

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124217.5 + phase: 0 /pseudo
(1307 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
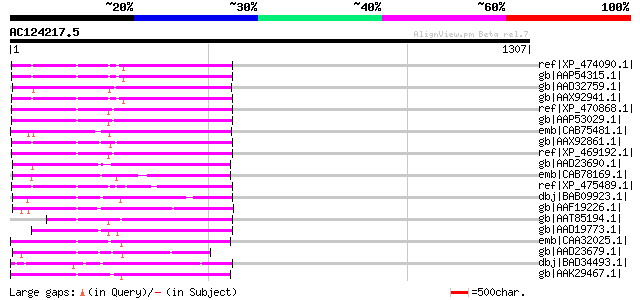
ref|XP_474090.1| OSJNBa0033G05.13 [Oryza sativa (japonica cultiv... 404 e-111
gb|AAP54315.1| putative polyprotein [Oryza sativa (japonica cult... 390 e-106
gb|AAD32759.1| putative retroelement pol polyprotein [Arabidopsi... 388 e-106
gb|AAX92941.1| retrotransposon protein, putative, Ty1-copia sub-... 387 e-105
ref|XP_470868.1| Putative retroelement pol polyprotein [Oryza sa... 385 e-105
gb|AAP53029.1| putative retrotransposon-related protein [Oryza s... 378 e-103
emb|CAB75481.1| copia-like polyprotein [Arabidopsis thaliana] gi... 377 e-102
gb|AAX92861.1| retrotransposon protein, putative, Ty1-copia sub-... 376 e-102
ref|XP_469192.1| putative polyprotein [Oryza sativa (japonica cu... 374 e-101
gb|AAD23690.1| putative retroelement pol polyprotein [Arabidopsi... 372 e-101
emb|CAB78169.1| putative retrotransposon [Arabidopsis thaliana] ... 369 e-100
ref|XP_475489.1| putative polyprotein [Oryza sativa (japonica cu... 358 5e-97
dbj|BAB09923.1| copia-like retrotransposable element [Arabidopsi... 353 2e-95
gb|AAF19226.1| Highly similar to Ta1-3 polyprotein [Arabidopsis ... 352 5e-95
gb|AAT85194.1| putative polyprotein [Oryza sativa (japonica cult... 351 8e-95
gb|AAD19773.1| putative retroelement pol polyprotein [Arabidopsi... 350 2e-94
emb|CAA32025.1| unnamed protein product [Nicotiana tabacum] gi|1... 349 3e-94
gb|AAD23679.1| putative retroelement pol polyprotein [Arabidopsi... 345 4e-93
dbj|BAD34493.1| Gag-Pol [Ipomoea batatas] 340 1e-91
gb|AAK29467.1| polyprotein-like [Lycopersicon chilense] 338 7e-91
>ref|XP_474090.1| OSJNBa0033G05.13 [Oryza sativa (japonica cultivar-group)]
gi|38344889|emb|CAD41912.2| OSJNBa0033G05.13 [Oryza
sativa (japonica cultivar-group)]
Length = 1181
Score = 404 bits (1039), Expect = e-111
Identities = 238/583 (40%), Positives = 341/583 (57%), Gaps = 36/583 (6%)
Query: 5 KWDIEKFTGSNDFGLWKVKMQAVLTQQKCVEALKGEAAMPATLTQEEKREMIDKAKSAIV 64
K+D+ F LW+VKM+AVL QQ+ +AL G + +EK+ KA S I
Sbjct: 5 KYDLPLLDRDTRFSLWQVKMRAVLAQQELDDALSGFDKRTQDWSNDEKKRD-RKAMSYIH 63
Query: 65 LCLGDKVLRDVAREATAASM*AKLESLYMTKSLAHRQLLKQQLYSFKMVESISISEQLTE 124
L L + +L++V +E TAA + KLE + MTK L + LKQ+L+ K+ + S+ + L+
Sbjct: 64 LHLSNNILQEVLKEETAAGLWLKLEQICMTKDLTSKMHLKQKLFLHKLQDDGSVMDHLSA 123
Query: 125 FNKILVDLANIEVNTEDEDKALLLLCSLPKSFEHFKDTILYGKEGTTTLEEVQAALRTKE 184
F +I+ DL ++EV +++D AL+LLCSLP S+ +F+DTILY ++ T TL+EV AL KE
Sbjct: 124 FKEIVADLESMEVKYDEKDLALILLCSLPSSYANFRDTILYSRD-TLTLKEVYDALHAKE 182
Query: 185 LTKFKDLKVDEGS----EGLNVARGRNEHRGKGKGKSRSKSRSKGFDKSK--YK-CFLCH 237
K K + EGS EGL V + E K + +S S +G KS+ YK C C
Sbjct: 183 --KMKKMVPSEGSNSQAEGLVVRGSQQEKNTNNKSRDKSSSSYRGRSKSRGRYKSCKYCK 240
Query: 238 KQGHFKKDC---PDKGGDGSPSVQVAEASNEEGYESTGALVVTSWKSEKS---------- 284
+ GH C DK D + + EE + A VVT KS+
Sbjct: 241 RDGHDISKCWKLQDK--DKRTGKYIPKGKKEEEGK---AAVVTDEKSDAELLVAYAGCAQ 295
Query: 285 ----WVLDSGCSYHMCPRKEYFETLTLKEGGVVRLGNNKACKVQGMGNVRLKMFDGREFL 340
W+LD+ C+YHMCP +++F T + +GG V +G++ C+V G+G V++KMFDG
Sbjct: 296 TSDQWILDTACTYHMCPNRDWFATYEVVQGGTVLMGDDTPCEVAGIGTVQIKMFDGCIRT 355
Query: 341 LRDVRFVPELKRNLISLSMFDGLGYCTRIEHGVCKISHGALITVKGS-KMNGLYILDGSI 399
L DVR +P LKR+LISL D GY G+ K++ G+L+ +K S K LY L G+
Sbjct: 356 LSDVRHIPNLKRSLISLCTLDRKGYKYSGGDGILKVTKGSLVVMKASIKSANLYHLQGTT 415
Query: 400 VIGNASVASVVPHNN--SELWHLRLGHVSERGLVELAKQGLLGKDKLDKLEFCEHCILGK 457
++GN + S N+ + LWH+RLGH+SE GL EL+K+GLL + KL+FCEHCI GK
Sbjct: 416 ILGNVATVSDSLSNSDATNLWHMRLGHMSEIGLAELSKRGLLDGQSISKLKFCEHCIFGK 475
Query: 458 QHRVKFGSGMHHSSRLFEYVHSDLLGPSKTPTHGGGSYFLSIIDDYSRRVWVFVLKKKSD 517
RVKF + H + + +YVHSDL GP++ + GG Y ++I+DDYSR+VW + LK K
Sbjct: 476 HKRVKFNTSTHTTEGILDYVHSDLWGPARKTSFGGARYMMTIVDDYSRKVWPYFLKHKYQ 535
Query: 518 TF*KFKE*HTLIENQMGTKLKGLRTDNGLEFVSEQFNDFLQVE 560
F FKE T++E Q K+K LRTDNG+EF S+ F + + E
Sbjct: 536 AFNVFKEWKTMVERQTERKVKILRTDNGMEFCSKIFKSYCKSE 578
>gb|AAP54315.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
gi|37535452|ref|NP_922028.1| putative polyprotein [Oryza
sativa (japonica cultivar-group)]
gi|22094359|gb|AAM91886.1| putative polyprotein [Oryza
sativa (japonica cultivar-group)]
Length = 1280
Score = 390 bits (1003), Expect = e-106
Identities = 235/584 (40%), Positives = 336/584 (57%), Gaps = 38/584 (6%)
Query: 5 KWDIEKFTGSNDFGLWKVKMQAVLTQQKCVEALKGEAAMPATLTQEEKREMIDKAKSAIV 64
K+D+ F LW+VKM+AVL QQ +AL G + +EK++ KA S I
Sbjct: 40 KYDLPLLDRDTRFSLWQVKMRAVLAQQDLDDALSGFDKRTQDWSNDEKKKD-RKAMSYIH 98
Query: 65 LCLGDKVLRDVAREATAASM*AKLESLYMTKSLAHRQLLKQQLYSFKMVESISISEQLTE 124
L L + +L++V +E TAA + KLE + MTK L + LKQ+L+ K+ + S+ + L+
Sbjct: 99 LHLSNNILQEVLKEETAAGLWLKLEQICMTKDLTSKMHLKQKLFLHKLQDDGSVMDHLST 158
Query: 125 FNKILVDLANIEVNTEDEDKALLLLCSLPKSFEHFKDTILYGKEGTTTLEEVQAALRTKE 184
F +I+ DL +IEV ++ED L+LLCSLP S+ +F+DTILY + T L+EV AL KE
Sbjct: 159 FKEIVADLESIEVKYDEEDLGLILLCSLPSSYANFRDTILYSHD-TLILKEVYDALHAKE 217
Query: 185 LTKFKDLKVDEGS----EGLNVARGRNEHRG---KGKGKSRSKSRSKGFDKSKYK-CFLC 236
K K + EGS EGL V RGR + + + + KS S R + + +YK C C
Sbjct: 218 --KMKKMVPSEGSNSQAEGL-VVRGRQQEKNTKNQSRDKSSSSYRGRSKSRGRYKSCKYC 274
Query: 237 HKQGHFKKDC---PDKGGDGSPSVQVAEASNEEGYESTGALVVTSWKSEKS--------- 284
+ GH +C DK D + + EE + A VVT KS+
Sbjct: 275 KRDGHDISECWKLQDK--DKRTGKYIPKGKKEEEGK---AAVVTDEKSDTELLVAYAGCA 329
Query: 285 -----WVLDSGCSYHMCPRKEYFETLTLKEGGVVRLGNNKACKVQGMGNVRLKMFDGREF 339
W+LD+ +YHMCP +++F T +GG V +G++ C+V G+G V++KMFDG
Sbjct: 330 QTSDQWILDTAWTYHMCPNRDWFATYEALQGGTVLMGDDTPCEVAGIGTVQIKMFDGYIR 389
Query: 340 LLRDVRFVPELKRNLISLSMFDGLGYCTRIEHGVCKISHGALITVKGS-KMNGLYILDGS 398
L DVR +P LKR+LISL D GY G+ K++ G+L+ +K K LY L G+
Sbjct: 390 TLSDVRHIPNLKRSLISLCTLDRKGYKYSGGDGILKVTKGSLVVMKADIKSANLYHLRGT 449
Query: 399 IVIGNASVASVVPHNN--SELWHLRLGHVSERGLVELAKQGLLGKDKLDKLEFCEHCILG 456
++GN + S N+ + LWH+RLGH+SE GL EL+K+ LL + KL+FCEHCI G
Sbjct: 450 TILGNVAAVSDSLSNSDATNLWHMRLGHMSEIGLAELSKRELLDGQSIGKLKFCEHCIFG 509
Query: 457 KQHRVKFGSGMHHSSRLFEYVHSDLLGPSKTPTHGGGSYFLSIIDDYSRRVWVFVLKKKS 516
K RVKF + H + + +YVHSDL GP+ + GG Y ++I+DDYSR+VW + LK K
Sbjct: 510 KHKRVKFNTSTHTTEGILDYVHSDLWGPACKTSFGGARYMMTIVDDYSRKVWPYFLKHKY 569
Query: 517 DTF*KFKE*HTLIENQMGTKLKGLRTDNGLEFVSEQFNDFLQVE 560
F FKE T++E Q K+K LRTDNG+EF S+ F + + E
Sbjct: 570 QAFDVFKEWKTMVERQTEKKVKILRTDNGMEFCSKIFKSYCKSE 613
>gb|AAD32759.1| putative retroelement pol polyprotein [Arabidopsis thaliana]
gi|25301696|pir||F84486 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana
Length = 1356
Score = 388 bits (996), Expect = e-106
Identities = 227/584 (38%), Positives = 342/584 (57%), Gaps = 40/584 (6%)
Query: 7 DIEKFTGSNDFGLWKVKMQAVLTQQKCVEALK-----GEAAMPATLTQEEKREMIDK--- 58
++EKF G D+ +WK K+ A + ALK GE + E+ E ++K
Sbjct: 7 EVEKFDGRGDYTMWKEKLLAHMDILGLNTALKESESTGEKKSVLDESDEDYEEKLEKFEA 66
Query: 59 -------AKSAIVLCLGDKVLRDVAREATAASM*AKLESLYMTKSLAHRQLLKQQLYSFK 111
A+SAIVL + D+VLR + +E+TAA+M L+ LYM+K+L +R KQ+LYSFK
Sbjct: 67 LEEKKKKARSAIVLSVTDRVLRKIKKESTAAAMLLALDKLYMSKALPNRIYPKQKLYSFK 126
Query: 112 MVESISISEQLTEFNKILVDLANIEVNTEDEDKALLLLCSLPKSFEHFKDTILYGK-EGT 170
M E++S+ + EF +I+ DL N+ V DED+A+LLL +LPK+F+ KDT+ Y +
Sbjct: 127 MSENLSVEGNIDEFLQIITDLENMNVIISDEDQAILLLTALPKAFDQLKDTLKYSSGKSI 186
Query: 171 TTLEEVQAALRTKEL---TKFKDLKVDEGSEGLNVARGRNEHRGKGKGKSRSKSRSKGFD 227
TL+EV AA+ +KEL + K +KV +EGL V + +NE++GKG+ K + K + KG
Sbjct: 187 LTLDEVAAAIYSKELELGSVKKSIKVQ--AEGLYV-KDKNENKGKGEQKGKGKGK-KGKS 242
Query: 228 KSKYKCFLCHKQGHFKKDCPD------------KGGDGSPSVQVAEASNEEGYESTGALV 275
K K C+ C ++GHF+ CP+ KG +AEA+ GY + AL
Sbjct: 243 KKKPGCWTCGEEGHFRSSCPNQNKPQFKQSQVVKGESSGGKGNLAEAA---GYYVSEALS 299
Query: 276 VTSWKSEKSWVLDSGCSYHMCPRKEYFETLTLKEGGVVRLGNNKACKVQGMGNVRLKMFD 335
T E W+LD+GCSYHM ++E+F GG VR+GN +V+G+G +R+K D
Sbjct: 300 STEVHLEDEWILDTGCSYHMTYKREWFHEFNEDAGGSVRMGNKTVSRVRGVGTIRVKNSD 359
Query: 336 GREFLLRDVRFVPELKRNLISLSMFDGLGYCTRIEHGVCKISHGALITVKGSKMNGLYIL 395
G +L +VR++P++ RNL+SL F+ GY E G+ +I G + + G + + LY+L
Sbjct: 360 GLTIVLTNVRYIPDMDRNLLSLGTFEKAGYKFESEDGILRIKAGNQVLLTGRRYDTLYLL 419
Query: 396 DGSIVIGNASVASVVPHNNSELWHLRLGHVSERGLVELAKQGLLGKDKLDKLEFCEHCIL 455
+ + + S+A V +++ LWH RL H+S++ + L ++G L K K+ L+ CE CI
Sbjct: 420 NWK-PVASESLAVVKRADDTVLWHQRLCHMSQKNMEILVRKGFLDKKKVSSLDVCEDCIY 478
Query: 456 GKQHRVKFGSGMHHSSRLFEYVHSDLLGPSKTP-THGGGSYFLSIIDDYSRRVWVFVLKK 514
GK R F H + EY+HSDL G P + G YF+SIIDD++R+VWV+ +K
Sbjct: 479 GKAKRKSFSLAHHDTKEKLEYIHSDLWGAPFVPLSLGKCQYFMSIIDDFTRKVWVYFMKT 538
Query: 515 KSDTF*KFKE*HTLIENQMGTKLKGLRTDNGLEFVSEQFNDFLQ 558
K + F KF E L+ENQ ++K LRTDNGLEF ++ F+ F +
Sbjct: 539 KDEAFEKFVEWVNLVENQTDRRVKTLRTDNGLEFCNKLFDGFCE 582
>gb|AAX92941.1| retrotransposon protein, putative, Ty1-copia sub-class [Oryza
sativa (japonica cultivar-group)]
Length = 2340
Score = 387 bits (994), Expect = e-105
Identities = 234/583 (40%), Positives = 335/583 (57%), Gaps = 36/583 (6%)
Query: 5 KWDIEKFTGSNDFGLWKVKMQAVLTQQKCVEALKGEAAMPATLTQEEKREMIDKAKSAIV 64
K+D+ F LW+VKM+AVL QQ +AL G + +EK+ KA S I
Sbjct: 212 KYDLPLLYRDTRFSLWQVKMRAVLAQQDLDDALSGFDKRTHDWSNDEKKRD-RKAMSYIH 270
Query: 65 LCLGDKVLRDVAREATAASM*AKLESLYMTKSLAHRQLLKQQLYSFKMVESISISEQLTE 124
L L + +L++V +E AA + KLE + MTK L + LKQ L+ K+ + S+ + L+
Sbjct: 271 LHLSNNILQEVLKEEIAAGLWLKLEQICMTKDLTSKMHLKQTLFLHKLQDDGSVMDHLSA 330
Query: 125 FNKILVDLANIEVNTEDEDKALLLLCSLPKSFEHFKDTILYGKEGTTTLEEVQAALRTKE 184
F +I+ DL ++EV ++ED L+LLCSLP S+ +F+DTILY ++ T TL+EV AL KE
Sbjct: 331 FKEIIADLESMEVKYDEEDLGLILLCSLPSSYANFRDTILYSRD-TLTLKEVYDALHVKE 389
Query: 185 LTKFKDLKVDEGS----EGLNVARGRNEHRGKGKGKSRSKSRSKGFDKSK--YK-CFLCH 237
K K + EGS EGL V + E K + + +S S +G KS+ YK C C
Sbjct: 390 --KMKKMVPSEGSNSQAEGLIVWGRQQEKNTKNQSRDKSSSSYRGRSKSRGRYKSCKYCK 447
Query: 238 KQGHFKKDC---PDKGGDGSPSVQVAEASNEEGYESTGALVVTSWKSEKS---------- 284
+ GH +C DK D V + EE + A VVT KS+
Sbjct: 448 RDGHDIFECWKLHDK--DKRTGKYVPKGKKEEEGK---AAVVTDEKSDAELLVAYAGCAQ 502
Query: 285 ----WVLDSGCSYHMCPRKEYFETLTLKEGGVVRLGNNKACKVQGMGNVRLKMFDGREFL 340
W+L++ C YHMCP +++F T + G V +G++ C+V G+G V++KMFDG
Sbjct: 503 TSDQWILNTACIYHMCPNRDWFATYEAVQVGTVLMGDDTPCEVAGIGTVQIKMFDGCIRT 562
Query: 341 LRDVRFVPELKRNLISLSMFDGLGYCTRIEHGVCKISHGALITVKGS-KMNGLYILDGSI 399
L DVR +P LKR+LISL D GY G+ K++ G+L+ +K K LY L G+
Sbjct: 563 LSDVRHIPNLKRSLISLCTLDRKGYKYSGGDGILKVTKGSLVVMKADIKSANLYHLRGTT 622
Query: 400 VIGNASVASVVPHNN--SELWHLRLGHVSERGLVELAKQGLLGKDKLDKLEFCEHCILGK 457
++GN + S N+ + LWH+RLGH++E GL EL+K+GLL + KL+FCEHCI GK
Sbjct: 623 ILGNVAAVSDSLSNSDATNLWHMRLGHMTEIGLAELSKRGLLDGQSIGKLKFCEHCIFGK 682
Query: 458 QHRVKFGSGMHHSSRLFEYVHSDLLGPSKTPTHGGGSYFLSIIDDYSRRVWVFVLKKKSD 517
RVKF + H + + +YVHSDL GP++ + GG Y ++I+DDYSR+VW + LK K
Sbjct: 683 HKRVKFNTSTHTTEGILDYVHSDLWGPARKTSFGGTRYMMTIVDDYSRKVWPYFLKHKYQ 742
Query: 518 TF*KFKE*HTLIENQMGTKLKGLRTDNGLEFVSEQFNDFLQVE 560
F FKE T++E Q K+K LRTDNG+EF S+ F + + E
Sbjct: 743 AFDVFKEWKTMVERQTERKVKILRTDNGMEFCSKIFKSYCKSE 785
>ref|XP_470868.1| Putative retroelement pol polyprotein [Oryza sativa]
gi|14029020|gb|AAK52561.1| Putative retroelement pol
polyprotein [Oryza sativa]
Length = 1326
Score = 385 bits (989), Expect = e-105
Identities = 229/574 (39%), Positives = 337/574 (57%), Gaps = 25/574 (4%)
Query: 5 KWDIEKFTGSNDFGLWKVKMQAVLTQQKCV-EALKGEAAMPATLTQEEKREMIDKAKSAI 63
K+D+ F LW+VKM+A+L Q + EAL+ +T E++ KA I
Sbjct: 5 KYDLPLLDYKTRFSLWQVKMRAILAQTSDLDEALESFGKKKSTEWTAEEKRKDRKALLLI 64
Query: 64 VLCLGDKVLRDVAREATAASM*AKLESLYMTKSLAHRQLLKQQLYSFKMVESISISEQLT 123
L L + +L++V +E TAA + KLES+ M+K L + +K +L+S K+ ES S+ ++
Sbjct: 65 QLHLSNDILQEVLQEKTAAELWLKLESICMSKDLTSKMHIKMKLFSHKLQESGSVLNHIS 124
Query: 124 EFNKILVDLANIEVNTEDEDKALLLLCSLPKSFEHFKDTILYGKEGTTTLEEVQAALRTK 183
F +I+VDL +IEV +DED LLLLCSLP S+ +F+DTIL ++ TL EV AL+ +
Sbjct: 125 VFKEIVVDLVSIEVQFDDEDLGLLLLCSLPSSYANFRDTILLSRD-ELTLAEVYEALQNR 183
Query: 184 ELTKFKDLKVDEGSEGLNVA---RGRNEHRGKGKGKSRSKSRSKGFDKSKYK--CFLCHK 238
E K K + + S A RGR+E R R KS+S+G KS+ K C C K
Sbjct: 184 E--KMKGMVQSDASSSKGEALQVRGRSEQRTYNDSSDRDKSQSRGRSKSRGKKFCKYCKK 241
Query: 239 QGHFKKDC------PDKGGDGSPSVQVAEASNEEGYESTGALVVTSW--KSEKSWVLDSG 290
+ HF ++C + DG SV ++ E +S LVV + S W+LD+
Sbjct: 242 KNHFIEECWKLQNKEKRKSDGKASV----VTSAENSDSGDCLVVFAGCVASHDEWILDTA 297
Query: 291 CSYHMCPRKEYFETL-TLKEGGVVRLGNNKACKVQGMGNVRLKMFDGREFLLRDVRFVPE 349
CS+H+C +++F + +++ G VVR+G++ ++ G+G+V++K DG L+DVR +P
Sbjct: 298 CSFHICINRDWFSSYKSVQNGDVVRMGDDNPREIVGIGSVQIKTHDGMTRTLKDVRHIPG 357
Query: 350 LKRNLISLSMFDGLGYCTRIEHGVCKISHGALITVKGSKMNG-LYILDGSIVIGNASVAS 408
+ RNLISLS D GY GV K+S G+L+ + G + LY+L GS + G+ + A+
Sbjct: 358 MARNLISLSTLDAEGYKYSSSGGVVKVSKGSLVYMIGDMNSANLYVLRGSTLHGSVTAAA 417
Query: 409 VVPHN--NSELWHLRLGHVSERGLVELAKQGLLGKDKLDKLEFCEHCILGKQHRVKFGSG 466
V + LWH+RLGH+SE G+ EL K+ LL K++FCEHC+ GK RVKF +
Sbjct: 418 VSKDEPIKTNLWHMRLGHMSELGMAELMKRNLLDGCTQGKMKFCEHCVFGKHKRVKFNTS 477
Query: 467 MHHSSRLFEYVHSDLLGPSKTPTHGGGSYFLSIIDDYSRRVWVFVLKKKSDTF*KFKE*H 526
+H + + +YVH+DL GPS+ GG Y L+IIDDYSR+VW + LK K DTF FKE
Sbjct: 478 VHRTKGILDYVHTDLWGPSRKAYLGGARYMLTIIDDYSRKVWPYFLKHKDDTFAAFKEWK 537
Query: 527 TLIENQMGTKLKGLRTDNGLEFVSEQFNDFLQVE 560
IE Q ++K LRTDNG EF S+ F+D+ + E
Sbjct: 538 VRIERQTEKEVKVLRTDNGGEFCSDAFDDYCRKE 571
>gb|AAP53029.1| putative retrotransposon-related protein [Oryza sativa (japonica
cultivar-group)] gi|37532880|ref|NP_920742.1| putative
retrotransposon-related protein [Oryza sativa (japonica
cultivar-group)] gi|22655747|gb|AAN04164.1| Putative
retrotransposon protein [Oryza sativa (japonica
cultivar-group)] gi|16905223|gb|AAL31093.1| putative
retrotransposon-related protein [Oryza sativa]
Length = 1229
Score = 378 bits (971), Expect = e-103
Identities = 227/574 (39%), Positives = 334/574 (57%), Gaps = 25/574 (4%)
Query: 5 KWDIEKFTGSNDFGLWKVKMQAVLTQQKCV-EALKGEAAMPATLTQEEKREMIDKAKSAI 63
K+D+ F LW+VKM+AVL Q + EAL+ T E++ KA S I
Sbjct: 2 KYDLPLQDYKTRFSLWQVKMRAVLAQTSDLDEALESFGKKKTTEWTAEEKRKDRKALSLI 61
Query: 64 VLCLGDKVLRDVAREATAASM*AKLESLYMTKSLAHRQLLKQQLYSFKMVESISISEQLT 123
L L + +L+ V +E TAA + KLES+ M+K L + +K +L+S K+ ES S+ ++
Sbjct: 62 QLHLSNDILQKVLQEKTAAELWFKLESICMSKDLTSKMHIKMKLFSHKLQESGSVLNHIS 121
Query: 124 EFNKILVDLANIEVNTEDEDKALLLLCSLPKSFEHFKDTILYGKEGTTTLEEVQAALRTK 183
F +I+ DL ++EV +DED LLLLCSLP + +F+DTIL ++ TL EV AL+ +
Sbjct: 122 VFKEIIADLVSMEVQFDDEDLGLLLLCSLPSLYANFRDTILLSRD-ELTLAEVYEALQNR 180
Query: 184 ELTKFKDLKVDEGSEGLNVA---RGRNEHRGKGKGKSRSKSRSKGFDKSKYK--CFLCHK 238
E K K + + S A RGR+E R R KS+S+G KS+ K C C K
Sbjct: 181 E--KMKGMVQSDASSSKGKALQVRGRSEQRTYNDSNDRDKSQSRGRSKSRGKKFCKYCKK 238
Query: 239 QGHFKKDC------PDKGGDGSPSVQVAEASNEEGYESTGALVVTSW--KSEKSWVLDSG 290
+ HF ++C + DG SV ++ E +S LV + S W+LD+
Sbjct: 239 KNHFIEECWKLQNKEKRKSDGKASV----VTSAENSDSADCLVFFAGCVASHDEWILDTA 294
Query: 291 CSYHMCPRKEYFET-LTLKEGGVVRLGNNKACKVQGMGNVRLKMFDGREFLLRDVRFVPE 349
C + +C +++F + +++ G VVR+G+N ++ G+G+V++K DG L+DVR +P
Sbjct: 295 CLFLICINRDWFSSHKSVQNGDVVRMGDNNPREIMGIGSVQIKTHDGMTRTLKDVRHIPG 354
Query: 350 LKRNLISLSMFDGLGYCTRIEHGVCKISHGALITVKGSKMNG-LYILDGSIVIGNASVAS 408
+ RNLISLS D GY GV K+S G+L+ + G + LY+L GS + G+ + A+
Sbjct: 355 MARNLISLSTLDAEGYKYSGSGGVVKVSKGSLVYMIGDMNSANLYVLRGSTLHGSLTAAA 414
Query: 409 VVPHNNSE--LWHLRLGHVSERGLVELAKQGLLGKDKLDKLEFCEHCILGKQHRVKFGSG 466
V S+ LWH+RLGH+SE G+ EL K+ LL ++FCEHC+ GK RVKF +
Sbjct: 415 VSKDEPSKTNLWHMRLGHMSELGMAELMKRNLLDGCTQGNMKFCEHCVFGKHKRVKFNTS 474
Query: 467 MHHSSRLFEYVHSDLLGPSKTPTHGGGSYFLSIIDDYSRRVWVFVLKKKSDTF*KFKE*H 526
+H + + +YVH+DL GPS+ P+ GG Y L+IIDDYSR+VW + LK K DTF FKE
Sbjct: 475 VHRTKGILDYVHADLWGPSRKPSLGGACYMLTIIDDYSRKVWPYFLKHKDDTFAAFKEWK 534
Query: 527 TLIENQMGTKLKGLRTDNGLEFVSEQFNDFLQVE 560
+IE Q ++K LRTDNG EF S+ F+D+ + E
Sbjct: 535 VMIERQAEKEVKVLRTDNGGEFCSDAFDDYCRKE 568
>emb|CAB75481.1| copia-like polyprotein [Arabidopsis thaliana]
gi|11278366|pir||T47492 copia-like polyprotein -
Arabidopsis thaliana
Length = 1363
Score = 377 bits (968), Expect = e-102
Identities = 227/597 (38%), Positives = 342/597 (57%), Gaps = 51/597 (8%)
Query: 3 GSKWDIEKFTGSNDFGLWKVKMQAVLTQQKCVEALKGEAAMP---------ATLTQEEKR 53
G++ ++EKF G D+ +WK K+ A + L+ E+ P + ++E+R
Sbjct: 3 GARIEVEKFDGRGDYTMWKEKLLAHIDMLGLSAVLR-ESETPMGKERDSEKSDEDEKEER 61
Query: 54 EMID-------KAKSAIVLCLGDKVLRDVAREATAASM*AKLESLYMTKSLAHRQLLKQQ 106
E ++ KA+S IVL + D+VLR + +E +AA+M L+ LYM+K+L +R LKQ+
Sbjct: 62 EKMEAFEEKKRKARSTIVLSVSDRVLRKIKKETSAAAMLEALDRLYMSKALPNRIYLKQK 121
Query: 107 LYSFKMVESISISEQLTEFNKILVDLANIEVNTEDEDKALLLLCSLPKSFEHFKDTILYG 166
LYSFKM E++SI + EF I+ DL N+ V DED+A+LLL SLPK F+ KDT+ Y
Sbjct: 122 LYSFKMSENLSIEGNIDEFLHIVADLENLNVLVSDEDQAILLLMSLPKPFDQLKDTLKYS 181
Query: 167 KEGTT-TLEEVQAALRTKELTKFKDLK--VDEGSEGLNVA-----RGRNEHRGKGKGKSR 218
T +L+EV AA+ ++EL +F +K + +EGL V RGR+E + KGKGK R
Sbjct: 182 SGKTVLSLDEVAAAIYSREL-EFGSVKKSIKGQAEGLYVKDKAENRGRSEQKDKGKGK-R 239
Query: 219 SKSRSKGFDKSKYKCFLCHKQGHFKKDCPDK----------------GGDGSPSVQVAEA 262
SKS KSK C++C + GH K CP+K GG G+
Sbjct: 240 SKS------KSKRGCWICGEDGHLKSTCPNKNKPQFKNQGSNKGESSGGKGNLVEGSVNF 293
Query: 263 SNEEGYESTGALVVTSWKSEKSWVLDSGCSYHMCPRKEYFETLTLKEGGVVRLGNNKACK 322
G + AL T E W++D+GC YHM ++E+ E + GG VR+GN +
Sbjct: 294 VESAGMFVSEALSSTDIHLEDEWIMDTGCIYHMTHKREWLEDFDEEAGGSVRMGNKSISR 353
Query: 323 VQGMGNVRLKMFDGREFLLRDVRFVPELKRNLISLSMFDGLGYCTRIEHGVCKISHGALI 382
V+G+G VR+ +G L++VR++P++ RNL+SL F+ G+ E+G+ +I G +
Sbjct: 354 VKGVGTVRIVNDNGLTVTLQNVRYIPDMDRNLLSLGTFEKAGHKFESENGMLRIKSGNQV 413
Query: 383 TVKGSKMNGLYILDGSIVIGNASVASVVPHNNSELWHLRLGHVSERGLVELAKQGLLGKD 442
++G + + LYIL G + S+A ++++ LWH RL H+S++ + L K+G L K
Sbjct: 414 LLEGRRYDTLYILHGKPAT-DESLAVARANDDTVLWHRRLCHMSQKNMSLLIKKGFLDKK 472
Query: 443 KLDKLEFCEHCILGKQHRVKFGSGMHHSSRLFEYVHSDLLGPSKTP-THGGGSYFLSIID 501
K+ L+ CE CI G+ ++ F H + + EYVHSDL G P + G YF+S ID
Sbjct: 473 KVSMLDTCEDCIYGRAKKIGFNLAQHDTKKKLEYVHSDLWGAPTVPMSLGNCQYFISFID 532
Query: 502 DYSRRVWVFVLKKKSDTF*KFKE*HTLIENQMGTKLKGLRTDNGLEFVSEQFNDFLQ 558
DY+R+VWV+ LK K + F KF +L+ENQ G ++K LRTDNGLEF + F+ F +
Sbjct: 533 DYTRKVWVYFLKTKDEAFEKFVSWISLVENQSGERVKTLRTDNGLEFCNRMFDGFCE 589
>gb|AAX92861.1| retrotransposon protein, putative, Ty1-copia sub-class [Oryza
sativa (japonica cultivar-group)]
Length = 1373
Score = 376 bits (966), Expect = e-102
Identities = 232/580 (40%), Positives = 336/580 (57%), Gaps = 35/580 (6%)
Query: 5 KWDIEKFTGSNDFGLWKVKMQAVLTQQKCV-EALKGEAAMPATLTQEEKREMIDKAKSAI 63
K+D+ F LW+VKM+ +L Q EAL A T EE R+ KA + I
Sbjct: 2 KFDLPLLNYDTRFSLWQVKMRGILAQTHDYDEALDNFGKRRAEWTAEEIRKD-QKALALI 60
Query: 64 VLCLGDKVLRDVAREATAASM*AKLESLYMTKSLAHRQLLKQQLYSFKMVESISISEQLT 123
L L + +L++ E T+A + KLES+ M+K L + +K +L++ KM E S+ +
Sbjct: 61 QLHLHNDILQECLTEKTSAELWLKLESICMSKDLTSKMQMKMKLFTLKMKEEDSVITHMA 120
Query: 124 EFNKILVDLANIEVNTEDEDKALLLLCSLPKSFEHFKDTILYGKEGTTTLEEVQAALRTK 183
EF KI+ DL ++EV +DED LLLLCSLP S+ +F+DTIL ++ TL+EV AL+ K
Sbjct: 121 EFKKIVADLVSMEVKYDDEDLGLLLLCSLPNSYANFRDTILLSRD-ELTLKEVYDALQNK 179
Query: 184 ELTKFKDLKVDEGS-----EGLNVARGRNEHRGKGKG----KSRSKSRSKGFDKSKYKCF 234
E K K + ++GS E L+V RGR E+R + + RSKS+ G +K C
Sbjct: 180 E--KMKIMVQNDGSSSSKGEALHV-RGRTENRTSNEKNYDRRGRSKSKPPG---NKKFCV 233
Query: 235 LCHKQGHFKKDCPDKGG-------DGSPSVQVAEASNEEGYESTGALVVTSW--KSEKSW 285
C + H +C DG SV A AS+++ S LVV + W
Sbjct: 234 YCKLKNHNIDECKKVQAKERKNKKDGKVSVASAAASDDD---SGDCLVVFAGCVAGHDEW 290
Query: 286 VLDSGCSYHMCPRKEYFETLT-LKEGGVVRLGNNKACKVQGMGNVRLKMFDGREFLLRDV 344
+LDS CS+H+C ++ +F + +++G VVR+G++ C + G+G+V++K DG L++V
Sbjct: 291 ILDSACSFHICTKRNWFSSYKPVQKGDVVRMGDDNPCAIVGIGSVQIKTDDGMTRTLKNV 350
Query: 345 RFVPELKRNLISLSMFDGLGYCTRIEHGVCKISHGALITVKGSKMNG-LYILDGSIVIGN 403
R++P + RNLISLS D GY GV K+S G+L+ +KG + LY+L G + G+
Sbjct: 351 RYIPGMSRNLISLSTLDAEGYKYSGSDGVLKVSKGSLVCLKGDVNSAKLYVLRGCTLTGS 410
Query: 404 ASVASVVPHNN---SELWHLRLGHVSERGLVELAKQGLLGKDKLDKLEFCEHCILGKQHR 460
S A+ + ++ + LWH+RLGH+S G+ EL K+ LL K++FCEHCI GK R
Sbjct: 411 DSAAAAITNDEPSKTNLWHMRLGHMSHLGMTELMKRNLLKGCTSSKIKFCEHCIFGKHKR 470
Query: 461 VKFGSGMHHSSRLFEYVHSDLLGPSKTPTHGGGSYFLSIIDDYSRRVWVFVLKKKSDTF* 520
V+F + +H + +YVH+DL GPSK P+ GG Y L+IIDDYSR+VW + LK K DTF
Sbjct: 471 VQFNTSVHTTKGTLDYVHADLWGPSKKPSLGGARYMLTIIDDYSRKVWPYFLKHKDDTFT 530
Query: 521 KFKE*HTLIENQMGTKLKGLRTDNGLEFVSEQFNDFLQVE 560
FK +IE Q K+K LRTDNG EF S FND+ + E
Sbjct: 531 AFKNWKVMIERQTERKVKLLRTDNGGEFCSHAFNDYCRQE 570
>ref|XP_469192.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
gi|53370655|gb|AAU89150.1| integrase core domain
containing protein [Oryza sativa (japonica
cultivar-group)] gi|40538906|gb|AAR87163.1| putative
polyprotein [Oryza sativa (japonica cultivar-group)]
Length = 1322
Score = 374 bits (960), Expect = e-101
Identities = 223/572 (38%), Positives = 333/572 (57%), Gaps = 21/572 (3%)
Query: 5 KWDIEKFTGSNDFGLWKVKMQAVLTQQKCV-EALKGEAAMPATLTQEEKREMIDKAKSAI 63
K+D+ F LW+VKM+AVL Q + EAL+ T E++ KA S I
Sbjct: 5 KYDLPLLDYKTRFSLWQVKMRAVLAQTSDLDEALESFGKKKTTEWTAEEKRKDRKALSLI 64
Query: 64 VLCLGDKVLRDVAREATAASM*AKLESLYMTKSLAHRQLLKQQLYSFKMVESISISEQLT 123
L L + +L++V ++ TAA + KLES+ M+K L + +K +L+S K+ ES S+ ++
Sbjct: 65 QLHLSNDILQEVLQKKTAAELWLKLESICMSKDLTSKMHIKMKLFSHKLHESGSVLNHIS 124
Query: 124 EFNKILVDLANIEVNTEDEDKALLLLCSLPKSFEHFKDTILYGKEGTTTLEEVQAALRTK 183
F +I+ DL ++EV +DED LLLLCSLP S+ +F+ TIL ++ TL EV AL+ +
Sbjct: 125 VFKEIVADLVSMEVQFDDEDLGLLLLCSLPSSYANFRHTILLSRD-ELTLAEVYEALQNR 183
Query: 184 ELTKFKDLKVDEGSEGLNV-ARGRNEHRGKGKGKSRSKSRSKGFDKSKYK--CFLCHKQG 240
E K S+G + RGR+E R KS+S+G KS+ K C C K+
Sbjct: 184 EKMKGMVQSYASSSKGEALQVRGRSEQRTYNDSNDHDKSQSRGRSKSRGKKFCKYCKKKN 243
Query: 241 HFKKDC------PDKGGDGSPSVQVAEASNEEGYESTGALVVTSW--KSEKSWVLDSGCS 292
HF ++C + DG SV ++ E +S LVV + S W+LD+ CS
Sbjct: 244 HFIEECWKLQNKEKRKSDGKASV----VTSAENSDSGDCLVVFAGYVASHDEWILDTACS 299
Query: 293 YHMCPRKEYFETL-TLKEGGVVRLGNNKACKVQGMGNVRLKMFDGREFLLRDVRFVPELK 351
+H+C +++F + +++ VVR+G++ ++ G+G+V++K DG L+DVR +P +
Sbjct: 300 FHICINRDWFSSYKSVQNEDVVRMGDDNPREIVGIGSVQIKTHDGMTRTLKDVRHIPGMA 359
Query: 352 RNLISLSMFDGLGYCTRIEHGVCKISHGALITVKGSKMNG-LYILDGSIVIGNASVASVV 410
RNLISLS D GY GV K+S G+L+ + G + LY+L GS + G+ + A+V
Sbjct: 360 RNLISLSTLDAEGYKYSGSGGVVKVSKGSLVYMIGDMNSANLYVLRGSTLHGSVTAAAVT 419
Query: 411 PHNNSE--LWHLRLGHVSERGLVELAKQGLLGKDKLDKLEFCEHCILGKQHRVKFGSGMH 468
S+ LWH+RLGH+SE G+ EL K+ LL ++FCEHC+ GK RVKF + +H
Sbjct: 420 KDEPSKTNLWHMRLGHMSELGMAELMKRNLLDGCTQGNMKFCEHCVFGKHKRVKFNTSVH 479
Query: 469 HSSRLFEYVHSDLLGPSKTPTHGGGSYFLSIIDDYSRRVWVFVLKKKSDTF*KFKE*HTL 528
+ + +YVH+DL GPS+ P+ GG Y L+IIDDYSR+ W + LK K DTF FKE +
Sbjct: 480 RTKGILDYVHADLWGPSRKPSLGGARYMLTIIDDYSRKEWPYFLKHKDDTFAAFKERKVM 539
Query: 529 IENQMGTKLKGLRTDNGLEFVSEQFNDFLQVE 560
IE Q ++K L TDNG EF S+ F+D+ + E
Sbjct: 540 IERQTEKEVKVLCTDNGGEFCSDAFDDYCRKE 571
>gb|AAD23690.1| putative retroelement pol polyprotein [Arabidopsis thaliana]
gi|25301702|pir||E84601 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana
Length = 1333
Score = 372 bits (954), Expect = e-101
Identities = 229/573 (39%), Positives = 325/573 (55%), Gaps = 44/573 (7%)
Query: 7 DIEKFTGSNDFGLWKVKMQAVLTQQKCVEALKGEAAM-----PATLTQEEKREMI----- 56
++EKF G D+ +WK K+ A L ALK E + LT+EE++E +
Sbjct: 7 EVEKFDGRGDYTMWKEKLMAHLDILGLSVALKEEDDLVEKVAEMQLTEEEEKEEVLRREL 66
Query: 57 -----DKAKSAIVLCLGDKVLRDVAREATAASM*AKLESLYMTKSLAHRQLLKQQLYSFK 111
KA+SAIVL + D+VLR + +E +AA+M L+ LYM+K+L +R KQ+LYSFK
Sbjct: 67 LEEKRRKARSAIVLSVTDRVLRKIKKEQSAAAMLGVLDKLYMSKALPNRIYQKQKLYSFK 126
Query: 112 MVESISISEQLTEFNKILVDLANIEVNTEDEDKALLLLCSLPKSFEHFKDTILYG-KEGT 170
M E++SI + EF +I+ DL N V DED+A+LLL SLPK F+ +DT+ YG T
Sbjct: 127 MSENLSIEGNIDEFLRIIADLENTNVLVSDEDQAILLLMSLPKPFDQLRDTLKYGLGRVT 186
Query: 171 TTLEEVQAALRTKELTKFKDLKVDEG-SEGLNV-----ARGRNEHRGKGKGKSRSKSRSK 224
+L+EV AA+ +KEL + K +G +EGL V RGR E RG +S+S+S
Sbjct: 187 LSLDEVVAAIYSKELELGSNKKSIKGQAEGLFVKEKTETRGRTEQRGNNNNNKKSRSKS- 245
Query: 225 GFDKSKYKCFLCHKQGHFKKDCPDKGGDGSPSVQVAEASNEEGYESTGALVVTSWKSEKS 284
+SK C++C G S + S G + AL T E
Sbjct: 246 ---RSKKGCWIC----------------GESSNGSSNYSEANGLYVSEALSSTDIHLEDE 286
Query: 285 WVLDSGCSYHMCPRKEYFETLTLKEGGVVRLGNNKACKVQGMGNVRLKMFDGREFLLRDV 344
WV+D+GCSYHM ++E+FE L GG VR+GN KV+G+G +R+K G L +V
Sbjct: 287 WVMDTGCSYHMTYKREWFEDLNEDAGGSVRMGNKTVSKVRGIGTIRVKNEAGMVVRLTNV 346
Query: 345 RFVPELKRNLISLSMFDGLGYCTRIEHGVCKISHGALITVKGSKMNGLYILDGSIVIGNA 404
R++PE+ RNL+SL F+ GY ++E+G I G + + + LY+L V
Sbjct: 347 RYIPEMDRNLLSLGTFEKSGYSFKLENGTLSIIAGDSVLLTVRRCYTLYLLQWRPVT-EE 405
Query: 405 SVASVVPHNNSELWHLRLGHVSERGLVELAKQGLLGKDKLDKLEFCEHCILGKQHRVKFG 464
S++ V +++ LWH RLGH+S++ + L K+GLL K K+ KLE CE CI GK R+ F
Sbjct: 406 SLSVVKRQDDTILWHRRLGHMSQKNMDLLLKKGLLDKKKVSKLETCEDCIYGKAKRIGFN 465
Query: 465 SGMHHSSRLFEYVHSDLLGPSKTP-THGGGSYFLSIIDDYSRRVWVFVLKKKSDTF*KFK 523
H + EYVHSDL G P + G YF+S IDDY+R+V ++ LK K + F KF
Sbjct: 466 LAQHDTREKLEYVHSDLWGAPSVPFSLGKCQYFISFIDDYTRKVRIYFLKTKDEAFDKFV 525
Query: 524 E*HTLIENQMGTKLKGLRTDNGLEFVSEQFNDF 556
E L+ENQ ++K LRTDNGLEF + F++F
Sbjct: 526 EWANLVENQTDKRIKTLRTDNGLEFCNRSFDEF 558
>emb|CAB78169.1| putative retrotransposon [Arabidopsis thaliana]
gi|4539406|emb|CAB40039.1| putative retrotransposon
[Arabidopsis thaliana] gi|7444416|pir||T04181
hypothetical protein F7L13.40 - Arabidopsis thaliana
Length = 1230
Score = 369 bits (947), Expect = e-100
Identities = 225/583 (38%), Positives = 321/583 (54%), Gaps = 58/583 (9%)
Query: 7 DIEKFTGSNDFGLWKVKMQAVLTQQKCVEALKGEAAMPATLTQEEK-------------R 53
++EKF G D+ LWK K+ A + AL+ ++ L EE+
Sbjct: 7 EMEKFDGHGDYTLWKEKLMAHMDLLGLTVALRETQSVSDPLESEEEGKESEKGDKEALME 66
Query: 54 EMIDKAKSAIVLCLGDKVLRDVAREATAASM*AKLESLYMTKSLAHRQLLKQQLYSFKMV 113
E KA+S IVL + D+VLR +E TA SM L+ LYM+K+L +R LKQ+LYS+KM
Sbjct: 67 EKRQKARSTIVLSVSDQVLRKSKKEKTAPSMLEALDKLYMSKALPNRIYLKQKLYSYKMQ 126
Query: 114 ESISISEQLTEFNKILVDLANIEVNTEDEDKALLLLCSLPKSFEHFKDTILYGKEGTT-T 172
E++S+ + EF +++ DL N V DED+A+LLL SLPK F+ KDT+ YG TT +
Sbjct: 127 ENLSVEGNIDEFLRLIADLENTNVLVSDEDQAILLLMSLPKQFDQLKDTLKYGSGRTTLS 186
Query: 173 LEEVQAALRTKELTKFKDLKVDEG-SEGLNV-----ARGRNEHRGKGKGKSRSKSRSKGF 226
++EV AA+ +KEL + K G +EGL V RG +E + KG K RS+SRSKG+
Sbjct: 187 VDEVVAAIYSKELELGSNKKSIRGQAEGLYVKDKPETRGMSEQKEKG-NKGRSRSRSKGW 245
Query: 227 DKSKYKCFLCHKQGHFKKDCPDKGGDGSPSVQVAEASNEE------------GYESTGAL 274
C++C ++GHFK CP+KG + A S E GY + AL
Sbjct: 246 K----GCWICGEEGHFKTSCPNKGKQQNKGKDQASGSKGEAATIKGNTSEGSGYYVSEAL 301
Query: 275 VVTSWKSEKSWVLDSGCSYHMCPRKEYFETLTLKEGGVVRLGNNKACKVQGMGNVRLKMF 334
T WV+D+GC+YHM +KE+FE L+ GG VR+GN K +
Sbjct: 302 HSTDVNLGNEWVMDTGCNYHMTHKKEWFEELSEDAGGTVRMGNKSTSKFR---------- 351
Query: 335 DGREFLLRDVRFVPELKRNLISLSMFDGLGYCTRIEHGVCKISHGALITVKGSKMNGLYI 394
V+++P++ RNL+S+ + GY ++GV + G + GS+ LY+
Sbjct: 352 ---------VKYIPDMDRNLLSMGTLEEHGYSFESKNGVLVVKEGTRTLLIGSRHEKLYL 402
Query: 395 LDGSIVIGNASVASVVPHNNSELWHLRLGHVSERGLVELAKQGLLGKDKLDKLEFCEHCI 454
L G + + S+ ++++ LWH RLGH+S++ + L K+G L K+ KLE CE CI
Sbjct: 403 LQGKPEVSH-SMTVERRNDDTVLWHRRLGHISQKNMDILVKKGYLDGKKVSKLELCEDCI 461
Query: 455 LGKQHRVKFGSGMHHSSRLFEYVHSDLLG-PSKTPTHGGGSYFLSIIDDYSRRVWVFVLK 513
GK R+ F H++ YVHSDL G PS + G YF+S ID YSR+ WV+ LK
Sbjct: 462 YGKARRLSFVVATHNTEDKLNYVHSDLWGAPSVPLSLGKCQYFISFIDVYSRKTWVYFLK 521
Query: 514 KKSDTF*KFKE*HTLIENQMGTKLKGLRTDNGLEFVSEQFNDF 556
K + F F E ++ENQ G K+K LR DNGLEF ++QFNDF
Sbjct: 522 HKDEAFGTFAEWSVMVENQTGRKIKILRIDNGLEFCNQQFNDF 564
>ref|XP_475489.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
gi|48475213|gb|AAT44282.1| putative polyprotein [Oryza
sativa (japonica cultivar-group)]
Length = 1243
Score = 358 bits (920), Expect = 5e-97
Identities = 222/570 (38%), Positives = 327/570 (56%), Gaps = 38/570 (6%)
Query: 5 KWDIEKFTGSNDFGLWKVKMQAVLTQQKCVEALKGEAAMPATLTQEEKREMIDKAKSAIV 64
K+D+ F LW+VKM+AVL QQ +AL G + +EK+ KA S I
Sbjct: 5 KYDLPLLDRDTRFSLWQVKMRAVLAQQDLDDALSGFDKRTQDWSNDEKKRD-RKAISYIH 63
Query: 65 LCLGDKVLRDVAREATAASM*AKLESLYMTKSLAHRQLLKQQLYSFKMVESISISEQLTE 124
L L + +L++V +E TAA + KLE + MTK L + LKQ+L+ K+ + S+ + L+
Sbjct: 64 LHLSNNILQEVLKEETAAGLWLKLEQICMTKDLTSKMHLKQKLFLHKLQDDESVMDHLSA 123
Query: 125 FNKILVDLANIEVNTEDEDKALLLLCSLPKSFEHFKDTILYGKEGTTTLEEVQAALRTKE 184
F +I+ DL ++EV +++D L+LLCSLP S+ +F+ TILY ++ T TL+EV A KE
Sbjct: 124 FKEIVADLESMEVKYDEDDLGLILLCSLPSSYANFRGTILYSRD-TLTLKEVYDAFHAKE 182
Query: 185 LTKFKDLKVDEGS----EGLNVARGRNEHRG-KGKGKSRSKSRSKGFDKSK--YK-CFLC 236
K K + EGS EGL V RGR + + K + + +S S +G KS+ YK C C
Sbjct: 183 --KMKKMVTSEGSNSQAEGL-VVRGRQQKKNTKNQSRDKSSSSYRGRTKSRGRYKSCKYC 239
Query: 237 HKQGHFKKDC---PDKGGDGSPSVQVAEASNEEGYESTGALVVTSWKSEKSWVLDSGCSY 293
+ GH +C DK D + + EE E A+V + V +GC+
Sbjct: 240 KRDGHDISECWKLQDK--DKRTGKYIPKGKKEE--EGKAAVVTDEKSDAELLVAYAGCA- 294
Query: 294 HMCPRKEYFETLTLKEGGVVRLGNNKACKVQGMGNVRLKMFDGREFLLRDVRFVPELKRN 353
+++F T +GG V +G++ C+V G+G V++KMFDG L DV+ +P LKR+
Sbjct: 295 -QTSDQDWFATYEALQGGTVLMGDDTPCEVAGIGTVQIKMFDGCIRTLSDVQHIPNLKRS 353
Query: 354 LISLSMFDGLGYCTRIEHGVCKISHGALITVKGS-KMNGLYILDGSIVIGNASVA--SVV 410
LISL +G+ K++ G+L+ +K K LY L G+ ++GN + S+
Sbjct: 354 LISL-------------YGILKVTKGSLVVMKVDIKSANLYHLRGTTILGNVAAVFDSLS 400
Query: 411 PHNNSELWHLRLGHVSERGLVELAKQGLLGKDKLDKLEFCEHCILGKQHRVKFGSGMHHS 470
+ + LWH+RLGH+SE GL EL+K+GLL + KL+FCEHCI GK RVKF + H +
Sbjct: 401 NSDATNLWHMRLGHMSEIGLAELSKRGLLDGQSIRKLKFCEHCIFGKHKRVKFNTSTHTT 460
Query: 471 SRLFEYVHSDLLGPSKTPTHGGGSYFLSIIDDYSRRVWVFVLKKKSDTF*KFKE*HTLIE 530
+ +YVHSDL GP+ + GG Y ++I+DDYSR+VW + LK K F FKE T++E
Sbjct: 461 EGILDYVHSDLWGPAHKTSFGGARYMMTIVDDYSRKVWPYFLKHKYQAFDGFKEWKTMVE 520
Query: 531 NQMGTKLKGLRTDNGLEFVSEQFNDFLQVE 560
Q K+K LRTDNG+EF S+ F + + E
Sbjct: 521 RQTERKVKILRTDNGMEFCSKIFKSYCKSE 550
>dbj|BAB09923.1| copia-like retrotransposable element [Arabidopsis thaliana]
Length = 1342
Score = 353 bits (907), Expect = 2e-95
Identities = 219/586 (37%), Positives = 329/586 (55%), Gaps = 53/586 (9%)
Query: 7 DIEKFTGSNDFGLWKVKMQAVLTQQKCVEALKGEAAM----------------PATLTQ- 49
++EKF G D+ LWK K+ A + +E L E P T T
Sbjct: 7 EVEKFDGDGDYILWKEKLLAHMEMLGLLEGLGEEEEAVVEDSTTEISDGGNQDPETATSK 66
Query: 50 -EEK--REMIDKAKSAIVLCLGDKVLRDVAREATAASM*AKLESLYMTKSLAHRQLLKQQ 106
E+K +E KA+S I+L LG+ VLR V ++ TAA M L+ L+M KSL +R LKQ+
Sbjct: 67 LEDKILKEKRGKARSTIILSLGNNVLRKVIKQKTAAGMIKVLDQLFMAKSLPNRIYLKQR 126
Query: 107 LYSFKMVESISISEQLTEFNKILVDLANIEVNTEDEDKALLLLCSLPKSFEHFKDTILYG 166
LY +KM E++++ E + +F K++ DL N++V DED+A++LL SLP+ F+ K+T+ Y
Sbjct: 127 LYGYKMSENMTMEENVNDFFKLISDLENVKVVVPDEDQAIVLLMSLPRQFDQLKETLKYC 186
Query: 167 KEGTTTLEEVQAALRTKELTKFKDLK-VDEGSEGLNVA-RGRNEHRGKGKGKSRSKSRSK 224
K T LEE+ +A+R+K L K + S+GL V RGR+E RGKG K++S+S+SK
Sbjct: 187 KT-TLHLEEITSAIRSKILELGASGKLLKNNSDGLFVQDRGRSETRGKGPNKNKSRSKSK 245
Query: 225 GFDKSKYKCFLCHKQGHFKKDC---PDKGGDGSPSVQVAEASNEEGYESTGALVVT---- 277
G K+ C++C K+GHFKK C ++ GS S + ++ ALVV+
Sbjct: 246 GAGKT---CWICGKEGHFKKQCYVWKERNKQGSTSERGEASTVTARVTDAAALVVSRALL 302
Query: 278 --SWKSEKSWVLDSGCSYHMCPRKEYFETLTLKEGGVVRLGNNKACKVQGMGNVRLKMFD 335
+ + +W+LD+GCS+HM RK++ G VR+GN+ +V+G+G+VR+K D
Sbjct: 303 GFAEVTPDTWILDTGCSFHMTCRKDWIIDFKETASGKVRMGNDTYSEVKGIGDVRIKNED 362
Query: 336 GREFLLRDVRFVPELKRNLISLSMFDGLGYCTRIEHGVCKISHGALITVKGSKMNGLYIL 395
G LL DVR++PE+ +NLISL + G + G+ I L + G K + LY L
Sbjct: 363 GSTILLTDVRYIPEMSKNLISLGTLEDKGCWFESKKGILTIFKNDLTVLTGKKESTLYFL 422
Query: 396 DGSIVIGNASVASVVPHNNSELWHLRLGHVSERGLVELAKQGLLGKDKLDKLEFCEHCIL 455
G+ + G A+V + + LWH RLGH+ +GL L +G L K+ +
Sbjct: 423 QGTTLAGEANVID-KEKDETSLWHSRLGHIGAKGLQVLVSKGHLDKNIM----------- 470
Query: 456 GKQHRVKFGSGMHHSSRLFEYVHSDLLGPSKTP-THGGGSYFLSIIDDYSRRVWVFVLKK 514
+ FG+ H + +YVHSDL G + P + G YF++ IDD++RR W++ ++
Sbjct: 471 -----ISFGAAKHVTKDKLDYVHSDLWGSTNVPFSIGKCQYFITFIDDFTRRTWIYFIRT 525
Query: 515 KSDTF*KFKE*HTLIENQMGTKLKGLRTDNGLEFVSEQFNDFLQVE 560
K + F KF E T IENQ KLK L TDNGLEF +++F+ F + E
Sbjct: 526 KDEAFSKFVEWKTQIENQQDKKLKILITDNGLEFCNQEFDSFCRKE 571
>gb|AAF19226.1| Highly similar to Ta1-3 polyprotein [Arabidopsis thaliana]
gi|25301707|pir||E86490 hypothetical protein F28L22.3 -
Arabidopsis thaliana
Length = 1356
Score = 352 bits (903), Expect = 5e-95
Identities = 216/592 (36%), Positives = 326/592 (54%), Gaps = 46/592 (7%)
Query: 7 DIEKFTGSNDFGLWKVKMQAVL------------TQQKCVEALKGEAAMPA--------- 45
+I+ F G DF LWK+++QA L + K V K EA +
Sbjct: 9 EIKVFNGDRDFSLWKIRIQAQLGVLGLKDTLTDFSLTKTVPLTKSEAKQESGDGESSGTK 68
Query: 46 TLTQEEKREMIDKAKSAIVLCLGDKVLRDVAREATAASM*AKLESLYMTKSLAHRQLLKQ 105
+ K E ++AK+ I+ + D VL V AT A + A L YM SL +R +
Sbjct: 69 EVPDPVKIEQSEQAKNIIINHISDVVLLKVNHYATTADLWATLNKKYMETSLPNRIYTQL 128
Query: 106 QLYSFKMVESISISEQLTEFNKILVDLANIEVNTEDEDKALLLLCSLPKSFEHFKDTILY 165
+LYSFKMV +++I + + EF +I+ +L ++E+ ++E +A+L+L SLP S K T+ Y
Sbjct: 129 KLYSFKMVSTMTIDQNVDEFLRIVAELGSLEIQVDEEVQAILILNSLPASHIQLKHTLKY 188
Query: 166 GKEGTTTLEEVQAALRT--KELTKFKDLKVDEGSEGLNVARGR-----NEHRGKGKGKSR 218
G + T T+++V ++ ++ +EL + DL + + RGR N+ G+GKG+SR
Sbjct: 189 GNK-TLTVQDVTSSAKSLERELAEAVDLDKGQAAVLYTTERGRPLVRNNQKGGQGKGRSR 247
Query: 219 SKSRSKGFDKSKYKCFLCHKQGHFKKDCPDKGGDGSPSVQVAEASNEEGYESTGALVVTS 278
S S K+K C+ C K+GH KKDC + Q E + AL V
Sbjct: 248 SNS------KTKVPCWYCKKEGHVKKDCYSRKKKMESEGQGEAGVITEKLVFSEALSVNE 301
Query: 279 WKSEKSWVLDSGCSYHMCPRKEYFETLTLKEGGVVRLGNNKACKVQGMGNVRLKMFDGRE 338
+ W+LDSGC+ HM R+++F + K + LG++ + + QG G +R+ G
Sbjct: 302 QMVKDLWILDSGCTSHMTSRRDWFISFQEKGNTTILLGDDHSVESQGQGTIRIDTHGGTI 361
Query: 339 FLLRDVRFVPELKRNLISLSMFDGLGYCTRIEHGVCKISHGALITVKGSKMNGLYILDGS 398
+L +V++VP L+RNLIS D LGY G + ++GS NGLY+LDGS
Sbjct: 362 KILENVKYVPHLRRNLISTGTLDKLGYRHEGGEGKVRYFKNNKTALRGSLSNGLYVLDGS 421
Query: 399 IVIG---NASVASVVPHNNSELWHLRLGHVSERGLVELAKQGLLGKDKLDKLEFCEHCIL 455
V+ NA V + LWH RLGH+S L LA +GL+ + ++++LEFCEHC++
Sbjct: 422 TVMSELCNAETDKV----KTALWHSRLGHMSMNNLKVLAGKGLIDRKEINELEFCEHCVM 477
Query: 456 GKQHRVKFGSGMHHSSRLFEYVHSDLLG-PSKTPTHGGGSYFLSIIDDYSRRVWVFVLKK 514
GK +V F G H S YVH+DL G P+ TP+ G YFLSIIDD +R+VW++ LK
Sbjct: 478 GKSKKVSFNVGKHTSEDALSYVHADLWGSPNVTPSISGKQYFLSIIDDKTRKVWLYFLKS 537
Query: 515 KSDTF*KFKE*HTLIENQMGTKLKGLRTDNGLEFVSEQFNDFLQ---VERNQ 563
K +TF KF E +L+ENQ+ K+K LRTDNGLEF + +F+ + + +ER++
Sbjct: 538 KDETFDKFCEWKSLVENQVNKKVKCLRTDNGLEFCNSRFDSYCKEHGIERHR 589
>gb|AAT85194.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
Length = 1241
Score = 351 bits (901), Expect = 8e-95
Identities = 201/495 (40%), Positives = 288/495 (57%), Gaps = 35/495 (7%)
Query: 93 MTKSLAHRQLLKQQLYSFKMVESISISEQLTEFNKILVDLANIEVNTEDEDKALLLLCSL 152
MTK L + LKQ+L+ K+ + S+ + L+ F +I+ DL ++EV ++ED L+LLCSL
Sbjct: 1 MTKDLTSKMHLKQKLFLHKLQDDGSVMDHLSAFKEIVADLESMEVKYDEEDLGLILLCSL 60
Query: 153 PKSFEHFKDTILYGKEGTTTLEEVQAALRTKELTKFKDLKVDEGS----EGLNVARGRNE 208
P S+ +F+DTILY ++ T TL+EV AL KE K K + EGS EGL V RGR +
Sbjct: 61 PSSYANFRDTILYSRD-TLTLKEVYDALHAKE--KMKKMVPSEGSNSQAEGL-VVRGRQQ 116
Query: 209 HRG---KGKGKSRSKSRSKGFDKSKYK-CFLCHKQGHFKKDC----------------PD 248
+ K + KS S R + + +YK C C + GH +C
Sbjct: 117 EKNTNNKSRDKSSSIYRGRSKSRGRYKSCKYCKRDGHDISECWKLQDKDKRTRKYIPKGK 176
Query: 249 KGGDGSPSVQVAEASNEEGYESTGALVVTSWKSEKSWVLDSGCSYHMCPRKEYFETLTLK 308
K +G +V E S+ E + TS W+LD+ C+YHMCP +++F T
Sbjct: 177 KEEEGKAAVVTDEKSDAELLVAYAGCAQTS----DQWILDTACTYHMCPNRDWFATYEAV 232
Query: 309 EGGVVRLGNNKACKVQGMGNVRLKMFDGREFLLRDVRFVPELKRNLISLSMFDGLGYCTR 368
+GG V +G++ C+V G+G V++KMFDG L DVR +P LKR+LISL D GY
Sbjct: 233 QGGTVLMGDDTPCEVAGIGTVQIKMFDGCIRTLLDVRHIPNLKRSLISLCTLDRKGYKYS 292
Query: 369 IEHGVCKISHGALITVKGS-KMNGLYILDGSIVIGNASVASVVPHNN--SELWHLRLGHV 425
G+ K++ G+L+ +K K LY L G+ ++GN + S N+ + LWH+RLGH+
Sbjct: 293 GGDGILKVTKGSLVVMKADIKYANLYHLRGTTILGNVAAVSDSLSNSDATNLWHMRLGHM 352
Query: 426 SERGLVELAKQGLLGKDKLDKLEFCEHCILGKQHRVKFGSGMHHSSRLFEYVHSDLLGPS 485
SE GL EL+K+GLL + KL+FCEHCI GK RVKF + H + + +YVHSDL GP+
Sbjct: 353 SEIGLAELSKRGLLDGQSIGKLKFCEHCIFGKHKRVKFNTSTHTTEGILDYVHSDLWGPA 412
Query: 486 KTPTHGGGSYFLSIIDDYSRRVWVFVLKKKSDTF*KFKE*HTLIENQMGTKLKGLRTDNG 545
+ + GG Y ++I+DDYSR+VW + LK K F FKE T++E Q K+K LRTDNG
Sbjct: 413 RKTSFGGARYMMTIVDDYSRKVWPYFLKHKYQAFDVFKEWKTMVERQTERKVKILRTDNG 472
Query: 546 LEFVSEQFNDFLQVE 560
+E S+ F + + E
Sbjct: 473 MELCSKIFKSYCKSE 487
>gb|AAD19773.1| putative retroelement pol polyprotein [Arabidopsis thaliana]
gi|25301697|pir||B84512 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana
Length = 1335
Score = 350 bits (897), Expect = 2e-94
Identities = 200/523 (38%), Positives = 296/523 (56%), Gaps = 22/523 (4%)
Query: 54 EMIDKAKSAIVLCLGDKVLRDVAREATAASM*AKLESLYMTKSLAHRQLLKQQLYSFKMV 113
E DKAK+ I L + DKVLR + TAA L+ L+M +SL HR + Y+FKM
Sbjct: 44 ERCDKAKNVIFLNVADKVLRKIELCKTAAEAWETLDRLFMIRSLPHRVYTQLSFYTFKMQ 103
Query: 114 ESISISEQLTEFNKILVDLANIEVNTEDEDKALLLLCSLPKSFEHFKDTILYGKEGTTT- 172
E+ I E + +F KI+ DL +++++ DE +A+LLL SLP ++ +T+ Y
Sbjct: 104 ENKKIDENIDDFLKIVADLNHLQIDVTDEVQAILLLSSLPARYDGLVETMKYSNSREKLR 163
Query: 173 LEEVQAALRTKELTKFKDLK-VDEGSEGLNVARGRNEHRG-KGKGKSRSKSRSKGFDKSK 230
L++V A R KE ++ + V EG G+N ++G KGK +SRSKS K
Sbjct: 164 LDDVMVAARDKERELSQNNRPVVEGHFARGRPDGKNNNQGNKGKNRSRSKSAD-----GK 218
Query: 231 YKCFLCHKQGHFKKDC------PDKGGDGSPSVQVAEASNEEGYE------STGALVVTS 278
C++C K+GHFKK C GS + + + A + E + +T +V +
Sbjct: 219 RVCWICGKEGHFKKQCYKWIERNKSKQQGSDNGESSLAKSTEAFNPAMVLLATDETLVVT 278
Query: 279 WKSEKSWVLDSGCSYHMCPRKEYFETLTLKEGGVVRLGNNKACKVQGMGNVRLKMFDGRE 338
WVLD+GCS+HM PRK++F+ G V++GN+ V+G+G+++++ DG +
Sbjct: 279 DSIANEWVLDTGCSFHMTPRKDWFKDFKELSSGYVKMGNDTYSPVKGIGSIKIRNSDGSQ 338
Query: 339 FLLRDVRFVPELKRNLISLSMFDGLGYCTRIEHGVCKISHGALITVKGSKMNGLYILDGS 398
+L DVR++P + RNLISL + G + + G+ KI G +KG K + LYILDG
Sbjct: 339 VILTDVRYMPNMTRNLISLGTLEDRGCWFKSQDGILKIVKGCSTILKGQKRDTLYILDGV 398
Query: 399 IVIGNASVASVVPHNNSELWHLRLGHVSERGLVELAKQGLLGKDKLDKLEFCEHCILGKQ 458
G S +S + + LWH RLGH+S++G+ L K+G L ++ + +LEFCE C+ GKQ
Sbjct: 399 TEEGE-SHSSAEVKDETALWHSRLGHMSQKGMEILVKKGCLRREVIKELEFCEDCVYGKQ 457
Query: 459 HRVKFGSGMHHSSRLFEYVHSDLLGPSKTPTH-GGGSYFLSIIDDYSRRVWVFVLKKKSD 517
HRV F H + YVHSDL G P G YF+S +DDYSR+VW++ L+KK +
Sbjct: 458 HRVSFAPAQHVTKEKLAYVHSDLWGSPHNPASLGNSQYFISFVDDYSRKVWIYFLRKKDE 517
Query: 518 TF*KFKE*HTLIENQMGTKLKGLRTDNGLEFVSEQFNDFLQVE 560
F KF E ++ENQ K+K LRTDNGLE+ + F F + E
Sbjct: 518 AFEKFVEWKKMVENQSDRKVKKLRTDNGLEYCNHYFEKFCKEE 560
>emb|CAA32025.1| unnamed protein product [Nicotiana tabacum]
gi|130582|sp|P10978|POLX_TOBAC Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
; Reverse transcriptase ; Endonuclease]
Length = 1328
Score = 349 bits (896), Expect = 3e-94
Identities = 199/572 (34%), Positives = 321/572 (55%), Gaps = 29/572 (5%)
Query: 3 GSKWDIEKFTGSNDFGLWKVKMQAVLTQQKCVEALKGEAAMPATLTQEEKREMIDKAKSA 62
G K+++ KF G N F W+ +M+ +L QQ + L ++ P T+ E+ ++ ++A SA
Sbjct: 3 GVKYEVAKFNGDNGFSTWQRRMRDLLIQQGLHKVLDVDSKKPDTMKAEDWADLDERAASA 62
Query: 63 IVLCLGDKVLRDVAREATAASM*AKLESLYMTKSLAHRQLLKQQLYSFKMVESISISEQL 122
I L L D V+ ++ E TA + +LESLYM+K+L ++ LK+QLY+ M E + L
Sbjct: 63 IRLHLSDDVVNNIIDEDTARGIWTRLESLYMSKTLTNKLYLKKQLYALHMSEGTNFLSHL 122
Query: 123 TEFNKILVDLANIEVNTEDEDKALLLLCSLPKSFEHFKDTILYGKEGTTTLEEVQAALRT 182
FN ++ LAN+ V E+EDKA+LLL SLP S+++ TIL+GK T L++V +AL
Sbjct: 123 NVFNGLITQLANLGVKIEEEDKAILLLNSLPSSYDNLATTILHGKT-TIELKDVTSALLL 181
Query: 183 KELTKFKDLKVDEGSEGLNVARGRNEHRGKGK-GKSRSKSRSKGFDKSKYK-CFLCHKQG 240
E K + ++G + RGR+ R G+S ++ +SK KS+ + C+ C++ G
Sbjct: 182 NE--KMRKKPENQGQALITEGRGRSYQRSSNNYGRSGARGKSKNRSKSRVRNCYNCNQPG 239
Query: 241 HFKKDCPDKGGDGSPSVQVAEASNEEGYESTGALVVTSWK----------------SEKS 284
HFK+DCP+ P E S ++ ++T A+V + E
Sbjct: 240 HFKRDCPN------PRKGKGETSGQKNDDNTAAMVQNNDNVVLFINEEEECMHLSGPESE 293
Query: 285 WVLDSGCSYHMCPRKEYFETLTLKEGGVVRLGNNKACKVQGMGNVRLKMFDGREFLLRDV 344
WV+D+ S+H P ++ F + G V++GN K+ G+G++ +K G +L+DV
Sbjct: 294 WVVDTAASHHATPVRDLFCRYVAGDFGTVKMGNTSYSKIAGIGDICIKTNVGCTLVLKDV 353
Query: 345 RFVPELKRNLISLSMFDGLGYCTRIEHGVCKISHGALITVKGSKMNGLYILDGSIVIGNA 404
R VP+L+ NLIS D GY + + +++ G+L+ KG LY + I G
Sbjct: 354 RHVPDLRMNLISGIALDRDGYESYFANQKWRLTKGSLVIAKGVARGTLYRTNAEICQGEL 413
Query: 405 SVASVVPHNNSELWHLRLGHVSERGLVELAKQGLLGKDKLDKLEFCEHCILGKQHRVKFG 464
+ A + +LWH R+GH+SE+GL LAK+ L+ K ++ C++C+ GKQHRV F
Sbjct: 414 NAAQ--DEISVDLWHKRMGHMSEKGLQILAKKSLISYAKGTTVKPCDYCLFGKQHRVSFQ 471
Query: 465 SGMHHSSRLFEYVHSDLLGPSKTPTHGGGSYFLSIIDDYSRRVWVFVLKKKSDTF*KFKE 524
+ + + V+SD+ GP + + GG YF++ IDD SR++WV++LK K F F++
Sbjct: 472 TSSERKLNILDLVYSDVCGPMEIESMGGNKYFVTFIDDASRKLWVYILKTKDQVFQVFQK 531
Query: 525 *HTLIENQMGTKLKGLRTDNGLEFVSEQFNDF 556
H L+E + G KLK LR+DNG E+ S +F ++
Sbjct: 532 FHALVERETGRKLKRLRSDNGGEYTSREFEEY 563
>gb|AAD23679.1| putative retroelement pol polyprotein [Arabidopsis thaliana]
gi|25412027|pir||G84599 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana
Length = 838
Score = 345 bits (886), Expect = 4e-93
Identities = 199/525 (37%), Positives = 310/525 (58%), Gaps = 30/525 (5%)
Query: 7 DIEKFTGSNDFGLWKVKMQAV---------LTQQKCVEALKGEAAMPATLTQEEKR---E 54
++EK G D+ LWK K+ A L + + +E + A + LT+ E + E
Sbjct: 7 EVEKLDGEGDYVLWKEKLLAHIELLGLLEGLEEDEAIEEEESTAETDSLLTKTEDKVLKE 66
Query: 55 MIDKAKSAIVLCLGDKVLRDVAREATAASM*AKLESLYMTKSLAHRQLLKQQLYSFKMVE 114
KA+S ++L LG+ VLR V +E TAA M L+ L+M KSL +R LKQ+LY +KM +
Sbjct: 67 KRGKARSTVILSLGNHVLRKVIKEKTAAGMIRVLDKLFMAKSLPNRIYLKQRLYGYKMSD 126
Query: 115 SISISEQLTEFNKILVDLANIEVNTEDEDKALLLLCSLPKSFEHFKDTILYGKEGTTTLE 174
S++I E + +F K++ DL N++V+ DED+A++LL SLPK F+ KDT+ YGK T L+
Sbjct: 127 SMTIEENVNDFFKLISDLENVKVSVPDEDQAIVLLMSLPKQFDQLKDTLKYGKT-TLALD 185
Query: 175 EVQAALRTKELTKFKDLK-VDEGSEGLNVA-RGRNEHRGKGKGKSRSKSRSKGFDKSKYK 232
E+ A+R+K L K + S+ L V RGR+E R K +++S+SRSK + K
Sbjct: 186 EITGAIRSKVLELGASGKMLKNSSDALFVQDRGRSEKRDKSSERNKSQSRSK--SREKKV 243
Query: 233 CFLCHKQGHFKKDC---PDKGGDGSPSVQVAEASNEEGYESTGALVVTSWKS-------E 282
C++C K+GHFKK C +K G+ S + E+SN G + A + +S +
Sbjct: 244 CWVCGKEGHFKKQCYVWKEKNKKGNNS-EKGESSNVIGQAADAAALAVREESNADNQEVD 302
Query: 283 KSWVLDSGCSYHMCPRKEYFETLTLKEGGVVRLGNNKACKVQGMGNVRLKMFDGREFLLR 342
W++D+GCS+HM PR+++F + G V++ N +++G+G++R++ D LL+
Sbjct: 303 NEWIMDTGCSFHMTPRRDWFVEFDESQTGRVKMANQTYSEIKGIGSIRIQNDDNTTVLLK 362
Query: 343 DVRFVPELKRNLISLSMFDGLGYCTRIEHGVCKISHGALITVKGSKMNGLYILDGSIVIG 402
+VR+VP + +NLIS+ + G + + G K+ G + +KG K+ LY+L G +V G
Sbjct: 363 NVRYVPSMSKNLISMGTLEDQGCWFQSKAGTLKVVKGCMTLLKGKKVGTLYLLQGVVVTG 422
Query: 403 NASVASVVPHNNSELWHLRLGHVSERGLVELAKQGLLGKDKLDKLEFCEHCILGKQHRVK 462
NA+ A + S++WH RL H+S+R + L K+G L +K++ LEFCE C+ GK HRV
Sbjct: 423 NAN-AVTSSKDESKIWHSRLCHMSQRNIDVLIKKGCLQAEKINGLEFCEDCVYGKTHRVG 481
Query: 463 FGSGMHHSSRLFEYVHSDLLG-PSKTPTHGGGSYFLSIIDDYSRR 506
FGS H + EY+HSDL G PS + G YF++ IDD +R+
Sbjct: 482 FGSAKHVTREKLEYIHSDLWGAPSVPNSLGNCQYFITFIDDLTRK 526
>dbj|BAD34493.1| Gag-Pol [Ipomoea batatas]
Length = 1298
Score = 340 bits (873), Expect = 1e-91
Identities = 210/574 (36%), Positives = 319/574 (54%), Gaps = 32/574 (5%)
Query: 2 MGSKWDIEKFTGSNDFGLWKVKMQAVLTQQKCVEALKGEAAMPATLTQEEK-REMIDKAK 60
M +K++IEKF G N F LWK+K++A+L + C+ A+ + P T ++K EM + A
Sbjct: 1 MAAKFEIEKFNGKN-FSLWKLKVKAILRKDNCLAAI---SERPVDFTDDKKWSEMNEDAM 56
Query: 61 SAIVLCLGDKVLRDVAREATAASM*AKLESLYMTKSLAHRQLLKQQLYSFKMVESISISE 120
+ + L + D VL + + TA + L LY KSL ++ LK++LY+ +M ES S++E
Sbjct: 57 ADLYLSIADGVLSSIEEKKTANEIWDHLNRLYEAKSLHNKIFLKRKLYTLRMSESTSVTE 116
Query: 121 QLTEFNKILVDLANIEVNTEDEDKALLLLCSLPKSFEHF---------KDTILYGKEGTT 171
L N + L ++ E +++A LLL SLP S++ D +++
Sbjct: 117 HLNTLNTLFSQLTSLSCKIEPQERAELLLQSLPDSYDQLIINLTNNILTDYLVFDDVAAA 176
Query: 172 TLEEVQAALRTKELTKFKDLKVD-EGSEGLNVARGRNEHRGKGKGKSRSKSRSKGFDKSK 230
LEE ++ + KE D +V+ + +E L V RGR+ RG+ G+ RSKS K
Sbjct: 177 VLEE-ESRRKNKE-----DRQVNLQQAEALTVMRGRSTERGQSSGRGRSKSSKKNLT--- 227
Query: 231 YKCFLCHKQGHFKKDCPDKGGDGSPSVQVAEASNEEGYESTGALVVTSWKSEKS--WVLD 288
C+ C K+GH KKDC + + +P VA S++ A + + + W++D
Sbjct: 228 --CYNCGKKGHLKKDCWNLAQNSNPQGNVASTSDDGSALCCEASIAREGRKRFADIWLID 285
Query: 289 SGCSYHMCPRKEYFETLTLKEGGVVRLGNNKACKVQGMGNVRLKMFDGREFLLRDVRFVP 348
SG +YHM RKE+F GG V ++ A ++ G+G ++LKM+DG ++DVR V
Sbjct: 286 SGATYHMTSRKEWFHHYEPISGGSVYSCDDHALEIIGIGTIKLKMYDGTVQTVQDVRHVK 345
Query: 349 ELKRNLISLSMFDGLGYCTRIEHGVCKISHGALITVKGSKM-NGLYILDG-SIVIGNASV 406
LK+NL+S + D + GV KI GAL+ +KG K+ LY+L G ++ ASV
Sbjct: 346 GLKKNLLSYGILDNSATQIETQKGVMKIFQGALVVMKGEKIAANLYMLKGETLQEAEASV 405
Query: 407 ASVVPHNNSELWHLRLGHVSERGLVELAKQGLLGKDKLDKLEFCEHCILGKQHRVKFGSG 466
A+ P +++ LWH +LGH+S++G+ L +Q L+ L CEHCI KQHR+KF +
Sbjct: 406 AACSP-DSTLLWHQKLGHMSDQGMKILVEQKLIPGLTKVSLPLCEHCITSKQHRLKFSTS 464
Query: 467 MHHSSRLFEYVHSDLLGPSKTPTHGGGSYFLSIIDDYSRRVWVFVLKKKSDTF*KFKE*H 526
+ E VHSD + + P+ GG YF+S IDDYSRR WV+ +KKKSD F FK
Sbjct: 465 NSRGKVVLELVHSD-VWQAPVPSLGGAKYFVSFIDDYSRRCWVYPIKKKSDVFATFKAFK 523
Query: 527 TLIENQMGTKLKGLRTDNGLEFVSEQFNDFLQVE 560
+E G K+K RTDNG E+ SE+F+DF + E
Sbjct: 524 ARVELDSGKKIKCFRTDNGGEYTSEEFDDFCKKE 557
>gb|AAK29467.1| polyprotein-like [Lycopersicon chilense]
Length = 1328
Score = 338 bits (867), Expect = 7e-91
Identities = 199/574 (34%), Positives = 320/574 (55%), Gaps = 32/574 (5%)
Query: 3 GSKWDIEKFTGSND-FGLWKVKMQAVLTQQKCVEALKGEAAMPATLTQEEKREMIDKAKS 61
G K+++ KF G F +W+ +M+ +L QQ +AL G++ P ++ E+ E+ +KA S
Sbjct: 3 GVKYEVAKFNGDKPVFSMWQRRMKDLLIQQGLHKALGGKSKKPESMKLEDWEELDEKAAS 62
Query: 62 AIVLCLGDKVLRDVAREATAASM*AKLESLYMTKSLAHRQLLKQQLYSFKMVESISISEQ 121
AI L L D V+ ++ E +A + KLE+LYM+K+L ++ LK+QLY+ M E +
Sbjct: 63 AIRLHLTDDVVNNIVDEESACGIWTKLENLYMSKTLTNKLYLKKQLYTLHMDEGTNFLSH 122
Query: 122 LTEFNKILVDLANIEVNTEDEDKALLLLCSLPKSFEHFKDTILYGKEGTTTLEEVQAALR 181
L N ++ LAN+ V E+EDK ++LL SLP S++ TIL+GK+ + L++V +AL
Sbjct: 123 LNVLNGLITQLANLGVKIEEEDKRIVLLNSLPSSYDTLSTTILHGKD-SIQLKDVTSALL 181
Query: 182 TKELTKFKDLKVDEGSEGLNVARGRNEHRGKGK-GKSRSKSRSKGFDKSKYK-CFLCHKQ 239
E K + + G + +RGR+ R G+S ++ +SK KSK + C+ C +
Sbjct: 182 LNE--KMRKKPENHGQVFITESRGRSYQRSSSNYGRSGARGKSKVRSKSKARNCYNCDQP 239
Query: 240 GHFKKDCPD-KGGDGSPSVQVAEASNEEGYESTGALVVTSWK----------------SE 282
GHFK+DCP+ K G G E+S ++ ++T A+V + +E
Sbjct: 240 GHFKRDCPNPKRGKG-------ESSGQKNDDNTAAMVQNNDDVVLLINEEEECMHLAGTE 292
Query: 283 KSWVLDSGCSYHMCPRKEYFETLTLKEGGVVRLGNNKACKVQGMGNVRLKMFDGREFLLR 342
WV+D+ SYH P ++ F + G V++GN K+ G+G++ K G +L+
Sbjct: 293 SEWVVDTAASYHATPVRDLFCRYVAGDYGNVKMGNTSYSKIAGIGDICFKTNVGCTLVLK 352
Query: 343 DVRFVPELKRNLISLSMFDGLGYCTRIEHGVCKISHGALITVKGSKMNGLYILDGSIVIG 402
DVR VP+L+ NLIS D GY + +++ GAL+ KG LY + I G
Sbjct: 353 DVRHVPDLRMNLISGIALDQDGYENYFANQKWRLTKGALVIAKGVARGTLYRTNAEICQG 412
Query: 403 NASVASVVPHNNSELWHLRLGHVSERGLVELAKQGLLGKDKLDKLEFCEHCILGKQHRVK 462
+ A N+++LWH R+GH SE+GL L+K+ L+ K ++ C + + GKQHRV
Sbjct: 413 ELNAAH--EENSADLWHKRMGHTSEKGLQILSKKSLISFTKGTTIKPCNYWLFGKQHRVS 470
Query: 463 FGSGMHHSSRLFEYVHSDLLGPSKTPTHGGGSYFLSIIDDYSRRVWVFVLKKKSDTF*KF 522
F + S + + V+SD+ GP + + GG YF++ IDD SR++WV++ + K F F
Sbjct: 471 FQTSSERKSNILDLVYSDVCGPMEIESMGGNKYFVTFIDDASRKLWVYIFRAKDQVFQVF 530
Query: 523 KE*HTLIENQMGTKLKGLRTDNGLEFVSEQFNDF 556
++ H L+E + G K K LRTDNG E+ S +F ++
Sbjct: 531 QKFHALVERETGRKRKRLRTDNGGEYTSREFEEY 564
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.346 0.154 0.537
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,043,683,341
Number of Sequences: 2540612
Number of extensions: 81477081
Number of successful extensions: 293933
Number of sequences better than 10.0: 1076
Number of HSP's better than 10.0 without gapping: 736
Number of HSP's successfully gapped in prelim test: 340
Number of HSP's that attempted gapping in prelim test: 290172
Number of HSP's gapped (non-prelim): 2003
length of query: 1307
length of database: 863,360,394
effective HSP length: 140
effective length of query: 1167
effective length of database: 507,674,714
effective search space: 592456391238
effective search space used: 592456391238
T: 11
A: 40
X1: 15 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.7 bits)
S2: 81 (35.8 bits)
Medicago: description of AC124217.5


