

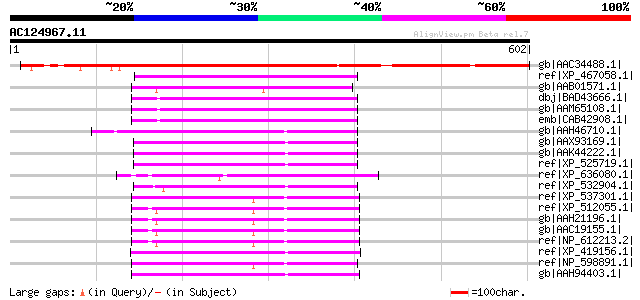
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124967.11 + phase: 0
(602 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAC34488.1| similar to late embryogenesis abundant proteins [... 556 e-157
ref|XP_467058.1| putative late embryonic abundant protein EMB8 [... 169 2e-40
gb|AAB01571.1| EMB8 [Picea glauca] gi|7446463|pir||T09290 late e... 167 9e-40
dbj|BAD43666.1| putative LEA protein [Arabidopsis thaliana] 157 7e-37
gb|AAM65108.1| putative LEA protein [Arabidopsis thaliana] 157 9e-37
emb|CAB42908.1| putative LEA protein [Arabidopsis thaliana] gi|1... 157 1e-36
gb|AAH46710.1| Abhd1-prov protein [Xenopus laevis] 132 2e-29
gb|AAX93169.1| unknown [Homo sapiens] gi|23397655|ref|NP_115993.... 127 1e-27
gb|AAK44222.1| lung alpha/beta hydrolase protein 1 [Homo sapiens... 125 4e-27
ref|XP_525719.1| PREDICTED: hypothetical protein XP_525719 [Pan ... 123 1e-26
ref|XP_636080.1| hypothetical protein DDB0188543 [Dictyostelium ... 117 8e-25
ref|XP_532904.1| PREDICTED: hypothetical protein XP_532904 [Cani... 117 1e-24
ref|XP_537301.1| PREDICTED: similar to alpha/beta hydrolase doma... 116 2e-24
ref|XP_512055.1| PREDICTED: similar to alpha/beta hydrolase doma... 115 3e-24
gb|AAH21196.1| Alpha/beta hydrolase domain containing protein 3 ... 115 3e-24
gb|AAC19155.1| unknown [Homo sapiens] 115 3e-24
ref|NP_612213.2| alpha/beta hydrolase domain containing protein ... 115 3e-24
ref|XP_419156.1| PREDICTED: similar to lung alpha/beta hydrolase... 111 8e-23
ref|NP_598891.1| abhydrolase domain containing 3 [Mus musculus] ... 110 1e-22
gb|AAH94403.1| Unknown (protein for MGC:84955) [Xenopus laevis] 110 2e-22
>gb|AAC34488.1| similar to late embryogenesis abundant proteins [Arabidopsis
thaliana] gi|7487147|pir||T02712 similar to late
embryogenesis abundant proteins [imported] - Arabidopsis
thaliana gi|15227541|ref|NP_178413.1| CAAX amino
terminal protease family protein [Arabidopsis thaliana]
Length = 1805
Score = 556 bits (1434), Expect = e-157
Identities = 314/621 (50%), Positives = 412/621 (65%), Gaps = 61/621 (9%)
Query: 13 AKPFPSRQFRL----YKR--RRLKIKASFPVPPPSPFENLFNTLISQCSSVNSIDFIAPS 66
A PF R+ R +KR R L + S PPP F+ + SS +APS
Sbjct: 22 ASPFILRRSRKRGARFKRQSRNLVLVNSSIFPPP------FDGSVPLDSS------LAPS 69
Query: 67 L-GFASGSALFFS-RF----------KSSQNSDVGEWILFASPTPFNRFVLLRCPSISFK 114
L G ASG A++ S RF K + VGEWILF +PTPFNRFVLLRC +SF
Sbjct: 70 LAGIASGLAVYLSSRFFGKSLEKISDKIVDDVVVGEWILFTTPTPFNRFVLLRCSLLSFD 129
Query: 115 DN------ERLIKDEKHY-----GRIRVNKREKDLEEELKYQRVCLSASDGGVVSLDWPV 163
D+ +RL+ +E+H+ G+I + D + L+YQRVC++ DGGVVSLDWP
Sbjct: 130 DDSEKSLSDRLVTEERHFVTLDSGKIVRDGAVTDEKTPLEYQRVCITMEDGGVVSLDWPA 189
Query: 164 ELDLEEERGLDSTLLIVPGTPQGSMDDNIRVFVIDALKRGFFPVVMNPRGCASSPITTPR 223
LD+ EERGLD+T++ +PGTP+GSM++ +R FV +AL+RG FPVVMNPRGCA SP+TTPR
Sbjct: 190 NLDIREERGLDTTVVFIPGTPEGSMEEGVRSFVCEALRRGVFPVVMNPRGCAGSPLTTPR 249
Query: 224 LFTAADSDDICTAITYINKARPWTTLMGVGWGYGANMLTKYLAEVGERTPLTAATCIDNP 283
LFTA DSDDI TA+ +++K RPWTTL VG GYGANMLTKYLAE GERTPLTAA CIDNP
Sbjct: 250 LFTAGDSDDISTALRFLSKTRPWTTLTAVGRGYGANMLTKYLAEAGERTPLTAAVCIDNP 309
Query: 284 FDLDEATRTFPYHHVTDQKLTRGLINILQTNKALFQGKAKGXDVGKALLAKSVRDFEEAI 343
FDL+E TRT PY DQ+LTRGL+ IL NK LFQG+AK DVGKAL +KSVR+F++A+
Sbjct: 310 FDLEEITRTSPYSTSLDQQLTRGLVEILLANKELFQGRAKAFDVGKALCSKSVREFDKAL 369
Query: 344 SMVSYGFVDIEDFYTKASTRNMIKDIKIPVLFIQSDNGMVPAFSVPRNLIAENPFTSLLL 403
SMV+YG IEDFY+ +TR +I ++K+P+LFIQ+D+ +VP +++PR+ IAENPFTSLLL
Sbjct: 370 SMVTYGCESIEDFYSSCATREVIGEVKVPLLFIQNDD-VVPPYTIPRSSIAENPFTSLLL 428
Query: 404 CSCLSSSVMDTDTSALSWCQ-LVTVEVAI*LSIFYMCNFGMHMQWLAAVELGLLKGRHPL 462
CS S +++D T A+SWCQ L ++ + + MQWL AVELGLLKGRHPL
Sbjct: 429 CSS-SPNLIDGRTVAVSWCQDLAKIDFPM-----------LAMQWLTAVELGLLKGRHPL 476
Query: 463 LTDIDLTIIPSKGLTLVEDARTDKNPKVGKLLELARSDAYNGYSIDPSEDLLEGSKNDAG 522
L D+D+T+ PSKGL E +K+ KL++ A NGY +DP + LE S
Sbjct: 477 LEDVDVTVNPSKGLVFSEARAPEKSIGAKKLVQAAHEKTVNGYHLDPFRETLEDSDMTPN 536
Query: 523 LHFGPQQDVQQNFEQGDMSLQVKDGPLQQTSSSGRALVGEEDAASV-DSEHGHVMQTAQV 581
+ P+ D+++N + + + S R E++ ++V +S+ G V+QTA+V
Sbjct: 537 SNLSPETDLEKN-----VKIDYGSDETENNIVSTRVESIEDNESNVEESDRGQVLQTAEV 591
Query: 582 VTNMLDVTMPGTLTEEQKKKV 602
V +MLDVTMPGTL E+KKKV
Sbjct: 592 VVSMLDVTMPGTLKAEEKKKV 612
>ref|XP_467058.1| putative late embryonic abundant protein EMB8 [Oryza sativa
(japonica cultivar-group)] gi|49388448|dbj|BAD25578.1|
putative late embryonic abundant protein EMB8 [Oryza
sativa (japonica cultivar-group)]
Length = 469
Score = 169 bits (429), Expect = 2e-40
Identities = 94/262 (35%), Positives = 153/262 (57%), Gaps = 4/262 (1%)
Query: 145 QRVCLSASDGGVVSLDWPVELDLEEERGLDSTLLIVPGTPQGSMDDNIRVFVIDALKRGF 204
+R CL D G V+LDW D R L+++PG GS D +R ++ A +G+
Sbjct: 120 RRECLRTPDDGAVALDWVSGDDRALPRDAP-VLILLPGLTGGSDDTYVRHMLLRARNKGW 178
Query: 205 FPVVMNPRGCASSPITTPRLFTAADSDDICTAITYINKARPWTTLMGVGWGYGANMLTKY 264
VV N RGCA SP+TT + ++A+ + D+ + ++ P + + VGW GAN+L +Y
Sbjct: 179 RVVVFNSRGCAGSPVTTAKFYSASFTGDLRQVVDHVLGRFPQSNVYAVGWSLGANILVRY 238
Query: 265 LAEVGERTPLTAATCIDNPFDLDEATRTF--PYHHVTDQKLTRGLINILQTNKALFQGKA 322
L E ++ L+ A + NPFDL A F +++V D+ L + L NI + + LF+G
Sbjct: 239 LGEETDKCVLSGAVSLCNPFDLVIADEDFHKGFNNVYDRALAKALRNIFKKHALLFEGLE 298
Query: 323 KGXDVGKALLAKSVRDFEEAISMVSYGFVDIEDFYTKASTRNMIKDIKIPVLFIQSDNG- 381
++ KA A+SVRDF+E ++ VS+GF ++D+Y+ +S+ + IK++ IP+L IQ+DN
Sbjct: 299 GEYNIPKAANARSVRDFDEGLTRVSFGFKSVDDYYSNSSSSDSIKNVSIPLLCIQADNDP 358
Query: 382 MVPAFSVPRNLIAENPFTSLLL 403
+ P+ +PR I NP L++
Sbjct: 359 IAPSRGIPREDIKANPNCLLIV 380
>gb|AAB01571.1| EMB8 [Picea glauca] gi|7446463|pir||T09290 late embryonic abundant
protein EMB8 - white spruce
gi|2501572|sp|Q40863|EMB8_PICGL Embryogenesis-associated
protein EMB8
Length = 457
Score = 167 bits (423), Expect = 9e-40
Identities = 96/266 (36%), Positives = 151/266 (56%), Gaps = 10/266 (3%)
Query: 142 LKYQRVCLSASDGGVVSLDWPVELDLEE----ERGLDSTLLIV-PGTPQGSMDDNIRVFV 196
+K +R CL DGG V LDWP+E + E E ++S +LI+ PG GS D ++ +
Sbjct: 113 IKSRRECLRMEDGGTVELDWPLEGEDAELWNGELPVNSPVLILLPGLTGGSDDSYVKHML 172
Query: 197 IDALKRGFFPVVMNPRGCASSPITTPRLFTAADSDDICTAITYINKARPWTTLMGVGWGY 256
+ A K G+ VV N RGCA SP+TTP+ ++A+ + D+C + ++ + + VGW
Sbjct: 173 LRARKHGWHSVVFNSRGCADSPVTTPQFYSASFTKDLCQVVKHVAVRFSESNIYAVGWSL 232
Query: 257 GANMLTKYLAEVGERTPLTAATCIDNPFDLDEATRTF----PYHHVTDQKLTRGLINILQ 312
GAN+L +YL EV PL+ A + NPF+L A F +++V D+ L RGL I
Sbjct: 233 GANILVRYLGEVAGNCPLSGAVSLCNPFNLVIADEDFHKGLGFNNVYDKALARGLRQIFP 292
Query: 313 TNKALFQGKAKGXDVGKALLAKSVRDFEEAISMVSYGFVDIEDFYTKASTRNMIKDIKIP 372
+ LF+G ++ A+SVRDF+ ++ VS+GF + D+Y+ +S+ IK ++
Sbjct: 293 KHTRLFEGIEGEYNIPTVAKARSVRDFDGGLTRVSFGFQSVGDYYSNSSSSLSIKYVQTS 352
Query: 373 VLFIQSDNG-MVPAFSVPRNLIAENP 397
+L IQ+ N + P+ +P I ENP
Sbjct: 353 LLCIQASNDPIAPSRGIPWEDIKENP 378
>dbj|BAD43666.1| putative LEA protein [Arabidopsis thaliana]
Length = 408
Score = 157 bits (398), Expect = 7e-37
Identities = 93/266 (34%), Positives = 146/266 (53%), Gaps = 6/266 (2%)
Query: 142 LKYQRVCLSASDGGVVSLDWPVELDLEEERGLDSTLLIV-PGTPQGSMDDNIRVFVIDAL 200
++ +R CL D G V+LDW V D DS +LI+ PG GS D +R ++ A
Sbjct: 91 VRLRRECLRTKDNGSVALDWVVGEDRHFPP--DSPILILLPGLTGGSQDSYVRHMLLRAQ 148
Query: 201 KRGFFPVVMNPRGCASSPITTPRLFTAADSDDICTAITYINKARPWTTLMGVGWGYGANM 260
+ VV N RGC SP+TTP+ ++A+ DI I ++ P L GW G N+
Sbjct: 149 SEKWRCVVFNSRGCGDSPVTTPQFYSASFLGDIGEVIDHVVDKFPKANLYAAGWSLGGNI 208
Query: 261 LTKYLAEVGERTPLTAATCIDNPFDLDEATRTF--PYHHVTDQKLTRGLINILQTNKALF 318
L YL + PLTAA + NPFDL A F +++V D+ L++ L I + LF
Sbjct: 209 LVNYLGQESHNCPLTAAVSLCNPFDLVIADEDFHKGFNNVYDKALSKSLRRIFSKHSLLF 268
Query: 319 QGKAKGXDVGKALLAKSVRDFEEAISMVSYGFVDIEDFYTKASTRNMIKDIKIPVLFIQS 378
+ ++ A A++VRDF++ ++ VS+GF ++++Y+K+S+ IK ++IP+L IQ+
Sbjct: 269 EDIGGEFNIPLAANAETVRDFDDGLTRVSFGFKSVDEYYSKSSSSKAIKHVRIPLLCIQA 328
Query: 379 DNG-MVPAFSVPRNLIAENPFTSLLL 403
N + P +PR+ I NP L++
Sbjct: 329 ANDPIAPERGIPRDDIKANPNCVLIV 354
>gb|AAM65108.1| putative LEA protein [Arabidopsis thaliana]
Length = 408
Score = 157 bits (397), Expect = 9e-37
Identities = 92/266 (34%), Positives = 146/266 (54%), Gaps = 6/266 (2%)
Query: 142 LKYQRVCLSASDGGVVSLDWPVELDLEEERGLDSTLLIV-PGTPQGSMDDNIRVFVIDAL 200
++ +R CL D G V+LDW D DS +LI+ PG GS D +R ++ A
Sbjct: 91 VRLRRECLRTKDNGSVALDWVAGEDRHFPP--DSPILILLPGLTGGSQDSYVRHMLLRAQ 148
Query: 201 KRGFFPVVMNPRGCASSPITTPRLFTAADSDDICTAITYINKARPWTTLMGVGWGYGANM 260
+ + VV N RGC SP+TTP+ ++A+ DI I ++ P L GW G N+
Sbjct: 149 SKKWRCVVFNSRGCGDSPVTTPQFYSASFLGDIGEVIDHVVDKFPKANLYAAGWSLGGNI 208
Query: 261 LTKYLAEVGERTPLTAATCIDNPFDLDEATRTF--PYHHVTDQKLTRGLINILQTNKALF 318
L YL + PLTAA + NPFDL A F +++V D+ L++ L I + LF
Sbjct: 209 LVNYLGQESHNCPLTAAVSLCNPFDLVIADEDFHKGFNNVYDKALSKSLRRIFSKHSLLF 268
Query: 319 QGKAKGXDVGKALLAKSVRDFEEAISMVSYGFVDIEDFYTKASTRNMIKDIKIPVLFIQS 378
+ ++ A A++VRDF++ ++ VS+GF ++++Y+K+S+ IK ++IP+L IQ+
Sbjct: 269 EDIGGEFNIPLAANAETVRDFDDGLTRVSFGFKSVDEYYSKSSSSKAIKHVRIPLLCIQA 328
Query: 379 DNG-MVPAFSVPRNLIAENPFTSLLL 403
N + P +PR+ I NP L++
Sbjct: 329 ANDPIAPERGIPRDDIKSNPNCVLIV 354
>emb|CAB42908.1| putative LEA protein [Arabidopsis thaliana]
gi|15230305|ref|NP_190648.1| late embryogenesis abundant
protein, putative / LEA protein, putative [Arabidopsis
thaliana] gi|7487939|pir||T08400 late embryonic abundant
protein EMB8 homolog F18B3.70 - Arabidopsis thaliana
Length = 408
Score = 157 bits (396), Expect = 1e-36
Identities = 92/266 (34%), Positives = 146/266 (54%), Gaps = 6/266 (2%)
Query: 142 LKYQRVCLSASDGGVVSLDWPVELDLEEERGLDSTLLIV-PGTPQGSMDDNIRVFVIDAL 200
++ +R CL D G V+LDW D DS +LI+ PG GS D +R ++ A
Sbjct: 91 VRLRRECLRTKDNGSVALDWVAGEDRHFPP--DSPILILLPGLTGGSQDSYVRHMLLRAQ 148
Query: 201 KRGFFPVVMNPRGCASSPITTPRLFTAADSDDICTAITYINKARPWTTLMGVGWGYGANM 260
+ + VV N RGC SP+TTP+ ++A+ DI I ++ P L GW G N+
Sbjct: 149 SKKWRCVVFNSRGCGDSPVTTPQFYSASFLGDIGEVIDHVVDKFPKANLYAAGWSLGGNI 208
Query: 261 LTKYLAEVGERTPLTAATCIDNPFDLDEATRTF--PYHHVTDQKLTRGLINILQTNKALF 318
L YL + PLTAA + NPFDL A F +++V D+ L++ L I + LF
Sbjct: 209 LVNYLGQESHNCPLTAAVSLCNPFDLVIADEDFHKGFNNVYDKALSKSLRRIFSKHSLLF 268
Query: 319 QGKAKGXDVGKALLAKSVRDFEEAISMVSYGFVDIEDFYTKASTRNMIKDIKIPVLFIQS 378
+ ++ A A++VRDF++ ++ VS+GF ++++Y+K+S+ IK ++IP+L IQ+
Sbjct: 269 EDIGGEFNIPLAANAETVRDFDDGLTRVSFGFKSVDEYYSKSSSSKAIKHVRIPLLCIQA 328
Query: 379 DNG-MVPAFSVPRNLIAENPFTSLLL 403
N + P +PR+ I NP L++
Sbjct: 329 ANDPIAPERGIPRDDIKANPNCVLIV 354
>gb|AAH46710.1| Abhd1-prov protein [Xenopus laevis]
Length = 411
Score = 132 bits (333), Expect = 2e-29
Identities = 92/315 (29%), Positives = 158/315 (49%), Gaps = 13/315 (4%)
Query: 96 SPTPFNRFVLLRCPSISFKDNERLIKDEKHYGRIRVNKREKDLEEE-LKYQRVCLSASDG 154
S PF F+ CP + K + GRI+ R L + + Y++ L +DG
Sbjct: 42 SDLPFQLFLERYCPIVKEKFRPTMWC---FGGRIQTILRVILLSKPPVSYRKEILKTTDG 98
Query: 155 GVVSLDWPVELDLEEERG--LDSTLLIVPGTPQGSMDDNIRVFVIDALKRGFFPVVMNPR 212
G +SLDW + + G L T++ +PG S I V A + G+ VV N R
Sbjct: 99 GQISLDWVDNNESSQFPGASLRPTVIFLPGLTGNSRQSYILHLVHQASRDGYRSVVFNNR 158
Query: 213 GCASSPITTPRLFTAADSDDICTAITYINKARPWTTLMGVGWGYGANMLTKYLAEVGERT 272
G + TPR F AA++DD+ ++++++ P ++ VG G ML YLA G
Sbjct: 159 GFGGEELLTPRTFCAANTDDLSAVVSHVHRQFPDAPVLAVGVSLGGMMLLNYLAAQGSSV 218
Query: 273 PLTAATCIDNPFDLDEATRTF--PYHHVT-DQKLTRGLINILQTNKALFQGKAKGXDVGK 329
PL AA C P+++ E+TR+ P +++ + L + L + ++ + K +V
Sbjct: 219 PLWAALCFSTPWNVFESTRSLEEPLNYLLFNYSLNKSLRRATEQHRHVI---GKAVNVDH 275
Query: 330 ALLAKSVRDFEEAISMVSYGFVDIEDFYTKASTRNMIKDIKIPVLFIQ-SDNGMVPAFSV 388
L ++S+R+F+E + V +GF +D+YT+AS + I+ PVL + +D+ P+ ++
Sbjct: 276 ILQSRSIREFDERYTSVVWGFPSCDDYYTQASPDFKLSHIRTPVLCLNAADDPFSPSAAI 335
Query: 389 PRNLIAENPFTSLLL 403
P + + NP +LL+
Sbjct: 336 PVSKASSNPHVALLI 350
>gb|AAX93169.1| unknown [Homo sapiens] gi|23397655|ref|NP_115993.2| alpha/beta
hydrolase domain containing protein 1 isoform 1 [Homo
sapiens]
Length = 405
Score = 127 bits (318), Expect = 1e-27
Identities = 79/266 (29%), Positives = 137/266 (50%), Gaps = 9/266 (3%)
Query: 144 YQRVCLSASDGGVVSLDWPVELDLEEERG--LDSTLLIVPGTPQGSMDDNIRVFVIDALK 201
YQ L DGG + LDW + D ++ +L++PG S D + V AL+
Sbjct: 90 YQSDILQTPDGGQLLLDWAKQPDSSQDPDPTTQPIVLLLPGITGSSQDTYVLHLVNQALR 149
Query: 202 RGFFPVVMNPRGCASSPITTPRLFTAADSDDICTAITYINKARPWTTLMGVGWGYGANML 261
G+ VV N RGC + T R F A++++D+ T + +I P L+ VG +G ++
Sbjct: 150 DGYQAVVFNNRGCRGEELRTHRAFCASNTEDLETVVNHIKHRYPQAPLLAVGISFGGILV 209
Query: 262 TKYLAEVGERTPLTAATCIDNPFDLDEATRTF--PYHHVT-DQKLTRGLINILQTNKALF 318
+LA+ + L AA + +D E TR+ P + + +Q LT GL +++ N+ +
Sbjct: 210 LNHLAQARQAAGLVAALTLSACWDSFETTRSLETPLNSLLFNQPLTAGLCQLVERNRKVI 269
Query: 319 QGKAKGXDVGKALLAKSVRDFEEAISMVSYGFVDIEDFYTKASTRNMIKDIKIPVLFIQ- 377
+ K D+ L A+++R F+E + V++G+ D +Y AS R I I+IPVL++
Sbjct: 270 E---KVVDIDFVLQARTIRQFDERYTSVAFGYQDCVTYYKAASPRTKIDAIRIPVLYLSA 326
Query: 378 SDNGMVPAFSVPRNLIAENPFTSLLL 403
+D+ P ++P +P+ +LL+
Sbjct: 327 ADDPFSPVCALPIQAAQHSPYVALLI 352
>gb|AAK44222.1| lung alpha/beta hydrolase protein 1 [Homo sapiens]
gi|37572291|gb|AAH39576.2| Alpha/beta hydrolase domain
containing protein 1, isoform 1 [Homo sapiens]
Length = 405
Score = 125 bits (314), Expect = 4e-27
Identities = 78/266 (29%), Positives = 137/266 (51%), Gaps = 9/266 (3%)
Query: 144 YQRVCLSASDGGVVSLDWPVELDLEEERG--LDSTLLIVPGTPQGSMDDNIRVFVIDALK 201
YQ L DGG + LDW + D ++ +L++PG S + + V AL+
Sbjct: 90 YQSDILQTPDGGQLLLDWAKQPDSSQDPDPTTQPIVLLLPGITGSSQETYVLHLVNQALR 149
Query: 202 RGFFPVVMNPRGCASSPITTPRLFTAADSDDICTAITYINKARPWTTLMGVGWGYGANML 261
G+ VV N RGC + T R F A++++D+ T + +I P L+ VG +G ++
Sbjct: 150 DGYQAVVFNNRGCRGEELRTHRAFCASNTEDLETVVNHIKHRYPQAPLLAVGISFGGILV 209
Query: 262 TKYLAEVGERTPLTAATCIDNPFDLDEATRTF--PYHHVT-DQKLTRGLINILQTNKALF 318
+LA+ + L AA + +D E TR+ P + + +Q LT GL +++ N+ +
Sbjct: 210 LNHLAQARQAAGLVAALTLSACWDSFETTRSLETPLNSLLFNQPLTAGLCQLVERNRKVI 269
Query: 319 QGKAKGXDVGKALLAKSVRDFEEAISMVSYGFVDIEDFYTKASTRNMIKDIKIPVLFIQ- 377
+ K D+ L A+++R F+E + V++G+ D +Y AS R I I+IPVL++
Sbjct: 270 E---KVVDIDFVLQARTIRQFDERYTSVAFGYQDCVTYYKAASPRTKIDAIRIPVLYLSA 326
Query: 378 SDNGMVPAFSVPRNLIAENPFTSLLL 403
+D+ P ++P +P+ +LL+
Sbjct: 327 ADDPFSPVCALPIQAAQHSPYVALLI 352
>ref|XP_525719.1| PREDICTED: hypothetical protein XP_525719 [Pan troglodytes]
Length = 402
Score = 123 bits (309), Expect = 1e-26
Identities = 78/266 (29%), Positives = 136/266 (50%), Gaps = 9/266 (3%)
Query: 144 YQRVCLSASDGGVVSLDWPVELDLEEERG--LDSTLLIVPGTPQGSMDDNIRVFVIDALK 201
YQ L DGG + LDW + D ++ +L++PG S + + V AL+
Sbjct: 87 YQSDILQTPDGGQLLLDWAKQPDSSQDPDPTTQPIVLLLPGITGSSQETYVLHLVNQALR 146
Query: 202 RGFFPVVMNPRGCASSPITTPRLFTAADSDDICTAITYINKARPWTTLMGVGWGYGANML 261
G+ VV N RGC + T R F A++++D+ T + +I P L+ VG +G ++
Sbjct: 147 DGYQAVVFNNRGCRGEELRTHRAFCASNTEDLETVVNHIKHRYPQAPLLAVGISFGGILV 206
Query: 262 TKYLAEVGERTPLTAATCIDNPFDLDEATRTF--PYHHVT-DQKLTRGLINILQTNKALF 318
+LA+ G+ L AA + +D E TR+ P + + +Q LT L +++ N+ +
Sbjct: 207 LNHLAQAGQAAGLVAALTLSACWDSFETTRSLETPLNSLLFNQPLTARLCQLVERNRKVI 266
Query: 319 QGKAKGXDVGKALLAKSVRDFEEAISMVSYGFVDIEDFYTKASTRNMIKDIKIPVLFIQ- 377
+ K D+ L A+++R F+E + V++G+ D +Y AS R I I+IPVL +
Sbjct: 267 E---KVVDIDFVLQARTIRQFDERYTSVAFGYQDCVTYYKAASPRTKIDAIRIPVLCLNA 323
Query: 378 SDNGMVPAFSVPRNLIAENPFTSLLL 403
+D+ P ++P +P+ +LL+
Sbjct: 324 TDDPFSPVCALPIQAAQHSPYVALLI 349
>ref|XP_636080.1| hypothetical protein DDB0188543 [Dictyostelium discoideum]
gi|60464426|gb|EAL62573.1| hypothetical protein
DDB0188543 [Dictyostelium discoideum]
Length = 395
Score = 117 bits (294), Expect = 8e-25
Identities = 87/312 (27%), Positives = 145/312 (45%), Gaps = 19/312 (6%)
Query: 125 HYGRIRVNKREKDLEEELKYQRVCLSASDGGVVSLDWPVELDLEEERGLDSTLLIVPGTP 184
+YG ++ K E + ++ DGG +SLD+ +L E + T++I G
Sbjct: 75 YYGSYKIPKLNLKTRRE-----ILVNPIDGGTISLDF---FELGEFKEDTPTIVINHGLT 126
Query: 185 QGSMDDNIRVFVIDALK-RGFFPVVMNPRGCASSPITTPRLFTAADSDDICTAITYINK- 242
GS + ++ F A K +GF VV N RGCA +PIT R ++A DDI Y+ K
Sbjct: 127 GGSHERYVQYFAQRAYKEKGFRSVVFNYRGCAGNPITADRAYSAVQLDDIKFVTEYLTKT 186
Query: 243 ----ARPWTTLMGVGWGYGANMLTKYLAEVGERTPLTAATCIDNPFDLDEATRTFPYHHV 298
+ W VG+ G+ +L Y+A+ G+ +P A I NP ++ E T+ ++
Sbjct: 187 ALPLVKKWFL---VGFSLGSAILVNYMADAGKDSPYLAHVSISNPMNMVECTKNLSSTYI 243
Query: 299 TDQKLTRGLINILQTNKALFQGKA-KGXDVGKALLAKSVRDFEEAISMVSYGFVDIEDFY 357
+ +GL N L+ F G+ K + + A+++ DF++ I+ +GF D+Y
Sbjct: 244 NNLIYNKGLANNLKRLFRKFDGRLDKYATKEQIMAAQTIADFDDLITSKMFGFETAHDYY 303
Query: 358 TKASTRNMIKDIKIPVLFIQS-DNGMVPAFSVPRNLIAENPFTSLLLCSCLSSSVMDTDT 416
AS+ I+++ P+LFI + D+ + P P NP T L + +
Sbjct: 304 LAASSSKSIRNLVKPILFINAIDDPIAPTSGFPWKDFKSNPNTILCVSRWGGHLGFISYE 363
Query: 417 SALSWCQLVTVE 428
+SW VE
Sbjct: 364 DHMSWSDKAAVE 375
>ref|XP_532904.1| PREDICTED: hypothetical protein XP_532904 [Canis familiaris]
Length = 429
Score = 117 bits (293), Expect = 1e-24
Identities = 80/268 (29%), Positives = 133/268 (48%), Gaps = 13/268 (4%)
Query: 144 YQRVCLSASDGGVVSLDWPVELDLEEERGLDST----LLIVPGTPQGSMDDNIRVFVIDA 199
Y L DGG LDW + D +G D T +L++PG S D I V A
Sbjct: 114 YWSEVLQTPDGGQFLLDWACQHD--RSQGPDPTTQPIVLLLPGITGSSQDSYILQLVNQA 171
Query: 200 LKRGFFPVVMNPRGCASSPITTPRLFTAADSDDICTAITYINKARPWTTLMGVGWGYGAN 259
L+ G+ VV N RGC + T R F+A +++D+ + +I P L+ VG G
Sbjct: 172 LRDGYRAVVFNNRGCRGEELLTHRAFSAGNTEDLEIVVKHIKHHYPRAPLLAVGISLGGI 231
Query: 260 MLTKYLAEVGERTPLTAATCIDNPFDLDEATRTF--PYHHVT-DQKLTRGLINILQTNKA 316
++ +LA G+ + L AA + +D E TR+ P + + +Q+LT L +I+ N+
Sbjct: 232 LVLNHLARTGQASGLVAALTLSACWDSSETTRSLETPLNSLLFNQRLTTELCHIVNRNRK 291
Query: 317 LFQGKAKGXDVGKALLAKSVRDFEEAISMVSYGFVDIEDFYTKASTRNMIKDIKIPVLFI 376
+ + K +V L A+++R F+E + V++G+ D +Y AS R + I+ PVL +
Sbjct: 292 VME---KVVNVDFVLQARTIRQFDERYTTVAFGYQDCVTYYQTASPRTKVDAIQTPVLCL 348
Query: 377 Q-SDNGMVPAFSVPRNLIAENPFTSLLL 403
+D+ P ++P +P +LL+
Sbjct: 349 NAADDPFSPVHALPLQAAQHSPHVALLI 376
>ref|XP_537301.1| PREDICTED: similar to alpha/beta hydrolase domain containing
protein 3 [Canis familiaris]
Length = 403
Score = 116 bits (291), Expect = 2e-24
Identities = 76/270 (28%), Positives = 136/270 (50%), Gaps = 9/270 (3%)
Query: 142 LKYQRVCLSASDGGVVSLDWPVELDLE--EERGLDSTLLIVPGTPQGSMDDNIRVFVIDA 199
++Y+ + +DGG +SLDW + + + T+L++PG S + I + +
Sbjct: 97 VQYRNELIKTADGGQISLDWFDNYNSKCYMDATTRPTILLLPGLTGTSKESYILHMIHLS 156
Query: 200 LKRGFFPVVMNPRGCASSPITTPRLFTAADSDDICTAITYINKARPWTTLMGVGWGYGAN 259
+ G+ VV N RG A + TPR + A+++D+ T I +++ P + G G
Sbjct: 157 EELGYRCVVFNNRGVAGENLLTPRTYCCANTEDLETVIHHVHSLYPSAPFLAAGVSMGGM 216
Query: 260 MLTKYLAEVGERTPLTAATCID---NPFDLDEATRTFPYHHVTDQKLTRGLINILQTNKA 316
+L YL ++G +TPL AA N F E+ + + LT L + + ++
Sbjct: 217 LLLNYLGKIGPKTPLKAAATFSVGWNTFACSESLEKPLNWLLFNYYLTTCLQSSVNKHRH 276
Query: 317 LFQGKAKGXDVGKALLAKSVRDFEEAISMVSYGFVDIEDFYTKASTRNMIKDIKIPVLFI 376
+F K D+ + AKS+R+F++ + V +G+ I+D+YT AS +K + IPVL +
Sbjct: 277 MF---VKQIDMDHVMKAKSIREFDKRFTSVMFGYQTIDDYYTDASPNRRLKSVGIPVLCL 333
Query: 377 QS-DNGMVPAFSVPRNLIAENPFTSLLLCS 405
S D+ P+ ++P +NP +L+L S
Sbjct: 334 NSVDDVFSPSHAIPIETAKQNPNVALVLTS 363
>ref|XP_512055.1| PREDICTED: similar to alpha/beta hydrolase domain containing
protein 3; lung alpha/beta hydrolase 3 [Pan troglodytes]
Length = 620
Score = 115 bits (289), Expect = 3e-24
Identities = 77/273 (28%), Positives = 136/273 (49%), Gaps = 15/273 (5%)
Query: 142 LKYQRVCLSASDGGVVSLDWPVELDLEE-----ERGLDSTLLIVPGTPQGSMDDNIRVFV 196
++Y+ + +DGG +SLDW D + + T+L++PG S + I +
Sbjct: 316 VQYRNELIKTADGGQISLDW---FDNDNSTCYMDASTRPTILLLPGLTGTSKESYILHMI 372
Query: 197 IDALKRGFFPVVMNPRGCASSPITTPRLFTAADSDDICTAITYINKARPWTTLMGVGWGY 256
+ + G+ VV N RG A + TPR + A+++D+ T I +++ P + G
Sbjct: 373 HLSEELGYRCVVFNNRGVAGENLLTPRTYCCANTEDLETVIHHVHSLYPSAPFLAAGVSM 432
Query: 257 GANMLTKYLAEVGERTPLTAATCID---NPFDLDEATRTFPYHHVTDQKLTRGLINILQT 313
G +L YL ++G +TPL AA N F E+ + + LT L + +
Sbjct: 433 GGMLLLNYLGKIGSKTPLMAAATFSVGWNTFACSESLEKPLNWLLFNYYLTTCLQSSVNK 492
Query: 314 NKALFQGKAKGXDVGKALLAKSVRDFEEAISMVSYGFVDIEDFYTKASTRNMIKDIKIPV 373
++ +F K D+ + AKS+R+F++ + V +G+ I+D+YT AS +K + IPV
Sbjct: 493 HRHMF---VKQVDMDHVMKAKSIREFDKRFTSVMFGYQTIDDYYTDASPSPRLKSVGIPV 549
Query: 374 LFIQS-DNGMVPAFSVPRNLIAENPFTSLLLCS 405
L + S D+ P+ ++P +NP +L+L S
Sbjct: 550 LCLNSVDDVFSPSHAIPVETAKQNPNVALVLTS 582
>gb|AAH21196.1| Alpha/beta hydrolase domain containing protein 3 [Homo sapiens]
Length = 409
Score = 115 bits (289), Expect = 3e-24
Identities = 77/273 (28%), Positives = 136/273 (49%), Gaps = 15/273 (5%)
Query: 142 LKYQRVCLSASDGGVVSLDWPVELDLEE-----ERGLDSTLLIVPGTPQGSMDDNIRVFV 196
++Y+ + +DGG +SLDW D + + T+L++PG S + I +
Sbjct: 105 VQYRNELIKTADGGQISLDW---FDNDNSTCYMDASTRPTILLLPGLTGTSKESYILHMI 161
Query: 197 IDALKRGFFPVVMNPRGCASSPITTPRLFTAADSDDICTAITYINKARPWTTLMGVGWGY 256
+ + G+ VV N RG A + TPR + A+++D+ T I +++ P + G
Sbjct: 162 HLSEELGYRCVVFNNRGVAGENLLTPRTYCCANTEDLETVIHHVHSLYPSAPFLAAGVSM 221
Query: 257 GANMLTKYLAEVGERTPLTAATCID---NPFDLDEATRTFPYHHVTDQKLTRGLINILQT 313
G +L YL ++G +TPL AA N F E+ + + LT L + +
Sbjct: 222 GGMLLLNYLGKIGSKTPLMAAATFSVGWNTFACSESLEKPLNWLLFNYYLTTCLQSSVNK 281
Query: 314 NKALFQGKAKGXDVGKALLAKSVRDFEEAISMVSYGFVDIEDFYTKASTRNMIKDIKIPV 373
++ +F K D+ + AKS+R+F++ + V +G+ I+D+YT AS +K + IPV
Sbjct: 282 HRHMF---VKQVDMDHVMKAKSIREFDKRFTSVMFGYQTIDDYYTDASPSPRLKSVGIPV 338
Query: 374 LFIQS-DNGMVPAFSVPRNLIAENPFTSLLLCS 405
L + S D+ P+ ++P +NP +L+L S
Sbjct: 339 LCLNSVDDVFSPSHAIPIETAKQNPNVALVLTS 371
>gb|AAC19155.1| unknown [Homo sapiens]
Length = 442
Score = 115 bits (289), Expect = 3e-24
Identities = 77/273 (28%), Positives = 136/273 (49%), Gaps = 15/273 (5%)
Query: 142 LKYQRVCLSASDGGVVSLDWPVELDLEE-----ERGLDSTLLIVPGTPQGSMDDNIRVFV 196
++Y+ + +DGG +SLDW D + + T+L++PG S + I +
Sbjct: 138 VQYRNELIKTADGGQISLDW---FDNDNSTCYMDASTRPTILLLPGLTGTSKESYILHMI 194
Query: 197 IDALKRGFFPVVMNPRGCASSPITTPRLFTAADSDDICTAITYINKARPWTTLMGVGWGY 256
+ + G+ VV N RG A + TPR + A+++D+ T I +++ P + G
Sbjct: 195 HLSEELGYRCVVFNNRGVAGENLLTPRTYCCANTEDLETVIHHVHSLYPSAPFLAAGVSM 254
Query: 257 GANMLTKYLAEVGERTPLTAATCID---NPFDLDEATRTFPYHHVTDQKLTRGLINILQT 313
G +L YL ++G +TPL AA N F E+ + + LT L + +
Sbjct: 255 GGMLLLNYLGKIGSKTPLMAAATFSVGWNTFACSESLEKPLNWLLFNYYLTTCLQSSVNK 314
Query: 314 NKALFQGKAKGXDVGKALLAKSVRDFEEAISMVSYGFVDIEDFYTKASTRNMIKDIKIPV 373
++ +F K D+ + AKS+R+F++ + V +G+ I+D+YT AS +K + IPV
Sbjct: 315 HRHMF---VKQVDMDHVMKAKSIREFDKRFTSVMFGYQTIDDYYTDASPSPRLKSVGIPV 371
Query: 374 LFIQS-DNGMVPAFSVPRNLIAENPFTSLLLCS 405
L + S D+ P+ ++P +NP +L+L S
Sbjct: 372 LCLNSVDDVFSPSHAIPIETAKQNPNVALVLTS 404
>ref|NP_612213.2| alpha/beta hydrolase domain containing protein 3 [Homo sapiens]
Length = 409
Score = 115 bits (289), Expect = 3e-24
Identities = 77/273 (28%), Positives = 136/273 (49%), Gaps = 15/273 (5%)
Query: 142 LKYQRVCLSASDGGVVSLDWPVELDLEE-----ERGLDSTLLIVPGTPQGSMDDNIRVFV 196
++Y+ + +DGG +SLDW D + + T+L++PG S + I +
Sbjct: 105 VQYRNELIKTADGGQISLDW---FDNDNSTCYMDASTRPTILLLPGLTGTSKESYILHMI 161
Query: 197 IDALKRGFFPVVMNPRGCASSPITTPRLFTAADSDDICTAITYINKARPWTTLMGVGWGY 256
+ + G+ VV N RG A + TPR + A+++D+ T I +++ P + G
Sbjct: 162 HLSEELGYRCVVFNNRGVAGENLLTPRTYCCANTEDLETVIHHVHSLYPSAPFLAAGVSM 221
Query: 257 GANMLTKYLAEVGERTPLTAATCID---NPFDLDEATRTFPYHHVTDQKLTRGLINILQT 313
G +L YL ++G +TPL AA N F E+ + + LT L + +
Sbjct: 222 GGMLLLNYLGKIGSKTPLMAAATFSVGWNTFACSESLEKPLNWLLFNYYLTTCLQSSVNK 281
Query: 314 NKALFQGKAKGXDVGKALLAKSVRDFEEAISMVSYGFVDIEDFYTKASTRNMIKDIKIPV 373
++ +F K D+ + AKS+R+F++ + V +G+ I+D+YT AS +K + IPV
Sbjct: 282 HRHMF---VKQVDMDHVMKAKSIREFDKRFTSVMFGYQTIDDYYTDASPSPRLKSVGIPV 338
Query: 374 LFIQS-DNGMVPAFSVPRNLIAENPFTSLLLCS 405
L + S D+ P+ ++P +NP +L+L S
Sbjct: 339 LCLNSVDDVFSPSHAIPIETAKQNPNVALVLTS 371
>ref|XP_419156.1| PREDICTED: similar to lung alpha/beta hydrolase fold protein 3
[Gallus gallus]
Length = 468
Score = 111 bits (277), Expect = 8e-23
Identities = 75/272 (27%), Positives = 135/272 (49%), Gaps = 9/272 (3%)
Query: 141 ELKYQRVCLSASDGGVVSLDWPVELD--LEEERGLDSTLLIVPGTPQGSMDDNIRVFVID 198
+++Y+ + +DGG +SLDW D + T+L++PG S + I +
Sbjct: 161 QVQYRNELIRTADGGQISLDWFDNNDSLYYPDASTRPTVLLLPGLTGTSKESYILHMIHQ 220
Query: 199 ALKRGFFPVVMNPRGCASSPITTPRLFTAADSDDICTAITYINKARPWTTLMGVGWGYGA 258
+ G+ VV N RG A + TPR + AA+++D+ I +I+ P M G G
Sbjct: 221 SETLGYRCVVFNNRGIAGEELLTPRTYCAANTEDLEAVIHHIHNLHPSAPFMAAGVSMGG 280
Query: 259 NMLTKYLAEVGERTPLTAATCIDNPFDLDEATRTF--PYHHVT-DQKLTRGLINILQTNK 315
+L YL + G TPL AA +++ E+ + P + + + LT L + + ++
Sbjct: 281 MLLLNYLGKTGRDTPLMAAAIFSAGWNVFESVESLEKPLNWLLFNYYLTTCLQSSISRHR 340
Query: 316 ALFQGKAKGXDVGKALLAKSVRDFEEAISMVSYGFVDIEDFYTKASTRNMIKDIKIPVLF 375
+ + K D+ + A++VR+F++ + V +G+ I+D+Y AS +K + IPVL
Sbjct: 341 QMLE---KLFDMDLVMKARTVREFDKQFTSVMFGYRTIDDYYEDASPCRKLKSVGIPVLC 397
Query: 376 IQS-DNGMVPAFSVPRNLIAENPFTSLLLCSC 406
+ S D+ P ++P +N +L+L SC
Sbjct: 398 LNSVDDVFSPGHAIPVETAKQNANVALVLTSC 429
>ref|NP_598891.1| abhydrolase domain containing 3 [Mus musculus]
gi|16660465|gb|AAL27565.1| lung alpha/beta hydrolase
fold protein 3 [Mus musculus] gi|20071306|gb|AAH26770.1|
Abhydrolase domain containing 3 [Mus musculus]
Length = 411
Score = 110 bits (276), Expect = 1e-22
Identities = 70/268 (26%), Positives = 135/268 (50%), Gaps = 9/268 (3%)
Query: 142 LKYQRVCLSASDGGVVSLDWPVELDLEE--ERGLDSTLLIVPGTPQGSMDDNIRVFVIDA 199
++Y+ + +DGG +SLDW + + T+L++PG S + I + +
Sbjct: 105 VQYRNELIKTADGGQISLDWFDNNNSAYYVDASTRPTILLLPGLTGTSKESYILHMIHLS 164
Query: 200 LKRGFFPVVMNPRGCASSPITTPRLFTAADSDDICTAITYINKARPWTTLMGVGWGYGAN 259
+ G+ VV N RG A + TPR + A+++D+ + +++ P + G G
Sbjct: 165 EELGYRCVVFNNRGVAGESLLTPRTYCCANTEDLEAVVHHVHSLYPGAPFLAAGVSMGGM 224
Query: 260 MLTKYLAEVGERTPLTAATCID---NPFDLDEATRTFPYHHVTDQKLTRGLINILQTNKA 316
+L YL ++G +TPL AA N F E+ + + LT L + ++ ++
Sbjct: 225 LLLNYLGKIGSKTPLMAAATFSVGWNTFACSESLERPLNWLLFNYYLTTCLQSSVKKHRH 284
Query: 317 LFQGKAKGXDVGKALLAKSVRDFEEAISMVSYGFVDIEDFYTKASTRNMIKDIKIPVLFI 376
+F + D+ + + AKS+R+F++ + V +G+ ++D+YT AS +K + IPVL +
Sbjct: 285 MF---VEQIDMDQVMKAKSIREFDKRFTAVMFGYRTLDDYYTDASPNRRLKSVGIPVLCL 341
Query: 377 Q-SDNGMVPAFSVPRNLIAENPFTSLLL 403
+D+ P+ ++P +NP +L+L
Sbjct: 342 NATDDVFSPSHAIPIETAKQNPNVALVL 369
>gb|AAH94403.1| Unknown (protein for MGC:84955) [Xenopus laevis]
Length = 410
Score = 110 bits (274), Expect = 2e-22
Identities = 75/270 (27%), Positives = 133/270 (48%), Gaps = 9/270 (3%)
Query: 142 LKYQRVCLSASDGGVVSLDWPVELD--LEEERGLDSTLLIVPGTPQGSMDDNIRVFVIDA 199
+ Y + +DGG +SLDW D L + T+L++PG S + I + +
Sbjct: 104 VNYNNELIKTADGGQISLDWFNNDDNTLYPDTSSRPTILLLPGLTGTSRESYILHMIKHS 163
Query: 200 LKRGFFPVVMNPRGCASSPITTPRLFTAADSDDICTAITYINKARPWTTLMGVGWGYGAN 259
G+ VV N RG + + TPR + AA+++D+ I +++ P +LM G G
Sbjct: 164 EALGYRCVVFNNRGVSGEKLLTPRTYCAANTEDLEAVINHVHAMYPEASLMAAGVSMGGM 223
Query: 260 MLTKYLAEVGERTPLTAATCIDNPFDLDEATRTF--PYHHVT-DQKLTRGLINILQTNKA 316
+L YL ++G +TPL A ++ E+ T P + + + LT L + ++
Sbjct: 224 LLVNYLGKMGRQTPLKGAAVFSAGWEAFESAITLEKPINWMMFNYYLTTCLQAAIVRHRH 283
Query: 317 LFQGKAKGXDVGKALLAKSVRDFEEAISMVSYGFVDIEDFYTKASTRNMIKDIKIPVLFI 376
+ + K D+ L AKS+R+F+ + V +G+ E +Y AS + +K ++IPVL +
Sbjct: 284 ILE---KHFDIDHILRAKSIREFDTRFTSVMFGYPTNEHYYRDASPCHKVKSVEIPVLCL 340
Query: 377 QS-DNGMVPAFSVPRNLIAENPFTSLLLCS 405
+ D+ P ++P +NP +L+L S
Sbjct: 341 NALDDVFSPGHAIPVETARQNPNVALVLTS 370
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.320 0.136 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,016,366,111
Number of Sequences: 2540612
Number of extensions: 44533096
Number of successful extensions: 97102
Number of sequences better than 10.0: 188
Number of HSP's better than 10.0 without gapping: 149
Number of HSP's successfully gapped in prelim test: 39
Number of HSP's that attempted gapping in prelim test: 96670
Number of HSP's gapped (non-prelim): 215
length of query: 602
length of database: 863,360,394
effective HSP length: 134
effective length of query: 468
effective length of database: 522,918,386
effective search space: 244725804648
effective search space used: 244725804648
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 78 (34.7 bits)
Medicago: description of AC124967.11


