

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124967.12 - phase: 0
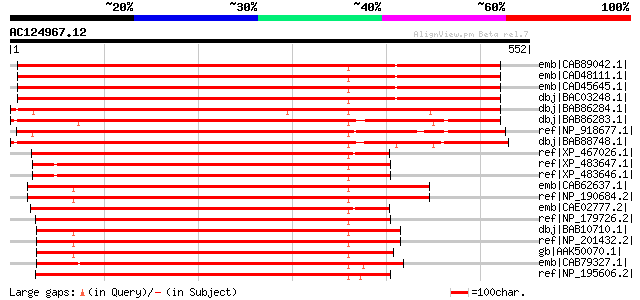
(552 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAB89042.1| kinesin-like protein [Arabidopsis thaliana] gi|1... 624 e-177
emb|CAD48111.1| putative kinesin protein [Arabidopsis thaliana] ... 624 e-177
emb|CAD45645.1| putative kinesin-like protein [Arabidopsis thali... 624 e-177
dbj|BAC03248.1| kinesin-like protein [Arabidopsis thaliana] gi|3... 624 e-177
dbj|BAB86284.1| kinesin-like protein NACK2 [Nicotiana tabacum] 618 e-175
dbj|BAB86283.1| kinesin-like protein NACK1 [Nicotiana tabacum] 564 e-159
ref|NP_918677.1| putative kinesin [Oryza sativa (japonica cultiv... 546 e-154
dbj|BAB88748.1| AtNACK1 kinesin-like protein [Arabidopsis thalia... 538 e-151
ref|XP_467026.1| putative kinesin heavy chain [Oryza sativa (jap... 421 e-116
ref|XP_483647.1| putative kinesin heavy chain [Oryza sativa (jap... 418 e-115
ref|XP_483646.1| putative kinesin heavy chain [Oryza sativa (jap... 418 e-115
emb|CAB62637.1| putative protein [Arabidopsis thaliana] gi|11357... 414 e-114
ref|NP_190684.2| kinesin motor family protein [Arabidopsis thali... 414 e-114
emb|CAE02777.2| OSJNBa0011L07.1 [Oryza sativa (japonica cultivar... 413 e-114
ref|NP_179726.2| kinesin motor family protein [Arabidopsis thali... 406 e-111
dbj|BAB10710.1| kinesin heavy chain DNA binding protein-like [Ar... 403 e-111
ref|NP_201432.2| kinesin motor family protein [Arabidopsis thali... 403 e-111
gb|AAK50070.1| AT5g66310/K1L20_9 [Arabidopsis thaliana] 402 e-110
emb|CAB79327.1| putative protein [Arabidopsis thaliana] gi|56686... 401 e-110
ref|NP_195606.2| kinesin motor family protein [Arabidopsis thali... 401 e-110
>emb|CAB89042.1| kinesin-like protein [Arabidopsis thaliana] gi|11358478|pir||T49235
kinesin-like protein - Arabidopsis thaliana
Length = 932
Score = 624 bits (1609), Expect = e-177
Identities = 332/535 (62%), Positives = 409/535 (76%), Gaps = 22/535 (4%)
Query: 9 TPSSKIRRKLADTPGGSKIREEKIRVTVRMRPLNRKEQAMYDLIAWDCLDDKTIVFKNPN 68
TP SKI + TP GSK+ EEKI VTVRMRPLN +E A YDLIAW+C DD+TIVFKNPN
Sbjct: 6 TPLSKIDKSNPYTPCGSKVTEEKILVTVRMRPLNWREHAKYDLIAWECPDDETIVFKNPN 65
Query: 69 QERPSTSYTFDRVFPPACSTQKVYDEGAKDVALSALSGINATIFAYGQTSSGKTFTMRGI 128
++ T Y+FD+VF P C+TQ+VY+ G++DVALSAL+G NATIFAYGQTSSGKTFTMRG+
Sbjct: 66 PDKAPTKYSFDKVFEPTCATQEVYEGGSRDVALSALAGTNATIFAYGQTSSGKTFTMRGV 125
Query: 129 TENAIRDIYECIKNTPDRDFVLKISALEIYNETVIDLLNRESGPLRILDDTEKVTVVENL 188
TE+ ++DIYE I+ T +R FVLK+SALEIYNETV+DLLNR++GPLR+LDD EK T+VENL
Sbjct: 126 TESVVKDIYEHIRKTQERSFVLKVSALEIYNETVVDLLNRDTGPLRLLDDPEKGTIVENL 185
Query: 189 FEEVARDAQHLRHLIGICEAHRQVGETTLNDKSSRSHQIIRLTVESFHRESPDHVKSYIA 248
EEV QHL+HLI ICE RQVGET LNDKSSRSHQIIRLT+ S RE V+S++A
Sbjct: 186 VEEVVESRQHLQHLISICEDQRQVGETALNDKSSRSHQIIRLTIHSSLREIAGCVQSFMA 245
Query: 249 SLNFVDLAGSERASQTNTCGTRLKEGSHINKSLLQLALVIRQLSSG-KSGHISYRTSKLT 307
+LN VDLAGSERA QTN G RLKEGSHIN+SLL L VIR+LSSG K H+ YR SKLT
Sbjct: 246 TLNLVDLAGSERAFQTNADGLRLKEGSHINRSLLTLTTVIRKLSSGRKRDHVPYRDSKLT 305
Query: 308 RILQSSLGGNARTAIICTVSPSLSHVEQTRNTLSFATNAKEVINTARVNMV--------- 358
RILQ+SLGGNARTAIICT+SP+LSHVEQT+ TLSFA +AKEV N A+VNMV
Sbjct: 306 RILQNSLGGNARTAIICTISPALSHVEQTKKTLSFAMSAKEVTNCAKVNMVVSEKKLLKH 365
Query: 359 --------EGELRNPEPEHAG-LRSLLAEKELKIQQMEKDMEDLRRQRDLAQCQLDLERR 409
E ELR+PEP + L+SLL EKE+KIQQME +M++L+RQRD+AQ +LDLER+
Sbjct: 366 LQQKVAKLESELRSPEPSSSTCLKSLLIEKEMKIQQMESEMKELKRQRDIAQSELDLERK 425
Query: 410 ANKVQKGSSDYGPSSQVVRCLSFAEENELAIGKHTP--ERTETVSRQAMLKNLLASPDPS 467
A K +KGSS+ P SQV RCLS+ + E K P RT R+ ++ L S DP+
Sbjct: 426 A-KERKGSSECEPFSQVARCLSYHTKEESIPSKSVPSSRRTARDRRKDNVRQSLTSADPT 484
Query: 468 ILVDEIQKLEHRQLQLCEDANRALEVLHKDFATHNLGNQETAETMSKVLSEIKDL 522
LV EI+ LE Q +L E+AN+AL+++HK+ +H LG+Q+ AE ++K+LSEI+D+
Sbjct: 485 ALVQEIRLLEKHQKKLGEEANQALDLIHKEVTSHKLGDQQAAEKVAKMLSEIRDM 539
>emb|CAD48111.1| putative kinesin protein [Arabidopsis thaliana]
gi|22796151|emb|CAD42234.1| kinesin-like protein
[Arabidopsis thaliana]
Length = 937
Score = 624 bits (1609), Expect = e-177
Identities = 332/535 (62%), Positives = 409/535 (76%), Gaps = 22/535 (4%)
Query: 9 TPSSKIRRKLADTPGGSKIREEKIRVTVRMRPLNRKEQAMYDLIAWDCLDDKTIVFKNPN 68
TP SKI + TP GSK+ EEKI VTVRMRPLN +E A YDLIAW+C DD+TIVFKNPN
Sbjct: 6 TPLSKIDKSNPYTPCGSKVTEEKILVTVRMRPLNWREHAKYDLIAWECPDDETIVFKNPN 65
Query: 69 QERPSTSYTFDRVFPPACSTQKVYDEGAKDVALSALSGINATIFAYGQTSSGKTFTMRGI 128
++ T Y+FD+VF P C+TQ+VY+ G++DVALSAL+G NATIFAYGQTSSGKTFTMRG+
Sbjct: 66 PDKAPTKYSFDKVFEPTCATQEVYEGGSRDVALSALAGTNATIFAYGQTSSGKTFTMRGV 125
Query: 129 TENAIRDIYECIKNTPDRDFVLKISALEIYNETVIDLLNRESGPLRILDDTEKVTVVENL 188
TE+ ++DIYE I+ T +R FVLK+SALEIYNETV+DLLNR++GPLR+LDD EK T+VENL
Sbjct: 126 TESVVKDIYEHIRKTQERSFVLKVSALEIYNETVVDLLNRDTGPLRLLDDPEKGTIVENL 185
Query: 189 FEEVARDAQHLRHLIGICEAHRQVGETTLNDKSSRSHQIIRLTVESFHRESPDHVKSYIA 248
EEV QHL+HLI ICE RQVGET LNDKSSRSHQIIRLT+ S RE V+S++A
Sbjct: 186 VEEVVESRQHLQHLISICEDQRQVGETALNDKSSRSHQIIRLTIHSSLREIAGCVQSFMA 245
Query: 249 SLNFVDLAGSERASQTNTCGTRLKEGSHINKSLLQLALVIRQLSSG-KSGHISYRTSKLT 307
+LN VDLAGSERA QTN G RLKEGSHIN+SLL L VIR+LSSG K H+ YR SKLT
Sbjct: 246 TLNLVDLAGSERAFQTNADGLRLKEGSHINRSLLTLTTVIRKLSSGRKRDHVPYRDSKLT 305
Query: 308 RILQSSLGGNARTAIICTVSPSLSHVEQTRNTLSFATNAKEVINTARVNMV--------- 358
RILQ+SLGGNARTAIICT+SP+LSHVEQT+ TLSFA +AKEV N A+VNMV
Sbjct: 306 RILQNSLGGNARTAIICTISPALSHVEQTKKTLSFAMSAKEVTNCAKVNMVVSEKKLLKH 365
Query: 359 --------EGELRNPEPEHAG-LRSLLAEKELKIQQMEKDMEDLRRQRDLAQCQLDLERR 409
E ELR+PEP + L+SLL EKE+KIQQME +M++L+RQRD+AQ +LDLER+
Sbjct: 366 LQQKVAKLESELRSPEPSSSTCLKSLLIEKEMKIQQMESEMKELKRQRDIAQSELDLERK 425
Query: 410 ANKVQKGSSDYGPSSQVVRCLSFAEENELAIGKHTP--ERTETVSRQAMLKNLLASPDPS 467
A K +KGSS+ P SQV RCLS+ + E K P RT R+ ++ L S DP+
Sbjct: 426 A-KERKGSSECEPFSQVARCLSYHTKEESIPSKSVPSSRRTARDRRKDNVRQSLTSADPT 484
Query: 468 ILVDEIQKLEHRQLQLCEDANRALEVLHKDFATHNLGNQETAETMSKVLSEIKDL 522
LV EI+ LE Q +L E+AN+AL+++HK+ +H LG+Q+ AE ++K+LSEI+D+
Sbjct: 485 ALVQEIRLLEKHQKKLGEEANQALDLIHKEVTSHKLGDQQAAEKVAKMLSEIRDM 539
>emb|CAD45645.1| putative kinesin-like protein [Arabidopsis thaliana]
gi|21954474|emb|CAD42658.1| kinesin-like protein
[Arabidopsis thaliana]
Length = 937
Score = 624 bits (1609), Expect = e-177
Identities = 332/535 (62%), Positives = 409/535 (76%), Gaps = 22/535 (4%)
Query: 9 TPSSKIRRKLADTPGGSKIREEKIRVTVRMRPLNRKEQAMYDLIAWDCLDDKTIVFKNPN 68
TP SKI + TP GSK+ EEKI VTVRMRPLN +E A YDLIAW+C DD+TIVFKNPN
Sbjct: 6 TPLSKIDKSNPYTPCGSKVTEEKILVTVRMRPLNWREHAKYDLIAWECPDDETIVFKNPN 65
Query: 69 QERPSTSYTFDRVFPPACSTQKVYDEGAKDVALSALSGINATIFAYGQTSSGKTFTMRGI 128
++ T Y+FD+VF P C+TQ+VY+ G++DVALSAL+G NATIFAYGQTSSGKTFTMRG+
Sbjct: 66 PDKAPTKYSFDKVFEPTCATQEVYEGGSRDVALSALAGTNATIFAYGQTSSGKTFTMRGV 125
Query: 129 TENAIRDIYECIKNTPDRDFVLKISALEIYNETVIDLLNRESGPLRILDDTEKVTVVENL 188
TE+ ++DIYE I+ T +R FVLK+SALEIYNETV+DLLNR++GPLR+LDD EK T+VENL
Sbjct: 126 TESVVKDIYEHIRKTQERSFVLKVSALEIYNETVVDLLNRDTGPLRLLDDPEKGTIVENL 185
Query: 189 FEEVARDAQHLRHLIGICEAHRQVGETTLNDKSSRSHQIIRLTVESFHRESPDHVKSYIA 248
EEV QHL+HLI ICE RQVGET LNDKSSRSHQIIRLT+ S RE V+S++A
Sbjct: 186 VEEVVESRQHLQHLISICEDQRQVGETALNDKSSRSHQIIRLTIHSSLREIAGCVQSFMA 245
Query: 249 SLNFVDLAGSERASQTNTCGTRLKEGSHINKSLLQLALVIRQLSSG-KSGHISYRTSKLT 307
+LN VDLAGSERA QTN G RLKEGSHIN+SLL L VIR+LSSG K H+ YR SKLT
Sbjct: 246 TLNLVDLAGSERAFQTNADGLRLKEGSHINRSLLTLTTVIRKLSSGRKRDHVPYRDSKLT 305
Query: 308 RILQSSLGGNARTAIICTVSPSLSHVEQTRNTLSFATNAKEVINTARVNMV--------- 358
RILQ+SLGGNARTAIICT+SP+LSHVEQT+ TLSFA +AKEV N A+VNMV
Sbjct: 306 RILQNSLGGNARTAIICTISPALSHVEQTKKTLSFAMSAKEVTNCAKVNMVVSEKKLLKH 365
Query: 359 --------EGELRNPEPEHAG-LRSLLAEKELKIQQMEKDMEDLRRQRDLAQCQLDLERR 409
E ELR+PEP + L+SLL EKE+KIQQME +M++L+RQRD+AQ +LDLER+
Sbjct: 366 LQQKVAKLESELRSPEPSSSTCLKSLLIEKEMKIQQMESEMKELKRQRDIAQSELDLERK 425
Query: 410 ANKVQKGSSDYGPSSQVVRCLSFAEENELAIGKHTP--ERTETVSRQAMLKNLLASPDPS 467
A K +KGSS+ P SQV RCLS+ + E K P RT R+ ++ L S DP+
Sbjct: 426 A-KERKGSSECEPFSQVARCLSYHTKEESIPSKSVPSSRRTARDRRKDNVRQSLTSADPT 484
Query: 468 ILVDEIQKLEHRQLQLCEDANRALEVLHKDFATHNLGNQETAETMSKVLSEIKDL 522
LV EI+ LE Q +L E+AN+AL+++HK+ +H LG+Q+ AE ++K+LSEI+D+
Sbjct: 485 ALVQEIRLLEKHQKKLGEEANQALDLIHKEVTSHKLGDQQAAEKVAKMLSEIRDM 539
>dbj|BAC03248.1| kinesin-like protein [Arabidopsis thaliana]
gi|30690898|ref|NP_189907.2| kinesin motor family
protein (NACK2) [Arabidopsis thaliana]
Length = 938
Score = 624 bits (1609), Expect = e-177
Identities = 332/535 (62%), Positives = 409/535 (76%), Gaps = 22/535 (4%)
Query: 9 TPSSKIRRKLADTPGGSKIREEKIRVTVRMRPLNRKEQAMYDLIAWDCLDDKTIVFKNPN 68
TP SKI + TP GSK+ EEKI VTVRMRPLN +E A YDLIAW+C DD+TIVFKNPN
Sbjct: 7 TPLSKIDKSNPYTPCGSKVTEEKILVTVRMRPLNWREHAKYDLIAWECPDDETIVFKNPN 66
Query: 69 QERPSTSYTFDRVFPPACSTQKVYDEGAKDVALSALSGINATIFAYGQTSSGKTFTMRGI 128
++ T Y+FD+VF P C+TQ+VY+ G++DVALSAL+G NATIFAYGQTSSGKTFTMRG+
Sbjct: 67 PDKAPTKYSFDKVFEPTCATQEVYEGGSRDVALSALAGTNATIFAYGQTSSGKTFTMRGV 126
Query: 129 TENAIRDIYECIKNTPDRDFVLKISALEIYNETVIDLLNRESGPLRILDDTEKVTVVENL 188
TE+ ++DIYE I+ T +R FVLK+SALEIYNETV+DLLNR++GPLR+LDD EK T+VENL
Sbjct: 127 TESVVKDIYEHIRKTQERSFVLKVSALEIYNETVVDLLNRDTGPLRLLDDPEKGTIVENL 186
Query: 189 FEEVARDAQHLRHLIGICEAHRQVGETTLNDKSSRSHQIIRLTVESFHRESPDHVKSYIA 248
EEV QHL+HLI ICE RQVGET LNDKSSRSHQIIRLT+ S RE V+S++A
Sbjct: 187 VEEVVESRQHLQHLISICEDQRQVGETALNDKSSRSHQIIRLTIHSSLREIAGCVQSFMA 246
Query: 249 SLNFVDLAGSERASQTNTCGTRLKEGSHINKSLLQLALVIRQLSSG-KSGHISYRTSKLT 307
+LN VDLAGSERA QTN G RLKEGSHIN+SLL L VIR+LSSG K H+ YR SKLT
Sbjct: 247 TLNLVDLAGSERAFQTNADGLRLKEGSHINRSLLTLTTVIRKLSSGRKRDHVPYRDSKLT 306
Query: 308 RILQSSLGGNARTAIICTVSPSLSHVEQTRNTLSFATNAKEVINTARVNMV--------- 358
RILQ+SLGGNARTAIICT+SP+LSHVEQT+ TLSFA +AKEV N A+VNMV
Sbjct: 307 RILQNSLGGNARTAIICTISPALSHVEQTKKTLSFAMSAKEVTNCAKVNMVVSEKKLLKH 366
Query: 359 --------EGELRNPEPEHAG-LRSLLAEKELKIQQMEKDMEDLRRQRDLAQCQLDLERR 409
E ELR+PEP + L+SLL EKE+KIQQME +M++L+RQRD+AQ +LDLER+
Sbjct: 367 LQQKVAKLESELRSPEPSSSTCLKSLLIEKEMKIQQMESEMKELKRQRDIAQSELDLERK 426
Query: 410 ANKVQKGSSDYGPSSQVVRCLSFAEENELAIGKHTP--ERTETVSRQAMLKNLLASPDPS 467
A K +KGSS+ P SQV RCLS+ + E K P RT R+ ++ L S DP+
Sbjct: 427 A-KERKGSSECEPFSQVARCLSYHTKEESIPSKSVPSSRRTARDRRKDNVRQSLTSADPT 485
Query: 468 ILVDEIQKLEHRQLQLCEDANRALEVLHKDFATHNLGNQETAETMSKVLSEIKDL 522
LV EI+ LE Q +L E+AN+AL+++HK+ +H LG+Q+ AE ++K+LSEI+D+
Sbjct: 486 ALVQEIRLLEKHQKKLGEEANQALDLIHKEVTSHKLGDQQAAEKVAKMLSEIRDM 540
>dbj|BAB86284.1| kinesin-like protein NACK2 [Nicotiana tabacum]
Length = 955
Score = 618 bits (1593), Expect = e-175
Identities = 341/551 (61%), Positives = 415/551 (74%), Gaps = 30/551 (5%)
Query: 1 MVKTPMAATPSSKIRRKLADTPGG-----SKIREEKIRVTVRMRPLNRKEQAMYDLIAWD 55
++ TP+ TP SKI R + PG SKIREEKI VT+R+RPL+ KEQA YDLIAWD
Sbjct: 2 VIGTPVT-TPLSKIVRTPSRVPGSRRTTPSKIREEKILVTIRVRPLSPKEQAAYDLIAWD 60
Query: 56 CLDDKTIVFKNPNQERPSTSYTFDRVFPPACSTQKVYDEGAKDVALSALSGINATIFAYG 115
D++TIV KN N ER + Y+FD VF P CST KVY++GA+DVALSAL+GINATIFAYG
Sbjct: 61 FPDEQTIVSKNLNHERHTGPYSFDYVFDPTCSTSKVYEQGARDVALSALNGINATIFAYG 120
Query: 116 QTSSGKTFTMRGITENAIRDIYECIKNTPDRDFVLKISALEIYNETVIDLLNRESGPLRI 175
QTSSGKTFTMRGITE+A+ DIY IK T +RDFVLK SALEIYNETV+DLLNRES LR+
Sbjct: 121 QTSSGKTFTMRGITESAVNDIYGRIKLTTERDFVLKFSALEIYNETVVDLLNRESVSLRL 180
Query: 176 LDDTEKVTVVENLFEEVARDAQHLRHLIGICEAHRQVGETTLNDKSSRSHQIIRLTVESF 235
LDD EK +VE EE+ +D +HL+ LIG EAHRQVGET LNDKSSRSHQIIRLT+ES
Sbjct: 181 LDDPEKGVIVEKQVEEIVKDEEHLKTLIGTVEAHRQVGETALNDKSSRSHQIIRLTIESS 240
Query: 236 HRESPDHVKSYIASLNFVDLAGSERASQTNTCGTRLKEGSHINKSLLQLALVIRQLSSG- 294
RE+ VKS++A+LN VDLAGSERASQT+ GTRLKEGSHIN+SLL + VIR+LS
Sbjct: 241 IRENSGCVKSFLATLNLVDLAGSERASQTSADGTRLKEGSHINRSLLTVTNVIRKLSCSG 300
Query: 295 --KSGHISYRTSKLTRILQSSLGGNARTAIICTVSPSLSHVEQTRNTLSFATNAKEVINT 352
+SGHI YR SKLTRILQ+SLGGN+RTAIICT+SP+LSH+EQ+RNTL FAT+AKEV T
Sbjct: 301 GKRSGHIPYRDSKLTRILQASLGGNSRTAIICTLSPALSHLEQSRNTLCFATSAKEVTTT 360
Query: 353 ARVNMV-----------------EGELRNPEPEHAG-LRSLLAEKELKIQQMEKDMEDLR 394
A+VNMV E ELR+P+P + LRSLL EKE KIQ+ME++M +L+
Sbjct: 361 AQVNMVVAEKQLLKHLQKEVSRLEAELRSPDPAASPCLRSLLIEKERKIQKMEEEMNELK 420
Query: 395 RQRDLAQCQLDLERRANKVQKGSSDYGPSSQVVRCLSFAEENELAIGKHTPE---RTETV 451
RQRDLAQ QL+LERR+ K KGS +GPS QVV+CLSF E+E G R +
Sbjct: 421 RQRDLAQSQLELERRSKKELKGSDHHGPSRQVVKCLSFTPEDEEVSGASLSTNLGRKSLL 480
Query: 452 SRQAMLKNLLASPDPSILVDEIQKLEHRQLQLCEDANRALEVLHKDFATHNLGNQETAET 511
RQA ++ S +PS+LV EI+KLE RQ QL ++AN AL++LHK+FA+H +G+Q ET
Sbjct: 481 ERQAAIRRSTNSTNPSMLVHEIRKLEMRQRQLGDEANHALQLLHKEFASHRIGSQGATET 540
Query: 512 MSKVLSEIKDL 522
++K+ SEIK+L
Sbjct: 541 IAKLFSEIKEL 551
>dbj|BAB86283.1| kinesin-like protein NACK1 [Nicotiana tabacum]
Length = 959
Score = 564 bits (1453), Expect = e-159
Identities = 322/548 (58%), Positives = 396/548 (71%), Gaps = 40/548 (7%)
Query: 2 VKTPMAATPSSKIRRKLADTPGGSKIREEKIRVTVRMRPLNRKEQAMYDLIAWDCLDDKT 61
V+TP TP+SKI + A TP G + REEKI VTVR+RPLN++E + D AW+C+DD T
Sbjct: 3 VRTP--GTPASKIDKTPATTPNGHRGREEKIVVTVRLRPLNKRELSAKDHAAWECIDDHT 60
Query: 62 IVFKNPNQER---PSTSYTFDRVFPPACSTQKVYDEGAKDVALSALSGINATIFAYGQTS 118
I+++ QER P++S+TFD+VF P T+ VY+EG K+VALS+L GINATIFAYGQTS
Sbjct: 61 IIYRPVPQERAAQPASSFTFDKVFGPDSITEAVYEEGVKNVALSSLMGINATIFAYGQTS 120
Query: 119 SGKTFTMRGITENAIRDIYECIKNTPDRDFVLKISALEIYNETVIDLLNRESG-PLRILD 177
SGKT+TMRGITE A+ DIY I +TP+R+F ++IS LEIYNE V DLLN ESG L++LD
Sbjct: 121 SGKTYTMRGITEKAVNDIYAHIMSTPEREFRIRISGLEIYNENVRDLLNSESGRSLKLLD 180
Query: 178 DTEKVTVVENLFEEVARDAQHLRHLIGICEAHRQVGETTLNDKSSRSHQIIRLTVESFHR 237
D EK TVVE L EE A + QHLRHLI ICEA RQVGET LND SSRSHQIIRLT+ES R
Sbjct: 181 DPEKGTVVEKLVEETASNDQHLRHLISICEAQRQVGETALNDTSSRSHQIIRLTIESTLR 240
Query: 238 ESPDHVKSYIASLNFVDLAGSERASQTNTCGTRLKEGSHINKSLLQLALVIRQLSSGK-S 296
ES D V+SY+ASLNFVDLAGSERASQTN G RL+EG HIN SL+ L VIR+LS GK S
Sbjct: 241 ESSDCVRSYVASLNFVDLAGSERASQTNADGARLREGCHINLSLMTLTTVIRKLSVGKRS 300
Query: 297 GHISYRTSKLTRILQSSLGGNARTAIICTVSPSLSHVEQTRNTLSFATNAKEVINTARVN 356
GHI YR SKLTRILQ SLGGNARTAIICT+SP+ SHVEQ+RNTL FAT AKEV N A+VN
Sbjct: 301 GHIPYRDSKLTRILQHSLGGNARTAIICTLSPASSHVEQSRNTLYFATRAKEVTNNAQVN 360
Query: 357 MV-----------------EGELRNPEPEHAGLRSLLAEKELKIQQMEKDMEDLRRQRDL 399
MV E ELR P+P + EK+ KIQQME ++E+L+RQRDL
Sbjct: 361 MVVSDKQLVKHLQKEVARLEAELRTPDPAN--------EKDWKIQQMEMEIEELKRQRDL 412
Query: 400 AQCQLDLERRANKVQKGSSDYGPSSQVV-RCLSFAEENELAIGKHTPERTE----TVSRQ 454
AQ Q+D RR + ++G S VV +CLSF+ + + P R+E T+ RQ
Sbjct: 413 AQSQVDELRRKLQEEQGPKPSESVSPVVKKCLSFSGTLSPNLEEKAPVRSERTRNTMGRQ 472
Query: 455 AMLKNLLASPDPSILVDEIQKLEHRQLQLCEDANRALEVLHKDFATHNLGNQETAETMSK 514
+M ++L A P L+ EI+KLEH Q QL ++ANRALEVL K+ A H LGNQ+ AET++K
Sbjct: 473 SMRQSLAA---PFTLMHEIRKLEHLQEQLGDEANRALEVLQKEVACHRLGNQDAAETIAK 529
Query: 515 VLSEIKDL 522
+ +EI+++
Sbjct: 530 LQAEIREM 537
>ref|NP_918677.1| putative kinesin [Oryza sativa (japonica cultivar-group)]
gi|13161377|dbj|BAB32972.1| putative KIF3 protein [Oryza
sativa (japonica cultivar-group)]
Length = 954
Score = 546 bits (1408), Expect = e-154
Identities = 305/543 (56%), Positives = 394/543 (72%), Gaps = 35/543 (6%)
Query: 8 ATPSSKIRRKLADTP---GGSKIREEKIRVTVRMRPLNRKEQAMYDLIAWDCLDDKTIVF 64
+TP+SKI R TP G ++++EEKI VTVR+RPL++KE A+ D +AW+C D++TI++
Sbjct: 8 STPASKIERTPMSTPTPGGSTRVKEEKIFVTVRVRPLSKKELALKDQVAWECDDNQTILY 67
Query: 65 KNPNQERPS-TSYTFDRVFPPACSTQKVYDEGAKDVALSALSGINATIFAYGQTSSGKTF 123
K P Q+R + TSYTFD+VF PA T+ VY+EGAKDVA+SAL+GINATIFAYGQTSSGKTF
Sbjct: 68 KGPPQDRAAPTSYTFDKVFGPASQTEVVYEEGAKDVAMSALTGINATIFAYGQTSSGKTF 127
Query: 124 TMRGITENAIRDIYECIKNTPDRDFVLKISALEIYNETVIDLLNRESGPLRILDDTEKVT 183
TMRG+TE+A+ DIY I+NTP+RDF++KISA+EIYNE V DLL ES LR+LDD EK T
Sbjct: 128 TMRGVTESAVNDIYRHIENTPERDFIIKISAMEIYNEIVKDLLRPESTNLRLLDDPEKGT 187
Query: 184 VVENLFEEVARDAQHLRHLIGICEAHRQVGETTLNDKSSRSHQIIRLTVESFHRESPDHV 243
+VE L EE+A+D+QHLRHLI ICE RQVGET LND SSRSHQIIRLTVES RE V
Sbjct: 188 IVEKLEEEIAKDSQHLRHLISICEEQRQVGETALNDTSSRSHQIIRLTVESRLREVSGCV 247
Query: 244 KSYIASLNFVDLAGSERASQTNTCGTRLKEGSHINKSLLQLALVIRQLSSGK-SGHISYR 302
KS++A+LNFVDLAGSERA+QT+ G RLKEG HIN+SLL L VIR+LSS K SGHI YR
Sbjct: 248 KSFVANLNFVDLAGSERAAQTHAVGARLKEGCHINRSLLTLTTVIRKLSSDKRSGHIPYR 307
Query: 303 TSKLTRILQSSLGGNARTAIICTVSPSLSHVEQTRNTLSFATNAKEVINTARVNMV---- 358
SKLTRILQ SLGGNARTAIICT+SP+ +HVEQ+RNTL FAT AKEV N A+VNMV
Sbjct: 308 DSKLTRILQLSLGGNARTAIICTMSPAQTHVEQSRNTLFFATCAKEVTNNAKVNMVVSDK 367
Query: 359 -------------EGELRNPEPEHAGLRSLLAEKELKIQQMEKDMEDLRRQRDLAQCQL- 404
E ELR P+ + ++ E++ KI+QMEK+ME+L++QRD AQ +L
Sbjct: 368 QLVKHLQMEVARLEAELRTPD-RASSSEIIIMERDRKIRQMEKEMEELKKQRDNAQLKLE 426
Query: 405 DLERRANKVQKGSSDYGPSSQVVRCLSFAEENELAIGKHTPERTETVSRQAMLKNLLASP 464
+L+++ Q G + + + +CL+++ G P + R ++ ++ A
Sbjct: 427 ELQKKMGDNQPGWNPFDSPQRTRKCLTYS-------GSLQPSNKMKI-RSSIRQSATA-- 476
Query: 465 DPSILVDEIQKLEHRQLQLCEDANRALEVLHKDFATHNLGNQETAETMSKVLSEIKDLVA 524
P +L EI+KLE Q QL +ANRA+EVLHK+ H GNQ+ AET++K+ +EI+ + +
Sbjct: 477 -PFMLKHEIRKLEQLQQQLEVEANRAIEVLHKEVECHKHGNQDAAETIAKLQAEIRGMQS 535
Query: 525 ASS 527
S
Sbjct: 536 VRS 538
>dbj|BAB88748.1| AtNACK1 kinesin-like protein [Arabidopsis thaliana]
gi|22329653|ref|NP_173273.2| kinesin motor family
protein (NACK1) [Arabidopsis thaliana]
Length = 974
Score = 538 bits (1385), Expect = e-151
Identities = 313/560 (55%), Positives = 386/560 (68%), Gaps = 43/560 (7%)
Query: 2 VKTPMAATPSSKIRRKLADTPGGS-KIREEKIRVTVRMRPLNRKEQAMYDLIAWDCLDDK 60
+KTP TP SK+ R A TPGGS + REEKI VTVR+RP+N++E D +AW+C++D
Sbjct: 3 IKTP--GTPVSKMDRTPAVTPGGSSRSREEKIVVTVRLRPMNKRELLAKDQVAWECVNDH 60
Query: 61 TIVFKNPNQER--PSTSYTFDRVFPPACSTQKVYDEGAKDVALSALSGINATIFAYGQTS 118
TIV K QER +S+TFD+VF P T+ VY++G K+VALSAL GINATIFAYGQTS
Sbjct: 61 TIVSKPQVQERLHHQSSFTFDKVFGPESLTENVYEDGVKNVALSALMGINATIFAYGQTS 120
Query: 119 SGKTFTMRGITENAIRDIYECIKNTPDRDFVLKISALEIYNETVIDLLNRESG-PLRILD 177
SGKT+TMRG+TE A+ DIY I TP+RDF +KIS LEIYNE V DLLN +SG L++LD
Sbjct: 121 SGKTYTMRGVTEKAVNDIYNHIIKTPERDFTIKISGLEIYNENVRDLLNSDSGRALKLLD 180
Query: 178 DTEKVTVVENLFEEVARDAQHLRHLIGICEAHRQVGETTLNDKSSRSHQIIRLTVESFHR 237
D EK TVVE L EE A + HLRHLI ICEA RQVGET LND SSRSHQIIRLT++S HR
Sbjct: 181 DPEKGTVVEKLVEETANNDNHLRHLISICEAQRQVGETALNDTSSRSHQIIRLTIQSTHR 240
Query: 238 ESPDHVKSYIASLNFVDLAGSERASQTNTCGTRLKEGSHINKSLLQLALVIRQLSSGK-S 296
E+ D V+SY+ASLNFVDLAGSERASQ+ GTRL+EG HIN SL+ L VIR+LS GK S
Sbjct: 241 ENSDCVRSYMASLNFVDLAGSERASQSQADGTRLREGCHINLSLMTLTTVIRKLSVGKRS 300
Query: 297 GHISYRTSKLTRILQSSLGGNARTAIICTVSPSLSHVEQTRNTLSFATNAKEVINTARVN 356
GHI YR SKLTRILQ SLGGNARTAIICT+SP+L+HVEQ+RNTL FA AKEV N A VN
Sbjct: 301 GHIPYRDSKLTRILQHSLGGNARTAIICTLSPALAHVEQSRNTLYFANRAKEVTNNAHVN 360
Query: 357 MV-----------------EGELRNPEPEHAGLRSLLAEKELKIQQMEKDMEDLRRQRDL 399
MV E E R P P EK+ KIQQME ++ +LRRQRD
Sbjct: 361 MVVSDKQLVKHLQKEVARLEAERRTPGPS--------TEKDFKIQQMEMEIGELRRQRDD 412
Query: 400 AQCQLDLERRA----NKVQKGSSDY-GPSSQVVRCLSFAEENELAIGKHTPERTE----T 450
AQ QL+ R+ + KG + + P V +CLS++ + T R E T
Sbjct: 413 AQIQLEELRQKLQGDQQQNKGLNPFESPDPPVRKCLSYSVAVTPSSENKTLNRNERARKT 472
Query: 451 VSRQAMLKNLLASPDPSILVDEIQKLEHRQLQLCEDANRALEVLHKDFATHNLGNQETAE 510
RQ+M++ +S P L+ EI+KLEH Q QL E+A +ALEVL K+ A H LGNQ+ A+
Sbjct: 473 TMRQSMIRQ--SSTAPFTLMHEIRKLEHLQEQLGEEATKALEVLQKEVACHRLGNQDAAQ 530
Query: 511 TMSKVLSEIKDLVAASSTAL 530
T++K+ +EI+++ +A+
Sbjct: 531 TIAKLQAEIREMRTVKPSAM 550
>ref|XP_467026.1| putative kinesin heavy chain [Oryza sativa (japonica
cultivar-group)] gi|49387615|dbj|BAD25811.1| putative
kinesin heavy chain [Oryza sativa (japonica
cultivar-group)]
Length = 884
Score = 421 bits (1083), Expect = e-116
Identities = 226/402 (56%), Positives = 293/402 (72%), Gaps = 22/402 (5%)
Query: 24 GSKIREEKIRVTVRMRPLNRKEQAMYDLIAWDCLDDKTIVFKNPNQERPS--TSYTFDRV 81
G + ++I+V VR+RPL+ KE A + W+C++D T++F++ +RP+ T+YTFDRV
Sbjct: 26 GGAGKMDRIQVLVRLRPLSEKEVARREPAEWECINDSTVMFRSTFPDRPTAPTAYTFDRV 85
Query: 82 FPPACSTQKVYDEGAKDVALSALSGINATIFAYGQTSSGKTFTMRGITENAIRDIYECIK 141
F CST++VY+EG K+VALS +SGIN++IFAYGQTSSGKT+TM G+TE + DIY+ I
Sbjct: 86 FHSDCSTKEVYEEGVKEVALSVVSGINSSIFAYGQTSSGKTYTMTGVTEYTVADIYDYIN 145
Query: 142 NTPDRDFVLKISALEIYNETVIDLLNRESGPLRILDDTEKVTVVENLFEEVARDAQHLRH 201
+R FVLK SA+EIYNE + DLL+ E+ PLR+ DD EK T VENL E V RD HL+
Sbjct: 146 KHEERAFVLKFSAIEIYNEVIRDLLSAENTPLRLWDDAEKGTYVENLTEVVLRDWNHLKG 205
Query: 202 LIGICEAHRQVGETTLNDKSSRSHQIIRLTVESFHRE--SPDHVKSYIASLNFVDLAGSE 259
LI +CEA R+ GET LN+KSSRSHQI+RLTVES RE D + +AS NFVDLAGSE
Sbjct: 206 LISVCEAQRRTGETFLNEKSSRSHQILRLTVESSAREFLGKDKSTTLVASANFVDLAGSE 265
Query: 260 RASQTNTCGTRLKEGSHINKSLLQLALVIRQLSSGKSGHISYRTSKLTRILQSSLGGNAR 319
RASQ + GTRLKEG HIN+SLL L VIR+LS G + HI YR SKLTRILQ SLGGNAR
Sbjct: 266 RASQALSAGTRLKEGCHINRSLLALGTVIRKLSMGSNAHIPYRDSKLTRILQPSLGGNAR 325
Query: 320 TAIICTVSPSLSHVEQTRNTLSFATNAKEVINTARVNMV-----------------EGEL 362
TAIICT+SP+ SH+EQ+RNTL F + AKEV+ A+VN+V E EL
Sbjct: 326 TAIICTLSPATSHIEQSRNTLLFGSCAKEVVTNAQVNVVMSDKALVKHLQKELARLESEL 385
Query: 363 RNPEPEHAGLRSLLAEKELKIQQMEKDMEDLRRQRDLAQCQL 404
R+P + + L +LL EK+ +I++MEK++++L+ QRDLAQ +L
Sbjct: 386 RHP-VQSSSLETLLKEKDNQIRKMEKEIKELKSQRDLAQSRL 426
>ref|XP_483647.1| putative kinesin heavy chain [Oryza sativa (japonica
cultivar-group)] gi|42408720|dbj|BAD09938.1| putative
kinesin heavy chain [Oryza sativa (japonica
cultivar-group)]
Length = 986
Score = 418 bits (1075), Expect = e-115
Identities = 220/403 (54%), Positives = 294/403 (72%), Gaps = 24/403 (5%)
Query: 25 SKIREEKIRVTVRMRPLNRKEQAMYDLIAWDCLDDKTIVFKNPNQERPS--TSYTFDRVF 82
++ +EE+I V+VR+RPLN +E D W+C+ T++F++ ER T+YT+DRVF
Sbjct: 15 AEAKEERIMVSVRLRPLNGREAG--DSCDWECISPTTVMFRSTVPERAMFPTAYTYDRVF 72
Query: 83 PPACSTQKVYDEGAKDVALSALSGINATIFAYGQTSSGKTFTMRGITENAIRDIYECIKN 142
P ST++VY+EGAK+VALS +SGIN++IFAYGQTSSGKT+TM GITE ++ DIY+ I+
Sbjct: 73 GPDSSTRQVYEEGAKEVALSVVSGINSSIFAYGQTSSGKTYTMTGITEYSVLDIYDYIEK 132
Query: 143 TPDRDFVLKISALEIYNETVIDLLNRESGPLRILDDTEKVTVVENLFEEVARDAQHLRHL 202
P+R+F+L+ SA+EIYNE V DLL+ ++ PLR+LDD EK T VE L EE RD HLR+L
Sbjct: 133 HPEREFILRFSAIEIYNEAVRDLLSHDTTPLRLLDDPEKGTTVEKLTEETLRDKDHLRNL 192
Query: 203 IGICEAHRQVGETTLNDKSSRSHQIIRLTVESFHRE--SPDHVKSYIASLNFVDLAGSER 260
+ +CEA RQ+GET LN+ SSRSHQI+RLT+ES R+ + + +A +NFVDLAGSER
Sbjct: 193 LAVCEAQRQIGETALNETSSRSHQILRLTIESSTRQYLGRGNSSTLVACVNFVDLAGSER 252
Query: 261 ASQTNTCGTRLKEGSHINKSLLQLALVIRQLSSGKSGHISYRTSKLTRILQSSLGGNART 320
ASQT + G RLKEGSHIN+SLL L V+RQLS G++GHI YR SKLTRILQSSLGGNART
Sbjct: 253 ASQTASAGVRLKEGSHINRSLLTLGKVVRQLSKGRNGHIPYRDSKLTRILQSSLGGNART 312
Query: 321 AIICTVSPSLSHVEQTRNTLSFATNAKEVINTARVNMV-----------------EGELR 363
AIICT+SP+ SH+EQ+RNTL FAT AKEV+ A+VN+V + E++
Sbjct: 313 AIICTMSPARSHIEQSRNTLLFATCAKEVVTNAQVNVVMSDKALVKHLQRELERLQSEIK 372
Query: 364 NPEPEHAGLRS-LLAEKELKIQQMEKDMEDLRRQRDLAQCQLD 405
P P + L EK+ +I+++EK +++L +RD + QLD
Sbjct: 373 FPAPASCTTHAEALREKDAQIKKLEKQLKELMEERDTVKSQLD 415
>ref|XP_483646.1| putative kinesin heavy chain [Oryza sativa (japonica
cultivar-group)] gi|42408719|dbj|BAD09937.1| putative
kinesin heavy chain [Oryza sativa (japonica
cultivar-group)]
Length = 1003
Score = 418 bits (1075), Expect = e-115
Identities = 220/403 (54%), Positives = 294/403 (72%), Gaps = 24/403 (5%)
Query: 25 SKIREEKIRVTVRMRPLNRKEQAMYDLIAWDCLDDKTIVFKNPNQERPS--TSYTFDRVF 82
++ +EE+I V+VR+RPLN +E D W+C+ T++F++ ER T+YT+DRVF
Sbjct: 31 AEAKEERIMVSVRLRPLNGREAG--DSCDWECISPTTVMFRSTVPERAMFPTAYTYDRVF 88
Query: 83 PPACSTQKVYDEGAKDVALSALSGINATIFAYGQTSSGKTFTMRGITENAIRDIYECIKN 142
P ST++VY+EGAK+VALS +SGIN++IFAYGQTSSGKT+TM GITE ++ DIY+ I+
Sbjct: 89 GPDSSTRQVYEEGAKEVALSVVSGINSSIFAYGQTSSGKTYTMTGITEYSVLDIYDYIEK 148
Query: 143 TPDRDFVLKISALEIYNETVIDLLNRESGPLRILDDTEKVTVVENLFEEVARDAQHLRHL 202
P+R+F+L+ SA+EIYNE V DLL+ ++ PLR+LDD EK T VE L EE RD HLR+L
Sbjct: 149 HPEREFILRFSAIEIYNEAVRDLLSHDTTPLRLLDDPEKGTTVEKLTEETLRDKDHLRNL 208
Query: 203 IGICEAHRQVGETTLNDKSSRSHQIIRLTVESFHRE--SPDHVKSYIASLNFVDLAGSER 260
+ +CEA RQ+GET LN+ SSRSHQI+RLT+ES R+ + + +A +NFVDLAGSER
Sbjct: 209 LAVCEAQRQIGETALNETSSRSHQILRLTIESSTRQYLGRGNSSTLVACVNFVDLAGSER 268
Query: 261 ASQTNTCGTRLKEGSHINKSLLQLALVIRQLSSGKSGHISYRTSKLTRILQSSLGGNART 320
ASQT + G RLKEGSHIN+SLL L V+RQLS G++GHI YR SKLTRILQSSLGGNART
Sbjct: 269 ASQTASAGVRLKEGSHINRSLLTLGKVVRQLSKGRNGHIPYRDSKLTRILQSSLGGNART 328
Query: 321 AIICTVSPSLSHVEQTRNTLSFATNAKEVINTARVNMV-----------------EGELR 363
AIICT+SP+ SH+EQ+RNTL FAT AKEV+ A+VN+V + E++
Sbjct: 329 AIICTMSPARSHIEQSRNTLLFATCAKEVVTNAQVNVVMSDKALVKHLQRELERLQSEIK 388
Query: 364 NPEPEHAGLRS-LLAEKELKIQQMEKDMEDLRRQRDLAQCQLD 405
P P + L EK+ +I+++EK +++L +RD + QLD
Sbjct: 389 FPAPASCTTHAEALREKDAQIKKLEKQLKELMEERDTVKSQLD 431
>emb|CAB62637.1| putative protein [Arabidopsis thaliana] gi|11357626|pir||T45746
hypothetical protein F24M12.190 - Arabidopsis thaliana
Length = 968
Score = 414 bits (1064), Expect = e-114
Identities = 224/445 (50%), Positives = 307/445 (68%), Gaps = 18/445 (4%)
Query: 20 DTPGGSKIREEKIRVTVRMRPLNRKEQAMYDLIAWDCLDDKTIVFKN----PNQERPSTS 75
D GS REEKI V+VR+RPLN +E+A D+ W+C++D+T+++++ + T+
Sbjct: 6 DQMQGSSGREEKIFVSVRLRPLNVRERARNDVADWECINDETVIYRSHLSISERSMYPTA 65
Query: 76 YTFDRVFPPACSTQKVYDEGAKDVALSALSGINATIFAYGQTSSGKTFTMRGITENAIRD 135
YTFDRVF P CST++VYD+GAK+VALS +SG++A++FAYGQTSSGKT+TM GIT+ A+ D
Sbjct: 66 YTFDRVFGPECSTREVYDQGAKEVALSVVSGVHASVFAYGQTSSGKTYTMIGITDYALAD 125
Query: 136 IYECIKNTPDRDFVLKISALEIYNETVIDLLNRESGPLRILDDTEKVTVVENLFEEVARD 195
IY+ I+ +R+F+LK SA+EIYNE+V DLL+ + PLR+LDD EK TVVE L EE RD
Sbjct: 126 IYDYIEKHNEREFILKFSAMEIYNESVRDLLSTDISPLRVLDDPEKGTVVEKLTEETLRD 185
Query: 196 AQHLRHLIGICEAHRQVGETTLNDKSSRSHQIIRLTVESFHRE--SPDHVKSYIASLNFV 253
H + L+ IC A RQ+GET LN+ SSRSHQI+RLTVES RE + D + A++NF+
Sbjct: 186 WNHFKELLSICIAQRQIGETALNEVSSRSHQILRLTVESTAREYLAKDKFSTLTATVNFI 245
Query: 254 DLAGSERASQTNTCGTRLKEGSHINKSLLQLALVIRQLSSGKSGHISYRTSKLTRILQSS 313
DLAGSERASQ+ + GTRLKEG HIN+SLL L VIR+LS GK+GHI +R SKLTRILQ+S
Sbjct: 246 DLAGSERASQSLSAGTRLKEGGHINRSLLTLGTVIRKLSKGKNGHIPFRDSKLTRILQTS 305
Query: 314 LGGNARTAIICTVSPSLSHVEQTRNTLSFATNAKEVINTARVNMV----------EGELR 363
LGGNART+IICT+SP+ HVEQ+RNTL FA+ AKEV A+VN+V + EL
Sbjct: 306 LGGNARTSIICTLSPARVHVEQSRNTLLFASCAKEVTTNAQVNVVMSDKALVRHLQRELA 365
Query: 364 NPEPEHAGLRSLLAEKELKIQQMEKDMEDLRRQRDLAQCQLDLERRANKVQKGSSDYG-- 421
E E + R L + EKD++ + +++ Q +LER ++++ G
Sbjct: 366 KLESELSSPRQALVVSDTTALLKEKDLQIEKLNKEVFQLAQELERAYSRIEDLQQIIGEA 425
Query: 422 PSSQVVRCLSFAEENELAIGKHTPE 446
P +++ S + +G+ P+
Sbjct: 426 PQQEILSTDSEQTNTNVVLGRQYPK 450
>ref|NP_190684.2| kinesin motor family protein [Arabidopsis thaliana]
Length = 1025
Score = 414 bits (1064), Expect = e-114
Identities = 224/445 (50%), Positives = 307/445 (68%), Gaps = 18/445 (4%)
Query: 20 DTPGGSKIREEKIRVTVRMRPLNRKEQAMYDLIAWDCLDDKTIVFKN----PNQERPSTS 75
D GS REEKI V+VR+RPLN +E+A D+ W+C++D+T+++++ + T+
Sbjct: 6 DQMQGSSGREEKIFVSVRLRPLNVRERARNDVADWECINDETVIYRSHLSISERSMYPTA 65
Query: 76 YTFDRVFPPACSTQKVYDEGAKDVALSALSGINATIFAYGQTSSGKTFTMRGITENAIRD 135
YTFDRVF P CST++VYD+GAK+VALS +SG++A++FAYGQTSSGKT+TM GIT+ A+ D
Sbjct: 66 YTFDRVFGPECSTREVYDQGAKEVALSVVSGVHASVFAYGQTSSGKTYTMIGITDYALAD 125
Query: 136 IYECIKNTPDRDFVLKISALEIYNETVIDLLNRESGPLRILDDTEKVTVVENLFEEVARD 195
IY+ I+ +R+F+LK SA+EIYNE+V DLL+ + PLR+LDD EK TVVE L EE RD
Sbjct: 126 IYDYIEKHNEREFILKFSAMEIYNESVRDLLSTDISPLRVLDDPEKGTVVEKLTEETLRD 185
Query: 196 AQHLRHLIGICEAHRQVGETTLNDKSSRSHQIIRLTVESFHRE--SPDHVKSYIASLNFV 253
H + L+ IC A RQ+GET LN+ SSRSHQI+RLTVES RE + D + A++NF+
Sbjct: 186 WNHFKELLSICIAQRQIGETALNEVSSRSHQILRLTVESTAREYLAKDKFSTLTATVNFI 245
Query: 254 DLAGSERASQTNTCGTRLKEGSHINKSLLQLALVIRQLSSGKSGHISYRTSKLTRILQSS 313
DLAGSERASQ+ + GTRLKEG HIN+SLL L VIR+LS GK+GHI +R SKLTRILQ+S
Sbjct: 246 DLAGSERASQSLSAGTRLKEGGHINRSLLTLGTVIRKLSKGKNGHIPFRDSKLTRILQTS 305
Query: 314 LGGNARTAIICTVSPSLSHVEQTRNTLSFATNAKEVINTARVNMV----------EGELR 363
LGGNART+IICT+SP+ HVEQ+RNTL FA+ AKEV A+VN+V + EL
Sbjct: 306 LGGNARTSIICTLSPARVHVEQSRNTLLFASCAKEVTTNAQVNVVMSDKALVRHLQRELA 365
Query: 364 NPEPEHAGLRSLLAEKELKIQQMEKDMEDLRRQRDLAQCQLDLERRANKVQKGSSDYG-- 421
E E + R L + EKD++ + +++ Q +LER ++++ G
Sbjct: 366 KLESELSSPRQALVVSDTTALLKEKDLQIEKLNKEVFQLAQELERAYSRIEDLQQIIGEA 425
Query: 422 PSSQVVRCLSFAEENELAIGKHTPE 446
P +++ S + +G+ P+
Sbjct: 426 PQQEILSTDSEQTNTNVVLGRQYPK 450
>emb|CAE02777.2| OSJNBa0011L07.1 [Oryza sativa (japonica cultivar-group)]
Length = 945
Score = 413 bits (1062), Expect = e-114
Identities = 227/403 (56%), Positives = 291/403 (71%), Gaps = 22/403 (5%)
Query: 23 GGSKIREEKIRVTVRMRPLNRKEQAMYDLIAWDCLDDKTIVFKNPNQERPS--TSYTFDR 80
GG + E+I V+VR+RPL+ KE A D W+C++D TI+ ++ +RPS T+Y+FDR
Sbjct: 26 GGGVGKLERILVSVRLRPLSDKEIARGDPSEWECINDTTIISRSTFPDRPSAPTAYSFDR 85
Query: 81 VFPPACSTQKVYDEGAKDVALSALSGINATIFAYGQTSSGKTFTMRGITENAIRDIYECI 140
VF C T +VY +GAK+VALS +SGIN++IFAYGQTSSGKT+TM GITE + DIY+ I
Sbjct: 86 VFRSDCDTNEVYKQGAKEVALSVVSGINSSIFAYGQTSSGKTYTMTGITEYTVADIYDYI 145
Query: 141 KNTPDRDFVLKISALEIYNETVIDLLNRESGPLRILDDTEKVTVVENLFEEVARDAQHLR 200
+R FVLK SA+EIYNE V DLL+ E+ PLR+ DD EK T VENL E V RD HL+
Sbjct: 146 GKHEERAFVLKFSAIEIYNEVVRDLLSAENTPLRLWDDAEKGTYVENLTEVVLRDWNHLK 205
Query: 201 HLIGICEAHRQVGETTLNDKSSRSHQIIRLTVESFHRE--SPDHVKSYIASLNFVDLAGS 258
LI +CEA R+ GET LN+ SSRSHQI++LT+ES RE D + +AS+NFVDLAGS
Sbjct: 206 ELISVCEAQRKTGETYLNENSSRSHQILKLTIESSAREFLGKDKSTTLVASVNFVDLAGS 265
Query: 259 ERASQTNTCGTRLKEGSHINKSLLQLALVIRQLSSGKSGHISYRTSKLTRILQSSLGGNA 318
ERASQ + G RLKEG HIN+SLL L VIR+LS ++GHI YR SKLTRILQ SLGGNA
Sbjct: 266 ERASQALSAGARLKEGCHINRSLLTLGTVIRKLSKVRNGHIPYRDSKLTRILQPSLGGNA 325
Query: 319 RTAIICTVSPSLSHVEQTRNTLSFATNAKEVINTARVNMV-----------------EGE 361
RTAIICT+SP+ SH+EQ+RNTL FA+ AKEV+ A+VN+V E E
Sbjct: 326 RTAIICTMSPARSHMEQSRNTLLFASCAKEVVTNAQVNVVMSDKALVKQLQKELARLESE 385
Query: 362 LRNPEPEHAGLRSLLAEKELKIQQMEKDMEDLRRQRDLAQCQL 404
LR P ++ L SL+ EK+ +I++MEK++++L+ QRDLAQ +L
Sbjct: 386 LRCP-ASYSSLESLVKEKDNQIRKMEKEIKELKLQRDLAQSRL 427
>ref|NP_179726.2| kinesin motor family protein [Arabidopsis thaliana]
Length = 862
Score = 406 bits (1043), Expect = e-111
Identities = 216/401 (53%), Positives = 287/401 (70%), Gaps = 23/401 (5%)
Query: 28 REEKIRVTVRMRPLNRKEQAMYDLIAWDCLDDKTIVFKNPNQERPS--TSYTFDRVFPPA 85
REEKI V VR+RPLN KE + W+C++D T++++N +E + ++Y+FDRV+
Sbjct: 21 REEKILVLVRLRPLNEKEILANEAADWECINDTTVLYRNTLREGSTFPSAYSFDRVYRGE 80
Query: 86 CSTQKVYDEGAKDVALSALSGINATIFAYGQTSSGKTFTMRGITENAIRDIYECIKNTPD 145
C T++VY++G K+VALS + GIN++IFAYGQTSSGKT+TM GITE A+ DI++ I D
Sbjct: 81 CPTRQVYEDGPKEVALSVVKGINSSIFAYGQTSSGKTYTMSGITEFAVADIFDYIFKHED 140
Query: 146 RDFVLKISALEIYNETVIDLLNRESGPLRILDDTEKVTVVENLFEEVARDAQHLRHLIGI 205
R FV+K SA+EIYNE + DLL+ +S PLR+ DD EK VE EE RD HL+ LI +
Sbjct: 141 RAFVVKFSAIEIYNEAIRDLLSPDSTPLRLRDDPEKGAAVEKATEETLRDWNHLKELISV 200
Query: 206 CEAHRQVGETTLNDKSSRSHQIIRLTVESFHRE--SPDHVKSYIASLNFVDLAGSERASQ 263
CEA R++GET+LN++SSRSHQII+LTVES RE ++ + +AS+NF+DLAGSERASQ
Sbjct: 201 CEAQRKIGETSLNERSSRSHQIIKLTVESSAREFLGKENSTTLMASVNFIDLAGSERASQ 260
Query: 264 TNTCGTRLKEGSHINKSLLQLALVIRQLSSGKSGHISYRTSKLTRILQSSLGGNARTAII 323
+ G RLKEG HIN+SLL L VIR+LS+G+ GHI+YR SKLTRILQ LGGNARTAI+
Sbjct: 261 ALSAGARLKEGCHINRSLLTLGTVIRKLSNGRQGHINYRDSKLTRILQPCLGGNARTAIV 320
Query: 324 CTVSPSLSHVEQTRNTLSFATNAKEVINTARVNMV-----------------EGELRNPE 366
CT+SP+ SHVEQTRNTL FA AKEV A++N+V E ELRNP
Sbjct: 321 CTLSPARSHVEQTRNTLLFACCAKEVTTKAQINVVMSDKALVKQLQRELARLESELRNPA 380
Query: 367 PEHAGLRS--LLAEKELKIQQMEKDMEDLRRQRDLAQCQLD 405
P + L +K+L+IQ+MEK + ++ +QRD+AQ +L+
Sbjct: 381 PATSSCDCGVTLRKKDLQIQKMEKQLAEMTKQRDIAQSRLE 421
>dbj|BAB10710.1| kinesin heavy chain DNA binding protein-like [Arabidopsis thaliana]
Length = 1037
Score = 403 bits (1035), Expect = e-111
Identities = 216/412 (52%), Positives = 295/412 (71%), Gaps = 25/412 (6%)
Query: 29 EEKIRVTVRMRPLNRKEQAMYDLIAWDCLDDKTIVFKN----PNQERPSTSYTFDRVFPP 84
+EKI V+VRMRPLN KE+ D+ W+C+++ TI++++ + ++YTFDRVF P
Sbjct: 16 QEKIYVSVRMRPLNDKEKFRNDVPDWECINNTTIIYRSHLSISERSMYPSAYTFDRVFSP 75
Query: 85 ACSTQKVYDEGAKDVALSALSGINATIFAYGQTSSGKTFTMRGITENAIRDIYECIKNTP 144
C T++VY++GAK+VA S +SG+NA++FAYGQTSSGKT+TM GIT+ A+ DIY I
Sbjct: 76 ECCTRQVYEQGAKEVAFSVVSGVNASVFAYGQTSSGKTYTMSGITDCALVDIYGYIDKHK 135
Query: 145 DRDFVLKISALEIYNETVIDLLNRESGPLRILDDTEKVTVVENLFEEVARDAQHLRHLIG 204
+R+F+LK SA+EIYNE+V DLL+ ++ PLR+LDD EK TVVE L EE RD H + L+
Sbjct: 136 EREFILKFSAMEIYNESVRDLLSTDTSPLRLLDDPEKGTVVEKLTEETLRDWNHFKELLS 195
Query: 205 ICEAHRQVGETTLNDKSSRSHQIIRLTVESFHRE--SPDHVKSYIASLNFVDLAGSERAS 262
+C+A RQ+GET LN+ SSRSHQI+RLTVES RE + D + A++NF+DLAGSERAS
Sbjct: 196 VCKAQRQIGETALNEVSSRSHQILRLTVESIAREFSTNDKFSTLTATVNFIDLAGSERAS 255
Query: 263 QTNTCGTRLKEGSHINKSLLQLALVIRQLSSGKSGHISYRTSKLTRILQSSLGGNARTAI 322
Q+ + GTRLKEG HIN+SLL L VIR+LS K+GHI +R SKLTRILQSSLGGNARTAI
Sbjct: 256 QSLSAGTRLKEGCHINRSLLTLGTVIRKLSKEKTGHIPFRDSKLTRILQSSLGGNARTAI 315
Query: 323 ICTVSPSLSHVEQTRNTLSFATNAKEVINTARVNMV-----------------EGELRNP 365
ICT+SP+ HVEQ+RNTL FA+ AKEV A+VN+V E ELR+P
Sbjct: 316 ICTMSPARIHVEQSRNTLLFASCAKEVTTNAQVNVVMSDKALVKHLQRELAKLESELRSP 375
Query: 366 EPEH--AGLRSLLAEKELKIQQMEKDMEDLRRQRDLAQCQLDLERRANKVQK 415
+ +LL EK+L++++++K++ L +Q + A+ ++ RR + +K
Sbjct: 376 SQASIVSDTTALLTEKDLEVEKLKKEVFQLAQQLEQARSEIKDLRRMVEEEK 427
>ref|NP_201432.2| kinesin motor family protein [Arabidopsis thaliana]
Length = 1063
Score = 403 bits (1035), Expect = e-111
Identities = 216/412 (52%), Positives = 295/412 (71%), Gaps = 25/412 (6%)
Query: 29 EEKIRVTVRMRPLNRKEQAMYDLIAWDCLDDKTIVFKN----PNQERPSTSYTFDRVFPP 84
+EKI V+VRMRPLN KE+ D+ W+C+++ TI++++ + ++YTFDRVF P
Sbjct: 16 QEKIYVSVRMRPLNDKEKFRNDVPDWECINNTTIIYRSHLSISERSMYPSAYTFDRVFSP 75
Query: 85 ACSTQKVYDEGAKDVALSALSGINATIFAYGQTSSGKTFTMRGITENAIRDIYECIKNTP 144
C T++VY++GAK+VA S +SG+NA++FAYGQTSSGKT+TM GIT+ A+ DIY I
Sbjct: 76 ECCTRQVYEQGAKEVAFSVVSGVNASVFAYGQTSSGKTYTMSGITDCALVDIYGYIDKHK 135
Query: 145 DRDFVLKISALEIYNETVIDLLNRESGPLRILDDTEKVTVVENLFEEVARDAQHLRHLIG 204
+R+F+LK SA+EIYNE+V DLL+ ++ PLR+LDD EK TVVE L EE RD H + L+
Sbjct: 136 EREFILKFSAMEIYNESVRDLLSTDTSPLRLLDDPEKGTVVEKLTEETLRDWNHFKELLS 195
Query: 205 ICEAHRQVGETTLNDKSSRSHQIIRLTVESFHRE--SPDHVKSYIASLNFVDLAGSERAS 262
+C+A RQ+GET LN+ SSRSHQI+RLTVES RE + D + A++NF+DLAGSERAS
Sbjct: 196 VCKAQRQIGETALNEVSSRSHQILRLTVESIAREFSTNDKFSTLTATVNFIDLAGSERAS 255
Query: 263 QTNTCGTRLKEGSHINKSLLQLALVIRQLSSGKSGHISYRTSKLTRILQSSLGGNARTAI 322
Q+ + GTRLKEG HIN+SLL L VIR+LS K+GHI +R SKLTRILQSSLGGNARTAI
Sbjct: 256 QSLSAGTRLKEGCHINRSLLTLGTVIRKLSKEKTGHIPFRDSKLTRILQSSLGGNARTAI 315
Query: 323 ICTVSPSLSHVEQTRNTLSFATNAKEVINTARVNMV-----------------EGELRNP 365
ICT+SP+ HVEQ+RNTL FA+ AKEV A+VN+V E ELR+P
Sbjct: 316 ICTMSPARIHVEQSRNTLLFASCAKEVTTNAQVNVVMSDKALVKHLQRELAKLESELRSP 375
Query: 366 EPEH--AGLRSLLAEKELKIQQMEKDMEDLRRQRDLAQCQLDLERRANKVQK 415
+ +LL EK+L++++++K++ L +Q + A+ ++ RR + +K
Sbjct: 376 SQASIVSDTTALLTEKDLEVEKLKKEVFQLAQQLEQARSEIKDLRRMVEEEK 427
>gb|AAK50070.1| AT5g66310/K1L20_9 [Arabidopsis thaliana]
Length = 425
Score = 402 bits (1034), Expect = e-110
Identities = 216/406 (53%), Positives = 293/406 (71%), Gaps = 26/406 (6%)
Query: 29 EEKIRVTVRMRPLNRKEQAMYDLIAWDCLDDKTIVFKN----PNQERPSTSYTFDRVFPP 84
+EKI V+VRMRPLN KE+ D+ W+C+++ TI++++ + ++YTFDRVF P
Sbjct: 16 QEKIYVSVRMRPLNDKEKFRNDVPDWECINNTTIIYRSHLSISERSMYPSAYTFDRVFSP 75
Query: 85 ACSTQKVYDEGAKDVALSALSGINATIFAYGQTSSGKTFTMRGITENAIRDIYECIKNTP 144
C T++VY++GAK+VA S +SG+NA++FAYGQTSSGKT+TM GIT+ A+ DIY I
Sbjct: 76 ECCTRQVYEQGAKEVAFSVVSGVNASVFAYGQTSSGKTYTMSGITDCALVDIYGYIDKHK 135
Query: 145 DRDFVLKISALEIYNETVIDLLNRESGPLRILDDTEKVTVVENLFEEVARDAQHLRHLIG 204
+R+F+LK SA+EIYNE+V DLL+ ++ PLR+LDD EK TVVE L EE RD H + L+
Sbjct: 136 EREFILKFSAMEIYNESVRDLLSTDTSPLRLLDDPEKGTVVEKLTEETLRDWNHFKELLS 195
Query: 205 ICEAHRQVGETTLNDKSSRSHQIIRLTVESFHRE--SPDHVKSYIASLNFVDLAGSERAS 262
+C+A RQ+GET LN+ SSRSHQI+RLTVES RE + D + A++NF+DLAGSERAS
Sbjct: 196 VCKAQRQIGETALNEVSSRSHQILRLTVESIAREFSTNDKFSTLTATVNFIDLAGSERAS 255
Query: 263 QTNTCGTRLKEGSHINKSLLQLALVIRQLSSGKSGHISYRTSKLTRILQSSLGGNARTAI 322
Q+ + GTRLKEG HIN+SLL L VIR+LS K+GHI +R SKLTRILQSSLGGNARTAI
Sbjct: 256 QSLSAGTRLKEGCHINRSLLTLGTVIRKLSKEKTGHIPFRDSKLTRILQSSLGGNARTAI 315
Query: 323 ICTVSPSLSHVEQTRNTLSFATNAKEVINTARVNMV-----------------EGELRNP 365
ICT+SP+ HVEQ+RNTL FA+ AKEV A+VN+V E ELR+P
Sbjct: 316 ICTMSPARIHVEQSRNTLLFASCAKEVTTNAQVNVVMSDKALVKHLQRELAKLESELRSP 375
Query: 366 EPEH--AGLRSLLAEKELKIQQMEKDMEDLRRQRDLAQCQL-DLER 408
+ +LL EK+L++++++K++ L +Q + A+ ++ DL R
Sbjct: 376 SQASIVSDTTALLTEKDLEVEKLKKEVFQLAQQLEQARSEIKDLRR 421
>emb|CAB79327.1| putative protein [Arabidopsis thaliana] gi|5668645|emb|CAB51660.1|
putative protein [Arabidopsis thaliana]
gi|7487172|pir||T13465 hypothetical protein T19F6.160 -
Arabidopsis thaliana
Length = 1263
Score = 401 bits (1031), Expect = e-110
Identities = 229/415 (55%), Positives = 292/415 (70%), Gaps = 26/415 (6%)
Query: 29 EEKIRVTVRMRPLNRKEQAMYDLIAWDCLDDKTIVFKNPNQERPSTSYTFDRVFPPACST 88
EEKI V+VR+RPLN KE+ D W+C++D TI+ K N S SYTFD+VF C T
Sbjct: 4 EEKILVSVRVRPLNEKEKTRNDRCDWECINDTTIICKFHNLPDKS-SYTFDKVFGFECPT 62
Query: 89 QKVYDEGAKDVALSALSGINATIFAYGQTSSGKTFTMRGITENAIRDIYECI-KNTPDRD 147
++VYD+GAK+VAL LSGIN++IFAYGQTSSGKT+TM GITE A+ DI+ I K+ +R
Sbjct: 63 KQVYDDGAKEVALCVLSGINSSIFAYGQTSSGKTYTMSGITEFAMDDIFAYIDKHKQERK 122
Query: 148 FVLKISALEIYNETVIDLLNRESG-PLRILDDTEKVTVVENLFEEVARDAQHLRHLIGIC 206
F LK SA+EIYNE V DLL +S PLR+LDD E+ TVVE L EE RD HL L+ IC
Sbjct: 123 FTLKFSAMEIYNEAVRDLLCEDSSTPLRLLDDPERGTVVEKLREETLRDRSHLEELLSIC 182
Query: 207 EAHRQVGETTLNDKSSRSHQIIRLTVESFHRE-SPDHVKSYIASLNFVDLAGSERASQTN 265
E R++GET+LN+ SSRSHQI+RLT+ES ++ SP+ + AS+ FVDLAGSERASQT
Sbjct: 183 ETQRKIGETSLNEISSRSHQILRLTIESSSQQFSPESSATLAASVCFVDLAGSERASQTL 242
Query: 266 TCGTRLKEGSHINKSLLQLALVIRQLSSGKSGHISYRTSKLTRILQSSLGGNARTAIICT 325
+ G+RLKEG HIN+SLL L VIR+LS GK+GHI YR SKLTRILQ+SLGGNARTAIICT
Sbjct: 243 SAGSRLKEGCHINRSLLTLGTVIRKLSKGKNGHIPYRDSKLTRILQNSLGGNARTAIICT 302
Query: 326 VSPSLSHVEQTRNTLSFATNAKEVINTARVNMV-----------------EGELRNPEPE 368
+SP+ SH+EQ+RNTL FAT AKEV A+VN+V E EL+N P
Sbjct: 303 MSPARSHLEQSRNTLLFATCAKEVTTNAQVNLVVSEKALVKQLQRELARMENELKNLGPA 362
Query: 369 HAGLRS-----LLAEKELKIQQMEKDMEDLRRQRDLAQCQLDLERRANKVQKGSS 418
A S +L +KE I +ME+ + +L+ QRD+AQ +++ ++ ++ SS
Sbjct: 363 SASSTSDFYALMLKQKEELIAKMEEQIHELKWQRDVAQSRVENLLKSTAEERSSS 417
>ref|NP_195606.2| kinesin motor family protein [Arabidopsis thaliana]
Length = 836
Score = 401 bits (1031), Expect = e-110
Identities = 216/402 (53%), Positives = 287/402 (70%), Gaps = 24/402 (5%)
Query: 28 REEKIRVTVRMRPLNRKEQAMYDLIAWDCLDDKTIVFKNPNQERPS--TSYTFDRVFPPA 85
REEKI V VR+RPLN+KE A + W+C++D TI+++N +E + ++Y+FD+V+
Sbjct: 10 REEKILVLVRLRPLNQKEIAANEAADWECINDTTILYRNTLREGSNFPSAYSFDKVYRGE 69
Query: 86 CSTQKVYDEGAKDVALSALSGINATIFAYGQTSSGKTFTMRGITENAIRDIYECIKNTPD 145
C T++VY++G K++ALS + GIN +IFAYGQTSSGKT+TM GITE A+ DI++ I +
Sbjct: 70 CPTRQVYEDGTKEIALSVVKGINCSIFAYGQTSSGKTYTMTGITEFAVADIFDYIFQHEE 129
Query: 146 RDFVLKISALEIYNETVIDLLNRESGPLRILDDTEKVTVVENLFEEVARDAQHLRHLIGI 205
R F +K SA+EIYNE + DLL+ + LR+ DD EK TVVE EE RD HL+ L+ I
Sbjct: 130 RAFSVKFSAIEIYNEAIRDLLSSDGTSLRLRDDPEKGTVVEKATEETLRDWNHLKELLSI 189
Query: 206 CEAHRQVGETTLNDKSSRSHQIIRLTVESFHRE--SPDHVKSYIASLNFVDLAGSERASQ 263
CEA R++GET+LN++SSRSHQ+IRLTVES RE ++ + +AS+NF+DLAGSERASQ
Sbjct: 190 CEAQRKIGETSLNERSSRSHQMIRLTVESSAREFLGKENSTTLMASVNFIDLAGSERASQ 249
Query: 264 TNTCGTRLKEGSHINKSLLQLALVIRQLSSGKSGHISYRTSKLTRILQSSLGGNARTAII 323
+ GTRLKEG HIN+SLL L VIR+LS G+ GHI++R SKLTRILQ LGGNARTAII
Sbjct: 250 AMSAGTRLKEGCHINRSLLTLGTVIRKLSKGRQGHINFRDSKLTRILQPCLGGNARTAII 309
Query: 324 CTVSPSLSHVEQTRNTLSFATNAKEVINTARVNMV-----------------EGELRNPE 366
CT+SP+ SHVE T+NTL FA AKEV AR+N+V E ELRNP
Sbjct: 310 CTLSPARSHVELTKNTLLFACCAKEVTTKARINVVMSDKALLKQLQRELARLETELRNPA 369
Query: 367 PEHAG---LRSLLAEKELKIQQMEKDMEDLRRQRDLAQCQLD 405
A + +K+L+IQ+MEK++ +LR+QRDLAQ +L+
Sbjct: 370 SSPASNCDCAMTVRKKDLQIQKMEKEIAELRKQRDLAQSRLE 411
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.315 0.130 0.357
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 846,964,248
Number of Sequences: 2540612
Number of extensions: 32827951
Number of successful extensions: 133075
Number of sequences better than 10.0: 1902
Number of HSP's better than 10.0 without gapping: 1527
Number of HSP's successfully gapped in prelim test: 384
Number of HSP's that attempted gapping in prelim test: 124858
Number of HSP's gapped (non-prelim): 2952
length of query: 552
length of database: 863,360,394
effective HSP length: 133
effective length of query: 419
effective length of database: 525,458,998
effective search space: 220167320162
effective search space used: 220167320162
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 78 (34.7 bits)
Medicago: description of AC124967.12


