

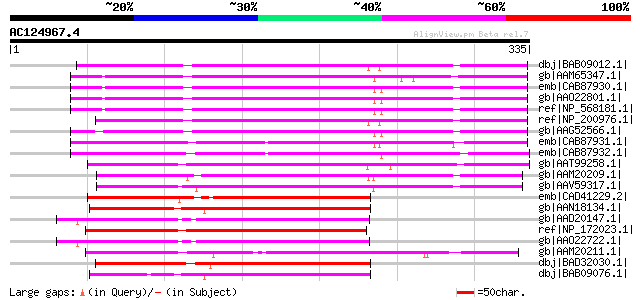
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124967.4 - phase: 0 /pseudo
(335 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAB09012.1| pectin methylesterase-like protein [Arabidopsis ... 252 1e-65
gb|AAM65347.1| pectin methyl-esterase-like protein [Arabidopsis ... 245 2e-63
emb|CAB87930.1| pectin methyl-esterase-like protein [Arabidopsis... 244 3e-63
gb|AAO22801.1| putative pectinesterase [Arabidopsis thaliana] gi... 244 3e-63
ref|NP_568181.1| pectinesterase family protein [Arabidopsis thal... 244 3e-63
ref|NP_200976.1| pectinesterase family protein [Arabidopsis thal... 243 4e-63
gb|AAG52566.1| putative pectin methylesterase; 8433-9798 [Arabid... 238 1e-61
emb|CAB87931.1| pectin methyl-esterase-like protein [Arabidopsis... 220 5e-56
emb|CAB87932.1| pectin methyl-esterase-like protein [Arabidopsis... 214 4e-54
gb|AAT99258.1| pectin-methyltransferase precursor [Salsola kali] 210 4e-53
gb|AAM20209.1| putative pectin methylesterase [Arabidopsis thali... 157 6e-37
gb|AAV59317.1| putative pectin methylesterase [Oryza sativa (jap... 156 9e-37
emb|CAD41229.2| OSJNBa0010H02.16 [Oryza sativa (japonica cultiva... 155 2e-36
gb|AAN18134.1| At5g47500/MNJ7_9 [Arabidopsis thaliana] gi|208568... 155 2e-36
gb|AAD20147.1| putative pectinesterase [Arabidopsis thaliana] gi... 152 1e-35
ref|NP_172023.1| pectinesterase family protein [Arabidopsis thal... 151 3e-35
gb|AAO22722.1| putative pectinesterase family protein [Arabidops... 149 9e-35
gb|AAM20211.1| putative pectinesterase [Arabidopsis thaliana] gi... 149 1e-34
dbj|BAD32030.1| pectin methylesterase-like protein [Oryza sativa... 147 3e-34
dbj|BAB09076.1| pectin methylesterase-like [Arabidopsis thaliana] 145 1e-33
>dbj|BAB09012.1| pectin methylesterase-like protein [Arabidopsis thaliana]
Length = 364
Score = 252 bits (643), Expect = 1e-65
Identities = 134/313 (42%), Positives = 186/313 (58%), Gaps = 31/313 (9%)
Query: 44 LTAAETTVRVVRVRKDGTGDFTTVTDAVKSIPSGNKRRVVVWIGMGEYREKITVDRSKRF 103
L AE R+++V+++G G F T+T+A+ S+ +GN RRV++ IG G Y+EK+T+DRSK F
Sbjct: 59 LLTAEAKPRIIKVKQNGRGHFKTITEAINSVRAGNTRRVIIKIGPGVYKEKVTIDRSKPF 118
Query: 104 VTFYGERNGKDNDMMPIITYDATALRYGTLDSATVAVDADYFVAVNVAFVNSSPMPDENS 163
+T YG N MP++T+D TA +YGT+DSAT+ V +DYF+AVN+ NS+PMPD
Sbjct: 119 ITLYGHPNA-----MPVLTFDGTAAQYGTVDSATLIVLSDYFMAVNIILKNSAPMPDGKR 173
Query: 164 VGGQALAMRISGDKAAFYNCKFIGFQDTLCDDYGKHFFKDCFIQGTYDFIFGNGKSIYLR 223
G QAL+MRISG+KAAFYNCKF G+QDT+CDD G HFFKDC+I+GT+DFIFG+G+S+YL
Sbjct: 174 KGAQALSMRISGNKAAFYNCKFYGYQDTICDDTGNHFFKDCYIEGTFDFIFGSGRSLYLG 233
Query: 224 TTIESVA-----------------KGFECHH----STR*GKNVGRHWIHFCSLQHNWK-W 261
T + V G+ H T G +GR W+ + + +
Sbjct: 234 TQLNVVGDGIRVITAHAGKSAAEKSGYSFVHCKVTGTGTGIYLGRSWMSHPKVVYAYTDM 293
Query: 262 SQEHLPWSWLEEEP*GCICLYLYGLCRQFQRMVPPWVQ*AVRLNYKRILSDEEAKHFLSM 321
S P W E G YG + R+ Y + + D EAK+F+S+
Sbjct: 294 SSVVNPSGWQENREAGRDKTVFYGE----YKCTGTGSHKEKRVKYTQDIDDIEAKYFISL 349
Query: 322 AYIHGEQWVRPPP 334
YI G W+ PPP
Sbjct: 350 GYIQGSSWLLPPP 362
>gb|AAM65347.1| pectin methyl-esterase-like protein [Arabidopsis thaliana]
Length = 342
Score = 245 bits (625), Expect = 2e-63
Identities = 132/317 (41%), Positives = 186/317 (58%), Gaps = 32/317 (10%)
Query: 40 LEEHLTAAETTVRVVRVRKDGTGDFTTVTDAVKSIPSGNKRRVVVWIGMGEYREKITVDR 99
L+ L AAE R++ V G G+F T+TDA+KS+P+GN +RV++ + GEYREK+T+DR
Sbjct: 34 LDPALVAAEAAPRIINVNPKG-GEFKTLTDAIKSVPAGNTKRVIIKMAPGEYREKVTIDR 92
Query: 100 SKRFVTFYGERNGKDNDMMPIITYDATALRYGTLDSATVAVDADYFVAVNVAFVNSSPMP 159
+K F+T G+ N MP+ITYD TA +YGT+DSA++ + +DYF+AVN+ N++P P
Sbjct: 93 NKPFITLMGQPNA-----MPVITYDGTAAKYGTVDSASLIILSDYFMAVNIVVKNTAPAP 147
Query: 160 DENSVGGQALAMRISGDKAAFYNCKFIGFQDTLCDDYGKHFFKDCFIQGTYDFIFGNGKS 219
D + G QAL+MRISG+ AAFYNCKF GFQDT+CDD G HFFKDC+++GT+DFIFG+G S
Sbjct: 148 DGKTKGAQALSMRISGNFAAFYNCKFYGFQDTICDDTGNHFFKDCYVEGTFDFIFGSGTS 207
Query: 220 IYLRTTIESVAKGFE---CHHSTR*GKNVGRHWIH--------FCSLQHNW--------- 259
+YL T + V G H +N G ++H L W
Sbjct: 208 MYLGTQLHVVGDGIRVIAAHAGKSAEENSGYSFVHCKVTGTGGVIYLGRAWMSHPKVVYA 267
Query: 260 --KWSQEHLPWSWLEEEP*GCICLYLYGLCRQFQRMVPPWVQ*AVRLNYKRILSDEEAKH 317
+ + P W E + YG + P A R+ + + + D+EA
Sbjct: 268 YTEMTSVVNPTGWQENKTPAHDKTVFYG----EYKCSGPGSHKAKRVPFTQDIDDKEANR 323
Query: 318 FLSMAYIHGEQWVRPPP 334
FLS+ YI G +W+ PPP
Sbjct: 324 FLSLGYIQGSKWLLPPP 340
>emb|CAB87930.1| pectin methyl-esterase-like protein [Arabidopsis thaliana]
gi|11358603|pir||T49880 pectin methyl-esterase-like
protein - Arabidopsis thaliana
Length = 342
Score = 244 bits (622), Expect = 3e-63
Identities = 132/317 (41%), Positives = 189/317 (58%), Gaps = 32/317 (10%)
Query: 40 LEEHLTAAETTVRVVRVRKDGTGDFTTVTDAVKSIPSGNKRRVVVWIGMGEYREKITVDR 99
L+ L AAE R++ V G G+F T+TDA+KS+P+GN +RV++ + GEYREK+T+DR
Sbjct: 34 LDPALVAAEAAPRIINVNPKG-GEFKTLTDAIKSVPAGNTKRVIIKMAHGEYREKVTIDR 92
Query: 100 SKRFVTFYGERNGKDNDMMPIITYDATALRYGTLDSATVAVDADYFVAVNVAFVNSSPMP 159
+K F+T G+ N MP+ITYD TA +YGT+DSA++ + +DYF+AVN+ N++P P
Sbjct: 93 NKPFITLMGQPNA-----MPVITYDGTAAKYGTVDSASLIILSDYFMAVNIVVKNTAPAP 147
Query: 160 DENSVGGQALAMRISGDKAAFYNCKFIGFQDTLCDDYGKHFFKDCFIQGTYDFIFGNGKS 219
D + G QAL+MRISG+ AAFYNCKF GFQDT+CDD G HFFKDC+++GT+DFIFG+G S
Sbjct: 148 DGKTKGAQALSMRISGNFAAFYNCKFYGFQDTICDDTGNHFFKDCYVEGTFDFIFGSGTS 207
Query: 220 IYLRTTIESVAKGFE--CHHS-------------------TR*GKNVGRHWIHFCSLQHN 258
+YL T + V G H+ T G +GR W+ + +
Sbjct: 208 MYLGTQLHVVGDGIRVIAAHAGKSAEEKSGYSFVHCKVTGTGGGIYLGRAWMSHPKVVYA 267
Query: 259 W-KWSQEHLPWSWLEEEP*GCICLYLYGLCRQFQRMVPPWVQ*AVRLNYKRILSDEEAKH 317
+ + + P W E + YG + P A R+ + + + D+EA
Sbjct: 268 YTEMTSVVNPTGWQENKTPAHDKTVFYGE----YKCSGPGSHKAKRVPFTQDIDDKEANC 323
Query: 318 FLSMAYIHGEQWVRPPP 334
FLS+ YI G +W+ PPP
Sbjct: 324 FLSLGYIQGSKWLLPPP 340
>gb|AAO22801.1| putative pectinesterase [Arabidopsis thaliana]
gi|30697951|ref|NP_177152.2| pectinesterase family
protein [Arabidopsis thaliana]
Length = 361
Score = 244 bits (622), Expect = 3e-63
Identities = 131/317 (41%), Positives = 189/317 (59%), Gaps = 32/317 (10%)
Query: 40 LEEHLTAAETTVRVVRVRKDGTGDFTTVTDAVKSIPSGNKRRVVVWIGMGEYREKITVDR 99
L+ L AAE R++ V G G+F T+TDA+KS+P+GN +RV++ + GEY+EK+T+DR
Sbjct: 53 LDPALVAAEAAPRIINVNPKG-GEFKTLTDAIKSVPAGNTKRVIIKMAPGEYKEKVTIDR 111
Query: 100 SKRFVTFYGERNGKDNDMMPIITYDATALRYGTLDSATVAVDADYFVAVNVAFVNSSPMP 159
+K F+T G+ N MP+ITYD TA +YGT+DSA++ + +DYF+AVN+ N++P P
Sbjct: 112 NKPFITLMGQPNA-----MPVITYDGTAAKYGTVDSASLIILSDYFMAVNIVVKNTAPAP 166
Query: 160 DENSVGGQALAMRISGDKAAFYNCKFIGFQDTLCDDYGKHFFKDCFIQGTYDFIFGNGKS 219
D + G QAL+MRISG+ AAFYNCKF GFQDT+CDD G HFFKDC+++GT+DFIFG+G S
Sbjct: 167 DGKTKGAQALSMRISGNFAAFYNCKFYGFQDTICDDTGNHFFKDCYVEGTFDFIFGSGTS 226
Query: 220 IYLRTTIESVAKGFE--CHHS-------------------TR*GKNVGRHWIHFCSLQHN 258
+YL T + V G H+ T G +GR W+ + +
Sbjct: 227 MYLGTQLHVVGDGIRVIAAHAGKSAEEKSGYSFVHCKVTGTGGGIYLGRAWMSHPKVVYA 286
Query: 259 W-KWSQEHLPWSWLEEEP*GCICLYLYGLCRQFQRMVPPWVQ*AVRLNYKRILSDEEAKH 317
+ + + P W E + YG + P A R+ + + + D+EA
Sbjct: 287 YTEMTSVVNPTGWQENKTPAHDKTVFYGE----YKCSGPGSHKAKRVPFTQDIDDKEANR 342
Query: 318 FLSMAYIHGEQWVRPPP 334
FLS+ YI G +W+ PPP
Sbjct: 343 FLSLGYIQGSKWLLPPP 359
>ref|NP_568181.1| pectinesterase family protein [Arabidopsis thaliana]
Length = 361
Score = 244 bits (622), Expect = 3e-63
Identities = 132/317 (41%), Positives = 189/317 (58%), Gaps = 32/317 (10%)
Query: 40 LEEHLTAAETTVRVVRVRKDGTGDFTTVTDAVKSIPSGNKRRVVVWIGMGEYREKITVDR 99
L+ L AAE R++ V G G+F T+TDA+KS+P+GN +RV++ + GEYREK+T+DR
Sbjct: 53 LDPALVAAEAAPRIINVNPKG-GEFKTLTDAIKSVPAGNTKRVIIKMAHGEYREKVTIDR 111
Query: 100 SKRFVTFYGERNGKDNDMMPIITYDATALRYGTLDSATVAVDADYFVAVNVAFVNSSPMP 159
+K F+T G+ N MP+ITYD TA +YGT+DSA++ + +DYF+AVN+ N++P P
Sbjct: 112 NKPFITLMGQPNA-----MPVITYDGTAAKYGTVDSASLIILSDYFMAVNIVVKNTAPAP 166
Query: 160 DENSVGGQALAMRISGDKAAFYNCKFIGFQDTLCDDYGKHFFKDCFIQGTYDFIFGNGKS 219
D + G QAL+MRISG+ AAFYNCKF GFQDT+CDD G HFFKDC+++GT+DFIFG+G S
Sbjct: 167 DGKTKGAQALSMRISGNFAAFYNCKFYGFQDTICDDTGNHFFKDCYVEGTFDFIFGSGTS 226
Query: 220 IYLRTTIESVAKGFE--CHHS-------------------TR*GKNVGRHWIHFCSLQHN 258
+YL T + V G H+ T G +GR W+ + +
Sbjct: 227 MYLGTQLHVVGDGIRVIAAHAGKSAEEKSGYSFVHCKVTGTGGGIYLGRAWMSHPKVVYA 286
Query: 259 W-KWSQEHLPWSWLEEEP*GCICLYLYGLCRQFQRMVPPWVQ*AVRLNYKRILSDEEAKH 317
+ + + P W E + YG + P A R+ + + + D+EA
Sbjct: 287 YTEMTSVVNPTGWQENKTPAHDKTVFYGE----YKCSGPGSHKAKRVPFTQDIDDKEANC 342
Query: 318 FLSMAYIHGEQWVRPPP 334
FLS+ YI G +W+ PPP
Sbjct: 343 FLSLGYIQGSKWLLPPP 359
>ref|NP_200976.1| pectinesterase family protein [Arabidopsis thaliana]
Length = 338
Score = 243 bits (621), Expect = 4e-63
Identities = 130/301 (43%), Positives = 179/301 (59%), Gaps = 31/301 (10%)
Query: 56 VRKDGTGDFTTVTDAVKSIPSGNKRRVVVWIGMGEYREKITVDRSKRFVTFYGERNGKDN 115
V+++G G F T+T+A+ S+ +GN RRV++ IG G Y+EK+T+DRSK F+T YG N
Sbjct: 45 VKQNGRGHFKTITEAINSVRAGNTRRVIIKIGPGVYKEKVTIDRSKPFITLYGHPNA--- 101
Query: 116 DMMPIITYDATALRYGTLDSATVAVDADYFVAVNVAFVNSSPMPDENSVGGQALAMRISG 175
MP++T+D TA +YGT+DSAT+ V +DYF+AVN+ NS+PMPD G QAL+MRISG
Sbjct: 102 --MPVLTFDGTAAQYGTVDSATLIVLSDYFMAVNIILKNSAPMPDGKRKGAQALSMRISG 159
Query: 176 DKAAFYNCKFIGFQDTLCDDYGKHFFKDCFIQGTYDFIFGNGKSIYLRTTIESVA----- 230
+KAAFYNCKF G+QDT+CDD G HFFKDC+I+GT+DFIFG+G+S+YL T + V
Sbjct: 160 NKAAFYNCKFYGYQDTICDDTGNHFFKDCYIEGTFDFIFGSGRSLYLGTQLNVVGDGIRV 219
Query: 231 ------------KGFECHH----STR*GKNVGRHWIHFCSLQHNWK-WSQEHLPWSWLEE 273
G+ H T G +GR W+ + + + S P W E
Sbjct: 220 ITAHAGKSAAEKSGYSFVHCKVTGTGTGIYLGRSWMSHPKVVYAYTDMSSVVNPSGWQEN 279
Query: 274 EP*GCICLYLYGLCRQFQRMVPPWVQ*AVRLNYKRILSDEEAKHFLSMAYIHGEQWVRPP 333
G YG + R+ Y + + D EAK+F+S+ YI G W+ PP
Sbjct: 280 REAGRDKTVFYGE----YKCTGTGSHKEKRVKYTQDIDDIEAKYFISLGYIQGSSWLLPP 335
Query: 334 P 334
P
Sbjct: 336 P 336
>gb|AAG52566.1| putative pectin methylesterase; 8433-9798 [Arabidopsis thaliana]
gi|25404855|pir||H96721 probable pectin methylesterase
T17F3.3 [imported] - Arabidopsis thaliana
Length = 338
Score = 238 bits (608), Expect = 1e-61
Identities = 129/317 (40%), Positives = 187/317 (58%), Gaps = 36/317 (11%)
Query: 40 LEEHLTAAETTVRVVRVRKDGTGDFTTVTDAVKSIPSGNKRRVVVWIGMGEYREKITVDR 99
L+ L AAE R++ G+F T+TDA+KS+P+GN +RV++ + GEY+EK+T+DR
Sbjct: 34 LDPALVAAEAAPRIIN-----GGEFKTLTDAIKSVPAGNTKRVIIKMAPGEYKEKVTIDR 88
Query: 100 SKRFVTFYGERNGKDNDMMPIITYDATALRYGTLDSATVAVDADYFVAVNVAFVNSSPMP 159
+K F+T G+ N MP+ITYD TA +YGT+DSA++ + +DYF+AVN+ N++P P
Sbjct: 89 NKPFITLMGQPNA-----MPVITYDGTAAKYGTVDSASLIILSDYFMAVNIVVKNTAPAP 143
Query: 160 DENSVGGQALAMRISGDKAAFYNCKFIGFQDTLCDDYGKHFFKDCFIQGTYDFIFGNGKS 219
D + G QAL+MRISG+ AAFYNCKF GFQDT+CDD G HFFKDC+++GT+DFIFG+G S
Sbjct: 144 DGKTKGAQALSMRISGNFAAFYNCKFYGFQDTICDDTGNHFFKDCYVEGTFDFIFGSGTS 203
Query: 220 IYLRTTIESVAKGFE--CHHS-------------------TR*GKNVGRHWIHFCSLQHN 258
+YL T + V G H+ T G +GR W+ + +
Sbjct: 204 MYLGTQLHVVGDGIRVIAAHAGKSAEEKSGYSFVHCKVTGTGGGIYLGRAWMSHPKVVYA 263
Query: 259 W-KWSQEHLPWSWLEEEP*GCICLYLYGLCRQFQRMVPPWVQ*AVRLNYKRILSDEEAKH 317
+ + + P W E + YG + P A R+ + + + D+EA
Sbjct: 264 YTEMTSVVNPTGWQENKTPAHDKTVFYGE----YKCSGPGSHKAKRVPFTQDIDDKEANR 319
Query: 318 FLSMAYIHGEQWVRPPP 334
FLS+ YI G +W+ PPP
Sbjct: 320 FLSLGYIQGSKWLLPPP 336
>emb|CAB87931.1| pectin methyl-esterase-like protein [Arabidopsis thaliana]
gi|21555245|gb|AAM63813.1| pectin methyl-esterase-like
protein [Arabidopsis thaliana]
gi|28827496|gb|AAO50592.1| putative pectinesterase
[Arabidopsis thaliana] gi|27754286|gb|AAO22596.1|
putative pectinesterase [Arabidopsis thaliana]
gi|15240781|ref|NP_196359.1| pectinesterase family
protein [Arabidopsis thaliana] gi|11358604|pir||T49881
pectin methyl-esterase-like protein - Arabidopsis
thaliana
Length = 361
Score = 220 bits (560), Expect = 5e-56
Identities = 128/319 (40%), Positives = 180/319 (56%), Gaps = 36/319 (11%)
Query: 40 LEEHLTAAETTVRVVRVRKDGTGDFTTVTDAVKSIPSGNKRRVVVWIGMGEYREKITVDR 99
L+ L AAE + RV+ V ++G GDF T+ A+KSIP NK RV++ + G Y EK+TVD
Sbjct: 53 LDPELEAAEASRRVIIVNQNGGGDFKTINAAIKSIPLANKNRVIIKLAPGIYHEKVTVDV 112
Query: 100 SKRFVTFYGERNGKDNDMMPIITYDATALRYGTLDSATVAVDADYFVAVNVAFVNSSPMP 159
+ +VT G+ + N +TY TA +YGT++SAT+ V A F+A N+ +N+SPMP
Sbjct: 113 GRPYVTLLGKPGAETN-----LTYAGTAAKYGTVESATLIVWATNFLAANLNIINTSPMP 167
Query: 160 DENSVGGQALAMRISGDKAAFYNCKFIGFQDTLCDDYGKHFFKDCFIQGTYDFIFGNGKS 219
+ G QALAMRI+GDKAAFYNC+F GFQDTLCDD G HFFK+C+I+GTYDFIFG G S
Sbjct: 168 KPGTQG-QALAMRINGDKAAFYNCRFYGFQDTLCDDRGNHFFKNCYIEGTYDFIFGRGAS 226
Query: 220 IYLRTTIESVAKGFE---CHH------------------STR*GKNVGRHWIHFCSLQHN 258
+YL T + +V G H+ G +GR W+ + ++
Sbjct: 227 LYLTTQLHAVGDGLRVIAAHNRQSTTEQNGYSFVHCKVTGVGTGIYLGRAWMSHPKVVYS 286
Query: 259 W-KWSQEHLPWSWLEEEP*GCICLYLYG--LCRQFQRMVPPWVQ*AVRLNYKRILSDEEA 315
+ + S P W E YG +C P A R+ + + + ++EA
Sbjct: 287 YTEMSSVVNPSGWQENRVRAHDKTVFYGEYMC------TGPGSHKAKRVAHTQDIDNKEA 340
Query: 316 KHFLSMAYIHGEQWVRPPP 334
FL++ YI G +W+ PPP
Sbjct: 341 SQFLTLGYIKGSKWLLPPP 359
>emb|CAB87932.1| pectin methyl-esterase-like protein [Arabidopsis thaliana]
gi|15240792|ref|NP_196360.1| pectinesterase family
protein [Arabidopsis thaliana] gi|11358605|pir||T49882
pectin methyl-esterase-like protein - Arabidopsis
thaliana
Length = 361
Score = 214 bits (544), Expect = 4e-54
Identities = 122/318 (38%), Positives = 176/318 (54%), Gaps = 32/318 (10%)
Query: 40 LEEHLTAAETTVRVVRVRKDGTGDFTTVTDAVKSIPSGNKRRVVVWIGMGEYREKITVDR 99
L+ L AAE +++ V + G +F T+ +A+KSIP+GNK RV++ + G Y EK+T+D
Sbjct: 53 LDPALEAAEAARQIITVNQKGGANFKTLNEAIKSIPTGNKNRVIIKLAPGVYNEKVTIDI 112
Query: 100 SKRFVTFYGERNGKDNDMMPIITYDATALRYGTLDSATVAVDADYFVAVNVAFVNSSPMP 159
++ F+T G+ + ++TY TA +YGT++SAT+ V A+YF A ++ N++PMP
Sbjct: 113 ARPFITLLGQPGAET-----VLTYHGTAAQYGTVESATLIVWAEYFQAAHLTIKNTAPMP 167
Query: 160 DENSVGGQALAMRISGDKAAFYNCKFIGFQDTLCDDYGKHFFKDCFIQGTYDFIFGNGKS 219
S G QALAMRI+ DKAAFY+C+F GFQDTLCDD G HFFKDC+I+GTYDFIFG G S
Sbjct: 168 KPGSQG-QALAMRINADKAAFYSCRFHGFQDTLCDDKGNHFFKDCYIEGTYDFIFGRGAS 226
Query: 220 IYLRTTIESVAKGFECHHS---------------------TR*GKNVGRHWIHFCSLQHN 258
+YL T + +V G + T G +GR W+ + +
Sbjct: 227 LYLNTQLHAVGDGLRVITAQGRQSATEQNGYTFVHCKVTGTGTGIYLGRSWMSHPKVVYA 286
Query: 259 W-KWSQEHLPWSWLEEEP*GCICLYLYGLCRQFQRMVPPWVQ*AVRLNYKRILSDEEAKH 317
+ + + P W E G YG + F P R+ Y + + E
Sbjct: 287 FTEMTSVVNPSGWRENLNRGYDKTVFYGEYKCFG----PGSHLEKRVPYTQDIDKNEVTP 342
Query: 318 FLSMAYIHGEQWVRPPPK 335
FL++ YI G W+ PPPK
Sbjct: 343 FLTLGYIKGSTWLLPPPK 360
>gb|AAT99258.1| pectin-methyltransferase precursor [Salsola kali]
Length = 362
Score = 210 bits (535), Expect = 4e-53
Identities = 119/308 (38%), Positives = 164/308 (52%), Gaps = 31/308 (10%)
Query: 51 VRVVRVRKDGTGDFTTVTDAVKSIPSGNKRRVVVWIGMGEYREKITVDRSKRFVTFYGER 110
V + VR+DG+G F T++DAVK + GN +RV++ IG GEYREK+ ++R ++T YG
Sbjct: 62 VETIEVRQDGSGKFKTISDAVKHVKVGNTKRVIITIGPGEYREKVKIERLHPYITLYGI- 120
Query: 111 NGKDNDMMPIITYDATALRYGTLDSATVAVDADYFVAVNVAFVNSSPMPDENSVGGQALA 170
D P IT+ TA +GT+DSATV V++DY V ++ NS+P PD G QA A
Sbjct: 121 ---DPKNRPTITFAGTAAEFGTVDSATVIVESDYSVGAHLIVTNSAPRPDGKRKGAQAGA 177
Query: 171 MRISGDKAAFYNCKFIGFQDTLCDDYGKHFFKDCFIQGTYDFIFGNGKSIYLRTTIESV- 229
+RISGD+AAFYNCKF GFQDT+CDD G HFF DC+ +GT DFIFG +S+YL T + V
Sbjct: 178 LRISGDRAAFYNCKFTGFQDTVCDDKGNHFFTDCYTEGTVDFIFGEARSLYLNTELHVVP 237
Query: 230 -----------------AKGFECHHSTR*GKN----VGRHWIHFCSLQHNW-KWSQEHLP 267
G+ H G +GR W + ++ S P
Sbjct: 238 GDPMAMITAHARKNADGVGGYSFVHCKVTGTGGTALLGRAWFDAARVVFSYCNLSDAAKP 297
Query: 268 WSWLEEEP*GCICLYLYGLCRQFQRMVPPWVQ*AVRLNYKRILSDEEAKHFLSMAYIHGE 327
W + L+G + P R Y + L++ +AK F S+ YI
Sbjct: 298 EGWSDNNKPEAQKTILFGEYKN----TGPGAAPDKRAPYTKQLTEADAKTFTSLEYIEAA 353
Query: 328 QWVRPPPK 335
+W+ PPPK
Sbjct: 354 KWLLPPPK 361
>gb|AAM20209.1| putative pectin methylesterase [Arabidopsis thaliana]
gi|17529290|gb|AAL38872.1| putative pectin
methylesterase [Arabidopsis thaliana]
gi|15241163|ref|NP_197474.1| pectinesterase family
protein [Arabidopsis thaliana]
Length = 383
Score = 157 bits (396), Expect = 6e-37
Identities = 100/302 (33%), Positives = 153/302 (50%), Gaps = 35/302 (11%)
Query: 57 RKDGTGDFTTVTDAVKSIPSGNKRRVVVWIGMGEYREKITVDRSKRFVTFYGERNGK--- 113
+K GDFT + DA+ S+P N RVV+ + G Y+EK+++ K F+T GE K
Sbjct: 89 KKSNKGDFTKIQDAIDSLPLINFVRVVIKVHAGVYKEKVSIPPLKAFITIEGEGAEKTTV 148
Query: 114 ---DNDMMPIITYDATALRYGTLDSATVAVDADYFVAVNVAFVNSSPMPDENSVGGQALA 170
D P D+ GT +SA+ AV++ +FVA N+ F N++P+P +VG QA+A
Sbjct: 149 EWGDTAQTP----DSKGNPMGTYNSASFAVNSPFFVAKNITFRNTTPVPLPGAVGKQAVA 204
Query: 171 MRISGDKAAFYNCKFIGFQDTLCDDYGKHFFKDCFIQGTYDFIFGNGKSIYLRTTIESVA 230
+R+S D AAF+ C+ +G QDTL D G+H++KDC+I+G+ DFIFGN S+Y + ++A
Sbjct: 205 LRVSADNAAFFGCRMLGAQDTLYDHLGRHYYKDCYIEGSVDFIFGNALSLYEGCHVHAIA 264
Query: 231 -----------------KGF---ECHHSTR*GKNVGRHWIHFCSLQHNWKWSQE-HLPWS 269
GF +C + +GR W F + + + LP
Sbjct: 265 DKLGAVTAQGRSSVLEDTGFSFVKCKVTGTGVLYLGRAWGPFSRVVFAYTYMDNIILPRG 324
Query: 270 WLEEEP*GCICLYLYGLCRQFQRMVPPWVQ*AVRLNYKRILSDEEAKHFLSMAYIHGEQW 329
W YG + R+ + R L+DEEAK FLS+ +I G +W
Sbjct: 325 WYNWGDPSREMTVFYGQ----YKCTGAGANYGGRVAWARELTDEEAKPFLSLTFIDGSEW 380
Query: 330 VR 331
++
Sbjct: 381 IK 382
>gb|AAV59317.1| putative pectin methylesterase [Oryza sativa (japonica
cultivar-group)]
Length = 398
Score = 156 bits (394), Expect = 9e-37
Identities = 97/300 (32%), Positives = 150/300 (49%), Gaps = 31/300 (10%)
Query: 57 RKDGTGDFTTVTDAVKSIPSGNKRRVVVWIGMGEYREKITVDRSKRFVTFYGERNGKDND 116
+ G+FT++ AV SIP N RVV+ + G Y EK+T+ + FVT G G D
Sbjct: 104 KNPAAGNFTSIQAAVDSIPLINLARVVIKVNAGTYTEKVTISPLRAFVTIEGA--GADKT 161
Query: 117 MMP----IITYDATALRYGTLDSATVAVDADYFVAVNVAFVNSSPMPDENSVGGQALAMR 172
++ T +GT SAT AV+A +FVA N+ F N++P+P ++G Q +A+R
Sbjct: 162 VVQWGDTADTVGPLGRPFGTFASATFAVNAQFFVAKNITFKNTAPVPRPGALGKQGVALR 221
Query: 173 ISGDKAAFYNCKFIGFQDTLCDDYGKHFFKDCFIQGTYDFIFGNGKSIYLRTTIESVAKG 232
IS D AAF C F+G QDTL D G+H+++DC+I+G+ DFIFGN S+Y + ++A+
Sbjct: 222 ISADNAAFLGCNFLGAQDTLYDHLGRHYYRDCYIEGSVDFIFGNALSLYEGCHVHAIARN 281
Query: 233 F--------------------ECHHSTR*GKNVGRHWIHFCSLQHNWKWSQE-HLPWSWL 271
+ C + +GR W F + + + +P W
Sbjct: 282 YGALTAQNRMSILEDTGFSFVNCRVTGSGALYLGRAWGTFSRVVFAYTYMDNIIIPRGWY 341
Query: 272 EEEP*GCICLYLYGLCRQFQRMVPPWVQ*AVRLNYKRILSDEEAKHFLSMAYIHGEQWVR 331
YG + P A R+ + R L+D+EAK F+S+++I G +WV+
Sbjct: 342 NWGDPTREMTVFYGQ----YKCTGPGSNYAGRVAWSRELTDQEAKPFISLSFIDGLEWVK 397
>emb|CAD41229.2| OSJNBa0010H02.16 [Oryza sativa (japonica cultivar-group)]
gi|50927829|ref|XP_473442.1| OSJNBa0010H02.16 [Oryza
sativa (japonica cultivar-group)]
Length = 344
Score = 155 bits (392), Expect = 2e-36
Identities = 82/191 (42%), Positives = 117/191 (60%), Gaps = 14/191 (7%)
Query: 51 VRVVRVRKDGTGDFTTVTDAVKSIPSGNKRRVVVWIGMGEYREKITVDRSKRFVTFYG-- 108
VR + V G GDF ++ AV S+P N RV++ I G Y EK+ V +K ++TF G
Sbjct: 41 VRRIVVDASGGGDFLSIQQAVNSVPENNTVRVIMQINAGSYIEKVVVPATKPYITFQGAG 100
Query: 109 ------ERNGKDNDMMPIITYDATALRYGTLDSATVAVDADYFVAVNVAFVNSSPMPDEN 162
E + + +D P D LR T ++A+V V ++YF A N++F N++P P
Sbjct: 101 RDVTVVEWHDRASDRGP----DGQQLR--TYNTASVTVLSNYFTAKNISFKNTAPAPMPG 154
Query: 163 SVGGQALAMRISGDKAAFYNCKFIGFQDTLCDDYGKHFFKDCFIQGTYDFIFGNGKSIYL 222
G QA+A RISGDKA F+ C F G QDTLCDD G+H+F+DC+I+G+ DF+FGNG+S+Y
Sbjct: 155 MQGWQAVAFRISGDKAFFFGCGFYGAQDTLCDDAGRHYFRDCYIEGSIDFVFGNGRSLYK 214
Query: 223 RTTIESVAKGF 233
+ S A+ F
Sbjct: 215 DCELHSTAQRF 225
>gb|AAN18134.1| At5g47500/MNJ7_9 [Arabidopsis thaliana] gi|20856815|gb|AAM26686.1|
AT5g47500/MNJ7_9 [Arabidopsis thaliana]
gi|15238111|ref|NP_199561.1| pectinesterase family
protein [Arabidopsis thaliana]
Length = 362
Score = 155 bits (391), Expect = 2e-36
Identities = 81/188 (43%), Positives = 113/188 (60%), Gaps = 10/188 (5%)
Query: 52 RVVRVRKDGTGDFTTVTDAVKSIPSGNKRRVVVWIGMGEYREKITVDRSKRFVTFYGERN 111
+V+ V +G F +V DAV SIP N + + + I G YREK+ V +K ++TF
Sbjct: 59 KVITVSLNGHAQFRSVQDAVDSIPKNNNKSITIKIAPGFYREKVVVPATKPYITF----K 114
Query: 112 GKDNDMMPIITYD------ATALRYGTLDSATVAVDADYFVAVNVAFVNSSPMPDENSVG 165
G D+ I +D A + T +A+V V A+YF A N++F N++P P G
Sbjct: 115 GAGRDVTAIEWHDRASDLGANGQQLRTYQTASVTVYANYFTARNISFTNTAPAPLPGMQG 174
Query: 166 GQALAMRISGDKAAFYNCKFIGFQDTLCDDYGKHFFKDCFIQGTYDFIFGNGKSIYLRTT 225
QA+A RISGDKA F C F G QDTLCDD G+H+FK+C+I+G+ DFIFGNG+S+Y
Sbjct: 175 WQAVAFRISGDKAFFSGCGFYGAQDTLCDDAGRHYFKECYIEGSIDFIFGNGRSMYKDCE 234
Query: 226 IESVAKGF 233
+ S+A F
Sbjct: 235 LHSIASRF 242
>gb|AAD20147.1| putative pectinesterase [Arabidopsis thaliana]
gi|25408502|pir||G84783 probable pectinesterase
[imported] - Arabidopsis thaliana
gi|15228023|ref|NP_181209.1| pectinesterase family
protein [Arabidopsis thaliana]
Length = 407
Score = 152 bits (384), Expect = 1e-35
Identities = 86/206 (41%), Positives = 119/206 (57%), Gaps = 9/206 (4%)
Query: 31 FNRHREITLLEE---HLTAAETTVRVVRVRKDGTGDFTTVTDAVKSIPSGNKRRVVVWIG 87
++ H I E+ L T V+ V G G+F+ V A+ +P + + ++ +
Sbjct: 65 YHHHEPIKCCEKWTSRLRHQYKTSLVLTVDLHGCGNFSNVQSAIDVVPDLSSSKTLIIVN 124
Query: 88 MGEYREKITVDRSKRFVTFYGERNGKDNDMMPIITYDATALRYG-TLDSATVAVDADYFV 146
G YREK+TV+ +K + G G N I ++ TA G T DS + V A F
Sbjct: 125 SGSYREKVTVNENKTNLVIQGR--GYQNTS---IEWNDTAKSAGNTADSFSFVVFAANFT 179
Query: 147 AVNVAFVNSSPMPDENSVGGQALAMRISGDKAAFYNCKFIGFQDTLCDDYGKHFFKDCFI 206
A N++F N++P PD QA+A+RI GD+AAFY C F G QDTL DD G+HFFK+CFI
Sbjct: 180 AYNISFKNNAPEPDPGEADAQAVALRIEGDQAAFYGCGFYGAQDTLLDDKGRHFFKECFI 239
Query: 207 QGTYDFIFGNGKSIYLRTTIESVAKG 232
QG+ DFIFGNG+S+Y TI S+AKG
Sbjct: 240 QGSIDFIFGNGRSLYQDCTINSIAKG 265
>ref|NP_172023.1| pectinesterase family protein [Arabidopsis thaliana]
Length = 393
Score = 151 bits (381), Expect = 3e-35
Identities = 80/181 (44%), Positives = 112/181 (61%), Gaps = 4/181 (2%)
Query: 50 TVRVVRVRKDGTGDFTTVTDAVKSIPSGNKRRVVVWIGMGEYREKITVDRSKRFVTFYGE 109
T + V K+G +FTTV AV ++ + ++RR V+WI G Y EK+ + ++K +T G+
Sbjct: 87 TTSYLCVDKNGCCNFTTVQSAVDAVGNFSQRRNVIWINSGMYYEKVVIPKTKPNITLQGQ 146
Query: 110 RNGKDNDMMPIITYDATALRYGTLDSATVAVDADYFVAVNVAFVNSSPMPDENSVGGQAL 169
D+ I D GT ATV V FVA N++F+N +P+P VG QA+
Sbjct: 147 ----GFDITAIAWNDTAYSANGTFYCATVQVFGSQFVAKNISFMNVAPIPKPGDVGAQAV 202
Query: 170 AMRISGDKAAFYNCKFIGFQDTLCDDYGKHFFKDCFIQGTYDFIFGNGKSIYLRTTIESV 229
A+RI+GD++AF C F G QDTL DD G+H+FKDC+IQG+ DFIFGN KS+Y I S+
Sbjct: 203 AIRIAGDESAFVGCGFFGAQDTLHDDRGRHYFKDCYIQGSIDFIFGNAKSLYQDCRIISM 262
Query: 230 A 230
A
Sbjct: 263 A 263
>gb|AAO22722.1| putative pectinesterase family protein [Arabidopsis thaliana]
Length = 407
Score = 149 bits (377), Expect = 9e-35
Identities = 85/206 (41%), Positives = 118/206 (57%), Gaps = 9/206 (4%)
Query: 31 FNRHREITLLEE---HLTAAETTVRVVRVRKDGTGDFTTVTDAVKSIPSGNKRRVVVWIG 87
++ H I E+ L T V+ V G G+F+ V A+ +P + + ++ +
Sbjct: 65 YHHHEPIKCCEKWTSRLRHQYKTSLVLTVDLHGCGNFSNVQSAIDVVPDLSSSKTLIIVN 124
Query: 88 MGEYREKITVDRSKRFVTFYGERNGKDNDMMPIITYDATALRYG-TLDSATVAVDADYFV 146
G YREK+TV+ +K + G G N I ++ TA G T DS + V A F
Sbjct: 125 SGSYREKVTVNENKTNLVIQGR--GYQNTS---IEWNDTAKSAGNTADSFSFVVFAANFT 179
Query: 147 AVNVAFVNSSPMPDENSVGGQALAMRISGDKAAFYNCKFIGFQDTLCDDYGKHFFKDCFI 206
A N++F N++P PD QA+A+RI GD+AAFY C F G QDTL DD G+HFFK+CFI
Sbjct: 180 AYNISFKNNAPEPDPGEADAQAVALRIEGDQAAFYGCGFYGAQDTLLDDKGRHFFKECFI 239
Query: 207 QGTYDFIFGNGKSIYLRTTIESVAKG 232
QG+ FIFGNG+S+Y TI S+AKG
Sbjct: 240 QGSIGFIFGNGRSLYQDCTINSIAKG 265
>gb|AAM20211.1| putative pectinesterase [Arabidopsis thaliana]
gi|17979179|gb|AAL49785.1| putative pectinesterase
[Arabidopsis thaliana] gi|21536660|gb|AAM60992.1|
putative pectinesterase [Arabidopsis thaliana]
gi|9294028|dbj|BAB01985.1| pectin methylesterase-like
protein [Arabidopsis thaliana]
gi|18406048|ref|NP_566842.1| pectinesterase family
protein [Arabidopsis thaliana]
Length = 317
Score = 149 bits (376), Expect = 1e-34
Identities = 99/315 (31%), Positives = 157/315 (49%), Gaps = 53/315 (16%)
Query: 50 TVRVVRVRKDGTGDFTTVTDAVKSIPSGNKRRVVVWIGMGEYREKITVDRSKRFVTFYGE 109
T R+VRV +DG+GD+ +V DA+ S+P GN R V+ + G YR+ + V + K F+TF G
Sbjct: 3 TTRMVRVSQDGSGDYCSVQDAIDSVPLGNTCRTVIRLSPGIYRQPVYVPKRKNFITFAGI 62
Query: 110 RNGKDNDMMPIITYDATALRY-----------GTLDSATVAVDADYFVAVNVAFVNSSPM 158
+ + ++T++ TA + GT +V V+ + F+A N+ F NS+P
Sbjct: 63 -----SPEITVLTWNNTASKIEHHQASRVIGTGTFGCGSVIVEGEDFIAENITFENSAP- 116
Query: 159 PDENSVGGQALAMRISGDKAAFYNCKFIGFQDTLCDDYGKHFFKDCFIQGTYDFIFGNGK 218
E S GQA+A+R++ D+ AFYNC+F+G+QDTL +GK + KDC+I+G+ DFIFGN
Sbjct: 117 --EGS--GQAVAIRVTADRCAFYNCRFLGWQDTLYLHHGKQYLKDCYIEGSVDFIFGNST 172
Query: 219 SIYLRTTIESVAKGFECHHSTR*GKNVGRHWIHFCSLQHNWKWSQEHL--PW-------- 268
++ I ++GF S + + + C + N + +L PW
Sbjct: 173 ALLEHCHIHCKSQGFITAQSRKSSQESTGYVFLRCVITGNGQSGYMYLGRPWGPFGRVVL 232
Query: 269 ---------------SWLEEEP*GCICLYLYGLCRQFQRMVPPWVQ*AVRLNYKRILSDE 313
+W E C Y Y R P + R+ + R L D+
Sbjct: 233 AYTYMDACIRNVGWHNWGNAENERSACFYEY-------RCFGPGSCSSERVPWSRELMDD 285
Query: 314 EAKHFLSMAYIHGEQ 328
EA HF+ +++ EQ
Sbjct: 286 EAGHFVHHSFVDPEQ 300
>dbj|BAD32030.1| pectin methylesterase-like protein [Oryza sativa (japonica
cultivar-group)] gi|50508665|dbj|BAD31151.1| pectin
methylesterase-like protein [Oryza sativa (japonica
cultivar-group)]
Length = 324
Score = 147 bits (372), Expect = 3e-34
Identities = 80/185 (43%), Positives = 112/185 (60%), Gaps = 12/185 (6%)
Query: 56 VRKDGTGDFTTVTDAVKSIPSGNKRRVVVWIGMGEYREKITVDRSKRFVTFYGERNGKDN 115
V DGTG TV AV +P+GN RRV + + G YREK+TV +K FV+ G G
Sbjct: 77 VSPDGTGHSRTVQGAVDMVPAGNTRRVKIVVRPGVYREKVTVPITKPFVSLIGMGTGHT- 135
Query: 116 DMMPIITYDATAL-------RYGTLDSATVAVDADYFVAVNVAFVNSSPMPDENSVGGQA 168
+IT+ + A + GT SA+VAV+ADYF A ++ F NS+ +VG QA
Sbjct: 136 ----VITWHSRASDVGASGHQVGTFYSASVAVEADYFCASHITFENSAAAAAPGAVGQQA 191
Query: 169 LAMRISGDKAAFYNCKFIGFQDTLCDDYGKHFFKDCFIQGTYDFIFGNGKSIYLRTTIES 228
+A+R+SGDK Y C+ +G QDTL D+ G+H+ +C IQG+ DFIFGN +S+Y T+ +
Sbjct: 192 VALRLSGDKTVLYKCRILGTQDTLFDNIGRHYLYNCDIQGSIDFIFGNARSLYQGCTLHA 251
Query: 229 VAKGF 233
VA +
Sbjct: 252 VATSY 256
>dbj|BAB09076.1| pectin methylesterase-like [Arabidopsis thaliana]
Length = 359
Score = 145 bits (367), Expect = 1e-33
Identities = 79/188 (42%), Positives = 111/188 (59%), Gaps = 12/188 (6%)
Query: 52 RVVRVRKDGTGDFTTVTDAVKSIPSGNKRRVVVWIGMGEYREKITVDRSKRFVTFYGERN 111
+V+ V +G F +V DAV SIP N + + + I G EK+ V +K ++TF
Sbjct: 59 KVITVSLNGHAQFRSVQDAVDSIPKNNNKSITIKIAPG--LEKVVVPATKPYITF----K 112
Query: 112 GKDNDMMPIITYD------ATALRYGTLDSATVAVDADYFVAVNVAFVNSSPMPDENSVG 165
G D+ I +D A + T +A+V V A+YF A N++F N++P P G
Sbjct: 113 GAGRDVTAIEWHDRASDLGANGQQLRTYQTASVTVYANYFTARNISFTNTAPAPLPGMQG 172
Query: 166 GQALAMRISGDKAAFYNCKFIGFQDTLCDDYGKHFFKDCFIQGTYDFIFGNGKSIYLRTT 225
QA+A RISGDKA F C F G QDTLCDD G+H+FK+C+I+G+ DFIFGNG+S+Y
Sbjct: 173 WQAVAFRISGDKAFFSGCGFYGAQDTLCDDAGRHYFKECYIEGSIDFIFGNGRSMYKDCE 232
Query: 226 IESVAKGF 233
+ S+A F
Sbjct: 233 LHSIASRF 240
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.328 0.141 0.457
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 594,067,666
Number of Sequences: 2540612
Number of extensions: 24274386
Number of successful extensions: 60104
Number of sequences better than 10.0: 303
Number of HSP's better than 10.0 without gapping: 287
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 59346
Number of HSP's gapped (non-prelim): 386
length of query: 335
length of database: 863,360,394
effective HSP length: 128
effective length of query: 207
effective length of database: 538,162,058
effective search space: 111399546006
effective search space used: 111399546006
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 75 (33.5 bits)
Medicago: description of AC124967.4


