

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126786.6 - phase: 0
(228 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
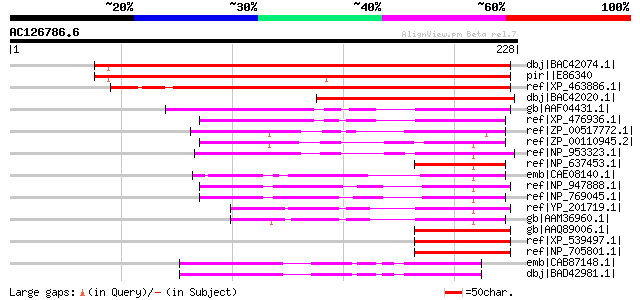
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAC42074.1| unknown protein [Arabidopsis thaliana] gi|288275... 280 2e-74
pir||E86340 protein F2D10.32 [imported] - Arabidopsis thaliana g... 276 4e-73
ref|XP_463886.1| putative immunophilin / FKBP-type peptidyl-prol... 259 3e-68
dbj|BAC42020.1| unknown protein [Arabidopsis thaliana] 138 1e-31
gb|AAF04431.1| unknown protein [Arabidopsis thaliana] gi|2168961... 106 6e-22
ref|XP_476936.1| immunophilin / FKBP-type peptidyl-prolyl cis-tr... 84 4e-15
ref|ZP_00517772.1| Peptidylprolyl isomerase [Crocosphaera watson... 65 1e-09
ref|ZP_00110945.2| COG0545: FKBP-type peptidyl-prolyl cis-trans ... 63 7e-09
ref|NP_953323.1| FKBP-type peptidyl-prolyl cis-trans isomerase [... 57 4e-07
ref|NP_637453.1| FKBP-type peptidyl-prolyl cis-trans isomerase [... 54 3e-06
emb|CAE08140.1| FKBP-type peptidyl-prolyl cis-trans isomerase (P... 53 7e-06
ref|NP_947888.1| FKBP-type peptidyl-prolyl cis-trans isomerase [... 53 7e-06
ref|NP_769045.1| Peptidylprolyl isomerase [Bradyrhizobium japoni... 52 1e-05
ref|YP_201719.1| FKBP-type peptidyl-prolyl cis-trans isomerase [... 52 1e-05
gb|AAM36960.1| FKBP-type peptidyl-prolyl cis-trans isomerase [Xa... 52 1e-05
gb|AAQ89006.1| FKBP14 [Homo sapiens] gi|51105873|gb|EAL24457.1| ... 51 3e-05
ref|XP_539497.1| PREDICTED: similar to Secernin 1 [Canis familia... 51 3e-05
ref|NP_705801.1| FK506 binding protein 14 [Mus musculus] gi|2213... 49 8e-05
emb|CAB87148.1| putative protein [Arabidopsis thaliana] gi|15240... 49 8e-05
dbj|BAD42981.1| unknown protein [Arabidopsis thaliana] 49 8e-05
>dbj|BAC42074.1| unknown protein [Arabidopsis thaliana] gi|28827556|gb|AAO50622.1|
putative FKBP-type peptidyl-prolyl cis-trans isomerase
[Arabidopsis thaliana] gi|15218039|ref|NP_173504.1|
immunophilin / FKBP-type peptidyl-prolyl cis-trans
isomerase family protein [Arabidopsis thaliana]
gi|18203241|sp|Q9LM71|FKBP1_ARATH Probable FKBP-type
peptidyl-prolyl cis-trans isomerase 1, chloroplast
precursor (PPIase) (Rotamase)
Length = 232
Score = 280 bits (717), Expect = 2e-74
Identities = 138/190 (72%), Positives = 159/190 (83%), Gaps = 3/190 (1%)
Query: 39 VVALS---QSRRSTAILISSLPFTFVFLSPPPPAEARERRKKKNIPIDDYITSPDGLKYY 95
VVA S R ++ IL+SS+P T F+ P +EARERR +K IP+++Y T P+GLK+Y
Sbjct: 36 VVAFSVPISRRDASIILLSSIPLTSFFVLTPSSSEARERRSRKVIPLEEYSTGPEGLKFY 95
Query: 96 DFLEGKGPIAEKGSTVQVHFDCLYRGITAVSSRESKLLAGNRVIAQPYEFKVGAPPGKER 155
D EGKGP+A +GST QVHFDC YR ITA+S+RESKLLAGNR IAQPYEFKVG+ PGKER
Sbjct: 96 DIEEGKGPVATEGSTAQVHFDCRYRSITAISTRESKLLAGNRSIAQPYEFKVGSTPGKER 155
Query: 156 KREFVDNPNGLFSAQAAPKPPQAMYTIVEGMRVGGKRTVIVPPEKGYGKKGMNEIPPGAT 215
KREFVDNPNGLFSAQAAPKPP AMY I EGM+VGGKRTVIVPPE GYG+KGMNEIPPGAT
Sbjct: 156 KREFVDNPNGLFSAQAAPKPPPAMYFITEGMKVGGKRTVIVPPEAGYGQKGMNEIPPGAT 215
Query: 216 FELNIELLQL 225
FELNIELL++
Sbjct: 216 FELNIELLRV 225
>pir||E86340 protein F2D10.32 [imported] - Arabidopsis thaliana
gi|8886936|gb|AAF80622.1| F2D10.32 [Arabidopsis
thaliana]
Length = 233
Score = 276 bits (705), Expect = 4e-73
Identities = 138/191 (72%), Positives = 159/191 (82%), Gaps = 4/191 (2%)
Query: 39 VVALS---QSRRSTAILISSLPFTFVFLSPPPPAEARERRKKKNIPIDDYITSPDGLKYY 95
VVA S R ++ IL+SS+P T F+ P +EARERR +K IP+++Y T P+GLK+Y
Sbjct: 36 VVAFSVPISRRDASIILLSSIPLTSFFVLTPSSSEARERRSRKVIPLEEYSTGPEGLKFY 95
Query: 96 DFLEGKGPIAEKGSTVQVHFDCLYRGITAVSSRESKLLAGNRVIAQ-PYEFKVGAPPGKE 154
D EGKGP+A +GST QVHFDC YR ITA+S+RESKLLAGNR IAQ PYEFKVG+ PGKE
Sbjct: 96 DIEEGKGPVATEGSTAQVHFDCRYRSITAISTRESKLLAGNRSIAQQPYEFKVGSTPGKE 155
Query: 155 RKREFVDNPNGLFSAQAAPKPPQAMYTIVEGMRVGGKRTVIVPPEKGYGKKGMNEIPPGA 214
RKREFVDNPNGLFSAQAAPKPP AMY I EGM+VGGKRTVIVPPE GYG+KGMNEIPPGA
Sbjct: 156 RKREFVDNPNGLFSAQAAPKPPPAMYFITEGMKVGGKRTVIVPPEAGYGQKGMNEIPPGA 215
Query: 215 TFELNIELLQL 225
TFELNIELL++
Sbjct: 216 TFELNIELLRV 226
>ref|XP_463886.1| putative immunophilin / FKBP-type peptidyl-prolyl cis-trans
isomerase [Oryza sativa (japonica cultivar-group)]
gi|41052837|dbj|BAD07728.1| putative immunophilin /
FKBP-type peptidyl-prolyl cis-trans isomerase [Oryza
sativa (japonica cultivar-group)]
Length = 242
Score = 259 bits (663), Expect = 3e-68
Identities = 130/180 (72%), Positives = 151/180 (83%), Gaps = 4/180 (2%)
Query: 46 RRSTAILISSLPFTFVFLSPPPPAEARERRKKKNIPIDDYITSPDGLKYYDFLEGKGPIA 105
RR+ A L+S+ F + +SPP A RR + + ++DY+TSPDGLKYYD +EGKGP A
Sbjct: 61 RRAAAQLLSAAGF-LIAVSPPSLAA---RRGRMVVSLEDYVTSPDGLKYYDLVEGKGPTA 116
Query: 106 EKGSTVQVHFDCLYRGITAVSSRESKLLAGNRVIAQPYEFKVGAPPGKERKREFVDNPNG 165
EKGSTVQVHFDC+YRGITAVSSRE+KLLAGNR IAQPYEF VG+ PGKERKREFVD+ NG
Sbjct: 117 EKGSTVQVHFDCIYRGITAVSSREAKLLAGNRSIAQPYEFSVGSLPGKERKREFVDSANG 176
Query: 166 LFSAQAAPKPPQAMYTIVEGMRVGGKRTVIVPPEKGYGKKGMNEIPPGATFELNIELLQL 225
L+SAQA+PKPP AMYTI EGM+VGGKR VIVPPE GYGKKGMNEIPP A FEL+IELL++
Sbjct: 177 LYSAQASPKPPAAMYTITEGMKVGGKRRVIVPPELGYGKKGMNEIPPDAPFELDIELLEV 236
>dbj|BAC42020.1| unknown protein [Arabidopsis thaliana]
Length = 95
Score = 138 bits (348), Expect = 1e-31
Identities = 67/89 (75%), Positives = 74/89 (82%)
Query: 139 IAQPYEFKVGAPPGKERKREFVDNPNGLFSAQAAPKPPQAMYTIVEGMRVGGKRTVIVPP 198
+ QPYEFKVG+ PGKERKREFVDNPNGLFSAQAAPKPP AMY I EGM+VGGKRTVIVPP
Sbjct: 7 LEQPYEFKVGSTPGKERKREFVDNPNGLFSAQAAPKPPPAMYFITEGMKVGGKRTVIVPP 66
Query: 199 EKGYGKKGMNEIPPGATFELNIELLQLAS 227
E GYG+KGMNEIP ++L + L L S
Sbjct: 67 EAGYGQKGMNEIPVRTYYKLYMSLELLLS 95
>gb|AAF04431.1| unknown protein [Arabidopsis thaliana] gi|21689613|gb|AAM67428.1|
AT3g10060/T22K18_11 [Arabidopsis thaliana]
gi|20453145|gb|AAM19814.1| AT3g10060/T22K18_11
[Arabidopsis thaliana] gi|15232828|ref|NP_187617.1|
immunophilin, putative / FKBP-type peptidyl-prolyl
cis-trans isomerase, putative [Arabidopsis thaliana]
Length = 230
Score = 106 bits (264), Expect = 6e-22
Identities = 66/155 (42%), Positives = 82/155 (52%), Gaps = 24/155 (15%)
Query: 71 ARERRKKKNIPIDDYITSPDGLKYYDFLEGKGPIAEKGSTVQVHFDCLYRGITAVSSRES 130
+R + +P D+ T P+GLKYYD G G A KGS V VH+ ++GIT ++SR+
Sbjct: 86 SRRALRASKLPESDFTTLPNGLKYYDIKVGNGAEAVKGSRVAVHYVAKWKGITFMTSRQG 145
Query: 131 KLLAGNRVIAQPYEFKVGAPPGKERKREFVDNPNGLFSAQAAPKPPQAMYTIVEGMRVGG 190
+ G PY F VG ER G VEGMRVGG
Sbjct: 146 LGVGGGT----PYGFDVGQ---SERGNVLKGLDLG-----------------VEGMRVGG 181
Query: 191 KRTVIVPPEKGYGKKGMNEIPPGATFELNIELLQL 225
+R VIVPPE YGKKG+ EIPP AT EL+IELL +
Sbjct: 182 QRLVIVPPELAYGKKGVQEIPPNATIELDIELLSI 216
>ref|XP_476936.1| immunophilin / FKBP-type peptidyl-prolyl cis-trans isomerase-like
[Oryza sativa (japonica cultivar-group)]
gi|34394609|dbj|BAC83911.1| immunophilin / FKBP-type
peptidyl-prolyl cis-trans isomerase-like [Oryza sativa
(japonica cultivar-group)] gi|50508941|dbj|BAD31845.1|
immunophilin / FKBP-type peptidyl-prolyl cis-trans
isomerase-like [Oryza sativa (japonica cultivar-group)]
Length = 236
Score = 83.6 bits (205), Expect = 4e-15
Identities = 54/138 (39%), Positives = 70/138 (50%), Gaps = 24/138 (17%)
Query: 86 ITSPDGLKYYDFLEGKGPIAEKGSTVQVHFDCLYRGITAVSSRESKLLAGNRVIAQPYEF 145
+TS +YYD G G A KGS V VH+ ++GIT ++SR+ + G PY F
Sbjct: 118 VTSEALSRYYDITVGSGLKAVKGSRVAVHYVAKWKGITFMTSRQGLGVGGGT----PYGF 173
Query: 146 KVGAPPGKERKREFVDNPNGLFSAQAAPKPPQAMYTIVEGMRVGGKRTVIVPPEKGYGKK 205
+G ER G VEGM+VGG+R +IVPPE YGKK
Sbjct: 174 DIG---NSERGNVLKGLDLG-----------------VEGMKVGGQRLIIVPPELAYGKK 213
Query: 206 GMNEIPPGATFELNIELL 223
G+ EIPP AT E++ LL
Sbjct: 214 GVQEIPPNATIEVDCHLL 231
>ref|ZP_00517772.1| Peptidylprolyl isomerase [Crocosphaera watsonii WH 8501]
gi|67853825|gb|EAM49151.1| Peptidylprolyl isomerase
[Crocosphaera watsonii WH 8501]
Length = 175
Score = 65.1 bits (157), Expect = 1e-09
Identities = 50/144 (34%), Positives = 67/144 (45%), Gaps = 34/144 (23%)
Query: 82 IDDYITSPDGLKYYDFLEGKGPIAEKGSTVQVHF-DCLYRGITAVSSRESKLLAGNRVIA 140
ID+ +T+ GLKY D EG G +KG TV V + L G SSR+ K
Sbjct: 61 IDNAVTTESGLKYIDLKEGDGESPQKGQTVTVDYTGTLENGKKFDSSRDRK--------- 111
Query: 141 QPYEFKVGAPPGKERKREFVDNPNGLFSAQAAPKPPQAMYTIVEGMRVGGKRTVIVPPEK 200
QP+ FK+G G+ K V M+VGG+R +I+PPE
Sbjct: 112 QPFSFKIGV--GQVIK---------------------GWDEGVASMKVGGQRILIIPPEL 148
Query: 201 GYGKKGMNEIPPG-ATFELNIELL 223
GYG +G + PG AT ++ELL
Sbjct: 149 GYGSRGAGGVIPGNATLIFDVELL 172
>ref|ZP_00110945.2| COG0545: FKBP-type peptidyl-prolyl cis-trans isomerases 1 [Nostoc
punctiforme PCC 73102]
Length = 163
Score = 62.8 bits (151), Expect = 7e-09
Identities = 46/140 (32%), Positives = 64/140 (44%), Gaps = 34/140 (24%)
Query: 86 ITSPDGLKYYDFLEGKGPIAEKGSTVQVHF-DCLYRGITAVSSRESKLLAGNRVIAQPYE 144
+T+P GLKY + EG G + G TV+VH+ L G SSR+ QP+
Sbjct: 53 VTTPSGLKYVELKEGTGATPQPGQTVEVHYVGTLEDGTKFDSSRDR---------GQPFS 103
Query: 145 FKVGAPPGKERKREFVDNPNGLFSAQAAPKPPQAMYTIVEGMRVGGKRTVIVPPEKGYGK 204
FK+G Q + + TI +VGG+R +I+P E GYG
Sbjct: 104 FKIGV-------------------GQVIKGWDEGVSTI----KVGGRRKLIIPSELGYGA 140
Query: 205 KGM-NEIPPGATFELNIELL 223
+G IPP AT ++ELL
Sbjct: 141 RGAGGVIPPNATLIFDVELL 160
>ref|NP_953323.1| FKBP-type peptidyl-prolyl cis-trans isomerase [Geobacter
sulfurreducens PCA] gi|39984263|gb|AAR35650.1| FKBP-type
peptidyl-prolyl cis-trans isomerase [Geobacter
sulfurreducens PCA]
Length = 138
Score = 57.0 bits (136), Expect = 4e-07
Identities = 43/145 (29%), Positives = 59/145 (40%), Gaps = 32/145 (22%)
Query: 84 DYITSPDGLKYYDFLEGKGPIAEKGSTVQVHFDCLYRGITAVSSRESKLLAGNRVIAQPY 143
D +T+ GL Y D G G G V+VH+ T S + +P+
Sbjct: 25 DAVTTASGLSYVDLAAGSGAAPVAGKPVKVHYTGWLENGTKFDSSVDR--------GEPF 76
Query: 144 EFKVGAPPGKERKREFVDNPNGLFSAQAAPKPPQAMYTIVEGMRVGGKRTVIVPPEKGYG 203
F +GA + P + V M+VGGKR +IVPP+ GYG
Sbjct: 77 VFTIGA-------------------GEVIPGWDEG----VMSMKVGGKRRLIVPPQLGYG 113
Query: 204 KKGM-NEIPPGATFELNIELLQLAS 227
G IPP AT +ELL +A+
Sbjct: 114 AAGAGGVIPPNATLIFEVELLDVAT 138
>ref|NP_637453.1| FKBP-type peptidyl-prolyl cis-trans isomerase [Xanthomonas
campestris pv. campestris str. ATCC 33913]
gi|21113218|gb|AAM41377.1| FKBP-type peptidyl-prolyl
cis-trans isomerase [Xanthomonas campestris pv.
campestris str. ATCC 33913] gi|66768407|ref|YP_243169.1|
FKBP-type peptidyl-prolyl cis-trans isomerase
[Xanthomonas campestris pv. campestris str. 8004]
gi|66573739|gb|AAY49149.1| FKBP-type peptidyl-prolyl
cis-trans isomerase [Xanthomonas campestris pv.
campestris str. 8004]
Length = 132
Score = 54.3 bits (129), Expect = 3e-06
Identities = 26/42 (61%), Positives = 32/42 (75%), Gaps = 1/42 (2%)
Query: 183 VEGMRVGGKRTVIVPPEKGYGKKGM-NEIPPGATFELNIELL 223
V GMRVGGKRT+++PPE GYG KG IPPGA+ ++ELL
Sbjct: 86 VAGMRVGGKRTLMIPPEFGYGDKGAGGVIPPGASLVFDVELL 127
>emb|CAE08140.1| FKBP-type peptidyl-prolyl cis-trans isomerase (PPIase)
[Synechococcus sp. WH 8102] gi|33866159|ref|NP_897718.1|
FKBP-type peptidyl-prolyl cis-trans isomerase (PPIase)
[Synechococcus sp. WH 8102]
Length = 208
Score = 52.8 bits (125), Expect = 7e-06
Identities = 45/142 (31%), Positives = 60/142 (41%), Gaps = 33/142 (23%)
Query: 83 DDYITSPDGLKYYDFLEGKGPIAEKGSTVQVHFDCLYRGITAVSSRESKLLAGNRVIAQP 142
D IT+ GLK + G+G A G TV VH YRG + K + P
Sbjct: 96 DTQITA-SGLKIIELQVGEGAEAASGQTVSVH----YRG----TLENGKQFDASYDRGTP 146
Query: 143 YEFKVGAPPGKERKREFVDNPNGLFSAQAAPKPPQAMYTIVEGMRVGGKRTVIVPPEKGY 202
+ F +GA + E VD GM+VGGKR +++PP+ Y
Sbjct: 147 FTFPLGAGRVIKGWDEGVD-----------------------GMKVGGKRKLVIPPDLAY 183
Query: 203 GKKGM-NEIPPGATFELNIELL 223
G +G IPP AT +ELL
Sbjct: 184 GSRGAGGVIPPNATLVFEVELL 205
>ref|NP_947888.1| FKBP-type peptidyl-prolyl cis-trans isomerase [Rhodopseudomonas
palustris CGA009] gi|39649465|emb|CAE27987.1| FKBP-type
peptidyl-prolyl cis-trans isomerase [Rhodopseudomonas
palustris CGA009]
Length = 152
Score = 52.8 bits (125), Expect = 7e-06
Identities = 41/141 (29%), Positives = 62/141 (43%), Gaps = 28/141 (19%)
Query: 86 ITSPDGLKYYDFLEGKGPIAEKGSTVQVHFDCLYRGITAVSSRESKLLAGNRVIAQPYEF 145
+T+P GL+ D G G +G +H Y G + +++ + +P+EF
Sbjct: 38 VTTPSGLQIVDTQVGTGASPARGQICVMH----YTGWLYENGAKTRKFDSSVDRNEPFEF 93
Query: 146 KVGAPPGKERKREFVDNPNGLFSAQAAPKPPQAMYTIVEGMRVGGKRTVIVPPEKGYGKK 205
+G R G+ S M+VGGKRT+I+PP+ GYG +
Sbjct: 94 PIGMG------RVIKGWDEGVAS-----------------MKVGGKRTLIIPPDLGYGAR 130
Query: 206 GM-NEIPPGATFELNIELLQL 225
G IPP AT ++ELL L
Sbjct: 131 GAGGVIPPNATLVFDVELLGL 151
>ref|NP_769045.1| Peptidylprolyl isomerase [Bradyrhizobium japonicum USDA 110]
gi|27350660|dbj|BAC47670.1| Peptidylprolyl isomerase
[Bradyrhizobium japonicum USDA 110]
Length = 154
Score = 52.4 bits (124), Expect = 1e-05
Identities = 40/139 (28%), Positives = 61/139 (43%), Gaps = 28/139 (20%)
Query: 86 ITSPDGLKYYDFLEGKGPIAEKGSTVQVHFDCLYRGITAVSSRESKLLAGNRVIAQPYEF 145
+T+ GL+ D G G + G +H Y G + ++ K + +P+EF
Sbjct: 40 MTTASGLQIIDTAVGTGASPQPGQICVMH----YTGWLYENGQKGKKFDSSVDRKEPFEF 95
Query: 146 KVGAPPGKERKREFVDNPNGLFSAQAAPKPPQAMYTIVEGMRVGGKRTVIVPPEKGYGKK 205
+G + R G+ S M+VGGKRT+I+PP+ GYG +
Sbjct: 96 PIG------KGRVIAGWDEGVAS-----------------MKVGGKRTLIIPPQLGYGAR 132
Query: 206 GM-NEIPPGATFELNIELL 223
G IPP AT ++ELL
Sbjct: 133 GAGGVIPPNATLMFDVELL 151
>ref|YP_201719.1| FKBP-type peptidyl-prolyl cis-trans isomerase [Xanthomonas oryzae
pv. oryzae KACC10331] gi|58427297|gb|AAW76334.1|
FKBP-type peptidyl-prolyl cis-trans isomerase
[Xanthomonas oryzae pv. oryzae KACC10331]
Length = 107
Score = 52.4 bits (124), Expect = 1e-05
Identities = 40/127 (31%), Positives = 59/127 (45%), Gaps = 25/127 (19%)
Query: 100 GKGPIAEKGSTVQVHFDCLYRGITAVSSRESKLLAGNRVIAQPYEFKVGAPPGKERKREF 159
G G A G+ V VH+ A + + K + A+P++F +G G + R +
Sbjct: 2 GTGAEATPGAMVTVHYTGWLYDENA-ADKHGKKFDSSLDRAEPFQFVLG---GHQVIRGW 57
Query: 160 VDNPNGLFSAQAAPKPPQAMYTIVEGMRVGGKRTVIVPPEKGYGKKGM-NEIPPGATFEL 218
D V GMRVGGKRT+++PP+ GYG G IPPGA+
Sbjct: 58 DDG--------------------VAGMRVGGKRTLMIPPDYGYGDNGAGGVIPPGASLVF 97
Query: 219 NIELLQL 225
++ELL +
Sbjct: 98 DVELLDV 104
>gb|AAM36960.1| FKBP-type peptidyl-prolyl cis-trans isomerase [Xanthomonas
axonopodis pv. citri str. 306]
gi|21242842|ref|NP_642424.1| FKBP-type peptidyl-prolyl
cis-trans isomerase [Xanthomonas axonopodis pv. citri
str. 306]
Length = 132
Score = 52.0 bits (123), Expect = 1e-05
Identities = 42/126 (33%), Positives = 59/126 (46%), Gaps = 27/126 (21%)
Query: 100 GKGPIAEKGSTVQVHFD-CLYRGITAVSSRESKLLAGNRVIAQPYEFKVGAPPGKERKRE 158
G G A G+ V VH+ LY A + K + A+P++F +G G + R
Sbjct: 27 GTGAEATPGAMVTVHYTGWLYDEKAA--DKHGKKFDSSLDRAEPFQFVLG---GHQVIRG 81
Query: 159 FVDNPNGLFSAQAAPKPPQAMYTIVEGMRVGGKRTVIVPPEKGYGKKGM-NEIPPGATFE 217
+ D V GMRVGGKRT+++PP+ GYG G IPPGA+
Sbjct: 82 WDDG--------------------VAGMRVGGKRTLMIPPDYGYGDNGAGGVIPPGASLV 121
Query: 218 LNIELL 223
++ELL
Sbjct: 122 FDVELL 127
>gb|AAQ89006.1| FKBP14 [Homo sapiens] gi|51105873|gb|EAL24457.1| FK506 binding
protein 14, 22 kDa [Homo sapiens]
gi|7021013|dbj|BAA91351.1| unnamed protein product [Homo
sapiens] gi|55729987|emb|CAH91719.1| hypothetical
protein [Pongo pygmaeus] gi|13528810|gb|AAH05206.1|
FK506 binding protein 14, 22 kDa [Homo sapiens]
gi|8923659|ref|NP_060416.1| FK506 binding protein 14, 22
kDa [Homo sapiens] gi|23821568|sp|Q9NWM8|FKB14_HUMAN
FK506 binding protein 14 precursor (Peptidyl-prolyl
cis-trans isomerase) (PPIase) (Rotamase) (22 kDa
FK506-binding protein) (FKBP-22)
Length = 211
Score = 50.8 bits (120), Expect = 3e-05
Identities = 22/43 (51%), Positives = 32/43 (74%)
Query: 183 VEGMRVGGKRTVIVPPEKGYGKKGMNEIPPGATFELNIELLQL 225
++GM VG KR +I+PP GYGK+G +IPP +T NI+LL++
Sbjct: 92 LKGMCVGEKRKLIIPPALGYGKEGKGKIPPESTLIFNIDLLEI 134
>ref|XP_539497.1| PREDICTED: similar to Secernin 1 [Canis familiaris]
Length = 892
Score = 50.8 bits (120), Expect = 3e-05
Identities = 22/43 (51%), Positives = 32/43 (74%)
Query: 183 VEGMRVGGKRTVIVPPEKGYGKKGMNEIPPGATFELNIELLQL 225
++GM VG KR +I+PP GYGK+G +IPP +T NI+LL++
Sbjct: 218 LKGMCVGEKRKLIIPPALGYGKEGKGKIPPESTLIFNIDLLEI 260
>ref|NP_705801.1| FK506 binding protein 14 [Mus musculus] gi|22137713|gb|AAH29109.1|
FK506 binding protein 14 [Mus musculus]
gi|23396585|sp|P59024|FKB14_MOUSE FK506 binding protein
14 precursor (Peptidyl-prolyl cis-trans isomerase)
(PPIase) (Rotamase) gi|26349311|dbj|BAC38295.1| unnamed
protein product [Mus musculus]
Length = 211
Score = 49.3 bits (116), Expect = 8e-05
Identities = 22/43 (51%), Positives = 31/43 (71%)
Query: 183 VEGMRVGGKRTVIVPPEKGYGKKGMNEIPPGATFELNIELLQL 225
++GM VG KR + VPP GYGK+G +IPP +T NI+LL++
Sbjct: 92 LKGMCVGEKRKLTVPPALGYGKEGKGKIPPESTLIFNIDLLEI 134
>emb|CAB87148.1| putative protein [Arabidopsis thaliana]
gi|15240623|ref|NP_196845.1| immunophilin / FKBP-type
peptidyl-prolyl cis-trans isomerase family protein
[Arabidopsis thaliana] gi|46931302|gb|AAT06455.1|
At5g13410 [Arabidopsis thaliana] gi|11358206|pir||T48588
hypothetical protein T22N19.60 - Arabidopsis thaliana
Length = 256
Score = 49.3 bits (116), Expect = 8e-05
Identities = 43/136 (31%), Positives = 57/136 (41%), Gaps = 20/136 (14%)
Query: 77 KKNIPIDDYITSPDGLKYYDFLEGKGPIAEKGSTVQVHFDCLYRGITAVSSRESKLLAGN 136
K + DY + GL+Y D G GPIA+KG V V +D G
Sbjct: 104 KTKMKYPDYTETQSGLQYKDLRVGTGPIAKKGDKVVVDWDGYTIGYY------------G 151
Query: 137 RVIAQPYEFKVGAPPGKERKREFVDNPNGLFSAQAAPKPPQAMYTIVEGMRVGGKRTVIV 196
R+ + K G+ G + +EF G S + P A V GM +GG R +IV
Sbjct: 152 RIFEARNKTKGGSFEGDD--KEFFKFTLG--SNEVIP----AFEEAVSGMALGGIRRIIV 203
Query: 197 PPEKGYGKKGMNEIPP 212
PPE GY N+ P
Sbjct: 204 PPELGYPDNDYNKSGP 219
>dbj|BAD42981.1| unknown protein [Arabidopsis thaliana]
Length = 255
Score = 49.3 bits (116), Expect = 8e-05
Identities = 43/136 (31%), Positives = 57/136 (41%), Gaps = 20/136 (14%)
Query: 77 KKNIPIDDYITSPDGLKYYDFLEGKGPIAEKGSTVQVHFDCLYRGITAVSSRESKLLAGN 136
K + DY + GL+Y D G GPIA+KG V V +D G
Sbjct: 103 KTKMKYPDYTETQSGLQYKDLRVGTGPIAKKGDKVVVDWDGYTIGYY------------G 150
Query: 137 RVIAQPYEFKVGAPPGKERKREFVDNPNGLFSAQAAPKPPQAMYTIVEGMRVGGKRTVIV 196
R+ + K G+ G + +EF G S + P A V GM +GG R +IV
Sbjct: 151 RIFEARNKTKGGSFEGDD--KEFFKFTLG--SNEVIP----AFEEAVSGMALGGIRRIIV 202
Query: 197 PPEKGYGKKGMNEIPP 212
PPE GY N+ P
Sbjct: 203 PPELGYPDNDYNKSGP 218
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.317 0.136 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 422,607,857
Number of Sequences: 2540612
Number of extensions: 18427663
Number of successful extensions: 46382
Number of sequences better than 10.0: 378
Number of HSP's better than 10.0 without gapping: 248
Number of HSP's successfully gapped in prelim test: 130
Number of HSP's that attempted gapping in prelim test: 45838
Number of HSP's gapped (non-prelim): 596
length of query: 228
length of database: 863,360,394
effective HSP length: 124
effective length of query: 104
effective length of database: 548,324,506
effective search space: 57025748624
effective search space used: 57025748624
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 73 (32.7 bits)
Medicago: description of AC126786.6


