

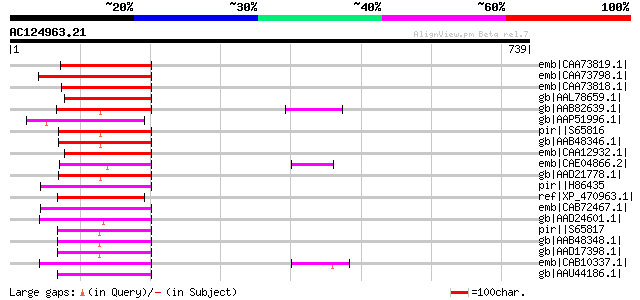
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124963.21 + phase: 0 /pseudo
(739 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAA73819.1| reverse transcriptase [Antirrhinum majus] gi|748... 141 7e-32
emb|CAA73798.1| reverse transcriptase [Beta vulgaris subsp. vulg... 131 9e-29
emb|CAA73818.1| reverse transcriptase [Antirrhinum majus] gi|748... 127 1e-27
gb|AAL78659.1| reverse transcriptase [Fagus sylvatica] 110 1e-22
gb|AAB82639.1| putative non-LTR retroelement reverse transcripta... 102 3e-20
gb|AAP51996.1| putative non-LTR retroelement reverse transcripta... 101 1e-19
pir||S65816 RNA-directed DNA polymerase (EC 2.7.7.49) (clone B2)... 99 7e-19
gb|AAB48346.1| reverse transcriptase 99 7e-19
emb|CAA12932.1| reverse transcriptase [Pinus elliottii] 98 9e-19
emb|CAE04866.2| OSJNBa0086O06.14 [Oryza sativa (japonica cultiva... 98 1e-18
gb|AAD21778.1| putative non-LTR retroelement reverse transcripta... 97 2e-18
pir||H86435 protein F17F8.5 [imported] - Arabidopsis thaliana gi... 94 1e-17
ref|XP_470963.1| OSJNBa0094O15.6 [Oryza sativa (japonica cultiva... 94 1e-17
emb|CAB72467.1| putative protein [Arabidopsis thaliana] gi|11358... 94 2e-17
gb|AAD24601.1| putative non-LTR retroelement reverse transcripta... 92 5e-17
pir||S65817 RNA-directed DNA polymerase (EC 2.7.7.49) (clone B3)... 92 6e-17
gb|AAB48348.1| reverse transcriptase 92 6e-17
gb|AAD17398.1| putative non-LTR retroelement reverse transcripta... 92 8e-17
emb|CAB10337.1| reverse transcriptase like protein [Arabidopsis ... 91 1e-16
gb|AAU44186.1| putative polyprotein [Oryza sativa (japonica cult... 91 1e-16
>emb|CAA73819.1| reverse transcriptase [Antirrhinum majus] gi|7489309|pir||T17102
RNA-directed DNA polymerase (EC 2.7.7.49) (clone AmLi2)
- garden snapdragon (fragment)
Length = 136
Score = 141 bits (356), Expect = 7e-32
Identities = 70/130 (53%), Positives = 97/130 (73%)
Query: 73 NDFCLISLVESMYKILAKVLANRLRYVIGMVISDSQSTFVKGRQILDGILVANEVVDDAH 132
N F ISL+ MYK+LAK+LANR+ V+G +IS+ Q+ F++GRQI+D IL+ANEV+DD
Sbjct: 1 NKFRPISLIGCMYKMLAKLLANRVSKVMGPIISNEQAAFIEGRQIMDSILIANEVLDDLK 60
Query: 133 RCKKELLLFKVDFEEAYDIVDWSYLDEVMYKMGFPVL*QK*IKECIGTATTSMLVNGSPT 192
R + L+FKVDFE+A+D V+W+YLD +M +GF +K I EC+ + T S+LVNGSPT
Sbjct: 61 RKRIPSLVFKVDFEKAFDCVNWNYLDSMMDNLGFCATWRKWISECLHSGTISVLVNGSPT 120
Query: 193 DEFPLKRGCK 202
DEF +RG +
Sbjct: 121 DEFRAQRGLR 130
>emb|CAA73798.1| reverse transcriptase [Beta vulgaris subsp. vulgaris]
gi|7484649|pir||T14619 reverse transcriptase - beet
retrotransposon (fragment)
Length = 507
Score = 131 bits (329), Expect = 9e-29
Identities = 64/162 (39%), Positives = 109/162 (66%), Gaps = 1/162 (0%)
Query: 42 LLVNFIGMGGLPKESIALSLLLFLRLR-ARRLNDFCLISLVESMYKILAKVLANRLRYVI 100
++ +F G LP + + L + + R N+F IS+V +YKI+AK+LA RL+ V+
Sbjct: 340 IVEDFWASGSLPHGCNSAFIALIPKTQHPRGFNEFRPISMVGCLYKIIAKILARRLQRVM 399
Query: 101 GMVISDSQSTFVKGRQILDGILVANEVVDDAHRCKKELLLFKVDFEEAYDIVDWSYLDEV 160
+I QS+F+KGRQILDG+L+A+E++D R K E ++ K+DF +A+D + W+YL+ V
Sbjct: 400 DHLIGPYQSSFIKGRQILDGVLIASELIDSCRRMKNEAVVLKLDFHKAFDSISWNYLEWV 459
Query: 161 MYKMGFPVL*QK*IKECIGTATTSMLVNGSPTDEFPLKRGCK 202
+ +M FP++ + I+ C+ +A+ ++L+NGSP+ F L+RG +
Sbjct: 460 LCQMNFPLVWRSWIRSCVMSASAAVLINGSPSSVFKLQRGLR 501
>emb|CAA73818.1| reverse transcriptase [Antirrhinum majus] gi|7489310|pir||T17095
RNA-directed DNA polymerase (EC 2.7.7.49) - garden
snapdragon (fragment)
Length = 136
Score = 127 bits (319), Expect = 1e-27
Identities = 66/129 (51%), Positives = 89/129 (68%)
Query: 74 DFCLISLVESMYKILAKVLANRLRYVIGMVISDSQSTFVKGRQILDGILVANEVVDDAHR 133
+F ISL+ +YK+LAK+LA RL V+ VIS+ QS F+ GRQI + I++ANEVVD+ R
Sbjct: 2 EFRPISLIGCIYKVLAKLLAKRLSKVMHSVISEEQSAFIGGRQITESIVIANEVVDELRR 61
Query: 134 CKKELLLFKVDFEEAYDIVDWSYLDEVMYKMGFPVL*QK*IKECIGTATTSMLVNGSPTD 193
KKE + K+DFE+AYD V W +LD +M K G +K + C TAT S+L+NGSPT+
Sbjct: 62 KKKEGFILKIDFEKAYDRVSWEFLDHMMDKYGVQREMEKVDRACNETATASVLINGSPTE 121
Query: 194 EFPLKRGCK 202
EF L RG +
Sbjct: 122 EFSLHRGLR 130
>gb|AAL78659.1| reverse transcriptase [Fagus sylvatica]
Length = 168
Score = 110 bits (276), Expect = 1e-22
Identities = 60/125 (48%), Positives = 84/125 (67%)
Query: 78 ISLVESMYKILAKVLANRLRYVIGMVISDSQSTFVKGRQILDGILVANEVVDDAHRCKKE 137
ISL+ S+YK+LAKVLANRL+ V+ VIS+SQ++FV GRQILD +L+ANE +D +
Sbjct: 4 ISLIGSIYKLLAKVLANRLKLVLDSVISESQNSFVGGRQILDSVLIANECLDSRIKSGIP 63
Query: 138 LLLFKVDFEEAYDIVDWSYLDEVMYKMGFPVL*QK*IKECIGTATTSMLVNGSPTDEFPL 197
L+ K+D E+AYD V+W L ++ +MGF +K C+ T S++VNGSPT F
Sbjct: 64 GLICKLDIEKAYDHVNWDCLFFILDRMGFGRKWVCWMKACVSTVRYSVIVNGSPTGFFDS 123
Query: 198 KRGCK 202
RG +
Sbjct: 124 SRGLR 128
>gb|AAB82639.1| putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana] gi|25408936|pir||A84888 hypothetical protein
At2g45230 [imported] - Arabidopsis thaliana
Length = 1374
Score = 102 bits (255), Expect = 3e-20
Identities = 51/139 (36%), Positives = 87/139 (61%), Gaps = 3/139 (2%)
Query: 67 LRARRLNDFCLISLVESMYKILAKVLANRLRYVIGMVISDSQSTFVKGRQILDGILVANE 126
L+A ++ DF ISL +YK++ K++ANRL+ ++ +IS++Q+ FVKGR I D IL+A+E
Sbjct: 496 LKAEKMTDFRPISLCNVIYKVIGKLMANRLKKILPSLISETQAAFVKGRLISDNILIAHE 555
Query: 127 V---VDDAHRCKKELLLFKVDFEEAYDIVDWSYLDEVMYKMGFPVL*QK*IKECIGTATT 183
+ + ++C +E + K D +AYD V+W +L++ M +GF + I EC+ +
Sbjct: 556 LLHALSSNNKCSEEFIAIKTDISKAYDRVEWPFLEKAMRGLGFADHWIRLIMECVKSVRY 615
Query: 184 SMLVNGSPTDEFPLKRGCK 202
+L+NG+P E RG +
Sbjct: 616 QVLINGTPHGEIIPSRGLR 634
Score = 53.9 bits (128), Expect = 2e-05
Identities = 29/84 (34%), Positives = 43/84 (50%), Gaps = 2/84 (2%)
Query: 393 FFCEGSDNHKKVTWVDWNFVCLSKEVGGYGVRRIREFNSALLGKWCWQLLIEKESLWYRV 452
F+ + + + W W + K VGG G + I FN ALLGK W+++ EK+SL +V
Sbjct: 825 FWWKNKKEGRGLHWKAWCHLSRPKAVGGLGFKEIEAFNIALLGKQLWRMITEKDSLMAKV 884
Query: 453 LSARYNEEGGRLLD--GGRSTSAW 474
+RY + L G R + AW
Sbjct: 885 FKSRYFSKSDPLNAPLGSRPSFAW 908
>gb|AAP51996.1| putative non-LTR retroelement reverse transcriptase [Oryza sativa
(japonica cultivar-group)] gi|37530814|ref|NP_919709.1|
putative non-LTR retroelement reverse transcriptase
[Oryza sativa (japonica cultivar-group)]
gi|21326501|gb|AAM47629.1| Putative non-LTR retroelement
reverse transcriptase [Oryza sativa (japonica
cultivar-group)]
Length = 1345
Score = 101 bits (251), Expect = 1e-19
Identities = 63/171 (36%), Positives = 97/171 (55%), Gaps = 4/171 (2%)
Query: 25 LASLKIFGIFCVIMLCVL-LVNFIGMGG---LPKESIALSLLLFLRLRARRLNDFCLISL 80
+ SL +F + +I + +L V F G G L + A L+ + A ++ DF ISL
Sbjct: 749 MVSLGLFSSWDIIKIDMLQAVTFFGDLGYSRLQDLNSAHICLIPKKTDASKVEDFRPISL 808
Query: 81 VESMYKILAKVLANRLRYVIGMVISDSQSTFVKGRQILDGILVANEVVDDAHRCKKELLL 140
+ S KI+AK+LANRL +G ++S SQS F++ R I D +L ++ HR KK L
Sbjct: 809 IHSFAKIIAKLLANRLAPKLGDLVSQSQSAFIRKRAIHDNVLFVQNMIKACHRAKKPTLF 868
Query: 141 FKVDFEEAYDIVDWSYLDEVMYKMGFPVL*QK*IKECIGTATTSMLVNGSP 191
KVD +A+D V+W YL EV+ GF I +GT+++++L+NG+P
Sbjct: 869 LKVDISKAFDTVNWPYLLEVLQNFGFGQKWCNWISNLLGTSSSAILLNGTP 919
>pir||S65816 RNA-directed DNA polymerase (EC 2.7.7.49) (clone B2) - Arabidopsis
thaliana retrotransposon Ta23 (fragment)
Length = 184
Score = 98.6 bits (244), Expect = 7e-19
Identities = 51/136 (37%), Positives = 83/136 (60%), Gaps = 3/136 (2%)
Query: 70 RRLNDFCLISLVESMYKILAKVLANRLRYVIGMVISDSQSTFVKGRQILDGILVANEV-- 127
+ ++D+ ISL + YKI++K+L RL+ +G VISDSQ+ FV GR I D +LVA+E+
Sbjct: 49 KHMSDYRPISLCTASYKIISKILIKRLKQCLGDVISDSQAAFVPGRNISDNVLVAHELLH 108
Query: 128 -VDDAHRCKKELLLFKVDFEEAYDIVDWSYLDEVMYKMGFPVL*QK*IKECIGTATTSML 186
+ C+ + K D +AYD V+W++L++VM ++GF K I C+ + + +L
Sbjct: 109 SLKSRRECQSGYVAVKTDISKAYDRVEWNFLEKVMIQLGFAPRWVKWIMTCVTSVSYEVL 168
Query: 187 VNGSPTDEFPLKRGCK 202
+NGSP + RG +
Sbjct: 169 INGSPYGKIFPSRGIR 184
>gb|AAB48346.1| reverse transcriptase
Length = 199
Score = 98.6 bits (244), Expect = 7e-19
Identities = 51/136 (37%), Positives = 83/136 (60%), Gaps = 3/136 (2%)
Query: 70 RRLNDFCLISLVESMYKILAKVLANRLRYVIGMVISDSQSTFVKGRQILDGILVANEV-- 127
+ ++D+ ISL + YKI++K+L RL+ +G VISDSQ+ FV GR I D +LVA+E+
Sbjct: 55 KHMSDYRPISLCTASYKIISKILIKRLKQCLGDVISDSQAAFVPGRNISDNVLVAHELLH 114
Query: 128 -VDDAHRCKKELLLFKVDFEEAYDIVDWSYLDEVMYKMGFPVL*QK*IKECIGTATTSML 186
+ C+ + K D +AYD V+W++L++VM ++GF K I C+ + + +L
Sbjct: 115 SLKSRRECQSGYVAVKTDISKAYDRVEWNFLEKVMIQLGFAPRWVKWIMTCVTSVSYEVL 174
Query: 187 VNGSPTDEFPLKRGCK 202
+NGSP + RG +
Sbjct: 175 INGSPYGKIFPSRGIR 190
>emb|CAA12932.1| reverse transcriptase [Pinus elliottii]
Length = 194
Score = 98.2 bits (243), Expect = 9e-19
Identities = 51/125 (40%), Positives = 79/125 (62%)
Query: 78 ISLVESMYKILAKVLANRLRYVIGMVISDSQSTFVKGRQILDGILVANEVVDDAHRCKKE 137
I+L +YKI++KV+ANRL+ ++ +IS QS +V+GRQILD IL+A E++ H K
Sbjct: 63 IALCNVIYKIISKVIANRLKIILPGIISQEQSGYVEGRQILDNILLAQEMIHSLHSRKVA 122
Query: 138 LLLFKVDFEEAYDIVDWSYLDEVMYKMGFPVL*QK*IKECIGTATTSMLVNGSPTDEFPL 197
+L ++D +AYD V W+YL+ ++ GF K + E I + S+LVNG+P+ F
Sbjct: 123 GMLIQLDLSKAYDKVSWTYLEAILKAFGFSRPWIKWVLELIKSMRYSVLVNGTPSTPFSP 182
Query: 198 KRGCK 202
RG +
Sbjct: 183 SRGIR 187
>emb|CAE04866.2| OSJNBa0086O06.14 [Oryza sativa (japonica cultivar-group)]
gi|50928373|ref|XP_473714.1| OSJNBa0086O06.14 [Oryza
sativa (japonica cultivar-group)]
Length = 1205
Score = 97.8 bits (242), Expect = 1e-18
Identities = 52/134 (38%), Positives = 78/134 (57%), Gaps = 3/134 (2%)
Query: 72 LNDFCLISLVESMYKILAKVLANRLRYVIGMVISDSQSTFVKGRQILDGILVANEVVDDA 131
+ DF +SL +YK++AK L NRLR ++ +IS++QS FV GR I D LVA E
Sbjct: 489 MKDFRPVSLCNVIYKVVAKCLVNRLRPLLQEIISETQSAFVPGRMITDNALVAFECFHSI 548
Query: 132 HRCKKE---LLLFKVDFEEAYDIVDWSYLDEVMYKMGFPVL*QK*IKECIGTATTSMLVN 188
H+C +E K+D +AYD VDW +LD + K+GF + +K I C+ + S+ +N
Sbjct: 549 HKCTRESQDFCALKLDLSKAYDRVDWGFLDGALQKLGFGNIWRKWIMSCVTSVRYSVRLN 608
Query: 189 GSPTDEFPLKRGCK 202
G+ + F RG +
Sbjct: 609 GNMLEPFYPTRGLR 622
Score = 54.3 bits (129), Expect = 1e-05
Identities = 27/60 (45%), Positives = 35/60 (58%)
Query: 402 KKVTWVDWNFVCLSKEVGGYGVRRIREFNSALLGKWCWQLLIEKESLWYRVLSARYNEEG 461
KKV W+ W + K +GG G R IR FN ALL + W+L+ +SL RVL A+Y G
Sbjct: 822 KKVHWIAWEKLTSPKLLGGLGFRDIRCFNQALLARQAWRLIESPDSLCARVLKAKYYPNG 881
>gb|AAD21778.1| putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana] gi|25410938|pir||G84429 hypothetical protein
At2g01840 [imported] - Arabidopsis thaliana
Length = 1715
Score = 97.1 bits (240), Expect = 2e-18
Identities = 50/136 (36%), Positives = 83/136 (60%), Gaps = 3/136 (2%)
Query: 70 RRLNDFCLISLVESMYKILAKVLANRLRYVIGMVISDSQSTFVKGRQILDGILVANEV-- 127
+ ++D+ ISL + YKI++K+L RL+ +G VISDSQ+ FV G+ I D +LVA+E+
Sbjct: 861 KHMSDYRPISLCTASYKIISKILIKRLKQCLGDVISDSQAAFVPGQNISDNVLVAHELLH 920
Query: 128 -VDDAHRCKKELLLFKVDFEEAYDIVDWSYLDEVMYKMGFPVL*QK*IKECIGTATTSML 186
+ C+ + K D +AYD V+W++L++VM ++GF K I C+ + + +L
Sbjct: 921 SLKSRRECQSGYVAVKTDISKAYDRVEWNFLEKVMIQLGFAPRWVKWIMTCVTSVSYEVL 980
Query: 187 VNGSPTDEFPLKRGCK 202
+NGSP + RG +
Sbjct: 981 INGSPYGKIFPSRGIR 996
>pir||H86435 protein F17F8.5 [imported] - Arabidopsis thaliana
gi|9755374|gb|AAF98181.1| F17F8.5 [Arabidopsis thaliana]
Length = 872
Score = 94.4 bits (233), Expect = 1e-17
Identities = 57/161 (35%), Positives = 91/161 (56%), Gaps = 4/161 (2%)
Query: 45 NFIGMGGLPK--ESIALSLLLFLRLRARRLNDFCLISLVESMYKILAKVLANRLRYVIGM 102
+F G LPK SI L+L+ +L A+ + D+ IS +YK+++K++ANRL+ ++
Sbjct: 145 SFFQKGFLPKGINSIILALIP-KKLAAKEMRDYRPISCCNVLYKVISKIIANRLKLLLPR 203
Query: 103 VISDSQSTFVKGRQILDGILVANEVVDDAHRCK-KELLLFKVDFEEAYDIVDWSYLDEVM 161
I+++QS FVK R +++ +L+A E+V D H+ K+D +A+D V WS+L +
Sbjct: 204 FIAENQSAFVKDRLLIENLLLATELVKDYHKDSISARCAIKIDISKAFDSVQWSFLTNTL 263
Query: 162 YKMGFPVL*QK*IKECIGTATTSMLVNGSPTDEFPLKRGCK 202
M F I CI TA+ S+ VNG F KRG +
Sbjct: 264 VAMNFSPTFIHWINLCITTASFSVQVNGDLVGYFQSKRGLR 304
Score = 42.7 bits (99), Expect = 0.042
Identities = 25/92 (27%), Positives = 40/92 (43%), Gaps = 2/92 (2%)
Query: 393 FFCEGSDNHKKVTWVDWNFVCLSKEVGGYGVRRIREFNSALLGKWCWQLLIEKESLWYR- 451
F GS+ + W+ VC K GG G+R ++E N K W+++ SLW +
Sbjct: 493 FLWSGSEMSSHKAKISWDIVCKPKAEGGLGLRNLKEANDVSCLKLVWRIISNSNSLWTKW 552
Query: 452 VLSARYNEEGGRLLDGGRSTSAW-WRVIATLQ 482
V ++ L S +W WR I ++
Sbjct: 553 VAEYLIRKKSIWSLKQSTSMGSWIWRKILKIR 584
>ref|XP_470963.1| OSJNBa0094O15.6 [Oryza sativa (japonica cultivar-group)]
gi|21741576|emb|CAD39338.1| OSJNBa0094O15.6 [Oryza
sativa (japonica cultivar-group)]
Length = 699
Score = 94.4 bits (233), Expect = 1e-17
Identities = 50/123 (40%), Positives = 76/123 (61%)
Query: 69 ARRLNDFCLISLVESMYKILAKVLANRLRYVIGMVISDSQSTFVKGRQILDGILVANEVV 128
A R+ DF LISL+ S KILAK+LANRL + ++S +QS F++ R I D L ++
Sbjct: 75 ATRVEDFRLISLMHSFAKILAKMLANRLAPKLHELVSPNQSAFIRKRAIHDNFLYVQNMI 134
Query: 129 DDAHRCKKELLLFKVDFEEAYDIVDWSYLDEVMYKMGFPVL*QK*IKECIGTATTSMLVN 188
+ +R KK +L KVD +A+DIV+W YL EV+ GF + I +G +++ +++N
Sbjct: 135 KNFNRKKKPMLFIKVDISKAFDIVNWPYLLEVLQNFGFGQRWRNWISNILGNSSSRIILN 194
Query: 189 GSP 191
G P
Sbjct: 195 GIP 197
>emb|CAB72467.1| putative protein [Arabidopsis thaliana] gi|11358077|pir||T47440
hypothetical protein T18B22.50 - Arabidopsis thaliana
Length = 762
Score = 94.0 bits (232), Expect = 2e-17
Identities = 55/160 (34%), Positives = 90/160 (55%), Gaps = 2/160 (1%)
Query: 45 NFIGMGGLPKESIALSLLLF-LRLRARRLNDFCLISLVESMYKILAKVLANRLRYVIGMV 103
+F +G LPK + L L +L ++ + D+ IS MYK+++K+LANRL+ ++
Sbjct: 51 SFFALGFLPKGVNSTILALIPKKLESKEMKDYRPISCCNVMYKVISKILANRLKLLLPQF 110
Query: 104 ISDSQSTFVKGRQILDGILVANEVVDDAHRCK-KELLLFKVDFEEAYDIVDWSYLDEVMY 162
I+ +QS+FVK R +++ +L+A ++V D H+ E K+D +A D V WS+L +
Sbjct: 111 IAGNQSSFVKDRLLIENVLLATDLVKDYHKDSISERCAIKIDISKASDSVQWSFLINTLT 170
Query: 163 KMGFPVL*QK*IKECIGTATTSMLVNGSPTDEFPLKRGCK 202
M FP + I+ CI T + S+ VNG F RG +
Sbjct: 171 AMHFPEMFIHWIRLCITTPSFSVQVNGELAGFFQSSRGLR 210
Score = 46.6 bits (109), Expect = 0.003
Identities = 29/106 (27%), Positives = 49/106 (45%), Gaps = 10/106 (9%)
Query: 346 GKLNIYLLVVIWFC*NLSCHLSLFMLFLSSRLLQVLSPLLNLF*NVFFFCEGSDNHKKVT 405
G+ N+ + +IW SC+ L L +Q + L + F G++ + K
Sbjct: 362 GRFNL-ISSIIWS----SCNFWLSAFQLPRACIQEIEKLCSSF-----LWSGTNLNSKKA 411
Query: 406 WVDWNFVCLSKEVGGYGVRRIREFNSALLGKWCWQLLIEKESLWYR 451
+ WN VC K GG G+R ++E N K W+++ +SLW +
Sbjct: 412 KISWNQVCKPKSEGGLGLRSLKEANDVCCLKLVWRIISHGDSLWVK 457
>gb|AAD24601.1| putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana] gi|25411729|pir||H84542 hypothetical protein
At2g16680 [imported] - Arabidopsis thaliana
Length = 1319
Score = 92.4 bits (228), Expect = 5e-17
Identities = 55/164 (33%), Positives = 90/164 (54%), Gaps = 4/164 (2%)
Query: 43 LVNFIGMGGLPKESIALSLLLFLRL-RARRLNDFCLISLVESMYKILAKVLANRLRYVIG 101
++ F G LP+E L L + +R++D ISL +YKI++K+L+ +L+ +
Sbjct: 423 ILGFFETGVLPQEWNHTHLYLIPKFTNPQRMSDIRPISLCSVLYKIISKILSFKLKKHLP 482
Query: 102 MVISDSQSTFVKGRQILDGILVANEVVDDAH---RCKKELLLFKVDFEEAYDIVDWSYLD 158
++S SQS F R I D IL+A+E+V + KE ++FK D +AYD V+WS+L
Sbjct: 483 SIVSPSQSAFFAERLISDNILIAHEIVHSLRTNDKISKEFMVFKTDMSKAYDRVEWSFLQ 542
Query: 159 EVMYKMGFPVL*QK*IKECIGTATTSMLVNGSPTDEFPLKRGCK 202
E++ +GF I C+ + T S+L+NG +RG +
Sbjct: 543 EILVALGFNDKWISWIMGCVTSVTYSVLINGQHFGHITPERGIR 586
Score = 46.6 bits (109), Expect = 0.003
Identities = 23/88 (26%), Positives = 44/88 (49%), Gaps = 2/88 (2%)
Query: 393 FFCEGSDNHKKVTWVDWNFVCLSKEVGGYGVRRIREFNSALLGKWCWQLLIEKESLWYRV 452
F+ + K+ W+ + L K +GG+G + ++ FN ALL K W+L + +S+ ++
Sbjct: 777 FWWNSMEFSNKIHWIGGKKLTLPKSLGGFGFKDLQCFNQALLAKQAWRLFSDSKSIVSQI 836
Query: 453 LSARY--NEEGGRLLDGGRSTSAWWRVI 478
+RY N + G R + W ++
Sbjct: 837 FKSRYFMNTDFLNARQGTRPSYTWRSIL 864
>pir||S65817 RNA-directed DNA polymerase (EC 2.7.7.49) (clone B3) - Arabidopsis
thaliana retrotransposon Ta25 (fragment)
Length = 184
Score = 92.0 bits (227), Expect = 6e-17
Identities = 52/138 (37%), Positives = 80/138 (57%), Gaps = 3/138 (2%)
Query: 68 RARRLNDFCLISLVESMYKILAKVLANRLRYVIGMVISDSQSTFVKGRQILDGILVANE- 126
R RR+ +F ISL YK+++KVL++RL+ ++ +IS++QS FV GR I D ILVA E
Sbjct: 47 RPRRMAEFRPISLCNVSYKVISKVLSSRLKKILPDLISETQSAFVAGRLITDNILVAQEN 106
Query: 127 --VVDDAHRCKKELLLFKVDFEEAYDIVDWSYLDEVMYKMGFPVL*QK*IKECIGTATTS 184
+ C+K+ + K D +AY V+WS+L +M KMGF I C+ + +
Sbjct: 107 FHALRTNPACRKKFMAIKTDMSKAYGRVEWSFLRALMLKMGFAQKWVDLIMFCVSSVSYK 166
Query: 185 MLVNGSPTDEFPLKRGCK 202
+L+NG+P RG +
Sbjct: 167 VLLNGAPRGFIKPTRGIR 184
>gb|AAB48348.1| reverse transcriptase
Length = 199
Score = 92.0 bits (227), Expect = 6e-17
Identities = 52/138 (37%), Positives = 80/138 (57%), Gaps = 3/138 (2%)
Query: 68 RARRLNDFCLISLVESMYKILAKVLANRLRYVIGMVISDSQSTFVKGRQILDGILVANE- 126
R RR+ +F ISL YK+++KVL++RL+ ++ +IS++QS FV GR I D ILVA E
Sbjct: 53 RPRRMAEFRPISLCNVSYKVISKVLSSRLKKILPDLISETQSAFVAGRLITDNILVAQEN 112
Query: 127 --VVDDAHRCKKELLLFKVDFEEAYDIVDWSYLDEVMYKMGFPVL*QK*IKECIGTATTS 184
+ C+K+ + K D +AY V+WS+L +M KMGF I C+ + +
Sbjct: 113 FHALRTNPACRKKFMAIKTDMSKAYGRVEWSFLRALMLKMGFAQKWVDLIMFCVSSVSYK 172
Query: 185 MLVNGSPTDEFPLKRGCK 202
+L+NG+P RG +
Sbjct: 173 VLLNGAPRGFIKPTRGIR 190
>gb|AAD17398.1| putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana] gi|25411644|pir||C84530 hypothetical protein
At2g15540 [imported] - Arabidopsis thaliana
Length = 1225
Score = 91.7 bits (226), Expect = 8e-17
Identities = 53/138 (38%), Positives = 80/138 (57%), Gaps = 3/138 (2%)
Query: 68 RARRLNDFCLISLVESMYKILAKVLANRLRYVIGMVISDSQSTFVKGRQILDGILVANE- 126
R R++ +F ISL YK+++KVL++RL+ ++ +IS++QS FV R I D IL+A E
Sbjct: 444 RPRKMAEFRPISLCNVSYKVISKVLSSRLKRLLPELISETQSAFVAERLITDNILIAQEN 503
Query: 127 --VVDDAHRCKKELLLFKVDFEEAYDIVDWSYLDEVMYKMGFPVL*QK*IKECIGTATTS 184
+ CKK+ + K D +AYD V+WS+L +M KMGF I CI + +
Sbjct: 504 FHALRTNPACKKKYMAIKTDMSKAYDRVEWSFLRALMLKMGFAQKWVDWIIFCISSVSYK 563
Query: 185 MLVNGSPTDEFPLKRGCK 202
+L+NGSP RG +
Sbjct: 564 ILLNGSPKGFIKPSRGIR 581
Score = 47.8 bits (112), Expect = 0.001
Identities = 23/65 (35%), Positives = 33/65 (50%)
Query: 393 FFCEGSDNHKKVTWVDWNFVCLSKEVGGYGVRRIREFNSALLGKWCWQLLIEKESLWYRV 452
F+ + + + W+ W+ +C GG G R + EFN LL K W+L+ SL RV
Sbjct: 705 FWWKTREESNGIHWIAWDKLCTPFSDGGLGFRTLEEFNLVLLAKQLWRLIRFPNSLLSRV 764
Query: 453 LSARY 457
L RY
Sbjct: 765 LRGRY 769
>emb|CAB10337.1| reverse transcriptase like protein [Arabidopsis thaliana]
gi|7268307|emb|CAB78601.1| reverse transcriptase like
protein [Arabidopsis thaliana] gi|7485171|pir||G71420
hypothetical protein - Arabidopsis thaliana
Length = 929
Score = 91.3 bits (225), Expect = 1e-16
Identities = 55/163 (33%), Positives = 87/163 (52%), Gaps = 3/163 (1%)
Query: 43 LVNFIGMGGLPKESIALSLLLFLRL-RARRLNDFCLISLVESMYKILAKVLANRLRYVIG 101
++ F G LPK + + L+L ++ + R+ F +SL ++KI+ K++ RL+ VI
Sbjct: 287 VMEFFESGVLPKSTNDVLLVLLAKVAKPERITQFRPVSLCNVLFKIITKMMVIRLKNVIS 346
Query: 102 MVISDSQSTFVKGRQILDGILVANEVVDDAHRCK--KELLLFKVDFEEAYDIVDWSYLDE 159
+I +Q++F+ GR D I+V E V R K K +L K+D E+AYD + W +L E
Sbjct: 347 KLIGPAQASFIPGRLSFDNIVVVQEAVHSMRRKKGRKGWMLLKLDLEKAYDRIRWDFLAE 406
Query: 160 VMYKMGFPVL*QK*IKECIGTATTSMLVNGSPTDEFPLKRGCK 202
+ G K I EC+ S+L NG TD F +RG +
Sbjct: 407 TLEAAGLSEGWIKRIMECVAGPEMSLLWNGEKTDSFTPERGLR 449
Score = 52.4 bits (124), Expect = 5e-05
Identities = 32/86 (37%), Positives = 43/86 (49%), Gaps = 3/86 (3%)
Query: 402 KKVTWVDWNFVCLSKEVGGYGVRRIREFNSALLGKWCWQLLIEKESLWYRVLSARY---N 458
+K + W VC K GG G+R ++ N ALL K W+LL +K SLW RVL +Y +
Sbjct: 632 RKQHLLSWKKVCRPKAAGGLGLRASKDMNRALLAKVGWRLLNDKVSLWARVLRRKYKVTD 691
Query: 459 EEGGRLLDGGRSTSAWWRVIATLQRE 484
L + S+ WR I RE
Sbjct: 692 VHDSSWLVPKATWSSTWRSIGVGLRE 717
>gb|AAU44186.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
Length = 1023
Score = 91.3 bits (225), Expect = 1e-16
Identities = 50/134 (37%), Positives = 79/134 (58%)
Query: 69 ARRLNDFCLISLVESMYKILAKVLANRLRYVIGMVISDSQSTFVKGRQILDGILVANEVV 128
A ++D+ ISL+ S+ KI AK+LA RLR + +IS +QS F+KGR I D L ++
Sbjct: 344 AESVSDYRPISLIHSIAKIFAKMLALRLRPHMHELISVNQSAFIKGRSIHDNYLFVRNMI 403
Query: 129 DDAHRCKKELLLFKVDFEEAYDIVDWSYLDEVMYKMGFPVL*QK*IKECIGTATTSMLVN 188
HR ++ +LLFK+D +A+D V W YL ++ + GFP + + ++T+ +L+N
Sbjct: 404 RRYHRLRRAMLLFKLDITKAFDSVRWDYLLALLQRKGFPTRWIDWLGALLSSSTSQVLLN 463
Query: 189 GSPTDEFPLKRGCK 202
GSP RG +
Sbjct: 464 GSPGQRIKHGRGLR 477
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.350 0.157 0.564
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,118,297,470
Number of Sequences: 2540612
Number of extensions: 43238572
Number of successful extensions: 146338
Number of sequences better than 10.0: 666
Number of HSP's better than 10.0 without gapping: 419
Number of HSP's successfully gapped in prelim test: 248
Number of HSP's that attempted gapping in prelim test: 145118
Number of HSP's gapped (non-prelim): 1066
length of query: 739
length of database: 863,360,394
effective HSP length: 136
effective length of query: 603
effective length of database: 517,837,162
effective search space: 312255808686
effective search space used: 312255808686
T: 11
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.9 bits)
S2: 79 (35.0 bits)
Medicago: description of AC124963.21


