

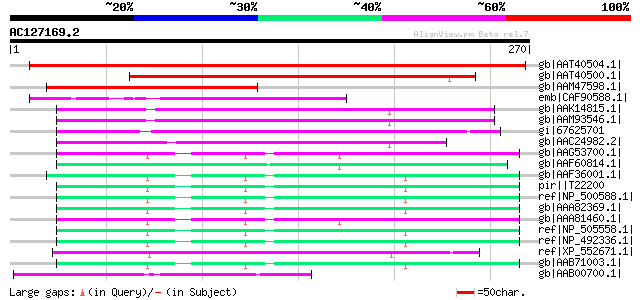
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC127169.2 - phase: 0
(270 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAT40504.1| putative polyprotein [Solanum demissum] 290 4e-77
gb|AAT40500.1| putative reverse transcriptase [Solanum demissum] 204 2e-51
gb|AAM47598.1| NBS/LRR resistance protein-like protein [Capsicum... 130 4e-29
emb|CAF90588.1| unnamed protein product [Tetraodon nigroviridis] 87 5e-16
gb|AAK14815.1| polyprotein [Schistosoma japonicum] 80 6e-14
gb|AAM93546.1| polyprotein [Schistosoma japonicum] 80 6e-14
gi|67625701 TPA: endonuclease-reverse transcriptase [Schistosoma... 77 7e-13
gb|AAC24982.2| reverse transcriptase [synthetic construct] 70 6e-11
gb|AAG53700.1| Hypothetical protein Y32G9A.9 [Caenorhabditis ele... 69 2e-10
gb|AAF60814.1| Hypothetical protein Y58G8A.2 [Caenorhabditis ele... 69 2e-10
gb|AAF36001.1| Hypothetical protein Y71F9AL.3 [Caenorhabditis el... 68 2e-10
pir||T22200 hypothetical protein F44G3.3 - Caenorhabditis elegans 67 4e-10
ref|NP_500588.1| reverse transcriptase family member (4F322) [Ca... 67 5e-10
gb|AAA82369.1| Hypothetical protein B0302.3 [Caenorhabditis eleg... 67 5e-10
gb|AAA81460.1| Hypothetical protein C24A3.7 [Caenorhabditis eleg... 67 5e-10
ref|NP_505558.1| predicted CDS, reverse transcriptase family mem... 67 5e-10
ref|NP_492336.1| reverse transcriptase family member, possibly N... 67 7e-10
ref|XP_552671.1| ENSANGP00000005174 [Anopheles gambiae str. PEST... 67 7e-10
gb|AAB71003.1| Hypothetical protein K10F12.5 [Caenorhabditis ele... 66 9e-10
gb|AAB00700.1| Hypothetical protein C34D4.5 [Caenorhabditis eleg... 66 9e-10
>gb|AAT40504.1| putative polyprotein [Solanum demissum]
Length = 832
Score = 290 bits (741), Expect = 4e-77
Identities = 139/258 (53%), Positives = 181/258 (69%)
Query: 11 VTEHIQELAPRCMLFADDVVLVGESREEVNGRLETWRQALEAYGFRLSRSKTEYIECNFS 70
+T IQE P CMLFADD+VL+ E+R+ VN RLE WRQ LE+ GFRLSR+KTEY+ C FS
Sbjct: 411 LTRSIQETVPWCMLFADDIVLIDETRDRVNARLEVWRQTLESKGFRLSRTKTEYLGCKFS 470
Query: 71 GRRSRSTLEVKVEDHIIPQVTRSKYLGSFVQNDGEIEADVSHRIQAGWLKWRRDSDVLCD 130
+ +EV++ +IP+ +YLG+ +Q G+I+ DV+HR+ A W+KWR S VLCD
Sbjct: 471 DGLDETDVEVRLAAQVIPKKESFRYLGAVIQGSGDIDDDVTHRVGAAWMKWRLASGVLCD 530
Query: 131 KKVPLKLKGKFYRPAFRPALMYGRECWAVKSQHENQVSVAEMRMLRWMSSKTRHDRTRND 190
KK+ KLKGKFYR RPAL+YG ECW VK+ H +++ VAEMRMLRWM TR D+ RN+
Sbjct: 531 KKISPKLKGKFYRVVVRPALLYGAECWPVKNAHVHKMHVAEMRMLRWMCGHTRSDKIRNE 590
Query: 191 TIRKRVEVAPIVEKLVENRLRWFGHVERRPVDAVVRRVYQMEDSQVKRGRGRPRKTIRET 250
IR++V VA +V+KL E RLRWFGHV+RR DA VRR M +RGRGRP+K E
Sbjct: 591 VIREKVGVASVVDKLREARLRWFGHVKRRSADAPVRRCEVMVVEGTRRGRGRPKKYWEEV 650
Query: 251 IRKDLEVNELDPNLVYDR 268
IR+DL + + ++ DR
Sbjct: 651 IRQDLAMLHITEDMTLDR 668
>gb|AAT40500.1| putative reverse transcriptase [Solanum demissum]
Length = 213
Score = 204 bits (519), Expect = 2e-51
Identities = 99/182 (54%), Positives = 128/182 (69%), Gaps = 2/182 (1%)
Query: 63 EYIECNFSGRRSRSTLEVKVEDHIIPQVTRSKYLGSFVQNDGEIEADVSHRIQAGWLKWR 122
EY+ C FS + +EV++ +IP+ KYLG+ +Q G+I+ DV+HR+ A W+KWR
Sbjct: 7 EYLGCKFSDVLDETDVEVRLAAQVIPKKESFKYLGAVIQGSGDIDDDVTHRVGAAWMKWR 66
Query: 123 RDSDVLCDKKVPLKLKGKFYRPAFRPALMYGRECWAVKSQHENQVSVAEMRMLRWMSSKT 182
S VLCDKK+PLKLKGKFYR RPAL+YG ECW VK+ H +++ VAEMRMLRWM T
Sbjct: 67 LASGVLCDKKIPLKLKGKFYRVVVRPALLYGAECWPVKNAHVHKMHVAEMRMLRWMCGHT 126
Query: 183 RHDRTRNDTIRKRVEVAPIVEKLVENRLRWFGHVERRPVDAVVRR--VYQMEDSQVKRGR 240
R D+ RN+ IR++V VA +V+KL E RLRWFGHV+RR DA VRR V +E S RG
Sbjct: 127 RSDKIRNEVIREKVGVASVVDKLREARLRWFGHVKRRSADAPVRRCEVMVVEASLPPRGS 186
Query: 241 GR 242
G+
Sbjct: 187 GK 188
>gb|AAM47598.1| NBS/LRR resistance protein-like protein [Capsicum annuum]
Length = 122
Score = 130 bits (327), Expect = 4e-29
Identities = 60/110 (54%), Positives = 82/110 (74%)
Query: 20 PRCMLFADDVVLVGESREEVNGRLETWRQALEAYGFRLSRSKTEYIECNFSGRRSRSTLE 79
P CMLFADDVVL+ E+R VN +LE WRQ LE+ GFR+SR+KTEY+EC F+ R + +
Sbjct: 10 PWCMLFADDVVLIDETRGGVNDKLELWRQTLESKGFRVSRTKTEYVECKFNDVRRENEVV 69
Query: 80 VKVEDHIIPQVTRSKYLGSFVQNDGEIEADVSHRIQAGWLKWRRDSDVLC 129
V++E + + + KYLGS +Q++GEI+ DVSHRI AGW+KW+ S V+C
Sbjct: 70 VRLEAQEVKKRDKFKYLGSVIQSNGEIDEDVSHRIGAGWMKWKLASGVMC 119
>emb|CAF90588.1| unnamed protein product [Tetraodon nigroviridis]
Length = 183
Score = 87.0 bits (214), Expect = 5e-16
Identities = 58/165 (35%), Positives = 92/165 (55%), Gaps = 17/165 (10%)
Query: 11 VTEHIQELAPRCMLFADDVVLVGESREEVNGRLETWRQALEAYGFRLSRSKTEYIECNFS 70
+T+ +++ +P +FA D+V+ SRE+V +LE R ALE+ SKTE E + S
Sbjct: 29 LTDEVRQESPWTTMFAGDIVMC--SREQVEEKLEERRFALES-------SKTEN-ERDLS 78
Query: 71 GRRSRSTLEVKVEDHIIPQVTRSKYLGSFVQNDGEIEADVSHRIQAGWLKWRRDSDVLCD 130
G V+++ I + K LGS VQ GE +V R+QAGW W + S V+CD
Sbjct: 79 G-------SVRLQGEEIKKGEDLKNLGSTVQTSGECGKEVKKRVQAGWNWWGKVSGVMCD 131
Query: 131 KKVPLKLKGKFYRPAFRPALMYGRECWAVKSQHENQVSVAEMRML 175
+ V K+K K + RPA+++G E ++ + E ++ +AEM+ L
Sbjct: 132 RGVSAKIKRKVDKTVARPAIIFGLETVPLRKRQEAELELAEMKAL 176
>gb|AAK14815.1| polyprotein [Schistosoma japonicum]
Length = 1091
Score = 80.1 bits (196), Expect = 6e-14
Identities = 62/232 (26%), Positives = 110/232 (46%), Gaps = 8/232 (3%)
Query: 25 FADDVVLVGESREEVNGRLETWRQALEAYGFRLSRSKTEYIECNFSGRRSRSTLEVKVED 84
+ADD+VL+ E +++ L T + G R S SK + + ++ S ++ +
Sbjct: 812 YADDIVLLSEDADKMQDFLTTLNMNVSMLGMRFSPSKCKMLLQDWLN----SAPKLVIGR 867
Query: 85 HIIPQVTRSKYLGSFVQNDGEIEADVSHRIQAGWLKWRRDSDVLCDKKVPLKLKGKFYRP 144
I V R YLGS + +G + ++S RI + + + + L KG+ Y
Sbjct: 868 ETIECVNRFTYLGSLISPNGLVSDEISARIHKARSAFANLRHLWRRRDIRLMTKGRVYCA 927
Query: 145 AFRPALMYGRECWAVKSQHENQVSVAEMRMLRWMSSKTRHDRTRNDTIRKRV--EVAPIV 202
A R L YG E W ++ + ++ V + R LR ++ +R N +R RV + +
Sbjct: 928 AVRSVLPYGCETWPLRVEDIRRILVFDHRCLRNIARVCWDNRVSNAWVRNRVLGKYGKSI 987
Query: 203 EKLVE-NRLRWFGHVERRPVDAVVRR-VYQMEDSQVKRGRGRPRKTIRETIR 252
+++V +RLRW GHV R P + RR + + K+ RG KT ++++
Sbjct: 988 DEVVNLHRLRWLGHVLRMPDHRLPRRAMLSVVGVGWKKARGGQTKTWHQSMK 1039
>gb|AAM93546.1| polyprotein [Schistosoma japonicum]
Length = 976
Score = 80.1 bits (196), Expect = 6e-14
Identities = 62/232 (26%), Positives = 110/232 (46%), Gaps = 8/232 (3%)
Query: 25 FADDVVLVGESREEVNGRLETWRQALEAYGFRLSRSKTEYIECNFSGRRSRSTLEVKVED 84
+ADD+VL+ E +++ L T + G R S SK + + ++ S ++ +
Sbjct: 697 YADDIVLLSEDADKMQDFLTTLNMNVSMLGMRFSPSKCKMLLQDWLN----SAPKLVIGR 752
Query: 85 HIIPQVTRSKYLGSFVQNDGEIEADVSHRIQAGWLKWRRDSDVLCDKKVPLKLKGKFYRP 144
I V R YLGS + +G + ++S RI + + + + L KG+ Y
Sbjct: 753 ETIECVNRFTYLGSLISPNGLVSDEISARIHKARSAFANLRHLWRRRDIRLMTKGRVYCA 812
Query: 145 AFRPALMYGRECWAVKSQHENQVSVAEMRMLRWMSSKTRHDRTRNDTIRKRV--EVAPIV 202
A R L YG E W ++ + ++ V + R LR ++ +R N +R RV + +
Sbjct: 813 AVRSVLPYGCETWPLRVEDIRRILVFDHRCLRNIARVCWDNRVSNAWVRNRVLGKYGKSI 872
Query: 203 EKLVE-NRLRWFGHVERRPVDAVVRR-VYQMEDSQVKRGRGRPRKTIRETIR 252
+++V +RLRW GHV R P + RR + + K+ RG KT ++++
Sbjct: 873 DEVVNLHRLRWLGHVLRMPDHRLPRRAMLSVVGVGWKKARGGQTKTWHQSMK 924
>gi|67625701 TPA: endonuclease-reverse transcriptase [Schistosoma mansoni]
Length = 992
Score = 76.6 bits (187), Expect = 7e-13
Identities = 51/231 (22%), Positives = 101/231 (43%), Gaps = 6/231 (2%)
Query: 25 FADDVVLVGESREEVNGRLETWRQALEAYGFRLSRSKTEYIECNFSGRRSRSTLEVKVED 84
FADD+ L+ ++++++ + + A A G +++ K++ + N + T + ++
Sbjct: 732 FADDLALLSQTQQQMQEKTTSVAAASAAVGLNINKGKSKTLRYN-----TICTNPITLDG 786
Query: 85 HIIPQVTRSKYLGSFVQNDGEIEADVSHRIQAGWLKWRRDSDVLCDKKVPLKLKGKFYRP 144
+ V YLGS + G +ADV RI + + ++ K++ K + +
Sbjct: 787 EALEDVEIFTYLGSIIDEHGGSDADVRARIGKARAAYLQLKNIWSSKQLSTNTKVRIFNT 846
Query: 145 AFRPALMYGRECWAVKSQHENQVSVAEMRMLRWMSSKTRHDRTRNDTIRKRVEVAPIVEK 204
+ L+YG E W ++ V LR + D N + + P E+
Sbjct: 847 NVKTVLLYGAETWRTTKAIIQKIQVFINSCLRKILRIRWPDTISNKLLWETTNQIPAEEE 906
Query: 205 LVENRLRWFGHVERRPVDAVVRRVYQMEDSQVKRGRGRPRKTIRETIRKDL 255
+ + R +W H R+ + V R+ + +R RGRP+ T+R I D+
Sbjct: 907 IRKKRWKWIWHTLRKSPNCVTRQALTWNPERQRR-RGRPKNTLRREIETDM 956
>gb|AAC24982.2| reverse transcriptase [synthetic construct]
Length = 1016
Score = 70.1 bits (170), Expect = 6e-11
Identities = 51/207 (24%), Positives = 95/207 (45%), Gaps = 8/207 (3%)
Query: 25 FADDVVLVGESREEVNGRLETWRQALEAYGFRLSRSKTEYIECNFSGRRSRSTLEVKVED 84
+ADD+VL+ ++ + + L ++ YG + SK + + ++ TL+ +
Sbjct: 737 YADDIVLLCDNAQGMQSALNQLAISVRRYGMCFAPSKCKVLLQDWQDSHPVLTLDGEQ-- 794
Query: 85 HIIPQVTRSKYLGSFVQNDGEIEADVSHRIQAGWLKWRRDSDVLCDKKVPLKLKGKFYRP 144
I V + YLGS++ G + ++ RI + + + V L +KG+ Y
Sbjct: 795 --IEVVEKFVYLGSYISAGGGVSDEIDARIMKARAAYANLGHLWRLRDVSLAVKGRIYNA 852
Query: 145 AFRPALMYGRECWAVKSQHENQVSVAEMRMLRWMSSKTRHDRTRNDTIRKRV----EVAP 200
+ R L+Y E W ++ + ++SV + R LR ++ N +R RV +
Sbjct: 853 SVRAVLLYACETWPLRVEDVRRLSVFDHRCLRRIADIQWQHHVSNAEVRHRVFGRRDDNS 912
Query: 201 IVEKLVENRLRWFGHVERRPVDAVVRR 227
I ++++RLRW GHV R + RR
Sbjct: 913 IGVTILKHRLRWLGHVLRMSSQRLPRR 939
>gb|AAG53700.1| Hypothetical protein Y32G9A.9 [Caenorhabditis elegans]
gi|17565388|ref|NP_503526.1| predicted CDS, reverse
transcriptase family member (5C378) [Caenorhabditis
elegans]
Length = 417
Score = 68.6 bits (166), Expect = 2e-10
Identities = 57/255 (22%), Positives = 103/255 (40%), Gaps = 26/255 (10%)
Query: 25 FADDVVLVGESREEVNGRLETWRQALEAYGFRLSRSKTEYIECNFS-------GRRSRST 77
FADD+VL+ + L+ Q G ++ KT+ + F+ G S +T
Sbjct: 151 FADDIVLIANHPNTASKMLQELVQKCSEVGLEINTGKTKVLRNRFADPSKVYFGNPSSTT 210
Query: 78 LEVKVEDHIIPQVTRSKYLGSFVQNDGEIEADVSHRIQAGWLKW---RRDSDVLCDKKVP 134
V+++I YLG + + ++ R +A W + + +D + DKK+
Sbjct: 211 QLDDVDEYI--------YLGRQINAQNNLMPEIHRRRRAAWAAFNGIKNTTDSITDKKI- 261
Query: 135 LKLKGKFYRPAFRPALMYGRECWAVKSQHENQVSVA----EMRMLRWMSSKTRHDRTRND 190
+ + PAL YG E W +V + E R++ ++ R +
Sbjct: 262 ---RANLFDSIVLPALTYGSEAWTFTKALSERVRITHASLERRLVGITLTQQRERDLHRE 318
Query: 191 TIRKRVEVAPIVEKLVENRLRWFGHVERRPVDAVVRRVYQMEDSQVKRGRGRPRKTIRET 250
IRK V + + + +L W GHV RR + + KR GRP ++
Sbjct: 319 DIRKMSLVRDPLNFVKKRKLGWAGHVARRKDGRWTTLMTEWRPYGWKRPVGRPPMRWTDS 378
Query: 251 IRKDLEVNELDPNLV 265
+RK++ + D ++
Sbjct: 379 LRKEITTRDADGEVI 393
>gb|AAF60814.1| Hypothetical protein Y58G8A.2 [Caenorhabditis elegans]
gi|17566042|ref|NP_503166.1| predicted CDS, reverse
transcriptase family member (5A739) [Caenorhabditis
elegans]
Length = 769
Score = 68.6 bits (166), Expect = 2e-10
Identities = 50/239 (20%), Positives = 97/239 (39%), Gaps = 5/239 (2%)
Query: 25 FADDVVLVGESREEVNGRLETWRQALEAYGFRLSRSKTEYIECNFSGRRSRSTLEVKVED 84
FADD+VLV + + L + + G +++ KT+ + F+ +R +
Sbjct: 519 FADDIVLVAHNPRTASQMLTELVEKCSSVGLKINTGKTKVLRNRFAYKRKVEIRCPNTTN 578
Query: 85 HIIPQVTRSKYLGSFVQNDGEIEADVSHRIQAGWLKWRRDSDVLCDKKVPLKLKGKFYRP 144
II V YLG + + + ++ R +A W + P +L+ +
Sbjct: 579 IIIDDVNEYIYLGRQINDSNNLLPELHRRRRAAWAAFTNIKSTTDQITCP-RLRANLFDS 637
Query: 145 AFRPALMYGRECWAVKSQHENQVSVA----EMRMLRWMSSKTRHDRTRNDTIRKRVEVAP 200
PAL YG E W + +V V E +++ ++ R + +R++ ++
Sbjct: 638 TVLPALTYGSEAWTFTKELAERVRVTHAALERKLVGLTLTEQRERNIHREEVREKSKLRD 697
Query: 201 IVEKLVENRLRWFGHVERRPVDAVVRRVYQMEDSQVKRGRGRPRKTIRETIRKDLEVNE 259
+ + + +L W GHV RR + KR GRP +++RK++ +
Sbjct: 698 PLIHIKKKKLGWAGHVARRTDGRWTTLMNDWCPRDEKRPVGRPPMRWNDSLRKEVTTRD 756
>gb|AAF36001.1| Hypothetical protein Y71F9AL.3 [Caenorhabditis elegans]
gi|17510457|ref|NP_491073.1| reverse transcriptase
family member (1D477) [Caenorhabditis elegans]
Length = 423
Score = 68.2 bits (165), Expect = 2e-10
Identities = 56/260 (21%), Positives = 103/260 (39%), Gaps = 26/260 (10%)
Query: 20 PRCMLFADDVVLVGESREEVNGRLETWRQALEAYGFRLSRSKTEYIECNFS-------GR 72
P + FADD+VL+ + L+ Q G ++ KT+ + F+ G
Sbjct: 152 PTNLRFADDIVLIANHPNTASKMLQELVQKCSEVGLEINTGKTKVLRNRFADPSKVYFGS 211
Query: 73 RSRSTLEVKVEDHIIPQVTRSKYLGSFVQNDGEIEADVSHRIQAGWLKW---RRDSDVLC 129
S +T V+++I YLG + + ++ R +A W + + +D +
Sbjct: 212 PSPTTQLDDVDEYI--------YLGRQINAQNNLMPEIHRRRRAAWAAFNGIKNTADSIT 263
Query: 130 DKKVPLKLKGKFYRPAFRPALMYGRECWAVKSQHENQVSVAEMRMLRWMSSKTRHDRTRN 189
DKK+ + + PAL YG E W +V + + R + T +
Sbjct: 264 DKKI----RANLFDSIVLPALTYGSEAWTFTKALSERVRITHASLERRLVGITLTQQRER 319
Query: 190 DTIRKRVEVAPIVEK----LVENRLRWFGHVERRPVDAVVRRVYQMEDSQVKRGRGRPRK 245
D R+ + +V + + +L W GHV RR + + KR GRP
Sbjct: 320 DLHREDIRTMSLVRDPLNFVKKRKLGWAGHVARRKDGRWTTLMTEWRPYGWKRPVGRPPM 379
Query: 246 TIRETIRKDLEVNELDPNLV 265
+++RK++ + D ++
Sbjct: 380 RWTDSLRKEITTRDADGEVI 399
>pir||T22200 hypothetical protein F44G3.3 - Caenorhabditis elegans
Length = 775
Score = 67.4 bits (163), Expect = 4e-10
Identities = 55/255 (21%), Positives = 101/255 (39%), Gaps = 26/255 (10%)
Query: 25 FADDVVLVGESREEVNGRLETWRQALEAYGFRLSRSKTEYIECNFS-------GRRSRST 77
FADD+VL+ + L+ Q G ++ KT+ + F+ G S +T
Sbjct: 509 FADDIVLIANHPNTASKMLQELVQKCSEVGLEINTGKTKVLRNRFADPSKVYFGSPSSTT 568
Query: 78 LEVKVEDHIIPQVTRSKYLGSFVQNDGEIEADVSHRIQAGWLKW---RRDSDVLCDKKVP 134
V+++I YLG + + ++ R +A W + + +D + DKK+
Sbjct: 569 QLDDVDEYI--------YLGRQINAQNNLMPEIHRRRRAAWAAFNGIKNTTDSITDKKI- 619
Query: 135 LKLKGKFYRPAFRPALMYGRECWAVKSQHENQVSVAEMRMLRWMSSKTRHDRTRNDTIRK 194
+ + PAL YG E W +V + + R + T + D R+
Sbjct: 620 ---RANLFDSIVLPALTYGSEAWTFTKALSERVRITHASLERRLVGITLTQQRERDLHRE 676
Query: 195 RVEVAPIVEK----LVENRLRWFGHVERRPVDAVVRRVYQMEDSQVKRGRGRPRKTIRET 250
+ +V + + +L W GHV RR + + KR GRP ++
Sbjct: 677 DIRTMSLVRDPLNFVKKRKLGWAGHVARRKDGRWTTLMTEWRPYGWKRPVGRPPMRWTDS 736
Query: 251 IRKDLEVNELDPNLV 265
+RK++ + D ++
Sbjct: 737 LRKEITTRDADGEVI 751
>ref|NP_500588.1| reverse transcriptase family member (4F322) [Caenorhabditis
elegans] gi|25396035|pir||H88671 protein F28E10.3
[imported] - Caenorhabditis elegans
Length = 553
Score = 67.0 bits (162), Expect = 5e-10
Identities = 55/255 (21%), Positives = 101/255 (39%), Gaps = 26/255 (10%)
Query: 25 FADDVVLVGESREEVNGRLETWRQALEAYGFRLSRSKTEYIECNFS-------GRRSRST 77
FADD+VL+ + L+ Q G ++ KT+ + F+ G S +T
Sbjct: 287 FADDIVLIANHPNTASKMLQELVQKCSEVGLEINTGKTKVLRNRFADPSKVYFGSPSPTT 346
Query: 78 LEVKVEDHIIPQVTRSKYLGSFVQNDGEIEADVSHRIQAGWLKW---RRDSDVLCDKKVP 134
V+++I YLG + + ++ R +A W + + +D + DKK+
Sbjct: 347 QLDDVDEYI--------YLGRQINAQNNLMPEIHRRRRAAWAAFNGIKNTTDSITDKKI- 397
Query: 135 LKLKGKFYRPAFRPALMYGRECWAVKSQHENQVSVAEMRMLRWMSSKTRHDRTRNDTIRK 194
+ + PAL YG E W +V + + R + T + D R+
Sbjct: 398 ---RANLFDSIVLPALTYGSEAWTFTKALSERVRITHASLERRLVGITLTQQRERDLHRE 454
Query: 195 RVEVAPIVEK----LVENRLRWFGHVERRPVDAVVRRVYQMEDSQVKRGRGRPRKTIRET 250
+ +V + + +L W GHV RR + + KR GRP ++
Sbjct: 455 DIRTMSLVRDPLNFVKKRKLGWAGHVARRKDGRWTTLMTEWRPYGWKRPVGRPPMRWTDS 514
Query: 251 IRKDLEVNELDPNLV 265
+RK++ + D ++
Sbjct: 515 LRKEITTRDADGEVI 529
>gb|AAA82369.1| Hypothetical protein B0302.3 [Caenorhabditis elegans]
gi|17549935|ref|NP_510780.1| predicted CDS, reverse
transcriptase family member (XR781) [Caenorhabditis
elegans] gi|7459643|pir||T15313 hypothetical protein
B0302.3 - Caenorhabditis elegans
Length = 279
Score = 67.0 bits (162), Expect = 5e-10
Identities = 55/255 (21%), Positives = 101/255 (39%), Gaps = 26/255 (10%)
Query: 25 FADDVVLVGESREEVNGRLETWRQALEAYGFRLSRSKTEYIECNFS-------GRRSRST 77
FADD+VL+ + L+ Q G ++ KT+ + F+ G S +T
Sbjct: 13 FADDIVLIANHPNTASKMLQELVQKCSEVGLEINTGKTKVLRNRFADPSKVYFGSPSPTT 72
Query: 78 LEVKVEDHIIPQVTRSKYLGSFVQNDGEIEADVSHRIQAGWLKW---RRDSDVLCDKKVP 134
V+++I YLG + + ++ R +A W + + +D + DKK+
Sbjct: 73 QLDDVDEYI--------YLGRQINAQNNLMPEIHRRRRAAWAAFNGIKNTTDSITDKKI- 123
Query: 135 LKLKGKFYRPAFRPALMYGRECWAVKSQHENQVSVAEMRMLRWMSSKTRHDRTRNDTIRK 194
+ + PAL YG E W +V + + R + T + D R+
Sbjct: 124 ---RANLFDSIVLPALTYGSEAWTFTKALSERVRITHASLERRLVGITLTQQRERDLHRE 180
Query: 195 RVEVAPIVEK----LVENRLRWFGHVERRPVDAVVRRVYQMEDSQVKRGRGRPRKTIRET 250
+ +V + + +L W GHV RR + + KR GRP ++
Sbjct: 181 DIRTMSLVRDPLNFVKKRKLGWAGHVARRKDGRWTTLMTEWRPYGWKRPVGRPPMRWTDS 240
Query: 251 IRKDLEVNELDPNLV 265
+RK++ + D ++
Sbjct: 241 LRKEITTRDADGEVI 255
>gb|AAA81460.1| Hypothetical protein C24A3.7 [Caenorhabditis elegans]
gi|17550592|ref|NP_509520.1| predicted CDS, reverse
transcriptase family member (XJ618) [Caenorhabditis
elegans] gi|7496376|pir||T15589 hypothetical protein
C24A3.7 - Caenorhabditis elegans
Length = 448
Score = 67.0 bits (162), Expect = 5e-10
Identities = 57/255 (22%), Positives = 103/255 (40%), Gaps = 26/255 (10%)
Query: 25 FADDVVLVGESREEVNGRLETWRQALEAYGFRLSRSKTEYIECNFS-------GRRSRST 77
FADD+VL+ + L+ Q G ++ KT+ + F+ G S +T
Sbjct: 182 FADDIVLIANHPNTASKMLQELVQKCSEVGLEINTGKTKVLRNRFADPSKVYFGSPSPTT 241
Query: 78 LEVKVEDHIIPQVTRSKYLGSFVQNDGEIEADVSHRIQAGWLKW---RRDSDVLCDKKVP 134
V+++I YLG + + ++ R +A W + + +D + DKK+
Sbjct: 242 QLDDVDEYI--------YLGRQINAQNNLMPEIHRRRRAAWAAFNGIKNTTDSITDKKI- 292
Query: 135 LKLKGKFYRPAFRPALMYGRECWAVKSQHENQVSVA----EMRMLRWMSSKTRHDRTRND 190
+ + PAL YG E WA +V + E R++ ++ R +
Sbjct: 293 ---RANLFDSIVLPALTYGSEAWAFTKALSERVRITHASLERRLVGITLTQQRERDLHQE 349
Query: 191 TIRKRVEVAPIVEKLVENRLRWFGHVERRPVDAVVRRVYQMEDSQVKRGRGRPRKTIRET 250
IR V + + + +L W GHV RR + + KR GRP ++
Sbjct: 350 DIRTMSLVRDPLNFVKKRKLGWAGHVARRKDGRWTTLMTEWRPYGWKRPVGRPPMRWTDS 409
Query: 251 IRKDLEVNELDPNLV 265
+RK++ + D ++
Sbjct: 410 LRKEITTRDADGEVI 424
>ref|NP_505558.1| predicted CDS, reverse transcriptase family member (5K419)
[Caenorhabditis elegans] gi|7507352|pir||T24639
hypothetical protein T07C12.2 - Caenorhabditis elegans
Length = 553
Score = 67.0 bits (162), Expect = 5e-10
Identities = 55/255 (21%), Positives = 101/255 (39%), Gaps = 26/255 (10%)
Query: 25 FADDVVLVGESREEVNGRLETWRQALEAYGFRLSRSKTEYIECNFS-------GRRSRST 77
FADD+VL+ + L+ Q G ++ KT+ + F+ G S +T
Sbjct: 287 FADDIVLIANHPNTASKMLQELVQKCSEVGLEINTGKTKVLRNRFADPSKVYFGSPSPTT 346
Query: 78 LEVKVEDHIIPQVTRSKYLGSFVQNDGEIEADVSHRIQAGWLKW---RRDSDVLCDKKVP 134
V+++I YLG + + ++ R +A W + + +D + DKK+
Sbjct: 347 QLDDVDEYI--------YLGRQINAQNNLMPEIHRRRRAAWAAFNGIKNTTDSITDKKI- 397
Query: 135 LKLKGKFYRPAFRPALMYGRECWAVKSQHENQVSVAEMRMLRWMSSKTRHDRTRNDTIRK 194
+ + PAL YG E W +V + + R + T + D R+
Sbjct: 398 ---RANLFDSIVLPALTYGSEAWTFTKALSERVRITHASLERRLVGITLTQQRERDLHRE 454
Query: 195 RVEVAPIVEK----LVENRLRWFGHVERRPVDAVVRRVYQMEDSQVKRGRGRPRKTIRET 250
+ +V + + +L W GHV RR + + KR GRP ++
Sbjct: 455 DIRTLSLVRDPLNFVKKRKLGWAGHVARRKDGRWTTLMTEWRPYGWKRPVGRPPMRWTDS 514
Query: 251 IRKDLEVNELDPNLV 265
+RK++ + D ++
Sbjct: 515 LRKEITTRDADGEVI 529
>ref|NP_492336.1| reverse transcriptase family member, possibly N-myristoylated
(1J197) [Caenorhabditis elegans] gi|25395320|pir||C87861
protein F43G9.7 [imported] - Caenorhabditis elegans
Length = 805
Score = 66.6 bits (161), Expect = 7e-10
Identities = 55/255 (21%), Positives = 101/255 (39%), Gaps = 26/255 (10%)
Query: 25 FADDVVLVGESREEVNGRLETWRQALEAYGFRLSRSKTEYIECNFS-------GRRSRST 77
FADD+VL+ + L+ Q G ++ KT+ + F+ G S +T
Sbjct: 539 FADDIVLIANHPNTASKMLQELVQKCSEVGLEINTGKTKALRNRFADPSKVYFGSPSPTT 598
Query: 78 LEVKVEDHIIPQVTRSKYLGSFVQNDGEIEADVSHRIQAGWLKW---RRDSDVLCDKKVP 134
V+++I YLG + + ++ R +A W + + +D + DKK+
Sbjct: 599 QLDDVDEYI--------YLGRQINAQNNLMPEIHRRRRAAWAAFNGIKNTTDSITDKKI- 649
Query: 135 LKLKGKFYRPAFRPALMYGRECWAVKSQHENQVSVAEMRMLRWMSSKTRHDRTRNDTIRK 194
+ + PAL YG E W +V + + R + T + D R+
Sbjct: 650 ---RANLFDSIVLPALTYGSEAWTFTKALSERVRITHASLERRLVGITLTQQRERDLHRE 706
Query: 195 RVEVAPIVEK----LVENRLRWFGHVERRPVDAVVRRVYQMEDSQVKRGRGRPRKTIRET 250
+ +V + + +L W GHV RR + + KR GRP ++
Sbjct: 707 DIRTMSLVRDPLNFVKKRKLGWAGHVARRKDGRWTTLMTEWRPYGWKRPVGRPPMRWTDS 766
Query: 251 IRKDLEVNELDPNLV 265
+RK++ + D ++
Sbjct: 767 LRKEITTRDADGEVI 781
>ref|XP_552671.1| ENSANGP00000005174 [Anopheles gambiae str. PEST]
gi|55235101|gb|EAL38938.1| ENSANGP00000005174 [Anopheles
gambiae str. PEST]
Length = 329
Score = 66.6 bits (161), Expect = 7e-10
Identities = 54/228 (23%), Positives = 96/228 (41%), Gaps = 7/228 (3%)
Query: 23 MLFADDVVLVGESREEVNGRLETWRQALEAYGFRLSRSKTEYIECNFSG----RRSRSTL 78
+ +ADD+ ++ V + QA E+ G +++ +KT+ + +G ++
Sbjct: 76 LAYADDIDIIDLRLSYVAEAYQGIEQAAESLGLQINEAKTKLMVATSAGLPINNQNLRRR 135
Query: 79 EVKVEDHIIPQVTRSKYLGSFVQNDGEIEADVSHRIQAGWLKWRRDSDVLCDKKVPLKLK 138
+V++ + V LGS V ND +EA++ R+ A + K + + K
Sbjct: 136 DVQIGERTFEVVPEFTCLGSKVSNDNSMEAELRARMLAANRSFYSLKKQFTSKNLSRRTK 195
Query: 139 GKFYRPAFRPALMYGRECWAVKSQHENQVSVAEMRMLRWMSSKTRHDRTRNDTIRKRVE- 197
Y P Y E W + E ++ E +MLR + + I
Sbjct: 196 LGLYSTYIVPVFTYASETWTLSKSDETLLAAFERKMLRRILAPYVWKDNEGAAIMTSYTR 255
Query: 198 -VAPIVEKLVENRLRWFGHVERRPVDAVVRRVYQMEDSQVKRGRGRPR 244
A ++++ RLRW GHV R D R+V+ + Q +R RGRP+
Sbjct: 256 CTATSLQRIKLARLRWAGHVVRMETDDPARKVF-LGRPQGQRRRGRPK 302
>gb|AAB71003.1| Hypothetical protein K10F12.5 [Caenorhabditis elegans]
gi|17554144|ref|NP_497194.1| reverse transcriptase
family member, possibly N-myristoylated (3B12)
[Caenorhabditis elegans] gi|7505736|pir||T32377
hypothetical protein K10F12.5 - Caenorhabditis elegans
Length = 805
Score = 66.2 bits (160), Expect = 9e-10
Identities = 55/255 (21%), Positives = 101/255 (39%), Gaps = 26/255 (10%)
Query: 25 FADDVVLVGESREEVNGRLETWRQALEAYGFRLSRSKTEYIECNFS-------GRRSRST 77
FADD+VL+ + L+ Q G ++ KT+ + F+ G S +T
Sbjct: 539 FADDIVLIANHPNIASKMLQELVQKCSEVGLEINTGKTKVLRNRFADPSKVYFGSPSPTT 598
Query: 78 LEVKVEDHIIPQVTRSKYLGSFVQNDGEIEADVSHRIQAGWLKW---RRDSDVLCDKKVP 134
V+++I YLG + + ++ R +A W + + +D + DKK+
Sbjct: 599 QLDDVDEYI--------YLGRQINAQNNLMPEIHRRRRAAWAAFNGIKNTTDSITDKKI- 649
Query: 135 LKLKGKFYRPAFRPALMYGRECWAVKSQHENQVSVAEMRMLRWMSSKTRHDRTRNDTIRK 194
+ + PAL YG E W +V + + R + T + D R+
Sbjct: 650 ---RANLFDSIVLPALTYGSEAWTFTKALSERVRITHASLERRLVGITLTQQRERDLHRE 706
Query: 195 RVEVAPIVEK----LVENRLRWFGHVERRPVDAVVRRVYQMEDSQVKRGRGRPRKTIRET 250
+ +V + + +L W GHV RR + + KR GRP ++
Sbjct: 707 DIRTMSLVRDPLNFVKKRKLGWAGHVARRKDGRWTTLMTEWRPYGWKRPVGRPPMRWTDS 766
Query: 251 IRKDLEVNELDPNLV 265
+RK++ + D ++
Sbjct: 767 LRKEITTRDADGEVI 781
>gb|AAB00700.1| Hypothetical protein C34D4.5 [Caenorhabditis elegans]
gi|17539028|ref|NP_501121.1| predicted CDS, reverse
transcriptase family member (4H911) [Caenorhabditis
elegans] gi|7497012|pir||T29286 hypothetical protein
C34D4.5 - Caenorhabditis elegans
Length = 624
Score = 66.2 bits (160), Expect = 9e-10
Identities = 49/155 (31%), Positives = 78/155 (49%), Gaps = 7/155 (4%)
Query: 3 NCFNRNMSVTEHIQELAPRCMLFADDVVLVGESREEVNGRLETWRQALEAYGFRLSRSKT 62
+C N V ++ R + FADDVVL+ + EEV RLE + YG ++++SKT
Sbjct: 340 DCDNEFAGVGIKVEGRHIRRLEFADDVVLICSTPEEVQERLEILDRISSIYGLKINQSKT 399
Query: 63 EYIECNFSGRRSRSTLEVKVEDHIIPQVTRSKYLGSFVQNDGEIEADVSHRIQAGWLKWR 122
++ F RS+ +V I V +YLG ++ G I+ ++S RI+AGW
Sbjct: 400 VLLKNKFC--RSQ---DVFFNGSPIIPVPGCRYLGRWIDISGSIDEEISRRIRAGWGALV 454
Query: 123 RDSDVLCDKKVPLKLKGKFYRPAFRPALMYGRECW 157
+VL + +P K + ++ PAL+Y E W
Sbjct: 455 GIKEVL--RIMPNKERIILFKQNVLPALLYASETW 487
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.322 0.136 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 441,360,028
Number of Sequences: 2540612
Number of extensions: 17435399
Number of successful extensions: 39113
Number of sequences better than 10.0: 93
Number of HSP's better than 10.0 without gapping: 27
Number of HSP's successfully gapped in prelim test: 66
Number of HSP's that attempted gapping in prelim test: 38986
Number of HSP's gapped (non-prelim): 98
length of query: 270
length of database: 863,360,394
effective HSP length: 126
effective length of query: 144
effective length of database: 543,243,282
effective search space: 78227032608
effective search space used: 78227032608
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 74 (33.1 bits)
Medicago: description of AC127169.2


