSGSDE 332
Query: 414 -------------------------------TITYRFGSVGQDTQLLLWELEMDEIV--V 440
++TYRFGSVGQDTQL LW+L D +
Sbjct: 333 DFQDLLHFGRDRANSTQSRLSKRNSTDSRPVSVTYRFGSVGQDTQLCLWDLTEDILFPHQ 392
Query: 441 PLRR-----------GPPGGS-----PTFG---------------SPTFSAGSQSSHWDN 469
PL R PP GS T G S +AGS+SS D
Sbjct: 393 PLSRARTHTNVMNATSPPAGSNGNSVTTPGNSVPPPLPRSNSLPHSAVSNAGSKSSVMDG 452
Query: 470 AVPLGTLQPA-------------------------------------------------- 479
A+ G + A
Sbjct: 453 AIASGVSKFATLSLHDRKERHHEKDHKRNHSMGHISSKSSDKLNLVTKTKTDPAKTLGTP 512
Query: 480 --PSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTRPG 527
P M DVP + PL+ ++ E L+ LIF ++ ++TAC+EG I W RPG
Sbjct: 513 LCPRMEDVPLLEPLICKKIAHERLTVLIFLEDCIVTACQEGFICTWGRPG 562
>emb|CAI46118.1| hypothetical protein [Homo sapiens]
Length = 581
Score = 233 bits (593), Expect = 2e-59
Identities = 176/542 (32%), Positives = 238/542 (43%), Gaps = 154/542 (28%)
Query: 131 SIGSSSLSSSVANPNFD-GKGSYLVFNAGDAILISDLNSQDK-----------------D 172
S GS+ + S N N G G L FN G + K
Sbjct: 42 SQGSNPVRVSFVNLNDQSGNGDRLCFNVGRELYFYIYKGVRKVPTRASPEPASGAADLSK 101
Query: 173 PIKSIHFSNSNPVCHAFDQ--DAKDGHDLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHY 230
PI + + P CH F+ + LL+G +G V QL D KK + +
Sbjct: 102 PIDKRIYKGTQPTCHDFNHLTATAESVSLLVGFSAGQV-------QLIDPIKKET-SKLF 153
Query: 231 NKDGILNNSRCTCISWVPGGDGAFVVAHADGNLYVYE-KNKDGAGESSFPILKDQTLFSV 289
N++ +++ SR TC+ WVPG + F+VAH+ GN+Y+Y ++ G + +LK F+V
Sbjct: 154 NEERLIDKSRVTCVKWVPGSESLFLVAHSSGNMYLYNVEHTCGTTAPHYQLLKQGESFAV 213
Query: 290 AHARYSKS--NPIARWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKS 347
H SKS NP+ +W + +G++N +FS DG +LA V +DG+LRVF++ L KS
Sbjct: 214 -HTCKSKSTRNPLLKWTVGEGALNEFAFSPDGKFLACVSQDGFLRVFNFDSVELHGTMKS 272
Query: 348 YYGGLLCCAWSMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPN 407
Y+GGLLC WS DGKYI+TGGEDDLV VWS D +V+A G GH SWVS VAFD Y +S
Sbjct: 273 YFGGLLCVCWSPDGKYIVTGGEDDLVTVWSFVDCRVIARGHGHKSWVSVVAFDPYTTSVE 332
Query: 408 SNDNGE-------------------------------------TITYRFGSVGQDTQLLL 430
D E ++TYRFGSVGQDTQL L
Sbjct: 333 EGDPMEFSGSDEDFQDLLHFGRDRANSTQSRLSKRNSTDSRPVSVTYRFGSVGQDTQLCL 392
Query: 431 WELEMDEIV--VPLRR-----------GPPGGS-----PTFG---------------SPT 457
W+L D + PL R PP GS T G S
Sbjct: 393 WDLTEDILFPHQPLSRARTHTNVMNATSPPAGSNGNSVTTPGNSVPPPLPRSNSLPHSAV 452
Query: 458 FSAGSQSSHWDNAVPLGTLQPA-------------------------------------- 479
+AGS+SS D A+ G + A
Sbjct: 453 SNAGSKSSVMDGAIASGVSKFATLSLHDRKERHHEKDHKRNHSMGHISSKSSDKLNLVTK 512
Query: 480 --------------PSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTR 525
P M DVP + PL+ ++ E L+ LIF ++ ++TAC+EG I W R
Sbjct: 513 TKTDPAKTLGTPLCPRMEDVPLLEPLICKKIAHERLTVLIFLEDCIVTACQEGFICTWGR 572
Query: 526 PG 527
PG
Sbjct: 573 PG 574
>gb|AAF63188.1| CreC [Emericella nidulans] gi|25090015|sp|Q9P4R5|CREC_EMENI
Catabolite repression protein creC
Length = 630
Score = 221 bits (563), Expect = 5e-56
Identities = 165/540 (30%), Positives = 255/540 (46%), Gaps = 82/540 (15%)
Query: 40 EGRYKLQFD---KTHPSGLLQFNHGKTVSMVTLAHLKEKPAPLTPTASSSSFSASSGVRS 96
EG Y+LQ D T P H ++ L P P PT+ S G+R+
Sbjct: 87 EGTYQLQDDLHLATPPP------HPSEAPIINPNPLATVPNP--PTSGVKLSLISLGLRN 138
Query: 97 ---------AAARLLGGSNGNRALSFVGGNGSSKSNGGASRIGSIGSSSLSSSVANP--- 144
AR G +GN AL+ SK + I SS +S + +
Sbjct: 139 KTAFPSKVQVTARPFG--DGNSALAAAPVKDPSKKRKPKNNIIKSSSSFVSRVIIHEATT 196
Query: 145 ----NFDGKGSYLVFNAGDAILISDLNSQDKD-PIKSIHFSNSNPVCHAFDQDAKDGH-- 197
+ D +G + N A DL+S+ KD P+ I F+ ++ + H ++ K
Sbjct: 197 KRLNDRDPEGLFAFANINRAFQWLDLSSKHKDEPLSKILFTKAHMISHDVNEITKSSAHI 256
Query: 198 DLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCTCISWVPGGDGAFVVA 257
D+++G +GD++ + + +K NK+GI+N+S T I W+PG + F+ A
Sbjct: 257 DVIMGSSAGDIF------WYEPISQKYA---RINKNGIINSSPVTHIKWIPGSENFFIAA 307
Query: 258 HADGNLYVYEKNKDGAGESSFPILKDQTLFSVAHARYS------------KSNPIARWHI 305
H +G L VY+K K+ A P L +Q+ SV +R+S K+NP+A W +
Sbjct: 308 HENGQLVVYDKEKEDA--LFIPELPEQSAESVKPSRWSLQVLKSVNSKNQKANPVAVWRL 365
Query: 306 CQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYIL 365
I +FS D +LA V DG LR+ DY +E ++ +SYYGG C WS DGKYI+
Sbjct: 366 ANQKITQFAFSPDHRHLAVVLEDGTLRLMDYLQEEVLDVFRSYYGGFTCVCWSPDGKYIV 425
Query: 366 TGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQD 425
TGG+DDLV +WS+ +RK++A +GH+SWVS VAFD + + TYR GSVG D
Sbjct: 426 TGGQDDLVTIWSLPERKIIARCQGHDSWVSAVAFDPW--------RCDERTYRIGSVGDD 477
Query: 426 TQLLLWELEM-----------------DEIVVPLRRGPPGGSPTFGSPTFSAGSQSSHWD 468
LLLW+ + ++ P + P + SQ + D
Sbjct: 478 CNLLLWDFSVGMLHRPKVHHQTSARHRTSLIAPSSQQPNRHRADSSGNRMRSDSQRTAAD 537
Query: 469 N-AVPLGTLQ-PAPSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTRP 526
+ + P +Q P S + P+++ V +P+ L F +++++T+ EGHI+ W RP
Sbjct: 538 SESAPDQPVQHPVESRARTALLPPIMSKAVGEDPICWLGFQEDTIMTSSLEGHIRTWDRP 597
>ref|NP_081890.1| WD repeat domain 20 [Mus musculus]
gi|28380207|sp|Q9D5R2|WDR20_MOUSE WD-repeat protein 20
gi|12853185|dbj|BAB29671.1| unnamed protein product [Mus
musculus]
Length = 567
Score = 214 bits (545), Expect = 6e-54
Identities = 146/406 (35%), Positives = 201/406 (48%), Gaps = 63/406 (15%)
Query: 131 SIGSSSLSSSVANPNFD-GKGSYLVFNAGDAILISDLNSQDK-----DPIKSIHFSNSNP 184
S GS+ + S N N G G+ L F+ G + + K PI + + P
Sbjct: 42 SQGSNPVRVSFVNLNDQTGNGNRLCFSVGRELYVYIYKGVRKAADLSKPIDKRIYKGTQP 101
Query: 185 VCHAFDQ--DAKDGHDLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCT 242
CH F+ + LL+G +G V QL D KK + +N++ +++ SR T
Sbjct: 102 TCHDFNHLTATAESVSLLVGFSAGQV-------QLIDPIKKET-SKLFNEERLIDKSRVT 153
Query: 243 CISWVPGGDGAFVVAHADGNLYVYE-KNKDGAGESSFPILKDQTLFSVAHARYSKSNPIA 301
C+ WVPG D F+VAH+ GN+Y Y ++ G + +LK F+V + + SKS
Sbjct: 154 CVKWVPGSDSLFLVAHSSGNMYSYNVEHTYGTTAPHYQLLKQGESFAVHNCK-SKSTRDL 212
Query: 302 RWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDG 361
+W + +G++N +FS DG +LA V +DG+LRVF++ L KSY+GGLLC WS DG
Sbjct: 213 KWTVGEGALNEFAFSPDGKFLACVSQDGFLRVFNFDSAELHGTMKSYFGGLLCLCWSPDG 272
Query: 362 KYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGE-------- 413
KYI+TGGEDDLV VWS D +V+A G GH SWVS VAFD Y +S +D E
Sbjct: 273 KYIVTGGEDDLVTVWSFLDCRVIARGRGHKSWVSVVAFDPYTTSVEESDPMEFSDSDKNF 332
Query: 414 -----------------------------TITYRFGSVGQDTQLLLWELEMDEIV--VPL 442
++TYRFGSVGQDTQL LW+L D + PL
Sbjct: 333 QDLLHFGRDRANSTQSRLSKQNSTDSRPVSVTYRFGSVGQDTQLCLWDLTEDILFPHQPL 392
Query: 443 RRGPPGGS--PTFGSPTFSAGSQSSHWDNAVPLGTLQPAPSMRDVP 486
R + G P S GS + N+VP P P +P
Sbjct: 393 SRARAHANVMNATGLPAGSNGSAVTTPGNSVP----PPLPRSNSLP 434
Score = 56.2 bits (134), Expect = 3e-06
Identities = 29/67 (43%), Positives = 42/67 (62%), Gaps = 2/67 (2%)
Query: 468 DNAVPLGTLQPAPSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTRPG 527
D A LGT P M DVP + PL+ ++ E L+ L+F ++ ++TAC+EG I W RPG
Sbjct: 502 DPAKTLGT-SLCPRMEDVPLLEPLICKKIAHERLTVLVFLEDCIVTACQEGFICTWARPG 560
Query: 528 -VAESQP 533
V++ QP
Sbjct: 561 KVSKFQP 567
>ref|NP_081425.2| WD repeat domain 20 [Mus musculus]
Length = 569
Score = 213 bits (543), Expect = 1e-53
Identities = 135/354 (38%), Positives = 183/354 (51%), Gaps = 57/354 (16%)
Query: 131 SIGSSSLSSSVANPNFD-GKGSYLVFNAGDAILISDLNSQDK-----DPIKSIHFSNSNP 184
S GS+ + S N N G G L FN G + K PI + + P
Sbjct: 42 SQGSNPVRVSFVNLNDQSGNGDRLCFNVGRELYFYIYKGVRKAADLSKPIDKRIYKGTQP 101
Query: 185 VCHAFD--QDAKDGHDLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCT 242
CH F+ + LL+G +G V QL D KK + +N++ +++ SR T
Sbjct: 102 TCHDFNLLTATAESVSLLVGFSAGQV-------QLIDPIKKET-SKLFNEERLIDKSRVT 153
Query: 243 CISWVPGGDGAFVVAHADGNLYVYE-KNKDGAGESSFPILKDQTLFSVAHARYSKS--NP 299
C+ WVPG + F+VAH+ GN+Y+Y ++ G + +LK F+V H SKS NP
Sbjct: 154 CVKWVPGSESLFLVAHSSGNMYLYNVEHTCGTTAPHYQLLKQGESFAV-HTCKSKSTRNP 212
Query: 300 IARWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSM 359
+ +W + +G++N +FS DG +LA V +DG+LRVF++ L KSY+GGLLC WS
Sbjct: 213 LLKWTVGEGALNEFAFSPDGKFLACVSQDGFLRVFNFDSVELHGTMKSYFGGLLCVCWSP 272
Query: 360 DGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGE------ 413
DGKYI+TGGEDDLV VWS D +V+A G GH SWVS VAFD Y +S +D E
Sbjct: 273 DGKYIVTGGEDDLVTVWSFLDCRVIARGHGHKSWVSVVAFDPYTTSVEESDPMEFSGSDE 332
Query: 414 -------------------------------TITYRFGSVGQDTQLLLWELEMD 436
++TYRFGSVGQDTQL LW+L D
Sbjct: 333 DFQDLLHFGRDRANSTQSRLSKRNSTDSRPVSVTYRFGSVGQDTQLCLWDLTED 386
Score = 54.7 bits (130), Expect = 8e-06
Identities = 26/60 (43%), Positives = 37/60 (61%), Gaps = 1/60 (1%)
Query: 468 DNAVPLGTLQPAPSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTRPG 527
D A LGT P M DVP + PL+ ++ E L+ L+F ++ ++TAC+EG I W RPG
Sbjct: 504 DPAKTLGT-SLCPRMEDVPLLEPLICKKIAHERLTVLVFLEDCIVTACQEGFICTWARPG 562
>gb|AAH50778.1| WD repeat domain 20 [Mus musculus]
Length = 567
Score = 213 bits (542), Expect = 1e-53
Identities = 146/406 (35%), Positives = 200/406 (48%), Gaps = 63/406 (15%)
Query: 131 SIGSSSLSSSVANPNFD-GKGSYLVFNAGDAILISDLNSQDK-----DPIKSIHFSNSNP 184
S GS+ + S N N G G+ L F+ G + + K PI + + P
Sbjct: 42 SQGSNPVRVSFVNLNDQTGNGNRLCFSVGRELYVYIYKGVRKAADLSKPIDKRIYKGTQP 101
Query: 185 VCHAFDQ--DAKDGHDLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCT 242
CH F+ + LL+G G V QL D KK + +N++ +++ SR T
Sbjct: 102 TCHDFNHLTATAESVSLLVGFSVGQV-------QLIDPIKKET-SKLFNEERLIDKSRVT 153
Query: 243 CISWVPGGDGAFVVAHADGNLYVYE-KNKDGAGESSFPILKDQTLFSVAHARYSKSNPIA 301
C+ WVPG D F+VAH+ GN+Y Y ++ G + +LK F+V + + SKS
Sbjct: 154 CVKWVPGSDSLFLVAHSSGNMYSYNVEHTYGTTAPHYQLLKQGESFAVHNCK-SKSTRDL 212
Query: 302 RWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDG 361
+W + +G++N +FS DG +LA V +DG+LRVF++ L KSY+GGLLC WS DG
Sbjct: 213 KWTVGEGALNEFAFSPDGKFLACVSQDGFLRVFNFDSAELHGTMKSYFGGLLCLCWSPDG 272
Query: 362 KYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGE-------- 413
KYI+TGGEDDLV VWS D +V+A G GH SWVS VAFD Y +S +D E
Sbjct: 273 KYIVTGGEDDLVTVWSFLDCRVIARGRGHKSWVSVVAFDPYTTSVEESDPMEFSDSDKNF 332
Query: 414 -----------------------------TITYRFGSVGQDTQLLLWELEMDEIV--VPL 442
++TYRFGSVGQDTQL LW+L D + PL
Sbjct: 333 QDLLHFGRDRANSTQSRLSKQNSTDSRPVSVTYRFGSVGQDTQLCLWDLTEDILFPHQPL 392
Query: 443 RRGPPGGS--PTFGSPTFSAGSQSSHWDNAVPLGTLQPAPSMRDVP 486
R + G P S GS + N+VP P P +P
Sbjct: 393 SRARAHANVMNATGLPAGSNGSAVTTPGNSVP----PPLPRSNSLP 434
Score = 56.2 bits (134), Expect = 3e-06
Identities = 29/67 (43%), Positives = 42/67 (62%), Gaps = 2/67 (2%)
Query: 468 DNAVPLGTLQPAPSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTRPG 527
D A LGT P M DVP + PL+ ++ E L+ L+F ++ ++TAC+EG I W RPG
Sbjct: 502 DPAKTLGT-SLCPRMEDVPLLEPLICKKIAHERLTVLVFLEDCIVTACQEGFICTWARPG 560
Query: 528 -VAESQP 533
V++ QP
Sbjct: 561 KVSKFQP 567
>emb|CAG10418.1| unnamed protein product [Tetraodon nigroviridis]
Length = 626
Score = 211 bits (538), Expect = 4e-53
Identities = 136/365 (37%), Positives = 187/365 (50%), Gaps = 60/365 (16%)
Query: 131 SIGSSSLSSSVANPNFD-GKGSYLVFNAGDAILISDLNSQDK-----DPIKSIHFSNSNP 184
S GS+ + S N N G G + FN G + K PI + + P
Sbjct: 42 SQGSNPVKVSFVNVNDQSGNGDRICFNVGRELYFYIYKGVRKAADLSKPIDKRIYKGTQP 101
Query: 185 VCHAFDQ--DAKDGHDLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCT 242
CH F+ + LL+G +G V QL D KK + +N++ +++ SR T
Sbjct: 102 TCHDFNHLTATAESVSLLVGFSAGQV-------QLIDPIKKET-SKLFNEERLIDKSRVT 153
Query: 243 CISWVPGGDGAFVVAHADGNLYVYE-KNKDGAGESSFPILKDQTLFSVAHARYSKS--NP 299
C+ WVPG + F+V+HA GN+Y+Y ++ G + +LK +SV H SKS NP
Sbjct: 154 CVKWVPGSESLFLVSHASGNMYLYNVEHTCGTTAPHYQLLKQGESYSV-HTCKSKSTRNP 212
Query: 300 IARWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSM 359
+ +W + +G++N +FS DG +LA V +DG+LRVF++ L KSY+GGLLC WS
Sbjct: 213 LLKWTVGEGALNEFAFSPDGKFLACVSQDGFLRVFNFDAVELHGTMKSYFGGLLCVCWSP 272
Query: 360 DGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGE------ 413
DGKYI+ GGEDDLV VWS D +V+A G GH SWVS VAFD Y +S +D E
Sbjct: 273 DGKYIVAGGEDDLVTVWSFLDCRVIARGHGHKSWVSVVAFDHYTTSVEESDPMEFSGSDE 332
Query: 414 --------------------------------TITYRFGSVGQDTQLLLWELEMDEIV-- 439
++TYRFGSVGQDTQL LW+L D +
Sbjct: 333 DFQDHVIHFGRDRANSTQSRLSKRNSTDSRPVSVTYRFGSVGQDTQLCLWDLTEDILFPH 392
Query: 440 VPLRR 444
+PL R
Sbjct: 393 LPLSR 397
Score = 56.2 bits (134), Expect = 3e-06
Identities = 28/60 (46%), Positives = 38/60 (62%), Gaps = 1/60 (1%)
Query: 468 DNAVPLGTLQPAPSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTRPG 527
D A LGT Q P M DVP + PL+ ++ E L+ LIF ++ ++TAC+EG I W RPG
Sbjct: 549 DPAKTLGT-QLCPRMEDVPLLEPLICKKIAHERLTVLIFLEDCLVTACQEGFICTWARPG 607
>ref|NP_001011447.1| hypothetical protein LOC496935 [Xenopus tropicalis]
gi|56971909|gb|AAH88535.1| Hypothetical LOC496935
[Xenopus tropicalis]
Length = 574
Score = 211 bits (537), Expect = 5e-53
Identities = 150/417 (35%), Positives = 204/417 (47%), Gaps = 76/417 (18%)
Query: 131 SIGSSSLSSSVANPNFD-GKGSYLVFNAGDAILISDLNSQDK-----DPIKSIHFSNSNP 184
S GS+ + S N N G G + FN G + K PI + + P
Sbjct: 42 SQGSNPVKVSFVNVNDQSGNGDRICFNVGRELYFYIYKGVRKAADLSKPIDKRIYKGTQP 101
Query: 185 VCHAFDQ--DAKDGHDLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCT 242
CH F+ D LL+G +G V QL D KK + +N++ +++ SR T
Sbjct: 102 TCHDFNHLTATADSVSLLVGFSAGQV-------QLIDPIKKET-SKLFNEERLIDKSRVT 153
Query: 243 CISWVPGGDGAFVVAHADGNLYVYE-KNKDGAGESSFPILKDQTLFSVAHARYSKS--NP 299
+ WVPG + F+VAH+ GN+Y+Y ++ G + +LK F+V H SKS NP
Sbjct: 154 SVKWVPGSESLFLVAHSSGNMYLYNVEHTCGTTAPHYQLLKQGESFAV-HTCKSKSTRNP 212
Query: 300 IARWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSM 359
+ +W + +G++N +FS DG +LA V +DG+LRVF++ L KSY+GGLLC WS
Sbjct: 213 LLKWTVGEGALNEFAFSPDGKFLACVSQDGFLRVFNFDSVELHGTMKSYFGGLLCVCWSP 272
Query: 360 DGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFD------------------- 400
DGKYI+TGGEDDLV VWS D +V+A G+GH SWVS VAFD
Sbjct: 273 DGKYIVTGGEDDLVTVWSFVDCRVIARGQGHKSWVSVVAFDPNTTSVEETDPMEFSGSDE 332
Query: 401 -----------------SYWSSPNSNDNGE-TITYRFGSVGQDTQLLLWELEMDEIV--V 440
S S NS D+ ++TYRFGSVGQDTQL LW+L D +
Sbjct: 333 DFQELISFGRDRANSTQSRLSKRNSTDSRPVSVTYRFGSVGQDTQLCLWDLTEDILFPHQ 392
Query: 441 PLRR-----------GPPGGSPTFGSPTFSAGSQSSHWDNAVPLGTLQPAPSMRDVP 486
PL R PP GS G S G+ + N++P P P +P
Sbjct: 393 PLSRTRTHTNVMNATSPPAGSG--GPNPGSNGNSITTPGNSIP----PPLPRSNSLP 443
Score = 54.7 bits (130), Expect = 8e-06
Identities = 27/60 (45%), Positives = 37/60 (61%), Gaps = 1/60 (1%)
Query: 468 DNAVPLGTLQPAPSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTRPG 527
D A LGT P M DVP + PL+ ++ E L+ LIF ++ ++TAC+EG I W RPG
Sbjct: 509 DPAKTLGT-PLCPRMEDVPLLEPLICKKIAHERLTVLIFLEDCIVTACQEGFICTWARPG 567
>gb|EAL88639.1| catabolite repressor protein (CreC), putative [Aspergillus
fumigatus Af293]
Length = 523
Score = 202 bits (513), Expect = 3e-50
Identities = 131/403 (32%), Positives = 203/403 (49%), Gaps = 59/403 (14%)
Query: 149 KGSYLVFNAGDAILISDLNSQDK-DPIKSIHFSNSNPVCHAFDQDAKDGH--DLLIGLFS 205
+G + N A DL+S+ K DP+ I F+ ++ +CH ++ K D+++G +
Sbjct: 144 EGLFAFANINRAFQWLDLSSKQKEDPLAKILFTKAHMLCHDINEVTKSPSHIDVVMGSSA 203
Query: 206 GDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCTCISWVPGGDGAFVVAHADGNLYV 265
GD+ + + +K NK+G++ NS T I W+PG + F+ +HA+G L V
Sbjct: 204 GDII------WYEPISQKYA---RINKNGVVCNSPVTHIKWIPGSENLFMASHANGQLVV 254
Query: 266 YEKNKDGAGESSFPILKDQTLFSV-------------AHARYSKSNPIARWHICQGSINS 312
Y+K K+ A + P ++DQ+ +V ++R K+NP+A W + I+
Sbjct: 255 YDKEKEDALFT--PEIQDQSAEAVKASSTQPLQVLKSVNSRNQKTNPVALWKLANQRISH 312
Query: 313 ISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGEDDL 372
+FS D +LA V DG LRV DY KE YYGGL+C WS DGKYI+TGG+DDL
Sbjct: 313 FAFSPDQRHLAVVLEDGSLRVMDYLKE-------DYYGGLICVCWSPDGKYIVTGGQDDL 365
Query: 373 VQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDTQLLLWE 432
V +WS +RK+VA +GHNSWVS VAFD + TYRFGSVG D +LLLW+
Sbjct: 366 VTIWSFPERKIVARCQGHNSWVSAVAFDPWQCDER--------TYRFGSVGDDCRLLLWD 417
Query: 433 LEMDEIVVP--------LRRGPPGGSPTF---------GSPTFSAGSQSSHWDNAVPLGT 475
+ + P R GS + G+ S ++++ +
Sbjct: 418 FSVGMLHRPKVHQTSARQRTSMVAGSSQYGNRHRADSVGNRMRSDSQRTANTYESCDQAV 477
Query: 476 LQPAPSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREG 518
P + P+++ V T+P+ L F ++ ++T+ EG
Sbjct: 478 RHPVEPRARTALLPPIMSKVVGTDPICWLGFQEDCIMTSSLEG 520
>gb|AAH26761.1| 2310040A13Rik protein [Mus musculus]
Length = 539
Score = 196 bits (498), Expect = 2e-48
Identities = 123/311 (39%), Positives = 169/311 (53%), Gaps = 25/311 (8%)
Query: 131 SIGSSSLSSSVANPNFD-GKGSYLVFNAGDAILISDLNSQDK-----DPIKSIHFSNSNP 184
S GS+ + S N N G G L FN G + K PI + + P
Sbjct: 42 SQGSNPVRVSFVNLNDQSGNGDRLCFNVGRELYFYIYKGVRKAADLSKPIDKRIYKGTQP 101
Query: 185 VCHAFD--QDAKDGHDLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCT 242
CH F+ + LL+G +G V QL D KK + +N++ +++ SR T
Sbjct: 102 TCHDFNLLTATAESVSLLVGFSAGQV-------QLIDPIKKET-SKLFNEERLIDKSRVT 153
Query: 243 CISWVPGGDGAFVVAHADGNLYVYE-KNKDGAGESSFPILKDQTLFSVAHARYSKS--NP 299
C+ WVPG + F+VAH+ GN+Y+Y ++ G + +LK F+V H SKS NP
Sbjct: 154 CVKWVPGSESLFLVAHSSGNMYLYNVEHTCGTTAPHYQLLKQGESFAV-HTCKSKSTRNP 212
Query: 300 IARWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSM 359
+ +W + +G++N +FS DG +LA V +DG+LRVF++ L KSY+GGLLC WS
Sbjct: 213 LLKWTVGEGALNEFAFSPDGKFLACVSQDGFLRVFNFDSVELHGTMKSYFGGLLCVCWSP 272
Query: 360 DGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRF 419
DGKYI+TGGEDDLV VWS D +V+A G GH SWVS VAFD Y +S +D E F
Sbjct: 273 DGKYIVTGGEDDLVTVWSFLDCRVIARGHGHKSWVSVVAFDPYTTSVEESDPME-----F 327
Query: 420 GSVGQDTQLLL 430
+D Q LL
Sbjct: 328 SGSDEDFQDLL 338
Score = 54.7 bits (130), Expect = 8e-06
Identities = 26/60 (43%), Positives = 37/60 (61%), Gaps = 1/60 (1%)
Query: 468 DNAVPLGTLQPAPSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTRPG 527
D A LGT P M DVP + PL+ ++ E L+ L+F ++ ++TAC+EG I W RPG
Sbjct: 474 DPAKTLGT-SLCPRMEDVPLLEPLICKKIAHERLTVLVFLEDCIVTACQEGFICTWARPG 532
>ref|XP_234540.3| PREDICTED: similar to 2310040A13Rik protein [Rattus norvegicus]
Length = 539
Score = 194 bits (494), Expect = 5e-48
Identities = 122/311 (39%), Positives = 169/311 (54%), Gaps = 25/311 (8%)
Query: 131 SIGSSSLSSSVANPNFD-GKGSYLVFNAGDAILISDLNSQDK-----DPIKSIHFSNSNP 184
S GS+ + S N N G G L FN G + K PI + + P
Sbjct: 42 SQGSNPVRVSFVNLNDQSGNGDRLCFNVGRELYFYIYKGVRKAADLSKPIDKRIYKGTQP 101
Query: 185 VCHAFDQ--DAKDGHDLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCT 242
CH F+ + LL+G +G V QL D KK + +N++ +++ SR T
Sbjct: 102 TCHDFNHLTATAESVSLLVGFSAGQV-------QLIDPIKKET-SKLFNEERLIDKSRVT 153
Query: 243 CISWVPGGDGAFVVAHADGNLYVYE-KNKDGAGESSFPILKDQTLFSVAHARYSKS--NP 299
C+ WVPG + F+VAH+ G++Y+Y ++ G + +LK F+V H SKS NP
Sbjct: 154 CVKWVPGSESLFLVAHSSGSMYLYNVEHTCGTTAPHYQLLKQGESFAV-HTCKSKSTRNP 212
Query: 300 IARWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSM 359
+ +W + +G++N +FS DG +LA V +DG+LRVF++ L KSY+GGLLC WS
Sbjct: 213 LLKWTVGEGALNEFAFSPDGKFLACVSQDGFLRVFNFDSVELHGTMKSYFGGLLCVCWSP 272
Query: 360 DGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRF 419
DGKYI+TGGEDDLV VWS D +V+A G GH SWVS VAFD Y +S +D E F
Sbjct: 273 DGKYIVTGGEDDLVTVWSFLDCRVIARGHGHKSWVSVVAFDPYTTSVEESDPME-----F 327
Query: 420 GSVGQDTQLLL 430
+D Q LL
Sbjct: 328 SGSDEDFQDLL 338
Score = 53.9 bits (128), Expect = 1e-05
Identities = 26/60 (43%), Positives = 37/60 (61%), Gaps = 1/60 (1%)
Query: 468 DNAVPLGTLQPAPSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWTRPG 527
D A LGT P M DVP + PL+ ++ E L+ L+F ++ ++TAC+EG I W RPG
Sbjct: 474 DPAKTLGT-SLCPRMEDVPLLEPLICKKIAHERLTVLVFLEDCLVTACQEGFICTWARPG 532
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.313 0.131 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,086,698,862
Number of Sequences: 2540612
Number of extensions: 52774820
Number of successful extensions: 188852
Number of sequences better than 10.0: 3245
Number of HSP's better than 10.0 without gapping: 1381
Number of HSP's successfully gapped in prelim test: 1907
Number of HSP's that attempted gapping in prelim test: 168531
Number of HSP's gapped (non-prelim): 14450
length of query: 561
length of database: 863,360,394
effective HSP length: 133
effective length of query: 428
effective length of database: 525,458,998
effective search space: 224896451144
effective search space used: 224896451144
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 78 (34.7 bits)
Medicago: description of AC128638.4


