

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126780.8 - phase: 0 /pseudo
(172 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
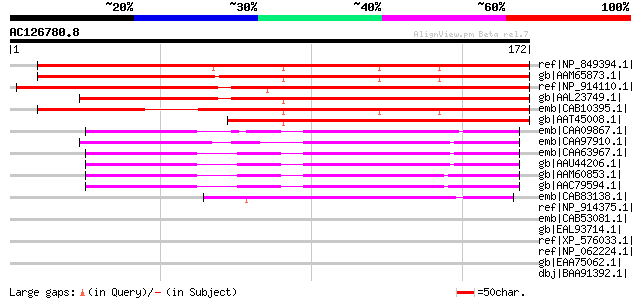
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_849394.1| mitochondrial import inner membrane translocase... 206 2e-52
gb|AAM65873.1| pore protein homolog [Arabidopsis thaliana] gi|18... 202 3e-51
ref|NP_914110.1| B1146F03.20 [Oryza sativa (japonica cultivar-gr... 180 1e-44
gb|AAL23749.1| stress-inducible membrane pore protein [Bromus in... 169 3e-41
emb|CAB10395.1| pore protein homolog [Arabidopsis thaliana] gi|7... 165 5e-40
gb|AAT45008.1| stress-inducible membrane pore-like protein [Xero... 130 2e-29
emb|CAA09867.1| amino acid selective channel protein [Hordeum vu... 84 2e-15
emb|CAA97910.1| core protein [Pisum sativum] gi|7488792|pir||T06... 83 3e-15
emb|CAA63967.1| pom14 [Solanum tuberosum] 82 6e-15
gb|AAU44206.1| putative amino acid selective channel protein [Or... 81 1e-14
gb|AAM60853.1| putative membrane channel protein [Arabidopsis th... 74 2e-12
gb|AAC79594.1| putative membrane channel protein [Arabidopsis th... 74 2e-12
emb|CAB83138.1| putative protein [Arabidopsis thaliana] gi|26450... 49 4e-05
ref|NP_914375.1| P0698H10.23 [Oryza sativa (japonica cultivar-gr... 39 0.045
emb|CAB53081.1| SPCC16A11.09c [Schizosaccharomyces pombe] gi|190... 38 0.099
gb|EAL93714.1| Mitochondrial import inner membrane translocase s... 38 0.13
ref|XP_576033.1| PREDICTED: similar to translocator of inner mit... 37 0.29
ref|NP_062224.1| translocator of inner mitochondrial membrane 17... 36 0.38
gb|EAA75062.1| hypothetical protein FG06120.1 [Gibberella zeae P... 35 0.64
dbj|BAA91392.1| unnamed protein product [Homo sapiens] gi|128030... 35 0.84
>ref|NP_849394.1| mitochondrial import inner membrane translocase subunit
Tim17/Tim22/Tim23 family protein [Arabidopsis thaliana]
Length = 178
Score = 206 bits (525), Expect = 2e-52
Identities = 109/171 (63%), Positives = 131/171 (75%), Gaps = 8/171 (4%)
Query: 10 RTLLDELSDFNKGGLFDFGHPLVNRIAESFVKAAGIGAVQAVSREAYFTVIEGTGID-NA 68
R ++DE+ F K LFD GHPL+NRIA+SFVKAAG+GA+QAVSREAYFTV++G G D N
Sbjct: 7 RIVMDEIRSFEKAHLFDLGHPLLNRIADSFVKAAGVGALQAVSREAYFTVVDGAGFDSNN 66
Query: 69 GGMPPEISGAKKNRFHGLRDQ-----Q*IY*GNGKESFQWGLAAGLYSGLTYGMKEAR-G 122
G P EI+G KK+RF LR + + GKES QWGLAAGLYSG+TYGM E R G
Sbjct: 67 VGPPSEITGNKKHRFPNLRGESSKSLDALVKNTGKESLQWGLAAGLYSGITYGMTEVRGG 126
Query: 123 THDWKNSAVAGAITGAALA-CTSDNTSHEQIAQCAITGAAISTAANLLTGI 172
HDW+NSAVAGA+TGAA+A TS+ TSHEQ+ Q A+TGAAISTAANLL+ +
Sbjct: 127 AHDWRNSAVAGALTGAAMAMTTSERTSHEQVVQSALTGAAISTAANLLSSV 177
>gb|AAM65873.1| pore protein homolog [Arabidopsis thaliana]
gi|18414605|ref|NP_567488.1| mitochondrial import inner
membrane translocase subunit Tim17/Tim22/Tim23 family
protein [Arabidopsis thaliana]
Length = 176
Score = 202 bits (514), Expect = 3e-51
Identities = 107/170 (62%), Positives = 130/170 (75%), Gaps = 8/170 (4%)
Query: 10 RTLLDELSDFNKGGLFDFGHPLVNRIAESFVKAAGIGAVQAVSREAYFTVIEGTGIDNAG 69
R ++DE+ F K LFD GHPL+NRIA+SFVKAAG+GA+QAVSREAYFTV++G +N
Sbjct: 7 RIVMDEIRSFEKAHLFDLGHPLLNRIADSFVKAAGVGALQAVSREAYFTVVDGFDSNNV- 65
Query: 70 GMPPEISGAKKNRFHGLRDQ-----Q*IY*GNGKESFQWGLAAGLYSGLTYGMKEAR-GT 123
G P EI+G KK+RF LR + + GKES QWGLAAGLYSG+TYGM E R G
Sbjct: 66 GPPSEITGNKKHRFPNLRGESSKSLDALVKNTGKESLQWGLAAGLYSGITYGMTEVRGGA 125
Query: 124 HDWKNSAVAGAITGAALA-CTSDNTSHEQIAQCAITGAAISTAANLLTGI 172
HDW+NSAVAGA+TGAA+A TS+ TSHEQ+ Q A+TGAAISTAANLL+ +
Sbjct: 126 HDWRNSAVAGALTGAAMAMTTSERTSHEQVVQSALTGAAISTAANLLSSV 175
>ref|NP_914110.1| B1146F03.20 [Oryza sativa (japonica cultivar-group)]
gi|21104571|dbj|BAB93165.1| putative stress-inducible
membrane pore protein [Oryza sativa (japonica
cultivar-group)] gi|12060507|dbj|BAB20636.1| putative
stress-inducible membrane pore protein [Oryza sativa
(japonica cultivar-group)]
Length = 263
Score = 180 bits (457), Expect = 1e-44
Identities = 94/174 (54%), Positives = 124/174 (71%), Gaps = 8/174 (4%)
Query: 3 SNSNLETRTLLDELSDFNKGGLFDFGHPLVNRIAESFVKAAGIGAVQAVSREAYFTVIEG 62
S+S L+T TL +E++ +K L D GHPLVNR+A+SF++AAG+GA +AVSREAYF +EG
Sbjct: 93 SSSRLDTWTLKEEVASMDKRWLVDLGHPLVNRVADSFIRAAGVGAARAVSREAYFVTVEG 152
Query: 63 TGIDNAGGMPPEISGAKKNRFH----GLRDQQ*IY*GNGKESFQWGLAAGLYSGLTYGMK 118
G D AG + K++ F G + + GKE+FQWGLAAG+YSGLTYG++
Sbjct: 153 LGGDTAG----LDNAVKRSNFSRGDDGQKSLDAVVKSAGKEAFQWGLAAGVYSGLTYGLR 208
Query: 119 EARGTHDWKNSAVAGAITGAALACTSDNTSHEQIAQCAITGAAISTAANLLTGI 172
EARG HDWKNSAVAGAI G A+A T D + + + AITGAA+S+AA+LL+GI
Sbjct: 209 EARGCHDWKNSAVAGAIAGVAVALTGDTGNADHMVHFAITGAALSSAASLLSGI 262
>gb|AAL23749.1| stress-inducible membrane pore protein [Bromus inermis]
Length = 157
Score = 169 bits (428), Expect = 3e-41
Identities = 85/155 (54%), Positives = 111/155 (70%), Gaps = 10/155 (6%)
Query: 24 LFDFGHPLVNRIAESFVKAAGIGAVQAVSREAYFTVIEGTGIDNAGGMPPEISGAKKNRF 83
L D GHPL+NR+A+SF++AAG+GA +AVSREAYF +EG G D+ G S AK++ F
Sbjct: 6 LLDLGHPLLNRVADSFIRAAGVGAARAVSREAYFVTVEGMGGDSTG----LDSNAKRSHF 61
Query: 84 HGLRDQ------Q*IY*GNGKESFQWGLAAGLYSGLTYGMKEARGTHDWKNSAVAGAITG 137
R + + GKE+FQWGLAAG+YSGLTYG++EARG HDWKNSA+AGA+ G
Sbjct: 62 SSARGDDGQKSFEVVVKSAGKEAFQWGLAAGVYSGLTYGLREARGCHDWKNSAIAGALAG 121
Query: 138 AALACTSDNTSHEQIAQCAITGAAISTAANLLTGI 172
AA+A T DN + + AITGAA+S+AA +L+GI
Sbjct: 122 AAVALTGDNGHSDHVVHFAITGAALSSAATMLSGI 156
>emb|CAB10395.1| pore protein homolog [Arabidopsis thaliana]
gi|7268365|emb|CAB78658.1| pore protein homolog
[Arabidopsis thaliana] gi|7485023|pir||A71428
hypothetical protein - Arabidopsis thaliana
Length = 160
Score = 165 bits (417), Expect = 5e-40
Identities = 93/170 (54%), Positives = 113/170 (65%), Gaps = 24/170 (14%)
Query: 10 RTLLDELSDFNKGGLFDFGHPLVNRIAESFVKAAGIGAVQAVSREAYFTVIEGTGIDNAG 69
R ++DE+ F K LFD GHPL+NRIA+SFVKAAG+ + N
Sbjct: 7 RIVMDEIRSFEKAHLFDLGHPLLNRIADSFVKAAGV-----------------SFDSNNV 49
Query: 70 GMPPEISGAKKNRFHGLRDQ-----Q*IY*GNGKESFQWGLAAGLYSGLTYGMKEAR-GT 123
G P EI+G KK+RF LR + + GKES QWGLAAGLYSG+TYGM E R G
Sbjct: 50 GPPSEITGNKKHRFPNLRGESSKSLDALVKNTGKESLQWGLAAGLYSGITYGMTEVRGGA 109
Query: 124 HDWKNSAVAGAITGAALA-CTSDNTSHEQIAQCAITGAAISTAANLLTGI 172
HDW+NSAVAGA+TGAA+A TS+ TSHEQ+ Q A+TGAAISTAANLL+ +
Sbjct: 110 HDWRNSAVAGALTGAAMAMTTSERTSHEQVVQSALTGAAISTAANLLSSV 159
>gb|AAT45008.1| stress-inducible membrane pore-like protein [Xerophyta humilis]
Length = 110
Score = 130 bits (326), Expect = 2e-29
Identities = 64/106 (60%), Positives = 80/106 (75%), Gaps = 6/106 (5%)
Query: 73 PEISGAKKNRFHGLRDQ------Q*IY*GNGKESFQWGLAAGLYSGLTYGMKEARGTHDW 126
P+++ K NRF LR + + GKE+FQWGLAAG+YSGLTYG+KEARGTHDW
Sbjct: 4 PDMTAGKNNRFPNLRGDNSSKSLEEMVKATGKEAFQWGLAAGVYSGLTYGLKEARGTHDW 63
Query: 127 KNSAVAGAITGAALACTSDNTSHEQIAQCAITGAAISTAANLLTGI 172
KNSA+AGAITGAALA T++ ++ + + Q A+TGAAIST ANLL GI
Sbjct: 64 KNSAIAGAITGAALALTTEKSNSDHVVQSAVTGAAISTVANLLRGI 109
>emb|CAA09867.1| amino acid selective channel protein [Hordeum vulgare subsp.
vulgare]
Length = 144
Score = 83.6 bits (205), Expect = 2e-15
Identities = 50/144 (34%), Positives = 76/144 (52%), Gaps = 21/144 (14%)
Query: 26 DFGHPLVNRIAESFVKAAGIGAVQAVSREAYFTVIEGTGIDNAGGMPPEISGAKKNRFHG 85
D G+PL+NR + F+K +GA + V+ +A+ + G +IS K+
Sbjct: 18 DLGNPLLNRTVDGFLKIGAVGACRVVAEDAFDCLHRG-----------DIS--KRQLEET 64
Query: 86 LRDQQ*IY*GNGKESFQWGLAAGLYSGLTYGMKEARGTHDWKNSAVAGAITGAALACTSD 145
L+ KE WG AG+Y G+ YG++ RG DWKN+ + G TGA ++ S+
Sbjct: 65 LKKMC-------KEGAYWGAVAGVYVGMEYGVERVRGDRDWKNALIGGIATGALVSAASN 117
Query: 146 NTSHEQIAQCAITGAAISTAANLL 169
N + +IAQ AITG AI+TA +
Sbjct: 118 NKGN-KIAQDAITGGAIATAVEFI 140
>emb|CAA97910.1| core protein [Pisum sativum] gi|7488792|pir||T06471 core protein -
garden pea
Length = 146
Score = 83.2 bits (204), Expect = 3e-15
Identities = 47/146 (32%), Positives = 76/146 (51%), Gaps = 21/146 (14%)
Query: 24 LFDFGHPLVNRIAESFVKAAGIGAVQAVSREAYFTVIEGTGIDNAGGMPPEISGAKKNRF 83
+ D G+P +N + F+K + A ++V+ + + + +G+ N + + K
Sbjct: 18 VIDMGNPFLNLTVDGFLKIGAVAATRSVAEDTFHIIRKGSISSN------DFEKSLKKMC 71
Query: 84 HGLRDQQ*IY*GNGKESFQWGLAAGLYSGLTYGMKEARGTHDWKNSAVAGAITGAALACT 143
KE WG AG+Y G+ YG++ RGT DWKN+ GA+TGA ++
Sbjct: 72 --------------KEGAYWGAIAGVYVGMEYGVERIRGTRDWKNAMFGGAVTGALVSAA 117
Query: 144 SDNTSHEQIAQCAITGAAISTAANLL 169
S+N ++IA AITGAAI+TAA +
Sbjct: 118 SNN-KKDKIAVDAITGAAIATAAEFI 142
>emb|CAA63967.1| pom14 [Solanum tuberosum]
Length = 146
Score = 82.0 bits (201), Expect = 6e-15
Identities = 46/144 (31%), Positives = 73/144 (49%), Gaps = 21/144 (14%)
Query: 26 DFGHPLVNRIAESFVKAAGIGAVQAVSREAYFTVIEGTGIDNAGGMPPEISGAKKNRFHG 85
D G+P +N ++F+ + A + V+ E Y V G S + N
Sbjct: 20 DMGNPFLNHTVDAFLNIGTVAATKTVAEETYGMVTRG-------------SVSSHNFEKS 66
Query: 86 LRDQQ*IY*GNGKESFQWGLAAGLYSGLTYGMKEARGTHDWKNSAVAGAITGAALACTSD 145
L+ KE WG AG+Y+G+ YG + RGT+DWKN+ + GA+TGA ++ +
Sbjct: 67 LKKMC-------KEGAYWGTVAGVYAGMEYGAERIRGTNDWKNAMIGGALTGALISAACN 119
Query: 146 NTSHEQIAQCAITGAAISTAANLL 169
N + ++I AITG A++TA+ L
Sbjct: 120 N-NRDKIVMDAITGGAVATASEFL 142
>gb|AAU44206.1| putative amino acid selective channel protein [Oryza sativa
(japonica cultivar-group)]
Length = 146
Score = 80.9 bits (198), Expect = 1e-14
Identities = 44/144 (30%), Positives = 73/144 (50%), Gaps = 21/144 (14%)
Query: 26 DFGHPLVNRIAESFVKAAGIGAVQAVSREAYFTVIEGTGIDNAGGMPPEISGAKKNRFHG 85
D G+P +NR + F+K +GA + + + + + G +K H
Sbjct: 20 DMGNPFLNRTVDGFLKIGAVGACKVAAEDTFDCLHRG-------------DVSKHKLEHM 66
Query: 86 LRDQQ*IY*GNGKESFQWGLAAGLYSGLTYGMKEARGTHDWKNSAVAGAITGAALACTSD 145
L+ KE WG AG+Y G+ YG++ RG HDWKN+ + GA++GA ++ S+
Sbjct: 67 LKKMC-------KEGAYWGTVAGVYVGMEYGVERIRGRHDWKNAMIGGALSGALISAASN 119
Query: 146 NTSHEQIAQCAITGAAISTAANLL 169
N ++I + AITG A++TA +
Sbjct: 120 N-HKDKIIKDAITGGAVATAVEFI 142
>gb|AAM60853.1| putative membrane channel protein [Arabidopsis thaliana]
Length = 148
Score = 73.6 bits (179), Expect = 2e-12
Identities = 43/144 (29%), Positives = 70/144 (47%), Gaps = 21/144 (14%)
Query: 26 DFGHPLVNRIAESFVKAAGIGAVQAVSREAYFTVIEGTGIDNAGGMPPEISGAKKNRFHG 85
D G+P +N ++F+K +G ++++ + Y + +G S +K H
Sbjct: 20 DMGNPFLNLTVDAFLKIGAVGVTKSLAEDTYKAIEKG-------------SLSKSTLEHA 66
Query: 86 LRDQQ*IY*GNGKESFQWGLAAGLYSGLTYGMKEARGTHDWKNSAVAGAITGAALACTSD 145
L+ KE WG A G+Y G YG++ RG+ DWKN+ +AGA TGA L+
Sbjct: 67 LKKLC-------KEGVYWGAAGGVYIGTEYGIERIRGSRDWKNAMLAGAATGAVLSAVG- 118
Query: 146 NTSHEQIAQCAITGAAISTAANLL 169
+ I AI G A++TA+ +
Sbjct: 119 KKGKDTIVIDAILGGALATASQFV 142
>gb|AAC79594.1| putative membrane channel protein [Arabidopsis thaliana]
gi|20147377|gb|AAM10398.1| At2g28900/F8N16.19
[Arabidopsis thaliana] gi|15010584|gb|AAK73951.1|
At2g28900/F8N16.19 [Arabidopsis thaliana]
gi|15226998|ref|NP_180456.1| mitochondrial import inner
membrane translocase subunit Tim17/Tim22/Tim23 family
protein [Arabidopsis thaliana] gi|25354609|pir||C84690
probable membrane channel protein [imported] -
Arabidopsis thaliana
Length = 148
Score = 73.6 bits (179), Expect = 2e-12
Identities = 43/144 (29%), Positives = 70/144 (47%), Gaps = 21/144 (14%)
Query: 26 DFGHPLVNRIAESFVKAAGIGAVQAVSREAYFTVIEGTGIDNAGGMPPEISGAKKNRFHG 85
D G+P +N ++F+K +G ++++ + Y + +G S +K H
Sbjct: 20 DMGNPFLNLTVDAFLKIGAVGVTKSLAEDTYKAIDKG-------------SLSKSTLEHA 66
Query: 86 LRDQQ*IY*GNGKESFQWGLAAGLYSGLTYGMKEARGTHDWKNSAVAGAITGAALACTSD 145
L+ KE WG A G+Y G YG++ RG+ DWKN+ +AGA TGA L+
Sbjct: 67 LKKLC-------KEGVYWGAAGGVYIGTEYGIERIRGSRDWKNAMLAGAATGAVLSAVG- 118
Query: 146 NTSHEQIAQCAITGAAISTAANLL 169
+ I AI G A++TA+ +
Sbjct: 119 KKGKDTIVIDAILGGALATASQFV 142
>emb|CAB83138.1| putative protein [Arabidopsis thaliana] gi|26450248|dbj|BAC42241.1|
unknown protein [Arabidopsis thaliana]
gi|28827678|gb|AAO50683.1| unknown protein [Arabidopsis
thaliana] gi|15229339|ref|NP_191847.1| mitochondrial
import inner membrane translocase subunit
Tim17/Tim22/Tim23 family protein [Arabidopsis thaliana]
gi|11357679|pir||T48077 hypothetical protein F26K9.310 -
Arabidopsis thaliana
Length = 136
Score = 49.3 bits (116), Expect = 4e-05
Identities = 33/106 (31%), Positives = 55/106 (51%), Gaps = 5/106 (4%)
Query: 65 IDNAGGMPPEISG---AKKNRFHGLRDQQ*IY*GNGKESFQWGLAAGLYSGLTYGMKEAR 121
+ AGG+ +G A+K G+ + G+ FQ GL +G+++ G++ R
Sbjct: 22 VATAGGLYGLCAGPRDARKIGLSGVSQASFVAKSIGRFGFQCGLVSGVFTMTHCGLQRYR 81
Query: 122 GTHDWKNSAVAGAITGAALACTSDNTSHEQIAQCAITGAAISTAAN 167
G +DW N+ V GA+ GAA+A ++ N + Q+ A +A S AN
Sbjct: 82 GKNDWVNALVGGAVAGAAVAISTRNWT--QVVGMAGLVSAFSVLAN 125
>ref|NP_914375.1| P0698H10.23 [Oryza sativa (japonica cultivar-group)]
Length = 84
Score = 39.3 bits (90), Expect = 0.045
Identities = 20/43 (46%), Positives = 29/43 (66%), Gaps = 1/43 (2%)
Query: 127 KNSAVAGAITGAALACTSDNTSHEQIAQCAITGAAISTAANLL 169
KN+ V GA+TGA ++ S N+ + + + AITG AI+TAA L
Sbjct: 39 KNAMVGGAVTGALVSAAS-NSHRQNVVKNAITGGAIATAAEFL 80
>emb|CAB53081.1| SPCC16A11.09c [Schizosaccharomyces pombe]
gi|19075496|ref|NP_587996.1| hypothetical protein
SPCC16A11.09c [Schizosaccharomyces pombe 972h-]
gi|7522504|pir||T41082 probable mitochondrial import
inner membrane translocase subunit [imported] - fission
yeast (Schizosaccharomyces pombe)
gi|18203510|sp|Q9USM7|TIM23_SCHPO Mitochondrial import
inner membrane translocase subunit tim23
Length = 210
Score = 38.1 bits (87), Expect = 0.099
Identities = 22/67 (32%), Positives = 35/67 (51%), Gaps = 6/67 (8%)
Query: 104 GLAAGLYSGLTYGMKEARGTHDWKNSAVAGAITGAALACTSDNTSHEQIAQCAITGAAIS 163
G+ A +Y+G+ + R H W+NS AGA+TGA T + AI+ + ++
Sbjct: 139 GVLALVYNGINSLIGYKRQKHGWENSVAAGALTGALYKST------RGLRAMAISSSLVA 192
Query: 164 TAANLLT 170
TAA + T
Sbjct: 193 TAAGIWT 199
>gb|EAL93714.1| Mitochondrial import inner membrane translocase subunit TIM23,
putative [Aspergillus fumigatus Af293]
Length = 212
Score = 37.7 bits (86), Expect = 0.13
Identities = 18/40 (45%), Positives = 24/40 (60%)
Query: 104 GLAAGLYSGLTYGMKEARGTHDWKNSAVAGAITGAALACT 143
G+ A +Y+G G+ ARG HD NS VAGA++G T
Sbjct: 138 GVVAMVYNGFNSGLGYARGKHDAANSIVAGALSGMLFKST 177
>ref|XP_576033.1| PREDICTED: similar to translocator of inner mitochondrial membrane
17a [Rattus norvegicus]
Length = 171
Score = 36.6 bits (83), Expect = 0.29
Identities = 27/71 (38%), Positives = 36/71 (50%), Gaps = 8/71 (11%)
Query: 103 WGLAAGLYSGLTYGMKEARGTHDWKNSAVAGAITGAALACTSDNTSHEQIAQCAITG--- 159
WG GL+S + GM + RG D NS +GA+TGA LA + + + A+ G
Sbjct: 69 WG---GLFSTIDCGMVQIRGKEDRWNSITSGALTGAILAARTGPVA--MVGSAAMGGILL 123
Query: 160 AAISTAANLLT 170
A I A LLT
Sbjct: 124 ALIEGAGILLT 134
>ref|NP_062224.1| translocator of inner mitochondrial membrane 17a [Rattus
norvegicus] gi|3219807|sp|O35092|TI17A_RAT Mitochondrial
import inner membrane translocase subunit Tim17 A (Inner
membrane preprotein translocase Tim17a)
gi|2335037|dbj|BAA21818.1| Tim17 [Rattus norvegicus]
Length = 171
Score = 36.2 bits (82), Expect = 0.38
Identities = 28/71 (39%), Positives = 36/71 (50%), Gaps = 8/71 (11%)
Query: 103 WGLAAGLYSGLTYGMKEARGTHDWKNSAVAGAITGAALACTSDNTSHEQIAQCAITG--- 159
WG GL+S + GM + RG D NS +GA+TGA LA + N + A+ G
Sbjct: 69 WG---GLFSTIDCGMVQIRGKEDPWNSITSGALTGAILA--ARNGPVAMVGSAAMGGILL 123
Query: 160 AAISTAANLLT 170
A I A LLT
Sbjct: 124 ALIEGAGILLT 134
>gb|EAA75062.1| hypothetical protein FG06120.1 [Gibberella zeae PH-1]
gi|46123485|ref|XP_386296.1| hypothetical protein
FG06120.1 [Gibberella zeae PH-1]
Length = 178
Score = 35.4 bits (80), Expect = 0.64
Identities = 22/63 (34%), Positives = 30/63 (46%), Gaps = 3/63 (4%)
Query: 103 WGLAAGLYSGLTYGMKEARGTHDWKNSAVAGAITGAALACTSDNTSHEQIAQCAITGAAI 162
+G LYSG+ G++ R +D NS AG +TG LA N + A + AA
Sbjct: 107 FGKVGALYSGVECGIEGLRAKNDLTNSVAAGCLTGGILA---KNAGPQAAAGGCLAFAAF 163
Query: 163 STA 165
S A
Sbjct: 164 SAA 166
>dbj|BAA91392.1| unnamed protein product [Homo sapiens] gi|12803047|gb|AAH02324.1|
Translocase of inner mitochondrial membrane 22 homolog
[Homo sapiens] gi|24638462|sp|Q9Y584|TIM22_HUMAN
Mitochondrial import inner membrane translocase subunit
Tim22 gi|56606061|ref|NP_037469.2| translocase of inner
mitochondrial membrane 22 homolog [Homo sapiens]
Length = 194
Score = 35.0 bits (79), Expect = 0.84
Identities = 14/38 (36%), Positives = 23/38 (59%)
Query: 103 WGLAAGLYSGLTYGMKEARGTHDWKNSAVAGAITGAAL 140
+ + ++S ++ RGT DWKNS ++G ITG A+
Sbjct: 129 FAIVGAMFSCTECLIESYRGTSDWKNSVISGCITGGAI 166
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.322 0.137 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 282,113,374
Number of Sequences: 2540612
Number of extensions: 10919063
Number of successful extensions: 21270
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 55
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 21189
Number of HSP's gapped (non-prelim): 71
length of query: 172
length of database: 863,360,394
effective HSP length: 119
effective length of query: 53
effective length of database: 561,027,566
effective search space: 29734460998
effective search space used: 29734460998
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 70 (31.6 bits)
Medicago: description of AC126780.8


