

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC125480.15 - phase: 0
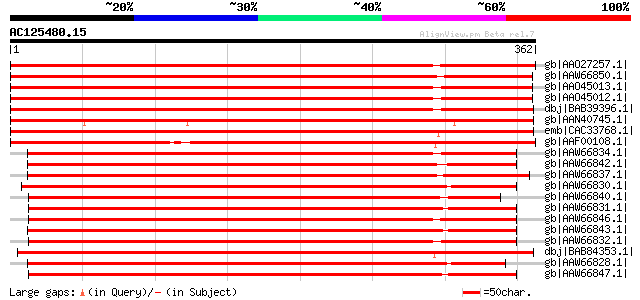
(362 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAO27257.1| putative S-adenosyl-L-methionine:salicylic acid c... 449 e-125
gb|AAW66850.1| SAMT [Nicotiana tabacum] 446 e-124
gb|AAO45013.1| S-adenosyl-L-methionine:benzoic acid/salicylic ac... 444 e-123
gb|AAO45012.1| S-adenosyl-L-methionine:benzoic acid/salicylic ac... 444 e-123
dbj|BAB39396.1| S-adenosyl-L-methionine:salicylic acid carboxyl ... 429 e-119
gb|AAN40745.1| S-adenosyl-L-methionine:salicylic acid methyltran... 426 e-118
emb|CAC33768.1| S-adenosyl-L-methionine:salicylic acid carboxyl ... 419 e-116
gb|AAF00108.1| S-adenosyl-L-methionine:salicylic acid carboxyl m... 416 e-115
gb|AAW66834.1| SAMT [Petunia nyctaginiflora] 407 e-112
gb|AAW66842.1| SAMT [Streptosolen jamesonii] 405 e-111
gb|AAW66837.1| SAMT [Brunfelsia americana] 405 e-111
gb|AAW66830.1| SAMT [Cestrum nocturnum] 402 e-111
gb|AAW66840.1| SAMT [Capsicum annuum] 400 e-110
gb|AAW66831.1| SAMT [Brugmansia sp. Robadey 027] 398 e-109
gb|AAW66846.1| SAMT [Physalis virginiana] 396 e-109
gb|AAW66843.1| SAMT [Juanulloa aurantiaca] 394 e-108
gb|AAW66832.1| SAMT [Hyoscyamus albus] 393 e-108
dbj|BAB84353.1| S-adenosyl-L-methionine:salicylic acid calboxyl ... 392 e-107
gb|AAW66828.1| SAMT [Cestrum elegans] 391 e-107
gb|AAW66847.1| SAMT [Solandra maxima] 390 e-107
>gb|AAO27257.1| putative S-adenosyl-L-methionine:salicylic acid carboxyl
methyltransferase [Pisum sativum]
Length = 360
Score = 449 bits (1154), Expect = e-125
Identities = 219/363 (60%), Positives = 284/363 (77%), Gaps = 5/363 (1%)
Query: 1 MEVAKVLHMNGGVGETSYANNSLV-QRKVIYLTKPLRDEAITSMYNNTLSKSLTIADLGC 59
MEVA+ +HM GG GE SYANNS++ Q VI TK +R+EAITS+Y++TL +SL IADLGC
Sbjct: 1 MEVAQGMHMKGGDGEESYANNSIIFQGNVISSTKLIREEAITSLYSSTLPRSLAIADLGC 60
Query: 60 SSGSNTLLVILDIIKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDE 119
S G NTL V+ ++I VVE LC+KLNH SPEY IYLNDL GNDFN++F SLD FKEKL DE
Sbjct: 61 SCGPNTLSVVSEVIHVVENLCKKLNHSSPEYKIYLNDLAGNDFNSVFRSLDSFKEKLRDE 120
Query: 120 MGTEMGPCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTSP 179
TE+ C+F GVPGSF+GR+FP +SLHFVHSSYS+HWLSKVPEG +N+KG IYI+ TSP
Sbjct: 121 TKTEIDRCYFFGVPGSFYGRVFPDRSLHFVHSSYSVHWLSKVPEGIENSKGAIYINETSP 180
Query: 180 LNVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCYIWELLAMAL 239
NV+KAYY+QF+ DFS+F+KCRAEEIVEGG M+++ +GR+ D+P SKE C + +LLA AL
Sbjct: 181 SNVIKAYYEQFERDFSVFIKCRAEEIVEGGRMVLSILGRRGDDPFSKESCDLLDLLATAL 240
Query: 240 NDMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWNARDDFESE 299
N MVL+G+I+E+K+NTFNIP YYPS SEVK ++ EGSF +N +E SEV+ N E
Sbjct: 241 NHMVLKGLIEEDKVNTFNIPNYYPSRSEVKSSILEEGSFAINHVEFSEVDLNN----SGE 296
Query: 300 SFGDDGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRTKFINITILLA 359
S D GYNVAQ +RA+ EPL+VSHFGE ++ ++F RY + + D+ S+ + + + TI L
Sbjct: 297 SLHDSGYNVAQTIRAVFEPLMVSHFGEAIIDDVFQRYHEIVVDQMSREKMQTVYFTISLT 356
Query: 360 KRS 362
+++
Sbjct: 357 RKA 359
>gb|AAW66850.1| SAMT [Nicotiana tabacum]
Length = 358
Score = 446 bits (1146), Expect = e-124
Identities = 210/360 (58%), Positives = 291/360 (80%), Gaps = 4/360 (1%)
Query: 1 MEVAKVLHMNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLSKSLTIADLGCS 60
M+V +VLHMNGG+G+ SYA NSLVQ+KVI +TKP+ ++AIT +Y + ++L IADLGCS
Sbjct: 1 MKVVEVLHMNGGIGDISYAKNSLVQQKVILMTKPITEQAITDLYCSLFPQNLCIADLGCS 60
Query: 61 SGSNTLLVILDIIKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEM 120
SG+NT +V+ ++IK+VEK +K +SPE+ NDLPGNDFNTIF SLDIF++ L ++
Sbjct: 61 SGANTFIVVSELIKIVEKERKKHGFQSPEFHFNFNDLPGNDFNTIFQSLDIFQQDLRKQI 120
Query: 121 GTEMGPCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTSPL 180
G E GPCFFSGV GSF+ R+FP SLHFVHSSYSL WLS+VP+ +NNKGNIY++STSP
Sbjct: 121 GEEFGPCFFSGVSGSFYTRLFPSNSLHFVHSSYSLMWLSQVPDAVENNKGNIYMASTSPP 180
Query: 181 NVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCYIWELLAMALN 240
+V+KAYYKQ++ DFS FLK R+EE+++GG M++TF+GR+S++PTSKECCYIWELLAMALN
Sbjct: 181 SVIKAYYKQYEKDFSNFLKYRSEELMKGGKMVLTFLGRESEDPTSKECCYIWELLAMALN 240
Query: 241 DMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWNARDDFESES 300
++V+EG+I+EEK+++FNIP Y PSP++VK V EGSF +NQLE + V+WNA +D
Sbjct: 241 ELVVEGLIEEEKVDSFNIPQYTPSPADVKYVVEKEGSFTINQLEATRVHWNACND----K 296
Query: 301 FGDDGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRTKFINITILLAK 360
+ + GY+V++CMRA+AEPLLVS FGE ++ +F++Y++ +++ SK +T+F N+ + L K
Sbjct: 297 YKNVGYSVSRCMRAVAEPLLVSQFGEELMDLVFHKYEQIISECMSKAQTEFTNVIVSLTK 356
>gb|AAO45013.1| S-adenosyl-L-methionine:benzoic acid/salicylic acid carboxyl
methyltransferase [Petunia x hybrida]
Length = 357
Score = 444 bits (1143), Expect = e-123
Identities = 212/360 (58%), Positives = 286/360 (78%), Gaps = 5/360 (1%)
Query: 1 MEVAKVLHMNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLSKSLTIADLGCS 60
MEV +VLHMNGG G++SYANNSLVQ+KVI +TKP+ ++A+ +Y++ ++L IADLGCS
Sbjct: 1 MEVVEVLHMNGGNGDSSYANNSLVQQKVILMTKPITEQAMIDLYSSLFPETLCIADLGCS 60
Query: 61 SGSNTLLVILDIIKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEM 120
G+NT LV+ ++K+VEK +K KSPE+ + NDLPGNDFNT+F SL F+E L +
Sbjct: 61 LGANTFLVVSQLVKIVEKERKKHGFKSPEFYFHFNDLPGNDFNTLFQSLGAFQEDLRKHI 120
Query: 121 GTEMGPCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTSPL 180
G GPCFFSGVPGSF+ R+FP +SLHFV+SSYSL WLS+VP G +NNKGNIY++ TSPL
Sbjct: 121 GESFGPCFFSGVPGSFYTRLFPSKSLHFVYSSYSLMWLSQVPNGIENNKGNIYMARTSPL 180
Query: 181 NVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCYIWELLAMALN 240
+V+KAYYKQ++IDFS FLK R+EE+++GG M++T +GR+S++PTSKECCYIWELLAMALN
Sbjct: 181 SVIKAYYKQYEIDFSNFLKYRSEELMKGGKMVLTLLGRESEDPTSKECCYIWELLAMALN 240
Query: 241 DMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWNARDDFESES 300
+V EG+IKEEK++ FNIP Y PSP+EVK V EGSF +N+LE S V+WNA S +
Sbjct: 241 KLVEEGLIKEEKVDAFNIPQYTPSPAEVKYIVEKEGSFTINRLETSRVHWNA-----SNN 295
Query: 301 FGDDGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRTKFINITILLAK 360
+ GYNV++CMRA+AEPLLVSHF + ++ +F++Y++ ++D SK +T+FIN+ + L K
Sbjct: 296 EKNGGYNVSRCMRAVAEPLLVSHFDKELMDLVFHKYEEIISDCMSKEKTEFINVIVSLTK 355
>gb|AAO45012.1| S-adenosyl-L-methionine:benzoic acid/salicylic acid carboxyl
methyltransferase [Petunia x hybrida]
Length = 357
Score = 444 bits (1141), Expect = e-123
Identities = 213/360 (59%), Positives = 285/360 (79%), Gaps = 5/360 (1%)
Query: 1 MEVAKVLHMNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLSKSLTIADLGCS 60
MEV +VLHMNGG G++SYANNSLVQ+KVI +TKP+ ++A+ +Y++ ++L IADLGCS
Sbjct: 1 MEVVEVLHMNGGNGDSSYANNSLVQQKVILMTKPITEQAMIDLYSSLFPETLCIADLGCS 60
Query: 61 SGSNTLLVILDIIKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEM 120
G+NT LV+ ++K+VEK +K KSPE+ + NDLPGNDFNT+F SL F+E L +
Sbjct: 61 LGANTFLVVSQLVKIVEKERKKHGFKSPEFYFHFNDLPGNDFNTLFQSLGAFQEDLRKHI 120
Query: 121 GTEMGPCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTSPL 180
G GPCFFSGVPGSF+ R+FP +SLHFV+SSYSL WLS+VP G +NNKGNIY++ TSPL
Sbjct: 121 GESFGPCFFSGVPGSFYTRLFPSKSLHFVYSSYSLMWLSQVPNGIENNKGNIYMARTSPL 180
Query: 181 NVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCYIWELLAMALN 240
+V+KAYYKQ++IDFS FLK R+EE+++GG M++T +GR+S++PTSKECCYIWELLAMALN
Sbjct: 181 SVIKAYYKQYEIDFSNFLKYRSEELMKGGKMVLTLLGRESEDPTSKECCYIWELLAMALN 240
Query: 241 DMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWNARDDFESES 300
+V EG+IKEEK++ FNIP Y PSP+EVK V EGSF +N+LE S V+WNA S +
Sbjct: 241 KLVEEGLIKEEKVDAFNIPQYTPSPAEVKYIVEKEGSFTINRLETSRVHWNA-----SNN 295
Query: 301 FGDDGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRTKFINITILLAK 360
+ GYNV++CMRA+AEPLLVSHF + ++ +F++Y++ ++D SK T+FIN+ I L K
Sbjct: 296 EKNGGYNVSRCMRAVAEPLLVSHFDKELMDLVFHKYEEIVSDCMSKENTEFINVIISLTK 355
>dbj|BAB39396.1| S-adenosyl-L-methionine:salicylic acid carboxyl methyltransferase
[Atropa belladonna]
Length = 357
Score = 429 bits (1102), Expect = e-119
Identities = 207/361 (57%), Positives = 285/361 (78%), Gaps = 5/361 (1%)
Query: 1 MEVAKVLHMNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLSKSLTIADLGCS 60
M+V +VLHMNGG G+ SYANNSLVQRKVI +TKP+ ++AI+ +Y + ++L IADLGCS
Sbjct: 1 MKVVEVLHMNGGNGDISYANNSLVQRKVILMTKPITEQAISDLYCSFFPETLCIADLGCS 60
Query: 61 SGSNTLLVILDIIKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEM 120
SG+NT LV+ +++K+VEK + N +S + + NDLPGNDFNTIF SL F++ L ++
Sbjct: 61 SGANTFLVVSELVKIVEKERKIHNLQSAGNLFHFNDLPGNDFNTIFQSLGKFQQDLRKQI 120
Query: 121 GTEMGPCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTSPL 180
G E GPCFFSGVPGSF+ R+FP +SLHFVHSSYSL WLS+VP+ + NK NIYI+STSP
Sbjct: 121 GEEFGPCFFSGVPGSFYTRLFPSESLHFVHSSYSLMWLSQVPDLIEKNKENIYIASTSPP 180
Query: 181 NVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCYIWELLAMALN 240
+V+KAYYKQ++ DFS FLK R+EE+++GG M++TF+GR+S++P+SKECCYIWELL+MALN
Sbjct: 181 SVIKAYYKQYEKDFSNFLKYRSEELMKGGKMVLTFLGRESEDPSSKECCYIWELLSMALN 240
Query: 241 DMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWNARDDFESES 300
++VLEG+I+EEK+++FNIP Y PSP EVK V EGSF++N+LE + V+WN S
Sbjct: 241 ELVLEGLIEEEKVDSFNIPQYTPSPEEVKYIVEKEGSFIINRLEATRVHWNV-----SNE 295
Query: 301 FGDDGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRTKFINITILLAK 360
+ GYNVA+CMRA+AEPLLVS F + ++ +F +Y++ ++D SK +T+FIN+ + L K
Sbjct: 296 GINGGYNVAKCMRAVAEPLLVSQFDQKLMNLVFQKYEEIISDCISKEKTEFINVIVSLTK 355
Query: 361 R 361
+
Sbjct: 356 K 356
>gb|AAN40745.1| S-adenosyl-L-methionine:salicylic acid methyltransferase
[Antirrhinum majus]
Length = 383
Score = 426 bits (1096), Expect = e-118
Identities = 214/371 (57%), Positives = 287/371 (76%), Gaps = 10/371 (2%)
Query: 1 MEVAKVLHMNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLSK--SLTIADLG 58
M++A+VLHMNGG+G++SYA+NSLVQRKVI +TKP+ +EA+T Y L +++IADLG
Sbjct: 12 MKLAQVLHMNGGLGKSSYASNSLVQRKVISITKPIIEEAMTEFYTRMLPSPHTISIADLG 71
Query: 59 CSSGSNTLLVILDIIKVVEKLCRKLNHKSP-EYMIYLNDLPGNDFNTIFTSL-DIFKEKL 116
CS G NTLLV +++K++ KL +KL+ + P E+ I+LNDLPGNDFN+IF L +F+E+L
Sbjct: 72 CSCGPNTLLVAAELVKIIVKLRQKLDREPPPEFQIHLNDLPGNDFNSIFRYLLPMFREEL 131
Query: 117 LDEMG--TEMGP-CFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIY 173
+E+G E G CF SGVPGSF+GR+FP +SLHFVHSSYSL WLSKVPEG NK NIY
Sbjct: 132 REEIGGGEEAGRRCFVSGVPGSFYGRLFPTKSLHFVHSSYSLMWLSKVPEGVKMNKENIY 191
Query: 174 ISSTSPLNVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCYIWE 233
I+STSP NV+ AYY+QFQ DFS FL CR+EE++ GG M++TF+GRKS + SKECCYIWE
Sbjct: 192 IASTSPQNVINAYYEQFQRDFSSFLICRSEEVIGGGRMVLTFLGRKSASARSKECCYIWE 251
Query: 234 LLAMALNDMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNW-NA 292
LL++AL +VLEG+I++EKL++F+IP Y PSP+EVK EV EGSF +N+LE+SE+ W +
Sbjct: 252 LLSLALKQLVLEGVIEKEKLHSFHIPQYTPSPTEVKAEVEKEGSFTVNRLEVSEITWASC 311
Query: 293 RDDFESESFGDDG--YNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRTK 350
+DF G YNVA+CMR++AEPLL+ HFGE V+ +F +Y++ + DR S+ TK
Sbjct: 312 GNDFHLPELVASGNEYNVAKCMRSVAEPLLIEHFGESVIDRLFEKYREIIFDRMSREETK 371
Query: 351 FINITILLAKR 361
F N+TI + +R
Sbjct: 372 FFNVTISMTRR 382
>emb|CAC33768.1| S-adenosyl-L-methionine:salicylic acid carboxyl methyltransferase
[Stephanotis floribunda]
Length = 366
Score = 419 bits (1078), Expect = e-116
Identities = 203/365 (55%), Positives = 274/365 (74%), Gaps = 4/365 (1%)
Query: 1 MEVAKVLHMNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLSKSLTIADLGCS 60
MEV +VLHMNGG G+ SYA+NSL+Q+KVI LTKP+ +EAIT +Y KS+ IAD+GCS
Sbjct: 1 MEVVEVLHMNGGTGDASYASNSLLQKKVILLTKPITEEAITELYTRLFPKSICIADMGCS 60
Query: 61 SGSNTLLVILDIIKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEM 120
SG NT L + ++IK VEK L H+SPEY I+LNDLP NDFNTIF SL F++ +M
Sbjct: 61 SGPNTFLAVSELIKNVEKKRTSLGHESPEYQIHLNDLPSNDFNTIFRSLPSFQKSFSKQM 120
Query: 121 GTEMGPCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTSPL 180
G+ G CFF+GVPGSF+GR+FP +SLHFVHSSYSL WLS+VP+ + NKGNIY+SSTSPL
Sbjct: 121 GSGFGHCFFTGVPGSFYGRLFPNKSLHFVHSSYSLMWLSRVPDLEEVNKGNIYLSSTSPL 180
Query: 181 NVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCYIWELLAMALN 240
+V++AY KQFQ DF+ FL+CRAEE+V GG M++T +GRK ++ + KE Y ELLA ALN
Sbjct: 181 SVIRAYLKQFQRDFTTFLQCRAEELVPGGVMVLTLMGRKGEDHSGKESGYALELLARALN 240
Query: 241 DMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWNARD----DF 296
++V EG I+EE+L+ FN+P Y PSP+EVK V EGSF + +LE + ++W A D
Sbjct: 241 ELVSEGQIEEEQLDCFNVPQYTPSPAEVKYFVEEEGSFSITRLEATTIHWTAYDHDHVTG 300
Query: 297 ESESFGDDGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRTKFINITI 356
+F D GY+++ C+RA+ EPLLV HFGE ++ E+F+RY++ LT+ +K + +FIN+T+
Sbjct: 301 HHHAFKDGGYSLSNCVRAVVEPLLVRHFGEAIMDEVFHRYREILTNCMTKEKIEFINVTV 360
Query: 357 LLAKR 361
+ +R
Sbjct: 361 SMKRR 365
>gb|AAF00108.1| S-adenosyl-L-methionine:salicylic acid carboxyl methyltransferase
[Clarkia breweri] gi|34809619|pdb|1M6E|X Chain X,
Crystal Structure Of Salicylic Acid Carboxyl
Methyltransferase (Samt)
Length = 359
Score = 416 bits (1068), Expect = e-115
Identities = 201/366 (54%), Positives = 281/366 (75%), Gaps = 12/366 (3%)
Query: 1 MEVAKVLHMNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSMYN-NTLSKSLTIADLGC 59
M+V +VLHM GG GE SYA NS +QR+VI +TKP+ + AIT++Y+ +T++ L IADLGC
Sbjct: 1 MDVRQVLHMKGGAGENSYAMNSFIQRQVISITKPITEAAITALYSGDTVTTRLAIADLGC 60
Query: 60 SSGSNTLLVILDIIKVVEKLCRKLNHK-SPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLD 118
SSG N L + ++IK VE+L +K+ + SPEY I+LNDLPGNDFN IF SL I E +D
Sbjct: 61 SSGPNALFAVTELIKTVEELRKKMGRENSPEYQIFLNDLPGNDFNAIFRSLPI--ENDVD 118
Query: 119 EMGTEMGPCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTS 178
G CF +GVPGSF+GR+FP +LHF+HSSYSL WLS+VP G ++NKGNIY+++T
Sbjct: 119 ------GVCFINGVPGSFYGRLFPRNTLHFIHSSYSLMWLSQVPIGIESNKGNIYMANTC 172
Query: 179 PLNVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCYIWELLAMA 238
P +V+ AYYKQFQ D +LFL+CRA+E+V GG M++T +GR+S++ S ECC IW+LLAMA
Sbjct: 173 PQSVLNAYYKQFQEDHALFLRCRAQEVVPGGRMVLTILGRRSEDRASTECCLIWQLLAMA 232
Query: 239 LNDMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWNA--RDDF 296
LN MV EG+I+EEK++ FNIP Y PSP+EV+ E++ EGSF+++ +E SE+ W++ +D
Sbjct: 233 LNQMVSEGLIEEEKMDKFNIPQYTPSPTEVEAEILKEGSFLIDHIEASEIYWSSCTKDGD 292
Query: 297 ESESFGDDGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRTKFINITI 356
S ++GYNVA+CMRA+AEPLL+ HFGE +++++F+RYK + +R SK +TKFIN+ +
Sbjct: 293 GGGSVEEEGYNVARCMRAVAEPLLLDHFGEAIIEDVFHRYKLLIIERMSKEKTKFINVIV 352
Query: 357 LLAKRS 362
L ++S
Sbjct: 353 SLIRKS 358
>gb|AAW66834.1| SAMT [Petunia nyctaginiflora]
Length = 332
Score = 407 bits (1046), Expect = e-112
Identities = 193/337 (57%), Positives = 265/337 (78%), Gaps = 5/337 (1%)
Query: 13 VGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLSKSLTIADLGCSSGSNTLLVILDI 72
+G+ SYANNSLVQ KVI +TKP+ ++A+ +Y++ ++L IADLGCSSG+NT LV+ ++
Sbjct: 1 IGDMSYANNSLVQAKVILMTKPIIEQAMKDLYSSLFPETLCIADLGCSSGANTFLVVSEL 60
Query: 73 IKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEMGTEMGPCFFSGV 132
+K++EK + KSPE+ + NDLPGNDFNTIF SL F+E L ++G GPCFFSGV
Sbjct: 61 VKIIEKERKNHGFKSPEFYFHFNDLPGNDFNTIFQSLGPFQEDLTKQIGESFGPCFFSGV 120
Query: 133 PGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTSPLNVVKAYYKQFQI 192
PGSF+ R+FP SL+F+HSSYSL WLS+VP ++NKGNIY++ TSP +V+KAYYKQ++I
Sbjct: 121 PGSFYTRLFPSNSLNFIHSSYSLMWLSQVPVAVESNKGNIYMARTSPPSVIKAYYKQYEI 180
Query: 193 DFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCYIWELLAMALNDMVLEGIIKEEK 252
DFS FLK R+EE+++GG M++T +GR+S++PTSKECCYIWELLAMALN++V EG+I+EEK
Sbjct: 181 DFSNFLKYRSEELMKGGRMVLTLLGRESEDPTSKECCYIWELLAMALNELVEEGLIEEEK 240
Query: 253 LNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWNARDDFESESFGDDGYNVAQCM 312
L+ FNIP Y PSP EVK V EGSF +N+LE S V+WNA S + + GYNV++CM
Sbjct: 241 LDAFNIPQYTPSPEEVKYVVEKEGSFTINRLETSRVHWNA-----SNNVKNGGYNVSRCM 295
Query: 313 RALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRT 349
RA+AEPLLVSHF + ++ +F++Y++ ++D SK +T
Sbjct: 296 RAVAEPLLVSHFDKELMDLVFHKYEEIVSDCMSKEKT 332
>gb|AAW66842.1| SAMT [Streptosolen jamesonii]
Length = 331
Score = 405 bits (1041), Expect = e-111
Identities = 194/337 (57%), Positives = 262/337 (77%), Gaps = 6/337 (1%)
Query: 13 VGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLSKSLTIADLGCSSGSNTLLVILDI 72
+G+ SYANNS VQ+ VI +TKP+ ++AI +YN + L +ADLGCSSG+NT LV+ ++
Sbjct: 1 IGDVSYANNSSVQQTVILMTKPITEQAIADLYNTLFPEILQVADLGCSSGANTFLVVSEL 60
Query: 73 IKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEMGTEMGPCFFSGV 132
+KVVEK +K +SPE+ +LNDL GNDFNTIF SL F+E L E+G +GPCFFSGV
Sbjct: 61 VKVVEKERKKHGFESPEFHFHLNDLSGNDFNTIFRSLGAFQEDLSKEIGEGLGPCFFSGV 120
Query: 133 PGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTSPLNVVKAYYKQFQI 192
PGSF+ R+FP +SLHFVHSSYSL WLS+VPE + NKGNIY++S+SP +V+KAYYKQ++
Sbjct: 121 PGSFYTRLFPSKSLHFVHSSYSLMWLSQVPEVIETNKGNIYMASSSPASVIKAYYKQYEK 180
Query: 193 DFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCYIWELLAMALNDMVLEGIIKEEK 252
DFS FLK R EE+++GG M++TF+GR+S++P SKECCYIWELLAMALN++V E +I+EEK
Sbjct: 181 DFSSFLKYRREELMKGGKMVLTFLGRESEDPCSKECCYIWELLAMALNELVAERLIEEEK 240
Query: 253 LNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWNARDDFESESFGDDGYNVAQCM 312
+++FNIP Y PSP+EV+ V EGSF +N+LE S V+W+ ++ + GYNV++CM
Sbjct: 241 MDSFNIPQYTPSPAEVRCIVEKEGSFTVNRLESSRVHWDVSNN------SNGGYNVSRCM 294
Query: 313 RALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRT 349
RA+AEPLLVSHFGE + +F +Y+ +TDR S +T
Sbjct: 295 RAVAEPLLVSHFGEKHMDLVFQKYQDIITDRMSNEKT 331
>gb|AAW66837.1| SAMT [Brunfelsia americana]
Length = 344
Score = 405 bits (1041), Expect = e-111
Identities = 192/346 (55%), Positives = 269/346 (77%), Gaps = 3/346 (0%)
Query: 13 VGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLSKSLTIADLGCSSGSNTLLVILDI 72
+GE SYANNSLVQ+KVI +TK + + AI +Y + ++L +ADLGCSSG+NT LV+ ++
Sbjct: 1 IGEISYANNSLVQQKVILMTKQITEXAIKDLYXSLFPETLCMADLGCSSGANTFLVVSEL 60
Query: 73 IKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEMGTEMGPCFFSGV 132
K VEK + KSPE+ + NDLPGNDFN+IF SL F+E L ++G E GPCFFSGV
Sbjct: 61 XKXVEKERKXHGFKSPEFHFHFNDLPGNDFNSIFQSLGXFQEDLRKQIGGEFGPCFFSGV 120
Query: 133 PGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTSPLNVVKAYYKQFQI 192
PGSF+ R++P SLHFVHSSYSL WLS+VP+ +NNKGNIY++ TSP +V+KAYY+Q++
Sbjct: 121 PGSFYTRLYPTNSLHFVHSSYSLMWLSQVPDATENNKGNIYLARTSPPSVIKAYYEQYER 180
Query: 193 DFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCYIWELLAMALNDMVLEGIIKEEK 252
DFS FLK R+EE+++GG M++TF+GR+S++PT++ECCYIWELLA+ALN++V EG+I+E K
Sbjct: 181 DFSNFLKYRSEELMKGGRMVLTFLGRESEDPTNRECCYIWELLAVALNELVEEGLIEEXK 240
Query: 253 LNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWNARDDFESESFGDDGYNVAQCM 312
+++FNIP Y PSP+EVK V EGSF +N LE S V+W+A B+ + GYNVA+CM
Sbjct: 241 VDSFNIPQYTPSPAEVKYXVEKEGSFTINHLESSRVHWSACBE---NCVNNGGYNVARCM 297
Query: 313 RALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRTKFINITILL 358
RA+AEPLLVS F + ++ +F +Y++ +++ SK +T+FIN+T+ L
Sbjct: 298 RAVAEPLLVSQFDKKLMDLVFQKYEEIISNCMSKEKTEFINVTVSL 343
>gb|AAW66830.1| SAMT [Cestrum nocturnum]
Length = 340
Score = 402 bits (1034), Expect = e-111
Identities = 194/342 (56%), Positives = 266/342 (77%), Gaps = 3/342 (0%)
Query: 9 MNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTL-SKSLTIADLGCSSGSNTLL 67
MNGG+G+ SYANNSLVQ+KVI +TKP+ ++AIT +YN+ + ++L IADLGCSSG+NT L
Sbjct: 1 MNGGIGDISYANNSLVQQKVILMTKPITEQAITDLYNSLIFPQTLHIADLGCSSGANTFL 60
Query: 68 VILDIIKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEMGTEMGPC 127
VI + +K++EK + +SPE+ NDLPGNDFNTIF SL F+E L ++G +GPC
Sbjct: 61 VISEFVKIIEKQXKIHGFESPEFNFNFNDLPGNDFNTIFRSLGAFEEDLRMQVGENLGPC 120
Query: 128 FFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTSPLNVVKAYY 187
FF GVPGSF+ R+FP +SLHFVHSSYSL WLS+VPE + NK NIY++STSP +V+KAYY
Sbjct: 121 FFKGVPGSFYXRLFPSKSLHFVHSSYSLMWLSQVPEMTETNKXNIYMASTSPPSVIKAYY 180
Query: 188 KQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCYIWELLAMALNDMVLEGI 247
KQ++ DF+ FLK R+EE+++GG M++TF+GR+S++ SKECCYIWELLA ALN++V EG+
Sbjct: 181 KQYESDFTSFLKYRSEELMKGGKMVLTFLGRESEDACSKECCYIWELLAKALNELVQEGL 240
Query: 248 IKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWNARDDFESESFGDDGYN 307
+E KL++FNIP Y PSP+EVK V GSF +N+LE S V+WN ++ + G GYN
Sbjct: 241 XEEXKLDSFNIPQYTPSPAEVKYIVGKHGSFAVNRLESSRVHWNVTNNNNNSING--GYN 298
Query: 308 VAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRT 349
V++CMRA+AEPLLVSHFGE ++ +F +Y+ +++ +K +T
Sbjct: 299 VSRCMRAVAEPLLVSHFGEDLMDLVFQKYEGIISECMAKEKT 340
>gb|AAW66840.1| SAMT [Capsicum annuum]
Length = 323
Score = 400 bits (1028), Expect = e-110
Identities = 192/325 (59%), Positives = 259/325 (79%), Gaps = 5/325 (1%)
Query: 14 GETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLSKSLTIADLGCSSGSNTLLVILDII 73
G+ SYANNSLVQRKVI +TKP+ +EAI+ +Y + L ++L IADLGCSSG+NT LV+ +++
Sbjct: 2 GDVSYANNSLVQRKVILMTKPITEEAISDLYCSLLPETLCIADLGCSSGANTFLVVSELV 61
Query: 74 KVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEMGTEMGPCFFSGVP 133
KVVEK +K +SPE+ NDLPGNDFN IF SL F++ L ++ G E+GPCFFSGVP
Sbjct: 62 KVVEKERKKHKLQSPEFYFRFNDLPGNDFNVIFQSLGEFEQDLRNQTGEELGPCFFSGVP 121
Query: 134 GSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTSPLNVVKAYYKQFQID 193
GSF+ R+FP +SLHFVHSSYSL WLS+VP + NKGNIY+SSTSP +V+KAYYKQ+ D
Sbjct: 122 GSFYTRLFPSKSLHFVHSSYSLMWLSQVPNLIEKNKGNIYMSSTSPPSVIKAYYKQYGKD 181
Query: 194 FSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCYIWELLAMALNDMVLEGIIKEEKL 253
F+ FLK R+EE+++GG M++TF+GR++++P+SKECCYIWELL+MALN++V+EG+I+EEKL
Sbjct: 182 FTNFLKYRSEELMKGGKMVLTFLGRENEDPSSKECCYIWELLSMALNELVVEGLIEEEKL 241
Query: 254 NTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWNARDDFESESFGDDGYNVAQCMR 313
+ FNIP Y PSP+EVK V E SF +N+LE + ++WNA +D + GYNV++CMR
Sbjct: 242 DAFNIPQYTPSPAEVKYIVEKENSFTINRLEATRIHWNASNDHI-----NGGYNVSRCMR 296
Query: 314 ALAEPLLVSHFGEGVVKEIFNRYKK 338
A+AEPLLVS FG ++ +F +Y+K
Sbjct: 297 AVAEPLLVSQFGPKLMDLVFQKYEK 321
>gb|AAW66831.1| SAMT [Brugmansia sp. Robadey 027]
Length = 335
Score = 398 bits (1023), Expect = e-109
Identities = 192/337 (56%), Positives = 266/337 (77%), Gaps = 4/337 (1%)
Query: 14 GETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLSKSLTIADLGCSSGSNTLLVILDII 73
G+ SYANNSLVQRKVI +TKP+ ++AI+ +Y++ ++L +ADLGCSSG+NT LV+ +++
Sbjct: 2 GDISYANNSLVQRKVILMTKPITEQAISDLYSSLFPETLYVADLGCSSGANTFLVVTELV 61
Query: 74 KVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEMGTEMGPCFFSGVP 133
K++EK +K N +SPE+ NDLPGNDFNTIF SL F++ L ++G G CFFSGV
Sbjct: 62 KIIEKERKKHNLQSPEFHFNFNDLPGNDFNTIFQSLGKFQQDLRKQIGEGFGSCFFSGVA 121
Query: 134 GSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTSPLNVVKAYYKQFQID 193
GSF+ R+FP +SLHFVHSSYSL WLS+VP ++ NKGNIYI+STSP +V+KAYYKQ++ D
Sbjct: 122 GSFYNRLFPSKSLHFVHSSYSLMWLSQVPNLSEKNKGNIYIASTSPPSVIKAYYKQYEKD 181
Query: 194 FSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCYIWELLAMALNDMVLEGIIKEEKL 253
FS FLK R+EE+++GG M++TF+GR+S++P+SKECC IWELL++ALN++V+EG+I+EEK+
Sbjct: 182 FSNFLKYRSEELMKGGKMVLTFLGRESEDPSSKECCCIWELLSIALNELVVEGLIEEEKV 241
Query: 254 NTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNW-NARDDFESESFGDDGYNVAQCM 312
N+FNIP Y PSP EVK V EGSF++NQLE + V+W NA +D E+ + GYNV++CM
Sbjct: 242 NSFNIPQYTPSPGEVKYIVEKEGSFIINQLETTRVHWNNASNDHENI---NGGYNVSKCM 298
Query: 313 RALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRT 349
RA+AEPLLVS FG + +F +Y+ ++D K +T
Sbjct: 299 RAVAEPLLVSQFGPKFMDLVFQKYEDNISDCMGKEKT 335
>gb|AAW66846.1| SAMT [Physalis virginiana]
Length = 333
Score = 396 bits (1017), Expect = e-109
Identities = 189/336 (56%), Positives = 263/336 (78%), Gaps = 4/336 (1%)
Query: 14 GETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLSKSLTIADLGCSSGSNTLLVILDII 73
G+ SYANNSLVQRKVI +TKP+ ++AI+ +Y + ++L IADLGCSSG+NT LV+ +++
Sbjct: 2 GDISYANNSLVQRKVILMTKPITEQAISDLYCSLFPETLCIADLGCSSGTNTXLVVSEVV 61
Query: 74 KVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEMGTEMGPCFFSGVP 133
KVVE +K N KSPE+ NDLPGNDFN IF SL F++ L +++G +GPCFFSGVP
Sbjct: 62 KVVENERKKHNLKSPEFYFRFNDLPGNDFNAIFRSLGEFEQXLRNQIGEGLGPCFFSGVP 121
Query: 134 GSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTSPLNVVKAYYKQFQID 193
GSF+ R+FP SLHFVHSSYSL WLS+VP + NKGNIY++STSP +V+KAYYKQ++ D
Sbjct: 122 GSFYTRLFPSNSLHFVHSSYSLMWLSQVPNLIEKNKGNIYMASTSPASVIKAYYKQYEND 181
Query: 194 FSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCYIWELLAMALNDMVLEGIIKEEKL 253
F+ FLK R+EE+++GG M++TF+GR+S++P+SKECCYIWELL+MALN++V+EG+I++EKL
Sbjct: 182 FTXFLKYRSEELMKGGKMVLTFLGRESEDPSSKECCYIWELLSMALNELVIEGLIEKEKL 241
Query: 254 NTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWNARDDFESESFGDDGYNVAQCMR 313
++FNIP Y PSP+EV V + SF +N LE + V+WN S + + GYNV++CMR
Sbjct: 242 DSFNIPQYTPSPAEVXYIVEKQDSFTINXLETTRVHWNN----ASNNDNNGGYNVSKCMR 297
Query: 314 ALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRT 349
A+AEPLLVS FG ++ +F +Y++ ++D +K +T
Sbjct: 298 AVAEPLLVSQFGPKLMDLVFQKYEEIVSDCMAKEKT 333
>gb|AAW66843.1| SAMT [Juanulloa aurantiaca]
Length = 334
Score = 394 bits (1013), Expect = e-108
Identities = 190/337 (56%), Positives = 262/337 (77%), Gaps = 3/337 (0%)
Query: 13 VGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLSKSLTIADLGCSSGSNTLLVILDI 72
+G+ SYANNSLVQRKVI +TKP+ ++AI+ +Y ++L IADLGCSSG+NT LV+ ++
Sbjct: 1 IGDISYANNSLVQRKVILMTKPITEQAISDLYCTLFPETLCIADLGCSSGANTFLVVSEL 60
Query: 73 IKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEMGTEMGPCFFSGV 132
IK++EK +K N +SPE+ NDLPGNDFNTIF SL F++ L ++G G CFFSGV
Sbjct: 61 IKIIEKERKKHNLQSPEFHFNFNDLPGNDFNTIFQSLGGFEQDLRKQIGEGFGSCFFSGV 120
Query: 133 PGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTSPLNVVKAYYKQFQI 192
GSF+ R+FP SLHFVHSSYSL WLS+VP+ + NKGNIY++STSP +V+KAYY+Q++
Sbjct: 121 AGSFYTRLFPSNSLHFVHSSYSLMWLSQVPDLIEKNKGNIYMASTSPASVIKAYYEQYEK 180
Query: 193 DFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCYIWELLAMALNDMVLEGIIKEEK 252
DFS FLK R+EE+++GG M++TF+GR+S++P+SKECCYIWELL+MALN++V+EG+I+E K
Sbjct: 181 DFSNFLKYRSEELMKGGKMVLTFLGRESEDPSSKECCYIWELLSMALNELVVEGLIEEGK 240
Query: 253 LNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWNARDDFESESFGDDGYNVAQCM 312
++ FNIP Y PSP+EVK V EGSF +N+LE + V+WN + E+ + GYNV++CM
Sbjct: 241 VDAFNIPQYTPSPTEVKYVVEKEGSFTINRLEATRVHWNNASNDENI---NGGYNVSRCM 297
Query: 313 RALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRT 349
RA+AEPLL S FG ++ +F +YK+ + D SK +T
Sbjct: 298 RAVAEPLLASQFGPKLMDLVFQKYKEIIFDCMSKEKT 334
>gb|AAW66832.1| SAMT [Hyoscyamus albus]
Length = 332
Score = 393 bits (1009), Expect = e-108
Identities = 189/336 (56%), Positives = 263/336 (78%), Gaps = 5/336 (1%)
Query: 14 GETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLSKSLTIADLGCSSGSNTLLVILDII 73
G+ SYANNSLVQRKVI +TK + ++AI+ +Y + ++L IADLGCS G+NT LV+ +++
Sbjct: 2 GDFSYANNSLVQRKVILMTKSITEQAISDLYCSFFPETLCIADLGCSLGANTFLVVSELV 61
Query: 74 KVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEMGTEMGPCFFSGVP 133
K+VEK + N +SPE + + NDLPGNDFNTIF SL F++ L ++G E GPCFF GVP
Sbjct: 62 KIVEKERKLHNIQSPEIVFHFNDLPGNDFNTIFQSLGKFQQDLRKQIGEEFGPCFFXGVP 121
Query: 134 GSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTSPLNVVKAYYKQFQID 193
GSF+ R+FP +SLHFVHSSYSL WLS+VPE +NNKGNIY+++TSP +V+KAYYKQ++ D
Sbjct: 122 GSFYTRLFPSKSLHFVHSSYSLMWLSQVPELIENNKGNIYMANTSPASVIKAYYKQYEKD 181
Query: 194 FSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCYIWELLAMALNDMVLEGIIKEEKL 253
FS FLK R+EE+++GG M++TF+GR+S++P SKECCYIWELL+MALN++V EG+I+EEK+
Sbjct: 182 FSNFLKYRSEELMKGGKMVLTFLGRESEDPCSKECCYIWELLSMALNELVFEGLIEEEKV 241
Query: 254 NTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWNARDDFESESFGDDGYNVAQCMR 313
++FNIP Y PSP+EVK V EGSF +N+LE + V+WNA S + GY+V++CMR
Sbjct: 242 DSFNIPQYTPSPAEVKNIVEKEGSFTINRLEATRVHWNA-----SNEGINGGYDVSRCMR 296
Query: 314 ALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRT 349
A+AEPLL++ F ++ +F +Y+ ++D SK +T
Sbjct: 297 AVAEPLLLTQFDHKLMDLVFQKYQHIISDCMSKEKT 332
>dbj|BAB84353.1| S-adenosyl-L-methionine:salicylic acid calboxyl
methyltransferase-like protein [Cucumis sativus]
Length = 370
Score = 392 bits (1006), Expect = e-107
Identities = 195/368 (52%), Positives = 269/368 (72%), Gaps = 12/368 (3%)
Query: 6 VLHMNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLSKSLTIADLGCSSGSNT 65
+LH+NGG+G TSYANNS +QR++I +T + EA+T+ YN + S+TIADLGCSSG NT
Sbjct: 1 MLHVNGGMGNTSYANNSRLQREIISMTCSIAKEALTNFYNQHIPTSITIADLGCSSGQNT 60
Query: 66 LLVILDIIKVVEKLCRKLNHKSP-EYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEMGTEM 124
L+++ +IK VE++ +KL+ + P EY I+LNDL GNDFN +FTSL F E L ++G +
Sbjct: 61 LMLVSYLIKQVEEIRQKLHQRLPLEYQIFLNDLHGNDFNAVFTSLPRFLEDLGTQIGGDF 120
Query: 125 GPCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTSPLNVVK 184
GPCFF+GVPGSF+ R+FP +S+HF HSS SLHWLS+VP G +NNKGNIYI STSP +V +
Sbjct: 121 GPCFFNGVPGSFYARLFPTKSVHFFHSSSSLHWLSRVPVGIENNKGNIYIGSTSPKSVGE 180
Query: 185 AYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCYIWELLAMALNDMVL 244
AYYKQFQ DFS+FLKCRAEE+V GG M++T VGR S++P+ YIWELL +ALN MV
Sbjct: 181 AYYKQFQKDFSMFLKCRAEELVMGGGMVLTLVGRTSEDPSKSGGYYIWELLGLALNTMVA 240
Query: 245 EGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWN---------ARDD 295
EGI++E+K ++FNIP Y PSP EV+ EV+ EGSF++NQL+ S +N N +
Sbjct: 241 EGIVEEKKADSFNIPYYIPSPKEVEAEVVKEGSFILNQLKASSINLNHTVHKTEEESSTP 300
Query: 296 FESESFGD-DGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGR-TKFIN 353
+ S D Y+ A+C+++++EPLL+ HFGE ++ E+F R++ + +K R + IN
Sbjct: 301 LINNSLADATDYDFAKCIQSVSEPLLIRHFGEAIMDELFIRHRNIVAGCMAKHRIMECIN 360
Query: 354 ITILLAKR 361
+TI L K+
Sbjct: 361 LTISLTKK 368
>gb|AAW66828.1| SAMT [Cestrum elegans]
Length = 332
Score = 391 bits (1004), Expect = e-107
Identities = 188/331 (56%), Positives = 258/331 (77%), Gaps = 3/331 (0%)
Query: 13 VGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTL-SKSLTIADLGCSSGSNTLLVILD 71
+G+ SYANNSLVQ+KVI +TKP+ ++AIT +YN+ + ++L IADLGCSSG+NT LVI +
Sbjct: 1 IGDISYANNSLVQQKVILMTKPITEQAITDLYNSLIFPQTLHIADLGCSSGANTFLVISE 60
Query: 72 IIKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEMGTEMGPCFFSG 131
+K++EK + +SPE+ Y NDLPGNDFNTIF SL F+E L ++G +GPCFF G
Sbjct: 61 FVKIIEKQRKIHGFESPEFNFYFNDLPGNDFNTIFRSLGAFEEDLRMQVGENLGPCFFKG 120
Query: 132 VPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTSPLNVVKAYYKQFQ 191
VPGSF+ R+FP +SLHFVHSSYSL WLS+VPE + NK NIY++STSP +V+KAYYKQ++
Sbjct: 121 VPGSFYTRLFPSKSLHFVHSSYSLMWLSQVPEMTETNKRNIYMASTSPPSVIKAYYKQYE 180
Query: 192 IDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCYIWELLAMALNDMVLEGIIKEE 251
DF+ FLK R+EE+++GG M++TF+GR+S++ SKECCYIWELLA AL ++V EG+ +E
Sbjct: 181 SDFTSFLKYRSEELMKGGKMVLTFLGRESEDACSKECCYIWELLAKALXELVQEGLXEEX 240
Query: 252 KLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWNARDDFESESFGDDGYNVAQC 311
KL++FNIP Y PSP+EVK V GSF +N+LE S V+WN ++ + G GYNV++C
Sbjct: 241 KLDSFNIPQYTPSPAEVKYIVGKHGSFAVNRLESSRVHWNVTNNNNNSING--GYNVSRC 298
Query: 312 MRALAEPLLVSHFGEGVVKEIFNRYKKYLTD 342
MRA+AEPLLVSHFGE ++ +F +Y+ +++
Sbjct: 299 MRAVAEPLLVSHFGEDLMDLVFQKYEGIISE 329
>gb|AAW66847.1| SAMT [Solandra maxima]
Length = 333
Score = 390 bits (1002), Expect = e-107
Identities = 188/336 (55%), Positives = 263/336 (77%), Gaps = 4/336 (1%)
Query: 14 GETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLSKSLTIADLGCSSGSNTLLVILDII 73
G+ SYANNSLVQRKVI +TKP+ ++AI+ +Y + ++L IADLGCSSG+NT LV+ +++
Sbjct: 2 GDISYANNSLVQRKVILMTKPIIEQAISDLYCSLFPETLCIADLGCSSGANTFLVVSELV 61
Query: 74 KVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEMGTEMGPCFFSGVP 133
K+VEK +K N +SPE+ NDLPGNDFNTIF SL F++ L ++G GP FFSGV
Sbjct: 62 KMVEKERKKHNLQSPEFHFNFNDLPGNDFNTIFQSLGEFEQDLRKQIGEGFGPYFFSGVA 121
Query: 134 GSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTSPLNVVKAYYKQFQID 193
GSF+ R+FP +SLHFVHS YSL WLS+VP+ + NKGNIY++ TSP +V+ AYYKQ++ D
Sbjct: 122 GSFYTRLFPSKSLHFVHSXYSLMWLSQVPDLTEKNKGNIYMAXTSPPSVINAYYKQYEKD 181
Query: 194 FSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCYIWELLAMALNDMVLEGIIKEEKL 253
FS FLK R+EE+++GG M++TF+GR+S++P+SKECCYIWELL+MALN++V+EG+I+EEK+
Sbjct: 182 FSNFLKYRSEELMKGGKMVLTFLGRESEDPSSKECCYIWELLSMALNELVVEGLIEEEKV 241
Query: 254 NTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWNARDDFESESFGDDGYNVAQCMR 313
++FNIP Y PSP+EVK V EGSF +N+LE + V+WN + E+ + YNV++CMR
Sbjct: 242 DSFNIPQYTPSPAEVKYIVEKEGSFTINRLEATRVHWNNASNNEN----NGSYNVSRCMR 297
Query: 314 ALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRT 349
A+AEPLLVS FG ++ +F +Y++ ++D SK +T
Sbjct: 298 AVAEPLLVSQFGPKLMDLVFQKYEEIISDCMSKEKT 333
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.137 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 620,497,920
Number of Sequences: 2540612
Number of extensions: 26566179
Number of successful extensions: 60993
Number of sequences better than 10.0: 173
Number of HSP's better than 10.0 without gapping: 160
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 60217
Number of HSP's gapped (non-prelim): 182
length of query: 362
length of database: 863,360,394
effective HSP length: 129
effective length of query: 233
effective length of database: 535,621,446
effective search space: 124799796918
effective search space used: 124799796918
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 76 (33.9 bits)
Medicago: description of AC125480.15


