

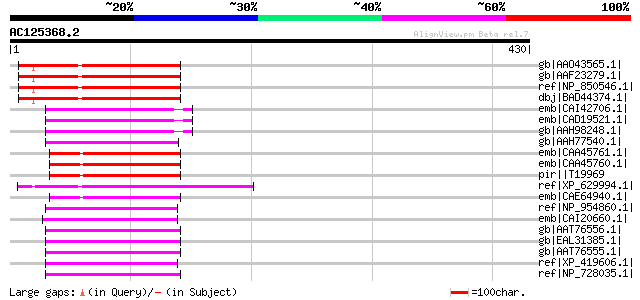
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC125368.2 - phase: 0
(430 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAO43565.1| At3g09470/F11F8_4 [Arabidopsis thaliana] gi|15146... 177 4e-43
gb|AAF23279.1| hypothetical protein [Arabidopsis thaliana] 177 4e-43
ref|NP_850546.1| expressed protein [Arabidopsis thaliana] gi|519... 177 4e-43
dbj|BAD44374.1| unnamed protein product [Arabidopsis thaliana] g... 177 4e-43
emb|CAI42706.1| unc-93 homolog A (C. elegans) [Homo sapiens] 87 1e-15
emb|CAD19521.1| HUNC-93A protein [Homo sapiens] gi|29336077|ref|... 87 1e-15
gb|AAH98248.1| Unknown (protein for MGC:119394) [Homo sapiens] 87 1e-15
gb|AAH77540.1| MGC83353 protein [Xenopus laevis] gi|67462040|sp|... 84 1e-14
emb|CAA45761.1| unc-93 [Caenorhabditis elegans] gi|58081858|emb|... 82 4e-14
emb|CAA45760.1| unc-93 [Caenorhabditis elegans] gi|61855544|emb|... 82 4e-14
pir||T19969 hypothetical protein C46F11.1 - Caenorhabditis elegans 82 4e-14
ref|XP_629994.1| hypothetical protein DDB0184030 [Dictyostelium ... 79 2e-13
emb|CAE64940.1| Hypothetical protein CBG09771 [Caenorhabditis br... 79 2e-13
ref|NP_954860.1| unc-93 homolog A [Mus musculus] gi|40067228|emb... 79 4e-13
emb|CAI20660.1| novel protein similar to vertebrate unc-93 homol... 79 4e-13
gb|AAT76556.1| CG4928 [Drosophila pseudoobscura] 78 5e-13
gb|EAL31385.1| GA18532-PA [Drosophila pseudoobscura] 78 5e-13
gb|AAT76555.1| CG4928 [Drosophila lutescens] 78 6e-13
ref|XP_419606.1| PREDICTED: similar to unc-93 homolog A; unc93 (... 78 6e-13
ref|NP_728035.1| CG4928-PA, isoform A [Drosophila melanogaster] ... 77 8e-13
>gb|AAO43565.1| At3g09470/F11F8_4 [Arabidopsis thaliana] gi|15146218|gb|AAK83592.1|
AT3g09470/F11F8_4 [Arabidopsis thaliana]
gi|67462074|sp|Q94AA1|UN93L_ARATH UNC93-like protein
gi|30680871|ref|NP_187558.2| expressed protein
[Arabidopsis thaliana]
Length = 464
Score = 177 bits (450), Expect = 4e-43
Identities = 89/136 (65%), Positives = 115/136 (84%), Gaps = 4/136 (2%)
Query: 8 HDEETPLVVES--PIQLQPHKSHSRDVHILSIAFVLIFLAYGAAQNLQSTLNTEEDLGTT 65
+DEE PL+ S +++ K ++RDVHILSI+F+LIFLAYGAAQNL++T+N +DLGT
Sbjct: 5 NDEEAPLISASGEDRKVRAGKCYTRDVHILSISFLLIFLAYGAAQNLETTVN--KDLGTI 62
Query: 66 SLGILYLSFTFFSVFASLVVRILGSKNALIIGTSGYWLYVAANLKPNWYTLVPASVYLGF 125
SLGILY+SF F S+ ASLVVR++GSKNAL++GT+GYWL+VAANLKP+W+T+VPAS+YLGF
Sbjct: 63 SLGILYVSFMFCSMVASLVVRLMGSKNALVLGTTGYWLFVAANLKPSWFTMVPASLYLGF 122
Query: 126 CASIIWVGQITISTKV 141
ASIIWVGQ T T +
Sbjct: 123 AASIIWVGQGTYLTSI 138
>gb|AAF23279.1| hypothetical protein [Arabidopsis thaliana]
Length = 285
Score = 177 bits (450), Expect = 4e-43
Identities = 89/136 (65%), Positives = 115/136 (84%), Gaps = 4/136 (2%)
Query: 8 HDEETPLVVES--PIQLQPHKSHSRDVHILSIAFVLIFLAYGAAQNLQSTLNTEEDLGTT 65
+DEE PL+ S +++ K ++RDVHILSI+F+LIFLAYGAAQNL++T+N +DLGT
Sbjct: 5 NDEEAPLISASGEDRKVRAGKCYTRDVHILSISFLLIFLAYGAAQNLETTVN--KDLGTI 62
Query: 66 SLGILYLSFTFFSVFASLVVRILGSKNALIIGTSGYWLYVAANLKPNWYTLVPASVYLGF 125
SLGILY+SF F S+ ASLVVR++GSKNAL++GT+GYWL+VAANLKP+W+T+VPAS+YLGF
Sbjct: 63 SLGILYVSFMFCSMVASLVVRLMGSKNALVLGTTGYWLFVAANLKPSWFTMVPASLYLGF 122
Query: 126 CASIIWVGQITISTKV 141
ASIIWVGQ T T +
Sbjct: 123 AASIIWVGQGTYLTSI 138
>ref|NP_850546.1| expressed protein [Arabidopsis thaliana]
gi|51971813|dbj|BAD44571.1| unnamed protein product
[Arabidopsis thaliana]
Length = 437
Score = 177 bits (450), Expect = 4e-43
Identities = 89/136 (65%), Positives = 115/136 (84%), Gaps = 4/136 (2%)
Query: 8 HDEETPLVVES--PIQLQPHKSHSRDVHILSIAFVLIFLAYGAAQNLQSTLNTEEDLGTT 65
+DEE PL+ S +++ K ++RDVHILSI+F+LIFLAYGAAQNL++T+N +DLGT
Sbjct: 5 NDEEAPLISASGEDRKVRAGKCYTRDVHILSISFLLIFLAYGAAQNLETTVN--KDLGTI 62
Query: 66 SLGILYLSFTFFSVFASLVVRILGSKNALIIGTSGYWLYVAANLKPNWYTLVPASVYLGF 125
SLGILY+SF F S+ ASLVVR++GSKNAL++GT+GYWL+VAANLKP+W+T+VPAS+YLGF
Sbjct: 63 SLGILYVSFMFCSMVASLVVRLMGSKNALVLGTTGYWLFVAANLKPSWFTMVPASLYLGF 122
Query: 126 CASIIWVGQITISTKV 141
ASIIWVGQ T T +
Sbjct: 123 AASIIWVGQGTYLTSI 138
>dbj|BAD44374.1| unnamed protein product [Arabidopsis thaliana]
gi|51971130|dbj|BAD44257.1| unnamed protein product
[Arabidopsis thaliana]
Length = 437
Score = 177 bits (450), Expect = 4e-43
Identities = 89/136 (65%), Positives = 115/136 (84%), Gaps = 4/136 (2%)
Query: 8 HDEETPLVVES--PIQLQPHKSHSRDVHILSIAFVLIFLAYGAAQNLQSTLNTEEDLGTT 65
+DEE PL+ S +++ K ++RDVHILSI+F+LIFLAYGAAQNL++T+N +DLGT
Sbjct: 5 NDEEAPLISASGEDRKVRAGKCYTRDVHILSISFLLIFLAYGAAQNLETTVN--KDLGTI 62
Query: 66 SLGILYLSFTFFSVFASLVVRILGSKNALIIGTSGYWLYVAANLKPNWYTLVPASVYLGF 125
SLGILY+SF F S+ ASLVVR++GSKNAL++GT+GYWL+VAANLKP+W+T+VPAS+YLGF
Sbjct: 63 SLGILYVSFMFCSMVASLVVRLMGSKNALVLGTTGYWLFVAANLKPSWFTMVPASLYLGF 122
Query: 126 CASIIWVGQITISTKV 141
ASIIWVGQ T T +
Sbjct: 123 AASIIWVGQGTYLTSI 138
>emb|CAI42706.1| unc-93 homolog A (C. elegans) [Homo sapiens]
Length = 452
Score = 87.0 bits (214), Expect = 1e-15
Identities = 48/123 (39%), Positives = 70/123 (56%), Gaps = 8/123 (6%)
Query: 30 RDVHILSIAFVLIFLAYGAAQNLQSTLNTEEDLGTTSLGILYLSFTFFSVF-ASLVVRIL 88
R+V ++S F+L+F AYG Q+LQS+L +EE LG T+L LY S+F L++ L
Sbjct: 6 RNVLVVSFGFLLLFTAYGGLQSLQSSLYSEEGLGVTALSTLYGGMLLSSMFLPPLLIERL 65
Query: 89 GSKNALIIGTSGYWLYVAANLKPNWYTLVPASVYLGFCASIIWVGQITISTKVL*LVTST 148
G K +I+ GY + N +WYTL+P S+ LG A+ +W Q T +T T
Sbjct: 66 GCKGTIILSMCGYVAFSVGNFFASWYTLIPTSILLGLGAAPLWSAQCT-------YLTIT 118
Query: 149 GNS 151
GN+
Sbjct: 119 GNT 121
>emb|CAD19521.1| HUNC-93A protein [Homo sapiens] gi|29336077|ref|NP_061847.2| unc-93
homolog A [Homo sapiens]
gi|67462066|sp|Q86WB7|UN93A_HUMAN UNC93 homolog A
(UNC-93A protein) (HmUNC-93A)
gi|29170388|emb|CAD48541.1| UNC93A protein [Homo
sapiens]
Length = 457
Score = 87.0 bits (214), Expect = 1e-15
Identities = 48/123 (39%), Positives = 70/123 (56%), Gaps = 8/123 (6%)
Query: 30 RDVHILSIAFVLIFLAYGAAQNLQSTLNTEEDLGTTSLGILYLSFTFFSVF-ASLVVRIL 88
R+V ++S F+L+F AYG Q+LQS+L +EE LG T+L LY S+F L++ L
Sbjct: 6 RNVLVVSFGFLLLFTAYGGLQSLQSSLYSEEGLGVTALSTLYGGMLLSSMFLPPLLIERL 65
Query: 89 GSKNALIIGTSGYWLYVAANLKPNWYTLVPASVYLGFCASIIWVGQITISTKVL*LVTST 148
G K +I+ GY + N +WYTL+P S+ LG A+ +W Q T +T T
Sbjct: 66 GCKGTIILSMCGYVAFSVGNFFASWYTLIPTSILLGLGAAPLWSAQCT-------YLTIT 118
Query: 149 GNS 151
GN+
Sbjct: 119 GNT 121
>gb|AAH98248.1| Unknown (protein for MGC:119394) [Homo sapiens]
Length = 415
Score = 87.0 bits (214), Expect = 1e-15
Identities = 48/123 (39%), Positives = 70/123 (56%), Gaps = 8/123 (6%)
Query: 30 RDVHILSIAFVLIFLAYGAAQNLQSTLNTEEDLGTTSLGILYLSFTFFSVF-ASLVVRIL 88
R+V ++S F+L+F AYG Q+LQS+L +EE LG T+L LY S+F L++ L
Sbjct: 6 RNVLVVSFGFLLLFTAYGGLQSLQSSLYSEEGLGVTALSTLYGGMLLSSMFLPPLLIERL 65
Query: 89 GSKNALIIGTSGYWLYVAANLKPNWYTLVPASVYLGFCASIIWVGQITISTKVL*LVTST 148
G K +I+ GY + N +WYTL+P S+ LG A+ +W Q T +T T
Sbjct: 66 GCKGTIILSMCGYVAFSVGNFFASWYTLIPTSILLGLGAAPLWSAQCT-------YLTIT 118
Query: 149 GNS 151
GN+
Sbjct: 119 GNT 121
>gb|AAH77540.1| MGC83353 protein [Xenopus laevis] gi|67462040|sp|Q6DDL7|UN93A_XENLA
UNC93 homolog A
Length = 460
Score = 83.6 bits (205), Expect = 1e-14
Identities = 40/112 (35%), Positives = 67/112 (59%), Gaps = 1/112 (0%)
Query: 30 RDVHILSIAFVLIFLAYGAAQNLQSTLNTEEDLGTTSLGILYLSFTFFSVFA-SLVVRIL 88
+++ I+S F+L+F A+G Q+LQS+LN++E LG SL ++Y + SVF +V++ +
Sbjct: 5 KNILIVSFGFLLLFTAFGGLQSLQSSLNSDEGLGVASLSVIYAALIVSSVFVPPIVIKKI 64
Query: 89 GSKNALIIGTSGYWLYVAANLKPNWYTLVPASVYLGFCASIIWVGQITISTK 140
G K ++ Y Y N +WYTL+P S+ LGF + +W + T T+
Sbjct: 65 GCKWTIVASMCCYITYSLGNFYASWYTLIPTSLILGFGGAPLWAAKCTYLTE 116
>emb|CAA45761.1| unc-93 [Caenorhabditis elegans] gi|58081858|emb|CAB03760.3|
Hypothetical protein C46F11.1b [Caenorhabditis elegans]
Length = 700
Score = 81.6 bits (200), Expect = 4e-14
Identities = 43/109 (39%), Positives = 67/109 (61%), Gaps = 3/109 (2%)
Query: 34 ILSIAFVLIFLAYGAAQNLQSTLNTEEDLGTTSLGILYLSFTFFSVFA-SLVVRILGSKN 92
ILS+AF+ +F A+ QNLQ+++N DLG+ SL LYLS S+F S ++ LG K
Sbjct: 247 ILSVAFLFLFTAFNGLQNLQTSVNG--DLGSDSLVALYLSLAISSLFVPSFMINRLGCKL 304
Query: 93 ALIIGTSGYWLYVAANLKPNWYTLVPASVYLGFCASIIWVGQITISTKV 141
+I Y+LY+ NL+P + +++PAS++ G AS IW + T++
Sbjct: 305 TFLIAIFVYFLYIVINLRPTYSSMIPASIFCGIAASCIWGAKCAYITEM 353
>emb|CAA45760.1| unc-93 [Caenorhabditis elegans] gi|61855544|emb|CAI70399.1|
Hypothetical protein C46F11.1a [Caenorhabditis elegans]
gi|62511220|sp|Q93380|UNC93_CAEEL Putative potassium
channel regulatory protein unc-93 (Uncoordinated protein
93) gi|25144062|ref|NP_497738.2| UNCoordinated
locomotion UNC-93, putative protein, with at least 11
transmembrane domains, of eukaryotic origin (unc-93)
[Caenorhabditis elegans] gi|102475|pir||S23352 gene
unc-93 protein 1 - Caenorhabditis elegans
Length = 705
Score = 81.6 bits (200), Expect = 4e-14
Identities = 43/109 (39%), Positives = 67/109 (61%), Gaps = 3/109 (2%)
Query: 34 ILSIAFVLIFLAYGAAQNLQSTLNTEEDLGTTSLGILYLSFTFFSVFA-SLVVRILGSKN 92
ILS+AF+ +F A+ QNLQ+++N DLG+ SL LYLS S+F S ++ LG K
Sbjct: 252 ILSVAFLFLFTAFNGLQNLQTSVNG--DLGSDSLVALYLSLAISSLFVPSFMINRLGCKL 309
Query: 93 ALIIGTSGYWLYVAANLKPNWYTLVPASVYLGFCASIIWVGQITISTKV 141
+I Y+LY+ NL+P + +++PAS++ G AS IW + T++
Sbjct: 310 TFLIAIFVYFLYIVINLRPTYSSMIPASIFCGIAASCIWGAKCAYITEM 358
>pir||T19969 hypothetical protein C46F11.1 - Caenorhabditis elegans
Length = 708
Score = 81.6 bits (200), Expect = 4e-14
Identities = 43/109 (39%), Positives = 67/109 (61%), Gaps = 3/109 (2%)
Query: 34 ILSIAFVLIFLAYGAAQNLQSTLNTEEDLGTTSLGILYLSFTFFSVFA-SLVVRILGSKN 92
ILS+AF+ +F A+ QNLQ+++N DLG+ SL LYLS S+F S ++ LG K
Sbjct: 252 ILSVAFLFLFTAFNGLQNLQTSVNG--DLGSDSLVALYLSLAISSLFVPSFMINRLGCKL 309
Query: 93 ALIIGTSGYWLYVAANLKPNWYTLVPASVYLGFCASIIWVGQITISTKV 141
+I Y+LY+ NL+P + +++PAS++ G AS IW + T++
Sbjct: 310 TFLIAIFVYFLYIVINLRPTYSSMIPASIFCGIAASCIWGAKCAYITEM 358
>ref|XP_629994.1| hypothetical protein DDB0184030 [Dictyostelium discoideum]
gi|60463369|gb|EAL61557.1| hypothetical protein
DDB0184030 [Dictyostelium discoideum]
Length = 447
Score = 79.3 bits (194), Expect = 2e-13
Identities = 52/197 (26%), Positives = 100/197 (50%), Gaps = 4/197 (2%)
Query: 7 HHDEETPLVVESPIQLQPHKSHSRDVHILSIAFVLIFLAYGAAQNLQSTLNTEEDLGTTS 66
HH EE S +Q ++ ++ IL + F ++F A+ Q LQ+T+N ++LG +
Sbjct: 17 HHQEENDYYSHSK-SIQNKNNNKINIIILGLCFCILFSAFSPTQILQTTIN--QNLGYYT 73
Query: 67 LGILYLSFTFFSVFASLVVRILGSKNALIIGTSGYWLYVAANLKPNWYTLVPASVYLGFC 126
L +LY S + + + +V G K +LIIGT Y +Y+ +N+ +L +S+ +GF
Sbjct: 74 LAVLYSSLSISNFISPFIVSKFGEKISLIIGTLSYAIYIGSNIYVTQPSLYISSILVGFG 133
Query: 127 ASIIWVGQITISTKVL*LVTSTGNSGLFMHSISLLEILLHL-LYSVMGRGEVPTAPLYCL 185
+I+W Q ++ K T N+GLF +I+ ++ +++ + + + L+ +
Sbjct: 134 GAILWNAQGSLIIKYSTEETIGANTGLFFALFQTDQIIGNIGSATLINKAGLSNSILFTI 193
Query: 186 LYSFSL*PLVQY*CSSC 202
SL P++ + C
Sbjct: 194 FMGISLMPIIGFLFLKC 210
>emb|CAE64940.1| Hypothetical protein CBG09771 [Caenorhabditis briggsae]
Length = 716
Score = 79.3 bits (194), Expect = 2e-13
Identities = 41/109 (37%), Positives = 66/109 (59%), Gaps = 3/109 (2%)
Query: 34 ILSIAFVLIFLAYGAAQNLQSTLNTEEDLGTTSLGILYLSFTFFSVFA-SLVVRILGSKN 92
ILS+A + +F A+ QNLQ+++N DLG+ SL LYLS S+F S ++ LG K
Sbjct: 247 ILSVALLFLFTAFNGLQNLQTSVNG--DLGSDSLVALYLSLAISSLFVPSFMINRLGCKM 304
Query: 93 ALIIGTSGYWLYVAANLKPNWYTLVPASVYLGFCASIIWVGQITISTKV 141
++ Y+LY+ NL+P + +++PAS++ G AS IW + T++
Sbjct: 305 TFLVAIFVYFLYIVINLRPTYSSMIPASIFCGIAASCIWGAKCAYITEM 353
>ref|NP_954860.1| unc-93 homolog A [Mus musculus] gi|40067228|emb|CAD19523.1| UNC-93A
protein [Mus musculus] gi|67462048|sp|Q710D3|UN93A_MOUSE
UNC93 homolog A (UNC-93A protein)
Length = 458
Score = 78.6 bits (192), Expect = 4e-13
Identities = 41/111 (36%), Positives = 64/111 (56%), Gaps = 1/111 (0%)
Query: 30 RDVHILSIAFVLIFLAYGAAQNLQSTLNTEEDLGTTSLGILYLSFTFFSVF-ASLVVRIL 88
++V ++S F+L+F AYG QNLQS+L +E+ LG +L LY S S+F ++++
Sbjct: 6 KNVLVVSCGFLLLFTAYGGLQNLQSSLYSEQGLGVATLSTLYASVLLSSMFLPPILIKKC 65
Query: 89 GSKNALIIGTSGYWLYVAANLKPNWYTLVPASVYLGFCASIIWVGQITIST 139
G K ++ Y ++ N NWYTL+P S+ LG A+ +W Q T T
Sbjct: 66 GCKWTIVGSMCCYVVFSLGNFHANWYTLIPTSILLGLGAAPLWSAQGTYLT 116
>emb|CAI20660.1| novel protein similar to vertebrate unc-93 homolog A (C. elegans)
(UNC93A) [Danio rerio] gi|67462027|sp|Q5SPF7|UN93A_BRARE
UNC93 homolog A
Length = 465
Score = 78.6 bits (192), Expect = 4e-13
Identities = 39/113 (34%), Positives = 65/113 (57%), Gaps = 1/113 (0%)
Query: 28 HSRDVHILSIAFVLIFLAYGAAQNLQSTLNTEEDLGTTSLGILYLSFTFFSVF-ASLVVR 86
++++V ++S F+L+F AYG Q+LQS+LN EE +G SL ++Y + S+F ++++
Sbjct: 4 NTKNVLVVSFGFLLLFTAYGGLQSLQSSLNAEEGMGVISLSVIYAAIILSSMFLPPIMIK 63
Query: 87 ILGSKNALIIGTSGYWLYVAANLKPNWYTLVPASVYLGFCASIIWVGQITIST 139
LG K +++ Y Y NL P W +L+ S LG S +W + T T
Sbjct: 64 NLGCKWTIVVSMGCYVAYSFGNLAPGWASLMSTSAILGMGGSPLWSAKCTYLT 116
>gb|AAT76556.1| CG4928 [Drosophila pseudoobscura]
Length = 514
Score = 78.2 bits (191), Expect = 5e-13
Identities = 41/113 (36%), Positives = 68/113 (59%), Gaps = 1/113 (0%)
Query: 30 RDVHILSIAFVLIFLAYGAAQNLQSTLNTEEDLGTTSLGILYLSFTFFSVF-ASLVVRIL 88
+++ I+SIAF++ F A+ NLQS++N+++ LGT SL +Y + +F +L++R L
Sbjct: 35 KNISIISIAFMVQFTAFQGTANLQSSINSKDGLGTVSLSAIYAALVVSCIFLPTLIIRKL 94
Query: 89 GSKNALIIGTSGYWLYVAANLKPNWYTLVPASVYLGFCASIIWVGQITISTKV 141
K L+ Y Y+A L P +YTLVPA + +G A+ +W + T T+V
Sbjct: 95 TVKWTLVFSMLCYAPYIAFQLFPRFYTLVPAGILVGMGAAPMWASKATYLTQV 147
>gb|EAL31385.1| GA18532-PA [Drosophila pseudoobscura]
Length = 538
Score = 78.2 bits (191), Expect = 5e-13
Identities = 41/113 (36%), Positives = 68/113 (59%), Gaps = 1/113 (0%)
Query: 30 RDVHILSIAFVLIFLAYGAAQNLQSTLNTEEDLGTTSLGILYLSFTFFSVF-ASLVVRIL 88
+++ I+SIAF++ F A+ NLQS++N+++ LGT SL +Y + +F +L++R L
Sbjct: 44 KNISIISIAFMVQFTAFQGTANLQSSINSKDGLGTVSLSAIYAALVVSCIFLPTLIIRKL 103
Query: 89 GSKNALIIGTSGYWLYVAANLKPNWYTLVPASVYLGFCASIIWVGQITISTKV 141
K L+ Y Y+A L P +YTLVPA + +G A+ +W + T T+V
Sbjct: 104 TVKWTLVFSMLCYAPYIAFQLFPRFYTLVPAGILVGMGAAPMWASKATYLTQV 156
>gb|AAT76555.1| CG4928 [Drosophila lutescens]
Length = 515
Score = 77.8 bits (190), Expect = 6e-13
Identities = 41/113 (36%), Positives = 67/113 (59%), Gaps = 1/113 (0%)
Query: 30 RDVHILSIAFVLIFLAYGAAQNLQSTLNTEEDLGTTSLGILYLSFTFFSVF-ASLVVRIL 88
+++ I+SIAF++ F A+ NLQS++N ++ LGT SL +Y + +F +L++R L
Sbjct: 35 KNISIISIAFMVQFTAFQGTANLQSSINAQDGLGTVSLSAIYAALVVSCIFLPTLIIRKL 94
Query: 89 GSKNALIIGTSGYWLYVAANLKPNWYTLVPASVYLGFCASIIWVGQITISTKV 141
K L+ Y Y+A L P +YTLVPA + +G A+ +W + T T+V
Sbjct: 95 TVKWTLVCSMLCYAPYIAFQLFPRFYTLVPAGILVGMGAAPMWASKATYLTQV 147
>ref|XP_419606.1| PREDICTED: similar to unc-93 homolog A; unc93 (C.elegans) homolog A
[Gallus gallus]
Length = 458
Score = 77.8 bits (190), Expect = 6e-13
Identities = 40/111 (36%), Positives = 64/111 (57%), Gaps = 1/111 (0%)
Query: 30 RDVHILSIAFVLIFLAYGAAQNLQSTLNTEEDLGTTSLGILYLSFTFFSVF-ASLVVRIL 88
++V +++ F+L+F AYG Q+LQS+LN+EE LG SL +LY + S+F ++++ L
Sbjct: 6 KNVLVIAFGFLLLFTAYGGLQSLQSSLNSEEGLGVASLSVLYAALIVSSMFLPPILIKKL 65
Query: 89 GSKNALIIGTSGYWLYVAANLKPNWYTLVPASVYLGFCASIIWVGQITIST 139
G K + Y + N +WYTL+P SV LG + +W + T T
Sbjct: 66 GCKWTIAGSMCCYIAFSLGNFYASWYTLIPTSVILGLGGAPLWSAKCTYLT 116
>ref|NP_728035.1| CG4928-PA, isoform A [Drosophila melanogaster]
gi|18859663|ref|NP_573179.1| CG4928-PB, isoform B
[Drosophila melanogaster] gi|7293301|gb|AAF48682.1|
CG4928-PB, isoform B [Drosophila melanogaster]
gi|22832423|gb|AAN09429.1| CG4928-PA, isoform A
[Drosophila melanogaster]
gi|67462083|sp|Q9Y115|UN93L_DROME UNC93-like protein
gi|5052604|gb|AAD38632.1| BcDNA.GH10120 [Drosophila
melanogaster]
Length = 538
Score = 77.4 bits (189), Expect = 8e-13
Identities = 41/113 (36%), Positives = 67/113 (59%), Gaps = 1/113 (0%)
Query: 30 RDVHILSIAFVLIFLAYGAAQNLQSTLNTEEDLGTTSLGILYLSFTFFSVF-ASLVVRIL 88
+++ I+SIAF++ F A+ NLQS++N ++ LGT SL +Y + +F +L++R L
Sbjct: 44 KNISIISIAFMVQFTAFQGTANLQSSINAKDGLGTVSLSAIYAALVVSCIFLPTLIIRKL 103
Query: 89 GSKNALIIGTSGYWLYVAANLKPNWYTLVPASVYLGFCASIIWVGQITISTKV 141
K L+ Y Y+A L P +YTLVPA + +G A+ +W + T T+V
Sbjct: 104 TVKWTLVCSMLCYAPYIAFQLFPRFYTLVPAGILVGMGAAPMWASKATYLTQV 156
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.344 0.150 0.524
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 682,591,926
Number of Sequences: 2540612
Number of extensions: 25619262
Number of successful extensions: 98488
Number of sequences better than 10.0: 118
Number of HSP's better than 10.0 without gapping: 88
Number of HSP's successfully gapped in prelim test: 30
Number of HSP's that attempted gapping in prelim test: 98315
Number of HSP's gapped (non-prelim): 147
length of query: 430
length of database: 863,360,394
effective HSP length: 131
effective length of query: 299
effective length of database: 530,540,222
effective search space: 158631526378
effective search space used: 158631526378
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.6 bits)
S2: 77 (34.3 bits)
Medicago: description of AC125368.2


