

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC125475.9 - phase: 0 /pseudo
(932 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
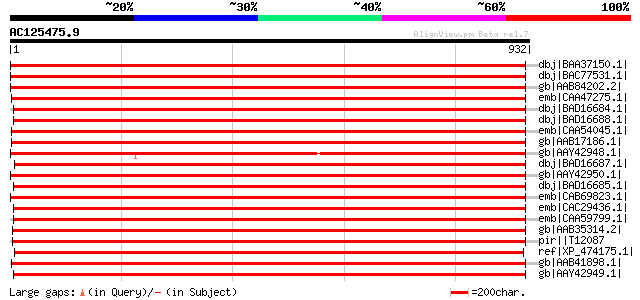
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAA37150.1| p-type H+-ATPase [Vicia faba] 1754 0.0
dbj|BAC77531.1| plasma membrane H+-ATPase [Sesbania rostrata] 1694 0.0
gb|AAB84202.2| plasma membrane proton ATPase [Kosteletzkya virgi... 1666 0.0
emb|CAA47275.1| plasma membrane H+-ATPase [Nicotiana plumbaginif... 1659 0.0
dbj|BAD16684.1| plasma membrane H+-ATPase [Daucus carota] 1657 0.0
dbj|BAD16688.1| plasma membrane H+-ATPase [Daucus carota] 1654 0.0
emb|CAA54045.1| H(+)-transporting ATPase [Solanum tuberosum] gi|... 1652 0.0
gb|AAB17186.1| plasma membrane H+-ATPase [Lycopersicon esculentum] 1651 0.0
gb|AAY42948.1| plasma membrane H+ ATPase [Lupinus albus] 1643 0.0
dbj|BAD16687.1| plasma membrane H+-ATPase [Daucus carota] 1642 0.0
gb|AAY42950.1| plasma membrane H+ ATPase [Lupinus albus] 1640 0.0
dbj|BAD16685.1| plasma membrane H+-ATPase [Daucus carota] 1639 0.0
emb|CAB69823.1| plasma membrane H+ ATPase [Prunus persica] 1634 0.0
emb|CAC29436.1| P-type H+-ATPase [Vicia faba] 1616 0.0
emb|CAA59799.1| H(+)-transporting ATPase [Phaseolus vulgaris] gi... 1611 0.0
gb|AAB35314.2| plasma membrane H(+)-ATPase precursor [Vicia faba] 1608 0.0
pir||T12087 H+-exporting ATPase (EC 3.6.3.6), plasma membrane - ... 1608 0.0
ref|XP_474175.1| OSJNBa0071I13.11 [Oryza sativa (japonica cultiv... 1604 0.0
gb|AAB41898.1| H+-transporting ATPase [Mesembryanthemum crystall... 1604 0.0
gb|AAY42949.1| plasma membrane H+ ATPase [Lupinus albus] 1598 0.0
>dbj|BAA37150.1| p-type H+-ATPase [Vicia faba]
Length = 952
Score = 1754 bits (4544), Expect = 0.0
Identities = 886/926 (95%), Positives = 906/926 (97%)
Query: 1 MAESKSISLEQIKNETVDLERIPVEEVFEQLKCTKEGLSSEEGANRLQIFGPNKLEEKKD 60
MA +KSI+LE+IKNETVDLE IPVEEVFEQLKCTKEGLS EEGANRLQIFGPNKLEEKK+
Sbjct: 1 MATNKSITLEEIKNETVDLEHIPVEEVFEQLKCTKEGLSLEEGANRLQIFGPNKLEEKKE 60
Query: 61 SKILKFLGFMWNPLSWVMEAAALMAIGLANGNGKPPDWQDFVGIICLLVINSTISFIEEN 120
SK+LKFLGFMWNPLSWVMEAAALMAIGLANG+GKPPDWQDFVGI+CLLVINSTISFIEEN
Sbjct: 61 SKLLKFLGFMWNPLSWVMEAAALMAIGLANGDGKPPDWQDFVGIVCLLVINSTISFIEEN 120
Query: 121 NAGNAAAALMAGLAPKTKVLRDGKWSEQEAAILVPGDIISIKLGDIVPADARLLEGDPLK 180
NAGNAAAALMAGLAP+TKVLRDGKWSEQEAAILVPGDIISIKLGDIVPADARLLEGDPLK
Sbjct: 121 NAGNAAAALMAGLAPETKVLRDGKWSEQEAAILVPGDIISIKLGDIVPADARLLEGDPLK 180
Query: 181 IDQSALTGESLPVTRNPGDEVYSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQVGH 240
IDQSALTGESLPVTR+PG EV+SGSTCKQGE+EAVVIATGVHTFFGKAAHLVDSTNQVGH
Sbjct: 181 IDQSALTGESLPVTRSPGQEVFSGSTCKQGEIEAVVIATGVHTFFGKAAHLVDSTNQVGH 240
Query: 241 FQKVLTAIGNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSV 300
FQKVLTAIGNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAM TVLSV
Sbjct: 241 FQKVLTAIGNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMSTVLSV 300
Query: 301 TMAIGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDK 360
T AIGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDK
Sbjct: 301 TTAIGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDK 360
Query: 361 EHVMLLAARASRVENQDAIDAAIVGTLADPKEARAGVREIHFLPFNPVDKRTALTYIDGN 420
EHVMLLAARASR+ENQDAIDAAIVGTLADPKEARAGVRE+HFLPFNPVDKRTALTYID N
Sbjct: 361 EHVMLLAARASRIENQDAIDAAIVGTLADPKEARAGVREVHFLPFNPVDKRTALTYIDSN 420
Query: 421 GNWHRASKGAPEQIMDLCKLREDTKRNIHAIIDKFAERGLRSLAVARQEVPEKTKESPGA 480
GNWHRASKGAPEQIM+LC LRED KRNIHAIIDKFAERGLRSLAV+RQEVPEKTKES G
Sbjct: 421 GNWHRASKGAPEQIMNLCNLREDAKRNIHAIIDKFAERGLRSLAVSRQEVPEKTKESAGG 480
Query: 481 PWQFVGLLSLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSA 540
PWQFVGLLSLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSA
Sbjct: 481 PWQFVGLLSLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSA 540
Query: 541 TLLGQDKDANIAALPVEELIEKADGFAGVFPEHKYEIVRKLQERKHICGMTGDGVNDAPA 600
TLLGQDKDA+IAALPVEELIEKADGFAGVFPEHKYEIV+KLQERKHICGMTGDGVNDAPA
Sbjct: 541 TLLGQDKDASIAALPVEELIEKADGFAGVFPEHKYEIVKKLQERKHICGMTGDGVNDAPA 600
Query: 601 LKRADIGIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIR 660
LK+ADIGIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIR
Sbjct: 601 LKKADIGIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIR 660
Query: 661 IVFGFMFIALIWKFDFSPFMVLIIAILNDGTIMTISKDRVVPSPLPDSWKLKEIFATGIV 720
IVFGFMFIALIWKFDFSPFMVLIIAILNDGTIMTISKDRVVPSPLPDSWKL EIFATGIV
Sbjct: 661 IVFGFMFIALIWKFDFSPFMVLIIAILNDGTIMTISKDRVVPSPLPDSWKLNEIFATGIV 720
Query: 721 LGGYLALMTVIFFWAMKENDFFPDKFGVRKLNHDEMMSALYLQVSIVSQALIFVTRSRGW 780
LGGYLALMTVIFFWA+KE FFPDKFGVR L HDEMMSALYLQVSIVSQALIFVTRSRGW
Sbjct: 721 LGGYLALMTVIFFWAIKETHFFPDKFGVRHLIHDEMMSALYLQVSIVSQALIFVTRSRGW 780
Query: 781 SFLERPGALLVIAFFIAQLIATIIAVYANWGFAKVQGIGWGWAGVIWLYSIVFYIPLDVM 840
SFLERPGALLVIAF IAQLIAT+IAVYANWGFAKVQGIGWGWAGVIWLYSIVFYIPLDVM
Sbjct: 781 SFLERPGALLVIAFLIAQLIATLIAVYANWGFAKVQGIGWGWAGVIWLYSIVFYIPLDVM 840
Query: 841 KFAIRYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPESSGIFNEK 900
KFAIRYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPESSGIFNEK
Sbjct: 841 KFAIRYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPESSGIFNEK 900
Query: 901 SSYRELSEIAEQAKRRAEVARLIPFH 926
SSYRELSEIAEQAKRRAEVARL H
Sbjct: 901 SSYRELSEIAEQAKRRAEVARLRELH 926
>dbj|BAC77531.1| plasma membrane H+-ATPase [Sesbania rostrata]
Length = 954
Score = 1694 bits (4388), Expect = 0.0
Identities = 848/928 (91%), Positives = 894/928 (95%), Gaps = 2/928 (0%)
Query: 1 MAESKSISLEQIKNETVDLERIPVEEVFEQLKCTKEGLSSEEGANRLQIFGPNKLEEKKD 60
MA SI+LE+IKNETVDLERIPVEEVFEQLKCT+EGLSSEEGANRLQIFGPNKLEEKK+
Sbjct: 1 MAAKGSITLEEIKNETVDLERIPVEEVFEQLKCTREGLSSEEGANRLQIFGPNKLEEKKE 60
Query: 61 SKILKFLGFMWNPLSWVMEAAALMAIGLANGNGKPPDWQDFVGIICLLVINSTISFIEEN 120
SKILKFLGFMWNPLSWVMEAAA+MAI LANG+GKPPDWQDFVGI+CLL+INSTISFIEEN
Sbjct: 61 SKILKFLGFMWNPLSWVMEAAAIMAIALANGDGKPPDWQDFVGIVCLLLINSTISFIEEN 120
Query: 121 NAGNAAAALMAGLAPKTKVLRDGKWSEQEAAILVPGDIISIKLGDIVPADARLLEGDPLK 180
NAGNAAAALMAGL PKTKVLRDG+WSEQEAAILVPGDIISIKLGDI+PADARLLEGDPLK
Sbjct: 121 NAGNAAAALMAGLTPKTKVLRDGQWSEQEAAILVPGDIISIKLGDIIPADARLLEGDPLK 180
Query: 181 IDQSALTGESLPVTRNPGDEVYSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQVGH 240
+DQSALTGESLPV +NPGDEV+SGSTCKQGE+EAVVIATGVHTFFGKAAHLVDSTNQVGH
Sbjct: 181 VDQSALTGESLPVNKNPGDEVFSGSTCKQGEIEAVVIATGVHTFFGKAAHLVDSTNQVGH 240
Query: 241 FQKVLTAIGNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSV 300
FQKVLTAIGNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSV
Sbjct: 241 FQKVLTAIGNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSV 300
Query: 301 TMAIGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDK 360
TMAIGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKL+VDKNLIEVF KGVDK
Sbjct: 301 TMAIGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLTVDKNLIEVFAKGVDK 360
Query: 361 EHVMLLAARASRVENQDAIDAAIVGTLADPKEARAGVREIHFLPFNPVDKRTALTYIDGN 420
EHV+LLAARASR ENQDAIDAA+VGTLADPKEARAG+RE+HF PFNPVDKRTALTYID +
Sbjct: 361 EHVLLLAARASRTENQDAIDAAVVGTLADPKEARAGIREVHFFPFNPVDKRTALTYIDSD 420
Query: 421 GNWHRASKGAPEQIMDLCKLREDTKRNIHAIIDKFAERGLRSLAVARQEVPEKTKESPGA 480
GNWHRASKGAPEQIM LC LR+D K+ IHAIIDKFAERGLRSLAVARQEVPEK+K+S G
Sbjct: 421 GNWHRASKGAPEQIMTLCNLRDDAKKKIHAIIDKFAERGLRSLAVARQEVPEKSKDSAGG 480
Query: 481 PWQFVGLLSLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSA 540
PWQFVGLLSLFDPPRHDSAETIRRAL+LGVNVKMITGDQLAIAKETGRRLGMGTNMYPSA
Sbjct: 481 PWQFVGLLSLFDPPRHDSAETIRRALNLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSA 540
Query: 541 TLLGQDKDANIAALPVEELIEKADGFAGVFPEHKYEIVRKLQERKHICGMTGDGVNDAPA 600
+LLGQDKDA+IAALP+EELIEKADGFAGVFPEHKYEIV+KLQERKHICGMTGDGVNDAPA
Sbjct: 541 SLLGQDKDASIAALPIEELIEKADGFAGVFPEHKYEIVKKLQERKHICGMTGDGVNDAPA 600
Query: 601 LKRADIGIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIR 660
LK+ADIGIAVADATDAAR ASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIR
Sbjct: 601 LKKADIGIAVADATDAARSASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIR 660
Query: 661 IVFGFMFIALIWKFDFSPFMVLIIAILNDGTIMTISKDRVVPSPLPDSWKLKEIFATGIV 720
IVFGFMFIALIWKFDFSPFMVLIIAILNDGTIMTISKDRV PSP+PDSWKLKEIFATG+V
Sbjct: 661 IVFGFMFIALIWKFDFSPFMVLIIAILNDGTIMTISKDRVKPSPMPDSWKLKEIFATGVV 720
Query: 721 LGGYLALMTVIFFWAMKENDFFPDKFGVRKLNH--DEMMSALYLQVSIVSQALIFVTRSR 778
LGGYLALMTVIFFWAMKE FF DKFGVR L+ DEM++ALYLQVSIVSQALIFVTRSR
Sbjct: 721 LGGYLALMTVIFFWAMKETTFFSDKFGVRSLHDSPDEMIAALYLQVSIVSQALIFVTRSR 780
Query: 779 GWSFLERPGALLVIAFFIAQLIATIIAVYANWGFAKVQGIGWGWAGVIWLYSIVFYIPLD 838
WS++ERPG LL+ AF IAQLIAT+IAVYANWGFA+++GIGWGWAGVIWLYSIVFY+PLD
Sbjct: 781 SWSYVERPGLLLMSAFVIAQLIATLIAVYANWGFARIKGIGWGWAGVIWLYSIVFYVPLD 840
Query: 839 VMKFAIRYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPESSGIFN 898
+MKFAIRYILSGKAW NLL+NKTAFTTKKDYGKEEREAQWA AQRTLHGLQPPE+SGIFN
Sbjct: 841 IMKFAIRYILSGKAWLNLLENKTAFTTKKDYGKEEREAQWALAQRTLHGLQPPETSGIFN 900
Query: 899 EKSSYRELSEIAEQAKRRAEVARLIPFH 926
EKSSYRELSEIAEQAKRRAEVARL H
Sbjct: 901 EKSSYRELSEIAEQAKRRAEVARLRELH 928
>gb|AAB84202.2| plasma membrane proton ATPase [Kosteletzkya virginica]
Length = 954
Score = 1666 bits (4315), Expect = 0.0
Identities = 832/928 (89%), Positives = 884/928 (94%), Gaps = 2/928 (0%)
Query: 1 MAESKSISLEQIKNETVDLERIPVEEVFEQLKCTKEGLSSEEGANRLQIFGPNKLEEKKD 60
MA SK ISLE+I+NETVDLE+IP+EEVFEQLKCTKEGLSSEEGANRLQIFGPNKLEEKK+
Sbjct: 1 MASSKGISLEEIRNETVDLEKIPIEEVFEQLKCTKEGLSSEEGANRLQIFGPNKLEEKKE 60
Query: 61 SKILKFLGFMWNPLSWVMEAAALMAIGLANGNGKPPDWQDFVGIICLLVINSTISFIEEN 120
SKILKFLGFMWNPLSWVMEAAA+MAI LANG GKPPDWQDFVGI+CLLVINSTISFIEEN
Sbjct: 61 SKILKFLGFMWNPLSWVMEAAAIMAIALANGEGKPPDWQDFVGIVCLLVINSTISFIEEN 120
Query: 121 NAGNAAAALMAGLAPKTKVLRDGKWSEQEAAILVPGDIISIKLGDIVPADARLLEGDPLK 180
NAGNAAAALMAGLAPKTKVLRDGKWSEQEAAILVPGDIIS+KLGDI+PADARLLEGDPLK
Sbjct: 121 NAGNAAAALMAGLAPKTKVLRDGKWSEQEAAILVPGDIISVKLGDIIPADARLLEGDPLK 180
Query: 181 IDQSALTGESLPVTRNPGDEVYSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQVGH 240
+DQSALTGESLPVT++PG EV+SGSTCKQGE+EAVVIATGVHTFFGKAAHLVDSTNQVGH
Sbjct: 181 VDQSALTGESLPVTKHPGQEVFSGSTCKQGEIEAVVIATGVHTFFGKAAHLVDSTNQVGH 240
Query: 241 FQKVLTAIGNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSV 300
FQKVLTAIGNFCICSIAVGML EIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSV
Sbjct: 241 FQKVLTAIGNFCICSIAVGMLVEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSV 300
Query: 301 TMAIGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDK 360
TMAIGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKL+VDKNLIEVF K DK
Sbjct: 301 TMAIGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLTVDKNLIEVFVKDGDK 360
Query: 361 EHVMLLAARASRVENQDAIDAAIVGTLADPKEARAGVREIHFLPFNPVDKRTALTYIDGN 420
+HV+LLAARASRVENQDAIDAAIVGTLADP+EARA + E+HFLPFNPVDKRTA+TYID N
Sbjct: 361 DHVLLLAARASRVENQDAIDAAIVGTLADPREARASITEVHFLPFNPVDKRTAITYIDSN 420
Query: 421 GNWHRASKGAPEQIMDLCKLREDTKRNIHAIIDKFAERGLRSLAVARQEVPEKTKESPGA 480
GNWHRASKGAPEQI+ LC +ED K+ +H+IIDKFAERGLRSLAV+RQ+VPEK+KES GA
Sbjct: 421 GNWHRASKGAPEQILALCNAKEDFKKKVHSIIDKFAERGLRSLAVSRQQVPEKSKESAGA 480
Query: 481 PWQFVGLLSLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSA 540
PWQFVGLLSLFDPPRHDSAETIR+ LHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSA
Sbjct: 481 PWQFVGLLSLFDPPRHDSAETIRQTLHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSA 540
Query: 541 TLLGQDKDANIAALPVEELIEKADGFAGVFPEHKYEIVRKLQERKHICGMTGDGVNDAPA 600
+LLGQDKDANIAALPVEELIEKADGFAGVFPEHKYEIV+KLQERKHICGMTGDGVNDAPA
Sbjct: 541 SLLGQDKDANIAALPVEELIEKADGFAGVFPEHKYEIVKKLQERKHICGMTGDGVNDAPA 600
Query: 601 LKRADIGIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIR 660
LK+ADIGIAVADATDAAR ASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIR
Sbjct: 601 LKKADIGIAVADATDAARSASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIR 660
Query: 661 IVFGFMFIALIWKFDFSPFMVLIIAILNDGTIMTISKDRVVPSPLPDSWKLKEIFATGIV 720
IVFGF+FIALIWKFDFSPFMVLIIAILNDGTIMTISKDRV PSPLPDSWKLKEIFATGIV
Sbjct: 661 IVFGFLFIALIWKFDFSPFMVLIIAILNDGTIMTISKDRVKPSPLPDSWKLKEIFATGIV 720
Query: 721 LGGYLALMTVIFFWAMKENDFFPDKFGVRKL--NHDEMMSALYLQVSIVSQALIFVTRSR 778
LGGYLALMTVIFFWAM + DFF +KF VR L + +EMM ALYLQVSIVSQALIFVTRSR
Sbjct: 721 LGGYLALMTVIFFWAMHDTDFFSEKFSVRSLRGSENEMMGALYLQVSIVSQALIFVTRSR 780
Query: 779 GWSFLERPGALLVIAFFIAQLIATIIAVYANWGFAKVQGIGWGWAGVIWLYSIVFYIPLD 838
WS+ ERPG LL+ AF IAQL+AT+IAVYANWGFA+++GIGWGWAGVIWLYSIVFY+PLD
Sbjct: 781 SWSYAERPGLLLLSAFIIAQLVATLIAVYANWGFARIKGIGWGWAGVIWLYSIVFYVPLD 840
Query: 839 VMKFAIRYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPESSGIFN 898
+KFAIRYILSGKAW L +NKTAFTTKKDYGKEEREAQWA AQRTLHGLQPPE+S +F+
Sbjct: 841 FIKFAIRYILSGKAWLTLFENKTAFTTKKDYGKEEREAQWALAQRTLHGLQPPETSNLFH 900
Query: 899 EKSSYRELSEIAEQAKRRAEVARLIPFH 926
EK+SYRELSEIAEQAKRRAEVARL H
Sbjct: 901 EKNSYRELSEIAEQAKRRAEVARLRELH 928
>emb|CAA47275.1| plasma membrane H+-ATPase [Nicotiana plumbaginifolia]
gi|416664|sp|Q03194|PMA4_NICPL Plasma membrane ATPase 4
(Proton pump 4)
Length = 952
Score = 1659 bits (4296), Expect = 0.0
Identities = 821/925 (88%), Positives = 886/925 (95%), Gaps = 2/925 (0%)
Query: 4 SKSISLEQIKNETVDLERIPVEEVFEQLKCTKEGLSSEEGANRLQIFGPNKLEEKKDSKI 63
+K+ISLE+IKNETVDLE+IP+EEVFEQLKCT+EGLS++EGA+RLQIFGPNKLEEK +SKI
Sbjct: 2 AKAISLEEIKNETVDLEKIPIEEVFEQLKCTREGLSADEGASRLQIFGPNKLEEKNESKI 61
Query: 64 LKFLGFMWNPLSWVMEAAALMAIGLANGNGKPPDWQDFVGIICLLVINSTISFIEENNAG 123
LKFLGFMWNPLSWVMEAAA+MAI LANG+GKPPDWQDF+GIICLLVINSTISFIEENNAG
Sbjct: 62 LKFLGFMWNPLSWVMEAAAVMAIALANGDGKPPDWQDFIGIICLLVINSTISFIEENNAG 121
Query: 124 NAAAALMAGLAPKTKVLRDGKWSEQEAAILVPGDIISIKLGDIVPADARLLEGDPLKIDQ 183
NAAAALMAGLAPKTKVLRDG+WSEQEAAILVPGDIIS+KLGDI+PADARLLEGDPLKIDQ
Sbjct: 122 NAAAALMAGLAPKTKVLRDGRWSEQEAAILVPGDIISVKLGDIIPADARLLEGDPLKIDQ 181
Query: 184 SALTGESLPVTRNPGDEVYSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQVGHFQK 243
SALTGESLPVT+NPGDEV+SGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTN VGHFQK
Sbjct: 182 SALTGESLPVTKNPGDEVFSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNNVGHFQK 241
Query: 244 VLTAIGNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMA 303
VLTAIGNFCICSIA+GML EIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMA
Sbjct: 242 VLTAIGNFCICSIAIGMLVEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMA 301
Query: 304 IGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDKEHV 363
IGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVD+NL+EVF KGVDKE+V
Sbjct: 302 IGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDRNLVEVFAKGVDKEYV 361
Query: 364 MLLAARASRVENQDAIDAAIVGTLADPKEARAGVREIHFLPFNPVDKRTALTYIDGNGNW 423
+LLAARASRVENQDAIDA +VG LADPKEARAG+RE+HFLPFNPVDKRTALTYID N NW
Sbjct: 362 LLLAARASRVENQDAIDACMVGMLADPKEARAGIREVHFLPFNPVDKRTALTYIDNNNNW 421
Query: 424 HRASKGAPEQIMDLCKLREDTKRNIHAIIDKFAERGLRSLAVARQEVPEKTKESPGAPWQ 483
HRASKGAPEQI+DLC +ED +R +H+++DK+AERGLRSLAVAR+ VPEK+KESPG W+
Sbjct: 422 HRASKGAPEQILDLCNAKEDVRRKVHSMMDKYAERGLRSLAVARRTVPEKSKESPGGRWE 481
Query: 484 FVGLLSLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSATLL 543
FVGLL LFDPPRHDSAETIRRAL+LGVNVKMITGDQLAIAKETGRRLGMGTNMYPSA+LL
Sbjct: 482 FVGLLPLFDPPRHDSAETIRRALNLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSASLL 541
Query: 544 GQDKDANIAALPVEELIEKADGFAGVFPEHKYEIVRKLQERKHICGMTGDGVNDAPALKR 603
GQDKD+ IA+LP+EELIEKADGFAGVFPEHKYEIV+KLQERKHI GMTGDGVNDAPALK+
Sbjct: 542 GQDKDSAIASLPIEELIEKADGFAGVFPEHKYEIVKKLQERKHIVGMTGDGVNDAPALKK 601
Query: 604 ADIGIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVF 663
ADIGIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVF
Sbjct: 602 ADIGIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVF 661
Query: 664 GFMFIALIWKFDFSPFMVLIIAILNDGTIMTISKDRVVPSPLPDSWKLKEIFATGIVLGG 723
GFMFIALIWK+DFS FMVLIIAILNDGTIMTISKDRV PSP+PDSWKLKEIFATG+VLGG
Sbjct: 662 GFMFIALIWKYDFSAFMVLIIAILNDGTIMTISKDRVKPSPMPDSWKLKEIFATGVVLGG 721
Query: 724 YLALMTVIFFWAMKENDFFPDKFGVRKLNH--DEMMSALYLQVSIVSQALIFVTRSRGWS 781
Y ALMTV+FFWAM + DFF DKFGV+ L + +EMMSALYLQVSI+SQALIFVTRSR WS
Sbjct: 722 YQALMTVVFFWAMHDTDFFSDKFGVKSLRNSDEEMMSALYLQVSIISQALIFVTRSRSWS 781
Query: 782 FLERPGALLVIAFFIAQLIATIIAVYANWGFAKVQGIGWGWAGVIWLYSIVFYIPLDVMK 841
FLERPG LLVIAF IAQL+AT+IAVYANW FA+V+G GWGWAGVIWLYSI+FY+PLD+MK
Sbjct: 782 FLERPGMLLVIAFMIAQLVATLIAVYANWAFARVKGCGWGWAGVIWLYSIIFYLPLDIMK 841
Query: 842 FAIRYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPESSGIFNEKS 901
FAIRYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWA AQRTLHGLQPPE++ +FNEK+
Sbjct: 842 FAIRYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWALAQRTLHGLQPPEATNLFNEKN 901
Query: 902 SYRELSEIAEQAKRRAEVARLIPFH 926
SYRELSEIAEQAKRRAE+ARL H
Sbjct: 902 SYRELSEIAEQAKRRAEMARLRELH 926
>dbj|BAD16684.1| plasma membrane H+-ATPase [Daucus carota]
Length = 950
Score = 1657 bits (4292), Expect = 0.0
Identities = 827/922 (89%), Positives = 878/922 (94%), Gaps = 2/922 (0%)
Query: 7 ISLEQIKNETVDLERIPVEEVFEQLKCTKEGLSSEEGANRLQIFGPNKLEEKKDSKILKF 66
+SLE+IKNETVDLE+IP+EEVFEQLKCT+EGLS++EGANRLQIFGPNKLEEKK+SK+LKF
Sbjct: 3 LSLEEIKNETVDLEKIPIEEVFEQLKCTREGLSADEGANRLQIFGPNKLEEKKESKLLKF 62
Query: 67 LGFMWNPLSWVMEAAALMAIGLANGNGKPPDWQDFVGIICLLVINSTISFIEENNAGNAA 126
LGFMWNPLSWVMEAAA+MAI LANGNGKPPDWQDFVGIICLLVINSTISFIEENNAGNAA
Sbjct: 63 LGFMWNPLSWVMEAAAIMAIALANGNGKPPDWQDFVGIICLLVINSTISFIEENNAGNAA 122
Query: 127 AALMAGLAPKTKVLRDGKWSEQEAAILVPGDIISIKLGDIVPADARLLEGDPLKIDQSAL 186
AALMAGLAPKTKVLRDG+WSEQ+AAILVPGDIISIKLGDIVPADARLLEGDPLKIDQSAL
Sbjct: 123 AALMAGLAPKTKVLRDGRWSEQDAAILVPGDIISIKLGDIVPADARLLEGDPLKIDQSAL 182
Query: 187 TGESLPVTRNPGDEVYSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQVGHFQKVLT 246
TGESLPVTRNP DEV+SGSTCKQGE+EAVVIATGVHTFFGKAAHLVDSTNQVGHFQKVLT
Sbjct: 183 TGESLPVTRNPYDEVFSGSTCKQGEIEAVVIATGVHTFFGKAAHLVDSTNQVGHFQKVLT 242
Query: 247 AIGNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGS 306
AIGNFCICSIAVGML E++VMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGS
Sbjct: 243 AIGNFCICSIAVGMLVELVVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGS 302
Query: 307 HRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDKEHVMLL 366
HRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVF KG DKEHV+L
Sbjct: 303 HRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFAKGFDKEHVLLC 362
Query: 367 AARASRVENQDAIDAAIVGTLADPKEARAGVREIHFLPFNPVDKRTALTYIDGNGNWHRA 426
AARASR ENQDAIDAAIVGTLADPKEARAG+RE+HFLPFNPVDKRTALTYID +GNWHR
Sbjct: 363 AARASRTENQDAIDAAIVGTLADPKEARAGIREVHFLPFNPVDKRTALTYIDSDGNWHRT 422
Query: 427 SKGAPEQIMDLCKLREDTKRNIHAIIDKFAERGLRSLAVARQEVPEKTKESPGAPWQFVG 486
SKGAPEQI+ LC +ED K+ +HA+IDKFAERGLRSL VA Q VPEK+K+S G PWQFVG
Sbjct: 423 SKGAPEQILTLCNCKEDLKKKVHAMIDKFAERGLRSLGVASQVVPEKSKDSAGGPWQFVG 482
Query: 487 LLSLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSATLLGQD 546
LLSLFDPPRHDSAETIRRAL+LGVNVKMITGDQLAIAKETGRRLGMGTNMYPSA+LLGQD
Sbjct: 483 LLSLFDPPRHDSAETIRRALNLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSASLLGQD 542
Query: 547 KDANIAALPVEELIEKADGFAGVFPEHKYEIVRKLQERKHICGMTGDGVNDAPALKRADI 606
KDA+IA+LPVEELIEKADGFAGVFPEHKYEIV+KLQERKHICGMTGDGVNDAPALK+ADI
Sbjct: 543 KDASIASLPVEELIEKADGFAGVFPEHKYEIVKKLQERKHICGMTGDGVNDAPALKKADI 602
Query: 607 GIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFM 666
GIAVADATDAAR ASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFM
Sbjct: 603 GIAVADATDAARSASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFM 662
Query: 667 FIALIWKFDFSPFMVLIIAILNDGTIMTISKDRVVPSPLPDSWKLKEIFATGIVLGGYLA 726
FIALIWKFDFSPFMVLIIAILNDGTIMTISKDRV PSPLPDSWKLKEIFATGIVLGGYLA
Sbjct: 663 FIALIWKFDFSPFMVLIIAILNDGTIMTISKDRVKPSPLPDSWKLKEIFATGIVLGGYLA 722
Query: 727 LMTVIFFWAMKENDFFPDKFGVRKLNH--DEMMSALYLQVSIVSQALIFVTRSRGWSFLE 784
L+TVIFFW MK+ D+ P+ FGVR + + DEMM+ALYLQVSIVSQALIFVTRSR WSF+E
Sbjct: 723 LLTVIFFWLMKDTDWLPNTFGVRSIRNKPDEMMAALYLQVSIVSQALIFVTRSRSWSFVE 782
Query: 785 RPGALLVIAFFIAQLIATIIAVYANWGFAKVQGIGWGWAGVIWLYSIVFYIPLDVMKFAI 844
RPG LL+ AF IAQLIAT+IAVYANWGFA++QG GWGWAGVIWLYSIVFY PLD+MKFA
Sbjct: 783 RPGFLLLGAFLIAQLIATLIAVYANWGFARIQGCGWGWAGVIWLYSIVFYFPLDIMKFAT 842
Query: 845 RYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPESSGIFNEKSSYR 904
RY LS KAW +++DN+TAFTTKKDYGKEEREAQWA AQRTLHGLQPPE+S IFNEKSSYR
Sbjct: 843 RYALSNKAWQSMIDNRTAFTTKKDYGKEEREAQWALAQRTLHGLQPPEASNIFNEKSSYR 902
Query: 905 ELSEIAEQAKRRAEVARLIPFH 926
ELSEIAEQAKRRAEVARL H
Sbjct: 903 ELSEIAEQAKRRAEVARLRELH 924
>dbj|BAD16688.1| plasma membrane H+-ATPase [Daucus carota]
Length = 950
Score = 1654 bits (4282), Expect = 0.0
Identities = 820/922 (88%), Positives = 878/922 (94%), Gaps = 2/922 (0%)
Query: 7 ISLEQIKNETVDLERIPVEEVFEQLKCTKEGLSSEEGANRLQIFGPNKLEEKKDSKILKF 66
+SLE+IKNETVDLE+IP+EEVFEQLKCT+EGLS++EG NRL+IFGPNKLEEKK+SK+LKF
Sbjct: 3 LSLEEIKNETVDLEKIPIEEVFEQLKCTREGLSADEGTNRLEIFGPNKLEEKKESKLLKF 62
Query: 67 LGFMWNPLSWVMEAAALMAIGLANGNGKPPDWQDFVGIICLLVINSTISFIEENNAGNAA 126
LGFMWNPLSWVMEAAA+MAI LANGNGKPPDWQDFVGI+CLLVINSTISFIEENNAGNAA
Sbjct: 63 LGFMWNPLSWVMEAAAIMAIALANGNGKPPDWQDFVGIMCLLVINSTISFIEENNAGNAA 122
Query: 127 AALMAGLAPKTKVLRDGKWSEQEAAILVPGDIISIKLGDIVPADARLLEGDPLKIDQSAL 186
AALMAGLAPKTKVLRDG+WSEQ+AAILVPGDIISIKLGDIVPADARLLEGDPLKIDQSAL
Sbjct: 123 AALMAGLAPKTKVLRDGRWSEQDAAILVPGDIISIKLGDIVPADARLLEGDPLKIDQSAL 182
Query: 187 TGESLPVTRNPGDEVYSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQVGHFQKVLT 246
TGESLPVTRNP DEV+SGSTCKQGE+EAVVIATGVHTFFGKAAHLVDSTNQVGHFQKVLT
Sbjct: 183 TGESLPVTRNPYDEVFSGSTCKQGEIEAVVIATGVHTFFGKAAHLVDSTNQVGHFQKVLT 242
Query: 247 AIGNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGS 306
AIGNFCICSIA+GML EI+VMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGS
Sbjct: 243 AIGNFCICSIAIGMLVEIVVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGS 302
Query: 307 HRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDKEHVMLL 366
HRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKL+VDKNLIEVF KG DKE+V+L
Sbjct: 303 HRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLTVDKNLIEVFAKGFDKENVLLC 362
Query: 367 AARASRVENQDAIDAAIVGTLADPKEARAGVREIHFLPFNPVDKRTALTYIDGNGNWHRA 426
AARASRVENQDAIDAAIVGTLADPKEARAG+RE+HFLPFNPVDKRTALTYID +GNWHRA
Sbjct: 363 AARASRVENQDAIDAAIVGTLADPKEARAGIREVHFLPFNPVDKRTALTYIDSDGNWHRA 422
Query: 427 SKGAPEQIMDLCKLREDTKRNIHAIIDKFAERGLRSLAVARQEVPEKTKESPGAPWQFVG 486
SKGAPEQI+ LC +ED K+ +HAIIDKFAERGLRSL VA Q VPEK+K+S G PWQFVG
Sbjct: 423 SKGAPEQILTLCNCKEDQKKKVHAIIDKFAERGLRSLGVASQVVPEKSKDSAGGPWQFVG 482
Query: 487 LLSLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSATLLGQD 546
LLSLFDPPRHDSAETIRRAL+LGVNVKMITGDQLAIAKETGRRLGMGTNMYPS++LLGQ
Sbjct: 483 LLSLFDPPRHDSAETIRRALNLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSSSLLGQH 542
Query: 547 KDANIAALPVEELIEKADGFAGVFPEHKYEIVRKLQERKHICGMTGDGVNDAPALKRADI 606
KD +IAALP+EELIEKADGFAGVFPEHKYEIV+KLQERKHICGMTGDGVNDAPALK+ADI
Sbjct: 543 KDESIAALPIEELIEKADGFAGVFPEHKYEIVKKLQERKHICGMTGDGVNDAPALKKADI 602
Query: 607 GIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFM 666
GIAVADATDAAR ASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFM
Sbjct: 603 GIAVADATDAARSASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFM 662
Query: 667 FIALIWKFDFSPFMVLIIAILNDGTIMTISKDRVVPSPLPDSWKLKEIFATGIVLGGYLA 726
FIALIWKFDFSPFMVLIIAILNDGTIMTISKDRV PSPLPDSWKLKEIFATG+VLGGYLA
Sbjct: 663 FIALIWKFDFSPFMVLIIAILNDGTIMTISKDRVKPSPLPDSWKLKEIFATGVVLGGYLA 722
Query: 727 LMTVIFFWAMKENDFFPDKFGVRKLNH--DEMMSALYLQVSIVSQALIFVTRSRGWSFLE 784
L+TVIFFW +K+ DFFPDKFGVR + H +EMM+ LYLQVSIVSQALIFVTRSR WSF+E
Sbjct: 723 LLTVIFFWLIKDTDFFPDKFGVRSIRHNPEEMMAVLYLQVSIVSQALIFVTRSRSWSFVE 782
Query: 785 RPGALLVIAFFIAQLIATIIAVYANWGFAKVQGIGWGWAGVIWLYSIVFYIPLDVMKFAI 844
RPG LL+ AF IAQL+AT+IAVYANWGFA++ G GWGWAGV+WLYSIVFY PLD+MKFA
Sbjct: 783 RPGFLLLGAFMIAQLLATVIAVYANWGFARIHGCGWGWAGVVWLYSIVFYFPLDIMKFAT 842
Query: 845 RYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPESSGIFNEKSSYR 904
RY LSGKAW N++DN+TAF+TKKDYGKEEREAQWA AQRTLHGLQPPE+S IFN+KSSYR
Sbjct: 843 RYALSGKAWQNMIDNRTAFSTKKDYGKEEREAQWALAQRTLHGLQPPEASTIFNDKSSYR 902
Query: 905 ELSEIAEQAKRRAEVARLIPFH 926
ELSEIAEQAKRRAEVARL H
Sbjct: 903 ELSEIAEQAKRRAEVARLRELH 924
>emb|CAA54045.1| H(+)-transporting ATPase [Solanum tuberosum] gi|1076663|pir||S50752
H+-exporting ATPase (EC 3.6.3.6) (clone PHA2) - potato
Length = 952
Score = 1652 bits (4278), Expect = 0.0
Identities = 822/925 (88%), Positives = 883/925 (94%), Gaps = 2/925 (0%)
Query: 4 SKSISLEQIKNETVDLERIPVEEVFEQLKCTKEGLSSEEGANRLQIFGPNKLEEKKDSKI 63
+K+ISLE+IKNETVDLE+IP+EEVFEQLKC++EGL+S+EGANRLQIFGPNKLEEKK+SKI
Sbjct: 2 AKAISLEEIKNETVDLEKIPIEEVFEQLKCSREGLTSDEGANRLQIFGPNKLEEKKESKI 61
Query: 64 LKFLGFMWNPLSWVMEAAALMAIGLANGNGKPPDWQDFVGIICLLVINSTISFIEENNAG 123
LKFLGFMWNPLSWVMEAAA+MAI LANGNGKPPDWQDFVGI+CLLVINSTISFIEENNAG
Sbjct: 62 LKFLGFMWNPLSWVMEAAAIMAIALANGNGKPPDWQDFVGIVCLLVINSTISFIEENNAG 121
Query: 124 NAAAALMAGLAPKTKVLRDGKWSEQEAAILVPGDIISIKLGDIVPADARLLEGDPLKIDQ 183
NAAAALMAGLAPKTKVLRDG+WSEQEAAILVPGDIIS+KLGDIVPADARLLEGDPLKIDQ
Sbjct: 122 NAAAALMAGLAPKTKVLRDGRWSEQEAAILVPGDIISVKLGDIVPADARLLEGDPLKIDQ 181
Query: 184 SALTGESLPVTRNPGDEVYSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQVGHFQK 243
SALTGESLPVT+NPGDEV+SGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTN VGHFQK
Sbjct: 182 SALTGESLPVTKNPGDEVFSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNNVGHFQK 241
Query: 244 VLTAIGNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMA 303
VLTAIGNFCICSIAVGML EIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMA
Sbjct: 242 VLTAIGNFCICSIAVGMLIEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMA 301
Query: 304 IGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDKEHV 363
IGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDK L+EVF KGVDKE+V
Sbjct: 302 IGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKTLVEVFVKGVDKEYV 361
Query: 364 MLLAARASRVENQDAIDAAIVGTLADPKEARAGVREIHFLPFNPVDKRTALTYIDGNGNW 423
+LL ARASRVENQDAIDA +VG LADPKEARAG+RE+HFLPFNPVDKRTALTYID NGNW
Sbjct: 362 LLLPARASRVENQDAIDACMVGMLADPKEARAGIREVHFLPFNPVDKRTALTYIDNNGNW 421
Query: 424 HRASKGAPEQIMDLCKLREDTKRNIHAIIDKFAERGLRSLAVARQEVPEKTKESPGAPWQ 483
HRASKGAPEQI+DLC +ED +R +H++IDK+AE GLRSLAVARQEVPEK+KES G PWQ
Sbjct: 422 HRASKGAPEQILDLCNCKEDVRRKVHSMIDKYAEAGLRSLAVARQEVPEKSKESAGGPWQ 481
Query: 484 FVGLLSLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSATLL 543
FVGLL LFDPPRHDSAETIRRAL+LGVNVKMITGDQLAIAKETGRRLGMGTNMYPSA+LL
Sbjct: 482 FVGLLPLFDPPRHDSAETIRRALNLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSASLL 541
Query: 544 GQDKDANIAALPVEELIEKADGFAGVFPEHKYEIVRKLQERKHICGMTGDGVNDAPALKR 603
GQDKD++IA+LPVEELIEKADGFAGVFPEHKYEIV+KLQERKHI GMTGDGVNDAPALK+
Sbjct: 542 GQDKDSSIASLPVEELIEKADGFAGVFPEHKYEIVKKLQERKHIVGMTGDGVNDAPALKK 601
Query: 604 ADIGIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVF 663
ADIGIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVF
Sbjct: 602 ADIGIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVF 661
Query: 664 GFMFIALIWKFDFSPFMVLIIAILNDGTIMTISKDRVVPSPLPDSWKLKEIFATGIVLGG 723
GFM IALIWK+DFS FMVLIIAILNDGTIMTISKDRV PSP+PDSWKL EIFATG+VLGG
Sbjct: 662 GFMLIALIWKYDFSAFMVLIIAILNDGTIMTISKDRVKPSPMPDSWKLNEIFATGVVLGG 721
Query: 724 YLALMTVIFFWAMKENDFFPDKFGVRKL--NHDEMMSALYLQVSIVSQALIFVTRSRGWS 781
Y ALMTV+FFWAM + FF DKFGV+ + + +EMMSALYLQVSI+SQALIFVTRSR WS
Sbjct: 722 YQALMTVLFFWAMHDTKFFSDKFGVKDIRESDEEMMSALYLQVSIISQALIFVTRSRSWS 781
Query: 782 FLERPGALLVIAFFIAQLIATIIAVYANWGFAKVQGIGWGWAGVIWLYSIVFYIPLDVMK 841
F+ERPGALL+IAF IAQL+AT+IAVYA+W FA+V+G GWGWAGVIW++SIV Y PLD+MK
Sbjct: 782 FVERPGALLMIAFLIAQLVATLIAVYADWTFARVKGCGWGWAGVIWIFSIVTYFPLDIMK 841
Query: 842 FAIRYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPESSGIFNEKS 901
FAIRYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWA AQRTLHGLQPPE+S +FNEK+
Sbjct: 842 FAIRYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWALAQRTLHGLQPPEASNLFNEKN 901
Query: 902 SYRELSEIAEQAKRRAEVARLIPFH 926
SYRELSEIAEQAKRRAE+ARL H
Sbjct: 902 SYRELSEIAEQAKRRAEMARLRELH 926
>gb|AAB17186.1| plasma membrane H+-ATPase [Lycopersicon esculentum]
Length = 952
Score = 1651 bits (4276), Expect = 0.0
Identities = 820/925 (88%), Positives = 884/925 (94%), Gaps = 2/925 (0%)
Query: 4 SKSISLEQIKNETVDLERIPVEEVFEQLKCTKEGLSSEEGANRLQIFGPNKLEEKKDSKI 63
+K+ISLE+IKNETVDLE+IP+EEVFEQLKC++EGL+S+EGANRLQIFGPNKLEEKK+SKI
Sbjct: 2 AKAISLEEIKNETVDLEKIPIEEVFEQLKCSREGLTSDEGANRLQIFGPNKLEEKKESKI 61
Query: 64 LKFLGFMWNPLSWVMEAAALMAIGLANGNGKPPDWQDFVGIICLLVINSTISFIEENNAG 123
LKFLGFMWNPLSWVME AA+MAI LANG+GKPPDWQDFVGI+CLLVINSTISFIEENNAG
Sbjct: 62 LKFLGFMWNPLSWVMEMAAIMAIALANGDGKPPDWQDFVGIVCLLVINSTISFIEENNAG 121
Query: 124 NAAAALMAGLAPKTKVLRDGKWSEQEAAILVPGDIISIKLGDIVPADARLLEGDPLKIDQ 183
NAAAALMAGLAPKTKVLRDG+WSEQEAAILVPGDIIS+KLGDIVPADARLLEGDPLKIDQ
Sbjct: 122 NAAAALMAGLAPKTKVLRDGRWSEQEAAILVPGDIISVKLGDIVPADARLLEGDPLKIDQ 181
Query: 184 SALTGESLPVTRNPGDEVYSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQVGHFQK 243
SALTGESLPVT+NPGDEV+SGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTN VGHFQK
Sbjct: 182 SALTGESLPVTKNPGDEVFSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNNVGHFQK 241
Query: 244 VLTAIGNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMA 303
VLTAIGNFCICSIA+GML EIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMA
Sbjct: 242 VLTAIGNFCICSIAIGMLVEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMA 301
Query: 304 IGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDKEHV 363
IGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVD++L+EVF KGVDKE+V
Sbjct: 302 IGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDRSLVEVFTKGVDKEYV 361
Query: 364 MLLAARASRVENQDAIDAAIVGTLADPKEARAGVREIHFLPFNPVDKRTALTYIDGNGNW 423
+LLAARASRVENQDAIDA +VG LADPKEARAG+RE+HFLPFNPVDKRTALTYID NGNW
Sbjct: 362 LLLAARASRVENQDAIDACMVGMLADPKEARAGIREVHFLPFNPVDKRTALTYIDSNGNW 421
Query: 424 HRASKGAPEQIMDLCKLREDTKRNIHAIIDKFAERGLRSLAVARQEVPEKTKESPGAPWQ 483
HRASKGAPEQI+DLC +ED +R +H++IDK+AERGLRSLAVARQEVPEK+KES G PWQ
Sbjct: 422 HRASKGAPEQILDLCNCKEDVRRKVHSMIDKYAERGLRSLAVARQEVPEKSKESTGGPWQ 481
Query: 484 FVGLLSLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSATLL 543
FVGLL LFDPPRHDSAETIRRAL+LGVNVKMITGDQLAIAKETGRRLGMGTNMYPSA+LL
Sbjct: 482 FVGLLPLFDPPRHDSAETIRRALNLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSASLL 541
Query: 544 GQDKDANIAALPVEELIEKADGFAGVFPEHKYEIVRKLQERKHICGMTGDGVNDAPALKR 603
GQDKD++IA+LPVEELIEKADGFAGVFPEHKYEIV+KLQERKHI GMTGDGVNDAPALK+
Sbjct: 542 GQDKDSSIASLPVEELIEKADGFAGVFPEHKYEIVKKLQERKHIVGMTGDGVNDAPALKK 601
Query: 604 ADIGIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVF 663
ADIGIAVADATDAARG SDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVF
Sbjct: 602 ADIGIAVADATDAARGRSDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVF 661
Query: 664 GFMFIALIWKFDFSPFMVLIIAILNDGTIMTISKDRVVPSPLPDSWKLKEIFATGIVLGG 723
GFM IALIWK+DFS FMVLIIAILNDGTIMTISKDRV PSP+PDSWKL EIFATG+VLGG
Sbjct: 662 GFMLIALIWKYDFSAFMVLIIAILNDGTIMTISKDRVKPSPMPDSWKLNEIFATGVVLGG 721
Query: 724 YLALMTVIFFWAMKENDFFPDKFGVRKL--NHDEMMSALYLQVSIVSQALIFVTRSRGWS 781
Y ALMTVIFFWAM + FF DKFGV+ + + +EMMSALYLQVSI+SQALIFVTRSR WS
Sbjct: 722 YQALMTVIFFWAMHDTSFFTDKFGVKDIRESDEEMMSALYLQVSIISQALIFVTRSRSWS 781
Query: 782 FLERPGALLVIAFFIAQLIATIIAVYANWGFAKVQGIGWGWAGVIWLYSIVFYIPLDVMK 841
F+ERPGALL+IAF IAQL+AT+IAVYA+W FA+V+G GWGWAGVIW++SIV Y PLD+MK
Sbjct: 782 FVERPGALLMIAFLIAQLVATLIAVYADWTFARVKGCGWGWAGVIWIFSIVTYFPLDIMK 841
Query: 842 FAIRYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPESSGIFNEKS 901
FAIRYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWA AQRTLHGLQPPE+S +FNEK+
Sbjct: 842 FAIRYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWALAQRTLHGLQPPEASNLFNEKN 901
Query: 902 SYRELSEIAEQAKRRAEVARLIPFH 926
SYRELSEIAEQAKRRAE+ARL H
Sbjct: 902 SYRELSEIAEQAKRRAEMARLRELH 926
>gb|AAY42948.1| plasma membrane H+ ATPase [Lupinus albus]
Length = 956
Score = 1643 bits (4254), Expect = 0.0
Identities = 834/934 (89%), Positives = 881/934 (94%), Gaps = 12/934 (1%)
Query: 1 MAESKSISLEQIKNETVDLERIPVEEVFEQLKCTKEGLSSEEGANRLQIFGPNKLEEKKD 60
MA K I LE+IKNE VDLERIPVEEVFEQLKCTKEGLS++EGA+RL+IFGPNKLEEKK+
Sbjct: 1 MASGKGIPLEEIKNENVDLERIPVEEVFEQLKCTKEGLSTQEGASRLEIFGPNKLEEKKE 60
Query: 61 SKILKFLGFMWNPLSWVMEAAALMAIGLANGNGKPPDWQDFVGIICLLVINSTISFIEEN 120
SK LKFLGFMWNPLSWVME+AA+MAI LANG+GKPPDWQDFVGIICLLVINSTISF+EEN
Sbjct: 61 SKFLKFLGFMWNPLSWVMESAAIMAIALANGDGKPPDWQDFVGIICLLVINSTISFVEEN 120
Query: 121 NAGNAAAALMAGLAPKTKVLRDGKWSEQEAAILVPGDIISIKLGDIVPADARLLEGDPLK 180
NAGNAAAALMAGLAPKTKVLRDGKW E+EAAILVPGDIISIKLGDIVPADARLLEGDPLK
Sbjct: 121 NAGNAAAALMAGLAPKTKVLRDGKWCEEEAAILVPGDIISIKLGDIVPADARLLEGDPLK 180
Query: 181 IDQSALTGESLPVTRNPGDEVYSGSTCKQGELEAVVIATGVHT-----FFGKAAHLVDST 235
IDQSALTGESLPVT+NPGDEV+SGSTCKQGE+EAVVIATGVHT FFGKAAHLVDST
Sbjct: 181 IDQSALTGESLPVTKNPGDEVFSGSTCKQGEIEAVVIATGVHTTGVHTFFGKAAHLVDST 240
Query: 236 NQVGHFQKVLTAIGNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMP 295
NQVGHFQKVLTAIGNFCI SIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMP
Sbjct: 241 NQVGHFQKVLTAIGNFCIVSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMP 300
Query: 296 TVLSVTMAIGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFE 355
TVLSVTMAIGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVF
Sbjct: 301 TVLSVTMAIGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFT 360
Query: 356 KGVDKEHVMLLAARASRVENQDAIDAAIVGTLADPKEARAGVREIHFLPFNPVDKRTALT 415
KGVDK+HVMLLAARASRVENQDAIDAAIVG LADPKEARAGVRE+HFLPFNPVDKRTALT
Sbjct: 361 KGVDKDHVMLLAARASRVENQDAIDAAIVGMLADPKEARAGVREVHFLPFNPVDKRTALT 420
Query: 416 YIDGNGNWHRASKGAPEQIMDLCKLREDTKRNIHAIIDKFAERGLRSLAVARQEVPEKTK 475
YID +G WHRASKGAPEQIM LC L+ED K+ +HAIIDKFAERGLRSLAVARQEVPEK K
Sbjct: 421 YIDTDGIWHRASKGAPEQIMILCGLKEDAKKKVHAIIDKFAERGLRSLAVARQEVPEKAK 480
Query: 476 ESPGAPWQFVGLLSLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTN 535
ES G PW+FVGLLSLFDPPRHDSAETIR+AL+LGVNVKMITGDQLAIAKETGRRLGMGTN
Sbjct: 481 ESAGGPWEFVGLLSLFDPPRHDSAETIRKALNLGVNVKMITGDQLAIAKETGRRLGMGTN 540
Query: 536 MYPSATLLGQDKDANIAALPVEELIEKADGFAGVFPEHKYEIVRKLQERKHICGMTGDGV 595
MYPS++LLGQ KD +I + VEELIEKADGFAGVFPEHKYEIV+KLQER+HICGMTGDGV
Sbjct: 541 MYPSSSLLGQHKDESIGS--VEELIEKADGFAGVFPEHKYEIVKKLQERRHICGMTGDGV 598
Query: 596 NDAPALKRADIGIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAV 655
NDAPALK+ADIGIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAV
Sbjct: 599 NDAPALKKADIGIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAV 658
Query: 656 SITIRIVFGFMFIALIWKFDFSPFMVLIIAILNDGTIMTISKDRVVPSPLPDSWKLKEIF 715
SITIRIVFGFMFIALIWKFDFSPFMVLIIAILNDGTIMTISKDRV PSPLPDSWKLKEIF
Sbjct: 659 SITIRIVFGFMFIALIWKFDFSPFMVLIIAILNDGTIMTISKDRVKPSPLPDSWKLKEIF 718
Query: 716 ATGIVLGGYLALMTVIFFWAMKENDFFPDKFGVRKLNHD---EMMSALYLQVSIVSQALI 772
ATGIVLGGY+ALMTVIFFWAMK+ FFP KFGVR + HD EM +ALYLQVS VSQALI
Sbjct: 719 ATGIVLGGYMALMTVIFFWAMKDTTFFPRKFGVRPI-HDSPYEMTAALYLQVSTVSQALI 777
Query: 773 FVTRSRGWSFLERPGALLVIAFFIAQLIATIIAVYANWGFAKVQGIGWGWAGVIWLYSIV 832
FVTRSR WSF+ERPG LL+ AF IAQLIATIIAVYANWGFAK+QG+GWGWAGVIWLYS+V
Sbjct: 778 FVTRSRSWSFVERPGMLLMGAFVIAQLIATIIAVYANWGFAKIQGVGWGWAGVIWLYSVV 837
Query: 833 FYIPLDVMKFAIRYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPE 892
FY PLD++KFAIRY+LSGKAW N ++NKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPE
Sbjct: 838 FYFPLDLLKFAIRYVLSGKAWVN-IENKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPE 896
Query: 893 SSGIFNEKSSYRELSEIAEQAKRRAEVARLIPFH 926
+S IFNE ++YRELSEIAEQAKRRAEVARL H
Sbjct: 897 TSNIFNESNNYRELSEIAEQAKRRAEVARLRELH 930
>dbj|BAD16687.1| plasma membrane H+-ATPase [Daucus carota]
Length = 949
Score = 1642 bits (4253), Expect = 0.0
Identities = 816/920 (88%), Positives = 873/920 (94%), Gaps = 2/920 (0%)
Query: 9 LEQIKNETVDLERIPVEEVFEQLKCTKEGLSSEEGANRLQIFGPNKLEEKKDSKILKFLG 68
LE IKNETVDLE+IP+EEVFEQLKCT+EGLS +EGANRLQIFGPNKLEEKK+SK+LKFLG
Sbjct: 4 LEDIKNETVDLEKIPIEEVFEQLKCTREGLSGDEGANRLQIFGPNKLEEKKESKLLKFLG 63
Query: 69 FMWNPLSWVMEAAALMAIGLANGNGKPPDWQDFVGIICLLVINSTISFIEENNAGNAAAA 128
FMWNPLSWVMEAAA+MAI LANG+GKPPDWQDFVGIICLL+INSTISF EENNAGNAAAA
Sbjct: 64 FMWNPLSWVMEAAAIMAIVLANGDGKPPDWQDFVGIICLLLINSTISFWEENNAGNAAAA 123
Query: 129 LMAGLAPKTKVLRDGKWSEQEAAILVPGDIISIKLGDIVPADARLLEGDPLKIDQSALTG 188
LMAGLAPKTKVLRDG+WSEQ+AAILVPGDIISIKLGDIVPADARLLEGDPLKIDQSALTG
Sbjct: 124 LMAGLAPKTKVLRDGRWSEQDAAILVPGDIISIKLGDIVPADARLLEGDPLKIDQSALTG 183
Query: 189 ESLPVTRNPGDEVYSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQVGHFQKVLTAI 248
ESLPVTRNP DEV+SGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQVGHFQ VLTAI
Sbjct: 184 ESLPVTRNPHDEVFSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQVGHFQTVLTAI 243
Query: 249 GNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGSHR 308
GNFCICSIAVGM+ EIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGSHR
Sbjct: 244 GNFCICSIAVGMVVEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGSHR 303
Query: 309 LSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDKEHVMLLAA 368
LS+QGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKL+VDKNLIEVF KGVDKE+V+L AA
Sbjct: 304 LSEQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLTVDKNLIEVFAKGVDKEYVLLCAA 363
Query: 369 RASRVENQDAIDAAIVGTLADPKEARAGVREIHFLPFNPVDKRTALTYIDGNGNWHRASK 428
RASR ENQDAIDAAIVGTLADPKEARAG+RE+HF PFNPVDKRTALT+ID GNWHRASK
Sbjct: 364 RASRTENQDAIDAAIVGTLADPKEARAGIREVHFFPFNPVDKRTALTFIDSEGNWHRASK 423
Query: 429 GAPEQIMDLCKLREDTKRNIHAIIDKFAERGLRSLAVARQEVPEKTKESPGAPWQFVGLL 488
GAPEQI+ LC +ED K+ +HAIIDKFAERGLRSLAVARQEVP+K+K+S G PWQFVGLL
Sbjct: 424 GAPEQILTLCNCKEDQKKKVHAIIDKFAERGLRSLAVARQEVPQKSKDSEGGPWQFVGLL 483
Query: 489 SLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSATLLGQDKD 548
SLFDPPRHDS+ETIRRAL+LGVNVKMITGDQLAIAKETGRRLGMGTNMYPSA LLGQ+KD
Sbjct: 484 SLFDPPRHDSSETIRRALNLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSAALLGQNKD 543
Query: 549 ANIAALPVEELIEKADGFAGVFPEHKYEIVRKLQERKHICGMTGDGVNDAPALKRADIGI 608
A+IA+LPV+ELIEKADGFAGVFPEHKYEIV++LQE+KHICGMTGDGVNDAPALK+ADIGI
Sbjct: 544 ASIASLPVDELIEKADGFAGVFPEHKYEIVKRLQEKKHICGMTGDGVNDAPALKKADIGI 603
Query: 609 AVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFMFI 668
AVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFMFI
Sbjct: 604 AVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFMFI 663
Query: 669 ALIWKFDFSPFMVLIIAILNDGTIMTISKDRVVPSPLPDSWKLKEIFATGIVLGGYLALM 728
ALIWKFDFSPFMVLIIAILNDGTIMTISKDRV PSP+PDSWKLKEIFATGIVLGGYLAL+
Sbjct: 664 ALIWKFDFSPFMVLIIAILNDGTIMTISKDRVKPSPMPDSWKLKEIFATGIVLGGYLALL 723
Query: 729 TVIFFWAMKENDFFPDKFGVRKLNH--DEMMSALYLQVSIVSQALIFVTRSRGWSFLERP 786
TVIFFW +K+ DFFP+KFGVR + + DEMM+ LYLQVSIVSQALIFVTRSR WSF+ERP
Sbjct: 724 TVIFFWLIKDTDFFPEKFGVRPIRNKPDEMMAVLYLQVSIVSQALIFVTRSRSWSFMERP 783
Query: 787 GALLVIAFFIAQLIATIIAVYANWGFAKVQGIGWGWAGVIWLYSIVFYIPLDVMKFAIRY 846
G LLV AF +AQLIAT +AVYANW FA++ G GWGWAGVIW+YSIVFYIPLD++KF RY
Sbjct: 784 GLLLVAAFLVAQLIATFVAVYANWDFARIHGCGWGWAGVIWIYSIVFYIPLDILKFGTRY 843
Query: 847 ILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPESSGIFNEKSSYREL 906
LSGKAW NLL+NKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPP S IF++K+SYREL
Sbjct: 844 ALSGKAWLNLLENKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPADSNIFDDKNSYREL 903
Query: 907 SEIAEQAKRRAEVARLIPFH 926
SEIAEQAKRRAEVARL H
Sbjct: 904 SEIAEQAKRRAEVARLRELH 923
>gb|AAY42950.1| plasma membrane H+ ATPase [Lupinus albus]
Length = 953
Score = 1640 bits (4248), Expect = 0.0
Identities = 831/928 (89%), Positives = 874/928 (93%), Gaps = 3/928 (0%)
Query: 1 MAESKSISLEQIKNETVDLERIPVEEVFEQLKCTKEGLSSEEGANRLQIFGPNKLEEKKD 60
MA K ISLE+IKNETVDLER+P+EEVF+QLKCTKEGLSS EGANRL+IFGPNKLEEKKD
Sbjct: 1 MASGKGISLEEIKNETVDLERVPIEEVFQQLKCTKEGLSSGEGANRLEIFGPNKLEEKKD 60
Query: 61 SKILKFLGFMWNPLSWVMEAAALMAIGLANGNGKPPDWQDFVGIICLLVINSTISFIEEN 120
SK LKFLGFMWNPLSWVME AA+MA+ LANG GKPPDWQDFVGIICLLVINSTISFIEEN
Sbjct: 61 SKFLKFLGFMWNPLSWVMELAAIMAVALANGEGKPPDWQDFVGIICLLVINSTISFIEEN 120
Query: 121 NAGNAAAALMAGLAPKTKVLRDGKWSEQEAAILVPGDIISIKLGDIVPADARLLEGDPLK 180
NAGNAAAALMAGLAPKTKVLRDGKW E+EAAILVPGDIISIKLGDIVPADARLLEGDPLK
Sbjct: 121 NAGNAAAALMAGLAPKTKVLRDGKWCEEEAAILVPGDIISIKLGDIVPADARLLEGDPLK 180
Query: 181 IDQSALTGESLPVTRNPGDEVYSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQVGH 240
IDQSALTGESLPVT+NPGDEV+SGSTCKQGE+EAVVIATGVHTFFGKAAHLVDSTNQVGH
Sbjct: 181 IDQSALTGESLPVTKNPGDEVFSGSTCKQGEIEAVVIATGVHTFFGKAAHLVDSTNQVGH 240
Query: 241 FQKVLTAIGNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSV 300
FQKVLTAIGNFCI SIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSV
Sbjct: 241 FQKVLTAIGNFCIVSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSV 300
Query: 301 TMAIGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDK 360
TMAIGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVF KGVDK
Sbjct: 301 TMAIGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFTKGVDK 360
Query: 361 EHVMLLAARASRVENQDAIDAAIVGTLADPKEARAGVREIHFLPFNPVDKRTALTYIDGN 420
+HVMLLAARASRVENQDAIDAAIVG LADPKEARAGVRE+HFLPFNPVDKRTALTYID +
Sbjct: 361 DHVMLLAARASRVENQDAIDAAIVGMLADPKEARAGVREVHFLPFNPVDKRTALTYIDTD 420
Query: 421 GNWHRASKGAPEQIMDLCKLREDTKRNIHAIIDKFAERGLRSLAVARQEVPEKTKESPGA 480
G WHRASKGAPEQIM LC L+ED K+ +HAIIDKFAERGLRSLAVARQEVPEK KES G
Sbjct: 421 GIWHRASKGAPEQIMILCGLKEDAKKKVHAIIDKFAERGLRSLAVARQEVPEKAKESAGG 480
Query: 481 PWQFVGLLSLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSA 540
P QFVGLLSLFDPPRHDSAETI +AL+LGVNVKMITGDQLAIAKETGRRLGMGTNMYPS+
Sbjct: 481 PRQFVGLLSLFDPPRHDSAETISKALNLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSS 540
Query: 541 TLLGQDKDANIAALPVEELIEKADGFAGVFPEHKYEIVRKLQERKHICGMTGDGVNDAPA 600
+LLGQ KD +IA++PVEELIEKADGFAGVFPEHKYEI +KLQERKHICGMTGDGVNDAPA
Sbjct: 541 SLLGQHKDESIASIPVEELIEKADGFAGVFPEHKYEIDKKLQERKHICGMTGDGVNDAPA 600
Query: 601 LKRADIGIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIR 660
LK+ADIGIAVADATDAARGAS IVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIR
Sbjct: 601 LKKADIGIAVADATDAARGASXIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIR 660
Query: 661 IVFGFMFIALIWKFDFSPFMVLIIAILNDGTIMTISKDRVVPSPLPDSWKLKEIFATGIV 720
IVFGFMFIALIWKFDFSPFMVLIIAILNDGTIMTISKDRV PSPLPDSWKLKEIFATGIV
Sbjct: 661 IVFGFMFIALIWKFDFSPFMVLIIAILNDGTIMTISKDRVKPSPLPDSWKLKEIFATGIV 720
Query: 721 LGGYLALMTVIFFWAMKENDFFPDKFGV--RKLNHDEMMSALYLQVSIVSQALIFVTRSR 778
LGGY+ALMTVIFFWAMK+ +F KFGV DEM +ALYLQVS VSQALIFVTRSR
Sbjct: 721 LGGYMALMTVIFFWAMKDTNFLSRKFGVDPYMTAPDEMTAALYLQVSTVSQALIFVTRSR 780
Query: 779 GWSFLERPGALLVIAFFIAQLIATIIAVYANWGFAKVQGIGWGWAGVIWLYSIVFYIPLD 838
WSF+ERPG LL+ AF IAQLIATIIAVYANWGFAK+QG+GWGWAGVIWLYS+VFY PLD
Sbjct: 781 SWSFVERPGMLLMGAFVIAQLIATIIAVYANWGFAKIQGVGWGWAGVIWLYSVVFYFPLD 840
Query: 839 VMKFAIRYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPESSGIFN 898
++KFAIRY+LSGKAW N ++NKTAFTTKKD GKEEREAQWAHAQRTLHGLQPPE+S IFN
Sbjct: 841 LLKFAIRYVLSGKAWVN-IENKTAFTTKKDCGKEEREAQWAHAQRTLHGLQPPETSNIFN 899
Query: 899 EKSSYRELSEIAEQAKRRAEVARLIPFH 926
E ++YRELSEIAEQAKRRAEVARL H
Sbjct: 900 ESNNYRELSEIAEQAKRRAEVARLRELH 927
>dbj|BAD16685.1| plasma membrane H+-ATPase [Daucus carota]
Length = 951
Score = 1639 bits (4245), Expect = 0.0
Identities = 814/923 (88%), Positives = 879/923 (95%), Gaps = 3/923 (0%)
Query: 7 ISLEQIKNETVDLERIPVEEVFEQLKCTKEGLSSEEGANRLQIFGPNKLEEKKDSKILKF 66
ISLE+IKNETVDLE+IP+EEVFEQLKCT+EGLS++EGANRL+IFGPNKLEEKK+SK LKF
Sbjct: 3 ISLEEIKNETVDLEKIPIEEVFEQLKCTREGLSTDEGANRLEIFGPNKLEEKKESKFLKF 62
Query: 67 LGFMWNPLSWVMEAAALMAIGLANGNGKPPDWQDFVGIICLLVINSTISFIEENNAGNAA 126
LGFMWNPLSWVMEAAA+MAI LANG+GKPPDWQDFVGIICLLVINSTISF+EENNAGNAA
Sbjct: 63 LGFMWNPLSWVMEAAAIMAIALANGSGKPPDWQDFVGIICLLVINSTISFVEENNAGNAA 122
Query: 127 AALMAGLAPKTKVLRDGKWSEQEAAILVPGDIISIKLGDIVPADARLLEGDPLKIDQSAL 186
AALMAGLAPKTKVLRDG+WSEQ+AAILVPGDIISIKLGDIVPADARLLEGDPLKIDQSAL
Sbjct: 123 AALMAGLAPKTKVLRDGRWSEQDAAILVPGDIISIKLGDIVPADARLLEGDPLKIDQSAL 182
Query: 187 TGESLPVTRNPGDEVYSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQVGHFQKVLT 246
TGESLPVTR+P DEV+SGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQVGHFQKVLT
Sbjct: 183 TGESLPVTRHPYDEVFSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQVGHFQKVLT 242
Query: 247 AIGNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGS 306
AIGNFCICSIAVGM+ EIIVMYPIQHRKYR+GIDNLLVLLIGGIPIAMPTVLSVTMAIGS
Sbjct: 243 AIGNFCICSIAVGMIVEIIVMYPIQHRKYRNGIDNLLVLLIGGIPIAMPTVLSVTMAIGS 302
Query: 307 HRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDKEHVMLL 366
HRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKL+VDKNLIEVF KG+DK+ V+L
Sbjct: 303 HRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLTVDKNLIEVFAKGMDKDFVLLC 362
Query: 367 AARASRVENQDAIDAAIVGTLADPKEARAGVREIHFLPFNPVDKRTALTYIDGNGNWHRA 426
AARASR ENQDAIDAAIVGTLADPKEARAG++E+HF PFNPVDKRTALT+ID +GNWHRA
Sbjct: 363 AARASRTENQDAIDAAIVGTLADPKEARAGIKEVHFFPFNPVDKRTALTFIDADGNWHRA 422
Query: 427 SKGAPEQIMDLCKLREDTKRNIHAIIDKFAERGLRSLAVARQEVPEKTKESPGAPWQFVG 486
SKGAPEQI+ LC +ED K+ +HAIIDKFAERGLRSL VARQ VP+K+K+S G PW+FVG
Sbjct: 423 SKGAPEQILTLCNCKEDLKKKVHAIIDKFAERGLRSLGVARQVVPQKSKDSAGGPWEFVG 482
Query: 487 LLSLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSATLLGQD 546
LLSLFDPPRHDSAETIRRAL+LGVNVKMITGDQLAIAKETGRRLGMGTNMYPSA LLGQ+
Sbjct: 483 LLSLFDPPRHDSAETIRRALNLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSAALLGQN 542
Query: 547 KDANIAALPVEELIEKADGFAGVFPEHKYEIVRKLQERKHICGMTGDGVNDAPALKRADI 606
KDA+IA+LPV+ELIEKADGFAGVFPEHKYEIV+KLQE+KHICGMTGDGVNDAPALK+ADI
Sbjct: 543 KDASIASLPVDELIEKADGFAGVFPEHKYEIVKKLQEKKHICGMTGDGVNDAPALKKADI 602
Query: 607 GIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFM 666
GIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFM
Sbjct: 603 GIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFM 662
Query: 667 FIALIWKFDFSPFMVLIIAILNDGTIMTISKDRVVPSPLPDSWKLKEIFATGIVLGGYLA 726
FIALIWKFDFSPFMVLIIAILNDGTIMTISKDRV PSPLPDSWKLKEIFATGIVLGGYLA
Sbjct: 663 FIALIWKFDFSPFMVLIIAILNDGTIMTISKDRVKPSPLPDSWKLKEIFATGIVLGGYLA 722
Query: 727 LMTVIFFWAMKENDFFPDKFGVRKL--NHDEMMSALYLQVSIVSQALIFVTRSRGWSFLE 784
L+TVIFFW MK+ DFFP+KFGVR + + DEMM+ALYLQVSIVSQALIFVTRSR WSF+E
Sbjct: 723 LLTVIFFWLMKDTDFFPNKFGVRPIRDSPDEMMAALYLQVSIVSQALIFVTRSRSWSFVE 782
Query: 785 RPGALLVIAFFIAQLIATIIAVYANWGFAKVQGIGWGWAGVIWLYSIVFYIPLDVMKFAI 844
RPG LL+ AF IAQLIAT+IAVYANWGFA+++G GWGWAGVIW+YS+VFY PLD+MKF
Sbjct: 783 RPGFLLLGAFLIAQLIATLIAVYANWGFARIEGCGWGWAGVIWIYSVVFYFPLDIMKFGT 842
Query: 845 RYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPESSGIFNEK-SSY 903
RY LSGKAWNN+++ + AFTTKKDYGKEEREAQWAH QRTLHGLQPPE++ IFN+K S+Y
Sbjct: 843 RYALSGKAWNNMIEQRVAFTTKKDYGKEEREAQWAHVQRTLHGLQPPEATNIFNDKNSNY 902
Query: 904 RELSEIAEQAKRRAEVARLIPFH 926
RELSEIAEQAKRRAEVARL H
Sbjct: 903 RELSEIAEQAKRRAEVARLRELH 925
>emb|CAB69823.1| plasma membrane H+ ATPase [Prunus persica]
Length = 954
Score = 1634 bits (4230), Expect = 0.0
Identities = 810/928 (87%), Positives = 878/928 (94%), Gaps = 2/928 (0%)
Query: 1 MAESKSISLEQIKNETVDLERIPVEEVFEQLKCTKEGLSSEEGANRLQIFGPNKLEEKKD 60
M K+ISLE+IKNE+VDLERIP+EEVFEQLKCT+EGL+ +EGANRLQ+FGPNKLEEKK+
Sbjct: 1 MGGDKAISLEEIKNESVDLERIPIEEVFEQLKCTREGLTGDEGANRLQVFGPNKLEEKKE 60
Query: 61 SKILKFLGFMWNPLSWVMEAAALMAIGLANGNGKPPDWQDFVGIICLLVINSTISFIEEN 120
SK+LKFLGFMWNPLSWVMEAAA+MAI LANG G+PPDWQDFVGI+ LLVINSTISFIEEN
Sbjct: 61 SKLLKFLGFMWNPLSWVMEAAAVMAIALANGGGRPPDWQDFVGIVVLLVINSTISFIEEN 120
Query: 121 NAGNAAAALMAGLAPKTKVLRDGKWSEQEAAILVPGDIISIKLGDIVPADARLLEGDPLK 180
NAGNAAAALMAGLAPKTKVLRDG+W+EQEA+ILVPGDIISIKLGDIVPADARLLEGDPLK
Sbjct: 121 NAGNAAAALMAGLAPKTKVLRDGRWTEQEASILVPGDIISIKLGDIVPADARLLEGDPLK 180
Query: 181 IDQSALTGESLPVTRNPGDEVYSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQVGH 240
IDQSALTGESLPVT+NP +EV+SGSTCKQGE+EAVVIATGVHTFFGKAAHLVDSTNQVGH
Sbjct: 181 IDQSALTGESLPVTKNPSEEVFSGSTCKQGEIEAVVIATGVHTFFGKAAHLVDSTNQVGH 240
Query: 241 FQKVLTAIGNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSV 300
FQKVLTAIGNFCICSIAVG+L E+IVMYPIQ RKYRDGIDNLLVLLIGGIPIAMPTVLSV
Sbjct: 241 FQKVLTAIGNFCICSIAVGILIELIVMYPIQKRKYRDGIDNLLVLLIGGIPIAMPTVLSV 300
Query: 301 TMAIGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDK 360
TMAIGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVD+NLIEVF KGV+K
Sbjct: 301 TMAIGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDRNLIEVFAKGVEK 360
Query: 361 EHVMLLAARASRVENQDAIDAAIVGTLADPKEARAGVREIHFLPFNPVDKRTALTYIDGN 420
EHVMLLAARASR ENQDAIDAAIVG LADPKEAR G+RE+HFLPFNPVDKRTALTYID +
Sbjct: 361 EHVMLLAARASRTENQDAIDAAIVGMLADPKEARVGIREVHFLPFNPVDKRTALTYIDSD 420
Query: 421 GNWHRASKGAPEQIMDLCKLREDTKRNIHAIIDKFAERGLRSLAVARQEVPEKTKESPGA 480
GNWHRASKGAPEQI+ LC +ED K+ +HA+IDKFAERGLRSLAVARQ+VPEKTKESPG
Sbjct: 421 GNWHRASKGAPEQILALCNCKEDFKKRVHAVIDKFAERGLRSLAVARQQVPEKTKESPGT 480
Query: 481 PWQFVGLLSLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSA 540
PWQFVGLL LFDPPRHDSAETIRRAL+LGVNVKMITGDQLAI KETGRRLGMGTNMYPS+
Sbjct: 481 PWQFVGLLPLFDPPRHDSAETIRRALNLGVNVKMITGDQLAIGKETGRRLGMGTNMYPSS 540
Query: 541 TLLGQDKDANIAALPVEELIEKADGFAGVFPEHKYEIVRKLQERKHICGMTGDGVNDAPA 600
LLGQDKDA+IA+LPV+ELIEKADGFAGVFPEHKYEIV++LQERKHICGMTGDGVNDAPA
Sbjct: 541 ALLGQDKDASIASLPVDELIEKADGFAGVFPEHKYEIVKRLQERKHICGMTGDGVNDAPA 600
Query: 601 LKRADIGIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIR 660
LK+ADIGIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIR
Sbjct: 601 LKKADIGIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIR 660
Query: 661 IVFGFMFIALIWKFDFSPFMVLIIAILNDGTIMTISKDRVVPSPLPDSWKLKEIFATGIV 720
IVFGFMFIALIWKFDF+PFMVLIIAILNDGTIMTISKDRV PSP PDSWKLKEIFATGIV
Sbjct: 661 IVFGFMFIALIWKFDFAPFMVLIIAILNDGTIMTISKDRVKPSPQPDSWKLKEIFATGIV 720
Query: 721 LGGYLALMTVIFFWAMKENDFFPDKFGVRKLNH--DEMMSALYLQVSIVSQALIFVTRSR 778
LGGY+ALMTV+FFW MK+ FF + F VR L ++MM+ALYLQVSIVSQALIFVTRSR
Sbjct: 721 LGGYMALMTVVFFWLMKDTKFFSNTFNVRHLGDRPEQMMAALYLQVSIVSQALIFVTRSR 780
Query: 779 GWSFLERPGALLVIAFFIAQLIATIIAVYANWGFAKVQGIGWGWAGVIWLYSIVFYIPLD 838
WSF+ERPG LL+ AF +AQL+AT+IAVYANW FA+++G GWGWAGVIWL+S+V Y PLD
Sbjct: 781 SWSFVERPGLLLLGAFMVAQLVATLIAVYANWAFARIEGCGWGWAGVIWLFSVVTYFPLD 840
Query: 839 VMKFAIRYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPESSGIFN 898
++KFAIRYILSGKAW+NLL+NKTAFTTKKDYGKEEREAQWA AQRTLHGLQPPE++ +F+
Sbjct: 841 LLKFAIRYILSGKAWDNLLENKTAFTTKKDYGKEEREAQWAAAQRTLHGLQPPETNNLFS 900
Query: 899 EKSSYRELSEIAEQAKRRAEVARLIPFH 926
EK+SYRELSEIAEQAKRRAEVARL H
Sbjct: 901 EKNSYRELSEIAEQAKRRAEVARLRELH 928
>emb|CAC29436.1| P-type H+-ATPase [Vicia faba]
Length = 951
Score = 1616 bits (4184), Expect = 0.0
Identities = 799/922 (86%), Positives = 870/922 (93%), Gaps = 2/922 (0%)
Query: 7 ISLEQIKNETVDLERIPVEEVFEQLKCTKEGLSSEEGANRLQIFGPNKLEEKKDSKILKF 66
ISLE+IKNE VDLERIPVEEVFEQLKC+KEGLSS+EGANRLQ+FGPNKLEEKK+SK LKF
Sbjct: 4 ISLEEIKNENVDLERIPVEEVFEQLKCSKEGLSSDEGANRLQVFGPNKLEEKKESKFLKF 63
Query: 67 LGFMWNPLSWVMEAAALMAIGLANGNGKPPDWQDFVGIICLLVINSTISFIEENNAGNAA 126
LGFMWNPLSWVMEAAA+MAI LANG+G+PPDWQDFVGII LLVINSTISFIEENNAGNAA
Sbjct: 64 LGFMWNPLSWVMEAAAIMAIALANGSGRPPDWQDFVGIISLLVINSTISFIEENNAGNAA 123
Query: 127 AALMAGLAPKTKVLRDGKWSEQEAAILVPGDIISIKLGDIVPADARLLEGDPLKIDQSAL 186
AALMAGLAPKT+VLRDG+WSE++ AILVPGDIISIKLGDI+PADARLLEGD L +DQSAL
Sbjct: 124 AALMAGLAPKTRVLRDGRWSEEDTAILVPGDIISIKLGDIIPADARLLEGDALSVDQSAL 183
Query: 187 TGESLPVTRNPGDEVYSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQVGHFQKVLT 246
TGESLP T+NP DE +SGST K+GE+EAVVIATGVHTFFGKAAHLVD+TNQVGHFQKVLT
Sbjct: 184 TGESLPATKNPSDESFSGSTVKKGEIEAVVIATGVHTFFGKAAHLVDNTNQVGHFQKVLT 243
Query: 247 AIGNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGS 306
AIGNFCICSIA+G+L E++VMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGS
Sbjct: 244 AIGNFCICSIALGILIELVVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGS 303
Query: 307 HRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDKEHVMLL 366
HRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVF K V+K++V+LL
Sbjct: 304 HRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFAKNVEKDYVILL 363
Query: 367 AARASRVENQDAIDAAIVGTLADPKEARAGVREIHFLPFNPVDKRTALTYIDGNGNWHRA 426
AARASR ENQDAIDAAIVG LA+PKEARAGVREIHF PFNPVDKRTALTYID +GNWHR+
Sbjct: 364 AARASRTENQDAIDAAIVGMLANPKEARAGVREIHFFPFNPVDKRTALTYIDSDGNWHRS 423
Query: 427 SKGAPEQIMDLCKLREDTKRNIHAIIDKFAERGLRSLAVARQEVPEKTKESPGAPWQFVG 486
SKGAPEQI++LC +ED ++ H++IDKFAERGLRSL VARQEVPEK K+SPGAPWQFVG
Sbjct: 424 SKGAPEQILNLCNCKEDVRKKAHSVIDKFAERGLRSLGVARQEVPEKNKDSPGAPWQFVG 483
Query: 487 LLSLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSATLLGQD 546
LL LFDPPRHDSAETI RAL+LGVNVKMITGDQLAIAKETGRRLGMGTNMYPS++LLGQ
Sbjct: 484 LLPLFDPPRHDSAETITRALNLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSSSLLGQS 543
Query: 547 KDANIAALPVEELIEKADGFAGVFPEHKYEIVRKLQERKHICGMTGDGVNDAPALKRADI 606
KDA +AALPV+ELIEKADGFAGVFPEHKYEIV++LQERKHICGMTGDGVNDAPALKRADI
Sbjct: 544 KDAAVAALPVDELIEKADGFAGVFPEHKYEIVKRLQERKHICGMTGDGVNDAPALKRADI 603
Query: 607 GIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFM 666
GIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFM
Sbjct: 604 GIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFM 663
Query: 667 FIALIWKFDFSPFMVLIIAILNDGTIMTISKDRVVPSPLPDSWKLKEIFATGIVLGGYLA 726
FIALIWKFDF+PFMVLIIAILNDGTIMTISKDRV PSPLPDSWKLKEIFATG+VLG Y+A
Sbjct: 664 FIALIWKFDFAPFMVLIIAILNDGTIMTISKDRVKPSPLPDSWKLKEIFATGVVLGSYMA 723
Query: 727 LMTVIFFWAMKENDFFPDKFGVRKL--NHDEMMSALYLQVSIVSQALIFVTRSRGWSFLE 784
LMTV+FFW MK+ DFF DKFGVR + N DEMM+ALYLQVSI+SQALIFVTRSR WSFLE
Sbjct: 724 LMTVVFFWLMKDTDFFSDKFGVRSIRKNPDEMMAALYLQVSIISQALIFVTRSRSWSFLE 783
Query: 785 RPGALLVIAFFIAQLIATIIAVYANWGFAKVQGIGWGWAGVIWLYSIVFYIPLDVMKFAI 844
RPG LL+ AF IAQL+AT IAVYANWGFA+++G+GWGWAGVIW+YS+V Y PLD++KF I
Sbjct: 784 RPGLLLLGAFMIAQLVATFIAVYANWGFARIKGMGWGWAGVIWVYSLVTYFPLDILKFVI 843
Query: 845 RYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPESSGIFNEKSSYR 904
RY+LSGKAW+NLL+NKTAFTTKKDYGKEEREAQWA AQRTLHGLQ PE++ +FN+K+SYR
Sbjct: 844 RYVLSGKAWDNLLENKTAFTTKKDYGKEEREAQWATAQRTLHGLQSPETTNLFNDKNSYR 903
Query: 905 ELSEIAEQAKRRAEVARLIPFH 926
ELSEIAEQAKRRAEVARL H
Sbjct: 904 ELSEIAEQAKRRAEVARLRELH 925
>emb|CAA59799.1| H(+)-transporting ATPase [Phaseolus vulgaris]
gi|1076511|pir||S52728 H+-exporting ATPase (EC 3.6.3.6)
- kidney bean
Length = 951
Score = 1611 bits (4172), Expect = 0.0
Identities = 796/922 (86%), Positives = 872/922 (94%), Gaps = 2/922 (0%)
Query: 7 ISLEQIKNETVDLERIPVEEVFEQLKCTKEGLSSEEGANRLQIFGPNKLEEKKDSKILKF 66
ISLE+IKNE VDLERIP+EEVFEQLKC++ GL+S+EGANRLQ+FGPNKLEEKK+SK LKF
Sbjct: 4 ISLEEIKNENVDLERIPIEEVFEQLKCSRAGLTSDEGANRLQVFGPNKLEEKKESKFLKF 63
Query: 67 LGFMWNPLSWVMEAAALMAIGLANGNGKPPDWQDFVGIICLLVINSTISFIEENNAGNAA 126
LGFMWNPLSWVMEAAA+MAI LANG G+PPDWQDFVGII LLVINSTISFIEENNAGNAA
Sbjct: 64 LGFMWNPLSWVMEAAAIMAIALANGGGRPPDWQDFVGIIALLVINSTISFIEENNAGNAA 123
Query: 127 AALMAGLAPKTKVLRDGKWSEQEAAILVPGDIISIKLGDIVPADARLLEGDPLKIDQSAL 186
AALMAGLAPKTKVLRDG+WSEQ+AAILVPGDIISIKLGDI+ ADARLLEGDPL +DQSAL
Sbjct: 124 AALMAGLAPKTKVLRDGRWSEQDAAILVPGDIISIKLGDIIAADARLLEGDPLSVDQSAL 183
Query: 187 TGESLPVTRNPGDEVYSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQVGHFQKVLT 246
TGESLPVT++ DEV+SGST K+GE+EAVVIATGVHTFFGKAAHLVDSTNQVGHFQKVLT
Sbjct: 184 TGESLPVTKSSSDEVFSGSTVKKGEIEAVVIATGVHTFFGKAAHLVDSTNQVGHFQKVLT 243
Query: 247 AIGNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGS 306
AIGNFCICSIA+G+ E+IVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGS
Sbjct: 244 AIGNFCICSIAIGIAIELIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGS 303
Query: 307 HRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDKEHVMLL 366
HRLSQQGAITKRMTAIEEM GMDVLCSDKTGTLTLNKLSVD+NLIEVF KGV+KE+V+LL
Sbjct: 304 HRLSQQGAITKRMTAIEEMDGMDVLCSDKTGTLTLNKLSVDRNLIEVFAKGVEKEYVILL 363
Query: 367 AARASRVENQDAIDAAIVGTLADPKEARAGVREIHFLPFNPVDKRTALTYIDGNGNWHRA 426
AARASR ENQDAIDAAIVG LADPKEAR+G+RE+HFLPFNPVDKRTALTYID +GNWHR+
Sbjct: 364 AARASRTENQDAIDAAIVGMLADPKEARSGIREVHFLPFNPVDKRTALTYIDSDGNWHRS 423
Query: 427 SKGAPEQIMDLCKLREDTKRNIHAIIDKFAERGLRSLAVARQEVPEKTKESPGAPWQFVG 486
SKGAPEQI+ LC +ED ++ +HA+IDKFAERGLRSL VARQEVPEK+K+ G PWQFVG
Sbjct: 424 SKGAPEQIITLCNCKEDVRKKVHAVIDKFAERGLRSLGVARQEVPEKSKDGAGGPWQFVG 483
Query: 487 LLSLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSATLLGQD 546
LL LFDPPRHDSAETIRRAL+LGVNVKMITGDQLAI KETGRRLGMGTNMYPS+ LLGQD
Sbjct: 484 LLPLFDPPRHDSAETIRRALNLGVNVKMITGDQLAIGKETGRRLGMGTNMYPSSALLGQD 543
Query: 547 KDANIAALPVEELIEKADGFAGVFPEHKYEIVRKLQERKHICGMTGDGVNDAPALKRADI 606
KDA+I+ALPV+ELI+KADGFAGVFPEHKYEIV++LQERKHICGMTGDGVNDAPALK+ADI
Sbjct: 544 KDASISALPVDELIDKADGFAGVFPEHKYEIVKRLQERKHICGMTGDGVNDAPALKKADI 603
Query: 607 GIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFM 666
GIAVADATDAAR ASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGF+
Sbjct: 604 GIAVADATDAARSASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFL 663
Query: 667 FIALIWKFDFSPFMVLIIAILNDGTIMTISKDRVVPSPLPDSWKLKEIFATGIVLGGYLA 726
FIALIWKFDF+PFMVLIIAILNDGTIMTISKDRV PSPLPDSWKL+EIFATG+VLG Y+A
Sbjct: 664 FIALIWKFDFAPFMVLIIAILNDGTIMTISKDRVKPSPLPDSWKLREIFATGVVLGSYMA 723
Query: 727 LMTVIFFWAMKENDFFPDKFGVR--KLNHDEMMSALYLQVSIVSQALIFVTRSRGWSFLE 784
LMTVIFFWAMK+ +FF +KFGVR +L+ +EMM+ALYLQVSI+SQALIFVTRSR WSF E
Sbjct: 724 LMTVIFFWAMKDTNFFSNKFGVRSLRLSPEEMMAALYLQVSIISQALIFVTRSRSWSFAE 783
Query: 785 RPGALLVIAFFIAQLIATIIAVYANWGFAKVQGIGWGWAGVIWLYSIVFYIPLDVMKFAI 844
RPG LL+ AF IAQL+AT IAVYANWGFA+++G+GWGWAGVIWLYS+V YIPLD++KFAI
Sbjct: 784 RPGLLLLGAFLIAQLVATFIAVYANWGFARIKGMGWGWAGVIWLYSVVTYIPLDILKFAI 843
Query: 845 RYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPESSGIFNEKSSYR 904
RYILSGKAW+NLL+NKTAFTTKKDYGKEEREAQWA AQRTLHGLQPPE+S +FN+K+SYR
Sbjct: 844 RYILSGKAWDNLLENKTAFTTKKDYGKEEREAQWAAAQRTLHGLQPPETSNLFNDKNSYR 903
Query: 905 ELSEIAEQAKRRAEVARLIPFH 926
ELSEIAEQAKRRAEVARL H
Sbjct: 904 ELSEIAEQAKRRAEVARLRELH 925
>gb|AAB35314.2| plasma membrane H(+)-ATPase precursor [Vicia faba]
Length = 956
Score = 1608 bits (4165), Expect = 0.0
Identities = 805/924 (87%), Positives = 871/924 (94%), Gaps = 4/924 (0%)
Query: 6 SISLEQIKNETVDLERIPVEEVFEQLKCTKEGLSSEEGANRLQIFGPNKLEEKKDSKILK 65
+ISLEQIKNE+VDLE+IP+EEVF QLKCT+EGLSS EG +R+QIFGPNKLEEKK+SK LK
Sbjct: 3 AISLEQIKNESVDLEKIPIEEVFAQLKCTREGLSSTEGESRIQIFGPNKLEEKKESKFLK 62
Query: 66 FLGFMWNPLSWVMEAAALMAIGLANGNGKPPDWQDFVGIICLLVINSTISFIEENNAGNA 125
FLGFMWNPLSWVMEAAA+MAI LANG G+PPDWQDFVGI+CLLVINSTISFIEENNAGNA
Sbjct: 63 FLGFMWNPLSWVMEAAAVMAIALANGGGQPPDWQDFVGIVCLLVINSTISFIEENNAGNA 122
Query: 126 AAALMAGLAPKTKVLRDGKWSEQEAAILVPGDIISIKLGDIVPADARLLEGDPLKIDQSA 185
AAALMAGLAPKTKVLRDGKWSEQEAAILVPGDIISIKLGDI+PADARLLEGDPLK+DQ+A
Sbjct: 123 AAALMAGLAPKTKVLRDGKWSEQEAAILVPGDIISIKLGDIIPADARLLEGDPLKVDQAA 182
Query: 186 LTGESLPVTRNPGDEVYSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQVGHFQKVL 245
LTGESLPVTR+PG EV+SGSTCKQGE+EAVVIATGVHTFFGKAAHLVD+TN VGHFQ VL
Sbjct: 183 LTGESLPVTRHPGQEVFSGSTCKQGEIEAVVIATGVHTFFGKAAHLVDNTNNVGHFQMVL 242
Query: 246 TAIGNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIG 305
+IGNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIG
Sbjct: 243 KSIGNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIG 302
Query: 306 SHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDKEHVML 365
SH+LSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVD+NLIEVF KG+DKEHV+L
Sbjct: 303 SHKLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDRNLIEVFIKGMDKEHVIL 362
Query: 366 LAARASRVENQDAIDAAIVGTLADPKEARAGVREIHFLPFNPVDKRTALTYIDG-NGNWH 424
LAARA+R ENQDAIDAAIVG LADPKEARA + E+HFLPFNP DKRTALTYID +G WH
Sbjct: 363 LAARAARTENQDAIDAAIVGMLADPKEARAEITEVHFLPFNPNDKRTALTYIDNKDGTWH 422
Query: 425 RASKGAPEQIMDLCKLREDTKRNIHAIIDKFAERGLRSLAVARQEVPEKTKESPGAPWQF 484
RASKGAPEQI++LC +RED ++ IH++I+KFAERGLRSL VARQEVPEKTKES GAPWQF
Sbjct: 423 RASKGAPEQIIELCNMREDAQKKIHSMIEKFAERGLRSLGVARQEVPEKTKESAGAPWQF 482
Query: 485 VGLLSLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSATLLG 544
VGLLS+FDPPRHDSAETIR+AL+LGVNVKMITGDQLAIAKETGRRLGMGTNMYPSATLLG
Sbjct: 483 VGLLSVFDPPRHDSAETIRQALNLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSATLLG 542
Query: 545 QDKDANIAALPVEELIEKADGFAGVFPEHKYEIVRKLQERKHICGMTGDGVNDAPALKRA 604
DKD+++A++PVEELIEKADGFAGVFPEHKYEIV+KLQERKHICGMTGDGVNDAPALK+A
Sbjct: 543 LDKDSSVASMPVEELIEKADGFAGVFPEHKYEIVKKLQERKHICGMTGDGVNDAPALKKA 602
Query: 605 DIGIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFG 664
DIGIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFG
Sbjct: 603 DIGIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFG 662
Query: 665 FMFIALIWKFDFSPFMVLIIAILNDGTIMTISKDRVVPSPLPDSWKLKEIFATGIVLGGY 724
FMFIALIWKFDFSPFM+LIIAILNDGTIMTISKDRV PSPLPDSWKLKEIFATG++LGGY
Sbjct: 663 FMFIALIWKFDFSPFMILIIAILNDGTIMTISKDRVKPSPLPDSWKLKEIFATGVMLGGY 722
Query: 725 LALMTVIFFWAMKENDFFPDKFGVRKL--NHDEMMSALYLQVSIVSQALIFVTRSRGWSF 782
ALMTVIFFW ++ FFPD+FGVR + N DE+ +ALYLQVSIVSQALIFVTRSR
Sbjct: 723 QALMTVIFFWIVQGTKFFPDRFGVRHIHDNPDELTAALYLQVSIVSQALIFVTRSRSGLM 782
Query: 783 LERPGALLVIAFFIAQLIATIIAVYANWGFAKVQGIGWGWAGVIWLYSIVFYIPLDVMKF 842
L PG LL+ AF IAQLIAT+IAVYANW FA++QGIGWGWAGVIWLYSI+FYIPLD++KF
Sbjct: 783 LNAPGLLLLGAFLIAQLIATLIAVYANWAFARIQGIGWGWAGVIWLYSIIFYIPLDIIKF 842
Query: 843 AIRYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPESSGIFNEKSS 902
A RY LSGKAW+N L+NKTAFTTKKDYGK EREAQWAHAQRTLHGL+PPESSGIF+EK+S
Sbjct: 843 ATRYFLSGKAWSN-LENKTAFTTKKDYGKGEREAQWAHAQRTLHGLEPPESSGIFHEKNS 901
Query: 903 YRELSEIAEQAKRRAEVARLIPFH 926
YRELSEIAEQAKRRAEVARL H
Sbjct: 902 YRELSEIAEQAKRRAEVARLRELH 925
>pir||T12087 H+-exporting ATPase (EC 3.6.3.6), plasma membrane - fava bean
Length = 963
Score = 1608 bits (4165), Expect = 0.0
Identities = 805/924 (87%), Positives = 871/924 (94%), Gaps = 4/924 (0%)
Query: 6 SISLEQIKNETVDLERIPVEEVFEQLKCTKEGLSSEEGANRLQIFGPNKLEEKKDSKILK 65
+ISLEQIKNE+VDLE+IP+EEVF QLKCT+EGLSS EG +R+QIFGPNKLEEKK+SK LK
Sbjct: 10 AISLEQIKNESVDLEKIPIEEVFAQLKCTREGLSSTEGESRIQIFGPNKLEEKKESKFLK 69
Query: 66 FLGFMWNPLSWVMEAAALMAIGLANGNGKPPDWQDFVGIICLLVINSTISFIEENNAGNA 125
FLGFMWNPLSWVMEAAA+MAI LANG G+PPDWQDFVGI+CLLVINSTISFIEENNAGNA
Sbjct: 70 FLGFMWNPLSWVMEAAAVMAIALANGGGQPPDWQDFVGIVCLLVINSTISFIEENNAGNA 129
Query: 126 AAALMAGLAPKTKVLRDGKWSEQEAAILVPGDIISIKLGDIVPADARLLEGDPLKIDQSA 185
AAALMAGLAPKTKVLRDGKWSEQEAAILVPGDIISIKLGDI+PADARLLEGDPLK+DQ+A
Sbjct: 130 AAALMAGLAPKTKVLRDGKWSEQEAAILVPGDIISIKLGDIIPADARLLEGDPLKVDQAA 189
Query: 186 LTGESLPVTRNPGDEVYSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQVGHFQKVL 245
LTGESLPVTR+PG EV+SGSTCKQGE+EAVVIATGVHTFFGKAAHLVD+TN VGHFQ VL
Sbjct: 190 LTGESLPVTRHPGQEVFSGSTCKQGEIEAVVIATGVHTFFGKAAHLVDNTNNVGHFQMVL 249
Query: 246 TAIGNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIG 305
+IGNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIG
Sbjct: 250 KSIGNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIG 309
Query: 306 SHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDKEHVML 365
SH+LSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVD+NLIEVF KG+DKEHV+L
Sbjct: 310 SHKLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDRNLIEVFIKGMDKEHVIL 369
Query: 366 LAARASRVENQDAIDAAIVGTLADPKEARAGVREIHFLPFNPVDKRTALTYIDG-NGNWH 424
LAARA+R ENQDAIDAAIVG LADPKEARA + E+HFLPFNP DKRTALTYID +G WH
Sbjct: 370 LAARAARTENQDAIDAAIVGMLADPKEARAEITEVHFLPFNPNDKRTALTYIDNKDGTWH 429
Query: 425 RASKGAPEQIMDLCKLREDTKRNIHAIIDKFAERGLRSLAVARQEVPEKTKESPGAPWQF 484
RASKGAPEQI++LC +RED ++ IH++I+KFAERGLRSL VARQEVPEKTKES GAPWQF
Sbjct: 430 RASKGAPEQIIELCNMREDAQKKIHSMIEKFAERGLRSLGVARQEVPEKTKESAGAPWQF 489
Query: 485 VGLLSLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSATLLG 544
VGLLS+FDPPRHDSAETIR+AL+LGVNVKMITGDQLAIAKETGRRLGMGTNMYPSATLLG
Sbjct: 490 VGLLSVFDPPRHDSAETIRQALNLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSATLLG 549
Query: 545 QDKDANIAALPVEELIEKADGFAGVFPEHKYEIVRKLQERKHICGMTGDGVNDAPALKRA 604
DKD+++A++PVEELIEKADGFAGVFPEHKYEIV+KLQERKHICGMTGDGVNDAPALK+A
Sbjct: 550 LDKDSSVASMPVEELIEKADGFAGVFPEHKYEIVKKLQERKHICGMTGDGVNDAPALKKA 609
Query: 605 DIGIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFG 664
DIGIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFG
Sbjct: 610 DIGIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFG 669
Query: 665 FMFIALIWKFDFSPFMVLIIAILNDGTIMTISKDRVVPSPLPDSWKLKEIFATGIVLGGY 724
FMFIALIWKFDFSPFM+LIIAILNDGTIMTISKDRV PSPLPDSWKLKEIFATG++LGGY
Sbjct: 670 FMFIALIWKFDFSPFMILIIAILNDGTIMTISKDRVKPSPLPDSWKLKEIFATGVMLGGY 729
Query: 725 LALMTVIFFWAMKENDFFPDKFGVRKL--NHDEMMSALYLQVSIVSQALIFVTRSRGWSF 782
ALMTVIFFW ++ FFPD+FGVR + N DE+ +ALYLQVSIVSQALIFVTRSR
Sbjct: 730 QALMTVIFFWIVQGTKFFPDRFGVRHIHDNPDELTAALYLQVSIVSQALIFVTRSRSGLM 789
Query: 783 LERPGALLVIAFFIAQLIATIIAVYANWGFAKVQGIGWGWAGVIWLYSIVFYIPLDVMKF 842
L PG LL+ AF IAQLIAT+IAVYANW FA++QGIGWGWAGVIWLYSI+FYIPLD++KF
Sbjct: 790 LNAPGLLLLGAFLIAQLIATLIAVYANWAFARIQGIGWGWAGVIWLYSIIFYIPLDIIKF 849
Query: 843 AIRYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPESSGIFNEKSS 902
A RY LSGKAW+N L+NKTAFTTKKDYGK EREAQWAHAQRTLHGL+PPESSGIF+EK+S
Sbjct: 850 ATRYFLSGKAWSN-LENKTAFTTKKDYGKGEREAQWAHAQRTLHGLEPPESSGIFHEKNS 908
Query: 903 YRELSEIAEQAKRRAEVARLIPFH 926
YRELSEIAEQAKRRAEVARL H
Sbjct: 909 YRELSEIAEQAKRRAEVARLRELH 932
>ref|XP_474175.1| OSJNBa0071I13.11 [Oryza sativa (japonica cultivar-group)]
gi|39545733|emb|CAE03410.3| OSJNBa0071I13.11 [Oryza
sativa (japonica cultivar-group)]
Length = 951
Score = 1604 bits (4154), Expect = 0.0
Identities = 797/918 (86%), Positives = 861/918 (92%), Gaps = 4/918 (0%)
Query: 9 LEQIKNETVDLERIPVEEVFEQLKCTKEGLSSEEGANRLQIFGPNKLEEKKDSKILKFLG 68
LE+IKNE VDLE IP+EEVFEQLKCT+EGLSSEEG R+++FGPNKLEEKK+SKILKFLG
Sbjct: 4 LEEIKNEAVDLENIPIEEVFEQLKCTREGLSSEEGNRRIEMFGPNKLEEKKESKILKFLG 63
Query: 69 FMWNPLSWVMEAAALMAIGLANGNGKPPDWQDFVGIICLLVINSTISFIEENNAGNAAAA 128
FMWNPLSWVME AA+MAI LANG GKPPDW+DFVGII LLVINSTISFIEENNAGNAAAA
Sbjct: 64 FMWNPLSWVMEMAAIMAIALANGGGKPPDWEDFVGIIVLLVINSTISFIEENNAGNAAAA 123
Query: 129 LMAGLAPKTKVLRDGKWSEQEAAILVPGDIISIKLGDIVPADARLLEGDPLKIDQSALTG 188
LMA LAPKTKVLRDG+W EQEAAILVPGDIISIKLGDIVPADARLLEGDPLKIDQSALTG
Sbjct: 124 LMANLAPKTKVLRDGRWGEQEAAILVPGDIISIKLGDIVPADARLLEGDPLKIDQSALTG 183
Query: 189 ESLPVTRNPGDEVYSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQVGHFQKVLTAI 248
ESLPVT+NPGDEV+SGSTCKQGE+EAVVIATGVHTFFGKAAHLVDSTNQVGHFQ VLTAI
Sbjct: 184 ESLPVTKNPGDEVFSGSTCKQGEIEAVVIATGVHTFFGKAAHLVDSTNQVGHFQTVLTAI 243
Query: 249 GNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGSHR 308
GNFCICSIAVG++ EIIVM+PIQHR YR GI+NLLVLLIGGIPIAMPTVLSVTMAIGSH+
Sbjct: 244 GNFCICSIAVGIVIEIIVMFPIQHRAYRSGIENLLVLLIGGIPIAMPTVLSVTMAIGSHK 303
Query: 309 LSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDKEHVMLLAA 368
LSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNL+EVF KGVDK+HV+LLAA
Sbjct: 304 LSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLVEVFTKGVDKDHVLLLAA 363
Query: 369 RASRVENQDAIDAAIVGTLADPKEARAGVREIHFLPFNPVDKRTALTYIDGNGNWHRASK 428
RASR ENQDAIDAA+VG LADPKEARAG+RE+HFLPFNPVDKRTALTYID +GNWHRASK
Sbjct: 364 RASRTENQDAIDAAMVGMLADPKEARAGIREVHFLPFNPVDKRTALTYIDADGNWHRASK 423
Query: 429 GAPEQIMDLCKLREDTKRNIHAIIDKFAERGLRSLAVARQEVPEKTKESPGAPWQFVGLL 488
GAPEQI+ LC +ED KR +HA+IDK+AERGLRSLAVARQEVPEK+KES G PWQFVGLL
Sbjct: 424 GAPEQILTLCNCKEDVKRKVHAVIDKYAERGLRSLAVARQEVPEKSKESAGGPWQFVGLL 483
Query: 489 SLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSATLLGQDKD 548
LFDPPRHDSAETIR+ALHLGVNVKMITGDQLAI KETGRRLGMGTNMYPS+ LLGQ+KD
Sbjct: 484 PLFDPPRHDSAETIRKALHLGVNVKMITGDQLAIGKETGRRLGMGTNMYPSSALLGQNKD 543
Query: 549 ANIAALPVEELIEKADGFAGVFPEHKYEIVRKLQERKHICGMTGDGVNDAPALKRADIGI 608
A++ ALPV+ELIEKADGFAGVFPEHKYEIV++LQE+KHI GMTGDGVNDAPALK+ADIGI
Sbjct: 544 ASLEALPVDELIEKADGFAGVFPEHKYEIVKRLQEKKHIVGMTGDGVNDAPALKKADIGI 603
Query: 609 AVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFMFI 668
AVADATDAAR ASDIVLTEPGLSVIISAVLTSR IFQRMKNYTIYAVSITIRIV GF+ I
Sbjct: 604 AVADATDAARSASDIVLTEPGLSVIISAVLTSRCIFQRMKNYTIYAVSITIRIVLGFLLI 663
Query: 669 ALIWKFDFSPFMVLIIAILNDGTIMTISKDRVVPSPLPDSWKLKEIFATGIVLGGYLALM 728
ALIWK+DFSPFMVLIIAILNDGTIMTISKDRV PSPLPDSWKLKEIFATGIVLG YLALM
Sbjct: 664 ALIWKYDFSPFMVLIIAILNDGTIMTISKDRVKPSPLPDSWKLKEIFATGIVLGSYLALM 723
Query: 729 TVIFFWAMKENDFFPDKFGVRKLNHD--EMMSALYLQVSIVSQALIFVTRSRGWSFLERP 786
TVIFFWAM + DFF DKFGVR + + EMMSALYLQVSIVSQALIFVTRSR WSF+ERP
Sbjct: 724 TVIFFWAMHKTDFFTDKFGVRSIRNSEHEMMSALYLQVSIVSQALIFVTRSRSWSFIERP 783
Query: 787 GALLVIAFFIAQLIATIIAVYANWGFAKVQGIGWGWAGVIWLYSIVFYIPLDVMKFAIRY 846
G LLV AF +AQL+AT +AVYANWGFA+++GIGWGWAGVIWLYSIVFY PLD+ KF IR+
Sbjct: 784 GLLLVTAFMLAQLVATFLAVYANWGFARIKGIGWGWAGVIWLYSIVFYFPLDIFKFFIRF 843
Query: 847 ILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPE--SSGIFNEKSSYR 904
+LSG+AW+NLL+NK AFTTKKDYG+EEREAQWA AQRTLHGLQPPE S+ +FN+KSSYR
Sbjct: 844 VLSGRAWDNLLENKIAFTTKKDYGREEREAQWATAQRTLHGLQPPEVASNTLFNDKSSYR 903
Query: 905 ELSEIAEQAKRRAEVARL 922
ELSEIAEQAKRRAE+ARL
Sbjct: 904 ELSEIAEQAKRRAEIARL 921
>gb|AAB41898.1| H+-transporting ATPase [Mesembryanthemum crystallinum]
gi|7436370|pir||T12577 H+-exporting ATPase (EC 3.6.3.6)
- common ice plant
Length = 953
Score = 1604 bits (4154), Expect = 0.0
Identities = 791/923 (85%), Positives = 866/923 (93%)
Query: 4 SKSISLEQIKNETVDLERIPVEEVFEQLKCTKEGLSSEEGANRLQIFGPNKLEEKKDSKI 63
+ S+SL++IK+E VDLE+IP+EEVF+ LKC++EGLSS EGANRLQIFGPNKLEEKKDSK
Sbjct: 5 NSSMSLQEIKDEKVDLEKIPIEEVFDSLKCSREGLSSAEGANRLQIFGPNKLEEKKDSKF 64
Query: 64 LKFLGFMWNPLSWVMEAAALMAIGLANGNGKPPDWQDFVGIICLLVINSTISFIEENNAG 123
LKFLGFMWNPLSWVMEAAALMAI LANG+ KPPDWQDFVGII LLVINSTISFIEENNAG
Sbjct: 65 LKFLGFMWNPLSWVMEAAALMAIVLANGDHKPPDWQDFVGIIILLVINSTISFIEENNAG 124
Query: 124 NAAAALMAGLAPKTKVLRDGKWSEQEAAILVPGDIISIKLGDIVPADARLLEGDPLKIDQ 183
NAAAALMA LAPKTKVLRDG+W EQEA+ILVPGDIISIKLGDIVPADARLLEGD LKIDQ
Sbjct: 125 NAAAALMANLAPKTKVLRDGRWGEQEASILVPGDIISIKLGDIVPADARLLEGDALKIDQ 184
Query: 184 SALTGESLPVTRNPGDEVYSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQVGHFQK 243
SALTGES+PVT+NPG+EV+SGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQVGHFQK
Sbjct: 185 SALTGESMPVTKNPGEEVFSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQVGHFQK 244
Query: 244 VLTAIGNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMA 303
VLT+IGNFCICSIA+GML EIIVMYPIQ RKYRDGIDNLLVLLIGGIPIAMPTVLSVTMA
Sbjct: 245 VLTSIGNFCICSIAIGMLIEIIVMYPIQRRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMA 304
Query: 304 IGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDKEHV 363
IGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKL+VD+NL+EVF KGV+KE+V
Sbjct: 305 IGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLTVDRNLVEVFAKGVEKEYV 364
Query: 364 MLLAARASRVENQDAIDAAIVGTLADPKEARAGVREIHFLPFNPVDKRTALTYIDGNGNW 423
+LLAARASR ENQDAIDAAIVG LADPKEARAG+RE+HFLPFNPVDKRTALTYID +GNW
Sbjct: 365 ILLAARASRTENQDAIDAAIVGMLADPKEARAGIREVHFLPFNPVDKRTALTYIDADGNW 424
Query: 424 HRASKGAPEQIMDLCKLREDTKRNIHAIIDKFAERGLRSLAVARQEVPEKTKESPGAPWQ 483
HRASKGAPEQI+ LC+ +ED K+ H +I+KFA+RGLRSLAVARQEVPEKTKESPG PWQ
Sbjct: 425 HRASKGAPEQILTLCRCKEDVKKKAHGVIEKFADRGLRSLAVARQEVPEKTKESPGGPWQ 484
Query: 484 FVGLLSLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSATLL 543
FVGLL LFDPPRHDSAETI+RAL+LGVNVKMITGDQLAI KETGRRLGMGTNMYPS++LL
Sbjct: 485 FVGLLPLFDPPRHDSAETIKRALNLGVNVKMITGDQLAIGKETGRRLGMGTNMYPSSSLL 544
Query: 544 GQDKDANIAALPVEELIEKADGFAGVFPEHKYEIVRKLQERKHICGMTGDGVNDAPALKR 603
GQDKD+N+A LPV+ELIEKADGFAGVFPEHKYEIV++LQ+RKHICGMTGDGVNDAPALKR
Sbjct: 545 GQDKDSNVAGLPVDELIEKADGFAGVFPEHKYEIVKRLQDRKHICGMTGDGVNDAPALKR 604
Query: 604 ADIGIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVF 663
ADIGIAVADATDAAR ASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIR+VF
Sbjct: 605 ADIGIAVADATDAARSASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRVVF 664
Query: 664 GFMFIALIWKFDFSPFMVLIIAILNDGTIMTISKDRVVPSPLPDSWKLKEIFATGIVLGG 723
GFMFIALIWKFDF+PFMVLIIAILNDGTIMTISKDRV PSPLPDSWKLKEIFATGIVLGG
Sbjct: 665 GFMFIALIWKFDFAPFMVLIIAILNDGTIMTISKDRVKPSPLPDSWKLKEIFATGIVLGG 724
Query: 724 YLALMTVIFFWAMKENDFFPDKFGVRKLNHDEMMSALYLQVSIVSQALIFVTRSRGWSFL 783
Y A+MTV+FFW +++ FF DKF V+ L +MM+ALYLQVS +SQALIFVTRSR WSF
Sbjct: 725 YQAIMTVVFFWLVRDTTFFVDKFHVKPLTDGQMMAALYLQVSAISQALIFVTRSRSWSFA 784
Query: 784 ERPGALLVIAFFIAQLIATIIAVYANWGFAKVQGIGWGWAGVIWLYSIVFYIPLDVMKFA 843
ERPG +L+ AF +AQLIAT+IAVYANW FAK++G+GWGWA +W+Y++V YIPLD++KF
Sbjct: 785 ERPGLMLLGAFVVAQLIATLIAVYANWSFAKIEGMGWGWALAVWIYTLVTYIPLDILKFT 844
Query: 844 IRYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPESSGIFNEKSSY 903
IRY LSG+AWNNLLDNKTAFTTKKDYGKEEREAQWA AQRT+HGLQPPE++ +F EKS+Y
Sbjct: 845 IRYALSGRAWNNLLDNKTAFTTKKDYGKEEREAQWAAAQRTMHGLQPPETTNLFPEKSNY 904
Query: 904 RELSEIAEQAKRRAEVARLIPFH 926
RELSEIAEQAKRRAEVARL H
Sbjct: 905 RELSEIAEQAKRRAEVARLRELH 927
>gb|AAY42949.1| plasma membrane H+ ATPase [Lupinus albus]
Length = 951
Score = 1598 bits (4139), Expect = 0.0
Identities = 791/922 (85%), Positives = 867/922 (93%), Gaps = 2/922 (0%)
Query: 7 ISLEQIKNETVDLERIPVEEVFEQLKCTKEGLSSEEGANRLQIFGPNKLEEKKDSKILKF 66
ISLE+IKNE VDLERIPV+EVFEQLKC++EGL+S+EGA+RLQ+FGPNKLEEKK+SK+LKF
Sbjct: 4 ISLEEIKNENVDLERIPVDEVFEQLKCSREGLTSDEGASRLQVFGPNKLEEKKESKLLKF 63
Query: 67 LGFMWNPLSWVMEAAALMAIGLANGNGKPPDWQDFVGIICLLVINSTISFIEENNAGNAA 126
LGFMWNPLSWVMEAAA+MAI LANG+G+PPDWQDFVGII LLVINSTISFIEENNAGNAA
Sbjct: 64 LGFMWNPLSWVMEAAAIMAIALANGDGRPPDWQDFVGIIVLLVINSTISFIEENNAGNAA 123
Query: 127 AALMAGLAPKTKVLRDGKWSEQEAAILVPGDIISIKLGDIVPADARLLEGDPLKIDQSAL 186
AALMAGLAPKTKVLRDG+WSEQ+AAILVPGDIISIKLGDI+PADARLLEGD L +DQSAL
Sbjct: 124 AALMAGLAPKTKVLRDGRWSEQDAAILVPGDIISIKLGDIIPADARLLEGDALSVDQSAL 183
Query: 187 TGESLPVTRNPGDEVYSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQVGHFQKVLT 246
TGESLP T+ P DEV+SGST K+GE+EAVVIATGVHTFFGKAAHLVDSTNQVGHFQKVLT
Sbjct: 184 TGESLPATKKPHDEVFSGSTVKKGEIEAVVIATGVHTFFGKAAHLVDSTNQVGHFQKVLT 243
Query: 247 AIGNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGS 306
AIGNFCICSIAVG++ E+IVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGS
Sbjct: 244 AIGNFCICSIAVGIVIELIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGS 303
Query: 307 HRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDKEHVMLL 366
HRLSQQGAITKRMTAIEEMAGMDVLCSDK GTLTLNKLSVDKNL+EVF KGV+K++V+LL
Sbjct: 304 HRLSQQGAITKRMTAIEEMAGMDVLCSDKAGTLTLNKLSVDKNLVEVFAKGVEKDYVILL 363
Query: 367 AARASRVENQDAIDAAIVGTLADPKEARAGVREIHFLPFNPVDKRTALTYIDGNGNWHRA 426
AARASR ENQDAIDAAIVG LADPKEARAG+RE+HFLPFNPVDKRTALTYID +GNWHR+
Sbjct: 364 AARASRTENQDAIDAAIVGMLADPKEARAGIREVHFLPFNPVDKRTALTYIDSDGNWHRS 423
Query: 427 SKGAPEQIMDLCKLREDTKRNIHAIIDKFAERGLRSLAVARQEVPEKTKESPGAPWQFVG 486
SKGAPEQI++LC +ED ++ HA IDKFAERGLRSL VARQEVPE+TKES GAPWQFVG
Sbjct: 424 SKGAPEQILNLCNCKEDVRKRAHATIDKFAERGLRSLGVARQEVPERTKESLGAPWQFVG 483
Query: 487 LLSLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSATLLGQD 546
LL LFDPPRHDSAETI RAL+LGVNVKMITGDQLAIAKETGRRLGMGTNMYPS++LLGQ
Sbjct: 484 LLPLFDPPRHDSAETITRALNLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSSSLLGQH 543
Query: 547 KDANIAALPVEELIEKADGFAGVFPEHKYEIVRKLQERKHICGMTGDGVNDAPALKRADI 606
KD I +LPV+ELIEKADGFAGVFPEHKYEIV++LQ+RKHICGMTGDGVNDAPALKRADI
Sbjct: 544 KDPAIESLPVDELIEKADGFAGVFPEHKYEIVKRLQDRKHICGMTGDGVNDAPALKRADI 603
Query: 607 GIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFM 666
GIAVADATDAAR ASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFM
Sbjct: 604 GIAVADATDAARSASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFM 663
Query: 667 FIALIWKFDFSPFMVLIIAILNDGTIMTISKDRVVPSPLPDSWKLKEIFATGIVLGGYLA 726
FIAL+W+FDF+PFMVLIIAILNDGTIMTISKDRV PSP+PDSWKLKEIFATG+VLG Y+A
Sbjct: 664 FIALLWRFDFAPFMVLIIAILNDGTIMTISKDRVKPSPMPDSWKLKEIFATGVVLGSYMA 723
Query: 727 LMTVIFFWAMKENDFFPDKFGVRKL--NHDEMMSALYLQVSIVSQALIFVTRSRGWSFLE 784
LMTVIFFW +K+ DFF DKFGVR L N EMM+ALYLQVSI+SQALIFVTRSR WS++E
Sbjct: 724 LMTVIFFWLIKDTDFFSDKFGVRSLRNNPAEMMAALYLQVSIISQALIFVTRSRSWSYVE 783
Query: 785 RPGALLVIAFFIAQLIATIIAVYANWGFAKVQGIGWGWAGVIWLYSIVFYIPLDVMKFAI 844
RPG LL+ AF IAQL+AT +AVYANW FA+++G+GWGWAGVIWLYS+V Y+PLD++KFAI
Sbjct: 784 RPGFLLMGAFLIAQLVATFLAVYANWSFARIKGMGWGWAGVIWLYSLVTYVPLDILKFAI 843
Query: 845 RYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPESSGIFNEKSSYR 904
Y LSGKAWN LL+NKTAFTTKKDYGKEEREAQWA AQRTLHGLQPPE++ +FN+K+SYR
Sbjct: 844 AYALSGKAWNTLLENKTAFTTKKDYGKEEREAQWATAQRTLHGLQPPETTNLFNDKNSYR 903
Query: 905 ELSEIAEQAKRRAEVARLIPFH 926
ELSEIAEQAKRRAEVARL H
Sbjct: 904 ELSEIAEQAKRRAEVARLRELH 925
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.321 0.138 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,543,763,101
Number of Sequences: 2540612
Number of extensions: 66468851
Number of successful extensions: 174419
Number of sequences better than 10.0: 3015
Number of HSP's better than 10.0 without gapping: 2773
Number of HSP's successfully gapped in prelim test: 242
Number of HSP's that attempted gapping in prelim test: 159415
Number of HSP's gapped (non-prelim): 6904
length of query: 932
length of database: 863,360,394
effective HSP length: 138
effective length of query: 794
effective length of database: 512,755,938
effective search space: 407128214772
effective search space used: 407128214772
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 80 (35.4 bits)
Medicago: description of AC125475.9


