

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124966.6 + phase: 0
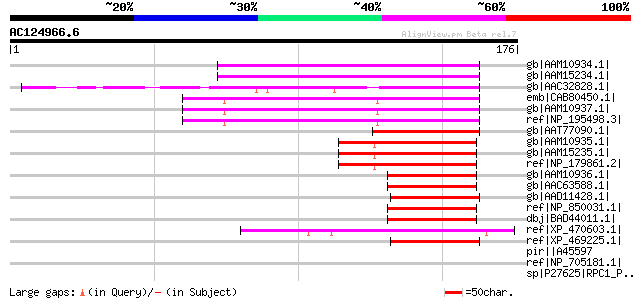
(176 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM10934.1| putative bHLH transcription factor [Arabidopsis t... 60 2e-08
gb|AAM15234.1| putative bHLH transcription factor [Arabidopsis t... 60 2e-08
gb|AAC32828.1| symbiotic ammonium transporter; nodulin [Glycine ... 59 4e-08
emb|CAB80450.1| putative protein [Arabidopsis thaliana] gi|44907... 56 5e-07
gb|AAM10937.1| putative bHLH transcription factor [Arabidopsis t... 56 5e-07
ref|NP_195498.3| basic helix-loop-helix (bHLH) family protein [A... 56 5e-07
gb|AAT77090.1| putative transcription factor [Oryza sativa (japo... 50 4e-05
gb|AAM10935.1| putative bHLH transcription factor [Arabidopsis t... 49 6e-05
gb|AAM15235.1| putative bHLH transcription factor [Arabidopsis t... 49 6e-05
ref|NP_179861.2| basic helix-loop-helix (bHLH) family protein [A... 49 6e-05
gb|AAM10936.1| putative bHLH transcription factor [Arabidopsis t... 48 1e-04
gb|AAC63588.1| putative bHLH transcription factor [Arabidopsis t... 48 1e-04
gb|AAD11428.1| transporter homolog [Mesembryanthemum crystallinum] 48 1e-04
ref|NP_850031.1| basic helix-loop-helix (bHLH) family protein [A... 48 1e-04
dbj|BAD44011.1| putative bHLH transcription factor (bHLH020) [Ar... 48 1e-04
ref|XP_470603.1| Putative bHLH transcription factor [Oryza sativ... 47 3e-04
ref|XP_469225.1| putative ammonium transporter [Oryza sativa (ja... 46 5e-04
pir||A45597 DNA-directed RNA polymerase (EC 2.7.7.6) III largest... 43 0.003
ref|NP_705181.1| DNA-directed RNA polymerase 3 largest subunit [... 43 0.003
sp|P27625|RPC1_PLAFA DNA-directed RNA polymerase III largest sub... 43 0.003
>gb|AAM10934.1| putative bHLH transcription factor [Arabidopsis thaliana]
gi|30681807|ref|NP_179860.2| basic helix-loop-helix
(bHLH) family protein [Arabidopsis thaliana]
Length = 304
Score = 60.5 bits (145), Expect = 2e-08
Identities = 36/91 (39%), Positives = 51/91 (55%)
Query: 73 PSQYILSFENSTMQPSPNSDIATCSSIMVQETTTLNNNVSSELPKIIKKRTKNLRSSCEM 132
PS ILSFE + + ++ S +E + + + + K+ RS
Sbjct: 65 PSSRILSFEKTGLHVMNHNSPNLIFSPKDEEIGLPEHKKAELIIRGTKRAQSLTRSQSNA 124
Query: 133 QDHIMAERKRRQVLTERFIALSATIPGLKKV 163
QDHI+AERKRR+ LT+RF+ALSA IPGLKK+
Sbjct: 125 QDHILAERKRREKLTQRFVALSALIPGLKKM 155
>gb|AAM15234.1| putative bHLH transcription factor [Arabidopsis thaliana]
gi|3738089|gb|AAC63586.1| putative bHLH transcription
factor [Arabidopsis thaliana] gi|25412135|pir||D84616
probable bHLH transcription factor [imported] -
Arabidopsis thaliana
Length = 288
Score = 60.5 bits (145), Expect = 2e-08
Identities = 36/91 (39%), Positives = 51/91 (55%)
Query: 73 PSQYILSFENSTMQPSPNSDIATCSSIMVQETTTLNNNVSSELPKIIKKRTKNLRSSCEM 132
PS ILSFE + + ++ S +E + + + + K+ RS
Sbjct: 65 PSSRILSFEKTGLHVMNHNSPNLIFSPKDEEIGLPEHKKAELIIRGTKRAQSLTRSQSNA 124
Query: 133 QDHIMAERKRRQVLTERFIALSATIPGLKKV 163
QDHI+AERKRR+ LT+RF+ALSA IPGLKK+
Sbjct: 125 QDHILAERKRREKLTQRFVALSALIPGLKKM 155
>gb|AAC32828.1| symbiotic ammonium transporter; nodulin [Glycine max]
gi|7488716|pir||T06329 symbiotic ammonium transport
protein SAT1 - soybean
Length = 347
Score = 59.3 bits (142), Expect = 4e-08
Identities = 55/174 (31%), Positives = 83/174 (47%), Gaps = 36/174 (20%)
Query: 5 KSHFYLDEDKFQREIVQHQPSFSFESGNSQSSTIQYMNNIVDNVCVTNTVHENIAQEYTW 64
+ H + D F P S E+ SQ+ +Y + DN + + N +Q+ T
Sbjct: 49 QKHSFSDNSNFN-------PKTSMET--SQTGIERYAKQLGDN-----SWNHNKSQQQTP 94
Query: 65 ATNSQQLPPSQYILSFENST-------MQPS-----PNSDIATCSSIMVQETTTLNNNV- 111
T Q +LSF N+ ++P P D + +++ + T N N
Sbjct: 95 ET---QFASCSNLLSFVNTNYTSELGLVKPKVEMACPKIDNNALADMLISQGTLGNQNYI 151
Query: 112 --SSELPKIIKKRTKNLRSSCEMQDHIMAERKRRQVLTERFIALSATIPGLKKV 163
+S+ K IK R K + QDHI+AERKRR+ L++RFIALSA +PGLKK+
Sbjct: 152 FKASQETKKIKTRPK----LSQPQDHIIAERKRREKLSQRFIALSALVPGLKKM 201
>emb|CAB80450.1| putative protein [Arabidopsis thaliana] gi|4490730|emb|CAB38933.1|
putative protein [Arabidopsis thaliana]
gi|7487499|pir||T06032 hypothetical protein T28I19.130 -
Arabidopsis thaliana
Length = 314
Score = 55.8 bits (133), Expect = 5e-07
Identities = 40/118 (33%), Positives = 56/118 (46%), Gaps = 15/118 (12%)
Query: 61 EYTWATNSQQLPP-----------SQYILSFENSTMQPSPNSDIATCSSIMVQETTTLNN 109
E T+ + S LPP S ILSFE+ + T + +
Sbjct: 40 ETTYISPSSHLPPNSKPHHIHRHSSSRILSFEDYGSNDMEHEYSPTYLNSIFSPKLEAQV 99
Query: 110 NVSSELPKIIKKRTKNL----RSSCEMQDHIMAERKRRQVLTERFIALSATIPGLKKV 163
+ + +K TK R+ QDHI+AERKRR+ LT+RF+ALSA +PGLKK+
Sbjct: 100 QPHQKSDEFNRKGTKRAQPFSRNQSNAQDHIIAERKRREKLTQRFVALSALVPGLKKM 157
>gb|AAM10937.1| putative bHLH transcription factor [Arabidopsis thaliana]
Length = 304
Score = 55.8 bits (133), Expect = 5e-07
Identities = 40/118 (33%), Positives = 56/118 (46%), Gaps = 15/118 (12%)
Query: 61 EYTWATNSQQLPP-----------SQYILSFENSTMQPSPNSDIATCSSIMVQETTTLNN 109
E T+ + S LPP S ILSFE+ + T + +
Sbjct: 40 ETTYISPSSHLPPNSKPHHIHRHSSSRILSFEDYGSNDMEHEYSPTYLNSIFSPKLEAQV 99
Query: 110 NVSSELPKIIKKRTKNL----RSSCEMQDHIMAERKRRQVLTERFIALSATIPGLKKV 163
+ + +K TK R+ QDHI+AERKRR+ LT+RF+ALSA +PGLKK+
Sbjct: 100 QPHQKSDEFNRKGTKRAQPFSRNQSNAQDHIIAERKRREKLTQRFVALSALVPGLKKM 157
>ref|NP_195498.3| basic helix-loop-helix (bHLH) family protein [Arabidopsis thaliana]
Length = 328
Score = 55.8 bits (133), Expect = 5e-07
Identities = 40/118 (33%), Positives = 56/118 (46%), Gaps = 15/118 (12%)
Query: 61 EYTWATNSQQLPP-----------SQYILSFENSTMQPSPNSDIATCSSIMVQETTTLNN 109
E T+ + S LPP S ILSFE+ + T + +
Sbjct: 64 ETTYISPSSHLPPNSKPHHIHRHSSSRILSFEDYGSNDMEHEYSPTYLNSIFSPKLEAQV 123
Query: 110 NVSSELPKIIKKRTKNL----RSSCEMQDHIMAERKRRQVLTERFIALSATIPGLKKV 163
+ + +K TK R+ QDHI+AERKRR+ LT+RF+ALSA +PGLKK+
Sbjct: 124 QPHQKSDEFNRKGTKRAQPFSRNQSNAQDHIIAERKRREKLTQRFVALSALVPGLKKM 181
>gb|AAT77090.1| putative transcription factor [Oryza sativa (japonica
cultivar-group)] gi|41469283|gb|AAS07165.1| putative
symbiotic ammonium transport protein [Oryza sativa
(japonica cultivar-group)]
Length = 359
Score = 49.7 bits (117), Expect = 4e-05
Identities = 22/37 (59%), Positives = 31/37 (83%)
Query: 127 RSSCEMQDHIMAERKRRQVLTERFIALSATIPGLKKV 163
R + + Q+HI+AERKRR+ L++RFIALS +PGLKK+
Sbjct: 174 RPASQNQEHILAERKRREKLSQRFIALSKIVPGLKKM 210
>gb|AAM10935.1| putative bHLH transcription factor [Arabidopsis thaliana]
Length = 295
Score = 48.9 bits (115), Expect = 6e-05
Identities = 25/54 (46%), Positives = 37/54 (68%), Gaps = 6/54 (11%)
Query: 115 LPKIIKKRTKN------LRSSCEMQDHIMAERKRRQVLTERFIALSATIPGLKK 162
+ K++ + TK RS ++H++AERKRR+ L+E+FIALSA +PGLKK
Sbjct: 94 MDKLVGRGTKRKTCSHGTRSPVLAKEHVLAERKRREKLSEKFIALSALLPGLKK 147
>gb|AAM15235.1| putative bHLH transcription factor [Arabidopsis thaliana]
gi|3738090|gb|AAC63587.1| putative bHLH transcription
factor [Arabidopsis thaliana] gi|25412136|pir||E84616
probable bHLH transcription factor [imported] -
Arabidopsis thaliana
Length = 284
Score = 48.9 bits (115), Expect = 6e-05
Identities = 25/54 (46%), Positives = 37/54 (68%), Gaps = 6/54 (11%)
Query: 115 LPKIIKKRTKN------LRSSCEMQDHIMAERKRRQVLTERFIALSATIPGLKK 162
+ K++ + TK RS ++H++AERKRR+ L+E+FIALSA +PGLKK
Sbjct: 94 MDKLVGRGTKRKTCSHGTRSPVLAKEHVLAERKRREKLSEKFIALSALLPGLKK 147
>ref|NP_179861.2| basic helix-loop-helix (bHLH) family protein [Arabidopsis thaliana]
Length = 295
Score = 48.9 bits (115), Expect = 6e-05
Identities = 25/54 (46%), Positives = 37/54 (68%), Gaps = 6/54 (11%)
Query: 115 LPKIIKKRTKN------LRSSCEMQDHIMAERKRRQVLTERFIALSATIPGLKK 162
+ K++ + TK RS ++H++AERKRR+ L+E+FIALSA +PGLKK
Sbjct: 94 MDKLVGRGTKRKTCSHGTRSPVLAKEHVLAERKRREKLSEKFIALSALLPGLKK 147
>gb|AAM10936.1| putative bHLH transcription factor [Arabidopsis thaliana]
Length = 320
Score = 47.8 bits (112), Expect = 1e-04
Identities = 21/31 (67%), Positives = 27/31 (86%)
Query: 132 MQDHIMAERKRRQVLTERFIALSATIPGLKK 162
+++H++AERKRRQ L ER IALSA +PGLKK
Sbjct: 130 LKEHVLAERKRRQKLNERLIALSALLPGLKK 160
>gb|AAC63588.1| putative bHLH transcription factor [Arabidopsis thaliana]
gi|25412138|pir||F84616 probable bHLH transcription
factor [imported] - Arabidopsis thaliana
Length = 322
Score = 47.8 bits (112), Expect = 1e-04
Identities = 21/31 (67%), Positives = 27/31 (86%)
Query: 132 MQDHIMAERKRRQVLTERFIALSATIPGLKK 162
+++H++AERKRRQ L ER IALSA +PGLKK
Sbjct: 130 LKEHVLAERKRRQKLNERLIALSALLPGLKK 160
>gb|AAD11428.1| transporter homolog [Mesembryanthemum crystallinum]
Length = 300
Score = 47.8 bits (112), Expect = 1e-04
Identities = 20/31 (64%), Positives = 28/31 (89%)
Query: 133 QDHIMAERKRRQVLTERFIALSATIPGLKKV 163
Q+H++AERKRR+ +T+RF ALSA +PGLKK+
Sbjct: 117 QEHVLAERKRREKMTQRFHALSALVPGLKKM 147
>ref|NP_850031.1| basic helix-loop-helix (bHLH) family protein [Arabidopsis thaliana]
Length = 314
Score = 47.8 bits (112), Expect = 1e-04
Identities = 21/31 (67%), Positives = 27/31 (86%)
Query: 132 MQDHIMAERKRRQVLTERFIALSATIPGLKK 162
+++H++AERKRRQ L ER IALSA +PGLKK
Sbjct: 124 LKEHVLAERKRRQKLNERLIALSALLPGLKK 154
>dbj|BAD44011.1| putative bHLH transcription factor (bHLH020) [Arabidopsis thaliana]
Length = 320
Score = 47.8 bits (112), Expect = 1e-04
Identities = 21/31 (67%), Positives = 27/31 (86%)
Query: 132 MQDHIMAERKRRQVLTERFIALSATIPGLKK 162
+++H++AERKRRQ L ER IALSA +PGLKK
Sbjct: 130 LKEHVLAERKRRQKLNERLIALSALLPGLKK 160
>ref|XP_470603.1| Putative bHLH transcription factor [Oryza sativa (japonica
cultivar-group)] gi|20532320|gb|AAM27466.1| Putative
bHLH transcription factor [Oryza sativa (japonica
cultivar-group)]
Length = 451
Score = 46.6 bits (109), Expect = 3e-04
Identities = 33/103 (32%), Positives = 51/103 (49%), Gaps = 8/103 (7%)
Query: 81 ENSTMQPSPNSDIATCSSIMVQ-ETTTLNNN-----VSSELPKIIKKRTKNLRSSCEMQD 134
+ QPSP+S + VQ TTTL+ + + + + R Q+
Sbjct: 228 KEQVQQPSPSSSNVLSFAGQVQGSTTTLDFSGRGWQQDDGVGVFQQPPERRSRPPANAQE 287
Query: 135 HIMAERKRRQVLTERFIALSATIPGLKKVS--FLYKPTISLIK 175
H++AERKRR+ L ++F+AL+ +PGLKK L TI +K
Sbjct: 288 HVIAERKRREKLQQQFVALATIVPGLKKTDKISLLGSTIDYVK 330
>ref|XP_469225.1| putative ammonium transporter [Oryza sativa (japonica
cultivar-group)] gi|29788848|gb|AAP03394.1| putative
ammonium transporter [Oryza sativa (japonica
cultivar-group)]
Length = 353
Score = 45.8 bits (107), Expect = 5e-04
Identities = 20/31 (64%), Positives = 26/31 (83%)
Query: 133 QDHIMAERKRRQVLTERFIALSATIPGLKKV 163
Q+HI+AERKRR+ + +RFI LS IPGLKK+
Sbjct: 191 QEHIIAERKRREKINQRFIELSTVIPGLKKM 221
>pir||A45597 DNA-directed RNA polymerase (EC 2.7.7.6) III largest chain - malaria
parasite (Plasmodium falciparum)
Length = 2339
Score = 43.1 bits (100), Expect = 0.003
Identities = 33/144 (22%), Positives = 68/144 (46%), Gaps = 16/144 (11%)
Query: 5 KSHFYLDEDKFQREIVQHQPSFSFESGNSQSSTIQYMN---NIVDNVCVTNTVHENIAQE 61
K+ F++++ K + H+ + E N+Q IQY N N + + + NT H + +
Sbjct: 1337 KAKFFIEKKKGK----MHECNDDIEYNNTQYDNIQYNNISCNYIKSQNLENT-HHQVNND 1391
Query: 62 YTWATNSQQLPPSQY--ILSFENSTMQPSPNSDIATCSSIMVQETTTLNNNVSSELPKII 119
++ N+ LPP +Y I F N ++ ++M ++ LNN+ + +
Sbjct: 1392 LSFIKNNVILPPKEYHSIFHFVNDYR------NVVEIKNLMDKKKIFLNNSEKNVVQSKY 1445
Query: 120 KKRTKNLRSSCEMQDHIMAERKRR 143
+ +KNL+ E+ ++I K++
Sbjct: 1446 NRMSKNLKKKIEIINNIYRNEKKK 1469
>ref|NP_705181.1| DNA-directed RNA polymerase 3 largest subunit [Plasmodium falciparum
3D7] gi|23615426|emb|CAD52417.1| DNA-directed RNA
polymerase 3 largest subunit [Plasmodium falciparum 3D7]
Length = 2356
Score = 43.1 bits (100), Expect = 0.003
Identities = 33/144 (22%), Positives = 68/144 (46%), Gaps = 16/144 (11%)
Query: 5 KSHFYLDEDKFQREIVQHQPSFSFESGNSQSSTIQYMN---NIVDNVCVTNTVHENIAQE 61
K+ F++++ K + H+ + E N+Q IQY N N + + + NT H + +
Sbjct: 1347 KAKFFIEKKKGK----MHECNDDIEYNNTQYDNIQYNNISCNYIKSQNLENT-HHQVNND 1401
Query: 62 YTWATNSQQLPPSQY--ILSFENSTMQPSPNSDIATCSSIMVQETTTLNNNVSSELPKII 119
++ N+ LPP +Y I F N ++ ++M ++ LNN+ + +
Sbjct: 1402 LSFIKNNVILPPKEYHSIFHFVNDYR------NVVEIKNLMDKKKIFLNNSEKNVVQSKY 1455
Query: 120 KKRTKNLRSSCEMQDHIMAERKRR 143
+ +KNL+ E+ ++I K++
Sbjct: 1456 NRMSKNLKKKIEIINNIYRNEKKK 1479
>sp|P27625|RPC1_PLAFA DNA-directed RNA polymerase III largest subunit
gi|160596|gb|AAA29729.1| RNA polymerase III largest
subunit
Length = 2339
Score = 43.1 bits (100), Expect = 0.003
Identities = 33/144 (22%), Positives = 68/144 (46%), Gaps = 16/144 (11%)
Query: 5 KSHFYLDEDKFQREIVQHQPSFSFESGNSQSSTIQYMN---NIVDNVCVTNTVHENIAQE 61
K+ F++++ K + H+ + E N+Q IQY N N + + + NT H + +
Sbjct: 1337 KAKFFIEKKKGK----MHECNDDIEYNNTQYDNIQYNNISCNYIKSQNLENT-HHQVNND 1391
Query: 62 YTWATNSQQLPPSQY--ILSFENSTMQPSPNSDIATCSSIMVQETTTLNNNVSSELPKII 119
++ N+ LPP +Y I F N ++ ++M ++ LNN+ + +
Sbjct: 1392 LSFIKNNVILPPKEYHSIFHFVNDYR------NVVEIKNLMDKKKIFLNNSEKNVVQSKY 1445
Query: 120 KKRTKNLRSSCEMQDHIMAERKRR 143
+ +KNL+ E+ ++I K++
Sbjct: 1446 NRMSKNLKKKIEIINNIYRNEKKK 1469
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.314 0.127 0.354
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 269,581,525
Number of Sequences: 2540612
Number of extensions: 9425635
Number of successful extensions: 34551
Number of sequences better than 10.0: 209
Number of HSP's better than 10.0 without gapping: 104
Number of HSP's successfully gapped in prelim test: 106
Number of HSP's that attempted gapping in prelim test: 34419
Number of HSP's gapped (non-prelim): 228
length of query: 176
length of database: 863,360,394
effective HSP length: 119
effective length of query: 57
effective length of database: 561,027,566
effective search space: 31978571262
effective search space used: 31978571262
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 71 (32.0 bits)
Medicago: description of AC124966.6


