

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126006.5 + phase: 0
(652 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
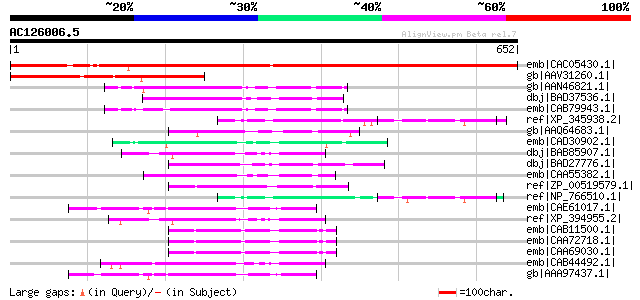
Score E
Sequences producing significant alignments: (bits) Value
emb|CAC05430.1| ankyrin-repeat containing protein [Arabidopsis t... 945 0.0
gb|AAV31260.1| hypothetical protein [Oryza sativa (japonica cult... 234 5e-60
gb|AAN46821.1| At4g32250/F10M6_110 [Arabidopsis thaliana] gi|214... 138 6e-31
dbj|BAD37536.1| protein kinase-like [Oryza sativa (japonica cult... 131 6e-29
emb|CAB79943.1| putative protein [Arabidopsis thaliana] gi|28646... 124 1e-26
ref|XP_345938.2| PREDICTED: similar to ankyrin repeat and kinase... 87 1e-15
gb|AAQ64683.1| NIMA-related kinase 2 [Chlamydomonas reinhardtii] 87 1e-15
emb|CAD30902.1| putative bifunctional protein [Streptomyces coel... 86 3e-15
dbj|BAB85907.1| p90 ribosomal S6 kinase [Asterina pectinifera] 86 5e-15
dbj|BAD27776.1| putative MAP3K alpha 1 protein kinase [Oryza sat... 84 2e-14
emb|CAA55382.1| cdc7 [Schizosaccharomyces pombe] gi|4455770|emb|... 83 2e-14
ref|ZP_00519579.1| Protein kinase:Serine/threonine protein kinas... 83 3e-14
ref|NP_766510.1| ankyrin repeat and kinase domain containing 1 [... 83 3e-14
emb|CAE61017.1| Hypothetical protein CBG04756 [Caenorhabditis br... 83 3e-14
ref|XP_394955.2| PREDICTED: similar to Rsk-2 [Apis mellifera] 82 4e-14
emb|CAB11500.1| wis4 [Schizosaccharomyces pombe] gi|19114469|ref... 82 5e-14
emb|CAA72718.1| Wak1 protein [Schizosaccharomyces pombe] 82 5e-14
emb|CAA69030.1| protein kinase [Schizosaccharomyces pombe] 82 5e-14
emb|CAB44492.1| ribosomal protein S6 kinase 3 [Mus musculus] gi|... 82 7e-14
gb|AAA97437.1| serine/threonine kinase 82 7e-14
>emb|CAC05430.1| ankyrin-repeat containing protein [Arabidopsis thaliana]
gi|15240646|ref|NP_196857.1| protein kinase family
protein / ankyrin repeat family protein [Arabidopsis
thaliana]
Length = 834
Score = 945 bits (2443), Expect = 0.0
Identities = 467/661 (70%), Positives = 541/661 (81%), Gaps = 24/661 (3%)
Query: 1 MKIPCCSVCQTRYNEEERVPLLLQCGHGFCKECLSRMFSSSSDANLTCPRCRHVSTVGNS 60
+K+PCCSVC TRYNE+ERVPLLLQCGHGFCK+CLS+MFS+SSD LTCPRCRHVS VGNS
Sbjct: 5 VKVPCCSVCHTRYNEDERVPLLLQCGHGFCKDCLSKMFSTSSDTTLTCPRCRHVSVVGNS 64
Query: 61 VQALRKNYAVLSLILSAADSAAAAGGGGGGDCDFTDDDEDRDDSEVDDGDDQKLDCRKNS 120
VQ LRKNYA+L+LI AA GG DCD+TDD++D D+ +DG D+ D + +
Sbjct: 65 VQGLRKNYAMLALI-------HAASGGANFDCDYTDDEDDDDE---EDGSDE--DGARAA 112
Query: 121 RGSQASSSGG--CAPVIEVGVHQDLKLVRRIGE----GRRAGVEMWSAVI--GGGRCKHQ 172
RG ASSS C PVIEVG H ++KLVR+IGE G GVEMW A + GGGRCKH+
Sbjct: 113 RGFHASSSINSLCGPVIEVGAHPEMKLVRQIGEESSSGGFGGVEMWDATVAGGGGRCKHR 172
Query: 173 VAVKKVVLNEGMDLDWMLGKLEDLRRTSMWCRNVCTFHGAMKVDEGLCLVMDKCFGSVQS 232
VAVKK+ L E MD++WM G+LE LRR SMWCRNVCTFHG +K+D LCL+MD+CFGSVQS
Sbjct: 173 VAVKKMTLTEDMDVEWMQGQLESLRRASMWCRNVCTFHGVVKMDGSLCLLMDRCFGSVQS 232
Query: 233 EMLRNEGRLTLEQVLRYGADIARGVVELHAAGVVCMSLKPSNLLLDANGHAVVSDYGLAT 292
EM RNEGRLTLEQ+LRYGAD+ARGV ELHAAGV+CM++KPSNLLLDA+G+AVVSDYGLA
Sbjct: 233 EMQRNEGRLTLEQILRYGADVARGVAELHAAGVICMNIKPSNLLLDASGNAVVSDYGLAP 292
Query: 293 ILKKPSCWKARPECDSAKIHSCMECIMLSPHYTAPEAWEPVKKSLNLFWDDGIGISPESD 352
ILKKP+C K RPE DS+K+ +C+ LSPHYTAPEAW PVKK LFW+D G+SPESD
Sbjct: 293 ILKKPTCQKTRPEFDSSKVTLYTDCVTLSPHYTAPEAWGPVKK---LFWEDASGVSPESD 349
Query: 353 AWSFGCTLVEMCTGAIPWAGLSAEEIYRQVVKAKKQPPQYASVVGGGIPRELWKMIGECL 412
AWSFGCTLVEMCTG+ PW GLS EEI++ VVKA+K PPQY +VG GIPRELWKMIGECL
Sbjct: 350 AWSFGCTLVEMCTGSTPWDGLSREEIFQAVVKARKVPPQYERIVGVGIPRELWKMIGECL 409
Query: 413 QFKPSKRPTFNAMLAIFLRHLQEIPRSPPASPDNDLVKGSVSNVTEASPVPELEIPQD-P 471
QFKPSKRPTFNAMLA FLRHLQEIPRSP ASPDN + K N+ +A + + QD P
Sbjct: 410 QFKPSKRPTFNAMLATFLRHLQEIPRSPSASPDNGIAKICEVNIVQAPRATNIGVFQDNP 469
Query: 472 NRLHRLVSEGDVTGVRDFLAKAASENESNFISSLLEAQNADGQTALHLACRRGSAELVET 531
N LHR+V EGD GVR+ LAKAA+ + + SLLEAQNADGQ+ALHLACRRGSAELVE
Sbjct: 470 NNLHRVVLEGDFEGVRNILAKAAAGGGGSSVRSLLEAQNADGQSALHLACRRGSAELVEA 529
Query: 532 ILDYPEANVDVLDKDGDPPLVFALAAGSHECVCSLIKRNANVTSRLRDGLGPSVAHVCAY 591
IL+Y EANVD++DKDGDPPLVFALAAGS +CV LIK+ ANV SRLR+G GPSVAHVC+Y
Sbjct: 530 ILEYGEANVDIVDKDGDPPLVFALAAGSPQCVHVLIKKGANVRSRLREGSGPSVAHVCSY 589
Query: 592 HGQPDCMRELLLAGADPNAVDDEGESVLHRAIAKKFTDCALVIVENGGCRSMAISNSKNL 651
HGQPDCMRELL+AGADPNAVDDEGE+VLHRA+AKK+TDCA+VI+ENGG RSM +SN+K L
Sbjct: 590 HGQPDCMRELLVAGADPNAVDDEGETVLHRAVAKKYTDCAIVILENGGSRSMTVSNAKCL 649
Query: 652 T 652
T
Sbjct: 650 T 650
Score = 40.8 bits (94), Expect = 0.14
Identities = 42/150 (28%), Positives = 57/150 (38%), Gaps = 49/150 (32%)
Query: 496 ENESNFISSLLEAQNAD--------GQTALHLACRRGSAELVETILDYPEANVDVLDKDG 547
E E + +L A AD G+TALH A + ELV ILD N ++ +
Sbjct: 701 EKEGRELVQILLAAGADPTAQDAQHGRTALHTAAMANNVELVRVILD-AGVNANIRNVHN 759
Query: 548 DPPLVFALAAGSHECVCSLIKRNANVTSRLRDGLGPSVAHVCAYHGQPDCMRELLLAGAD 607
PL ALA G++ C+ LL +G+D
Sbjct: 760 TIPLHMALARGANS-----------------------------------CVSLLLESGSD 784
Query: 608 PNAVDDEGESVLHRAIAKKFTDCALVIVEN 637
N DDEG++ H A D A +I EN
Sbjct: 785 CNIQDDEGDNAFHIA-----ADAAKMIREN 809
>gb|AAV31260.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 264
Score = 234 bits (598), Expect = 5e-60
Identities = 127/261 (48%), Positives = 165/261 (62%), Gaps = 15/261 (5%)
Query: 1 MKIPCCSVCQTRYNEEERVPLLLQCGHGFCKECLSRMFSSSSDANLTCPRCRHVSTVGNS 60
M++PCCS+C RY+EEER PLLL CGHGFC+ CL+RM ++++ A L CPRCRH + VGNS
Sbjct: 1 MRVPCCSLCHVRYDEEERAPLLLHCGHGFCRACLARMLANAAGAVLACPRCRHPTAVGNS 60
Query: 61 VQALRKNYAVLSLILSAADSAAAAGGGGGGDCDFTDDDEDRDDSEVDDGDDQKLDCRKNS 120
V ALRKN+ +LSL+ S+ S + GG + D D DD + ++
Sbjct: 61 VSALRKNFPILSLLSSSPSSPSFLHSDSGGS---SSDGSDDDDDDFFGRPSRRSSAAGAG 117
Query: 121 RGSQASS--SGGCAPVIEVGVHQDLKLVRRIGEG--RRAGVEMWSAVI--GGG-----RC 169
G+ A S GCA ++ H DLKL RRIG G AG E+WS + GGG RC
Sbjct: 118 AGAAAPSLQPAGCAS-FDLASHPDLKLARRIGSGPPGPAGQEVWSGTLSRGGGGGGAKRC 176
Query: 170 KHQVAVKKVVLNEGMDLDWMLGKLEDLRRTSMWCRNVCTFHGAMKVDEGLCLVMDKCFGS 229
KH VAVK+V + G L+ + ++E LRR + WCRNV TFHGA++V LC VMD+ GS
Sbjct: 177 KHPVAVKRVPVTAGDVLEGVQEEVERLRRAATWCRNVTTFHGAVRVGGHLCFVMDRYAGS 236
Query: 230 VQSEMLRNEGRLTLEQVLRYG 250
VQ+EM +N GRLTLEQ+LR G
Sbjct: 237 VQTEMRQNGGRLTLEQILRCG 257
>gb|AAN46821.1| At4g32250/F10M6_110 [Arabidopsis thaliana]
gi|21436453|gb|AAM51427.1| unknown protein [Arabidopsis
thaliana] gi|20259492|gb|AAM13866.1| unknown protein
[Arabidopsis thaliana] gi|21703136|gb|AAM74508.1|
AT4g32250/F10M6_110 [Arabidopsis thaliana]
gi|30689316|ref|NP_849560.1| protein kinase family
protein [Arabidopsis thaliana]
gi|22329080|ref|NP_194952.2| protein kinase family
protein [Arabidopsis thaliana]
Length = 611
Score = 138 bits (347), Expect = 6e-31
Identities = 102/317 (32%), Positives = 157/317 (49%), Gaps = 36/317 (11%)
Query: 123 SQASSSGGCAPVIEVGVHQDLKLVRRIGEGRRAGVEMWSAVIGGGRCK----HQVAVKKV 178
S+++ + G +P + LKL RIG G V W A H+VA+K +
Sbjct: 22 SESALAAGTSPWMNSST---LKLRHRIGRGPFGDV--WLATHHQSTEDYDEHHEVAIKML 76
Query: 179 VLNEGMDLDWMLGKLEDLRRTSMWCRNVCTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNE 238
+ ++ K EDL NVC G ++ +C+VM GS+ +M R +
Sbjct: 77 YPIKEDQRRVVVDKFEDLFSKCQGLENVCLLRGVSSINGKICVVMKFYEGSLGDKMARLK 136
Query: 239 G-RLTLEQVLRYGADIARGVVELHAAGVVCMSLKPSNLLLDANGHAVVSDYGLATILKKP 297
G +L+L VLRYG D+A G++ELH+ G + ++LKPSN LL N A++ D G+ +L
Sbjct: 137 GGKLSLPDVLRYGVDLATGILELHSKGFLILNLKPSNFLLSDNDKAILGDVGIPYLLLSI 196
Query: 298 SCWKARPECDSAKIHSCMECIMLSPHYTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFG 357
P D M + +P+Y APE W+P D +S E+D+W FG
Sbjct: 197 PL----PSSD-------MTERLGTPNYMAPEQWQP---------DVRGPMSFETDSWGFG 236
Query: 358 CTLVEMCTGAIPWAGLSAEEIYRQVVKAKKQPPQYASVVGGGIPRELWKMIGECLQFKPS 417
C++VEM TG PW+G SA+EIY VV+ +++ + IP L ++ C +
Sbjct: 237 CSIVEMLTGVQPWSGRSADEIYDLVVRKQEK-----LSIPSSIPPPLENLLRGCFMYDLR 291
Query: 418 KRPTFNAMLAIFLRHLQ 434
RP+ +L + L+ LQ
Sbjct: 292 SRPSMTDILLV-LKSLQ 307
>dbj|BAD37536.1| protein kinase-like [Oryza sativa (japonica cultivar-group)]
gi|51536357|dbj|BAD37488.1| protein kinase-like [Oryza
sativa (japonica cultivar-group)]
Length = 630
Score = 131 bits (330), Expect = 6e-29
Identities = 81/260 (31%), Positives = 134/260 (51%), Gaps = 26/260 (10%)
Query: 171 HQVAVKKVVLNEGMDLDWMLGKLEDLRRTSMWCRNVCTFHGAMKVDEGLCLVMDKCFGSV 230
H+VAVK + L + +++ NVC HG + +C+ M GS+
Sbjct: 75 HEVAVKMLHPIREDQLQAFSVRFDEIFSKCQGLSNVCFLHGISTQNGRICIAMKFYEGSI 134
Query: 231 QSEMLRNEG-RLTLEQVLRYGADIARGVVELHAAGVVCMSLKPSNLLLDANGHAVVSDYG 289
+M R +G R+ L VLRYGAD+ARG+++LH+ G++ ++LKP N LLD + HAV+ D+G
Sbjct: 135 GDKMARLKGGRIPLSDVLRYGADLARGIIDLHSRGILILNLKPCNFLLDEHDHAVLGDFG 194
Query: 290 LATILKKPSCWKARPECDSAKIHSCMECIMLSPHYTAPEAWEPVKKSLNLFWDDGIGISP 349
+ ++L S P D + + +P+Y APE W+P + IS
Sbjct: 195 IPSLLFGLSL----PNPDLIQ-------RLGTPNYMAPEQWQPSIRG---------PISY 234
Query: 350 ESDAWSFGCTLVEMCTGAIPWAGLSAEEIYRQVVKAKKQPPQYASVVGGGIPRELWKMIG 409
E+D+W F +++EM +G PW G S +E+Y+ VV K++P + +P + ++
Sbjct: 235 ETDSWGFAWSILEMLSGIQPWRGKSPDEVYQLVVLKKEKP-----IFPYNLPPAIENVLS 289
Query: 410 ECLQFKPSKRPTFNAMLAIF 429
C ++ RP +L F
Sbjct: 290 GCFEYDFRDRPQMTDILDAF 309
>emb|CAB79943.1| putative protein [Arabidopsis thaliana] gi|2864618|emb|CAA16965.1|
putative protein [Arabidopsis thaliana]
gi|7485349|pir||T05403 hypothetical protein F10M6.110 -
Arabidopsis thaliana
Length = 593
Score = 124 bits (311), Expect = 1e-26
Identities = 97/313 (30%), Positives = 152/313 (47%), Gaps = 46/313 (14%)
Query: 123 SQASSSGGCAPVIEVGVHQDLKLVRRIGEGRRAGVEMWSAVIGGGRCKHQVAVKKVVLNE 182
S+++ + G +P + LKL RIG G V W A H + + +
Sbjct: 22 SESALAAGTSPWMNSST---LKLRHRIGRGPFGDV--WLAT-------HHQSTEDYDEHH 69
Query: 183 GMDLDWMLGKLEDLRRTSMWCRNVCTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNEG-RL 241
+ + + ED RR V G ++ +C+VM GS+ +M R +G +L
Sbjct: 70 EVAIKMLYPIKEDQRR-------VVVDKGVSSINGKICVVMKFYEGSLGDKMARLKGGKL 122
Query: 242 TLEQVLRYGADIARGVVELHAAGVVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWK 301
+L VLRYG D+A G++ELH+ G + ++LKPSN LL N A++ D G+ +L
Sbjct: 123 SLPDVLRYGVDLATGILELHSKGFLILNLKPSNFLLSDNDKAILGDVGIPYLLLSIPL-- 180
Query: 302 ARPECDSAKIHSCMECIMLSPHYTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLV 361
P D M + +P+Y APE W+P D +S E+D+W FGC++V
Sbjct: 181 --PSSD-------MTERLGTPNYMAPEQWQP---------DVRGPMSFETDSWGFGCSIV 222
Query: 362 EMCTGAIPWAGLSAEEIYRQVVKAKKQPPQYASVVGGGIPRELWKMIGECLQFKPSKRPT 421
EM TG PW+G SA+EIY VV+ +++ + IP L ++ C + RP+
Sbjct: 223 EMLTGVQPWSGRSADEIYDLVVRKQEK-----LSIPSSIPPPLENLLRGCFMYDLRSRPS 277
Query: 422 FNAMLAIFLRHLQ 434
+L + L+ LQ
Sbjct: 278 MTDILLV-LKSLQ 289
>ref|XP_345938.2| PREDICTED: similar to ankyrin repeat and kinase domain containing 1
[Rattus norvegicus]
Length = 762
Score = 87.4 bits (215), Expect = 1e-15
Identities = 96/383 (25%), Positives = 164/383 (42%), Gaps = 38/383 (9%)
Query: 268 MSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDSAKIHSCMECIMLSPHYTAP 327
+ LKP N+LLD N H +SD+GL S W E + K + + + Y P
Sbjct: 156 LDLKPGNILLDNNMHVKISDFGL-------SKWM---EQSTQKQYIERSALRGTLSYIPP 205
Query: 328 EAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIYRQVVKAKK 387
E +F ++ PE D +SFG + E+ T P+AGL+ I QV +
Sbjct: 206 E----------MFLENNKAPGPEYDVYSFGIVVWEILTQKKPYAGLNMMTIIIQVAAGMR 255
Query: 388 QPPQYASVVGGGIPRELWKMIGECLQFKPSKRPTFNAMLAIFLRHLQEIPRSPPASPDND 447
S ++ ++ C P KRP F ++ I L + ++P A P+ +
Sbjct: 256 PSLHDVSAEWPEEAHQMVDLMKLCWDQDPKKRPCFLNVI-IETDMLLSLFQTPVADPEYE 314
Query: 448 LVKGSVS-----NVTEASPVPE-----LEIPQDPNRLHRLVSEGDVTGVRDFLAKAASEN 497
+ VS + +++ + L++ + + + E VT + +A+ + E
Sbjct: 315 ALTRKVSKEVSQEIADSADSDDSLKWILQLSDSKSLVPSDIYENKVTPLHFLVARGSLEQ 374
Query: 498 ESNFISSL--LEAQNADGQTALHLACRRGSAELVETILDYPEANVDVLDKDGDPPLVFAL 555
+S ++ Q A G T L +A + +L +L + A+ ++ D+DG PL FA
Sbjct: 375 VRLLLSHEVDVDCQTASGYTPLLIATQDQQPDLCALLLAH-GADTNLADEDGWAPLHFAA 433
Query: 556 AAGSHECVCSLIKRNANVTSRLRDGLGPSVAHVCAYHGQPDCMRELLLAGADPNAVDDEG 615
+G L+ A V +R +G P H+ A + + R L+ AD N + EG
Sbjct: 434 QSGDDHTARLLLDHGALVNAREHEGWTP--LHLAAQNNFENIARLLVSRQADLNPCEAEG 491
Query: 616 ESVLHRAIAKKFTDCALVIVENG 638
++ LH +A F LV + G
Sbjct: 492 KTPLH--VAAYFGHIGLVKLLTG 512
Score = 53.9 bits (128), Expect = 2e-05
Identities = 43/155 (27%), Positives = 74/155 (47%), Gaps = 19/155 (12%)
Query: 474 LHRLVSEGDVTGVRDFLAKAASENESNFISSLLEAQNADGQTALHLACRRGSAELVETIL 533
LH V +G V ++ L + A L + + +G + LH+A +G + + +L
Sbjct: 528 LHLAVEKGKVRAIQHLLKRGA----------LPDVLDHNGYSPLHIAAAKGKYLIFKMLL 577
Query: 534 DYPEANVDVLDKDGDPPLVFALAAGSHECVCSLIKRNANVTSRLRDGLGP---SVAHVCA 590
+ A++++ + G PL A G E + L K +A++ D LG + H+ A
Sbjct: 578 RHG-ASLELCTQQGWAPLHLATYKGHLEIIHLLAKSHADL-----DALGSMQWTPLHLAA 631
Query: 591 YHGQPDCMRELLLAGADPNAVDDEGESVLHRAIAK 625
+ G+ M LL GA+PNA + G + LH A+ K
Sbjct: 632 FCGEEGMMLALLQCGANPNAAEQSGWTPLHLAVHK 666
>gb|AAQ64683.1| NIMA-related kinase 2 [Chlamydomonas reinhardtii]
Length = 653
Score = 87.4 bits (215), Expect = 1e-15
Identities = 63/255 (24%), Positives = 114/255 (44%), Gaps = 42/255 (16%)
Query: 205 NVCTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNEGR----LTLEQVLRYGADIARGVVEL 260
NV ++ A LC++M+ +++++ + L + + +Y + G+ L
Sbjct: 69 NVVCYNEAFLDGNRLCIIMEYAADGDLAKVIKKQQMMKRPLPEDMIWKYFIQVVMGLQAL 128
Query: 261 HAAGVVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDSAKIHSCMECIML 320
H+ ++ +KP N+++ NG A + D G+A +L K + K + +
Sbjct: 129 HSMKILHRDIKPGNIMVFDNGVAKIGDLGIAKLLTKTAAAKTQ---------------IG 173
Query: 321 SPHYTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIYR 380
+PHY PE W+ S SD W+ GC L E+ A+P+ S E+
Sbjct: 174 TPHYMGPEIWKNRPYSYT------------SDTWAIGCLLYELAALAVPFEARSMSELRY 221
Query: 381 QVVKAKKQPPQYASVVGGGIPRELWKMIGECLQFKPSKRPTFNAML--AIFLRHLQEIP- 437
+V++ P + R+L +M+ ECL P KRPT + +L A + +P
Sbjct: 222 KVLRGTYPP------IPNTFSRDLQQMVRECLDPNPDKRPTMDQILASAAVASRAKLVPH 275
Query: 438 --RSPPASPDNDLVK 450
R PPA+ ++LV+
Sbjct: 276 ESRHPPATAGSNLVE 290
>emb|CAD30902.1| putative bifunctional protein [Streptomyces coelicolor A3(2)]
gi|21223285|ref|NP_629064.1| putative bifunctional
protein [Streptomyces coelicolor A3(2)]
Length = 670
Score = 86.3 bits (212), Expect = 3e-15
Identities = 90/371 (24%), Positives = 149/371 (39%), Gaps = 49/371 (13%)
Query: 133 PVIEVGVHQDLKLVRRIGEGRRAGVEMWSAVIGGGRCKHQVAVKKVVLNEGMDLDWMLGK 192
PV + +L+ IG G E+W A R + VAVK ++ M D
Sbjct: 23 PVPHTVIEGRYELLEPIGSGGMG--EVWKA--HDRRLRRFVAVKGLLDRRAMTPDTQKAA 78
Query: 193 LEDLRRTS-----MWCRNVCTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNEGRLTLEQVL 247
++ RR + + +NV T H ++ + + +VM G +++L + L + +
Sbjct: 79 MQRARREAEALAKIEHQNVVTVHDQIETADQVWIVMKLLEGRSLADLLSRDRVLGVPRAA 138
Query: 248 RYGADIARGVVELHAAGVVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECD 307
G +A+G+ +H A V+ +KP N+L+ G V+ D+G+AT D
Sbjct: 139 EIGLQMAQGLRAVHEASVLHRDVKPGNVLVRDGGQVVLVDFGIATF----------EGAD 188
Query: 308 SAKIHSCMECIMLSPHYTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGA 367
H I+ +P Y APE + P G + SD W+ G TL EM G
Sbjct: 189 RVTRHG---GIIGTPPYLAPELFAPAAP----------GPTSASDLWALGVTLYEMVEGR 235
Query: 368 IPWAGLSAEEIYRQVVKAKKQPPQYASVVGGGIPRELW-------------KMIGECLQF 414
+P+ G E+ + +A +YA +G I L +M+ + L
Sbjct: 236 LPFGGNEVWEVQANIQQAPDPVLRYAGPLGPVIQGLLTTDPDRRLDAAGAEEMLRDVLAD 295
Query: 415 KPSKRPTFNAMLAIFLRHLQEIPRSPPASPDNDLVKGSVSNVTEASPVPELEIPQDPNRL 474
P A A H +PPAS V +V + + PVP +P +P+ +
Sbjct: 296 PGGPTPARAATPA----HPPTARPTPPASGPVPAVTAAVPAASSSGPVPSAAVPSEPSPV 351
Query: 475 HRLVSEGDVTG 485
G ++G
Sbjct: 352 ASGGGRGRLSG 362
>dbj|BAB85907.1| p90 ribosomal S6 kinase [Asterina pectinifera]
Length = 738
Score = 85.5 bits (210), Expect = 5e-15
Identities = 76/274 (27%), Positives = 119/274 (42%), Gaps = 50/274 (18%)
Query: 144 KLVRRIGEGRRAGVEMWSAVIG--GGRCKHQVAVKKVVLNEGMDLDWMLGKLEDLRRTSM 201
+L++ +G+G V + V G G +KK L K+ D RT M
Sbjct: 62 ELLKVLGQGSFGKVFLVRKVFGEDSGTLYAMKVLKKATL-----------KVRDRMRTKM 110
Query: 202 WCRNVCT---------FHGAMKVDEGLCLVMDKCFGSVQSEMLRNEGRLTLEQVLRYGAD 252
RN+ H A + + L L++D G L E T + Y A+
Sbjct: 111 E-RNILVDVNHPFIVKLHYAFQTEGKLYLILDFLRGGDLFTRLSKEVMFTEDDAKLYPAE 169
Query: 253 IARGVVELHAAGVVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDSAKIH 312
+A + LH+ G++ LKP N+LLDA+GH ++D+GL ++ D++K +
Sbjct: 170 LALALDHLHSLGIIYRDLKPENILLDASGHIKLTDFGL-----------SKESVDNSKTY 218
Query: 313 SCMECIMLSPHYTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAG 372
S + Y APE V + G + +D WSFG + EM TGA+P+ G
Sbjct: 219 SFCGTV----EYMAPEV---VNRR---------GHTTAADWWSFGVLMFEMLTGALPFQG 262
Query: 373 LSAEEIYRQVVKAKKQPPQYASVVGGGIPRELWK 406
+E + KAK PQ+ S + R+L+K
Sbjct: 263 QHRKETMAMIPKAKLGMPQFLSPEAQSLLRQLFK 296
Score = 41.2 bits (95), Expect = 0.11
Identities = 57/259 (22%), Positives = 107/259 (41%), Gaps = 41/259 (15%)
Query: 205 NVCTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNEGRLTLEQVLRYGADIARGVVELHAAG 264
N+ T + + +VM+ G + + + L+ + + + V LH
Sbjct: 473 NIITLRDVYDDGQNVYMVMELMKGGELLDKILKKKCLSEREACEIMHVVTKTVDYLHQQR 532
Query: 265 VVCMSLKPSNLL-LDANGHAV---VSDYGLATILKKPSCWKARPECDSAKIHSCMECIML 320
VV LKPSN+L D +G+ +SD+G A ++ + +++
Sbjct: 533 VVHRDLKPSNILYADDSGNPSSLRISDFGFAKQMRADN------------------GMLM 574
Query: 321 SPHYTAP-EAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIY 379
+P YTA A E +K+ G D WS G L M G P+A E
Sbjct: 575 TPCYTANFVAPEVLKRQ---------GYDAACDIWSLGVLLYTMLAGYTPFA--HGPEDS 623
Query: 380 RQVVKAKKQPPQYASVVGGG----IPRELWKMIGECLQFKPSKRPTFNAMLA-IFLRHLQ 434
+++ A+ ++ + GG + +++ L P KRP+ +L +++H +
Sbjct: 624 AEIILARIGEGKFP--LNGGNWDHVSPMAEELVRWMLHVDPFKRPSAAKVLQHPWMQHCE 681
Query: 435 EIPRSPPASPDNDLVKGSV 453
+ ++ + D DLVKG++
Sbjct: 682 SLSQNRLTTQDYDLVKGAM 700
>dbj|BAD27776.1| putative MAP3K alpha 1 protein kinase [Oryza sativa (japonica
cultivar-group)] gi|50251372|dbj|BAD28399.1| putative
MAP3K alpha 1 protein kinase [Oryza sativa (japonica
cultivar-group)]
Length = 894
Score = 83.6 bits (205), Expect = 2e-14
Identities = 70/278 (25%), Positives = 118/278 (42%), Gaps = 41/278 (14%)
Query: 205 NVCTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNEGRLTLEQVLRYGADIARGVVELHAAG 264
N+ ++G+ VD+ L + ++ G ++L+ G+ + Y I G+ LHA
Sbjct: 470 NIVRYYGSEMVDDKLYIYLEYVSGGSIHKLLQEYGQFGEPAIRSYTKQILLGLAYLHAKN 529
Query: 265 VVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDSAKIHSCMECIMLSPHY 324
V +K +N+L+D NG ++D+G+A + C SP++
Sbjct: 530 TVHRDIKGANILVDPNGRVKLADFGMAKHING---------------QQCAFSFKGSPYW 574
Query: 325 TAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIYRQVVK 384
APE V K+ N G + D WS GCT++EM T PW+ ++
Sbjct: 575 MAPE----VIKNSN-------GCNLAVDIWSLGCTVLEMATSKPPWSQYEGIAAVFKIGN 623
Query: 385 AKKQPPQYASVVGGGIPRELWKMIGECLQFKPSKRPTFNAMLA-IFLRHLQEIPRSPPAS 443
+K+ PP + + E I +CLQ PS RPT +L F+R+ AS
Sbjct: 624 SKELPP-----IPDHLSEEGRDFIRQCLQRNPSSRPTAVDLLQHSFIRN---------AS 669
Query: 444 PDNDLVKGSVSNVTEASPVPELEIPQDPNRLHRLVSEG 481
P + + ++ S P+L++ + L EG
Sbjct: 670 PLEKSLSDPLLQLSTTSCKPDLKVVGHARNMSSLGLEG 707
>emb|CAA55382.1| cdc7 [Schizosaccharomyces pombe] gi|4455770|emb|CAB36886.1| cdc7
[Schizosaccharomyces pombe] gi|19113132|ref|NP_596340.1|
hypothetical protein SPBC21.06c [Schizosaccharomyces
pombe 972h-] gi|1076912|pir||S46367 protein kinase CDC7
(EC 2.7.1.-) - fission yeast (Schizosaccharomyces
pombe) gi|1168817|sp|P41892|CDC7_SCHPO Cell division
control protein 7
Length = 1062
Score = 83.2 bits (204), Expect = 2e-14
Identities = 61/247 (24%), Positives = 115/247 (45%), Gaps = 34/247 (13%)
Query: 173 VAVKKVVLNEGMDLDWMLGKLEDLRRTSMWCRNVCTFHGAMKVDEGLCLVMDKCFGSVQS 232
VAVKKV L++ + D + K+E ++ N+ + G+ + ++ LC++++ C
Sbjct: 35 VAVKKVKLSKMLKSDLSVIKMEIDLLKNLDHPNIVKYRGSYQTNDSLCIILEYCENGSLR 94
Query: 233 EMLRNEGRLTLEQVLRYGADIARGVVELHAAGVVCMSLKPSNLLLDANGHAVVSDYGLAT 292
+ +N G++ V Y + +G++ LH GV+ +K +N+L +G ++D+G+AT
Sbjct: 95 SICKNFGKIPENLVALYTFQVLQGLLYLHNQGVIHRDIKGANILTTKDGTIKLADFGVAT 154
Query: 293 ILKKPSCWKARPECDSAKIHSCMECIMLSPHYTAPEAWEPVKKSLNLFWDDGIGISPESD 352
+ ++ + HS ++ SP++ APE E V G + SD
Sbjct: 155 ------------KINALEDHS----VVGSPYWMAPEVIELV------------GATTASD 186
Query: 353 AWSFGCTLVEMCTGAIPWAGLSAEEIYRQVVKAKKQPPQYASVVGGGIPRELWKMIGECL 412
WS GCT++E+ G P+ L ++VK + P + I + +C
Sbjct: 187 IWSVGCTVIELLDGNPPYYDLDPTSALFRMVKDEHPP------LPSNISSAAKSFLMQCF 240
Query: 413 QFKPSKR 419
Q P+ R
Sbjct: 241 QKDPNLR 247
>ref|ZP_00519579.1| Protein kinase:Serine/threonine protein kinase, active site
[Solibacter usitatus Ellin6076]
gi|67866139|gb|EAM61137.1| Protein
kinase:Serine/threonine protein kinase, active site
[Solibacter usitatus Ellin6076]
Length = 912
Score = 82.8 bits (203), Expect = 3e-14
Identities = 67/233 (28%), Positives = 105/233 (44%), Gaps = 18/233 (7%)
Query: 205 NVCTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNEGRLTLEQVLRYGADIARGVVELHAAG 264
N+ H + D + +VM+ G S+++ +G + L + L +IA + HAAG
Sbjct: 68 NIIVVHEIGEHDRQIYIVMELVDGKPLSQLIPKKG-MRLTEALHIAVEIADALAAAHAAG 126
Query: 265 VVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDSA--KIHSCMECIMLSP 322
+V LKP+NL++D G A V D+GLA + + A ++ + IM S
Sbjct: 127 IVHRDLKPANLMVDRLGRAKVLDFGLAKMTASAAVATDEETRTLAVTQLQTEEGVIMGSI 186
Query: 323 HYTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIYRQV 382
Y +PE E G + SD +SFG L EM TG + G S V
Sbjct: 187 PYMSPEQAE------------GRPVDARSDIFSFGAVLYEMITGRRAFGGESRVSTLAAV 234
Query: 383 VKAKKQPPQYASVVGGGIPRELWKMIGECLQFKPSKRPTFNAMLAIFLRHLQE 435
V+ PP S + G P EL ++I CL+ + +R A + + L L++
Sbjct: 235 VEKDPVPP---SEIAAGTPPELERLIARCLRKEVGRRSQSMADVKLALEELRD 284
>ref|NP_766510.1| ankyrin repeat and kinase domain containing 1 [Mus musculus]
gi|26331766|dbj|BAC29613.1| unnamed protein product [Mus
musculus]
Length = 745
Score = 82.8 bits (203), Expect = 3e-14
Identities = 96/375 (25%), Positives = 149/375 (39%), Gaps = 48/375 (12%)
Query: 268 MSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDSAKIHSCMECIMLSPHYTAP 327
+ LKP N+LLD N H +SD+GL S W E + K + + + Y P
Sbjct: 156 LDLKPGNILLDNNMHVKISDFGL-------SKWM---EQSTQKQYIERSALRGTLSYIPP 205
Query: 328 EAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIYRQVVKAKK 387
E +F ++ PE D +SF + E+ T P+AGL+ I +V +
Sbjct: 206 E----------MFLENNKAPGPEYDVYSFAIVIWEILTQKKPYAGLNMMTIIIRVAAGMR 255
Query: 388 QPPQYASVVGGGIPRELWKMIGECLQFKPSKRPTFNAMLAIFLRHLQEIPRSPPASPDND 447
Q S ++ ++ C P KRP F +A+ L + +SP P
Sbjct: 256 PSLQDVSDEWPEEVHQMVNLMKRCWDQDPKKRPCF-LNVAVETDMLLSLFQSPMTDP--- 311
Query: 448 LVKGSVSNVTEASPVPELEIPQDPNRLHRLVSEGDVTGVRDFLAKAASENESNFISSLLE 507
G + + S P L P H++ E V +A + S + +I L +
Sbjct: 312 ---GCEALTQKVSCKPSLSQP------HKVSKE-----VNQEIADSVSSDSLKWILQLSD 357
Query: 508 AQN-------ADGQTALHLACRRGSAELVETILDYPEANVDVLDKDGDPPLVFALAAGSH 560
+++ + T LH GS E V +L + + +VD G PL+ A
Sbjct: 358 SKSLVASDVYENRVTPLHFLVAGGSLEQVRLLLSH-DVDVDCQTASGYTPLLIATQDQQP 416
Query: 561 ECVCSLIKRNANVTSRLRDGLGPSVAHVCAYHGQPDCMRELLLAGADPNAVDDEGESVLH 620
+ L+ A+ DG P H A +G R LL GA NA + EG + LH
Sbjct: 417 DLCALLLAHGADTNLADEDGWAP--LHFAAQNGDDHTARLLLDHGALVNAREHEGWTPLH 474
Query: 621 RAIAKKFTDCALVIV 635
A F + A ++V
Sbjct: 475 LAAQNNFENVARLLV 489
Score = 55.8 bits (133), Expect = 4e-06
Identities = 45/155 (29%), Positives = 71/155 (45%), Gaps = 19/155 (12%)
Query: 474 LHRLVSEGDVTGVRDFLAKAASENESNFISSLLEAQNADGQTALHLACRRGSAELVETIL 533
LH V G V ++ L A L +A + G + LH+A RG + + +L
Sbjct: 539 LHLAVERGKVRAIQHLLKCGA----------LPDALDHSGYSPLHIAAARGKDLIFKMLL 588
Query: 534 DYPEANVDVLDKDGDPPLVFALAAGSHECVCSLIKRNANVTSRLRDGLGP---SVAHVCA 590
Y A++++ + G PL A G E + L K + ++ D LG + H+ A
Sbjct: 589 RYG-ASLELRTQQGWTPLHLATYKGHLEIIHQLAKSHVDL-----DALGSMQWTPLHLAA 642
Query: 591 YHGQPDCMRELLLAGADPNAVDDEGESVLHRAIAK 625
+ G+ M LL GA+PNA + G + LH A+ K
Sbjct: 643 FQGEEGVMLALLQCGANPNAAEQSGWTPLHLAVHK 677
>emb|CAE61017.1| Hypothetical protein CBG04756 [Caenorhabditis briggsae]
Length = 1088
Score = 82.8 bits (203), Expect = 3e-14
Identities = 78/323 (24%), Positives = 135/323 (41%), Gaps = 41/323 (12%)
Query: 76 SAADSAAAAGGGGGGDCDFTDDDEDRDDSEVDDGDDQKLDCRKNSRGSQASSSGGCAPVI 135
+ S GGG ++ R G S G SS A
Sbjct: 64 TTTSSGVPTASTGGGSARYSSSSSGRSHPTTGGGSSS----HARSSGQSGMSSRSAARRN 119
Query: 136 EVGVHQD-LKLVRRIGEGRRAGVEMWSAVIGGGRCKHQVAVK---KVVLNEGMDLDWMLG 191
+ VH KL++ IG+G A V++ VI G H+VA+K K LN L +
Sbjct: 120 DQDVHVGKYKLLKTIGKGNFAKVKLAKHVITG----HEVAIKIIDKTALNPS-SLQKLFR 174
Query: 192 KLEDLRRTSMWCRNVCTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNEGRLTLEQVLRYGA 251
+++ +++ N+ + M+ ++ L LV++ G + L GR+ ++
Sbjct: 175 EVKIMKQLDH--PNIVKLYQVMETEQTLYLVLEYASGGEVFDYLVAHGRMKEKEARAKFR 232
Query: 252 DIARGVVELHAAGVVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDSAKI 311
I V LH+ ++ LK NLLLD + + ++D+G + ++ +
Sbjct: 233 QIVSAVQYLHSKNIIHRDLKAENLLLDQDMNIKIADFGFS---------------NTFSL 277
Query: 312 HSCMECIMLSPHYTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWA 371
+ ++ SP Y APE + K DG PE D WS G L + +G++P+
Sbjct: 278 GNKLDTFCGSPPYAAPELFSGKKY-------DG----PEVDVWSLGVILYTLVSGSLPFD 326
Query: 372 GLSAEEIYRQVVKAKKQPPQYAS 394
G + +E+ +V++ K + P Y S
Sbjct: 327 GQNLKELRERVLRGKYRIPFYMS 349
>ref|XP_394955.2| PREDICTED: similar to Rsk-2 [Apis mellifera]
Length = 746
Score = 82.4 bits (202), Expect = 4e-14
Identities = 83/302 (27%), Positives = 126/302 (41%), Gaps = 62/302 (20%)
Query: 128 SGGCAPVIEVGVHQ------------DLKLVRRIGEGRRAGVEMWSAVIG--GGRCKHQV 173
S GC+ E+ V + +L++ +G+G V + V+G G
Sbjct: 44 SNGCSESHEIEVREVVRDGHEKADPSQFELLKVLGQGSFGKVFLVRKVVGKDSGTLYAMK 103
Query: 174 AVKKVVLNEGMDLDWMLGKLEDLRRTSMWCRNVCT---------FHGAMKVDEGLCLVMD 224
+KK L K+ D RT M RN+ H A + + L L++D
Sbjct: 104 VLKKATL-----------KVRDRVRTKME-RNILVDVEHPFIVRLHYAFQTEGNLYLILD 151
Query: 225 KCFGSVQSEMLRNEGRLTLEQVLRYGADIARGVVELHAAGVVCMSLKPSNLLLDANGHAV 284
G L E T + V Y A++A + +H G++ LKP N+LLD GH
Sbjct: 152 FLRGGDLFSRLAKELMFTEDDVKFYLAELALALDHIHKLGIIYRDLKPENILLDTEGHIA 211
Query: 285 VSDYGLATILKKPSCWKARPECDSAKIHSCMECIMLSPHYTAPEAWEPVKKSLNLFWDDG 344
++D+GL+ K+P D K +S + Y APE +K
Sbjct: 212 LTDFGLS---KQP--------LDDCKAYSFCGTV----EYMAPEI--VTRK--------- 245
Query: 345 IGISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIYRQVVKAKKQPPQYASVVGGGIPREL 404
G S +D WSFG + EM TGA+P+ G + +E Q++KAK P S + R L
Sbjct: 246 -GHSFAADWWSFGVLMFEMLTGALPFQGANRKETMTQILKAKLGMPLNISPDAQALLRVL 304
Query: 405 WK 406
+K
Sbjct: 305 FK 306
Score = 42.7 bits (99), Expect = 0.037
Identities = 59/233 (25%), Positives = 88/233 (37%), Gaps = 44/233 (18%)
Query: 205 NVCTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNEGRLTLEQVLRYGADIARGVVELHAAG 264
++ T + D+ LV++ G + L LT ++ I V LH G
Sbjct: 478 HIVTLRAVHEDDKRAYLVLELLRGGELLDRLLQRRNLTEKEAAEVMYTIVSVVNYLHENG 537
Query: 265 VVCMSLKPSNLLLDANG----HAVVSDYGLATILKKPSCWKARPECDSAKIHSCMECIML 320
VV LKPSN+L G + D G A L+ + +++
Sbjct: 538 VVHRDLKPSNILYSKLGADPTTLCLCDLGFAKQLRAEN------------------GLLM 579
Query: 321 SPHYTAP-EAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPW---AGLSAE 376
+P YTA A E +K+ G D WS G L M G P+ G SA
Sbjct: 580 TPCYTANFVAPEVLKRQ---------GYDAACDIWSLGVLLYIMLAGYTPFRNSPGDSAT 630
Query: 377 EIYRQVVKAKKQPPQYASVVGG---GIPRELWKMIGECLQFKPSKRPTFNAML 426
+I ++ P Y V G I E +++ L P++RPT A+L
Sbjct: 631 DILDRI------GPGYIDVESGIWCQISNEAKELVKRMLHVDPNRRPTAAAIL 677
>emb|CAB11500.1| wis4 [Schizosaccharomyces pombe] gi|19114469|ref|NP_593557.1|
hypothetical protein SPAC9G1.02 [Schizosaccharomyces
pombe 972h-] gi|18201962|sp|O14299|WIS4_SCHPO MAP kinase
kinase kinase wis4 (MAP kinase kinase kinase wak1) (MAP
kinase kinase kinase wik1)
Length = 1401
Score = 82.0 bits (201), Expect = 5e-14
Identities = 63/219 (28%), Positives = 101/219 (45%), Gaps = 23/219 (10%)
Query: 205 NVCTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNEGRLTLEQVLR-YGADIARGVVELHAA 263
NV T++G E + + M+ C G +++L + GR+ E VL+ Y + G+ +H+
Sbjct: 1097 NVVTYYGVEVHREKVYIFMEFCQGGSLADLLAH-GRIEDENVLKVYVVQLLEGLAYIHSQ 1155
Query: 264 GVVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDSAKIHSCMECIMLSPH 323
++ +KP+N+LLD G SD+G A + P+ PE I ++ + +P
Sbjct: 1156 HILHRDIKPANILLDHRGMIKYSDFGSALYVSPPT----DPEVRYEDIQPELQHLAGTPM 1211
Query: 324 YTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIYRQVV 383
Y APE KK G D WS GC ++EM TG+ PW+ + E V
Sbjct: 1212 YMAPEIILGTKK----------GDFGAMDIWSLGCVILEMMTGSTPWSEMDNEWAIMYHV 1261
Query: 384 KAKKQP--PQYASVVGGGIPRELWKMIGECLQFKPSKRP 420
A P PQ + + R+ I +C + P +RP
Sbjct: 1262 AAMHTPSIPQNEKI--SSLARD---FIEQCFERDPEQRP 1295
>emb|CAA72718.1| Wak1 protein [Schizosaccharomyces pombe]
Length = 1306
Score = 82.0 bits (201), Expect = 5e-14
Identities = 63/219 (28%), Positives = 101/219 (45%), Gaps = 23/219 (10%)
Query: 205 NVCTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNEGRLTLEQVLR-YGADIARGVVELHAA 263
NV T++G E + + M+ C G +++L + GR+ E VL+ Y + G+ +H+
Sbjct: 1002 NVVTYYGVEVHREKVYIFMEFCQGGSLADLLAH-GRIEDENVLKVYVVQLLEGLAYIHSQ 1060
Query: 264 GVVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDSAKIHSCMECIMLSPH 323
++ +KP+N+LLD G SD+G A + P+ PE I ++ + +P
Sbjct: 1061 HILHRDIKPANILLDHRGMIKYSDFGSALYVSPPT----DPEVRYEDIQPELQHLAGTPM 1116
Query: 324 YTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIYRQVV 383
Y APE KK G D WS GC ++EM TG+ PW+ + E V
Sbjct: 1117 YMAPEIILGTKK----------GDFGAMDIWSLGCVILEMMTGSTPWSEMDNEWAIMYHV 1166
Query: 384 KAKKQP--PQYASVVGGGIPRELWKMIGECLQFKPSKRP 420
A P PQ + + R+ I +C + P +RP
Sbjct: 1167 AAMHTPSIPQNEKI--SSLARD---FIEQCFERDPEQRP 1200
>emb|CAA69030.1| protein kinase [Schizosaccharomyces pombe]
Length = 1275
Score = 82.0 bits (201), Expect = 5e-14
Identities = 63/219 (28%), Positives = 101/219 (45%), Gaps = 23/219 (10%)
Query: 205 NVCTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNEGRLTLEQVLR-YGADIARGVVELHAA 263
NV T++G E + + M+ C G +++L + GR+ E VL+ Y + G+ +H+
Sbjct: 971 NVVTYYGVEVHREKVYIFMEFCQGGSLADLLAH-GRIEDENVLKVYVVQLLEGLAYIHSQ 1029
Query: 264 GVVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDSAKIHSCMECIMLSPH 323
++ +KP+N+LLD G SD+G A + P+ PE I ++ + +P
Sbjct: 1030 HILHRDIKPANILLDHRGMIKYSDFGSALYVSPPT----DPEVRYEDIQPELQHLAGTPM 1085
Query: 324 YTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIYRQVV 383
Y APE KK G D WS GC ++EM TG+ PW+ + E V
Sbjct: 1086 YMAPEIILGTKK----------GDFGAMDIWSLGCVILEMMTGSTPWSEMDNEWAIMYHV 1135
Query: 384 KAKKQP--PQYASVVGGGIPRELWKMIGECLQFKPSKRP 420
A P PQ + + R+ I +C + P +RP
Sbjct: 1136 AAMHTPSIPQNEKI--SSLARD---FIEQCFERDPEQRP 1169
>emb|CAB44492.1| ribosomal protein S6 kinase 3 [Mus musculus]
gi|29835158|gb|AAH51079.1| Ribosomal protein S6 kinase,
polypeptide 2 [Mus musculus] gi|34785208|gb|AAH56946.1|
Ribosomal protein S6 kinase, polypeptide 2 [Mus
musculus] gi|27696717|gb|AAH43064.1| Ribosomal protein
S6 kinase, polypeptide 2 [Mus musculus]
gi|11133183|sp|Q9WUT3|KS6A2_MOUSE Ribosomal protein S6
kinase alpha 2 (S6K-alpha 2) (90 kDa ribosomal protein
S6 kinase 2) (p90-RSK 2) (Ribosomal S6 kinase 3) (RSK-3)
(pp90RSK3) (Protein-tyrosine kinase Mpk-9) (MAP
kinase-activated protein kinase 1c) (MAPKAPK1C)
gi|6755374|ref|NP_035429.1| ribosomal protein S6 kinase,
polypeptide 2 [Mus musculus]
Length = 733
Score = 81.6 bits (200), Expect = 7e-14
Identities = 81/306 (26%), Positives = 125/306 (40%), Gaps = 45/306 (14%)
Query: 117 RKNSRGSQASSSG----GCAPVIEVGVH----------QDLKLVRRIGEGRRAGVEMWSA 162
RK SR +S S G I++ H +L++ +G+G V +
Sbjct: 19 RKKSRSKSSSLSRLEEEGIVKEIDISNHVKEGFEKADPSQFELLKVLGQGSYGKVFLVRK 78
Query: 163 VIGG--GRCKHQVAVKKVVLNEGMDLDWMLGKLEDLRRTSMWCRNVCTFHGAMKVDEGLC 220
V G G+ +KK L D + K+E + + H A + + L
Sbjct: 79 VTGSDAGQLYAMKVLKKATLKVR---DRVRSKMERDILAEVNHPFIVKLHYAFQTEGKLY 135
Query: 221 LVMDKCFGSVQSEMLRNEGRLTLEQVLRYGADIARGVVELHAAGVVCMSLKPSNLLLDAN 280
L++D G L E T E V Y A++A + LH G++ LKP N+LLD
Sbjct: 136 LILDFLRGGDLFTRLSKEVMFTEEDVKFYLAELALALDHLHGLGIIYRDLKPENILLDEE 195
Query: 281 GHAVVSDYGLATILKKPSCWKARPECDSAKIHSCMECIMLSPHYTAPEAWEPVKKSLNLF 340
GH ++D+GL+ K + D C + Y APE V +
Sbjct: 196 GHIKITDFGLS---------KEATDHDKRAYSFCG-----TIEYMAPEV---VNRR---- 234
Query: 341 WDDGIGISPESDAWSFGCTLVEMCTGAIPWAGLSAEEIYRQVVKAKKQPPQYASVVGGGI 400
G + +D WSFG + EM TG++P+ G +E ++KAK PQ+ S +
Sbjct: 235 -----GHTQSADWWSFGVLMFEMLTGSLPFQGKDRKETMALILKAKLGMPQFLSAEAQSL 289
Query: 401 PRELWK 406
R L+K
Sbjct: 290 LRALFK 295
Score = 38.5 bits (88), Expect = 0.69
Identities = 55/213 (25%), Positives = 91/213 (41%), Gaps = 43/213 (20%)
Query: 253 IARGVVELHAAGVVCMSLKPSNLL-LDANGHAV---VSDYGLATILKKPSCWKARPECDS 308
IAR + LH+ GVV LKPSN+L +D +G+ + D+G A L+ +
Sbjct: 516 IARTMDYLHSQGVVHRDLKPSNILYMDESGNPESIRICDFGFAKQLRAEN---------- 565
Query: 309 AKIHSCMECIMLSPHYTAP-EAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGA 367
++++P YTA A E +K+ G D WS G L M G
Sbjct: 566 --------GLLMTPCYTANFVAPEVLKRQ---------GYDAACDVWSLGILLYTMLAGF 608
Query: 368 IPWAG---LSAEEIYRQVVKAKKQPPQYASVVGG--GIPRELWKMIGECLQFKPSKRPTF 422
P+A + EEI ++ K YA G I ++ + L P +R T
Sbjct: 609 TPFANGPDDTPEEILARIGSGK-----YALSGGNWDSISDAAKDVVSKMLHVDPQQRLTA 663
Query: 423 NAMLA-IFLRHLQEIPRSPPASPDNDLVKGSVS 454
+L ++ + + + ++ + D LVKG+++
Sbjct: 664 VQVLKHPWIVNREYLSQNQLSRQDVHLVKGAMA 696
>gb|AAA97437.1| serine/threonine kinase
Length = 1192
Score = 81.6 bits (200), Expect = 7e-14
Identities = 79/323 (24%), Positives = 136/323 (41%), Gaps = 44/323 (13%)
Query: 76 SAADSAAAAGGGGGGDCDFTDDDEDRDDSEVDDGDDQKLDCRKNSRGSQASSSGGCAPVI 135
+ A S A A GG ++ S S G SS A
Sbjct: 109 TTASSGAPAASSGGSSARYSSSGRSHPTSGSSSS-------HARSTGQSGMSSRSAARRN 161
Query: 136 EVGVHQD-LKLVRRIGEGRRAGVEMWSAVIGGGRCKHQVAVK---KVVLNEGMDLDWMLG 191
+ VH KL++ IG+G A V++ VI G H+VA+K K LN L +
Sbjct: 162 DQDVHVGKYKLLKTIGKGNFAKVKLAKHVITG----HEVAIKIIDKTALNPS-SLQKLFR 216
Query: 192 KLEDLRRTSMWCRNVCTFHGAMKVDEGLCLVMDKCFGSVQSEMLRNEGRLTLEQVLRYGA 251
+++ +++ N+ + M+ ++ L LV++ G + L GR+ ++
Sbjct: 217 EVKIMKQLDH--PNIVKLYQVMETEQTLYLVLEYASGGEVFDYLVAHGRMKEKEARAKFR 274
Query: 252 DIARGVVELHAAGVVCMSLKPSNLLLDANGHAVVSDYGLATILKKPSCWKARPECDSAKI 311
I V LH+ ++ LK NLLLD + + ++D+G + ++ +
Sbjct: 275 QIVSAVQYLHSKNIIHRDLKAENLLLDQDMNIKIADFGFS---------------NTFSL 319
Query: 312 HSCMECIMLSPHYTAPEAWEPVKKSLNLFWDDGIGISPESDAWSFGCTLVEMCTGAIPWA 371
+ ++ SP Y APE + K DG PE D WS G L + +G++P+
Sbjct: 320 GNKLDTFCGSPPYAAPELFSGKKY-------DG----PEVDVWSLGVILYTLVSGSLPFD 368
Query: 372 GLSAEEIYRQVVKAKKQPPQYAS 394
G + +E+ +V++ K + P Y S
Sbjct: 369 GQNLKELRERVLRGKYRIPFYMS 391
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.318 0.135 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,176,244,599
Number of Sequences: 2540612
Number of extensions: 52404744
Number of successful extensions: 295628
Number of sequences better than 10.0: 19843
Number of HSP's better than 10.0 without gapping: 7970
Number of HSP's successfully gapped in prelim test: 11938
Number of HSP's that attempted gapping in prelim test: 252290
Number of HSP's gapped (non-prelim): 37059
length of query: 652
length of database: 863,360,394
effective HSP length: 135
effective length of query: 517
effective length of database: 520,377,774
effective search space: 269035309158
effective search space used: 269035309158
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 78 (34.7 bits)
Medicago: description of AC126006.5


