

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126009.1 - phase: 0
(326 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
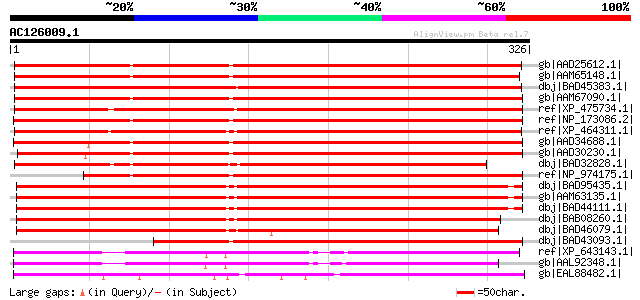
Score E
Sequences producing significant alignments: (bits) Value
gb|AAD25612.1| Unknown protein [Arabidopsis thaliana] gi|1738611... 413 e-114
gb|AAM65148.1| unknown [Arabidopsis thaliana] gi|20258911|gb|AAM... 412 e-114
dbj|BAD45383.1| LEM3-like [Oryza sativa (japonica cultivar-group)] 409 e-113
gb|AAM67090.1| unknown [Arabidopsis thaliana] gi|18412377|ref|NP... 408 e-112
ref|XP_475734.1| putative membrane protein [Oryza sativa (japoni... 407 e-112
ref|NP_173086.2| LEM3 (ligand-effect modulator 3) family protein... 407 e-112
ref|XP_464311.1| LEM3 (ligand-effect modulator 3)-like [Oryza sa... 397 e-109
gb|AAD34688.1| >F3O9.16 [Arabidopsis thaliana] gi|25347303|pir||... 396 e-109
gb|AAD30230.1| T8K14.13 [Arabidopsis thaliana] gi|25347301|pir||... 395 e-109
dbj|BAD32828.1| hypothetical protein [Lotus corniculatus var. ja... 366 e-100
ref|NP_974175.1| LEM3 (ligand-effect modulator 3) family protein... 355 8e-97
dbj|BAD95435.1| hypothetical protein [Arabidopsis thaliana] gi|3... 354 2e-96
gb|AAM63135.1| unknown [Arabidopsis thaliana] 353 4e-96
dbj|BAD44111.1| unknown protein [Arabidopsis thaliana] 353 5e-96
dbj|BAB08260.1| unnamed protein product [Arabidopsis thaliana] 343 4e-93
dbj|BAD46079.1| LEM3 (ligand-effect modulator 3)-like [Oryza sat... 338 1e-91
dbj|BAD43093.1| unknown protein [Arabidopsis thaliana] 325 1e-87
ref|XP_643143.1| hypothetical protein DDB0217816 [Dictyostelium ... 235 1e-60
gb|AAL92348.1| similar to membrane protein common family; protei... 212 1e-53
gb|EAL88482.1| LEM3/CDC50 family protein [Aspergillus fumigatus ... 207 3e-52
>gb|AAD25612.1| Unknown protein [Arabidopsis thaliana] gi|17386112|gb|AAL38602.1|
At1g54320/F20D21_50 [Arabidopsis thaliana]
gi|18404877|ref|NP_564656.1| LEM3 (ligand-effect
modulator 3) family protein / CDC50 family protein
[Arabidopsis thaliana] gi|15450729|gb|AAK96636.1|
At1g54320/F20D21_50 [Arabidopsis thaliana]
gi|15010742|gb|AAK74030.1| At1g54320/F20D21_50
[Arabidopsis thaliana] gi|25347305|pir||G96584
hypothetical protein F20D21.14 [imported] - Arabidopsis
thaliana
Length = 349
Score = 413 bits (1061), Expect = e-114
Identities = 196/318 (61%), Positives = 240/318 (74%), Gaps = 3/318 (0%)
Query: 4 SRFSQQELHAWQPILTPGWVIAIFTFIGLVFIPIGVASLFASEQVVEVPLRYDDQCLPSL 63
S+F+QQEL A +PILTPGWVI+ F + ++FIP+GV SLFAS+ VVE+ RYD +C+P+
Sbjct: 31 SKFTQQELPACKPILTPGWVISTFLIVSVIFIPLGVISLFASQDVVEIVDRYDTECIPAP 90
Query: 64 YKDDAMTYIKGNRISKTCTKKLTVKSKMKAPIYVYYQLSNFYQNHRHYVKSRDHKQLRSK 123
+ + + YI+G+ K C + L V +MK PIYVYYQL NFYQNHR YVKSR QLRS
Sbjct: 91 ARTNKVAYIQGDG-DKVCNRDLKVTKRMKQPIYVYYQLENFYQNHRRYVKSRSDSQLRST 149
Query: 124 ADENDVGKCFPEDYTANGYLPVVPCGLAAWSLFNDTYRFSNNNKDLVINKKNIAWKSDQK 183
EN + C PED G P+VPCGL AWSLFNDTY S NN L +NKK IAWKSD++
Sbjct: 150 KYENQISACKPEDDV--GGQPIVPCGLIAWSLFNDTYALSRNNVSLAVNKKGIAWKSDKE 207
Query: 184 AKFGSDVYPKNFQTGSLIGGARLNESIPLSEQEDLIVWMRTAALPTFRKLYGKIEVDLEA 243
KFG+ V+PKNFQ G++ GGA L+ IPLSEQEDLIVWMRTAALPTFRKLYGKIE DLE
Sbjct: 208 HKFGNKVFPKNFQKGNITGGATLDPRIPLSEQEDLIVWMRTAALPTFRKLYGKIESDLEM 267
Query: 244 NDEITVVIENNYNTYQFGGTKSVILSTTTWIGGKNDFLGIAYILIGGLSLVYSLVFLLMY 303
D I V + NNYNTY F G K ++LSTT+W+GGKNDFLGIAY+ +GG+ + +L F +MY
Sbjct: 268 GDTIHVKLNNNYNTYSFNGKKKLVLSTTSWLGGKNDFLGIAYLTVGGICFILALAFTIMY 327
Query: 304 LMKPRPLGDPRYLTWNKN 321
L+KPR LGDP YL+WN+N
Sbjct: 328 LVKPRRLGDPSYLSWNRN 345
>gb|AAM65148.1| unknown [Arabidopsis thaliana] gi|20258911|gb|AAM14149.1| unknown
protein [Arabidopsis thaliana]
gi|15028095|gb|AAK76578.1| unknown protein [Arabidopsis
thaliana] gi|11994416|dbj|BAB02418.1| unnamed protein
product [Arabidopsis thaliana]
gi|15294236|gb|AAK95295.1| AT3g12740/MBK21_10
[Arabidopsis thaliana] gi|18399730|ref|NP_566435.1| LEM3
(ligand-effect modulator 3) family protein / CDC50
family protein [Arabidopsis thaliana]
Length = 350
Score = 412 bits (1058), Expect = e-114
Identities = 197/317 (62%), Positives = 242/317 (76%), Gaps = 3/317 (0%)
Query: 4 SRFSQQELHAWQPILTPGWVIAIFTFIGLVFIPIGVASLFASEQVVEVPLRYDDQCLPSL 63
S+F+QQEL A +PILTPGWVI+ F I ++FIP+GV SLFAS+ VVE+ RYD C+P
Sbjct: 32 SKFTQQELPACKPILTPGWVISTFLIISVIFIPLGVISLFASQDVVEIVDRYDSACIPLS 91
Query: 64 YKDDAMTYIKGNRISKTCTKKLTVKSKMKAPIYVYYQLSNFYQNHRHYVKSRDHKQLRSK 123
+ + + YI+G +K+CT+ L V +MK PIYVYYQL NFYQNHR YVKSR QLRS
Sbjct: 92 DRANKVAYIQGTG-NKSCTRTLIVPKRMKQPIYVYYQLENFYQNHRRYVKSRSDSQLRSV 150
Query: 124 ADENDVGKCFPEDYTANGYLPVVPCGLAAWSLFNDTYRFSNNNKDLVINKKNIAWKSDQK 183
DEN + C PED G P+VPCGL AWSLFNDTY S NN+ L +NKK IAWKSD++
Sbjct: 151 KDENQIDACKPEDDF--GGQPIVPCGLIAWSLFNDTYVLSRNNQGLTVNKKGIAWKSDKE 208
Query: 184 AKFGSDVYPKNFQTGSLIGGARLNESIPLSEQEDLIVWMRTAALPTFRKLYGKIEVDLEA 243
KFG +V+PKNFQ G+L GGA L+ + PLS+QEDLIVWMRTAALPTFRKLYGKIE DLE
Sbjct: 209 HKFGKNVFPKNFQKGNLTGGASLDPNKPLSDQEDLIVWMRTAALPTFRKLYGKIESDLEK 268
Query: 244 NDEITVVIENNYNTYQFGGTKSVILSTTTWIGGKNDFLGIAYILIGGLSLVYSLVFLLMY 303
+ I V ++NNYNTY F G K ++LSTT+W+GGKNDFLGIAY+ +GG+ V +L F +MY
Sbjct: 269 GENIQVTLQNNYNTYSFSGKKKLVLSTTSWLGGKNDFLGIAYLTVGGICFVLALAFTVMY 328
Query: 304 LMKPRPLGDPRYLTWNK 320
L+KPR LGDP YL+WN+
Sbjct: 329 LVKPRRLGDPTYLSWNR 345
>dbj|BAD45383.1| LEM3-like [Oryza sativa (japonica cultivar-group)]
Length = 358
Score = 409 bits (1051), Expect = e-113
Identities = 188/318 (59%), Positives = 241/318 (75%), Gaps = 1/318 (0%)
Query: 4 SRFSQQELHAWQPILTPGWVIAIFTFIGLVFIPIGVASLFASEQVVEVPLRYDDQCLPSL 63
S+F+QQEL A +PILTP WV+++F +G++F+P+GV SL A++ VVE+ RYDD C+P+
Sbjct: 38 SKFTQQELPACKPILTPKWVVSVFFLVGVIFVPVGVVSLLAAQNVVEIVDRYDDACVPAN 97
Query: 64 YKDDAMTYIKGNRISKTCTKKLTVKSKMKAPIYVYYQLSNFYQNHRHYVKSRDHKQLRSK 123
D+ + YI+ ISK CT+ LT+ M PI+VYYQL NFYQNHR YVKSR+ QLR
Sbjct: 98 MTDNKLAYIQNPNISKECTRTLTITEDMNQPIFVYYQLDNFYQNHRRYVKSRNDGQLRDA 157
Query: 124 ADENDVGKCFPEDYTANGYLPVVPCGLAAWSLFNDTYRFSNNNKDLVINKKNIAWKSDQK 183
A N C PE TA+G P+VPCGL AWSLFNDTY F+ N++L ++KK+I+WKSD++
Sbjct: 158 AKANQTSACEPEKTTADGK-PIVPCGLIAWSLFNDTYSFTRGNENLTVDKKDISWKSDRE 216
Query: 184 AKFGSDVYPKNFQTGSLIGGARLNESIPLSEQEDLIVWMRTAALPTFRKLYGKIEVDLEA 243
KFG +VYP NFQ G L GG L+ +IPLSEQEDLIVWMRTAALPTFRKLYG+I VDL+
Sbjct: 217 HKFGKNVYPSNFQNGLLKGGGTLDPAIPLSEQEDLIVWMRTAALPTFRKLYGRIYVDLKK 276
Query: 244 NDEITVVIENNYNTYQFGGTKSVILSTTTWIGGKNDFLGIAYILIGGLSLVYSLVFLLMY 303
ND ITV + NNYNTY FGG K ++LST TW+GGKNDFLG AY+++GG+ + F L+Y
Sbjct: 277 NDTITVKLSNNYNTYNFGGKKKLVLSTATWLGGKNDFLGFAYVIVGGVCFFLAFAFTLLY 336
Query: 304 LMKPRPLGDPRYLTWNKN 321
L+KPR LGD YL+WN++
Sbjct: 337 LIKPRKLGDHNYLSWNRH 354
>gb|AAM67090.1| unknown [Arabidopsis thaliana] gi|18412377|ref|NP_565210.1| LEM3
(ligand-effect modulator 3) family protein / CDC50
family protein [Arabidopsis thaliana]
Length = 350
Score = 408 bits (1048), Expect = e-112
Identities = 194/319 (60%), Positives = 244/319 (75%), Gaps = 3/319 (0%)
Query: 4 SRFSQQELHAWQPILTPGWVIAIFTFIGLVFIPIGVASLFASEQVVEVPLRYDDQCLPSL 63
SRF+QQEL A +PILTP WVI F G+VFIP+GV LFAS+ VVE+ RYD C+P+
Sbjct: 31 SRFTQQELPACKPILTPRWVILTFLVAGVVFIPLGVICLFASQGVVEIVDRYDTDCIPTS 90
Query: 64 YKDDAMTYIKGNRISKTCTKKLTVKSKMKAPIYVYYQLSNFYQNHRHYVKSRDHKQLRSK 123
+++ + YI+G K C + +TV MK P+YVYYQL NFYQNHR YVKSR+ QLRS
Sbjct: 91 SRNNMVAYIQGEG-DKICKRTITVTKAMKHPVYVYYQLENFYQNHRRYVKSRNDAQLRSP 149
Query: 124 ADENDVGKCFPEDYTANGYLPVVPCGLAAWSLFNDTYRFSNNNKDLVINKKNIAWKSDQK 183
+E+DV C PED G P+VPCGL AWSLFNDTY FS N++ L++NKK I+WKSD++
Sbjct: 150 KEEHDVKTCAPEDNV--GGEPIVPCGLVAWSLFNDTYSFSRNSQQLLVNKKGISWKSDRE 207
Query: 184 AKFGSDVYPKNFQTGSLIGGARLNESIPLSEQEDLIVWMRTAALPTFRKLYGKIEVDLEA 243
KFG +V+PKNFQ G+ IGG LN S PLSEQEDLIVWMRTAALPTFRKLYGKIE DL A
Sbjct: 208 NKFGKNVFPKNFQKGAPIGGGTLNISKPLSEQEDLIVWMRTAALPTFRKLYGKIETDLHA 267
Query: 244 NDEITVVIENNYNTYQFGGTKSVILSTTTWIGGKNDFLGIAYILIGGLSLVYSLVFLLMY 303
D ITV+++NNYNTY F G K ++LSTT+W+GG+NDFLGIAY+ +G + L ++ F ++Y
Sbjct: 268 GDTITVLLQNNYNTYSFNGQKKLVLSTTSWLGGRNDFLGIAYLTVGSICLFLAVTFAVLY 327
Query: 304 LMKPRPLGDPRYLTWNKNS 322
L+KPR LGDP YL+WN+++
Sbjct: 328 LVKPRQLGDPSYLSWNRSA 346
>ref|XP_475734.1| putative membrane protein [Oryza sativa (japonica cultivar-group)]
gi|45642720|gb|AAS72348.1| putative membrane protein
[Oryza sativa (japonica cultivar-group)]
Length = 345
Score = 407 bits (1046), Expect = e-112
Identities = 192/319 (60%), Positives = 247/319 (77%), Gaps = 4/319 (1%)
Query: 4 SRFSQQELHAWQPILTPGWVIAIFTFIGLVFIPIGVASLFASEQVVEVPLRYDDQCLPSL 63
S+F+QQEL AW+P+ TPG VI F+ IG++FIPIG+ S+ AS++VVE+ +YD +C+ +
Sbjct: 26 SKFTQQELPAWKPLYTPGIVIGAFSLIGIIFIPIGLVSIAASQEVVELVDKYDGECVTA- 84
Query: 64 YKDDAMTYIKGNRISKTCTKKLTVKSKMKAPIYVYYQLSNFYQNHRHYVKSRDHKQLRSK 123
+D + +I+ + K CT+ +TV MK PI VYYQL NFYQNHR YVKSR KQLRSK
Sbjct: 85 --NDKVGFIQDTKTDKACTRTITVPKPMKGPIQVYYQLENFYQNHRRYVKSRSDKQLRSK 142
Query: 124 ADENDVGKCFPEDYTANGYLPVVPCGLAAWSLFNDTYRFSNNNKDLVINKKNIAWKSDQK 183
+ + C PE + G P+VPCGL AWSLFNDT+ FS N K + +NKKNIAW SD+
Sbjct: 143 EFSSVIKTCDPEAISEGG-APIVPCGLIAWSLFNDTFTFSVNKKTVQVNKKNIAWSSDRT 201
Query: 184 AKFGSDVYPKNFQTGSLIGGARLNESIPLSEQEDLIVWMRTAALPTFRKLYGKIEVDLEA 243
KFGSDV+P+NFQ G LIGG +LNE +PLSEQEDLIVWMRTAALPTFRKLYG+IE D+ A
Sbjct: 202 IKFGSDVFPENFQKGGLIGGGQLNEKLPLSEQEDLIVWMRTAALPTFRKLYGRIETDIMA 261
Query: 244 NDEITVVIENNYNTYQFGGTKSVILSTTTWIGGKNDFLGIAYILIGGLSLVYSLVFLLMY 303
+DEITVVI+NNYNTY FGGTK+++LSTT+WIGGKN+F+G AY+ IG +S + +L F+ +
Sbjct: 262 SDEITVVIQNNYNTYSFGGTKALVLSTTSWIGGKNNFIGFAYVAIGTISFLIALAFVGLN 321
Query: 304 LMKPRPLGDPRYLTWNKNS 322
++KPR LGDP YL+WNK +
Sbjct: 322 MVKPRTLGDPSYLSWNKEN 340
>ref|NP_173086.2| LEM3 (ligand-effect modulator 3) family protein / CDC50 family
protein [Arabidopsis thaliana]
Length = 336
Score = 407 bits (1046), Expect = e-112
Identities = 192/320 (60%), Positives = 244/320 (76%), Gaps = 3/320 (0%)
Query: 3 DSRFSQQELHAWQPILTPGWVIAIFTFIGLVFIPIGVASLFASEQVVEVPLRYDDQCLPS 62
DSRF+QQEL A +PILTP WVI F G+VFIP+GV LFAS+ V+E+ RYD C+P
Sbjct: 16 DSRFTQQELPACKPILTPKWVILTFLVSGVVFIPLGVICLFASQGVIEIVDRYDTDCIPL 75
Query: 63 LYKDDAMTYIKGNRISKTCTKKLTVKSKMKAPIYVYYQLSNFYQNHRHYVKSRDHKQLRS 122
+D+ + YI+G K C + +TV MK P+YVYYQL N+YQNHR YVKSR QLRS
Sbjct: 76 SSRDNKVRYIQGLE-DKRCNRTITVTKTMKNPVYVYYQLENYYQNHRRYVKSRQDGQLRS 134
Query: 123 KADENDVGKCFPEDYTANGYLPVVPCGLAAWSLFNDTYRFSNNNKDLVINKKNIAWKSDQ 182
DE++ C PED G P+VPCGL AWSLFNDTY F+ NN+ L +NKK+I+WKSD+
Sbjct: 135 PKDEHETKSCAPEDTL--GGQPIVPCGLVAWSLFNDTYDFTRNNQKLPVNKKDISWKSDR 192
Query: 183 KAKFGSDVYPKNFQTGSLIGGARLNESIPLSEQEDLIVWMRTAALPTFRKLYGKIEVDLE 242
++KFG +V+PKNFQ GSLIGG L++ IPLSEQEDLIVWMRTAALPTFRKLYGKI+ DL+
Sbjct: 193 ESKFGKNVFPKNFQKGSLIGGKSLDQDIPLSEQEDLIVWMRTAALPTFRKLYGKIDTDLQ 252
Query: 243 ANDEITVVIENNYNTYQFGGTKSVILSTTTWIGGKNDFLGIAYILIGGLSLVYSLVFLLM 302
A D I V+++NNYNTY F G K ++LSTT+W+GG+NDFLGIAY+ +G + L ++ F ++
Sbjct: 253 AGDTIKVLLQNNYNTYSFNGKKKLVLSTTSWLGGRNDFLGIAYLTVGSICLFLAVSFSVL 312
Query: 303 YLMKPRPLGDPRYLTWNKNS 322
YL KPR LGDP YL+WN+++
Sbjct: 313 YLAKPRQLGDPSYLSWNRSA 332
>ref|XP_464311.1| LEM3 (ligand-effect modulator 3)-like [Oryza sativa (japonica
cultivar-group)] gi|49388971|dbj|BAD26188.1| LEM3
(ligand-effect modulator 3)-like [Oryza sativa (japonica
cultivar-group)]
Length = 350
Score = 397 bits (1021), Expect = e-109
Identities = 190/318 (59%), Positives = 238/318 (74%), Gaps = 3/318 (0%)
Query: 4 SRFSQQELHAWQPILTPGWVIAIFTFIGLVFIPIGVASLFASEQVVEVPLRYDDQCLPSL 63
S+F+QQEL A +PILTP WVI++F +G++F+PIG+ SL AS +VVE+ RYDD C+P+
Sbjct: 32 SKFTQQELPACKPILTPKWVISVFVLVGVIFVPIGLVSLKASRKVVEIVDRYDDACVPA- 90
Query: 64 YKDDAMTYIKGNRISKTCTKKLTVKSKMKAPIYVYYQLSNFYQNHRHYVKSRDHKQLRSK 123
D + YI+ ISK C + L V M API+VYYQL NFYQNHR YVKSR QLR
Sbjct: 91 NTTDKLAYIQNPTISKNCRRTLKVPKDMDAPIFVYYQLDNFYQNHRRYVKSRSDAQLRDP 150
Query: 124 ADENDVGKCFPEDYTANGYLPVVPCGLAAWSLFNDTYRFSNNNKDLVINKKNIAWKSDQK 183
ND C PE TANG + +VPCGL AWS+FNDTY F N+K+L ++KKNI+WKSD++
Sbjct: 151 KKANDTSTCDPEG-TANG-MAIVPCGLIAWSIFNDTYGFVRNSKNLPVDKKNISWKSDRE 208
Query: 184 AKFGSDVYPKNFQTGSLIGGARLNESIPLSEQEDLIVWMRTAALPTFRKLYGKIEVDLEA 243
KFG DV+PKNFQ GSLIGG L+ + LS+QEDLIVWMRTAALPTFRKLYG+I DL+
Sbjct: 209 HKFGRDVFPKNFQNGSLIGGKTLDPNKSLSKQEDLIVWMRTAALPTFRKLYGRIHTDLKK 268
Query: 244 NDEITVVIENNYNTYQFGGTKSVILSTTTWIGGKNDFLGIAYILIGGLSLVYSLVFLLMY 303
D ITV +ENNYNTY F G K ++LST+TW+GGKNDFLG+AY+ +GGL + F L+Y
Sbjct: 269 GDTITVTLENNYNTYSFSGKKKLVLSTSTWLGGKNDFLGLAYLSVGGLCFFLAFAFTLLY 328
Query: 304 LMKPRPLGDPRYLTWNKN 321
L+KPR +GD YL+WN+N
Sbjct: 329 LIKPRKMGDNNYLSWNRN 346
>gb|AAD34688.1| >F3O9.16 [Arabidopsis thaliana] gi|25347303|pir||G86298 protein
F3O9.16 [imported] - Arabidopsis thaliana
Length = 353
Score = 396 bits (1018), Expect = e-109
Identities = 192/337 (56%), Positives = 244/337 (71%), Gaps = 20/337 (5%)
Query: 3 DSRFSQQELHAWQPILTPGWVIAIFTFIGLVFIPIGVASLFASEQV-------------- 48
DSRF+QQEL A +PILTP WVI F G+VFIP+GV LFAS+ V
Sbjct: 16 DSRFTQQELPACKPILTPKWVILTFLVSGVVFIPLGVICLFASQGVCFKSFLTFLSYNYI 75
Query: 49 ---VEVPLRYDDQCLPSLYKDDAMTYIKGNRISKTCTKKLTVKSKMKAPIYVYYQLSNFY 105
+E+ RYD C+P +D+ + YI+G K C + +TV MK P+YVYYQL N+Y
Sbjct: 76 YFVIEIVDRYDTDCIPLSSRDNKVRYIQGLE-DKRCNRTITVTKTMKNPVYVYYQLENYY 134
Query: 106 QNHRHYVKSRDHKQLRSKADENDVGKCFPEDYTANGYLPVVPCGLAAWSLFNDTYRFSNN 165
QNHR YVKSR QLRS DE++ C PED G P+VPCGL AWSLFNDTY F+ N
Sbjct: 135 QNHRRYVKSRQDGQLRSPKDEHETKSCAPEDTL--GGQPIVPCGLVAWSLFNDTYDFTRN 192
Query: 166 NKDLVINKKNIAWKSDQKAKFGSDVYPKNFQTGSLIGGARLNESIPLSEQEDLIVWMRTA 225
N+ L +NKK+I+WKSD+++KFG +V+PKNFQ GSLIGG L++ IPLSEQEDLIVWMRTA
Sbjct: 193 NQKLPVNKKDISWKSDRESKFGKNVFPKNFQKGSLIGGKSLDQDIPLSEQEDLIVWMRTA 252
Query: 226 ALPTFRKLYGKIEVDLEANDEITVVIENNYNTYQFGGTKSVILSTTTWIGGKNDFLGIAY 285
ALPTFRKLYGKI+ DL+A D I V+++NNYNTY F G K ++LSTT+W+GG+NDFLGIAY
Sbjct: 253 ALPTFRKLYGKIDTDLQAGDTIKVLLQNNYNTYSFNGKKKLVLSTTSWLGGRNDFLGIAY 312
Query: 286 ILIGGLSLVYSLVFLLMYLMKPRPLGDPRYLTWNKNS 322
+ +G + L ++ F ++YL KPR LGDP YL+WN+++
Sbjct: 313 LTVGSICLFLAVSFSVLYLAKPRQLGDPSYLSWNRSA 349
>gb|AAD30230.1| T8K14.13 [Arabidopsis thaliana] gi|25347301|pir||F96825 T8K14.13
[imported] - Arabidopsis thaliana
Length = 335
Score = 395 bits (1015), Expect = e-109
Identities = 192/333 (57%), Positives = 243/333 (72%), Gaps = 19/333 (5%)
Query: 6 FSQQELHAWQPILTPGWVIAIFTFIGLVFIPIGVASLFASE----------------QVV 49
F+QQEL A +PILTP WVI F G+VFIP+GV LFAS+ +VV
Sbjct: 2 FTQQELPACKPILTPRWVILTFLVAGVVFIPLGVICLFASQGLRFCGFIEDMFHSYLKVV 61
Query: 50 EVPLRYDDQCLPSLYKDDAMTYIKGNRISKTCTKKLTVKSKMKAPIYVYYQLSNFYQNHR 109
E+ RYD C+P+ +++ + YI+G K C + +TV MK P+YVYYQL NFYQNHR
Sbjct: 62 EIVDRYDTDCIPTSSRNNMVAYIQGEG-DKICKRTITVTKAMKHPVYVYYQLENFYQNHR 120
Query: 110 HYVKSRDHKQLRSKADENDVGKCFPEDYTANGYLPVVPCGLAAWSLFNDTYRFSNNNKDL 169
YVKSR+ QLRS +E+DV C PED G P+VPCGL AWSLFNDTY FS N++ L
Sbjct: 121 RYVKSRNDAQLRSPKEEHDVKTCAPEDNV--GGEPIVPCGLVAWSLFNDTYSFSRNSQQL 178
Query: 170 VINKKNIAWKSDQKAKFGSDVYPKNFQTGSLIGGARLNESIPLSEQEDLIVWMRTAALPT 229
++NKK I+WKSD++ KFG +V+PKNFQ G+ IGG LN S PLSEQEDLIVWMRTAALPT
Sbjct: 179 LVNKKGISWKSDRENKFGKNVFPKNFQKGAPIGGGTLNISKPLSEQEDLIVWMRTAALPT 238
Query: 230 FRKLYGKIEVDLEANDEITVVIENNYNTYQFGGTKSVILSTTTWIGGKNDFLGIAYILIG 289
FRKLYGKIE DL A D ITV+++NNYNTY F G K ++LSTT+W+GG+NDFLGIAY+ +G
Sbjct: 239 FRKLYGKIETDLHAGDTITVLLQNNYNTYSFNGQKKLVLSTTSWLGGRNDFLGIAYLTVG 298
Query: 290 GLSLVYSLVFLLMYLMKPRPLGDPRYLTWNKNS 322
+ L ++ F ++YL+KPR LGDP YL+WN+++
Sbjct: 299 SICLFLAVTFAVLYLVKPRQLGDPSYLSWNRSA 331
>dbj|BAD32828.1| hypothetical protein [Lotus corniculatus var. japonicus]
Length = 337
Score = 366 bits (940), Expect = e-100
Identities = 184/296 (62%), Positives = 221/296 (74%), Gaps = 5/296 (1%)
Query: 4 SRFSQQELHAWQPILTPGWVIAIFTFIGLVFIPIGVASLFASEQVVEVPLRYDDQCLPSL 63
S+F+QQEL A +PILTP VI+ F + +VF+PIGVASL AS +VVE+ RY+ CL +
Sbjct: 27 SKFTQQELPACKPILTPRAVISAFLLVSVVFVPIGVASLIASRKVVEIVHRYESSCLKGV 86
Query: 64 YKDDAMTYIKGNRISKTCTKKLTVKSKMKAPIYVYYQLSNFYQNHRHYVKSRDHKQLRSK 123
D+ + YI+ + KTC L V MK+PIYVYYQL NFYQNHR YVKSR +QLR
Sbjct: 87 --DNKIAYIQSSA-DKTCKITLKVDKHMKSPIYVYYQLDNFYQNHRRYVKSRSDQQLRDP 143
Query: 124 ADENDVGKCFPEDYTANGYLPVVPCGLAAWSLFNDTYRFSNNNKDLVINKKNIAWKSDQK 183
+E+ C PED ANG +VPCGL AWSLFNDTY FS NK L +NKK IAWKSD++
Sbjct: 144 KEESSTSACKPEDI-ANGRA-IVPCGLIAWSLFNDTYSFSYKNKSLTVNKKGIAWKSDRE 201
Query: 184 AKFGSDVYPKNFQTGSLIGGARLNESIPLSEQEDLIVWMRTAALPTFRKLYGKIEVDLEA 243
KFG +V PKNFQ GS+IGGA LNESI LSEQEDLIVWMRTAALPTFRKLYGKIEVDL+
Sbjct: 202 HKFGKNVLPKNFQNGSIIGGAHLNESIALSEQEDLIVWMRTAALPTFRKLYGKIEVDLDE 261
Query: 244 NDEITVVIENNYNTYQFGGTKSVILSTTTWIGGKNDFLGIAYILIGGLSLVYSLVF 299
+ I+V ++NNYNTY F G K ++LSTT+W+GGKNDFLGIAY+ +GGL +L F
Sbjct: 262 GENISVKLQNNYNTYSFNGKKKLVLSTTSWLGGKNDFLGIAYLTVGGLCFFLALAF 317
>ref|NP_974175.1| LEM3 (ligand-effect modulator 3) family protein / CDC50 family
protein [Arabidopsis thaliana]
Length = 283
Score = 355 bits (912), Expect = 8e-97
Identities = 166/276 (60%), Positives = 212/276 (76%), Gaps = 3/276 (1%)
Query: 47 QVVEVPLRYDDQCLPSLYKDDAMTYIKGNRISKTCTKKLTVKSKMKAPIYVYYQLSNFYQ 106
+VVE+ RYD C+P+ +++ + YI+G K C + +TV MK P+YVYYQL NFYQ
Sbjct: 7 KVVEIVDRYDTDCIPTSSRNNMVAYIQGEG-DKICKRTITVTKAMKHPVYVYYQLENFYQ 65
Query: 107 NHRHYVKSRDHKQLRSKADENDVGKCFPEDYTANGYLPVVPCGLAAWSLFNDTYRFSNNN 166
NHR YVKSR+ QLRS +E+DV C PED G P+VPCGL AWSLFNDTY FS N+
Sbjct: 66 NHRRYVKSRNDAQLRSPKEEHDVKTCAPEDNV--GGEPIVPCGLVAWSLFNDTYSFSRNS 123
Query: 167 KDLVINKKNIAWKSDQKAKFGSDVYPKNFQTGSLIGGARLNESIPLSEQEDLIVWMRTAA 226
+ L++NKK I+WKSD++ KFG +V+PKNFQ G+ IGG LN S PLSEQEDLIVWMRTAA
Sbjct: 124 QQLLVNKKGISWKSDRENKFGKNVFPKNFQKGAPIGGGTLNISKPLSEQEDLIVWMRTAA 183
Query: 227 LPTFRKLYGKIEVDLEANDEITVVIENNYNTYQFGGTKSVILSTTTWIGGKNDFLGIAYI 286
LPTFRKLYGKIE DL A D ITV+++NNYNTY F G K ++LSTT+W+GG+NDFLGIAY+
Sbjct: 184 LPTFRKLYGKIETDLHAGDTITVLLQNNYNTYSFNGQKKLVLSTTSWLGGRNDFLGIAYL 243
Query: 287 LIGGLSLVYSLVFLLMYLMKPRPLGDPRYLTWNKNS 322
+G + L ++ F ++YL+KPR LGDP YL+WN+++
Sbjct: 244 TVGSICLFLAVTFAVLYLVKPRQLGDPSYLSWNRSA 279
>dbj|BAD95435.1| hypothetical protein [Arabidopsis thaliana]
gi|30694892|ref|NP_851139.1| LEM3 (ligand-effect
modulator 3) family protein / CDC50 family protein
[Arabidopsis thaliana] gi|18422638|ref|NP_568657.1| LEM3
(ligand-effect modulator 3) family protein / CDC50
family protein [Arabidopsis thaliana]
gi|51971485|dbj|BAD44407.1| unknown protein [Arabidopsis
thaliana] gi|51970926|dbj|BAD44155.1| unknown protein
[Arabidopsis thaliana] gi|51968732|dbj|BAD43058.1|
unknown protein [Arabidopsis thaliana]
Length = 343
Score = 354 bits (909), Expect = 2e-96
Identities = 170/318 (53%), Positives = 228/318 (71%), Gaps = 5/318 (1%)
Query: 5 RFSQQELHAWQPILTPGWVIAIFTFIGLVFIPIGVASLFASEQVVEVPLRYDDQCLPSLY 64
+F QQ+L A +P+LTP VI +F +G VFIPIG+ +L AS +E+ RYD +C+P Y
Sbjct: 28 QFKQQKLPACKPVLTPISVITVFMLMGFVFIPIGLITLRASRDAIEIIDRYDVECIPEEY 87
Query: 65 KDDAMTYIKGNRISKTCTKKLTVKSKMKAPIYVYYQLSNFYQNHRHYVKSRDHKQLRSKA 124
+ + + YI + I K CT+ L V+ MKAPI++YYQL N+YQNHR YVKSR +QL
Sbjct: 88 RTNKLLYITDSSIPKNCTRYLKVQKYMKAPIFIYYQLDNYYQNHRRYVKSRSDQQLLHGL 147
Query: 125 DENDVGKCFPEDYTANGYLPVVPCGLAAWSLFNDTYRFSNNNKDLVINKKNIAWKSDQKA 184
+ + C PE+ ++NG LP+VPCGL AWS+FNDT+ FS L +++ NIAWKSD++
Sbjct: 148 EYSHTSSCEPEE-SSNG-LPIVPCGLIAWSMFNDTFTFSRERTKLNVSRNNIAWKSDREH 205
Query: 185 KFGSDVYPKNFQTGSLIGGARLNESIPLSEQEDLIVWMRTAALPTFRKLYGKIEVDLEAN 244
KFG +VYP NFQ G+LIGGA+L+ IPLS+QED IVWMR AAL +FRKLYG+IE DLE
Sbjct: 206 KFGKNVYPINFQNGTLIGGAKLDPKIPLSDQEDFIVWMRAAALLSFRKLYGRIEEDLEPG 265
Query: 245 DEITVVIENNYNTYQFGGTKSVILSTTTWIGGKNDFLGIAYILIGGLSLVYSLVFLLMYL 304
+ V + NNYNTY F G K +ILST+ W+GG+NDFLGI Y+++G S+V S++F+L++L
Sbjct: 266 KVVEVNLMNNYNTYSFSGQKKLILSTSNWLGGRNDFLGITYLVVGSSSIVISIIFMLLHL 325
Query: 305 MKPRPLGDPRYLTWNKNS 322
PRP GD +WNK S
Sbjct: 326 KNPRPYGDN---SWNKKS 340
>gb|AAM63135.1| unknown [Arabidopsis thaliana]
Length = 336
Score = 353 bits (906), Expect = 4e-96
Identities = 169/318 (53%), Positives = 228/318 (71%), Gaps = 5/318 (1%)
Query: 5 RFSQQELHAWQPILTPGWVIAIFTFIGLVFIPIGVASLFASEQVVEVPLRYDDQCLPSLY 64
+F QQ+L A +P+LTP VI +F +G VFIPIG+ +L AS +E+ RYD +C+P Y
Sbjct: 21 QFKQQKLPACKPVLTPISVITVFMLMGFVFIPIGLITLRASRDAIEIIDRYDVECIPEEY 80
Query: 65 KDDAMTYIKGNRISKTCTKKLTVKSKMKAPIYVYYQLSNFYQNHRHYVKSRDHKQLRSKA 124
+ + + YI + I K CT+ L V+ MKAPI++YYQL N+YQNHR YVKSR +QL
Sbjct: 81 RTNKLLYITDSSIPKNCTRYLKVQKYMKAPIFIYYQLDNYYQNHRRYVKSRSDQQLLHGL 140
Query: 125 DENDVGKCFPEDYTANGYLPVVPCGLAAWSLFNDTYRFSNNNKDLVINKKNIAWKSDQKA 184
+ + C PE+ ++NG LP+VPCGL AWS+FNDT+ FS L +++ NIAWKSD++
Sbjct: 141 EYSHTSSCEPEE-SSNG-LPIVPCGLIAWSMFNDTFTFSRERTKLNVSRNNIAWKSDREH 198
Query: 185 KFGSDVYPKNFQTGSLIGGARLNESIPLSEQEDLIVWMRTAALPTFRKLYGKIEVDLEAN 244
KFG +VYP NFQ G+LIGGA+L+ +PLS+QED IVWMR AAL +FRKLYG+IE DLE
Sbjct: 199 KFGKNVYPINFQNGTLIGGAKLDPKLPLSDQEDFIVWMRAAALLSFRKLYGRIEEDLEPG 258
Query: 245 DEITVVIENNYNTYQFGGTKSVILSTTTWIGGKNDFLGIAYILIGGLSLVYSLVFLLMYL 304
+ V + NNYNTY F G K +ILST+ W+GG+NDFLGI Y+++G S+V S++F+L++L
Sbjct: 259 KVVEVNLMNNYNTYSFSGQKKLILSTSNWLGGRNDFLGITYLVVGSSSVVISIIFMLLHL 318
Query: 305 MKPRPLGDPRYLTWNKNS 322
PRP GD +WNK S
Sbjct: 319 KNPRPYGDN---SWNKKS 333
>dbj|BAD44111.1| unknown protein [Arabidopsis thaliana]
Length = 343
Score = 353 bits (905), Expect = 5e-96
Identities = 170/318 (53%), Positives = 227/318 (70%), Gaps = 5/318 (1%)
Query: 5 RFSQQELHAWQPILTPGWVIAIFTFIGLVFIPIGVASLFASEQVVEVPLRYDDQCLPSLY 64
+F QQ+L A +P+LTP VI +F +G VFIPIG+ +L AS +E+ RYD +C+P Y
Sbjct: 28 QFKQQKLPACKPVLTPISVITVFMLMGFVFIPIGLITLRASRDAIEIIDRYDVECIPEEY 87
Query: 65 KDDAMTYIKGNRISKTCTKKLTVKSKMKAPIYVYYQLSNFYQNHRHYVKSRDHKQLRSKA 124
+ + + YI + I K CT+ L V+ MKAPI++YYQL N+YQNHR YVKSR +QL
Sbjct: 88 RTNKLLYITDSSIPKNCTRYLKVQKYMKAPIFIYYQLDNYYQNHRRYVKSRSDQQLLHGL 147
Query: 125 DENDVGKCFPEDYTANGYLPVVPCGLAAWSLFNDTYRFSNNNKDLVINKKNIAWKSDQKA 184
+ + C PE+ ++NG LP+VPCGL AWS+FNDT+ FS L +++ NIAWKSD++
Sbjct: 148 EYSHTSSCEPEE-SSNG-LPIVPCGLIAWSMFNDTFTFSRERTKLNVSRNNIAWKSDREH 205
Query: 185 KFGSDVYPKNFQTGSLIGGARLNESIPLSEQEDLIVWMRTAALPTFRKLYGKIEVDLEAN 244
KFG +VYP NFQ G+LIGGA+L+ IPLS+QED IVWMR AAL +FRKLYG IE DLE
Sbjct: 206 KFGKNVYPINFQNGTLIGGAKLDPKIPLSDQEDFIVWMRAAALLSFRKLYGGIEEDLEPG 265
Query: 245 DEITVVIENNYNTYQFGGTKSVILSTTTWIGGKNDFLGIAYILIGGLSLVYSLVFLLMYL 304
+ V + NNYNTY F G K +ILST+ W+GG+NDFLGI Y+++G S+V S++F+L++L
Sbjct: 266 KVVEVNLMNNYNTYSFSGQKKLILSTSNWLGGRNDFLGITYLVVGSSSIVISIIFMLLHL 325
Query: 305 MKPRPLGDPRYLTWNKNS 322
PRP GD +WNK S
Sbjct: 326 KNPRPYGDN---SWNKKS 340
>dbj|BAB08260.1| unnamed protein product [Arabidopsis thaliana]
Length = 329
Score = 343 bits (880), Expect = 4e-93
Identities = 163/304 (53%), Positives = 220/304 (71%), Gaps = 2/304 (0%)
Query: 5 RFSQQELHAWQPILTPGWVIAIFTFIGLVFIPIGVASLFASEQVVEVPLRYDDQCLPSLY 64
+F QQ+L A +P+LTP VI +F +G VFIPIG+ +L AS +E+ RYD +C+P Y
Sbjct: 28 QFKQQKLPACKPVLTPISVITVFMLMGFVFIPIGLITLRASRDAIEIIDRYDVECIPEEY 87
Query: 65 KDDAMTYIKGNRISKTCTKKLTVKSKMKAPIYVYYQLSNFYQNHRHYVKSRDHKQLRSKA 124
+ + + YI + I K CT+ L V+ MKAPI++YYQL N+YQNHR YVKSR +QL
Sbjct: 88 RTNKLLYITDSSIPKNCTRYLKVQKYMKAPIFIYYQLDNYYQNHRRYVKSRSDQQLLHGL 147
Query: 125 DENDVGKCFPEDYTANGYLPVVPCGLAAWSLFNDTYRFSNNNKDLVINKKNIAWKSDQKA 184
+ + C PE+ ++NG LP+VPCGL AWS+FNDT+ FS L +++ NIAWKSD++
Sbjct: 148 EYSHTSSCEPEE-SSNG-LPIVPCGLIAWSMFNDTFTFSRERTKLNVSRNNIAWKSDREH 205
Query: 185 KFGSDVYPKNFQTGSLIGGARLNESIPLSEQEDLIVWMRTAALPTFRKLYGKIEVDLEAN 244
KFG +VYP NFQ G+LIGGA+L+ IPLS+QED IVWMR AAL +FRKLYG+IE DLE
Sbjct: 206 KFGKNVYPINFQNGTLIGGAKLDPKIPLSDQEDFIVWMRAAALLSFRKLYGRIEEDLEPG 265
Query: 245 DEITVVIENNYNTYQFGGTKSVILSTTTWIGGKNDFLGIAYILIGGLSLVYSLVFLLMYL 304
+ V + NNYNTY F G K +ILST+ W+GG+NDFLGI Y+++G S+V S++F+L++L
Sbjct: 266 KVVEVNLMNNYNTYSFSGQKKLILSTSNWLGGRNDFLGITYLVVGSSSIVISIIFMLLHL 325
Query: 305 MKPR 308
PR
Sbjct: 326 KNPR 329
>dbj|BAD46079.1| LEM3 (ligand-effect modulator 3)-like [Oryza sativa (japonica
cultivar-group)] gi|52076953|dbj|BAD45964.1| LEM3
(ligand-effect modulator 3)-like [Oryza sativa (japonica
cultivar-group)]
Length = 351
Score = 338 bits (868), Expect = 1e-91
Identities = 167/305 (54%), Positives = 217/305 (70%), Gaps = 4/305 (1%)
Query: 5 RFSQQELHAWQPILTPGWVIAIFTFIGLVFIPIGVASLFASEQVVEVPLRYDDQCLPSLY 64
RF+QQEL A +PIL P VI + F+GL+FIPIG+A + AS +VVE+ RYD +C+P
Sbjct: 32 RFTQQELQACKPILIPQTVILVLVFVGLIFIPIGLACIAASNKVVELVDRYDTKCVPRNM 91
Query: 65 KDDAMTYIKGNRISKTCTKKLTVKSKMKAPIYVYYQLSNFYQNHRHYVKSRDHKQLRSKA 124
+ + +I+ + I KTCT+ V MK PIY+YYQL FYQNHR YVKS + QLR+
Sbjct: 92 LRNKVAFIQNSSIDKTCTRVFKVPKDMKKPIYIYYQLDKFYQNHRRYVKSLNDMQLRNPK 151
Query: 125 DENDVGKCFPEDYTANGYLPVVPCGLAAWSLFNDTYRFS--NNNKDLVINKKNIAWKSDQ 182
D C PE TANG P+VPCGL AWSLFNDTY F+ + N+ L +NK I+WKS++
Sbjct: 152 KVADTQYCSPEA-TANGR-PIVPCGLIAWSLFNDTYSFTRGHGNETLRVNKDGISWKSER 209
Query: 183 KAKFGSDVYPKNFQTGSLIGGARLNESIPLSEQEDLIVWMRTAALPTFRKLYGKIEVDLE 242
+FG +VYPKNFQ G+LIGG +LN S PLSEQEDLIVWMR AALPTFRKLYG+I++DL+
Sbjct: 210 NRRFGKNVYPKNFQNGTLIGGGQLNPSKPLSEQEDLIVWMRIAALPTFRKLYGRIDMDLQ 269
Query: 243 ANDEITVVIENNYNTYQFGGTKSVILSTTTWIGGKNDFLGIAYILIGGLSLVYSLVFLLM 302
A D + V ++NNYN+Y F G KS++LST W+GGKN FLG AY ++G + +L+ L+
Sbjct: 270 AGDRVEVTMQNNYNSYSFNGKKSLVLSTAGWLGGKNAFLGRAYAIVGLACFLLALLLALL 329
Query: 303 YLMKP 307
Y + P
Sbjct: 330 YFVFP 334
>dbj|BAD43093.1| unknown protein [Arabidopsis thaliana]
Length = 234
Score = 325 bits (832), Expect = 1e-87
Identities = 148/232 (63%), Positives = 186/232 (79%), Gaps = 2/232 (0%)
Query: 91 MKAPIYVYYQLSNFYQNHRHYVKSRDHKQLRSKADENDVGKCFPEDYTANGYLPVVPCGL 150
MK P+YVYYQL N+YQNHR YVKSR QLRS DE++ C PED G P+VPCGL
Sbjct: 1 MKNPVYVYYQLENYYQNHRRYVKSRQDGQLRSPKDEHETKSCAPEDTL--GGQPIVPCGL 58
Query: 151 AAWSLFNDTYRFSNNNKDLVINKKNIAWKSDQKAKFGSDVYPKNFQTGSLIGGARLNESI 210
AWSLFNDTY F+ NN+ L +NKK+I+WKSD+++KFG +V+PKNFQ GSLIGG L++ I
Sbjct: 59 VAWSLFNDTYDFTRNNQKLPVNKKDISWKSDRESKFGKNVFPKNFQKGSLIGGKSLDQDI 118
Query: 211 PLSEQEDLIVWMRTAALPTFRKLYGKIEVDLEANDEITVVIENNYNTYQFGGTKSVILST 270
PLSEQEDLIVWMRTAALPTFRKLYGKI+ DL+A D I V+++NNYNTY F G K ++LST
Sbjct: 119 PLSEQEDLIVWMRTAALPTFRKLYGKIDTDLQAGDTIKVLLQNNYNTYSFNGKKKLVLST 178
Query: 271 TTWIGGKNDFLGIAYILIGGLSLVYSLVFLLMYLMKPRPLGDPRYLTWNKNS 322
T+W+GG+NDFLGIAY+ +G + L ++ F ++YL KPR LGDP YL+WN+++
Sbjct: 179 TSWLGGRNDFLGIAYLTVGSICLFLAVSFSVLYLAKPRQLGDPSYLSWNRSA 230
>ref|XP_643143.1| hypothetical protein DDB0217816 [Dictyostelium discoideum]
gi|60471274|gb|EAL69237.1| hypothetical protein
DDB0217816 [Dictyostelium discoideum]
Length = 312
Score = 235 bits (600), Expect = 1e-60
Identities = 140/325 (43%), Positives = 193/325 (59%), Gaps = 32/325 (9%)
Query: 3 DSRFSQQELHAWQPILTPGWVIAIFTFIGLVFIPIGVASLFASEQVVEVPLRYDDQCLPS 62
++ F QQ L AW+PILTPG VI F IG+VFIPIG A + +S +V+E +RYDD
Sbjct: 13 NTAFKQQRLKAWEPILTPGPVIIAFIVIGIVFIPIGAAIINSSNKVLEHVIRYDDN---- 68
Query: 63 LYKDDAMTYIKGNRISKTCTKKLTVKSKMKAPIYVYYQLSNFYQNHRHYVKSRDHKQLRS 122
++ + CT T+ ++M++P+Y+YY+L NFYQNHR YVKSR+ QLR
Sbjct: 69 ----------PNCQLGQKCTINYTLPTEMESPVYLYYKLDNFYQNHRRYVKSRNDDQLRG 118
Query: 123 --KADENDVGKCFP---EDYTANGYLPVVPCGLAAWSLFNDTYRFSNNNKDLV-INKKNI 176
D + + C P D + + +VPCGL A S+FND+ ++ + +L+ + KK I
Sbjct: 119 IEVTDFSKLKDCEPLITTDGSEDVNKILVPCGLIANSVFNDSISLADASGNLINLTKKGI 178
Query: 177 AWKSDQKAKFGSDVYPKNFQTGSLIGGARLNESIPLSEQEDLIVWMRTAALPTFRKLYGK 236
AW+SD KF + YP+N G +N S E ED IVWMRTAALP F+KLY
Sbjct: 179 AWQSDIDKKFKN--YPEN-------GVGVINFSN--FEDEDFIVWMRTAALPDFKKLYRI 227
Query: 237 IE-VDLEANDEITVVIENNYNTYQFGGTKSVILSTTTWIGGKNDFLGIAYILIGGLSLVY 295
+ + I + I+N Y F G K V+LSTT+WIGGKN FLG AYI++G + V
Sbjct: 228 YHGQNGPLSGTIQIRIDNKYPVASFDGKKYVVLSTTSWIGGKNPFLGYAYIIVGIICFVQ 287
Query: 296 SLVFLLMYLMKPRPLGDPRYLTWNK 320
+VF + + + PR LG P+YL WNK
Sbjct: 288 GVVFAIKHKISPRVLGSPKYLEWNK 312
>gb|AAL92348.1| similar to membrane protein common family; protein id: At3g12740.1,
supported by cDNA: 37019., supported by cDNA:
gi_15028094, supported by cDNA: gi_15294235, supported
by cDNA: gi_20258910 [Arabidopsis thaliana]
[Dictyostelium discoideum]
Length = 374
Score = 212 bits (540), Expect = 1e-53
Identities = 132/313 (42%), Positives = 185/313 (58%), Gaps = 33/313 (10%)
Query: 3 DSRFSQQELHAWQPILTPGWVIAIFTFIGLVFIPIGVASLFASEQVVEVPLRYDDQCLPS 62
++ F QQ L AW+PILTPG VI F IG+VFIPIG A + +S +V+E +RYDD
Sbjct: 13 NTAFKQQRLKAWEPILTPGPVIIAFIVIGIVFIPIGAAIINSSNKVLEHVIRYDDN---- 68
Query: 63 LYKDDAMTYIKGNRISKTCTKKLTVKSKMKAPIYVYYQLSNFYQNHRHYVKSRDHKQLR- 121
++ + CT T+ ++M++P+Y+YY+L NFYQNHR YVKSR+ QLR
Sbjct: 69 ----------PNCQLGQKCTINYTLPTEMESPVYLYYKLDNFYQNHRRYVKSRNDDQLRG 118
Query: 122 -SKADENDVGKCFP---EDYTANGYLPVVPCGLAAWSLFNDTYRFSNNNKDLV-INKKNI 176
D + + C P D + + +VPCGL A S+FND+ ++ + +L+ + KK I
Sbjct: 119 IEVTDFSKLKDCEPLITTDGSEDVNKILVPCGLIANSVFNDSISLADASGNLINLTKKGI 178
Query: 177 AWKSDQKAKFGSDVYPKNFQTGSLIGGARLNESIPLSEQEDLIVWMRTAALPTFRKLYGK 236
AW+SD KF + YP+N G +N S E ED IVWMRTAALP F+KLY
Sbjct: 179 AWQSDIDKKFKN--YPEN-------GVGVINFS--NFEDEDFIVWMRTAALPDFKKLYRI 227
Query: 237 IE-VDLEANDEITVVIENNYNTYQFGGTKSVILSTTTWIGGKNDFLGIAYILIGGLSLVY 295
+ + I + I+N Y F G K V+LSTT+WIGGKN FLG AYI++G + V
Sbjct: 228 YHGQNGPLSGTIQIRIDNKYPVASFDGKKYVVLSTTSWIGGKNPFLGYAYIIVGIICFVQ 287
Query: 296 SLVFL-LMYLMKP 307
+VF+ ++ L+ P
Sbjct: 288 GVVFVSILKLIDP 300
>gb|EAL88482.1| LEM3/CDC50 family protein [Aspergillus fumigatus Af293]
Length = 400
Score = 207 bits (528), Expect = 3e-52
Identities = 136/356 (38%), Positives = 184/356 (51%), Gaps = 41/356 (11%)
Query: 3 DSRFSQQELHAWQPILTPGWVIAIFTFIGLVFIPIGVASLFASEQVVEVPLRYDD----- 57
++ F QQ L AWQPILTP V+ IF G++F PIG L+AS QV E+ + Y +
Sbjct: 34 NTAFRQQRLKAWQPILTPRSVLPIFFVFGIIFAPIGGLLLWASSQVQEISIDYSECAEKA 93
Query: 58 QCLPSLYKDDAMTYIKGNRISKT-------------CTKKLTVKSKMKAPIYVYYQLSNF 104
P D + K + T C V + P+++YY+L+NF
Sbjct: 94 PSYPVSIADRVKSSFKSSTGQSTPTWERRIENGTTICRLSFEVPDDLGPPVFLYYRLTNF 153
Query: 105 YQNHRHYVKSRDHKQLRSKADEN---DVGKCFPE--DYTANGYLPVVPCGLAAWSLFNDT 159
YQNHR YVKS D QL+ KA +N D G C P D T Y P CGL A S FNDT
Sbjct: 154 YQNHRRYVKSLDIDQLKGKAVDNKTIDGGSCDPLKLDPTGKAYYP---CGLIANSQFNDT 210
Query: 160 YRFSNNNKDL-----VINKKNIAWKSDQKA------KFGSDVYPKNFQTGSLIGGARLNE 208
DL + K IAW SD++ K V P N+ G +
Sbjct: 211 IHSPELLSDLNPTVYFMTNKGIAWDSDKELIKTTQYKPWEVVPPPNWHDRYPNGYI---D 267
Query: 209 SIP-LSEQEDLIVWMRTAALPTFRKLYGKIEVDLEANDEITVVIENNYNTYQFGGTKSVI 267
IP L E ED +VWMRTAALP F KL + + + IE+ + ++GGTKS++
Sbjct: 268 GIPDLHEDEDFMVWMRTAALPAFSKLSRRNDSAPMKAGSYRLDIEDRFPVTEYGGTKSIL 327
Query: 268 LSTTTWIGGKNDFLGIAYILIGGLSLVYSLVFLLMYLMKPRPLGDPRYLTWNKNSK 323
+ST T IGG+N F+GIAY+++GG+ ++ +F L +L++PR LGD YLTWN +
Sbjct: 328 ISTRTVIGGQNPFMGIAYVVVGGICVLLGALFTLAHLVRPRKLGDHTYLTWNNEQE 383
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.320 0.138 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 601,986,767
Number of Sequences: 2540612
Number of extensions: 26490500
Number of successful extensions: 58397
Number of sequences better than 10.0: 161
Number of HSP's better than 10.0 without gapping: 127
Number of HSP's successfully gapped in prelim test: 34
Number of HSP's that attempted gapping in prelim test: 57806
Number of HSP's gapped (non-prelim): 215
length of query: 326
length of database: 863,360,394
effective HSP length: 128
effective length of query: 198
effective length of database: 538,162,058
effective search space: 106556087484
effective search space used: 106556087484
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 75 (33.5 bits)
Medicago: description of AC126009.1


