

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126790.7 - phase: 0 /pseudo
(625 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
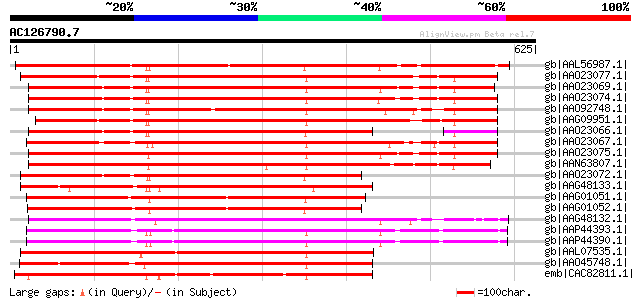
Score E
Sequences producing significant alignments: (bits) Value
gb|AAL56987.1| functional candidate resistance protein KR1 [Glyc... 505 e-141
gb|AAO23077.1| R 8 protein [Glycine max] 483 e-135
gb|AAO23069.1| R 4 protein [Glycine max] 476 e-133
gb|AAO23074.1| R 10 protein [Glycine max] 476 e-132
gb|AAO92748.1| candidate disease-resistance protein SR1 [Glycine... 472 e-131
gb|AAG09951.1| resistance protein LM6 [Glycine max] 466 e-129
gb|AAO23066.1| R 3 protein [Glycine max] 461 e-128
gb|AAO23067.1| R 12 protein [Glycine max] 455 e-126
gb|AAO23075.1| R 5 protein [Glycine max] 454 e-126
gb|AAN63807.1| resistance protein KR3 [Glycine max] 412 e-113
gb|AAO23072.1| R 14 protein [Glycine max] 410 e-113
gb|AAG48133.1| putative resistance protein [Glycine max] 400 e-110
gb|AAG01051.1| resistance protein LM12 [Glycine max] 391 e-107
gb|AAG01052.1| resistance protein MG23 [Glycine max] 387 e-106
gb|AAG48132.1| putative resistance protein [Glycine max] 384 e-105
gb|AAP44393.1| nematode resistance-like protein [Solanum tuberosum] 352 2e-95
gb|AAP44390.1| nematode resistance protein [Solanum tuberosum] 349 2e-94
gb|AAL07535.1| resistance gene analog PU3 [Helianthus annuus] 337 5e-91
gb|AAO45748.1| MRGH5 [Cucumis melo] 330 1e-88
emb|CAC82811.1| resistance gene-like [Solanum tuberosum subsp. a... 321 4e-86
>gb|AAL56987.1| functional candidate resistance protein KR1 [Glycine max]
Length = 1124
Score = 505 bits (1301), Expect = e-141
Identities = 308/626 (49%), Positives = 400/626 (63%), Gaps = 50/626 (7%)
Query: 8 MQLPTSSSSLSYDFNFDVFISFRGTDTRFGFTGNLYKALSDKGIHTFIDDKELPTGDEIT 67
M +SSSS SY F+ DVF+SFRG DTR GFTGNLYKALSD+GIHTF+DDK++P GD+IT
Sbjct: 1 MAKQSSSSSFSYRFSNDVFLSFRGEDTRRGFTGNLYKALSDRGIHTFMDDKKIPRGDQIT 60
Query: 68 PSLRKSIEESRIAIIIFSKNYATSSFCLDELVHIIHCFREKVTKVIPVFYGTEPSHVRKL 127
L K+IEESRI II+ S+NYA+SSFCL+EL +I+ + K ++PVFY +PS VR
Sbjct: 61 SGLEKAIEESRIFIIVLSENYASSSFCLNELDYILKFIKGKGILILPVFYKVDPSDVRNH 120
Query: 128 EDSYGEALAKHEVEFQNDMENMERLLKWKEALHQ------FHSW---------------- 165
S+G+AL HE +F++ +ME+L WK AL++ +H +
Sbjct: 121 TGSFGKALTNHEKKFKS-TNDMEKLETWKMALNKVANLSGYHHFKHGEEYEYEFIQRIVE 179
Query: 166 -----INRCHLHVAEYLVGLESRISEVNSLLDLGCTDGVYIIGILGTGGLGKTTLAEAVY 220
INR LHVA+Y VGLESRI EV +LLD+G D V+++GI G GG+GKTTLA AVY
Sbjct: 180 LVSKKINRAPLHVADYPVGLESRIQEVKALLDVGSDDVVHMLGIHGLGGVGKTTLAAAVY 239
Query: 221 NSIVNQFECRCFLYNVRENSFKHSLKYLQEQLLSKSIGYDTPLEHDNEGIEIIKQRLCRK 280
NSI + FE CFL NVRE S KH L++LQ LLS+ G D L +GI II+ RL +K
Sbjct: 240 NSIADHFEALCFLQNVRETSKKHGLQHLQRNLLSEMAGED-KLIGVKQGISIIEHRLRQK 298
Query: 281 KVLLILDDVDKPNQLEKLVGEPGWFGQGSRVIITTRDRYLLSCHGITKIYEADSLNKEES 340
KVLLILDDVDK QL+ L G P FG GSRVIITTRD+ LL+CHG+ + YE + LN+E +
Sbjct: 299 KVLLILDDVDKREQLQALAGRPDLFGPGSRVIITTRDKQLLACHGVERTYEVNELNEEYA 358
Query: 341 LELLRKMTF---KNDSSYDYILNRAVEYASGLPLALKVVGSNLFGKSIADCESTLDKYER 397
LELL F K D Y +LNRA YASGLPLAL+V+GSNL GK+I S LD+Y+R
Sbjct: 359 LELLNWKAFKLEKVDPFYKDVLNRAATYASGLPLALEVIGSNLSGKNIEQWISALDRYKR 418
Query: 398 IPPEDIQKILKVSFDTLEEEQQSVFLDIACCFKGCDWQKFQ----RHFNFIMAIQ*HIML 453
IP ++IQ+ILKVS+D LEE++QS+FLDIACCFK D + Q H M +++
Sbjct: 419 IPNKEIQEILKVSYDALEEDEQSIFLDIACCFKKYDLAEVQDILHAHHGHCMKHHIGVLV 478
Query: 454 ECWLMNLS*RLVLQIHTTLLILLHYMT**SIWV*K*SDKNQLKSLESALGCGVMMI*LMF 513
E L+ +S L + TL L+ M + + Q S L ++ ++
Sbjct: 479 EKSLIKIS----LDGYVTLHDLIEDMGKEIVR----KESPQEPGKRSRLWLPTDIVQVLE 530
Query: 514 *NK-THKIEMIYLN-CSSMEPINI--NEKAFKKMKKLKTLIIEKGYFSKGLKYLPKSLIV 569
NK T I +I +N SS E + I + AFKKMK LKTLII G+FSKG K+ PKSL V
Sbjct: 531 ENKGTSHIGIICMNFYSSFEEVEIQWDGDAFKKMKNLKTLIIRSGHFSKGPKHFPKSLRV 590
Query: 570 LKWKGFTSEPLSFCFSFKASKIIVFS 595
L+W + S F + F+ K+ +F+
Sbjct: 591 LEWWRYPSH--YFPYDFQMEKLAIFN 614
>gb|AAO23077.1| R 8 protein [Glycine max]
Length = 892
Score = 483 bits (1243), Expect = e-135
Identities = 293/605 (48%), Positives = 381/605 (62%), Gaps = 48/605 (7%)
Query: 13 SSSSLSYDFNFDVFISFRGTDTRFGFTGNLYKALSDKGIHTFIDDKELPTGDEITPSLRK 72
++++ S +N+DVF+SF G DTR GFTG LYKAL D+GI+TFIDD+EL GDEI P+L
Sbjct: 2 AATTRSLAYNYDVFLSFTGQDTRQGFTGYLYKALCDRGIYTFIDDQELRRGDEIKPALSN 61
Query: 73 SIEESRIAIIIFSKNYATSSFCLDELVHIIHCFREKVTKVIPVFYGTEPSHVRKLEDSYG 132
+I+ESRIAI + S+NYA+SSFCLDELV I+HC + + VIPVFY +PSHVR + SYG
Sbjct: 62 AIQESRIAITVLSQNYASSSFCLDELVTILHC-KSQGLLVIPVFYKVDPSHVRHQKGSYG 120
Query: 133 EALAKHEVEFQNDMENMERLLKWKEALHQ------FH--------------------SWI 166
EA+AKH+ F+ N E+L KW+ ALHQ +H
Sbjct: 121 EAMAKHQKRFK---ANKEKLQKWRMALHQVADLSGYHFKDGDSYEYEFIGSIVEEISRKF 177
Query: 167 NRCHLHVAEYLVGLESRISEVNSLLDLGCTDGVYIIGILGTGGLGKTTLAEAVYNSIVNQ 226
+R LHVA+Y VGLES ++EV LLD+G D V+IIGI G GGLGKTTLA AV+N I
Sbjct: 178 SRASLHVADYPVGLESEVTEVMKLLDVGSHDVVHIIGIHGMGGLGKTTLALAVHNFIALH 237
Query: 227 FECRCFLYNVRENSFKHSLKYLQEQLLSKSIG-YDTPLEHDNEGIEIIKQRLCRKKVLLI 285
F+ CFL NVRE S KH LK+LQ LLSK +G D L EG +I+ RL RKKVLLI
Sbjct: 238 FDESCFLQNVREESNKHGLKHLQSILLSKLLGEKDITLTSWQEGASMIQHRLQRKKVLLI 297
Query: 286 LDDVDKPNQLEKLVGEPGWFGQGSRVIITTRDRYLLSCHGITKIYEADSLNKEESLELLR 345
LDDVDK QL+ +VG P WFG GSRVIITTRD++LL H + + YE LN+ +L+LL
Sbjct: 298 LDDVDKRQQLKAIVGRPDWFGPGSRVIITTRDKHLLKYHEVERTYEVKVLNQSAALQLLT 357
Query: 346 KMTFKN---DSSYDYILNRAVEYASGLPLALKVVGSNLFGKSIADCESTLDKYERIPPED 402
FK D SY+ +LNR V YASGLPLAL+V+GSNLF K++A+ ES ++ Y+RIP ++
Sbjct: 358 WNAFKREKIDPSYEDVLNRVVTYASGLPLALEVIGSNLFEKTVAEWESAMEHYKRIPSDE 417
Query: 403 IQKILKVSFDTLEEEQQSVFLDIACCFKGCDWQKFQRHFNFIM--AIQ*HI--MLECWLM 458
IQ+ILKVSFD L EEQ++VFLDIACCFKG +W + + + HI ++E L+
Sbjct: 418 IQEILKVSFDALGEEQKNVFLDIACCFKGYEWTEVDNILRDLYGNCTKHHIGVLVEKSLV 477
Query: 459 NLS*RLVLQIHTTLLILLHYMT**SIWV*K*SDKNQLKSLESALGCGVMMI*LMF*NKTH 518
+S +++H + + + + S + K L ++ + L T
Sbjct: 478 KVSCCDTVEMHDMIQDMGREIE------RQRSPEEPGKCKRLLLPKDIIQV-LKDNTGTS 530
Query: 519 KIEMIYLNCS---SMEPININEKAFKKMKKLKTLIIEKGYFSKGLKYLPKSLIVLKWKGF 575
KIE+I L+ S E + NE AF KMK LK LII FSKG Y P+ L VL+W +
Sbjct: 531 KIEIICLDFSISDKEETVEWNENAFMKMKNLKILIIRNCKFSKGPNYFPEGLRVLEWHRY 590
Query: 576 TSEPL 580
S L
Sbjct: 591 PSNCL 595
>gb|AAO23069.1| R 4 protein [Glycine max]
Length = 895
Score = 476 bits (1225), Expect = e-133
Identities = 287/592 (48%), Positives = 373/592 (62%), Gaps = 49/592 (8%)
Query: 23 FDVFISFRGTDTRFGFTGNLYKALSDKGIHTFIDDKELPTGDEITPSLRKSIEESRIAII 82
+DVF+SFRG DTR GFTGNLYKAL D+GI+T IDD+ELP GDEITP+L K+I+ESRIAI
Sbjct: 12 YDVFLSFRGLDTRHGFTGNLYKALDDRGIYTSIDDQELPRGDEITPALSKAIQESRIAIT 71
Query: 83 IFSKNYATSSFCLDELVHIIHCFREKVTKVIPVFYGTEPSHVRKLEDSYGEALAKHEVEF 142
+ S+NYA+SSFCLDELV I+HC E + VIPVFY +PS VR + SYGEA+AKH+ F
Sbjct: 72 VLSQNYASSSFCLDELVTILHCKSEGLL-VIPVFYKVDPSDVRHQKGSYGEAMAKHQKRF 130
Query: 143 QNDMENMERLLKWKEALHQ------FH--------------------SWINRCHLHVAEY 176
+ E+L KW+ AL Q +H I+R LHVA+Y
Sbjct: 131 K---AKKEKLQKWRMALKQVADLSGYHFEDGDAYEYKFIGSIVEEVSRKISRASLHVADY 187
Query: 177 LVGLESRISEVNSLLDLGCTDGVYIIGILGTGGLGKTTLAEAVYNSIVNQFECRCFLYNV 236
VGLES+++EV LLD+G D V+IIGI G GGLGKTTLA VYN I F+ CFL NV
Sbjct: 188 PVGLESQVTEVMKLLDVGSDDLVHIIGIHGMGGLGKTTLALEVYNLIALHFDESCFLQNV 247
Query: 237 RENSFKHSLKYLQEQLLSKSIGY-DTPLEHDNEGIEIIKQRLCRKKVLLILDDVDKPNQL 295
RE S KH LK+LQ LLSK +G D L EG I+ RL RKKVLLILDDV+K QL
Sbjct: 248 REESNKHGLKHLQSILLSKLLGEKDITLTSWQEGASTIQHRLQRKKVLLILDDVNKREQL 307
Query: 296 EKLVGEPGWFGQGSRVIITTRDRYLLSCHGITKIYEADSLNKEESLELLRKMTFKN---D 352
+ +VG P WFG GSRVIITTRD++LL CH + + YE LN +L+LL FK D
Sbjct: 308 KAIVGRPDWFGPGSRVIITTRDKHLLKCHEVERTYEVKVLNHNAALQLLTWNAFKREKID 367
Query: 353 SSYDYILNRAVEYASGLPLALKVVGSNLFGKSIADCESTLDKYERIPPEDIQKILKVSFD 412
SY+ +LNR V YASGLPLAL+++GSN+FGKS+A ES ++ Y+RIP ++I +ILKVSFD
Sbjct: 368 PSYEDVLNRVVTYASGLPLALEIIGSNMFGKSVAGWESAVEHYKRIPNDEILEILKVSFD 427
Query: 413 TLEEEQQSVFLDIACCFKGCDWQKFQRH----FNFIMAIQ*HIMLECWLMNLS*RLVLQI 468
L EEQ++VFLDIA C KGC + + ++ M ++++ L+ + ++++
Sbjct: 428 ALGEEQKNVFLDIAFCLKGCKLTEVEHMLCSLYDNCMKHHIDVLVDKSLIKVK-HGIVEM 486
Query: 469 HTTLLILLHYMT**SIWV*K*SDKNQLKSLESALGCGVMMI*LMF*NKTHKIEMIYLNCS 528
H + ++ + + S + K L ++ + L T KIE+I L+ S
Sbjct: 487 HDLIQVVGREIE------RQRSPEEPGKRKRLWLPKDIIHV-LKDNTGTSKIEIICLDFS 539
Query: 529 ---SMEPININEKAFKKMKKLKTLIIEKGYFSKGLKYLPKSLIVLKWKGFTS 577
E + NE AF KM+ LK LII G FSKG Y P+ L VL+W + S
Sbjct: 540 ISYKEETVEFNENAFMKMENLKILIIRNGKFSKGPNYFPEGLRVLEWHRYPS 591
>gb|AAO23074.1| R 10 protein [Glycine max]
Length = 901
Score = 476 bits (1224), Expect = e-132
Identities = 288/602 (47%), Positives = 372/602 (60%), Gaps = 60/602 (9%)
Query: 23 FDVFISFRGTDTRFGFTGNLYKALSDKGIHTFIDDKELPTGDEITPSLRKSIEESRIAII 82
+DVF++FRG DTR+GFTGNLY+AL DKGIHTF D+K+L G+EITP+L K+I+ESRIAI
Sbjct: 12 YDVFLNFRGGDTRYGFTGNLYRALCDKGIHTFFDEKKLHRGEEITPALLKAIQESRIAIT 71
Query: 83 IFSKNYATSSFCLDELVHIIHCFREKVTKVIPVFYGTEPSHVRKLEDSYGEALAKHEVEF 142
+ SKNYA+SSFCLDELV I+HC E + VIPVFY +PS VR + SYG +AKH+ F
Sbjct: 72 VLSKNYASSSFCLDELVTILHCKSEGLL-VIPVFYNVDPSDVRHQKGSYGVEMAKHQKRF 130
Query: 143 QNDMENMERLLKWKEALHQ------FH--------------------SWINRCHLHVAEY 176
+ E+L KW+ AL Q +H INR LHVA+Y
Sbjct: 131 K---AKKEKLQKWRIALKQVADLCGYHFKDGDAYEYKFIQSIVEQVSREINRAPLHVADY 187
Query: 177 LVGLESRISEVNSLLDLGCTDGVYIIGILGTGGLGKTTLAEAVYNSIVNQFECRCFLYNV 236
VGL S++ EV LLD+G D V+IIGI G GGLGKTTLA AVYN I F+ CFL NV
Sbjct: 188 PVGLGSQVIEVRKLLDVGSHDVVHIIGIHGMGGLGKTTLALAVYNLIALHFDESCFLQNV 247
Query: 237 RENSFKHSLKYLQEQLLSKSIGY-DTPLEHDNEGIEIIKQRLCRKKVLLILDDVDKPNQL 295
RE S KH LK+LQ LLSK +G D L EG +I+ RL RKKVLLILDDVDK QL
Sbjct: 248 REESNKHGLKHLQSILLSKLLGEKDITLTSWQEGASMIQHRLQRKKVLLILDDVDKREQL 307
Query: 296 EKLVGEPGWFGQGSRVIITTRDRYLLSCHGITKIYEADSLNKEESLELLRKMTFKN---D 352
+ +VG P WFG GSRVIITTRD++LL H + + YE LN+ +L+LL+ FK D
Sbjct: 308 KAIVGRPDWFGPGSRVIITTRDKHLLKYHEVERTYEVKVLNQSAALQLLKWNAFKREKID 367
Query: 353 SSYDYILNRAVEYASGLPLALKVVGSNLFGKSIADCESTLDKYERIPPEDIQKILKVSFD 412
SY+ +LNR V YASGLPLAL+V+GSNLFGK++A+ ES ++ Y+RIP ++I +ILKVSFD
Sbjct: 368 PSYEDVLNRVVTYASGLPLALEVIGSNLFGKTVAEWESAMEHYKRIPSDEILEILKVSFD 427
Query: 413 TLEEEQQSVFLDIACCFKGCDWQKF-----------QRHFNFIMAIQ*HIMLECWLMNLS 461
L EEQ++VFLDIACCF+G W + ++H ++ + I L C+ +
Sbjct: 428 ALGEEQKNVFLDIACCFRGYKWTEVDDILRALYGNCKKHHIGVLVEKSLIKLNCYGTD-- 485
Query: 462 *RLVLQIHTTLLILLHYMT**SIWV*K*SDKNQLKSLESALGCGVMMI*LMF*NKTHKIE 521
+++H + + + K S + K L ++ + T KIE
Sbjct: 486 ---TVEMHDLIQDMAREIE------RKRSPQEPGKCKRLWLPKDIIQV-FKDNTGTSKIE 535
Query: 522 MIYLNCS---SMEPININEKAFKKMKKLKTLIIEKGYFSKGLKYLPKSLIVLKWKGFTSE 578
+I L+ S E + NE AF KM+ LK LII FSKG Y P+ L VL+W + S
Sbjct: 536 IICLDSSISDKEETVEWNENAFMKMENLKILIIRNDKFSKGPNYFPEGLRVLEWHRYPSN 595
Query: 579 PL 580
L
Sbjct: 596 CL 597
>gb|AAO92748.1| candidate disease-resistance protein SR1 [Glycine max]
Length = 1137
Score = 472 bits (1215), Expect = e-131
Identities = 293/607 (48%), Positives = 371/607 (60%), Gaps = 73/607 (12%)
Query: 23 FDVFISFRGTDTRFGFTGNLYKALSDKGIHTFIDDKELPTGDEITPSLRKSIEESRIAII 82
+DVF+SF G DTR GFTG LYKAL D+GI+TFIDD+ELP GDEI P+L +I+ SRIAI
Sbjct: 12 YDVFLSFTGQDTRHGFTGYLYKALDDRGIYTFIDDQELPRGDEIKPALSDAIQGSRIAIT 71
Query: 83 IFSKNYATSSFCLDELVHIIHCFREKVTKVIPVFYGTEPSHVRKLEDSYGEALAKHEVEF 142
+ S+NYA S+FCLDELV I+HC E + VIPVFY +PSHVR + SYGEA+AKH+ F
Sbjct: 72 VLSQNYAFSTFCLDELVTILHCKSEGLL-VIPVFYKVDPSHVRHQKGSYGEAMAKHQKRF 130
Query: 143 QNDMENMERLLKWKEALHQ------FH--------------------SWINRCHLHVAEY 176
+ N E+L KW+ AL Q +H INR LHVA+Y
Sbjct: 131 K---ANKEKLQKWRMALQQVADLSGYHFKDGDAYEYKFIQSIVEQVSREINRAPLHVADY 187
Query: 177 LVGLESRISEVNSLLDLGCTDGVYIIGILGTGGLGKTTLAEAVYNSIVNQFECRCFLYNV 236
VGL S++ EV LLD+G D V+IIGI G GGLGKTTLA AVYN I F+ CFL NV
Sbjct: 188 PVGLGSQVIEVRKLLDVGSDDVVHIIGIHGMGGLGKTTLAVAVYNLIAPHFDESCFLQNV 247
Query: 237 RENSFKHSLKYLQEQLLSKSIGY-DTPLEHDNEGIEIIKQRLCRKKVLLILDDVDKPNQL 295
RE S +LK+LQ LLSK +G D L EG +I+ RL RKKVLLILDDVDK QL
Sbjct: 248 REES---NLKHLQSSLLSKLLGEKDITLTSWQEGASMIQHRLRRKKVLLILDDVDKREQL 304
Query: 296 EKLVGEPGWFGQGSRVIITTRDRYLLSCHGITKIYEADSLNKEESLELLRKMTFKN---D 352
+ +VG+P WFG GSRVIITTRD++LL H + + YE LN +L LL FK D
Sbjct: 305 KAIVGKPDWFGPGSRVIITTRDKHLLKYHEVERTYEVKVLNHNAALHLLTWNAFKREKID 364
Query: 353 SSYDYILNRAVEYASGLPLALKVVGSNLFGKSIADCESTLDKYERIPPEDIQKILKVSFD 412
YD +LNR V YASGLPLAL+V+GSNL+GK++A+ ES L+ Y+RIP +I KIL+VSFD
Sbjct: 365 PIYDDVLNRVVTYASGLPLALEVIGSNLYGKTVAEWESALETYKRIPSNEILKILQVSFD 424
Query: 413 TLEEEQQSVFLDIACCFKGCDWQKFQRHFNFIMA----IQ*HIMLECWLM--NLS*RLVL 466
LEEEQQ+VFLDIACCFKG +W + F + +++E L+ N + R +
Sbjct: 425 ALEEEQQNVFLDIACCFKGHEWTEVDDIFRALYGNGKKYHIGVLVEKSLIKYNRNNRGTV 484
Query: 467 QIHTTLLILLHYM----------T**SIWV*K*SDKNQLKSLESALGCGVMMI*LMF*NK 516
Q+H + + + +W S K+ ++ L+ G
Sbjct: 485 QMHNLIQDMGREIERQRSPEEPGKRKRLW----SPKDIIQVLKHNTG------------- 527
Query: 517 THKIEMIYLNCS---SMEPININEKAFKKMKKLKTLIIEKGYFSKGLKYLPKSLIVLKWK 573
T KIE+I L+ S E + NE AF KM+ LK LII G FS G Y+P+ L VL+W
Sbjct: 528 TSKIEIICLDSSISDKEETVEWNENAFMKMENLKILIIRNGKFSIGPNYIPEGLRVLEWH 587
Query: 574 GFTSEPL 580
+ S L
Sbjct: 588 RYPSNCL 594
>gb|AAG09951.1| resistance protein LM6 [Glycine max]
Length = 863
Score = 466 bits (1199), Expect = e-129
Identities = 284/587 (48%), Positives = 366/587 (61%), Gaps = 55/587 (9%)
Query: 31 GTDTRFGFTGNLYKALSDKGIHTFIDDKELPTGDEITPSLRKSIEESRIAIIIFSKNYAT 90
G DTR GFTG LYKAL D+GI+TFIDD+EL GDEI P+L +I+ESRIAI + S+NYA+
Sbjct: 3 GQDTRQGFTGYLYKALCDRGIYTFIDDQELRRGDEIKPALSNAIQESRIAITVLSQNYAS 62
Query: 91 SSFCLDELVHIIHCFREKVTKVIPVFYGTEPSHVRKLEDSYGEALAKHEVEFQNDMENME 150
SSFCLDELV I+HC + + VIPVFY +PSHVR + SYGEA+AKH+ F+ N E
Sbjct: 63 SSFCLDELVTILHC-KSQGLLVIPVFYKVDPSHVRHQKGSYGEAMAKHQKRFK---ANKE 118
Query: 151 RLLKWKEALHQ------FH--------------------SWINRCHLHVAEYLVGLESRI 184
+L KW+ ALHQ +H +R LHVA+Y VGLES +
Sbjct: 119 KLQKWRMALHQVADLSGYHFKDGDSYEYEFIGSIVEEISRKFSRASLHVADYPVGLESEV 178
Query: 185 SEVNSLLDLGCTDGVYIIGILGTGGLGKTTLAEAVYNSIVNQFECRCFLYNVRENSFKHS 244
+EV LLD+G D V+IIGI G GGLGKTTLA AV+N I F+ CFL NVRE S KH
Sbjct: 179 TEVMKLLDVGSHDVVHIIGIHGMGGLGKTTLALAVHNFIALHFDESCFLQNVREESNKHG 238
Query: 245 LKYLQEQLLSKSIG-YDTPLEHDNEGIEIIKQRLCRKKVLLILDDVDKPNQLEKLVGEPG 303
LK+LQ LLSK +G D L EG +I+ RL RKKVLLILDDVDK QL+ +VG P
Sbjct: 239 LKHLQSILLSKLLGEKDITLTSWQEGASMIQHRLQRKKVLLILDDVDKRQQLKAIVGRPD 298
Query: 304 WFGQGSRVIITTRDRYLLSCHGITKIYEADSLNKEESLELLRKMTFKN---DSSYDYILN 360
WFG GSRVIITTRD++LL H + + YE LN+ +L+LL FK D SY+ +LN
Sbjct: 299 WFGPGSRVIITTRDKHLLKYHEVERTYEVKVLNQSAALQLLTWNAFKREKIDPSYEDVLN 358
Query: 361 RAVEYASGLPLALKVVGSNLFGKSIADCESTLDKYERIPPEDIQKILKVSFDTLEEEQQS 420
R V YASGLPLAL+V+GSNLF K++A+ ES ++ Y+RIP ++IQ+ILKVSFD L EEQ++
Sbjct: 359 RVVTYASGLPLALEVIGSNLFEKTVAEWESAMEHYKRIPSDEIQEILKVSFDALGEEQKN 418
Query: 421 VFLDIACCFKGCDWQKFQRHFNFIM--AIQ*HI--MLECWLMNLS*RLVLQIHTTLLILL 476
VFLDIACCFKG +W + + + HI ++E L+ +S +++H + +
Sbjct: 419 VFLDIACCFKGYEWTEVDNILRDLYGNCTKHHIGVLVEKSLVKVSCCDTVEMHDMIQDMG 478
Query: 477 HYMT**SIWV*K*SDKNQLKSLESALGCGVMMI*LMF*NKTHKIEMIYLNCS---SMEPI 533
+ + +S E C +++ + KIE+I L+ S E +
Sbjct: 479 R-------------EIERQRSPEEPGKCKRLLLPKDI-IQVFKIEIICLDFSISDKEETV 524
Query: 534 NINEKAFKKMKKLKTLIIEKGYFSKGLKYLPKSLIVLKWKGFTSEPL 580
NE AF KMK LK LII FSKG Y P+ L VL+W + S L
Sbjct: 525 EWNENAFMKMKNLKILIIRNCKFSKGPNYFPEGLRVLEWHRYPSNCL 571
>gb|AAO23066.1| R 3 protein [Glycine max]
Length = 897
Score = 461 bits (1185), Expect = e-128
Identities = 250/440 (56%), Positives = 312/440 (70%), Gaps = 34/440 (7%)
Query: 23 FDVFISFRGTDTRFGFTGNLYKALSDKGIHTFIDDKELPTGDEITPSLRKSIEESRIAII 82
+DVF+SFRG DTR GFTGNLYKAL D+GI+TFIDD+ELP GDEITP+L K+I+ESRIAI
Sbjct: 12 YDVFLSFRGLDTRHGFTGNLYKALDDRGIYTFIDDQELPRGDEITPALSKAIQESRIAIT 71
Query: 83 IFSKNYATSSFCLDELVHIIHCFREKVTKVIPVFYGTEPSHVRKLEDSYGEALAKHEVEF 142
+ S+NYA+SSFCLDELV ++ C R+ + VIPVFY +PS VR+ + SYGEA+AKH+ F
Sbjct: 72 VLSQNYASSSFCLDELVTVLLCKRKGLL-VIPVFYNVDPSDVRQQKGSYGEAMAKHQKRF 130
Query: 143 QNDMENMERLLKWKEALHQ------FH--------------------SWINRCHLHVAEY 176
+ E+L KW+ ALHQ +H INR LHVA+Y
Sbjct: 131 K---AKKEKLQKWRMALHQVADLSGYHFKDGDAYEYKFIQSIVEQVSREINRTPLHVADY 187
Query: 177 LVGLESRISEVNSLLDLGCTDGVYIIGILGTGGLGKTTLAEAVYNSIVNQFECRCFLYNV 236
VGL S++ EV LLD+G D V+IIGI G GGLGKTTLA AVYN I F+ CFL NV
Sbjct: 188 PVGLGSQVIEVRKLLDVGSHDVVHIIGIHGMGGLGKTTLALAVYNLIALHFDESCFLQNV 247
Query: 237 RENSFKHSLKYLQEQLLSKSIGY-DTPLEHDNEGIEIIKQRLCRKKVLLILDDVDKPNQL 295
RE S KH LK+LQ +LSK +G D L EG +I+ RL RKKVLLILDDVDK QL
Sbjct: 248 REESNKHGLKHLQSIILSKLLGEKDINLTSWQEGASMIQHRLQRKKVLLILDDVDKRQQL 307
Query: 296 EKLVGEPGWFGQGSRVIITTRDRYLLSCHGITKIYEADSLNKEESLELLRKMTFK---ND 352
+ +VG P WFG GSRVIITTRD+++L H + + YE LN+ +L+LL+ FK ND
Sbjct: 308 KAIVGRPDWFGPGSRVIITTRDKHILKYHEVERTYEVKVLNQSAALQLLKWNAFKREKND 367
Query: 353 SSYDYILNRAVEYASGLPLALKVVGSNLFGKSIADCESTLDKYERIPPEDIQKILKVSFD 412
SY+ +LNR V YASGLPLAL+++GSNLFGK++A+ ES ++ Y+RIP ++I +ILKVSFD
Sbjct: 368 PSYEDVLNRVVTYASGLPLALEIIGSNLFGKTVAEWESAMEHYKRIPSDEILEILKVSFD 427
Query: 413 TLEEEQQSVFLDIACCFKGC 432
L EEQ++VFLDIACC KGC
Sbjct: 428 ALGEEQKNVFLDIACCLKGC 447
Score = 56.6 bits (135), Expect = 2e-06
Identities = 32/67 (47%), Positives = 40/67 (58%), Gaps = 3/67 (4%)
Query: 517 THKIEMIYLNCS---SMEPININEKAFKKMKKLKTLIIEKGYFSKGLKYLPKSLIVLKWK 573
T KIE+IY++ S E + NE AF KM+ LK LII G FSKG Y P+ L VL+W
Sbjct: 528 TSKIEIIYVDFSISDKEETVEWNENAFMKMENLKILIIRNGKFSKGPNYFPQGLRVLEWH 587
Query: 574 GFTSEPL 580
+ S L
Sbjct: 588 RYPSNCL 594
>gb|AAO23067.1| R 12 protein [Glycine max]
Length = 893
Score = 455 bits (1170), Expect = e-126
Identities = 284/605 (46%), Positives = 369/605 (60%), Gaps = 68/605 (11%)
Query: 21 FNFDVFISFRGTDTRFGFTGNLYKALSDKGIHTFIDDKELPTGDEITPSLRKSIEESRIA 80
F +DVF+SF DT GFT LYKAL+D+GI+TF D+ELP E+TP L K+I SR+A
Sbjct: 10 FIYDVFLSFIREDTHRGFTFYLYKALNDRGIYTFFYDQELPRETEVTPGLYKAILASRVA 69
Query: 81 IIIFSKNYATSSFCLDELVHIIHCFREKVTKVIPVFYGTEPSHVRKLEDSYGEALAKHEV 140
II+ S+NYA SSFCLDELV I+HC RE VIPVF+ +PS VR + SYGEA+AKH+
Sbjct: 70 IIVLSENYAFSSFCLDELVTILHCERE----VIPVFHNVDPSDVRHQKGSYGEAMAKHQK 125
Query: 141 EFQNDMENMERLLKWKEALHQF---------------HSWINR-----------CHLHVA 174
F+ ++L KW+ AL Q + I R LHVA
Sbjct: 126 RFK-----AKKLQKWRMALKQVANLCGYHFKDGGSYEYMLIGRIVKQVSRMFGLASLHVA 180
Query: 175 EYLVGLESRISEVNSLLDLGCTDGVYIIGILGTGGLGKTTLAEAVYNSIVNQFECRCFLY 234
+Y VGLES+++EV LLD+G D V+IIGI G GGLGKTTLA AVYN I F+ CFL
Sbjct: 181 DYPVGLESQVTEVMKLLDVGSDDVVHIIGIHGMGGLGKTTLAMAVYNFIAPHFDESCFLQ 240
Query: 235 NVRENSFKHSLKYLQEQLLSKSIGY-DTPLEHDNEGIEIIKQRLCRKKVLLILDDVDKPN 293
NVRE S KH LK+LQ LLSK +G D L EG +I+ RL KK+LLILDDVDK
Sbjct: 241 NVREESNKHGLKHLQSVLLSKLLGEKDITLTSWQEGASMIQHRLRLKKILLILDDVDKRE 300
Query: 294 QLEKLVGEPGWFGQGSRVIITTRDRYLLSCHGITKIYEADSLNKEESLELLRKMTFKN-- 351
QL+ +VG+P WFG GSRVIITTRD++LL H + + YE + LN +++ +LL FK
Sbjct: 301 QLKAIVGKPDWFGPGSRVIITTRDKHLLKYHEVERTYEVNVLNHDDAFQLLTWNAFKREK 360
Query: 352 -DSSYDYILNRAVEYASGLPLALKVVGSNLFGKSIADCESTLDKYERIPPEDIQKILKVS 410
D SY +LNR V YASGLPLAL+V+GSNL+GK++A+ ES L+ Y+RIP +I KIL+VS
Sbjct: 361 IDPSYKDVLNRVVTYASGLPLALEVIGSNLYGKTVAEWESALETYKRIPSNEILKILEVS 420
Query: 411 FDTLEEEQQSVFLDIACCFKGCDWQKFQRHFNFIMA-IQ*H----IMLECWLMNLS*RLV 465
FD LEEEQ++VFLDIACCFKG W + F + + + H ++ + L+ +S R
Sbjct: 421 FDALEEEQKNVFLDIACCFKGYKWTEVYDIFRALYSNCKMHHIGVLVEKSLLLKVSWRDN 480
Query: 466 LQIHTTLLILLHYMT**SIWV*K*SDKNQLKSLESALGC-------GVMMI*LMF*NKTH 518
+++H L+ M D + +S E C ++ + L T
Sbjct: 481 VEMHD----LIQDMG---------RDIERQRSPEEPGKCKRLWSPKDIIQV-LKHNTGTS 526
Query: 519 KIEMIYLNCS---SMEPININEKAFKKMKKLKTLIIEKGYFSKGLKYLPKSLIVLKWKGF 575
K+E+I L+ S E + NE AF KM+ LK LII G FSKG Y P+ L VL+W +
Sbjct: 527 KLEIICLDSSISDKEETVEWNENAFMKMENLKILIIRNGKFSKGPNYFPEGLRVLEWHRY 586
Query: 576 TSEPL 580
S L
Sbjct: 587 PSNCL 591
>gb|AAO23075.1| R 5 protein [Glycine max]
Length = 907
Score = 454 bits (1167), Expect = e-126
Identities = 266/590 (45%), Positives = 365/590 (61%), Gaps = 40/590 (6%)
Query: 23 FDVFISFRGTDTRFGFTGNLYKALSDKGIHTFIDDKELPTGDEITPSLRKSIEESRIAII 82
+DVF+SFRG DTR+GFTGNLYKAL DKGIHTF D+ +L +G+EITP+L K+I++SRIAI
Sbjct: 12 YDVFLSFRGEDTRYGFTGNLYKALCDKGIHTFFDEDKLHSGEEITPALLKAIQDSRIAIT 71
Query: 83 IFSKNYATSSFCLDELVHIIHCFREKVTKVIPVFYGTEPSHVRKLEDSYGEALAKHEVEF 142
+ S+++A+SSFCLDEL I+ C + VIPVFY P VR + +YGEALAKH+ F
Sbjct: 72 VLSEDFASSSFCLDELATILFCAQYNGMMVIPVFYKVYPCDVRHQKGTYGEALAKHKKRF 131
Query: 143 QNDMENMERLLKWKEALHQFH--------------------SWINRCHLHVAEYLVGLES 182
+ ++ ER L+ L H IN LHVA+ VGLES
Sbjct: 132 PDKLQKWERALRQVANLSGLHFKDRDEYEYKFIGRIVASVSEKINPASLHVADLPVGLES 191
Query: 183 RISEVNSLLDLGCTDGVYIIGILGTGGLGKTTLAEAVYNSIV--NQFECRCFLYNVRENS 240
++ EV LLD+G DGV +IGI G GG+GK+TLA AVYN ++ F+ CFL NVRE+S
Sbjct: 192 KVQEVRKLLDVGNHDGVCMIGIHGMGGIGKSTLARAVYNDLIITENFDGLCFLENVRESS 251
Query: 241 FKHSLKYLQEQLLSKSIGYDTPLEHDNEGIEIIKQRLCRKKVLLILDDVDKPNQLEKLVG 300
H L++LQ LLS+ +G D + +GI I+ L KKVLLILDDVDKP QL+ + G
Sbjct: 252 NNHGLQHLQSILLSEILGEDIKVRSKQQGISKIQSMLKGKKVLLILDDVDKPQQLQTIAG 311
Query: 301 EPGWFGQGSRVIITTRDRYLLSCHGITKIYEADSLNKEESLELLRKMTFKN---DSSYDY 357
WFG GS +IITTRD+ LL+ HG+ K YE + LN+ +L+LL FK D SY+
Sbjct: 312 RRDWFGPGSIIIITTRDKQLLAPHGVKKRYEVEVLNQNAALQLLTWNAFKREKIDPSYED 371
Query: 358 ILNRAVEYASGLPLALKVVGSNLFGKSIADCESTLDKYERIPPEDIQKILKVSFDTLEEE 417
+LNR V YASGLPLAL+V+GSN+FGK +A+ +S ++ Y+RIP ++I +ILKVSFD L EE
Sbjct: 372 VLNRVVTYASGLPLALEVIGSNMFGKRVAEWKSAVEHYKRIPNDEILEILKVSFDALGEE 431
Query: 418 QQSVFLDIACCFKGCDWQKFQRH----FNFIMAIQ*HIMLECWLMNLS*RLVLQIHTTLL 473
Q++VFLDIACCFKGC + + +N M ++++ L+ + + +H +
Sbjct: 432 QKNVFLDIACCFKGCKLTEVEHMLRGLYNNCMKHHIDVLVDKSLIKVR-HGTVNMHDLIQ 490
Query: 474 ILLHYMT**SIWV*K*SDKNQLKSLESALGCGVMMI*LMF*NKTHKIEMIYLNCS---SM 530
++ + + S + K L ++ + L T KIE+I L+ S
Sbjct: 491 VVGREIE------RQISPEEPGKCKRLWLPKDIIQV-LKHNTGTSKIEIICLDFSISDKE 543
Query: 531 EPININEKAFKKMKKLKTLIIEKGYFSKGLKYLPKSLIVLKWKGFTSEPL 580
+ + N+ AF KM+ LK LII G FSKG Y P+ L VL+W + S+ L
Sbjct: 544 QTVEWNQNAFMKMENLKILIIRNGKFSKGPNYFPEGLRVLEWHRYPSKCL 593
>gb|AAN63807.1| resistance protein KR3 [Glycine max]
Length = 636
Score = 412 bits (1058), Expect = e-113
Identities = 249/585 (42%), Positives = 358/585 (60%), Gaps = 41/585 (7%)
Query: 23 FDVFISFRGTDTRFGFTGNLYKALSDKGIHTFIDDKELPTGDEITPSLRKSIEESRIAII 82
+DVFI+FRG DTRF FTG+L+KAL +KGI F+D+ ++ GDEI +L ++I+ SRIAI
Sbjct: 35 YDVFINFRGEDTRFAFTGHLHKALCNKGIRAFMDENDIKRGDEIRATLEEAIKGSRIAIT 94
Query: 83 IFSKNYATSSFCLDELVHIIHCFREKVTKVIPVFYGTEPSHVRKLEDSYGEALAKHEVEF 142
+FSK+YA+SSFCLDEL I+ C+REK VIPVFY +PS VR+L+ SY E LA+ E F
Sbjct: 95 VFSKDYASSSFCLDELATILGCYREKTLLVIPVFYKVDPSDVRRLQGSYAEGLARLEERF 154
Query: 143 QNDMENMERLLKWKEALHQFH--------------------SWINRCH--LHVAEYLVGL 180
+MEN ++ L+ L H IN+ ++VA++ VGL
Sbjct: 155 HPNMENWKKALQKVAELAGHHFKDGAGYEFKFIRKIVDDVFDKINKAEASIYVADHPVGL 214
Query: 181 ESRISEVNSLLDLGCTDGVYIIGILGTGGLGKTTLAEAVYNSIVNQFECRCFLYNVRENS 240
+ ++ LL+ G +D + +IGI G GG+GK+TLA AVYN + F+ CFL NVRE S
Sbjct: 215 HLEVEKIRKLLEAGSSDAISMIGIHGMGGVGKSTLARAVYNLHTDHFDDSCFLQNVREES 274
Query: 241 FKHSLKYLQEQLLSKSIGYDTPLEHDNEGIEIIKQRLCRKKVLLILDDVDKPNQLEKLVG 300
+H LK LQ LLS+ + + L + +G +IK +L KKVLL+LDDVD+ QL+ +VG
Sbjct: 275 NRHGLKRLQSILLSQILKKEINLASEQQGTSMIKNKLKGKKVLLVLDDVDEHKQLQAIVG 334
Query: 301 EPGW----FGQGSRVIITTRDRYLLSCHGITKIYEADSLNKEESLELLRKMTFKN----D 352
+ W FG +IITTRD+ LL+ +G+ + +E L+K+++++LL++ FK D
Sbjct: 335 KSVWSESEFGTRLVLIITTRDKQLLTSYGVKRTHEVKELSKKDAIQLLKRKAFKTYDEVD 394
Query: 353 SSYDYILNRAVEYASGLPLALKVVGSNLFGKSIADCESTLDKYERIPPEDIQKILKVSFD 412
SY+ +LN V + SGLPLAL+V+GSNLFGKSI + ES + +Y+RIP ++I KILKVSFD
Sbjct: 395 QSYNQVLNDVVTWTSGLPLALEVIGSNLFGKSIKEWESAIKQYQRIPNKEILKILKVSFD 454
Query: 413 TLEEEQQSVFLDIACCFKGCDWQKFQ--RHFNFIMAIQ*HIMLECWLMNLS*RLVLQIHT 470
LEEE++SVFLDI CC KG ++ + H + ++ HI + L++ S +
Sbjct: 455 ALEEEEKSVFLDITCCLKGYKCREIEDILHSLYDNCMKYHIGV---LVDKSLIQISDDRV 511
Query: 471 TLLILLHYMT**SIWV*K*SDKNQLKSLESALGCGVMMI*LMF*NKTHKIEMIYLN---C 527
TL L+ M + S K K L ++ + L + T ++++I L+
Sbjct: 512 TLHDLIENMG--KEIDRQKSPKETGKRRRLWLLKDIIQV-LKDNSGTSEVKIICLDFPIS 568
Query: 528 SSMEPININEKAFKKMKKLKTLIIEKGYFSKGLKYLPKSLIVLKW 572
E I N AFK+MK LK LII G S+G YLP+SL +L+W
Sbjct: 569 DKQETIEWNGNAFKEMKNLKALIIRNGILSQGPNYLPESLRILEW 613
>gb|AAO23072.1| R 14 protein [Glycine max]
Length = 641
Score = 410 bits (1054), Expect = e-113
Identities = 229/437 (52%), Positives = 295/437 (67%), Gaps = 34/437 (7%)
Query: 13 SSSSLSYDFNFDVFISFRGTDTRFGFTGNLYKALSDKGIHTFIDDKELPTGDEITPSLRK 72
++++ S F +DVF+SFRG DTR+GFTGNLY+AL +KGIHTF D+++L GDEITP+L K
Sbjct: 2 AATTRSLPFIYDVFLSFRGEDTRYGFTGNLYRALCEKGIHTFFDEEKLHGGDEITPALSK 61
Query: 73 SIEESRIAIIIFSKNYATSSFCLDELVHIIHCFREKVTKVIPVFYGTEPSHVRKLEDSYG 132
+I+ESRIAI + S+NYA SSFCLDELV I+HC E + VIPVFY +PS +R + SYG
Sbjct: 62 AIQESRIAITVLSQNYAFSSFCLDELVTILHCKSEGLL-VIPVFYNVDPSDLRHQKGSYG 120
Query: 133 EALAKHEVEFQNDMENMERLLKWKEALHQF-----HSW---------------------I 166
EA+ KH+ F++ ME+L KW+ AL Q H + I
Sbjct: 121 EAMIKHQKRFES---KMEKLQKWRMALKQVADLSGHHFKDGDAYEYKFIGSIVEEVSRKI 177
Query: 167 NRCHLHVAEYLVGLESRISEVNSLLDLGCTDGVYIIGILGTGGLGKTTLAEAVYNSIVNQ 226
NR LHV +Y VGLES+++++ LLD+G D V+IIGI G GLGKTTL+ AVYN I
Sbjct: 178 NRASLHVLDYPVGLESQVTDLMKLLDVGSDDVVHIIGIHGMRGLGKTTLSLAVYNLIALH 237
Query: 227 FECRCFLYNVRENSFKHSLKYLQEQLLSKSIG-YDTPLEHDNEGIEIIKQRLCRKKVLLI 285
F+ CFL NVRE S KH LK+LQ LL K +G D L EG +I+ RL RKKVLLI
Sbjct: 238 FDESCFLQNVREESNKHGLKHLQSILLLKLLGEKDINLTSWQEGASMIQHRLRRKKVLLI 297
Query: 286 LDDVDKPNQLEKLVGEPGWFGQGSRVIITTRDRYLLSCHGITKIYEADSLNKEESLELLR 345
LDD D+ QL+ +VG P WFG GSRVIITTRD++LL HGI + YE LN +L+LL
Sbjct: 298 LDDADRHEQLKAIVGRPDWFGPGSRVIITTRDKHLLKYHGIERTYEVKVLNDNAALQLLT 357
Query: 346 KMTF---KNDSSYDYILNRAVEYASGLPLALKVVGSNLFGKSIADCESTLDKYERIPPED 402
F K D SY+++LNR V YASGLPLAL+V+GS+LF K++A+ E ++ Y RIP ++
Sbjct: 358 WNAFRREKIDPSYEHVLNRVVAYASGLPLALEVIGSHLFEKTVAEWEYAVEHYSRIPIDE 417
Query: 403 IQKILKVSFDTLEEEQQ 419
I ILKVSFD ++E Q
Sbjct: 418 IVDILKVSFDATKQETQ 434
>gb|AAG48133.1| putative resistance protein [Glycine max]
Length = 522
Score = 400 bits (1027), Expect = e-110
Identities = 217/444 (48%), Positives = 299/444 (66%), Gaps = 29/444 (6%)
Query: 14 SSSLSYDFNFDVFISFRGTDTRFGFTGNLYKALSDKGIHTFIDDKELPTGDEITPS---L 70
SSS + + FDVF+SFRG DTR GF GNLYKAL++KG HTF +K L G+EI S +
Sbjct: 7 SSSRVWHYEFDVFLSFRGEDTRLGFVGNLYKALTEKGFHTFFREK-LVRGEEIAASPSVV 65
Query: 71 RKSIEESRIAIIIFSKNYATSSFCLDELVHIIHCFREKVTKVIPVFYGTEPSHVRKLEDS 130
K+I+ SR+ +++FS+NYA+S+ CL+EL+ I+ ++ V+PVFY +PS V
Sbjct: 66 EKAIQHSRVFVVVFSQNYASSTRCLEELLSILRFSQDNRRPVLPVFYYVDPSDVGLQTGM 125
Query: 131 YGEALAKHEVEFQNDMENMERLLKWKEALHQ---FHSW------------INRCHLHVAE 175
YGEALA HE F ++ + +++KW++AL + W I + V++
Sbjct: 126 YGEALAMHEKRFNSESD---KVMKWRKALCEAAALSGWPFKHGDGYEYELIEKIVEGVSK 182
Query: 176 YL---VGLESRISEVNSLLDLGCTDGVYIIGILGTGGLGKTTLAEAVYNSIVNQFECRCF 232
+ VGL+ R+ E+N LLD GV++IGI G GG+GKTTLA A+Y+S+ QF+ CF
Sbjct: 183 KINRPVGLQYRMLELNGLLDAASLSGVHLIGIYGVGGIGKTTLARALYDSVAVQFDALCF 242
Query: 233 LYNVRENSFKHSLKYLQEQLLSKSIGY-DTPLEHDNEGIEIIKQRLCRKKVLLILDDVDK 291
L VREN+ KH L +LQ+ +L++++G D L +GI ++KQRL K+VLL+LDD+++
Sbjct: 243 LDEVRENAMKHGLVHLQQTILAETVGEKDIRLPSVKQGITLLKQRLQEKRVLLVLDDINE 302
Query: 292 PNQLEKLVGEPGWFGQGSRVIITTRDRYLLSCHGITKIYEADSLNKEESLELLRKMTFKN 351
QL+ LVG PGWFG GSRVIITTRDR LL HG+ KIYE ++L E+LELL FK
Sbjct: 303 SEQLKALVGSPGWFGPGSRVIITTRDRQLLESHGVEKIYEVENLADGEALELLCWKAFKT 362
Query: 352 DSSYDYILN---RAVEYASGLPLALKVVGSNLFGKSIADCESTLDKYERIPPEDIQKILK 408
D Y +N RA+ YASGLPLAL+V+GSNLFG+ I + + TLD YE+I +DIQKILK
Sbjct: 363 DKVYPDFINKIYRALTYASGLPLALEVIGSNLFGREIVEWQYTLDLYEKIHDKDIQKILK 422
Query: 409 VSFDTLEEEQQSVFLDIACCFKGC 432
+SFD L+E ++ +FLDIAC FKGC
Sbjct: 423 ISFDALDEHEKDLFLDIACFFKGC 446
>gb|AAG01051.1| resistance protein LM12 [Glycine max]
Length = 438
Score = 391 bits (1004), Expect = e-107
Identities = 222/436 (50%), Positives = 287/436 (64%), Gaps = 36/436 (8%)
Query: 21 FNFDVFISFRGTDTRFGFTGNLYKALSDKGIHTFIDDKELPTGDEITPSLRKSIEESRIA 80
F++DVF+SFRG DTR+GFTG LY L ++GIHTFIDD E GDEIT +L +IE+S+I
Sbjct: 6 FSYDVFLSFRGEDTRYGFTGYLYNVLRERGIHTFIDDDEPQEGDEITTALEAAIEKSKIF 65
Query: 81 IIIFSKNYATSSFCLDELVHIIHCFREKV-TKVIPVFYGTEPSHVRKLEDSYGEALAKHE 139
II+ S+NYA+SSFCL+ L HI++ +E V+PVFY PS VR S+GEALA HE
Sbjct: 66 IIVLSENYASSSFCLNSLTHILNFTKENNDVLVLPVFYRVNPSDVRHHRGSFGEALANHE 125
Query: 140 VEFQNDMENMERLLKWKEALHQFHSW---------------------------INRCHLH 172
+ ++ NME+L WK ALHQ + N HLH
Sbjct: 126 KK--SNSNNMEKLETWKMALHQVSNISGHHFQHDGNKYEYKFIKEIVESVSNKFNHDHLH 183
Query: 173 VAEYLVGLESRISEVNSLLDLGCTDGVYIIGILGTGGLGKTTLAEAVYNSIVNQFECRCF 232
V++ LVGLES + EV SLLD+G D V+++GI G G+GKTTLA AVYNSI FE CF
Sbjct: 184 VSDVLVGLESPVLEVKSLLDVGRDDVVHMVGIHGLAGVGKTTLAIAVYNSIAGHFEASCF 243
Query: 233 LYNVRENSFK-HSLKYLQEQLLSKSIGYDTPLEHDNEGIEIIKQRLCRKKVLLILDDVDK 291
L NV+ S + L+ LQ LLSK+ G + L + EGI IIK++L +KKVLLILDDVD+
Sbjct: 244 LENVKRTSNTINGLEKLQSFLLSKTAG-EIKLTNWREGIPIIKRKLKQKKVLLILDDVDE 302
Query: 292 PNQLEKLVGEPGWFGQGSRVIITTRDRYLLSCHGITKIYEADSLNKEESLELLRKMTFK- 350
QL+ L+G P WFG GSR+IITTRD +LL+ H + Y+ LN++ +L+LL + F+
Sbjct: 303 DKQLQALIGSPDWFGLGSRIIITTRDEHLLALHNVKITYKVRELNEKHALQLLTQKAFEL 362
Query: 351 ---NDSSYDYILNRAVEYASGLPLALKVVGSNLFGKSIADCESTLDKYERIPPEDIQKIL 407
D SY ILNRAV YASGLP L+V+GSNLFGKSI + +S LD YERIP + IL
Sbjct: 363 EKGIDPSYHDILNRAVTYASGLPFVLEVIGSNLFGKSIEEWKSALDGYERIPHKKNLCIL 422
Query: 408 KVSFDTLEEEQQSVFL 423
KVS+D L E+++S+FL
Sbjct: 423 KVSYDALNEDEKSIFL 438
>gb|AAG01052.1| resistance protein MG23 [Glycine max]
Length = 435
Score = 387 bits (993), Expect = e-106
Identities = 219/432 (50%), Positives = 288/432 (65%), Gaps = 37/432 (8%)
Query: 22 NFDVFISFRGTDTRFGFTGNLYKALSDKGIHTFIDDKELPTGDEITPSLRKSIEESRIAI 81
++DVF+SFRG DTR GFTGNLY L ++GI TFIDD+EL G EIT +L ++IE+S+I I
Sbjct: 7 SYDVFLSFRGEDTRHGFTGNLYNVLRERGIDTFIDDEELQKGHEITKALEEAIEKSKIFI 66
Query: 82 IIFSKNYATSSFCLDELVHIIHCFREKVTK-VIPVFYGTEPSHVRKLEDSYGEALAKHEV 140
I+ S+NYA+SSFCL+EL HI++ + K + ++PVFY +PS VR S+GEALA HE
Sbjct: 67 IVLSENYASSSFCLNELTHILNFTKGKSDRSILPVFYKVDPSDVRYHRGSFGEALANHEK 126
Query: 141 EFQNDMENMERLLKWKEALHQFHSW---------------------------INRCHLHV 173
+ +++ ME+L WK AL Q ++ NR L+V
Sbjct: 127 KLKSNY--MEKLQIWKMALQQVSNFSGHHFQPDGDKYEYDFIKEIVESVPSKFNRNLLYV 184
Query: 174 AEYLVGLESRISEVNSLLDLGCTDGVYIIGILGTGGLGKTTLAEAVYNSIVNQFECRCFL 233
++ LVGL+S + V SLLD+G D V+++GI G GG+GKTTLA AVYNSI FE CFL
Sbjct: 185 SDVLVGLKSPVLAVKSLLDVGADDVVHMVGIHGLGGVGKTTLAVAVYNSIACHFEACCFL 244
Query: 234 YNVRENSFKHSLKYLQEQLLSKSIGYDTPLEHDN--EGIEIIKQRLCRKKVLLILDDVDK 291
NVRE S K L+ LQ LLSK++G D +E N EG +IIK++L KKVLL+LDDV++
Sbjct: 245 ENVRETSNKKGLESLQNILLSKTVG-DMKIEVTNSREGTDIIKRKLKEKKVLLVLDDVNE 303
Query: 292 PNQLEKLVGEPGWFGQGSRVIITTRDRYLLSCHGITKIYEADSLNKEESLELLRKMTF-- 349
QL+ ++ P WFG+GSRVIITTRD LL H + + Y+ LN++ +L+LL + F
Sbjct: 304 HEQLQAIIDSPDWFGRGSRVIITTRDEQLLVLHNVKRTYKVRELNEKHALQLLTQKAFGL 363
Query: 350 --KNDSSYDYILNRAVEYASGLPLALKVVGSNLFGKSIADCESTLDKYERIPPEDIQKIL 407
K D SY ILNRAV YASGLPLALKV+GSNLFGKSI + ES LD YER P + I L
Sbjct: 364 EKKVDPSYHDILNRAVTYASGLPLALKVIGSNLFGKSIEEWESVLDGYERSPDKSIYMTL 423
Query: 408 KVSFDTLEEEQQ 419
KVS+D L E+++
Sbjct: 424 KVSYDALNEDEK 435
>gb|AAG48132.1| putative resistance protein [Glycine max]
Length = 1093
Score = 384 bits (987), Expect = e-105
Identities = 251/615 (40%), Positives = 347/615 (55%), Gaps = 68/615 (11%)
Query: 23 FDVFISFRGTDTRFGFTGNLYKALSDKGIHTFIDDKELPTGDEITPSLRKSIEESRIAII 82
+DVF+SFRG DTR FTGNLY L +GIHTFI D + +G+EI SL ++IE SR+ +I
Sbjct: 14 YDVFLSFRGEDTRRSFTGNLYNCLEKRGIHTFIGDYDFESGEEIKASLSEAIEHSRVFVI 73
Query: 83 IFSKNYATSSFCLDELVHIIHCFREKVTKVIPVFYGTEPSHVRKLEDSYGEALAKHEVEF 142
+FS+NYA+SS+CLD LV I+ + VIPVF+ EPSHVR + YGEALA HE
Sbjct: 74 VFSENYASSSWCLDGLVRILDFTEDNHRPVIPVFFDVEPSHVRHQKGIYGEALAMHERRL 133
Query: 143 QNDMENMERLLKWKEALHQFHSWINRCHLH-------------------------VAEYL 177
+ +++KW+ AL Q + H V +
Sbjct: 134 NPE---SYKVMKWRNALRQAANLSGYAFKHGDGYEYKLIEKIVEDISNKIKISRPVVDRP 190
Query: 178 VGLESRISEVNSLLDLGCTDGVYIIGILGTGGLGKTTLAEAVYNSIVNQFECRCFLYNVR 237
VGLE R+ EV+ LLD GV++IGI G GG+GKTTLA AVY+S F+ CFL NVR
Sbjct: 191 VGLEYRMLEVDWLLDATSLAGVHMIGICGIGGIGKTTLARAVYHSAAGHFDTSCFLGNVR 250
Query: 238 ENSFKHSLKYLQEQLLSKSIGYDT-PLEHDNEGIEIIKQRLCRKKVLLILDDVDKPNQLE 296
EN+ KH L +LQ+ LL++ + L +GI +IK+ L RK++LL+LDDV + + L
Sbjct: 251 ENAMKHGLVHLQQTLLAEIFRENNIRLTSVEQGISLIKKMLPRKRLLLVLDDVCELDDLR 310
Query: 297 KLVGEPGWFGQGSRVIITTRDRYLLSCHGITKIYEADSLNKEESLELLRKMTFKNDSSY- 355
LVG P WFG GSRVIITTRDR+LL HG+ K+YE + L E+LELL F+ D +
Sbjct: 311 ALVGSPDWFGPGSRVIITTRDRHLLKAHGVDKVYEVEVLANGEALELLCWKAFRTDRVHP 370
Query: 356 DYI--LNRAVEYASGLPLALKVVGSNLFGKSIADCESTLDKYERIPPEDIQKILKVSFDT 413
D+I LNRA+ +ASG+PLAL+++GS+L+G+ I + ESTLD+YE+ PP DI LK+SFD
Sbjct: 371 DFINKLNRAITFASGIPLALELIGSSLYGRGIEEWESTLDQYEKNPPRDIHMALKISFDA 430
Query: 414 LEEEQQSVFLDIACCFKGCDWQKFQR----HFNFIMAIQ*HIMLECWLMNLS*RLVLQIH 469
L ++ VFLDIAC F G + + + H + ++E L+ + +Q+H
Sbjct: 431 LGYLEKEVFLDIACFFNGFELAEIEHILGAHHGCCLKFHIGALVEKSLIMIDEHGRVQMH 490
Query: 470 TTLLIL----------LHYMT**SIWV*K*SDKNQLKSLESALGCGVMMI*LMF*NKTHK 519
+ + H +W S ++ + LE G T K
Sbjct: 491 DLIQQMGREIVRQESPEHPGKRSRLW----STEDIVHVLEDNTG-------------TCK 533
Query: 520 IEMIYLNCSSMEP-ININEKAFKKMKKLKTLIIEKGYFSKGLKYLPKSLIVLKWKGFTSE 578
I+ I L+ S E + + AF KM L+TLII K FSKG K + L +L+W G S+
Sbjct: 534 IQSIILDFSKSEKVVQWDGMAFVKMISLRTLIIRK-MFSKGPKNF-QILKMLEWWGCPSK 591
Query: 579 PLSFCFSFKASKIIV 593
L FK K+ +
Sbjct: 592 SLP--SDFKPEKLAI 604
>gb|AAP44393.1| nematode resistance-like protein [Solanum tuberosum]
Length = 1136
Score = 352 bits (904), Expect = 2e-95
Identities = 233/608 (38%), Positives = 338/608 (55%), Gaps = 53/608 (8%)
Query: 21 FNFDVFISFRGTDTRFGFTGNLYKALSDKGIHTFIDDKELPTGDEITPSLRKSIEESRIA 80
+++DVF+SFRG D R F +LY AL K I+TF DD++L G I+P L SIEESRIA
Sbjct: 16 WSYDVFLSFRGEDVRKTFVDHLYLALQQKCINTFKDDEKLEKGKFISPELVSSIEESRIA 75
Query: 81 IIIFSKNYATSSFCLDELVHIIHCFREKVTKVIPVFYGTEPSHVRKLEDSYGEALAKHEV 140
+IIFSKNYA S++CLDEL I+ C K V+PVFY +PS VRK + +GEA +KHE
Sbjct: 76 LIIFSKNYANSTWCLDELTKIMECKNVKGQIVVPVFYDVDPSTVRKQKSIFGEAFSKHEA 135
Query: 141 EFQNDMENMERLLKWKEALHQ---FHSWI-------------------------NRCHLH 172
FQ D ++ KW+ AL + W ++ H
Sbjct: 136 RFQED-----KVQKWRAALEEAANISGWDLPNTSNGHEARVMEKIAEDIMARLGSQRHAS 190
Query: 173 VAEYLVGLESRISEVNSLLDLGCTDGVYIIGILGTGGLGKTTLAEAVYNSIVNQFECRCF 232
A LVG+ES + +V +L +G + GV+ +GILG G+GKTTLA +Y++I +QF+ CF
Sbjct: 191 NARNLVGMESHMHQVYKMLGIG-SGGVHFLGILGMSGVGKTTLARVIYDNIRSQFQGACF 249
Query: 233 LYNVRENSFKHSLKYLQEQLLSKSIGYDTPLEHDN-EGIEIIKQRLCRKKVLLILDDVDK 291
L+ VR+ S K L+ LQE LLS+ + +D+ EG + KQRL KKVLL+LDDVD
Sbjct: 250 LHEVRDRSAKQGLERLQEILLSEILVVKKLRINDSFEGANMQKQRLQYKKVLLVLDDVDH 309
Query: 292 PNQLEKLVGEPGWFGQGSRVIITTRDRYLLSCHGITKIYEADSLNKEESLELLRKMTFKN 351
+QL L GE WFG GSR+IITT+D++LL + KIY +LN ESL+L ++ FK
Sbjct: 310 IDQLNALAGEREWFGDGSRIIITTKDKHLLVKYETEKIYRMKTLNNYESLQLFKQHAFKK 369
Query: 352 D---SSYDYILNRAVEYASGLPLALKVVGSNLFGKSIADCESTLDKYERIPPEDIQKILK 408
+ ++ + + +++ GLPLALKV+GS L+G+ + + S +++ ++IP +I K L+
Sbjct: 370 NRPTKEFEDLSAQVIKHTDGLPLALKVLGSFLYGRGLDEWISEVERLKQIPENEILKKLE 429
Query: 409 VSFDTLEEEQQSVFLDIACCFKGCDWQKFQR---HFNFIMAIQ*HIMLECWLMNLS*RLV 465
SF L +Q +FLDIAC F G R F+F I +++E L+
Sbjct: 430 QSFTGLHNTEQKIFLDIACFFSGKKKDSVTRILESFHFCPVIGIKVLMEKCLIT-----T 484
Query: 466 LQIHTTLLILLHYMT**SIWV*K*SDKNQLKSLESALG-CGVMMI*LMF*NKTHKIEMIY 524
LQ T+ L+ M + D L C V+ L T KIE +
Sbjct: 485 LQGRITIHQLIQDMGWHIVRREATDDPRMCSRLWKREDICPVLERNL----GTDKIEGMS 540
Query: 525 LNCSSMEPININEKAFKKMKKLKTLIIEKGYFSKGLKYLPKSLIVLKWKGFTSEPLSFCF 584
L+ ++ E +N KAF +M +L+ L + Y +G ++LP L L W G+ S+ L
Sbjct: 541 LHLTNEEEVNFGGKAFMQMTRLRFLKFQNAYVCQGPEFLPDELRWLDWHGYPSKSLP--N 598
Query: 585 SFKASKII 592
SFK +++
Sbjct: 599 SFKGDQLV 606
>gb|AAP44390.1| nematode resistance protein [Solanum tuberosum]
Length = 1136
Score = 349 bits (895), Expect = 2e-94
Identities = 231/608 (37%), Positives = 337/608 (54%), Gaps = 53/608 (8%)
Query: 21 FNFDVFISFRGTDTRFGFTGNLYKALSDKGIHTFIDDKELPTGDEITPSLRKSIEESRIA 80
+++DVF+SFRG D R F +LY AL K I+TF DD++L G I+P L SIEESRIA
Sbjct: 16 WSYDVFLSFRGEDVRKTFVDHLYLALEQKCINTFKDDEKLEKGKFISPELVSSIEESRIA 75
Query: 81 IIIFSKNYATSSFCLDELVHIIHCFREKVTKVIPVFYGTEPSHVRKLEDSYGEALAKHEV 140
+IIFSKNYA S++CLDEL I+ C K V+PVFY +PS VRK + +GEA +KHE
Sbjct: 76 LIIFSKNYANSTWCLDELTKIMECKNVKGQIVVPVFYDVDPSTVRKQKSIFGEAFSKHEA 135
Query: 141 EFQNDMENMERLLKWKEALHQ---FHSWI-------------------------NRCHLH 172
FQ D ++ KW+ AL + W ++ H
Sbjct: 136 RFQED-----KVQKWRAALEEAANISGWDLPNTANGHEARVMEKIAEDIMARLGSQRHAS 190
Query: 173 VAEYLVGLESRISEVNSLLDLGCTDGVYIIGILGTGGLGKTTLAEAVYNSIVNQFECRCF 232
A LVG+ES + +V +L +G + GV+ +GILG G+GKTTLA +Y++I +QF+ CF
Sbjct: 191 NARNLVGMESHMHKVYKMLGIG-SGGVHFLGILGMSGVGKTTLARVIYDNIRSQFQGACF 249
Query: 233 LYNVRENSFKHSLKYLQEQLLSKSIGYDTPLEHDN-EGIEIIKQRLCRKKVLLILDDVDK 291
L+ VR+ S K L+ LQE LLS+ + +D+ EG + KQRL KKVLL+LDDVD
Sbjct: 250 LHEVRDRSAKQGLERLQEILLSEILVVKKLRINDSFEGANMQKQRLQYKKVLLVLDDVDH 309
Query: 292 PNQLEKLVGEPGWFGQGSRVIITTRDRYLLSCHGITKIYEADSLNKEESLELLRKMTFKN 351
+QL L GE WFG GSR+IITT+D++LL + KIY +LN ESL+L ++ FK
Sbjct: 310 IDQLNALAGEREWFGDGSRIIITTKDKHLLVKYETEKIYRMKTLNNYESLQLFKQHAFKK 369
Query: 352 D---SSYDYILNRAVEYASGLPLALKVVGSNLFGKSIADCESTLDKYERIPPEDIQKILK 408
+ ++ + + +++ GLPLALKV+GS L+G+ + + S +++ ++IP +I K L+
Sbjct: 370 NRPTKEFEDLSAQVIKHTDGLPLALKVLGSFLYGRGLDEWISEVERLKQIPENEILKKLE 429
Query: 409 VSFDTLEEEQQSVFLDIACCFKGCDWQKFQR---HFNFIMAIQ*HIMLECWLMNLS*RLV 465
SF L +Q +FLDIAC F G R F+F I +++E L+ +
Sbjct: 430 QSFTGLHNTEQKIFLDIACFFSGKKKDSVTRILESFHFCPVIGIKVLMEKCLIT-----I 484
Query: 466 LQIHTTLLILLHYMT**SIWV*K*SDKNQLKSLESALG-CGVMMI*LMF*NKTHKIEMIY 524
LQ T+ L+ M + D + C V+ L T K E +
Sbjct: 485 LQGRITIHQLIQDMGWHIVRREATDDPRMCSRMWKREDICPVLERNL----GTDKNEGMS 540
Query: 525 LNCSSMEPININEKAFKKMKKLKTLIIEKGYFSKGLKYLPKSLIVLKWKGFTSEPLSFCF 584
L+ ++ E +N KAF +M +L+ L Y +G ++LP L L W G+ S+ L
Sbjct: 541 LHLTNEEEVNFGGKAFMQMTRLRFLKFRNAYVCQGPEFLPDELRWLDWHGYPSKSLP--N 598
Query: 585 SFKASKII 592
SFK +++
Sbjct: 599 SFKGDQLV 606
>gb|AAL07535.1| resistance gene analog PU3 [Helianthus annuus]
Length = 770
Score = 337 bits (865), Expect = 5e-91
Identities = 194/443 (43%), Positives = 273/443 (60%), Gaps = 25/443 (5%)
Query: 14 SSSLSYDFNFDVFISFRGTDTRFGFTGNLYKALSDKGIHTFIDDKELPTGDEITPSLRKS 73
S+S S +N DVF+SFRG DTR F +LY AL +GI T+ DD+ LP G+ I P+L K+
Sbjct: 74 SASTSQSWNHDVFLSFRGEDTRNSFVDHLYAALVQQGIQTYKDDQTLPRGERIGPALLKA 133
Query: 74 IEESRIAIIIFSKNYATSSFCLDELVHIIHCFREKVTKVIPVFYGTEPSHVRKLEDSYGE 133
I+ESRIA+++FS+NYA SS+CLDEL HI+ C + VIP+FY +PS VRK + YG+
Sbjct: 134 IQESRIAVVVFSQNYADSSWCLDELAHIMECMDTRGQIVIPIFYFVDPSDVRKQKGKYGK 193
Query: 134 ALAKHEVEFQNDMENMERLLK-------W------------KEALHQFHSWINRCHLHVA 174
A KH+ E + +E+ + L+ W KE + S + +V
Sbjct: 194 AFRKHKRENKQKVESWRKALEKAGNLSGWVINENSHEAKCIKEIVATISSRLPTLSTNVN 253
Query: 175 EYLVGLESRISEVNSLLDLGCTDGVYIIGILGTGGLGKTTLAEAVYNSIVNQFECRCFLY 234
+ L+G+E+R+ ++ S L + D V IIGI G GG GKTTLA A Y I ++FE C L
Sbjct: 254 KDLIGIETRLQDLKSKLKMESGD-VRIIGIWGVGGGGKTTLASAAYAEISHRFEAHCLLQ 312
Query: 235 NVRENSFKHSLKYLQEQLLSKSI-GYDTPLEHDNEGIEIIKQRLCRKKVLLILDDVDKPN 293
N+RE S KH L+ LQE++LS + D + + EG +I++RL K VL++LDDVD
Sbjct: 313 NIREESNKHGLEKLQEKILSLVLKTKDVVVGSEIEGRSMIERRLRNKSVLVVLDDVDDLK 372
Query: 294 QLEKLVGEPGWFGQGSRVIITTRDRYLLSCHGITKIYEADSLNKEESLELLRKMTFKND- 352
QLE L G WFG+GSR+IITTRD +LL+ H IYE L+ +E++EL K ++ D
Sbjct: 373 QLEALAGSHAWFGKGSRIIITTRDEHLLTRHA-DMIYEVSLLSDDEAMELFNKHAYREDE 431
Query: 353 --SSYDYILNRAVEYASGLPLALKVVGSNLFGKSIADCESTLDKYERIPPEDIQKILKVS 410
Y + V YASGLPLAL+++GS L+ K+ D +S L K + IP ++ + LK+S
Sbjct: 432 LIEDYGMLSKDVVSYASGLPLALEILGSFLYDKNKDDWKSALAKLKCIPNVEVTERLKIS 491
Query: 411 FDTLEEEQQSVFLDIACCFKGCD 433
+D LE E Q +FLDIAC ++ D
Sbjct: 492 YDGLEPEHQKLFLDIACFWRRRD 514
>gb|AAO45748.1| MRGH5 [Cucumis melo]
Length = 1092
Score = 330 bits (845), Expect = 1e-88
Identities = 194/450 (43%), Positives = 284/450 (63%), Gaps = 37/450 (8%)
Query: 12 TSSSSLSYDFNFDVFISFRGTDTRFGFTGNLYKALSDKGIHTFIDDKELPTGDEITPSLR 71
+SSSS +++++DVF+SFRG DTR FTG+LY L KG++ FIDD L G++I+ +L
Sbjct: 10 SSSSSPIFNWSYDVFLSFRGEDTRSNFTGHLYMFLRQKGVNVFIDDG-LERGEQISETLF 68
Query: 72 KSIEESRIAIIIFSKNYATSSFCLDELVHIIHCFREKVTKVIPVFYGTEPSHVRKLEDSY 131
K+I+ S I+I+IFS+NYA+S++CLDELV I+ C + K KV+P+FY +PS VRK +
Sbjct: 69 KTIQNSLISIVIFSENYASSTWCLDELVEIMECKKSKGQKVLPIFYKVDPSDVRKQNGWF 128
Query: 132 GEALAKHEVEFQNDMENMERLLKWKEAL------------------------HQFHSWIN 167
E LAKHE F ME++ W++AL + S +N
Sbjct: 129 REGLAKHEANF------MEKIPIWRDALTTAANLSGWHLGARKEAHLIQDIVKEVLSILN 182
Query: 168 RCH-LHVAEYLVGLESRISEVNSLLDLGCTDGVYIIGILGTGGLGKTTLAEAVYNSIVNQ 226
L+ E+LVG++S+I + ++ ++ V ++GI G GG+GKTTLA+A+Y+ + +Q
Sbjct: 183 HTKPLNANEHLVGIDSKIEFLYRKEEMYKSECVNMLGIYGIGGIGKTTLAKALYDKMASQ 242
Query: 227 FECRCFLYNVRENS-FKHSLKYLQEQLLSKSIGYDTPLEHDNEGIEIIKQRLCRKKVLLI 285
FE C+L +VRE S L LQ++LL + + YD + + GI IIK RL KKVL++
Sbjct: 243 FEGCCYLRDVREASKLFDGLTQLQKKLLFQILKYDLEVVDLDWGINIIKNRLRSKKVLIL 302
Query: 286 LDDVDKPNQLEKLVGEPGWFGQGSRVIITTRDRYLLSCHGITKIYEADSLNKEESLELLR 345
LDDVDK QL+ LVG WFGQG+++I+TTR++ LL HG K+YE L+K E++EL R
Sbjct: 303 LDDVDKLEQLQALVGGHDWFGQGTKIIVTTRNKQLLVSHGFDKMYEVQGLSKHEAIELFR 362
Query: 346 KMTFKN---DSSYDYILNRAVEYASGLPLALKVVGSNLFGKS-IADCESTLDKYERIPPE 401
+ FKN S+Y + RA Y +G PLAL V+GS L +S +A+ LD +E +
Sbjct: 363 RHAFKNLQPSSNYLDLSERATRYCTGHPLALIVLGSFLCDRSDLAEWSGILDGFENSLRK 422
Query: 402 DIQKILKVSFDTLEEEQQSVFLDIACCFKG 431
DI+ IL++SFD LE+E + +FLDI+C G
Sbjct: 423 DIKDILQLSFDGLEDEVKEIFLDISCLLVG 452
Score = 41.6 bits (96), Expect = 0.078
Identities = 21/56 (37%), Positives = 32/56 (56%), Gaps = 1/56 (1%)
Query: 533 ININEKAFKKMKKLKTLIIEKGY-FSKGLKYLPKSLIVLKWKGFTSEPLSFCFSFK 587
I+++ +AF+ MK L+ L+++ F K +KYLP L +KW F L CF K
Sbjct: 550 IDLDPEAFRSMKNLRILMVDGNVRFCKKIKYLPNGLKWIKWHRFAHPSLPSCFITK 605
>emb|CAC82811.1| resistance gene-like [Solanum tuberosum subsp. andigena]
Length = 1126
Score = 321 bits (823), Expect = 4e-86
Identities = 185/459 (40%), Positives = 279/459 (60%), Gaps = 38/459 (8%)
Query: 6 MAMQLPTSSSSLSYD-----FNFDVFISFRGTDTRFGFTGNLYKALSDKGIHTFIDDKEL 60
MA +S S+ Y + +DVF+SFRG DTR FT +LY+ L ++GI TF+DDK L
Sbjct: 1 MASSSSSSESNSQYSCPQRKYKYDVFLSFRGKDTRRNFTSHLYERLDNRGIFTFLDDKRL 60
Query: 61 PTGDEITPSLRKSIEESRIAIIIFSKNYATSSFCLDELVHIIHCFREKVTKVIPVFYGTE 120
GD ++ L K+I+ES++A+IIFSKNYATS +CL+E+V I+ C E VIPVFY +
Sbjct: 61 ENGDSLSKELVKAIKESQVAVIIFSKNYATSRWCLNEVVKIMECKEENGQLVIPVFYDVD 120
Query: 121 PSHVRKLEDSYGEALAKHEVEFQNDMENMERLLKWKEALHQ-----------------FH 163
PS VRK S+ EA A+HE +++D+E M+++ +W+ AL +
Sbjct: 121 PSDVRKQTKSFAEAFAEHESRYKDDVEGMQKVQRWRTALSEAADLKGYDIRERIESECIG 180
Query: 164 SWINRCHLHVAE----YL---VGLESRISEVNSLLDLGCTDGVYIIGILGTGGLGKTTLA 216
+N + E YL VG+++ + +VNSLL++ D V I+ I G GG+GKTT+A
Sbjct: 181 ELVNEISPKLCETSLSYLTDVVGIDAHLKKVNSLLEMKIDD-VRIVWIWGMGGVGKTTIA 239
Query: 217 EAVYNSIVNQFECRCFLYNVRENSFKHSLKYLQEQLLSKSIGYDTPLEHDNE-GIEIIKQ 275
A+++ + ++F+ CFL + +EN K+ + LQ LLSK +G HD E G ++ +
Sbjct: 240 RAIFDILSSKFDGACFLPDNKEN--KYEIHSLQSILLSKLVGEKENCVHDKEDGRHLMAR 297
Query: 276 RLCRKKVLLILDDVDKPNQLEKLVGEPGWFGQGSRVIITTRDRYLLSCHGITKIYEADSL 335
RL KKVL++LD++D +QL+ L G+ GWFG G+R+I TTRD++ + + +Y +L
Sbjct: 298 RLRLKKVLVVLDNIDHEDQLKYLAGDLGWFGNGTRIIATTRDKHFIRKNDA--VYPVTTL 355
Query: 336 NKEESLELLRKMTFKN---DSSYDYILNRAVEYASGLPLALKVVGSNLFGKSIADCESTL 392
+ ++++L + FKN D ++ I V +A GLPLALKV GS+L K I S +
Sbjct: 356 LEHDAVQLFNQYAFKNEVPDKCFEEITLEVVSHAEGLPLALKVWGSSLHKKDIHVWRSAV 415
Query: 393 DKYERIPPEDIQKILKVSFDTLEEEQQSVFLDIACCFKG 431
D+ +R P + + LKVS+D LE E Q +FLDIAC +G
Sbjct: 416 DRIKRNPSSKVVENLKVSYDGLEREDQEIFLDIACFLRG 454
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.331 0.145 0.443
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 998,319,194
Number of Sequences: 2540612
Number of extensions: 41216072
Number of successful extensions: 174018
Number of sequences better than 10.0: 2309
Number of HSP's better than 10.0 without gapping: 1172
Number of HSP's successfully gapped in prelim test: 1137
Number of HSP's that attempted gapping in prelim test: 168910
Number of HSP's gapped (non-prelim): 2847
length of query: 625
length of database: 863,360,394
effective HSP length: 134
effective length of query: 491
effective length of database: 522,918,386
effective search space: 256752927526
effective search space used: 256752927526
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 78 (34.7 bits)
Medicago: description of AC126790.7


