

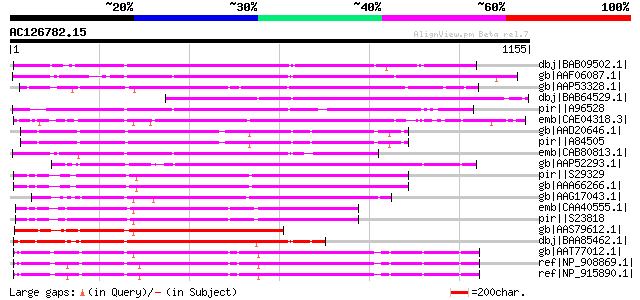
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126782.15 + phase: 0
(1155 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAB09502.1| transposon protein-like [Arabidopsis thaliana] 827 0.0
gb|AAF06087.1| Similar to gi|4325351 T25H8.2 TNP2 protein homolo... 756 0.0
gb|AAP53328.1| putative TNP2-like transposon protein [Oryza sati... 694 0.0
dbj|BAB64529.1| TdcA1-ORF2 [Daucus carota] 677 0.0
pir||A96528 protein F27J15.14 [imported] - Arabidopsis thaliana ... 655 0.0
emb|CAE04318.3| OSJNBb0016D16.9 [Oryza sativa (japonica cultivar... 646 0.0
gb|AAD20646.1| putative TNP2-like transposon protein [Arabidopsi... 640 0.0
pir||A84505 probable TNP2-like transposon protein [imported] - A... 640 0.0
emb|CAB80813.1| putative transposon protein [Arabidopsis thalian... 627 e-178
gb|AAP52293.1| putative transposable element [Oryza sativa (japo... 626 e-177
pir||S29329 hypothetical protein 1 - maize transposon En-1 gi|22... 612 e-173
gb|AAA66266.1| unknown protein 612 e-173
gb|AAG17043.1| transposase [Zea mays] 605 e-171
emb|CAA40555.1| TNP2 [Antirrhinum majus] 588 e-166
pir||S23818 hypothetical protein Tnp2 - garden snapdragon transp... 588 e-166
gb|AAS79612.1| putative tnp2 transposase [Ipomoea trifida] 572 e-161
dbj|BAA85462.1| transposon-like ORF [Brassica rapa] 568 e-160
gb|AAT77012.1| putative Transposase family tnp2 protein [Oryza s... 544 e-153
ref|NP_908869.1| B1151H08.16 [Oryza sativa (japonica cultivar-gr... 544 e-153
ref|NP_915890.1| putative retrotransposable elements TNP2 [Oryza... 544 e-153
>dbj|BAB09502.1| transposon protein-like [Arabidopsis thaliana]
Length = 1089
Score = 827 bits (2137), Expect = 0.0
Identities = 461/1058 (43%), Positives = 624/1058 (58%), Gaps = 62/1058 (5%)
Query: 8 RRWMYDRTFPGRTGLKPQFVEGVDGFISWAWEQEICRDEGGIRCPCLKCRCRHIITDPGD 67
R WMY+R P + F EG+D F+ +A Q + + + CPC+KC +
Sbjct: 11 RHWMYERIDPTTNRVSQAFYEGLDNFLKFAKSQPLFLENKKLFCPCIKCGNGGRQQEEHI 70
Query: 68 VKKHLKKVGFMPNYWVWTYNGEMFQNFGAGVNAQASTSHGGTNVETNVEYNEDVNTDQFN 127
V HL GFM NYWVWT +GE + N N++ T ++ E Y E
Sbjct: 71 VATHLFNKGFMLNYWVWTSHGEDY-NMLINNNSEEDTVRFESHTEPVDPYVE-------- 121
Query: 128 MMDDMVADALGVELSYGEDVEDDEEEQPPNEKAQRFYRLLSESNTPLYEGSSH--SKLSM 185
MV+DA G E D E+ PN +A++FY +L + P+Y+G SKLS+
Sbjct: 122 ----MVSDAFGST----ESEFDQMREEDPNFEAKKFYDILDAAKQPIYDGCKEGLSKLSL 173
Query: 186 CVWLLSHKSNYLNPDDGMDDITKMLKAVTPFKDNLPMNYHAAKRLVMKLGLSVKKIDCCR 245
L+S K++ + MD I ++++ P +N P +Y+ K+L+ LGL +KID C+
Sbjct: 174 AARLMSLKTDNNLSQNCMDSIAQIMQEYLPEGNNSPKSYYEIKKLMRSLGLPYQKIDVCQ 233
Query: 246 NGCMLFYDNEFGINDGPLEECKFCQSPRYGISK---QKRVAVKSMFYLPIIPRLQRLFAS 302
+ CM+F+ E C FC+ RY ++ QK + + MFYLPI RL+RL+ S
Sbjct: 234 DNCMIFWKET-----EKEEYCLFCKKDRYRPTQKIGQKSIPYRQMFYLPIADRLKRLYQS 288
Query: 303 MKTASQMTWHHSNGIS-GVMRHPSDGEAWKHFDRVHPDFAADPRNVRLGLCSDGFQPYVQ 361
TA M WH + S G M HPSDGEAWKHF +VHPDFA+ PRNV LGLC+DGF P+
Sbjct: 289 HNTAKHMRWHAEHLASDGEMGHPSDGEAWKHFHKVHPDFASKPRNVYLGLCTDGFNPFGM 348
Query: 362 SSAKPYSCWPIIVTPYNLPPEMCMTKPYLFLSCIVPGPDNPKAGIDVFLQPLIDDLKRLW 421
S YS WP+I+TPYNLPPEMCM + ++FL+ +VPGP++PK +D+FLQPLI++LK LW
Sbjct: 349 SGHN-YSLWPVILTPYNLPPEMCMKQEFMFLTILVPGPNHPKRSLDIFLQPLIEELKDLW 407
Query: 422 V-GELTYDISRKQNFKLRAALMWTINDFPAYGMLSGWSTHGRFACPHCMENTKAFTLESG 480
V G YDIS KQNF L+ LMWTI+DFPAYGMLSGW+THGR ACP+C + T AF L++G
Sbjct: 408 VNGVEAYDISTKQNFLLKVVLMWTISDFPAYGMLSGWTTHGRLACPYCSDQTGAFWLKNG 467
Query: 481 GKSSWFDCHRPFLPHNHPFRRLKNGFKKDERVFVGPPPKITSEEVWTRVCDYPKVTDY-G 539
K+ WFDCHR FLP NH +R K FKK + V P +T EE++ VC PK D G
Sbjct: 468 RKTCWFDCHRCFLPVNHSYRGNKKDFKKGKVVEDSKPEILTGEELYNEVCCLPKTVDCGG 527
Query: 540 RAVPIPRYGVDHHWTKRSIFWDLPYWKDNLLRHNLDVMHIEKNVFDNIFNTVMDVKGKTK 599
+ YG H+W K+SI W+L YWKD LRHNLDVMHIEKNV DN T+++V+GKTK
Sbjct: 528 NHGRLEGYGKTHNWHKQSILWELSYWKDLKLRHNLDVMHIEKNVLDNFIKTLLNVQGKTK 587
Query: 600 DNEKARKDLQIYCKRKDLELKPQPNGKLLKPKANYNLSAQEAKLVCQWL-KDLRMPDGYS 658
DN K+R DLQ C RKDL L P+ GK PK + L ++ +WL KD++ DGYS
Sbjct: 588 DNIKSRLDLQENCNRKDLHLTPE--GKAPIPK--FRLKPDAKEIFLRWLEKDVKFSDGYS 643
Query: 659 SNIGRCADVNTGRLHGMKSHDSHIFMENFIPIAFSSL-PKHVLDPLIEISQFFKNLCAST 717
S++ C D+ +L GMKSHD H+ M+ PIAF+ L K V D L +S FF++LCA T
Sbjct: 644 SSLANCVDLPGRKLTGMKSHDCHVLMQRLFPIAFAELMDKSVHDALSSVSLFFRDLCART 703
Query: 718 LRVDELEKMEKNMPIIICQLEQIFPPGFFDSMEHLPVHLAYEAILGGPVQYRWMYPFERF 777
+ D +E ++KN+ +++C LE+IFPP FFD MEHL +HL +EA LGGPVQ+RWMY FER+
Sbjct: 704 IHKDGIEMLQKNICMVLCNLEKIFPPSFFDVMEHLLIHLPHEASLGGPVQFRWMYCFERY 763
Query: 778 MGDSKRAVKNLAKVEGSIVAIYTAKETTYFIGHYFVDRLLTPSNTRNEVQVNDLRDPST- 836
MG K+ VKN AKVEGSIV Y +E + F +YF DR + N + + T
Sbjct: 764 MGHLKKKVKNKAKVEGSIVEQYINEEISTFCTYYF-DRHIKTKNRAGDRHYDGGNQEDTH 822
Query: 837 -----LSVFNLDGRHAGKVLQYWMVDQKEMRSAHVHVLINCAEVKPYLEEFIAFYAEFGE 891
+F+ GR +GK + W+ D K+ AH ++L NC +++P+ F
Sbjct: 823 EFDGVPDIFSQAGRDSGKESEIWLQD-KDYHIAHRYILRNCDQLRPFERLFDESLIAANP 881
Query: 892 GSTVGSIHE----YFPAWFKQRVYNAEPTDEVIRLRQLSQGPLQCANEFHTYFVNGYKFH 947
G + ++E + +W K + N+ P L + GP+ + YF GY FH
Sbjct: 882 GISEKDLNELREKQYSSWLK-KYDNSYPE----WLLSIVHGPMVKVTCWPMYFCRGYIFH 936
Query: 948 THSWTEGKKTINSGVYVKGVTDGG---EDDFYGVIKHIYELSYN---DNKVVLFYCDWFD 1001
T+ + KK N GV VKG T E DFYG+++ IYEL Y D KVV+F CDW+D
Sbjct: 937 TYDHGKNKKNANYGVCVKGTTSSSSNEEADFYGILREIYELHYPGHVDLKVVVFKCDWYD 996
Query: 1002 PS-PRWTKINKLCNTVDIRVDKKYKEYDPFIMAHNVRQ 1038
R + NK +DI + Y+EYDPF++A Q
Sbjct: 997 SKVGRGIRRNK-SGIIDINAKRHYEEYDPFVLASQADQ 1033
>gb|AAF06087.1| Similar to gi|4325351 T25H8.2 TNP2 protein homolog from Arabidopsis
thaliana BAC gb|AF128394 gi|25518667|pir||F86485
hypothetical protein F28J9.11 [imported] - Arabidopsis
thaliana
Length = 1121
Score = 756 bits (1953), Expect = 0.0
Identities = 448/1164 (38%), Positives = 628/1164 (53%), Gaps = 95/1164 (8%)
Query: 6 YYRRWMYDRTFPGRTGLKPQFVEGVDGFISWAWEQEICRDEGG-IRCPCLKCRCRHIITD 64
+YR WMY R L ++V GV+ F+++A Q I + G CPC C+ I
Sbjct: 13 FYRDWMYKRFDEVTGNLSAEYVAGVEQFMTFANSQPIVQSSRGKFHCPCSVCKNEKHIIS 72
Query: 65 PGDVKKHLKKVGFMPNYWVWTYNGEMFQNFGAGVNAQASTSHGGTNVETNVEYNEDVNTD 124
V HL GFMP+Y+VW +GE +N TS+ TN E +E+V
Sbjct: 73 GKRVSSHLFSHGFMPDYYVWYKHGEE-------MNMDIGTSY--TNRTYFSENHEEVGNI 123
Query: 125 QFNMMDDMVADALGVELSYGEDVEDDEE----EQPPNEKAQRFYRLLSESNTPLYEGSSH 180
+ DMV DA + Y ++ D+ E+P + +FY LL ++N PLY+
Sbjct: 124 VEDPYVDMVNDAFNFNVGYDDNYRHDDSYQNVEEPVRNHSNKFYDLLEDANNPLYD---- 179
Query: 181 SKLSMCVWLLSHKSNYLNPDDGMDDITKMLKAVTPFKDNLPMNYHAAKRLVMKLGLSVKK 240
D + + +A T +++ ++L+ LGL
Sbjct: 180 -----------------------DFLPEGNQATT--------SHYQTEKLMRNLGLPYHT 208
Query: 241 IDCCRNGCMLFYDNEFGINDGPLEECKFCQSPRYGISKQKR---VAVKSMFYLPIIPRLQ 297
ID C+N CML + D ++C+FC + R+ +R V M+YLPI RL+
Sbjct: 209 IDVCQNNCMLSWKE-----DEKEDQCRFCGAKRWKPKDDRRRTKVPYSRMWYLPIGDRLK 263
Query: 298 RLFASMKTASQMTWHHSN-GISGVMRHPSDGEAWKHFDRVHPDFAADPRNVRLGLCSDGF 356
R++ S KTA+ M WH + G M H SD W++F +HP+FA +PRNV LGLC+DGF
Sbjct: 264 RMYQSHKTAAAMRWHAEHQSKEGEMNHLSDAAEWRYFQGLHPEFAEEPRNVYLGLCTDGF 323
Query: 357 QPYVQSSAKPYSCWPIIVTPYNLPPEMCMTKPYLFLSCIVPGPDNPKAGIDVFLQPLIDD 416
P+ S + +S WP+I+TPYNLPP MCM YLFL+ + GP++P+A +DVFLQPLI++
Sbjct: 324 NPFGMS--RNHSLWPVILTPYNLPPGMCMNTEYLFLTILNSGPNHPRASLDVFLQPLIEE 381
Query: 417 LKRLW-VGELTYDISRKQNFKLRAALMWTINDFPAYGMLSGWSTHGRFACPHCMENTKAF 475
LK W G YD+S QNF L+A L+WTI+DFPAY ML GW+THG+ +CP CME+TK+F
Sbjct: 382 LKEFWSTGVDAYDVSLSQNFNLKAVLLWTISDFPAYNMLLGWTTHGKLSCPVCMESTKSF 441
Query: 476 TLESGGKSSWFDCHRPFLPHNHPFRRLKNGFKKDERVFVG-PPPKITSEEVWTR---VCD 531
L +G K+ WFDCHR FLPH HP RR + F K PP +T E+V+ +
Sbjct: 442 YLPNGRKTCWFDCHRRFLPHGHPSRRNRKDFLKGRDASSEYPPESLTGEQVYYERLASVN 501
Query: 532 YPKVTDYG---RAVPIPRYGVDHHWTKRSIFWDLPYWKDNLLRHNLDVMHIEKNVFDNIF 588
PK D G + YG +H+W K SI W+L YWKD LRHN+DVMH EKN DNI
Sbjct: 502 PPKTKDVGGNGHEKKMRGYGKEHNWHKESILWELSYWKDLNLRHNIDVMHTEKNFLDNIM 561
Query: 589 NTVMDVKGKTKDNEKARKDLQIYCKRKDLELKPQPNGKLLKPKANYNLSAQEAKLVCQWL 648
NT+M VKGK+KDN +R D++ +C R L + +GK P Y L+ + + + Q +
Sbjct: 562 NTLMRVKGKSKDNIMSRLDIEKFCSRPGLHI--DSSGKA--PFPAYTLTEEAKQSLLQCV 617
Query: 649 K-DLRMPDGYSSNIGRCADVNTGRLHGMKSHDSHIFMENFIPIAFSSL-PKHVLDPLIEI 706
K D+R PDGYSS++ C D++ G+ GMKSHD H+FME +P F+ L ++V L I
Sbjct: 618 KYDIRFPDGYSSDLASCVDLDNGKFSGMKSHDCHVFMERLLPFIFAELLDRNVHLALSGI 677
Query: 707 SQFFKNLCASTLRVDELEKMEKNMPIIICQLEQIFPPGFFDSMEHLPVHLAYEAILGGPV 766
FF++LC+ TL+ ++ +++N+ +IIC E+IFPP FFD MEHLP+HL YEA LGGPV
Sbjct: 678 GAFFRDLCSRTLQTSRVQILKQNIVLIICNFEKIFPPSFFDVMEHLPIHLPYEAELGGPV 737
Query: 767 QYRWMYPFERFMGDSKRAVKNLAKVEGSIVAIYTAKETTYFIGHYFVDRLLTPSN-TR-N 824
QYRWMYPFERF K KN GSIV Y E YF HYF D + T S TR N
Sbjct: 738 QYRWMYPFERFFKKLKGKAKNKRYAAGSIVESYINDEIAYFSEHYFADHIQTKSRLTRFN 797
Query: 825 EVQVNDLRDPSTLSVFNLDGRHAGKVLQYWMVDQKEMRSAHVHVLINCAEVKPY---LEE 881
E +V ++F GR +G++ W++ +K+ +SAH +VL NC KP+ E+
Sbjct: 798 EGEVPVYHVSGVPNIFMHVGRPSGEMHVDWLL-EKDYQSAHAYVLRNCDYFKPFESMFED 856
Query: 882 FI-AFYAEFGEGSTVGSIHEYFPAWFKQRVYNAEPTDEV-IRLRQLSQGPLQCANEFHTY 939
++ A Y E E + W K+ V T ++++ QGPL + Y
Sbjct: 857 YLSAKYPCLPEKELYARRAEEYHLWVKEYVTYWNTTSPFPTWVQEIVQGPLNKVKTWPMY 916
Query: 940 FVNGYKFHTHSWTEGKKTINSGVYVKG---VTDGGEDDFYGVIKHIYELSYN---DNKVV 993
F GY FHT + G+KT N G+ VKG E DFYG + I EL Y + ++
Sbjct: 917 FTRGYLFHTQNHGAGRKTCNYGICVKGENYADSSDEADFYGTLTDIIELEYEGIVNLRIT 976
Query: 994 LFYCDWFDPSPRWTKINKLCNTVDIRVDKKYKEYDPFIMAHNVRQVYYVPYPPTLPRKRG 1053
LF C W+DP VDI +KY +Y+PFI+ QV Y+PYP T K
Sbjct: 977 LFKCKWYDPKIGRGTRRSHSGVVDILSTRKYNKYEPFILGSQADQVCYIPYPYTKKPKNI 1036
Query: 1054 WSVAIKTKPRGRIETE-ERNEEVAYQADD-----MTHVNEIIEVDQVTSWQDSQVEGDQV 1107
W +K PRG I E E N+ Q ++ +T + +++ V + ++ D
Sbjct: 1037 WLNVLKVNPRGNISGEYENNDPTLLQTENDDDVLLTTIEDLVLETPVANLNPIILDYDVG 1096
Query: 1108 DPSIL-EMRNEVEDENEDVVDDNN 1130
D E R + +ED V+D +
Sbjct: 1097 DAEPEDEFRCNLSSLDEDEVEDED 1120
>gb|AAP53328.1| putative TNP2-like transposon protein [Oryza sativa (japonica
cultivar-group)] gi|37533478|ref|NP_921041.1| putative
TNP2-like transposon protein [Oryza sativa (japonica
cultivar-group)] gi|20279447|gb|AAM18727.1| putative
TNP2-like transposon protein [Oryza sativa (japonica
cultivar-group)]
Length = 1571
Score = 694 bits (1790), Expect = 0.0
Identities = 411/1056 (38%), Positives = 598/1056 (55%), Gaps = 75/1056 (7%)
Query: 22 LKPQFVEGVDGFISWAWEQEICRDEGGIRCPCLKCRCRHIITDPGDVKKHLKKVGFMPNY 81
L +++EGV+ FI +A+ + IRCPC+KC C V HLK G + +Y
Sbjct: 15 LNHEYLEGVEKFIDYAFSK--LDGVQVIRCPCIKC-CNTYSLPHHIVSSHLKAYGILRSY 71
Query: 82 WVWTYNGEMFQNFGAGVNAQASTSHGGTNVETNVEYNEDVNTDQFNMMDDMVADAL---- 137
W ++GE+ + ++E + +++ N + +N M D++ D
Sbjct: 72 TFWYHHGEVL-------------AEQENDIEVDEYDSQEFNGEGYNGMQDLMEDLFPQCN 118
Query: 138 ---GVELSYGEDVEDDEEEQPPNEKAQRFYRLLSESNTPLYEGSSHSKLSMCVWLLSHKS 194
G E ++ + EEE PN A +FY LL + N PLYEGS SKLS V LL K+
Sbjct: 119 TPGGDEATHESTDQGPEEE--PNTDAAKFYALLDKYNQPLYEGSRISKLSALVNLLHVKN 176
Query: 195 NYLNPDDGMDDITKML-KAVTPFKDNLPMNYHAAKRLVMKLGLSVKKIDCCRNGCMLFYD 253
+ + ++L K P LP +Y+ AK+ + +LGLS KID C+N CML++
Sbjct: 177 LGKWSNKSFTALLELLRKDFLPHDSTLPNSYYEAKKTIRELGLSYIKIDACKNDCMLYWK 236
Query: 254 NEFGINDGPLEECKFCQSPRYG---------ISKQKRVAVKSMFYLPIIPRLQRLFASMK 304
+ E CK C S R+ S K++ K+M Y PI PRLQRLF K
Sbjct: 237 EDIDA-----ESCKVCGSSRWKEKHTREIKHSSAGKKIPHKTMRYFPIKPRLQRLFMCRK 291
Query: 305 TASQMTWHHSNGISG-VMRHPSDGEAWKHFDRVHPDFAADPRNVRLGLCSDGFQPYVQSS 363
TA+ WH + G VMRHP+D +AWK FD++H FA++PRNVRL L SDGFQP+
Sbjct: 292 TAAYAKWHKECRVDGGVMRHPADSKAWKEFDKIHSSFASEPRNVRLCLASDGFQPFANMR 351
Query: 364 AKPYSCWPIIVTPYNLPPEMCMTKPYLFLSCIVPGPDNPKAGIDVFLQPLIDDLKRLWV- 422
YS WP+ + P NLPP MC+ + + LS ++PGPD P IDV+LQPLI++L LW
Sbjct: 352 TS-YSIWPVFLVPLNLPPWMCIKQQNVMLSMLLPGPDGPGDAIDVYLQPLIEELIELWED 410
Query: 423 GELTYDISRKQNFKLRAALMWTINDFPAYGMLSGWSTHGRFACPHCMENTKAFTLESGGK 482
G TYD S K NFKLRAAL+WT++DFPAYG +SG ST G+ ACP C + T + L+ G K
Sbjct: 411 GVDTYDSSTKTNFKLRAALLWTVHDFPAYGNISGHSTKGKLACPVCHKETNSMWLKKGQK 470
Query: 483 SSWFDCHRPFLPHNHPFRRLKNGFK--KDERVFVGPPPKITSEEVWTRVCDYPKVT---D 537
+ HR FL +H +R + F K+ R GPP +++ ++V +V D + D
Sbjct: 471 FCYMG-HRCFLRRSHRWRNDRTSFDGTKESR---GPPMELSGDDVLKQVQDLQGIILSKD 526
Query: 538 YGRAVPIPRYGVDHHWTKRSIFWDLPYWKDNLLRHNLDVMHIEKNVFDNIFNTVMDVKGK 597
+ + R +W KRSIF++LPY+K LLRHNLDVMHIEKN+ D+I T+M+VKGK
Sbjct: 527 MSKKTKVSRSDRGDNWKKRSIFFELPYFKTLLLRHNLDVMHIEKNICDSIIGTLMNVKGK 586
Query: 598 TKDNEKARKDLQIYCKRKDLELKPQPNGKLLKPKANYNLSAQEAKLVCQWLKDLRMPDGY 657
TKDN R DL++ R DL GK++ P A Y LS + K + + K+L++PDG+
Sbjct: 587 TKDNINTRSDLKLMNLRPDLH-PVDDGGKVVLPPAPYTLSPDQKKALLSFFKELKVPDGF 645
Query: 658 SSNIGRCADVNTGRLHGMKSHDSHIFMENFIPIAFSSL-PKHVLDPLIEISQFFKNLCAS 716
SSNI RC ++ ++ G+KSHD H+ + + +P+A L P+++ +PL+E+SQFFK L +
Sbjct: 646 SSNISRCVNMKERKMSGLKSHDCHVILNHILPLALRGLLPENIYEPLVELSQFFKKLNSK 705
Query: 717 TLRVDELEKMEKNMPIIICQLEQIFPPGFFDSMEHLPVHLAYEAILGGPVQYRWMYPFER 776
L V++LE+M+ +P+I+C+LE+ FPP FFD M HLPVHLA EA++GGP YRWMY FER
Sbjct: 706 ALSVEQLEEMDAQIPVILCKLEKEFPPSFFDVMMHLPVHLAREALIGGPTMYRWMYLFER 765
Query: 777 FMGDSKRAVKNLAKVEGSIVAIYTAKETTYFIGHYFVDRLLTPSNTRNEVQVNDLRDPST 836
+ K V+N+A+ EGSI Y A E Y +D + T N + + +
Sbjct: 766 QIHYLKSLVRNMARPEGSIAEGYIADEFMTLSSRY-LDDVETKHNRPGRIHDTSIGNKFN 824
Query: 837 LSVFNLDGRHAGKVLQYWMVDQKEMRSAHVHVLINCAEVKPYLEEFIAFYAEFGEGSTV- 895
LS+F+ GR G + +D E AH+++L NC EV+ Y+ E Y EGS++
Sbjct: 825 LSIFSCAGRPIGS-RKTRDLDMLESEQAHIYILRNCDEVQAYISE----YCNSTEGSSMV 879
Query: 896 ---GSIHEYFPAWFKQRVYNAEPTD--EVIR-LRQLSQGPLQCANEFHTYFVNGYKFHTH 949
+ ++ F WFK+++Y D EV+ L LS+GPL+ F Y +NG+++ T
Sbjct: 880 QFTSTWNKKFINWFKEKIYQQHQHDKSEVMEDLLSLSRGPLKNITCFTGYDMNGFRYRTE 939
Query: 950 SWTEGKKTINSGVYVKGVTDGGEDDFYGVIKHIYELSY-NDNKVVLFYCDWFD--PSPRW 1006
+ T NSGV V G + ++YGV+ I E+ Y +V LF C+W D R
Sbjct: 940 RRDSRRCTQNSGVMVL----GDDVEYYGVLTEILEIQYLGGRRVPLFRCNWVDVFNKDRG 995
Query: 1007 TKINKLCNTVDIRVDKKYKEYDPFIMAHNVRQVYYV 1042
+I+K + + + + +PF++A QV++V
Sbjct: 996 IRIDKY-GLTSVNLQRLLQTDEPFVLASQAAQVFFV 1030
>dbj|BAB64529.1| TdcA1-ORF2 [Daucus carota]
Length = 812
Score = 677 bits (1746), Expect = 0.0
Identities = 370/828 (44%), Positives = 495/828 (59%), Gaps = 41/828 (4%)
Query: 348 RLGLCSDGFQPYVQSSAKPYSCWPIIVTPYNLPPEMCMTKPYLFLSCIVPGPDNPKAGID 407
RLGL +DGFQP+ SS K YS WPIIVTPYNLPP MC + Y+FLS +VPGP NPK ID
Sbjct: 1 RLGLSADGFQPF-GSSGKQYSSWPIIVTPYNLPPWMCSKEEYMFLSILVPGPRNPKQKID 59
Query: 408 VFLQPLIDDLKRLW-VGELTYDISRKQNFKLRAALMWTINDFPAYGMLSGWSTHGRFACP 466
VFLQPLI +LK LW VG T+D S KQNF++RAALMWTI+DFPAY MLSGW T G ACP
Sbjct: 60 VFLQPLISELKMLWEVGVETWDTSLKQNFQMRAALMWTISDFPAYSMLSGWKTAGHLACP 119
Query: 467 HCMENTKAFTLESGGKSSWFDCHRPFLPHNHPFRRLKNGFKKDERVFVGPPPKITSEEVW 526
HC A+ L+ GGK +WFD HR FLP NHPFR+ KN F K + V PPP T E+V
Sbjct: 120 HCAHEHDAYNLKHGGKPTWFDNHRKFLPANHPFRKNKNWFTKGKVVSEFPPPIRTGEDVL 179
Query: 527 TRV--CDYPKVTDYG----RAVPIPRYGVDHHWTKRSIFWDLPYWKDNLLRHNLDVMHIE 580
+ K+T+ G A I Y W KRSIFWDLPYW+ +RHNLDVMHIE
Sbjct: 180 QEIESLGLMKITELGSEEHNAKIIKTYNCG--WKKRSIFWDLPYWRTLSIRHNLDVMHIE 237
Query: 581 KNVFDNIFNTVMDVKGKTKDNEKARKDLQIYCKRKDLELKPQPNGKLLKPKANYNLSAQE 640
KNVF+NIFNT+M ++GKTKDN KAR D+ + C+R +L + PKA Y+L +
Sbjct: 238 KNVFENIFNTIMAIEGKTKDNAKARADIALLCRRPELAIDESTRKY---PKACYSLDKKG 294
Query: 641 AKLVCQWLKDLRMPDGYSSNIGRCADVNTGRLHGMKSHDSHIFMENFIPIAFSS-LPKHV 699
+ VC+WL+DL+ PDGY SN+GRC D+ +L GMKSHD H+FM+ +PIAF LP +V
Sbjct: 295 KEAVCKWLQDLKFPDGYVSNMGRCIDMKKYKLFGMKSHDCHVFMQRLMPIAFREFLPNNV 354
Query: 700 LDPLIEISQFFKNLCASTLRVDELEKMEKNMPIIICQLEQIFPPGFFDSMEHLPVHLAYE 759
+ + E+S FFK+L ++TL+V ++ ++E+ +P+I+C+LE+IFPP FDSMEHL VHL YE
Sbjct: 355 WEAVTELSLFFKDLTSATLKVSDMCRLEEQIPVILCKLERIFPPALFDSMEHLLVHLPYE 414
Query: 760 AILGGPVQYRWMYPFERFMGDSKRAVKNLAKVEGSIVAIYTAKETTYFIGHYFVDRLLTP 819
A + GPVQYRWMYPFERF+ K +KN A+VEGS+ Y +E + F HYF + T
Sbjct: 415 ARIAGPVQYRWMYPFERFLRHLKNNIKNKARVEGSMCNAYLVEEASTFCSHYFEQHVQTK 474
Query: 820 SN--TRNEVQVNDLRDPSTLSVFNLDGRHAGKVLQYWMVDQKEMRSAHVHVLINCAEVKP 877
RN+ S+F+ GR +G +D +E +AH +VL+NC EV P
Sbjct: 475 HRKVPRNDSSGGGENFEGNFSIFSHPGRASG-CANVRYLDDREYMAAHNYVLLNCPEVAP 533
Query: 878 YLEEFIAFYAEFGEGSTVGSIHEY----FPAWFKQRVYNAEPTDEVIRLRQLSQGPLQCA 933
Y E FI E + T I + F WFKQ N I +R ++ GPL+
Sbjct: 534 YTEIFINMVRENNQSITDAEIDKCLESDFALWFKQYAQNPSLVPNEI-VRDIASGPLRSV 592
Query: 934 NEFHTYFVNGYKFHTHSWTEGKKTINSGVYVKGVT-DGGEDDFYGVIKHIYELSYND--- 989
++VNGYKFHT + + T NSGV +KG +D++G+I I L Y
Sbjct: 593 RSVPIFYVNGYKFHTRKYGANRSTFNSGVCIKGSNYSETSNDYFGIIDEILILEYPRLPI 652
Query: 990 NKVVLFYCDWFDPSPR-WTKINKLCNTVDIRVDKKYKEYDPFIMAHNVRQVYYVPYPPTL 1048
K LF C+WFDP+P T+++ V++ KK Y+PFI+A QVY+ YP
Sbjct: 653 KKTTLFKCEWFDPTPSVGTRVHLRFKMVEVNRKKKLSVYEPFILASQAMQVYFCNYPSLR 712
Query: 1049 PRKRGWSVAIKTKPRGRIETEE--RNEEVAYQADDMTHVNEIIEVDQVTSWQDSQVEGDQ 1106
K W V K K R +E + ++ + YQ + ++N I D T D++
Sbjct: 713 RDKMDWLVVCKIKARPLVELSQASQSHQEPYQDETPENLNRIDIRDIPTHLNDNE----- 767
Query: 1107 VDPSILEMRNEVEDENEDVVDDNNEGDHQDNQVDDGDVDEYNYASDNH 1154
IL++ + E++ D++ + + +Q D D +NY SD++
Sbjct: 768 ---GILDLDDGEGSPEEEIELDSDSEEGRSSQSD----DIHNYGSDSN 808
>pir||A96528 protein F27J15.14 [imported] - Arabidopsis thaliana
gi|7770347|gb|AAF69717.1| F27J15.14 [Arabidopsis
thaliana]
Length = 1526
Score = 655 bits (1691), Expect = 0.0
Identities = 392/1065 (36%), Positives = 577/1065 (53%), Gaps = 118/1065 (11%)
Query: 6 YYRRWMYDRTFPGRTG-LKPQFVEGVDGFISWAWEQEICRDEGGIRCPCLKCRCRHIITD 64
+ R WMY R F TG L ++V GV+ F+++A Q I + CR
Sbjct: 13 FNRDWMYKR-FDEMTGNLSAEYVAGVEEFLTFANSQPIVQS------------CR----- 54
Query: 65 PGDVKKHLKKVGFMPNYWVWTYNGEMFQNFGAGVNAQASTSHGGTNVETNVEYNEDVNTD 124
+Y+VW +GE F N +++ N E+ ED D
Sbjct: 55 ---------------DYYVWYMHGEDFNMNVGTSNYVIDSTYLRENYESVGNVVEDPYVD 99
Query: 125 QFNMMDD----MVADA----LGVELSYGEDVEDDEEEQPPNEKAQRFYRLLSESNTPLYE 176
N+++D MV DA +G + +Y +D + E+P + +++FY LL + PLY+
Sbjct: 100 VGNVVEDPYVDMVNDAFRYNVGFDDNYHQDGTNQNVEEPVHNHSKKFYDLLEGAQNPLYD 159
Query: 177 G--SSHSKLSMCVWLLSHKSNYLNPDDGMDDITKMLKAVTPFKDNLPMNYHAAKRLVMKL 234
G S+LS+ ++ +K+++ + +D + +ML P + +++ ++L+ L
Sbjct: 160 GCRQGQSQLSLAARVMQNKADHNMSERCVDSVYQMLTDFLPEGNQATDSHYKTEKLMCNL 219
Query: 235 GLSVKKIDCCRNGCMLFYDNEFGINDGPLEECKFCQSPRY----GISKQKRVAVKSMFYL 290
GL ID C N CM+F+ D ++C+FC + R+ ++ +V M YL
Sbjct: 220 GLPYYTIDVCINNCMIFWKE-----DEKEDKCRFCSAQRWKPMDDYRRRTKVPYSRMRYL 274
Query: 291 PIIPRLQRLFASMKTASQMTWHHSN-GISGVMRHPSDGEAWKHFDRVHPDFAADPRNVRL 349
PI RL+R++ S KTA+ M WH + M HPSD W++F +HP FA +PRNV L
Sbjct: 275 PIGDRLKRMYQSHKTAAAMRWHAEHQSKEEEMNHPSDAAEWRYFQELHPMFAEEPRNVYL 334
Query: 350 GLCSDGFQPYVQSSAKPYSCWPIIVTPYNLPPEMCMTKPYLFLSCIVPGPDNPKAGIDVF 409
GLC+DGF P+ S + +S WP+I+TPYNLPP MCM YLFL+ + GP++P+ +DVF
Sbjct: 335 GLCTDGFNPFGMS--RNHSLWPVILTPYNLPPGMCMNIEYLFLTILNSGPNHPRGSLDVF 392
Query: 410 LQPLIDDLKRLW-VGELTYDISRKQNFKLRAALMWTINDFPAYGMLSGWSTHGRFACPHC 468
LQPLI++LK LW G YD+S QNF L+A L+WTI+DFPAY MLSGW+THG+ +CP C
Sbjct: 393 LQPLIEELKELWSTGIDAYDVSLNQNFNLKAVLLWTISDFPAYSMLSGWTTHGKLSCPIC 452
Query: 469 MENTKAFTLESGGKSSWFDCHRPFLPHNHPFRRLKNGFKKDERVFVG-PPPKITSEEVWT 527
ME+T +F L +G K+ WFDCHR FL H HP R+ K F+K + PP +T E+++
Sbjct: 453 MESTNSFYLPNGRKTCWFDCHRRFLSHGHPLRKNKKDFRKGKDASTEYPPESLTGEQIYY 512
Query: 528 RVC---DYPKVTDYG---RAVPIPRYGVDHHWTKRSIFWDLPYWKDNLLRHNLDVMHIEK 581
+ P+ D G +P YG +H+W K SI W+LPYWKD LRH +DVMH EK
Sbjct: 513 ERLSGVNPPRTKDVGGNGHEKKMPGYGKEHNWHKESILWELPYWKDLNLRHCIDVMHTEK 572
Query: 582 NVFDNIFNTVMDVKGKTKDNEKARKDLQIYCKRKDLELKPQPNGKLLKPKANYNLSAQEA 641
N DNI NT+M VKGK+KDN AR D++ +C R DL + + K P Y L+ +
Sbjct: 573 NFLDNIMNTLMSVKGKSKDNIMARMDIERFCSRPDLHI----DSKRKAPFPAYTLTNEAK 628
Query: 642 KLVCQWLKD-LRMPDGYSSNIGRCADVNTGRLHGMKSHDSHIFMENFIPIAFSSLPKHVL 700
+ Q +K ++ PDGYSS++ C D+ G+L + + H+ +
Sbjct: 629 MSLLQCVKHAIKFPDGYSSDLSSCVDMENGKLSELLDRNVHLALSG-------------- 674
Query: 701 DPLIEISQFFKNLCASTLRVDELEKMEKNMPIIICQLEQIFPPGFFDSMEHLPVHLAYEA 760
+ FF++LC+ TL+ ++ +++N+ +IIC LE+IFPP FFD MEHLP+HL YEA
Sbjct: 675 -----VGAFFRDLCSRTLQKSHVQILKQNIVLIICNLEKIFPPSFFDVMEHLPIHLPYEA 729
Query: 761 ILGGPVQYRWMYPFERFMGDSKRAVKNLAKVEGSIVAIYTAKETTYFIGHYFVDRLLTPS 820
LGGPVQYRWMYPFERF K KN GSIV Y E +YF HYF D + T S
Sbjct: 730 ELGGPVQYRWMYPFERFFKKLKGKAKNKRYAAGSIVESYINDEISYFSEHYFADDIQTKS 789
Query: 821 N-TR-NEVQVNDLRDPSTLSVFNLDGRHAGKVLQYWMVDQKEMRSAHVHVLINC---AEV 875
TR NE +V P ++F+ GR +G++ + W+ +++ + AH +V+ NC +
Sbjct: 790 RLTRFNEGEVPVYHVPGVPTIFSSVGRPSGEIREVWL-SEEDYQCAHGYVIRNCDYFQVI 848
Query: 876 KPYLEEFIAF-YAEFGEGSTVGSIHEYFPAWFKQ-RVYNAEPTDEVIRLRQLSQGPLQCA 933
+ E+F++ Y E +E + W N PT ++++ GPL
Sbjct: 849 ESMFEDFLSIKYPGLNEKELFVKRNEEYHVWVTYWNSSNPFPT----WVQEIVNGPL--- 901
Query: 934 NEFHTYFVNGYKFHTHSWTEGKKTINSGVYVKG--VTDGGE-DDFYGVIKHIYELSYN-- 988
++ T+ ++G G+KT N GV VKG TD + DFYG + I EL Y
Sbjct: 902 HKVKTWPIHG---------AGRKTCNYGVCVKGENYTDASDAADFYGNLTDIIELEYEGV 952
Query: 989 -DNKVVLFYCDWFDPSPRWTKINKLCNTVDIRVDKKYKEYDPFIM 1032
K+ LF C W+DP VD+ +KY +Y+PFI+
Sbjct: 953 VSLKITLFKCSWYDPKLGRGTRRSNSGVVDVLSSRKYNKYEPFIL 997
>emb|CAE04318.3| OSJNBb0016D16.9 [Oryza sativa (japonica cultivar-group)]
gi|50928325|ref|XP_473690.1| OSJNBb0016D16.9 [Oryza
sativa (japonica cultivar-group)]
Length = 1097
Score = 646 bits (1667), Expect = 0.0
Identities = 410/1180 (34%), Positives = 597/1180 (49%), Gaps = 128/1180 (10%)
Query: 8 RRWMYDRTFPGRTGLKPQFVEGVDGFISWAWEQEICRDEGGIRCPCLKCRCRHIITDP-- 65
R WMY+ R + ++ F+ A+ ++ D G+ CP + C H T
Sbjct: 5 RSWMYNGRRK-RNQVSADWILKTTDFLDRAFSRDTSAD--GVMCPFVTCE--HTKTQAHT 59
Query: 66 ---GDVKKHLKKVGFMPNYWVWTYNGEMFQNFGAGVNAQASTSHGGTNVETNVEYNEDVN 122
+ KHL K GF P Y VW Y+GE + + +V ++ED +
Sbjct: 60 KQWDTMMKHLFKHGFRPEYTVWVYHGE------------SDSDPTRDDVLRQRTFDEDGD 107
Query: 123 TDQFNMMDDMVADALGVELSYGEDVEDDEEEQPPNEKAQRFYRLLSESNTPLYEGSSHSK 182
+ + MDDMV D +S EE+ P A+ FY +L+ SN L+ + S+
Sbjct: 108 AE-VHRMDDMVDDVRDAHISV--------EEEEPEPIARAFYEMLTASNQSLHAHTEVSQ 158
Query: 183 LSMCVWLLSHKSNYLNPDDGMDDITKMLKAVTPFKDNLPMNYHAAKRLVMKLGLSVKKID 242
L LL+ KS + G D + + A+ P LP+N + AK+ + L + +KID
Sbjct: 159 LDAITRLLAVKSQFSISIAGFDALLNVFGALLPQGHKLPLNLYEAKKFLSALTMPYEKID 218
Query: 243 CCRNGCMLFYDNEFGINDGPLEECKFCQSPRY-------GISKQKRVAVKSMFYLPIIPR 295
C CMLF + + C C RY G KQ + +K + Y+P IPR
Sbjct: 219 VCPKYCMLFREE-----NSEKTHCDKCGESRYVEVQNSYGEKKQLNIPMKVLRYIPFIPR 273
Query: 296 LQRLFASMKTASQMTWH------HSNGISGVMRHPSDGEAWKHFDRVHPDFAADPRNVRL 349
LQRL+ S A QMTWH H N I H +D EAWK FDR +FA+D RNVR+
Sbjct: 274 LQRLYMSEPQAKQMTWHKYGHRYHPNKIV----HTADAEAWKQFDRDFSEFASDARNVRI 329
Query: 350 GLCSDGFQPYVQSSAKPYSCWPIIVTPYNLPPEMCMTKPYLFLSCIVPGPDNPKAGIDVF 409
+ +DGF P+ +A Y+CWP+ V P NLPP +CM K +FLS I+PGPD P I ++
Sbjct: 330 AIATDGFNPFGMGAAS-YTCWPVFVIPLNLPPGVCMQKHNMFLSLIIPGPDYPGKKISMY 388
Query: 410 LQPLIDDLKRLWV-GELTYDISRKQNFKLRAALMWTINDFPAYGMLSGWSTHGRFACPHC 468
++PL+DDL W G TYD + KQNF +R + +++ +D PAYG+ GW HG+ CP C
Sbjct: 389 MEPLVDDLLHAWEHGVQTYDRATKQNFNMRVSYLFSFHDLPAYGIFCGWCVHGKMPCPVC 448
Query: 469 MENTKAFTLESGGKSSWFDCHRPFLPHNHPFRRLKNGFKKDERVFVGPPPKITSEEVWTR 528
ME K L+ GGK S+FDCHR FLPH H FR N F + V PP + +EEV R
Sbjct: 449 MEVLKGRRLKFGGKYSFFDCHRQFLPHGHIFRNDPNSFLANTTVTTEPPHRFKTEEVHVR 508
Query: 529 VCDYPKVTDYGRAVPIPRYGVDHHWTKRSIFWDLPYWKDNLLRHNLDVMHIEKNVFDNIF 588
+ ++ YG DH+WT W LPY+ +L HN+DVMH EKNV + IF
Sbjct: 509 L---QRLQPAANGEGFEGYGEDHNWTHIPGLWRLPYFHKLVLPHNIDVMHNEKNVAEAIF 565
Query: 589 NTVMDVKGKTKDNEKARKDLQIYCKRKDLEL-KPQPNGKLLKPKANYNLSAQEAKLVCQW 647
NT D+ KTKDN KAR D I C R DL + + Q +G+ LKP+A++ L+ + K + +W
Sbjct: 566 NTCFDIPEKTKDNVKARLDQAILCNRPDLNMIRRQTSGQWLKPRADFCLNRTQKKEILEW 625
Query: 648 LKDLRMPDGYSSNIGRCADVNTGRLHGMKSHDSHIFMENFIPIAFSS-LPKHVLDPLIEI 706
+ L+ PDGY SN+ R + R++G+KSHD HI ME +P+ F ++ + + E+
Sbjct: 626 FQTLKFPDGYGSNLRRGVNFKKMRINGLKSHDYHIMMERLLPVMFRGYFQPYLWEVIAEL 685
Query: 707 SQFFKNLCASTLRVDELEKMEKNMPIIICQLEQIFPPGFFDSMEHLPVHLAYEAILGGPV 766
S F++ LCA + ELE ME +P+++C+LE IFP GFF+ M+HL +HL YEA +GGPV
Sbjct: 686 SFFYRKLCAKEVDPIELESMELQVPVLLCKLEMIFPSGFFNPMQHLILHLPYEARMGGPV 745
Query: 767 QYRWMYPFERFMGDSKRAVKNLAKVEGSIVAIYTAKETTYFIGHYFVDRLLTPSNTRNEV 826
QYRWMYP ER D K VKN A+VE SI Y E F YF D++ T N
Sbjct: 746 QYRWMYPGERDQKDLKSKVKNRARVEASIAEAYILDEIANFTTIYFADQVYTVHNPVPRY 805
Query: 827 QVNDLRDPSTLSVFNLDGRHAGKVLQYWMVDQKEMRSAHVHVLINCAEVKPYLEEFIAFY 886
V +LS+F++ G + + + + E +A ++VL N EV Y+ +F
Sbjct: 806 NVTVESRECSLSLFSIKGDSTSRGVTRHLT-EVEWEAAMLYVLTNLPEVDDYVGKF---- 860
Query: 887 AEFGEGSTVGSIHEYFPAWFKQRVYNAEPTDEVIRLRQLSQGPLQCANEFHTYFVNGYKF 946
+HE W ++ EPT + + L G F T+F
Sbjct: 861 -----------LHE---EWSRR----GEPTRQQ-KENLLRNGARNGCPNFVTWF------ 895
Query: 947 HTHSWTEGKKTINSGVYVKGVTDGGEDDFYGVIKHIYELSYNDNK---VVLFYCDWFDPS 1003
+ + TI G + D+YG IK I ELS++ + +VLF C WFDP+
Sbjct: 896 ----YQQCLVTIGQG------ENSDITDYYGYIKEIIELSFHGDSELTLVLFNCHWFDPA 945
Query: 1004 PRWTKINKLCNTVDIRVDKKYKEYDPFIMAHNVRQVYYVPYP-PTLPRKRGWSVAIKTKP 1062
T+ V++ Y+PF++AH QVYY+PYP ++P W V K +P
Sbjct: 946 Q--TRYTPQYGLVEVAHSSTLAVYEPFVVAHQATQVYYIPYPCKSVPALIDWWVVYKVQP 1003
Query: 1063 RGRIETE--------ERNEEVAYQADD---MTHVNEIIE----VDQVTSWQDSQVEGDQV 1107
G+I N+ V Y +D T V +++E V + T+ + D+
Sbjct: 1004 TGKIAAPVDKDYDFLPNNDNVHYFQEDGLQGTFVVDLMEGLHNVSEATTRDAEGIVNDK- 1062
Query: 1108 DPSILEMRNEVEDENEDVVDDNNEGDHQDNQVDDGDVDEY 1147
D +L N ++ D+++ + +++ + +DEY
Sbjct: 1063 DLQLLNGSNVLQQ------DESDSSEFDEDEEEGPIIDEY 1096
>gb|AAD20646.1| putative TNP2-like transposon protein [Arabidopsis thaliana]
Length = 1040
Score = 640 bits (1651), Expect = 0.0
Identities = 357/890 (40%), Positives = 523/890 (58%), Gaps = 64/890 (7%)
Query: 25 QFVEGVDGFISWAWEQEICRDEGGIRCPCLKCRCRHIITDPGDVKKHLKKVGFMPNYWVW 84
++ + + GF+ + R+ G I CPC KC + + VKKHL GFMPNY+VW
Sbjct: 187 EYYDELRGFMYQTTNEPFARENGTILCPCRKCLNEKYL-EFNVVKKHLYNRGFMPNYYVW 245
Query: 85 TYNGEMFQNFGAGVNAQASTSHGGTNVETNVEYNED---VNTDQFNMMDDMVADALGVEL 141
+G++ + V +S+ + + D +Q N DMV +A +
Sbjct: 246 LRHGDVVEIAHELVIQIMVSSNLVILIMMRIMQVPDQFGYGYNQENRFHDMVTNAFHETI 305
Query: 142 -SYGEDVEDDEEEQPPNEKAQRFYRLLSESNTPLYEG--SSHSKLSMCVWLLSHKSNYLN 198
S+ E++ ++ PN AQ FY +L +N P+YEG +SKLS+ +++ K++
Sbjct: 306 ASFPENISEE-----PNVDAQHFYDMLDAANQPIYEGCREGNSKLSLASRMMTIKADNNL 360
Query: 199 PDDGMDDITKMLKAVTPFKDNLPMNYHAAKRLVMKLGLSVKKIDCCRNGCMLFYDNEFGI 258
+ MD +++K P + +Y+ ++LV GL + ID C + CM+F+ ++
Sbjct: 361 SEKCMDSWAELIKEYLPPDNISAESYYEIQKLVSSHGLPSEMIDVCIDYCMIFWGDDVN- 419
Query: 259 NDGPLEECKFCQSPRYGISKQK-RVAVKSMFYLPIIPRLQRLFASMKTASQMTWHHSNGI 317
L+EC+FC PRY + + RV K M+YLPI RL+RL+ S +TA++M WH +
Sbjct: 420 ----LQECRFCGKPRYQTTGGRTRVPYKRMWYLPITDRLKRLYQSERTAAKMRWHAQHTT 475
Query: 318 S-GVMRHPSDGEAWKHFDRVHPDFAADPRNVRLGLCSDGFQPYVQSSAKPYSCWPIIVTP 376
+ G + HPSD +AWKHF V+PDFA + RNV LGLC+DGF P+ + YS WP+I+TP
Sbjct: 476 ADGEITHPSDAKAWKHFQTVYPDFANECRNVYLGLCTDGFSPF-GLCGRQYSLWPVILTP 534
Query: 377 YNLPPEMCMTKPYLFLSCIVPGPDNPKAGIDVFLQPLIDDLKRLWV-GELTYDISRKQNF 435
Y LPP+MCM ++FLS +VPGP +PK +DVFLQPLI +LK+LW G T+D S KQNF
Sbjct: 535 YKLPPDMCMQSEFMFLSILVPGPKHPKRALDVFLQPLIHELKKLWYEGVHTFDYSSKQNF 594
Query: 436 KLRAALMWTINDFPAYGMLSGWSTHGRFACPHCMENTKAFTLESGGKSSWFDCHRPFLPH 495
+RA LMWTI+DFPAY MLSGW+ HGR P K WF+CHR FLP
Sbjct: 595 NMRAMLMWTISDFPAYRMLSGWTMHGRLYVP-----------TKDRKPCWFNCHRRFLPL 643
Query: 496 NHPFRRLKNGFKKDERVFVGPPPKITSEEVWTRVCDY------PKVTDYGRAVPIP-RYG 548
+HP+RR K F+K++ V + PP ++ +++ ++ Y + ++ +P YG
Sbjct: 644 HHPYRRNKKLFRKNKIVRLLPPSYVSGTDLFEQIDYYGAQETCKRGGNWHTTANMPDEYG 703
Query: 549 VDHHWTKRSIFWDLPYWKDNLLRHNLDVMHIEKNVFDNIFNTVMDVKGKTKDNEKARKDL 608
H+W K+SIFW LPYWKD LLRHNL VMHIEKN FDNI NT+++++GKTKD K+R DL
Sbjct: 704 STHNWYKQSIFWQLPYWKDLLLRHNLYVMHIEKNFFDNIMNTLLNLQGKTKDTLKSRLDL 763
Query: 609 QIYCKRKDLELKPQPNGKLLKPKANYNLSAQEAKLVCQWL-KDLRMPDGYSSNIGRCADV 667
C + +L + +GKL P + + LS + K + +W+ +++ PDGY S RC +
Sbjct: 764 AEICSKPELHVTR--DGKL--PVSKFRLSNEAKKALFEWVVAEVKFPDGYVSKFSRCVEQ 819
Query: 668 NTGRLHGMKSHDSHIFMENFIP-IAFSSLPKHVLDPLIEISQFFKNLCASTLRVDELEKM 726
+ GMKSHD H+FM+ +P + LP ++ + + I FFK+LC TL D +E++
Sbjct: 820 GQ-KFSGMKSHDCHVFMQRLLPFVLVELLPTNIHEAIAGIGVFFKDLCTRTLTTDTIEQL 878
Query: 727 EKNMPIIICQLEQIFPPGFFDSMEHLPVHLAYEAILGGPVQYRWMYPFERFMGDSKRAVK 786
+KN+P+++C LE++F P FFD MEHLP+HL +E LGGPVQ+RWMYPFERFM K K
Sbjct: 879 DKNIPVLLCNLEKLFSPAFFDVMEHLPIHLPHEVALGGPVQFRWMYPFERFMKSLKGKAK 938
Query: 787 NLAKVEGSIVAIYTAKETTYFIGHYFVDRLLTPSNTRNEVQVNDLRDPSTLSVFNLD--- 843
NLA+VEGSIVA +ET++F +YF +PS + + D +N+
Sbjct: 939 NLARVEGSIVAGSLTEETSHFTSYYF-----SPSVRTKKTRPRCYDDGGVAPTYNVPDVP 993
Query: 844 ------GRHAGKVLQYWMVDQKEMRSAHVHVLINCAEVKPYLEEFIAFYA 887
GR AGK+ + W D R+AH ++L N Y+++F + A
Sbjct: 994 DTFAQIGRLAGKLKEVWWDDHTLSRAAHNYILRNV----DYIQKFERYKA 1039
>pir||A84505 probable TNP2-like transposon protein [imported] - Arabidopsis
thaliana
Length = 1040
Score = 640 bits (1651), Expect = 0.0
Identities = 357/890 (40%), Positives = 523/890 (58%), Gaps = 64/890 (7%)
Query: 25 QFVEGVDGFISWAWEQEICRDEGGIRCPCLKCRCRHIITDPGDVKKHLKKVGFMPNYWVW 84
++ + + GF+ + R+ G I CPC KC + + VKKHL GFMPNY+VW
Sbjct: 187 EYYDELRGFMYQTTNEPFARENGTILCPCRKCLNEKYL-EFNVVKKHLYNRGFMPNYYVW 245
Query: 85 TYNGEMFQNFGAGVNAQASTSHGGTNVETNVEYNED---VNTDQFNMMDDMVADALGVEL 141
+G++ + V +S+ + + D +Q N DMV +A +
Sbjct: 246 LRHGDVVEIAHELVIQIMVSSNLVILIMMRIMQVPDQFGYGYNQENRFHDMVTNAFHETI 305
Query: 142 -SYGEDVEDDEEEQPPNEKAQRFYRLLSESNTPLYEG--SSHSKLSMCVWLLSHKSNYLN 198
S+ E++ ++ PN AQ FY +L +N P+YEG +SKLS+ +++ K++
Sbjct: 306 ASFPENISEE-----PNVDAQHFYDMLDAANQPIYEGCREGNSKLSLASSMMTIKADNNL 360
Query: 199 PDDGMDDITKMLKAVTPFKDNLPMNYHAAKRLVMKLGLSVKKIDCCRNGCMLFYDNEFGI 258
+ MD +++K P + +Y+ ++LV GL + ID C + CM+F+ ++
Sbjct: 361 SEKCMDSWAELIKEYLPPDNISAESYYEIQKLVSSHGLPSEMIDVCIDYCMIFWGDDVN- 419
Query: 259 NDGPLEECKFCQSPRYGISKQK-RVAVKSMFYLPIIPRLQRLFASMKTASQMTWHHSNGI 317
L+EC+FC PRY + + RV K M+YLPI RL+RL+ S +TA++M WH +
Sbjct: 420 ----LQECRFCGKPRYQTTGGRTRVPYKRMWYLPITDRLKRLYQSERTAAKMRWHAQHTT 475
Query: 318 S-GVMRHPSDGEAWKHFDRVHPDFAADPRNVRLGLCSDGFQPYVQSSAKPYSCWPIIVTP 376
+ G + HPSD +AWKHF V+PDFA + RNV LGLC+DGF P+ + YS WP+I+TP
Sbjct: 476 ADGEITHPSDAKAWKHFQTVYPDFANECRNVYLGLCTDGFSPF-GLCGRQYSLWPVILTP 534
Query: 377 YNLPPEMCMTKPYLFLSCIVPGPDNPKAGIDVFLQPLIDDLKRLWV-GELTYDISRKQNF 435
Y LPP+MCM ++FLS +VPGP +PK +DVFLQPLI +LK+LW G T+D S KQNF
Sbjct: 535 YKLPPDMCMQSEFMFLSILVPGPKHPKRALDVFLQPLIHELKKLWYEGVHTFDYSSKQNF 594
Query: 436 KLRAALMWTINDFPAYGMLSGWSTHGRFACPHCMENTKAFTLESGGKSSWFDCHRPFLPH 495
+RA LMWTI+DFPAY MLSGW+ HGR P K WF+CHR FLP
Sbjct: 595 NMRAMLMWTISDFPAYRMLSGWTMHGRLYVP-----------TKDRKPCWFNCHRRFLPL 643
Query: 496 NHPFRRLKNGFKKDERVFVGPPPKITSEEVWTRVCDY------PKVTDYGRAVPIP-RYG 548
+HP+RR K F+K++ V + PP ++ +++ ++ Y + ++ +P YG
Sbjct: 644 HHPYRRNKKLFRKNKIVRLLPPSYVSGTDLFEQIDYYGAQETCKRGGNWHTTANMPDEYG 703
Query: 549 VDHHWTKRSIFWDLPYWKDNLLRHNLDVMHIEKNVFDNIFNTVMDVKGKTKDNEKARKDL 608
H+W K+SIFW LPYWKD LLRHNL VMHIEKN FDNI NT+++++GKTKD K+R DL
Sbjct: 704 STHNWYKQSIFWQLPYWKDLLLRHNLYVMHIEKNFFDNIMNTLLNLQGKTKDTLKSRLDL 763
Query: 609 QIYCKRKDLELKPQPNGKLLKPKANYNLSAQEAKLVCQWL-KDLRMPDGYSSNIGRCADV 667
C + +L + +GKL P + + LS + K + +W+ +++ PDGY S RC +
Sbjct: 764 AEICSKPELHVTR--DGKL--PVSKFRLSNEAKKALFEWVVAEVKFPDGYVSKFSRCVEQ 819
Query: 668 NTGRLHGMKSHDSHIFMENFIP-IAFSSLPKHVLDPLIEISQFFKNLCASTLRVDELEKM 726
+ GMKSHD H+FM+ +P + LP ++ + + I FFK+LC TL D +E++
Sbjct: 820 GQ-KFSGMKSHDCHVFMQRLLPFVLVELLPTNIHEAIAGIGVFFKDLCTRTLTTDTIEQL 878
Query: 727 EKNMPIIICQLEQIFPPGFFDSMEHLPVHLAYEAILGGPVQYRWMYPFERFMGDSKRAVK 786
+KN+P+++C LE++F P FFD MEHLP+HL +E LGGPVQ+RWMYPFERFM K K
Sbjct: 879 DKNIPVLLCNLEKLFSPAFFDVMEHLPIHLPHEVALGGPVQFRWMYPFERFMKSLKGKAK 938
Query: 787 NLAKVEGSIVAIYTAKETTYFIGHYFVDRLLTPSNTRNEVQVNDLRDPSTLSVFNLD--- 843
NLA+VEGSIVA +ET++F +YF +PS + + D +N+
Sbjct: 939 NLARVEGSIVAGSLTEETSHFTSYYF-----SPSVRTKKTRPRCYDDGGVAPTYNVPDVP 993
Query: 844 ------GRHAGKVLQYWMVDQKEMRSAHVHVLINCAEVKPYLEEFIAFYA 887
GR AGK+ + W D R+AH ++L N Y+++F + A
Sbjct: 994 DTFAQIGRLAGKLKEVWWDDHTLSRAAHNYILRNV----DYIQKFERYKA 1039
>emb|CAB80813.1| putative transposon protein [Arabidopsis thaliana]
gi|4325351|gb|AAD17349.1| similar to Antirrhinum majus
(garden snapdragon) TNP2 protein (GB:X57297)
[Arabidopsis thaliana] gi|25407250|pir||D85049 probable
transposon protein [imported] - Arabidopsis thaliana
Length = 817
Score = 627 bits (1618), Expect = e-178
Identities = 351/842 (41%), Positives = 478/842 (56%), Gaps = 65/842 (7%)
Query: 6 YYRRWMYDRTFPGRTGLKPQFVEGVDGFISWAWEQEICRD-EGGIRCPCLKCRCRHIITD 64
+YR WMY R L ++V GV+ F+++A Q I + G CPC C+ I
Sbjct: 13 FYRDWMYKRFDEVTGNLSAEYVAGVEEFMTFANSQPIVQSCRGKFYCPCSVCKNEKHIIS 72
Query: 65 PGDVKKHLKKVGFMPNYWVWTYNGEMFQNFGAGVNAQASTSHGGTNVETNVEYNEDVNTD 124
V HL FMP+Y+VW +GE +N TS+ T+ E +E+V
Sbjct: 73 GRRVSSHLFSHEFMPDYYVWYKHGEE-------LNMDIGTSY--TDRTYFSENHEEVGNV 123
Query: 125 QFNMMDDMVADALGVELSYGEDVEDDEE----------EQPPNEKAQRFYRLLSESNTPL 174
+ DMV DA + Y ++V D+ E+P + +FY LL +N PL
Sbjct: 124 VEDPYVDMVNDAFNFNVGYDDNVGHDDNYHHDGSYQNVEEPVRNHSNKFYDLLEGANNPL 183
Query: 175 YEG--SSHSKLSMCVWLLSHKSNYLNPDDGMDDITKMLKAVTPFKDNLPMNYHAAKRLVM 232
Y+G S+LS+ L+ +K+ Y + +D + +M A P + +++ ++L+
Sbjct: 184 YDGCREGQSQLSLASRLMHNKAEYNMSEKLVDSVCEMFTAFLPEGNQATTSHYQTEKLMR 243
Query: 233 KLGLSVKKIDCCRNGCMLFYDNEFGINDGPLEECKFCQSPRYGISKQKR---VAVKSMFY 289
LGL ID +N CMLF+ D ++C+FC + R+ +R V M+Y
Sbjct: 244 NLGLPYHTIDVYKNNCMLFWKE-----DEKEDQCRFCGAQRWKPKDDRRRTKVPYSRMWY 298
Query: 290 LPIIPRLQRLFASMKTASQMTWHHSN-GISGVMRHPSDGEAWKHFDRVHPDFAADPRNVR 348
LPI RL+R++ S KTA+ M WH + G M HPSD W++F +HP FA +P NV
Sbjct: 299 LPIADRLKRMYQSHKTAAAMRWHAEHQSKEGEMNHPSDAAEWRYFQELHPRFAEEPHNVY 358
Query: 349 LGLCSDGFQPYVQSSAKPYSCWPIIVTPYNLPPEMCMTKPYLFLSCIVPGPDNPKAGIDV 408
LGL +DGF P+ S + +S WP+I+TPYNLPP MCM YLFL+ + GP++P+A +DV
Sbjct: 359 LGLYTDGFNPFGMS--RNHSLWPVILTPYNLPPGMCMNTEYLFLTILNSGPNHPRASLDV 416
Query: 409 FLQPLIDDLKRLW-VGELTYDISRKQNFKLRAALMWTINDFPAYGMLSGWSTHGRFACPH 467
FLQPLI++LK LW G YD+S QNF L+A L+WTI+DFPAY MLSGW+THG+ +CP
Sbjct: 417 FLQPLIEELKELWCTGVDAYDVSLSQNFNLKAVLLWTISDFPAYSMLSGWTTHGKLSCPV 476
Query: 468 CMENTKAFTLESGGKSSWFDCHRPFLPHNHPFRRLKNGFKKDERVFVG-PPPKITSEEVW 526
CME+TK+F L +G K+ WFDCHR FLPH HP RR K F K PP +T E+V+
Sbjct: 477 CMESTKSFYLPNGRKTCWFDCHRRFLPHGHPSRRNKKDFLKGRDASSEYPPESLTGEQVY 536
Query: 527 TR---VCDYPKVTDYG---RAVPIPRYGVDHHWTKRSIFWDLPYWKDNLLRHNLDVMHIE 580
+ PK D G + YG +H+W K SI W+L YWKD LRHN+DVMH E
Sbjct: 537 YERLASVNPPKTKDVGGNGHEKKMRGYGKEHNWHKESILWELSYWKDLNLRHNIDVMHTE 596
Query: 581 KNVFDNIFNTVMDVKGKTKDNEKARKDLQIYCKRKDLELKPQPNGKLLKPKANYNLSAQE 640
KN DNI NT++ VKGK+KDN +R D++ YC R L + GK P Y L+ +
Sbjct: 597 KNFLDNIMNTLLGVKGKSKDNIMSRLDIEKYCSRPGLHI--DSTGKA--PFPPYKLTEEA 652
Query: 641 AKLVCQWLK-DLRMPDGYSSNIGRCADVNTGRLHGMKSHDSHIFMENFIPIAFSSL-PKH 698
+ + Q +K D+R PDG MKSHD H+FME +P F+ L ++
Sbjct: 653 KQSLFQCVKHDVRFPDG------------------MKSHDCHVFMERLLPFIFAELLDRN 694
Query: 699 VLDPLIEISQFFKNLCASTLRVDELEKMEKNMPIIICQLEQIFPPGFFDSMEHLPVHLAY 758
V L I FF++LC+ TL+ ++ +++N+ +IIC LE+IFPP FFD MEHLP+HL Y
Sbjct: 695 VHLALSGIRAFFRDLCSRTLQTSRVQILKQNIVLIICNLEKIFPPSFFDVMEHLPIHLPY 754
Query: 759 EAILGGPVQYRWMYPFERFMGDSKRAVKNLAKVEGSIVAIYTAKETTYFIGHYFVDRLLT 818
EA LGGPVQYRWMYPFERF K KN GSIV Y E YF HYF D + T
Sbjct: 755 EAELGGPVQYRWMYPFERFFKKLKGKAKNKRYAAGSIVESYINDEIAYFSEHYFADHIQT 814
Query: 819 PS 820
S
Sbjct: 815 KS 816
>gb|AAP52293.1| putative transposable element [Oryza sativa (japonica
cultivar-group)] gi|37531408|ref|NP_920006.1| putative
transposable element [Oryza sativa (japonica
cultivar-group)] gi|22655757|gb|AAN04174.1| Putative
transposable element [Oryza sativa (japonica
cultivar-group)] gi|21672010|gb|AAM74372.1| Putative
transposable element [Oryza sativa (japonica
cultivar-group)]
Length = 992
Score = 626 bits (1614), Expect = e-177
Identities = 362/968 (37%), Positives = 538/968 (55%), Gaps = 68/968 (7%)
Query: 94 FGAGVNAQASTSHGGTNVETNVEYNEDVNTDQFNMMDDMVADALGVELSYGEDVEDDEEE 153
+G + + TSHG + E + + + M++M+ DA+ DV EE
Sbjct: 8 YGFQLGYERWTSHGEPEILEEDEAEQHIGL--VDQMNEMLTDAIAAH-----DVNPSEE- 59
Query: 154 QPPNEKAQRFYRLLSESNTPLYEGSSHSKLSMCVWLLSHKSNYLNPDDGMDDITKMLKAV 213
P A YRLL E +TP++E + ++LS+ L+S K+ + ++I +L
Sbjct: 60 --PTPTAAELYRLLEEVDTPVHENTRQTRLSIVARLMSIKTQNNMSEACYNEIMNLLHDS 117
Query: 214 TPFK--DNLPMNYHAAKRLVMKLGLSVKKIDCCRNGCMLFY-DNEFGINDGPLEECKFCQ 270
+ + LP NYH +K+LV LG+ KKI C N CMLFY +NE E C C
Sbjct: 118 LGDEAGNKLPKNYHRSKKLVHSLGMPYKKIHACTNNCMLFYKENEHK------ENCTVCG 171
Query: 271 SPRYGIS----KQKRVAVKSMFYLPIIPRLQRLFASMKTASQMTWHHSNGISGVMRHPSD 326
RY S + KRV K + YLPI PRLQRL+ S A M +H R S+
Sbjct: 172 EARYEESARGNRSKRVPRKVLRYLPITPRLQRLYMSETMAKHMRYHARP------REESN 225
Query: 327 GEAWKHFDRVHPDFAADPRNVRLGLCSDGFQPYVQSSAKPYSCWPIIVTPYNLPPEMCMT 386
+ RNVRL L +DGF P+ +S PYSCWP+ V PYNLPPEMC
Sbjct: 226 ----------------EVRNVRLCLATDGFTPFSIAST-PYSCWPVFVAPYNLPPEMCTK 268
Query: 387 KPYLFLSCIVPGPDNPKAGIDVFLQPLIDDLKRLWV-GELTYDISRKQNFKLRAALMWTI 445
+ LS ++PGP++P ++VF++PL+D+L LW+ G +TYD KQNF ++AA++WTI
Sbjct: 269 DNNILLSLVIPGPEHPGKNLNVFMEPLVDELLDLWMNGAVTYDRFLKQNFNMKAAVVWTI 328
Query: 446 NDFPAYGMLSGWSTHGRFACPHCMENTKAFTLESGGKSSWFDCHRPFLPHNHPFRRLKNG 505
+DFPAYG++S WSTHG+ ACP C TKAF L+ G K+ WFDCHR FLP NH FRR
Sbjct: 329 HDFPAYGLVSCWSTHGKLACPICGGGTKAFQLKHGHKACWFDCHRRFLPSNHEFRRSLKC 388
Query: 506 FKKDERVFVGPPPKITSEEVWTRVCDYPKVTDYGRAVPIPRYGVDHHWTKRSIFWDLPYW 565
F+K +V PP ++T +E++ ++ V D +G H+WT S W LPY+
Sbjct: 389 FRKKTKVLDPPPERLTGDEIYKQLMSL--VPDRTGKHTFEGFGETHNWTGISGLWKLPYF 446
Query: 566 KDNLLRHNLDVMHIEKNVFDNIFNTVMDVKGKTKDNEKARKDLQIYCKRKDLELKPQPNG 625
K +LRHN+DVMH EKNV + + +T +D++ KTKDN KAR D+ C R L + NG
Sbjct: 447 KKLMLRHNIDVMHNEKNVAEAVLSTCLDIQDKTKDNVKARLDMADICDRPTLNMTKSTNG 506
Query: 626 KLLKPKANYNLSAQEAKLVCQWLKDLRMPDGYSSNIGRCADVNTGRLHGMKSHDSHIFME 685
+ KP+A Y ++ ++ + +W KDL+ PD +++N+ + +++ + G+KSHD HIF+E
Sbjct: 507 RWQKPRAKYCVTKEDKLTIMKWFKDLKFPDRFAANLKKAVNLSQCKFIGLKSHDFHIFIE 566
Query: 686 NFIPIAFSS-LPKHVLDPLIEISQFFKNLCASTLRVDELEKMEKNMPIIICQLEQIFPPG 744
+P+A +P+ L E+S F++ LCA + +++ ++EK + +++C+LE++FP G
Sbjct: 567 RLLPVALRGFIPEKEWKDLSELSFFYRQLCAKEIDKEQMHRLEKEIVVLLCKLEKMFPLG 626
Query: 745 FFDSMEHLPVHLAYEAILGGPVQYRWMYPFERFMGDSKRAVKNLAKVEGSIVAIYTAKET 804
FF+SM+HL +HL YEA +GGPVQ+RWMYPFER M + V+N A+VEGSIV Y +ET
Sbjct: 627 FFNSMQHLILHLPYEARVGGPVQFRWMYPFERCMCKLRLKVRNKARVEGSIVEAYIVEET 686
Query: 805 TYFIGHYFVDRLLTPSNTRNEVQVNDL---RDPSTLSVFNLDGRHAGKVLQYWMVDQKEM 861
T F+ YF + T N N D TL VF G G+ + + Q E+
Sbjct: 687 TNFMSIYFNKDIRTTWNKPNRYNNGGTVVQNDGCTLDVFQEQGMLHGRGVPRDLSPQ-EL 745
Query: 862 RSAHVHVLINCAEVKPYLEEFI----AFYAEFGEGSTVGSIHEYFPAWFKQRVYNAEPTD 917
A ++VL NC+ V + E+F A + + E F WFK N D
Sbjct: 746 NVAMLYVLTNCSMVDKFQEKFSEEKHAMHNNLSVEGLDQLMREEFVDWFKNACRNDPSAD 805
Query: 918 EVIRLRQLSQGPLQCANEFHTYFVNGYKFHTHSWTEGKK---TINSGVYVKGVT-DGGED 973
+ L L+ G +++Y +NGY+F + + + ++ T+NSGV + T D +
Sbjct: 806 D--DLWNLATGCKPRVLSYNSYDINGYRFRSEKYEKSRRRLTTVNSGVCLSSFTSDDQQL 863
Query: 974 DFYGVIKHIYELSYNDNK---VVLFYCDWFDPSPRWTKINKLCNTVDIRVDKKYKEYDPF 1030
D+YGVI+ I +L+++ + +VL C WFDP R + ++I+ K ++PF
Sbjct: 864 DYYGVIEDIIKLTFDAGRKIEIVLLQCRWFDPI-RGKRSTPTLGMIEIKSTSKLTNFEPF 922
Query: 1031 IMAHNVRQ 1038
MAH Q
Sbjct: 923 AMAHQATQ 930
>pir||S29329 hypothetical protein 1 - maize transposon En-1
gi|225005|prf||1206239A ORF 1
Length = 905
Score = 612 bits (1578), Expect = e-173
Identities = 338/891 (37%), Positives = 489/891 (53%), Gaps = 60/891 (6%)
Query: 8 RRWMYDRTFPGRTGLKPQFVEGVDGFISWAWEQEICRDEGGIRCPCLKCRCRHIITDPGD 67
RR MYD G ++ D F++ A+ D RCPC+KCR + +
Sbjct: 14 RRAMYDGFDSVTHGHSDAWLRVADEFVALAF----VGDARLARCPCIKCR-NLVRLKKVE 68
Query: 68 VKKHLKKVGFMPNYWVWTYNGEMFQNFGAGVNAQASTSHGGTNVETNVEYNEDVNTDQFN 127
+ H+ K GFMPNY VW +GE V+ +E + D + D+
Sbjct: 69 LSYHIFKHGFMPNYLVWHEHGE---------------------VDHTIESDGDQDIDRME 107
Query: 128 MMDDMVADALGVELSYGEDVEDDEEEQPPNEKAQRFYRLLSESNTPLYEGSSHSKLSMCV 187
M D + + + D + Q E + FY+LL S ++EG++ S L +
Sbjct: 108 EMLDDIRN----------EYPDLQNNQAFPEDVREFYKLLEASEAKVHEGTNVSVLQVVT 157
Query: 188 WLLSHKSNYLNPDDGMDDITKMLKAVTPFKDNLPMNYHAAKRLVMKLGLSVKKIDCCRNG 247
L++ KS Y + +DI K++ ++P N+P + + K+LV LG++ +KID C +
Sbjct: 158 RLMAMKSKYTFSNKCYNDIVKLIIDISPPNHNMPKDLYHCKKLVAGLGMNYQKIDACEDN 217
Query: 248 CMLFYDNEFGINDGPLEECKFCQSPRYGISKQK-------RVAVKSMFYLPIIPRLQRLF 300
CMLF+ C C RY + + +V +K + Y+PI PRL+RLF
Sbjct: 218 CMLFWKEHENTT-----HCIHCSKSRYAVVLDEDGNEVTTKVPIKQLRYMPITPRLKRLF 272
Query: 301 ASMKTASQMTWHHSNGISG----VMRHPSDGEAWKHFDRVHPDFAADPRNVRLGLCSDGF 356
+ +TA QM WH G VM HPSDGEAW+ DR P+FA DPR+VRLGL +DGF
Sbjct: 273 LNQETAKQMRWHKEGDRQGQDPDVMVHPSDGEAWQALDRFDPEFARDPRSVRLGLSTDGF 332
Query: 357 QPYVQSSAKPYSCWPIIVTPYNLPPEMCMTKPYLFLSCIVPGPDNPKAGIDVFLQPLIDD 416
PY +S YSCWP+ + PYNLPP CM + +FL+ IVPGP +P I+VF++PLI++
Sbjct: 333 TPYSNNSTS-YSCWPVFMMPYNLPPNKCMKEEVMFLALIVPGPKDPVTKINVFMEPLIEE 391
Query: 417 LKRLWVGELTYDISRKQNFKLRAALMWTINDFPAYGMLSGWSTHGRFACPHCMENTKAFT 476
LK LW G YD K F LRAA +W+I+D AYG+ SGW HG CP CM +++A+
Sbjct: 392 LKMLWQGVEAYDSHLKCCFTLRAAYLWSIHDLLAYGIFSGWCVHGILRCPICMGDSQAYR 451
Query: 477 LESGGKSSWFDCHRPFLPHNHPFRRLKNGFKKDERVFVGPPPKITSEEVWTRVCDYPKVT 536
LE G K ++FD HR LP+NHPFR+ F+K +RV GPP + T E + + D
Sbjct: 452 LEHGKKETFFDVHRRLLPYNHPFRKDTKSFRKGKRVRDGPPKRQTGENIMRQHRDLKP-- 509
Query: 537 DYGRAVPIPRYGVDHHWTKRSIFWDLPYWKDNLLRHNLDVMHIEKNVFDNIFNTVMDVKG 596
G YG +H+WT S W+LPY K LL HN+D+MH E+NV ++I + D G
Sbjct: 510 --GVGGRFQGYGKEHNWTHISFIWELPYTKALLLPHNIDLMHQERNVAESIISMCFDFTG 567
Query: 597 KTKDNEKARKDLQIYCKRKDLELKPQPNGKLLKPKANYNLSAQEAKLVCQWLKDLRMPDG 656
+TKDN AR+DL C R LEL+ P+G +P+A Y L QE + + QWLK LR PD
Sbjct: 568 QTKDNMNARRDLAELCDRPHLELRKNPSGSESRPQAPYCLKRQEREEIFQWLKKLRFPDR 627
Query: 657 YSSNIGRCADVNTGRLHGMKSHDSHIFMENFIPIAFSS-LPKHVLDPLIEISQFFKNLCA 715
Y++NI R +++TG+L G+KSHD HI +E +P+ F V E+S F+K +CA
Sbjct: 628 YAANIKRAVNLDTGKLVGLKSHDYHILIERLVPVMFRGYFSPDVWKIFAELSYFYKQICA 687
Query: 716 STLRVDELEKMEKNMPIIICQLEQIFPPGFFDSMEHLPVHLAYEAILGGPVQYRWMYPFE 775
+ + + EK + +++C++E++FPPGFF+ M+HL VHL +EA++GGP Q+RWMY E
Sbjct: 688 KEISKKLMLRFEKEIVVLVCKMEKVFPPGFFNCMQHLLVHLPWEALVGGPAQFRWMYSQE 747
Query: 776 RFMGDSKRAVKNLAKVEGSIVAIYTAKETTYFIGHYFVDRLLTPSNTRNEVQVNDLRDPS 835
R + + V+N A+VEG I + A+E T F YF D + T V + +
Sbjct: 748 RELKKLRGMVRNKARVEGCIAEAFAAREITLFSSKYFSDTNNVNAQT-TRYHVAEQAPIT 806
Query: 836 TLSVFNLDGRHAGKVLQYWMVDQKEMRSAHVHVLINCAEVKPYLEEFIAFY 886
LS F DG+ G + +V E + + +N E+ PY + F + Y
Sbjct: 807 DLSAFKWDGKGVGAYTSH-LVGTIERNKTLLFLYVNMPELHPYFQIFDSIY 856
>gb|AAA66266.1| unknown protein
Length = 897
Score = 612 bits (1578), Expect = e-173
Identities = 338/891 (37%), Positives = 489/891 (53%), Gaps = 60/891 (6%)
Query: 8 RRWMYDRTFPGRTGLKPQFVEGVDGFISWAWEQEICRDEGGIRCPCLKCRCRHIITDPGD 67
RR MYD G ++ D F++ A+ D RCPC+KCR + +
Sbjct: 6 RRAMYDGFDSVTHGHSDAWLRVADEFVALAF----VGDARLARCPCIKCR-NLVRLKKVE 60
Query: 68 VKKHLKKVGFMPNYWVWTYNGEMFQNFGAGVNAQASTSHGGTNVETNVEYNEDVNTDQFN 127
+ H+ K GFMPNY VW +GE V+ +E + D + D+
Sbjct: 61 LSYHIFKHGFMPNYLVWHEHGE---------------------VDHTIESDGDQDIDRME 99
Query: 128 MMDDMVADALGVELSYGEDVEDDEEEQPPNEKAQRFYRLLSESNTPLYEGSSHSKLSMCV 187
M D + + + D + Q E + FY+LL S ++EG++ S L +
Sbjct: 100 EMLDDIRN----------EYPDLQNNQAFPEDVREFYKLLEASEAKVHEGTNVSVLQVVT 149
Query: 188 WLLSHKSNYLNPDDGMDDITKMLKAVTPFKDNLPMNYHAAKRLVMKLGLSVKKIDCCRNG 247
L++ KS Y + +DI K++ ++P N+P + + K+LV LG++ +KID C +
Sbjct: 150 RLMAMKSKYTFSNKCYNDIVKLIIDISPPNHNMPKDLYHCKKLVAGLGMNYQKIDACEDN 209
Query: 248 CMLFYDNEFGINDGPLEECKFCQSPRYGISKQK-------RVAVKSMFYLPIIPRLQRLF 300
CMLF+ C C RY + + +V +K + Y+PI PRL+RLF
Sbjct: 210 CMLFWKEHENTT-----HCIHCSKSRYAVVLDEDGNEVTTKVPIKQLRYMPITPRLKRLF 264
Query: 301 ASMKTASQMTWHHSNGISG----VMRHPSDGEAWKHFDRVHPDFAADPRNVRLGLCSDGF 356
+ +TA QM WH G VM HPSDGEAW+ DR P+FA DPR+VRLGL +DGF
Sbjct: 265 LNQETAKQMRWHKEGDRQGQDPDVMVHPSDGEAWQALDRFDPEFARDPRSVRLGLSTDGF 324
Query: 357 QPYVQSSAKPYSCWPIIVTPYNLPPEMCMTKPYLFLSCIVPGPDNPKAGIDVFLQPLIDD 416
PY +S YSCWP+ + PYNLPP CM + +FL+ IVPGP +P I+VF++PLI++
Sbjct: 325 TPYSNNSTS-YSCWPVFMMPYNLPPNKCMKEEVMFLALIVPGPKDPVTKINVFMEPLIEE 383
Query: 417 LKRLWVGELTYDISRKQNFKLRAALMWTINDFPAYGMLSGWSTHGRFACPHCMENTKAFT 476
LK LW G YD K F LRAA +W+I+D AYG+ SGW HG CP CM +++A+
Sbjct: 384 LKMLWQGVEAYDSHLKCCFTLRAAYLWSIHDLLAYGIFSGWCVHGILRCPICMGDSQAYR 443
Query: 477 LESGGKSSWFDCHRPFLPHNHPFRRLKNGFKKDERVFVGPPPKITSEEVWTRVCDYPKVT 536
LE G K ++FD HR LP+NHPFR+ F+K +RV GPP + T E + + D
Sbjct: 444 LEHGKKETFFDVHRRLLPYNHPFRKDTKSFRKGKRVRDGPPKRQTGENIMRQHRDLKP-- 501
Query: 537 DYGRAVPIPRYGVDHHWTKRSIFWDLPYWKDNLLRHNLDVMHIEKNVFDNIFNTVMDVKG 596
G YG +H+WT S W+LPY K LL HN+D+MH E+NV ++I + D G
Sbjct: 502 --GVGGRFQGYGKEHNWTHISFIWELPYTKALLLPHNIDLMHQERNVAESIISMCFDFTG 559
Query: 597 KTKDNEKARKDLQIYCKRKDLELKPQPNGKLLKPKANYNLSAQEAKLVCQWLKDLRMPDG 656
+TKDN AR+DL C R LEL+ P+G +P+A Y L QE + + QWLK LR PD
Sbjct: 560 QTKDNMNARRDLAELCDRPHLELRKNPSGSESRPQAPYCLKRQEREEIFQWLKKLRFPDR 619
Query: 657 YSSNIGRCADVNTGRLHGMKSHDSHIFMENFIPIAFSS-LPKHVLDPLIEISQFFKNLCA 715
Y++NI R +++TG+L G+KSHD HI +E +P+ F V E+S F+K +CA
Sbjct: 620 YAANIKRAVNLDTGKLVGLKSHDYHILIERLVPVMFRGYFSPDVWKIFAELSYFYKQICA 679
Query: 716 STLRVDELEKMEKNMPIIICQLEQIFPPGFFDSMEHLPVHLAYEAILGGPVQYRWMYPFE 775
+ + + EK + +++C++E++FPPGFF+ M+HL VHL +EA++GGP Q+RWMY E
Sbjct: 680 KEISKKLMLRFEKEIVVLVCKMEKVFPPGFFNCMQHLLVHLPWEALVGGPAQFRWMYSQE 739
Query: 776 RFMGDSKRAVKNLAKVEGSIVAIYTAKETTYFIGHYFVDRLLTPSNTRNEVQVNDLRDPS 835
R + + V+N A+VEG I + A+E T F YF D + T V + +
Sbjct: 740 RELKKLRGMVRNKARVEGCIAEAFAAREITLFSSKYFSDTNNVNAQT-TRYHVAEQAPIT 798
Query: 836 TLSVFNLDGRHAGKVLQYWMVDQKEMRSAHVHVLINCAEVKPYLEEFIAFY 886
LS F DG+ G + +V E + + +N E+ PY + F + Y
Sbjct: 799 DLSAFKWDGKGVGAYTSH-LVGTIERNKTLLFLYVNMPELHPYFQIFDSIY 848
>gb|AAG17043.1| transposase [Zea mays]
Length = 854
Score = 605 bits (1561), Expect = e-171
Identities = 326/818 (39%), Positives = 477/818 (57%), Gaps = 62/818 (7%)
Query: 49 IRCPCLKCRCRHIITDPGDVKKHLKKVGFMPNYWVWTYNGEMFQNFGAGVNAQASTSHGG 108
++CPC C + +++KHL K GFMPNY VW +GE ++
Sbjct: 52 VKCPCNHCENKWP-QKKEEMEKHLCKSGFMPNYLVWYRHGESIRH--------------- 95
Query: 109 TNVETNVEYNEDVNTDQFNMMDDMVADALGVELSYGEDVEDDEEE--QPPNEKAQRFYRL 166
V+ VE ++D + MDDM+ D G +VE + EE Q P + AQ F++L
Sbjct: 96 --VDAEVELDDD-----HDRMDDMLHDL-------GREVEMNAEEPGQLPRD-AQEFFQL 140
Query: 167 LSESNTPLYEGSSHSKLSMCVWLLSHKSNYLNPDDGMDDITKMLKAVTPFKDNLPMNYHA 226
L+ L+E + S L L++ KS + + +DI +++ V P LP N +
Sbjct: 141 LAAGEERLHEHTQMSVLGTLTRLMAIKSKHNISNSAYNDIVQLMGEVLPENHKLPKNMYF 200
Query: 227 AKRLVMKLGLSVKKIDCCRNGCMLFYDNEFGINDGPLEECKFCQSPRY-------GISKQ 279
K+++ LG++ +KID C N CMLF++ D L+ CK C++ RY G
Sbjct: 201 TKKMLAGLGMTYEKIDVCPNSCMLFFEE-----DDKLDRCKHCEASRYVEVTNDEGELVV 255
Query: 280 KRVAVKSMFYLPIIPRLQRLFASMKTASQMTWHHSNGIS-----GVMRHPSDGEAWKHFD 334
+VA K + LPIIPRL RLF + + A MTW NG+ +M HPSDG+AWK FD
Sbjct: 256 TKVAAKQLRRLPIIPRLSRLFLNKEIALHMTWP-KNGVRLVTDPDIMVHPSDGDAWKAFD 314
Query: 335 RVHPDFAADPRNVRLGLCSDGFQPYVQSSAKPYSCWPIIVTPYNLPPEMCMTKPYLFLSC 394
P+FA DPR+VRLGL +DGF P+ +SA PYSCWP+ + PYNLPPE+ + ++FL+
Sbjct: 315 EFDPEFANDPRSVRLGLSTDGFTPF-NTSASPYSCWPVFIVPYNLPPELVNKEEFMFLAL 373
Query: 395 IVPGPDNPKAGIDVFLQPLIDDLKRLWVGELTYDISRKQNFKLRAALMWTINDFPAYGML 454
++PGP++P +++F++PLI++LK+LW G YD ++ F +RAA +W+++D AYG
Sbjct: 374 VIPGPEHPGPKLNMFVRPLIEELKQLWRGVKAYDSHTEKEFTMRAAYLWSVHDLLAYGDW 433
Query: 455 SGWSTHGRFACPHCMENTKAFTLESGGKSSWFDCHRPFLPHNHPFRRLKNGFKKDERVFV 514
SGW HGR CP CM +T AF L+ GGK S+FD HR + P H FR F+ ++
Sbjct: 434 SGWCVHGRLCCPICMNDTDAFRLKHGGKVSFFDAHRRWTPFKHDFRNSLTAFRGGAKIRN 493
Query: 515 GPPPKITSEEVWTRVCDYPKVTDYGRAVPIPRYGVDHHWTKRSIFWDLPYWKDNLLRHNL 574
GPP + T+ ++ + G YG DH+WT S W+LPY K ++ HN+
Sbjct: 494 GPPKRQTAPQIMA----WHACLKQGENDRFQGYGEDHNWTHISSIWELPYAKALIMPHNI 549
Query: 575 DVMHIEKNVFDNIFNTVMDVKGKTKDNEKARKDLQIYCKRKDLELKPQPNGKLLKPKANY 634
D+MH E+NV ++I +T DV KTKDN KARKD+ CKR LELK G +P+A+Y
Sbjct: 550 DLMHQERNVAESIISTCFDVTDKTKDNMKARKDMAEICKRPMLELKVSDKGHESRPRADY 609
Query: 635 NLSAQEAKLVCQWLKDLRMPDGYSSNIGRCADVNTGRLHGMKSHDSHIFMENFIPIAFSS 694
L E K + +WLK+L+ PD Y++N+ R ++ TG+L G+KSHD HI ME +P+ F
Sbjct: 610 CLKPDERKEIFKWLKNLKFPDRYAANLKRSVNLKTGKLIGLKSHDYHIIMERLMPVMFRG 669
Query: 695 LPKHVLDPLI-EISQFFKNLCASTLRVDELEKMEKNMPIIICQLEQIFPPGFFDSMEHLP 753
K L + E+S F++ +CA T+ ++K EK +PI+IC+ E++FPPGFF+ M+HL
Sbjct: 670 YFKDELWSIFAELSYFYREVCAKTVSKRLMQKFEKEIPILICKFEKVFPPGFFNVMQHLI 729
Query: 754 VHLAYEAILGGPVQYRWMYPFERFMGDSKRAVKNLAKVEGSIVAIYTAKETTYFIGHYF- 812
VHL +EA++GGPVQ+RWMYP ER + + +V+N A+VEG I + KE + F YF
Sbjct: 730 VHLPWEALVGGPVQFRWMYPIERALKKLRASVRNKARVEGCIAEAFALKEISQFSTRYFA 789
Query: 813 -VDRLLTPSNTRNEVQVNDLRDPSTLSVFNLDGRHAGK 849
+ + PS + V + STL +F G+ GK
Sbjct: 790 RANNVFAPS---VRLHVENESPQSTLQIFANPGKAVGK 824
>emb|CAA40555.1| TNP2 [Antirrhinum majus]
Length = 752
Score = 588 bits (1515), Expect = e-166
Identities = 327/783 (41%), Positives = 461/783 (58%), Gaps = 56/783 (7%)
Query: 13 DRTFPGRTGLKPQFVEGVDGFISWAWEQEICRDEGGIRCPCLKCRCRHIITDPGDVKKHL 72
D+++ ++ L ++ G+D F+ +A++ +C + G I CPC +C+ + D K+HL
Sbjct: 7 DKSWMHKSRLTEEYENGLDQFLDFAFDN-VCLN-GKIICPCKQCK-NALWMDRETAKEHL 63
Query: 73 KKVGFMPNYWVWTYNGEMFQNFGAGVNAQASTSHGGTNVETNVEYNEDVNTDQFNMMDDM 132
GF+ Y W +GE G +N + N++ NE +NT F M ++
Sbjct: 64 IIDGFIKGYTHWVIHGE-----------------GSSNSQNNIQ-NERLNT--FEDMHNL 103
Query: 133 VADALGVELSYGEDVEDDEEEQPPNEKAQRFYRLLSESNTPLYEG-SSHSKLSMCVWLLS 191
V DA GVE E+ EE+ PNE+A+ FY+LL ++ LY G S+L V L
Sbjct: 104 VHDAFGVEDDNINSEEEPNEEEEPNEEAKTFYKLLDDAQQELYPGCKDFSRLRFIVRLFH 163
Query: 192 HKSNYLNPDDGMDDITKMLKAVTPFKDN-LPMNYHAAKRLVMKLGLSVKKIDCCRNGCML 250
K + + ++LK P + N LP +Y+ ++++ LGL+ KID C N CML
Sbjct: 164 LKCLGKWSNKTFTMLLELLKEAFPEQTNSLPKSYYEVQKIIGALGLTCVKIDACPNDCML 223
Query: 251 FYDNEFGINDGPLEECKFCQSPRY------------GISKQKRVAVKSMFYLPIIPRLQR 298
Y E +D C C + RY SK KRV K + Y P+ PRLQR
Sbjct: 224 -YRKEHANDDA----CHVCNTSRYIEGENNSVSEEASSSKCKRVPAKVLRYFPLKPRLQR 278
Query: 299 LFASMKTASQMTWHHSNGIS-GVMRHPSDGEAWKHFDRVHPDFAADPRNVRLGLCSDGFQ 357
LF S KTAS M WH G +RHPSD W FD HPDFA DPRN+RLGL +DGF
Sbjct: 279 LFTSSKTASFMRWHKEERTKDGQLRHPSDSPLWHAFDHQHPDFAEDPRNIRLGLAADGFN 338
Query: 358 PYVQSSAKPYSCWPIIVTPYNLPPEMCMTKPYLFLSCIVPGPDNPKAGIDVFLQPLIDDL 417
P+ ++ + +S WP+I+TPYN+PP MCM +P+ FL+ ++PGP P IDV++QPLI++L
Sbjct: 339 PF-RTMSVAHSTWPVILTPYNIPPWMCMKEPFFFLTLLIPGPSPPGNNIDVYMQPLIEEL 397
Query: 418 KRLWVGELTYDISRKQNFKLRAALMWTINDFPAYGMLSGWSTHGRFACPHCMENTKAFTL 477
+ LW G TYD S K+NF +RAAL+WTIND+PAY LSGWST G ACP C ++T + L
Sbjct: 398 QELWGGVNTYDASAKENFNVRAALLWTINDYPAYANLSGWSTKGELACPSCHKDTCSKYL 457
Query: 478 ESGGKSSWFDCHRPFLPHNHPFRRLKNGFKKDE---RVFVGPPPKITSEEVWTRVCDYPK 534
+ K + HR FL HP+R+ F +E R + + SEE+ + K
Sbjct: 458 QKSHKYCYMG-HRRFLNRYHPYRKDTKSFDGNEEYRRAPIALTGDMVSEEITGFNIKFGK 516
Query: 535 VTDYGRAVPIPRYGVDHHWTKRSIFWDLPYWKDNLLRHNLDVMHIEKNVFDNIFNTVMDV 594
D +P+ +W KRSIF+DLPYWKD+LLRHN DVMHIEKNV ++I T++++
Sbjct: 517 KVDDNPTLPL-------NWKKRSIFFDLPYWKDSLLRHNFDVMHIEKNVCESIIGTLLNL 569
Query: 595 KGKTKDNEKARKDLQIYCKRKDLELKPQPNGKLLKPKANYNLSAQEAKLVCQWLKDLRMP 654
+G+TKD+E +R DL+ R +L +GK P A Y++S +E ++V + LK +++P
Sbjct: 570 EGRTKDHENSRLDLKDMGIRSELHPISLESGKHYLPAACYSMSKKEKEIVFEILKTVKVP 629
Query: 655 DGYSSNIGRCADVNTGRLHGMKSHDSHIFMENFIPIAFSS-LPKHVLDPLIEISQFFKNL 713
DGY+SNI R + ++ G+KSHD HI M+ +PIA LPKHV PLI++ FF+ L
Sbjct: 630 DGYASNISRRVQLKPNKISGLKSHDHHILMQQLLPIALRKVLPKHVRTPLIKLCTFFREL 689
Query: 714 CASTLRVDELEKMEKNMPIIICQLEQIFPPGFFDSMEHLPVHLAYEAILGGPVQYRWMYP 773
C+ L +L +M K++ +C LE+IFPP FFD M HLP+HLAYEA + GPVQYRWMYP
Sbjct: 690 CSKVLNPQDLVRMGKDIAKTLCDLEKIFPPSFFDIMMHLPIHLAYEAQIAGPVQYRWMYP 749
Query: 774 FER 776
ER
Sbjct: 750 IER 752
>pir||S23818 hypothetical protein Tnp2 - garden snapdragon transposable element
Tam1
Length = 752
Score = 588 bits (1515), Expect = e-166
Identities = 327/783 (41%), Positives = 461/783 (58%), Gaps = 56/783 (7%)
Query: 13 DRTFPGRTGLKPQFVEGVDGFISWAWEQEICRDEGGIRCPCLKCRCRHIITDPGDVKKHL 72
D+++ ++ L ++ G+D F+ +A++ +C + G I CPC +C+ + D K+HL
Sbjct: 7 DKSWMHKSRLTEEYENGLDQFLDFAFDN-VCLN-GKIICPCKQCK-NALWMDRETAKEHL 63
Query: 73 KKVGFMPNYWVWTYNGEMFQNFGAGVNAQASTSHGGTNVETNVEYNEDVNTDQFNMMDDM 132
GF+ Y W +GE G +N + N++ NE +NT F M ++
Sbjct: 64 IIDGFIKGYTHWVIHGE-----------------GSSNSQNNIQ-NERLNT--FEDMHNL 103
Query: 133 VADALGVELSYGEDVEDDEEEQPPNEKAQRFYRLLSESNTPLYEG-SSHSKLSMCVWLLS 191
V DA GVE E+ EE+ PNE+A+ FY+LL ++ LY G S+L V L
Sbjct: 104 VHDAFGVEDDNINSEEEPNEEEEPNEEAKTFYKLLDDAQQELYPGCKDFSRLRFIVRLFH 163
Query: 192 HKSNYLNPDDGMDDITKMLKAVTPFKDN-LPMNYHAAKRLVMKLGLSVKKIDCCRNGCML 250
K + + ++LK P + N LP +Y+ ++++ LGL+ KID C N CML
Sbjct: 164 LKCLGKWSNKTFTMLLELLKEAFPEQTNSLPKSYYEVQKIIGALGLTCVKIDACPNDCML 223
Query: 251 FYDNEFGINDGPLEECKFCQSPRY------------GISKQKRVAVKSMFYLPIIPRLQR 298
Y E +D C C + RY SK KRV K + Y P+ PRLQR
Sbjct: 224 -YRKEHANDDA----CHVCNTSRYIEGENNSVSEEASSSKCKRVPAKVLRYFPLKPRLQR 278
Query: 299 LFASMKTASQMTWHHSNGIS-GVMRHPSDGEAWKHFDRVHPDFAADPRNVRLGLCSDGFQ 357
LF S KTAS M WH G +RHPSD W FD HPDFA DPRN+RLGL +DGF
Sbjct: 279 LFTSSKTASFMRWHKEERTKDGQLRHPSDSPLWHAFDHQHPDFAEDPRNIRLGLAADGFN 338
Query: 358 PYVQSSAKPYSCWPIIVTPYNLPPEMCMTKPYLFLSCIVPGPDNPKAGIDVFLQPLIDDL 417
P+ ++ + +S WP+I+TPYN+PP MCM +P+ FL+ ++PGP P IDV++QPLI++L
Sbjct: 339 PF-RTMSVAHSTWPVILTPYNIPPWMCMKEPFFFLTLLIPGPSPPGNNIDVYMQPLIEEL 397
Query: 418 KRLWVGELTYDISRKQNFKLRAALMWTINDFPAYGMLSGWSTHGRFACPHCMENTKAFTL 477
+ LW G TYD S K+NF +RAAL+WTIND+PAY LSGWST G ACP C ++T + L
Sbjct: 398 QELWGGVNTYDASAKENFNVRAALLWTINDYPAYANLSGWSTKGELACPSCHKDTCSKYL 457
Query: 478 ESGGKSSWFDCHRPFLPHNHPFRRLKNGFKKDE---RVFVGPPPKITSEEVWTRVCDYPK 534
+ K + HR FL HP+R+ F +E R + + SEE+ + K
Sbjct: 458 QKSHKYCYMG-HRRFLNRYHPYRKDTKSFDGNEEYRRAPIALTGDMVSEEITGFNIKFGK 516
Query: 535 VTDYGRAVPIPRYGVDHHWTKRSIFWDLPYWKDNLLRHNLDVMHIEKNVFDNIFNTVMDV 594
D +P+ +W KRSIF+DLPYWKD+LLRHN DVMHIEKNV ++I T++++
Sbjct: 517 KVDDNPTLPL-------NWKKRSIFFDLPYWKDSLLRHNFDVMHIEKNVCESIIGTLLNL 569
Query: 595 KGKTKDNEKARKDLQIYCKRKDLELKPQPNGKLLKPKANYNLSAQEAKLVCQWLKDLRMP 654
+G+TKD+E +R DL+ R +L +GK P A Y++S +E ++V + LK +++P
Sbjct: 570 EGRTKDHENSRLDLKDMGIRSELHPISLESGKHYLPAACYSMSKKEKEIVFEILKTVKVP 629
Query: 655 DGYSSNIGRCADVNTGRLHGMKSHDSHIFMENFIPIAFSS-LPKHVLDPLIEISQFFKNL 713
DGY+SNI R + ++ G+KSHD HI M+ +PIA LPKHV PLI++ FF+ L
Sbjct: 630 DGYASNISRRVQLKPNKISGLKSHDHHILMQQLLPIALRKVLPKHVRTPLIKLCTFFREL 689
Query: 714 CASTLRVDELEKMEKNMPIIICQLEQIFPPGFFDSMEHLPVHLAYEAILGGPVQYRWMYP 773
C+ L +L +M K++ +C LE+IFPP FFD M HLP+HLAYEA + GPVQYRWMYP
Sbjct: 690 CSKVLNPQDLVRMGKDIAKTLCDLEKIFPPSFFDIMMHLPIHLAYEAQIAGPVQYRWMYP 749
Query: 774 FER 776
ER
Sbjct: 750 IER 752
>gb|AAS79612.1| putative tnp2 transposase [Ipomoea trifida]
Length = 651
Score = 572 bits (1475), Expect = e-161
Identities = 303/611 (49%), Positives = 390/611 (63%), Gaps = 45/611 (7%)
Query: 11 MYDRTFPGRTGLKPQFVEGVDGFISWAWEQEICRDEGGIRCPCLKCRCR-HIITDPGDVK 69
MY+R PG+ G +F+ GV+ F+++A + IRCPC KC+ R H+ D +V+
Sbjct: 1 MYNRLSPGKRGYTDEFLAGVEEFVNYACTLPVYESTNTIRCPCSKCKNRVHLSAD--EVR 58
Query: 70 KHLKKVGFMPNYWVWTYNGEMFQNFGAGVNAQASTSHGGTNVETNVEYNEDVNTDQFNMM 129
HL K GF P YWVWT +GE N ++ E N + N
Sbjct: 59 VHLYKKGFEPYYWVWTCHGEPIPT-------------------VNEQFQETRN--ESNQY 97
Query: 130 DDMVADALGVELSYGEDVEDDEEEQPPNEKAQRFYRLLSESNTPLYEGS-SHSKLSMCVW 188
+ MV D G + + EE+ P+ A+ FY LLS + PL ++LS +
Sbjct: 98 ETMVFDVGGSSFIPFNVLNNQEED--PHTSAKDFYDLLSTARQPLSPSCVEETELSYALK 155
Query: 189 LLSHKSNYLNPDDGMDDITKMLKAVTPFKDNLPMNYHAAKRLVMKLGLSVKKIDCCRNGC 248
+++ K+ + P MD I + K + + +P Y+ +++L+ KLGL+ +KIDCC N C
Sbjct: 156 MMNIKATFNIPQLAMDQIFEYNKHIMGSDNRVPSRYYDSEKLISKLGLAHEKIDCCINSC 215
Query: 249 MLFYDNEFGINDGPLEECKFCQSPRY-----GISKQKRVAVKSMFYLPIIPRLQRLFASM 303
ML+Y ++ IND +CKFC PR+ ++ K V K M YLP+IPRLQRL+AS
Sbjct: 216 MLYYKSD--IND---RQCKFCGEPRFKPRSPNCTRGKEVPKKRMHYLPLIPRLQRLYASP 270
Query: 304 KTASQMTWHHSNGIS-GVMRHPSDGEAWKHFDRVHPDFAADPRNVRLGLCSDGFQPYVQS 362
+A M WH+ N GVM HPSDGEAWKHFD +PDF ADPRNVRLGLC+DGF P+
Sbjct: 271 SSAEHMRWHYENRRQPGVMCHPSDGEAWKHFDSQYPDFGADPRNVRLGLCADGFSPF-GL 329
Query: 363 SAKPYSCWPIIVTPYNLPPEMCMTKPYLFLSCIVPGPDNPKAGIDVFLQPLIDDLKRLW- 421
+AK YSCWP++V PYNLPP MCMT PY FL+CI+PG NPKA IDV+LQPLID+L+ LW
Sbjct: 330 NAKSYSCWPVMVVPYNLPPSMCMTPPYTFLTCIIPGEHNPKANIDVYLQPLIDELQLLWE 389
Query: 422 VGELTYDISRKQNFKLRAALMWTINDFPAYGMLSGWSTHGRFACPHCMENTKAFTLESGG 481
G LTYD+S +QNF +RA LMWTINDFPAYGMLSGW T G+ ACP+C TK+F L++G
Sbjct: 390 TGVLTYDVSLQQNFMMRAMLMWTINDFPAYGMLSGWQTAGKLACPYCNHYTKSFYLKNGR 449
Query: 482 KSSWFDCHRPFLPHNHPFRRLKNGFKKDERVFVGPPPKI-TSEEVWTRVCDYPKVTDYGR 540
K+ WFDCHR FLP +HPFRR ++ F K +RV PP+I T EE+ V +PK+TD G
Sbjct: 450 KTCWFDCHRQFLPMDHPFRRNRDSFIK-QRVEKSRPPRIWTGEELLANVLMFPKITD-GP 507
Query: 541 AVPIPRYGVDHHWTKRSIFWDLPYWKDNLLRHNLDVMHIEKNVFDNIFNTVMDVKG--KT 598
+ +G H+W KRSIFWDLPYWKDNLLRHNLDVMHIEKNVFDNIF TVMDVKG K
Sbjct: 508 IGRLEGFGCSHNWKKRSIFWDLPYWKDNLLRHNLDVMHIEKNVFDNIFYTVMDVKGTRKN 567
Query: 599 KDNEKARKDLQ 609
KD R D++
Sbjct: 568 KDTPATRLDIK 578
Score = 42.4 bits (98), Expect = 0.092
Identities = 20/51 (39%), Positives = 29/51 (56%), Gaps = 3/51 (5%)
Query: 971 GEDDFYGVIKHIYELSYND---NKVVLFYCDWFDPSPRWTKINKLCNTVDI 1018
G+ F+G +K I E+ Y + KVVLF C+WFDP+P + L + I
Sbjct: 586 GDSLFFGTLKEIVEVEYPNIPLKKVVLFNCEWFDPTPNTDQEEPLIMLISI 636
>dbj|BAA85462.1| transposon-like ORF [Brassica rapa]
Length = 703
Score = 568 bits (1465), Expect = e-160
Identities = 307/715 (42%), Positives = 441/715 (60%), Gaps = 44/715 (6%)
Query: 8 RRWMY-DRTFPGRTGLKPQFVEGVDGFISWAWEQEICRDEGGIRCPCLKCRCRHIITDPG 66
R WMY R GR + +++ G++ F+ A + ++ G + CPC KC + +
Sbjct: 11 RNWMYMHRDANGR--VTKEYLAGLETFMHQADSTPLAQESGKMFCPCRKCN-NSKLANRE 67
Query: 67 DVKKHLKKVGFMPNYWVWTYNGEMF---QNFGAGVNAQASTSHGGTNVETNVEYNEDVNT 123
+V KHL GF PNY++W +GE + QN + N+ ++ Y+++
Sbjct: 68 NVWKHLINRGFTPNYYIWFQHGEGYNYDQNEASSSNSNFQEEPVNHHLHNEHSYHQEEMV 127
Query: 124 DQFNMMDDMVADALGVELSYGEDVEDDEEEQPPNEKAQRFYRLLSESNTPLYEGSSH--S 181
D ++ + DMVADA V DE+E+P N A++FY +L+ +N PLY G S
Sbjct: 128 D-YDRVHDMVADAF---------VAHDEDEEP-NIDAKKFYGMLNAANQPLYSGCREGLS 176
Query: 182 KLSMCVWLLSHKSNYLNPDDGMDDITKMLKAVTPFKDNLPMNYHAAKRLVMKLGLSVKKI 241
KLS+ +++ K+++ P+ M++ + K P + +Y+ ++LV LGL + I
Sbjct: 177 KLSLAARMMNIKTDHNLPESCMNEWADLFKEYLPEDNVSADSYYEIQKLVYSLGLPSEMI 236
Query: 242 DCCRNGCMLFYDNEFGINDGPLEECKFCQSPRYGISKQ--KRVAVKSMFYLPIIPRLQRL 299
D C + CM+++ + D LE C+FC+ PR+ + RV + M+YLPI RL+RL
Sbjct: 237 DVCIDNCMIYWGD-----DEKLEACRFCKKPRFKPQGRGLNRVPYQRMWYLPITDRLKRL 291
Query: 300 FASMKTASQMTWHHSNG-ISGVMRHPSDGEAWKHFDRVHPDFAADPRNVRLGLCSDGFQP 358
+ S +TA+ M WH + G M HPSD AWKHF++VHPDFA++ RNV LGLC+DGF P
Sbjct: 292 YQSEQTAANMRWHAEHTQTDGEMTHPSDARAWKHFNKVHPDFASNSRNVYLGLCTDGFNP 351
Query: 359 YVQSSAKPYSCWPIIVTPYNLPPEMCMTKPYLFLSCIVPGPDNPKAGIDVFLQPLIDDLK 418
+ S + YS WP+ +TPYNLPPEMCM + LFL+ ++PGP++PK +DVFLQPLI +LK
Sbjct: 352 FGMSG-RQYSLWPVFLTPYNLPPEMCMQRELLFLTILIPGPNHPKRSLDVFLQPLIKELK 410
Query: 419 RLW-VGELTYDISRKQNFKLRAALMWTINDFPAYGMLSGWSTHGRFACPHCMENTKAFTL 477
LW G TYD S K NF +RA L+WTI+DFPAYGMLSGW+THGR ACP+C T AF L
Sbjct: 411 DLWSTGVRTYDCSTKTNFTMRAMLLWTISDFPAYGMLSGWTTHGRLACPYCNGTTDAFQL 470
Query: 478 ESGGKSSWFDCHRPFLPHNHPFRRLKNGFKKDERVFVGPPPKITSEEVWTRVCDY--PKV 535
++G K+SWFDCHR FLP HP+RR KN F+ V PP +T E++ ++ Y +
Sbjct: 471 KNGRKTSWFDCHRRFLPIGHPYRRNKNLFRHKRVVRDTSPPYLTGEQIEAQIDYYGANET 530
Query: 536 TDYGRAVPIPR-----YGVDHHWTKRSIFWDLPYWKDNLLRHNLDVMHIEKNVFDNIFNT 590
+G +PR YGV H+W K+SIFW+LP WKD LLRHNLDVMHIEKN F+NI NT
Sbjct: 531 VRWGGNWHVPRNMPDSYGVHHNWHKKSIFWELPCWKDLLLRHNLDVMHIEKNFFENIMNT 590
Query: 591 VMDVKGKTKDNEKARKDLQIYCKRKDLELKPQPNGKLLKPKANYNLSAQEAKLVCQWL-K 649
+++V GKTKDN K+R DL C R +L +K NG++ P + LS+++ ++ W+
Sbjct: 591 ILNVPGKTKDNIKSRLDLPDICSRSELHIK--SNGQV--PVPIFRLSSEKKSVLFNWVAS 646
Query: 650 DLRMPDGYSSNIGRCADVNTGRLHGMKSHDSHIFMENFIPIAFSS-LPKHVLDPL 703
+++ PDGY SN+ RC + + GMKSHD H+FM+ +P AF+ LP +V + L
Sbjct: 647 EVKFPDGYVSNLSRCVEKGQ-KFSGMKSHDCHVFMQRLLPFAFAELLPTNVHEAL 700
>gb|AAT77012.1| putative Transposase family tnp2 protein [Oryza sativa (japonica
cultivar-group)]
Length = 1095
Score = 544 bits (1402), Expect = e-153
Identities = 352/1076 (32%), Positives = 541/1076 (49%), Gaps = 83/1076 (7%)
Query: 8 RRWMYDRTFPGRTGLKPQFVEGVDGFISWAWEQEICRDEGGIRCPCLKCRCRHIITDPGD 67
RRWMY + R+ ++ EGV F+++A R + CPC C+ +I D +
Sbjct: 3 RRWMY---YAHRSST--EYREGVTEFVTFADNDRKSRMSMHMLCPCRDCKNEQMIEDKDE 57
Query: 68 VKKHLKKVGFMPNYWVWTYNGEMFQNFGAGVNAQASTSHGGTNVETNVEYNED--VNTDQ 125
V HL GFM Y WT +GE V A+ +VE E V +
Sbjct: 58 VHAHLIMNGFMKKYTCWTKHGEQ------EVPDVAAEEVLDQDVENTAAAREGMFVPSPL 111
Query: 126 FNMMDDMVADALGVELSYGEDVEDDEEEQPPNEKAQRFYRLLSESNTPLYEG--SSHSKL 183
D+ L L ED ED++ + ++F +L+ + PLY+G S HSKL
Sbjct: 112 GGETIDLDTQCLSTMLHDIEDAEDNDRDY------EKFSKLVEDCQMPLYDGCKSKHSKL 165
Query: 184 SMCVWLLSHKSNYLNPDDGMDDITKMLKAVTPFKDNLPMNYHAAKRLVMKLGLSVKKIDC 243
S + L+ K++ D ++ ++LK + P +NLP + AK+++ LGL V++I
Sbjct: 166 SCVLELMKLKASNGWSDKSFTELLELLKDLLPEGNNLPQTTYEAKQVLCPLGLEVRRIHA 225
Query: 244 CRNGCMLFYDNEFGINDGPLEECKFCQSPRY--------GISKQKRVAVKSMFYLPIIPR 295
C N C+L+Y L+ C C + RY G ++ VK ++YLPI R
Sbjct: 226 CPNDCILYYKEY-----ADLDVCPICGTSRYKRAKSEGEGSKSKRGGPVKVVWYLPIAER 280
Query: 296 LQRLFASMKTASQMTWH-HSNGISGVMRHPSDGEAWKHFDRVHPDFAADPRNVRLGLCSD 354
++R+FA+ + A + WH + ++RHP+D W+ DR++ +F+ DPRN+R +C+D
Sbjct: 281 MKRMFANKEQAKLVRWHAEERKVDTMLRHPADSVQWRTIDRIYQEFSDDPRNMRFAMCTD 340
Query: 355 GFQPYVQSSAKPYSCWPIIVTPYNLPPEMCMTKPYLFLSCIVPGPDNPKAGIDVFLQPLI 414
G P+ S++ +S WP+++ YNLPP +C + Y+ L+ ++ GP P IDVFL+P+I
Sbjct: 341 GINPFGDLSSR-HSTWPVLLVNYNLPPWLCFKRKYIMLAMLIQGPRQPGNDIDVFLEPII 399
Query: 415 DDLKRLW-VGELTYDISRKQNFKLRAALMWTINDFPAYGMLSGWSTHGRFACPHCMENTK 473
DD +RLW G T+D ++ F L A L TIND+PA G LSG + G++AC CM+ T+
Sbjct: 400 DDFERLWNEGTRTWDAYAQEYFNLHAMLFCTINDYPALGNLSGQTVKGKWACSECMKETR 459
Query: 474 AFTLESGGKSSWFDCHRPFLPHNHPFRRLKNGFKKDERVFVGPPPKITSEEVWTRVC--- 530
+ L+ K+ + HR FLP HP+R ++ F R GPP ++T EV V
Sbjct: 460 SKWLKHSHKTVYMG-HRRFLPRYHPYRNMRKNFN-GHRDTAGPPTELTGTEVHNLVMGIT 517
Query: 531 -DYPKVTDYGRAVP--IPRYGVDHH-----------WTKRSIFWDLPYWKDNLLRHNLDV 576
++ K G+ + + H W K+SIFW LPYWKD +RH +D+
Sbjct: 518 NEFGKKRKVGKRKEKSTSKEKTEEHVEKQKTKERSMWKKKSIFWRLPYWKDLEVRHCIDL 577
Query: 577 MHIEKNVFDNIFNTVMDVKGKTKDNEKARKDLQIYCKRKDLELKPQPNGKLLKPKANYNL 636
MH+EKNV +++ +++ G TKD AR+DL+ R +L +G++ P A Y L
Sbjct: 578 MHVEKNVCESLMGLLLN-PGTTKDGLNARRDLEDMGVRSELHPITTESGRVYLPPACYTL 636
Query: 637 SAQEAKLVCQWLKDLRMPDGYSSNIGRCADVNTGRLHGMKSHDSHIFMENFIPIAFSS-L 695
S +E + L +++P GYSS I R + +L GMKSHD H+ + +P+A + L
Sbjct: 637 SKEEKIDLLTCLSGIKVPSGYSSRISRLVSLQDLKLVGMKSHDCHVLITQLLPVAIRNIL 696
Query: 696 PKHVLDPLIEISQFFKNLCASTLRVDELEKMEKNMPIIICQLEQIFPPGFFDSMEHLPVH 755
P V + + FF + + + L++++ + +C LE FPP FFD MEHLPVH
Sbjct: 697 PPKVRHTIQRLCSFFHAIGQKIIDPEGLDELQAELVRTLCHLEMYFPPTFFDIMEHLPVH 756
Query: 756 LAYEAILGGPVQYRWMYPFERFMGDSKRAVKNLAKVEGSIVAIYTAKETTYFIGHYFVDR 815
L + GP MYP ER++G K V+N + EGSI+ YT +E F Y
Sbjct: 757 LVRQTKCCGPAFMTQMYPCERYLGILKGYVRNRSHPEGSIIESYTTEEAIEFCVDYM--- 813
Query: 816 LLTPSNTRNEVQVNDLRDPSTLSVFNLDGRHAGKVLQYWM-VDQKEMRSAHVHVLINCAE 874
+ + + P + LDG G V + + +D+K AH VL + E
Sbjct: 814 ----------SETSSIGLPRSHHEGRLDG--VGTVGRKTIRLDRKVYDKAHFTVLQHMTE 861
Query: 875 VKPYLEEFIAFYAEFGEGSTVGSIH----EYFPAWFKQRVYNAEPTDEVIRLRQLSQGPL 930
V PY++E +A + G + + F W K R+ + L+ LSQGP
Sbjct: 862 VVPYVDEHLAVIRQENPGRSESWVRNKDMSSFNEWLKNRIARLQNLSSE-TLQWLSQGPE 920
Query: 931 QCANEFHTYFVNGYKFHTHSWTEGKKTINSGVYVKGVTDGG-EDDFYGVIKHIYELSYND 989
A + Y +NGY FHT NSG+ ++ +DGG D +YG ++ I EL Y
Sbjct: 921 WSATTWQGYDINGYTFHTVKQDSKCTVQNSGLRIEAASDGGRRDQYYGKVEQILELDYLK 980
Query: 990 NKVVLFYCDWFDPSPRWTKINKLCNTVDIRVDKKYKEYDPFIMAHNVRQVYYVPYP 1045
KV LF C W D R K++ T + YK+ +PF++A V QV+Y+ P
Sbjct: 981 FKVPLFRCRWVD--LRNVKVDNEGFTTVNLANNAYKD-EPFVLAKQVVQVFYIVDP 1033
>ref|NP_908869.1| B1151H08.16 [Oryza sativa (japonica cultivar-group)]
Length = 1095
Score = 544 bits (1401), Expect = e-153
Identities = 354/1082 (32%), Positives = 545/1082 (49%), Gaps = 95/1082 (8%)
Query: 8 RRWMYDRTFPGRTGLKPQFVEGVDGFISWAWEQEICRDEGGIRCPCLKCRCRHIITDPGD 67
RRWMY + R+ ++ EGV F+++A R + CPC C+ +I D +
Sbjct: 3 RRWMY---YAHRSST--EYREGVTEFVTFADNDRKSRMSMHMLCPCRDCKNEQMIEDKDE 57
Query: 68 VKKHLKKVGFMPNYWVWTYNGEMFQNFGAGVNAQASTSHGGTNVETNVEYNEDVNTDQF- 126
V HL GFM Y WT +GE +A ++ +VE F
Sbjct: 58 VHAHLIMNGFMKKYTCWTKHGEQ----------EAPDVAAEEVLDQDVENTAAAREGMFV 107
Query: 127 -----NMMDDMVADALGVELSYGEDVEDDEEEQPPNEKAQRFYRLLSESNTPLYEG--SS 179
D+ L L ED ED++ + ++F +L+ + PLY+G S
Sbjct: 108 PSPLGGETIDLDTQCLSTMLHDIEDAEDNDRDY------EKFSKLVEDCQMPLYDGCKSK 161
Query: 180 HSKLSMCVWLLSHKSNYLNPDDGMDDITKMLKAVTPFKDNLPMNYHAAKRLVMKLGLSVK 239
H+KLS + L+ K++ D ++ ++LK + P +NLP + AK+++ LGL V+
Sbjct: 162 HNKLSCVLELMKLKASNGWSDKSFTELLELLKDLLPEGNNLPQTTYEAKQVLCPLGLEVR 221
Query: 240 KIDCCRNGCMLFYDNEFGINDGPLEECKFCQSPRYGISKQKRVAVKS--------MFYLP 291
+I C N C+L+Y L+ C C + RY +K + KS ++YLP
Sbjct: 222 RIHACPNDCILYYKEY-----ADLDVCPICGASRYKRAKSEGEGSKSKRGGPAKVVWYLP 276
Query: 292 IIPRLQRLFASMKTASQMTWH-HSNGISGVMRHPSDGEAWKHFDRVHPDFAADPRNVRLG 350
I R++R+FA+ + A + WH + ++RHP+D W+ DR++ +F+ DPRN+R
Sbjct: 277 IAERMKRMFANKEQAKLVRWHAEERKVDTMLRHPADSVQWRTIDRIYQEFSNDPRNMRFA 336
Query: 351 LCSDGFQPYVQSSAKPYSCWPIIVTPYNLPPEMCMTKPYLFLSCIVPGPDNPKAGIDVFL 410
+C+DG P+ S++ +S WP+++ YNLPP +C + Y+ L+ ++ GP P IDVFL
Sbjct: 337 MCTDGINPFGDLSSR-HSTWPVLLVNYNLPPWLCFKRKYIMLAMLIQGPRQPGNDIDVFL 395
Query: 411 QPLIDDLKRLW-VGELTYDISRKQNFKLRAALMWTINDFPAYGMLSGWSTHGRFACPHCM 469
+P+IDD +RLW G T+D ++ F L A L TIND+PA G LSG + G++AC CM
Sbjct: 396 EPIIDDFERLWNEGTRTWDAYAQEYFNLHAMLFCTINDYPALGNLSGQTVKGKWACSECM 455
Query: 470 ENTKAFTLESGGKSSWFDCHRPFLPHNHPFRRLKNGFKKDERVFVGPPPKITSEEVWTRV 529
E T++ L+ K+ + HR FLP HP+R ++ F R GPP ++T EV V
Sbjct: 456 EETRSKWLKHSHKTVYMG-HRRFLPRYHPYRNMRKNFN-GHRDTAGPPTELTGTEVHNLV 513
Query: 530 C----DYPKVTDYGRAVP--IPRYGVDHH-----------WTKRSIFWDLPYWKDNLLRH 572
++ K G+ + + H W K+SIFW LPYWKD +RH
Sbjct: 514 MGITNEFGKKRKVGKRKEKSTSKEKTEEHVEKQKTKERSMWKKKSIFWRLPYWKDLEVRH 573
Query: 573 NLDVMHIEKNVFDNIFNTVMDVKGKTKDNEKARKDLQIYCKRKDLELKPQPNGKLLKPKA 632
+D+MH+EKNV +++ +++ G TKD AR+DL+ R +L +G++ P A
Sbjct: 574 CIDLMHVEKNVCESLMGLLLN-PGTTKDGLNARRDLEDMGVRSELHPITTESGRVYLPPA 632
Query: 633 NYNLSAQEAKLVCQWLKDLRMPDGYSSNIGRCADVNTGRLHGMKSHDSHIFMENFIPIAF 692
Y LS +E + L +++P GYSS I R + +L GMKSHD H+ + +P+A
Sbjct: 633 CYTLSKEEKIDLLTCLSGIKVPSGYSSRISRLVSLQDLKLVGMKSHDCHVLITQLLPVAI 692
Query: 693 SS-LPKHVLDPLIEISQFFKNLCASTLRVDELEKMEKNMPIIICQLEQIFPPGFFDSMEH 751
+ LP V + + FF + + + L++++ + +C LE FPP FFD MEH
Sbjct: 693 RNILPPKVRHTIQRLCSFFHAIGQKIIDPEGLDELQAELVKTLCHLEMYFPPTFFDIMEH 752
Query: 752 LPVHLAYEAILGGPVQYRWMYPFERFMGDSKRAVKNLAKVEGSIVAIYTAKETTYFIGHY 811
LPVHL + GP MYP ER++G K V+N + EGSI+ YT +E F Y
Sbjct: 753 LPVHLVRQTKCCGPAFMTQMYPCERYLGILKGYVRNRSHPEGSIIESYTTEEAIEFCVDY 812
Query: 812 FVDRLLTPSNTRNEVQVNDLRDPSTLSVFNLDGRHAGKVLQYWM-VDQKEMRSAHVHVLI 870
+ + + P + LDG G V + + +D+K AH VL
Sbjct: 813 M-------------SETSSIGLPRSHHEGRLDG--VGTVGRKTIRLDRKVYDKAHFTVLQ 857
Query: 871 NCAEVKPYLEEFIAFYAEFGEG---STVGSIH-EYFPAWFKQRVYNAE--PTDEVIRLRQ 924
+ EV PY++E +A + G S V + H F W K R+ + P++ L+
Sbjct: 858 HMTEVVPYVDEHLAVIRQENPGRSESWVRNKHMSSFNEWLKNRIARLQNLPSE---TLQW 914
Query: 925 LSQGPLQCANEFHTYFVNGYKFHTHSWTEGKKTINSGVYVKGVTDGG-EDDFYGVIKHIY 983
LSQGP A + Y +NGY FHT NSG+ ++ +DGG D +YG ++ I
Sbjct: 915 LSQGPEWSATTWQGYDINGYTFHTVKQDSKCTVQNSGLCIEAASDGGRRDQYYGRVEQIL 974
Query: 984 ELSYNDNKVVLFYCDWFDPSPRWTKINKLCNTVDIRVDKKYKEYDPFIMAHNVRQVYYVP 1043
EL Y KV LF C W D R K++ T + YK+ +PF++A V QV+Y+
Sbjct: 975 ELDYLKFKVPLFRCRWVD--LRNVKVDNEAFTTVNLANNAYKD-EPFVLAKQVVQVFYIV 1031
Query: 1044 YP 1045
P
Sbjct: 1032 DP 1033
>ref|NP_915890.1| putative retrotransposable elements TNP2 [Oryza sativa (japonica
cultivar-group)]
Length = 1095
Score = 544 bits (1401), Expect = e-153
Identities = 354/1080 (32%), Positives = 542/1080 (49%), Gaps = 91/1080 (8%)
Query: 8 RRWMYDRTFPGRTGLKPQFVEGVDGFISWAWEQEICRDEGGIRCPCLKCRCRHIITDPGD 67
RRWMY + R+ ++ EGV F+++A R + CPC C+ +I D +
Sbjct: 3 RRWMY---YAHRSST--EYREGVTEFVTFADNDRKSRMSMHMLCPCRDCKNEQMIEDKDE 57
Query: 68 VKKHLKKVGFMPNYWVWTYNGEMFQNFGAGVNAQASTSHGGTNVETNVEYNEDVNTDQF- 126
V HL GFM Y WT +GE +A ++ +VE F
Sbjct: 58 VHAHLIMNGFMKKYTCWTKHGEQ----------EAPDVAAEEVLDQDVENTAAAREGMFV 107
Query: 127 -----NMMDDMVADALGVELSYGEDVEDDEEEQPPNEKAQRFYRLLSESNTPLYEG--SS 179
D+ L L ED ED++ + ++F +L+ + PLY+G S
Sbjct: 108 PSPLGGETIDLDTQCLSTMLHDIEDAEDNDRDY------EKFSKLVEDCQMPLYDGCKSK 161
Query: 180 HSKLSMCVWLLSHKSNYLNPDDGMDDITKMLKAVTPFKDNLPMNYHAAKRLVMKLGLSVK 239
HSKLS + L+ K++ D ++ ++LK + P +NLP + AK+++ LGL V+
Sbjct: 162 HSKLSCVLELMKLKASNGWSDKSFTELLELLKDLLPEGNNLPQTTYEAKQVLCPLGLEVR 221
Query: 240 KIDCCRNGCMLFYDNEFGINDGPLEECKFCQSPRYGISKQKRVAVKS--------MFYLP 291
+I C N C+L+Y L+ C C + RY +K + KS ++YLP
Sbjct: 222 RIHACPNDCILYYKEY-----ADLDVCPICGASRYKRAKSEGEGSKSKRGGPAKVVWYLP 276
Query: 292 IIPRLQRLFASMKTASQMTWH-HSNGISGVMRHPSDGEAWKHFDRVHPDFAADPRNVRLG 350
I R++R+FA+ + A + WH + ++RHP+D W+ DR++ +F+ DPRN+R
Sbjct: 277 IAERMKRMFANKEQAKLVRWHAEERKVDTMLRHPADSVQWRTIDRIYQEFSNDPRNMRFA 336
Query: 351 LCSDGFQPYVQSSAKPYSCWPIIVTPYNLPPEMCMTKPYLFLSCIVPGPDNPKAGIDVFL 410
+C+DG P+ S++ +S WP+++ YNLPP +C + Y+ L+ ++ GP P IDVFL
Sbjct: 337 MCTDGINPFGDLSSR-HSTWPVLLVNYNLPPWLCFKRKYIMLAMLIQGPRQPGNDIDVFL 395
Query: 411 QPLIDDLKRLW-VGELTYDISRKQNFKLRAALMWTINDFPAYGMLSGWSTHGRFACPHCM 469
+P+IDD +RLW G T+D ++ F L A L TIND+PA G LSG + G++AC CM
Sbjct: 396 EPIIDDFERLWNEGTRTWDAYAQEYFNLHAMLFCTINDYPALGNLSGQTVKGKWACSECM 455
Query: 470 ENTKAFTLESGGKSSWFDCHRPFLPHNHPFRRLKNGFKKDERVFVGPPPKITSEEVWTRV 529
E T++ L+ K+ + HR FLP HP+R ++ F R GPP ++T EV V
Sbjct: 456 EETRSKWLKHSHKTVYMG-HRRFLPRYHPYRNMRKNFN-GHRDTAGPPTELTGTEVHNLV 513
Query: 530 C----DYPKVTDYGRAVP--IPRYGVDHH-----------WTKRSIFWDLPYWKDNLLRH 572
++ K G+ + + H W K+SIFW LPYWKD +RH
Sbjct: 514 MGITNEFGKKRKVGKRKEKSTSKEKTEEHVEKQKTKERSMWKKKSIFWRLPYWKDLEVRH 573
Query: 573 NLDVMHIEKNVFDNIFNTVMDVKGKTKDNEKARKDLQIYCKRKDLELKPQPNGKLLKPKA 632
+D+MH+EKNV +++ +++ G TKD AR+DL+ R +L +G++ P A
Sbjct: 574 CIDLMHVEKNVCESLMGLLLN-PGTTKDGLNARRDLEDMGVRSELHPITTESGRVYLPPA 632
Query: 633 NYNLSAQEAKLVCQWLKDLRMPDGYSSNIGRCADVNTGRLHGMKSHDSHIFMENFIPIAF 692
Y LS +E + L +++P GYSS I R + +L GMKSHD H+ + +PIA
Sbjct: 633 CYTLSKEEKIDLLTCLSGIKVPSGYSSRISRLVSLQDLKLVGMKSHDCHVLITQLLPIAI 692
Query: 693 SS-LPKHVLDPLIEISQFFKNLCASTLRVDELEKMEKNMPIIICQLEQIFPPGFFDSMEH 751
+ LP V + + FF + + + L++++ + +C LE FPP FFD MEH
Sbjct: 693 RNILPPKVRHTIQRLCSFFHAIGQKIIDPEGLDELQAELVRTLCHLEMYFPPTFFDIMEH 752
Query: 752 LPVHLAYEAILGGPVQYRWMYPFERFMGDSKRAVKNLAKVEGSIVAIYTAKETTYFIGHY 811
LPVHL + GP MYP ER++G K V+N + EGSI+ YT +E F Y
Sbjct: 753 LPVHLVRQTKCCGPAFMTQMYPCERYLGILKGYVRNRSHPEGSIIESYTTEEAIEFCVDY 812
Query: 812 FVDRLLTPSNTRNEVQVNDLRDPSTLSVFNLDGRHAGKVLQYWM-VDQKEMRSAHVHVLI 870
+ + + P + LDG G V + + +D+K AH VL
Sbjct: 813 M-------------SETSSIGLPRSHHEGRLDG--VGTVGRKTIRLDRKVYDKAHFTVLQ 857
Query: 871 NCAEVKPYLEEFIAFYAEFGEG---STVGSIH-EYFPAWFKQRVYNAEPTDEVIRLRQLS 926
+ EV PY++E +A + G S V + H F W K R+ + L+ LS
Sbjct: 858 HMTEVVPYVDEHLAVIRQENPGRSESWVRNKHMSSFNEWLKNRIARLQNLSSE-TLQCLS 916
Query: 927 QGPLQCANEFHTYFVNGYKFHTHSWTEGKKTINSGVYVKGVTDGG-EDDFYGVIKHIYEL 985
QGP A + Y +NGY FHT NSG+ ++ +DGG D +YG ++ I EL
Sbjct: 917 QGPEWSATTWQGYDINGYTFHTVKQDSKCTVQNSGLRIEATSDGGRRDQYYGKVEQILEL 976
Query: 986 SYNDNKVVLFYCDWFDPSPRWTKINKLCNTVDIRVDKKYKEYDPFIMAHNVRQVYYVPYP 1045
Y K+ LF C W D R K++ T + YK+ +PF++A V QV+Y+ P
Sbjct: 977 DYLKFKIPLFRCRWVD--LRNVKVDNEGFTTVNLANNAYKD-EPFVLAKQVVQVFYIVDP 1033
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.320 0.138 0.441
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,257,393,104
Number of Sequences: 2540612
Number of extensions: 104911226
Number of successful extensions: 290941
Number of sequences better than 10.0: 886
Number of HSP's better than 10.0 without gapping: 624
Number of HSP's successfully gapped in prelim test: 273
Number of HSP's that attempted gapping in prelim test: 279500
Number of HSP's gapped (non-prelim): 4363
length of query: 1155
length of database: 863,360,394
effective HSP length: 139
effective length of query: 1016
effective length of database: 510,215,326
effective search space: 518378771216
effective search space used: 518378771216
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 81 (35.8 bits)
Medicago: description of AC126782.15


