

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126782.8 + phase: 0 /pseudo
(250 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
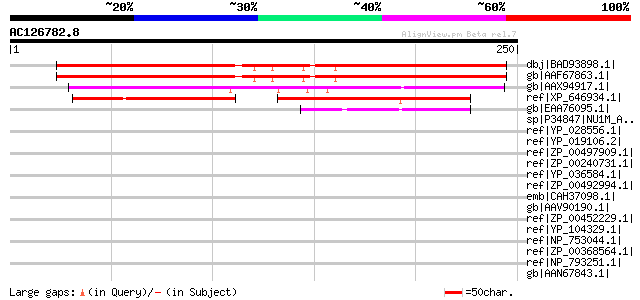
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAD93898.1| cyclopropyl isomerase [Arabidopsis thaliana] 268 1e-70
gb|AAF67863.1| cyclopropyl isomerase [Arabidopsis thaliana] gi|1... 267 2e-70
gb|AAX94917.1| Similar to cycloeucalenol cycloisomerase (ec 5.5.... 236 4e-61
ref|XP_646934.1| hypothetical protein DDB0202318 [Dictyostelium ... 96 7e-19
gb|EAA76095.1| hypothetical protein FG06622.1 [Gibberella zeae P... 63 7e-09
sp|P34847|NU1M_APILI NADH-ubiquinone oxidoreductase chain 1 (NAD... 38 0.23
ref|YP_028556.1| membrane protein, putative [Bacillus anthracis ... 36 1.1
ref|YP_019106.2| membrane protein, putative [Bacillus anthracis ... 36 1.1
ref|ZP_00497909.1| COG1835: Predicted acyltransferases [Burkhold... 35 2.5
ref|ZP_00240731.1| putative integral membrane protein [Bacillus ... 35 2.5
ref|YP_036584.1| possible membrane protein [Bacillus thuringiens... 34 4.3
ref|ZP_00492994.1| COG1835: Predicted acyltransferases [Burkhold... 33 5.6
emb|CAH37098.1| putative acyltransferase [Burkholderia pseudomal... 33 5.6
gb|AAV90190.1| conserved hypothetical protein [Zymomonas mobilis... 33 5.6
ref|ZP_00452229.1| COG1835: Predicted acyltransferases [Burkhold... 33 7.3
ref|YP_104329.1| acyltransferase family protein [Burkholderia ma... 33 7.3
ref|NP_753044.1| Cytochrome BD-II oxidase subunit I [Escherichia... 33 7.3
ref|ZP_00368564.1| sodium- and chloride-dependent transporter [C... 33 7.3
ref|NP_793251.1| monovalent cation:proton antiporter, putative [... 33 9.6
gb|AAN67843.1| multicomponent potassium-proton antiporter, subun... 33 9.6
>dbj|BAD93898.1| cyclopropyl isomerase [Arabidopsis thaliana]
Length = 280
Score = 268 bits (685), Expect = 1e-70
Identities = 150/283 (53%), Positives = 175/283 (61%), Gaps = 66/283 (23%)
Query: 24 SASPTPSLWFAPNLSKRWGELFFLLYTPFWLSLSLGIVVPYKLYENFTELEYLLLGLISA 83
S S +PSLW APN SKRWGELFFL YTPFWL+L LGIVVPYKLYE FTELEYLLL L+SA
Sbjct: 2 SGSSSPSLWLAPNPSKRWGELFFLFYTPFWLTLCLGIVVPYKLYETFTELEYLLLALVSA 61
Query: 84 VPAFLIPMLFVGKADKGISWQDRYWVKSMDNNLQLC---------WELFLDSLL------ 128
VPAF+IPML VGKAD+ + W+DRYWVK+ NL + W + +L
Sbjct: 62 VPAFVIPMLLVGKADRSLCWKDRYWVKA---NLWIIVFSYVGNYFWTHYFFKVLGASYTF 118
Query: 129 -------------FHSSGCILYLSFMEN---------------------EQSYLETLAIS 154
F + C L+ N E +++ LA+S
Sbjct: 119 PSWKMNNVPHTTFFLTHVCFLFYHVASNITLRRLRHSTADLPDSLKWCFEAAWI--LALS 176
Query: 155 NFPYY------------EFVDRESMYKVGSLFYAIYFIVSFPMFLRIDEKPGDKWDLPRV 202
F Y EFVDR +MY+VG LFYAIYFIVSFPMF R+DEK D+WDL RV
Sbjct: 177 YFIAYLETIAIANFPYYEFVDRSAMYRVGCLFYAIYFIVSFPMFFRVDEKSTDEWDLSRV 236
Query: 203 AVDALGAAMLVTIILDLWRIFLGPIVPVSEAKQCSPTGLPWFS 245
AVDALGAAMLVTIILDLWR+FLGPIVP+ E + C +GLPWFS
Sbjct: 237 AVDALGAAMLVTIILDLWRLFLGPIVPLPEGQNCLQSGLPWFS 279
>gb|AAF67863.1| cyclopropyl isomerase [Arabidopsis thaliana]
gi|18423143|ref|NP_568727.1| cyclopropyl isomerase
(CPI1) [Arabidopsis thaliana]
gi|24211551|sp|Q9M643|CCI1_ARATH Cycloeucalenol
cycloisomerase (Cycloeucalenol--obtusifoliol isomerase)
(Cyclopropyl sterol isomerase)
Length = 280
Score = 267 bits (683), Expect = 2e-70
Identities = 150/283 (53%), Positives = 175/283 (61%), Gaps = 66/283 (23%)
Query: 24 SASPTPSLWFAPNLSKRWGELFFLLYTPFWLSLSLGIVVPYKLYENFTELEYLLLGLISA 83
S S +PSLW APN SKRWGELFFL YTPFWL+L LGIVVPYKLYE FTELEYLLL L+SA
Sbjct: 2 SGSSSPSLWLAPNPSKRWGELFFLFYTPFWLTLCLGIVVPYKLYETFTELEYLLLALVSA 61
Query: 84 VPAFLIPMLFVGKADKGISWQDRYWVKSMDNNLQLC---------WELFLDSLL------ 128
VPAF+IPML VGKAD+ + W+DRYWVK+ NL + W + +L
Sbjct: 62 VPAFVIPMLLVGKADRSLCWKDRYWVKA---NLWIIVFSYVGNYFWTHYFFKVLGASYTF 118
Query: 129 -------------FHSSGCILYLSFMEN---------------------EQSYLETLAIS 154
F + C L+ N E +++ LA+S
Sbjct: 119 PSWKMNNVPHTTFFLTHVCFLFYHVASNITLRRLRHSTADLPDSLKWCFEAAWI--LALS 176
Query: 155 NFPYY------------EFVDRESMYKVGSLFYAIYFIVSFPMFLRIDEKPGDKWDLPRV 202
F Y EFVDR +MY+VG LFYAIYFIVSFPMF R+DEK D+WDL RV
Sbjct: 177 YFIAYLETIAIANFPYYEFVDRSAMYRVGCLFYAIYFIVSFPMFFRMDEKSTDEWDLSRV 236
Query: 203 AVDALGAAMLVTIILDLWRIFLGPIVPVSEAKQCSPTGLPWFS 245
AVDALGAAMLVTIILDLWR+FLGPIVP+ E + C +GLPWFS
Sbjct: 237 AVDALGAAMLVTIILDLWRLFLGPIVPLPEGQNCLQSGLPWFS 279
>gb|AAX94917.1| Similar to cycloeucalenol cycloisomerase (ec 5.5.1.9)
(cycloeucalenol--obtusifoliol isomerase) (cyclopropyl
sterol isomerase) [Oryza sativa (japonica
cultivar-group)]
Length = 326
Score = 236 bits (602), Expect = 4e-61
Identities = 139/299 (46%), Positives = 163/299 (54%), Gaps = 85/299 (28%)
Query: 30 SLWFAPNLSKRWGELFFLLYTPFWLSLSLGIVVPYKLYENFTELEYLLLGLISAVPAFLI 89
S W A + SKRWGE FFLLYTPFWL+L LG+VVP+KLYE FTELEYL++GL+S VPAF+I
Sbjct: 22 SRWMAADGSKRWGETFFLLYTPFWLTLCLGVVVPFKLYERFTELEYLVVGLVSTVPAFVI 81
Query: 90 PMLFVGKADKGISWQDRY------WVKSMDNNLQLCWELFLDSLLFHS------------ 131
P+ VGKAD S +DRY W+ W + ++L S
Sbjct: 82 PLFLVGKADSVRSLKDRYWVKANIWIIIFSYVGNYFWTHYFFTVLGASYTFPSWRMNNVP 141
Query: 132 -------SGCILYLSFMENEQ-------------------------------SYLETLAI 153
C L+ N +YLETLAI
Sbjct: 142 HTTFLLTHACFLFYHMTSNMSLRKLHHSTAHLPQFLRWSFEAAWVLALSYFIAYLETLAI 201
Query: 154 SN----------------------------FPYYEFVDRESMYKVGSLFYAIYFIVSFPM 185
+N FPYYEF+DR+ MYKVGSLFYAIYFIVSFPM
Sbjct: 202 ANKICGNAFQSGQIPLDRPSGYTTFEHWEKFPYYEFIDRDIMYKVGSLFYAIYFIVSFPM 261
Query: 186 FLRIDEKPGDKWDLPRVAVDALGAAMLVTIILDLWRIFLGPIVPVSEAKQCSPTGLPWF 244
F RIDE +KW L RVAVDALGAAMLVTIILDLWRIFLGPIVP+ E+++C GL WF
Sbjct: 262 FSRIDENE-EKWSLSRVAVDALGAAMLVTIILDLWRIFLGPIVPIPESRRCGQPGLAWF 319
>ref|XP_646934.1| hypothetical protein DDB0202318 [Dictyostelium discoideum]
gi|60475138|gb|EAL73074.1| hypothetical protein
DDB0202318 [Dictyostelium discoideum]
Length = 282
Score = 96.3 bits (238), Expect = 7e-19
Identities = 44/98 (44%), Positives = 64/98 (64%), Gaps = 3/98 (3%)
Query: 133 GCILYLSFMENEQSYLETLAISNFPYYEFVDRESMYKVGSLFYAIYFIVSFPMFLRIDE- 191
G IL + M +++E IS+ PYY DR MY +GS FY +YF++SFPMF RIDE
Sbjct: 170 GSILIVCAMAYFTAFMEAWTISSVPYYHIEDRTKMYTIGSTFYMLYFVISFPMFARIDET 229
Query: 192 --KPGDKWDLPRVAVDALGAAMLVTIILDLWRIFLGPI 227
K +KW++ R A++ LG+AM+V I+ D WR+ +G +
Sbjct: 230 GPKHNEKWNISRAALEGLGSAMIVFILCDFWRLAIGSL 267
Score = 75.9 bits (185), Expect = 1e-12
Identities = 33/80 (41%), Positives = 53/80 (66%), Gaps = 1/80 (1%)
Query: 32 WFAPNLSKRWGELFFLLYTPFWLSLSLGIVVPYKLYENFTELEYLLLGLISAVPAFLIPM 91
WF+ SKRW E FFL Y+P W+++ ++V Y++F ++E+ +LGL+ ++P + P+
Sbjct: 11 WFSKIPSKRWAEKFFLFYSPVWITV-FSLIVATGAYKSFYDVEFFILGLVVSLPCVIYPL 69
Query: 92 LFVGKADKGISWQDRYWVKS 111
LF ADKG +RYWVK+
Sbjct: 70 LFPCAADKGKPILERYWVKA 89
>gb|EAA76095.1| hypothetical protein FG06622.1 [Gibberella zeae PH-1]
gi|46124489|ref|XP_386798.1| hypothetical protein
FG06622.1 [Gibberella zeae PH-1]
Length = 269
Score = 63.2 bits (152), Expect = 7e-09
Identities = 33/84 (39%), Positives = 50/84 (59%), Gaps = 3/84 (3%)
Query: 144 EQSYLETLAISNFPYYEFVDRESMYKVGSLFYAIYFIVSFPMFLRIDEKPGDKWDLPRVA 203
E ++ + +S++ YYE DR M VGSL Y +FI PM RID K G+ W L RV
Sbjct: 189 ETFFMASPLLSDWFYYEKTDR--MMTVGSLGYMAFFITGLPMVGRIDAK-GEDWPLSRVV 245
Query: 204 VDALGAAMLVTIILDLWRIFLGPI 227
++ALG M + ++ ++W +GP+
Sbjct: 246 IEALGTFMSILVLFEVWAKVVGPL 269
Score = 45.1 bits (105), Expect = 0.002
Identities = 27/87 (31%), Positives = 43/87 (49%), Gaps = 7/87 (8%)
Query: 28 TPSLWFAPNLS----KRWGELFFLLYTPFWLSLSLGIVVPYKLYENFTELEYLLLGLISA 83
TPS++ A S KRW + L +PFW+ +++ + + ++ +++YLL + A
Sbjct: 6 TPSIYKATAQSIEAEKRWTQRLNLFLSPFWI-VAVAFAMLSGIMHSWNDIDYLLFSIAVA 64
Query: 84 VPAFLIPMLFVGKADKGISWQDRYWVK 110
P L P L K G W RYW K
Sbjct: 65 TPPALFPALLSKKI--GRPWYRRYWFK 89
>sp|P34847|NU1M_APILI NADH-ubiquinone oxidoreductase chain 1 (NADH dehydrogenase subunit
1) gi|829010|gb|AAB96810.1| NADH dehydrogenase subunit 1
[Apis mellifera ligustica]
gi|5834938|ref|NP_008094.1|ND1_10414 NADH dehydrogenase
subunit 1 [Apis mellifera ligustica]
Length = 305
Score = 38.1 bits (87), Expect = 0.23
Identities = 38/167 (22%), Positives = 69/167 (40%), Gaps = 33/167 (19%)
Query: 37 LSKRWGELFF-----LLYTP---FWLSLSLGIVVPYKLYENFTELEYLLLGLISAVPAFL 88
LSK W FF +Y+P F+LSL + I+ P+ + + E L + L+ + +
Sbjct: 57 LSKEW--FFFNYSNLFIYSPMLMFFLSLVMWILYPWFGFMYYIEFSILFMLLVLGLSVY- 113
Query: 89 IPMLFVGKADKGISWQDRYWVKSMDNNLQLCWELFLDSLLFHSSGCILYLSFMENEQSYL 148
P+LFVG W+ + + + L + F + L S M
Sbjct: 114 -PVLFVG------------WISNCNYAILGSMRLVSTMISFEINLFFLVFSLM------- 153
Query: 149 ETLAISNFPYYEFVDRESMYKVGSLFYAIYFIVSFPMFLRIDEKPGD 195
+ + +F + EF ++ K L Y +Y ++ M + ++ P D
Sbjct: 154 --MMVESFSFNEFFFFQNNIKFAILLYPLYLMMFTSMLIELNRTPFD 198
>ref|YP_028556.1| membrane protein, putative [Bacillus anthracis str. Sterne]
gi|49179231|gb|AAT54607.1| membrane protein, putative
[Bacillus anthracis str. Sterne]
Length = 182
Score = 35.8 bits (81), Expect = 1.1
Identities = 29/109 (26%), Positives = 55/109 (49%), Gaps = 15/109 (13%)
Query: 1 MVSSILFSKSLFCVSV------LEISSSSSASPTPSLWFAPNLSKRWGELFFLL-YTPFW 53
+V+S LF L V ++I S +P A N+ + +G+ +F+ +
Sbjct: 10 LVASYLFGNILTAYIVTKWRHNVDIRDEGSGNPG-----ARNMGRVYGKGYFVATFLGDA 64
Query: 54 LSLSLGIVVPYKLYENFTELEYLLLGLISAVPAFLIPMLFVGKADKGIS 102
+ ++ + + L+E+FT +++L L++ + + PMLF GK KGIS
Sbjct: 65 IKGAIVVSIAKYLFEDFT---FVMLTLLAVIMGHIYPMLFKGKGGKGIS 110
>ref|YP_019106.2| membrane protein, putative [Bacillus anthracis str. 'Ames
Ancestor'] gi|30262466|ref|NP_844843.1| membrane
protein, putative [Bacillus anthracis str. Ames]
gi|30257097|gb|AAP26329.1| membrane protein, putative
[Bacillus anthracis str. Ames]
gi|47551756|gb|AAT31581.2| membrane protein, putative
[Bacillus anthracis str. 'Ames Ancestor']
gi|65319760|ref|ZP_00392719.1| COG0344: Predicted
membrane protein [Bacillus anthracis str. A2012]
gi|46397155|sp|Q81QF8|Y2467_BACAN Hypothetical UPF0078
protein BA2467/GBAA2467/BAS2295
Length = 189
Score = 35.8 bits (81), Expect = 1.1
Identities = 29/109 (26%), Positives = 55/109 (49%), Gaps = 15/109 (13%)
Query: 1 MVSSILFSKSLFCVSV------LEISSSSSASPTPSLWFAPNLSKRWGELFFLL-YTPFW 53
+V+S LF L V ++I S +P A N+ + +G+ +F+ +
Sbjct: 17 LVASYLFGNILTAYIVTKWRHNVDIRDEGSGNPG-----ARNMGRVYGKGYFVATFLGDA 71
Query: 54 LSLSLGIVVPYKLYENFTELEYLLLGLISAVPAFLIPMLFVGKADKGIS 102
+ ++ + + L+E+FT +++L L++ + + PMLF GK KGIS
Sbjct: 72 IKGAIVVSIAKYLFEDFT---FVMLTLLAVIMGHIYPMLFKGKGGKGIS 117
>ref|ZP_00497909.1| COG1835: Predicted acyltransferases [Burkholderia pseudomallei S13]
Length = 372
Score = 34.7 bits (78), Expect = 2.5
Identities = 25/76 (32%), Positives = 34/76 (43%), Gaps = 6/76 (7%)
Query: 12 FCVSVLEISSSSSASPTPSLWFAPNLSKRWGELFFLLYTPFWLSLSLGIVVPYKLYENFT 71
FC+S + I SS +P A + FF LY FWLSL LG +V + L+
Sbjct: 61 FCISGMLIPSSLRGAP------AEGTRRFVVRRFFRLYPAFWLSLPLGYLVYWLLFGQRM 114
Query: 72 ELEYLLLGLISAVPAF 87
+ LL + AF
Sbjct: 115 DASGLLANVTMIPTAF 130
>ref|ZP_00240731.1| putative integral membrane protein [Bacillus cereus G9241]
gi|47553273|gb|EAL11664.1| putative integral membrane
protein [Bacillus cereus G9241]
Length = 182
Score = 34.7 bits (78), Expect = 2.5
Identities = 31/108 (28%), Positives = 52/108 (47%), Gaps = 13/108 (12%)
Query: 1 MVSSILFSKSLFCVSV------LEISSSSSASPTPSLWFAPNLSKRWGELFFLLYTPFWL 54
+V+S LF L V ++I S +P A N+ + +G+ +F+ T
Sbjct: 10 LVASYLFGNILTAYIVTKWRHNVDIRDEGSGNPG-----ARNMGRVYGKGYFVA-TFLGD 63
Query: 55 SLSLGIVVPYKLYENFTELEYLLLGLISAVPAFLIPMLFVGKADKGIS 102
++ IVV Y F + +L+L L++ + + P+LF GK KGIS
Sbjct: 64 AIKGAIVVSMAKYL-FEDSTFLMLALLAVIIGHIFPILFKGKGGKGIS 110
>ref|YP_036584.1| possible membrane protein [Bacillus thuringiensis serovar konkukian
str. 97-27] gi|49329266|gb|AAT59912.1| possible membrane
protein [Bacillus thuringiensis serovar konkukian str.
97-27]
Length = 182
Score = 33.9 bits (76), Expect = 4.3
Identities = 31/108 (28%), Positives = 52/108 (47%), Gaps = 13/108 (12%)
Query: 1 MVSSILFSKSLFCVSV------LEISSSSSASPTPSLWFAPNLSKRWGELFFLLYTPFWL 54
+V+S LF L V ++I S +P A N+ + +G+ +F+ T
Sbjct: 10 LVASYLFGNILTAYIVTKWRHNVDIRDEGSGNPG-----ARNMGRVYGKGYFVA-TFLGD 63
Query: 55 SLSLGIVVPYKLYENFTELEYLLLGLISAVPAFLIPMLFVGKADKGIS 102
++ IVV Y F + +L+L L++ + + P+LF GK KGIS
Sbjct: 64 AIKGAIVVSIAKYL-FEDSTFLMLTLLAVIMGHIYPILFKGKGGKGIS 110
>ref|ZP_00492994.1| COG1835: Predicted acyltransferases [Burkholderia pseudomallei
Pasteur] gi|67737990|ref|ZP_00488706.1| COG1835:
Predicted acyltransferases [Burkholderia pseudomallei
668] gi|67716007|ref|ZP_00485368.1| COG1835: Predicted
acyltransferases [Burkholderia pseudomallei 1710b]
gi|67681701|ref|ZP_00476026.1| COG1835: Predicted
acyltransferases [Burkholderia pseudomallei 1710a]
gi|67669810|ref|ZP_00466631.1| COG1835: Predicted
acyltransferases [Burkholderia pseudomallei 1655]
Length = 372
Score = 33.5 bits (75), Expect = 5.6
Identities = 24/76 (31%), Positives = 34/76 (44%), Gaps = 6/76 (7%)
Query: 12 FCVSVLEISSSSSASPTPSLWFAPNLSKRWGELFFLLYTPFWLSLSLGIVVPYKLYENFT 71
FC+S + I +S +P A + FF LY FWLSL LG +V + L+
Sbjct: 61 FCISGMLIPTSLRGAP------AEGTRRFVVRRFFRLYPAFWLSLPLGYLVYWLLFGQRM 114
Query: 72 ELEYLLLGLISAVPAF 87
+ LL + AF
Sbjct: 115 DASGLLANVTMIPTAF 130
>emb|CAH37098.1| putative acyltransferase [Burkholderia pseudomallei K96243]
gi|53720696|ref|YP_109682.1| putative acyltransferase
[Burkholderia pseudomallei K96243]
Length = 400
Score = 33.5 bits (75), Expect = 5.6
Identities = 24/76 (31%), Positives = 34/76 (44%), Gaps = 6/76 (7%)
Query: 12 FCVSVLEISSSSSASPTPSLWFAPNLSKRWGELFFLLYTPFWLSLSLGIVVPYKLYENFT 71
FC+S + I +S +P A + FF LY FWLSL LG +V + L+
Sbjct: 89 FCISGMLIPTSLRGAP------AEGTRRFVVRRFFRLYPAFWLSLPLGYLVYWLLFGQRM 142
Query: 72 ELEYLLLGLISAVPAF 87
+ LL + AF
Sbjct: 143 DASGLLANVTMIPTAF 158
>gb|AAV90190.1| conserved hypothetical protein [Zymomonas mobilis subsp. mobilis
ZM4] gi|59803117|sp|Q9RH13|Y1566_ZYMMO Hypothetical
UPF0060 protein ZMO1566 gi|56552462|ref|YP_163301.1|
hypothetical protein ZMO1566 [Zymomonas mobilis subsp.
mobilis ZM4]
Length = 107
Score = 33.5 bits (75), Expect = 5.6
Identities = 24/97 (24%), Positives = 48/97 (48%), Gaps = 5/97 (5%)
Query: 122 LFLDSLLFHSSGCILYLSFMENEQSYLETL-AISNFPYYE----FVDRESMYKVGSLFYA 176
L++ + L +GC + +++ +S L L I++ + F E+ K +++
Sbjct: 5 LYIPAALAEITGCFSFWAWIRLHKSPLWLLPGIASLLLFAWLLTFSPAENAGKAYAVYGG 64
Query: 177 IYFIVSFPMFLRIDEKPGDKWDLPRVAVDALGAAMLV 213
IY I+S +++ P D WDL A +GAA+++
Sbjct: 65 IYIIMSLLWSWKVEATPPDHWDLIGAAFCLVGAAIIL 101
>ref|ZP_00452229.1| COG1835: Predicted acyltransferases [Burkholderia mallei SAVP1]
gi|67645467|ref|ZP_00443755.1| COG1835: Predicted
acyltransferases [Burkholderia mallei NCTC 10247]
gi|67640667|ref|ZP_00439465.1| COG1835: Predicted
acyltransferases [Burkholderia mallei GB8 horse 4]
gi|67635074|ref|ZP_00434035.1| COG1835: Predicted
acyltransferases [Burkholderia mallei 10399]
gi|67628824|ref|ZP_00428684.1| COG1835: Predicted
acyltransferases [Burkholderia mallei 10229]
Length = 357
Score = 33.1 bits (74), Expect = 7.3
Identities = 24/76 (31%), Positives = 34/76 (44%), Gaps = 6/76 (7%)
Query: 12 FCVSVLEISSSSSASPTPSLWFAPNLSKRWGELFFLLYTPFWLSLSLGIVVPYKLYENFT 71
FC+S + I +S +P A + FF LY FWLSL LG +V + L+
Sbjct: 46 FCISGMLIPTSLRGAP------AEGTRRFVVRRFFRLYPAFWLSLLLGYLVYWLLFGQRM 99
Query: 72 ELEYLLLGLISAVPAF 87
+ LL + AF
Sbjct: 100 DASGLLANVTMIPTAF 115
>ref|YP_104329.1| acyltransferase family protein [Burkholderia mallei ATCC 23344]
gi|52427380|gb|AAU47973.1| acyltransferase family
protein [Burkholderia mallei ATCC 23344]
Length = 372
Score = 33.1 bits (74), Expect = 7.3
Identities = 24/76 (31%), Positives = 34/76 (44%), Gaps = 6/76 (7%)
Query: 12 FCVSVLEISSSSSASPTPSLWFAPNLSKRWGELFFLLYTPFWLSLSLGIVVPYKLYENFT 71
FC+S + I +S +P A + FF LY FWLSL LG +V + L+
Sbjct: 61 FCISGMLIPTSLRGAP------AEGTRRFVVRRFFRLYPAFWLSLLLGYLVYWLLFGQRM 114
Query: 72 ELEYLLLGLISAVPAF 87
+ LL + AF
Sbjct: 115 DASGLLANVTMIPTAF 130
>ref|NP_753044.1| Cytochrome BD-II oxidase subunit I [Escherichia coli CFT073]
gi|26107404|gb|AAN79587.1| Cytochrome BD-II oxidase
subunit I [Escherichia coli CFT073]
Length = 514
Score = 33.1 bits (74), Expect = 7.3
Identities = 31/149 (20%), Positives = 60/149 (39%), Gaps = 10/149 (6%)
Query: 36 NLSKRWGELF---FLLYTPFWLSLSLGIVVPYKLYENFTELEYLLLGLISAVPAFLIPML 92
++++ WG+LF F L L++ + Y N+ + + A+ AF +
Sbjct: 50 DMTRFWGKLFGINFALGVATGLTMEFQCGTNWSFYSNYVGDIFGAPLAMEALMAFFLEST 109
Query: 93 FVGKADKGISWQDRY------WVKSMDNNLQLCWELFLDSLLFHSSGCILYLSFMENEQS 146
FVG G ++Y W+ + +NL W L + + H +G + + E +
Sbjct: 110 FVGLFFFGWQRLNKYQHLLVTWLVAFGSNLSALWILNANGWMQHPTGAHFDIDTLRMEMT 169
Query: 147 YLETLAISNFPYYEFVDR-ESMYKVGSLF 174
L + +FV + Y G++F
Sbjct: 170 SFSELVFNPVSQVKFVHTVMAGYVTGAMF 198
>ref|ZP_00368564.1| sodium- and chloride-dependent transporter [Campylobacter lari
RM2100] gi|57018956|gb|EAL55729.1| sodium- and
chloride-dependent transporter [Campylobacter lari
RM2100]
Length = 450
Score = 33.1 bits (74), Expect = 7.3
Identities = 33/115 (28%), Positives = 49/115 (41%), Gaps = 5/115 (4%)
Query: 66 LYENFTELEYLLLGLISAVPAFLIPMLFVG--KADKGISWQDRYWVKSMDNNLQLCWELF 123
L+ N + +L LG I A+ AF I + F G A I Y + S N L
Sbjct: 291 LFSNIDPIGFLPLGNILAI-AFFIALFFAGITSAVSMIEPLTFYLINSF--NFTRKKALI 347
Query: 124 LDSLLFHSSGCILYLSFMENEQSYLETLAISNFPYYEFVDRESMYKVGSLFYAIY 178
+ + + G + LS +E + LE S F +F+ M +G LF AI+
Sbjct: 348 FIAFIVYFLGSLCILSGIEWSKDSLEFFGKSFFDILDFIASNLMMPLGGLFGAIF 402
>ref|NP_793251.1| monovalent cation:proton antiporter, putative [Pseudomonas syringae
pv. tomato str. DC3000] gi|28853880|gb|AAO56946.1|
monovalent cation:proton antiporter, putative
[Pseudomonas syringae pv. tomato str. DC3000]
Length = 972
Score = 32.7 bits (73), Expect = 9.6
Identities = 27/97 (27%), Positives = 46/97 (46%), Gaps = 7/97 (7%)
Query: 44 LFFLLYTPFWLSLSLGIVVPYKLYENFTELEYLLLGLISAVPAFLIPMLFVGKADKGISW 103
L LL PF + L ++P+ N E L GL++ + A + + F AD G+
Sbjct: 3 LIVLLLLPF-IGSCLAAILPH----NARNAESFLAGLVALIGAIQMALWFPQIADGGVIR 57
Query: 104 QDRYWVKSMDNNLQLCWELF--LDSLLFHSSGCILYL 138
++ +W+ S+ N L + F L S+L G ++ L
Sbjct: 58 EEYFWLPSLGMNFVLRIDGFAWLFSMLVLGIGALVSL 94
>gb|AAN67843.1| multicomponent potassium-proton antiporter, subunit A/B, putative
[Pseudomonas putida KT2440] gi|26988954|ref|NP_744379.1|
multicomponent potassium-proton antiporter, subunit A/B,
putative [Pseudomonas putida KT2440]
Length = 971
Score = 32.7 bits (73), Expect = 9.6
Identities = 29/97 (29%), Positives = 46/97 (46%), Gaps = 7/97 (7%)
Query: 44 LFFLLYTPFWLSLSLGIVVPYKLYENFTELEYLLLGLISAVPAFLIPMLFVGKADKGISW 103
L LL PF + L V+P+ N E +L GL++ V + +L+ A G+
Sbjct: 3 LIVLLLLPF-VGSCLAAVLPH----NARNAESILAGLVALVGTVQVALLYPQVAHGGVIR 57
Query: 104 QDRYWVKSMDNNLQLCWELF--LDSLLFHSSGCILYL 138
++ W+ S+ NL L + F L SLL G ++ L
Sbjct: 58 EEFLWLPSLGMNLVLRMDGFAWLFSLLVLGIGALVSL 94
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.326 0.141 0.449
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 442,215,989
Number of Sequences: 2540612
Number of extensions: 18646033
Number of successful extensions: 51881
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 18
Number of HSP's that attempted gapping in prelim test: 51857
Number of HSP's gapped (non-prelim): 37
length of query: 250
length of database: 863,360,394
effective HSP length: 125
effective length of query: 125
effective length of database: 545,783,894
effective search space: 68222986750
effective search space used: 68222986750
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 73 (32.7 bits)
Medicago: description of AC126782.8


