

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126782.9 + phase: 1 /pseudo
(871 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
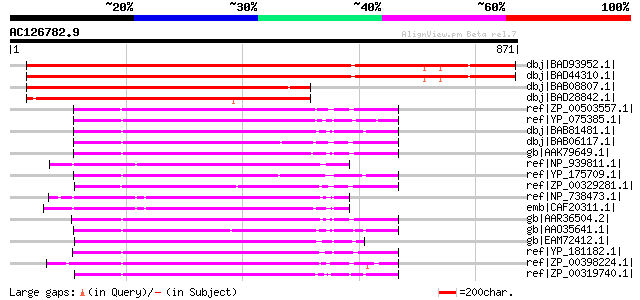
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAD93952.1| putative protein [Arabidopsis thaliana] gi|28393... 1186 0.0
dbj|BAD44310.1| putative protein [Arabidopsis thaliana] 1181 0.0
dbj|BAB08807.1| unnamed protein product [Arabidopsis thaliana] 785 0.0
dbj|BAD28842.1| putative metallo beta subunit lactamase [Oryza s... 762 0.0
ref|ZP_00503557.1| Conserved hypothetical protein MG423 [Clostri... 330 2e-88
ref|YP_075385.1| Zn-dependent hydrolase beta-lactamase superfami... 328 4e-88
dbj|BAB81481.1| conserved hypothetical protein [Clostridium perf... 327 1e-87
dbj|BAB06117.1| BH2398 [Bacillus halodurans C-125] gi|15614961|r... 322 4e-86
gb|AAK79649.1| Predicted metal-dependent hydrolase of metallo-be... 318 5e-85
ref|NP_939811.1| hypothetical protein DIP1463 [Corynebacterium d... 317 1e-84
ref|YP_175709.1| Zn-dependent hydrolase, beta-lactamase superfam... 316 2e-84
ref|ZP_00329281.1| COG0595: Predicted hydrolase of the metallo-b... 314 9e-84
ref|NP_738473.1| putative metallo-beta-lactamase superfamily pro... 314 9e-84
emb|CAF20311.1| HYDROLASE OF METALLO-BETA-LACTAMASE SUPERFAMILY ... 313 1e-83
gb|AAR36504.2| metallo-beta-lactamase family protein [Geobacter ... 313 1e-83
gb|AAO35641.1| putative Zn-dependent hydrolase (beta-lactamase s... 313 2e-83
gb|EAM72412.1| IMP dehydrogenase/GMP reductase:Beta-lactamase-li... 311 6e-83
ref|YP_181182.1| metallo-beta-lactamase family protein [Dehaloco... 311 6e-83
ref|ZP_00398224.1| Beta-lactamase-like:RNA-metabolising metallo-... 310 1e-82
ref|ZP_00319740.1| COG0595: Predicted hydrolase of the metallo-b... 309 3e-82
>dbj|BAD93952.1| putative protein [Arabidopsis thaliana] gi|28393617|gb|AAO42228.1|
unknown protein [Arabidopsis thaliana]
gi|42568733|ref|NP_201147.2| metallo-beta-lactamase
family protein [Arabidopsis thaliana]
Length = 911
Score = 1186 bits (3069), Expect = 0.0
Identities = 608/859 (70%), Positives = 702/859 (80%), Gaps = 27/859 (3%)
Query: 30 TKMATLTSLPPLPHSLLSLRSKPTRLSVSASALSASVGNDGSTSRVPQKRRRRIEGPRKS 89
+KMA ++L P++ +S + +VS S SA S+S+ P++R R+EG KS
Sbjct: 30 SKMAAFSALSLCPYTFTFRQSSRIKSTVSCSVTSAPASGTSSSSKTPRRRSGRLEGVGKS 89
Query: 90 MEDSVQRRMEQFYEGNDGPPLRVLPIGGLGEIGMNCMLVGNHDRYILIDAGIMFPDYDDL 149
MEDSV+R+MEQFYEG DGPPLR+LPIGGLGEIGMNCMLVGN+DRYILIDAGIMFPDYD+
Sbjct: 90 MEDSVKRKMEQFYEGTDGPPLRILPIGGLGEIGMNCMLVGNYDRYILIDAGIMFPDYDEP 149
Query: 150 GVQKIIPDTTFIRKWSHKIEALVITHGHEDHIGALPWVIPALDSNTPIFASSFTMELIKK 209
G+QKI+PDT FIR+W HKIEA+VITHGHEDHIGALPWVIPALD NTPIFASSFTMELIKK
Sbjct: 150 GIQKIMPDTGFIRRWKHKIEAVVITHGHEDHIGALPWVIPALDPNTPIFASSFTMELIKK 209
Query: 210 RLKEHGIFLPSRLKIFRTKNKFVAGPFEIEPIRVTHSIPDCCGLVLRCSDGTILHTGDWK 269
RLKEHGIF+ SRLK F T+ +F+AGPFEIEPI VTHSIPDC GL LRC+DG ILHTGDWK
Sbjct: 210 RLKEHGIFVQSRLKTFSTRRRFMAGPFEIEPITVTHSIPDCSGLFLRCADGNILHTGDWK 269
Query: 270 IDETPLDGKVFDREGLEELSKEGVTLMMSDSTNVLSPGRTTSESVVADSLLRHISASKGR 329
IDE PLDGKVFDRE LEELSKEGVTLMMSDSTNVLSPGRT SE VVAD+L+R++ A+KGR
Sbjct: 270 IDEAPLDGKVFDREALEELSKEGVTLMMSDSTNVLSPGRTISEKVVADALVRNVMAAKGR 329
Query: 330 VITTQFASNLHRIGSVKAAADLTGRKLVFVGMSLRTYLEAAWKDGKAPFDPSTLVKAEDI 389
VITTQFASN+HR+GS+KAAAD+TGRKLVFVGMSLRTYLEAAW+DGKAP DPS+L+K EDI
Sbjct: 330 VITTQFASNIHRLGSIKAAADITGRKLVFVGMSLRTYLEAAWRDGKAPIDPSSLIKVEDI 389
Query: 390 DAYAPKDLLIVTTGSQAEPRAALNLASFGSSHAFKLTKEDIVLYSAKVIPGNESRVMEMM 449
+AYAPKDLLIVTTGSQAEPRAALNLAS+GSSHAFKLTKEDI+LYSAKVIPGNESRVM+MM
Sbjct: 390 EAYAPKDLLIVTTGSQAEPRAALNLASYGSSHAFKLTKEDIILYSAKVIPGNESRVMKMM 449
Query: 450 NRISEIGSTIVMGRNENLHTSGHAYRGELEEVLRIVKPQHFLPVHGEYLFLKEHESLGKS 509
NRI++IG I+MG+NE LHTSGHAYRGELEEVL+IVKPQHFLP+HGE LFLKEHE LGKS
Sbjct: 450 NRIADIGPNIIMGKNEMLHTSGHAYRGELEEVLKIVKPQHFLPIHGELLFLKEHELLGKS 509
Query: 510 TGIRHTAVIKNGEMLGVSHLRNRRVLSNGFISLGKENLQLKYSDGDKAFGTSGELFLDER 569
TGIRHT VIKNGEMLGVSHLRNRRVLSNGF SLG+ENLQL YSDGDKAFGTS EL +DER
Sbjct: 510 TGIRHTTVIKNGEMLGVSHLRNRRVLSNGFSSLGRENLQLMYSDGDKAFGTSSELCIDER 569
Query: 570 MRIALDGIIVVSMEIFRPKNLESLAGNTLKGKIRITTRCLWLDKGKLLDALYKAAHAALS 629
+RI+ DGIIV+SMEI RP ++ NTLKGKIRITTRC+WLDKG+LLDAL+KAAHAALS
Sbjct: 570 LRISSDGIIVLSMEIMRP----GVSENTLKGKIRITTRCMWLDKGRLLDALHKAAHAALS 625
Query: 630 SCPVKSPLPHMERTVSEVLRKMVRKYSGKRPEVIAIAIENPGAVFADEINTKLSGKSQVG 689
SCPV PL HMERTVSEVLRK+VRKYSGKRPEVIAIA ENP AV ADE++ +LSG VG
Sbjct: 626 SCPVTCPLSHMERTVSEVLRKIVRKYSGKRPEVIAIATENPMAVRADEVSARLSGDPSVG 685
Query: 690 PGISTFRRSVDEHRKENQSTAL------------QIRDDGIDIEGLLVEIETITTAAEGD 737
G++ R+ V+ + K +++ + DD ID LL E ET + +
Sbjct: 686 SGVAALRKVVEGNDKRSRAKKAPSQEASPKEVDRTLEDDIIDSARLLAEEETAASTYTEE 745
Query: 738 L-----SDSGESDEFWKPFI--ASSVEKSIKANNGYVSRKEHKSNTKQDDSEDIDEAKSE 790
+ S S ESD+FWK FI +SS S N V+ E K+ K++ +D + A
Sbjct: 746 VDTPVGSSSEESDDFWKSFINPSSSPSPSETENMNKVADTEPKAEGKENSRDDDELA--- 802
Query: 791 EMSDSEPESS-KSEKKNKWKTEEVKKLIDLRSDLRDRFKVVKGRMALWEEISQSLLADGI 849
+ SDSE +SS K +KNKWK EE+KK+I +R +L RF+VVKGRMALWEEIS +L A+GI
Sbjct: 803 DASDSETKSSPKRVRKNKWKPEEIKKVIRMRGELHSRFQVVKGRMALWEEISSNLSAEGI 862
Query: 850 SRSPGQCKSLWTSLALKYE 868
+RSPGQCKSLW SL KYE
Sbjct: 863 NRSPGQCKSLWASLIQKYE 881
>dbj|BAD44310.1| putative protein [Arabidopsis thaliana]
Length = 911
Score = 1181 bits (3056), Expect = 0.0
Identities = 606/859 (70%), Positives = 701/859 (81%), Gaps = 27/859 (3%)
Query: 30 TKMATLTSLPPLPHSLLSLRSKPTRLSVSASALSASVGNDGSTSRVPQKRRRRIEGPRKS 89
+KMA ++L P++ +S + +VS S SA S+S+ P++R R+EG KS
Sbjct: 30 SKMAAFSALSLCPYTFTFRQSSRIKSTVSCSVTSAPASGTSSSSKTPRRRSGRLEGVGKS 89
Query: 90 MEDSVQRRMEQFYEGNDGPPLRVLPIGGLGEIGMNCMLVGNHDRYILIDAGIMFPDYDDL 149
MEDSV+R+MEQFYEG DGPPLR+LPIGGLGEIGMNCMLVGN+DRYILIDAGIMFPDYD+
Sbjct: 90 MEDSVKRKMEQFYEGTDGPPLRILPIGGLGEIGMNCMLVGNYDRYILIDAGIMFPDYDEP 149
Query: 150 GVQKIIPDTTFIRKWSHKIEALVITHGHEDHIGALPWVIPALDSNTPIFASSFTMELIKK 209
G+QKI+PDT FIR+W HKIEA+VITHGHEDHIGALPWVIPALD NTPIFASSFTMELIKK
Sbjct: 150 GIQKIMPDTGFIRRWKHKIEAVVITHGHEDHIGALPWVIPALDPNTPIFASSFTMELIKK 209
Query: 210 RLKEHGIFLPSRLKIFRTKNKFVAGPFEIEPIRVTHSIPDCCGLVLRCSDGTILHTGDWK 269
RLKEHGIF+ SRLK F T+ +F+AGPFEIEPI VTHSIPDC GL LRC+DG ILHTGDWK
Sbjct: 210 RLKEHGIFVQSRLKTFSTRRRFMAGPFEIEPITVTHSIPDCSGLFLRCADGNILHTGDWK 269
Query: 270 IDETPLDGKVFDREGLEELSKEGVTLMMSDSTNVLSPGRTTSESVVADSLLRHISASKGR 329
IDE PLDGKVFDRE LEELSKEGVTLMMSDSTNVLSPGRT SE VVAD+L+R++ A+K R
Sbjct: 270 IDEAPLDGKVFDREALEELSKEGVTLMMSDSTNVLSPGRTISEKVVADALVRNVMAAKER 329
Query: 330 VITTQFASNLHRIGSVKAAADLTGRKLVFVGMSLRTYLEAAWKDGKAPFDPSTLVKAEDI 389
VITTQFASN+HR+GS+KAAAD+TGRKLVFVGMSLRTYLEAAW+DGKAP DPS+L+K EDI
Sbjct: 330 VITTQFASNIHRLGSIKAAADITGRKLVFVGMSLRTYLEAAWRDGKAPIDPSSLIKVEDI 389
Query: 390 DAYAPKDLLIVTTGSQAEPRAALNLASFGSSHAFKLTKEDIVLYSAKVIPGNESRVMEMM 449
+AYAPKDLLIVTTGSQAEPRAALNLAS+GSSHAFKLTKEDI+LYSAKVIPGNESRVM+MM
Sbjct: 390 EAYAPKDLLIVTTGSQAEPRAALNLASYGSSHAFKLTKEDIILYSAKVIPGNESRVMKMM 449
Query: 450 NRISEIGSTIVMGRNENLHTSGHAYRGELEEVLRIVKPQHFLPVHGEYLFLKEHESLGKS 509
NRI++IG I+MG+NE LHTSGHAYRGELEEVL+IVKPQHFLP+HGE LFLKEHE LGKS
Sbjct: 450 NRIADIGPNIIMGKNEMLHTSGHAYRGELEEVLKIVKPQHFLPIHGELLFLKEHELLGKS 509
Query: 510 TGIRHTAVIKNGEMLGVSHLRNRRVLSNGFISLGKENLQLKYSDGDKAFGTSGELFLDER 569
TGIRHT VIKNGEMLGVSHLRNRRVLSNGF SLG+ENLQL YSDGDKAFGTS EL +DER
Sbjct: 510 TGIRHTTVIKNGEMLGVSHLRNRRVLSNGFSSLGRENLQLMYSDGDKAFGTSSELCIDER 569
Query: 570 MRIALDGIIVVSMEIFRPKNLESLAGNTLKGKIRITTRCLWLDKGKLLDALYKAAHAALS 629
+RI+ DGIIV+SMEI RP ++ NTLKGKIRITTRC+WLDKG+LLDAL+KAAHAALS
Sbjct: 570 LRISSDGIIVLSMEIMRP----GVSENTLKGKIRITTRCMWLDKGRLLDALHKAAHAALS 625
Query: 630 SCPVKSPLPHMERTVSEVLRKMVRKYSGKRPEVIAIAIENPGAVFADEINTKLSGKSQVG 689
SCPV PL HMERTVSEVLRK+VRKYSGKRPEVIAIA E+P AV ADE++ +LSG VG
Sbjct: 626 SCPVTCPLSHMERTVSEVLRKIVRKYSGKRPEVIAIATEDPMAVRADEVSARLSGDPSVG 685
Query: 690 PGISTFRRSVDEHRKENQSTAL------------QIRDDGIDIEGLLVEIETITTAAEGD 737
G++ R+ V+ + K +++ + DD ID LL E ET + +
Sbjct: 686 SGVAALRKVVEGNDKRSRAKKAPSQEASPKEVDRTLEDDIIDSARLLAEEETAASTYTEE 745
Query: 738 L-----SDSGESDEFWKPFI--ASSVEKSIKANNGYVSRKEHKSNTKQDDSEDIDEAKSE 790
+ S S ESD+FWK FI +SS S N V+ E K+ K++ +D + A
Sbjct: 746 VDTPVGSSSEESDDFWKSFINPSSSPSPSETENMNKVADTEPKAEGKENSRDDDELA--- 802
Query: 791 EMSDSEPESS-KSEKKNKWKTEEVKKLIDLRSDLRDRFKVVKGRMALWEEISQSLLADGI 849
+ SDSE +SS K +KNKWK EE+KK+I +R +L RF+VVKGRMALWEEIS +L A+GI
Sbjct: 803 DTSDSETKSSPKRVRKNKWKPEEIKKVIRMRGELHSRFQVVKGRMALWEEISSNLSAEGI 862
Query: 850 SRSPGQCKSLWTSLALKYE 868
+RSPGQCKSLW SL KYE
Sbjct: 863 NRSPGQCKSLWASLIQKYE 881
>dbj|BAB08807.1| unnamed protein product [Arabidopsis thaliana]
Length = 528
Score = 785 bits (2028), Expect = 0.0
Identities = 384/488 (78%), Positives = 433/488 (88%), Gaps = 2/488 (0%)
Query: 30 TKMATLTSLPPLPHSLLSLRSKPTRLSVSASALSASVGNDGSTSRVPQKRRRRIEGPRKS 89
+KMA ++L P++ +S + +VS S SA S+S+ P++R R+EG KS
Sbjct: 30 SKMAAFSALSLCPYTFTFRQSSRIKSTVSCSVTSAPASGTSSSSKTPRRRSGRLEGVGKS 89
Query: 90 MEDSVQRRMEQFYEGNDGPPLRVLPIGGLGEIGMNCMLVGNHDRYILIDAGIMFPDYDDL 149
MEDSV+R+MEQFYEG DGPPLR+LPIGGLGEIGMNCMLVGN+DRYILIDAGIMFPDYD+
Sbjct: 90 MEDSVKRKMEQFYEGTDGPPLRILPIGGLGEIGMNCMLVGNYDRYILIDAGIMFPDYDEP 149
Query: 150 GVQKIIPDTTFIRKWSHKIEALVITHGHEDHIGALPWVIPALDSNTPIFASSFTMELIKK 209
G+QKI+PDT FIR+W HKIEA+VITHGHEDHIGALPWVIPALD NTPIFASSFTMELIKK
Sbjct: 150 GIQKIMPDTGFIRRWKHKIEAVVITHGHEDHIGALPWVIPALDPNTPIFASSFTMELIKK 209
Query: 210 RLKEHGIFLPSRLKIFRTKNKFVAGPFEIEPIRVTHSIPDCCGLVLRCSDGTILHTGDWK 269
RLKEHGIF+ SRLK F T+ +F+AGPFEIEPI VTHSIPDC GL LRC+DG ILHTGDWK
Sbjct: 210 RLKEHGIFVQSRLKTFSTRRRFMAGPFEIEPITVTHSIPDCSGLFLRCADGNILHTGDWK 269
Query: 270 IDETPLDGKVFDREGLEELSKEGVTLMMSDSTNVLSPGRTTSESVVADSLLRHISASKGR 329
IDE PLDGKVFDRE LEELSKEGVTLMMSDSTNVLSPGRT SE VVAD+L+R++ A+KGR
Sbjct: 270 IDEAPLDGKVFDREALEELSKEGVTLMMSDSTNVLSPGRTISEKVVADALVRNVMAAKGR 329
Query: 330 VITTQFASNLHRIGSVKAAADLTGRKLVFVGMSLRTYLEAAWKDGKAPFDPSTLVKAEDI 389
VITTQFASN+HR+GS+KAAAD+TGRKLVFVGMSLRTYLEAAW+DGKAP DPS+L+K EDI
Sbjct: 330 VITTQFASNIHRLGSIKAAADITGRKLVFVGMSLRTYLEAAWRDGKAPIDPSSLIKVEDI 389
Query: 390 DAYAPKDLLIVTTGSQAEPRAALNLASFGSSHAFKLTKEDIVLYSAKVIPGNESRVMEMM 449
+AYAPKDLLIVTTGSQAEPRAALNLAS+GSSHAFKLTKEDI+LYSAKVIPGNESRVM+MM
Sbjct: 390 EAYAPKDLLIVTTGSQAEPRAALNLASYGSSHAFKLTKEDIILYSAKVIPGNESRVMKMM 449
Query: 450 NRISEIGSTIVMGRNENLHTSGHAYRGELEEVLRIVKPQHFLPVHGEYLFLKEHESLGKS 509
NRI++IG I+MG+NE LHTSGHAYRGEL VL+IVKPQHFLP+HGE LFLKEHE LGKS
Sbjct: 450 NRIADIGPNIIMGKNEMLHTSGHAYRGEL--VLKIVKPQHFLPIHGELLFLKEHELLGKS 507
Query: 510 TGIRHTAV 517
TGIRHT V
Sbjct: 508 TGIRHTTV 515
>dbj|BAD28842.1| putative metallo beta subunit lactamase [Oryza sativa (japonica
cultivar-group)]
Length = 506
Score = 762 bits (1968), Expect = 0.0
Identities = 379/501 (75%), Positives = 432/501 (85%), Gaps = 14/501 (2%)
Query: 29 LTKMATLTSLPPLPHSLLSLRSKPTRLSVSASALSASVGNDGST-SRVPQKRRRRIEGPR 87
+ +A+L+SL P L RS + S+S A GS SR P++R R+ EG
Sbjct: 1 MVALASLSSLSPC--GLARRRSASSAASISCCAAPPPPSAKGSQESRTPRRRVRKTEGAT 58
Query: 88 KSMEDSVQRRMEQFYEGNDGPPLRVLPIGGLGEIGMNCMLVGNHDRYILIDAGIMFPDYD 147
KS+EDSV+R+MEQFYEG DGPPLRVLPIGGLGEIGMNCMLVGN+DRYILIDAG+MFPD+D
Sbjct: 59 KSLEDSVKRKMEQFYEGLDGPPLRVLPIGGLGEIGMNCMLVGNYDRYILIDAGVMFPDFD 118
Query: 148 DLGVQKIIPDTTFIRKWSHKIEALVITHGHEDHIGALPWVIPALDSNTPIFASSFTMELI 207
+ GVQKIIPDTTFI+KWSHKIEA++ITHGHEDHIGALPWVIPALDS+TPIFASSFTMELI
Sbjct: 119 EFGVQKIIPDTTFIKKWSHKIEAVIITHGHEDHIGALPWVIPALDSSTPIFASSFTMELI 178
Query: 208 KKRLKEHGIFLPSRLKIFRTKNKFVAGPFEIEPIRVTHSIPDCCGLVLRCSDGTILHTGD 267
K+RLKE GIFL SRLK+FR + +F AGPFE+EP+RVTHSIPDCCGLVLRC+DG I HTGD
Sbjct: 179 KRRLKEFGIFLSSRLKVFRVRKRFQAGPFEVEPLRVTHSIPDCCGLVLRCADGIIFHTGD 238
Query: 268 WKIDETPLDGKVFDREGLEELSKEGVTLMMSDSTNVLSPGRTTSESVVADSLLRHISASK 327
WKIDE+P+DGK+FDR+ LEELSKEGVTLMMSDSTNVLSPGR+ SESVVA SLLRHIS +K
Sbjct: 239 WKIDESPVDGKIFDRQALEELSKEGVTLMMSDSTNVLSPGRSISESVVAGSLLRHISEAK 298
Query: 328 GRVITTQFASNLHRIGSVKAAADLTGRKLVFVGMSLRTYLEAAWKDGKAPFDPSTL---- 383
GRVITTQFASN+HRIGS+KAAADLTGRKLVFVGMSLRTYL+AA++DGK+P DPSTL
Sbjct: 299 GRVITTQFASNIHRIGSIKAAADLTGRKLVFVGMSLRTYLDAAFRDGKSPIDPSTLVHLI 358
Query: 384 -------VKAEDIDAYAPKDLLIVTTGSQAEPRAALNLASFGSSHAFKLTKEDIVLYSAK 436
VK ED+DAYAP DLL+VTTGSQAEPRAALNLASFG SHA KL+KED++LYSAK
Sbjct: 359 ESRFLLKVKVEDMDAYAPNDLLVVTTGSQAEPRAALNLASFGGSHALKLSKEDVLLYSAK 418
Query: 437 VIPGNESRVMEMMNRISEIGSTIVMGRNENLHTSGHAYRGELEEVLRIVKPQHFLPVHGE 496
VIPGNESRVM+M+NR++E+G IVMG++ LHTSGHAY ELEEVL+IVKPQHFLPVHGE
Sbjct: 419 VIPGNESRVMKMLNRLTELGPKIVMGKDAGLHTSGHAYHDELEEVLQIVKPQHFLPVHGE 478
Query: 497 YLFLKEHESLGKSTGIRHTAV 517
LFLKEHE LG+STGIRHT V
Sbjct: 479 LLFLKEHELLGRSTGIRHTTV 499
>ref|ZP_00503557.1| Conserved hypothetical protein MG423 [Clostridium thermocellum ATCC
27405] gi|67851674|gb|EAM47237.1| Conserved hypothetical
protein MG423 [Clostridium thermocellum ATCC 27405]
Length = 555
Score = 330 bits (845), Expect = 2e-88
Identities = 193/564 (34%), Positives = 314/564 (55%), Gaps = 22/564 (3%)
Query: 110 LRVLPIGGLGEIGMNCMLVGNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKWSHKIE 169
L+V+P+GGLGEIG N + D ++D GI FP+ D LG+ +IPD +++ K K+
Sbjct: 8 LKVIPLGGLGEIGKNITVFEYGDDIFVVDCGIAFPEDDMLGIDLVIPDISYLTKNREKVR 67
Query: 170 ALVITHGHEDHIGALPWVIPALDSNTPIFASSFTMELIKKRLKEHGIFLPSRLKIFRTKN 229
+V+THGHEDHIGALP+V+ D N P++ + T+ L++++L+EHG+ L + + +
Sbjct: 68 GIVLTHGHEDHIGALPYVLK--DLNVPVYGTKLTLGLLEQKLEEHGLLNNVVLNVVKHSD 125
Query: 230 KFVAGPFEIEPIRVTHSIPDCCGLVLRCSDGTILHTGDWKIDETPLDGKVFDREGLEELS 289
G F++E IR THSI D L + GTI HTGD+KID TP++G+ D L EL
Sbjct: 126 VIELGCFKVEFIRSTHSIADSTALAIFTPVGTIFHTGDFKIDYTPIEGEPIDLARLAELG 185
Query: 290 KEGVTLMMSDSTNVLSPGRTTSESVVADSLLRHISASKGRVITTQFASNLHRIGSVKAAA 349
K+GV L+M DSTNV G T SE V ++ +K R++ FASN+HRI + AA
Sbjct: 186 KKGVLLLMCDSTNVEREGYTMSEKTVGETFDEIFMNAKNRILVATFASNVHRIQQIVNAA 245
Query: 350 DLTGRKLVFVGMSLRTYLEAAWKDGKAPFDPSTLVKAEDIDAYAPKDLLIVTTGSQAEPR 409
GRK+ G S+ + A + G ++ + I+ Y P+ ++I+TTGSQ EP
Sbjct: 246 IKFGRKIAICGRSMVNVVNVAMELGYMNVPEGLIIDIDHINKYPPEKIVIITTGSQGEPM 305
Query: 410 AALNLASFGSSHAFKLTKEDIVLYSAKVIPGNESRVMEMMNRISEIGSTIVMGRNENLHT 469
+AL + G ++ D+V+ SA IPGNE V ++N + + G+ ++ ++H
Sbjct: 306 SALTRMASGDHKKVEIIPGDLVIISANPIPGNEKLVSRVVNDLFKKGAEVIYESLADIHV 365
Query: 470 SGHAYRGELEEVLRIVKPQHFLPVHGEYLFLKEHESLGKSTGI--RHTAVIKNGEMLGVS 527
SGHA + EL+ + R+++P++F+PVHGEY LK H +L G+ + ++ G++L
Sbjct: 366 SGHASQEELKLIHRLIRPKYFMPVHGEYRHLKRHANLAVELGMSPENIFIMDIGKVL--- 422
Query: 528 HLRNRRVLSNGFISLGKENLQLKYSDGDKAFGTSGELFLDERMRIALDGIIVVSMEIFRP 587
L N NG ++ G+ + DG G G + L +R ++ DG+IVV + I
Sbjct: 423 ELTNDSAKINGSVNAGRVLV-----DG-LGVGDVGNIVLRDRKHLSQDGLIVVVITI--- 473
Query: 588 KNLESLAGNTLKGKIRITTRCLWLDKGK-LLDALYKAAHAALSSC--PVKSPLPHMERTV 644
E GN + G I+ +++ + + L++ + + AAL C K+ + +
Sbjct: 474 ---EGDTGNVIAGPDVISRGFVYVRESEDLMEEIREVCKAALQKCNDKKKNDWSTKKSII 530
Query: 645 SEVLRKMVRKYSGKRPEVIAIAIE 668
+ LR + + + +RP ++ I +E
Sbjct: 531 RDALRDFLYERTKRRPMILPIIME 554
>ref|YP_075385.1| Zn-dependent hydrolase beta-lactamase superfamily [Symbiobacterium
thermophilum IAM 14863] gi|51856383|dbj|BAD40541.1|
Zn-dependent hydrolase beta-lactamase superfamily
[Symbiobacterium thermophilum IAM 14863]
Length = 558
Score = 328 bits (842), Expect = 4e-88
Identities = 196/565 (34%), Positives = 315/565 (55%), Gaps = 24/565 (4%)
Query: 110 LRVLPIGGLGEIGMNCMLVGNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKWSHKIE 169
L V+P+GGLGEIG NCM+V D I+IDAG+ FP+ + LG+ +IPD T++ + K+
Sbjct: 11 LYVIPLGGLGEIGKNCMVVQYGDDIIVIDAGLAFPEEEMLGIDIVIPDFTYLLENREKVR 70
Query: 170 ALVITHGHEDHIGALPWVIPALDSNTPIFASSFTMELIKKRLKEHGIFLPSRLKIFRTKN 229
++ITHGHEDHIGA+ +++ + N P++A+ T+ LI+ +L E G+ LP+ K R +
Sbjct: 71 GILITHGHEDHIGAVAYLLRNI--NIPVYATRLTLGLIEGKLAEMGMKLPAESKAVRAGD 128
Query: 230 KFVAGPFEIEPIRVTHSIPDCCGLVLRCSDGTILHTGDWKIDETPLDGKVFDREGLEELS 289
+ GPF E IRV HSIPD C + + GTILHTGD+K D+TP+DG+ D L EL
Sbjct: 129 QVKLGPFTCEFIRVNHSIPDACAIAVTTPVGTILHTGDFKFDQTPIDGEFADYRRLTELG 188
Query: 290 KEGVTLMMSDSTNVLSPGRTTSESVVADSLLRHISASKGRVITTQFASNLHRIGSVKAAA 349
K G+ ++SDSTN PG T SE V ++ R +KGR++ FASN+HRI A
Sbjct: 189 KRGILALLSDSTNAERPGYTPSERSVRPNMERVFREAKGRILMATFASNVHRIQQAIELA 248
Query: 350 DLTGRKLVFVGMSLRTYLEAAWKDGKAPFDPSTLVKAEDIDAYAPKDLLIVTTGSQAEPR 409
G+K+ VG S+ + A + G T+V +++ +I+TTGSQ EP
Sbjct: 249 AEMGKKVAVVGRSMENTVRIALELGYLNVPEGTMVTVDELRRVPLNQQVILTTGSQGEPM 308
Query: 410 AALNLASFGSSHAFKLTKEDIVLYSAKVIPGNESRVMEMMNRISEIGSTIVMGRNENLHT 469
AAL+ + A +L D VL++A IPGNE V +N + G+ ++ R+ +H
Sbjct: 309 AALSRMAVNDHKAIELYPGDTVLFAATAIPGNEKSVARTVNNLYRRGAEVIYDRSAGVHV 368
Query: 470 SGHAYRGELEEVLRIVKPQHFLPVHGEYLFLKEHESLGKSTGIRHTAVI--KNGEMLGVS 527
SGHA + E + ++ + +P+ F+P+HGEY L +H+ L ++ GI ++ +NG +
Sbjct: 369 SGHASQEEQKLMINLTRPKFFVPIHGEYRMLLKHKQLAEACGIPSENILIGENGTIF--E 426
Query: 528 HLRNRRVLSNGFISLGKENLQLKYSDGDKAFGTSGELFLDERMRIALDGIIVVSMEIFRP 587
RN ++ GK DG G G + L +R ++A +GI++V + + R
Sbjct: 427 FTRNSAQIT------GKVTSGQVLVDG-LGVGDVGNIVLRDRKQLAQEGILIVVVAVDRE 479
Query: 588 KNLESLAGNTLKGKIRITTRCLWLDKGK-LLDALYKAAHAALSSCPVKSPLPH---MERT 643
+ G + G ++ +++ + + LLD + AL+ ++ LP ++ +
Sbjct: 480 E------GTIVGGPDMVSRGFVYVRESEGLLDEAKERVKRALAD-QMQRGLPEWAVLKSS 532
Query: 644 VSEVLRKMVRKYSGKRPEVIAIAIE 668
+ +VL K + + +RP ++ I IE
Sbjct: 533 IRDVLSKYLYDKTHRRPMILPIIIE 557
>dbj|BAB81481.1| conserved hypothetical protein [Clostridium perfringens str. 13]
gi|18310757|ref|NP_562691.1| hypothetical protein
CPE1775 [Clostridium perfringens str. 13]
Length = 581
Score = 327 bits (838), Expect = 1e-87
Identities = 183/563 (32%), Positives = 313/563 (55%), Gaps = 20/563 (3%)
Query: 110 LRVLPIGGLGEIGMNCMLVGNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKWSHKIE 169
++++P+GGL EIG N ++ D I++D G+ FPD D G+ +IPD +++ K + K+
Sbjct: 34 VKIIPLGGLNEIGKNLTVIEFKDDIIVVDCGLKFPDEDMFGIDIVIPDVSYLVKNAEKVR 93
Query: 170 ALVITHGHEDHIGALPWVIPALDSNTPIFASSFTMELIKKRLKEHGIFLPSRLKIFRTKN 229
+ +THGHEDHIGALP+V+ L N P++ + T+ +++ +LKEHG+ + L + ++
Sbjct: 94 GIFLTHGHEDHIGALPYVLKNL--NVPVYGTKLTLGIVETKLKEHGLLSTTELIRVKPRD 151
Query: 230 KFVAGPFEIEPIRVTHSIPDCCGLVLRCSDGTILHTGDWKIDETPLDGKVFDREGLEELS 289
+E ++ HSI D + + G +LHTGD+K+D TP DG+V D EL
Sbjct: 152 IIKLKSSSVEFVKTNHSIADSVAIAVHTPLGVVLHTGDFKVDYTPTDGEVMDFARFAELG 211
Query: 290 KEGVTLMMSDSTNVLSPGRTTSESVVADSLLRHISASKGRVITTQFASNLHRIGSVKAAA 349
++GV MM+DSTNV PG T SE V ++L + +KGR+I FASN+HRI + AA
Sbjct: 212 RKGVLAMMADSTNVERPGYTMSERAVGENLKKIFVGAKGRIIIATFASNIHRIQQIVEAA 271
Query: 350 DLTGRKLVFVGMSLRTYLEAAWKDGKAPFDPSTLVKAEDIDAYAPKDLLIVTTGSQAEPR 409
++TGRK+ G S+ ++ A + G D + V + I+ Y + + I+TTGSQ EP
Sbjct: 272 EMTGRKIAVSGRSMENIVQVAIELGYLTIDKESFVSIDSINKYPNEQVTIITTGSQGEPM 331
Query: 410 AALNLASFGSSHAFKLTKEDIVLYSAKVIPGNESRVMEMMNRISEIGSTIVMGRNENLHT 469
+AL + + + D V+ SA IPGNE V +++N++ + G+ +V G+ ++H
Sbjct: 332 SALARMASSEHKKVNIIEGDTVILSATPIPGNEKLVSKVINQLFKKGAEVVYGKLADVHV 391
Query: 470 SGHAYRGELEEVLRIVKPQHFLPVHGEYLFLKEHESLGKSTGI--RHTAVIKNGEMLGVS 527
SGHA + EL+ + +VKP+ F+PVHGEY LK+H L G+ ++ + +NG+++ ++
Sbjct: 392 SGHACQEELKLMQALVKPKFFIPVHGEYRHLKQHAELAVDVGLSEKNFMIAENGDVIEIT 451
Query: 528 HLRNRRVLSNGFISLGKENLQLKYSDGDKAFGTSGELFLDERMRIALDGIIVVSMEIFRP 587
+ NG ++ G+ + DG G G + L +R ++ DGI+ V + I +
Sbjct: 452 ---RDSIKKNGSVTSGQ-----IFVDG-LGVGDVGNIVLRDRKHLSQDGILTVVVTISK- 501
Query: 588 KNLESLAGNTLKGKIRITTRCLWLDKGKLLDALYKAAHAALSSCPVKS--PLPHMERTVS 645
+ +AG + + + R + L+D + L C K M+ +
Sbjct: 502 ETASVVAGPDIISRGFVYVR----ESEDLMDEAKEIVKDVLRDCEKKGICDWATMKSNIR 557
Query: 646 EVLRKMVRKYSGKRPEVIAIAIE 668
+ LR + + + ++P ++ I +E
Sbjct: 558 DGLRSFLYEKTKRKPMILPIIME 580
>dbj|BAB06117.1| BH2398 [Bacillus halodurans C-125] gi|15614961|ref|NP_243264.1|
hypothetical protein BH2398 [Bacillus halodurans C-125]
gi|25304210|pir||F83949 hypothetical protein BH2398
[imported] - Bacillus halodurans (strain C-125)
Length = 555
Score = 322 bits (824), Expect = 4e-86
Identities = 188/564 (33%), Positives = 310/564 (54%), Gaps = 23/564 (4%)
Query: 110 LRVLPIGGLGEIGMNCMLVGNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKWSHKIE 169
+RV +GG+GEIG N +V D +IDAG+MFPD + LGV +IPD +++ + ++
Sbjct: 9 IRVFALGGVGEIGKNMYVVEVDDDLFVIDAGLMFPDDEMLGVDVVIPDISYLVENEERVR 68
Query: 170 ALVITHGHEDHIGALPWVIPALDSNTPIFASSFTMELIKKRLKEHGIFLPSRLKIFRTKN 229
A+++THGHEDHIG LP+V+ L N P++ + T+ L++++LKE G+ ++LK+ + +
Sbjct: 69 AILLTHGHEDHIGGLPYVLQKL--NVPVYGTKLTLGLVEEKLKEAGLIRSAKLKLIDSNS 126
Query: 230 KFVAGPFEIEPIRVTHSIPDCCGLVLRCSDGTILHTGDWKIDETPLDGKVFDREGLEELS 289
+ G + R HSIPD G+ ++ S G I+HTGD+K D+TP+DGK + + +
Sbjct: 127 RLKLGSTPVSFFRTNHSIPDSVGICIQTSQGFIVHTGDFKFDQTPVDGKQAEIGKMAAIG 186
Query: 290 KEGVTLMMSDSTNVLSPGRTTSESVVADSLLRHISASKGRVITTQFASNLHRIGSVKAAA 349
+GV ++SDSTN PG T SE+ V + +KGR+I T FASN+HR+ V AA
Sbjct: 187 HKGVLCLLSDSTNAERPGMTKSETEVGRGIAEAFEQTKGRIIVTTFASNVHRVQQVIHAA 246
Query: 350 DLTGRKLVFVGMSLRTYLEAAWKDGKAPFDPSTLVKAEDIDAYAPKDLLIVTTGSQAEPR 409
T RKL G S+ + A + G + E++ Y + + I+TTGSQ EP
Sbjct: 247 IATNRKLAVAGRSMVKVVSIAERLGYLEAPDDLFIDIEEVSKYDDERVAIITTGSQGEPM 306
Query: 410 AALNLASFGSSHAFKLTKEDIVLYSAKVIPGNESRVMEMMNRISEIGSTIVMGRNENLHT 469
+AL+ + G+ +T+ D V+ +A IPGNE V +++ + IG+ ++ G + +H
Sbjct: 307 SALSRMAKGAHRQITITENDTVIIAATPIPGNERSVSTIVDLLHRIGADVIFGHGK-VHA 365
Query: 470 SGHAYRGELEEVLRIVKPQHFLPVHGEYLFLKEHESLGKSTGIRHTAV--IKNGEMLGVS 527
SGH EL+ +L +++P+ F+P+HGE+ H+ L KS GIR A+ + GE V
Sbjct: 366 SGHGSAEELKLMLNLMRPKFFVPIHGEFRMQHAHKELAKSVGIREEAIFLVDKGE---VV 422
Query: 528 HLRNRRVLSNGFISLGKENLQLKYSDGDKAFGTSGELFLDERMRIALDGIIVVSMEIFRP 587
RN + G + G + DG G G + L +R ++ DGI+VV +
Sbjct: 423 EFRNGQGRKAGKVPSGNVLI-----DG-LGVGDVGNIVLRDRRLLSKDGILVVVV----- 471
Query: 588 KNLESLAGNTLKGKIRITTRCLWL-DKGKLLDALYKAAHAALSSCPVK--SPLPHMERTV 644
L +G L G I+ +++ + KL++ + L C + + ++ V
Sbjct: 472 -TLNKQSGTILSGPNIISRGFVYVRESEKLIEEANELVTETLKKCVTENVNEWSSLKSNV 530
Query: 645 SEVLRKMVRKYSGKRPEVIAIAIE 668
EVL + + + + +RP ++ I +E
Sbjct: 531 REVLSRFLFEKTKRRPMILPIIME 554
>gb|AAK79649.1| Predicted metal-dependent hydrolase of metallo-beta-lactamase
superfamily [Clostridium acetobutylicum ATCC 824]
gi|15894960|ref|NP_348309.1| Predicted metal-dependent
hydrolase of metallo-beta-lactamase superfamily
[Clostridium acetobutylicum ATCC 824]
gi|25304219|pir||F97107 probable metal-dependent
hydrolase of metallo-beta-lactamase superfamily
[imported] - Clostridium acetobutylicum
Length = 555
Score = 318 bits (815), Expect = 5e-85
Identities = 183/562 (32%), Positives = 307/562 (54%), Gaps = 18/562 (3%)
Query: 110 LRVLPIGGLGEIGMNCMLVGNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKWSHKIE 169
++++P+GG+ EIG N ++ + +ID G+ FPD + G+ +IPD T++ K K++
Sbjct: 8 IKIIPLGGVDEIGKNLTVIEYREEIAVIDCGLSFPDEEMFGIDLVIPDVTYLIKNREKVK 67
Query: 170 ALVITHGHEDHIGALPWVIPALDSNTPIFASSFTMELIKKRLKEHGIFLPSRLKIFRTKN 229
+ +THGHEDHIGALP+V+ L N P++ + T+ +++ +LKEH + LK+ + ++
Sbjct: 68 GIFLTHGHEDHIGALPYVLKEL--NVPVYGTKLTIGIVETKLKEHNLLSKVDLKVVKPRD 125
Query: 230 KFVAGPFEIEPIRVTHSIPDCCGLVLRCSDGTILHTGDWKIDETPLDGKVFDREGLEELS 289
IE IR +HSI D + + G +LHTGD+K+D TP+DG V D EL
Sbjct: 126 IIKLNSMSIEFIRQSHSIADSVAIAIHTPMGVVLHTGDFKVDYTPIDGCVADFARFAELG 185
Query: 290 KEGVTLMMSDSTNVLSPGRTTSESVVADSLLRHISASKGRVITTQFASNLHRIGSVKAAA 349
K GV MM DSTNV PG T SE+ V ++ + +KGR+I FASN+HRI + AA
Sbjct: 186 KRGVLAMMCDSTNVERPGYTMSETTVGETFNTVFTKAKGRIIVATFASNIHRIQQIVEAA 245
Query: 350 DLTGRKLVFVGMSLRTYLEAAWKDGKAPFDPSTLVKAEDIDAYAPKDLLIVTTGSQAEPR 409
+ RK+ G S+ + A + G L+ + I+ YA ++I+TTGSQ EP
Sbjct: 246 ERYNRKVAVSGRSMENIVAVAIELGYLKVKEGVLIGIDAINRYADNKIVIITTGSQGEPM 305
Query: 410 AALNLASFGSSHAFKLTKEDIVLYSAKVIPGNESRVMEMMNRISEIGSTIVMGRNENLHT 469
+AL + K+ + D+V+ SA IPGNE V ++N++ + G+ ++ ++H
Sbjct: 306 SALTRMAAQEHKKVKIQEGDMVVISANPIPGNEKLVSRVINQLFKKGAEVIYEALADVHV 365
Query: 470 SGHAYRGELEEVLRIVKPQHFLPVHGEYLFLKEHESLGKSTGIRHTAVIKNGEMLGVSHL 529
SGHA + EL+ + ++KP+ F+PVHGEY LK+H+ L + G+ V+ E+ GV L
Sbjct: 366 SGHACQEELKLMHTLIKPRFFIPVHGEYRHLKQHKELATNLGMPKANVV-IPEIGGVIEL 424
Query: 530 RNRRVLSNGFISLGKENLQLKYSDGDKAFGTSGELFLDERMRIALDGIIVVSMEIFRPKN 589
+ + GK + DG G G + L +R ++ DGI+ V + I
Sbjct: 425 TRSSIKKTDTVVAGK-----VFVDG-LGVGDVGNIVLRDRKHLSQDGIVTVVVTI----- 473
Query: 590 LESLAGNTLKGKIRITTRCLWLDKGK-LLDALYKAAHAALSSCPVK--SPLPHMERTVSE 646
E ++G + G I+ +++ + + L++ + AL C K + ++ + +
Sbjct: 474 -EKVSGTVIAGPDIISRGFVYVRESEDLMEESKEIVKTALLECEEKHITEWATIKSNIKD 532
Query: 647 VLRKMVRKYSGKRPEVIAIAIE 668
VLR + + + ++P ++ I +E
Sbjct: 533 VLRIFLYERTKRKPMILPIIME 554
>ref|NP_939811.1| hypothetical protein DIP1463 [Corynebacterium diphtheriae NCTC
13129] gi|38200306|emb|CAE49991.1| Conserved
hypothetical protein [Corynebacterium diphtheriae]
Length = 684
Score = 317 bits (812), Expect = 1e-84
Identities = 193/521 (37%), Positives = 290/521 (55%), Gaps = 18/521 (3%)
Query: 68 NDGSTSRVPQKRRRRIEGPRKSME--DSVQRRMEQFYEGNDGPPLRVLPIGGLGEIGMNC 125
N S S+ Q R P KSM+ D QR E +G LR+ +GG+ EIG N
Sbjct: 79 NRSSQSKNNQSNNRGRRTPVKSMQGADLTQRLPEPPKAPKNG--LRIYALGGISEIGRNM 136
Query: 126 MLVGNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKWSHKIEALVITHGHEDHIGALP 185
+ ++R +++D G++FP + GV I+PD I K++ALVITHGHEDHIGA+P
Sbjct: 137 TVFEYNNRLLIVDCGVLFPSSGEPGVDLILPDFGPIENKLDKVDALVITHGHEDHIGAIP 196
Query: 186 WVIPALDSNTPIFASSFTMELIKKRLKEHGIFLPSRLKIFRTKNKFVAGPFEIEPIRVTH 245
W++ L + PI AS FTM LI + +EH P +++ K+ GPF I V H
Sbjct: 197 WLLK-LRPDMPILASKFTMALIAAKCREHRQ-RPKLIEV-NEKSDINRGPFNIRFWAVNH 253
Query: 246 SIPDCCGLVLRCSDGTILHTGDWKIDETPLDGKVFDREGLEELSKEGVTLMMSDSTNVLS 305
SIPDC G+ ++ G ++HTGD K+D+TP DG+ D L EGV L++ +STN
Sbjct: 254 SIPDCLGVAIKTGAGLVIHTGDVKLDQTPPDGRPTDLPALSRFGDEGVDLLLCESTNATV 313
Query: 306 PGRTTSESVVADSLLRHISASKGRVITTQFASNLHRIGSVKAAADLTGRKLVFVGMSLRT 365
PG + SE+ VA +L R IS +K RVI FASN++R+ + AA GRK+ F G S+
Sbjct: 314 PGFSGSEADVAPTLKRLISDAKQRVILASFASNVYRVQAAVDAAVAAGRKVAFNGRSMIR 373
Query: 366 YLEAAWKDGKAPFDPSTLVKAEDIDAYAPKDLLIVTTGSQAEPRAALNLASFGSSHAFKL 425
+E A K G T+V +D AP ++++TTG+Q EP AAL+ + +
Sbjct: 374 NMEIAEKMGYLKAPRGTIVSMDDAAKMAPHKVVLITTGTQGEPMAALSRMARREHRQITV 433
Query: 426 TKEDIVLYSAKVIPGNESRVMEMMNRISEIGSTIVMGRNENLHTSGHAYRGELEEVLRIV 485
D+++ S+ ++PGNE V +MN +S+IG+T+V GR+ +HTSGH + GEL +
Sbjct: 434 RDGDLIILSSSLVPGNEEAVFGVMNMLSQIGATVVTGRDAKVHTSGHGFAGELLFLYNAA 493
Query: 486 KPQHFLPVHGEYLFLKEHESLGKSTGIRHTAVI--KNGEMLGVSHLRNRRVLSNGFISLG 543
+P++ +PVHGE+ L+ ++ L STG+ V+ +NG ++ + R R V G
Sbjct: 494 RPKNVMPVHGEWRHLRANKELAISTGVEADRVVLAQNGVVVDLVDGRARVV--------G 545
Query: 544 KENLQLKYSDGDKAFGTSGELFLDERMRIALDGIIVVSMEI 584
+ + Y DG E+ L+ER + G+I ++ I
Sbjct: 546 QMQVGHLYVDGVTMGDIDAEV-LEERTSLGEGGLIAITAVI 585
>ref|YP_175709.1| Zn-dependent hydrolase, beta-lactamase superfamily [Bacillus
clausii KSM-K16] gi|56910221|dbj|BAD64748.1|
Zn-dependent hydrolase, beta-lactamase superfamily
[Bacillus clausii KSM-K16]
Length = 555
Score = 316 bits (810), Expect = 2e-84
Identities = 183/566 (32%), Positives = 316/566 (55%), Gaps = 27/566 (4%)
Query: 110 LRVLPIGGLGEIGMNCMLVGNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKWSHKIE 169
+RV +GG+GEIG N + + I+IDAG+MFPD D LGV +IPD +++ + ++
Sbjct: 9 VRVFALGGVGEIGKNMYGIEVDEDIIVIDAGLMFPDDDMLGVDIVIPDVSYLVENEERVR 68
Query: 170 ALVITHGHEDHIGALPWVIPALDSNTPIFASSFTMELIKKRLKEHGIFLPSRLKIFRTKN 229
+++THGHEDHIG L +V+ + N P++A+ T+ L++++LKE GI + L++ + +
Sbjct: 69 GIILTHGHEDHIGGLSYVLKKI--NVPVYATKLTLGLVEEKLKEAGIHRHAELRLIDSNS 126
Query: 230 KFVAGPFEIEPIRVTHSIPDCCGLVLRCSDGTILHTGDWKIDETPLDGKVFDREGLEELS 289
+ G + RV+HSIPDC G+ + S G I+HTGD+K D+TP+DGK D + +
Sbjct: 127 RIKLGNTPVSFFRVSHSIPDCVGVCVETSQGAIVHTGDFKFDQTPVDGKHADIGKMAAIG 186
Query: 290 KEGVTLMMSDSTNVLSPGRTTSESVVADSLLRHISASKGRVITTQFASNLHRIGSVKAAA 349
++GV +MSDSTN PG T SE+ + + +KGR+I T FASN+HRI V AA
Sbjct: 187 QKGVLCLMSDSTNAERPGTTKSEAEAGKGIKDVFARTKGRLIVTSFASNVHRIQQVIQAA 246
Query: 350 DLTGRKLVFVGMSLRTYLEAAWKDGKAPFDPSTLVKAEDIDAYAPKDLLIVTTGSQAEPR 409
T RKL VG S+ + A K G L+ ++++ + P+ + I+TTGSQ EP
Sbjct: 247 SATNRKLAVVGKSMVRVISIAGKLGYLKIPKDLLIDMDEVNKHKPEKVAILTTGSQGEPM 306
Query: 410 AALNLASFGSSHAFKLTKEDIVLYSAKVIPGNESRVMEMMNRISEIGSTIVMGRNENLHT 469
+AL + G+ +T D V+ +A IPGNE+ V +++++ IG+ ++ G ++ +H
Sbjct: 307 SALTRMAKGAHRHITITPNDTVIIAATAIPGNETSVSAVIDQLYRIGAEVIYG-SQAVHA 365
Query: 470 SGHAYRGELEEVLRIVKPQHFLPVHGEYLFLKEHESLGKSTGIR--HTAVIKNGEMLGVS 527
SGH + EL+ +L +++P+ FLP+HGE H+ L GI+ +++ GE++ +
Sbjct: 366 SGHGSQEELKLMLNLMRPKFFLPIHGEIRMQHAHQQLALQVGIKKERIYIVEKGEVIEFT 425
Query: 528 HLRNRR--VLSNGFISLGKENLQLKYSDGDKAFGTSGELFLDERMRIALDGIIVVSMEIF 585
+ RR + +G + + DG G G + L +R ++ DGI+VV + +
Sbjct: 426 KQQARRGPKVPSGHVLI----------DG-LGVGDVGNIVLRDRRLLSKDGILVVVVTLN 474
Query: 586 RPKNLESLAGNTL-KGKIRITTRCLWLDKGKLLDALYKAAHAALSSCPVK--SPLPHMER 642
+ N N + +G + + + +LL+ + + L C + + ++
Sbjct: 475 KKTNTIVSGPNIISRGFVYVR------ESEELLEKSNELVTSILQKCVTENVNEWSSLKS 528
Query: 643 TVSEVLRKMVRKYSGKRPEVIAIAIE 668
+ E L + + + + +RP ++ I +E
Sbjct: 529 NIREGLSRYLFEKTKRRPMILPIIME 554
>ref|ZP_00329281.1| COG0595: Predicted hydrolase of the metallo-beta-lactamase
superfamily [Moorella thermoacetica ATCC 39073]
Length = 555
Score = 314 bits (804), Expect = 9e-84
Identities = 192/563 (34%), Positives = 314/563 (55%), Gaps = 24/563 (4%)
Query: 112 VLPIGGLGEIGMNCMLVGNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKWSHKIEAL 171
++P+GGLGEIG N M + + ++ID G+ FP+ + LGV +IPD T++ + ++ +
Sbjct: 10 LIPLGGLGEIGKNMMAIRYGNSILVIDCGLTFPEDELLGVDVVIPDYTYLLENRQMVKGI 69
Query: 172 VITHGHEDHIGALPWVIPALDSNTPIFASSFTMELIKKRLKEHGIFLPSRLKIFRTKNKF 231
++THGHEDHIGALP+V+ D N P++ + T+ LI+ +LKE G F RL+ + ++
Sbjct: 70 IVTHGHEDHIGALPYVLK--DLNVPVYGTKLTLALIQAKLKEQGNFNGVRLQQVKPRDTL 127
Query: 232 VAGPFEIEPIRVTHSIPDCCGLVLRCSDGTILHTGDWKIDETPLDGKVFDREGLEELSKE 291
GPF +E I V+HSI D L + GTI+HT D+KID TP+DG+VFD EL ++
Sbjct: 128 KIGPFRVEFIHVSHSIADTVALAIHTPVGTIVHTSDFKIDYTPIDGEVFDFYKFAELGEK 187
Query: 292 GVTLMMSDSTNVLSPGRTTSESVVADSLLRHISASKGRVITTQFASNLHRIGSVKAAADL 351
GV ++MSDSTNV PG T SE VV + ++ R+I FASN+HR+ + + A
Sbjct: 188 GVLVLMSDSTNVERPGFTMSERVVGGTFDEVFRRARERIIIASFASNIHRVQQIISTAYK 247
Query: 352 TGRKLVFVGMSLRTYLEAAWKDGKAPFDPSTLVKAEDIDAYAPKD-LLIVTTGSQAEPRA 410
RK+ VG S+ + A + G TLV+ ++ A+ PK+ +I++TGSQ EP +
Sbjct: 248 YNRKVAVVGRSMVNVVNIAQEIGYLNIPEGTLVELSEL-AHLPKNQTVIISTGSQGEPMS 306
Query: 411 ALNLASFGSSHAFKLTKEDIVLYSAKVIPGNESRVMEMMNRISEIGSTIVMGRNENLHTS 470
AL + ++ D V+ SA IPGNE V ++++ + G+ + E +H S
Sbjct: 307 ALTRIARNDHRQIEIVPGDTVIISALPIPGNEKLVARTVDQLFKQGADVYHEAVEGVHVS 366
Query: 471 GHAYRGELEEVLRIVKPQHFLPVHGEYLFLKEHESLGKSTGI--RHTAVIKNGEMLGVSH 528
GHA + EL+ VL +VKP+ F+PVHGEY L +H L + GI + V +NG+++ +
Sbjct: 367 GHASQEELKLVLSLVKPKFFVPVHGEYRMLIKHARLAEELGIPPENIFVAENGQVMEFTR 426
Query: 529 LRNRRVLSNGFISLGKENLQLKYSDGDKAFGTSGELFLDERMRIALDGIIVVSMEIFRPK 588
NG ++ G+ + DG G G + L +R ++A DG+++V +
Sbjct: 427 EEGN---FNGRVTAGRLLI-----DG-LGVGDVGNIVLRDRKQLAQDGLLIVVL------ 471
Query: 589 NLESLAGNTLKGKIRITTRCLWL-DKGKLLDALYKAAHAALSSCPVK--SPLPHMERTVS 645
L G+ + G I+ +++ + +LLD + AL C + + ++ +
Sbjct: 472 TLSKETGSVVAGPDIISRGFVYVRESEELLDEAKERVRQALDKCSERKVNDWSTIKGNIR 531
Query: 646 EVLRKMVRKYSGKRPEVIAIAIE 668
+ L K + + + +RP ++ I +E
Sbjct: 532 DNLSKFLYEKTRRRPMILPIIME 554
>ref|NP_738473.1| putative metallo-beta-lactamase superfamily protein
[Corynebacterium efficiens YS-314]
gi|23493704|dbj|BAC18673.1| putative
metallo-beta-lactamase superfamily protein
[Corynebacterium efficiens YS-314]
Length = 700
Score = 314 bits (804), Expect = 9e-84
Identities = 190/522 (36%), Positives = 295/522 (56%), Gaps = 21/522 (4%)
Query: 67 GNDGSTSRVPQKRRRRIEGPRKSME--DSVQRRMEQFYEGNDGPPLRVLPIGGLGEIGMN 124
GN G + Q RR + KSM+ D QR E +G LR+ +GG+ EIG N
Sbjct: 97 GNQGGRGQRNQGGRRNVV---KSMQGADLTQRLPEPPKAPANG--LRIYALGGISEIGRN 151
Query: 125 CMLVGNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKWSHKIEALVITHGHEDHIGAL 184
+ ++R +++D G++FP + GV I+PD I +++EALV+THGHEDHIGA+
Sbjct: 152 MTVFEYNNRLLIVDCGVLFPSSGEPGVDLILPDFGPIENHLNRVEALVVTHGHEDHIGAI 211
Query: 185 PWVIPALDSNTPIFASSFTMELIKKRLKEHGIFLPSRLKIFRTKNKFVAGPFEIEPIRVT 244
PW++ L + PI S FT+ LI + KEH + P +++ N+ GPF I V
Sbjct: 212 PWLLK-LRHDIPIIGSRFTLALIAAKCKEHRL-RPKLIEVNEQSNEN-RGPFNIRFWAVN 268
Query: 245 HSIPDCCGLVLRCSDGTILHTGDWKIDETPLDGKVFDREGLEELSKEGVTLMMSDSTNVL 304
HSIPDC GL ++ G ++HTGD K+D+TP DG+ D L EGV LM+ DSTN
Sbjct: 269 HSIPDCLGLAIKTPAGLVIHTGDIKLDQTPTDGRPTDLPALSRFGDEGVDLMLCDSTNAT 328
Query: 305 SPGRTTSESVVADSLLRHISASKGRVITTQFASNLHRIGSVKAAADLTGRKLVFVGMSLR 364
+PG + SE+ +A +L R +S +K RVI FASN++R+ + AA RK+ F G S+
Sbjct: 329 TPGVSGSEAEIAPTLKRLVSEAKQRVILASFASNVYRVQAAVDAAVAANRKVAFNGRSMI 388
Query: 365 TYLEAAWKDGKAPFDPSTLVKAEDIDAYAPKDLLIVTTGSQAEPRAALNLASFGSSHAFK 424
+E A K G T++ +D AP ++++TTG+Q EP AAL+ +
Sbjct: 389 RNMEIAEKMGYLKAPRGTIISMDDASRMAPHKVMLITTGTQGEPMAALSRMARREHRQIT 448
Query: 425 LTKEDIVLYSAKVIPGNESRVMEMMNRISEIGSTIVMGRNENLHTSGHAYRGELEEVLRI 484
+ D+++ S+ ++PGNE V ++N +++IG+T++ GR+ +HTSGH Y GEL +
Sbjct: 449 VRDGDLIILSSSLVPGNEEAVFGVINMLAQIGATVITGRDAKVHTSGHGYSGELLFLYNA 508
Query: 485 VKPQHFLPVHGEYLFLKEHESLGKSTGIRHTAVI--KNGEMLGVSHLRNRRVLSNGFISL 542
+P++ +PVHGE+ L+ ++ L STG++ V+ +NG ++ + + + R V G I +
Sbjct: 509 ARPKNAMPVHGEWRHLRANKELAISTGVQRENVVLAQNGVVVDLVNGQARVV---GQIPV 565
Query: 543 GKENLQLKYSDGDKAFGTSGELFLDERMRIALDGIIVVSMEI 584
G NL Y DG G L +R + G+I ++ I
Sbjct: 566 G--NL---YVDG-VTMGDIDADILADRTSLGEGGLISITCVI 601
>emb|CAF20311.1| HYDROLASE OF METALLO-BETA-LACTAMASE SUPERFAMILY [Corynebacterium
glutamicum ATCC 13032] gi|21324740|dbj|BAB99363.1|
Predicted hydrolase of the metallo-beta-lactamase
superfamily [Corynebacterium glutamicum ATCC 13032]
gi|11967652|emb|CAC19480.1| hypothetical protein
[Corynebacterium glutamicum]
gi|18266923|sp|P54122|Y1970_CORGL Hypothetical UPF0036
protein Cgl1970/cg2160 gi|62390810|ref|YP_226212.1|
HYDROLASE OF METALLO-BETA-LACTAMASE SUPERFAMILY
[Corynebacterium glutamicum ATCC 13032]
gi|19553174|ref|NP_601176.1| predicted hydrolase of the
metallo-beta-lactamase superfamily [Corynebacterium
glutamicum ATCC 13032]
Length = 718
Score = 313 bits (803), Expect = 1e-83
Identities = 196/534 (36%), Positives = 297/534 (54%), Gaps = 21/534 (3%)
Query: 58 SASALSASVGNDGSTSRVPQKRRRRIEGPR---KSME--DSVQRRMEQFYEGNDGPPLRV 112
S +A + N G+ +R R R G R KSM+ D QR E +G LR+
Sbjct: 100 SGNANEGANNNSGNQNRQGGNRGNRGGGRRNVVKSMQGADLTQRLPEPPKAPANG--LRI 157
Query: 113 LPIGGLGEIGMNCMLVGNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKWSHKIEALV 172
+GG+ EIG N + ++R +++D G++FP + GV I+PD I H+++ALV
Sbjct: 158 YALGGISEIGRNMTVFEYNNRLLIVDCGVLFPSSGEPGVDLILPDFGPIEDHLHRVDALV 217
Query: 173 ITHGHEDHIGALPWVIPALDSNTPIFASSFTMELIKKRLKEHGIFLPSRLKIFRTKNKFV 232
+THGHEDHIGA+PW++ L ++ PI AS FT+ LI + KEH P +++ N+
Sbjct: 218 VTHGHEDHIGAIPWLLK-LRNDIPILASRFTLALIAAKCKEHRQ-RPKLIEVNEQSNED- 274
Query: 233 AGPFEIEPIRVTHSIPDCCGLVLRCSDGTILHTGDWKIDETPLDGKVFDREGLEELSKEG 292
GPF I V HSIPDC GL ++ G ++HTGD K+D+TP DG+ D L EG
Sbjct: 275 RGPFNIRFWAVNHSIPDCLGLAIKTPAGLVIHTGDIKLDQTPPDGRPTDLPALSRFGDEG 334
Query: 293 VTLMMSDSTNVLSPGRTTSESVVADSLLRHISASKGRVITTQFASNLHRIGSVKAAADLT 352
V LM+ DSTN +PG + SE+ VA +L R + +K RVI FASN++R+ + AA +
Sbjct: 335 VDLMLCDSTNATTPGVSGSEADVAPTLKRLVGDAKQRVILASFASNVYRVQAAVDAAVAS 394
Query: 353 GRKLVFVGMSLRTYLEAAWKDGKAPFDPSTLVKAEDIDAYAPKDLLIVTTGSQAEPRAAL 412
RK+ F G S+ +E A K G T++ +D AP ++++TTG+Q EP AAL
Sbjct: 395 NRKVAFNGRSMIRNMEIAEKLGYLKAPRGTIISMDDASRMAPHKVMLITTGTQGEPMAAL 454
Query: 413 NLASFGSSHAFKLTKEDIVLYSAKVIPGNESRVMEMMNRISEIGSTIVMGRNENLHTSGH 472
+ + + D+++ S+ ++PGNE V ++N +++IG+T+V GR+ +HTSGH
Sbjct: 455 SRMARREHRQITVRDGDLIILSSSLVPGNEEAVFGVINMLAQIGATVVTGRDAKVHTSGH 514
Query: 473 AYRGELEEVLRIVKPQHFLPVHGEYLFLKEHESLGKSTGIRHTAVI--KNGEMLGVSHLR 530
Y GEL + +P++ +PVHGE+ L+ ++ L STG+ V+ +NG V +
Sbjct: 515 GYSGELLFLYNAARPKNAMPVHGEWRHLRANKELAISTGVNRDNVVLAQNGV---VVDMV 571
Query: 531 NRRVLSNGFISLGKENLQLKYSDGDKAFGTSGELFLDERMRIALDGIIVVSMEI 584
N R G I +G NL Y DG G L +R + G+I ++ I
Sbjct: 572 NGRAQVVGQIPVG--NL---YVDG-VTMGDIDADILADRTSLGEGGLISITAVI 619
>gb|AAR36504.2| metallo-beta-lactamase family protein [Geobacter sulfurreducens
PCA]
Length = 561
Score = 313 bits (803), Expect = 1e-83
Identities = 180/568 (31%), Positives = 309/568 (53%), Gaps = 22/568 (3%)
Query: 106 DGPPLRVLPIGGLGEIGMNCMLVGNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKWS 165
D LR++P+GGLGEIG+N M + + I+ D G+MFP+ LG+ +IPD T++R+ +
Sbjct: 10 DNGGLRIIPLGGLGEIGLNMMAYEHGEDIIVADCGLMFPEPQMLGIDLVIPDITYLRERA 69
Query: 166 HKIEALVITHGHEDHIGALPWVIPALDSNTPIFASSFTMELIKKRLKEHGIFLPSRLKIF 225
++ +++THGHEDHIGALP+V+ L + P++ ++ T+ I+++LKE + L +
Sbjct: 70 GRVRGILLTHGHEDHIGALPYVLQEL--SLPLYGTALTLGFIREKLKEFDLESEVELNVV 127
Query: 226 RTKNKFVAGPFEIEPIRVTHSIPDCCGLVLRCSDGTILHTGDWKIDETPLDGKVFDREGL 285
+ ++ G F +E IRV+HSI D C L +R +G ++HTGD+KID+TP+DG++ D
Sbjct: 128 KPRDVVTLGCFSVEFIRVSHSIVDGCALAIRTPEGVVIHTGDFKIDQTPVDGELTDLATF 187
Query: 286 EELSKEGVTLMMSDSTNVLSPGRTTSESVVADSLLRHISASKGRVITTQFASNLHRIGSV 345
EGV +++DSTNV G T SE VV ++ +GR+I F+SN+HR+
Sbjct: 188 SRYGDEGVLALLADSTNVEREGYTLSERVVGEAFDEIFPRCEGRIIVAAFSSNIHRVQQA 247
Query: 346 KAAADLTGRKLVFVGMSLRTYLEAAWKDGKAPFDPSTLVKAEDIDAYAPKDLLIVTTGSQ 405
AA GRK++ G S+ + A + G L +++ + + ++TTGSQ
Sbjct: 248 VDAAVRCGRKVLLNGRSMVANVAIARQLGYLRMPEGVLTDLKELQHLPKEQVCMITTGSQ 307
Query: 406 AEPRAALNLASFGSSHAFKLTKEDIVLYSAKVIPGNESRVMEMMNRISEIGSTIVMGRNE 465
EP ++L + KL D V+ S++ IPGNE + +++N + G+ + +
Sbjct: 308 GEPMSSLARIAMDDHKQIKLEAGDTVILSSRFIPGNEKTISDLINHLYRRGAEVFHEKVS 367
Query: 466 NLHTSGHAYRGELEEVLRIVKPQHFLPVHGEYLFLKEHESLGKSTGI--RHTAVIKNGEM 523
+H SGHA + EL+ +L + +P+HF+PVHGEY L +H L + G+ V NG++
Sbjct: 368 EVHVSGHASQEELKLMLNLTRPRHFVPVHGEYRHLVKHARLAQRVGVPAERCLVAVNGDV 427
Query: 524 LGVSHLRNRRVLSNGFISLGKENLQLKYSDGDKAFGTSGELFLDERMRIALDGIIVVSME 583
+ + R G + G+ Y DG K G GE+ L +R ++ DG++VV +
Sbjct: 428 IRFADGRGEIA---GAVESGR-----VYIDG-KGIGDVGEVVLKDRKHLSEDGMVVVIIA 478
Query: 584 IFRPKNLESLAGNTLKGKIRITTRCLWLDKG-KLLDALYKAAHAALSSCPVKS--PLPHM 640
I +G + G ++ ++ D+ + LD K L+ +++ +
Sbjct: 479 I------NQASGEVIYGPDIVSRGFVFEDESQQYLDETKKIVLDLLAGMSIETLGEWGEV 532
Query: 641 ERTVSEVLRKMVRKYSGKRPEVIAIAIE 668
++ V +LR+ K +RP ++ + +E
Sbjct: 533 KQEVRRILRRFFNKTIERRPVILPVILE 560
>gb|AAO35641.1| putative Zn-dependent hydrolase (beta-lactamase superfamily)
[Clostridium tetani E88] gi|28210760|ref|NP_781704.1|
putative Zn-dependent hydrolase (beta-lactamase
superfamily) [Clostridium tetani E88]
Length = 555
Score = 313 bits (802), Expect = 2e-83
Identities = 184/565 (32%), Positives = 308/565 (53%), Gaps = 24/565 (4%)
Query: 110 LRVLPIGGLGEIGMNCMLVGNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKWSHKIE 169
++V+P+GGL EIG N + ++ID G+ FPD + LG+ +IPD +++ K +++
Sbjct: 8 VKVIPLGGLNEIGKNITAFEYKNDIVVIDCGLKFPDEEMLGIDIVIPDISYLLKNKERVK 67
Query: 170 ALVITHGHEDHIGALPWVIPALDSNTPIFASSFTMELIKKRLKEHGIFLPSRLKIFRTKN 229
+ +THGHEDHIGALP+V+ L N P++ + T+ +++ +LKEH + +L + ++
Sbjct: 68 GIFLTHGHEDHIGALPYVLKEL--NVPVYGTKLTLGILETKLKEHNLLSSVKLNCIKPRD 125
Query: 230 KFVAGPFEIEPIRVTHSIPDCCGLVLRCSDGTILHTGDWKIDETPLDGKVFDREGLEELS 289
+E IR +HSI D + + G ILHTGD+KID TP+DG V D EL
Sbjct: 126 MIKLENCTVEFIRTSHSIADSVAIAIHTPVGAILHTGDFKIDYTPIDGHVADLSRFAELG 185
Query: 290 KEGVTLMMSDSTNVLSPGRTTSESVVADSLLRHISASKGRVITTQFASNLHRIGSVKAAA 349
++GV LM++DSTNV PG T SES V + + KGR+I FASN+HR+ + + A
Sbjct: 186 RKGVALMLADSTNVERPGYTMSESTVGKTFDNIFTKGKGRIIVATFASNIHRVQQIISTA 245
Query: 350 DLTGRKLVFVGMSLRTYLEAAWKDG--KAPFDPSTLVKAEDIDAYAPKDLLIVTTGSQAE 407
RK+ F G S+ +E A + G KAP + T + + I+ Y ++I+TTGSQ E
Sbjct: 246 VKFDRKVAFSGRSMENIVEVAKELGYIKAPLE--TFIDIDSINKYPDNKVVIITTGSQGE 303
Query: 408 PRAALNLASFGSSHAFKLTKEDIVLYSAKVIPGNESRVMEMMNRISEIGSTIVMGRNENL 467
P +AL+ + + D V+ SA IPGNE V ++N++ + G+ ++ ++
Sbjct: 304 PMSALSRMAASEHRKVNIVPGDTVVISATPIPGNEKLVSRVINQLFQKGADVIYEALADI 363
Query: 468 HTSGHAYRGELEEVLRIVKPQHFLPVHGEYLFLKEHESLGKSTGIRHTAVI--KNGEMLG 525
H SGHA + EL+ + +V+P++F+PVHGEY LK+H L + G+RH ++ NG+++
Sbjct: 364 HVSGHACQEELKLMHTLVRPKYFIPVHGEYRHLKQHGELAEKLGMRHENIVIAGNGDVIE 423
Query: 526 VSHLRNRRVLSNGFISLGKENLQLKYSDGDKAFGTSGELFLDERMRIALDGIIVVSMEIF 585
V+ + NG + G+ + DG G G + L +R ++ DGI+ V + I
Sbjct: 424 VA---RDCIKKNGTVMSGQ-----VFVDG-LGVGDVGNIVLRDRRHLSQDGILTVVVTIE 474
Query: 586 RPKNLESLAGNTLKGKIRITTRCLWLDKGKLLDALYKAAHAALSSCPVKSPL--PHMERT 643
R + +AG + + + R + L+ + L C + ++
Sbjct: 475 RGTGM-VIAGPDIISRGFVYVR----ESEYLMGEAREVVKEVLRKCEENHIVEWSTIKSN 529
Query: 644 VSEVLRKMVRKYSGKRPEVIAIAIE 668
+ + LR + + + +RP ++ I +E
Sbjct: 530 IKDALRAFLYEKTKRRPMILPIIME 554
>gb|EAM72412.1| IMP dehydrogenase/GMP
reductase:Beta-lactamase-like:RNA-metabolising
metallo-beta-lactamase [Desulfuromonas acetoxidans DSM
684]
Length = 547
Score = 311 bits (797), Expect = 6e-83
Identities = 177/500 (35%), Positives = 273/500 (54%), Gaps = 18/500 (3%)
Query: 112 VLPIGGLGEIGMNCMLVGNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKWSHKIEAL 171
+LP+GGLGEIG+N M + + +LID G+MFP+ LG+ ++PD + + + I L
Sbjct: 1 MLPLGGLGEIGLNMMALEYAGKIVLIDCGLMFPEAHMLGIDLVLPDVSCLEERREDICGL 60
Query: 172 VITHGHEDHIGALPWVIPALDSNTPIFASSFTMELIKKRLKEHGIFLPSRLKIFRTKNKF 231
VITHGHEDHIGA+P++ L P+FA+ T+ L+KK+L+E + L + + F
Sbjct: 61 VITHGHEDHIGAIPFLYEQL-GRPPLFATRLTLALLKKKLEEFELNLNQSMTQIVPRETF 119
Query: 232 VAGPFEIEPIRVTHSIPDCCGLVLRCSDGTILHTGDWKIDETPLDGKVFDREGLEELSKE 291
GPF IEP RV HS+PD GL +RC G ++H+GD+K+D TPLDG+ D L + +E
Sbjct: 120 DVGPFTIEPFRVAHSVPDGVGLAVRCGAGLVVHSGDFKLDATPLDGQSTDVARLAQYGEE 179
Query: 292 GVTLMMSDSTNVLSPGRTTSESVVADSLLRHISASKGRVITTQFASNLHRIGSVKAAADL 351
GV L+++DSTNV +PG SE V + + G V F+SN+HR+ AA
Sbjct: 180 GVLLLLADSTNVETPGFAGSERDVGPRFRDIMEQANGMVFVATFSSNIHRVQQAVDAALA 239
Query: 352 TGRKLVFVGMSLRTYLEAAWKDGKAPFDPSTLVKAEDIDAYAPKDLLIVTTGSQAEPRAA 411
GRK+ G S+ + A S L+ + Y + ++TTGSQ EPR+A
Sbjct: 240 CGRKVALYGRSMVSNCGVARVYDALTIPESELIDVRQLADYPRDQVAVITTGSQGEPRSA 299
Query: 412 LNLASFGSSHAFKLTKEDIVLYSAKVIPGNESRVMEMMNRISEIGSTIVMGRNENLHTSG 471
L + F + + D V+ S++ IPGNE + ++N++ +G+ + R +H SG
Sbjct: 300 LARLAADDHPQFAIEENDCVILSSRFIPGNEKAITSVINQLYRLGAEVHYQRTSGVHVSG 359
Query: 472 HAYRGELEEVLRIVKPQHFLPVHGEYLFLKEHESLGKSTGI--RHTAVIKNGEMLGVSHL 529
HA R EL+++L +V+PQ F+P+HGE+ L +H L + G+ + V++NG+ + VS
Sbjct: 360 HASREELKQLLALVRPQSFIPIHGEFRHLMQHARLAQQVGVAQERSLVVENGQPVLVS-- 417
Query: 530 RNRRVLSNGFISLGKENLQLKYSDGDKAFGTSGELFLDERMRIALDGIIVVSMEIFRPKN 589
++G L DG K G G + L +R R+A G ++ + + R K
Sbjct: 418 ------AHGMKLLEPVETGRVLVDG-KGVGDVGMMELRDRHRLARHGTVMAVLAVSRSK- 469
Query: 590 LESLAGNTLKGKIRITTRCL 609
G L G ++ CL
Sbjct: 470 -----GTLLHGPELVSRGCL 484
>ref|YP_181182.1| metallo-beta-lactamase family protein [Dehalococcoides ethenogenes
195] gi|57225208|gb|AAW40265.1| metallo-beta-lactamase
family protein [Dehalococcoides ethenogenes 195]
Length = 551
Score = 311 bits (797), Expect = 6e-83
Identities = 197/565 (34%), Positives = 305/565 (53%), Gaps = 22/565 (3%)
Query: 108 PPLRVLPIGGLGEIGMNCMLVGNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKWSHK 167
P L+++P+GGLGEIG N M V + ILID G+MFP+ + LG+ +IPD +++ + K
Sbjct: 4 PKLKIVPLGGLGEIGKNMMSVEYGEDIILIDCGLMFPEEEMLGIDLVIPDISYVLENREK 63
Query: 168 IEALVITHGHEDHIGALPWVIPALDSNTPIFASSFTMELIKKRLKEHGIFLPSRLKIFRT 227
+ +VITHGHEDHIGALP+++P + N PI+ + T LI +LKE + +++ + +
Sbjct: 64 VRGIVITHGHEDHIGALPYILPQI--NVPIYCTKLTQGLISVKLKEAKLLHQTKINVIPS 121
Query: 228 KNKFVAGPFEIEPIRVTHSIPDCCGLVLRCSDGTILHTGDWKIDETPLDGKVFDREGLEE 287
F G +++ V HS+PD GLV++ G ++H+GD+K+D TP+DGK + L
Sbjct: 122 DGTFKLGKLKVDFYPVCHSLPDSVGLVIQTPIGAVVHSGDFKLDYTPVDGKPTNLSRLAM 181
Query: 288 LSKEGVTLMMSDSTNVLSPGRTTSESVVADSLLRHISASKGRVITTQFASNLHRIGSVKA 347
L GV L+MSDST+V PG T SE+VV +++ R I A+ GRV+ T FAS + R V
Sbjct: 182 LGSRGVLLLMSDSTHVELPGYTPSETVVGENIDRIIGAATGRVLVTTFASLISRQQQVID 241
Query: 348 AADLTGRKLVFVGMSLRTYLEAAWKDGKAPFDPSTLVKAEDIDAYAPKDLLIVTTGSQAE 407
AA GRKL F G S+ + A G + + + P+ ++++TTGSQ E
Sbjct: 242 AAAKYGRKLFFAGRSMTEIHKMAADLGYLKVPEGLVCNIDQLQRVPPEKVVLMTTGSQGE 301
Query: 408 PRAALNLASFGSSHAFKLTKEDIVLYSAKVIPGNESRVMEMMNRISEIGSTIVMGRNENL 467
P +AL + + K D V+ SA IPGNES V ++ + G+ + R +
Sbjct: 302 PTSALVRIANRDFRHVHIMKGDTVIISASPIPGNESLVSRTIDSLFRQGAKVFYDRIAKV 361
Query: 468 HTSGHAYRGELEEVLRIVKPQHFLPVHGEYLFLKEHESLGKSTGI--RHTAVIKNGEMLG 525
H GHA + EL+ VL +VKP++F+PVHGEY L H L ++ G+ +T ++++G++L
Sbjct: 362 HVHGHASQEELKLVLNLVKPKYFVPVHGEYRHLTMHAELARTMGVAPENTFIMEDGDILE 421
Query: 526 VSHLRNRRVLSNGFISLGKENLQLKYSDGDKAFGTSGELFLDERMRIALDGIIVVSMEIF 585
++ R GK Y DG + G G + L R ++ DGI+V + I
Sbjct: 422 LNTKSGR--------ITGKVPSGNVYVDG-LSVGDVGGVVLRNRRMLSQDGIVVAIVAIN 472
Query: 586 RPKNLESLAGNTLKGKIRITTR-CLWLDKGK-LLDALYKAAHAALSSCPVKSPLPHMERT 643
+ + L G+ I +R + D+ K +LDA L P M
Sbjct: 473 KQTGM-------LVGRPDIVSRGFVDPDESKAMLDASRDLIIKLLDHDGKDIPEGAMYND 525
Query: 644 VSEVLRKMVRKYSGKRPEVIAIAIE 668
V E L K + + +RP VI + ++
Sbjct: 526 VKETLNKFYYEQTKRRPMVIPVMVK 550
>ref|ZP_00398224.1| Beta-lactamase-like:RNA-metabolising metallo-beta-lactamase
[Anaeromyxobacter dehalogenans 2CP-C]
gi|66779912|gb|EAL80987.1|
Beta-lactamase-like:RNA-metabolising
metallo-beta-lactamase [Anaeromyxobacter dehalogenans
2CP-C]
Length = 844
Score = 310 bits (795), Expect = 1e-82
Identities = 193/613 (31%), Positives = 320/613 (51%), Gaps = 34/613 (5%)
Query: 64 ASVGNDGSTSRVPQKRRRRIEGPRKSMEDSVQRRMEQFYEGNDGPPLRVLPIGGLGEIGM 123
A+ G G+ R EG R RR Y G P+R++P+GGLGEIGM
Sbjct: 257 AACGGHGAGRLPRSPGGRSPEGRRARAASGRDRR----YIGPVAAPVRIVPLGGLGEIGM 312
Query: 124 NCMLVGNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKWSHKIEALVITHGHEDHIGA 183
NCM V R ++D G++FP+ ++LGV I PD +++R+ ++ A+ +THGHEDH+GA
Sbjct: 313 NCMAVECDGRIAVVDCGVLFPN-ENLGVDLIAPDLSWLRERREQVGAVFLTHGHEDHLGA 371
Query: 184 LPWVIPALDSNTPIFASSFTMELIKKRLKEHGIFLPSR-LKIFRTKNKFVAGPFEIEPIR 242
LP+++ + P++ + FT+ L++ RL+E G+ R ++ + A P E +
Sbjct: 372 LPFLLREIP--VPVYGTRFTLALLRPRLEEAGVRADLREVRPGDVRPAGEASPIAAEFLA 429
Query: 243 VTHSIPDCCGLVLRCSDGTILHTGDWKIDETPLDGKVFDREGLEELSKEGVTLMMSDSTN 302
VTHSIPD CGL L GT+LH+GD+KID P+ G D + +E L ++GV L++SDSTN
Sbjct: 430 VTHSIPDACGLALSTPQGTLLHSGDFKIDPAPVRGPGMDLDRIEALGRKGVRLLLSDSTN 489
Query: 303 VLSPGRTTSESVVADSLLRHISASKGRVITTQFASNLHRIGSVKAAADLTGRKLVFVGMS 362
G + SES V +L + ++GRV FASN+HR+ + AA GR++ F+G S
Sbjct: 490 AEREGTSLSESEVGPALEQAFERARGRVFVACFASNVHRLQQIVDAARAFGRRVAFLGRS 549
Query: 363 LRTYLEAAWKDGKAPFDPSTLVKAEDIDAYAPKDLLIVTTGSQAEPRAALNLASFGSSHA 422
+ A + G V E+ P++L ++TTG+Q EPR+AL + G
Sbjct: 550 MEANARLAQELGYLELPAWMPVSPEEARTLPPRELCVLTTGTQGEPRSALARLARGEHPD 609
Query: 423 FKLTKEDIVLYSAKVIPGNESRVMEMMNRISEIGSTIVMGRNENLHTSGHAYRGELEEVL 482
+ D+V+ S++ IPGNE + +++N + G+ + LH SGHA E ++
Sbjct: 610 LAVAPGDLVVLSSRFIPGNEIAIGQVVNELCRRGAEVAYEGLAPLHVSGHAQADEQRRLI 669
Query: 483 RIVKPQHFLPVHGEYLFLKEHESLGKSTGIRHTAVIKNGEMLGVSHLRNRRVLSNGFISL 542
R+ +P HF+P+HGEY L H + + G+ V+ +G++L ++ R + ++
Sbjct: 670 RLARPAHFVPIHGEYRHLSRHAAHAAAEGVAGRDVLVDGQVLELADAGVR--VLEAPVAT 727
Query: 543 GKENLQLKYSDGDKAFGTS-GELFLDERMRIALDGIIVVSMEIFRPKNLESLAGNTLKGK 601
G+ Y+D D G G L + +R +A G+ + + I E G ++G
Sbjct: 728 GR-----VYTDRDALLGADMGALVVRDRRLLAETGLCIAVLAI------ERTTGAVVRGP 776
Query: 602 IRITTRCLWLD------KGKLLDALYKAAHAALSSCPVKSPLPHMERTVSEVLRKMVRKY 655
+ +G++L AL + + A ++ ++ V +R+ R+
Sbjct: 777 ELFARGVAGFEGAEEELRGEVLAALAELSPQA------RTDPAEVQEAVRLAVRRFFRRT 830
Query: 656 SGKRPEVIAIAIE 668
+GK+P V+ + +E
Sbjct: 831 TGKKPPVLPVVLE 843
>ref|ZP_00319740.1| COG0595: Predicted hydrolase of the metallo-beta-lactamase
superfamily [Oenococcus oeni PSU-1]
Length = 573
Score = 309 bits (791), Expect = 3e-82
Identities = 195/562 (34%), Positives = 304/562 (53%), Gaps = 21/562 (3%)
Query: 112 VLPIGGLGEIGMNCMLVGNHDRYILIDAGIMFPDYDDLGVQKIIPDTTFIRKWSHKIEAL 171
V+ +GGLGEIG N + D ++IDAGIMFP+ D LGV +IPD T++ + K++AL
Sbjct: 24 VMALGGLGEIGKNTYAIQYQDEIVVIDAGIMFPEDDLLGVDYVIPDYTYLVQNIEKVKAL 83
Query: 172 VITHGHEDHIGALPWVIPALDSNTPIFASSFTMELIKKRLKEHGIFLPSRLKIFRTKNKF 231
VITHGHEDHIGA+ + + A+ N P++A F + LIK +L+EH + + L + +
Sbjct: 84 VITHGHEDHIGAISYFLRAV--NVPVYAPPFALALIKGKLEEHNLLKKTELHEIKPDSFL 141
Query: 232 VAGPFEIEPIRVTHSIPDCCGLVLRCSDGTILHTGDWKIDETPLDGKVFDREGLEELSKE 291
G +E + THSIPD G+ + GTI+ TGD+K D TP+ + D + ++ +E
Sbjct: 142 DFGKLSVEFFQTTHSIPDTVGVAVHTPKGTIVETGDFKFDLTPVTNQYPDMLKMAKIGRE 201
Query: 292 GVTLMMSDSTNVLSPGRTTSESVVADSLLRHISA-SKGRVITTQFASNLHRIGSVKAAAD 350
GV L+MSDSTN P T SE+ V S+ R +KGR+I FASNL R+ +A
Sbjct: 202 GVLLLMSDSTNAERPEFTKSEAWVQKSIERIFDRITKGRIIFATFASNLSRVKMAMDSAV 261
Query: 351 LTGRKLVFVGMSLRTYLEAAWKDGKAPFDPSTLVKAEDIDAYAPKDLLIVTTGSQAEPRA 410
GRK+ G S+ T + G L++ D+ P + LI++TGSQ EP A
Sbjct: 262 RHGRKIAVFGRSMETAVNNGRDLGYLNISDEFLLEPSDLKNTPPDETLILSTGSQGEPMA 321
Query: 411 ALNLASFGSSHAFKLTKEDIVLYSAKVIPGNESRVMEMMNRISEIGSTIVMGRNENLHTS 470
AL + G+ KL D V++S+ IPGN + V +++N + E G+ ++ G N+HTS
Sbjct: 322 ALARIANGTHKQIKLQPNDTVVFSSNPIPGNTASVNKVINELEEGGANVIYGPLNNIHTS 381
Query: 471 GHAYRGELEEVLRIVKPQHFLPVHGEYLFLKEHESLGKSTGI--RHTAVIKNGEMLGVSH 528
GH + E + +L ++KP+ F+P+HGEY LK H L +STG+ H ++ NG++L
Sbjct: 382 GHGGQEEQKIMLELMKPKFFMPIHGEYRMLKIHSGLAQSTGVPSDHIFIMDNGDVLA--- 438
Query: 529 LRNRRVLSNGFISLGKENLQLKYSDGDKAFGTSGELFLDERMRIALDGIIVVSMEIFRPK 588
L G S E++ Y DG G G L ER +++ DG+++V+ I +
Sbjct: 439 LDGEHAHPAGHFS--SEDI---YVDGG-GIGDVGNAVLHERQKLSQDGLVIVTATI-NMQ 491
Query: 589 NLESLAGNTLKGKIRITTRCLWLDKGKLLDALYKAAHAALSSCPVKSPLPH--MERTVSE 646
E L+G L + + R + L++ + A + + S + + V +
Sbjct: 492 TKEILSGPDLLSRGFVYMR----ESVDLINDGRRVAFGTIRKAMMSSNADEGSIRQAVID 547
Query: 647 VLRKMVRKYSGKRPEVIAIAIE 668
L + K + ++P VI + I+
Sbjct: 548 ELSHYLYKKTARKPIVIPVLIQ 569
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.318 0.135 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,414,769,858
Number of Sequences: 2540612
Number of extensions: 60451984
Number of successful extensions: 255637
Number of sequences better than 10.0: 1100
Number of HSP's better than 10.0 without gapping: 388
Number of HSP's successfully gapped in prelim test: 727
Number of HSP's that attempted gapping in prelim test: 251522
Number of HSP's gapped (non-prelim): 2816
length of query: 871
length of database: 863,360,394
effective HSP length: 137
effective length of query: 734
effective length of database: 515,296,550
effective search space: 378227667700
effective search space used: 378227667700
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 80 (35.4 bits)
Medicago: description of AC126782.9


