

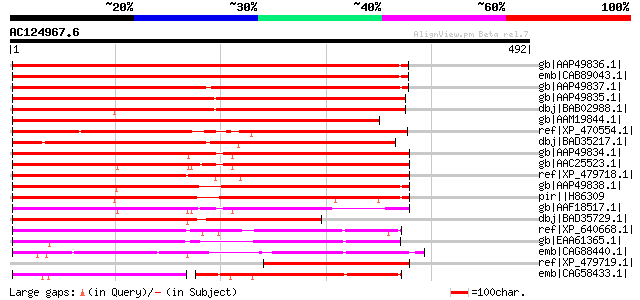
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124967.6 - phase: 0 /pseudo
(492 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAP49836.1| SAC domain protein 3 [Arabidopsis thaliana] gi|22... 529 e-149
emb|CAB89043.1| putative protein [Arabidopsis thaliana] gi|11357... 529 e-149
gb|AAP49837.1| SAC domain protein 4 [Arabidopsis thaliana] gi|30... 517 e-145
gb|AAP49835.1| SAC domain protein 2 [Arabidopsis thaliana] gi|20... 514 e-144
dbj|BAB02988.1| unnamed protein product [Arabidopsis thaliana] 507 e-142
gb|AAM19844.1| AT3g43220/F7K15_70 [Arabidopsis thaliana] 503 e-141
ref|XP_470554.1| Putative phosphoinositide phosphatase [Oryza sa... 452 e-125
dbj|BAD35217.1| putative Sac domain-containing inositol phosphat... 451 e-125
gb|AAP49834.1| SAC domain protein 1 [Arabidopsis thaliana] gi|22... 421 e-116
gb|AAC25523.1| Similar to hypothetical protein C34B7.2 gb|172950... 414 e-114
ref|XP_479718.1| putative sac domain-containing inositol phospha... 400 e-110
gb|AAP49838.1| SAC domain protein 5 [Arabidopsis thaliana] gi|20... 367 e-100
pir||H86309 F28G4.21 protein - Arabidopsis thaliana gi|9665125|g... 354 4e-96
gb|AAF18517.1| Hypothetical protein [Arabidopsis thaliana] 320 5e-86
dbj|BAD35729.1| Sac domain-containing inositol phosphatase-like ... 272 2e-71
ref|XP_640668.1| hypothetical protein DDB0205575 [Dictyostelium ... 254 3e-66
gb|EAA61365.1| hypothetical protein AN7314.2 [Aspergillus nidula... 248 3e-64
emb|CAG88440.1| unnamed protein product [Debaryomyces hansenii C... 211 5e-53
ref|XP_479719.1| sac domain-containing inositol phosphatase 3-li... 189 1e-46
emb|CAG58433.1| unnamed protein product [Candida glabrata CBS138... 142 2e-32
>gb|AAP49836.1| SAC domain protein 3 [Arabidopsis thaliana]
gi|22331537|ref|NP_189908.2| phosphoinositide
phosphatase family protein [Arabidopsis thaliana]
Length = 818
Score = 529 bits (1362), Expect = e-149
Identities = 263/376 (69%), Positives = 303/376 (79%), Gaps = 1/376 (0%)
Query: 3 RYLRRGVNEKGRVANDVETEQIVFEDVPEGLPIKISSVIQNRGSIPLFWSQETSRLNIKP 62
RYL+RGVN G VANDVETEQIV EDVPE P++ISSV+QNRGSIPLFWSQETSRLN+KP
Sbjct: 248 RYLKRGVNRNGDVANDVETEQIVSEDVPEDHPMQISSVVQNRGSIPLFWSQETSRLNLKP 307
Query: 63 DIILSKKDQSYQATKLHFENLVKRYGNPIIILNLIKTHEKKPREAILRQEFANAIDFINK 122
DI+LSKK+ +Y+AT+LHF+NLV+RYGNPIIILNLIKT E++PRE+ILR+EF NAIDFINK
Sbjct: 308 DIVLSKKEPNYEATRLHFDNLVERYGNPIIILNLIKTKERRPRESILREEFVNAIDFINK 367
Query: 123 DLSEENRLRFLHWDLHKHFQSKATNVLLLLGKVAAYALTLTGFFYCQVSSTLRPEDCLKW 182
DL EENRLRFLHWDLHKHF+SK NVL LL KVA AL LT FY QV+ + ED +
Sbjct: 368 DLPEENRLRFLHWDLHKHFRSKTKNVLALLCKVATCALMLTDLFYYQVTPAMTIEDSMSL 427
Query: 183 PSTDDVDKGSFSPTVHVEYDNEDANDLERKPSDEINVSNENHSAKPPRLQNGVLRTNCID 242
S+ D D G SP + DN D + LE+K S N++ KPPRLQ+GVLRTNCID
Sbjct: 428 SSSSDADTGDISPHTSSDDDNGDHDSLEKKSSRSKNIAYGKCDVKPPRLQSGVLRTNCID 487
Query: 243 CLDRTNVAQYAYGLAAIGHQLHSLGIIEHPKIDLDDPVANDLMQFYERMGDTLAHQYGGS 302
CLDRTNVAQYAYG AA+G QLH LGI + P I+LDDP+A LM YERMGDTLAHQYGGS
Sbjct: 488 CLDRTNVAQYAYGWAALGQQLHVLGIRDVPAIELDDPLAISLMGLYERMGDTLAHQYGGS 547
Query: 303 AAHNKIFSARRGQWRAATQSQEFFRTLQRYYSNAWMDAEKQDAINVFLGHFQPQQDKPAL 362
AAHNK+FS RRGQWRAATQSQEF RTLQRYY+NA+MDA+KQDAIN+FLG FQP+Q PA+
Sbjct: 548 AAHNKVFSERRGQWRAATQSQEFLRTLQRYYNNAYMDADKQDAINIFLGTFQPEQGMPAI 607
Query: 363 WELGSDQHYDTGRIGD 378
WEL S+ GR G+
Sbjct: 608 WELRSNS-LSNGRNGE 622
Score = 34.7 bits (78), Expect = 7.0
Identities = 16/32 (50%), Positives = 20/32 (62%), Gaps = 1/32 (3%)
Query: 456 SHHIYYSEHGDSFSCSNFVDLDWLSSSANSCE 487
S +Y+ D S SNFVD++WLSS N CE
Sbjct: 715 SDQVYFGGEEDMSSVSNFVDIEWLSSE-NPCE 745
>emb|CAB89043.1| putative protein [Arabidopsis thaliana] gi|11357847|pir||T49236
hypothetical protein F7K15.70 - Arabidopsis thaliana
Length = 794
Score = 529 bits (1362), Expect = e-149
Identities = 263/376 (69%), Positives = 303/376 (79%), Gaps = 1/376 (0%)
Query: 3 RYLRRGVNEKGRVANDVETEQIVFEDVPEGLPIKISSVIQNRGSIPLFWSQETSRLNIKP 62
RYL+RGVN G VANDVETEQIV EDVPE P++ISSV+QNRGSIPLFWSQETSRLN+KP
Sbjct: 231 RYLKRGVNRNGDVANDVETEQIVSEDVPEDHPMQISSVVQNRGSIPLFWSQETSRLNLKP 290
Query: 63 DIILSKKDQSYQATKLHFENLVKRYGNPIIILNLIKTHEKKPREAILRQEFANAIDFINK 122
DI+LSKK+ +Y+AT+LHF+NLV+RYGNPIIILNLIKT E++PRE+ILR+EF NAIDFINK
Sbjct: 291 DIVLSKKEPNYEATRLHFDNLVERYGNPIIILNLIKTKERRPRESILREEFVNAIDFINK 350
Query: 123 DLSEENRLRFLHWDLHKHFQSKATNVLLLLGKVAAYALTLTGFFYCQVSSTLRPEDCLKW 182
DL EENRLRFLHWDLHKHF+SK NVL LL KVA AL LT FY QV+ + ED +
Sbjct: 351 DLPEENRLRFLHWDLHKHFRSKTKNVLALLCKVATCALMLTDLFYYQVTPAMTIEDSMSL 410
Query: 183 PSTDDVDKGSFSPTVHVEYDNEDANDLERKPSDEINVSNENHSAKPPRLQNGVLRTNCID 242
S+ D D G SP + DN D + LE+K S N++ KPPRLQ+GVLRTNCID
Sbjct: 411 SSSSDADTGDISPHTSSDDDNGDHDSLEKKSSRSKNIAYGKCDVKPPRLQSGVLRTNCID 470
Query: 243 CLDRTNVAQYAYGLAAIGHQLHSLGIIEHPKIDLDDPVANDLMQFYERMGDTLAHQYGGS 302
CLDRTNVAQYAYG AA+G QLH LGI + P I+LDDP+A LM YERMGDTLAHQYGGS
Sbjct: 471 CLDRTNVAQYAYGWAALGQQLHVLGIRDVPAIELDDPLAISLMGLYERMGDTLAHQYGGS 530
Query: 303 AAHNKIFSARRGQWRAATQSQEFFRTLQRYYSNAWMDAEKQDAINVFLGHFQPQQDKPAL 362
AAHNK+FS RRGQWRAATQSQEF RTLQRYY+NA+MDA+KQDAIN+FLG FQP+Q PA+
Sbjct: 531 AAHNKVFSERRGQWRAATQSQEFLRTLQRYYNNAYMDADKQDAINIFLGTFQPEQGMPAI 590
Query: 363 WELGSDQHYDTGRIGD 378
WEL S+ GR G+
Sbjct: 591 WELRSNS-LSNGRNGE 605
Score = 34.7 bits (78), Expect = 7.0
Identities = 16/32 (50%), Positives = 20/32 (62%), Gaps = 1/32 (3%)
Query: 456 SHHIYYSEHGDSFSCSNFVDLDWLSSSANSCE 487
S +Y+ D S SNFVD++WLSS N CE
Sbjct: 691 SDQVYFGGEEDMSSVSNFVDIEWLSSE-NPCE 721
>gb|AAP49837.1| SAC domain protein 4 [Arabidopsis thaliana]
gi|30688003|ref|NP_197584.2| phosphoinositide
phosphatase family protein [Arabidopsis thaliana]
Length = 831
Score = 517 bits (1332), Expect = e-145
Identities = 257/376 (68%), Positives = 305/376 (80%), Gaps = 5/376 (1%)
Query: 3 RYLRRGVNEKGRVANDVETEQIVFEDVPEGLPIKISSVIQNRGSIPLFWSQETSRLNIKP 62
RYL+RG+NE G VANDVETEQIV EDVP P++ISSV+QNRGSIPLFWSQETSR+ +KP
Sbjct: 257 RYLKRGINESGNVANDVETEQIVSEDVPVDRPMQISSVVQNRGSIPLFWSQETSRMKVKP 316
Query: 63 DIILSKKDQSYQATKLHFENLVKRYGNPIIILNLIKTHEKKPREAILRQEFANAIDFINK 122
DI+LSK+D +Y+AT++HFENLV+RYG PIIILNLIKT+E+KPRE+ILR EFANAIDFINK
Sbjct: 317 DIVLSKRDLNYEATRVHFENLVERYGVPIIILNLIKTNERKPRESILRAEFANAIDFINK 376
Query: 123 DLSEENRLRFLHWDLHKHFQSKATNVLLLLGKVAAYALTLTGFFYCQVSSTLRPEDCLKW 182
DL EENRLRFLHWDLHKHF SK NVL LLGKVAA AL LTGFFY Q++ ++ E +
Sbjct: 377 DLPEENRLRFLHWDLHKHFHSKTENVLALLGKVAACALMLTGFFYYQLTPAMKLEGYMSL 436
Query: 183 PSTDDVDKGSFSPTVHVEYDNEDANDLERKPSDEINVSNENHSAKPPRLQNGVLRTNCID 242
S+D SP + D+ D + LE+ NV+N ++ KP RLQ+GVLRTNCID
Sbjct: 437 SSSD----ADTSPHNSSDDDSRDYDSLEKNCRPSKNVANGDYDVKPSRLQSGVLRTNCID 492
Query: 243 CLDRTNVAQYAYGLAAIGHQLHSLGIIEHPKIDLDDPVANDLMQFYERMGDTLAHQYGGS 302
CLDRTNVAQYAYG AA+G QLH+LGI + P I+LDDP+++ LM YERMGDTLA+QYGGS
Sbjct: 493 CLDRTNVAQYAYGWAALGQQLHALGIRDAPTIELDDPLSSTLMGLYERMGDTLAYQYGGS 552
Query: 303 AAHNKIFSARRGQWRAATQSQEFFRTLQRYYSNAWMDAEKQDAINVFLGHFQPQQDKPAL 362
AAHNK+FS RRGQWRAATQSQEF RTLQRYY+NA+MDA+KQDAIN+FLG F+P+Q A+
Sbjct: 553 AAHNKVFSERRGQWRAATQSQEFLRTLQRYYNNAYMDADKQDAINIFLGTFRPEQGSQAV 612
Query: 363 WELGSDQHYDTGRIGD 378
WEL SD H GR G+
Sbjct: 613 WELRSDSH-SNGRSGE 627
Score = 45.1 bits (105), Expect = 0.005
Identities = 21/34 (61%), Positives = 23/34 (66%)
Query: 456 SHHIYYSEHGDSFSCSNFVDLDWLSSSANSCEED 489
S IY SE D S SNFVD++WLSSS N CE D
Sbjct: 721 SSSIYLSEQEDMSSVSNFVDIEWLSSSENLCEND 754
>gb|AAP49835.1| SAC domain protein 2 [Arabidopsis thaliana]
gi|20334798|gb|AAM16260.1| at3g14201/at3g14201
[Arabidopsis thaliana] gi|15215806|gb|AAK91448.1|
At3g14201 [Arabidopsis thaliana]
gi|18400310|ref|NP_566481.1| phosphoinositide
phosphatase family protein [Arabidopsis thaliana]
Length = 808
Score = 514 bits (1325), Expect = e-144
Identities = 263/374 (70%), Positives = 298/374 (79%), Gaps = 2/374 (0%)
Query: 3 RYLRRGVNEKGRVANDVETEQIVFEDVPEGLPIKISSVIQNRGSIPLFWSQETSRLNIKP 62
RYL+RGVNEKGRVANDVETEQIVFE+ +G P +ISSV+QNRGSIPLFWSQETSRLNIKP
Sbjct: 253 RYLKRGVNEKGRVANDVETEQIVFEEAQDGNPGRISSVVQNRGSIPLFWSQETSRLNIKP 312
Query: 63 DIILSKKDQSYQATKLHFENLVKRYGNPIIILNLIKTHEKKPREAILRQEFANAIDFINK 122
DIILS KD +++AT+LHFENL +RYGNPIIILNLIKT EK+PRE ILR EFANAI FINK
Sbjct: 313 DIILSPKDPNFEATRLHFENLGRRYGNPIIILNLIKTREKRPRETILRAEFANAIRFINK 372
Query: 123 DLSEENRLRFLHWDLHKHFQSKATNVLLLLGKVAAYALTLTGFFYCQVSSTLRPEDCLKW 182
LS+E+RLR LHWDLHKH + K TNVL +LG++A YAL LT FYCQ++ LR E
Sbjct: 373 GLSKEDRLRPLHWDLHKHSRKKGTNVLAILGRLATYALNLTSIFYCQLTPDLRGEGFQNQ 432
Query: 183 -PSTDDVDKGSFSPTVHVEYDNEDANDLERKPSDEINVSNENHSAKPPRLQNGVLRTNCI 241
PST + D G S T +E A +L + ++ + E+ + LQ GVLRTNCI
Sbjct: 433 NPSTLENDDGECS-TYDPPSKDETAPNLVVENGNDSKDAKEDQQKEVTMLQKGVLRTNCI 491
Query: 242 DCLDRTNVAQYAYGLAAIGHQLHSLGIIEHPKIDLDDPVANDLMQFYERMGDTLAHQYGG 301
DCLDRTNVAQYAYGL A G QLH+LG+ E IDLD+P+A DLM YE MGDTLA QYGG
Sbjct: 492 DCLDRTNVAQYAYGLVAFGRQLHALGLTESTNIDLDNPLAEDLMGIYETMGDTLALQYGG 551
Query: 302 SAAHNKIFSARRGQWRAATQSQEFFRTLQRYYSNAWMDAEKQDAINVFLGHFQPQQDKPA 361
SAAHNKIF RRGQWRAATQSQEFFRTLQRYYSNA+MDAEKQDAINVFLG+FQPQ DKPA
Sbjct: 552 SAAHNKIFCERRGQWRAATQSQEFFRTLQRYYSNAYMDAEKQDAINVFLGYFQPQSDKPA 611
Query: 362 LWELGSDQHYDTGR 375
LWELGSDQHY+ R
Sbjct: 612 LWELGSDQHYNAAR 625
>dbj|BAB02988.1| unnamed protein product [Arabidopsis thaliana]
Length = 816
Score = 507 bits (1306), Expect = e-142
Identities = 263/382 (68%), Positives = 298/382 (77%), Gaps = 10/382 (2%)
Query: 3 RYLRRGVNEKGRVANDVETEQIVFEDVPEGLPIKISSVIQNRGSIPLFWSQETSRLNIKP 62
RYL+RGVNEKGRVANDVETEQIVFE+ +G P +ISSV+QNRGSIPLFWSQETSRLNIKP
Sbjct: 253 RYLKRGVNEKGRVANDVETEQIVFEEAQDGNPGRISSVVQNRGSIPLFWSQETSRLNIKP 312
Query: 63 DIILSKKDQSYQATKLHFENLVKRYGNPIIILNLIK--------THEKKPREAILRQEFA 114
DIILS KD +++AT+LHFENL +RYGNPIIILNLIK T EK+PRE ILR EFA
Sbjct: 313 DIILSPKDPNFEATRLHFENLGRRYGNPIIILNLIKVGILTPRQTREKRPRETILRAEFA 372
Query: 115 NAIDFINKDLSEENRLRFLHWDLHKHFQSKATNVLLLLGKVAAYALTLTGFFYCQVSSTL 174
NAI FINK LS+E+RLR LHWDLHKH + K TNVL +LG++A YAL LT FYCQ++ L
Sbjct: 373 NAIRFINKGLSKEDRLRPLHWDLHKHSRKKGTNVLAILGRLATYALNLTSIFYCQLTPDL 432
Query: 175 RPEDCLKW-PSTDDVDKGSFSPTVHVEYDNEDANDLERKPSDEINVSNENHSAKPPRLQN 233
R E PST + D G S T +E A +L + ++ + E+ + LQ
Sbjct: 433 RGEGFQNQNPSTLENDDGECS-TYDPPSKDETAPNLVVENGNDSKDAKEDQQKEVTMLQK 491
Query: 234 GVLRTNCIDCLDRTNVAQYAYGLAAIGHQLHSLGIIEHPKIDLDDPVANDLMQFYERMGD 293
GVLRTNCIDCLDRTNVAQYAYGL A G QLH+LG+ E IDLD+P+A DLM YE MGD
Sbjct: 492 GVLRTNCIDCLDRTNVAQYAYGLVAFGRQLHALGLTESTNIDLDNPLAEDLMGIYETMGD 551
Query: 294 TLAHQYGGSAAHNKIFSARRGQWRAATQSQEFFRTLQRYYSNAWMDAEKQDAINVFLGHF 353
TLA QYGGSAAHNKIF RRGQWRAATQSQEFFRTLQRYYSNA+MDAEKQDAINVFLG+F
Sbjct: 552 TLALQYGGSAAHNKIFCERRGQWRAATQSQEFFRTLQRYYSNAYMDAEKQDAINVFLGYF 611
Query: 354 QPQQDKPALWELGSDQHYDTGR 375
QPQ DKPALWELGSDQHY+ R
Sbjct: 612 QPQSDKPALWELGSDQHYNAAR 633
>gb|AAM19844.1| AT3g43220/F7K15_70 [Arabidopsis thaliana]
Length = 622
Score = 503 bits (1296), Expect = e-141
Identities = 249/348 (71%), Positives = 285/348 (81%)
Query: 3 RYLRRGVNEKGRVANDVETEQIVFEDVPEGLPIKISSVIQNRGSIPLFWSQETSRLNIKP 62
RYL+RGVN G VANDVETEQIV EDVPE P++ISSV+QNRGSIPLFWSQETSRLN+KP
Sbjct: 248 RYLKRGVNRNGDVANDVETEQIVSEDVPEDHPMQISSVVQNRGSIPLFWSQETSRLNLKP 307
Query: 63 DIILSKKDQSYQATKLHFENLVKRYGNPIIILNLIKTHEKKPREAILRQEFANAIDFINK 122
DI+LSKK+ +Y+AT+LHF+NLV+RYGNPIIILNLIKT E++PRE+ILR+EF NAIDFINK
Sbjct: 308 DIVLSKKEPNYEATRLHFDNLVERYGNPIIILNLIKTKERRPRESILREEFVNAIDFINK 367
Query: 123 DLSEENRLRFLHWDLHKHFQSKATNVLLLLGKVAAYALTLTGFFYCQVSSTLRPEDCLKW 182
DL EENRLRFLHWDLHKHF+SK NVL LL KVA AL LT FY QV+ + ED +
Sbjct: 368 DLPEENRLRFLHWDLHKHFRSKTKNVLALLCKVATCALMLTDLFYYQVTPAMTIEDSMSL 427
Query: 183 PSTDDVDKGSFSPTVHVEYDNEDANDLERKPSDEINVSNENHSAKPPRLQNGVLRTNCID 242
S+ D D G SP + DN D + LE+K S N++ KPPRLQ+GVLRTNCID
Sbjct: 428 SSSSDADTGDISPHTSSDDDNGDHDSLEKKSSRSKNIAYGKCDVKPPRLQSGVLRTNCID 487
Query: 243 CLDRTNVAQYAYGLAAIGHQLHSLGIIEHPKIDLDDPVANDLMQFYERMGDTLAHQYGGS 302
CLDRTNVAQYAYG AA+G QLH LGI + P I+LDDP+A LM YERMGDTLAHQYGGS
Sbjct: 488 CLDRTNVAQYAYGWAALGQQLHVLGIRDVPAIELDDPLAISLMGLYERMGDTLAHQYGGS 547
Query: 303 AAHNKIFSARRGQWRAATQSQEFFRTLQRYYSNAWMDAEKQDAINVFL 350
AAHNK+FS RRGQWRAATQSQEF RTLQRYY+NA+MDA+KQDAIN+FL
Sbjct: 548 AAHNKVFSERRGQWRAATQSQEFLRTLQRYYNNAYMDADKQDAINIFL 595
>ref|XP_470554.1| Putative phosphoinositide phosphatase [Oryza sativa]
gi|15217295|gb|AAK92639.1| Putative phosphoinositide
phosphatase [Oryza sativa]
Length = 779
Score = 452 bits (1162), Expect = e-125
Identities = 237/378 (62%), Positives = 282/378 (73%), Gaps = 29/378 (7%)
Query: 3 RYLRRGVNEKGRVANDVETEQIVFEDVPEGLPIKISSVIQNRGSIPLFWSQETSRLNIKP 62
R+L+RGVNEKGRVANDVETEQIVFED P+ +P +ISSV+Q+RGSIPL W QETSRLNI+P
Sbjct: 241 RFLKRGVNEKGRVANDVETEQIVFEDTPDEIPHQISSVVQHRGSIPLIWFQETSRLNIRP 300
Query: 63 DIILSKKDQSYQATKLHFENLVKRYGNPIIILNLIKTHEKKPREAILRQEFANAIDFINK 122
DIIL K D Y+ T+LHFENL RYGNPIIILNLIKT EKKPRE++LR EFA AI +INK
Sbjct: 301 DIIL-KPDVDYKTTRLHFENLALRYGNPIIILNLIKTREKKPRESLLRAEFAKAIHYINK 359
Query: 123 DLSEENRLRFLHWDLHKHFQSKATNVLLLLGKVAAYALTLTGFFYCQVSSTLRPEDCLKW 182
L ++ RL+FLH DL K + K TNVL LL KVA+ L LT F +C+++++
Sbjct: 360 GLPDDKRLKFLHMDLSKLSRRKGTNVLSLLNKVASDVLDLTDFLHCEITTS--------- 410
Query: 183 PSTDDVDKGSFSPTVHVEYDNEDANDLERKPSDEINVSNENHSAK---PPRLQNGVLRTN 239
+D G + ++ D+E N+ ++N A P LQ GVLRTN
Sbjct: 411 -KYEDASSGQGAVA--------NSGDIE-------NIQDQNLCATKLVPLLLQKGVLRTN 454
Query: 240 CIDCLDRTNVAQYAYGLAAIGHQLHSLGIIEHPKIDLDDPVANDLMQFYERMGDTLAHQY 299
CIDCLDRTNVAQ+AYGLAA+G QLH L + E P I+L P+A+DLM FYERMGDTLA QY
Sbjct: 455 CIDCLDRTNVAQFAYGLAALGRQLHVLQLNETPTIELHAPLADDLMDFYERMGDTLAIQY 514
Query: 300 GGSAAHNKIFSARRGQWRAATQSQEFFRTLQRYYSNAWMDAEKQDAINVFLGHFQPQQDK 359
GGSAAHNKIF +RGQW+AATQSQEF RTLQRYYSNA+ D EKQD+INVFLGHFQPQ+ K
Sbjct: 515 GGSAAHNKIFCEQRGQWKAATQSQEFLRTLQRYYSNAYTDPEKQDSINVFLGHFQPQEGK 574
Query: 360 PALWELGSDQHYDTGRIG 377
PALW+L SDQHY+ GR G
Sbjct: 575 PALWKLDSDQHYNIGRQG 592
Score = 38.1 bits (87), Expect = 0.64
Identities = 17/22 (77%), Positives = 19/22 (86%)
Query: 471 SNFVDLDWLSSSANSCEEDPYE 492
SNF+DLD LSSS NSCEE+ YE
Sbjct: 694 SNFLDLDLLSSSGNSCEEEIYE 715
>dbj|BAD35217.1| putative Sac domain-containing inositol phosphatase 3 [Oryza sativa
(japonica cultivar-group)]
Length = 803
Score = 451 bits (1160), Expect = e-125
Identities = 239/382 (62%), Positives = 279/382 (72%), Gaps = 24/382 (6%)
Query: 3 RYLRRGVNEKGRVANDVETEQIVFEDVPEGLPIKISSVIQNRGSIPLFWSQETSRLNIKP 62
RYL+RGVN++G VANDVETEQI+FED+ P +ISSV+QNRGSIPLFWSQETS+LN+KP
Sbjct: 254 RYLKRGVNDEGSVANDVETEQIIFEDMLG--PKQISSVVQNRGSIPLFWSQETSKLNLKP 311
Query: 63 DIILSKKDQSYQATKLHFENLVKRYGNPIIILNLIKTHEKKPREAILRQEFANAIDFINK 122
DIIL +KD++Y+AT+LHFENL RYGNPIIILNLIK E++PRE+ILR EF AI IN
Sbjct: 312 DIILHEKDKNYEATRLHFENLRIRYGNPIIILNLIKKRERRPRESILRSEFDKAIKIINN 371
Query: 123 DLSEENRLRFLHWDLHKHFQSKATNVLLLLGKVAAYALTLTGFFYCQVSSTLRPEDCLKW 182
DL EN LRFLHWDLHK+ Q K+TN L +L KVA AL LT FFY QV R E
Sbjct: 372 DLPGENHLRFLHWDLHKNSQRKSTNALQMLLKVAFEALNLTEFFYYQVPPARRAESSFNL 431
Query: 183 PSTDDVDKGSFSPTVHVEYDNEDANDLERKPSD-------------------EINVSNEN 223
+ K F P + +N+D D D +I N +
Sbjct: 432 HAPL---KNGFGPHECDDSNNDDITDCIDNIDDMSQEDTCGSSDTSGNGTAEDIAEGNGS 488
Query: 224 HSAKPPRLQNGVLRTNCIDCLDRTNVAQYAYGLAAIGHQLHSLGIIEHPKIDLDDPVAND 283
S KPP+ Q GVLRTNCIDCLDRTNVAQYAYGLAA+GHQLH+LG IE P++DLD P+A+
Sbjct: 489 ISVKPPKFQKGVLRTNCIDCLDRTNVAQYAYGLAALGHQLHALGSIESPELDLDSPLAHH 548
Query: 284 LMQFYERMGDTLAHQYGGSAAHNKIFSARRGQWRAATQSQEFFRTLQRYYSNAWMDAEKQ 343
LM FYERMGDTLA QYGGSAAHNKIFSA+RG + A QSQEFFRTLQRYYSNA+MDA KQ
Sbjct: 549 LMHFYERMGDTLAVQYGGSAAHNKIFSAKRGHLKFAIQSQEFFRTLQRYYSNAYMDAYKQ 608
Query: 344 DAINVFLGHFQPQQDKPALWEL 365
AIN+FLG+FQP + +PALWEL
Sbjct: 609 AAINLFLGYFQPCEGEPALWEL 630
>gb|AAP49834.1| SAC domain protein 1 [Arabidopsis thaliana]
gi|22329733|ref|NP_173676.2| phosphoinositide
phosphatase family protein [Arabidopsis thaliana]
gi|33337344|gb|AAQ13339.1| FIG4-like protein AtFIG4
[Arabidopsis thaliana]
Length = 912
Score = 421 bits (1083), Expect = e-116
Identities = 213/391 (54%), Positives = 276/391 (70%), Gaps = 19/391 (4%)
Query: 3 RYLRRGVNEKGRVANDVETEQIVFEDVPEGLPIKISSVIQNRGSIPLFWSQETSRLNIKP 62
RYL+RGVN++GRVANDVETEQ+V +D K+SSV+Q RGSIPLFWSQE SR + KP
Sbjct: 268 RYLKRGVNDRGRVANDVETEQLVLDDEAGSCKGKMSSVVQMRGSIPLFWSQEASRFSPKP 327
Query: 63 DIILSKKDQSYQATKLHFENLVKRYGNPIIILNLIKTHEKKPREAILRQEFANAIDFINK 122
DI L + D +Y++TK+HFE+LV RYGNPII+LNLIKT EK+PRE +LR+EFANA+ ++N
Sbjct: 328 DIFLQRYDPTYESTKMHFEDLVNRYGNPIIVLNLIKTVEKRPREMVLRREFANAVGYLNS 387
Query: 123 DLSEENRLRFLHWDLHKHFQSKATNVLLLLGKVAAYALTLTGFFYC-------QVSSTLR 175
EEN L+F+HWD HK +SK+ NVL +LG VA+ AL LTG ++ + +S L
Sbjct: 388 IFREENHLKFIHWDFHKFAKSKSANVLAVLGAVASEALDLTGLYFSGKPKIVKKKASQLS 447
Query: 176 PEDCLKWPSTDDVDKGSFSPTVHVEYDNEDANDL-------ERKPSDEINVSNENHSAKP 228
+ + PS D+ S + E AND+ E++ +E ++
Sbjct: 448 HANTAREPSLRDLRAYSAELS-----RGESANDILSALANREKEMKLTQQKKDEGTNSSA 502
Query: 229 PRLQNGVLRTNCIDCLDRTNVAQYAYGLAAIGHQLHSLGIIEHPKIDLDDPVANDLMQFY 288
PR Q+GVLRTNCIDCLDRTNVAQYAYGLAA+G QLH++G+ + PKID D +A LM Y
Sbjct: 503 PRYQSGVLRTNCIDCLDRTNVAQYAYGLAALGRQLHAMGLSDTPKIDPDSSIAAALMDMY 562
Query: 289 ERMGDTLAHQYGGSAAHNKIFSARRGQWRAATQSQEFFRTLQRYYSNAWMDAEKQDAINV 348
+ MGD LA QYGGSAAHN +F R+G+W+A TQS+EF ++++RYYSN + D EKQDAIN+
Sbjct: 563 QSMGDALAQQYGGSAAHNTVFPERQGKWKATTQSREFLKSIKRYYSNTYTDGEKQDAINL 622
Query: 349 FLGHFQPQQDKPALWELGSDQHYDTGRIGDD 379
FLG+FQPQ+ KPALWEL SD + IGDD
Sbjct: 623 FLGYFQPQEGKPALWELDSDYYLHVSGIGDD 653
>gb|AAC25523.1| Similar to hypothetical protein C34B7.2 gb|1729503 from C. elegans
cosmid gb|Z83220. [Arabidopsis thaliana]
gi|7487361|pir||T00781 hypothetical protein T22J18.20 -
Arabidopsis thaliana
Length = 925
Score = 414 bits (1063), Expect = e-114
Identities = 216/405 (53%), Positives = 280/405 (68%), Gaps = 34/405 (8%)
Query: 3 RYLRRGVNEKGRVANDVETEQIVFEDVPEGLPIKISSVIQNRGSIPLFWSQETSRLNIKP 62
RYL+RGVN++GRVANDVETEQ+V +D K+SSV+Q RGSIPLFWSQE SR + KP
Sbjct: 268 RYLKRGVNDRGRVANDVETEQLVLDDEAGSCKGKMSSVVQMRGSIPLFWSQEASRFSPKP 327
Query: 63 DIILSKKDQSYQATKLHFENLVKRYGNPIIILNLIKTH--------EKKPREAILRQEFA 114
DI L + D +Y++TK+HFE+LV RYGNPII+LNLIKT+ EK+PRE +LR+EFA
Sbjct: 328 DIFLQRYDPTYESTKMHFEDLVNRYGNPIIVLNLIKTNIHGNIQTVEKRPREMVLRREFA 387
Query: 115 NAIDFINKDLSEENRLRFLHWDLHKHFQSKATNVLLLLGKVAAYALTLTGFFYC------ 168
NA+ ++N EEN L+F+HWD HK +SK+ NVL +LG VA+ AL LTG ++
Sbjct: 388 NAVGYLNSIFREENHLKFIHWDFHKFAKSKSANVLAVLGAVASEALDLTGLYFSGKPKIV 447
Query: 169 -----QVS--STLRPEDCLKWPSTDDVDKGSFSPTVHVEYDNEDANDL-------ERKPS 214
Q+S +T R + C + PS D+ S + E AND+ E++
Sbjct: 448 KKKASQLSHANTARDKYCRE-PSLRDLRAYSAELS-----RGESANDILSALANREKEMK 501
Query: 215 DEINVSNENHSAKPPRLQNGVLRTNCIDCLDRTNVAQYAYGLAAIGHQLHSLGIIEHPKI 274
+E ++ PR Q+GVLRTNCIDCLDRTNVAQYAYGLAA+G QLH++G+ + PKI
Sbjct: 502 LTQQKKDEGTNSSAPRYQSGVLRTNCIDCLDRTNVAQYAYGLAALGRQLHAMGLSDTPKI 561
Query: 275 DLDDPVANDLMQFYERMGDTLAHQYGGSAAHNKIFSARRGQWRAATQSQEFFRTLQRYYS 334
D D +A LM Y+ MGD LA QYGGSAAHN +F R+G+W+A TQS+EF ++++RYYS
Sbjct: 562 DPDSSIAAALMDMYQSMGDALAQQYGGSAAHNTVFPERQGKWKATTQSREFLKSIKRYYS 621
Query: 335 NAWMDAEKQDAINVFLGHFQPQQDKPALWELGSDQHYDTGRIGDD 379
N + D EKQDAIN+FLG+FQPQ+ KPALWEL SD + IGDD
Sbjct: 622 NTYTDGEKQDAINLFLGYFQPQEGKPALWELDSDYYLHVSGIGDD 666
>ref|XP_479718.1| putative sac domain-containing inositol phosphatase 3 [Oryza sativa
(japonica cultivar-group)] gi|51964610|ref|XP_507089.1|
PREDICTED P0007D08.10-1 gene product [Oryza sativa
(japonica cultivar-group)] gi|42408372|dbj|BAD09523.1|
putative sac domain-containing inositol phosphatase 3
[Oryza sativa (japonica cultivar-group)]
Length = 889
Score = 400 bits (1027), Expect = e-110
Identities = 203/382 (53%), Positives = 271/382 (70%), Gaps = 6/382 (1%)
Query: 3 RYLRRGVNEKGRVANDVETEQIVFEDVPEGLPIKISSVIQNRGSIPLFWSQETSRLNIKP 62
RYL+RGVN+ G+VANDVETEQIVFE+ ++S+V+Q RGSIPLFWSQE RL+ KP
Sbjct: 248 RYLKRGVNDHGKVANDVETEQIVFEEEAGSWKGRMSAVVQMRGSIPLFWSQEAGRLSPKP 307
Query: 63 DIILSKKDQSYQATKLHFENLVKRYGNPIIILNLIKTHEKKPREAILRQEFANAIDFINK 122
DII+ + D +Y+ATKLHF+++ +RYG+PIIILNL KT EK+PRE +LR+E+ NA+ ++N+
Sbjct: 308 DIIVQRYDPTYEATKLHFDDVAQRYGHPIIILNLTKTFEKRPREMMLRREYFNAVGYLNQ 367
Query: 123 DLSEENRLRFLHWDLHKHFQSKATNVLLLLGKVAAYALTLTGFFYCQVSSTLRPEDCLKW 182
++ EE +LRF+HWD HK +SK+ NVL +LG VA+ AL LTGF+Y ++ + +
Sbjct: 368 NVPEEKKLRFIHWDFHKFAKSKSANVLGVLGGVASEALDLTGFYY-SGKPKVQKKRSTQL 426
Query: 183 PSTDDVDKGSF---SPTVHVEYDNEDANDLERKPSDEI--NVSNENHSAKPPRLQNGVLR 237
T GS + + + + +A+ L S +I + S + P Q GVLR
Sbjct: 427 SRTTTARDGSIDIRASSGDLSRLSSNADSLGPTASQDIRKDDSKQELLGDGPCYQTGVLR 486
Query: 238 TNCIDCLDRTNVAQYAYGLAAIGHQLHSLGIIEHPKIDLDDPVANDLMQFYERMGDTLAH 297
TNC+DCLDRTNVAQYAYGLAA+G QLH++G KI D +A+ LM Y+ MGD LAH
Sbjct: 487 TNCMDCLDRTNVAQYAYGLAALGRQLHAMGATNVSKIHPDSSIASALMDLYQSMGDALAH 546
Query: 298 QYGGSAAHNKIFSARRGQWRAATQSQEFFRTLQRYYSNAWMDAEKQDAINVFLGHFQPQQ 357
QYGGSAAHN +F R+G+W+A TQS+EF ++++RYYSNA+ D EKQDAIN+FLG+FQPQ
Sbjct: 547 QYGGSAAHNTVFPERQGKWKATTQSREFLKSIRRYYSNAYTDGEKQDAINLFLGYFQPQD 606
Query: 358 DKPALWELGSDQHYDTGRIGDD 379
KPALWEL SD + GDD
Sbjct: 607 GKPALWELDSDYYLHVTTYGDD 628
>gb|AAP49838.1| SAC domain protein 5 [Arabidopsis thaliana]
gi|20147341|gb|AAM10384.1| At1g17340/F28G4_6
[Arabidopsis thaliana] gi|22329625|ref|NP_173177.2|
phosphoinositide phosphatase family protein [Arabidopsis
thaliana] gi|25090443|gb|AAN72303.1| At1g17340/F28G4_6
[Arabidopsis thaliana]
Length = 785
Score = 367 bits (943), Expect = e-100
Identities = 196/379 (51%), Positives = 256/379 (66%), Gaps = 23/379 (6%)
Query: 3 RYLRRGVNEKGRVANDVETEQIVFEDVPEGLPIKISSVIQNRGSIPLFWSQETSRLNIKP 62
RYLRRGVN+ GRVANDVETEQIV + VP G I I+SV+Q RGSIPLFWSQE S N +P
Sbjct: 253 RYLRRGVNDIGRVANDVETEQIVSKVVPAGQKIPITSVVQVRGSIPLFWSQEASVFNPQP 312
Query: 63 DIILSKKDQSYQATKLHFENLVKRYGNPIIILNLIKT--HEKKPREAILRQEFANAIDFI 120
+IIL+KKD +Y+AT+ HF+NL +RYGN IIILNL+KT EKK RE ILR EFA I FI
Sbjct: 313 EIILNKKDANYEATQHHFQNLRQRYGNRIIILNLLKTVTGEKKHRETILRGEFAKTIRFI 372
Query: 121 NKDLSEENRLRFLHWDLHKHFQSKATNVLLLLGKVAAYALTLTGFFYCQVSSTLRPEDCL 180
NK + E+RL+ +H+DL KH++ A L + +L LT FYC+ S + E+ +
Sbjct: 373 NKGMDREHRLKAIHFDLSKHYKKGADGAFNHLCIFSRKSLELTDLFYCKAPSGVGAEEVI 432
Query: 181 KWPSTDDVDKGSFSPTVHVEYDNEDANDLERKPSDEINVSNENHSAKPPRLQNGVLRTNC 240
YD+ N + + + + E+ A LQNGVLRTNC
Sbjct: 433 --------------------YDSFFNNPIPSQDEEASSPEKEDMKADIFLLQNGVLRTNC 472
Query: 241 IDCLDRTNVAQYAYGLAAIGHQLHSLGIIEHPKIDLDDPVANDLMQFYERMGDTLAHQYG 300
IDCLDRTN AQYA+GL ++GHQL +LGI P +DL++P+A +LM Y++MG+TLA QYG
Sbjct: 473 IDCLDRTNFAQYAHGLVSLGHQLRTLGISGPPVVDLNNPLAIELMDAYQKMGNTLAMQYG 532
Query: 301 GSAAHNKIFSARRGQWRAATQSQEFFRTLQRYYSNAWMDAEKQDAINVFLGHFQPQQDKP 360
GS AH+K+F RG W + ++ F ++RYYSNA+ D++KQ+AINVFLGHF+P+ +P
Sbjct: 533 GSEAHSKMFCDLRGNWNMVMRQRDIFTAVRRYYSNAYQDSDKQNAINVFLGHFRPRLGRP 592
Query: 361 ALWELGSDQHYDTGRIGDD 379
ALWEL SDQH + GR G +
Sbjct: 593 ALWELDSDQH-NIGRSGSN 610
>pir||H86309 F28G4.21 protein - Arabidopsis thaliana gi|9665125|gb|AAF97309.1|
Unknown protein [Arabidopsis thaliana]
Length = 718
Score = 354 bits (908), Expect = 4e-96
Identities = 197/392 (50%), Positives = 256/392 (65%), Gaps = 36/392 (9%)
Query: 3 RYLRRGVNEKGRVANDVETEQIVFEDVPEGLPIKISSVIQNRGSIPLFWSQETSRLNIKP 62
RYLRRGVN+ GRVANDVETEQIV + VP G I I+SV+Q RGSIPLFWSQE S N +P
Sbjct: 157 RYLRRGVNDIGRVANDVETEQIVSKVVPAGQKIPITSVVQVRGSIPLFWSQEASVFNPQP 216
Query: 63 DIILSKKDQSYQATKLHFENLVKRYGNPIIILNLIK--THEKKPREAILRQEFANAIDFI 120
+IIL+KKD +Y+AT+ HF+NL +RYGN IIILNL+K T EKK RE ILR EFA I FI
Sbjct: 217 EIILNKKDANYEATQHHFQNLRQRYGNRIIILNLLKTVTGEKKHRETILRGEFAKTIRFI 276
Query: 121 NKDLSEENRLRFLHWDLHKHFQSKATNVLLLLGKVAAYALTLTGFFYCQVSSTLRPEDCL 180
NK + E+RL+ +H+DL KH++ A L + +L LT FYC+ S + E+
Sbjct: 277 NKGMDREHRLKAIHFDLSKHYKKGADGAFNHLCIFSRKSLELTDLFYCKAPSGVGAEE-- 334
Query: 181 KWPSTDDVDKGSFSPTVHVEYDNEDANDLERKPSDEINVSNENHSAKPPRLQNGVLRTNC 240
V YD+ N + + + + E+ A LQNGVLRTNC
Sbjct: 335 ------------------VIYDSFFNNPIPSQDEEASSPEKEDMKADIFLLQNGVLRTNC 376
Query: 241 IDCLDRTNVAQYAYGLAAIGHQLHSLGIIEHPKIDLDDPVANDLMQFYERMGDTLAHQYG 300
IDCLDRTN AQYA+GL ++GHQL +LGI P +DL++P+A +LM Y++MG+TLA QYG
Sbjct: 377 IDCLDRTNFAQYAHGLVSLGHQLRTLGISGPPVVDLNNPLAIELMDAYQKMGNTLAMQYG 436
Query: 301 GSAAHNK------IFSARRGQWRAATQSQEFFRTLQRYYSNAWMDAEKQDAINV------ 348
GS AH+K +F RG W + ++ F ++RYYSNA+ D++KQ+AINV
Sbjct: 437 GSEAHSKKVYGIQMFCDLRGNWNMVMRQRDIFTAVRRYYSNAYQDSDKQNAINVVKLIVD 496
Query: 349 -FLGHFQPQQDKPALWELGSDQHYDTGRIGDD 379
FLGHF+P+ +PALWEL SDQH + GR G +
Sbjct: 497 RFLGHFRPRLGRPALWELDSDQH-NIGRSGSN 527
>gb|AAF18517.1| Hypothetical protein [Arabidopsis thaliana]
Length = 876
Score = 320 bits (821), Expect = 5e-86
Identities = 186/405 (45%), Positives = 238/405 (57%), Gaps = 83/405 (20%)
Query: 3 RYLRRGVNEKGRVANDVETEQIVFEDVPEGLPIKISSVIQNRGSIPLFWSQETSRLNIKP 62
RYL+RGVN++GRVANDVETEQ+V +D K+SSV+Q RGSIPLFWSQE SR + KP
Sbjct: 268 RYLKRGVNDRGRVANDVETEQLVLDDEAGSCKGKMSSVVQMRGSIPLFWSQEASRFSPKP 327
Query: 63 DIILSKKDQSYQATKLHFENLVKRYGNPIIILNLIKTH--------EKKPREAILRQEFA 114
DI L + D +Y++TK+HFE+LV RYGNPII+LNLIKT+ EK+PRE +LR+EFA
Sbjct: 328 DIFLQRYDPTYESTKMHFEDLVNRYGNPIIVLNLIKTNIHGNIQTVEKRPREMVLRREFA 387
Query: 115 NAIDFINKDLSEENRLRFLHWDLHKHFQSKATNVLLLLGKVAAYALTLTGFFY------- 167
NA+ ++N EEN L+F+HWD HK +SK+ NVL +LG VA+ AL LTG ++
Sbjct: 388 NAVGYLNSIFREENHLKFIHWDFHKFAKSKSANVLAVLGAVASEALDLTGLYFSGKPKIV 447
Query: 168 ----CQVS--STLRPEDCLKWPSTDDVDKGSFSPTVHVEYDNEDANDL-------ERKPS 214
Q+S +T R + C + PS D+ S + E AND+ E++
Sbjct: 448 KKKASQLSHANTARDKYC-REPSLRDLRAYSAELS-----RGESANDILSALANREKEMK 501
Query: 215 DEINVSNENHSAKPPRLQNGVLRTNCIDCLDRTNVAQYAYGLAAIGHQLHSLGIIEHPKI 274
+E ++ PR Q+GVLRTNCIDCLDRTNVAQYAYGLAA+G QLH++G+ + PKI
Sbjct: 502 LTQQKKDEGTNSSAPRYQSGVLRTNCIDCLDRTNVAQYAYGLAALGRQLHAMGLSDTPKI 561
Query: 275 DLDDPVANDLMQFYERMGDTLAHQYGGSAAHNKIFSARRGQWRAATQSQEFFRTLQRYYS 334
D D +A LM Y+ MGD LA QYGGSAAHN
Sbjct: 562 DPDSSIAAALMDMYQSMGDALAQQYGGSAAHN---------------------------- 593
Query: 335 NAWMDAEKQDAINVFLGHFQPQQDKPALWELGSDQHYDTGRIGDD 379
+ KPALWEL SD + IGDD
Sbjct: 594 ---------------------TEGKPALWELDSDYYLHVSGIGDD 617
>dbj|BAD35729.1| Sac domain-containing inositol phosphatase-like [Oryza sativa
(japonica cultivar-group)] gi|51090870|dbj|BAD35418.1|
Sac domain-containing inositol phosphatase-like [Oryza
sativa (japonica cultivar-group)]
Length = 438
Score = 272 bits (696), Expect = 2e-71
Identities = 147/304 (48%), Positives = 196/304 (64%), Gaps = 18/304 (5%)
Query: 3 RYLRRGVNEKGRVANDVETEQIVFEDVPEGLPIKISSVIQNRGSIPLFWSQETSRLNIKP 62
RYL+RGVN+ G+VANDVETEQIVFE+ ++S+V+Q RGSIPLFW QE RL+ KP
Sbjct: 135 RYLKRGVNDHGKVANDVETEQIVFEEEAGSWKGRMSAVVQMRGSIPLFWWQEAGRLSPKP 194
Query: 63 DIILSKKDQSYQATKLHFENLVKRYGNPIIILNLIKTHEKKPREAILRQEFANAIDFINK 122
DII+ + D +Y+ATKLHF+++ +RYG+PIIILNL K EK+PRE +LR+E+ NA+ ++N+
Sbjct: 195 DIIVQRYDPTYEATKLHFDDVAQRYGHPIIILNLTKIFEKRPREMMLRREYFNAVGYLNQ 254
Query: 123 DLSEENRLRFLHWDLHKHFQSKATNVLLLLGKVAAYALTLTGFFY-----------CQVS 171
++ EE +LRF+HWD HK +SK+ NVL +LG A+ AL LTGF+Y Q+S
Sbjct: 255 NVPEEKKLRFIHWDFHKFAKSKSANVLGVLGGGASEALDLTGFYYNGKPKVQKKRSTQLS 314
Query: 172 STLRPEDCLKWPSTDDVDKGSFSPTVHVEYDNEDANDLERKPSDEINVSNENHSAKPPRL 231
T D +D + S + N D+ + S + P
Sbjct: 315 RTTFHRD-------GSIDTRASSGDLPRLSSNADSLGPTASQDIRKDDSKQELLGDGPCY 367
Query: 232 QNGVLRTNCIDCLDRTNVAQYAYGLAAIGHQLHSLGIIEHPKIDLDDPVANDLMQFYERM 291
Q GVLRTNC+DCLDRTNVAQYAYGLAA+G QLH++G KI D +A+ LM Y+ M
Sbjct: 368 QTGVLRTNCMDCLDRTNVAQYAYGLAALGRQLHAMGATNVSKIHPDSSIASALMDLYQSM 427
Query: 292 GDTL 295
GD L
Sbjct: 428 GDAL 431
>ref|XP_640668.1| hypothetical protein DDB0205575 [Dictyostelium discoideum]
gi|60468693|gb|EAL66695.1| hypothetical protein
DDB0205575 [Dictyostelium discoideum]
Length = 1391
Score = 254 bits (650), Expect = 3e-66
Identities = 151/382 (39%), Positives = 231/382 (59%), Gaps = 30/382 (7%)
Query: 3 RYLRRGVNEKGRVANDVETEQIVFEDVPE-GLPIKISSVIQNRGSIPLFWSQETSRLNIK 61
R+L+RG+NE G+VANDVETEQI+ E + K SS +Q RGSIPL+W Q+ + + K
Sbjct: 336 RFLKRGINENGQVANDVETEQILQEPLTGWSSKAKFSSFVQIRGSIPLYWEQDNNIMTPK 395
Query: 62 PDIILSKKDQSYQATKLHFENLVKRYGNPIIILNLIKTHEKKPREAILRQEFANAIDFIN 121
P I + + D +T LHF++L +R+G+PIIILNL+K++EKKPRE+ILR EF + I+ +N
Sbjct: 396 PPIKIQRMDPFLGSTILHFQDLFQRFGSPIIILNLVKSNEKKPRESILRNEFTDCINCLN 455
Query: 122 KDLSEENRLRFLHWDLHKHFQSKATNVLLLLGKVAAYALTLTGFFYCQVSSTLRPEDCLK 181
+ L E++R+ + WD H+ +SK + + L A ++ TG FY SS+ + L+
Sbjct: 456 EMLPEQHRIHYQAWDFHQAAKSKDEDHMAWLDSFANQSILKTGIFY---SSSKLYCNFLR 512
Query: 182 --WPSTDDVDKGSFSPT------VHVEYDNEDANDLERKPSDEINVSNENHSAKPPRLQN 233
+ S+ D + GS + T + NE N+L +EI Q
Sbjct: 513 ENYNSSSDDNGGSTTTTTTTTAATTISGLNEKENELFSYLFNEIGTE-----------QI 561
Query: 234 GVLRTNCIDCLDRTNVAQYAYGLAAIGHQLHSLGIIEHPKIDLDDPVANDLMQFYERMGD 293
GV+RTNCID LDRTN AQ+ G A+G+QL+S+G+++ P +D + + L+ YE G+
Sbjct: 562 GVVRTNCIDSLDRTNAAQFCIGKCALGYQLYSMGLLDSPSLDFNTGIVQVLIDMYEACGN 621
Query: 294 TLAHQYGGSAAHNKIFSARRGQWRAATQSQEFFRTLQRYYSNAWMDAEKQDAINVFLGHF 353
+A QYGGS N I + + ++QS++ ++RY SN+++DA+KQ +IN+FLG+F
Sbjct: 622 QIALQYGGSELANTIKTYTKHS--LSSQSRDLVNAIKRYISNSFIDADKQHSINLFLGYF 679
Query: 354 QPQQ----DKPALWELGSDQHY 371
P + +K LW L +D HY
Sbjct: 680 VPAKQIAIEKIPLWNLETD-HY 700
>gb|EAA61365.1| hypothetical protein AN7314.2 [Aspergillus nidulans FGSC A4]
gi|67900654|ref|XP_680583.1| hypothetical protein
AN7314_2 [Aspergillus nidulans FGSC A4]
gi|49106705|ref|XP_411451.1| hypothetical protein
AN7314.2 [Aspergillus nidulans FGSC A4]
Length = 1013
Score = 248 bits (633), Expect = 3e-64
Identities = 142/378 (37%), Positives = 210/378 (54%), Gaps = 65/378 (17%)
Query: 3 RYLRRGVNEKGRVANDVETEQIVFEDVPEGLPIK---------ISSVIQNRGSIPLFWSQ 53
R+L+RG N+ G VANDVETEQIV E + +S +Q+RGSIPL+W+Q
Sbjct: 394 RFLKRGANDLGYVANDVETEQIVSEMTTTSFHLAGPSLYANPLYTSYVQHRGSIPLYWTQ 453
Query: 54 ETSRLNIKPDIILSKKDQSYQATKLHFENLVKRYGNPIIILNLIKTHEKKPREAILRQEF 113
E S ++ KPDI L+ D Y A LHF+NL RYG PI +LNLIK+ E+ PRE+ L +EF
Sbjct: 454 ENSGVSPKPDIELNLVDPFYSAAALHFDNLFARYGAPIYVLNLIKSRERTPRESKLLREF 513
Query: 114 ANAIDFINKDLSEENRLRFLHWDLHKHFQSKATNVLLLLGKVAAYALTLTGFFYCQVSST 173
NA++++N+ L E+ +L + WD+ + +S+ +V+ L +A + TGFF +
Sbjct: 514 TNAVEYLNQFLPEDKKLIYKPWDMSRAAKSRDQDVIETLEDIAGEIIPKTGFF----QNG 569
Query: 174 LRPEDCLKWPSTDDVDKGSFSPTVHVEYDNEDANDLERKPSDEINVSNENHSAKPPRLQN 233
PE LK +QN
Sbjct: 570 QSPETGLK-------------------------------------------------VQN 580
Query: 234 GVLRTNCIDCLDRTNVAQYAYGLAAIGHQLHSLGIIEHPKIDLDDPVANDLMQFYERMGD 293
G+ RTNCIDCLDRTN AQ+ G A+G+QLH+LGIIE ++ D V N + GD
Sbjct: 581 GIARTNCIDCLDRTNAAQFVIGKRALGYQLHALGIIEGTTVEYDTDVVNLFTDMWHDHGD 640
Query: 294 TLAHQYGGSAAHNKIFSARR-GQWRAATQSQEFFRTLQRYYSNAWMDAEKQDAINVFLGH 352
+A QYGGS N + + R+ QW ++ S++ + +RYY+N+++DA++Q+A N+FLG+
Sbjct: 641 NIAIQYGGSHLVNTMATYRKINQW--SSHSRDMVESFKRYYNNSFLDAQRQEAYNLFLGN 698
Query: 353 FQPQQDKPALWELGSDQH 370
+ Q +P LW+L +D +
Sbjct: 699 YIFSQGQPMLWDLSTDYY 716
>emb|CAG88440.1| unnamed protein product [Debaryomyces hansenii CBS767]
gi|50423173|ref|XP_460167.1| unnamed protein product
[Debaryomyces hansenii]
Length = 1042
Score = 211 bits (536), Expect = 5e-53
Identities = 141/404 (34%), Positives = 215/404 (52%), Gaps = 77/404 (19%)
Query: 3 RYLRRGVNEKGRVANDVETEQIV-------FEDVPEGL--PIKISSVIQNRGSIPLFWSQ 53
R+L+RGVN+KG VAN+VETEQIV F D G + +S +Q+RGSIPL+W+Q
Sbjct: 343 RFLKRGVNDKGNVANEVETEQIVSDMLISSFHDTKHGFFNNPRYTSFVQHRGSIPLYWTQ 402
Query: 54 ETSRLNIKPDIILSKKDQSYQATKLHFENLVKRYGNPIIILNLIKTHEKKPREAILRQEF 113
+ +RL KP I ++ D YQ++ +HF L +RYG P+IILNLIKT EK+PRE L Q F
Sbjct: 403 DLNRLP-KPPIEINLPDPFYQSSAIHFNGLFRRYGLPVIILNLIKTKEKQPRELKLNQHF 461
Query: 114 ANAIDFINKDLSEENRLRFLHWDLHKHFQSKATNVLLLLGKVAAYALTLTGFFY--CQVS 171
N I ++N+ L EE++L++ +D+ KH K +V+ L K+A+ ++ GFF+ ++
Sbjct: 462 TNCIKYLNQFLPEEHKLQYHSFDMSKH-SKKNLDVITPLQKIASNSIDRIGFFHNGTDLA 520
Query: 172 STLRPEDCLKWPSTDDVDKGSFSPTVHVEYDNEDANDLERKPSDEINVSNENHSAKPPRL 231
ST + ++ D +D
Sbjct: 521 STKVQKGIIRTNCIDCLD------------------------------------------ 538
Query: 232 QNGVLRTNCIDCLDRTNVAQYAYGLAAIGHQLHSLGII-EHPKIDLDDPVANDLMQFYER 290
RTN AQ+ A+ +QL SL II E +D D + N L + +
Sbjct: 539 -----RTN---------AAQFIICKEALSYQLRSLNIISESTNLDYDSDLINILTEIFHD 584
Query: 291 MGDTLAHQYGGSAAHNKIFSARR-GQWRAATQSQEFFRTLQRYYSNAWMDAEKQDAINVF 349
GDT+A QYGGS N + S RR QW ++ +++ +++R YSN++MD +Q+AIN+F
Sbjct: 585 HGDTIAVQYGGSNLVNTMDSYRRINQW--SSHTRDILNSIKRIYSNSFMDLTRQEAINLF 642
Query: 350 LGHFQPQQDKPALWELGSDQHYDTGRIGDDDAS*VIFQKIFLRW 393
LG++Q DKP LWEL +D + + +D + + ++RW
Sbjct: 643 LGNYQYSPDKPKLWELQNDFYLHNDILIED----IRIKLSYIRW 682
>ref|XP_479719.1| sac domain-containing inositol phosphatase 3-like [Oryza sativa
(japonica cultivar-group)] gi|42408373|dbj|BAD09524.1|
sac domain-containing inositol phosphatase 3-like [Oryza
sativa (japonica cultivar-group)]
Length = 406
Score = 189 bits (481), Expect = 1e-46
Identities = 87/139 (62%), Positives = 107/139 (76%)
Query: 241 IDCLDRTNVAQYAYGLAAIGHQLHSLGIIEHPKIDLDDPVANDLMQFYERMGDTLAHQYG 300
+DCLDRTNVAQYAYGLAA+G QLH++G KI D +A+ LM Y+ MGD LAHQYG
Sbjct: 1 MDCLDRTNVAQYAYGLAALGRQLHAMGATNVSKIHPDSSIASALMDLYQSMGDALAHQYG 60
Query: 301 GSAAHNKIFSARRGQWRAATQSQEFFRTLQRYYSNAWMDAEKQDAINVFLGHFQPQQDKP 360
GSAAHN +F R+G+W+A TQS+EF ++++RYYSNA+ D EKQDAIN+FLG+FQPQ KP
Sbjct: 61 GSAAHNTVFPERQGKWKATTQSREFLKSIRRYYSNAYTDGEKQDAINLFLGYFQPQDGKP 120
Query: 361 ALWELGSDQHYDTGRIGDD 379
ALWEL SD + GDD
Sbjct: 121 ALWELDSDYYLHVTTYGDD 139
>emb|CAG58433.1| unnamed protein product [Candida glabrata CBS138]
gi|50286187|ref|XP_445522.1| unnamed protein product
[Candida glabrata]
Length = 871
Score = 142 bits (359), Expect = 2e-32
Identities = 80/206 (38%), Positives = 126/206 (60%), Gaps = 15/206 (7%)
Query: 177 EDCLKWPSTDDVDKGSFSPTVHVEYDNEDAND-------LERKPSDEINVSNENHSAKP- 228
E+C+K+ ++ D S T + E D LE D I + H++K
Sbjct: 410 EECIKYLNSLIYDNNMISYTSW-DMSRESKQDGQGVIEFLESYAIDTITSTGIFHNSKDF 468
Query: 229 --PRLQNGVLRTNCIDCLDRTNVAQYAYGLAAIGHQLHSLGIIEHPKIDLDDPVANDLMQ 286
++Q GV RTNCIDCLDRTN AQ+ G A+G QL SLGII+ ++ D + N L +
Sbjct: 469 KGTKIQQGVCRTNCIDCLDRTNAAQFVIGKRALGVQLKSLGIIDDSFLEYDSDIVNILTE 528
Query: 287 FYERMGDTLAHQYGGSAAHNKIFSARR-GQWRAATQSQEFFRTLQRYYSNAWMDAEKQDA 345
+ +GDT+A QYGGS N + + R+ QW+ + S++ +++R+YSN++MDA++QDA
Sbjct: 529 LFHDLGDTIALQYGGSHLVNTMETYRKINQWK--SHSRDMIESIKRFYSNSFMDAQRQDA 586
Query: 346 INVFLGHFQPQQDKPALWELGSDQHY 371
+N+FLG++ ++D+P+LWE+ +D HY
Sbjct: 587 VNLFLGNYVWKEDQPSLWEMNTD-HY 611
Score = 129 bits (323), Expect = 3e-28
Identities = 71/175 (40%), Positives = 103/175 (58%), Gaps = 10/175 (5%)
Query: 3 RYLRRGVNEKGRVANDVETEQIVFEDV-----PEGLPI----KISSVIQNRGSIPLFWSQ 53
R+L+RGVN G VAN+VETEQIV + V G+ + +S +Q+RGSIPLFW+Q
Sbjct: 289 RFLKRGVNNNGYVANEVETEQIVADMVLTAFHKSGMGYFDSNRYTSFVQHRGSIPLFWTQ 348
Query: 54 ETSRLNIKPDIILSKKDQSYQATKLHFENLVKRY-GNPIIILNLIKTHEKKPREAILRQE 112
E S L KP I ++ +D + A +HF+NL +RY G I +LNLIKT EK PRE L +E
Sbjct: 349 EASNLTAKPPIEITVRDPFFSAAAIHFDNLFQRYGGGRIDVLNLIKTKEKVPRETKLLKE 408
Query: 113 FANAIDFINKDLSEENRLRFLHWDLHKHFQSKATNVLLLLGKVAAYALTLTGFFY 167
+ I ++N + + N + + WD+ + + V+ L A +T TG F+
Sbjct: 409 YEECIKYLNSLIYDNNMISYTSWDMSRESKQDGQGVIEFLESYAIDTITSTGIFH 463
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.328 0.141 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 843,807,768
Number of Sequences: 2540612
Number of extensions: 35324966
Number of successful extensions: 98928
Number of sequences better than 10.0: 265
Number of HSP's better than 10.0 without gapping: 241
Number of HSP's successfully gapped in prelim test: 25
Number of HSP's that attempted gapping in prelim test: 97919
Number of HSP's gapped (non-prelim): 549
length of query: 492
length of database: 863,360,394
effective HSP length: 132
effective length of query: 360
effective length of database: 527,999,610
effective search space: 190079859600
effective search space used: 190079859600
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 77 (34.3 bits)
Medicago: description of AC124967.6


